Abstract
Chlorpyrifos (CP) is an organophosphate widely used as an insecticide and acaricide. Extensive application of CP contaminates ecosystems, polluting the environment and food products, creating health complications to humans due to its neurotoxicity. The study evaluated CP bioremediation by bacteria isolated from dairy farm soils in Nakuru County, Kenya, through enrichment culture technique. The growth response of the bacteria and degradation of chlorpyrifos was monitored every five days using UV-VIS Spectrophotometer (600nm). Enrichment culture technique led to the isolation of eighteen (MA1-MA18) potential CP degraders belonging to the genera Pseudomonas, Stenotrophomonas, Bacillus, Alicaligenes, and Achromobacter. The efficacy of four (4) strains was further investigated using Gas Chromatography-Mass Spectrometry (GC-MS) analysis. The results showed that all four strains significantly degraded chlorpyrifos in Minimum Salt Medium (MSM): Lysinibacillus sp.HBUM206408 (87.16 %), Stenotrophomonas maltophilia (82.04 %), Pseudomonas putida (89.52 %), and Achromobacter insuavis (91.08 %) within 16 days, producing 2-Hydroxy-3, 5, 6-trichloropyridine (TCP) as the main metabolite. Therefore, these strains can be used to degrade chlorpyrifos in contaminated soil. There is a need for further studies to determine the possible mechanisms and other metabolites of chlorpyrifos degradation by the isolates obtained in the study. Besides, future studies should explore the efficacy and survival of the organisms in the contaminated environment.
Keywords: Biodegradation, Bioremediation, Chlorpyrifos, Metabolite, Organophosphate, Spectrophotometer
Biodegradation, Bioremediation, Chlorpyrifos, Metabolite, Organophosphate, Spectrophotometer.
1. Introduction
Organophosphates (OPs) are widely used in agriculture as pesticides due to their high efficiency (Das and Adhya, 2015). Chlorpyrifos [O, Odiethyl-O-(3, 5, 6-trichloro-2-pyridyl) phosphorothioate] (CP) is among the leading acaricides used by dairy farmers in Kenya to control ticks (Mutavi et al., 2018). Although it has low solubility in water, it readily dissolves in most organic solvents (Barathidasan et al., 2014; Rayu et al., 2017). Excessive application of CP as an acaricide has caused contamination of animal products (Singh and Walker, 2006), including meat and milk (Barathidasan et al., 2014). In particular, milk retains acaricide residues due to its lipophilic nature.
In humans, CP residues interfere with the action of acetyl-cholinesterase (AChE-ase), thereby disrupting Central Nervous System (CNS) activity (Aswathi et al., 2019). Cases of severe poisoning in humans by CP are known to cause symptoms such as nausea, headache, muscle twitching, dizziness, increased sweating, weakness, and salivation (Ambreen and Yasmin, 2021). Although studies have documented the adverse impact of organophosphates on human and animal health, there is still a dearth of literature concerning the best ways of eliminating them from contaminated environments (Fatima, 2019; Jokar et al., 2021).
Often, pesticide residues in animal products originate from the contaminated environment (water, soil, and feed). In particular, the soil is a reservoir of CP residues and their metabolites (Rayu et al., 2017). Thus, there is a need to eliminate the residues from contaminated soils through biodegradation (Barathidasan et al., 2014; Das and Adhya, 2015). Although abiotic pathways can cause the degradation of xenobiotics in water and soil, the primary mechanism for breakdown and detoxification in soils is the action of microorganisms (Ortiz-Hernández and Sánchez-Salinas, 2010).
Various studies have shown that microbial activity is the most significant factor in xenobiotics degradation, although physical-chemical factors like moisture content, temperature, PH, pesticide formulation, and organic carbon content can also influence the process (Naphade et al., 2012; Kumar et al., 2018a, b). According to Barathidasan et al. (2014), microbial biodegradation is quite attractive for removing toxic chemicals from the environment. Bioremediation is a low-cost, environmentally friendly, and easy-to-use approach to eliminate CP from the soil and water (Rayu et al., 2017). Previous studies have reported a few chlorpyrifos-degrading (CP-degrading) bacteria, including Pseudomonas sp., Serratia, and Enterobacter, Alcaligenes, Sphingobacterium, Gordonia, Paracoccus, and Mesorhizobium (Barathidasan et al., 2014; Abdel-Wareth and Abd El-Hamid, 2016; Rayu et al., 2017; Kumar et al., 2018a, b; Akhdiya et al., 2020). The main products of CP degradation are 3, 5, 6-trichloro-2-pyridinol (TCP) diethyl thiophosphoric acid (DETP) (Rayu et al., 2017).
Despite the widespread application of OP acaricides in Kenya, including CP (Atego et al., 2021), few studies have isolated and characterized microorganisms with degradation potential. The current research aimed to isolate CP degrading bacteria from dairy farm soil, which ably, rapidly, and efficiently degrade CP. Thus, the isolation of native microorganisms would be favorable for in situ bioremediation because they are better adapted to the local environment. The study also aimed to characterize the isolated strains using biochemical and molecular techniques and monitor degradation by using GC-MS. Repetitive enrichment and successive culturing were used to examine their CP degrading potential for indigenous bacterial strains.
2. Materials and methods
2.1. Study site
The study was carried out in Nakuru Country in Kenya (Latitude: 0° 29' 59.99" N; Longitude: 36° 00' 0.00" E) (Figure 1). Nakuru is 1871 m above sea level, with a mild climate, generally temperate and warm. Annual rainfall is approximately 762 mm, and the average temperature is 17.5 °C.
Figure 1.
Map of the study area, Nakuru County, Kenya.
2.2. Pre-study survey
Before the actual study, a field survey was conducted on dairy farms in Nakuru County. The purpose of the survey was to identify dairy farms that applied organophosphate acaricides (Chimbevo et al., 2021). The survey findings were important in informing the subsequent phases of the study, including sample collection. Information sought in the survey included the history of pesticide application, type of acaricide used, the amount applied, and waste disposal.
2.3. Sample collection
Based on the survey findings that mapped dairy farms that frequently use organophosphates, fifteen soil and dip wash samples from dairy farms in Nakuru County, Kenya, suspected to be contaminated with chlorpyrifos, were collected. The soil was picked at 10 cm below the soil surface using a soil auger, transported in sterile sealable plastic bags, and kept in a refrigerator at 4 °C. The samples were air-dried and ground before being passed through 2 mm sieves according to (Das and Adhya, 2015). The soils were grouped into N1S, N2S, and N3S based on regions and mixed thoroughly to form three composite samples. Collected dip wash was also grouped into three composites as N1D, N2D, and N3D.
2.4. Enrichment culture technique
Enrichment culture technique using Minimum Salt Medium (MSM) (adapted from Barathidasan et al., 2014) was used to isolate potential chlorpyrifos bacteria. MSM was constituted as follows (in grams per liter of distilled water): Ammonium Nitrate ((NH4)2NO3) 1; Hydrated Calcium Nitrate (Ca(NO3)2.2H2O), 0.04; Hydrated Magnesium Sulphate (MgSO4.7H2O), 0.1; Potassium Chloride (KCl), 0.2; Hydrated Iron (ii) sulfate (FeSO4.7H2O), 0.001, Dipotassium phosphate (K2HPO4.12H2O), 1.5; and Potassium dihydrogen phosphate (KH2PO4), 4.8. Adjustment of pH was done to 7 by using the Benchtop pH meter. Commercial grade chlorpyrifos (Duodip 55% EC) was supplemented into the medium to serve as the only carbon source. The use of commercial grade acaricide was found appropriate because they resemble the active compound the microorganism is exposed to in the soil. Chlorpyrifos supplementation was done at three different concentrations: 5 ppm, 10 ppm, and 40 ppm. A gram of soil was dispersed in 10 ml of distilled water and serially diluted from each composite. The sixth dilution (10−6) was inoculated in MSM broth. The liquid media were incubated on a rotary shaker (150 rpm) and maintained at 30 °C and optimum pH for 21 days. Bacterial growth was assessed by measuring medium turbidity (OD600) changes on a UV-VIS Spectrophotometer. Samples of the liquid media were collected periodically (5 days) for Optical density measurement. Afterward, aliquots (1 ml) were inoculated onto MSM agar plates (Barathidasan et al., 2014). The plates were incubated at 30 °C for seven days. Bacterial colonies were then streaked into Nutrient Agar (supplemented with 10 ppm chlorpyrifos) plates and sequentially sub-cultured to obtain pure colonies.
2.5. Morphological and biochemical characterization of isolated CP-degrading bacteria
Cultural and morphological characteristics were used for preliminary characterization of the bacteria isolates. Colony characteristics used for identification include color, elevation, margin, and size. Besides, the isolates were viewed under a compound microscope, aided by the Gram Staining Technique (Rayu et al., 2017). Afterward, biochemical tests were done, including Indole Test, Methyl-Red Test, Catalase, Oxidase, Urease, Citrate Utilization, Lactose Utilization, and production of Hydrogen sulfide (Cao et al., 2018).
2.6. Molecular identification of bacterial isolates
2.6.1. Bacterial DNA extraction
The Zymo quick DNA™ Min Prep Kit (Zymo Research, USA) was used for genomic DNA extraction, with strict adherence to manufacturer instructions (Wei et al., 2020). In this regard, freshly cultured bacteria isolates were suspended in normal saline (500 μL) in Eppendorf tubes. Afterward, vertexing was done for 10 s, followed by 3 min of centrifugation at 13,000 revolutions per minute (rpm). Subsequently, the sodium chloride was discarded, and genomic lysis buffer (400 μL) was added to the mixture, followed by further 10 s of vertexing. Incubation of the contents was carried out for 30 min at room temperature, followed by a transfer to the Zymo-Spin column placed in collection tubes and subjected to centrifugation for a minute at 13,000 rpm. Afterward, a pre-wash buffer was added (200 μL) followed by another round of centrifugation (Daneshparvar et al., 2017). The next step was the addition of gDNA wash buffer (500 μL) and centrifugation (13,000 rpm). Afterward, contents were transferred to Eppendorf tubes, and DNA elution buffer (50 μL) was added. The elution was done through incubation at room temperature for 10 min, followed by centrifugation at 13,000 rpm for 30 s (Cao et al., 2018). Finally, the eluted DNA was kept at -20 °C.
2.6.2. PCR amplification of 16sRNA region of bacteria
The protocol described by Lorenz (2012) was used in the Polymerase Chain Reaction (PCR) to amplify rRNA in a 96 well thermocycler. After the initial denaturation at 95 °C for 5 min, automated sequential 30 cycles were done that included denaturing (95 °C; 45 s, primer annealing (62 °C; 45 s), extension (72 °C; 2 min). The final extension was set at 72 °C for 5 min. The obtained DNA was at 4 °C. The primers used in the amplification of the 16S rRNA gene were the 1492r (5'GTTACCTTGTTACGACTTC-3') and 27f (5'-AGTTTGATCCTGGCTCAG-3') (Daneshparvar et al., 2017). These are the most commonly used primers for bacterial 16 rRNA genes as they anneal to the highly conserved areas of the gene in specific bacteria (Lorenz, 2012).
2.6.3. Agarose gel electrophoresis
SYBR Green (BioLab, USA) was used for staining of the PCR products, and agarose gel electrophoresis was used for separation in 1.0 % (w/v) agarose gel, with 0.5X Tris Borate EDTA used as a buffer for 30 min at 80 V (Franco-Duarte et al., 2019). The gel was visualized using an Ultra Violet trans-illuminator (Seriennumber: UV 31208/070200004). A kilobase pair DNA (1000 base pair) ladder was used to estimate the band's molecular sizes. After that, a digital camera was used to photograph the gel image.
2.6.4. Molecular sequencing of amplified bacterial DNA
First, the QIAquick PCR purification kit was used to purify the PCR products by strictly following the instructions of the manufacturer (Qiagen, Tiangen, China). The primers 27F and 1492R were used for the sequencing (Johnson et al., 2019). In the PCR reaction, five picomoles of the forward and reverse primers were used in a 20 μl column with a DNA template of 2.5 μl. After PCR product purification, 500 bp sequencing was carried out at Macrogen Europe Laboratory in Amsterdam-Netherlands, where the Sanger sequencing method was used, which utilized the automated sequencer ABI 3700 DNA with Big Dye Terminator Kit V. 3.1, and the instructions of the manufacturer were followed in the reactions (Applied Biosystems, USA).
2.6.5. GC-MS analysis of degradation metabolites
Isolates that showed the greatest biodegradation potential were further analyzed using gas chromatography-mass spectrometry (GC-MS) to identify degradation metabolites. Liquid samples (10 ml) were added to 50 ml of deionized water and extracted using 20ml hexane on a rotary shaker for 1 h (El-Gawad, 2016). Dehydration of extracts was carried out using anhydrous Na2SO4 followed by drying by evaporation under Nitrogen at 45 °C in a rotary evaporator. Afterward, there was dilution with acetone to a final volume of 10 ml for analysis by chromatography. Samples were analyzed using Agilent 7890A gas chromatography (Agilent Technologies, Inc., Beijing, China) and a 5975 C inert mass detector under the following conditions: The injector temperature was set at 270 °C and the transfer line temperature at 280 °C. Initial temperature was set at 35 °C, maintained for 5 min, before an increase of 10 °C in the next 10.5 min, followed an increment of 50 ºC/minute in the next 30 min to 285 °C. Helium was used as the carrier gas, at a flow rate of 1.25 ml/min. The mass selective detector (Agilent 5973) operated an ion source temperature and a quadruple temperature of 250 °C and 180 °C respectively. A temperature of 230 °C was set as the temperature of the ion source (EL-Maali and Yehia Wahman, 2015). Moreover, an acceleration energy of 70 eV was used to obtain the electron impact (EI) mass spectra. Aliquots of 1.0 μL sample extract were injected directly by the auto-sampler under a quadruple technique (QMS) (Yin et al., 2021). Fragment ions were analyzed over the 40–550 m/z mass range in the full scan, with the filament delay time set at 3.3 min (El-Gawad, 2016).
2.6.6. Data management and analysis
The Spreadsheet Microsoft Excel was used for raw data entry. The Computer Software, Statistical Package for Social Sciences (SPSS) Version 20 was used for Analysis of Variance (ANOVA) for data obtained from GC-MS. The BLAST search program was used to compare the sequences of isolated bacterial strains with those in public databases on the National Center for Biotechnology Information (NCBI). Sequence similarity matrices and a phylogenetic tree were calculated and visualized using the Tree View of the MEGAX software (Kumar et al., 2018a, b). The sequences were deposited in the NCBI GenBank.
3. Results
3.1. Bacterial growth on Minimum Salt Medium
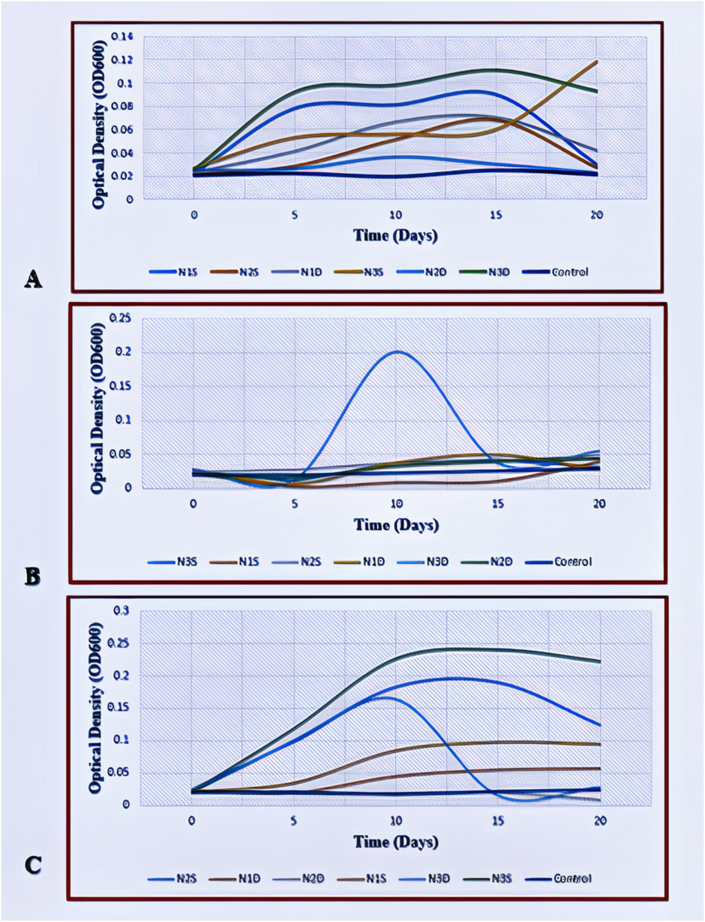
After incubation for 21 days and periodic monitoring of bacterial growth in MSM using UV-VIS Spectrophotometry, there was an increase in cell mass (OD600) in most flasks, indicative of bacterial growth. The OD levels in negative controls remained fairly constant (Figure 2). The bacterial growth curve in the media exhibited the typical four phases of lag, exponential, plateau, and decline.
Figure 2.
Microbial growth curves of chlorpyrifos-enriched MSM cultures (A- 5 ppm; B- 10 ppm; and C- 40 ppm). Abbreviations: N1S- Nakuru soil 1; N2S- Nakuru soil 2; N3S- Nakuru soil 3; N1D- Nakuru Dip wash 1; N2D-Nakuru Dip wash 2; and N3D- Nakuru Dip wash 3. Control- Uninoculated CP-supplemented MSM media.
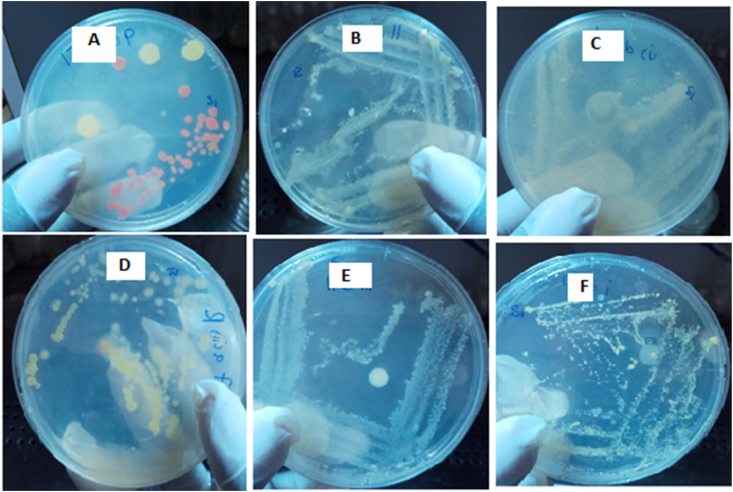
In MSM agar, colony formation occurred in 11 out of the 54 plates, representing a 20.4 % positivity rate. The colonies were sequentially cultured in nutrient agar until 18 pure colonies were obtained (Figure 3). The growth of bacteria in the soils and dip wash was not significantly different across the three regions.
Figure 3.
Nutrient Agar Plates of Isolated CP degrading bacteria (identified as A- Achromobacter insuavis, B- Lysinibacillus sp., C- Pseudomonas protegens, D- Alcaligenes faecalis, E- Stenotrophomonas maltophilia, and F- Pseudomonas putida).
3.2. Growth of bacteria in MSM media at different concentrations of chlorpyrifos
Table 1 summarizes MSM culture plates that showed colony formation at different concentrations of chlorpyrifos. At the concentration of 5 ppm, only two (2) plates had colony formation (11.1 %), five (5) plates at 10 ppm had growth (27.8 %), and four (4) at 40 ppm (20.4 %). A One Way ANOVA was carried out to determine if the concentration of CP affected the growth of bacteria isolates. The analysis found no statistically significant difference in growth across the three concentrations (p = 0.99).
Table 1.
Summary of MSM culture plates showing growth at different concentrations of the enriched chlorpyrifos.
| Concentration | Plates cultured | Number of plates showing growth | Number of plates without growth | Percent Growth |
|---|---|---|---|---|
| 5 ppm | 18 | 2 | 16 | 11,1% |
| 10 ppm | 18 | 5 | 13 | 27.8% |
| 40 ppm | 18 | 4 | 14 | 22.2% |
| Total | 54 | 11 | 43 | 20.4% |
Key: ppm-parts per million.
3.3. Morphological and biochemical characterization of isolated bacteria
The 18 isolated pure colonies of potential CP degrading bacteria were designated using MA1 to MA18. As viewed under a compound microscope, all the isolates were rod-shaped, 3 of them (16.7%) gram-positive (MA1, MA6, and MA10), and the rest were gram-negative. Isolates MA1 and MA8 were catalase-negative, while the other sixteen were positive. Results for the Oxidase test revealed that all the isolates were positive. None of the isolates was Urease or Methyl-red (MR) positive, while only one was Indole positive (MA1) and five lactose positive (MA1, MA13, MA16, MA17, and MA18). Only isolate MA10 and MA16 were Citrate positive, and Hydrogen Sulfide (H2S) production was only evident in isolate MA1 and MA16 (Table 2).
Table 2.
Summary of Biochemical Tests done for Preliminary Identification.
| Biochemical Test | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Isolate Code | Gram Stain | Catalase | Oxidase | Indole | Urease | MR | Citrate | Lactose | H2S |
| MA1 | + | - | + | + | - | - | + | + | + |
| MA2 | - | + | + | - | - | - | + | - | - |
| MA3 | - | + | + | - | - | - | + | - | - |
| MA4 | - | + | + | - | - | - | + | - | - |
| MA5 | - | + | + | - | - | - | + | - | - |
| MA6 | + | + | + | - | - | - | + | - | - |
| MA7 | - | + | + | - | - | - | + | - | - |
| MA8 | - | - | + | - | - | - | + | - | - |
| MA9 | - | + | + | - | - | - | + | - | - |
| MA10 | + | + | + | - | - | - | - | - | - |
| MA11 | - | + | + | - | - | - | + | - | - |
| MA12 | - | + | + | - | - | - | + | - | - |
| MA13 | - | + | + | - | - | - | + | + | - |
| MA14 | - | + | + | - | - | - | + | - | - |
| MA15 | - | + | + | - | - | - | + | - | - |
| MA16 | + | + | + | + | - | - | - | + | + |
| MA17 | - | - | + | - | - | - | + | + | - |
| MA18 | - | - | + | - | - | - | + | + | - |
Abbreviation: MR; Methyl-Red, H2S; Hydrogen Sulfide.
3.4. Molecular characterization of isolated bacteria
3.4.1. PCR amplification of 16sRNA regions
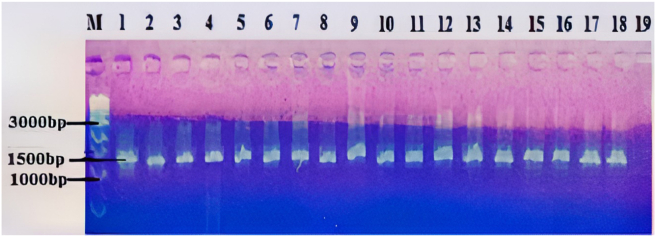
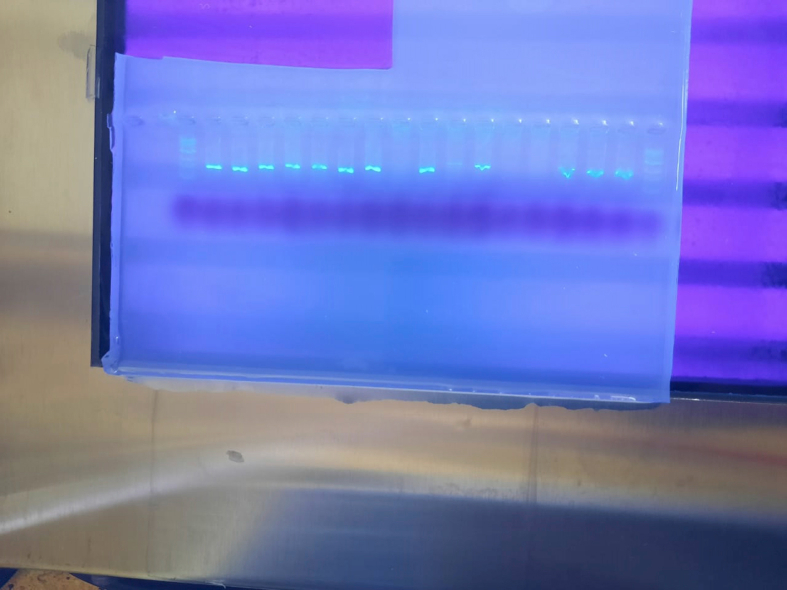
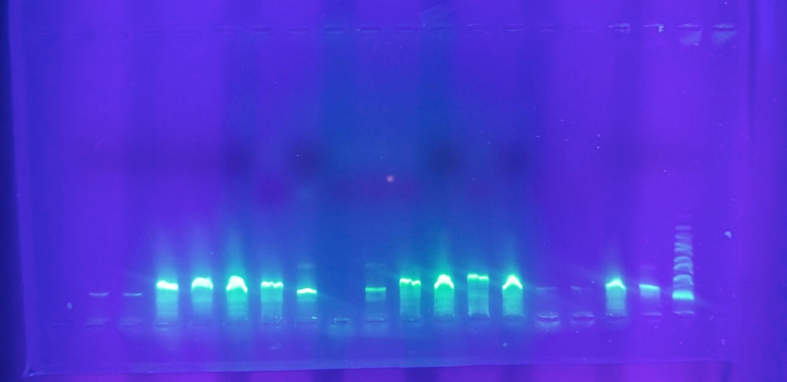
After PCR amplification, the isolates were shown to have fragments of the amplified DNA of around 1500 bp (Figure 4).
Figure 4.
PCR amplified 16S rDNA of the bacteria isolates in agarose gel. Lane M, 1kb DNA ladder; Lanes 1–18, genomic DNA of isolated strains (MA1 - MA18); Lane 19, Negative control. (The full, original gel images are provided in supplemental material as Agarose gel image of PCR products 1 and 2).
3.4.2. DNA sequencing and bacterial species identification
Isolates were identified based on the similarity to sequences in the GenBank. The isolates belonged to the genera Pseudomonas, Alcaligenes, Stenotrophomonas, Bacillus, and Achromobacter (Table 3).
Table 3.
Identity of Isolated Bacteria from soil samples based on similarity to the 16S rDNA sequence.
| Bacteria isolate | Accession | Species (16S rRNA gene analysis) | Accession | Identity (%) |
|---|---|---|---|---|
| MA1 | MZ314427.1 | Lysinibacillus sp. HBUM206408 | MT541001.1 | 100 |
| MA2 | MZ359883.1 | Stenotrophomonas maltophilia B9 | JQ900524.1 | 96.29 |
| MA3 | MZ310718.1 | Pseudomonas protegens L21 | MT505104.1 | 96.4 |
| MA4 | MZ310719.1 | Pseudomonas putida TCA1 | JQ782512.1 | 100 |
| MA5 | MZ359884.1 | Pseudomonas putida strain c275 | JQ782512.1 | 96.35 |
| MA6 | MZ310720.1 | Bacillus sp. C4P019a | FJ950565.1 | 96.56 |
| MA7 | MZ310721.1 | Pseudomonas sp. IAE245 | MN989109.1 | 92.9 |
| MA8 | MZ314428.1 | Achromobacter insuavis | MK414951.1 | 90.81 |
| MA9 | MZ359885.1 | Pseudomonas fluorescens strain psf4 | MF662233.1 | 98 |
| MA10 | MZ359886.1 | Uncultured bacterium WHENT_C1 | MN256390.1 | 89.53 |
| MA11 | MZ310722.1 | Pseudomonas vranovensis S3-4 | GU201990.1 | 98.8 |
| MA12 | MZ310723.1 | Pseudomonas aeruginosa 31 | LC425424.1 | 98.11 |
| MA13 | MZ310724.1 | Alcaligenes faecalis KWW 84 | MN732983.1 | 96.98 |
| MA14 | MZ310725.1 | Pseudomonas sp. FBF19 | LC425424.1 | 90.73 |
| MA15 | MZ314429.1 | Pseudomonas plecoglossicida B3 | LK391651.1 | 90 |
| MA16 | MZ310726.1 | Bacillus sp. FJAT-22078 | HG805697.1 | 93.54 |
| MA17 | MZ310727.1 | Alcaligenes faecalis H11 | MT071410.1 | 98.15 |
| MA18 | MZ310728.1 | Alcaligenes sp. A23 | KY949530.1 | 96.38 |
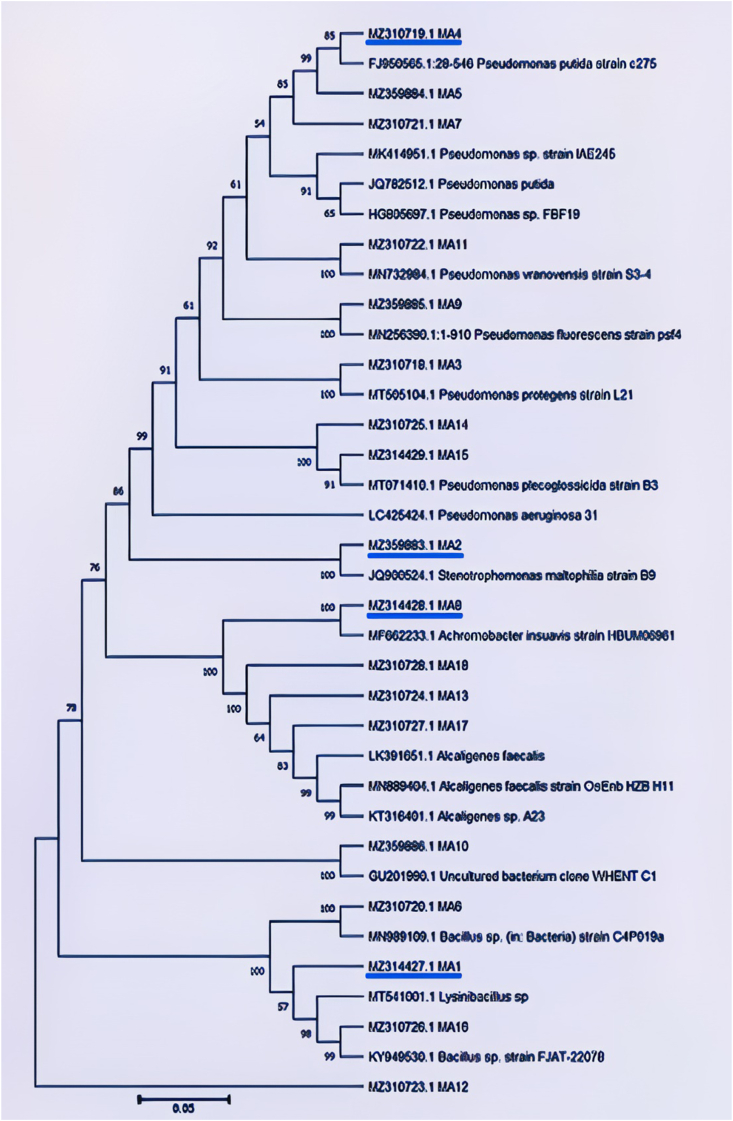
The phylogenetic grouping tree (Figure 5) was constructed using MEGAX Software (MEGA Software Company). Strains were clustered together based on the similarity of sequences. Moreover, 16S rRNA gene sequences formed the basis of the Maximum Likelihood phylogenetic tree in which closest relatives' bootstrap values were calculated and indicated (1000 replications). The tree's scale was 0.05 and the same unit for branch lengths as evolutionary distances. The Maximum Likelihood model was used to infer the evolutionary history, based on the Tamura-Nei model. In total, there were 36 nucleotide sequences in the analysis, which led to a total of 1804 positions in the dataset.
Figure 5.
Phylogenetic tree showing the relationship of the 18 isolates (MA1 to MA18) to closely related bacteria. Reference type strains of corresponding bacteria are involved in the tree. Four isolates that were subjected to GC-MS analysis (MA1, MA2, MA4, and MA8 are underlined in blue.
3.4.3. GC-MS analysis
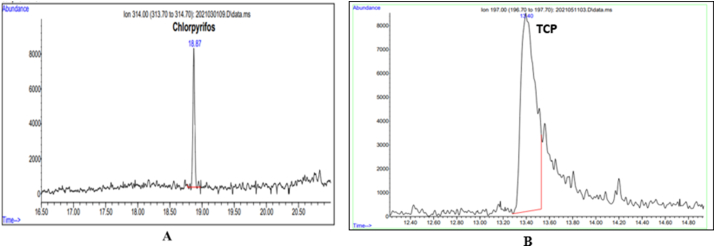
Isolates MA1, MA2, MA4, and MA8 were selected to assess their degradation potential, representing four bacteria genera, Lysinibacillus, Stenotrophomonas, Pseudomonas, and Achromobactor. GC-MS was used to analyze the degradation products of chlorpyrifos in the culture extracts. Based on the analysis, it was confirmed that TCP was the main degradation product (Figure 6).
Figure 6.
GC chromatograms showing the retention time of CP at 18.87 min (A) and TCP 13.40 min (B) and other smaller unidentified metabolites.
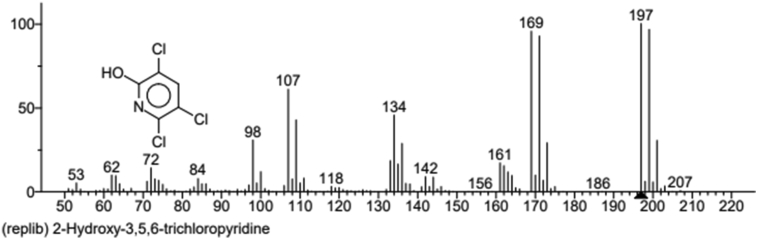
GC-MS confirmed the metabolic products of CP degradation through the retention times of peaks. Mass spectra and the National Institute of Standards and Technology (NIST) library identification program confirmed the metabolite to be TCP (Figure 7), based on the characteristic fragment ion peaks and molecular ion (m/z). At day zero, the sample showed m/z value of 350.76, indicating presence of chlorpyrifos in the medium. After incubation, TCP was formed as the metabolite of degradation for all strains, with m/z value of 197.
Figure 7.
Chemical structure and mass spectra of metabolite 3, 5, 6-trichloro-2-pyridinol (TCP) produced from chlorpyrifos degradation.
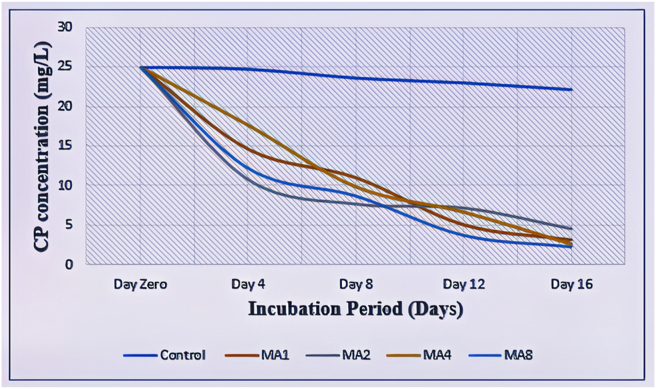
The rate of degradation of CP was very low in the uninoculated control during the 16 days of analysis. The percentage of CP degraded at the end of the period was only 11.56 %. In comparison, all the four isolates showed significant degradation, that is, MA1 (87.16 %), MA2 (82.04 %), MA4 (89.52% %), and MA8 (91.08 %) (Table 4). They were ranked in terms of their degradation ability from the highest to the lowest: MA8 (Achromobactor insuavis), MA4 (Pseudomonas putida), MA1 (Lysinibacillus sp.), and MA2 (Stenotrophomonas maltophilia). Conversely, only 11.56 % of CP degradation was observed in control after the 16 days of incubation (Figure 8).
Table 4.
Chlorpyrifos biodegradation in MSM medium (initial concentration of 25 ppm, temperature at 30 °C) for 12 days.
| Incubation Time (Days) |
||||||
|---|---|---|---|---|---|---|
| Initial | Day 4 | Day 8 | Day 12 | Day 16 | ||
| Control | Conc. (ppm) ± SD | 25 | 24.76 ± 0.07 | 23.61 ± 0.54 | 22.98 ± 0.66 | 22.11 ± 0.06 |
| % Degradation | 0 | 0.96 | 5.56 | 8.08 | 11.56 | |
| MA1 | Conc. | 25 | 14.64 ± 0.69 | 11.06 ± 0.93 | 5.12 ± 0.19 | 3.21 ± 0.11 |
| % | 0 | 41.44 | 55.76 | 79.52 | 87.16 | |
| MA2 | Conc. | 25 | 10.7 ± 0.08 | 7.62 ± 0.05 | 7.14 ± 0.06 | 4.49 ± 0.06 |
| % | 0 | 47.67 | 69.52 | 71.43 | 82.04 | |
| MA4 | Conc. | 25 | 17.65 ± 0.21 | 9.86 ± 0.06 | 6.67 ± 0.04 | 2.62 ± 0.07 |
| % | 0 | 29.4 | 60.56 | 73.32 | 89.52 | |
| MA8 | Conc | 25 | 12.16 ± 0.49 | 8.67 ± 0.47 | 3.69 ± 0.62 | 2.23 ± 0.06 |
| % | 0 | 42.8 | 54.43 | 71.03 | 91.08 | |
A two-way ANOVA found no significant difference in the rate of CP degradation by the four isolates (p = 0.442), but there was a significant difference between the isolates and the control (p < 0.05).
Figure 8.
Degradation kinetics of CP by Isolates MA1, MA2, MA4, and MA8 as compared to the control.
4. Discussion
The study's main aim was to isolate and characterize novel CP-degrading bacteria from dairy farm soils in Nakuru County, Kenya. Removal of CP residues from the soil is desired because the pesticide is toxic, and its extensive application impacts human health negatively (Aswathi et al., 2019). Bioremediation utilizes the degradation potential of microorganisms to provide a reliable and cost-effective approach for CP abatement (Akbar and Sultan, 2016; Jariyal et al., 2018). Biodegradation is the best option for in situ restoration operations (Farhan et al., 2021). In Nakuru County, Kenya, Chlorpyrifos is extensively used to control ticks in dairy farms, leading to the soil, water, and animal feed contamination (Chimbevo et al., 2021; Atego et al., 2021). The current study is among the first to explore the bioremediation of chlorpyrifos residues by native bacteria isolated from soils in dairy farms from the region. Besides, it is the first study to specifically assess biodegradation of Duodip, a commercial CP formulation that is widely used by dairy farmers in Kenya.
Isolation and characterization of indigenous microbial strains lead to an environmentally friendly method of in situ detoxification (Ortiz-Hernández and Sánchez-Salinas, 2010). Indigenous species are preferred because they do not negatively affect the soil micro-flora (Farhan et al., 2021). In contaminated environments, such as the dairy farm soils used in the present study, autochthonous bacteria populations have evolved to adapt to the contaminants. Therefore, soils from the sites are good ecological niches for isolating microorganisms capable of degrading the pesticides. In the present study, enrichment culture technique was used, a simple and straightforward method for isolating pesticide degrading microorganisms. The principle of the approach is that the provision of growth medium with a substrate only the microbe of interest utilizes prevents the growth of others, thereby serving as a selective medium for the organism of interest (Fatima, 2019; Verma et al., 2020). The use of Chlorpyrifos-supplemented Minimum Salt Medium (MSM) allowed the selection of bacteria that utilize CP and its intermediate products as the only carbon source. The results showed no statistically significant growth difference on the three concentrations (5 ppm, 10 ppm, and 40 ppm; p = 0.99). These findings corroborate earlier studies that had reported CP degradation at different concentrations (Mutua et al., 2015; Verma et al., 2020).
The MSM-isolated bacteria had genetic sequences homologous to Lysinibacillus sp. HBUM206408, Stenotrophomonas maltophilia, pseudomonas protegens L21, Pseudomonas putida TCA1, Pseudomonas putida strain c275, Bacillus sp. C4P019a, Pseudomonas sp. IAE245, Achromobacter insuavis, Pseudomonas fluorescens strain psf4, uncultured bacterium WHENT C1, Pseudomonas vranovensis S3-4, Pseudomonas auruginosa 31, Alcaligenes faecalis KWW 84, Pseudomonas sp. FBF19, Pseudomonas plecoglossicida B3, Bacillus sp. FJAT-22078, Alcaligenes faecalis H11, and Alcaligenes sp. A23. Perennial use of Chlorpyrifos as acaricide in Nakuru dairy farms led to repeated exposure of soil bacteria to the residues for an extended time. The prolonged exposure caused bacteria to develop ability to utilize the chemical as a source of carbon, as evidenced by the ability to grow in the media that contains CP as the only carbon source. Some previous studies have reported the degradation ability of Pseudomonas sp., Bacillus sp., and Alcaligenes faecalis against CP (Farhan et al., 2012; Akhdiya et al., 2020; Ambreen and Yasmin, 2021), but there is no documented knowledge that links the other isolated species to biodegradation of organophosphates.
GC-MS was used in the present study to monitor degradation and identify degradation products. The method was used because OPs are amenable to GC-MS due to its sensitivity (Farhan et al., 2021). The analysis showed that the four isolates MA1 (Lysinibacillus sp.), MA4 (Pseudomonas putida), MA2 (Stenotrophomonas maltophilia), and MA8 (Achromobactor insuavis) effectively degraded CP to produce TCP. The most efficient degrader, Achromobacter insuavis, degraded 91.08 % of the initial 25 ppm of CP within 16 days, while the least efficient was Stenotrophomonas maltophilia (82.04 %). The findings show that all four isolates can use CP as the sole energy source, without requiring additional nutrients to induce expression of OP-degrading enzymes. The variations in the degradation performance of the isolated strains can be attributed to differences in microbial activity and degradation pathways (Akhdiya et al., 2020; Ambreen and Yasmin, 2021). The current results are better compared to those reported in some earlier studies, especially concerning the time needed for maximum degradation. For instance, Hamsavathani et al. (2017) reported 30.78 % degradation of CP within 14 days by three isolates. Also, the results are more significant than those obtained by Kumar (2011), in which 77% of CP was degraded in 30 days.
The findings corroborate previous studies that isolated bacterial communities with the potential to degrade CP and TCP in the soil and liquid culture. Examples of species isolated in prior studies include Arthromobacter sp. Enterobacter strain B-14, Alcaligenes faecalis, Bacillus pumilus, Staphylococcus sp., Streptococcus sp., Achromobactor sp., Serratia marcescens, Bacillus cereus, Bacillus subtilis, Pseudomonas desmolyticum, and Pseudomonas aeruginosa (Singh et al., 2004; Anwar et al., 2009; Yadav et al., 2015; Hamsavathani et al., 2017; Anode et al., 2018). These bacterial communities have been shown to catabolize and co-metabolize TCP and CP. Yadav et al. (2015) reported co-metabolization of CP by Arthrobacter sp. and Flavobacterium sp. However, research has shown that the organism does not use CP as the energy source. Singh et al. (2004) reported that, instead, the bacterium hydrolyses CP to TCP and diethyl thiophosphate (DETP) and then uses the latter for growth and energy. Various other studies have confirmed TCP as the main metabolic product of CP degradation (Fawzy et al., 2014; Briceño et al., 2020; Bose et al., 2021). TCP is more mobile than CP, which adsorbs strongly unto organic matter and soil (Aswathi et al., 2019).
Microbial degradation is an effective and efficient method for eliminating harmful compounds from the environment (Abdel-Wareth and Abd El-Hamid, 2016). Bacteria and fungi are the main transformers, although only a few species have been shown to degrade the chemicals due to their complexity. Bioremediation, the use of microorganisms to immobilize or degrade wastes, is a prominent process of eliminating them. Biodegradation of OPs in the soil and water is important because of the potential of these chemicals to get into the livestock and eventually into human consumers of animal products. According to Yadav et al. (2015), bacteria can degrade pollutants efficiently because they have catabolic genes that enable them to survive in different ecological niches in various pH, temperature, and oxygen concentrations.
The bacterial consortium isolated from the samples contained a group of strains that significantly degraded the pesticide. However, it is important to note that the existing laboratory methods can only isolate around 10% of all bacteria. Therefore, many bacteria that interfere with the degradation process may not be isolated or cultured in the laboratory (Ortiz-Hernández and Sánchez-Salinas, 2010). Thus, metagenomics can be used to explore and exploit the enormous diversity of microorganisms in the soil. Successful detoxification of pesticides and other organic chemicals often requires bacteria consortia's concerted efforts (Akhdiya et al., 2020; Ambreen and Yasmin, 2021).
5. Conclusion
Acaricide production and application have become inevitable in agriculture to control pests. As shown by the GC-MS analysis, the four microbial isolates showed significant biodegradation of chlorpyrifos. Thus, the current study is useful in the practical application of bioremediation of soil contaminated by chlorpyrifos due to its efficiency, low cost, and less destruction to indigenous organisms. It is recommended that future studies should further explore and develop the strains isolated in the current study for enhanced bioremediation of CP in soils from dairy farms. Also, there is a need to develop a bacterial consortium that utilizes symbiotic associations among the strains for the degradation of CP. Moreover, further studies should explore other aspects, such as genes, enzymes, and various biochemical aspects, to better understand the biodegradation process.
Declarations
Author contribution statement
Paul Sifuna Oshule: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Suliman Essuman: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Ezekiel Njeru Mugendi: Conceived and designed the experiments.
Micah Nyabiba Asamba and Norbert Adum Atego: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Lenny Mwagandi Chimbevo: Conceived and designed the experiments; Analyzed and interpreted the data.
Funding statement
This work was supported by Kenya National Research Fund (grant number NRF-2017).
Data availability statement
Data associated with this study has been deposited at NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the accession numbers:
MZ314427.1; MZ359883.1; MZ310718.1; MZ310719.1; MZ359884.1; MZ310720.1; MZ310721.1; MZ314428.1; MZ359885.1; MZ359886.1; MZ310722.1; MZ310723.1; MZ310724.1; MZ310725.1; MZ314429.1; MZ310726.1; MZ310727.1; MZ310728.1
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Agarose Gel Image of PCR Products 1.
Agarose Gel Image of PCR Products 2.
:
References
- Abdel-Wareth M.T.A., Abd El-Hamid R.M. Mycoremediation of chlorpyrifos and lambda-cyhalothrin by two species of filamentous fungi. Int. J. Environ. Stud. 2016;73(6):974–987. [Google Scholar]
- Akbar S., Sultan S. Soil bacteria showing a potential of chlorpyrifos degradation and plant growth enhancement. Braz. J. Microbiol. 2016;47(3):563–570. doi: 10.1016/j.bjm.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhdiya A., Wartono, Sulaeman E. Degradation of chlorpyrifoschlorpyrifos and BPMC by the bacteria isolated from contaminated shallot farm soil. IOP Conf. Ser. Earth Environ. Sci. 2020;457 [Google Scholar]
- Ambreen S., Yasmin A. Novel degradation pathways for ChlorpyrifosChlorpyrifos and 3, 5, 6-Trichloro-2-pyridinol degradation by bacterial strain Bacillus thuringiensis MB497 isolated from agricultural fields of Mianwali, Pakistan. Pestic. Biochem. Physiol. 2021;172:104750. doi: 10.1016/j.pestbp.2020.104750. [DOI] [PubMed] [Google Scholar]
- Anode S.O., Magoma G., Onguso J., Abraha T. Bioremediation of xenobiotic pesticides by bacterial species isolated from flower farm soil around lake naivasha, Kenya. J. Biorem. Biodegrad. 2018;9(5) [Google Scholar]
- Anwar S., Liaquat F., Khan Q.M., Khalid Z.M., Iqbal S. Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol by Bacillus pumilus strain C2A1. J. Hazard Mater. 2009;168(1):400–405. doi: 10.1016/j.jhazmat.2009.02.059. [DOI] [PubMed] [Google Scholar]
- Aswathi A., Pandey A., Sukumaran R.K. Rapid degradation of the organophosphate pesticide – chlorpyrifos by a novel strain of Pseudomonas nitroreducens AR-3. Bioresour. Technol. 2019;292:122025. doi: 10.1016/j.biortech.2019.122025. [DOI] [PubMed] [Google Scholar]
- Atego A., Gicharu G., Chimbevo L., Oshule P., Essuman S., Asamba M. Detection and quantification of chlorpyrifos in soil, milk, dip wash, spray race residues using high performance liquid chromatography in selected dairy farms in Kenya. Sci. J. Anal. Chem. 2021;9(4):88–95. [Google Scholar]
- Barathidasan K., Reetha D., D J.M., N S., M G. Biodegradation of chlorpyrifoschlorpyrifos by co-culture of Cellulomonas fimi and Phanerochaete chrysosporium. Afr. J. Microbiol. Res. 2014;8(9):961–966. [Google Scholar]
- Bose S., Kumar P.S., Vo D.-V.N., Rajamohan N., Saravanan R. Microbial degradation of recalcitrant pesticides: a review. Environ. Chem. Lett. 2021;12(1) [Google Scholar]
- Briceño G., Lamilla C., Leiva B., Levio M., Donoso-Piñol P., Schalchli H., Gallardo F., Diez M.C. Pesticide-tolerant bacteria are isolated from a biopurification system to remove commonly used pesticides to protect water resources. PLoS One. 2020;15(6) doi: 10.1371/journal.pone.0234865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Shi W., Shu R., Pang J., Liu Y., Zhang X., Lei Y. Isolation and characterization of a bacterium able to degrade high concentrations of iprodione. Can. J. Microbiol. 2018;64(1):49–56. doi: 10.1139/cjm-2017-0185. [DOI] [PubMed] [Google Scholar]
- Chimbevo L.M., Atego N.A., Oshule P.S., Mapesa J., Essuman S., Nderitu J.H., Asamba M.N., Ngeny C. The role and sustainability of community-based county government funded agricultural infrastructure projects: a case of community cattle dips and Acaricides use in Kilifi, Kajiado and Nakuru counties. J. Agric. Ecol. Res. Int. 2021;22(4):26–36. [Google Scholar]
- Daneshparvar A., Mowlavi G., Mirjalali H., Hajjaran H., Mobedi I., Naddaf S.R., Shidfar M., Sadat Makki M. Molecular characterization and analysis of 16S ribosomal DNA in some isolates of demodex folicullorum. Iran. J. Parasitol. 2017;12(2):224–229. https://pubmed.ncbi.nlm.nih.gov/28761482/ [PMC free article] [PubMed] [Google Scholar]
- Das S., Adhya T.K. Degradation of chlorpyrifoschlorpyrifos in tropical rice soils. J. Environ. Manag. 2015;152:36–42. doi: 10.1016/j.jenvman.2015.01.025. [DOI] [PubMed] [Google Scholar]
- El-Gawad H.A. Validation method of organochlorine pesticides residues in water using gas chromatography–quadruple mass. Water Sci. 2016;30(2):96–107. [Google Scholar]
- EL-Maali N., Yehia Wahman A. Gas chromatography-mass spectrometric method for simultaneous separation and determination of several pops with health hazards effects. Mod. Chem. Appl. 2015;03(04) [Google Scholar]
- Farhan M., Ahmad M., Kanwal A., Butt Z.A., Khan Q.F., Raza S.A., Qayyum H., Wahid A. Biodegradation of chlorpyrifos using isolates from contaminated agricultural soil, its kinetic studies. Sci. Rep. 2021;11(1):10320. doi: 10.1038/s41598-021-88264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan M., Khan A.U., Wahid A., Ahmad M., Ahmad F. Biodegradation of chlorpyrifos using indigenous Pseudomonas sp. isolated from industrial drain. Pakistan J. Nutr. 2012;11(12):1183–1189. [Google Scholar]
- Fatima M. Isolation and characterization of chlorpyrifos degrading bacteria from contaminated agricultural lands. BioScient. Rev. 2019;1(2):53–65. [Google Scholar]
- Fawzy I., Eissa Khaled M., Ghanem Ibrahim M., Gomaa, Hend A., Mahmoud Osama N., Massoud Biodegradation of ChlorpyrifosChlorpyrifos by microbial strains isolated from agricultural wastewater. J. Am. Sci. 2014;10 http://free-journal.umm.ac.id/download-pdf-journal-2768--biodegradation-of-chlorpyrifoschlorpyrifos-by-microbial-strains-isolated-from-agricultural-wastewater---.pdf [Google Scholar]
- Franco-Duarte R., Černáková L., Kadam S., Kaushik S., Salehi B., Bevilacqua A., Corbo M.R., Antolak H., Dybka-Stępień K., Leszczewicz M., Relison Tintino S., Alexandrino de Souza V.C., Sharifi-Rad J., Melo Coutinho H.D., Martins N., Rodrigues C.F. Advances in chemical and biological methods to identify microorganisms—from past to present. Microorganisms. 2019;7(5):130. doi: 10.3390/microorganisms7050130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamsavathani V., Aysha O.S., Ajith A. Raughul. Isolation and identification of chlorpyrifoschlorpyrifos degrading bacteria from agricultural soil. Int. J. Adv. Res. 2017;5(5):1209–1221. [Google Scholar]
- Jariyal M., Jindal V., Mandal K., Gupta V.K., Singh B. Bioremediation of organophosphorus pesticide phorate in soil by microbial consortia. Ecotoxicol. Environ. Saf. 2018;159:310–316. doi: 10.1016/j.ecoenv.2018.04.063. [DOI] [PubMed] [Google Scholar]
- Johnson J.S., Spakowicz D.J., Hong B.-Y., Petersen L.M., Demkowicz P., Chen L., Leopold S.R., Hanson B.M., Agresta H.O., Gerstein M., Sodergren E., Weinstock G.M. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019;10(1):1–11. doi: 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokar S.H., Shavandi M., Haddadi A., Alaie E. Research Square; 2021. Impacts of Chlorpyrifos and Deltamethrin on Soil Bacterial Community Composition in Different Salinity Soils: Natural Attenuation Microcosm Studies. [Google Scholar]
- Kumar S. Bioremediation of chlorpyrifos by bacteria isolated from the cultivated soils. Int. J. Pharma Bio Sci. 2011;2:359–366. [Google Scholar]
- Kumar S., Kaushik G., Dar M.A., Mimesh S., López-Chuken U.J., Villarreal-Chiu J.F. Microbial degradation of Organophosphate Organophosphate pesticides: a review. Pedosphere. 2018;28(2):190–208. [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz T.C. Polymerase Chain reaction: basic protocol plus troubleshooting and optimization strategies. JoVE. 2012;5(63) doi: 10.3791/3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutavi F., Aarts N., Van Paassen A., Heitkönig I., Wieland B. Techne meets metis: knowledge and practices for tick control in Laikipia County, Kenya. NJAS - Wageningen J. Life Sci. 2018;86–87:136–145. [Google Scholar]
- Mutua G.K., Ngigi A.N., Getenga Z.M. Chlorpyrifos degradation in soils with different treatment regimes within Nzoia river drainage basin, Kenya. Bull. Environ. Contam. Toxicol. 2015;94(3):387–392. doi: 10.1007/s00128-015-1465-0. [DOI] [PubMed] [Google Scholar]
- Naphade S., Durve A., Bhot M., Varghese J., Ch N., Ra Isolation, characterization, and identification of pesticide tolerating bacteria from garden soil. Pelagia Res. Library. 2012;2(5) [Google Scholar]
- Ortiz-Hernández M.L., Sánchez-Salinas E. Biodegradation of the organophosphate organophosphate pesticide tetrachlorvinphos by bacteria isolated from agricultural soils in México. Rev. Int. Contam. Ambient. 2010;26(1):27–38. http://www.scielo.org.mx/scielo.php?script=sci_abstract&pid=S0188-49992010000100003&lng=es&nrm=iso&tlng=en [Google Scholar]
- Rayu S., Nielsen U.N., Nazaries L., Singh B.K. Isolation and molecular characterization of novel ChlorpyrifosChlorpyrifos and 3,5,6-trichloro-2-pyridinol-degrading bacteria from sugarcane farm soils. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B., Walker A. Microbial degradation of organophosphorus compounds. FEMS Microbiol. Rev. 2006:428–471. doi: 10.1111/j.1574-6976.2006.00018.x. [DOI] [PubMed] [Google Scholar]
- Singh B., Walker A., Morgan J., Wright D. Biodegradation of chlorpyrifos by enterobacter strain B-14 and its use in bioremediation of contaminated soils. Appl. Environ. Microbiol. 2004;70(8) doi: 10.1128/AEM.70.8.4855-4863.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Singh D., Chatterjee S. Biodegradation of organophosphorus pesticide chlorpyrifos by Sphingobacterium sp. C1B, a psychrotolerant bacterium isolated from apple orchard in Himachal Pradesh of India. Extremophiles. 2020;24(6):897–908. doi: 10.1007/s00792-020-01203-y. [DOI] [PubMed] [Google Scholar]
- Wei S., Levy B., Hoffman N., Cujar C., Rodney-Sandy R., Wapner R., D’AltonD'Alton M., Williams Z. A rapid and simple bead-bashing-based method for genomic DNA extraction from mammalian tissue. Biotechniques. 2020;68(5):240–244. doi: 10.2144/btn-2019-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav M., Shukla A.K., Srivastva N., Upadhyay S.N., Dubey S.K. Utilization of microbial community potential for removal of chlorpyrifos: a review. Crit. Rev. Biotechnol. 2015;36(4):727–742. doi: 10.3109/07388551.2015.1015958. [DOI] [PubMed] [Google Scholar]
- Yin X., Lv Y., Wen R., Wang Y., Chen Q., Kong B. Characterization of selected Harbin red sausages on the basis of their flavour profiles using HS-SPME-GC/MS combined with electronic nose and electronic tongue. Meat Sci. 2021;172:108345. doi: 10.1016/j.meatsci.2020.108345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with this study has been deposited at NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the accession numbers:
MZ314427.1; MZ359883.1; MZ310718.1; MZ310719.1; MZ359884.1; MZ310720.1; MZ310721.1; MZ314428.1; MZ359885.1; MZ359886.1; MZ310722.1; MZ310723.1; MZ310724.1; MZ310725.1; MZ314429.1; MZ310726.1; MZ310727.1; MZ310728.1