Abstract
A patient-specific airflow simulation was developed to help address the pressing need for an expansion of the ventilator capacity in response to the COVID-19 pandemic. The computational model provides guidance regarding how to split a ventilator between two or more patients with differing respiratory physiologies. To address the need for fast deployment and identification of optimal patient-specific tuning, there was a need to simulate hundreds of millions of different clinically relevant parameter combinations in a short time. This task, driven by the dire circumstances, presented unique computational and research challenges. We present here the guiding principles and lessons learned as to how a large-scale and robust cloud instance was designed and deployed within 24 hours and 800 000 compute hours were utilized in a 72-hour period. We discuss the design choices to enable a quick turnaround of the model, execute the simulation, and create an intuitive and interactive interface.
The ability to quickly spin-up cloud instances that strategically match the presented research question, alongside the increasing availability of high-performance computing resources, has resulted in an increased use of cloud computing for critical computational science research. A global pandemic, such as COVID-19, naturally amplifies the need for rapid development and deployment of tools that can have an impact. The dire circumstances evolving in spring of 2020 led to increased hospitalization rates and estimates of shortages of ventilators within the U.S. alone of between 45000 and 160 000.1 Ventilators are an essential component of life-preserving treatment for patients with respiratory failure and a shortage of ventilators was predicted,2 and in some cases realized,3 early in the global pandemic. As a result, considerable effort has been focused on methods to share one ventilator amongst multiple patients.1 4
4 5 However, due to safety concerns6 with preexisting ventilator splitting strategies, ventilator splitting in the past has not been recommended.7 In collaboration with restor3D (a local biotechnology company, https://www.restor3d.com/), a large team of engineers and clinicians at Duke University developed a ventilator splitter and resistor system (VSRS) aimed at increasing the safety profile of ventilator splitting by accurately predicting the delivered tidal volumes and pressures under the wide range of clinically relevant situations. To achieve this task, it was necessary to quickly develop and deploy new strategies of ventilator splitting, combining 3-D printed components designed to fit standard ventilator tubing (see Figure 1: top) with extensive computational modeling to ensure that each patient would receive a safe degree of ventilation (see Figure 1, bottom).
5 However, due to safety concerns6 with preexisting ventilator splitting strategies, ventilator splitting in the past has not been recommended.7 In collaboration with restor3D (a local biotechnology company, https://www.restor3d.com/), a large team of engineers and clinicians at Duke University developed a ventilator splitter and resistor system (VSRS) aimed at increasing the safety profile of ventilator splitting by accurately predicting the delivered tidal volumes and pressures under the wide range of clinically relevant situations. To achieve this task, it was necessary to quickly develop and deploy new strategies of ventilator splitting, combining 3-D printed components designed to fit standard ventilator tubing (see Figure 1: top) with extensive computational modeling to ensure that each patient would receive a safe degree of ventilation (see Figure 1, bottom).
Figure 1.
Top: Three-dimensional (3-D) printed splitter (in blue) connected to the resistor (silver). Airflow from the ventilator comes into the VSRS from the left and heads to the patient who requires the resistor (top right) or without the resistor (bottom right). Bottom: Example output from the numerical model without a resistor for a PIP of 30, PEEP of 8, and I:E of 1 (acronyms explained in Table 1), demonstrating how predicted tidal volumes vary greatly and nonlinearly from multiple parameters, specifically pulmonary compliance, respiratory rate, and endotracheal (ET) tube size. The result for two different ET tube sizes are displayed, with an 8 mm ET tube resulting in greater tidal volumes than a 6 mm ET tube. Red colors demonstrate potentially highly dangerous tidal volumes.
With the validated device in hand, the remaining question was how to tune it for any given patient pairing and, further, how to provide this decision support in an intuitive way to the clinicians. Addressing these questions presented unique challenges in an environment with significant time pressure. The entire process, from design of a new computational model, to architecting an appropriate computing environment, to creating a user interface, presented many challenging opportunities of interest to high-performance computing and biomedical researchers facing problems driven by short turnaround times. This article summarizes our design decisions and lessons learned during model design, HPC resource procurement and deployment, and execution of a massively parallel solution using over 800 000 compute hours in a 72 h period. The total time to create the initial model, validate it against benchtop data, deploy the model at scale, and to collate the simulation results into an easy-to-use mobile app was less than one month. We expect that these experiences can be applied by the computational science community to future research questions requiring rapid deployment.
DESIGN: MODEL DEVELOPMENT
Establishing a Numerical Model
A numerical model was established to aid clinicians in their use of the VSRS, by calculating the airflow characteristics and distribution through the system over a wide range of operating conditions. Under the dire circumstances of a global pandemic, not only was time-to-solution a driving design goal, but similarly was minimal development time. In conventional computational fluid dynamics research, time would be taken to optimize the code and the simulation setup so that the run-time of each simulation was minimized. Such an effort typically requires significant man-hours to establish a validated and optimized computational model. The challenges in developing the VSRS model were the competing needs of 1) creating a robust, well-validated, and accurate model to ensure that clinicians can deploy the VSRS while minimizing harm to their patients, and 2) the need to deliver results as quickly as possible.
Due to a lack of well-established numerical models available for ventilator splitting, we developed our own model to explore the general dynamics of a system where multiple patients are connected to one ventilator. Specifically, we approached the problem by using lumped parameter models to solve the governing equations of mass, energy, and momentum conservation in order to simulate airflow from a ventilator source to a patient's lungs. The lungs were modeled as a Hookean spring and viscous dashpot in parallel to represent the pulmonary compliance and resistance, respectively. The relevant inputs to the model are described in Table 1 and Figure 2, and the outputs are time-series of delivered tidal volumes (which can be condensed as displayed in Figure 1, bottom) and pressures.
Table 1. Input parameters to the numerical model.
| Parameter Name | Units | Min Value | Max Value | Step Size | # of Values |
|---|---|---|---|---|---|
| Peak Inspiratory Pressure (PIP) | cmH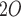
|
20 | 50 | 1 | 31 |
| Positive End-Expiratory Pressure (PEEP) | cmH
|
5 | 20 | 1 | 16 |
| Inspiratory to Expiratory Ratio (I:E) | ratio | 1:3 | 1:1 | fraction | 7 |
| Respiratory Rate (RR) |

|
10 | 30 | 1 | 21 |
| Pulmonary Compliance |

|
10 | 100 | 1-2 | 46-91 |
| Endotracheal Tube (ET) Diameter | mm | 6 | 8.5 | 0.5 | 6 |
| Resistor Radii | mm | 2.5 | 5.5 | 1 | 7 |
Final parameters, along with the minimum, maximum, and step size, and discrete values explored, that were required for the large parameter sweep. Step sizes were determined so that an incremental change resulted in less than a 5% change in delivered tidal volumes. The top four parameters are ventilator settings (PIP, PEEP, I:E, RR), the next two are patient-specific considerations (Compliances and ET tube size), and the final one is a circuit configuration parameter (Resistor size).
Figure 2.
Illustration of the directory structure, job types, and output file structure for the 7-D embarrassingly parallel parameter sweep. The number of parameters at each level are in parentheses. For each combination of PIP, PEEP, I:E, and RR, two jobs were submitted, for the cases without a resistor and one for cases with a resistor. Those jobs then swept through the parameters of ET Tube sizes, resistor sizes (if applicable), and pulmonary compliances, totaling over 270 million different simulations.
The ideal modeling tool for this task would be one which can be run locally, is highly flexible to allow different configurations and patient parameters, and is readily scalable and computationally efficient so that it could eventually be deployed for a massive parameter sweep. MATLABs Simscape has an easy-to-use GUI, which allows for rapid generation of highly flexible models that can run easily on a laptop. This allows one to experiment with the significance of different parameters and model configurations, however, using a higher-level proprietary software that requires a license to run potentially meant that we would encounter issues with scalability and efficiency when deploying the model at scale. While we chose MATLAB due to previous experience with the modeling software in an effort to decrease time to model generation, open source alternatives, such as OpenModelica or Scilab, could have been explored as well.
The initial models helped us make fundamental observations, which were key to designing our large-scale parameter sweep. Of note, we discovered that the one ventilator mode (pressure-controlled ventilation) was inherently safer for ventilator splitting than another (volume-controlled ventilation). Additionally, the preliminary models demonstrated that a decoupling occurs when multiple patients are placed on pressure-controlled ventilation, such that there is no interaction between patients connected to the same ventilator. This finding was important because it fundamentally changed our computational task. Namely, instead of simulating every possible combination of ventilator parameters as well as both patients’ individual characteristics, this permitted the more tractable task of solving for every possible combination of ventilator parameters and a single patients characteristics.
This difference ultimately reduced the number of simulations we had to perform by several orders of magnitude (270 million as compared to 125 billion) and turned an intractable task into a large, but achievable computational challenge given the time constraints.
A final important outcome from the initial testing was to understand the sensitivity of our outputs of interest, which are tidal volumes (see Figure 1 bottom) and delivered pressures, to the many input parameters (see Table 1). The minimum and maximum values for various parameters were known from clinical experience, but how finely we would have to sample parameter space was unknown. To overcome this, we performed sensitivity tests to determine the granularity with which we would have to sample parameter space in order to guarantee precise results within an acceptable level of uncertainty. With this knowledge, and initial speed tests of the numerical model on local resources, we were able to estimate the number of simulations and compute hours that would be required, assuming that we were able to efficiently scale the model to larger systems.
Lesson Learned 1: Using low overhead tools, such as MATLAB's Simscape, for initial modeling can enable rapid acquisition of baseline intuition needed for design of the large-scale study.
Embarrassingly Parallel Design
The goal of this project was to provide a decision support tool to aid clinicians in their use of the VSRS. To this end, we needed to precalculate the expected airflow for any potential patient- and ventilator-derived values, so that the resulting data could be made available in realtime to help direct the choice of resistor. As described in Table 1, there are seven parameters serving as input to the simulation. For the four ventilator settings in Table 1 (PIP, PEEP, I:E, RR) and ET tube diameters, the step size was chosen to allow clinicians to have an equal level of granularity as they would with a standard ventilator, with minimum and maximum values chosen based on clinical experience for the range of realistic clinical scenarios. The compliance step size was set such that a step change in compliance would lead to less than a 5% change in tidal volumes. Determining the number of values to explore was derived from discrete subtraction of the minimum from the maximum value, divided by the step size.
Figure 2 displays the chosen hierarchy for the parameter sweep. The batch submission script generates two jobs (one for the model with a resistor, one for the model without a resistor) for each combination of PIP, PEEP, I:E, and RR. Each job then sweeps through the different ET Tube sizes, Resistor sizes, and Compliance values. Due to the embarrassingly parallel nature of the parameter sweep, it was possible to break up the number of simulations into jobs in a variety of different ways. Ultimately, a balance was chosen such that an average job would last approximately 5 h and, therefore, resubmitting a failed job would not incur an undue strain on the allotment. Additionally, this results in a manageable number of writes to disk, with a tolerable overhead cost for each job.
With a cloud-based environment different node counts can be running, and generally speaking there is a lot of flexibility. This is well suited for an embarrassingly parallel framework. We further optimized the design by having each simulation save the time-series data for pressures and tidal volumes to the local disk. This was facilitated by a hierarchy that identified where in parameter space that simulation occurred, for ease of postprocessing and collating the results. In order to not overwhelm the storage capacity on-node, prior to completion the job would postprocess the results by: 1) determining the tidal volume, maximum pressures, and minimum pressures at the steady state, 2) delete the time-series data, and 3) create a reduced precision csv file with a row for each simulation that was included in the job. This configuration was designed to allow for easy debugging and resilience against network outages. Namely, one could look at the files on-node to determine which specific simulation failed by finding the last successful output in a parameter sweep. In total 146 000 intermediate csv files were created, for which subsequent scripts were deployed to concatenate them along the directory structure until the final data table was ultimately produced.
Lesson Learned 2: Relying on an embarrassingly parallel framework allowed us to match to a dynamic cloud-based environment, to best facilitate the required large-scale parameter sweep.
DEPLOYMENT: DYNAMIC CLOUD COMPUTING
With the initial modeling was complete, the next step was to develop a method for deploying the model at scale. In this section, we will discuss the unique opportunities provided by deploying on a cloud platform, unexpected challenges with maintaining an embarrassingly parallel code at scale, solutions we implemented, and how the hundreds of millions of simulations were synthesized into an intuitive and portable interface for clinicians.
Problem-Oriented Architecture Design
To obtain access to high-performance computing resources on a scale similar to what was required for this project, it is common practice to submit an architecture-targeted proposal requesting compute hours on a specific resource. While drafting such a proposal, care is taken to demonstrate feasibility, run-time, and parallel performance on the targeted system, since efficiency is paramount on these precious resources. For our project, however, the emphasis was the need for haste in solving the problem, which led to us seeking methods that were almost agnostic to the underlying hardware. The COVID-19 HPC Consortium (https://covid-19-hpc-consortium.org/) provided a unique opportunity for describing the needs and constraints posed by the problem, which could then be paired with an appropriate and available resource allocation. In this case, we had a working, validated code that could easily run on a variety of different platforms. The embarrassingly parallel nature of the setup meant that network connectivity was less of a consideration, and that our primary need was a large core count with sufficient memory (2–4 GB of memory-per-core) so that each core could independently complete a simulation without being bottlenecked by shared memory. Most importantly, we needed access to a high-throughput resource, where jobs could be submitted and executed quickly. Although time-to-solution was important, the constraints allowed some flexibility. For example, individual job run-time was not a concern, nor was the order of completion; if some jobs took longer to run than others, that was acceptable. The overriding need was to turnaround completion of all of the jobs within a few days so that the data were available for FDA review of the device and associated clinical support software. Rather than requesting a set number of core hours corresponding to jobs on set architecture and node sizes, the Consortium afforded us the ability to describe the problem, needs, and constraints. This flexible, problem-oriented design process facilitated a more efficient and effective matching of resources to problem that played a critical role in our ability to successfully accomplish our stated research goals.
The shift to a problem-driven approach was further enhanced through being paired with the Microsoft team for computing resource coordination and administration. The Consortium provided resources through the Microsoft cloud computing infrastructure, Azure. The use of a cloud-based architecture was a strong fit for our needs based on the ability to configure the infrastructural aggregate to suit the problem at hand. Thus, as we were leveraging an embarrassingly parallel setup, there was no necessity for the node count to remain fixed over the course of the simulations. Throughput, resilience, and resource availability could be balanced in a dynamic way as node counts assigned to the jobs could fluctuate throughout the execution duration. In the initial meetings between the Duke and Microsoft teams, we were able to outline the problem and associated resource needs to allow a cloud instance to be configured and tuned specifically to meet our need. As mentioned, we opted for an implementation that left each parameter-based simulation contained in an independent manner with minimal data collection and analysis handled between small groups of tasks. Therefore, rather than a tightly coupled, MPI-based model, we were able to employ a minimal communication, embarrassingly parallel framework. This design choice resulted in the processor selection being of far more consequence than choice of interconnect, so we searched for an Azure cloud configuration, which would allow for the largest core count at the lowest cost per core. The goal of maximizing throughput and minimizing wall-time could only be accomplished by taking advantage of as many nodes as was economically feasible. Shared storage requirements were relatively minimal, since individual compute nodes did not need to reference considerable shared data and postprocessing was designed to be performed on the local node; requirements were limited to those necessary for job submission—Slurm configuration and the users home directory. In order to scale the NFS filesystem on the head node to support hundreds of nodes connecting back to it, a managed premium disk was attached to the head node to use as the NFS export. With these considerations, the team was able to define a rough architecture using the Azure HB-series VMs, a basic NFS filesystem, and a Slurm front-end to manage job scheduling, all orchestrated by Azure CycleCloud. Slurm was chosen as the cloud scheduler to allow a seamless transition from our local cluster, which also used Slurm, to the cloud, yet another benefit of being able to fully customize the cloud architecture to meet the needs of the project. As a result, we were able to rapidly deploy our model with minimal time spent altering the code.
With an architecture defined, the next step was calculating the number of cores needed to meet the timeline. Based on this architecture and the desired timeframe, 24000 cores were estimated to be needed to complete the project in 2 days (24 000 cores × 48 h gave a total of 1152000 core hours). Alternatively, if we attempted to complete this task with only local resources (for example, by having unrestricted access to 1000 cores), the total time to solution (over a month) would have delayed our ability to combat the initial surge in COVID cases. Due to the computations not requiring communication between compute nodes and the capabilities of Azure, it would have been possible to secure the necessary number of cores by combining allocations from several geographically distinct locations—with some jobs running in Europe, for example, while others might run in South America. In order to simplify the debugging of failed jobs and minimize administrative overhead, however, all nodes were allocated within the same datacenter in Western Europe; thus, 24000 cores (using the Azure individual node type “HB60s”) were made available by Saturday.
Lesson Learned 3: By configuring the cloud architecture to match the needs of the problem (a problem-oriented approach) instead of manipulating the problem to fit the constraints of the platform (an architecture-oriented approach), we were able to be prepared to rapidly deploy our model.
Lesson Learned 4: By mimicking the feasibility testing environment, and thereby minimizing the need to rework scripts, we were able to rapidly implement our model at scale.
EXECUTION: COMPLETING 800 000 COMPUTE HOURS OF SIMULATION IN 72 H
In order to successfully complete the computing challenge, it was important to anticipate issues before they arose and to design the system architecture accordingly. However, some issues with underlying dependencies and hidden performance bottlenecks only arose when the model was deployed at the full scale. Solving these issues rapidly was paramount to being able to complete the computational task without depleting the allotment of compute hours.
Identify Underlying Dependencies
While initial performance testing on the cloud behaved as expected, significant slowdowns were observed while attempting to spin up all of the nodes and deploy the model. This eventually reached the point where job submission became infeasible and performance levels were significantly below expectations (both in terms of concurrently running jobs and completion rates).
To investigate this, we identified all communication channels, storage locations used, and potential hidden bottlenecks in the architecture and configuration of the supporting software. In this manner, a number of issues were addressed, including network addressing and Slurm scheduler tuning. The most significant issue identified was that file I/O from the head node slowed and eventually stopped. The cause was identified as the shared NFS filesystem being written to by all the running jobs, but only minimally; the I/O load was not commensurate with the slowdown being experienced. The underlying reason was a MATLAB-specific setting associated with the simulation execution, related to the location of the preferences directory. While unrelated to the simulation itself, this emphasizes the importance of running full-scale tests to identify hidden infrastructure interactions.
Lessons Learned 5: Beyond initial performance testing, full-scale tests deploying the model helped to identify hidden dependencies causing severe performance degradation.
Look for Hidden Performance Bottlenecks
Hidden performance bottlenecks at scale can create significant increases in necessary compute hours. For example, MATLAB is based on run-time compilation, and the time to compile the code was several factors larger than the time necessary to simulate the model for a given set of parameters. As a result, a primary consideration to maintain efficiency was to minimize time spent compiling. Fortunately, there is built-in functionality in MATLAB to perform a parameter sweep without recompiling the code for each iteration of a parameter sweep. This was only possible to implement, however, for sweeps over patient parameters; this was not easily achievable for sweeping through ventilator settings or changes in the circuit architecture. We took advantage of this by organizing the simulations into separate jobs for which only one compilation was required per job. This resulted in the 270 million simulations being handled by 146000 jobs, which drastically jminimized wall-time by reducing run-time recompilation.
Use of a folder directory architecture (see Figure 2) whereby files are stored across multiple directories, reduced slow-down associated with reading and writing to disk compared with single-directory storage. This was an important consideration given the significant I/O associated with the  files associated with the 146000 jobs. An additional advantage of storing both the submission scripts and output in this separated-directory structure was the triviality of generating auxiliary scripts for detecting failures in either the job submissions or model output generation; this allowed problematic jobs to be easily found and resubmitted.
files associated with the 146000 jobs. An additional advantage of storing both the submission scripts and output in this separated-directory structure was the triviality of generating auxiliary scripts for detecting failures in either the job submissions or model output generation; this allowed problematic jobs to be easily found and resubmitted.
To decrease data storage and transfer requirements, all postprocessing was performed on-node immediately after each simulation was completed, with intermediate results deleted. This converted the time-series output into three values (the steady-state tidal volume, maximum delivered pressure, and minimum delivered pressure), which were stored only at the clinically relevant precision. As a result, what would have required over 100 TB of storage space was decreased to approximately 10 GB.
Lesson Learned 6: Job organization with a run-time compilation code like MATLAB is important to minimize recompilation and, thus, maintain the embarrassingly parallel characteristic.
Lesson Learned 7: I/O, data storage, and transfers can be improved through specific usage of directory structures and on-node postprocessing.
TRANSLATION: ESTABLISHING AN INTUITIVE INTERFACE
With simulations completed, the remaining challenge was how to provide this data back to the clinician in a way that is simple, intuitive, up-to-date, and minimizes the chance of error. A mobile app, both for iOS and Android, that can run on low-end mobile phones as well as high-end tablets maximizes portability of the VSRS to global health scenarios as well as high-tech ICUs. Using a mobile app allows for a native, performant user interface (see Figure 3) and the ability to save and retrieve deidentified input value sets locally on the device. As the final data table, which stores all of the precomputed results, is larger than 10 GB, it is unlikely to be easily stored on low-end mobile phones. Consequently, it was decided to have the mobile app connect to a cloud-based API to receive the input values and return the corresponding results from the indexed database. While an Internet connection is necessary to retrieve new results, the installed app approach also leaves the door open to potentially precaching the data on high-capacity mobile devices should a no-Internet version be necessary in the future. An advantage of the cloud-based API is that it allows for clinicians to have the most up-to-date and accurate results at their fingertips. While the VSRS exclusively makes use of a mobile app, a remote webform is a reasonable alternative that could deliver similar functionality as the mobile app.
Figure 3.
Illustration of the mobile app's input (left) and output (right) displays. Note that the inputs are from a drop-down menu and are not free text fields to reduce the possibility of clinician error. The displayed output is for the case of the user querying the results for a 3 mm resistor.
As the data were precalculated, the end-user would need to query precalculated results based on a diverse but fixed number of input possibilities. Taking advantage of this and with an intent to reduce possible user-error, we followed a selection-based UI paradigm (akin to tabs and drop-down menus). This ensured that clinicians would only be able to input values that were consistent with the precomputed results and that no errors, such as confusion with unit conventions, would occur.
Lessons Learned 8: A cross-platform mobile app with a selection-based UI maximizes usability in various hospital settings, while minimizing user error.
CONCLUSION
Multiple lessons were learned in the process of rapid development and deployment of a parallel numerical model to support the clinical use of the VSRS in the event of ventilator shortages. Unlike conventional research projects that are designed and executed over months or years, a unique set of challenges arise for projects requiring rapid and agile development and deployment. The balance of developer time versus compute time under severe time-to-solution constraints leads to substantively different design choices. This article is an attempt to organize and articulate the valuable lessons learned in the process of generating the data necessary to guide resistor choice when using the VSRS to split ventilators between two or more patients.
For example, utilizing physical intuition gleaned from preliminary modeling can assist in the development of an embarrassingly parallel numerical model, which is advantageous for performing large-scale parameter sweeps in HPC environments. A cloud architecture as the HPC environment allows for platform customization to match the needs of the problem, instead of having to coerce the problem into functioning on the platform, which leads for rapid deployment. In order to avoid significant slowdowns when an embarrassingly parallel code is deployed at a massive scale, it is important to analyze the code and communication channels for hidden dependencies and performance bottlenecks. Last, for the results of the hundreds of millions of simulations to be utilized to help combat COVID-19, it is essential to create an intuitive user interface, with effort placed to minimize the potential user error. These lessons learned from rapidly deploying almost one million compute hours in a cloud-based infrastructure for a COVID-19 target are applicable to other situations where researchers have maximal motivation to minimize time-to-solution.
APPENDIX
Airflow Numerical Model
Air flow was simulated as a lumped parameter model using MathWork's Simscape pipe flow dynamics packages, which solve the laws of mass, momentum, and energy to determine the pressure, velocity, density, and temperature of gas as it travels through a network of pipes. The ventilator was modeled as either a volume or pressure source with a specified waveform representing the user-defined ventilator settings (PIP, PEEP, I:E, RR). Gas flow is then simulated for the travel though standard ventilator tubing, where it then interacts with the splitter, resistor of set diameter, endotracheal tube of a given size, and the patient's lungs. The lungs were modeled as a Hookean spring, representing the inverse of the compliance of the lungs, and a viscous dashpot, representing the resistance of the lungs, in parallel.8 The range of input parameters to the model were chosen based on simulating the wide range of ventilator settings, endotracheal tube sizes, and pulmonary characteristics that clinicians could encounter when treating patients with respiratory failure. Models were simulated until the gas flow to the lungs reached the steady state. Sensitivity tests were conducted to determine the granularity of the parameter sweep necessary to ensure that a change in tidal volume of less than 5% occurred for a given step size.
The model was validated against benchtop data. Using an anesthesia care station ventilator, standard tubing, the 3-D printed splitter and resistor system, standard endotracheal tubing, and artificial test lungs, experiments were performed to determine the delivered pressures and tidal volumes to the artificial test lungs for different ventilator settings. When these same settings were used as inputs to the numerical model, the predicted tidal volumes were found to be in excellent agreement with those from the benchtop experiments.
ACKNOWLEDGMENTS
The authors would like to thank many contributors who provided assistance, often at odd hours, to help complete this project with a fast turnaround. In particular, they would like to thank the COVID-19 HPC Consortium for both the compute hours and fast turnaround time and the broader Microsoft team for all of the support. They would like to thank T. Milledge for connecting us with the larger Research Computing team at OIT and assisting with MATLAB licensing issues, T. Lonergan for acting as the licensing liaison between Mathworks and Azure, and J. Hopkins and S. Rich for technical assistance with CycleCloud authorization tokens. The team would also like to thank the assistance of Microsoft and the Microsoft team. They provided assistance, insight, and funding all on short notice and with full commitment to the success of the project.
Biographies
Michael Kaplan is currently a medical student with Duke University, Durham, NC, USA, conducting research in the Biomedical Engineering Lab of Prof. A. Randles. He plans to start residency in Anesthesiology in 2021 and to continue to conduct research in basic and translational sciences, in fields ranging from computational hemodynamic modeling, machine learning, surgical optimization, vascular growth, and imaging of the microvasculature. He received the Ph.D. degree in geophysics from the University of Southern California, Los Angeles, CA, USA. Contact him at mike.kaplan@duke.edu.
Charles Kneifel is currently the Senior Technical Director with Duke OIT, Durham, NC, USA. He manages Dukes central technology infrastructure and Software Defined Networking Project. He has coordinated several technology grants with Duke including the National Science Foundations Data Infrastructure Building Blocks (DIBBS) grant to build campus cyberinfrastructures. Prior to Duke, he was a Chief Information Officer with the American Kennel Club for nine years. He has also held multiple technical positions at the NC State University. He received the Ph.D. degree in chemistry from the State University of New York at Stony Brook, Stony Brook, NY, USA. Contact him at charley.kneifel@duke.edu.
Victor Orlikowski is currently a Research Software Developer and Systems Administrator with Duke Research Computing, Stony Brook, NY, USA. He is a Senior IT Analyst focused on automation. He is experienced in networked storage devices and virtual machines. He has been a key member of research teams that have pioneered the tools regularly used by researchers at Duke. He has been with Duke for more than eight years in the Departments of Computer Science, Pratt School of Engineering and OIT. He regularly teaches Dukes Introduction to Linux seminars. Contact him at vjo@duke.edu.
James Dorff is currently an IT Senior Manager with Duke University, Durham, NC, USA. He specializes in Unix System Administration and computational physics. Contact him at jdorff@duke.edu.
Mike Newton is currently the Senior IT Analyst with Duke Universitys Office of Information Technology (OIT), Durham, NC, USA. He is the Primary System Administrator for OITs High Performance Computing (HPC) cluster. Contact him at jmnewton@duke.edu.
Andy Howard is currently a Senior Program Manager on the Azure Compute team focused on HPC and Azure CycleCloud. After spending almost a decade helping architect, build, and manage on-premises HPC systems for companies such as Purdue University, Cray, and the Department of Defense, he has spent the last seven years helping transition HPC workloads to the cloud. At Microsoft, he draws on this industry experience to help customers plan cloud HPC implementations and guide Microsoft's HPC product offerings. He received the B.S. degree in electrical and computer engineering technology from Purdue University, West Lafayette, IN, USA. Contact him at Howard.Andy@microsoft.com.
Don Shinn is currently the Founder and CEO of CrossComm—an award-winning mobile, web, and immersive app development studio with a 20+ year history of deploying innovative technologies to solve the toughest problems and challenges. Under Mr. Shin's leadership, the Durham, NC-based company has been recognized as one of the leading mobile and AR/VR app developers in the region by clutch.co, and has been nationally recognized as the Minority Technology Firm of the Year (2015) by the US Department of Commerce. He has been a passionate advocate for human-centric user interfaces throughout his career, and is currently interested in exploring the future of spatial computing and leveraging app innovation to advance social good. Contact him at don.shin@crosscomm.com.
Muath Bishawi is currently a Cardiothoracic Surgery resident with Duke University, Durham, NC, USA and a Ph.D. Research Scientist in biomedical engineering also at Duke. His translational and biomedical engineering research focuses on studying cardiovascular function and end-stage heart failure. He has experience in 3-D printing, novel catheter design, tissue engineered blood vessels, and ex-vivo modification of donor hearts to improve cardiac transplantation outcomes. His clinical research is focused on clinical outcomes after adult cardiac surgery with a focus on end-stage surgical heart failure and transplantation. He is a serial inventor and entrepreneur. Contact him at muath.bishawi@duke.edu.
Simbarashe Chidyagwai is currently working on the Ph.D. degree in biomedical engineering with Duke University, Durham, NC, USA. His research involves computational fluid dynamics modeling of blood flow in congenital heart diseases. He received the B.S.E. degree in mechanical engineering from Michigan State University, East Lansing, MI, USA. Contact him at simbarashe.chidyagwai@duke.edu.
Peter Balogh received the Ph.D. degree in mechanical engineering from Rutgers University in 2018. He developed a method for modeling blood cells flowing through complex capillary networks in 3D, for which he was awarded the Acrivos Dissertation Award in Fluid Dynamics from the American Physical Society. He is currently a postdoctoral associate in the Department of Biomedical Engineering at Duke University. His research interests include computational fluid dynamics modeling of biological flows, numerical methods for complex fluid-structure interfaces, and code development for high performance computing. His current research is focused on modeling cancer cells and their transport through the circulatory system, and on investigating the remodeling of microvasculatures through comparisons with experiments. Contact him at peter.balogh@duke.edu.
Amanda Randles is currently the Alfred Winborne Mordecai and Victoria Stover Mordecai Assistant Professor of biomedical engineering with Duke University, Durham, NC, USA. She was a Lawrence Fellow with the Lawrence Livermore National Laboratory from 2013 to 2015. Prior to graduate school, she worked with IBM on the Blue Gene supercomputer. She has been the recipient of the ACM Grace Murray Hopper Award, the LLNL Lawrence Fellowship, the NIH Early Independence Award, and named as a 2017 MIT TR35 Visionary. She is a Senior Member of the National Academy of Inventors. Her research in biomedical simulation and high-performance computing focuses on the development of new computational tools that she uses to provide insight into the localization and development of human disease. She has contributed more than 40 peer-reviewed papers, more than 120 granted US patents, and has more than 100 pending patent applications. She received the bachelor's degree in physics and computer science from Duke University, and the master's degree in computer science and the Ph.D. degree in applied physics and from Harvard University, Cambridge, MA, USA. Contact her at amanda.randles@duke.edu.
REFERENCES
- 1.Srinivasan S., et al. , “A rapidly deployable individualized system for augmenting ventilator capacity,” Sci. Trans. Med., vol. 9401, no. May, 2020, Art. no. eabb9401, doi: 10.1126/scitranslmed.abb9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenthal B. and Feuer A., “Coronavirus in N.Y.: Astronomical surge leads to quarantine warning,” pp. 3–7, 2020. [Online]. Available: https://www.nytimes.com/2020/03/24/nyregion/coronavirus-new-york-apex-a ndrew-cuomo.html
- 3.Beitler J. R., et al. , “Ventilator sharing during an acute shortage caused by the COVID-19 pandemic,” Amer. J. Respiratory Crit. Care Med., vol. 15, pp. 600–604, 2020. [Online]. Available: http://www.ncbi.nlm.nih.gov/pubmed/32515988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai B. K., Erian J. L., Pew S. H., and Eckmann M. S., “Emergency open-source three-dimensional printable ventilator circuit splitter and flow regulator during the COVID-19 pandemic,” Anesthesiology, vol. 133, pp. 246–248, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke A., Stephens A., Liao S., Byrne T., and Gregory S., “Coping with COVID-19: ventilator splitting with differential driving pressures using standard hospital equipment,” Anaesthesia, vol. 75, pp. 872–880, Apr. 2020, doi: 10.1111/anae.15078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherry A. D., Cappiello J., Bishawi M., Hollidge M. G., and MacLeod D. B., “Shared ventilation: Toward safer ventilator splitting in resource emergencies,” Anesthesiology, vol. 133, pp. 681–683, Sept. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.“Joint Statement on Multiple Patients Per Ventilator,” pp. 12–14, 2020. [Online]. Available: https://www.asahq.org/about-asa/newsroom/news-releases/2020/03/joint-st atement-on-multiple-patients-per-ventilator
- 8.Schmidt M., et al. , Computer Simulation Measured Respiratory Impedance Newborn Infants Effect Measurement Equipment, vol. 20. Amsterdam, The Netherlands: Elsevier, 1998, no. 3. [DOI] [PubMed] [Google Scholar]





