Abstract
This study evaluated the cytotoxic activity of Tamoxifen (TMX), an anti-estrogen drug, with microalgal crude extracts (MCEs) in single and synergistic application (TMX-MCEs) on MCF-7 and 4T1 breast cancer cells, and non-cancerous Vero cells. The MCEs of Nannochloropsis oculata, Tetraselmis suecica and Chlorella sp. from five different solvents (methanol, MET; ethanol, ETH; water, W; chloroform, CHL; and hexane, HEX) were developed. The TMX-MCEs-ETH and W at the 1:2 and 1:3 ratios, attained IC50 of 15.84–29.51 μg/mL against MCF-7; 13.8–31.62 μg/mL against 4T1; and 24.54–85.11 μg/mL against Vero cells. Higher late apoptosis was exhibited against MCF-7 by the TMX-N. oculata-ETH (41.15 %); and by the TMX-T. suecica-ETH (65.69 %) against 4T1 cells. The TMX-T. suecica-ETH also showed higher ADP/ATP ratios, but comparable Caspase activities to control. For Vero cells, overall apoptotic effects were lowered with synergistic application, and only early apoptosis was higher with TMX-T. suecica-ETH but at lower levels (29.84 %). The MCEs-W showed the presence of alanine, oleic acid, linoleic acid, lactic acid, and fumaric acid. Based on Principal Component Analysis (PCA), the spectral signals for polar solvents such as MET and ETH, were found in the same cluster, while the non-polar solvent CHL was with HEX, suggesting similar chemical profiles clustered for the same polarity. The CHL and HEX were more effective with N. oculata and T. suecica which were of the marine origin, while the ETH and MET were more effective with Chlorella sp., which was of the freshwater origin. The synergistic application of microalgal bioactive compounds with TMX can maintain the cytotoxicity against breast cancer cells whilst reducing the toxicity against non-cancerous Vero cells. These findings will benefit the biopharmaceutical, and functional and healthy food industries.
Keywords: Cytotoxicity, 1H NMR metabolomics, Tamoxifen, Microalgal extracts, Synergistic application
Cytotoxicity; 1H NMR metabolomics; Tamoxifen; Microalgal extracts; Synergistic application.
1. Introduction
The conventional treatments of breast cancer, as of today, face some drawbacks attributable to the associated side effects of the drugs, limited drug concentration and loss of specificity at the cancer site, and the development of chemo-resistance (Fanciullino et al., 2013; Singh et al., 2013; Hosseinia et al., 2017). Among the drugs commonly used to treat breast cancer are tamoxifen (TMX) (an anti-estrogen, hormone therapy drug), doxorubicin (a cytotoxic antibiotic), and paclitaxel (an anti-microtubule agent). These chemotherapy drugs are widely used to promote estrogen-dependent programmed breast cancer cell death (Cuzick et al., 2011). Paclitaxel causes mitotic arrest by stabilizing the cellular microtubule elements. Doxorubicin is used in a combination regimen where the cytotoxic activities have been attributed to free radical mechanism, lipid peroxidation, and direct membrane effects (Anjum et al., 2017). TMX or trans-1-[p-b-(dimethyl-amino) ethoxy- phenyl]1,2-diphenyl-1-butene is a substituted trans-isomer of triphenylethylene (Morrow and Jordan, 2000; Jordan, 2006), and a common drug to treat estrogen receptor positive breast cancer (ER+ve) (Jones and Buzdar, 2004), estrogen-receptor-negative breast cancers (ER-ve), and other unrelated cancers (Sun et al., 2012). TMX interacts strongly with bio-membranes, lipids and proteins, because of its lipophilic characteristic. The mitochondrial bioenergetics can also be affected by TMX (Marek et al., 2011). The main mechanism involves the binding of TMX to the estrogen receptor and inhibition of the proliferative activities of estrogen on the mammary epithelium. TMX stimulates DNA adduct formation, cytostatic (G0/G1 arrest) and cytotoxic (induction of apoptosis) effects, and may target the checkpoint between the cell cycle progression and apoptosis (Petinari et al., 2004). With the use of TMX, the risk of ER+ve breast cancer return is reduced to 50%, and the morbidity rate decreased to 28 % (Anjum et al., 2017). However, patients treated with TMX have been reported to develop resistance and severe side effects. The long-term use of TMX can cause aneuploidy, endometrial and hepatic cancer (Petinari et al., 2004).
It is becoming imperative to search for new alternatives, strategies and formulations of anti-breast cancer agents (Senthilraja and Kathiresan, 2015). The new chemo-preventive drugs against breast cancer need to consider the safety and efficacy aspects to improve the therapeutics management and reduce the high cost and pain of patients (Steward and Brown, 2013). There has been an increasing interest to develop novel therapeutic strategies against cancer using drugs or cytotoxic agents based on natural compounds (Gurunathan et al., 2013; Hussein et al., 2020a,b,c,d). Natural-based products including benzylpenicillin, didemnin B, girolline, lovastatin, bryostatin 1, dolastatin 10, cryptophycins, kahalalide F and bengamide derivative are utilized in clinical trials for cancer, cognitive diseases, analgesia and allergy (Newman and Cragg, 2004). Natural compounds derived from marine resources have great potentials to be developed as important components of functional food (Abdullah et al., 2017) and biopharmaceuticals, either as drugs directly or indirectly as precursors for biochemical drug synthesis (Molinski et al., 2009). In the market, there are seven drugs which are marine-based and four of these are for anticancer. With nearly twenty-six marine-based natural products in clinical trials, twenty-three are for cancer therapeutics (Jaspars et al., 2016; Martínez Andrade et al., 2018).
There is a promising future for the applications of microalgae in pharmaceuticals, nutraceutical, cosmetics, functional food, and aquaculture, apart from the applications for bioenergy and environmental remediation in a biorefinery set-up (Abdullah et al., 2015, 2016, 2017; Abdullah and Hussein, 2021). Microalgae could enhance the host defence mechanism by enhancing the activity of the natural killer cells (Yuan and Walsh, 2006), activating the immune system (Schumacher et al., 2012), and inhibiting cancer cells and carcinogenesis (Liu et al., 2012; El-hack et al., 2019). The microalgal bioactive compounds are also effective against many pathogens (El-hack et al., 2019). The co-applications of microalgal metabolites with silver nanoparticles have been proven to enhance cytotoxic effects against breast cancer cells, but with reduced toxicity against the non-cancerous Vero cells (Hussein et al., 2020a,b,d). Combination therapy has shown good potential in the treatment of cancer where the cytotoxicity and therapeutic activity can be increased by utilizing multiple drugs with various mechanisms of action delivered to the same disease site (Gao et al., 2015). In the case of paclitaxel administration, combination therapy is effective in lowering the drug doses to avoid the side effects of high-dose in patients. It acts in a synergistic manner by strengthening the drugs and increasing the anticancer activity, whilst reducing the toxicity on normal cells (Malhão et al., 2021).
Metabolite levels in any biosystems indicate their regulatory processes and responses to genetic or environmental perturbations (Fiehn, 2002). The responses may vary in time and space which make analyzing a huge array of compounds and chemical structures a daunting task. Metabolomics is an unbiased analysis of complex metabolite mixtures, which allows for estimation and identification of individual metabolite. It has found application in functional genomics to establish the molecular interaction global network within a biosystem (Gomez-casati et al., 2013). Other areas include systems biology, pharmaceutical research, drug discovery, toxicology, food and nutrition science and early disease detection (Pan and Raftery, 2007). The two analytical methods commonly used in metabolomics are Nuclear Magnetic Resonance (NMR) Spectroscopy and Liquid Chromatography-Mass Spectrometry (LC-MS). The 1H NMR-based metabolomics is a non-targeted analysis to evaluate the correlation between specific metabolites in 1H NMR spectra of a source, such as the plant extracts, with the bioactivities such as anti-inflammation, antioxidant, and antidiabetic properties (Azizan et al., 2018).
The aims of this study were to develop a new formulation of TMX with Microalgal Crude Extracts (MCEs) in single or synergistic applications and to optimize the concentrations, the ratios, and the duration of treatments for cytotoxic effects on MCF-7 and 4T1 breast cancer cells, with reduced toxicities against the non-cancerous Vero cells. Morphological changes, apoptosis (Annexin-V flow cytometry), cell cycle arrest, ADP/ATP ratio, and caspase 3/7 activities were determined. The metabolite profiles of the microalgae extracted by solvents of different polarity (W, MET, ETH, CHL and HEX) were established and the profiles were correlated to the biological activities using 1H NMR-based metabolomics, Principal Component Analysis, and relative quantification method.
2. Material and methods
2.1. Cultivation and preparation of MCEs
The microalgal species used in this study, Nannochloropsis oculata, Tetraselmis suecica, and Chlorella sp., were identified by Dr. Mohd Fariduddin Othman, from the Fisheries Research Institute (FRI), Kuala Muda, Kedah, Malaysia. The molecular identification of N. oculata carried out using partial 18S rRNA sequence, partial rbc1 gene, and internal transcribed spacer (ITS) region determination, exhibited 97–99 % similarity to N. oculata, based on sequence alignment and phylogenetic tree analysis (Shah, 2014; Abdullah et al., 2021). The protocol for microalgal culture maintenance and extraction was as reported earlier (Hussein et al., 2020a). The TMRL Enrichment medium was prepared by adding 1 mL of each nutrient (Table S1) (AQUACOPs, 1984) into 960 mL of sea water, pH adjusted to 7.6–7.8, topped up to 1 L, filtered by 0.22–1 micron, and then autoclaved at 121 °C for 15 min. All work was carried out aseptically in a laminar flow cabinet. The cultures were shaken at 130 rpm on a shaker, at 28 ± 2 °C, under white fluorescent light. After 14–16 days of cultivation, the cells were harvested, and centrifuged (3500 rpm, 10 min). The cells were dried in the oven overnight (50 °C), and kept until use in a freezer (4 °C). The MCEs preparation was as reported before (Hussein et al., 2020a).
2.2. Preparation of TMX:MCEs ratios
The stocks (10 mg/mL) of TMX (Sigma, USA) and MCEs were prepared by dissolving in Dimethyl sulfoxide (DMSO). For single application, the MCEs and TMX were prepared between 3.125-100 μg/mL. For synergistic application, each stock solution of TMX and MCEs was mixed at 1:2, 1:3, 1:4 and 1:5 ratios to yield 100 μg/mL final concentration (TMX: MCEs, w/w) (Table S2).
2.3. Determination of cytotoxicity
2.3.1. MCF-7, 4T1 and Vero cell-lines
The MCF-7 cells (American Tissue Culture Collection, ATCC® HTB-22™) are human breast adenocarcinoma cell-line, the 4T1 (ATCC® CRL-2539™) cells are mammary carcinoma that can metastasize, and the non-cancerous Vero cells (ATCC® CCL-81™) are the cell-line derived from the African green monkey kidney epithelial cells. The MCF-7 and Vero cells were cultured in Minimum Essential Medium (MEM), supplemented with 10 % Fetal Bovine Serum (FBS), 1 % non-essential amino acid, 1 % sodium pyruvate, and 1 % penicillin-streptomycin. The 4T1 cell-line was maintained in Roswell Park Memorial Institute (RPMI)-1640 medium supplemented with 10 % FBS, 2 mM L-glutamine, 100 unit/mL penicillin and 100 mg/mL streptomycin. The cell-lines were grown in 75 cm3 and 25 cm3 culture flasks, at 37 °C in a humidified atmosphere, with 5 % CO2. The cells were washed with Phosphate Buffered Saline (PBS) at pH 7.4, and the used media were replenished with the fresh media every 2–3 days.
2.3.2. Cytotoxic assay
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was carried out as a measure of the dehydrogenation enzyme in the mitochondria of the viable cells capable of cleaving the tetrazolium rings of the pale yellow coloured MTT, into purplish formazan crystal (Prabaharan et al., 2009). The number of viable cells is directly proportional to the level of the formazan crystal formed (Janitabar-Darzi et al., 2017). The cytotoxic effects of TMX (Sigma-Aldrich) and MCEs in single and synergistic application were evaluated after 24 h. One hundred μL of 5 × 104, 2 × 104 and 4 × 104 cells/mL for MCF-7, 4T1 and Vero cells respectively, were pipetted into each well of the 96-well plate, and incubated overnight for growth. Later, the MCEs and TMX were prepared at 3.125–100 μg/mL, for single and synergistic applications (Table S2) where 100 μL was loaded to each well and incubated for 24 h. Then, 5 mg/mL of the MTT reagent was dispensed into the well, incubated, and solubilized. The sample absorbance (Abs) was measured spectrophotometrically using a microplate reader at 570 nm. The percentage of cytotoxicity was calculated as described previously (Mosmann, 1983) by using the following formula in Eq. (1):
| Cell viability (%) = [(OD sample – OD blank)/OD Control] × 100 | (1) |
where OD sample = Abs of the treated sample, OD blank = Abs of media + DMSO, and OD Control = Abs of non-treated sample. The half maximal inhibitory concentration (IC50) was determined by using Graph Pad Prism (Version 6, CA, USA).
2.3.3. Morphological characterization
The cell-lines were seeded for 24 h. The media later was changed to a fresh media containing TMX, and the MCEs at the IC50 levels, for 24 h treatment. The negative Control was the cells without any treatment. The cell morphology was analysed using an inverted microscope.
2.4. Flow Cytometry
2.4.1. Annexin V
The cell lines were cultured in 75 cm3 flasks for 24 h. The fresh media was then loaded with the TMX and MCEs treatment (including Control culture without treatment). After 24 h, the cells were washed with the PBS three times, and harvested by adding 1 mL of trypsin and incubated for 7–10 min, for cell detachment. The cells were rinsed with the media and then centrifuged (1000 rpm, 5 min). The cell pellet was then suspended in 1 mL complete media. The cells at 106 cell number were washed in 1 mL Binding buffer and centrifuged (300×g, 10 min). The pellet was re-suspended in 100 μL of 1× Binding Buffer per 106 cells. Ten μL of Annexin V-FITC was added per 106 cells, and incubated in the dark (15 min, room temperature). The cells were washed with 1 mL of 1× Binding Buffer per 106 cells and centrifuged (300×g, 10 min). The cell pellet was then re-suspended in 500 μL of 1× Binding Buffer per 106 cells. Finally, 5 μL of Propidium iodide (PI) solution was added before flow cytometric analysis was carried out. The phosphatidylserine (PS) levels in the untreated control cells (in Relative Fluorescence Unit (RFU)), were used as the baseline indicator for normal PS levels (Sarojini et al., 2016).
2.4.2. Cell -cycle analysis
The cells were harvested after 24 h treatment. The cell number was determined and the cells were washed one time at 1 × 106 cells per tube. Later, PBS (1 mL) was added and the tube was centrifuged (1200 rpm, 4 °C). The pellet was re-suspended in PBS buffer (0.3 mL), and the cells were then fixed gently by adding dropwise 0.7 mL of 70 % cold ethanol into 0.3 mL of the cell suspension in PBS, and then vortexed. The cells were kept in ice for 1 h or in a freezer (4 °C), and then centrifuged, washed with cold PBS, re-centrifuged, and finally the pellet re-suspended in PBS (0.25 mL). Five μL of 10 mg/mL RNAse A was added to give 0.2–0.5 mg/mL final concentration and the cell suspension was incubated (30 min, 37 °C). Thereafter, ten μL of PI solution (1 mg/mL) was added to yield 10 μg/mL final concentration, and the sample was kept at 4 °C, in the dark, prior to flowcytometric analysis.
2.5. Biomarkers
2.5.1. ADP/ATP ratio
The cell-lines were cultured at 104 cells/well in a 96-well plate and incubated with the treatment to induce apoptosis, while the control culture was without the treatment. After 24 and 48 h, the culture medium was removed. The Nucleotide Releasing Buffer (NRB) (50 μL Buffer per 103–104 cells) was added, and incubated at room temperature with gentle shaking for 5 min. The NRB assists to loosen up the cell membrane to allow the ATP to leak out without cell lysis.
The ATP Monitoring Enzyme was added with Enzyme Reconstitution Buffer (2.1 mL), mixed gently, and the Reaction Mix added (100 μL), before the background luminescence (Data A) was read. Fifty μL of cells (103–104 cells) were then pipetted into the luminescence plate reader (luminometer) (GloMax® System, Promega, USA) and the NRB was added. After 5 min, the sample in the luminometer (Data B) was read. After 10 min incubation at room temperature, the sample was again read (Data C). Later, the 10X ADP-Converting enzyme, which was diluted (10-fold) with the NRB, was added, and after 5 min, the sample was read (Data D). The data was analyzed as follows in Eq. (2):
| ADP/ATP ratio = [Data D – Data C]/[Data B – Data A] | (2) |
Data D = Sample signal 5 min after the addition of 10 μL 1X ADP Converting Enzyme to the cells.
Data C = Sample signal prior to the addition of 1X ADP Converting Enzyme to the cells.
Data B = Sample signal 5 min after the addition of cells to the reaction mix.
Data A = Background signal of the reaction mix.
2.5.2. Caspase 3/7
The cell-lines were cultured at 2 ×104 cells/well for 24 h in a white 96-well plate. The cells were treated at the IC50 values of the TMX and MCEs, for 24 and 48 h treatments. The cells and Caspase-Glo® 3/7 Reagent were equilibrated to room temperature, before Caspase-Glo® 3/7 Reagent (100 μL) was added into each well containing 100 μL of the blank, negative Control and treated cells in the culture medium. The plate was gently mixed (300–500 rpm) for 30 s, and incubated (30 min-3 h) at room temperature. The sample luminescence was read using the luminometer as described in the manufacturer's protocol.
2.6. NMR analyses
2.6.1. Sample preparation
Approximately 10 mg of the MCEs, each from five solvents - methanol (MET), chloroform (CHL), hexane (HEX), ethanol (ETH), and water (W), was dissolved in 700 μL deuterated dimethyl sulfoxide (DMSO-d6) containing 0.03 % tetramethylsilane (TMS) in a 2 mL Eppendorf tube, in 6 replicates. The mixture was vortexed (1 min) and ultrasonicated (15 min) to solubilize the extract, and then centrifuged (13000 rpm, 10 min). The supernatant (approximately 600 μL) was pipetted into a 5 mm NMR tube for analyses.
2.6.2. NMR spectra pre-processing and Multivariate Data Analysis
The 1H NMR spectra was obtained on a 400 MHz Advance Π NMR spectrometer (Bruker, Germany), operating at 400 MHz and 25 °C. For each sample, the following parameters were used:- 64 number of scans, 2 s relaxation delay (Olive and van Genderen, 2000; Fox et al., 2018), and 8.49 min acquisition time. The spectral width was adjusted from −2 to 14 ppm. To reduce water signals, the presaturation (PRESAT) pulse sequence was applied.
The 1H NMR spectra of all samples were pre-processed (phasing, baseline correction and alignment) and binned automatically to ASCII files using Chenomx software (version 6.2, Edmonton, AB, Canada) at 0.04 ppm bin size. The spectra at 0.50–10.00 ppm was bucketed into 232 integrated regions, and the spectral ranges at δ 2.75–3.33 ppm and δ 2.5 ppm, representing residual signals for water and DMSO, respectively, were excluded. Metabolite identification of all the MCEs was performed by comparing the chemical shifts in the 1H-NMR spectrum with the references available in the Chenomx compound library, Human Metabolome Databases (HMDB) and published literature data. The chemical shifts were also further checked and compared with the values in the literature. The standardized bucketed data were pareto-scaled (PAR) and analysed with the Principal Component Analysis (PCA), using SIMCA-P+ software (version 12.0.1.0, Umetrics AB, Umea, Sweden). The 1H NMR signals for the metabolites of interest were used for relative quantification and the statistical analysis was carried out using Graph Pad Prism (Version 6, CA, USA).
2.7. Statistical analysis
All experiments were carried out in triplicate and the results were expressed as the means ± standard deviation (SD). The data were analyzed using GraphPad Prism software (version 6, CA, USA). The IC50 values were calculated from a dose response curve after 24 h treatment, based on the non-linear regression using GraphPad Prism software (Version 6, CA, USA).
3. Results
3.1. Cytotoxicity of TMX-MCEs
Table 1 shows the cytotoxic effects of TMX-MCEs-ETH and W at the ratios of 1:2, 1:3, 1:4, and 1:5, as compared to CHL, after 24 h treatments. At the 1:2 ratio, the highest cytotoxicity against MCF-7 cells was exhibited by TMX-N. oculata-W (15.8 μg/mL), TMX-N. oculata and TMX-T. suecica-ETH (16.98 μg/mL); and TMX-T. suecica-W (19.1 μg/mL), as compared to TMX-Chlorella sp.-CHL (13.5 μg/mL). However for 4T1 cells, the TMX-T. suecica-ETH 1:2 (13.8 μg/mL), and TMX-T. suecica-ETH 1:3 (14.1 μg/mL) exhibited higher or comparable cytotoxicity to TMX-T. suecica-CHL (14.12 μg/mL). For Vero cells, the TMX-T. suecica-ETH 1:2 (24.5 μg/mL) and TMX-Chlorella sp.-ETH 1:2 (30.2 μg/mL) exerted cytotoxicity higher than or comparable to the TMX-T. suecica-CHL (30.2 μg/mL). In general, the TMX:MCEs-ETH formulation at the 1:2 and 1:3 ratios showed the IC50 values of 16.98–26.9 μg/mL on MCF-7; 13.8–20.4 μg/mL on 4T1; and 24.5–38.9 μg/mL on Vero cells. For W, the IC50 values were 15.8–29.5 μg/mL on MCF-7; 21.4–31.6 μg/mL on 4T1; and much lower against Vero cells lines at 42.7–85.1 μg/mL. The CHL extracts exhibited higher cytotoxic effects on 4T1 cells with IC50 of 14.1–18.6 μg/mL, as compared to 13.5–36.3 μg/mL against MCF-7 cells, and 30.2–50.1 μg/mL on Vero cells.
Table 1.
IC50 values (μg/mL) of TMX-MCEs-ETH and W, in comparison to TMX-MCEs-CHL, on MCF-7, 4T1 and Vero cell-lines, after 24 h treatment (Tamoxifen is the positive control; Untreated cells are the negative control).
| Cell-lines /Formulation | MCF-7 |
4T1 |
Vero |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1:2 | 1:3 | 1:4 | 1:5 | 1:2 | 1:3 | 1:4 | 1:5 | 1:2 | 1:3 | 1:4 | 1:5 | |
| TMX | 12.02 ± 0.02∗∗∗ | 5.05 ± 0.01∗∗∗∗ | 11.22 ± 0.01∗∗∗ | |||||||||
| Negative Control | ˃100 | ˃100 | ˃100 | ˃100 | ˃100 | ˃100 | ˃100 | ˃100 | ˃100 | ˃100 | ˃100 | ˃100 |
| TMX -N. oculata-ETH | 16.98∗∗ ± 0.007 | 24.54∗∗ ± 0.02 | 26.3 ± 0.01 | 30.19 ± 0.02 | 18.62∗∗∗ ± 0.01 | 19.05∗ ± 0.01 | 21.37∗ ± 0.05 | 30.9 ± 0.05 | 25.11∗∗∗ ± 0.01 | 35.48∗ ± 0.02 | 60.25 ± 0.02 | 61.65 ± 0.02 |
| TMX- N. oculata-W | 15.84∗∗∗ ± 0.06 | 21.87∗∗ ± 0.05 | 30.19 ± 0.03 | 33.88 ± 0.06 | 30.9 ± 0.01 | 31.62 ± 0.02 | 38.01 ± 0.02 | 41.68 ± 0.01 | 56.23∗ ± 0.02 | 85.11 ± 0.01 | 87.09 ± 0.02 | ˃100 |
| TMX -N. oculata-CHL |
31.62∗ ± 0.02 |
36.3 ± 0.02 |
46.77 ± 0.05 |
53.7 ± 0.02 |
15.48∗∗∗ ± 0.06 |
17.78∗∗∗ ± 0.01 |
19.95∗∗ ± 0.05 |
22.38∗ ± 0.03 |
33.88∗∗ ± 0.01 |
47.86∗ ± 0.02 |
60.25 ± 0.02 |
66.06 ± 0.03 |
| TMX-T. suecica-ETH | 16.98∗∗ ± 0.01 | 21.37∗ ± 0.02 | 21.37∗ ± 0.01 | 33.11 ± 0.03 | 13.8∗∗∗∗ ± 0.02 | 14.12∗∗∗∗ ± 0.01 | 18.62∗∗ ± 0.03 | 24.54∗ ± 0.04 | 24.54∗∗ ± 0.01 | 33.88∗∗ ± 0.01 | 50.11 ± 0.01 | 58.88 ± 0.02 |
| TMX -T. suecica-W | 19.05∗∗ ± 0.03 | 26.91 ± 0.03 | 36.30 ± 0.04 | 38.90 ± 0.02 | 23.98∗∗ ± 0.02 | 30.19 ± 0.02 | 34.67 ± 0.02 | 43.65 ± 0.01 | 46.77∗ ± 0.02 | 56.23 ± 0.01 | 70.79 ± 0.02 | 85.11 ± 0.02 |
| TMX -T. suecica-CHL |
27.54∗ ± 0.01 |
33.11 ± 0.01 |
37.15 ± 0.01 |
43.65 ± 0.01 |
14.12∗∗∗ ± 0.01 |
15.13∗∗ ± 0.01 |
16.98∗∗ ± 0.03 |
28.18 ± 0.03 |
30.19∗∗ ± 0.01 |
44.66 ± 0.01 |
48.97 ± 0.01 |
66.06 ± 0.02 |
| TMX -Chlorella sp.-ETH | 22.38∗∗ ± 0.02 | 26.91 ± 0.02 | 38.9 ± 0.04 | 52.48 ± 0.02 | 16.98∗∗ ± 0.01 | 20.41∗ ± 0.01 | 21.37 ± 0.03 | 28.84 ± 0.06 | 30.19∗∗ ± 0.02 | 38.9 ± 0.01 | 56.23 ± 0.02 | 70.79 ± 0.02 |
| TMX -Chlorella sp.-W | 24.54∗ ± 0.04 | 29.51 ± 0.04 | 33.88 ± 0.02 | 39.81 ± 0.04 | 21.37∗ ± 0.02 | 26.30 ± 0.01 | 38.01 ± 0.02 | 39.81 ± 0.01 | 42.65 ± 0.02 | 56.23 ± 0.01 | 63.09 ± 0.01 | 85.11 ± 0.02 |
| TMX -Chlorella sp.-CHL | 13.48∗∗∗ ± 0.02 | 21.87∗∗ ± 0.02 | 44.66 ± 0.02 | 54.95 ± 0.03 | 16.98∗∗ ± 0.01 | 18.62∗ ± 0.03 | 19.95∗ ± 0.01 | 25.11 ± 0.03 | 31.62∗ ± 0.01 | 50.11 ± 0.03 | 60.25 ± 0.02 | 66.06 ± 0.02 |
Data expressed as mean ± standard deviation (n = 3). for statistically significant difference between microalgae extracts and the ratio. Significant level: 0.010 < p ≤ 0.05, significant ∗; 0.001 < p ≤ 0.010, very significant∗∗; and p ≤ 0.001, highly significant ∗∗∗. The symbol “- “indicates no IC50 estimated.
Synergistic application of silver nanoparticles (AgNPs) with T. suecica chloroform extracts (AgNPs-T. suecica-CHL at the ratio of 2:1) also achieves high cytotoxicity on MCF-7 (IC50 6.6 μg/mL), but low toxicity on 4T1 cells (IC50 53.7 μg/mL), and with no toxicity on Vero cells after 72 h treatment (Hussein et al., 2020a). For comparison, the single application of TMX was highly cytotoxic to both cancer and Vero cell-lines. The IC50 of TMX on MCF-7 at 12 μg/mL (Table 1) after 24 h, agrees well with the IC50 of 12 ± 0.52 μg/mL reported before (Nigjeh et al., 2013). Other values of TMX such as IC50 5 μg/mL after 24 h (Hassan et al., 2018), and IC50 ∼ 8 μM after 72 h, have also been reported (Sun et al., 2012). The single application of the MCEs however showed low cytotoxicity on MCF-7 cells based on Chlorella sp.-ETH (IC50 52.48 μg/mL), T. suecica-ETH (53.7 μg/mL) and N. oculata-CHL (57.5 μg/ml), after 24 h. The Chlorella sp.-ETH and T. suecica-ETH exhibited even lower cytotoxicity against 4T1 cells (91.2 μg/mL). All MCEs, regardless of the solvents used, do not show any cytotoxicity against the Vero cells (Hussein et al., 2020a,b).
Vero cells are widely used in research as established mammalian cell lines and have been used as a control normal cell line in the evaluation of cytotoxic effects (Fernández Freire et al., 2009; Sombatsri et al., 2019). This cell line is usually used in cancer studies (Siddiqui et al., 2019) and natural product screening tests (Mashjoor et al., 2015; Sit et al., 2018; Kumarihamy et al., 2019), because of their sensitivity to different types of microbes, toxins, and chemical complexes. It has also been used as a substrate for the production of vaccine (Mimeault et al., 2005). It does not cause tumors at the passage levels used for the production of the vaccine, although at higher passage levels it may cause a tumor (Szliszka et al., 2008). The Vero cell lineage is aneuploid and continuous, with abnormal chromosome number. A persistent cell lineage can be duplicated through many division cycles and it does not become aging (Sigma Aldrich, 2003). In contrast to the normal mammalian cells, Vero cells contain little interferon and do not produce interferon-α or β upon virus infection. However, they still have an α or β interferon receptor, such that they respond normally when complex interferon is added to the culture media (Desmyter et al., 1968). Our work proved that the presence of MCEs reduced the cytotoxicity of TMX or AgNPs against the Vero cells, whilst at the same time the TMX or AgNPs-MCEs synergistic formulations improved the cytotoxicity of MCEs on MCF-7 and 4T1 cells (Hussein et al., 2020a,b,d). This should be of great interest in cancer therapeutics as the cytotoxicity of TMX on the normal cells has been widely reported (Salih et al., 2017; Yusmazura et al., 2017).
3.2. Morphological characterization
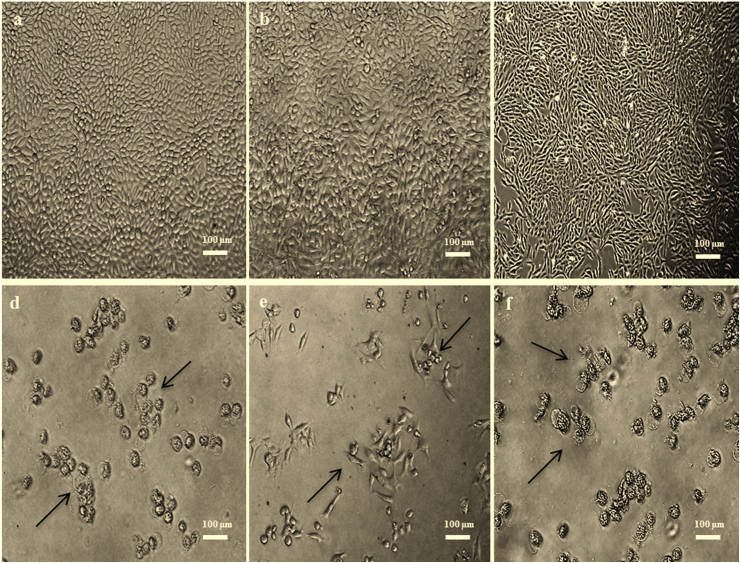
Cytotoxic agents have inhibitory effects against the proliferation of cancerous cells (Pantazis et al., 1995). The changes in the general shape of cancer cells under the influence of drugs are commonly used as the basis for evaluating the drug effects (Schempp et al., 2002). The TMX was investigated further at IC50 9 μg/mL for MCF-7 (Nigjeh et al., 2013), and at their respective IC50 values for 4T1 and Vero cells for morphological characterization. Figure 1a, b, and c show that the MCF-7, 4T1, and Vero control cells (untreated) appeared connected together, healthy-looking, and with no necrotic or apoptotic bodies. However, the treated MCF-7 cells with TMX (Figure 1d) and synergistic application (Figure 2) became lower in cell number, rounded up, smaller size, and detached with some cells exhibiting membranous blebbing, and apoptotic bodies seen as circular or oval masses of cytoplasm. The 4T1 (Figures 1e and S1) and Vero (Figures 1f and S2) cells treated with TMX and synergistic application also exhibited similar morphologies. These are unique changes in apoptosis where the nucleus becomes anaphase and dense, due to the nuclear chromatin condensation (Skerman et al., 2011).
Figure 1.
Morphology of Control cells for (a) MCF-7; (b); 4T1; (c) Vero; and cells treated with TMX for (d) MCF-7; (e) 4T1; and (f) Vero, after 24 h.
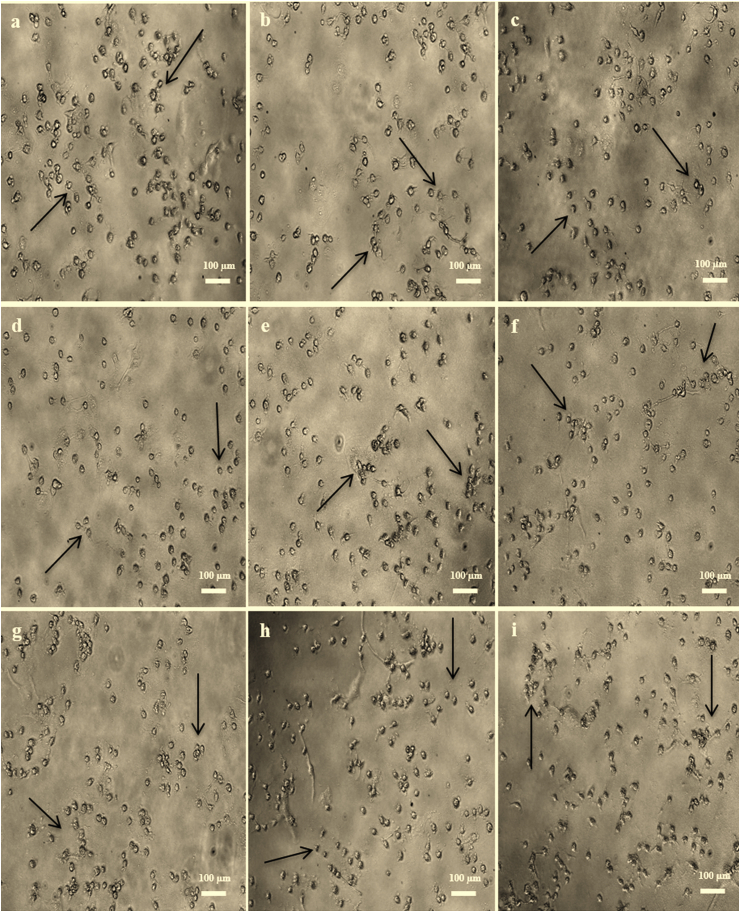
Figure 2.
Morphology of MCF-7 cells after 24 h synergistic application (1:3) at the IC50 levels for (a) TMX-N. oculata-CHL; (b) TMX-T. suecica-CHL; (c) TMX-Chlorella sp.-CHL; (d) TMX-N. oculata-ETH; (e) TMX-T. suecica-ETH; (f) TMX-Chlorella sp.-ETH; (g) TMX-N. oculata-W; (h) TMX-T. suecica-W; (i) TMX-Chlorella sp.-W.
3.3. Flow cytometric analyses
3.3.1. Flow Cytometry – Annexin V
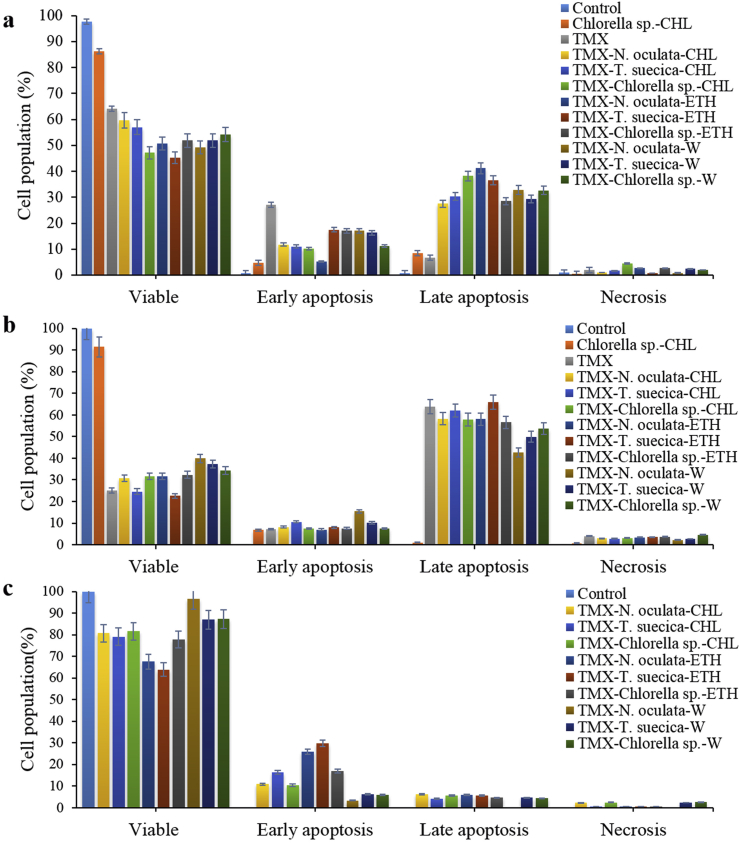
For analyses of apoptosis during the early and late apoptotic stage, Annexin-V attaches to the PS, the apoptotic biomarker. During the late stage of apoptosis or necrosis, PI penetrates the cells that have lost membrane integrity to allow DNA staining (Atasever-arslan et al., 2016). Four populations of cells are analyzed based on the staining:- unlabeled (viable cells, Annexin V−/PI−), labeled with Annexin V-FITC only (early apoptotic, Annexin V+/PI−), stained with PI (necrotic, Annexin V−/PI+), and bound Annexin V and stained with PI (late apoptotic/necrotic cells, Annexin V+/PI+) (Skerman et al., 2011; Hajiaghaalipour et al., 2017). Following the TMX-MCEs (1:3) synergistic treatment in MCF-7 cells, significant apoptotic activities were observed, especially the late apoptosis after 24 h with TMX-N. oculata-ETH (41.1 %), TMX-Chlorella sp.-CHL (36.8 %), TMX-T. suecica-ETH (36.6 %) and TMX-N. oculata-W (32.8 %). The TMX single application in contrast had resulted in much lower late apoptotic event (6.6 %). The highest early apoptosis was registered by the TMX (27.17 %), followed by the AgNPs (25.56 %) (Hussein et al., 2020a), as compared to the TMX-T. suecica-ETH (17.2 %), TMX-N. oculata-W (17.1 %), and TMX-Chlorella sp.-ETH (17 %) (Figure 3a).
Figure 3.
Flow cytometric analysis of Control, Chlorella sp.-CHL, TMX and TMX-MCEs synergistic application after 24 h treatment on (a) MCF-7; (b) 4T1; (c) Vero cell-lines. All treatments show significant differences between the control, single and synergistic application with p < 0.05 (n = 3).
For 4T1 cells (Figure 3b), significant increase in the late apoptosis with TMX-T. suecica-ETH (66 %) was recorded, which was much higher than the TMX alone. The apoptotic events with other synergistic applications were however slightly lower than the TMX, although still much higher than the MCEs alone. After 24 h, early apoptosis was the highest with TMX-N. oculata-W (15.7 %), followed by TMX-T. suecica-CHL (10.8 %). In Vero cell-lines (Figure 3c), the early apoptosis were registered lower with TMX-T. suecica-ETH (29.8 %) and TMX-N. oculata-ETH (25.9 %), and with no significant increase in the late apoptosis (less than 6.2 %), as compared to the TMX alone (38.4 %).
The synergistic applications of TMX and MCEs were capable of inducing high early and late apoptosis against MCF-7 and 4T, but not in Vero cells. These can be due to the capability of the synergistic treatment on the cells to initiate apoptotic mechanisms more readily through different apoptotic pathways. Other study has shown that the cells in the late apoptotic phase is significantly enhanced from 0.045 % in the Control to 45.7 % in the cells treated with TMX after 48 h, confirming apoptosis in the TMX-treated MCF-7 cells (Rouhimoghadam et al., 2018). The TMX at 5 μg/mL has been reported to stimulate the induction of intrinsic Caspase pathway in MCF-7 cells. Majority of anti-cancer drugs promote apoptosis by inducing the release of cytochrome c and the expression of Apaf-1 and caspase-9 activities in the mitochondria (Zhou et al., 2015).
3.3.2. Cell-cycle analyses
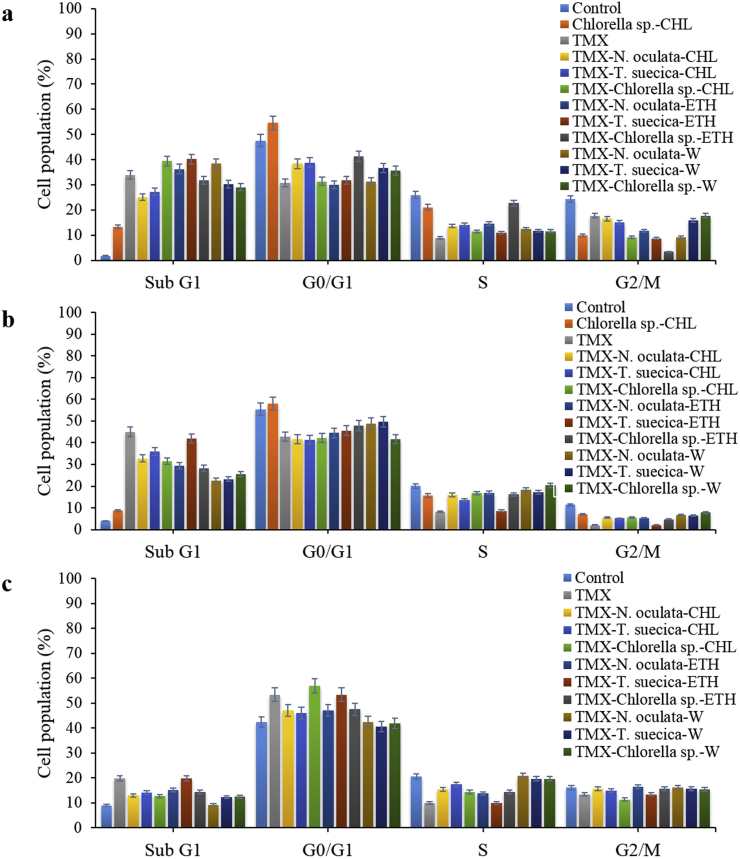
Most of anticancer drugs exert their cytotoxic effects at the G1 or G2 stage to inhibit the cell-cycle progression (Goh et al., 2010). If the check-point pathways detect problems in the cell DNA, the cell-cycle may halt, and the cell makes attempt to repair the damaged DNA or complete the DNA replication (Hwang et al., 2013). If the damage is beyond repair, this may lead to the cell-cycle arrest at G0/G1 and G2/M, which can trigger apoptosis (Kim et al., 2014). In our study, the TMX-MCEs synergistic application on MCF-7 cells showed significant increase in sub-G1 phase from 34 % with TMX, to 40.2 % with TMX-T. suecica-ETH, 39.4 % with TMX-Chlorella sp.-CHL, 38.4 % with TMX-N. oculata-W and 36.3 % with TMX-N. oculata-ETH at 1:3 ratios. There was also a significant reduction in the G1 and S and G2/M phases of all the treated cells (Figure 4a).
Figure 4.
Composition of sub-G1, G1, S and G2/M of Control, Chlorella sp.-CHL, TMX and TMX-MCEs synergistic application after 24 h treatment on (a) MCF-7; (b) 4T1; (c) Vero cell-lines. All treatments show significant differences between the control, single and synergistic application with p < 0.05 (n = 3).
For 4T1 cells, the TMX single application showed the highest sub-G1 phase (45 %), and a highly increased sub-G1 (22–41 %) in all the TMX-MCEs treated cells, as compared to 6–8 % with the MCEs as shown in Figure 4b. For Vero cells, the TMX-MCEs synergistic application showed only a slight increase in sub-G1 phase (9–19 %) suggesting lower apoptosis after 24 h, as compared to the TMX (51 %). There was also an increase in G1 (40.7–57.1 %), S (13.8–20.8 %), and G2/M phase (11.4–16.5 %), which were more than the TMX (35.9, 10.5, and 1.8 %, respectively). Only the TMX-T. suecica-ETH (1:3) (10 %) was slightly lower than the TMX in S phase (Figure 4c). Previous study shows that the TMX treatment of the MCF-7 cells after 48 h has resulted in no significant variations of the cell-cycle phase distribution (G1, S, G2), with an increase in the S (10 %) and the G2 (6 %) phase populations, when compared to Control. However, the drop in the viability of the cells to about 50 % after 48 h has been suggested as a result of the TMX mediating to reduce the cell viability, but not the cell cycle arrest (Rouhimoghadam et al., 2018).
3.3.3. Biomarkers
Significant enhancement of ADP/ATP ratios, after 24 and 48 h, respectively, was observed in the TMX-MCEs-CHL, ETH, and W treated cells between 1.5-1.9 and 3.5–3.9 fold, as compared to the TMX alone (Figure S3). The MCEs may play a role as an excellent carrier system for the TMX to the target cells resulting in enhanced ADP/ATP ratios, and therefore improved effectiveness of the TMX-MCEs application. The highest ADP/ATP ratio, after 24 and 48 h, respectively, was achieved with TMX-N. oculata-CHL (3.7, 6), TMX-T. suecica-ETH (3.6, 6) and TMX-T. suecica-CHL (3.5, 5.9) at 1:3 ratios, which was comparable to the TMX alone (3.4, 5.8). The other synergistic applications exhibited slightly lower ratios (2, 5.6) than the TMX alone, but still higher than the MCEs single application. For Vero cells, the significant reduction in ADP/ATP ratio with the TMX-MCEs (1:3) treatment at 1–2.4, 1.4–3.4 as compared to the TMX (3.2, 5.3) further proved the lower apoptotic events (Figure S3c).
The TMX-T. suecica-ETH showed significant increase, after 24 and 48 h, respectively, in the activity of Caspase (24 × 104, 33 × 104 RLU), followed by the TMX-N. oculata-W and TMX-Chlorella sp.-CHL (23 × 104, 32 × 104 RLU), which were however higher than the TMX alone. The other synergistic application attained comparable or slightly lower caspase activity than the TMX-treated cells (Figure S4a). For 4T1 cells (Figure S4b), the TMX-T. suecica-CHL and ETH (1:3) achieved the highest Caspase activity (29 × 104, 36 × 104 RLU), followed by TMX-N. oculata-CHL and ETH (25 × 104, 31 × 104 RLU) and TMX-Chlorella sp.-CHL (24 × 104, 29 × 104 RLU), respectively, as compared to the untreated cells, TMX and MCEs. For Vero cells (Figure S4c), the TMX-MCEs application showed improved Caspase activities as compared to the untreated cells, but lower than the TMX single application. These results indicated that the MCEs had reduced the toxicity of the TMX and the apoptosis in the non-cancerous Vero cells, with reduced Caspase activity.
3.4. NMR analyses
3.4.1. Assignments of metabolites by 1H NMR spectra
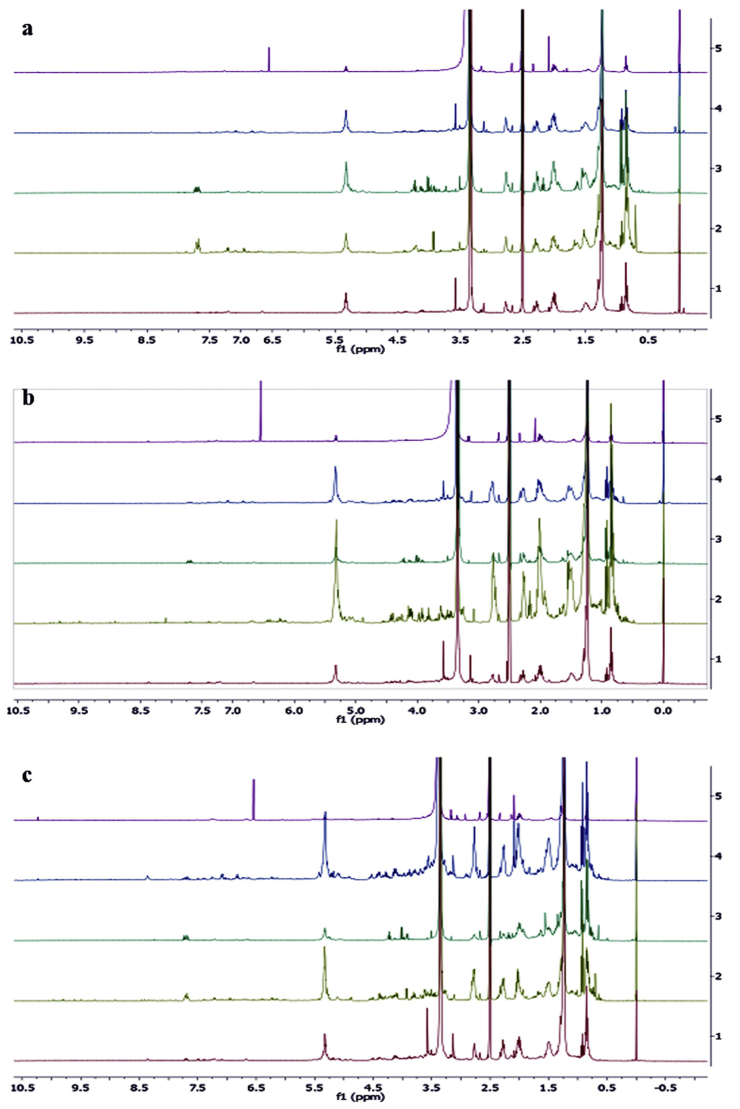
Several researches on marine microalgae have reported a large number of principal metabolites such as simple sugars, fatty acids, lipids, and amino acids. There is however incomplete information on microalgal secondary metabolites. Figure 5 shows the representative spectra of 1H NMR from different N. oculata, T. suecica, and Chlorella sp solvent extracts. In general, the spectra of 1H NMR exhibited that of carotenoids, fatty acids, amino acids, and organic acids. Significant differences were observed between different solvent extracts in the region δH 0.5–3.2 ppm, which mostly were related to the organic acids and amino acids. The signals δH 3.2–5.5 ppm were the characteristic signals of the anomeric and backbone-protons of carbohydrates. Some secondary metabolites such as the polyphenols can be recognized at δH 5.5–10.0 ppm (Aguilera-Sáez et al., 2019), as well as the characteristic signals of carotenoids and chlorophyll components at δH 6.0–7.0 ppm, 7.0–8 ppm and 8.2–10.0 ppm (Chauton et al., 2004). Tables S3 and S4 show the chemical shifts of 44 identified metabolites of the MCEs samples extracted with 5 different solvents (MET, CHL, HEX, ETH and W). The detected metabolites were 9 amino acids including alanine, threonine, aminolevulinic acid, valine, asparagine, tyrosine, leucine, isoleucine, and lysine; 1 vitamin (choline); 6 fatty acids (stearic acid, linoleic acid, oleic acid, glycerol, eicosapentaenoic acid (EPA C20:5) and triglyceride); 1 glucose; 4 carotenoids (fucoxanthin, lutein, β-carotene, and violaxanthin); 4 chlorophylls and chlorophyll transformation products including chlorophyll a, methyl pyropheophorbide, pheophorbide-a and isochlorin; 8 organic acid derivatives including lactic acid, formic acid, triethanolamine acid, fumaric acid, phosphatidylcholine, succinic acid, malonic acid and hydroxyphenyllactic acid; and other metabolite such as xanthine and unknown compounds.
-
1.
Amino acids
Figure 5.
Representative 400 MHz 1H Nuclear Magnetic Resonance (NMR) spectra of (a) N. oculata; (b) T. suecica (c) Chlorella sp., extracted by 1) MET; 2) CHL; 3) HEX; (4) ETH; (5) W.
Most of the 1H NMR signals were partially or completely overlapping because the signals of the metabolites were close to each other. The 1H NMR spectrum of the MCEs-W exhibited alanine at δH 1.48 ppm (d, J = 6.8 Hz), and 3.76 ppm (qt, overlapped). Both N. oculata and Chlorella-CHL showed threonine at δH 1.30 ppm (d, overlapped). The signals at δH 2.13 ppm (t, J = 2 Hz), 2.68 ppm (t, J = 2 Hz), and 4.09 ppm (s) suggested the presence of aminolevulinic acid in N. oculata-MET, HEX, W, as well as T. suecica-HEX, and Chlorella sp. HEX, ETH, W. The signals δH 2.29 ppm (m), 1.03 ppm (d, overlapped), and 0.98 ppm (d, overlapped) showed the presence of valine in MCEs-HEX. Asparagine produced a signal at δH 2.94 ppm (m), 4.04 ppm (dd, J = 7.6, 4 Hz), 2.84 ppm (m), and observed in MCEs-MET. Tyrosine was detected in MCEs-CHL and ETH at δH 6.84 ppm (d, J = 8.4 Hz), 7.15 ppm (d, J = 8.4 Hz), while isoleucine and leucine were detected in MCEs-HEX, and in MCEs-HEX at δH 0.94 ppm (t, J = 7.6 Hz); N. oculata-ETH at 1.04 ppm (d, overlapped); and Chlorella sp.-ETH of 0.942 ppm (t, J = 6.8 Hz), 1.7 ppm (m), 3.72 ppm (m). The 1H NMR spectrum of Chlorella sp.-W exhibited the presence of lysine at δH 3.08 ppm (t, J = 7.2 Hz), 1.5 ppm (m), 1.72 ppm (m), 1.9 ppm (m).
-
2.
Choline and Fatty acids
Choline produced a signal at 3.24 ppm (s), which was observed in Chlorella sp.-CHL. Choline is an aliphatic monoamine, associated with the phospholipid, the most popular being phosphotidylcholine (PC) in microalgae (Ma et al., 2018). The signals at δH 2.51 ppm (t, J = 7.2 Hz), 1.07 ppm (t, J = 6.8 Hz) showed the presence of stearic acid in N. oculata-CHL, Chlorella sp.-CHL, HEX, ETH, and T. suecica-ETH. Oleic acid was identified in the MCEs-MET, ETH, W; T. suecica- HEX, ETH and Chlorella sp.-HEX, ETH at δH 0.85 ppm (t, overlapped), 1.3 ppm (m), 1.60 ppm (m), 2.37 ppm (t, overlapped), and 5.32 ppm (m). Except for N. oculata-MET, linoleic acid was detected in all MCEs at δH 0.9 ppm (t, J = 6.8 Hz), 1.32 ppm (m), 1.64 ppm (m), 2.07 ppm (m), 2.36 ppm (t, overlapped), 2.78 ppm (t, J = 5.6 Hz), 5.37 ppm (m). The signals at δH 3.61 ppm (d, overlapped), 3.65 ppm (d, overlapped) were determined as glycerol, found in Chlorella sp.-CHL and T. suecica-ETH. Glycerol is an essential component of both neutral lipids and phospholipids, obtained from direct glycerol pathway or by starch fermentation (Singh et al., 2005; Sarpal et al., 2016). EPA C20:5 was detected at δH 1.69 ppm (s) found in MCEs-CHL, HEX, and Chlorella sp.-ETH; and the triglycerides at δH 4.3 ppm (dd, J = 8, 14.8 Hz), 4.16 ppm (dd, J = 6.8, 12.8 Hz), 5.36 ppm (m), were detected in MCEs-ETH, CHL. Triacylglycerides (TAG) are the essential lipid storage in microalgae (Zhang et al., 2016).
-
3.
Glucose
The 1H NMR spectra of MCEs-ETH, N. oculata-MET and Chlorella sp.-MET, CHL exhibited the presence of glucose signals at δH 5.18 ppm (d, J = 3.6 Hz), and 4.53 ppm (d, J = 8.4 Hz).
-
4.
Carotenoids
Four carotenoids were identified (Table S3). The signals at δH 6.44 ppm (d, J = 12 Hz), 6.55 ppm (m), 6.38 ppm (d, J = 16.4 Hz), 7.20 ppm (d, overlapped), 6.23 ppm (s), 6.29 ppm (d, J = 12 Hz), 6.12 ppm (d, overlapped) were those of fucoxanthin, found in MCEs CHL and ETH. Lutein was detected only in Chlorella sp.-CHL at δH 3.99 ppm (m), 6.67 ppm (m), 6.63 ppm (m), 6.24 ppm (d, J = 11 Hz), 6.38 ppm (d, J = 16 Hz), 6.16 ppm (m),1.68 ppm (s), 1.93 ppm (s). The 1H NMR spectrum of MCEs-CHL, HEX, and Chlorella sp.-ETH exhibited violaxanthin at δH 1.14 ppm (s), 1.94 ppm (s), 3.87 ppm (m), 6.23 ppm (d, J = 10 Hz), 6.58 ppm (d, overlapped). β-carotene was detected in MCEs-CHL, ETH; N. oculata-HEX, MET; T. suecica-MET and Chlorella sp.-HEX at δH 1.27 ppm (s), 1.86 ppm (s), 2.02 ppm (t, J = 7.2 Hz), 2,31 ppm (s), 6.26 ppm (d, overlapped), 6.63 ppm (d, overlapped).
-
5.
Chlorophylls
The chlorophyll was identified as chlorophyll a at δH 9.48 ppm (s), observed in MCEs-CHL and MET. The chlorophyll transformation products were determined in MCEs-CHL as methyl pyropheophorbide at δH 9.44 ppm (s), 9.35 ppm (s), 3.65 ppm (s), N. oculata and Chlorella sp.-CHL as pheophorbide –a at δH 9.57 ppm (s), 9.73 ppm (s), 8.71 ppm (s), 3.27 ppm (s), 3.73 ppm (s) and MCEs CHL, MET as isochlorin at δH 3.62 ppm (s), 9.58 ppm (s), 8.99 ppm (s). However, signals at δH 9.96 ppm (s), 9.91 ppm (s), 8.81 ppm (s), 8.30 ppm (s) which was assigned to chlorophyll c1, and at δH 10.02 ppm (s), 10.39 ppm (s) assigned to chlorophyll c2, as observed in MCEs-CHL and MET, were also detected. We could only speculate that these signals, suggesting the presence of chlorophyll c, were artefacts. Chlorophyll a is found in all higher plants, algae and cyanobacteria, while chlorophyll b is found in higher plants and specifically in green algae. However, chlorophyll c is unique to diatoms, dinoflagellates and brown algae, while chlorophyll d is only specific to red algae (Patel, 2011).
-
6.
Organic acids
The 1H NMR spectrum of MCEs-CHL, MCEs-W, T. suecica-MET, Chlorella sp.-MET, ETH; and, N. oculata and Chlorella sp.-CHL exhibited formic acid at δH 8.41 ppm (s) and lactic acid at δH 4.1 ppm (m), 1.33 ppm (d, J = 8.8), while the triethanolamine was identified at δH 3.88 ppm (s), 3.4 ppm (s), displayed in N. oculata-CHL and MCEs-MET. Fumaric acid was detected at δH 6.55 ppm (s) in MCEs-W; and N. oculata and T. suecica-CHL; and phosphatidylcholine at δH 3.17 ppm (m), 4.16 ppm (m) in MCEs-CHL, ETH, MET. Succinic acid was detected at δH 2.33 ppm (s) in T. suecica and Chlorella sp.-CHL; and N. oculata and Chlorella-ETH; malonic acid at δH 3 ppm (s) in MCEs-ETH, Chlorella sp.-HEX, and N. oculata and Chlorella sp.-CHL; and hydroxyphenyllactic acid at δH 7.22 ppm (d, J = 8) in N. oculata and T. suecica-CHL and Chlorella sp.-W.
-
7.
Unknown compounds
The 1H NMR spectrum of MCEs-ETH and Chlorella sp.-CHL showed xanthine at δH 7.89 ppm (s). The other unknown compounds were detected at δH 7.09 ppm (d, J = 7.6 Hz) which appeared in Chlorella sp.-CHL only; 7.74 ppm (m), 7.68 ppm (m) were displayed in MCEs-ETH and Chlorella sp.-CHL, MET; 9.87 ppm (s) found in MCEs-CHL, ETH; 1.56 ppm (d, J = 7.6 Hz) found only in Chlorella sp. and T. suecica-ETH; 1.82 ppm (s) detected in Chlorella sp. and T. suecica-ETH and N. oculata-MET; 1.24 ppm (s) found in Chlorella sp.-HEX, MET; N. oculata-MET and T. suecica-W; 10.22 (s) detected in MCEs-W, N. oculata and T. suecica-ETH, T. suecica and Chlorella sp.-CHL; and the last metabolite at δH 2.19 ppm (t, J = 7.6 Hz) identified only in Chlorella sp.-HEX. The 1H NMR spectrum of Ulva fasciata extracts exhibit a mix of fatty acids and triacylglycerols as the main compounds. Triacylglycerols 1H signals are observed at δH 5.36 ppm (m), 4.29 ppm (dd, J = 7.4; 14.6 Hz), and 4.16 ppm (dd, J = 6.0; 12.4 Hz). The mixture of fatty acids is detected at 0.88 ppm (t, J = 7.2 Hz), 2.31 ppm (t, J = 7.4 Hz), 1.60 ppm (m), 2.80 ppm, 2.02 ppm, 1.26 ppm (bs). The δH 0.65 ppm (s), 0.92 ppm (s) and 1.0 ppm (s) suggested a little amount of sterols in the MCEs (Mendes et al., 2010).
In general, there are some unique metabolites detected in specific microalgae, as shown in Table S5, which can be correlated to their bioactivities during synergistic application with TMX. The uniqueness was determined based on the spectral identification of the metabolite from Table S3 and S4, and summarized as shown in Table S5, based on the metabolites not found in all of the extracts and species. Some were detected only in one extract and one species like triethanolamine and threonine in N. oculata-CHL, malonic acid in Chlorella sp.-ETH, lysine in Chlorella sp.-W, and choline and lutein in Chlorella sp.-CHL. Other metabolites were detected only in one extract of all species like alanine and fumaric acid (W extracts of all species), or detected in one extract of two species such as isoleucine (N. oculata and Chlorella-ETH), methyl pyrophaeophoribide and hydroxyphenyllactic acid (N. oculata and T. suecica-CHL), or only in two extracts of two or three species such as tyrosine (N. oculata, Chlorella sp., T. suecica-CHL and ETH), fucoxanthin (N. oculata and T. suecica- ETH and CHL and Chlorella sp. ETH), triglycerides and tyrosine (N. oculata, Chlorella sp. and T. suecica-CHL and ETH).
3.4.2. Classification based on MCEs by Principal Component Analysis (PCA)
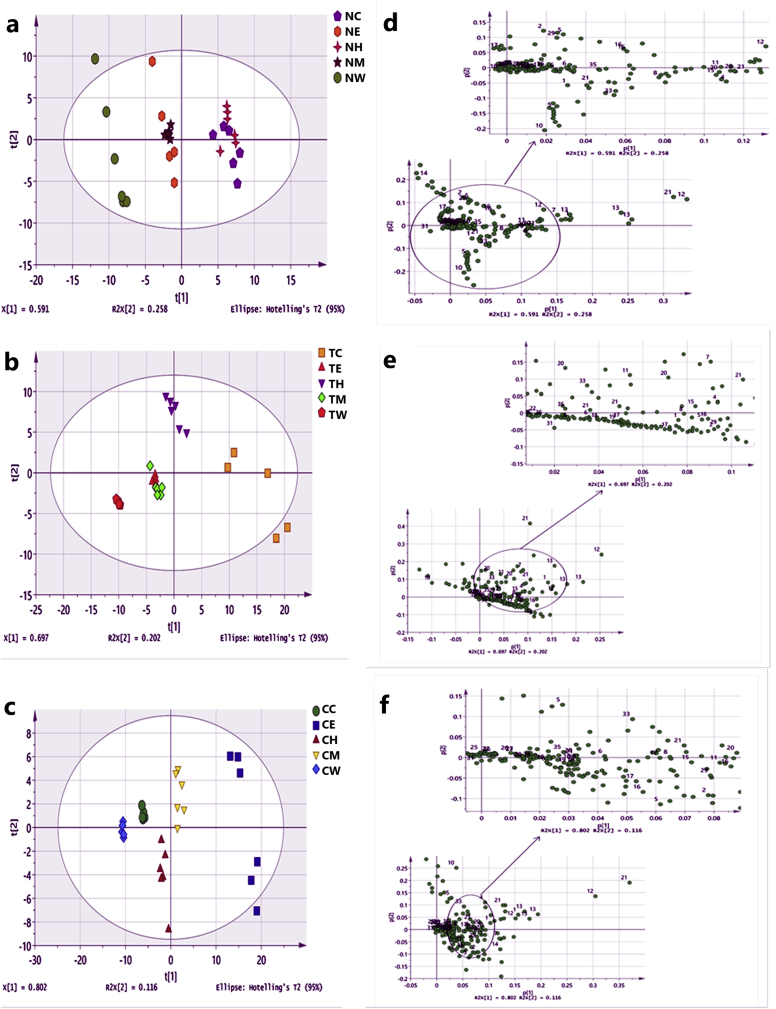
Multivariate Data Analysis analyses the differences in metabolites in different extracts from the three microalgae species. The PCA score plot (Figure 6a and b) shows that 36 samples of N. oculata and T. suecica were clustered into three and four groups, respectively, with R2X cum and Q2 cum values of 0.999 and 0.994; and 0.999 and 0.995, respectively (Table S6), and there was no indication of strong outliers. The metabolite profile of N. oculata and T. suecica was differentiated from each other, and the CHL and HEX were well detached based on the first principal component (PC1). The spectral signals for MET and ETH, as well as the CHL and HEX, were found in the same cluster due to the same polarity, suggesting similar chemical profiles.
Figure 6.
Score plot (a, b, c) and Loading plot (d, e, f) of Principal Component Analysis (PC1 versus PC2) of N. oculata, T. suecica, and Chlorella sp. The plot ellipse represents 95 % hotelling T2 confidence. C: CHL; E: ETH; H: HEX; M: MET; W: water.
The loading-plot with PC1 and PC2 for N. oculata (Figure 6d) highlights the possible marker metabolites responsible for aggregation. Table S7 shows the numbering and the assigned metabolites for the loading plot. The HEX and CHL had significantly higher amounts of fatty acids, carotenoids, and amino acids than the polar MET and ETH. The carotenoids were β-carotene and violaxanthin, and the amino acids were isoleucine, valine and leucine. The fatty acids identified included linoleic acid, oleic acid, EPA and triglyceride. The ETH, MET and W extracts consisted mainly of glycerol, glucose, fumaric acid and triethanolamine. The loading plot of T. suecica (Figure 6e) shows that the HEX and CHL had greater amounts of carotenoids (violaxanthin, fucoxanthin, lutein and β-carotene), chlorophyll (chlorophyll a), amino acids (alanine, tyrosine, valine, asparagine, threonine and leucine) and fatty acids (stearic acid, oleic acid, linoleic acid, EPA, triglyceride, and glycerol), as compared to the other extracts.
For Chlorella sp., the PCA score plot revealed the 36 samples grouped into five groups which attained R2X cum and Q2 cum values of 0.995 and 0.989, respectively (Table S6). The MET and ETH were well detached from the CHL, W and HEX by PC1 (Figure 6c). The loading plot (Figure 6f) shows that the ETH and MET had greater amounts of carotenoids (β-carotene, fucoxanthin, violaxanthin, and lutein), chlorophyll (chlorophyll a, and chlorophyll products (methyl pyrophaeophoribide, pheophorbide–a)), amino acids (tyrosine), organic acid (formic acid) and others (xanthine), as compared to the other extracts. The CHL and W were mainly having methyl pyrophaeophoribide and fumaric acid.
3.4.3. Relative quantification of metabolites
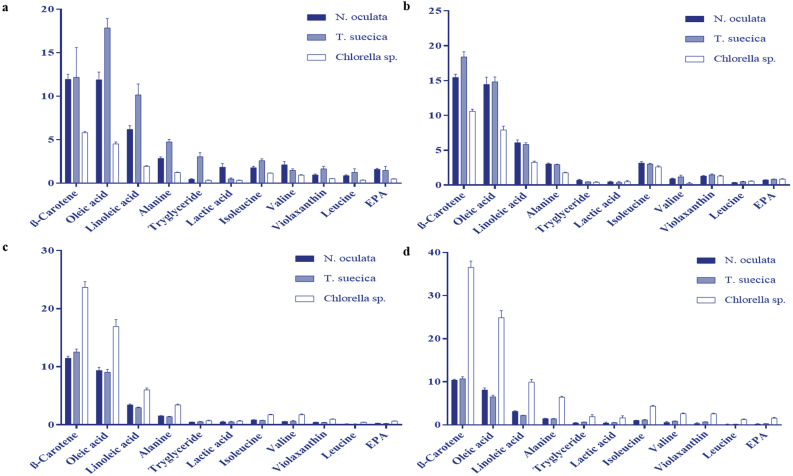
The detected metabolites (the peak area intensity) were further quantified by the Analysis of Variance (ANOVA) to evaluate the significance of differences in different species and extracting solvents. The metabolites with Variable-Importance-in-Projection (VIP) value (the metabolites with a higher rating) were checked if they could influence the assembly of the X-variable (the identified metabolites) in the PCA loading-plot (Ginsburg et al., 2011). Figure 7 shows the profile of the compounds and their relative amounts in different MCEs. For CHL (Figure 7a), T. suecica exhibited higher and more significant amount of all metabolites. Only lactic acid and valine were detected higher in N. oculata. Figure 7b shows that only β-carotene was higher in T. suecica-HEX, while the other metabolites were more comparable between N. oculata-HEX and T. suecica-HEX. For MET and ETH (Figures 7c & 7d), Chlorella sp. exhibited significantly higher amount of β-carotene (δH 1.22 ppm), oleic acid (δH 1.26 ppm), linoleic acid (δH 1.3 ppm), alanine (δH 1.5 ppm), triglycerides (δH 5.38 ppm), lactic acid (δH 4.1 ppm) isoleucine (δH 0.94 ppm), valine (δH 2.3 ppm), violaxanthin (δH 1.1 ppm), leucine (δH 1.7 ppm) and EPA (δH 1.66 ppm), in comparison to N. oculata and T. suecica.
Figure 7.
Relative quantification of the identified β-carotene, oleic acid, linolenic acid, alanine, lactic acid, isoleucine, valine, violaxanthin, triglyceride, leucine and EPA of microalgae species based on the mean peak area of 1H NMR signals for (a) CHL; (b) HEX; (c) MET; (d) ETH. The chemical shifts (ppm) used for the relative quantification are β-carotene (1.22), oleic acid (1.26), linolenic acid (1.3), alanine (1.5) triglyceride (5.38), lactic acid (4.1), isoleucine (0.94), valine (2.3), violaxanthin (1.1), leucine (1.7) and EPA (1.66). Data presented are based on the mean of six replicates for each of the microalgal species (N. oculata, T. suecica and Chlorella sp.) ± standard deviation (SD). Significant level: 0.010 < p < 0.05, significant ∗; 0.001 < p < 0.010, very significant∗∗; and p < 0.001, highly significant ∗∗∗.
Based on the relative quantification from different solvent systems (Figure S5), N. oculata-HEX was unique in having significantly higher quantities of β-carotene, oleic acid, and isoleucine, while the N. oculata-CHL contained only slightly higher levels of violaxanthin, and EPA, with comparable level of linoleic acid between N. oculata-CHL and HEX extracts (Figure S5a). For T. suecica (Figure S5b), the CHL extract showed significantly higher amount of oleic acid, linoleic acid, triglyceride and EPA in comparison to the other extracts, while the HEX extracts were significantly higher in β-carotene. For Chlorella sp. (Figure S5c), the ETH extract was characterized by significantly higher amounts of β-carotene, oleic acid, linoleic acid, isoleucine, violaxanthin, triglyceride, stearic acid and EPA. The MET extracts showed higher β-carotene, oleic acid and linoleic acid, in comparison to other extracts, although still lower than the ETH. However, the number of metabolites were lower in all W extracts. Based on the PCA, the W extracts (Figure S6) showed less variation in the constituents with similar metabolite profile in all microalgal species. The non-polar solvents such as CHL and HEX, and the polar solvents such as ETH and MET, had similar chemical profiling. It appears that the CHL and HEX were more effective with T. suecica and N. oculata which were of the marine origin, while the ETH and MET were more effective with Chlorella sp., which was of the freshwater origin.
Table S8 showed the significant difference in solvent extracts of N. oculata. β-carotene was highly significant (p < 0.0001) in all extracts, especially in comparison to the MET vs CHL extracts. Similarly, oleic acid, linolenic acids and isoleucine were significantly higher (p < 0.0001) in all extracts, while the violaxanthin were highly significant (p < 0.0001) in the CHL and ETH, and significant (p < 0.01) in the HEX and MET extracts. The triglyceride showed significant differences between HEX and W (p < 0.01) extracts, and when compared to the other extracts. Stearic acids were highly significant (p < 0.0001) in CHL, HEX, ETH and very significant (p < 0.001) in ETH and HEX, and the EPA was highly significant (p < 0.0001) in CHL and very significant (p < 0.01) in HEX. However, isoleucine and linolenic acids showed no significant differences between MET and ETH extracts. For T. suecica, β-carotene was highly significant (p < 0.0001) in all extracts, but the CHL vs ETH showed significant (p < 0.01) and the MET vs CHL extracts showed no significant differences. Oleic acid and linolenic acids were significantly higher (p < 0.0001) in all extracts, while the isoleucine exhibited no significant in MET vs ETH, MET vs W and CHL vs HEX. Violaxanthin was significant (p < 0.01) in the MET vs CHL and HEX vs W, and very significant (p < 0.001) in the HEX vs CHL extracts. The triglyceride showed higher significant differences (p < 0.0001) in MET and CHL extracts than the other extracts. Stearic acids were significant (p < 0.05) in CHL and HEX, while the EPA was significant (p < 0.01) with MET and CHL. For Chlorella sp., β-carotene, oleic acid and linolenic acids were highly significant (p < 0.0001) in all extracts. The isoleucine were significantly higher (p < 0.0001) in all extracts, but showed no significant differences between MET and CHL extracts. However, ETH extracts showed highly significant (p < 0.0001) amount of violaxanthin, triglyceride, stearic acids and EPA, in comparison to the other extracts.
4. Discussion
Cancer prevention through effective cancer chemotherapeutic is an important route to combat cancer (Sharif et al., 2014). Chemotherapeutic agents can be selectively toxic to cancer cells as they enhance the oxidative stress to the already exhausted cells beyond their limit (Moungjaroen et al., 2006). Table 2 shows the IC50 values of TMX single-application against various cell-types. At 50 μM TMX, the Chinese hamster lung fibroblasts (V79) shows 21.6 % of the cells exhibiting apoptosis (Petinari et al., 2004). TMX also achieves the lowest IC50 against HeLa and Vero cells, which explains the side-effects related to TMX treatment in cancer patients (Yusmazura et al., 2017). The systemic complications could be reduced through the use of lower TMX dose and a better delivery system to improve its efficiency in breast cancer treatment (Akim et al., 2013). The incorporation of TMX into Nanostructured Lipid Carrier (NLC), for example, decreases the peripheral distribution and increases the localization on the tumor site (How et al., 2013). The IC50 of TMX-NLC and TMX on MCF-7 cell-lines are 5.56 μg/mL and 2.72 μg/mL respectively; while on 4T1 cell lines are 5.19 μg/mL and 5.13 μg/mL, respectively. The low cytotoxic activity of blank NLC suggests that the cytotoxicity is principally due to the TMX in the NLC, and the TMX formulated into the NLC has similar overall performance against the MCF-7 and 4T1 cells (How et al., 2013). The major challenge is however in retaining the cytotoxic activity against the cancer cells, without causing any threat to the normal cells. Also challenging is in determining the cellular toxicity mechanism as to whether it is by apoptosis or necrosis (Reyna-Martinez et al., 2018).
Table 2.
IC50 values of TMX single application against different cell-lines.
| Treatments | IC50 | Treatment time (h) | Cell type | References |
|---|---|---|---|---|
| TMX | 12.59 μM | 48 | MCF-7 | Petinari et al. (2004) |
| ND | Vero | |||
| TMX | 2.72 μg/mL | 72 | MCF-7 | How et al. (2013) |
| TMX | 5.13 μg/mL | 4T1 | ||
| TMX-Nanostructured lipid carrier (NLC) | 5.56 μg/mL | MCF-7 | ||
| TMX-Nanostructured lipid carrier (NLC) | 5.19 μg/mL | 4T1 | ||
| TMX | 2.2 ± 0.029 μg/mL | 72 | Vero | Yusmazura et al. (2017) |
| TMX | 1.25 μg/mL | 44 | MCF-7 | Salih et al. (2017) |
| TMX | 2.3 μg/mL | Vero | ||
| TMX | 4.506 μg/mL | 24 | MCF-7 | Hassan et al. (2018) |
| TMX | 8.0 ± 0.7 μM | 24 | MCF-7 | Perry et al. (1995) |
Biocompounds from microalgal and plant extracts can be potentially used as complementary therapeutics against cancer and tumor cells with lower or no side-effects, and are generally regarded as safe and (Abdullah et al., 2016, 2017; Abdullah and Hussein, 2021). Microalgal species such as N. oculata (Ochrophyta, Eustigmatophyceae), T. suecica (Chlorophyceae) and Chlorella sp. (Chlorellaceae), have been developed for nutraceutical, functional food, pharmaceuticals, and biochemical resources, in addition to bioenergy, environmental remediation and animal feed applications (Abdullah et al., 2016, 2017; Abdullah and Hussein, 2021). The synergistic application of microalgal bioactive compounds with chemotherapeutic drugs is therefore highly relevant to be explored in clinical setting. Seaweed compounds in combination with anticancer drugs through various mechanisms have exhibited beneficial effects and is a promising strategy to treat breast cancer cells (Malhão et al., 2021). Apoptosis and cell cycle arrest are two essential mechanisms by which anticancer agents exert their anti-proliferative effects on cancer cells. While apoptosis could completely eliminate the cancer cells from the body, cell cycle arrest prevents the division of such cancerous cells. In our study, we have proven that the TMX-MCEs synergistic application triggered both apoptosis (Annexin V) and cell cycle arrest at sub-G1 in the breast cancer cells with lower toxicity in non-cancerous Vero cells. The apoptosis was confirmed by increased activities of Caspase 3/7 and ADP/ATP ratio.
The anti-tumor activity of microalgal metabolites can be attributed to the lipophilicity of some compounds to travel across the lipophilic membranes and potentially react with the apoptotic proteins. Combined therapy of drugs with a carrier or natural products such as MCEs is attractive as it may improve the penetration of drugs into the tumor cells, and enhance their ability to target the tumor site, with reduced side effects against the normal, healthy cells (Abdullah et al., 2014; Gul-e-Saba and Abdullah, 2015; Hussein and Abdullah, 2021). In this case, the newly developed TMX-MCEs formulation, not only retains the cytotoxicity against breast cancer cells, but also with potentially reduced side-effects. The new formulations based on ETH and W crude extracts of N. oculata, T. suecica and Chlorella sp. with TMX at the correct dosage, ratios and duration of treatment, showed strong cytotoxicity against MCF-7 and 4T1 cells, similar to or better than the single TMX and the MCEs-CHL, but with reduced cytotoxicity on the Vero (normal) cells (Table 1, Figure S7). The use of MCEs could achieve the optimum dosage and loading in a carrier for lower cytotoxicity of TMX against the normal healthy cells, whilst retaining the potency to kill cancer cells. These unique properties of the formulation can be harnessed and explored with different drugs for general and specific cancer therapeutic or other applications.
The different and unique metabolites in microalgae play major roles in conferring their specific bioactivities. Primary metabolites provide the needs of physiological activities and act as precursors for secondary metabolites synthesis (Gao et al., 2019). Secondary metabolites depend on environmental signals and a number of stimuli such as media culture, cultivation condition, ambient temperature, pH, carbon, nitrogen, and light (Keller et al., 2005; Chiang et al., 2009). These structurally different types of bioactive compounds have affinity/solubility with solvents of different polarity. Different solvent extracts result in the extraction of different compounds as well as the total extracts of varying quantities. Although methanol is a good organic solvent to extract most of the compounds, it is not as polar as water. Ethanol is highly polar, attributable to the OH group, and its high electro-negativity of oxygen allows hydrogen bonding especially to other ionic and polar molecules. As the ethyl group (C2H5) is nonpolar, ethanol could also attract the nonpolar molecules. Ethanol can therefore dissolve both non-polar and polar compounds (Easy Chem, 2013; Saleh, 2016). Hexane and chloroform are non-polar solvents which extract non-polar compounds, but as chloroform has some polarity, it can dissolve metabolites ranging from moderately polar to moderately non-polar (Saleh, 2016; Azizan et al., 2018).
Metabolite profiling via metabolomics approach is a great tool in identifying and selecting fractions/extracts for chemical identification and characterization (Wu et al., 2015). 1H NMR is an ideal and robust technique for metabolite profiling such as in the analyses of 45 metabolites (Arora et al., 2018), and 50 metabolites (Anderson et al., 2018), in different biosystems. PCA explores the metabolic differences in NMR spectra and compares the difference between samples (Costa et al., 2019). Low concentration and the presence of major number of metabolites however could make the spectrum overcrowded. Many metabolites are produced at low amount in microalgal species, and solvent extraction technique and the overlapping of these compounds with other compounds, or their possible involvement in other more complex metabolic processes either de novo synthesis or transformation from/into other metabolite, may render them undetectable. These metabolites could be important for environmental adaptation (Gao et al., 2019). It is this loss of spectral accuracy due to the sample homogeneity which limits the monitoring of the carotenoids in the vegetables (Sivathanu and Palaniswamy, 2012).
The fluctuation of metabolite levels in microalgae can be influenced by the product transformation and biocatalysis. Fatty acid such as oleic acid is considered as the principal bioactive compound extracted from C. vulgaris (De Morais et al., 2015). The transformation of stearic acid to linoleic acid by oleic acid has been reported to reduce the level of stearic acid in subsequent stages of C. saccharophila cultivation (Singh et al., 2013). In Chlorella pyrenoidosa, the amino acid profiles include tryptophan, phenylalanine, lysine, methionine, valine, leucine, isoleucine, threonine, and histidine. The variations in the level of proteins in C. pyrenoidosa and C. vulgaris could reduce the amount of lysine, asparagine, glutamine and the total amino acids (Safafar et al., 2016). In our study, the significant amount of carotenoids, chlorophyll, pigments, fatty acids and amino acids in the MCEs indicate their potential as the sources of antioxidant and anti-cancer compounds. Pigments such as carotenoids and chlorophyll are detected more in C. vulgaris than in Spirulina platensis (cyanobacteria). Chlorophyll a is known as the main pigments that transform photons into chemical energy. Chlorophyll is a polar compound, and when the amount of water is small or absent in the mobile phase, their separation will not be complete. The organic mobile phase is actually suitable for the separation of pure carotenoids, instead of carotenoids and chlorophyll. Hence, saponification of carotenoids and chlorophyll has to be carried out in the extraction. This is also the reason not all chlorophyll compounds are detected in some microalgal extracts (Hynstova et al., 2018).
Carotenoids (fucoxanthinol, halocynthiaxanthin, and fucoxanthin and its metabolites) exhibit anticancer, antioxidant, anti-inflammatory, anti-obesity and antidiabetic effects and can reduce heart disease (Martin, 2015; Chuyen and Eun, 2017). The antioxidant and anticancer activity of N. oculata and Chlorella sp.-CHL (Hussein et al., 2020b) may be attributable to the presence of choline, vioxanthine and hydroxyphenyllactatic acid. Many isolated compounds from algae, such as bromophenol, phlorotannins and sterols, have also shown high inhibitory activity (Ezzat et al., 2018). The pheophorbide a (PPB a) is a chlorophyll product with high inhibition activity and could promote apoptosis against human lymphoid leukemia Molt 4B cells better than chlorophyll a (Hibasami et al., 2000). Several flavonoid-like compounds such as 6-prenylapigenin 2, 6,8-diprenyleriodictyol (3), gancaonin Q 1, 4-hydroxylon-chocarpin (4), isolated from Dorstenia genus, have exhibited anti-proliferative effects against solid and leukemia cells, and also normal hepatocytes AML12, by cell-cycle arrest, and inducing apoptosis, caspase 3/7 and antiangiogenic activities (Kuete et al., 2011).
The diverse spectrum of metabolites such as carotenoids, phenols, protein, polysaccharide, flavonoids, and fatty acids, with varied bioactivities suggest the potential of microalgae to be used as the sources of adjuvants. Since the activity of TMX in the TMX-MCEs is well-preserved, the synergistic applications can be harnessed as a novel drug delivery system to treat breast cancer. The different metabolites detected in the MCEs may improve the effectiveness of TMX-MCEs and provide additional cytotoxicity to target cancer cells, with reduced toxicity against normal cells. Further study is needed to evaluate the effective concentrations of TMX-MCEs formulation in cancer-induced animal model, before implementation to attain clinically achievable concentrations of TMX and MCEs when administered in humans. The culturing of micro and macroalgae, and increased sampling efforts in extreme or diverse habitats open up ways for the discovery of new species or new anti-cancer agents (Abdullah et al., 2017; Shah and Abdullah, 2018). In combination with technological advancements in genome sequencing and transcription, identification of new species as a source of biocompounds, and assignment of unknown genes to specific metabolic pathways/or functions could be developed (Martínez Andrade et al., 2018). With increasing interest in biopharmaceuticals, and a healthy and functional food as a source of medication and therapeutics, this fundamental understanding may lead to a systems-wide optimization for efficient production, effective therapeutic strategies and safe applications in cancer treatment.
5. Conclusions
The use of ETH and W as green solvents for the production of MCEs and the incorporation in the TMX-MCEs formulation could pave the way for the green production route of natural product as an adjuvant to conventional drug formulation. The TMX:MCEs-ETH at 1:2 and 1:3 ratios showed the IC50 values of 16.98–26.91 μg/mL against MCF-7; 13.8–20.41 μg/mL against 4T1; and 24.54–38.9 μg/mL on Vero cells. For W, the IC50 values were 15.84–29.51 μg/mL against MCF-7; 21.37–31.62 μg/mL on 4T1; and 42.65–85.11 μg/mL on Vero cells. The TMX-MCEs synergistic application exhibited insignificant or lower apoptotic activity against non-cancerous Vero cells, but higher early and late apoptotic events against MCF-7 and 4T1 cells than the control and TMX single-applications. The cell-cycle analysis suggested apoptotic induction with significant enhancement of events in sub-G1 phase with increased Caspase 3/7 and ADP/ATP ratio, especially with TMX-T suecica-ETH. The 1H NMR metabolomics analyses identified forty-four metabolites of different classes including amino acids, vitamins, fatty acids, glucose, carotenoids, chlorophyll and chlorophyll transformation products, an organic acid derivative, and others such as xanthine and unknown compounds. The PCA showed pronounced samples clustering for the three microalgae species and different solvent systems with their respective metabolites, to support the association of the tentatively identified compounds with their anticancer activities. The T. suecica-CHL and HEX exhibited higher amount of metabolites, followed by N. oculata-CHL and HEX, and Chlorella sp.-ETH and MET. Some unique metabolite such as alanine was detected only in the MCEs-W; lysine only in Chlorella sp.-W; and isoleucine in MCEs-HEX, Chlorella sp.-ETH, and N. oculata. The tyrosine, triglycerides, and fucoxanthin were detected in the MCEs-ETH and CHL; glycerol in T. suecica-ETH and Chlorella sp.-CHL; xanthine in MCEs-ETH and Chlorella sp.-CHL; and succinic acid only in Chlorella sp. and N. oculata -ETH, and Chlorella sp. and T. suecica-CHL. The metabolites detected in the MCEs may improve the effectiveness of TMX-MCEs and provide not only cytotoxicity to the target cancer cells but also reduced toxicity on the non-cancerous cells. The toxic side-effects of the conventional drug such as TMX during cancer treatment can therefore be reduced without losing their therapeutic efficacy. This study could pave the way for increased use of microalgae for biopharmaceuticals and functional food industries.
Declarations
Author contribution statement
Hanaa Ali Hussein: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Murni Nur Islamiah Kassim; Faridah Abas: Contributed analysis tools.
M. Maulidiani: Contributed analysis tools, analyzed and interpreted the data.
Mohd Azmuddin Abdullah: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by the Fundamental Research Grant Scheme (FRGS/1/2015/SG05/UMT/02/4) under the Ministry of Higher Education, Malaysia.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors thank the Science Officers in the Institute of Marine Biotechnology, Universiti Malaysia Terengganu for their assistance with the facilities for the experiments, and Mr. Syed Tajudin and Mr. Haziq Hamid from the Laboratory of Animal Cell Culture in Universiti Sultan Zainal Abidin, Tembila Campus, Besut, Terengganu, for their assistance in flow cytometric analyses.
Contributor Information
M. Maulidiani, Email: maulidiani@umt.edu.my.
Mohd Azmuddin Abdullah, Email: azmuddin@sibcogroup.net, joule1602@gmail.com.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Figure S1. Morphology of 4T1 cells after 24 h synergistic application (1:3) at the IC50 levels for (a) TMX-N. oculata-CHL; (b) TMX-T. suecica-CHL; (c) TMX-Chlorella sp.-CHL; (d) TMX-N. oculata-ETH; (e) TMX-T. suecica-ETH; (f) TMX-Chlorella sp.-ETH; (g) TMX-N. oculata-W; (h) TMX-T. suecica-W; (i) TMX-Chlorella sp.-W.
Figure S2. Morphology of Vero cells after 24 h synergistic application (1:3) at the IC50 levels for (a) TMX-N. oculata-CHL; (b) TMX-T. suecica-CHL; (c) TMX-Chlorella sp.-CHL; (d) TMX-N. oculata-ETH; (e) TMX-T. suecica-ETH; (f) TMX-Chlorella sp-.ETH; (g) TMX-N. oculata-W; (h) TMX-T. suecica-W; (i) TMX-Chlorella sp.-W.
Figure S3. Effects of Chlorella sp.-CHL, TMX, TMX-MCEs-CHL, TMX:MCEs-ETH and W (1:3) at the IC50 levels on ADP/ATP ratio in (a) MCF-7; (b) 4T1; (c) Vero cell-lines, after 24 and 48 h treatments, in comparison to Control.
Figure S4. Effects of Chlorella sp.-CHL, TMX, TMX-MCEs-CHL, TMX:MCEs-ETH and W (1:3) at the IC50 levels on Caspase 3/7 activity in (a) MCF-7; (b) 4T1; (c) Vero cell-lines, after 24 and 48 h treatments, in comparison to Control.
Figure S5. Relative quantification of the identified β-carotene, oleic acid, linoleic acid, isoleucine, violaxanthin, triglyceride, stearic acid and EPA of different solvent of (a) N. oculata; (b) T. suecica; (c) Chlorella sp. based on the mean peak area of 1H NMR signals. The chemical shifts (ppm) used for the relative quantification are β-carotene (1.22), oleic acid (1.26), linoleic acid (1.3), isoleucine (0.94), violaxanthin (1.1), triglyceride (5.38), stearic acid (1.02) and EPA (1.66). Data presented are based on the mean of six replicates for each of the solvent systems (CHL, HEX, MET, ETH, W) ± standard deviation (SD).
Figure S6. Score plot of PCA (PC1 versus PC2) for W extracts. The symbol for CW: Chlorella sp.-W, NW: N. oculata-W, TW: T. suecica-W.
Figure S7 The newly developed TMX-MCEs formulation which retained the cytotoxicity against breast cancer cells, and with reduced cytotoxicity on Vero cells. The symbol: NE: N. oculata-ETH, NW: N. oculata-W, NC: N. oculata-CHL, TE: T. suecica-ETH, TW: T. suecica-W, CE: Chlorella sp.-ETH, CW: Chlorella sp.-W, CC: Chlorella sp.-CHL.
References
- Abdullah M.A., Hussein H.A. Integrated algal and oil palm biorefinery as a model system for bioenergy Co-generation with bioproducts and biopharmaceuticals. Bioresour. Bioproc. 2021;8:40. doi: 10.1186/s40643-021-00396-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdullah M.A., Wong H.L., Shah S.M.U., Loh P.C. In: Phycobiotechnology: Biodiversity and Biotechnology of Algae and Algal Products for Food, Feed and Fuel. Sangeetha J., Thangadurai D., Elumalai S., Thimmappa S.C., editors. Apple Academic Press/CRC Press; Burlington, Canada/Boca Raton, Florida, USA: 2021. Algal pathways and metabolic engineering for enhanced production of lipid, carbohydrate and bioactive compounds; pp. 363–430. [Google Scholar]
- Abdullah M.A., Shah S.M.U., Shanab S.M.M., Ali H.E.A. Integrated algal bioprocess engineering for enhanced productivity of lipid, carbohydrate and high-value bioactive compounds. Res. Rev. J. Microb. Biotechnol. 2017;6:61–92. [Google Scholar]
- Abdullah M.A., Ahmad A., Shah S.M.U., Shanab S.M.M., Ali H.E.A., Abo-State M.A.M., Othman M.F. Integrated algal engineering for bioenergy generation, effluent remediation, and production of high-value bioactive compounds. Biotechnol. Bioproc. Eng. 2016;21:236–249. [Google Scholar]
- Abdullah M.A., Ahmad A., Shah S.M.U., El-Sayed H. Biotechnology and Bioinformatics: Advances and Applications for Bioenergy, Bioremediation and Biopharmaceutical Research. CRC Press/Apple Academic Press; New Jersey, USA: 2015. Algal Biotechnology for bioenergy, environmental remediation and high-value biochemicals; pp. 301–344. [Google Scholar]
- Abdullah M.A., Gul-e-Saba, Abdah A. Cytotoxic effects of drug-loaded hyaluronan-glutaraldehyde cross-linked nanoparticles and the release kinetics modeling. J. Adv. Chem. Eng. 2014;1(104):1000104. [Google Scholar]
- Aguilera-Sáez L.M., et al. NMR metabolomics as an effective tool to unravel the effect of light intensity and temperature on the composition of the marine microalgae isochrysis galbana. J. Agric. Food Chem. 2019;67(14):3879–3889. doi: 10.1021/acs.jafc.8b06840. [DOI] [PubMed] [Google Scholar]
- Akim A.M., et al. 4th International Conference on Biomedical Engineering in Vietnam. Springer; Berlin, Heidelberg: 2013. Nanoparticle-encapsulated tamoxifen inducing cytotoxic effect on MCF-7 breast cancer cell lines; pp. 226–227. [Google Scholar]
- Anderson J.R., et al. 1H NMR metabolomics identifies underlying inflammatory pathology in osteoarthritis and rheumatoid arthritis synovial joints. J. Proteome Res. 2018;17(11):3780–3790. doi: 10.1021/acs.jproteome.8b00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjum F., Razvi N., Masood M.A. Breast cancer therapy: a mini review. MOJ Drug Des. Dev. Ther. 2017;1(2):35–38. [Google Scholar]
- AQUACOPs Review of ten years of experimental penaeid shrimp culture in Tahiti and New Caledonia (South Pacific) J. World Maricult. Soc. 1984;15:73–91. [Google Scholar]
- Arora N., et al. NMR-based metabolomic approach to elucidate the differential cellular responses during mitigation of arsenic (III, V) in a green microalga. ACS Omega. 2018;3:11847–11856. doi: 10.1021/acsomega.8b01692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasever-arslan B., et al. Screening of new antileukemic agents from essential oils of algae extracts and computational modeling of their interactions with intracellular signaling nodes. Eur. J. Pharmaceut. Sci. 2016;83:120–131. doi: 10.1016/j.ejps.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Azizan A., et al. Metabolite profiling of the microalgal diatom Chaetoceros calcitrans and correlation with antioxidant and nitric oxide inhibitory activities via 1H NMR-based metabolomics. Mar. Drugs. 2018;16(5):154. doi: 10.3390/md16050154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauton M.S., Størseth T.R., Krane J. High-resolution magic angle spinning NMR analysis of whole cells of chaetoceros muelleri (bacillariophyceae) and comparison with 13C-NMR and distortion less enhancement by polarization transfer 13C-NMR analysis of lipophilic extracts. J. Phycol. 2004;40:611–618. [Google Scholar]
- Chiang Y.M., et al. Unlocking fungal cryptic natural products. Nat. Prod. Commun. 2009;4(11):1505–1510. [PMC free article] [PubMed] [Google Scholar]
- Chuyen V.H., Eun J.B. Marine carotenoids: bioactivities and potential benefits to human health. Crit. Rev. Food Sci. Nutr. 2017;57(12):2600–2610. doi: 10.1080/10408398.2015.1063477. [DOI] [PubMed] [Google Scholar]
- Costa M., et al. Identification of cyanobacterial strains with potential for the treatment of obesity-related Co-morbidities by bioactivity, toxicity evaluation and metabolite profiling. Mar. Drugs. 2019;17(5):280. doi: 10.3390/md17050280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J., et al. Preventive therapy for breast cancer: a consensus statement. Lancet Oncol. 2011;12(5):496–503. doi: 10.1016/S1470-2045(11)70030-4. [DOI] [PubMed] [Google Scholar]
- De Morais M.B., da Silva Vaz de, Etiele Greque M., Vieira Costa J.A. Biologically active metabolites synthesized by microalgae. BioMed Res. Int. 2015;2015:1–15. doi: 10.1155/2015/835761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmyter J.A.N., Melnick J.L., Rawls W.E. Defectiveness of interferon production and of rubella virus interference in a line of Afri-can green monkey kidney cells (vero) J. Virol. 1968;2(10):955–961. doi: 10.1128/jvi.2.10.955-961.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easy Chem . 2013. Ethanol as a Solvent.http://www.easychem.com.au/production-of-materials/renewable- ethanol/ethanol-as-a-solvent [Google Scholar]
- El-hack M.E.A., et al. Microalgae in modern cancer therapy: current knowledge. Biomed. Pharmacother. 2019;111:42–50. doi: 10.1016/j.biopha.2018.12.069. [DOI] [PubMed] [Google Scholar]
- Ezzat S., et al. Looking at marine-derived bioactive molecules as upcoming anti-diabetic agents: a special emphasis on PTP1B inhibitors. Molecules. 2018;23(12):3334. doi: 10.3390/molecules23123334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanciullino R., Ciccolini J., Milano Challenges G. Expectations and limits for nanoparticles-based therapeutics in cancer: a focus on nano-albumin-bound drugs. Crit. Rev. Oncol. Hematol. 2013;88(3):504–513. doi: 10.1016/j.critrevonc.2013.06.010. [DOI] [PubMed] [Google Scholar]
- Fernández Freire P., et al. An integrated cellular model to evaluate cytotoxic effects in mammalian cell lines. Toxicol. Vitro. 2009;23(8):1553–1558. doi: 10.1016/j.tiv.2009.06.017. [DOI] [PubMed] [Google Scholar]
- Fiehn O. Metabolomics - the link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- Fox C.A., et al. Composition of dissolved organic matter in pore waters of anoxic marine sediments analyzed by 1H nuclear magnetic resonance spectroscopy. Front. Mar. Sci. 2018;5(May) [Google Scholar]
- Gao X., Liu B., Ji B. Profiling of small molecular metabolites in nostoc flagelliforme during periodic desiccation. Mar. Drugs. 2019;17(5):298. doi: 10.3390/md17050298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg S., Tiwari P., Kurhanewicz J., Madabhushi A. Variable ranking with PCA: finding multiparametric MR imaging markers for prostate cancer diagnosis and grading. Lect. Notes Comput. Sci.- LNCS. 2011;6963:146–157. [Google Scholar]
- Goh S.H., Yusoff F.M., Loh S.P. A comparison of the antioxidant properties and total phenolic content in a diatom, Chaetoceros sp. and a green Microalga, Nannochloropsis sp. J. Agric. Sci. 2010;2(3):123. [Google Scholar]
- Gomez-casati D.F., Zanor M.I., V Busi M. Metabolomics in plants and humans: applications in the prevention and diagnosis of diseases. BioMed Res. Int. 2013;2013:1–11. doi: 10.1155/2013/792527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Wang B., Wu Q., Wei X., Zheng F. Men et al., Combined delivery and anti-cancer activity of paclitaxel and curcumin using polymeric micelles. J. Biomed. Nanotechnol. 2015;11:578–589. doi: 10.1166/jbn.2015.1964. [DOI] [PubMed] [Google Scholar]
- Gul-e-Saba, Abdullah M.A. In: Biotechnology and Bioinformatics: Advances and Applications for Bioenergy, Bioremediation, and Biopharmaceutical Research. Thangadurai D., Sangeetha J., editors. CRC Press/Apple Academic Press; New Jersey, USA: 2015. Polymeric nanoparticle mediated targeted drug delivery to cancer cells; pp. 1–34. [Google Scholar]
- Gurunathan S., et al. Cytotoxicity of biologically synthesized silver nanoparticles in MDA-MB-231 human breast cancer cells. BioMed Res. Int. 2013;2013:10. doi: 10.1155/2013/535796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajiaghaalipour F., et al. Underlying mechanism for the modulation of apoptosis induced by a new benzoindole derivative on HT-29 colon cancer cells. RSC Adv. 2017;7(61):38257–38263. [Google Scholar]
- Hassan F., et al. Cytotoxic effects of tamoxifen in breast cancer cells. J. Unexplored Med. Data. 2018;3:3. [Google Scholar]
- Hibasami H., et al. Pheophorbide a, a moiety of chlorophyll a, induces apoptosis in human lymphoid leukemia Molt 4B cells. Int. J. Mol. Med. 2000;6(3):277–279. [PubMed] [Google Scholar]
- Hosseinia A., Bakhtiarib E., Mousavid S.H. Protective effect of Hibiscus sabdariffa on doxorubicin-induced cytotoxicity in H9c2 cardiomyoblast cells. Iran. J. Pharm. Res. (IJPR) 2017;16(2):708–713. [PMC free article] [PubMed] [Google Scholar]
- How C.W., Rasedee A., Manickam S., Rosli R. Tamoxifen-loaded nanostructured lipid carrier as a drug delivery system: characterization, stability assessment and cytotoxicity. Colloids Surf. B Biointerfaces. 2013;112:393–399. doi: 10.1016/j.colsurfb.2013.08.009. [DOI] [PubMed] [Google Scholar]
- Hussein H.A., Abdullah M.A. Novel drug delivery systems based on silver nanoparticles, hyaluronic acid, lipid nanoparticles and liposomes for cancer treatment. Appl. Nanosci. 2021 [Google Scholar]
- Hussein H.A., Mohamad H., Ghazaly M.M., Laith A.A., Abdullah M.A. Cytotoxic effects of Tetraselmis suecica chloroform extracts with silver nanoparticle co-application on MCF-7, 4 T1, and Vero cell lines. J. Appl. Phycol. 2020;32:127–143. [Google Scholar]
- Hussein H.A., Mohamad H., Ghazaly M.M., Laith A.A., Abdullah M.A. Anticancer and antioxidant activities of Nannochloropsis oculata and Chlorella sp. extracts in co-application with silver nanoparticle. J. King Saud Univ. Sci. 2020;32(8):3486–3494. [Google Scholar]
- Hussein H.A., Syamsumir D.F., Radzi S.A.M., Siong J.Y.F., Zin N.A.M., Abdullah M.A. Phytochemical screening, metabolite profiling and enhanced antimicrobial activities of microalgal crude extracts in co-application with silver nanoparticle. Bioresour. Bioproc. 2020;7:39. [Google Scholar]
- Hussein H.A., Maulidiani M., Abdullah M.A. Microalgal metabolites as anti-cancer/anti-oxidant agents reduce cytotoxicity of elevated silver nanoparticle levels against non-cancerous vero cells. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e05263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y.J., Lee E.J., Kim H.R., Hwang K.A. Molecular mechanisms of luteolin-7-o-glucoside-induced growth inhibition on human liver cancer cells: G2/M cell cycle arrest and caspase-independent apoptotic signaling pathways. BMB Rep. 2013;46(12):611–616. doi: 10.5483/BMBRep.2013.46.12.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynstova V., Sterbova D., Klejdus B., Hedbavny J., Huska D., Adam V. Separation, identification and quantification of carotenoids and chlorophylls in dietary supplements containing Chlorella vulgaris and Spirulina platensis using high performance thin layer chromatography. J. Pharm. Biomed. Anal. 2018;148:108–118. doi: 10.1016/j.jpba.2017.09.018. [DOI] [PubMed] [Google Scholar]
- Janitabar-Darzi S., Rezaei R., Yavari K. In vitro cytotoxicity effects of (197)Au/PAMAMG4 and (198)Au/PAMAMG4 nanocomposites against MCF7 and 4T1 breast cancer cell lines. Adv. Pharmaceut. Bull. 2017;7(1):87–95. doi: 10.15171/apb.2017.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspars M., et al. The marine biodiscovery pipeline and ocean medicines of tomorrow. J. Mar. Biol. Assoc. U. K. 2016;96(1):151–158. [Google Scholar]
- Jones K.L., Buzdar A.U. A review of adjuvant hormonal therapy in breast cancer. Endocr. Relat. Cancer. 2004;11(3):391–406. doi: 10.1677/erc.1.00594. [DOI] [PubMed] [Google Scholar]
- Jordan V.C. Tamoxifen (ICI46,474) as a targeted therapy to treat and prevent breast cancer. Br. J. Pharmacol. 2006;147(S1):S269–S276. doi: 10.1038/sj.bjp.0706399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller N.P., Turner G., Bennett J.W. Fungal secondary metabolism—from biochemistry to genomics. Nat. Rev. Microbiol. 2005;3(12):937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- Kim Y.S., et al. Stigmasterol isolated from marine microalgae navicula incerta induces apoptosis in human hepatoma HepG2 cells. BMB Rep. 2014;47(8):433–438. doi: 10.5483/BMBRep.2014.47.8.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuete V., et al. Cytotoxicity and mode of action of four naturally occurring flavonoids from the genus Dorstenia: gancaonin Q, 4-hydroxylonchocarpin, 6-prenylapigenin, and 6,8-diprenyleriodictyol. Planta Med. 2011;77(18):1984. doi: 10.1055/s-0031-1280023. –89. [DOI] [PubMed] [Google Scholar]
- Kumarihamy M., et al. Antiplasmodial and cytotoxic cytochalasins from an endophytic fungus, nemania sp. UM10M, isolated from a diseased Torreya taxifolia leaf. Molecules. 2019;24(4):777. doi: 10.3390/molecules24040777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Heinrich M., Myers S., Dworjanyn S.A. Towards a better understanding of medicinal uses of the Brown Seaweed Sargassum in traditional Chinese medicine: a phytochemical and pharmacological review. J. Ethnopharmacol. 2012;142(3):591–619. doi: 10.1016/j.jep.2012.05.046. [DOI] [PubMed] [Google Scholar]
- Ma N., et al. Metabolites Re-programming and physiological changes induced in scenedesmus regularis under nitrate treatment. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-018-27894-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhão F., Ramos A.A., Macedo A.C., Rocha E. Cytotoxicity of seaweed compounds, alone or combined to reference drugs, against breast cell lines cultured in 2D and 3D. Toxics. 2021;9(1):32. doi: 10.3390/toxics9020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek C.B., Peralta R.M., Itinose A.M., Bracht A. Influence of tamoxifen on gluconeogenesis and glycolysis in the perfused rat liver. Chem. Biol. Interact. 2011;193(1):22–33. doi: 10.1016/j.cbi.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Martin L.J. Fucoxanthin and its metabolite fucoxanthinol in cancer prevention and treatment. Mar. Drugs. 2015;13(8):4784–4798. doi: 10.3390/md13084784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez Andrade K., Lauritano C., Romano G., Ianora A. Marine microalgae with anti-cancer properties. Mar. Drugs. 2018;16(5):165. doi: 10.3390/md16050165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashjoor S., Yousefzadi M., Esmaeili M.A., Rafie R. Cytotoxicity and antimicrobial activity of marine macro algae (dictyotaceae and ulvaceae) from the Persian gulf. Cytotechnology. 2015;68(5):1717–1726. doi: 10.1007/s10616-015-9921-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes G.D.S., et al. Antiviral activity of the green marine alga Ulva fasciata on the replication of human metapneumovirus. Rev. Inst. Med. Trop. Sao Paulo. 2010;52(1) doi: 10.1590/s0036-46652010000100001. 03–10. [DOI] [PubMed] [Google Scholar]
- Mimeault M., Jouy N., Depreux P., Hénichart J.-P. Synergistic antiproliferative and apoptotic effects induced by mixed epidermal growth factor receptor inhibitor ZD1839 and nitric oxide donor in human prostatic cancer cell lines. Prostate. 2005;62:187–199. doi: 10.1002/pros.20138. [DOI] [PubMed] [Google Scholar]
- Molinski T.F., Dalisay D.S., Lievens S.L., Saludes J.P. Drug development from marine natural products. Nat. Rev. Drug Discov. 2009;8(1):69–85. doi: 10.1038/nrd2487. [DOI] [PubMed] [Google Scholar]
- Morrow M., Jordan V.C. Tamoxifen for the prevention of breast cancer in the high-risk woman. Ann. Surg Oncol. 2000;7(1):67–71. doi: 10.1007/s10434-000-0067-0. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65(1–2):55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Moungjaroen J., et al. Reactive oxygen species mediate caspase activation and apoptosis induced by lipoic acid in human lung epithelial cancer cells through Bcl-2 down-regulation. J. Pharmacol. Exp. Therapeut. 2006;319(3):1062–1069. doi: 10.1124/jpet.106.110965. [DOI] [PubMed] [Google Scholar]
- Newman D.J., Cragg G.M. Marine natural products and related compounds in clinical and advanced preclinical trials. J. Nat. Prod. 2004;67(8):1216–1238. doi: 10.1021/np040031y. [DOI] [PubMed] [Google Scholar]
- Nigjeh S.E., et al. Cytotoxic effect of ethanol extract of microalga, Chaetoceros calcitrans, and its mechanisms in inducing apoptosis in human breast cancer cell line. BioMed Res. Int. 2013;8 doi: 10.1155/2013/783690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive G., van Genderen M. A 1H, 13C, 31P and 15N NMR study of (Pyrrolidine-2,2-diyl)Bisphosphonic acid, tetraalkyl(pyrrolidine-2,2-diyl)Bisphosphonates and acyclic tetraethyl bisphosphonates. Magn. Reson. Chem. 2000;38(5):379–381. [Google Scholar]
- Pan Z., Raftery D. Comparing and combining NMR spectroscopy and mass Spectrometry in metabolomics. Anal. Bioanal. Chem. 2007;387(2):525–527. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- Pantazis P., et al. Sensitivity of camptothecin-resistant human leukemia cells and tumors to anticancer drugs with diverse mechanisms of action. Leuk. Res. 1995;19(1):43–55. doi: 10.1016/0145-2126(94)00060-n. [DOI] [PubMed] [Google Scholar]
- Patel B.H. In: Clark M., editor. Woodhead Publishing Ltd; Cambridge, UK: 2011. Natural dyes; pp. 395–424. (Handbook of textile and industrial dyeing, Principles, Processes and Types of Dyes). [Google Scholar]
- Perry R.R., Kang Y., Greaves B. Effects of tamoxifen on growth and apoptosis of estrogen-dependent and-independent human breast cancer cells. Ann. Surg Oncol. 1995;2(3):238–239. doi: 10.1007/BF02307030. [DOI] [PubMed] [Google Scholar]
- Petinari L., Kohn L.K., De Carvalho J.E., Genari S.C. Cytotoxicity of tamoxifen in normal and tumoral cell lines and its ability to induce cellular transformation in vitro. Cell Biol. Int. 2004;28(7):531–539. doi: 10.1016/j.cellbi.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Prabaharan M., et al. Folate-conjugated amphiphilic hyperbranched block copolymers based on Boltorn® H40, poly (l-Lactide) and poly (ethylene glycol) for tumor-targeted drug delivery. Biomaterials. 2009;30(16):3009–3019. doi: 10.1016/j.biomaterials.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Reyna-Martinez R., et al. Antitumor activity of Chlorella sorokiniana and scenedesmus sp. microalgae native of nuevo león state, México. PeerJ. 2018;6:e4358. doi: 10.7717/peerj.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhimoghadam M., et al. Tamoxifen-induced apoptosis of MCF-7 cells via GPR30/PI3K/MAPKs interactions: verification by ODE modeling and RNA sequencing. Front. Physiol. 2018;9:907. doi: 10.3389/fphys.2018.00907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safafar H., et al. Enhancement of protein and pigment content in two Chlorella species cultivated on industrial process water. J. Mar. Sci. Eng. 2016;4(4):84. [Google Scholar]
- Saleh F.R. Antibacterial activity of seeds of Iraqi dates. J. BioInnov. 2016;5(2):313–318. [Google Scholar]
- Salih G.A., Ahmad-Raus R., Shaban M.N., Abdullah N. Extraction and purification of cytotoxic compounds from Premna serratifolia L. (Bebuas) for human breast cancer treatment. Int. Food Res. J. 2017;24(Suppl):S281–S286. [Google Scholar]
- Sarojini S., Senthilkumaar P., Ramesh V. Impact of ethanolic extract of Mikania glomerata on human breast cancer (MCF 7) cell line. Int. J. Adv. Sci. Res. 2016;2(4):94–193. [Google Scholar]
- Sarpal S., et al. Compositional analyses of oil extracts of microalgae biomasses by NMR and chromatographic techniques. J. Anal. Bioanal. Sep. Tech. 2016;1(1):17–41. [Google Scholar]
- Schempp C.M., et al. Inhibition of tumour cell growth by hyperforin, a novel anticancer drug from st. John’s wort that acts by induction of apoptosis. Oncogene. 2002;6(8):1242–1250. doi: 10.1038/sj.onc.1205190. [DOI] [PubMed] [Google Scholar]
- Schumacher M., Kelkel M., Dicato M., Diederich M. Gold from the sea: marine compounds as inhibitors of the hallmarks of cancer. Biotechnol. Adv. 2012;29(5):531–547. doi: 10.1016/j.biotechadv.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Senthilraja P., Kathiresan K. In vitro cytotoxicity MTT assay in vero, HepG2 and MCF-7 cell lines study of marine yeast. J. Appl. Pharmaceut. Sci. 2015;5(3):80–84. [Google Scholar]
- Shah S.M.U. Universiti Teknologi PETRONAS, Seri Iskandar; Malaysia: 2014. Cell Culture Optimization and Reactor Studies of Green and Brown Microalgae for Enhanced Lipid Production. [Google Scholar]
- Shah S.M.U., Abdullah M.A. Effects of macro/micronutrients on green and Brown microalgal cell growth and fatty acids in photobioreactor and open-tank systems. Biocatal. Agric. Biotechnol. 2018;14:10–17. [Google Scholar]
- Sharif N., et al. Prolific anticancer bioactivity of algal extracts (review) Am. J. Drug Deliv. Therapeut. 2014;3(4):8. [Google Scholar]
- Siddiqui S., et al. Cytostatic and anti-tumor potential of ajwa date pulp against human hepatocellular carcinoma HepG2 cells. Sci. Rep. 2019;9(1):245. doi: 10.1038/s41598-018-36475-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigma Aldrich . 2003. Fundamental Techniques in Cell Culture: A Laboratory Handbook. [Google Scholar]
- Singh D., et al. Characterization of a new zeaxanthin producing strain of Chlorella saccharophila isolated from New Zealand marine waters. Bioresour. Technol. 2013;143:308–314. doi: 10.1016/j.biortech.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Singh S., Kate B.N., Banerjee U.C. Bioactive compounds from cyanobacteria and microalgae: an overview. Crit. Rev. Biotechnol. 2005;25(3):73–95. doi: 10.1080/07388550500248498. [DOI] [PubMed] [Google Scholar]
- Sit N.W., et al. In vitro antidermatophytic activity and cytotoxicity of extracts derived from medicinal plants and marine algae. J. Mycol. Med. 2018;28(3):561–567. doi: 10.1016/j.mycmed.2018.07.001. [DOI] [PubMed] [Google Scholar]
- Sivathanu B., Palaniswamy S. Purification and characterization of carotenoids from green algae Chlorococcum humicola by HPLC-NMR and LC-MS-APCI. Biomed. Prev. Nutr. 2012;2(4):276–282. [Google Scholar]
- Skerman N.B., Joubert A.M., Cronjé M.J. The apoptosis inducing effects of Sutherlandia spp. extracts on an oesophageal cancer cell line. J. Ethnopharmacol. 2011;137(3):1250–1260. doi: 10.1016/j.jep.2011.07.054. [DOI] [PubMed] [Google Scholar]
- Sombatsri A., et al. Atalantums H-K from the peels of Atalantia monophylla and their cytotoxicity. Nat. Prod. Res. 2019:1–7. doi: 10.1080/14786419.2019.1576042. [DOI] [PubMed] [Google Scholar]
- Steward W.P., Brown K. Cancer chemoprevention: a rapidly evolving field. Br. J. Cancer. 2013;109(1):1–7. doi: 10.1038/bjc.2013.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., et al. “Iron-saturated” bovine lactoferrin improves the chemotherapeutic effects of tamoxifen in the treatment of basal-like breast cancer in mice. BMC Cancer. 2012;12(1):591. doi: 10.1186/1471-2407-12-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szliszka E., Czuba Z.P., Jernas K., Król W. Dietary flavonoids sensitize HeLa cells to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) Int. J. Mol. Sci. 2008;9:56–64. doi: 10.3390/ijms9010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.H.K., Kim G.P., Van Wezel, Choi Y.H. Metabolomics in the natural products field–a gateway to novel antibiotics. Drug Discov. Today Technol. 2015;13:11–17. doi: 10.1016/j.ddtec.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Yuan Y.V., Walsh N.A. Antioxidant and antiproliferative activities of extracts from a variety of edible seaweeds. Food Chem. Toxicol. 2006;44:1144–1150. doi: 10.1016/j.fct.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Yusmazura Z., Lim W.Y., Nik Fakhuruddin N.H. Anti-cancer effects of clinacanthus nutans extract towards human cervical cancer cell. J. Biomed. Clin. Sci. 2017;2(1):11–19. [Google Scholar]
- Zhang Y., et al. Free amino acids and small molecular acids profiling of marine microalga Isochrysis zhangjiangensis under nitrogen deficiency. Algal Res. 2016;13:207–217. [Google Scholar]
- Zhou B., et al. Claudin 1 promotes migration and increases sensitivity to tamoxifen and anticancer drugs in luminal-like human breast cancer cells MCF7. Cancer Invest. 2015;33(9):429–439. doi: 10.3109/07357907.2015.1060996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Morphology of 4T1 cells after 24 h synergistic application (1:3) at the IC50 levels for (a) TMX-N. oculata-CHL; (b) TMX-T. suecica-CHL; (c) TMX-Chlorella sp.-CHL; (d) TMX-N. oculata-ETH; (e) TMX-T. suecica-ETH; (f) TMX-Chlorella sp.-ETH; (g) TMX-N. oculata-W; (h) TMX-T. suecica-W; (i) TMX-Chlorella sp.-W.
Figure S2. Morphology of Vero cells after 24 h synergistic application (1:3) at the IC50 levels for (a) TMX-N. oculata-CHL; (b) TMX-T. suecica-CHL; (c) TMX-Chlorella sp.-CHL; (d) TMX-N. oculata-ETH; (e) TMX-T. suecica-ETH; (f) TMX-Chlorella sp-.ETH; (g) TMX-N. oculata-W; (h) TMX-T. suecica-W; (i) TMX-Chlorella sp.-W.
Figure S3. Effects of Chlorella sp.-CHL, TMX, TMX-MCEs-CHL, TMX:MCEs-ETH and W (1:3) at the IC50 levels on ADP/ATP ratio in (a) MCF-7; (b) 4T1; (c) Vero cell-lines, after 24 and 48 h treatments, in comparison to Control.
Figure S4. Effects of Chlorella sp.-CHL, TMX, TMX-MCEs-CHL, TMX:MCEs-ETH and W (1:3) at the IC50 levels on Caspase 3/7 activity in (a) MCF-7; (b) 4T1; (c) Vero cell-lines, after 24 and 48 h treatments, in comparison to Control.
Figure S5. Relative quantification of the identified β-carotene, oleic acid, linoleic acid, isoleucine, violaxanthin, triglyceride, stearic acid and EPA of different solvent of (a) N. oculata; (b) T. suecica; (c) Chlorella sp. based on the mean peak area of 1H NMR signals. The chemical shifts (ppm) used for the relative quantification are β-carotene (1.22), oleic acid (1.26), linoleic acid (1.3), isoleucine (0.94), violaxanthin (1.1), triglyceride (5.38), stearic acid (1.02) and EPA (1.66). Data presented are based on the mean of six replicates for each of the solvent systems (CHL, HEX, MET, ETH, W) ± standard deviation (SD).
Figure S6. Score plot of PCA (PC1 versus PC2) for W extracts. The symbol for CW: Chlorella sp.-W, NW: N. oculata-W, TW: T. suecica-W.
Figure S7 The newly developed TMX-MCEs formulation which retained the cytotoxicity against breast cancer cells, and with reduced cytotoxicity on Vero cells. The symbol: NE: N. oculata-ETH, NW: N. oculata-W, NC: N. oculata-CHL, TE: T. suecica-ETH, TW: T. suecica-W, CE: Chlorella sp.-ETH, CW: Chlorella sp.-W, CC: Chlorella sp.-CHL.
Data Availability Statement
Data included in article/supplementary material/referenced in article.