Abstract
Background
Environmental health and other researchers can benefit from automated or semi-automated summaries of data within published studies as summarizing study methods and results is time and resource intensive. Automated summaries can be designed to identify and extract details of interest pertaining to the study design, population, testing agent/intervention, or outcome (etc.). Much of the data reported across existing publications lack unified structure, standardization and machine-readable formats or may be presented in complex tables which serve as barriers that impede the development of automated data extraction methodologies.
As full automation of data extraction seems unlikely soon, encouraging investigators to submit structured summaries of methods and results in standardized formats with meta-data tagging of content may be of value during the publication process. This would produce machine-readable content to facilitate automated data extraction, establish sharable data repositories, help make research data FAIR, and could improve reporting quality.
Objectives
A pilot study was conducted to assess the feasibility of asking participants to summarize study methods and results using a structured, web-based data extraction model as a potential workflow that could be implemented during the manuscript submission process.
Methods
Eight participants entered study details and data into the Health Assessment Workplace Collaborative (HAWC). Participants were surveyed after the extraction exercise to ascertain 1) whether this extraction exercise will impact their conducting and reporting of future research, 2) the ease of data extraction, including which fields were easiest and relatively more problematic to extract and 3) the amount of time taken to perform data extractions and other related tasks. Investigators then presented participants the potential benefits of providing structured data in the format they were extracting. After this, participants were surveyed about 1) their willingness to provide structured data during the publication process and 2) whether they felt the potential application of structured data entry approaches and their implementation during the journal submission process should continue to be further explored.
Conclusions
Routine provision of structured data that summarizes key information from research studies could reduce the amount of effort required for reusing that data in the future, such as in systematic reviews or agency scientific assessments. Our pilot study suggests that directly asking authors to provide that data, via structured templates, may be a viable approach to achieving this: participants were willing to do so, and the overall process was not prohibitively arduous. We also found some support for the hypothesis that use of study templates may have halo benefits in improving the conduct and completeness of reporting of future research. While limitations in the generalizability of our findings mean that the conditions of success of templates cannot be assumed, further research into how such templates might be designed and implemented does seem to have enough chance of success that it ought to be undertaken.
Keywords: Data extraction, Automated data extraction, Manuscript submission, Structured data, Data sharing, Author opinion, Author feedback, Author willingness, Data summary, Systematic review, Study evaluation template, Standardized data, Data templates, Author journal submission requirements, Science translation, Natural language Processing (NLP)
Data extraction; Automated data extraction; Electronic data extraction manuscript submission; Structured data; Data sharing; Natural language processing (NLP); Semantic authoring; Author opinion; Author feedback; Author willingness; Data summary; Systematic review; Study quality; Study quality reporting; Risk of bias reporting; Study evaluation; Data extraction tool; Data extraction software; Standardized data; Data templates; Author submission requirements; Machine readable; Knowledge translation; Science translation.
1. Introduction
The processes of identifying and noting salient details to summarize study methods and results (referred to as “data extraction”), and making judgements pertaining to study evaluation, are two of the most time- and resource-intensive aspects of conducting a systematic review (Nussbaumer-Streit et al., 2021; Wallace et al., 2016; Tsafnat et al., 2014). Thus, there is keen interest in assessing the extent to which this process could be automated or semi-automated by using natural language processing (NLP) methods (O'Connor et al., 2019). However, state-of-the-art methodologies for automated data extraction from text, figures and tables published in studies are to date only achieving limited success (Marshall and Wallace, 2019; Wallace et al., 2012, 2013, 2016; Tsafnat et al., 2014). Previously, such technologies obtained no more than moderately acceptable extraction results on controlled subsets of high-quality documents in narrowly defined content areas. Current technologies can more effectively utilize machine learning approaches to mine text [for extraction] but the success of such algorithms depends heavily on the availability of manually annotated data, which is limited in the biomedical health field (Ma et al., 2020; Jonnalagadda et al., 2015; Mishra et al., 2014). Other barriers to successful implementation of automated data extraction methods include the following: the heterogeneous way (e.g., inconsistencies in vocabulary/ontologies, annotations and reported data) in which information is reported in published manuscripts, that can be difficult to summarize even for experienced human reviewers (Tsafnat et al., 2014); and the frequent use of complex table structures and inconsistent or ambiguous schema for recording data in tabular format (Wolffe et al., 2020). It follows that dissemination of tabular content that is already in a machine-readable format would be beneficial for the efficiency of systematic review and data accessibility in general, and potentially contribute to making research data more Findable, Accessible, Interoperable, and Reusable (FAIR) in general (Wilkinson et al., 2016).
Published manuscripts would be more readily machine-readable if data within them were organized in a more consistent and standardized manner. The more scientific health studies present data that are similarly structured, the more readily data can be shared, mined, aggregated, extracted, and identified within and across related disciplines. In addition, provision of studies containing structured data is a prerequisite for the creation of data repositories. Use of a standardized format may also improve the reporting quality of manuscripts because authors would be prompted to enter key information about methods and results (Turner et al., 2012).
As a secondary benefit, it is also possible that asking authors to provide certain types of information when completing structured reporting templates could also encourage changes in research practices, as prompts to report data also function as prompts to conduct related elements of study practice (e.g., blinding of investigators) that might otherwise be overlooked.
One approach for obtaining structured data is to ask authors to enter their data into a structured format. An approach to increase provision of structured data on a large-scale basis would be encouragement or condition by journal publishers to require submitters to provide their data into a widely implemented, structured data format or data template. There are several related issues and challenges such as determining an agreed upon structured format, etc. In this pilot study, we were interested in ascertaining author willingness to enter their data into a given, structured format as a part of the journal submission process.
Encouraging structured data submission is not a novel concept (Sim and Detmer, 2005) but has been hindered by a lack of available software to support standardised formats and a lack of support from authors and publishers (Jin et al., 2015; Swan and Brown, 2008). With respect to software, new web-based tools used for structured data extraction in systematic review could serve as prototypes to assess the feasibility of structured data submission by authors during the publication process. The increasing use of protocol and study registries may also be acclimating authors to the process of structured data entry and reporting. For example, the International Committee of Medical Journal Editors (ICMJE) requires prospective registration of clinical trials as prerequisite for publication, providing formatting guidelines for preparing, sharing and reporting data summarized in tables and results for journal submission (ICMJE, 2019). Likewise, other initiatives such as the United States’ Clinical Trials registry (https://clinicaltrials.gov), containing over 395,000 U.S. and international trial registries, offers guidance for consistent style and formatting of reported data and flow-chart figures. Structured data submission has also been successfully implemented for genomics, proteomics and metabolomics data through the Gene Expression Omnibus (GEO), a public genomics data repository [(Barrett et al., 2011, 2013), http://www.ncbi.nlm.nih.gov/geo/].
Studies describing authors’ attitudes toward entering data into structured data templates are limited in number. Some have focused on physicians who enter data related to patient electronic health records (Bush et al., 2017; Doberne et al., 2017; Djalali et al., 2015).
Given the potential utility of author-entered data extraction for data standardization and sharing, a pilot study was conducted to assess the willingness of prospective authors to summarize study methods and results in a structured web-based format. In addition, we asked authors to evaluate reporting quality and internal validity (risk of bias) of the summarized study and asked the authors if this might alter how they would design and report future research.
2. Methods
2.1. Tool selection
HAWC (https://hawc.epa.gov/) (Shapiro et al., 2018) was selected to serve as the data template for the pilot because it is a web-accessible, free and open-source application increasingly used in environmentally-oriented systematic reviews (Supplemental Materials: HAWC FAQ). HAWC is designed to help structure data extraction and study evaluation for human (either randomized trial or observational), animal, and in vitro evidence. HAWC has detailed instructions to guide the data extraction process, that were developed and refined based on feedback from a broad base of over 100 scientists who regularly summarize studies for use in systematic reviews. Other applications have similar capabilities [(e.g., EPPI-Reviewer (Thomas et al., 2010), Table Builder (IARC, 2015), LitStream (ICF, 2014), SRDR (Li et al., 2015), and Covidence (https://www.covidence.org/home)], but none are all freely available, open-source, web-based, and designed to both collect and display information from human, animal and in vitro studies. Since the objective was to test the author's extraction of their own data, selection of only one suitable tool was considered sufficient for this pilot exercise.
2.2. Participant recruitment and study/endpoint selection
Prior to contacting and recruiting study participants, the National Institutes of Health (NIH), Office of Human Subjects Research (OHSR) reviewed and exempted this research (Exemption # 13366), stating “Federal regulations for the protection of human subjects do not apply” to this project.
Selected participants met the following criteria: unfamiliarity with HAWC; a current student or professional conducting health-related, scientific research; and had previous manuscript publishing experience. Volunteers were recruited by the team of study authors between January 2018–March 2019, primarily by word of mouth, e.g., advertising via a call with the NIEHS Superfund Research Program, contacting investigators familiar with systematic review, or simply by contacting researchers known to have a certain area of expertise, etc. A total of 17 people were contacted and showed initial interest, of which 15 attended an initial informational kick-off meeting. Eight people agreed to participate after the kick-off meeting. The reason given for not being involved was the time commitment, except for one participant who withdrew because they disagreed with the concept of providing additional structured summaries of study data. All participants had previously published manuscripts so were considered able to provide feedback on the pilot study from the perspective of an author.
Team members met and worked with participants to identify a published study with data to extract using the following study selection criteria:
-
•
Experimental animal study that included a control group, presented two or more dose level groups, and designed to evaluate effects in different dose groups.
-
•
The findings for the health outcome endpoint needed to be quantitatively presented in a table, text, or visuals (or by having access to the raw data). Specifically, animal numbers per group, a central estimate (e.g., mean), and variance estimate (standard deviation, standard error of the mean) needed to be presented to later calculate effect size information.
-
•
For simplicity, mixture studies and repeated measures were excluded because these study designs are more challenging to extract.
Initially, participants were encouraged to select a study for extraction among articles they had co-authored. However, in some cases the focus of the article was not a good match for the goals of the pilot and the criteria for study extraction selection (e.g., our pilot focused on experimental animal data and the participant had only published in vitro studies). In these cases, participants were permitted to extract data from another study of their choice that met the criteria above.
2.3. Participant orientation
Participants attended one of several 1-h kick-off meetings scheduled between November 2018 and March 2019 that included a brief overview of HAWC (including resources to set up an account and how to access training modules), schedule for completing pilot tasks, identification of data for extraction, and contacts for assistance. The meeting purposefully did not provide training on how to navigate HAWC because a goal was to assess the ability of participants to use the provided electronic instructional materials on their own. The kick-off meeting was recorded and later viewed by participants who were unable to attend the scheduled presentation.
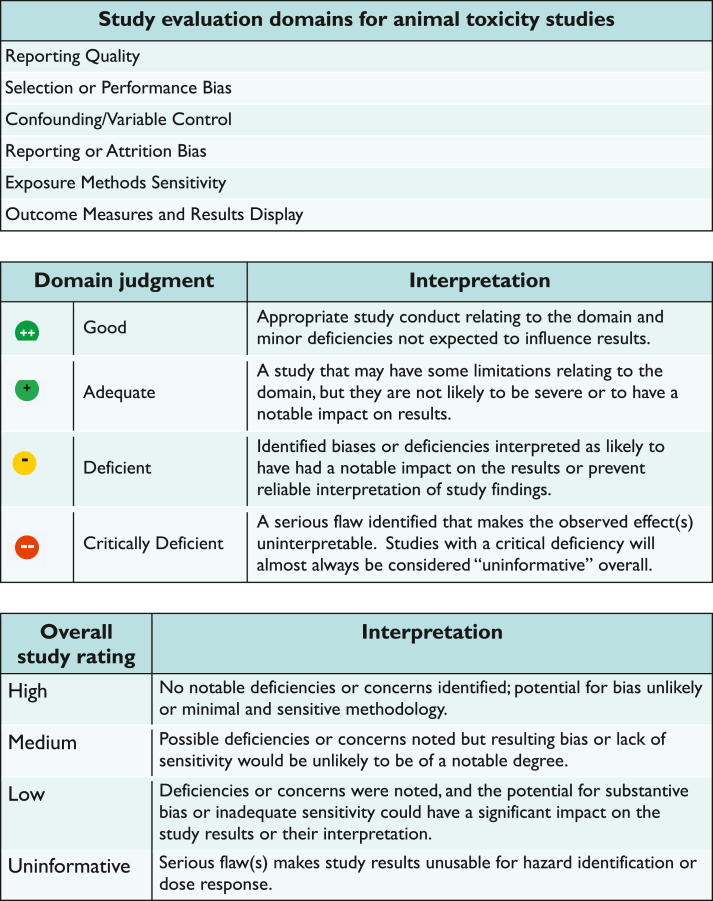
A HAWC Extraction Guide (Supplemental Materials: HAWC Extraction Guide) was shared to illustrate the data extraction fields participants were asked to complete to summarize study design and results, and to conduct study evaluation (risk of bias) using IRIS methodology (NASEM, 2018) (summarized schematically in Figure 1). Another resource, the ‘Instructions for Answering Study Evaluation Domains’, which also served as a checklist for judging study reporting and quality, was provided to assist participants with conducting study evaluation (Supplemental Materials: Instructions for Answering Study Evaluation Domains). Core questions are provided to prompt the reviewer in evaluating domains assessing different aspects of study design and conduct related to reporting, risk of bias and study sensitivity. Reviewers apply a judgment of Good, Adequate, Deficient, Not Reported or Critically Deficient and can include a rationale and study-specific information to support the judgment. Once all domains are evaluated, a confidence rating of High, Medium, or Low confidence or Uninformative is assigned for each endpoint/outcome from the study.
Figure 1.
Overview of IRIS Program study evaluation approach for animal toxicity studies.
2.4. Participant extraction activities
Participants followed the provided instructions to extract study details and results data, perform study evaluation and enter extracted data and evaluation information into appropriate sections of the HAWC template. Participants also noted the amount of time it took them to extract study details and results and to complete the study evaluation. Throughout the pilot, participants were able to contact team members for technical assistance or ask questions as issues arose.
2.5. Participants completed surveys and attended a small group meeting
Participants completed a post-extraction survey that was administered following participants' completion of the extraction and study evaluation exercise (Supplemental Materials: Supplemental Excel File, Tab 2). This survey consisted of nine questions (five Likert Scale questions and four open-ended responses) to query participants’ experience and attitudes regarding the extraction, study evaluation process, and amount of time taken to conduct the various pilot tasks (i.e., learn HAWC, extract data, perform risk of bias analyses, and complete surveys).
In addition, participants attended one of a series of small group meetings, to discuss general observations following the extraction and study evaluation activities. During these meetings, the research team presented a variety of interactive charts and plots displaying, where possible, the extracted data that participants entered into HAWC. The ability of HAWC to produce visualizations of data aggregated across different studies was demonstrated to participants (Supplemental Materials: Supplemental Excel File, Tab 1). This presentation was designed to illustrate how the steps taken by authors to structure their data within an individual journal article could eventually impact the future utility of large-scale, structured data extraction and be used to potentially create repositories of data and study findings. Such a repository would benefit the research community by furnishing data formatted to assess quantitative patterns more quickly across studies. Participants were asked to consider such visualizations and potential uses of structured data when assessing their willingness to submit data in a structured format during the publication submission process.
Following the small group meeting, participants completed a post-pilot survey consisting of three Likert Scale questions and one open-ended question to ascertain participants willingness to provide structured data as part of a publication submission process (Supplemental Materials: Supplemental Excel File, Tab 3).
3. Results
Survey data are summarized to report the frequency of participant responses while data indicating the time spent performing various pilot tasks is averaged across all the responses received in units of minutes. Narrative comments provided by participants are presented verbatim and in full when noted (i.e., Table 5), or otherwise summarized (i.e., Table 7). Participant narrative responses can be viewed in the non-summarized form from the below supplemental materials (Supplemental Materials: Supplemental Excel File). Table 1 summarizes information about the study participants including their career or educational position, pilot exercise tasks completed, and endpoints extracted.
Table 5.
Participant responses describing Pilot's impact on future research activities (from post-extraction survey).
| Impact Query | Impact | Participant Responses |
|---|---|---|
| What fields (if any) are you likely to change about the way you conduct, or report future research based on your experience with the reporting checklist used during this pilot? | Positive (71%) (5 of 7 responses) |
|
| Neutral (29%) (2 of 7 responses) |
|
|
| Negative (0%) (0 of 7 responses) |
• N/A | |
| What fields (if any) are you likely to change about the way you conduct or report research in the future based on your experience with the study quality (internal validity) assessment tool used during this pilot? | Positive (83%) (5 of 6 responses) |
|
| Neutral (0%) (0 of 6 responses) |
|
|
| Negative (17%) (1 of 6 responses) |
|
Reported after participant's 2nd extraction.
Table 7.
Easy and difficult data fields to extract.
| Query: Which data field(s) did you find easy to extract? Which field(s) of data extraction did you find problematic? | Participant Responses∗ | Number of Responses |
|---|---|---|
| Easy Fields to Extract | 13 | |
| Data Summary: Participants provided examples of easy fields to extract which approximately equalled 65% (13 out of 20) of the total number of responses received for this query. | Dose regimen/exposure | 5 |
| Animal husbandry | 3 | |
| Animal group and experiment set up | 3 | |
| Outcome/endpoint | 2 | |
| Difficult Fields to Extract | 7 | |
| Data Summary: Participants also provided details regarding fields that were difficult to extract which equalled 35% (7 out of 20) of the total number of responses received for this query. Of the total number of fields reported as difficult to extract (n = 7), 85.7% (6 of 7) were associated with entering endpoint or outcome details while 14.3% (1 of 7) were associated with the dosing regimen. |
Behavioral endpoint because there are many different types for each task/difficult to enter data by sex | 1 |
| Outcome | 2 | |
| Endpoint details: not sure how to enter data for system, organ effect and sub-type; not sure what frank effect means | 1 | |
| Reproductive end point entry | 1 | |
| Entering raw/dose response data was challenging [Note: participant had issues with entering response data associated with the dose] | 1 | |
| Dosing regimen: not clear on how to adjust doses; guess it may be done at set up | 1 | |
| 20 | ||
Reflects one or more responses indicated by a total of 7 participants.
Table 1.
Summary of study participants and selected studies.
| Participant Number | Educational Status | Data Extraction | Study Evaluation | Task Completion Time | Post-Extraction & Post-Pilot Survey | Selected Study and Extracted Endpoints |
|---|---|---|---|---|---|---|
| 1 | Associate dean | X | X | X | X | Neurobehavioral – male rat; open field test for anxiety related behaviors; oral exposure to Bisphenol A (BPA) (Study 1) |
| 2 | Graduate student | X | X | X | X | Neuromuscular – male and female rat; escape latency to visible platform, swimming speed; nicotine exposure via subcutaneous injection (Study 2) |
| 3∗ | Professor | X | X | X | X | Developmental/Neurobehavioral – adult male and female zebrafish; learning and memory (shock avoidance); exposed to benzo [a]pyrene via water habitat (Study 3) |
| 4 | Professor | X | X | Developmental/Neurobehavioral -weanling, male rat; Morris water maze; spatial learning and memory; lactational exposure from maternal dietary exposure to PCBs (Study 4) | ||
| 5 | Associate dean; Professor | X | X | X | X | Systemic (whole body) – male mouse; body weight; 14-day, oral exposure to valproic acid (Study 5) |
| 6 | Graduate student | X | X | X | X | Systemic (whole body)– male rats; oxidative stress (as measured by malondialdehyde (MDA) and hepatoxicity as measured by histopathology) via subchronic, oral exposure to arsenic (Study 6) |
| 7 | Graduate student | X | X | X | X | Systemic (whole body) – maternal and fetal weight change and placental markers of oxidative stress from maternal dietary TCE exposure and fetal placental exposure (Study 7)) |
| 8 | Graduate student | X | X | X | X | Developmental - Female reproduction and fertility: histopathology; body, organ, and ovary weights; prenatal exposure to benzo [a]pyrene and inorganic lead (Study 8) |
| Total n | 7 | 8 | 8 | 7 |
Participant performed two extractions (responses from the first extraction were used in the pilot results).
The pilot study was based on 8 participants for which 7 of the eight participants completed all tasks for this pilot. Four of the 8 participants were senior scientists (associate dean or professor), 4 were graduate students, and all had previously co-authored studies. Participants held diverse expertise including biochemistry, neurodevelopmental toxicology, microbiology, computational toxicology, biologist/behavioral sciences, general toxicology and molecular and cellular neurotoxicology and neurobiology. Most of the extracted endpoints were neurological in nature and others included body weight, histopathology, fertility, and biochemical responses (Table 1).
Table 2 presents participant Likert scale responses indicating their willingness to consider structured data entry during the publication process.
Table 2.
Willingness to consider structured data entry during publication process.
| Please use the scale below to rate your agreement with the statement (n = 7) | I am open to using structured data entry when submitting articles for publication. | Given the potential application of structured data entry, approaches to implement this during the journal submission process should continue to be explored. | My consideration of using structured data entry depends on the type of article being submitted for publication. |
|---|---|---|---|
| Strongly Agree | 43% (n = 3) | 86% (n = 6) | 43% (n = 3) |
| Moderately Agree | 43% (n = 3) | 14% (n = 1) | 14% (n = 1) |
| Agree | 14% (n = 1) | 0% (n = 0) | 43% (n = 3) |
| Neutral | 0% (n = 0) | 0% (n = 0) | 0% (n = 0) |
| Disagree | 0% (n = 0) | 0% (n = 0) | 0% (n = 0) |
| Moderately Disagree | 0% (n = 0) | 0% (n = 0) | 0% (n = 0) |
| Strongly Disagree | 0% (n = 0) | 0% (n = 0) | 0% (n = 0) |
Overall, all participants completing the post-pilot survey agreed that structured data entry during the journal submission process should be explored, with most expressing moderate or strong agreement (Table 2), although not necessarily for all types of articles.
Table 3 summarizes the time participants took to complete the various pilot tasks as shown in the first column.
Table 3.
Participant self-reported time spent performing various pilot tasks.
| Participants' Response to Time Spent Performing These Various Tasks (in minutes) | n | Average (Min) | Range (Min) |
|---|---|---|---|
| Orientation: account creation, becoming acquainted with the HAWC software, watching tutorial videos, etc. | 8 | 65 | 10–120 |
| Data Extraction: summarizing study methods and results into HAWC. | 8 | 83 | 30–120 |
| Receiving Technical Support: assistance required in addition to orientation and team provided written instructions (i.e., help with tool navigation, and rating study and reporting quality, etc. | 7 | 49 | 0–180 |
| Study Evaluation: determining risk of bias | 7 | 56 | 30–90 |
| Reporting Quality: rating how well results were reported and recorded within the study | 7 | 51 | 30–90 |
With respect to time spent completing pilot tasks, the process of ‘data extraction’ required the most time, with participants averaging 83 min per study. The time participants reported spending on ‘orientation’, ‘study evaluation’ and ‘reporting quality rating’ tasks averaged 65, 56 and 51 min respectively (Table 3). Of the 7 participants providing responses for time spent ‘receiving technical support’, 6 required extraction assistance (beyond the assistance offered during orientation and for tool access), by indicating more than 0 min were spent on this task. The range for technical support received by the 6 participants reporting a time greater than 0 min, is 10–180 min (a subset of Table 3 data not shown).
A summary of the technical assistance the team of study authors provided to study participants is presented in Table 4.
Table 4.
Summary of Technical Assistance Provided by Team (throughout duration of pilot).
| Summary of Team Provided Technical Assistance | n | Average Time in Minutes (Hours) | Range in Minutes (Hours) |
|---|---|---|---|
| Getting started | 8 | 47 min (0.78 h) | 15–60 min (0.25–1.0 h) |
| Data extraction assistance | 4 | 38 min (0.63 h) | 30–60 min (0.5–1.0 h) |
| Study evaluation assistance | 2 | 30 min (0.50 h) | 30–30 min (0.5–0.5 h) |
| Technical Assistance (provided after the ‘getting started’ phase) | 1 | 60 min (1 h) | 60–60 (1.0–1.0 h) |
Assistance offered for ‘getting started’ activities required an average of 47 min ‘Getting started’ activities included assistance setting up HAWC accounts, navigating the tool and providing emailed instructions (etc.), while ‘extraction assistance’ offered help in setting up experiments, animal groups (including those with several generations) and endpoints.
Table 5 includes participant responses to their likely change in conduct or the reporting of study details and quality in their future research activities. It also includes the study authors’ assessment of these qualitatively stated impacts as shown in Column 2.
Regarding the survey query on future research conduct and reporting impacts due to the reporting checklist, 71%, or 5 out of 7 responses, reflected positive resulting behaviors such as “[e]nsuring that my published methods contain all of the elements on the checklist to minimize risk of bias and maximize study quality”. The impact of 2 responses was not clear. For the survey query related to the impact on future conduct or reporting of study quality information, approximately 83%, or 5 out of 6 the responses indicated positive changes in associated behaviors, including “[b]eing specific about how data is blinded and randomized (instead of just stating that it was done).” One response “[c]orrecting errors and performing edits to the report could be made easier and straightforward” was assessed as a negative impact as it may indicate the process for making edits within the structure or tool, although possible, was not readily clear.
Table 6 highlights participants’ direct experience with extracting study data and information pertaining to the study evaluation (Supplemental Materials: Supplemental Excel File Tab 8).
Table 6.
Initial survey Likert scale results (from post-extraction survey).
| Question/Response (n = 7) | Easy | Relatively Easy | Neutral | Relatively Difficult | Difficult |
|---|---|---|---|---|---|
| Rate your experience entering study data | 29% (n = 2) | 43% (n = 3) | 14% (n = 1) | 14% (n = 1) | 0% (n = 0) |
| Rate your experience performing a study quality evaluation | 43% (n = 3) | 43% (n = 3) | 14% (n = 1) | 0% (n = 0) | 0% (n = 0) |
| Question/Response (n = 7) | Strongly Agree | Somewhat Agree | Neutral | Somewhat Disagree | Strongly Disagree |
| Completing the reporting quality checklist is likely to impact my future research activities | 57% (n = 4) | 29% (n = 2) | 0% (n = 0) | 0% (n = 0) | 14% (n = 1) |
| Completing the study quality evaluation is likely to impact my future research activities | 57% (n = 4) | 14% (n = 1) | 14% (n = 1) | 0% (n = 0) | 14% (n = 1) |
| I feel comfortable with applying the quality checklist and study quality tool to my other research | 57% (n = 4) | 29% (n = 2) | 14% (n = 1) | 0% (n = 0) | 0% (n = 0) |
In terms of ease of entering study data, most participants felt this data extraction task was “easy” or “relatively easy” (5 of 7 participants), with one person expressing a neutral response and another judging the task as relatively difficult (Table 6).
With respect to future design and reporting, many participants ‘strongly or ‘somewhat’ agreed that completing the reporting quality checklist (6 of 7) and study quality evaluation (5 of 7) will likely impact the way they conduct and report forthcoming research, with most expressing strong agreement (Table 6) and (Table 7).
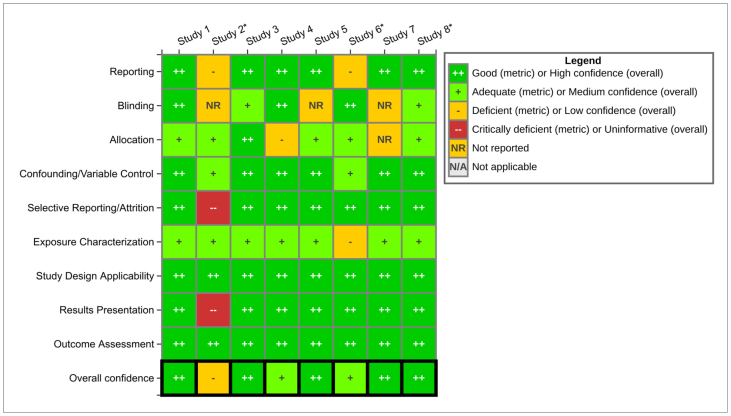
Figure 2 is a heat map, summarizing how participants assessed various study quality elements, aggregated across each of the 8 studies evaluated by all participants plus the repeated extraction (for a total of 9). Of the 8 participants, 5 extracted data from their own study while 3 extracted data from studies by other authors (see Limitations and Future Research for further discussion). Study authors prepared these visuals from the extracted data participants entered into HAWC.
Figure 2.
Participants Aggregated Study Evaluation Summary. ∗ Participant did not extract data from their own study. This heatmap can also be accessed from the following link: https://hawc.epa.gov/summary/visual/assessment/100500015/Table-7-Participant-Aggregated-Study-Evaluation-Su/ and (Supplemental Materials: Supplemental Excel File Tab 1, Figure 5).
Overall, with one exception, most studies were considered ‘high’ or ‘medium’ confidence with most limitations occurring in the reporting (i.e., selective reporting/attrition and results presentations), and blinding assessment domains.
Table 7 summarizes participant responses gaging the ease or difficulty in extracting data for various fields by listing the fields participants noted as easy or difficult to extract and a tally indicating the total number of times participants noted fields as easy or difficult to extract. Fields reported as easy to extract include dose regimen or exposure, animal husbandry, animal group and experiment set up, and outcome and endpoints. Such easy to extract fields accounted for approximately 65% (13 out of 20) of the total number of responses received for the query. Data fields noted as difficult to extract included those pertaining to behavioral and reproductive endpoints, outcomes, organ effect, and dose-response data and accounted for 35% (7 out of 20) of the total number of responses received for this query (Supplemental Materials: Supplemental Excel File Tab 10).
4. Discussion
We present three main themes for discussion in relation to our results: willingness of participants to provide structured data; potential impact on study design decisions by researchers; the arduousness of providing structured data, from both the perspective of author and provider; and the limitations of our study. This is followed by a discussion of study limitations and the impact of this pilot effort.
4.1. Willingness of participants to provide structured data
There was a general willingness among the study participants to provide structured data, with 6 out of the 7 participants either strongly or moderately agreeing with using structured data entry when submitting articles for publication, and 7 out of the 7 strongly or moderately supporting the exploration of applications and approaches for implementing structured data as part of the journal submission process (see Table 5). Only one of the 15 people who attended the kick-off meeting withdrew because they disagreed in principle with the concept being investigated in this pilot study. As this was a convenience sample of volunteers, it is likely to represent only researchers with an existing interest in this issue; however, the mere fact of anyone volunteering, plus the generally positive response of participants to the prospect of providing structured data in future, suggests that at least a portion of the research community is receptive to introduction of this practice.
4.2. Potential impact on study reporting and design decisions by researchers
Five (5) out of the 6 participants stated that, based on the information asked for in the templates used in this pilot study, they would likely change how they report studies in future, providing more detailed information useful for third-party evaluation. Four (4) of 7 participants reported that completing the reporting quality checklist and the study quality evaluation would likely impact their future research activities, increasing the likelihood of their introducing study design elements that would result in more favourable ratings in subsequent study appraisal processes. Again, being a convenience sample the generalizability of these findings cannot be assumed; however, there does seem to be grounds for believing that providing structured templates for study reporting could have halo benefits in terms of improving the design of future studies.
4.3. Arduousness of providing structured data
Five (5) out of the 7 participants stated that entering study data into a formatted template was easy or relatively easy to perform. However, at least on the first run-through, an average of approximately 3 h of technical support time was required (the one participant who performed the exercise twice completed the second run in 2 h rather than 6 (Supplemental Materials: Supplemental Excel File, Tab 9). There were also important differences in how easy participants found the reporting of various data items, with 6 out of the 7 describing endpoints and outcomes as difficult to report (qualitative feedback is shown in Table 7) - a finding that is consistent with the experience of programs that routinely conduct assessments such as EPA IRIS and other organizations (Mathes et al., 2017) and from the conduct of systematic reviews (Ganju and Heyman, 2018; Mathes et al., 2017; Leeflang, 2008).
These findings suggest that while time commitments for support may reduce over time, provision of initial support may be important, and its absence may be an obstacle to large-scale use. If such support cannot be provided, this may impact the willingness of researchers to use structured templates and/or lead to their incorrect use, limiting their potential use and effectiveness. Focusing on providing template elements that specifically address aspects that are difficult to report (e.g., integrating controlled vocabularies and semantic elements into templates that help authors correctly report information about their studies), may be important for usability and gaining acceptance for using such templates.
4.4. Limitations and Future Research
The main limitations of this pilot study are in its sampling strategy, sample size, and focus on simple study designs. These all limit its generalizability: we do not know how representative the sample population (a convenience sample of volunteers) is of the broader research population, although we did have representation from across career stages; the sample is also small, so we do not know if the proportion of responses we have in our sample would be reflected in a larger population; in focusing on simple study designs, we do not know if researchers would have the same degree of positive responses if they were presented with more complex templates for complex study designs; nor do we know how accurate researchers will be when applying templates to summarise their own studies. Nonetheless, we believe our results at least justify further examination of this topic, with larger studies from more backgrounds and career stages, more detailed use-case development and investigation, the development and testing of better template technology, and the accuracy of self-extraction of summary data all being candidates as valuable research.
5. Conclusions
Routine provision of structured data that summarizes key information from research studies could reduce the amount of effort required for reusing that data in the future, such as in systematic reviews or agency scientific assessments, and may contribute to making research data more FAIR. Our pilot study suggests that directly asking authors to provide that data, via structured templates, may be a viable approach to achieving this: participants were willing to do so, and the overall process was not prohibitively arduous. We also found some support for the hypothesis that use of study templates may have halo benefits in improving the conduct and completeness of reporting of future research. While limitations in the generalizability of our findings mean that the conditions of success of templates cannot be assumed, further research into how such templates might be designed and implemented does seem to have enough chance of success that it ought to be undertaken.
Disclaimer
The views expressed in this article are those of the author(s) and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency.
Declarations
Author contribution statement
Amanda S. Persad: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Ingrid L. Druwe and Janice S. Lee: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Michele M. Taylor and Kristina A. Thayer: Conceived and designed the experiments; Performed the experiments; Wrote the paper.
Natalie Blanton Southard and Courtney Lemeri: Conceived and designed the experiments; Wrote the paper.
Amina Wilkins: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Paul Whaley: Analyzed and interpreted the data; Wrote the paper.
Andrew J. Shapiro: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
The work was supported by way of staff salaries from the National Toxicology Program (NTP); and the Environmental Protection Agency (EPA). PW's time was covered under a consultancy agreement with Evidence-Based Toxicology Collaboration at Johns Hopkins University Bloomberg School of Public Health.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare the following conflict of interests: Mr. Whaley reports personal fees from Elsevier Ltd (Environment International), the Cancer Prevention and Education Society, the Evidence Based Toxicology Collaboration and Yordas Group, and grants from Lancaster University, which are outside the submitted work but relate to the development and promotion of systematic review and other evidence-based methods in environmental health research, delivering training around these methods, and providing editorial services.
Additional information
No additional information is available for this paper.
Acknowledgements
The co-authors would like to thank Michelle Angrish and Samuel Thacker for providing technical review and Ryan Jones for assisting with reference editing.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
The studies and data extracted by participants are contained in HAWC and are available for public viewing to become better acquainted with the tool and to view supplemental materials used in the pilot, such as surveys, participant responses, and HAWC FAQs and guides, which can be accessed in the ‘Attachments’ section from: https://hawc.epa.gov/assessment/100500015/
References
- Barrett T., Troup D.B., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Muertter R.N., Holko M., Ayanbule O., Yefanov A., Soboleva A. NCBI GEO: archive for functional genomics data sets--10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., Yefanov A., Lee H., Zhang N., Robertson C.L., Serova N., Davis S., Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush R.A., Kuelbs C., Ryu J., Jiang W., Chiang G. Structured data entry in the electronic medical record: perspectives of pediatric specialty physicians and surgeons. J. Med. Syst. 2017;41:75. doi: 10.1007/s10916-017-0716-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djalali S., Ursprung N., Rosemann T., Senn O., Tandjung R. Undirected health IT implementation in ambulatory care favors paper-based workarounds and limits health data exchange. Int. J. Med. Inf. 2015;84:920–932. doi: 10.1016/j.ijmedinf.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Doberne J.W., Redd T., Lattin D., Yackel T.R., Eriksson C.O., Mohan V., Gold J.A., Ash J.S., Chiang M.F. Perspectives and uses of the electronic health record among US pediatricians: a national survey. J. Ambul. Care Manag. 2017;40:59–68. doi: 10.1097/JAC.0000000000000167. [DOI] [PubMed] [Google Scholar]
- Ganju E., Heyman C. Wiley; Hoboken, NJ: 2018. Scaling Global Change: a Social Entrepreneur's Guide to Surviving the Start-Up Phase and Driving Impact.https://www.wiley.com/en-us/Scaling+Global+Change%3A+A+Social+Entrepreneur%27s+Guide+to+Surviving+the+Start+up+Phase+and+Driving+Impact-p-9781119483885 [Google Scholar]
- IARC (International Agency for Research on Cancer) 2015. IARC Monographs: Instructions to Authors; pp. 1–20.http://monographs.iarc.fr/ENG/Preamble/previous/Instructions_to_Authors.pdf (Updated 10 March 2017) [IARC Monograph] Lyon, France. [Google Scholar]
- ICF (ICF International) 2014. From Systematic Review to Assessment Development: Managing Big (And Small) Datasets with DRAGON.https://web.archive.org/web/20140811051047/www.icfi.com/insights/products-and-tools/dragon-dose-response Available online at. [Google Scholar]
- ICMJE (International Committee of Medical Journal Editors) 2019. Recommendations for the Conduct, Reporting, Editing, and Publication of Scholarly Work in Medical Journals.http://www.icmje.org/recommendations/ [PubMed] [Google Scholar]
- Jin X., Wah B.W., Cheng X., Wang Y. Significance and challenges of big data research. Big Data Res. 2015;2:59–64. [Google Scholar]
- Jonnalagadda S.R., Goyal P., Huffman M.D. Automating data extraction in systematic reviews: a systematic review [Review] Syst. Rev. 2015;4:78. doi: 10.1186/s13643-015-0066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeflang M.M.G. University of Amsterdam; Amsterdam, Netherlands: 2008. Systematic Reviews of Diagnostic Test Accuracy: Summary and Discussion.https://hdl.handle.net/11245/1.385458 (Doctoral Dissertation) Retrieved from. [Google Scholar]
- Li T., Vedula S.S., Hadar N., Parkin C., Lau J., Dickersin K. Innovations in data collection, management, and archiving for systematic reviews. Ann. Intern. Med. 2015;162:287–294. doi: 10.7326/M14-1603. [DOI] [PubMed] [Google Scholar]
- Ma X., Lu Y., Lu Y., Pei Z., Liu J. Biomedical event extraction using a new error detection learning approach based on neural network. Comput. Mater. Continua (CMC) 2020;63:923–941. [Google Scholar]
- Marshall I.J., Wallace B.C. Toward systematic review automation: a practical guide to using machine learning tools in research synthesis [Editorial] Syst. Rev. 2019;8:163. doi: 10.1186/s13643-019-1074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathes T., Klaßen P., Pieper D. Frequency of data extraction errors and methods to increase data extraction quality: a methodological review. BMC Med. Res. Methodol. 2017;17:152. doi: 10.1186/s12874-017-0431-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R., Bian J., Fiszman M., Weir C.R., Jonnalagadda S., Mostafa J., Del Fiol G. Text summarization in the biomedical domain: a systematic review of recent research [Review] J. Biomed. Inf. 2014;52:457–467. doi: 10.1016/j.jbi.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NASEM (National Academies of Sciences, Engineering, and Medicine) The National Academies Press; Washington, DC: 2018. Progress toward Transforming the Integrated Risk Information System (IRIS) Program. A 2018 Evaluation. [Google Scholar]
- Nussbaumer-Streit B., Ellen M., Klerings I., Sfetcu R., Riva N., MahmIc-Kaknjo, Poulentzas G., Martinez P., Baladia E., Ziganshina L.E., Marqués M.E., Aguilar L., Kassianos A.P., Frampton G., Silva A.G., Affengruber L., Spjker R., Thomas J., Berg R.C., Kontogiani M., Sousa M., Kontogiorgis C., Gartlehner G. Resource use during systematic review production varies widely: a scoping review [Review] J. Clin. Epidemiol. 2021;139:287–296. doi: 10.1016/j.jclinepi.2021.05.019. [DOI] [PubMed] [Google Scholar]
- O’Connor Annette M., Glasziou Paul, Taylor Michele, Thomas James, Spijker Rene, Wolfe Mary S., et al. A focus on cross-purpose tools, automated recognition of study design in multiple disciplines, and evaluation of automation tools: a summary of significant discussions at the fourth meeting of the International Collaboration for Automation of Systematic Reviews (ICASR) Syst. Rev. 2020;9(100) doi: 10.1186/s13643-020-01351-4. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro A.J., Antoni S., Guyton K.Z., Lunn R.M., Loomis D., Rusyn I., Jahnke G.D., Schwingl P.J., Mehta S.S., Addington J., Guha N. Software tools to facilitate systematic review used for cancer hazard identification. Environ. Health Perspect. 2018;126:104501. doi: 10.1289/EHP4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim I., Detmer D.E. Beyond trial registration: a global trial bank for clinical trial reporting. PLoS Med. 2005;2:e365. doi: 10.1371/journal.pmed.0020365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan A., Brown S. 2008. To Share or Not to Share: Publication and Quality Assurance of Research Data Outputs. A Report Commissioned by the Research Information Network.https://eprints.soton.ac.uk/266742/ Research Information Network. [Google Scholar]
- Thomas J., Brunton J., Graziosi S. Social Science Research Unit, UCL Institute of Education; London, UK: 2010. EPPI-reviewer 4: Software for Research Synthesis.https://eppi.ioe.ac.uk/cms/er4/Features/tabid/3396/Default.aspx Retrieved from. [Google Scholar]
- Tsafnat G., Glasziou P., Choong M.K., Dunn A., Galgani F., Coiera E. Systematic review automation technologies [Editorial] Syst. Rev. 2014;3:74. doi: 10.1186/2046-4053-3-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner L., Shamseer L., Altman D.G., Schulz K.F., Moher D. Does use of the CONSORT Statement impact the completeness of reporting of randomised controlled trials published in medical journals? A Cochrane review [Review] Syst. Rev. 2012;1:60. doi: 10.1186/2046-4053-1-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace B.C., Dahabreh I.J., Schmid C.H., Lau J., Trikalinos T.A. Modernizing the systematic review process to inform comparative effectiveness: tools and methods. J. Comp. Eff. Res. 2013;2:273–282. doi: 10.2217/cer.13.17. [DOI] [PubMed] [Google Scholar]
- Wallace B.C., Kuiper J., Sharma A., Zhu M.B., Marshall I.J. Extracting PICO sentences from clinical trial reports using supervised distant supervision. J. Mach. Learn. Res. 2016 (Print) 17. [PMC free article] [PubMed] [Google Scholar]
- Wallace B.C., Small K., Brodley C.E., Lau J., Schmid C.H., Bertram L., Lill C.M., Cohen J.T., Trikalinos T.A. Toward modernizing the systematic review pipeline in genetics: efficient updating via data mining. Genet. Med. 2012;14:663–669. doi: 10.1038/gim.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M.D., et al. Comment: the FAIR guiding principles for scientific data management and stewardship [Comment] Sci. Data. 2016;3:1–9. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe T.A.M., Vidler J., Halsall C., Hunt N., Whaley P. A survey of systematic evidence mapping practice and the case for knowledge graphs in environmental health & toxicology. Toxicol. Sci. 2020 doi: 10.1093/toxsci/kfaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The studies and data extracted by participants are contained in HAWC and are available for public viewing to become better acquainted with the tool and to view supplemental materials used in the pilot, such as surveys, participant responses, and HAWC FAQs and guides, which can be accessed in the ‘Attachments’ section from: https://hawc.epa.gov/assessment/100500015/
Data Availability Statement
Data included in article/supplementary material/referenced in article.