Abstract
Interaction of immune cells with the systemic environment is necessary for the coordinated development and execution of immune responses. Monocyte-macrophage lineage cells reside at the junction of innate and adaptive immunity. Previously we reported that the sialyltransferase ST6GAL1 in the extracellular milieu modulates B cell development and IgG production, granulocyte production, and attenuates acute airway inflammation to bacterial challenge in mouse models. Here, we report that extracellular ST6GAL1 also elicits profound responses in monocyte-macrophage lineage cells. We show that recombinant ST6GAL1 adheres to subsets of thioglycolate-elicited inflammatory cells in the mouse peritoneum and to cultured human monocyte THP-1 cells. Exposure of the inflammatory cells to recombinant ST6GAL1 elicited wholesale changes in the gene expression profile of primary mouse myeloid cells; most notable was the striking up-regulation of monocyte-macrophage and monocyte-derived dendritic cell development pathway signature genes and transcription factors PU.1, NFκB and their target genes, driving increased monocyte-macrophage population and survival ex vivo. In the cultured human monocyte cells, the essential cell surface receptor of the monocyte-macrophage lineage, the M-CSF receptor (M-CSF-R, Csfr1) was a target of extracellular ST6GAL1 catalytic activity. Extracellular ST6GAL1 activated the M-CSF-R and initiated intracellular signaling events, namely, the nuclear translocation of NFκB subunit p65, and phosphorylation of ERK 1/2 and AKT. The findings implicate extracellular ST6GAL1 in monocyte development by a mechanism initiated at the cell surface and support an emerging paradigm of an extracellular glycan-modifying enzyme as a central regulator coordinating immune hematopoietic cell development and function.
Keywords: inflammation, macrophage, monocyte, sialyltransferase, transcriptomics
Introduction
Maintaining immunity is critical for health throughout lifetime. The process requires producing the proper numbers of distinct functional immunocytes, delivery of these immunocytes to sites where they are needed and generating an appropriate response to neutralize the invading challenges. Cell-intrinsic developmental programs are required to generate functional leukocytes across all lineages. However, cell-extrinsic mechanisms are necessary to convey the systemic needs for these cells and to guide them to appropriate destinations. Glycans on the cell surface and in the extracellular milieu reside at the interface through which these cell-extrinsic cues are conveyed. It is well-recognized that selectin-mediated interactions with sialylated and fucosylated glycan motifs on counter-receptors are central to leukocyte trafficking along the vascular endothelium and delivery to distal sites (Tvaroska et al. 2020). Tumor cells and viruses bind selectins to metastasize and enter host cells, respectively (Kremsreiter et al. 2021). The Siglecs, a family of sialic acid-binding immunoglobin-like lectins, promote and regulate cell–cell interactions in innate and adaptive immunity through recognition of sialic acid glycan motifs (Bornhofft et al. 2018; Meyer et al. 2018; Laubli and Varki 2020). Targeted strategies to modulate Siglec interactions include antibodies, biologics, and nanoparticles (O'Sullivan et al. 2020). Likewise, the galectins, a family of galactose-recognizing proteins are involved in diverse physiologic functions including inflammation, immune responses, cell migration, autophagy, and signaling driven by their interactions with cytosolic and nuclear ligands (Johannes et al. 2018). Synthetic disaccharide galectin inhibitors have shown efficacy in animal models of vascular disease (Delaine et al. 2016; Chen et al. 2017), insulin resistance (Li et al. 2016), and clinical trials for idiopathic lung fibrosis (Hirani et al. 2021).
There is a resurgence of appreciation for the terminal α2,6-sialyl glycan epitope constructed by the sialyltransferase ST6GAL1. ST6GAL1-mediated addition of α2,6-sialic acids impacts a growing list of leukocyte cell-surface receptors. Examples of ST6GAL1-regulated receptor function include prolonged monocyte TLR4 inflammatory signaling cascades (Holdbrooks et al. 2020), Fas and TNFR1 sialylation preventing receptor internalization and subsequent apoptosis (Holdbrooks et al. 2018), α2,6-sialic acid addition to EGFR which activates tyrosine kinase activity (Britain et al. 2018), and α2,6-sialylation by ST6GAL1 which modulates the B-cell specific Siglec CD22, critical for B cell development and function (Irons et al. 2020). Over expression of ST6GAL1 is common in many human cancers; its tumorigenic effects include invasiveness and enhancement of the cancer stem cell phenotype (Garnham et al. 2019; Dorsett et al. 2021) .
Glycosyltransferases such as ST6GAL1 are traditionally viewed as occupants of the intracellular Golgi-ER secretory apparatus where they glycosylate nascent glycoconjugates in biosynthetic transit. However, catalytically active ST6GAL1, as well as numerous other glycosyltransferases, are also present in the extracellular milieu. Plasma levels of ST6GAL1 fluctuate with stress and disease states. ST6GAL1 levels and α2,6-sialylation are upregulated in several carcinomas and enzyme expression is correlated with more aggressive breast and prostate tumor grades (Garnham et al. 2019). A potential anti-inflammatory role for sialylated Fc is based on lower mRNA and enzyme levels of ST6GAL1 in IVIG therapy-resistant Kawasaki disease (Ogata et al. 2013).
Previously, we reported that extracellular ST6GAL1 modulates B cell development and IgG production (Irons and Lau 2018; Irons et al. 2019, 2020), as well as neutrophil (Nasirikenari et al. 2006; Dougher et al. 2017) and eosinophil (Nasirikenari et al. 2010) production. A role for extracellular ST6GAL1 in inflammation was demonstrated by the ability of intravenously infused recombinant ST6GAL1 to attenuate acute airway inflammation elicited by bacterial challenge in mouse models (Nasirikenari et al. 2019). Diminished inflammatory cytokine release in response to recombinant ST6GAL1 was attributed to altered airway macrophage function. Monocyte–macrophage lineage cells reside at the junction of innate and adaptive immunity. Here we report that extracellular ST6GAL1 elicits profound responses in monocyte–macrophage lineage cells, causing striking changes in transcription profiles and ultimately driving monocyte differentiation and survival. Together, this and the related reports implicate ST6GAL1 as a direct conveyer of systemic signals, affecting multiple immunocyte lineages and functions.
Results
Extracellular ST6GAL1 adheres to a subset of recruited murine inflammatory cells and human THP-1 monocytes
Freshly isolated primary inflammatory cells were examined for interaction with exogenously added rST6GAL1. Thioglycolate was used to recruit primary inflammatory cells to the peritoneum. Within 24 h of thioglycolate injection, the recruited cells consisted mostly of granulocytes with a significant subpopulation of monocyte–macrophage cells (Nasirikenari et al. 2006). These analyses were performed on only the adherent cells to avoid potential contamination by nonadherent granulocytes. In order to avoid confounding signal from natively expressed ST6GAL1, the St6gal1-null mouse unable to express functional ST6GAL1 was used. Figure 1A shows GFP-rST6G attachment to a subset of fixed, nonpermeabilized thioglycolate-elicited peritoneal cells within 3 min. Proteinase K digestion effectively abolished the GFP-rST6G signal, which further supports the localization of GFP-rST6G on cell surfaces. The adhesion of ST6GAL1 to inflammatory cell surfaces appears to be specific; GFP-huST3GAL1, and GFP-huβ4GALT1 did not attach to inflammatory cell surfaces under the same conditions.
Fig. 1.

Extracellular ST6GAL1 adheres to a subset of inflammatory cells. Panel A) ST6GAL1 binds a subset of fixed St6gal1-null primary inflammatory cells during a 3 min incubation. Thioglycolate-elicited peritoneal cells from St6gal1-null mice were fixed and then treated with GFP-rST6G (25 μg/mL) for 3 min only at room temperature, and washed once; alternatively, cells were further treated with Proteinase K (250 μg/mL) for 5 min at room temperature, washed once, and visualized by fluorescence microscopy at 200× magnification. Also, cells were treated with GFP-rST3GAL1 or GFP-rβ4GALT1 in lieu of GFP-rST6G. B) ST6GAL1 binds a subset of fixed C57BL/6 wild type primary inflammatory cells during a 30 min incubation. Thioglycolate-elicited peritoneal cells from wild type mice were fixed and then treated with GFP-rST6G (25 μg/mL) for 30 min at room temperature and washed once (100 μm magnification). C) ST6GAL1 binds human monocyte cell line THP-1. For the Western blot analysis (left), THP-1 cells in culture (3 × 10(6)/3 mL) were incubated for 24 h with buffer alone (control), CMP-Sia, (100 μM), recombinant soluble GFP-rST6GAL1 (GFP-rST6G, 10 μg/mL), or both CMP-Sia and GFP-rST6G. Cells were then lysed and visualized by Western blot with anti-GFP or anti-ST6GAL1. Actin as loading control is also shown. C, right) Alternatively, THP-1 cells in culture were incubated for only 30 min with buffer alone or with GFP-rST6G alone. Cells were washed once and observed for GFP fluorescence.
To address whether GFP-rST6GAL1 binding is due to lack of endogenous surface α2,6 sialic acids in St6gal1-null mice, GFP-rST6GAL1 was incubated with fixed thioglycolate peritoneal lavage cells from C57BL/6 WT mice (Fig. 1B). A 30-min incubation showed that wild type cells also significantly bind ST6GAL1 indicating that it is not preferentially targeting exposed galactose residues in the knockout cells.
ST6GAL1 binding was further evaluated in the human monocyte THP-1 cell line. GFP-rST6G added to the culture medium associated with the cells within 24 h, as revealed by Western blot analysis of cell lysates visualized with either anti-GFP or anti-ST6GAL1 (Fig. 1C). The strong signal with electrophoretic mobility on SDS-PAGE consistent with GFP-rST6G was absent in lysates prepared from cells not incubated with GFP-rST6G. Additionally, anti-ST6GAL1 visualized the cell-natively expressed ST6GAL1 similar in molecular size to the soluble ST6GAL1 (39–40 kDa) as well as, to a lesser degree, the full-length 49–50 kDa form. Addition of the sialyltransferase donor substrate, CMP-Neu5Ac (CMP-Sialic acid), did not augment or diminish GFP-rST6G association with the cells. The GFP-rST6G band on the Western blot remained intact after 24 h, suggesting that the added GFP-rST6 was not metabolically modified. Binding also was demonstrated in live THP-1 cells by fluorescence microscopy (Fig. 1C). To minimize the possibility of internalization of GFP-rST6G, incubation was reduced to 30 min. ST6GAL1 bound to a subset of cells within 30 min, and in many instances the interaction appears membrane associated. These data support the association of exogenous ST6GAL1 with monocytes in a rapid (3 min) and specific manner that can occur directly at the cell surface, although the possibility of binding through an endocytic pathway in live cells cannot be excluded. Additionally, it is possible that such interactions occur cell autonomously, non-cell autonomously, or both.
Extracellular ST6GAL1 upregulates gene expression in primary monocytes/macrophages
Peritoneal lavage was collected from St6gal1-null mice 24 h after thioglycolate elicitation. Monocyte/macrophage cells were enriched by magnetic bead depletion of neutrophils, lymphocytes, and erythroid cells as described in Materials and Methods, and then cultured for 24 h in the absence or presence of rST6G alone, CMP-Sia alone, or rST6G and CMP-Sia together. Bulk RNA-seq analysis was performed on only the adherent cells, which avoids potential contamination by any residual nonadherent granulocytes. Figure 2A is a representative portion of a Heatmap of the RNA-seq analysis, showing dramatic and wholesale changes in transcription profiles in response to extracellular ST6GAL1. Curiously, rST6G alone is sufficient to elicit changes in the transcription profile. Added sialyltransferase donor substrate, CMP-Sia, did not alter the transcription profile of the primary monocyte/macrophage cells. As noted earlier, our experiments cannot discriminate between a mechanism involving direct cell surface sialylation versus endocytosis and intracellular sialylation of receptors returning to the cell membrane. However, this does not diminish the critical finding that exogenous ST6GAL1 exerts a profound impact on gene expression and cell phenotype.
Fig. 2.

Extracellular ST6GAL1 exposure drives wholesale changes in cellular transcription profiles. A) Representative portion of a Heatmap analysis of thioglycolate-elicted St6gal1-null peritoneal inflammatory cells. Peritoneal lavage cells enriched for monocytes/macrophages were cultured 24 h with buffer alone (control), CMP-Sia (100 μM), recombinant soluble ST6GAL1 (rST6G, 25 μg/mL), or both CMP-Sia and rST6G. Triplicate culture wells were used for each condition. RNA isolation was performed as in Materials and Methods. RNA-seq methods are described in the Supplement. Analysis of bulk RNA-seq data is described in Materials and Methods. B) ST6GAL1-mediated extracellular α2,6-sialylation did not require added CMP-Sia. Mouse RAW264.7 cells were fixed and sialidase-treated as described in Materials and Methods, then incubated with rST6G (20 μg/mL) in the presence or absence of added CMP-Sia (250 μM) for 2 h and visualized for cell surface FITC-SNA reactivity at 400× magnification.
We hypothesize that cellular leakage of endogenous donor substrates might be sufficient to drive catalytic action of the extracellular rST6G, rendering superfluous additional CMP-Sia. To explore this hypothesis, the SNA lectin (Sambucus nigra agglutinin) that recognizes the α2,6-sialyl structures constructed by ST6GAL1 was used on primary cultured lavage cells and on the mouse RAW 264.7 monocyte/macrophage cell line treated with rST6 in the presence and absence of CMP-Sia. At 24 h post-treatment of primary cells in culture, sialylated targets may be long past the stage of endocytosis, no longer detectable on fixed cells with SNA. Indeed, we observed minimal sialylation 24 h after treatment of primary cells under any of the conditions tested. We chose to use RAW264.7 cells to validate our initial findings with primary cells because of their consistency and reproducibility (Fig. 2B). Fixation prior to rST6GAL1 treatment rendered them incapable of receptor endocytosis thereby making α2,6 modifications more apparent. Fixed RAW 264.7 cells also were sialidase-treated to render them SNA-negative before adding rST6G. Addition of rST6G alone to these fixed cells was sufficient to make them SNA-positive, indicating that cell surface α2,6-sialic acids could be constructed by rST6G without needing exogenously added CMP-Sia.
Figure 3 summarizes the results of the RNA-seq analysis between rST6-treated and mock buffer control. Red circles in the Dot-Plot (Fig. 3A) denote gene sets with high normalized enrichment scores (NES) and low false discovery rates, and the sizes of the circles denote the size of each gene set. Gene pathways for every major macrophage signature, M0, M1, and M2 were elevated, with high NES, large gene set sizes, and low false discovery rates. NES is the effect size of a gene set’s expression in the treatment group versus the control group. Higher positive values indicate up-regulation of genes contained within the given gene set in the treatment group. Lower negative values indicate down-regulation of the genes contained within the gene set within the treatment group. Monocyte/macrophage and monocyte/dendritic cell differentiation pathways also were highly enriched but had smaller gene sets.
Fig. 3.
Monocyte–Macrophage gene pathways and gene-based cell populations are profoundly impacted by extracellular ST6GAL1. Thioglycolate-recruited St6gal1-null inflammatory cells were prepared and cultured ex vivo as described for Fig. 2. Data in this figure (Fig. 3) represent rST6G-treated vs mock-treated cells. A) is a dot-plot summarizing gene set enrichment analysis statistics. The highly elevated pathways (represented as red circles) include those for M0, M1, and M2 macrophage subtypes. The size of each colored circle reflects the number of genes contributing to each signature pathway. B) shows the Gene Set Enrichment Analysis (GSEA), which reveals up-regulation of monocyte–macrophage and monocyte-dendritic cell pathway signature genes. Also highly enriched are targets of the monocyte/macrophage master regulator transcription factor PU.1 and target genes of NFκB, the transcription factor next in order of importance in this context. C) is qPCR confirmation of the RNA-seq dataset for elevation of 2 resident macrophage signature genes F4/80 and the resident peritoneal macrophage marker, serpin b2 (Summers et al. 2020). Error bars represent the standard deviation of triplicate measurements for each condition. The unpaired t-test yielded confidence for the difference between control and rST6G treated samples of P = 0.0054 and 0.0013 for F4/80 and serpin b2, respectively. D) is the predicted cell population content based on comparison with the Gene Expression Commons Database, which, similar to the GSEA profile reveals monocyte/macrophage lineage enrichment (in blue) in response to rST6G and rST6G + CMP-Sia.
Gene set enrichment analysis (GSEA) graphs of monocyte/macrophage and monocyte/dendritic cell pathway signature genes illustrate a high degree of correlation with ST6GAL1 addition (Fig. 3B). Values closer to zero indicate less difference between groups. The master transcriptional regulator of monocyte/macrophage differentiation, PU.1 and NFκB considered next in order of importance in this context had NES values indicating approximately 3-fold enrichment (Fig. 3B). Based on gene content, PU.1, followed in order by NFκB were the 2 most highly upregulated transcription factors in this data set. GSEA plots illustrate strong correlation between extrinsic ST6GAL1 and expression of PU.1 and NFκB target genes (Fig. 3A).
The stimulating effects of extracellular ST6GAL1 on monocyte/macrophage lineage genes were confirmed by quantitative PCR (qPCR) measurement demonstrating mature macrophage signature genes F4/80 and serpin b2 to be highly elevated (Summers et al. 2020) (Fig. 3C).
Cell populations based on gene expression were estimated using CIBERSORTx with the Gene Expression Commons Database as a reference (Choi et al. 2019) (Fig. 3D). The monocyte/macrophage population is the most significantly enriched fraction in response to extrinsic ST6GAL1 further supporting its critical influence on differentiation along this pathway. Extrinsic ST6GAL1 highly upregulated signature genes in the monocyte/dendritic cell pathway as well (Fig. 3B) apparently reflecting the positive influence of master regulator PU.1 and NFκB. The relevance of extrinsic ST6GAL1 function to these monocyte lineage pathways is underscored upon comparison with others such as p53 signaling, TGF-beta signaling, insulin receptor signaling, and cell cycle regulation (Fig. 3A).
Extracellular ST6GAL1 prolongs monocyte survival
Thioglycolate-elicited peritoneal inflammatory cells from the St6gal1-null mouse were cultured as in the RNA-seq study but without antibody-based depletion of specific cell types (Most monocytes are adherent; neutrophils were removed by washing). Cells were treated with rST6G and CMP-Sia added together to test for effects on survival. After 1 day in culture adherent cells were collected and the number of live cells and their phenotype determined by flow cytometry (Fig. 4). rST6G and CMP-Sia improved cell survival by roughly 40%–50%. The increase in live cells was primarily attributable to monocytes, whereas the low fraction of resident macrophages was unchanged. We conclude that extracellular ST6GAL1 combined with CMP-Sia has a pro-survival effect on monocytes.
Fig. 4.
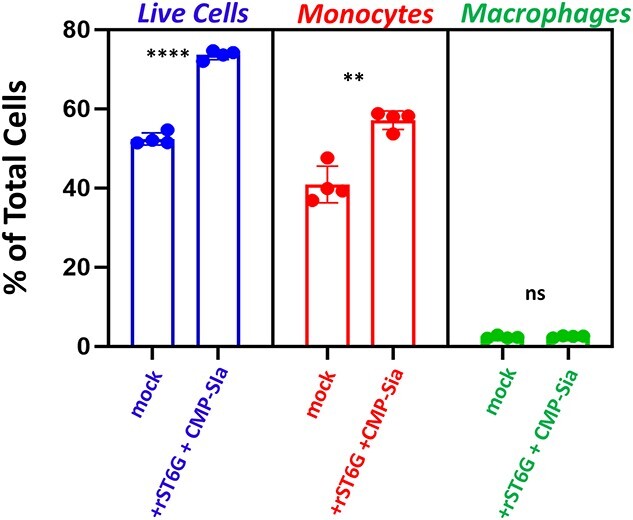
Extracellular ST6GAL1 combined with CMP-sialic acid enhances cell survival and increases monocyte abundance. Thioglycolate-recruited peritoneal lavage cells from St6gal1-null mice were cultured in the absence (control) and presence of rST6G + CMP-Sia for 24 h. Adherent cells were analyzed by flow cytometry in which live cells (left) were renumerated as % of total cells by DAPI, monocytes (middle) by Ly6C, and macrophages (right) by F4/80. Quadruplicates were used for each datapoint; error bars show standard deviation. The unpaired t-test yielded confidence for the difference between control and rST6G + CMP-Sia treated samples of P < 0.0001 and P = 0.0008 for % live cells and monocytes, respectively.
Extracellular ST6GAL1 targets the mature form of the M-CSF-receptor at the cell surface
The remarkable impact of extracellular ST6GAL1 on the monocyte/macrophage transcriptome prompted identification of a cell surface target, the M-CSF receptor selected based on its potential role in the effects we observed on gene expression and monocyte survival. Indeed, the extracellular domain of M-CSF-R contains 9 potential sites for N-linked glycans, 3 of which are occupied (https://www.ebi.ac.uk/ -UniProtKB-P09581 (CSF1R_MOUSE)). Here, we addressed the possibility of extracellular α2,6-sialylation of the human THP-1 monocyte M-CSF-R employing the One-Step Selective Exoenzymatic Labeling (SEEL) method (Sun et al. 2016). THP-1 cells were treated with rST6GAL1 and CMP–sialic acid–biotin conjugate (SEEL reagent) sugar donor followed by lysis and immunoprecipitation with anti-biotin antibody and M-CSF-R detection by Western blot. In Figure 5 (right panel), the pull-down of surface-localized mature receptor but not the precursor intracellular form demonstrated robust extracellular catalytic action of extracellular ST6GAL1 on the M-CSF-R in THP-1 monocytes.
Fig. 5.
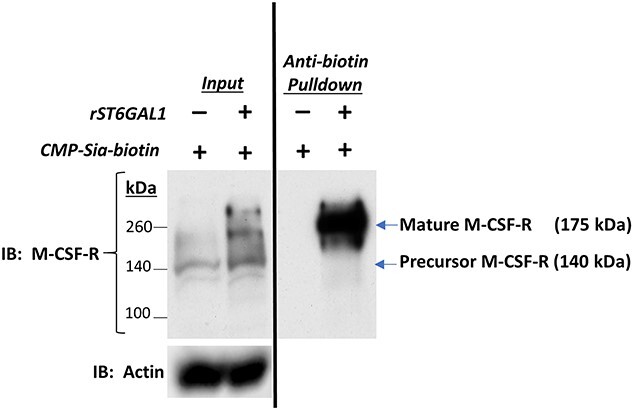
Cell-surface M-CSF receptor is sialylated by extracellular ST6GAL1. THP-1 cells were treated with rST6G and CMP-Sia(biotin) in culture for 2 h followed by immunoprecipitation of lysates with anti-biotin and Western blot using anti-M-CSF-R as described in Materials and Methods. Left Panel is a Western blot analysis of THP-1 lysates probed with anti-M-CSF-R showing enhanced formation of the precursor and mature receptor in the presence of rST6G. Right Panel is an anti-biotin pulldown of THP-1 homogenates, also probed with anti-M-CSF-R, showing sialylation by Sia(biotin) of the mature 175 kDa receptor but not the precursor 140 kDa form.
Extracellular ST6GAL1 initiates intracellular signaling
M-CSF-R signaling is initiated upon M-CSF/IL-34 ligand binding to its extracellular domain causing receptor dimerization and cytoplasmic domain tyrosine phosphorylation (Stanley and Chitu 2014). Downstream formation of P-AKT and P-ERK yields consequences for proliferation and differentiation. We hypothesize that sialylation of M-CSF-R by extracellular ST6GAL1 elicits an intracellular signaling cascade ultimately affecting gene expression and survival, and therefore we assessed the ability of extracellular rST6G to initiate intracellular signaling consistent with M-CSF-R activation in the absence of ligand. Extracellular ST6GAL1-induced phosphorylation of the M-CSF-R was evaluated in THP-1 cells (Fig. 6). While the absolute level of M-CSF-R was unaffected by the presence of extracellular ST6GAL1 (A, top panel), ST6GAL1 elicited phosphorylation of M-CSF-R, a feature of its activation, and it also increased P-ERK1/2 and P-AKT (A, middle panels). Remarkably Western blot reveals that rST6GAL1 becomes associated with cellular components contemporaneous with these events (Fig. 6A). This is consistent with the presence of GFP-rST6G in THP-1 lysates (Western blot, Fig. 1C).
Fig. 6.
Extracellular ST6GAL1 modulates intracellular signaling. THP-1 cells were treated with rST6G and CMP-Sia in culture for 24 h. Cellular lysates, nuclear extracts, and nuclei-free cytosolic extracts were prepared. A) Western blots showed showing similar expression for M-CSF-R in all conditions but increases in phospho-M-CSF-R, phospho-ERK1/2 and phospho-AKT, and ST6GAL1 in the presence of rST6G and CMP-Sia. Actin signal is shown (bottom) as loading control. B, left) shows a trend, although statistically not significant, for increased total phospho-p65 dependent on rST6 and rST6 + CMP-Sia, with actin as loading control. B, right) shows translocation of p65 into the nuclear fraction. (Fold = signal intensity of p65 relative to Histone H3 or GAPDH). The gel images are representative blots from at least 3 repeats, and the densitometric bar graphs of 3 or more repeats. Where there is statistical significance for the difference, the p value is shown.
NFκB exerts an anti-apoptotic/pro-survival effect on monocytes when its p65 subunit becomes phosphorylated and localized in the nucleus. Based on the pro-survival influence of extrinsic ST6GAL1 on monocytes (see Fig. 4), we tested the capacity of extracellular ST6GAL1 to cause phosphorylation and nuclear translocation of p65. ST6GAL1 induced an apparent trend in phosphorylation of p65 (Fig. 6B, left panel), although this was not statistically significant. However, it did cause the nuclear translocation of p65 (B, right panel). Together these observations implicate p65 in a mechanism for the pro-survival effect of extracellular ST6GAL1.
Discussion
Emerging data from recent years have pointed to the importance of circulating extracellular ST6GAL1 in diverse physiologic events in blood cell production, inflammation, and in the maintenance of the anti-inflammatory epitope in the Fc glycan of immunoglobulins (Jones et al. 2012, 2016; Nasirikenari et al. 2014; Dougher et al. 2017; Irons et al. 2019, 2020). In fact, ST6GAL1 is one of several hallmark genes upregulated during the inflammatory acute phase response (Baumann and Gauldie 1994). However, mechanistic insight on how extracellular ST6GAL1 acts on target cells has been lacking. This report is based on our earlier observation that recombinant ST6GAL1 infusion diminished inflammatory cytokine release from airway macrophages from an in vivo mouse model of acute airway inflammation (Nasirikenari et al. 2019). Our current results implicate extracellular ST6GAL1 in adhering to monocytes, catalytically modifying cell surfaces by transfer of α2–6 linked sialic acids and initiating an intracellular cascade of signaling events resulting in wholesale changes in transcription driving monocyte differentiation and survival. We have shown that exogenous ST6GAL1 interacts with monocytes by a rapid and specific mechanism, which can operate directly at the cell surface. We have not explored the possibility that exogenous ST6GAL1 binds and undergoes endocytosis in live cells. An added possibility is that ST6GAL1 binding interactions occur cell autonomously, noncell autonomously, or both. Regardless of the exact mechanism, circulatory ST6GAL1 has potential major consequences for myeloid cell biology.
Data from earlier studies have shown the ability of extracellular sialyltransferase to extrinsically sialylate target cell surfaces (Lee-Sundlov et al. 2017; Manhardt et al. 2017). In vivo, extrinsic sialylation is dynamically regulated by systemic triggers that involve the release of the sialic acid donor substrate necessary for ST6GAL1-mediated catalysis. Here, using an ST6GAL1 fusion protein (GFP-rST6G), we observed attachment of an ST6GAL1 adduct to specific inflammatory cell subsets and to cultured human monocytes using the fluorescent GFP tag or antibodies against either GFP or ST6GAL1. These data demonstrate cell surface adherence of ST6GAL1 by (i) microscopic cell surface localization of GFP-rST6G on nonpermeabilized fixed primary mouse ST6GAL1-null cells after only 3 min of binding; (ii) microscopic cell surface localization of GFP-rST6G on live human THP-1 monocytes; (iii) the absence of significant GFP-rST6G degradation by Western blot analysis even after 24 h of binding; and (iv) the ability of proteinase K to remove the GFP-rST6G signal from intact fixed cells. Curiously, we also observed that similar GFP-adducts of other glycosyltransferases, ST3GAL1 (GFP-huST3G) and β4GalT1 (GFP-huβ4GalT1), did not bind to cells under identical conditions. This latter observation, currently under investigation, implicates an unknown mechanism directing ST6GAL1 but not all extracellular glycosyltransferases to the cell surface. The critical finding regarding the binding of exogenous ST6GAL1 with monocytes is that the reaction is rapid and specific and can occur directly at the surface. The possibility of binding through an endocytic pathway in live cells cannot be excluded. Additionally, it is possible that such interactions occur cell autonomously, non-cell autonomously, or both.
The concentration of ST6GAL1 used in our study ranges from 20 to 42 ug/mL, higher than the extracellular ng/mL range reported in serum (Myojin et al. 2021). However, the effective in vivo concentration of ST6GAL1 in the local microenvironment at a surface receptor may be much higher than the average serum level. Additional possible factors for the requirement of higher amounts of recombinant enzyme might be the absence of still unknown enhancing co-factors and delivery mechanisms normally present in vivo, and it is these factors which could lower the effective dosage of ST6GAL1. For example, nanoparticle exomeres deliver membrane-anchored and cleaved, soluble ST6GAL1 to recipient cells where they sialylate β1-intergrin (Zhang et al. 2019).
Engagement of extracellular ST6GAL1 with primary mouse inflammatory monocyte–macrophage lineage cells resulted in profound changes in transcription profiles within 24 h (see Fig. 2A). We hypothesize that sialyltransferase recruitment to the cell surface initiates a signaling cascade beginning with cell surface sialylation ultimately driving changes in gene expression favoring monocyte/macrophage and monocyte/dendritic cell development. Curiously, we observed that rST6G alone, without addition of the canonical donor substrate CMP-sialic acid, was sufficient to elicit these transcriptional changes (see Fig. 2A). We had previously demonstrated that platelets contain a significant cargo of sialic acid donor substrate sufficient to drive ST6GAL1-mediated sialylation (Lee et al. 2014). Moreover, dying and perhaps healthy cells also leak their intracellular content of sugar nucleotides (Hoflack et al. 1979). To assess whether monocytes in culture were able to supply sufficient sialic acid donor substrate to the exogenous enzyme, we observed that rST6G efficiently sialylated the surface of mouse monocyte/macrophage-like RAW264.7 cells without requiring addition of CMP-sialic acid (see Fig. 2B). The RAW264.7 cells were lightly fixed and nonpermeabilized to minimize internalization of the exogenously added rST6G.
In human monocyte THP-1 cells, we observed robust sialylation of the mature M-CSF receptor in the presence of exogenously added rST6G (see Fig. 5). M-CSF-R is implicated in monocyte–macrophage differentiation, and we hypothesize that its sialylation elicits an intracellular signaling cascade driving monocyte–macrophage differentiation, activation, and survival. In support of this hypothesis, we show that rST6G-mediated sialylation of M-CSF-R and phosphorylation are accompanied by enhanced P-AKT and P-ERK formation, and by nuclear translocation of the p65 subunit of NFκB (see Fig. 6). Consistent with a pro-survival/anti-apoptotic impact due to nuclear translocation of p65, exogenous rST6GAL1 yielded a significant increase in the total fraction of live primary monocytes in culture (see Fig. 4). Although we cannot definitively conclude that the observed transcriptional changes are brought about directly by changes in the M-CSF-R, our observed activation of the PU.1 and related intracellular signal pathways associated with the extracellular M-CSF-R do strongly suggest this is how extracellular ST6GAL1 affects monocyte survival and differentiation. The mechanistic details by which sialylation of M-CSF-R might serve to sensitize M-CSF-R recognition by its putative ligand M-CSF, promote activation in the absence of ligand, or both, remain to be determined. Receptor tyrosine kinases including M-CSF-R undergo auto-phosphorylation brought about by conformational change (Du and Lovly 2018) suggesting that sialic acid glycan modification might cause altered M-CSF-R conformation and activation without added ligand. While our observations indicate that ST6GAL1-induced intracellular signaling occurs concomitantly with M-CSF-R sialylation and phosphorylation, our data do not exclude the possible involvement of other cell surface molecules. Indeed, M-CSF-R is one among many THP-1 cell surface components modified by rST6G, and all these remain to be formally and exhaustively identified and analyzed. Moreover, there is an intriguing possibility that ST6GAL1 engagement at the cell surface is sufficient to elicit intracellular signaling, which may occur without sialylation of the cell surface (Fig. 2A, see the similarity of the Heatmap for rST6 ± CMP-sialic acid; Fig. 2B, see surface SNA reactivity with rST6G alone; Fig. 6, see “+rST6G lanes”). The idea of extracellular glycosyltransferases such as ST6GAL1 directly functioning in receptor–counter–receptor signaling, such as in cellular adhesiveness, which was originally proposed by Saul Roseman (Roseman 1985) remains to be discounted.
Whatever the intermediate mechanisms, engagement of extracellular ST6GAL1 clearly drives a profound set of transcriptional changes manifest in pathways driving monocyte/macrophage and monocyte/dendritic cell differentiation, as well as gene signatures for M0, M1, and M2 macrophages (see Fig. 3). The most highly upregulated transcription factors identified, PU.1 (Spi1) and NFκB are the primary drivers of differentiation along these lineages and GSEA analyses support their impact on these pathways (see Fig. 3C). These data implicate an extracellular ST6GAL1-mediated gene expression program with fundamental consequences for myeloid cell development.
Elevated circulating ST6GAL1 is frequently associated with inflammatory states. Alternatively, α2,6 sialic acid is an anti-inflammatory indicator based on glycan profiles of polarized THP-1 and RAW264.7 cells where it is enhanced in the M2 anti-inflammatory phenotype and decreased for the M1 inflammatory state (Yang et al. 2021). Our observations differ from this slightly, and suggest that ST6GAL1 engagement promotes transcriptional activation not only in M2 but along all the major monocyte/macrophage/dendritic cell pathways (see Fig. 3A and B). While the precise mechanism(s) regulating pro- and anti-inflammatory responses to extracellular ST6GAL1 remain unclear, our current observations indicate that elevated extracellular ST6GAL1 has the capacity to set in motion a program of differentiation of monocytes into functional macrophages and dendritic cells bridging the gap between innate and adaptive immunity. Together with the other known correlations of circulatory ST6GAL1 in muting of inflammation and in promoting B cell maturation while suppressing granulocyte production, we speculate that circulatory ST6GAL1 might be a systemic signal coordinating the transition from inflammation to more productive immune responses.
Materials and methods
Animals
Mouse strains used were control C57BL/6 J and germline knockout strain St6gal1KO (Hennet et al. 1998) backcrossed 15 generations onto a C57BL/6 J background and maintained at Roswell Park’s Laboratory Animal Shared Resource facility. The Animal Care and Use Committee of Roswell Park Comprehensive Cancer Center approved all animal studies reported here.
Recombinant proteins, chemicals, and cell lines
Highly purified, acceptor-specific recombinant glycosyltransferases rST6GAL1 and fluorescent conjugates rST6GAL1-GFP, huST3GAL1-GFP, and huβ4GAL-T1-GFP were generated as previously described (Moremen et al. 2018). Homogeneity and acceptor specificity are documented in Supplementary Figure 1 and Supplementary Table 1. Sugar donor CMP-sialic acid disodium salt was purchased from Millipore Sigma, #233264 and CMP-sialic acid-biotin was generated as previously described (Sun et al. 2016). Mouse monocyte/macrophage cell line RAW264.7 was obtained from ATCC and maintained in DMEM with 10% FBS. Human monocyte THP-1 cells from ATCC were cultured in RPMI containing 10% FBS.
Binding of purified recombinant glycosyltransferase-GFP conjugates with fixed peritoneal lavage cells from the St6gal1-null and C57BL/6 wild-type mouse
Brewer’s yeast thioglycolate (Becton Dickinson Microbiology, Baltimore, MD) was injected intraperitoneally (1 mL, 4% sterile solution) to elicit leukocyte recruitment to the peritoneum and mice were sacrificed for collection of peritoneal lavage fluid 18 h later. Erythrocytes were removed by Ammonium–Chloride–Potassium (ACK) lysis and cells fixed in 2% paraformaldehyde (Electron Microscopy Sciences) in DPBS for 15 min at room temperature (RT). Fixed cells in DPBS were incubated 3 or 30 min at RT with 25 μg/mL recombinant GFP proteins and then washed once with DPBS. Proteinase K digestion (250 μg/mL, Invitrogen #25530049) for 5 min at RT was performed on an aliquot of the rST6GAL1-GFP sample followed by one DPBS wash. Approximately 2–5 × 10(5) cells were applied to a 1 cm spot encircled with a hydrophobic pen on Gold Plus slides (Electron Microscopy Sciences) for 1 h at RT and cells remaining after aspiration were mounted with Fluoroshield with DAPI (Sigma-Aldrich #F6057) for imaging on a Nikon Eclipse E600 fluorescence microscope.
Treatment of thioglycolate-recruited peritoneal lavage cells from the St6gal1KO mouse and RNA isolation for RNA-seq
Thioglycolate injection and preparation of peritoneal lavage fluid is described above. Monocytes/macrophages were enriched from lavage fluid using magnetic bead (BioMag goat anti-rat IgG, Qiagen #310107) depletion of antibody-bound B cells (Biotin rat anti-mouse CD19, BD Biosciences #553784; Biotin anti-mouse/human CD45R/B220 BioLegend #103204), neutrophils (Biotin anti-mouse Ly-6G, BioLegend #127604), lymphocytes (Biotin anti-mouse CD3, BioLegend #100244; Biotin anti-mouse CD5, BioLegend #100604) and erythroid cells (Biotin anti-mouse TER119, BioLegend #116204). To prepare cells for RNA-seq, the depleted cell population was cultured in DMEM with 10% FBS for approximately 24 h under 3 conditions: mock buffer (0.2 M NaCl, 0.02 M HEPES, 10% glycerol, pH 7.2), CMP-Sialic acid (100 μM), recombinant rST6GAL1 (25 μg/mL), and the combination of CMP-Sia and rST6GAL1 at the above concentrations. Triplicate culture wells were used for each of the 3 conditions. Following the 24 h incubation, cell culture medium was completely removed by aspiration and lysis solution directly added to adherent cells for total RNA isolation using the RNeasy Plus Micro Kit from Qiagen (Ref 74034). Details of the RNA-seq procedure are included in Supplementary Method for RNA-seq.
Analysis of bulk RNAseq data
Raw fastq sequences were aligned to the Mus musculus GRCm38.p4 reference genome (GENCODE M10 annotation) using STAR to allow for spliced alignments (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3530905/,https://pubmed.ncbi.nlm.nih.gov/30357393/). Data were assessed for quality using fastqc and RSeQC (https://pubmed.ncbi.nlm.nih.gov/22743226/). Reads were quantified at the gene level for each sample using the featureCounts function from Subread (https://pubmed.ncbi.nlm.nih.gov/24227677/). Gene level read counts were imported into R and samples were normalized using DESeq2 (https://pubmed.ncbi.nlm.nih.gov/25516281/). Gene level differential expression tests between treatment groups were calculated using Wald tests and P-values were adjusted using the Benjamini-Hochberg method. Preranked gene set enrichment analyses were calculated using the Wald test statistic provided by DESeq2 as the gene ranking metric (https://pubmed.ncbi.nlm.nih.gov/16199517/). The Broad Institute’s Molecular Signatures Database (MolSigDB) C2: Canonical Pathways and C3: Transcription Factor Targets databases were used to obtain gene sets for testing (https://pubmed.ncbi.nlm.nih.gov/21546393/). GSEA was conducted using fgsea and facilitated by clusterProfiler (https://pubmed.ncbi.nlm.nih.gov/22455463/). Cell type abundances within bulk RNAseq samples were estimated using CIBERSORTx (https://pubmed.ncbi.nlm.nih.gov/31960376/). Pure cell population microarray RNA expression data from the Gene Expression Commons were used as a reference for the CIBERSORTx composition analysis (https://pubmed.ncbi.nlm.nih.gov/22815738/).
Quantitative PCR
Thioglycolate-induced peritoneal inflammatory cells from the St6gal1KO mouse were lineage-depleted and cultured for RNA isolation as for the RNA-seq study. cDNA was prepared with the iScript cDNA Synthesis Kit (BIO-RAD # 1708890) and quantitative PCR carried out using iTaq Universal SYBR Green Super Mix (BIO-RAD # 1725121). Gene expression relative to mβ2-microglobulin was calculated by the 2-(ΔΔCT) method (Livak and Schmittgen 2001; Schmittgen and Livak 2008). PCR primer sequences are listed in Supplementary Table 2.
SNA reactivity detection by immunofluorescence
RAW264.7 cells were grown on sterile coverslips overnight in DMEM with 10% FBS, fixed in 5% formalin, treated with sialidase (20 U/mL, NEB P0722L) 2 h at 37 °C and incubated 2 h at 37 °C with mock buffer alone (0.2 M NaCl, 0.02 M HEPES, 10% glycerol, pH 7.2), CMP-sialic acid (250 μM), rST6GAL1 (20 μg/mL), or the 2 combined. Samples were stained with SNA-FITC (Vector Labs # FL-1301) and imaged on a Nikon Eclipse E600 fluorescence microscope.
Flow cytometry
Cells were resuspended in 1 mM EDTA with 0.05% BSA, 0.02% azide/DPBS. Nonspecific Fc receptor binding was blocked with CD16/32 IgG (anti-mouse CD16/32 BioLegend #101302) and DAPI staining provided live/dead cell discrimination. Cell populations were identified as myeloid (BV711 anti-mouse/human CD11b BioLegend #101242), macrophage (PE anti-mouse F4/80 BioLegend #123110) or monocyte (PE/Cyanine7 anti-mouse Ly-6C BioLegend #128018) with a LSRII-A Becton-Dickinson flow cytometer and data analysis using FlowJo software.
Selective exoenzymatic labeling
Sialylation of the M-CSF-R in THP-1 monocytes by rST6GAL1 was demonstrated using the SEEL approach (Sun et al. 2016). In this method, glycosylation targets are labeled with sugar donor CMP–sialic acid–biotin conjugate, immunoprecipitated with anti-biotin IgG and identified by Western blotting with anti-M-CSF-R. Briefly, THP-1 cells were incubated in serum-free medium for 2 h and then treated 2 h under serum-free conditions in a 37 °C CO2 incubator in open-capped Eppendorf tubes containing 300 μL rST6GAL1 (42 μg/mL), CMP–sialic acid–biotin (100 μM), BSA (13 μg/mL) and alkaline phosphatase (0.7 U/mL, Fast AP, Thermo EF0651). Cell lysis and immunoprecipitation were carried out as previously described (Hait et al. 2020). Antibodies for immunoprecipitation and Western blot identification of M-CSF-R are listed below under Western blot analysis.
Analysis of cell signaling in THP-1 cells
THP-1 cells [3 × 10(6)/mL] were grown 24 h in serum-free RPMI 1640 containing 0.05 mM β-mercaptoethanol. Cells were treated with vehicle alone (mock), CMP-Sia (10 μM), and rST6G (100 ng/mL) for 30 min, then harvested, washed for isolation of total protein extract or nuclear and nuclei-free cytosolic fractions. Protein lysates were run on Western blots (below) and experiments were repeated at least 3 times. P-65 and p65 blot bands were normalized to the respective loading controls (NIH Image J).
Western blot analysis
After resolution on 10% SDS-PAGE, the products were transferred to PVDF Immobilon membranes for Western blot analysis described previously (Hait et al. 2020). Briefly, membranes were blocked with 5% nonfat dry milk (Bio-Rad, # 170-G404) in Tris-buffered saline (TBS) containing 0.1% of Tween 20 for 1 h at RT. The following primary antibodies were used for SEEL pulldown and immunoblot analysis. Antibodies for SEEL pulldown assays: mouse monoclonal anti-biotin IgG (Jackson ImmunoResearch # 200-002-211). Primary antibodies for Western blot analysis: Goat anti-human ST6GAL1 (R&D Systems #AF5924), GFP (4B10) mouse mAb (Cell Signaling # 2955), rabbit polyclonal anti-M-CSF-R IgG (Cell Signaling # 3152), rabbit mAb NF-κB p65 (Cell Signaling # 8242), rabbit mAb Phospho-NF-κB p65 (Ser536, Cell Signaling # 3033), Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (E10) mouse mAb #9106, rabbit mAb Phospho-Akt (Ser473, Cell Signaling # 4060), β-actin mouse mAb (Santa Cruz sc-47,778), GAPDH (Cell Signaling #5174), and histone H3 (Cell Signaling #4499). Secondary antibodies were HRP-conjugated rabbit anti-goat IgG (R&D Systems #HAF109), HRP-conjugated goat anti-rabbit IgG (#111-035-045), and goat anti-mouse IgG (#115-035-062) from Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA. For some experiments, HRP-conjugated goat anti-rabbit IgG (Invitrogen #A16096) secondary antibodies were used. Protein bands were detected using Pierce ECL Western Blotting Substrate (ThermoFisher Scientific #32209). The membranes were imaged by exposing them to X-ray films (Thomas Scientific, LLC # 1141J51).
Statistical analysis
Statistical tests for were performed with GraphPad Prism 8 software. P < 0.05 was considered statistically significant (P < 0.05*, P < 0.01**, and P < 0.001***).
Supplementary Material
Acknowledgements
We would like to acknowledge the expert technical assistance of Valerie Andersen and Mary Kay Ellsworth.
Contributor Information
Michael E Rusiniak, Department of Molecular and Cellular Biology, Roswell Park Comprehensive Cancer Center, Elm and Carlton Streets, Buffalo, NY 14263, United States.
Patrick R Punch, Department of Molecular and Cellular Biology, Roswell Park Comprehensive Cancer Center, Elm and Carlton Streets, Buffalo, NY 14263, United States; Department of Oral Biology, University at Buffalo, Buffalo, NY 14215, United States.
Nitai C Hait, Department of Molecular and Cellular Biology, Roswell Park Comprehensive Cancer Center, Elm and Carlton Streets, Buffalo, NY 14263, United States; Department of Surgical Oncology, Roswell Park Comprehensive Cancer Center, Elm and Carlton Streets, Buffalo, NY 14263, United States.
Aparna Maiti, Department of Surgical Oncology, Roswell Park Comprehensive Cancer Center, Elm and Carlton Streets, Buffalo, NY 14263, United States.
Robert T Burns, Translational Glycomics Center, Blood Research Institute, 8727 W. Watertown Plank Rd, Milwaukee, WI 53226, United States.
Digantkumar Chapla, Complex Carbohydrate Research Center, University of Georgia, Athens, GA 30602, United States.
Kelley W Moremen, Complex Carbohydrate Research Center, University of Georgia, Athens, GA 30602, United States.
Peng Zhao, Complex Carbohydrate Research Center, University of Georgia, Athens, GA 30602, United States.
Lance Wells, Complex Carbohydrate Research Center, University of Georgia, Athens, GA 30602, United States.
Karin Hoffmeister, Translational Glycomics Center, Blood Research Institute, 8727 W. Watertown Plank Rd, Milwaukee, WI 53226, United States.
Joseph T Y Lau, Department of Molecular and Cellular Biology, Roswell Park Comprehensive Cancer Center, Elm and Carlton Streets, Buffalo, NY 14263, United States.
Funding
This work was supported by National Institute of Allergy and Infectious Diseases R01AI140736 and National Heart, Lung, and Blood Institute P01HL151333 to JTYL; and National Institute of General Medical Sciences P41GM103390 and R01GM130915 to KWM. The core facilities of Roswell Park Comprehensive Cancer Center used in this work, including the Genomics Shared Resource and Flow and Image Cytometry Shared Resource were supported in part by NIH National Cancer Institute Cancer Center Support Grant 5P30 CA016056.
Conflicts of interest statement: The authors declare that they have no conflicts of interest with the contents of this article.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files. The raw RNA-seq data will be uploaded to NCBI GEO and SRA upon the acceptance of the manuscript under the accession GSEXXXXX.
Ethics approval
This manuscript does not report studies involving human participation, human data, or human tissues.
Abbreviations
ST6GAL1: ST6 beta-galactoside alpha-2,6-sialyltransferase 1; rST6G: recombinant rat protein ST6 beta-galactoside alpha-2,6-sialyltransferase 1; PU.1: transcription factor PU.1 (SPi1); NFκB: transcription factor NB; M-CSF-R: macrophage colony-stimulating factor receptor; THP-1: human THP-1 monocyte cell line; RAW264.7; murine monocyte/macrophage cell line; p65: subunit of the transcription factor NFκB; p-ERK 1/2: phosphorylated extracellular signal-regulated kinase; p-AKT: phosphorylated Serine/Threonine protein kinase or phosphorylated protein kinase B.
References
- Baumann H, Gauldie J. The acute phase response. Immunol Today. 1994:15:74–80. [DOI] [PubMed] [Google Scholar]
- Bornhofft KF, Goldammer T, Rebl A, Galuska SP. Siglecs: a journey through the evolution of sialic acid-binding immunoglobulin-type lectins. Dev Comp Immunol. 2018:86:219–231. [DOI] [PubMed] [Google Scholar]
- Britain CM, Holdbrooks AT, Anderson JC, Willey CD, Bellis SL. Sialylation of EGFR by the ST6Gal-I sialyltransferase promotes EGFR activation and resistance to gefitinib-mediated cell death. J Ovarian Res. 2018:11:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WS, Cao Z, Leffler H, Nilsson UJ, Panjwani N. Galectin-3 inhibition by a small-molecule inhibitor reduces both pathological corneal neovascularization and fibrosis. Invest Ophthalmol Vis Sci. 2017:58:9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Baldwin TM, Wong M, Bolden JE, Fairfax KA, Lucas EC, Cole R, Biben C, Morgan C, Ramsay KA et al. Haemopedia RNA-seq: a database of gene expression during haematopoiesis in mice and humans. Nucleic Acids Res. 2019:47:D780–D785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaine T, Collins P, MacKinnon A, Sharma G, Stegmayr J, Rajput VK, Mandal S, Cumpstey I, Larumbe A, Salameh BA et al. Galectin-3-Binding glycomimetics that strongly reduce bleomycin-induced lung fibrosis and modulate intracellular glycan recognition. Chembiochem. 2016:17:1759–1770. [DOI] [PubMed] [Google Scholar]
- Dorsett KA, Marciel MP, Hwang J, Ankenbauer KE, Bhalerao N, Bellis SL. Regulation of ST6GAL1 sialyltransferase expression in cancer cells. Glycobiology. 2021:31:530–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougher CWL, Buffone A Jr, Nemeth MJ, Nasirikenari M, Irons EE, Bogner PN, Lau JTY. The blood-borne sialyltransferase ST6Gal-1 is a negative systemic regulator of granulopoiesis. J Leukoc Biol. 2017:102:507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Lovly CM. Mechanisms of receptor tyrosine kinase activation in cancer. Mol Cancer. 2018:17:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnham R, Scott E, Livermore KE, Munkley J. ST6GAL1: a key player in cancer. Oncol Lett. 2019:18:983–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hait NC, Maiti A, Xu P, Qi Q, Kawaguchi T, Okano M, Takabe K, Yan L, Luo C. Regulation of hypoxia-inducible factor functions in the nucleus by sphingosine-1-phosphate. FASEB J. 2020:34:4293–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennet T, Chui D, Paulson JC, Marth JD. Immune regulation by the ST6Gal sialyltransferase. Proc Natl Acad Sci U S A. 1998:95:4504–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirani N, MacKinnon AC, Nicol L, Ford P, Schambye H, Pedersen A, Nilsson UJ, Leffler H, Sethi T, Tantawi S et al. Target inhibition of galectin-3 by inhaled TD139 in patients with idiopathic pulmonary fibrosis. Eur Respir J. 2021:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoflack B, Cacan R, Montreuil J, Verbert A. Detection of ectosiallyltransferase activity using whole cells. Correction of misleading results due to the release of intracellular CMP-N-acetylneuraminic acid. Biochim Biophys Acta. 1979:568:348–356. [DOI] [PubMed] [Google Scholar]
- Holdbrooks AT, Britain CM, Bellis SL. ST6Gal-I sialyltransferase promotes tumor necrosis factor (TNF)-mediated cancer cell survival via sialylation of the TNF receptor 1 (TNFR1) death receptor. J Biol Chem. 2018:293:1610–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdbrooks AT, Ankenbauer KE, Hwang J, Bellis SL. Regulation of inflammatory signaling by the ST6Gal-I sialyltransferase. PLoS One. 2020:15:e0241850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons EE, Lau JTY. Systemic ST6Gal-1 Is a pro-survival factor for murine transitional B cells. Front Immunol. 2018:9:2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons EE, Lee-Sundlov MM, Zhu Y, Neelamegham S, Hoffmeister KM, Lau JT. B cells suppress medullary granulopoiesis by an extracellular glycosylation-dependent mechanism. Elife. 2019:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons EE, Punch PR, Lau JTY. Blood-borne ST6GAL1 regulates immunoglobulin production in B cells. Front Immunol. 2020:11:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L, Jacob R, Leffler H. Galectins at a glance. J Cell Sci. 2018:131. [DOI] [PubMed] [Google Scholar]
- Jones MB, Nasirikenari M, Lugade AA, Thanavala Y, Lau JT. Anti-inflammatory IgG production requires functional P1 promoter in beta-galactoside alpha2,6-sialyltransferase 1 (ST6Gal-1) gene. J Biol Chem. 2012:287:15365–15370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MB, Oswald DM, Joshi S, Whiteheart SW, Orlando R, Cobb BA. B-cell-independent sialylation of IgG. Proc Natl Acad Sci U S A. 2016:113:7207–7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremsreiter SM, Kroell AH, Weinberger K, Boehm H. Glycan-lectin interactions in cancer and viral infections and how to disrupt them. Int J Mol Sci. 2021:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubli H, Varki A. Sialic acid-binding immunoglobulin-like lectins (Siglecs) detect self-associated molecular patterns to regulate immune responses. Cell Mol Life Sci. 2020:77:593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Nasirikenari M, Manhardt CT, Ashline DJ, Hanneman AJ, Reinhold VN, Lau JT. Platelets support extracellular sialylation by supplying the sugar donor substrate. J Biol Chem. 2014:289:8742–8748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Sundlov MM, Ashline DJ, Hanneman AJ, Grozovsky R, Reinhold VN, Hoffmeister KM, Lau JT. Circulating blood and platelets supply glycosyltransferases that enable extrinsic extracellular glycosylation. Glycobiology. 2017:27:188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Liu S, Lu M, Bandyopadhyay G, Oh D, Imamura T, Johnson AMF, Sears D, Shen Z, Cui B et al. Hematopoietic-derived galectin-3 causes cellular and systemic insulin resistance. Cell. 2016:167:973, e912–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001:25:402–408. [DOI] [PubMed] [Google Scholar]
- Manhardt CT, Punch PR, Dougher CWL, Lau JTY. Extrinsic sialylation is dynamically regulated by systemic triggers in vivo. J Biol Chem. 2017:292:13514–13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer SJ, Linder AT, Brandl C, Nitschke L. B cell Siglecs-news on signaling and its interplay with ligand binding. Front Immunol. 2018:9:2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen KW, Ramiah A, Stuart M, Steel J, Meng L, Forouhar F, Moniz HA, Gahlay G, Gao Z, Chapla D et al. Expression system for structural and functional studies of human glycosylation enzymes. Nat Chem Biol. 2018:14:156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myojin Y, Kodama T, Maesaka K, Motooka D, Sato Y, Tanaka S, Abe Y, Ohkawa K, Mita E, Hayashi Y et al. ST6GAL1 is a novel serum biomarker for lenvatinib-susceptible FGF19-driven hepatocellular carcinoma. Clin Cancer Res. 2021:27:1150–1161. [DOI] [PubMed] [Google Scholar]
- Nasirikenari M, Segal BH, Ostberg JR, Urbasic A, Lau JT. Altered granulopoietic profile and exaggerated acute neutrophilic inflammation in mice with targeted deficiency in the sialyltransferase ST6Gal I. Blood. 2006:108:3397–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasirikenari M, Chandrasekaran EV, Matta KL, Segal BH, Bogner PN, Lugade AA, Thanavala Y, Lee JJ, Lau JT. Altered eosinophil profile in mice with ST6Gal-1 deficiency: an additional role for ST6Gal-1 generated by the P1 promoter in regulating allergic inflammation. J Leukoc Biol. 2010:87:457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasirikenari M, Veillon L, Collins CC, Azadi P, Lau JT. Remodeling of marrow hematopoietic stem and progenitor cells by non-self ST6Gal-1 sialyltransferase. J Biol Chem. 2014:289:7178–7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasirikenari M, Lugade AA, Neelamegham S, Gao Z, Moremen KW, Bogner PN, Thanavala Y, Lau JTY. Recombinant sialyltransferase infusion mitigates infection-driven acute lung inflammation. Front Immunol. 2019:10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata S, Shimizu C, Franco A, Touma R, Kanegaye JT, Choudhury BP, Naidu NN, Kanda Y, Hoang LT, Hibberd ML et al. Treatment response in Kawasaki disease is associated with sialylation levels of endogenous but not therapeutic intravenous immunoglobulin G. PLoS One. 2013:8:e81448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan JA, Chang AT, Youngblood BA, Bochner BS. Eosinophil and mast cell Siglecs: from biology to drug target. J Leukoc Biol. 2020:108:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman S. Studies on specific intercellular adhesion. J Biochem. 1985:97:709–718. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008:3:1101–1108. [DOI] [PubMed] [Google Scholar]
- Stanley ER, Chitu V. CSF-1 receptor signaling in myeloid cells. Cold Spring Harb Perspect Biol. 2014:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers KM, Bush SJ, Hume DA. Network analysis of transcriptomic diversity amongst resident tissue macrophages and dendritic cells in the mouse mononuclear phagocyte system. PLoS Biol. 2020:18:e3000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Yu SH, Zhao P, Meng L, Moremen KW, Wells L, Steet R, Boons GJ. One-step selective exoenzymatic labeling (SEEL) strategy for the biotinylation and identification of glycoproteins of living cells. J Am Chem Soc. 2016:138:11575–11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvaroska I, Selvaraj C, Koca J. Selectins—the two Dr. Jekyll and Mr. Hyde faces of adhesion molecules—a review. Molecules. 2020:25:2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhang Q, Lin L, Xu Y, Huang Y, Hu Z, Wang K, Zhang C, Yang P, Yu H. Microarray investigation of glycan remodeling during macrophage polarization reveals alpha2,6 sialic acid as an anti-inflammatory indicator. Mol Omics. 2021:17:565–571. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Higginbotham JN, Jeppesen DK, Yang YP, Li W, McKinley ET, Graves-Deal R, Ping J, Britain CM, Dorsett KA et al. Transfer of functional cargo in exomeres. Cell Rep. 2019:27:940, e946–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. The raw RNA-seq data will be uploaded to NCBI GEO and SRA upon the acceptance of the manuscript under the accession GSEXXXXX.




