Abstract
Little is known about the effects of Pseudomonas biocontrol inoculants on nontarget rhizosphere fungi. This issue was addressed using the biocontrol agent Pseudomonas fluorescens CHA0-Rif, which produces the antimicrobial polyketides 2,4-diacetylphloroglucinol (Phl) and pyoluteorin (Plt) and protects cucumber from several fungal pathogens, including Pythium spp., as well as the genetically modified derivative CHA0-Rif(pME3424). Strain CHA0-Rif(pME3424) overproduces Phl and Plt and displays improved biocontrol efficacy compared with CHA0-Rif. Cucumber was grown repeatedly in the same soil, which was left uninoculated, was inoculated with CHA0-Rif or CHA0-Rif(pME3424), or was treated with the fungicide metalaxyl (Ridomil). Treatments were applied to soil at the start of each 32-day-long cucumber growth cycle, and their effects on the diversity of the rhizosphere populations of culturable fungi were assessed at the end of the first and fifth cycles. Over 11,000 colonies were studied and assigned to 105 fungal species (plus several sterile morphotypes). The most frequently isolated fungal species (mainly belonging to the genera Paecilomyces, Phialocephala, Fusarium, Gliocladium, Penicillium, Mortierella, Verticillium, Trichoderma, Staphylotrichum, Coniothyrium, Cylindrocarpon, Myrothecium, and Monocillium) were common in the four treatments, and no fungal species was totally suppressed or found exclusively following one particular treatment. However, in each of the two growth cycles studied, significant differences were found between treatments (e.g., between the control and the other treatments and/or between the two inoculation treatments) using discriminant analysis. Despite these differences in the composition and/or relative abundance of species in the fungal community, treatments had no effect on species diversity indices, and species abundance distributions fit the truncated lognormal function in most cases. In addition, the impact of treatments at the 32-day mark of either growth cycle was smaller than the effect of growing cucumber repeatedly in the same soil.
Introduction of beneficial microorganisms into soil or the rhizosphere has been proposed for biological control of soilborne crop diseases (6, 20, 34). In certain cases, genetically modified (GM) strains with increased expression of biocontrol traits have been developed to improve biocontrol efficacy (8, 27, 53). However, the release of large populations of biocontrol agents into the environment raises important biosafety issues related to the possible ecological consequences of such introductions on resident populations and ecosystem functioning (14). This concern is of primary relevance in the case of GM inoculants (8, 19, 27, 61), and in many countries the existing regulatory framework makes their deliberate field release strictly dependent on detailed assessment of their potential environmental impact and associated risks (49).
Soilborne fungal pathogens cause considerable damage to crop plants (1), and they have been often targeted in biocontrol (20, 34, 60). However, the majority of soil fungi are not pathogenic, and a large number of them may even be beneficial to plants and/or contribute positively to ecosystem functioning. Indeed, nonpathogenic saprotrophic microfungi perform key ecological roles in the soil ecosystem through decomposition of organic matter, nutrient cycling, natural control of plant pathogens, and a myriad of other functions (9, 11, 17). Common rhizosphere fungi are well documented as decomposers of celluloses and hemicelluloses (Trichoderma, Penicillium, and Fusarium), as well as chitin (Mortierella) (18). The ability of certain saprotrophic Fusarium and Trichoderma strains to protect plants against pathogenic fungi through competition, parasitism, antagonism, and/or induced resistance is also well known (2, 5, 23). In this context, it is surprising that saprotrophic rhizosphere fungi have been largely neglected as nontarget, beneficial resident microorganisms potentially affected by bacterial biocontrol inoculants, especially when the latter produce or overproduce antifungal metabolites with a relatively broad range of action. Indeed, investigations of the ecological impact of biocontrol bacteria have focused mainly on the effects on crops, on nontarget resident bacteria, and on ecosystem functioning (15, 41, 45, 48).
The few studies dealing with nontarget fungi have mostly monitored the impact of GM inoculants with antifungal biocontrol traits on total fungal counts (reviewed in reference 65). These studies may allow the assessment of catastrophic effects on the resident fungi, but they do not address the possibility of specific changes in microfungal community organization, e.g., in terms of the relative abundance of fungal species. Such alterations in the composition and structure of fungal communities might have immediate or lasting effects on ecosystem functioning (35), as there is now experimental evidence of a link between microbial biodiversity and the maintenance and regulation of ecosystem functions (46).
Mathematical methods to analyze fungal diversity data are still the subject of considerable debate in mycological literature, especially in the case of soil microfungal communities and/or when ecological interpretation of community response to perturbation is attempted (16, 24, 71, 72). Species abundance distribution analysis may provide both a complete mathematical description of the data and information on resource-partitioning patterns among component species in a given assemblage (71, 72). For large, species-rich equilibrium communities, the species abundance distribution is usually lognormal, while for species-poor nonequilibrium communities under a harsh environmental regime a geometric series often pertains (40), thus making modeling a useful tool to examine the effects of disturbance. Species richness and dominance indices provide simpler information but may be useful when comparing treatments (40). Multivariate analysis techniques (especially ordination methods) have also been used to analyze soil fungal communities and generate hypotheses on the factors involved in community changes (see, e.g., references 66 to 68).
In this study, the ecological impacts of the biocontrol agent Pseudomonas fluorescens CHA0-Rif and its GM derivative CHA0-Rif(pME3424) on the diversity of the culturable microfungal assemblages in the rhizosphere of cucumber (Cucumis sativus L.) were examined. P. fluorescens CHA0-Rif produces several bioactive compounds, including the antimicrobial polyketides 2,4-diacetylphloroglucinol (Phl) and pyoluteorin (Plt), and can protect cucumber against Pythium ultimum Trow (32, 34, 63). P. ultimum rapidly infects seeds and causes both pre- and postemergence damping-off of cucumber seedlings, but it can produce root rots even at later plant growth stages (1). The GM strain P. fluorescens CHA0-Rif(pME3424) overproduces the antimicrobial compounds Phl and Plt and displays enhanced biocontrol activity against P. ultimum (53). Phl and Plt inhibit the growth of a broad spectrum of bacteria and fungi (21, 25, 32, 55, 60).
In the present work, a soil with low disease pressure was chosen, so that the potential negative impacts of inoculation on nontarget fungi could not be compensated for by the biocontrol effects of the inoculants. The inoculation treatments were compared with a control (no inoculation) and a chemical treatment, in which soil was treated with metalaxyl (Ridomil), a phenylamide fungicide with selective action almost exclusively against Peronosporales (Oomycetes) (12, 54). The chemical treatment served as positive control since (i) metalaxyl is one of the main chemical fungicides currently used against Pythium spp. and (ii) CHA0 and its derivatives are being studied as potential biocontrol agents against these fungal pathogens. Chemical fungicides may be applied several times within a given growing season and/or in successive growing seasons, and this is also relevant for biocontrol products. Therefore, several cucumber growth cycles were carried out in the same soil, and treatments (bacterial inoculum or metalaxyl) were applied to soil at the start of each cycle. Since the objective of this work was to assess whether treatments could have an impact on the composition and structure of rhizosphere microfungal assemblages, different approaches (species abundance distributions, diversity indices, and multivariate analysis) were followed for the description and characterization of the fungal community.
MATERIALS AND METHODS
Bacterial strains.
P. fluorescens CHA0-Rif (47) is a spontaneous rifampin-resistant mutant of the wild-type strain CHA0 (57). Strains CHA0 and CHA0-Rif display the same growth rate and produce the same amounts of Phl and Plt in laboratory media. The ecology of strain CHA0-Rif has been assessed in the field on several occasions (62). Strain CHA0-Rif(pME3424) carries a recombinant plasmid (i.e., pME3424 [53]) constructed by inserting a copy of the rpoD gene (coding for sigma factor 70) of CHA0 into the IncP vector pVK100 (36). Introduction of the plasmid into the pseudomonad caused a severalfold increase in the amounts of Phl and Plt produced in vitro and resulted in enhanced suppression of P. ultimum-mediated damping-off of cucumber in soil microcosms (53).
Experimental setup.
Soil was collected from the surface horizon of a fallow located at Eschikon (Zürich, Switzerland). The soil at the site corresponds to a cambisol and was described by Natsch et al. (47) and Guntli et al. (29). Disease pressure is usually low in this soil, unless it is experimentally inoculated with phytopathogenic fungi. In that case, Eschikon soil becomes disease conducive, but plant protection can be achieved using CHA0 as inoculant, both in the field and in soil microcosms (13, 69). This has been observed with several fungal pathogens, including P. ultimum. The soil was air-dried until friable and passed through a 5-mm-mesh screen.
The four treatments studied were as follows: (i) inoculation with CHA0-Rif, (ii) inoculation with CHA0-Rif(pME3424), (iii) addition of the chemical fungicide metalaxyl, and (iv) no treatment (control). At the start of the experiment, the soil was sprayed (17 ml kg of soil−1) with either a cell suspension of CHA0-Rif, a cell suspension of CHA0-Rif(pME3424), a metalaxyl solution (at 1 mg ml−1), or sterile distilled water. Inoculations resulted in 9 × 106 CFU of introduced pseudomonads per g of dry soil. The soil was mixed thoroughly to ensure an even distribution of inoculants and chemical fungicide and placed in pots. Each 3.8-dm3 pot contained the equivalent of 3.5 kg of dry soil.
Cucumbers seeds (cultivar Chinesische Schlange) were obtained from R. Geissler (Zürich, Switzerland). They were sown at a depth of 1 cm (four seeds per pot), and the pots were placed in a growth chamber set at 70% relative humidity with 16 h of light (160 mE m−2 s−1) at 22°C and 8 h of dark at 18°C. The soil was sprayed with 50 ml of sterile distilled water per pot per day on six consecutive days. The number of seedlings was adjusted to three per pot at 10 days after sowing. Each pot received 100 ml of sterile distilled water, poured in the plate under the pot, at 2, 3, and 4 weeks after sowing.
The pots were emptied at 32 days after sowing. The root systems were shaken to dislodge weakly adhering soil, which was mixed with bulk soil and put back into the pots. On the same day, each treatment was applied a second time on the same pots, at the same rates, except that volumes were sprayed onto the soil surface without mixing the soil (which mimics certain commercial protocols for inoculation). Cucumber was sown and a second 32-day-long cycle of plant growth was carried out. The same procedure was repeated three times for a total of five cycles of cucumber growth.
Sampling of rhizosphere, isolation of fungi, and identification at species level.
The effects of treatments on the culturable rhizosphere microfungi were investigated in the first and fifth cycles of cucumber growth. Strain CHA0-Rif was present at 7.6 and 5.6 log CFU g of root−1 and CHA0-Rif(pME3424) was present at 6.9 and 4.9 log CFU g of root−1 at the end of the first and fifth cycles, respectively. At the end of each of the two cycles, one plant was randomly chosen from each pot to study rhizosphere microfungi. The roots were excised and weighed after the excess soil had been removed. The roots were then washed under mechanical agitation (by vortexing) in sterile distilled water, blotted dry, and weighed again. Following determination of the amounts of suspended soil, the rhizosphere suspension was adjusted to reach a final dilution of 1:10,000. Aliquots (2 ml) of the final suspension were then plated on 2% malt agar (20 g of malt extract, 18 g of agar, 1 liter of distilled water) acidified to pH 5.5 (with HCl) and containing chloramphenicol (150 ppm). Plates were maintained at room temperature (average temperature, 22°C). Most isolates were obtained after a few days of incubation, but plates were checked over several weeks to allow isolation of slow-growing fungi.
Identification of fungal isolates at the species level was carried out on the basis of classical macro- and microscopic criteria (taking into account cultural features and morphology of vegetative and asexual or sexual reproductive structures), with the help of pertinent keys and literature (30, 64). Morphotypes were recognized for sterile isolates. More than 11,000 isolates were thus characterized.
Statistical design, data reduction, and analyses.
Each treatment was studied with 10 replicates. The experiment followed a randomized block design, with ten blocks and one pot (i.e., one replicate) per treatment per block. Each rhizosphere suspension was plated onto five plates, and data obtained from each plate were analyzed separately.
The soil dilution plate method is effective for detecting changes due to pollution, management practices, or environmental disturbances (22) but tends to favor the most-sporulating species (24, 72). Therefore, data were expressed with regard to fungal frequency (i.e., the number of plates in which the species occurred) rather than numbers of CFU in all data analyses, except when comparing fungal population levels or considering species abundance distribution (because this would have resulted in too few abundance classes).
Differences in CFU and species numbers (including sterile morphotypes) among treatments within each cucumber growth cycle, as well as between the two cycles within each treatment, were tested for significance by means of two-way analysis of variance with post hoc simple contrasts. SYSTAT (version 5.2; SYSTAT, Evanston, Ill.) was used to perform the test (P < 0.05).
Species abundance distributions.
Species abundance distribution analysis was performed by testing the fit to the four main species abundance distribution models (geometric series, logarithmic series, lognormal, and broken stick). The truncated form of the lognormal function was used. The procedure for fitting the models consisted of calculating the number of species expected in each abundance class and comparing it with the number of species actually observed. Expected species abundances for each model were derived as described by Magurran (40). Expected and observed values were compared using a chi-square goodness-of-fit test. The fit was considered statistically significant when P was >0.05.
Diversity indices.
Diversity indices represent a useful mean to quantify community diversity and have been instrumental in revealing the impact of biocontrol inoculants on resident populations (48). Here, several diversity indices were used to compare treatment effects. Both species richness indices (weighing towards uncommon species) and indices based on the proportional abundances of species (weighing towards abundant species) were computed.
Among species richness indices, Margalef's diversity measure (DMg), the log series α index, Shannon's index (H′), and Brillouin's index (HB) were chosen. Margalef's index was calculated from the formula DMg = (S − 1)/ln N (here and throughout, S is the number of taxa and N is the total number of individuals); Shannon's index was calculated from the formula H′ = −Spi(ln pi), where pi is the proportion of individuals found in the ith species (in a sample, the true value of pi is unknown but is estimated as ni/N, [here and throughout, ni is the number of individuals in the ith species]); and Brillouin's index was calculated from the formula HB = [ln N! − S(ln ni!)]/N. The log series α index is derived in the course of the calculations for fitting the log series model of species abundance distribution and was obtained from the equation α = [N (1 − x)]/x, in which x is estimated from the iterative solution of S/N = (1 − x)/x[− ln (1 − x)].
Among indices based on the proportional abundance of species, Simpson's index (D) and Berger-Parker's index (d) were computed. Simpson's index was calculated as D = S {[ni (ni − 1)]/[N (N − 1)]}. Berger-Parker's index expresses the proportional importance of the most abundant species and was obtained using the formula d = Nmax/N, where Nmax is the number of individuals in the most abundant species.
Treatments were compared within each cucumber growth cycle studied, using two-way analysis of variance with post hoc simple contrasts (P < 0.05). Similarly, each treatment was compared across the two cycles. SYSTAT version 5.2 was used.
Discriminant analysis.
Discriminant analysis (DA [also known as canonical variate analysis]) is an ordination technique based on an a priori partition of objects into groups: the new axes, called the canonical variates, are located so as to reveal the best discrimination among groups. In this study, the underlying assumption tested was that different treatments would result in different fungal assemblages.
Since the method is considerably robust against the violation of linear data structures, it can be used without knowing any property of the data set (50), and untransformed data (frequency values) were used in the analysis. In DA, the number of variables should never exceed the number of objects, because otherwise the analysis will not run due to singularity problems (50); therefore, the rarest fungal species (occurring with only one plant in one or several treatments in either cycle) were excluded from the analysis. With this lower bound, 34 fungal taxa (including sterile morphotypes) were retained and subjected to the analysis. The analysis was performed using the SYN-TAX 5.0 package subroutine “Canonical Variates” with the “Spherized scores of objects” (normalization of eigenvectors) option. Correlations with the original variables were also analyzed. DA biplots were taken into account to evaluate the relationships between the original variables and the new axes graphically. These biplots showed lines connecting the origin to the variable positions as well as isodensity circles representing each group (circles drawn around group centroids and expected to contain 95% of the observations within each group).
RESULTS
Effect on fungal propagule counts and species numbers.
Treatments had no effect on the number of fungal CFU within the first cycle (i.e., 3.0 × 105 to 3.3 × 105 CFU per g of rhizosphere soil) or the fifth cycle (i.e., 1.2 × 105 to 5.6 × 105 CFU per g of rhizosphere soil) of cucumber growth (Table 1). However, when treatments were compared over the two samplings, the numbers of CFU were significantly lower for the control in the fifth cycle than in the first one.
TABLE 1.
Occurrence of fungal taxa in the rhizosphere of cucumbera
| Taxon | Mean no. of plates ± SD (no. of plants)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| First growth cycle
|
Fifth growth cycle
|
|||||||
| Control | CHA0-Rif | CHA0-Rif (pME3424) | Metalaxyl | Control | CHA0-Rif | CHA0-Rif (pME3424) | Metalaxyl | |
| Paecilomyces marquandii (Massee) Hughes | 2.8 ± 1.7 (9) | 3.9 ± 1.0 (10) | 3.5 ± 1.8 (10) | 3.2 ± 1.6 (10) | 2.8 ± 1.6 (9) | 3.0 ± 1.2 (10) | 3.2 ± 1.5 (10) | 3.2 ± 1.1 (10) |
| Phialocephala humicola S. C. Jong & E. E. Davis | 1.9 ± 1.3 (9) | 3.7 ± 1.6 (9) | 2.5 ± 1.4 (10) | 3.2 ± 1.5 (10) | 2.0 ± 1.2 (9) | 2.1 ± 1.0 (10) | 2.0 ± 1.2 (10) | 2.6 ± 1.7 (9) |
| Fusarium oxysporum Snyder & Hansen | 4.1 ± 1.4 (10) | 4.5 ± 1.3 (10) | 3.1 ± 1.8 (9) | 3.6 ± 1.7 (10) | 1.5 ± 1.0 (8) | 2.2 ± 1.6 (8) | 1.8 ± 1.0 (9) | 1.4 ± 1.0 (8) |
| Gliocladium roseum Bainier | 2.5 ± 2.0 (8) | 2.8 ± 1.6 (9) | 3.6 ± 1.3 (10) | 3.1 ± 1.0 (10) | 1.6 ± 0.8 (9) | 2.1 ± 1.2 (9) | 1.5 ± 1.1 (8) | 1.3 ± 1.2 (8) |
| Penicillium rugulosum Thom | 2.3 ± 1.8 (7) | 3.5 ± 1.6 (10) | 3.4 ± 1.4 (10) | 3.0 ± 1.5 (10) | 1.7 ± 1.6 (8) | 2.1 ± 1.9 (8) | 2.2 ± 1.7 (8) | 1.8 ± 1.1 (9) |
| Mortierella alpina Peyronel | 2.3 ± 1.8 (7) | 3.4 ± 2.2 (8) | 3.4 ± 1.6 (9) | 3.2 ± 1.8 (9) | 1.6 ± 1.1 (9) | 1.7 ± 1.6 (8) | 2.2 ± 1.6 (9) | 2.1 ± 1.4 (8) |
| Verticillium chlamydosporium Goddard | 3.6 ± 1.8 (9) | 4.4 ± 0.8 (10) | 3.7 ± 1.5 (10) | 4.2 ± 0.9 (10) | 1.0 ± 0.9 (7) | 0.7 ± 0.7 (6) | 1.0 ± 1.2 (6) | 1.6 ± 1.4 (7) |
| Trichoderma hamatum (Bonord.) Bainier | 1.4 ± 1.1 (8) | 2.4 ± 1.6 (8) | 2.3 ± 1.8 (8) | 2.2 ± 1.3 (9) | 1.1 ± 0.6 (9) | 0.9 ± 1.0 (6) | 2.1 ± 1.3 (9) | 1.8 ± 1.5 (8) |
| Verticillium nigrescens Pethybr. | 1.6 ± 1.6 (6) | 2.5 ± 1.9 (8) | 1.4 ± 1.6 (5) | 2.3 ± 1.6 (9) | 3.3 ± 1.4 (10) | 2.1 ± 1.8 (8) | 1.8 ± 1.7 (7) | 2.5 ± 1.4 (9) |
| Fusarium tabacinum (Beyma) W. Gams | 2.6 ± 2.1 (7) | 2.6 ± 2.4 (7) | 2.2 ± 2.3 (7) | 2.4 ± 2.2 (7) | 2.5 ± 1.9 (9) | 1.6 ± 2.0 (6) | 3.8 ± 1.5 (10) | 2.2 ± 1.8 (8) |
| Staphylotrichum coccosporum J. Meyer & Nicot | 0.7 ± 1.3 (3) | 1.1 ± 1.3 (5) | 0.6 ± 0.7 (5) | 0.5 ± 0.7 (4) | 3.2 ± 1.5 (9) | 2.9 ± 2.0 (8) | 3.3 ± 1.5 (10) | 3.9 ± 1.9 (9) |
| Coniothyrium fuckelii Sacc. | 1.5 ± 1.4 (7) | 2.0 ± 1.9 (6) | 1.1 ± 1.4 (5) | 1.2 ± 1.0 (7) | 1.0 ± 0.9 (7) | 1.0 ± 1.2 (5) | 1.0 ± 1.2 (6) | 1.3 ± 1.1 (7) |
| Fusarium solani (Mart.) Sacc. | 1.5 ± 2.0 (6) | 2.4 ± 1.9 (8) | 1.3 ± 1.3 (7) | 1.8 ± 1.9 (7) | 0.7 ± 0.7 (6) | 0.9 ± 0.9 (7) | 0.2 ± 0.4 (2) | 0.6 ± 0.5 (6) |
| Trichoderma harzianum Rifai | 1.0 ± 0.8 (7) | 1.3 ± 1.2 (7) | 1.2 ± 1.1 (7) | 1.7 ± 0.8 (10) | 0.3 ± 0.5 (3) | 0.3 ± 0.5 (3) | 0.4 ± 0.5 (4) | 0.4 ± 0.7 (3) |
| Cylindrocarpon destructans (Zins.) Scholten | 0.7 ± 0.8 (5) | 0.5 ± 0.7 (4) | 0.5 ± 0.8 (3) | 0.8 ± 1.3 (4) | 0.7 ± 0.8 (5) | 0.7 ± 1.1 (4) | 0.5 ± 0.7 (4) | 0.4 ± 0.5 (4) |
| Myrothecium roridum Tode ex Steudel | 0.4 ± 0.7 (3) | 0.6 ± 0.7 (5) | 0.5 ± 1.0 (3) | 0.7 ± 0.7 (6) | 0.4 ± 0.7 (3) | 0.4 ± 0.7 (3) | 0.5 ± 0.5 (5) | 0.3 ± 0.7 (2) |
| Monocillium mucidum W. Gams | 0.8 ± 1.6 (3) | 0.3 ± 0.9 (1) | 0.9 ± 1.5 (3) | 0.6 ± 1.0 (4) | 0.5 ± 0.5 (5) | 0.5 ± 0.7 (4) | 0.6 ± 0.7 (5) | 0.3 ± 0.5 (3) |
| Mycelium sterile moniliaceum 1 | 1.5 ± 1.4 (7) | 1.6 ± 1.8 (6) | 2.2 ± 2.0 (6) | 1.8 ± 2.0 (6) | ||||
| Fusarium equiseti (Corda) Sacc. | 0.1 ± 0.3 (1) | 1.4 ± 1.7 (5) | 0.6 ± 1.3 (2) | 0.9 ± 1.7 (3) | 0.2 ± 0.4 (2) | 0.2 ± 0.4 (2) | 0.6 ± 0.5 (6) | 0.2 ± 0.4 (2) |
| Myrothecium verrucaria (Alb. & Schw.) Ditm. ex Steudel | 0.2 ± 0.4 (2) | 0.8 ± 1.2 (5) | 0.3 ± 0.7 (2) | 1.0 ± 1.3 (5) | 0.2 ± 0.4 (2) | 0.1 ± 0.3 (1) | 0.3 ± 0.5 (3) | 0.3 ± 0.5 (3) |
| Coniothyrium cerealis E. Müll. | 0.3 ± 0.7 (2) | 0.6 ± 1.0 (4) | 0.2 ± 0.4 (2) | 0.2 ± 0.4 (2) | 0.4 ± 0.5 (4) | 0.5 ± 1.0 (3) | 0.2 ± 0.4 (2) | 0.5 ± 0.8 (3) |
| Cladosporium cladosporioides (Fres.) de Vries | 0.1 ± 0.3 (1) | 0.7 ± 0.9 (4) | 0.7 ± 0.8 (5) | 0.7 ± 1.1 (4) | 0.4 ± 0.7 (3) | 0.1 ± 0.3 (1) | 0.4 ± 0.7 (3) | |
| Exophiala sp. | 0.2 ± 0.4 (2) | 0.2 ± 0.6 (1) | 0.3 ± 0.7 (2) | 1.1 ± 1.0 (7) | 0.3 ± 0.5 (3) | 0.6 ± 1.1 (3) | 0.4 ± 0.7 (3) | |
| Acremonium roseo-griseum (S. B. Saksena) W. Gams | 0.3 ± 0.7 (2) | 0.3 ± 0.7 (2) | 0.5 ± 0.7 (4) | 0.4 ± 0.5 (4) | 0.3 ± 0.5 (3) | 0.4 ± 0.7 (3) | ||
| Trichosporon beigelii (Küchenm. & Rabenh.) Vuill. | 0.4 ± 0.8 (2) | 0.7 ± 1.1 (4) | 0.9 ± 1.5 (5) | 0.2 ± 0.6 (1) | 0.1 ± 0.3 (1) | 0.2 ± 0.6 (1) | 0.3 ± 0.7 (2) | 0.1 ± 0.3 (1) |
| Mucor hiemalis Wehmer f. hiemalis | 0.9 ± 1.1 (5) | 0.4 ± 0.5 (4) | 0.6 ± 0.8 (4) | 0.2 ± 0.4 (2) | 0.1 ± 0.3 (1) | |||
| Mycelium sterile moniliaceum 2 | 0.3 ± 0.7 (2) | 0.1 ± 0.3 (1) | 0.4 ± 0.7 (3) | 0.4 ± 1.0 (2) | 0.2 ± 0.6 (1) | 0.3 ± 0.5 (3) | 0.2 ± 0.4 (2) | 0.2 ± 0.6 (1) |
| Truncatella angustata (Pers. ex Lk) Hughes | 0.4 ± 0.8 (2) | 0.8 ± 1.5 (3) | 0.4 ± 0.7 (3) | 0.2 ± 0.6 (1) | 0.3 ± 0.7 (2) | 0.3 ± 0.5 (3) | ||
| Stachybotrys chartarum (Ehrenb. ex Link) Hughes | 0.1 ± 0.3 (1) | 0.1 ± 0.3 (1) | 0.1 ± 0.3 (1) | 0.3 ± 0.7 (2) | 0.6 ± 0.8 (4) | 0.3 ± 0.5 (3) | ||
| Mycelium sterile dematiaceum 4 | 0.1 ± 0.3 (1) | 0.1 ± 0.3 (1) | 0.7 ± 1.6 (3) | 0.3 ± 0.5 (3) | 0.4 ± 0.7 (3) | |||
| Scedosporium apiospermum (Sacc.) Sacc. ex Castell. & Chalm. | 0.2 ± 0.4 (2) | 0.3 ± 0.7 (2) | 0.2 ± 0.4 (2) | 0.2 ± 0.6 (1) | 0.3 ± 0.9 (1) | 0.1 ± 0.3 (1) | ||
| Penicillium canescens Sopp | 0.2 ± 0.6 (1) | 0.1 ± 0.3 (1) | 0.2 ± 0.6 (1) | 0.1 ± 0.3 (1) | 0.1 ± 0.3 (1) | 0.3 ± 0.5 (3) | 0.1 ± 0.3 (1) | |
| Talaromyces luteus (Sacc.) Stolk & Samson | 0.1 ± 0.3 (1) | 0.1 ± 0.3 (1) | 0.1 ± 0.3 (1) | 0.2 ± 0.4 (2) | 0.1 ± 0.3 (1) | 0.2 ± 0.4 (2) | ||
| Mucor minutus (Baijal & B.R. Mehrotra) Schipper | 0.1 ± 0.3 (1) | 0.1 ± 0.3 (1) | 0.2 ± 0.4 (2) | 0.1 ± 0.3 (1) | 0.2 ± 0.4 (2) | |||
| Mucor racemosus Fres. f. racemosus | 0.5 ± 0.5 (5) | 0.3 ± 0.9 (1) | ||||||
| Humicola fuscoatra Traaen var. fuscoatra | 0.1 ± 0.3 (1) | 0.4 ± 0.7 (3) | 0.1 ± 0.3 (1) | 0.1 ± 0.3 (1) | ||||
| Phoma eupyrena Sacc. | 0.1 ± 0.3 (1) | 0.2 ± 0.4 (2) | 0.2 ± 0.4 (2) | 0.1 ± 0.3 (1) | ||||
| Gonytrichum macrocladum (Sacc.) Hughes | 0.1 ± 0.3 (1) | 0.2 ± 0.4 (2) | 0.1 ± 0.3 (1) | 0.1 ± 0.3 (1) | 0.1 ± 0.3 (1) | |||
| Stilbella aciculosa (Ellis & Everhart) Seifert | 0.1 ± 0.3 (1) | 0.3 ± 0.9 (1) | 0.6 ± 1.6 (2) | 0.2 ± 0.6 (1) | ||||
| Mortierella humilis Linnem. ex W. Gams | 0.2 ± 0.4 (2) | 0.4 ± 1.0 (2) | 0.1 ± 0.3 (1) | |||||
| Ramichloridium schulzeri (Sacc.) de Hoog | 0.1 ± 0.3 (1) | 0.2 ± 0.4 (2) | 0.1 ± 0.3 (1) | 0.1 ± 0.3 (1) | ||||
| Trichosporon sp. | 0.1 ± 0.3 (1) | 0.2 ± 0.4 (3) | 0.1 ± 0.3 (1) | |||||
| Fusarium sambucinum Fuckel | 0.1 ± 0.3 (1) | 0.2 ± 0.6 (1) | 0.3 ± 0.7 (2) | |||||
| Penicillium simplicissimum (Oudem.) Thom | 0.1 ± 0.3 (1) | 0.2 ± 0.4 (2) | 0.1 ± 0.3 (1) | |||||
| Chaetomium bostrychodes Zopf | 0.1 ± 0.3 (1) | 0.1 ± 0.3 (1) | 0.2 ± 0.4 (2) | |||||
| Fusarium culmorum (W. G. Sm.) Sacc. | 0.2 ± 0.4 (2) | 0.1 ± 0.3 (1) | 0.1 ± 0.3 (1) | |||||
| Stachybotrys cylindrospora Bainier & Sartory | 0.3 ± 0.5 (3) | 0.1 ± 0.3 (1) | ||||||
| Paecilomyces carneus (Duché & Heim.) A.H.S. Brown & G. Sm. | 0.3 ± 0.9 (1) | 0.2 ± 0.6 (1) | 0.1 ± 0.3 (1) | |||||
| Mycelium sterile dematiaceum 1 | 0.1 ± 0.3 (1) | 0.4 ± 1.0 (2) | ||||||
| Mycelium sterile dematiaceum 5 | 0.1 ± 0.3 (1) | 0.2 ± 0.4 (2) | ||||||
| Chaetomium sp. | 0.1 ± 0.3 (1) | |||||||
| Gliocladium sp. | 0.3 ± 0.9 (1) | 0.1 ± 0.3 (1) | ||||||
| Mycelium sterile dematiaceum 3 | 0.4 ± 1.0 (2) | |||||||
| Byssochlamys nivea Westling | 0.2 ± 0.6 (1) | 0.1 ± 0.3 (1) | ||||||
| Cladosporium cucumerinum Ellis & Arth. | 0.2 ± 0.6 (1) | 0.1 ± 0.3 (1) | ||||||
| Absidia spinosa Lendner | 0.2 ± 0.4 (2) | |||||||
| Aspergillus candidus Link ex Link | 0.2 ± 0.4 (2) | |||||||
| Aspergillus niger Tiegh. | 0.2 ± 0.4 (2) | |||||||
| Talaromyces sp. | 0.2 ± 0.4 (2) | |||||||
| Mycelium sterile moniliaceum 3 | 0.5 ± 1.6 (1) | |||||||
| Gliocladium solani (Harting) Petch | 0.4 ± 1.3 (1) | |||||||
| Penicillium chrysogenum Thom | 0.3 ± 0.9 (1) | |||||||
| Mycelium sterile dematiaceum 2 | 0.2 ± 0.6 (1) | |||||||
| Total no. of fungal taxab | 45 | 68 | 57 | 50 | 57 | 52 | 63 | 55 |
| Total no. of fungal propagules (105 CFU/g of rhizosphere soil)c | 3.24 | 3.31 | 2.99 | 2.95 | 1.18 | 5.62 | 1.81 | 1.31 |
The numbers of plates and individual plants in which each taxon was found are indicated. Each treatment was studied using 10 plants per cycle, and for each plant, five plates were used to recover fungi. Data are shown for fungal taxa recovered in at least two plates for one of the treatments in one cycle, and taxa are listed according to their overall occurrence. Fungal taxa occurring in no more than one plate for a given treatment in one cycle were as follows (treatment and cycle are specified in parentheses [c1, 1st cycle control; n1, 1st cycle CHA0-Rif; g1, 1st cycle CHA0-Rif(pME3424); r1, 1st cycle metalaxyl; c5, 5th cycle control; n5, 5th cycle CHA0-Rif; g5, 5th cycle CHA0-Rif(pME3424); r5, 5th cycle metalaxyl]): Acremonium sp. (g1, n1); Acremonium strictum W. Gams (g1, n1, g5); Arthrinium phaeospermum (Corda) M. B. Ellis (g5); Arthrobotrys oligospora Fres. (c1); Ascodesmis microscopica (Croulan) Le Gal (n5); Aspergillus fumigatus Fres. (n1, r5); Aspergillus speluneus Raper & Fennel (n1); Botryotrichum piluliferum Sacc. & March. (g1, n1); Chaetomium nozdrenkoae Serg. (n5); Chrysosporium ana. Renispora flavissima Sigler et al. (r5); Cladosporium herbarum (Pers.) Link ex Gray (r1); Coniothyrium minitans W. A. Campbell (n1); Curvularia inaequalis (Wakker) Boedijn (g1, n1, r5); Cylindrocarpon magnusianum (Sacc.) Wollenw. (r1); Cylindrocarpon obtusisporum (Cooke & Harkness) Wollenw. (g5, r5); Doratomyces stemonitis (Pers. ex Steud.) Morton & G. Sm. (n1); Embellisia chlamydospora (Hoes, Bruehl & Shaw) Simmons (n5); Emericellopsis terricola van Beyma (r1); Fusarium sp. (c5, n5); Geomyces pannorum (Link) Sigler & Carmich. var. pannorum (g5); Humicola grisea Traaen (n1); Leptodontium sp. (c5); Mariannea elegans (Corda) Samson var. punicea Samson (g5); Mortierella sp. (g1, n1); Mucor racemosus Fres. f. sphaerosporus (Haghem) Schipper (c1, n1); Paecilomyces sp. (n1); Penicillium commune Thom (g1); Penicillium olsonii Bain. & Sartory (c1); Penicillium purpurogenum Stoll (g1, n1, c5); Penicillium velutinum van Beyma (c1); Penicillium sp. (n1); Periconia macrospinosa Lefebvre & A. G. Johnson (g1, r1, n5); Petriellidium boydii (Shear) Malloch (g5); Phoma leveillei Boerema & Bollen (c5, n5); Phoma medicaginis Malbr. & Roum. var. pinodella (L.K. Jones) Boerema (c5, r5); Phoma sp. (g1, n1, r1, c5); Preussia terricola Cain (c5, g5); Pseudoeurotium sp. (c5, g5, n5); Rhinocladiella sp. (n1); Rhizopus stolonifer (Ehrenb. ex Link) Lind (g1, n1); Sordaria fimicola (Rob.) Ces. & de Not. (r5); Sporothrix schenckii Hektoen & Perkins (c5, n5, r5); Stachybotrys elegans (Pidopl.) W. Gams (r1, g5, r5); Teniolella sp. (n1); Trichocladium opacum (Corda) Hughes (g1, n5); Trichurus spiralis Hasselbr. (g1, r1); Verticillium lecanii (Zimm.) Viégas (g1, c5); Verticillium tenerum (Nees ex Pers.) Link (c1); Zygorrhinchus moelleri Vuill. (n1); M. sterile moniliaceum 4–25 (in either treatment or control); M. sterile dematiaceum 6–25 (in either treatment or control).
Including taxa listed in footnote a.
Treatments had no statistical influence on results within the first or the fifth cycle. When comparing each treatment over the first and the fifth cycles, the only significant difference was found in the case of the control (P < 0.05; two-way analysis of variance with post hoc simple contrasts).
In total, over 11,000 colonies were identified and assigned to 105 fungal species (plus 50 sterile morphotypes) (Table 1). Many fungal species, which were the most abundant, were found in all treatments in both cycles. The main exception was Mycelium sterile moniliaceum 1, which was absent from the first cycle. No statistically significant difference in species numbers (sterile morphotypes included) was found among treatments at either of the two cycles studied or when the treatments were compared across both cycles.
Effect on species abundance distributions.
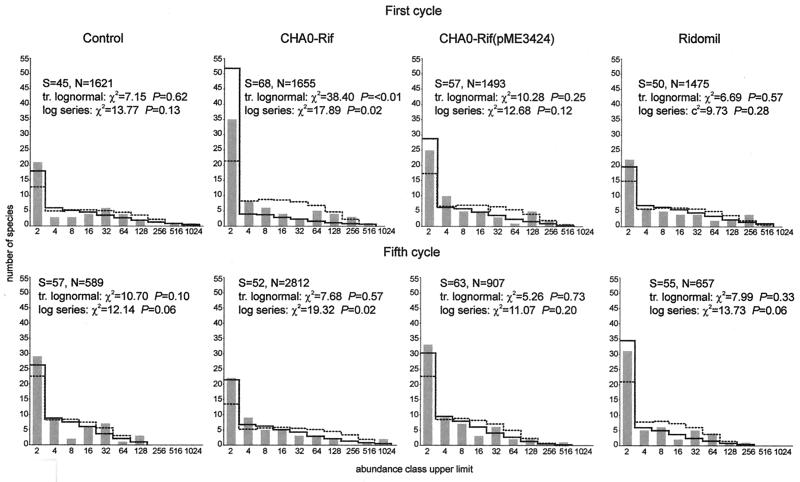
Distributions of fungal species in the rhizosphere following treatments with the control, CHA0-Rif(pME3424), and metalaxyl could be described both by the log series and the truncated lognormal models in each of the two growth cycles studied. However, results from the chi-square test indicated that the truncated lognormal function yielded a better fit in all cases. Instead, neither the log series nor the truncated lognormal model was appropriate for the CHA0-Rif treatment in the first cycle, mainly because the log series model predicted fewer rare species and the truncated lognormal model predicted fewer species of intermediate-high abundance than were actually recorded. In the fifth cycle, the fungal assemblage from the same treatment did not fit the log series, because of several discrepancies between actual and expected data for rare species and species with high abundances, but fit the truncated lognormal model (Fig. 1).
FIG. 1.
Species abundance distribution for rhizosphere fungal assemblages in the control, the CHA0-Rif, the CHA0-Rif(pME3424), and the metalaxyl treatments at the end of the first and the fifth cycles of cucumber growth. The abundance of each species is given based on CFU data. The x axis corresponds to the octave abundance classes (upper limit of each abundance class) where the different species identified have been ranked; the y axis corresponds to the actual or expected number of fungal isolates falling within each abundance class. Bars, observed values; solid line, expected values according to the truncated lognormal function; dotted line, expected values according to the log series function. S, number of taxa, N, number of CFU. The chi-square and associated probability values for the models are also indicated. Details of calculations are as described by Magurran (40). The fit is statistically significant when P is >0.05. For the first cycle, both the truncated lognormal model and the log series model were statistically significant for the control, the CHA0-Rif(pME3424), and the metalaxyl treatments. For the fifth cycle, the truncated lognormal model was statistically significant for each of the four treatments, and the log series model was statistically significant for the control, the CHA0-Rif(pME3424), and the metalaxyl treatments.
None of the species abundance distributions obtained from the different treatments in the first or the fifth cycle fit the broken stick model or the geometric series (P ≪ 0.05). For the broken stick model, this resulted from the fact that this function predicted fewer rare species than were recorded, except for the control in the fifth cycle (the number of species of intermediate abundance was too large to fit the model). Instead, the main reason that data did not conform to the geometric series was that the species with the highest rank (i.e., abundance) were more abundant than was predicted by the model.
Effect on diversity indices.
When diversity indices were computed (Table 2), no significant difference was detected between the treatments in either cycle. This was observed with indices based on the proportional abundance of species (Shannon's and Brillouin's indices), dominance measures (Simpson's and Berger-Parker's indices), and species richness indices (log series α and Margalef's indices). In a comparison between cycles, a significant difference was found for the control with log series α index, for the CHA0-Rif and the metalaxyl treatments with Berger-Parker's index.
TABLE 2.
Diversity indices for fungal assemblages in the cucumber rhizosphere
| Index | Valuea at end of:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| First growth cycle
|
Fifth growth cycle
|
|||||||
| Control | CHA0-Rif | CHA0-Rif (pME3424) | Metalaxyl | Control | CHA0-Rif | CHA0-Rif (pME3424) | Metalaxyl | |
| Log α | 10.5 ± 5.5 a | 13.4 ± 5.9 ab | 13.1 ± 6.6 ab | 12.9 ± 6.1 ab | 18.6 ± 6.0 b | 15.2 ± 5.4 ab | 19.6 ± 10.4 ab | 14.6 ± 5.2 ab |
| Margalef's DMg | 3.9 ± 1.3 | 5.0 ± 1.3 | 4.6 ± 1.4 | 4.7 ± 1.3 | 5.1 ± 1.0 | 4.6 ± 0.9 | 5.2 ± 1.0 | 4.7 ± 1.1 |
| Shannon's H′ | 2.5 ± 0.5 | 2.8 ± 0.3 | 2.7 ± 0.3 | 2.7 ± 0.3 | 2.8 ± 0.2 | 2.6 ± 0.2 | 2.8 ± 0.2 | 2.7 ± 0.3 |
| Brillouin's HB | 2.0 ± 0.4 | 2.4 ± 0.2 | 2.2 ± 0.2 | 2.3 ± 0.3 | 2.2 ± 0.2 | 2.1 ± 0.2 | 2.2 ± 0.2 | 2.1 ± 0.2 |
| Berger-Parker's d | 0.2 ± 0.1 abcd | 0.1 ± 0.0 ac | 0.1 ± 0.0 abcd | 0.1 ± 0.0 ad | 0.2 ± 0.0 abcd | 0.2 ± 0.0 bd | 0.1 ± 0.0 abcd | 0.1 ± 0.0 bc |
| Simpson's D | 0.1 ± 0.1 | 0.0 ± 0.0 | 0.1 ± 0.0 | 0.1 ± 0.0 | 0.1 ± 0.0 | 0.1 ± 0.0 | 0.0 ± 0.0 | 0.1 ± 0.0 |
Values followed by no common letter are significantly different within a row (P < 0.05; two-way analysis of variance with post hoc simple contrasts).
Discriminant analysis of treatment effects.
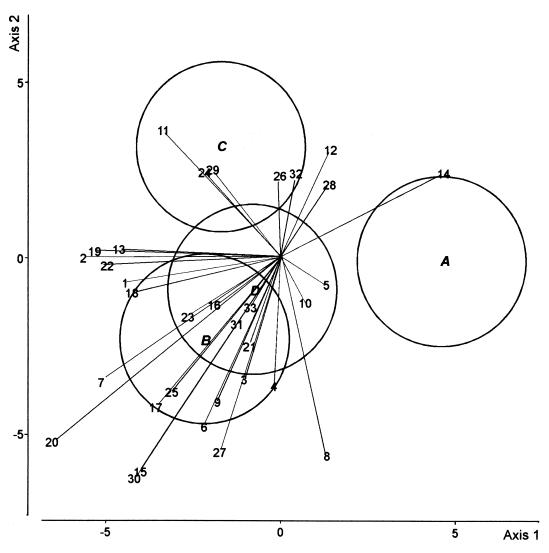
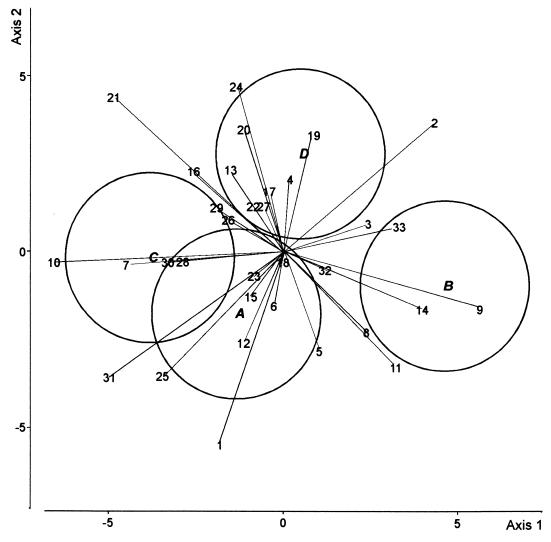
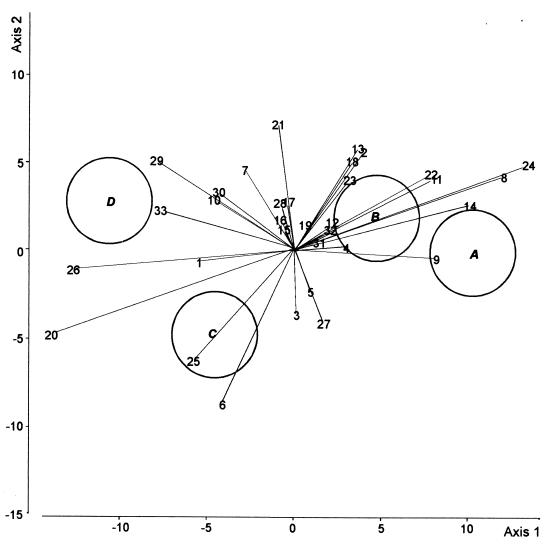
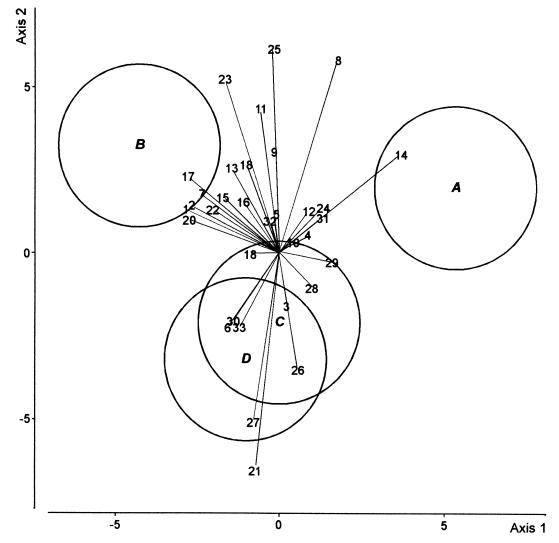
DA was carried out to compare the four treatments in the first cycle of cucumber growth (Fig. 2). The first axis (50% of the total variation) discriminated between the control and the other three treatments, but there was no difference between the latter along this axis. The second axis (27% of the total variation) distinguished the CHA0-Rif(pME3424) treatment from the CHA0-Rif treatment. In the fifth cycle of cucumber growth, the first DA axis (67% of the total variation) mainly discriminated between the CHA0-Rif and the CHA0-Rif(pME3424) treatments (Fig. 3). Axis 2 (20% of the total variation) failed to discriminate between treatments.
FIG. 2.
DA biplot showing group centroids, isodensity circles, and correlations of fungal species with canonical variates for fungal assemblages at the end of the first cycle of cucumber growth. Group A, control; group B, CHA0-Rif; group C, CHA0-Rif(pME3424); group D, metalaxyl. Fungal species names (values in parentheses are correlation with axis 1 and correlation with axis 2, respectively): 1, Acremonium roseo-griseum (−0.26, −0.04); 2, Cladosporium cladosporioides (−0.32, 0.004); 3, Coniothyrium cerealis (−0.06, −0.19); 4, C. fuckelii (−0.01, −0.21); 5, C. destructans (0.07, −0.04); 6, V. nigrescens (−0.13, −0.27); 7, Fusarium equiseti (−0.30, −0.20); 8, F. oxysporum (0.08, −0.32); 9, F. solani (−0.10, −0.23); 10, F. tabacinum (0.04, −0.07); 11, G. roseum (−0.19, 0.21); 12, Monocillium mucidum (0.08, 0.18); 13, Mortierella alpina (−0.26, 0.02); 14, Mucor hiemalis f. hiemalis (0.27, 0.14); 15, Mucor racemosus f. racemosus (−0.23, −0.35); 16, Myrothecium roridum (−0.11, −0.07); 17, Myrothecium verrucaria (−0.20, −0.24); 18, P. marquandii (−0.24, −0.06); 19, P. rugulosum (−0.31, 0.01); 20, P. humicola (−0.38, −0.30); 21, Staphylotrichum coccosporum (−0.05, −0.14); 22, Trichoderma hamatum (−0.29, −0.01); 23, Trichoderma harzianum (−0.15, −0.09); 24, Trichosporon beigelii (−0.12, 0.14); 25, Verticillium chlamydosporium (−0.18, −0.22); 26, Exophiala sp. (−0.003, 0.14);27, Humicola fuscoatra (−0.10, −0.31); 28, P. canescens (0.08, 0.12); 29, S. chartarum (−0.11, 0.15); 30, S. cylindrospora (−0.24, −0.36); 31, T. angustata (−0.07, − 0.11); 32, Mycelium sterile moniliaceum 1 (0.03, 0.14); 33, Mycelium sterile dematiaceum 4 (−0.05, −0.08). The three canonical variates accounted for 63.3, 14.7, and 7.3% of the total variation, respectively.
FIG. 3.
DA biplot showing group centroids, isodensity circles, and correlations of fungal species with canonical variates for fungal assemblages at the end of the fifth cycle of cucumber growth. Group A, control; group B, CHA0-Rif; group C, CHA0-Rif(pME3424); group D, metalaxyl. Fungal species names (values in parentheses are correlation with axis 1 and correlation with axis 2, respectively): 1, A. roseo-griseum (−0.12, −0.34); 2, C. cladosporioides (0.27, 0.23); 3, C. cerealis (0.16, 0.05); 4, C. fuckelii (0.01, 0.13); 5, C. destructans (0.07, −0.18); 6, V. nigrescens (−0.02, −0.09); 7, F. equiseti (−0.28, −0.02); 8, F. oxysporum (0.15, −0.14); 9, F. solani (0.36, −0.10); 10, F. tabacinum (−0.41, −0.02); 11, G. roseum (0.20, −0.20); 12, M. mucidum (−0.07, −0.16); 13, M. alpina (−0.09, 0.15); 14, M. hiemalis f. hiemalis (0.25, −0.10); 15, M. roridum (−0.06, −0.08); 16, M. verrucaria (−0.16, 0.14); 17, P. marquandii (−0.02, 0.11); 18, P. rugulosum (0.00, −0.02); 19, P. humicola (0.05, 0.21); 20, S. coccosporum (−0.07, 0.22); 21, T. hamatum (− 0.31, 0.28); 22, T. harzianum (−0.05, 0.08); 23, T. beigelii (−0.05, −0.04); 24, V. chlamydosporium (−0.08, 0.30); 25, Exophiala sp. (−0.22, −0.22); 26, Mycelium sterile moniliaceum 1 (−0.10, 0.06); 27, H. fuscoatra (−0.03, 0.08); 28, P. canescens (−0.18, −0.01); 29, S. chartarum (−0.12, 0.08); 30, S. cylindrospora (−0.21, −0.02); 31, T. angustata (−0.31, −0.22); 32, Mycelium sterile moniliaceum 2 (0.08, −0.03); 33, Mycelium sterile dematiaceum 4 (0.21, 0.05). The three canonical variates accounted for 66.9, 20.3, and 12.8% of the total variation, respectively.
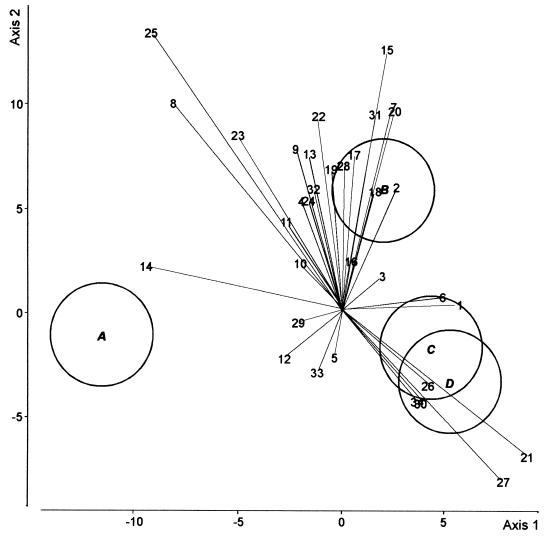
When the control and the CHA0-Rif treatments were compared in both cucumber growth cycles (Fig. 4), the first axis (74% of the total variation) distinguished the control in the first cycle from the other three situations. Whereas this axis clearly discriminated the CHA0-Rif treatment from the control in the first cycle, the two treatments could not be distinguished in the fifth cycle. The distance between the two controls was greater than the distance between the CHA0-Rif treatment and the respective control in either cycle. Axis 2 (20% of the total variation) discriminated the bacterial treatment in the first cycle from the other three situations.
FIG. 4.
DA biplot showing group centroids, isodensity circles, and correlations of fungal species for fungal assemblages in the control and the CHA0-Rif treatments, at the end of the first and the fifth cycles of cucumber growth. Group A, control in the first cycle; group B, CHA0-Rif in the first cycle; group C, control in the fifth cycle; group D, CHA0-Rif in the fifth cycle. Fungal species names (values in parentheses are correlation with axis 1 and correlation with axis 2, respectively): 1, A. roseo-griseum (0.30, 0.02); 2, C. cladosporioides (0.14, 0.32); 3, C. cerealis (0.10, 0.09); 4, C. fuckelii (−0.10, 0.28); 5, C. destructans (−0.02, −0.11); 6, V. nigrescens (0.26, 0.04); 7, F. equiseti (0.13, 0.52); 8, F. oxysporum (−0.42, 0.52); 9, F. solani (−0.12, 0.41); 10, F. tabacinum (−0.11, 0.12); 11, G. roseum (−0.14, 0.23); 12, M. mucidum (−0.14, −0.11); 13, M. alpina (−0.08, 0.40); 14, M. hiemalis f. hiemalis (−0.49, 0.12); 15, M. racemosus f. racemosus (0.11, 0.66); 16, M. roridum (0.02, 0.13); 17, M. verrucaria (0.04, 0.40); 18, P. marquandii (0.09, 0.31); 19, P. rugulosum (−0.03, 0.36); 20, P. humicola (0.13, 0.51); 21, S. coccosporum (0.47, −0.36); 22, T. hamatum (−0.06, 0.49); 23, T. harzianum (−0.26, 0.44); 24, T. beigelii (−0.09, 0.28); 25, V. chlamydosporium (−0.48, 0.70); 26, Exophiala sp. (0.22, −0.18); 27, Mycelium sterile moniliaceum 1 (0.41, −0.14); 28, H. fuscoatra (0.01, 0.37); 29, P. canescens (−0.11, − 0.02); 30, S. chartarum (0.20, −0.22); 31, S. cylindrospora (0.09, 0.50); 32, T. angustata (−0.07, 0.31); 33, Mycelium sterile moniliaceum 2 (−0.06, −0.42); 34, Mycelium sterile dematiaceum 4 (0.19, −0.22). The three canonical variates accounted for 74.0, 19.6, and 6.4% of the total variation, respectively.
When the control and the CHA0-Rif(pME3424) treatments of both cucumber growth cycles were compared (Fig. 5), each of the four situations was clearly different from the others along the first axis (82% of the total variation). The distance between the bacterial treatment and the respective control was greater in the fifth cycle than in the first cycle but was always smaller than the distance between the two controls. Axis 2 (only 11% of the total variation) mainly distinguished the control from the bacterial treatment in the fifth cycle.
FIG. 5.
DA biplot showing group centroids, isodensity circles, and correlations of fungal species for fungal assemblages in the control and the CHA0-Rif(pME3424) treatments, at the end of the first and the fifth cycles of cucumber growth. Group A, control in the first cycle; group B, CHA0-Rif(pME3424) in the first cycle; group C, control in the fifth cycle; group D, CHA0-Rif(pME3424) in the fifth cycle. Fungal species names (values in parentheses are correlation with axis 1 and correlation with axis 2, respectively): 1, A. roseo-griseum (−0.26, −0.03); 2, C. cladosporioides (0.20, 0.28); 3, C. cerealis (0.01, −0.17); 4, C. fuckelii (0.15, 0.01); 5, C. destructans (0.05, −0.11); 6, V. nigrescens (−0.20, −0.42); 7, F. equiseti (−0.14, 0.23); 8, F. oxysporum (0.59, 0.21); 9, F. solani (0.40, −0.02); 10, F. tabacinum (−0.23, 0.14); 11, G. roseum (0.40, 0.21); 12, M. mucidum (0.11, 0.08); 13, M. alpina (0.18, 0.29); 14, M. hiemalis f. hiemalis (0.50, 0.13); 15, M. roridum (−0.03, 0.06); 16, M. verrucaria (−0.04, 0.09); 17, P. marquandii (−0.02, 0.14); 18, P. rugulosum (0.17, 0.25); 19, P. humicola (0.03, 0.07); 20, S. coccosporum (−0.68, −0.23); 21, T. hamatum (−0.04, 0.36); 22, T. harzianum (0.39, 0.22); 23, T. beigelii (0.16, 0.20); 24, V. chlamydosporium (0.66, 0.25); 25, Exophiala sp. (−0.28, −0.31); 26, Mycelium sterile moniliaceum 1 (−0.61, −0.05); 27, H. fuscoatra (0.08, −0.20); 28, P. canescens (−0.04, 0.13); 29, S. chartarum (−0.39, 0.25); 30, S. cylindrospora (−0.21, 0.16); 31, T. angustata (0.07, 0.03); 32, Mycelium sterile moniliaceum 2 (0.10, 0.06); 33, Mycelium sterile dematiaceum 4 (−0.38, 0.11). The three canonical variates accounted for 81.6, 10.9, and 7.5% of the total variation, respectively.
When the control and the metalaxyl treatments of both cucumber growth cycles were compared (Fig. 6), the first axis (54% of the total variation) distinguished the control from the metalaxyl treatment for the first cycle but not for the fifth cycle. Along this axis the distance between the two controls was smaller. The second axis (33% of the total variation) distinguished the metalaxyl treatment of the first cycle from the two treatments of the fifth cycle.
FIG. 6.
DA biplot showing group centroids, isodensity circles, and correlations of fungal species for fungal assemblages in the control and the metalaxyl treatments, at the end of the first and the fifth cycles of cucumber growth. Group A, control in the first cycle; group B, metalaxyl in the first cycle; group C, control in the fifth cycle; group D, metalaxyl in the fifth cycle. Fungal species names (values in parentheses are correlation with axis 1 and correlation with axis 2, respectively): 1, A. roseo-griseum (−0.34, 0.16); 2, C. cladosporioides (−0.32, 0.18); 3, C. cerealis (0.03, −0.19); 4, C. fuckelii (0.11, 0.07); 5, C. destructans (−0.01, 0.14); 6, V. nigrescens (−0.19, −0.26); 7, F. equiseti (−0.28, 0.22); 8, F. oxysporum (0.21, 0.70); 9, F. solani (−0.02, 0.37); 10, F. tabacinum (0.05, 0.04); 11, G. roseum (−0.07, 0.52); 12, M. mucidum (0.11, 0.16); 13, M. alpina (−0.17, 0.31); 14, M. hiemalis f. hiemalis (0.44, 0.36); 15, M. racemosus f. racemosus (−0.21, 0.21); 16, M. roridum (−0.13, 0.19); 17, M. verrucaria (−0.33, 0.28); 18, P. marquandii (−0.11, 0.00); 19, P. rugulosum (−0.12, 0.32); 20, P. humicola (−0.33, 0.12); 21, S. coccosporum (−0.09, −0.78); 22, T. hamatum (−0.24, 0.16); 23, T. harzianum (−0.20, 0.63); 24, T. beigelii (0.16, 0.16); 25, V. chlamydosporium (−0.02, 0.74); 26, Exophiala sp. (0.07, −0.42); 27, Mycelium sterile moniliaceum 1 (− 0.10, −0.61), 28, H. fuscoatra (0.12, −0.12), 29, P. canescens (0.19, −0.03), 30, S. chartarum (−0.17, −0.25), 31, T. angustata (0.16, 0.13), 32, Mycelium sterile moniliaceum 2 (−0.04, 0.12), 33, Mycelium sterile dematiaceum 4 (−0.15, −0.27). The three canonical variates accounted for 54.3, 33.3, and 12.4% of the total variation, respectively.
Correlations between fungal species and canonical variates are shown in Fig. 2 to 6. Some species were found to display low correlations with canonical variates, i.e., to discriminate little among the treatments studied in either growth cycle (Fig. 2 and 3). These species were often among the commonest based on their overall occurrence, e.g., Fusarium tabacinum and Cylindrocarpon destructans in the first cycle and Penicillium rugulosum, Coniothyrium fuckelii, Verticillium nigrescens, Paecilomyces marquandii in the fifth cycle (Table 1). Some species consistently discriminated between the CHA0-Rif and the CHA0-Rif(pME3424) treatments in both cycles; e.g., Fusarium solani and Fusarium oxysporum were associated with the CHA0-Rif treatment, and Exophiala sp., Penicillium canescens, and Stachybotrys chartarum were associated with the CHA0-Rif(pME3424) treatment (Fig. 2 and 3). Other species instead appeared to be associated with one bacterial treatment in one cycle and with the other bacterial treatment in the other. For instance, Gliocladium roseum was associated with the CHA0-Rif(pME3424) treatment in the first cycle and the CHA0-Rif treatment in the fifth cycle, whereas Stachybotrys cylindrospora and Truncatella angustata were associated with the CHA0-Rif treatment in the first cycle and the CHA0-Rif(pME3424) treatment in the fifth cycle (Fig. 2 and 3).
DISCUSSION
Many biocontrol strains of fluorescent Pseudomonas spp. produce the antimicrobial polyketides Phl and/or Plt active against phytopathogenic fungi (33, 56). The ability of P. fluorescens CHA0-Rif to produce Phl and Plt was increased following the introduction of pME3424 in the strain, and the resulting derivative protected cucumber better against P. ultimum-mediated damping-off (53). In the current work, colony counts performed at the end of each cycle showed that the ability of the inoculants to colonize the rhizosphere of cucumber declined steadily with time, so that population levels of culturable cells of the inoculant strains present at the end of the fifth cucumber growth cycle were smaller than those found at the end of the first cycle. This suggests that growing cucumber repeatedly in the same soil favored resident microorganisms that were better adapted to the environmental conditions prevailing in the rhizosphere than CHA0-Rif or, especially, CHA0-Rif(pME3424) was. How this translated in terms of ecological impact on resident fungi is difficult to assess since (i) the biocontrol effect of CHA0 starts shortly after inoculation (i.e., at a time where population levels of the inoculants were still close to inoculation level) and (ii) the exact time at which interactions between indigenous fungi and introduced bacteria may take place is unknown. Population levels of the inoculants at the end of each growth cycle were still in the order of 105 CFU per g of root or higher, which is generally considered enough for disease suppression by biocontrol pseudomonads (51). However, it is possible that the impact on resident culturable fungi in each cycle could have been bigger at an earlier sampling, and that mostly resilience was being recorded at 32 days (48).
Isolation of microfungi from the rhizosphere of cucumber yielded a broad fungal spectrum (Table 1) dominated by genera and species rather widespread and frequently found in agricultural soils, rhizospheres, and roots of crop plants (the main exception being Phialocephala humicola, which is mostly a forest soil fungus) (3, 4, 18, 26). This fungal spectrum overlaps the one obtained by Hong (31), who found that rhizosphere fungi protect cucumber seedlings against damping-off caused by Fusarium oxysporum f. sp. cucumerinum. No Oomycetes were isolated in this study, despite the fact that they can grow on the laboratory medium used. This is in accordance with the fact that disease pressure is usually low in Eschikon soil. The choice of experimental conditions not conducive to plant disease in such a study enabled us (i) to assess potential negative ecological impacts of a biocontrol inoculant without the interference of the positive biocontrol effect on target pathogens (45) and (ii) to avoid large-scale plant mortality in the unprotected control treatment (56). Perhaps Oomycetes were present at population levels too low to be detected, or perhaps they were not competitive enough on the plates.
Soil and rhizosphere fungal communities have been shown to fit the lognormal species abundance distribution model (39), but Zak (71) found a logarithmic function, not a lognormal function, to be typical for root surface fungal assemblages, and Thomas and Shattock (59) observed that phylloplane fungal populations were best described by both the geometric and the log series model. In this study, most fungal assemblages fit both the log series function and the truncated lognormal model, but the latter provided a better fit (Fig. 1). Polluted or stressed communities are often characterized by a switch from the lognormal model (arising in response to many independent factors controlling the abundance of a heterogeneous collection of organisms) to the geometric series (associated with species-poor habitats, where dominant species preempt a significant portion of a limiting resource of the habitat and reach population levels proportional to the amount of resource utilized) (40, 71). Against this background, it appears that neither those treatments nor the fact of growing cucumber repeatedly in the same soil represented a significant perturbation for the fungal community of the rhizosphere. No conclusion could be drawn for the CHA0-Rif treatment in the first cucumber growth cycle, as none of the models tested were appropriate in this case.
These findings were strengthened by comparing fungal diversity levels (Table 2). Indeed, the majority of diversity indices did not highlight any difference between the fungal assemblages studied. Statistically significant differences were obtained only within treatments over the two cycles, fungal diversity being higher in the fifth cycle than in the first cycle. Instead, no significant effect of the different treatments was revealed in either cucumber growth cycle, indicating that the species richness and evenness of the fungal assemblages were unaffected by such treatments.
However, significant differences between treatments were found when fungal diversity was analyzed in more detail, using a DA approach (Fig. 2 to 6). Depending on the growth cycle, such differences were found between the control and each of the three other treatments and/or between the two inoculation treatments. Correlations were found between treatments and the occurrence of certain fungal species in both cucumber growth cycles, as illustrated by DA biplots. Seed inoculation with P. fluorescens E6 resulted in enhanced colonization of zinnia roots by Fusarium spp. and reduced colonization by Penicillium spp. at 3 weeks (70), which did not take place in this work with either inoculant (data not shown). The effects of the CHA0-Rif(pME3424) treatment were not greater in intensity than those of the CHA0-Rif treatment when compared with the effects of the control (Fig. 2 and 3). Some of the differences between the CHA0-Rif and CHA0-Rif(pME3424) treatments could already be anticipated by comparing the results for fungal species listed in Table 1, as, e.g., in the fifth cycle Fusarium solani was recovered from 7 of 10 plants in the CHA0-Rif treatment but from only 2 plants in the CHA0-Rif(pME3424) treatment. An effect of the metalaxyl treatment was also detected (Fig. 4), although this fungicide is thought to act almost exclusively against Peronosporales (12, 54). Peronosporales were not found in this work, but metalaxyl may display a wider specificity towards fungi considering that it inhibits rRNA synthesis. Indeed, there is evidence that arbuscular mycorrhizal fungi are sensitive to metalaxyl (58). Interestingly, treatment effects were greater in the fifth cycle than in the first cycle for the CHA0-Rif(pME3424) treatment (Fig. 5), while the effects of the CHA0-Rif treatment (Fig. 4) and the metalaxyl treatment (Fig. 6) had decreased by the fifth cycle. However, the effects of the bacterial treatments were less than those linked to cucumber monoculture (Fig. 4 to 6).
Whether the effects of the bacterial treatments resulted from the ability of the inoculants to produce (or overproduce) the antimicrobial compounds Phl and Plt remains to be ascertained. Under gnotobiotic conditions, it can be shown using a translational phlA′-′lacZ fusion that the biosynthetic gene phlA involved in Phl synthesis is expressed when CHA0 colonizes the cucumber rhizosphere (R. Notz, M. Maurhofer, U. Schnider-Keel, D. Haas, and G. Défago, unpublished). CHA0-Rif(pME3424) displayed enhanced protection of cucumber against P. ultimum compared with CHA0 (44), confirming previous disease suppression data obtained using the prototype plasmid pME3090 (42), from which pME3424 was constructed. Interestingly, high-pressure liquid chromatography analysis indicated that CHA0 can produce Phl and Plt in the rhizosphere and that the concentrations reached by these polyketides in the rhizosphere were higher when CHA0 contained pME3090 (44). It is considered that Phl-positive biocontrol pseudomonads should reach population levels of 105 CFU per g of root (or more) for effective production of Phl (52), which was the case here. Further work will assess whether repeated inoculations of CHA0-Rif or CHA0-Rif(pME3424) resulted in the enrichment of fungi resistant to Phl and/or Plt.
Interactions between CHA0-Rif or CHA0-Rif(pME3424) and the resident culturable fluorescent pseudomonads in the cucumber rhizosphere were not mediated by the production of antimicrobial polyketides (48). Likewise, some of the effects of the bacterial treatments were perhaps not mediated by Phl or Plt. For instance, positive interactions can take place between introduced biocontrol pseudomonads and resident antagonistic fungi, which may contribute to suppression of Fusarium wilt of radish (38). Indeed, interactions among component species have been recognized to play an important role in shaping fungal communities (66), and bacteria can have an indirect effect on saprotrophic microfungi by influencing interfungal interactions. This may generate great complexity and lead to the occurrence of unexpected phenomena in natural communities (10, 37). In addition, one of the mechanisms by which CHA0 can protect against disease corresponds to induced systemic resistance (43). Plant physiology is different when systemic resistance has been induced (23), which in turn may influence the way fungi can colonize the rhizosphere. This issue deserves further attention.
Comparable studies on the effects of introduced biocontrol bacteria on indigenous rhizosphere bacterial populations have shown that the impact of the inocula can vary depending on the experimental conditions (28). Therefore, it will be necessary to extend impact assessment of biocontrol bacterial inoculants on the indigenous fungal community to take into account the various environmental conditions prevailing in the field (e.g., from one year to the next), as well as crop rotation conditions. The effect of fungal disease pressure is another important issue that will have to be addressed in further work, as fungal pathogens can influence the growth and physiology of roots and the release of root exudates (1), which in turn will likely affect saprotrophic fungi.
In conclusion, DA revealed that inoculation with P. fluorescens CHA0-Rif or its GM derivative CHA0-Rif(pME3424) had a detectable influence on the fungal community of the cucumber rhizosphere at 32 days in the two cycles studied. The effects of the two inoculants were different, but in both cases the magnitude of the impact was small. The most frequently isolated fungal species were common to the different treatments, and no fungal species was totally suppressed or found exclusively in one particular treatment. No dramatic change in species abundance distribution or diversity level was observed following the various treatments, indicating that community organization was not profoundly altered, and treatment effects were equal to or smaller than the effects of growing cucumber repeatedly in the same soil.
ACKNOWLEDGMENTS
We thank Felipe Wettstein, Carsten Hase, Zensi Hopf, and Fabio Mascher (ETH) for technical help.
This work was supported by the EU IMPACT2 project (BIO4-CT96-0027) and by grant MURST (40%).
REFERENCES
- 1.Agrios G N. Plant pathology. 4th ed. San Diego, Calif: Academic Press; 1997. [Google Scholar]
- 2.Alabouvette C, Steinberg C. Suppressiveness of soils to invading micro-organisms. In: Hokkanen H M T, Lynch J M, editors. Plant and microbial biotechnology research series. 4. Biological control: benefits and risks. Cambridge, United Kingdom: Cambridge University Press; 1995. pp. 3–12. [Google Scholar]
- 3.Barron G L. The genera of Hyphomycetes from soil. Baltimore, Md: The Williams and Wilkins Company; 1968. [Google Scholar]
- 4.Booth C. The genus Fusarium. Kew, United Kingdom: Commonwealth Mycological Institute; 1971. [Google Scholar]
- 5.Chet I, Inbar J, Hadar Y. Fungal antagonists and mycoparasites. In: Wicklow D T, Söderström B, editors. The mycota. 4. Environmental and microbial relationships. Berlin, Germany: Springer-Verlag; 1997. pp. 165–184. [Google Scholar]
- 6.Cook R J. Making greater use of introduced microorganisms for biological control of plant pathogens. Annu Rev Phytopathol. 1993;31:53–80. doi: 10.1146/annurev.py.31.090193.000413. [DOI] [PubMed] [Google Scholar]
- 7.Cook R J. Assuring the safe use of microbial biocontrol agents—a need for policy based on real rather than perceived risks. Can J Plant Pathol. 1996;18:439–445. [Google Scholar]
- 8.Cook R J, Bruckart W L, Coulson J R, Goettel M S, Humber R A, Lumsden R D, Maddox J V, Memanus M L, Moore L, Meyer S F, Quimby P C, Stack J P, Vaughn J L. Safety of microorganisms intended for pest and plant disease control—a framework for scientific evaluation. Biol Control. 1996;7:333–351. [Google Scholar]
- 9.Cooke R C, Rayner A D M. Ecology of saprotrophic fungi. London, United Kingdom: Longman; 1984. [Google Scholar]
- 10.Culver D C. Introduction to the theory of species interactions. In: Carroll G C, Wicklow D T, editors. The fungal community: its organization and role in the ecosystem. 2nd ed. New York, N.Y: Marcel Dekker, Inc; 1992. pp. 229–241. [Google Scholar]
- 11.Curl E A, Truelove B. The rhizosphere. Berlin, Germany: Springer-Verlag; 1986. [Google Scholar]
- 12.Davidse L C. Phenylamide fungicides—biochemical action and resistance. In: Lyr H, editor. Modern selective fungicides: properties, applications, modalities of action. Iena, Germany: Gustav Fischer Verlag; 1995. pp. 347–354. [Google Scholar]
- 13.Défago G, Berling C-H, Henggeler S, Hungerbühler W, Kern H, Schleppi P, Stutz E W, Zürrer M. Survie d'un Pseudomonas fluorescens dans le sol et protection du blé contre les maladies d'origine fongique. Schweiz Landw Fo. 1987;26:155–160. [Google Scholar]
- 14.Défago G, Keel C, Moënne-Loccoz Y. Fate of released Pseudomonas bacteria in the soil profile: Implications for the use of genetically-modified microbial inoculants. In: Zelikoff J T, Lynch J M, Shepers J, editors. Ecotoxicology: responses, biomarkers and risk assessment. Fair Haven, N.J: SOS Publications; 1997. pp. 403–418. [Google Scholar]
- 15.de Leij F A A M, Sutton E J, Whipps J M, Lynch J M. Effect of a genetically modified Pseudomonas aureofaciens on indigenous microbial populations of wheat. FEMS Microbiol Ecol. 1994;13:249–257. [Google Scholar]
- 16.Dighton J. Analysis of micromycete communities in soil: a critique of methods. Mycol Res. 1994;98:796–798. [Google Scholar]
- 17.Dix N J, Webster J. Fungal ecology. London, United Kingdom: Chapman and Hall; 1995. [Google Scholar]
- 18.Domsch K H, Gams W, Anderson T H. Compendium of soil fungi. Eching, Germany: IHW-Verlag; 1980. [Google Scholar]
- 19.Doyle J D, Stotzky G. Methods for the detection of changes in the microbial ecology of soil caused by the introduction of microorganisms. Microb Releases. 1993;2:63–72. [Google Scholar]
- 20.Dunne C, Delany I, Fenton A, Lohrke S, Moënne-Loccoz Y, O'Gara F. The biotechnology and application of Pseudomonas inoculants for the biocontrol of phytopathogens. In: Stacey G, Mullin B, Gresshoff P M, editors. Biology of plant-microbe interactions. St. Paul, Minn: IS-MPMI; 1996. pp. 441–448. [Google Scholar]
- 21.Dutrecq A, Debras P, Stevaux J, Marlier M. Activity of 2,4-diacetylphloroglucinol isolated from a strain of Pseudomonas fluorescens to Gaeumannomyces graminis var. tritici. In: Beemster A B R, Bollen G J, Gerlagh M, Ruissen M A, Schippers B, Tempel A, editors. Biotic interactions and soil-borne diseases. Amsterdam, The Netherlands: Elsevier Science Publishers; 1991. pp. 252–257. [Google Scholar]
- 22.Fritze H, Bååth E. Microfungal species composition and fungal biomass in a coniferous forest soil polluted by alkaline deposition. Microb Ecol. 1993;25:83–92. doi: 10.1007/BF00182131. [DOI] [PubMed] [Google Scholar]
- 23.Fuchs J-G, Moënne-Loccoz Y, Défago G. Nonpathogenic Fusarium oxysporum strain Fo47 induces resistance to Fusarium wilt in tomato. Plant Dis. 1997;81:492–496. doi: 10.1094/PDIS.1997.81.5.492. [DOI] [PubMed] [Google Scholar]
- 24.Gams W. The analysis of communities of saprophytic microfungi with special reference to soil fungi. In: Winterhoff W, editor. Fungi in vegetation science. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1992. pp. 183–223. [Google Scholar]
- 25.Garagulya A D, Kirprianova E A, Boiko O I. Antibiotic effect of bacteria from the genus Pseudomonas on phytopathogenic fungi. Mikrobiol Zh. 1974;36:197–202. [PubMed] [Google Scholar]
- 26.Gerlach W, Nirenberg H. The genus Fusarium—a pictorial atlas. Berlin, Germany: Biologischen Bundesanstalt für Land und Forstwirtschaft; 1982. [Google Scholar]
- 27.Giddings G. Tansley review no. 99. The release of genetically engineered micro-organisms and viruses into the environment. New Phytol. 1998;140:173–184. doi: 10.1046/j.1469-8137.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- 28.Gilbert G S, Parke J L, Clayton M K, Handelsman J. Effects of an introduced bacterium on bacterial communities on roots. Ecology. 1993;74:840–854. [Google Scholar]
- 29.Guntli D, Burgos S, Moënne-Loccoz Y, Défago G. Calystegine degradation capacities of microbial rhizosphere communities of Zea mays (calystegine-negative) and Calystegia sepium (calystegine-positive) FEMS Microbiol Ecol. 1999;28:75–84. [Google Scholar]
- 30.Hawksworth D L, Kirk P M, Sutton B C, Pegler D N. Ainsworth and Bisby's dictionary of the fungi. 8th ed. Wallingford, United Kingdom: CAB International; 1995. [Google Scholar]
- 31.Hong C-Y. The relation between rhizosphere fungi and the occurrence of damping-off of cucumber seedlings. Ann Phytopathol Soc Jpn. 1969;25:308–314. [Google Scholar]
- 32.Keel C, Schnider U, Maurhofer M, Voisard C, Laville J, Burger U, Wirthner P, Haas D, Défago G. Suppression of root diseases by Pseudomonas fluorescens CHA0: Importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol Plant-Microbe Interact. 1992;5:4–13. [Google Scholar]
- 33.Keel C, Weller D M, Natsch A, Défago G, Cook R J, Thomashow L S. Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl Environ Microbiol. 1996;62:552–563. doi: 10.1128/aem.62.2.552-563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keel C, Défago G. Interactions between beneficial soil bacteria and root pathogens: mechanisms and ecological impact. In: Gange A C, Brown V K, editors. Multitrophic interactions in terrestrial systems. London, United Kingdom: Blackwell Scientific Publishers; 1997. pp. 27–46. [Google Scholar]
- 35.Kennedy A C, Smith K L. Soil microbial diversity and the sustainability of agricultural soils. Plant Soil. 1995;170:75–86. [Google Scholar]
- 36.Knauf V C, Nester E W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982;8:45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- 37.Lavelle P. Interactions, hiérarchies et régulations dans le sol: a la recherche d'une nouvelle approche conceptuelle. Rev Ecol Biol Sol. 1987;24:219–229. [Google Scholar]
- 38.Leeman M, Den Ouden F M, Van Pelt J A, Cornelissen C, Matamala Garros A, Bakker P A H M, Schippers B. Suppression of fusarium wilt of radish by co-inoculation of fluorescent Pseudomonas spp and root-colonizing fungi. Eur J Plant Pathol. 1996;102:21–31. [Google Scholar]
- 39.Lussenhop J. Analysis of microfungal component communities. In: Wicklow D T, Carroll G C, editors. The fungal community. Its organization and role in the ecosystem. New York, N.Y: Marcel Dekker, Inc; 1981. pp. 37–45. [Google Scholar]
- 40.Magurran A E. Ecological diversity and its measurement. London, United Kingdom: Chapman and Hall; 1988. [Google Scholar]
- 41.Mahaffee W F, Kloepper J W. Bacterial communities of the rhizosphere and endorhiza associated with field-grown cucumber plants inoculated with a plant growth-promoting Rhizobacterium or its genetically modified derivative. Can J Microbiol. 1997;43:344–353. doi: 10.1139/m97-048. [DOI] [PubMed] [Google Scholar]
- 42.Maurhofer M, Keel C, Schnider U, Voisard C, Haas D, Défago G. Influence of enhanced antibiotic production in Pseudomonas fluorescens strain CHA0 on its disease suppressive capacity. Phytopathology. 1992;82:190–195. [Google Scholar]
- 43.Maurhofer M, Hase C, Meuwly P, Métraux J-P, Défago G. Induction of systemic resistance of tobacco to tobacco necrosis virus by the root-colonizing Pseudomonas fluorescens strain CHA0: Influence of the gacA gene and of pyoverdine production. Phytopathology. 1994;84:139–146. [Google Scholar]
- 44.Maurhofer M, Keel C, Haas D, Défago Influence of plant species on disease suppression by Pseudomonas fluorescens strain CHA0 with enhanced antibiotic production. Plant Pathol. 1995;44:40–50. [Google Scholar]
- 45.Moënne-Loccoz Y, Powell J, Higgins P, Britton J, O'Gara F. Effect of the biocontrol agent Pseudomonas fluorescens F113 released as sugarbeet inoculant on the nutrient contents of soil and foliage of a red clover rotation crop. Biol Fertil Soils. 1998;27:380–385. [Google Scholar]
- 46.Naeem S, Thompson L J, Lawler S P, Lawton J H, Woodfin R M. Declining biodiversity can alter the performance of ecosystems. Nature. 1994;368:734–737. [Google Scholar]
- 47.Natsch A, Keel C, Pfister H A, Haas D, Défago G. Contribution of the global regulator gene gacA to persistence and dissemination of Pseudomonas fluorescens biocontrol strain CHA0 introduced into soil microcosms. Appl Environ Microbiol. 1994;60:2553–2560. doi: 10.1128/aem.60.7.2553-2560.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Natsch A, Keel C, Hebecker N, Laasik E, Défago G. Influence of biocontrol strain Pseudomonas fluorescens CHA0 and its antibiotic overproducing derivative on the diversity of resident root colonizing pseudomonads. FEMS Microbiol Ecol. 1997;23:341–352. [Google Scholar]
- 49.Nuti M P, Squartini A, Giacomini A. European Community regulation for the use and release of genetically modified organisms (GMOs) in the environment. In: O'Gara F, Dowling D, Boesten B, editors. Molecular ecology of rhizosphere microorganisms. Weinheim, Germany: VCH Publications; 1994. pp. 165–173. [Google Scholar]
- 50.Podani J. Multivariate data analysis in ecology and systematics. Ecological computations series. Vol. 6. The Hague, The Netherlands: SPB Academic Publishing; 1994. [Google Scholar]
- 51.Raaijmakers J M, Leeman M, van Oorschot M M P, van der Sluis I, Schippers B, Bakker P A H M. Dose-response relationships in biological control of Fusarium wilt of radish by Pseudomonas spp. Phytopathology. 1995;85:1075–1081. [Google Scholar]
- 52.Raaijmakers J M, Bonsall R F, Weller D M. Effect of population density of Pseudomonas fluorescens on production of 2,4-diacetylphloroglucinol in the rhizosphere of wheat. Phytopathology. 1999;89:470–475. doi: 10.1094/PHYTO.1999.89.6.470. [DOI] [PubMed] [Google Scholar]
- 53.Schnider U, Keel C, Blumer C, Troxler J, Défago G, Haas D. Amplification of the housekeeping sigma factor in Pseudomonas fluorescens CHA0 enhances antibiotic production and improves biocontrol abilities. J Bacteriol. 1995;177:5387–5392. doi: 10.1128/jb.177.18.5387-5392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwinn F, Staub T. Phenylamides and other fungicides against Oomycetes. In: Lyr H, editor. Modern selective fungicides. Properties, applications, modalities of action. Iena, Germany: Gustav Fischer Verlag; 1995. pp. 323–346. [Google Scholar]
- 55.Shanahan P, O'Sullivan D J, Simpson P, Glennon J D, O'Gara F. Isolation of 2,4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl Environ Microbiol. 1992;58:353–358. doi: 10.1128/aem.58.1.353-358.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharifi-Tehrani A, Zala M, Natsch A, Moënne-Loccoz Y, Défago G. Biocontrol of soil-borne fungal plant diseases by 2,4-diacetylphloroglucinol-producing fluorescent pseudomonads with different restriction profiles of amplified 16S rDNA. Eur J Plant Pathol. 1998;104:631–643. [Google Scholar]
- 57.Stutz E, Défago G, Kern H. Naturally occurring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology. 1986;76:181–185. [Google Scholar]
- 58.Sukarno N, Smith F A, Smith S E, Scott E S. The effects of fungicides on vesicular-arbuscular mycorrhizal symbiosis. 2. The effects on area of interface and efficiency of P uptake and transfer to plant. New Phytol. 1996;132:583–592. doi: 10.1111/j.1469-8137.1996.tb01877.x. [DOI] [PubMed] [Google Scholar]
- 59.Thomas M R, Shattock R C. Filamentous fungal associations in the phylloplane of Lolium perenne. Trans Br Mycol Soc. 1986;87:255–268. [Google Scholar]
- 60.Thomashow L S, Weller D M. Current concepts in the use of introduced bacteria for biological disease control: mechanisms and antifungal metabolites. In: Stacey G, Keen N T, editors. Plant-microbe interactions. Vol. 1. New York, N.Y: Chapman and Hall; 1996. pp. 187–235. [Google Scholar]
- 61.Tiedje J M, Colwell R K, Grossman Y L, Hodson R E, Lenski R E, Mack R N, Regal P J. The planned introduction of genetically engineered organisms: Ecological considerations and recommendations. Ecology. 1989;70:298–315. [Google Scholar]
- 62.Troxler J, Zala M, Moënne-Loccoz Y, Keel C, Défago G. Predominance of nonculturable cells of the biocontrol strain Pseudomonas fluorescens CHA0 in the surface horizon of large outdoor lysimeters. Appl Environ Microbiol. 1997;63:3776–3782. doi: 10.1128/aem.63.10.3776-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voisard C, Bull C, Keel C, Laville J, Maurhofer M, Schnider U, Défago G, Haas D. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches. In: O'Gara F, Dowling D, Boesten B, editors. Molecular ecology of rhizosphere microorganisms. Weinheim, Germany: VHC Publishers; 1994. pp. 67–89. [Google Scholar]
- 64.von Arx J A. The genera of fungi sporulating in pure culture. 3rd ed. Vaduz, Germany: J. Cramer; 1980. [Google Scholar]
- 65.Whipps J M, de Leij F A A M, Lynch J M, Bailey M J. Impact of genetically modified microorganisms on the terrestrial microbiota including fungi. In: Frankland J C, Magan N, Gadd G M, editors. Fungi and environmental change. Cambridge, United Kingdom: Cambridge University Press; 1996. pp. 299–316. [Google Scholar]
- 66.Widden P. Microfungal community structure from forest soils in southern Quebec, using discriminant function and factor analysis. Can J Bot. 1986;64:1402–1412. [Google Scholar]
- 67.Widden P. Seasonality of forest soil microfungi in southern Quebec. Can J Bot. 1986;64:1413–1423. [Google Scholar]
- 68.Widden P. Fungal communities in soils along an elevation gradient in northern England. Mycologia. 1987;79:298–309. [Google Scholar]
- 69.Wüthrich B, Défago G. Suppression of wheat take-all and black root rot of tobacco by Pseudomonas fluorescens strain CHA0: results of field and pot experiments. In: Keel C, Koller B, Défago G, editors. Plant growth-promoting rhizobacteria. Interlaken, Switzerland: Interlaken International Workshop; 1990. pp. 17–22. [Google Scholar]
- 70.Yuen G Y, Schroth M N. Interactions of Pseudomonas fluorescens strain E6 with ornamental plants and its effect on the composition of root-colonizing microflora. Phytopathology. 1986;76:176–180. [Google Scholar]
- 71.Zak J C. Response of soil fungal communities to disturbance. In: Carroll G C, Wicklow D T, editors. The fungal community: its organization and role in the ecosystem. 2nd ed. New York, N.Y: Marcel Dekker, Inc; 1992. pp. 403–425. [Google Scholar]
- 72.Zak J C, Rabatin S C. Organization and description of fungal communities. In: Wicklow D T, Söderström B, editors. The mycota. 4. Environmental and microbial relationships. Berlin, Germany: Springer-Verlag; 1997. pp. 33–46. [Google Scholar]