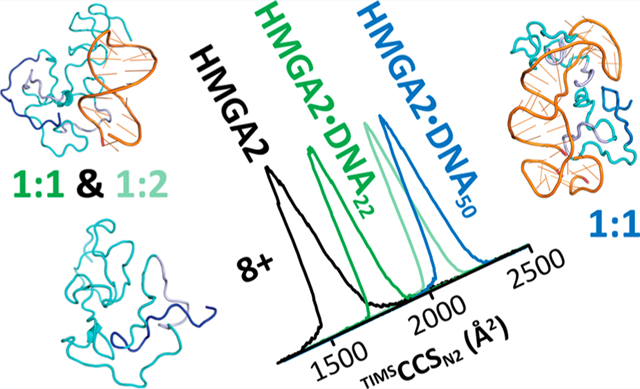
Abstract
The mammalian high mobility group protein AT-hook 2 (HMGA2) is an intrinsically disordered DNA-binding protein expressed during embryogenesis. In the present work, the conformational and binding dynamics of HMGA2 and HMGA2 in complex with a 22-nt (DNA22) and a 50-nt (DNA50) AT-rich DNA hairpin were investigated using trapped ion mobility spectrometry–mass spectrometry (TIMS–MS) under native starting solvent conditions (e.g., 100 mM aqueous NH4Ac) and collision-induced unfolding/dissociation (CIU/CID) as well as solution fluorescence anisotropy to assess the role of the DNA ligand when binding to the HMGA2 protein. CIU-TIMS–CID-MS/MS experiments showed a significant reduction of the conformational space and charge-state distribution accompanied by an energy stability increase of the native HMGA2 upon DNA binding. Fluorescence anisotropy experiments and CIU-TIMS–CID-MS/MS demonstrated for the first time that HMGA2 binds with high affinity to the minor groove of AT-rich DNA oligomers and with lower affinity to the major groove of AT-rich DNA oligomers (minor groove occupied by a minor groove binder Hoechst 33258). The HMGA2·DNA22 complex (18.2 kDa) 1:1 and 1:2 stoichiometry suggests that two of the AT-hook sites are accessible for DNA binding, while the other AT-hook site is probably coordinated by the C-terminal motif peptide (CTMP). The HMGA2 transition from disordered to ordered upon DNA binding is driven by the interaction of the three basic AT-hook residues with the minor and/or major grooves of AT-rich DNA oligomers.
Graphical Abstract
1. INTRODUCTION
The mammalian high mobility group protein AT-hook 2 (HMGA2) is an intrinsically disordered protein of 11.6 kDa that belongs to the nonhistone chromosomal HMGA protein family predominantly expressed during embryogenesis.1,2 This protein plays important roles in development as disruption of the HMGA2 expression pattern results in the deregulation of the cell growth,2,3 resulting in deficiency in human height4,5 and human intelligence.6 In addition, in vivo studies in mouse models have contributed to establishing the physiological role of HMGA2 by monitoring the effects of expression levels of the protein.7 These studies reported that aberrant expression of HMGA2 is also connected to tumorigenesis8–11 and adipogenesis.12,13 Such involvement in substantial physiological processes has implicated HMGA2 as potential therapeutic drug target for the treatment of cancer14 and obesity.15 The accumulating findings resulting in multiple functional roles demonstrate the broad importance of HMGA2 and make it one of the most investigated proteins to date.
HMGA2 is composed of a negatively charged C-terminal motif peptide (CTMP, highlighted in blue in Figure 1a) and three DNA binding motifs, each consisting of a positively charged “AT-hook” (highlighted in red in Figure 1a) peptide, which display specificity for binding to the minor groove of AT-rich DNA sequences.16 Each AT-hook is comprised of a conserved central Arg-Gly-Arg-Pro core sequence (underlined residues in Figure 1a) that deeply penetrates into the minor groove of DNA and forms a well-defined AT-hook:DNA complex.17 In addition, a recent study reported that synthesized AT-hooks also tightly bind to the major groove of DNA when the minor groove is occupied by a minor groove binder Hoechst 33258 (HOE).18 The particularity of HMGA2 to be able to transition from unstructured19 to a defined conformation, when bound to DNA,20 allows the protein to actively participate in diverse nuclear activities, including DNA replication, DNA repair, chromatin remodeling, and gene transcription and regulation.20–22 One main roadblock in the structural elucidation of these conformational changes is the lack of three-dimensional structures owing to the biological heterogeneity of conformations for HMGA2 and its DNA-bound forms. It is this conformational heterogeneity which impedes structural characterization using traditional methods such as nuclear magnetic resonance (NMR) and X-ray crystallography. These techniques have been only successful in the description of the intermolecular interactions that support the binding of the AT-hook motifs with DNA.23,24 Although the biological activities of HMGA2 have been demonstrated via in vivo and in vitro studies, structural information and appropriate models remain incomplete. HMGA2 and its DNA-bound forms do not necessarily have a unique ground state (folded state or native structure) representing a single free-energy minimum but rather a distribution of ground states with the same energy (conformational space). Their structural characterization requires the use of analytical tools capable of sampling each conformational state (or microstate) which is ultimately related to the protein functions. Native mass spectrometry (MS), particularly in combination with ion mobility spectrometry (IMS), has shown the potential for structural characterization as well as description of kinetic intermediates of biomolecules.25–28 IMS affords an additional dimension of separation of gas-phase conformation of ions and offers complementary insight relative to more conventional biophysical tools. Coupled with collision-induced unfolding (CIU), IMS can be used to determine the relative stability of gas-phase protein structures.29 The implementation of the trapped IMS (TIMS)30–32 technology has gained significant attention for the investigation of proteins due to the high trapping efficiency, higher ion mobility resolution (R up to ~425), and direct ion-neutral collision cross section measurements.32–34 In particular, the recent introduction of the convex electrode TIMS quadrupolar cell geometry enhanced the mobility range (K0 = 0.185–1.84 cm2·V−1·s−1), thus enabling the characterization of native macromolecular complexes (experimental data shown up to 1MDa).34 Moreover, TIMS has recently been successfully employed for the structural elucidation of peptide/protein DNA binding under native conditions.35,36
Figure 1.
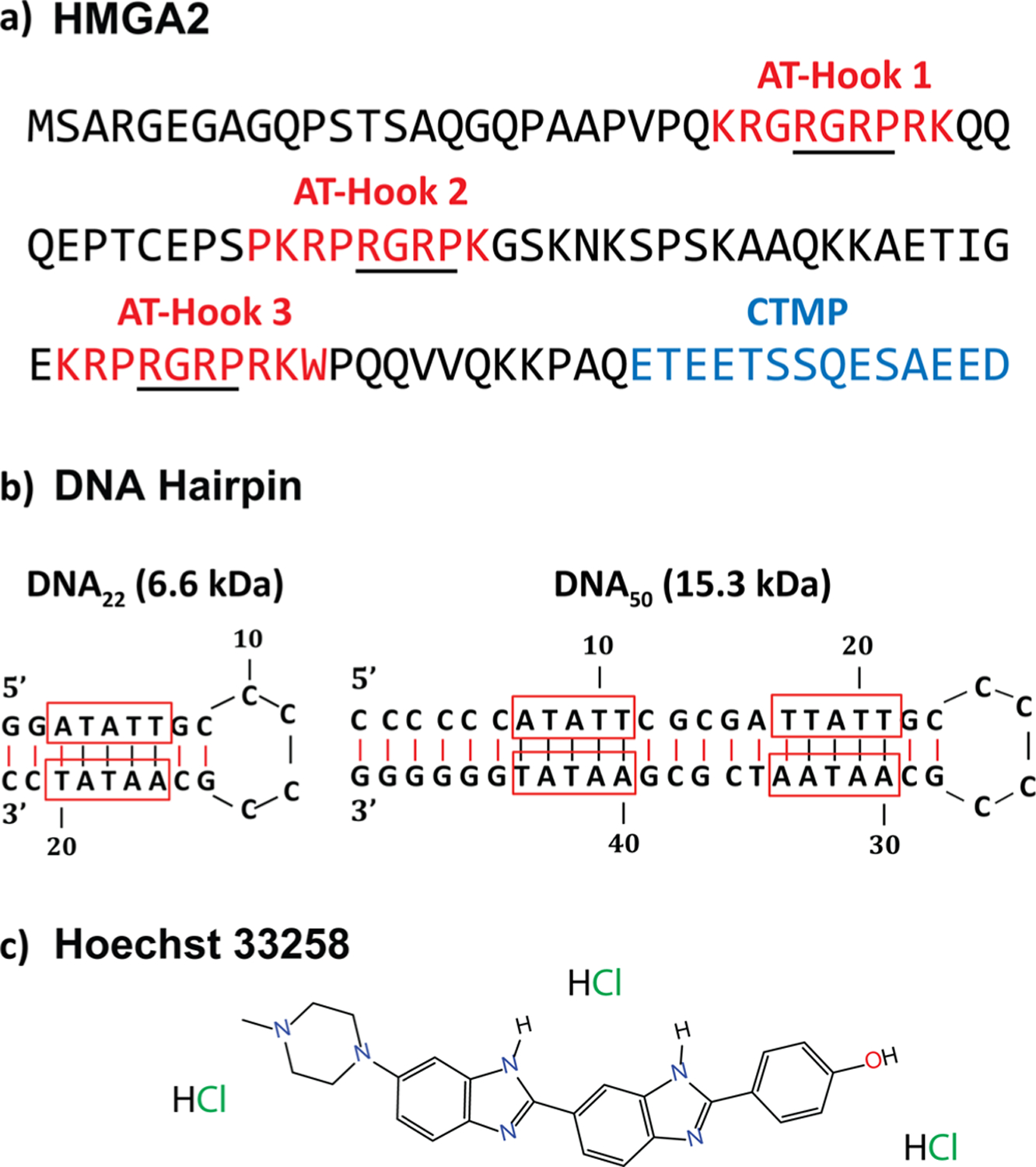
Sequences of (a) HMGA2 and (b) DNA hairpins, and (c) chemical structure of Hoechst 33258 minor groove binder. The three AT-hook DNA binding regions (red) and the negatively charged CTMP (blue) are highlighted. The underlined residues represent the conserved core of each AT-hook. The AT-rich domains of the DNA hairpins are outlined in red.
In the present work, the conformational and binding dynamics of free HMGA2 and in complex with a 22-nt (DNA22) or 50-nt (DNA50) AT-rich DNA hairpin (Figure 1b) were investigated using TIMS–MS under native starting solvent conditions (e.g., 100 mM aqueous NH4Ac) and with CIU/CID as well as solution fluorescence anisotropy to assess the role of the DNA ligand when binding to the HMGA2 protein. In the following discussion, a special emphasis is placed on the capability of TIMS to probe conformational changes upon DNA binding in native conditions and as a function of the collision energy to assess the stability for the free and the DNA-bound system. In addition, we report for the first-time experimental evidence of HMGA2 binding to the minor and/or major grooves of the AT-rich DNA oligomers (when the minor groove was occupied by HOE, Figure 1c).
2. EXPERIMENTAL SECTION
2.1. Materials and Reagents.
Details on HMGA2 expression and purification have been previously reported.37 A 22-nt (DNA22, 6.6 kDa) and a 50-nt (DNA50, 15.3 kDa) AT-rich DNA sequence (Figure 1b) were purchased from Eurofins Scientific (Luxembourg City, Luxembourg). These DNA hairpins contain a base pair stem and an AT-rich region in the middle of the stem. Hoechst 33258 was obtained from Sigma-Aldrich (St. Louis, MO). Ammonium acetate (NH4Ac) was purchased from Fisher Scientific (Pittsburgh, PA). Solutions of HMGA2 alone or in complex with DNA (1:1 ratio) and DNA/HOE (1:1:4 ratio) were analyzed at a concentration of 10 μM in 100 mM aqueous NH4Ac for native conditions. A low-concentration Tuning Mix standard (G1969–85000) was used to calibrate the TIMS instrument and obtained from Agilent Technologies (Santa Clara, CA).
2.2. HMGA2 Labeling and Fluorescence Anisotropy.
His-tagged mouse HMGA2 were labeled with the fluorescence reagent tetramethylrhodamine-5-maleimide (TMR) through the thiol group of the cysteine residue of HMGA2. Specifically, 100 μM of HMGA2 was incubated with 200 μM of TMR in the presence of 200 μM tris(2-carboxyethyl)phosphine hydro-chloride (TCEP) in 10 mM sodium phosphate buffer (pH 7.2) containing 20 mM NaCl for 2 h at room temperature to yield TMR-labeled HMGA2 (HMGA2-TMR). HMGA2-TMR was purified with a Ni-NTA column. An extinction coefficient of 95000 M−1 cm−1 at 541 nm in methanol was used to determine the HMGA2-TMR concentration. The purity of HMGA2-TMR was verified using size-exclusion chromatography (Superdex Peptide 10/300 GL) of AKTA FPLC.
Fluorescence anisotropy experiments were used to determine the dissociation constants (Kd) of HMGA2 binding to DNA50 oligomer in the absence or presence of HOE using a Varian Cary Eclipse fluorescence spectrophotometer with an excitation wavelength at 543 nm and an emission wavelength at 575 nm. Excitation and emission slits are 5 and 10 nm, respectively. For the HMGA2-TMR binding experiments, increasing concentrations of DNA50 or DNA50/HOE were titrated into 20 nM of HMGA2-TMR in a buffer solution containing 10 mM Tris–HCl (pH 8.0) and 200 mM NaCl at room temperature. The anisotropy values of HMGA2-TMR were determined and calculated by eq 138
| (1) |
where r is the calculated anisotropy, IVV is the observed polarized intensity corresponding to vertically polarized excitation and vertically polarized emission, IVH is the observed polarized intensity corresponding to vertically polarized excitation and horizontally polarized emission, and G is the ratio of the sensitivities of the detection system for vertically and horizontally polarized light. The binding data were fit using eq 239
| (2) |
where y is the binding ratio of A/Amax, for which A and Amax are the relative and maximum anisotropy, respectively, x is the titrate concentration, a is the HMGA2-TMR concentration, and Kd is the dissociation constant.
2.3. TIMS–MS Instrumentation.
Ion mobility experiments were performed on a custom built nESI–TIMS coupled to an Impact q-ToF mass spectrometer (Bruker Daltonics Inc., Billerica, MA, Figure S1).40 nESI emitters were pulled from quartz capillaries (O.D. = 1.0 mm and I.D. = 0.70 mm) using a Sutter Instruments Co. P2000 laser puller (Sutter Instruments, Novato, CA). Protein solutions were loaded in a pulled-tip capillary, housed in a mounted custom built XYZ stage in front of the MS inlet, and sprayed at 800–1100 V via a tungsten wire inserted inside the nESI emitters. Briefly, the ion mobility separation in a TIMS device is based on holding the ions stationary using an electric field (E) against a moving buffer gas (Figure S1).30 In the present design, the TIMS analyzer section is composed of convex electrode geometry, as described elsewhere.32,34 The TIMS unit is controlled by an in-house software in LabView (National Instruments) and synchronized with the MS platform controls.40 The general fundamentals of TIMS as well as the calibration procedure have been described in the literature.41–44
TIMS experiments were carried out using nitrogen (N2) as the buffer gas at ambient temperature (T). The gas velocity was kept constant between the funnel entrance (P1 = 2.6 mbar) and exit (P2 = 0.8 mbar, Figure S1) regardless of the protein solutions. An rf voltage of 250 Vpp at 450 kHz was applied to all electrodes. Ions were softly transferred and injected into the TIMS analyzer to avoid potential activation by keeping a low ΔV (ΔV = 50 V) between the deflector (Vdef) and the funnel entrance (Vfun) as well as between the funnel entrance and the TIMS analyzer to generate native-like ion mobility distributions (Figure S1). Here, a Vdef of −130 V with a Vfun of −180 V for a Vramp at −230 to −90 V and base voltage (Vout) of 60 V was used for HMGA2 and in complex with DNA and DNA/HOE oligomers. Ion-heating experiments (CIU) were performed prior to the TIMS by increasing the ΔV (ΔV = 50–250 V) between the deflector (Vdef) and the funnel entrance (Vfun). Collision-induced dissociation (CID) experiments were carried out by increasing the activation voltage at the entrance of the collision cell stepwise (6–40 V, with 2 V increment) without m/z selection (this setup is limited to m/z 3000 quadrupole isolation prior to CID events). All experiments were performed in triplicate and were reproducible across the replica measurements.
2.4. Structure Representation.
The HMGA2 protein as well as the DNA22 and DNA50 templates were built using Avogadro.45 Because HMGA2 contains three minor-groove binding AT-hook regions, docking was performed for each of these segments using HDOCK,46 which uses a hybrid algorithm of template-based modeling and ab initio for docking. For the HMGA2 docking to DNA22, each AT-hook region was first docked to the DNA minor-groove and the rest of the protein structure was connected to the docked segment. Also, while a specific AT-hook region was docked to DNA, the C-terminal tail of the HMGA2 was docked with either of the other two AT-hook regions at a time. In this way, six different HMGA2·DNA22 complexes were built. For docking to DNA50, two AT-hook segments were docked to two minor-groove sites at a time, while the remaining AT-hook region was docked to the C-terminal tail. This resulted in three different HMGA2 DNA50 complexes. Note that the generated structures are only for representation purposes and are not proposed candidate structures based on experimental collision cross-sections.
3. RESULTS AND DISCUSSION
3.1. Characterization of HMGA2 using nESI–(CIU)-TIMS-MS.
The native mass spectrometry analysis of the HMGA2 (11.6 kDa) resulted in a broad charge-state distribution, ranging from [M + 6H]6+ to [M + 12H]12+under native-like solution conditions (highlighted in black in Figure 2a). This broad charge state distribution was rationalized as a structural change in the folded protein (6+ to 8+) toward more extended conformations (9+ to 12+), which is typical of an intrinsically disordered protein. This assignment was further supported by the ion mobility distributions for which relatively narrow single ion mobility bands (~1500 Å2) were characteristic of the [M + 6H]6+ to [M + 8H]8+ ions, whereas significantly broader ion mobility bands at larger CCS values were observed for higher charge states (9+ to 12+, black traces in Figure 2b). The wider mobility bands suggest a higher structural heterogeneity for the 9+ to 12+ charge states.
Figure 2.
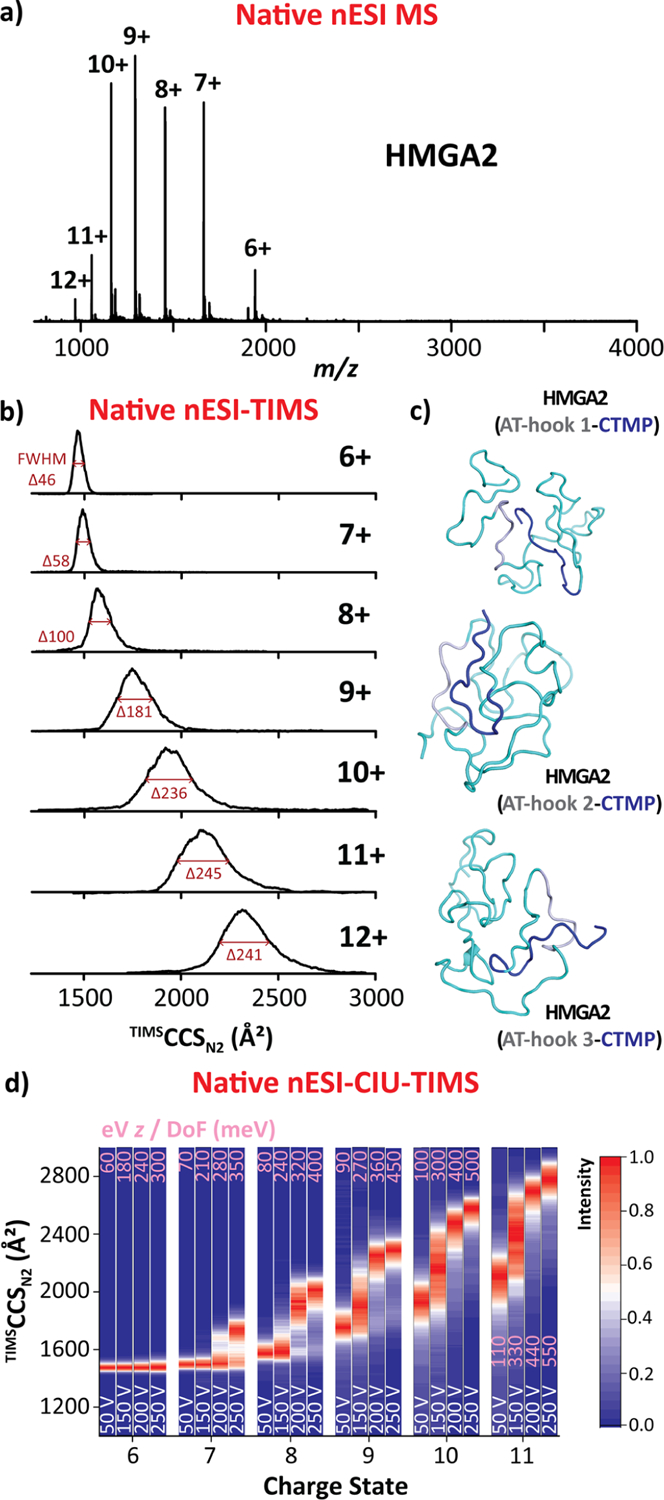
HMGA2 (11.6 kDa) native nESI generated mass spectrum (a), native ion mobility distributions (b), representative intramolecular interactions, for which AT-Hook segments and CTMP are colored in gray and blue, respectively (c), and CIU fingerprints (d). Voltage difference (ΔV) of the CIU is denoted in white and eV z/DoF (in meV) values in pink for each CIU profile. Note that individual TIMS spectra for each CIU conditions are available in Figure S2.
Recent experimental evidence showed that the negatively charged CTMP can effectively interact with each of the positively charged AT-hook sections (manuscript in preparation), thus increasing the structural heterogeneity. That is, at least multiple HMGA2 structural combinations should be considered based on the intramolecular CTMP interactions: (i) AT-hook 1 interacting with CTMP, (ii) AT-hook 2 interacting with CTMP, and (iii) AT-hook 3 interacting with CTMP, as illustrated in Figure 2c.
The conformational changes and stability of the HMGA2 were also studied using collision-induced unfolding (CIU) prior to the TIMS–MS analysis. During CIU, ion heating from energizing collisions with the buffer gas allows the molecular ions to overcome conformational barriers and sample other local, free-energy minima not initially accessible by native nESI–TIMS–MS.29,47 The TIMS settings were modified by increasing the potential difference (ΔV = 50–250 V) between the deflector (Vdef) and the funnel entrance (Vfun) to induce collisional unfolding (Figures 2d and S2). In the case of the free HMGA2, no major changes aside from a slight broadening were observed in the ion mobility band arising from the [M + 6H]6+ ions (Figure 2d), consistent with a more structured protein in the 6+ charge state. For higher charge states (7+ to 11+), CIU resulted in several unfolding transitions evidenced by the observation of additional ion mobility bands at higher TIMSCCSN2 values in agreement with an intrinsically disordered protein (Figures 2d and S2).
3.2. Characterization of HMGA2·DNA22 Complex Using nESI–(CIU)-TIMS–CID-MS of Complex.
Native nESI-MS analysis of the HMGA2·DNA22 complex (18.2 kDa) in an equimolar ratio resulted in a 1:1 and 1:2 binding stoichiometry (green squares and triangles in Figure 3a). A charge-state distribution, ranging from [M + 6H]6+ to [M +10H]10+, was observed for HMGA2·DNA22 1:1 complex (green squares), while a narrower charge state distribution (8+ and 9+) was observed for the HMGA2·DNA22 1:2 complex (green triangles). The TIMS spectra for HMGA2·DNA22 1:1 complex displayed similar TIMSCCSN2 values (~1750 Å2) for the [M + 6H]6+ to [M + 8H]8+ ions (green square traces in Figure 3b). This feature combined with narrower ion mobility distributions as compared to the free HMGA2 suggest that the structural heterogeneity of HMGA2 is restricted upon DNA22 binding. A slight increase in the CCS values was observed for the [M + 9H]9+ and [M + 10H]10+ ions relative to the lower charge states, suggesting that these structures are more extended and that the presence of the shorter DNA hairpin is not sufficient to stabilize multiple AT-rich binding motifs (i.e., AT-hooks). As the charge state increases, no major change in the width of the mobility bands was observed; these results suggest that the conformational rearrangement of the more extended structures upon DNA binding into a more collapsed distribution when compared to the 9+ to 12+ charge states of the free HMGA2 (black square traces in Figure 3b). The slight unfolding observed for the 9+ and 10+ charge states of HMGA2·DNA22 1:1 complex is attributed to the flexible domains of the protein rather than any disturbance in the intermolecular interaction network. Note that similar features were observed for the HMGA2·DNA22 1:2 complex, for which similar TIMSCCSN2 values (~2075 Å2) were obtained for the [M + 8H]8+ and [M + 9H]9+ ions (green triangle traces in Figure 3b). The narrow mobility bands observed for the HMGA2·DNA22 1:2 complex also suggest a collapse of the structural heterogeneity when compared to the free HMGA2 (black square traces in Figure 3b).
Figure 3.
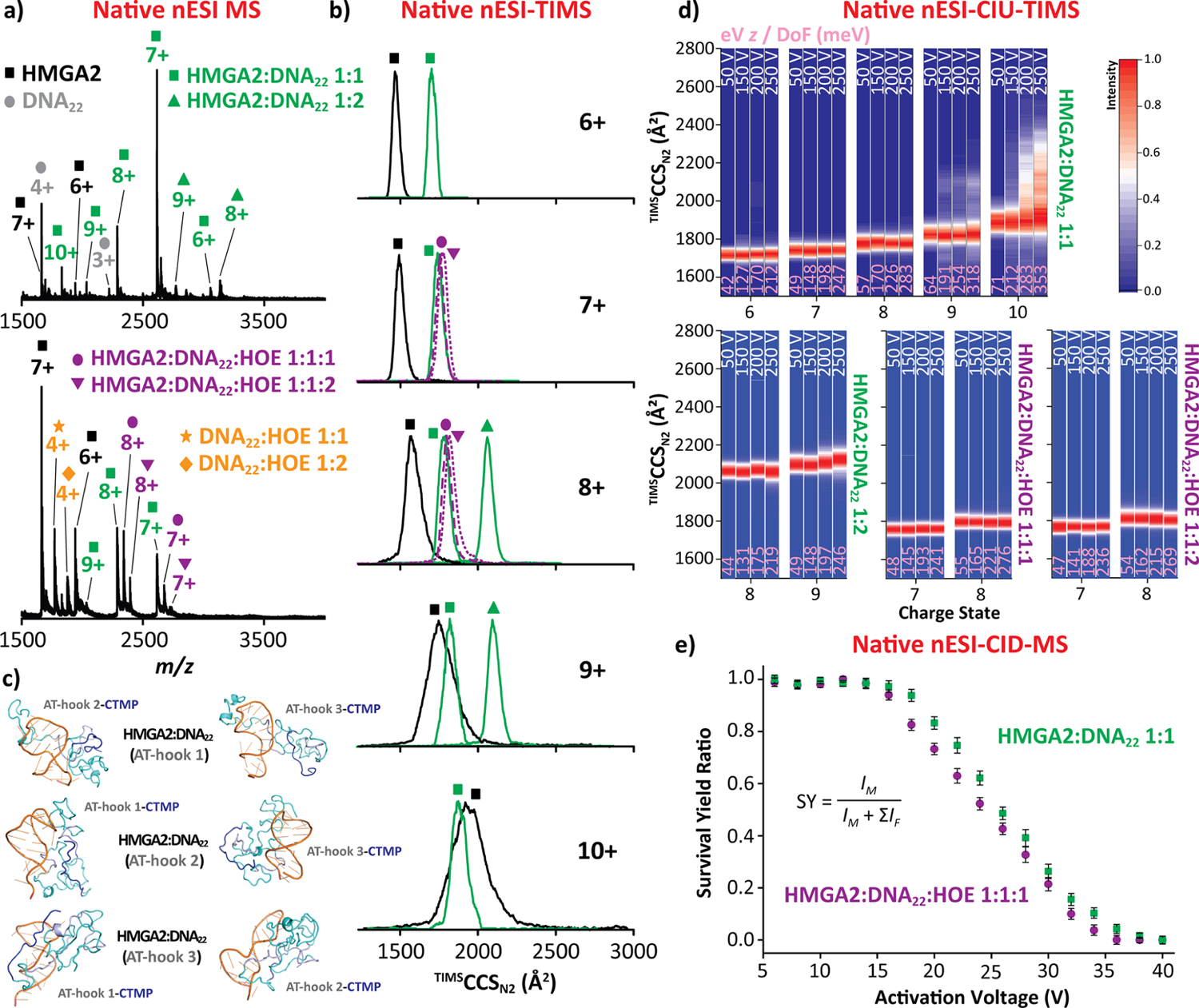
HMGA2·DNA22 (green) and HMGA2·DNA22·HOE (purple) native nESI-generated mass spectra (a), native ion mobility distributions (b), representative intra-and intermolecular interactions, for which DNA, AT-Hook segments, and CTMP are colored in orange, gray, and blue, respectively (c), CIU fingerprints (d), and collision-induced dissociations curves (e). Voltage difference (ΔV) of the CIU is denoted in white and the eV z/DoF (in meV) values are denoted in pink for each CIU profile. Note that individual TIMS spectra for each CIU conditions are shown in Figure S4. The CID data represent the mean value ± standard deviation (SD) of three independent experiments.
Interesting insight is derived by considering the conformational dynamics of HMGA2·DNA22 as compared to an analogous complex containing a truncated version of the protein and a 10-residue peptide containing a single AT hook (AT-Hook3).35 The conformational dynamics of AT-Hook3·DNA22 and HMGA2·DNA22 complexes exhibited significant differences. The AT-Hook3 peptide has only one RGRP core and fully interacts with the DNA22 substrate, restricting the complex conformational dynamics and thus yielding narrower IMS distribution. In the case of the HMGA2·DNA22 complex, the DNA22 substrate can interact with one out of the three RGRP cores from the AT-hook regions, as illustrated in Figure 3c, thus allowing the rest of the protein to adopt multiple conformations (broader IMS distribution). In addition, CTMP interacts with the positively charged AT-hook regions (manuscript in preparation).
For the HMGA2·DNA22 1:1 stoichiometry, multiple intra-and intermolecular interactions can be accessible: (i) AT-hook 1 and DNA22 while CTMP interacts with either AT-hook 2 or AT-hook 3, (ii) AT-hook 2 and DNA22 while CTMP interacts with either AT-hook 1 or AT-hook 3, and (iii) AT-hook 3 and DNA22 while CTMP interacts with either AT-hook 1 or AT-hook 2, as illustrated in Figure 3c. Moreover, the HMGA2·DNA22 1:2 stoichiometry was also observed using an equimolar starting solution ratio. In this case, two AT-hooks are interacting with the DNA22 while the third is likely interacting with the CTMP (three combinations). The hypothesis that one of the AT-hooks interacting with the CTMP (likely preformed structure/substrate) is also supported by the absence of the 1:3 stoichiometry in experiments with 1:3 excess DNA22 in solution (see the typical MS spectrum in Figure S3).
Individual AT-hooks exhibit specificity for binding to the minor groove of AT-rich DNA oligomers, but they can also interact with the DNA major groove when the minor groove is not available.18 Additional TIMS–MS experiments of incorporating a minor groove binder HOE48 to the DNA22 were performed to assess the potential of HMGA2 to bind to the major groove of the AT-rich DNA oligomers. HOE was added in excess (×4) to ensure the minor groove of DNA was fully occupied and restricted HMGA2 binding to the DNA major groove. Native nESI-MS analysis revealed that HMGA2 has the ability to bind to the DNA22·HOE complex, resulting in a 1:1:1 and 1:1:2 binding stoichiometry (purple circles and inverted triangles in Figure 3a), for which both HMGA2·DNA22·HOE complexes displayed a narrow charge state distribution (7+ and 8+). The TIMS spectra for both HMGA2·DNA22·HOE complexes showed TIMSCCSN2 values (~1750 Å2) and ion mobility widths comparable to those of HMGA2·DNA22 (purple circles and inverted triangle traces in Figure 3b). These results suggest that the conformational and binding dynamics are not disturbed when DNA22·HOE is added to the HMGA2 protein and will probably not alter the nuclear activities when bound to the major groove of the DNA oligomers.
The conformational changes and stability of the HMGA2·DNA22 complexes were studied using collision-induced unfolding (CIU) prior to the TIMS–MS analysis and collision-induced dissociation (Figures 3d,e and S4). Upon CIU of the HMGA2·DNA22 complexes, changes in the ion mobility distributions were only observed for the higher charge states (9+ and 10+) of HMGA2·DNA22 1:1 at high collisional energies (ΔV > 200 V), while no major changes in the ion mobility distributions were observed for all charge states (8+ and 9+) of the HMGA2·DNA22 1:2 complexes, supporting a transition from disordered to ordered upon DNA binding. Significant conformational changes were not observed for the HMGA2·DNA22·HOE 1:1:1 and 1:1:2 complexes. A direct comparison between the systems can be performed based on the energy per degrees of freedom (DoF, calculated as 3N – 6, where N is the number of atoms in each molecule) during the CIU process ((eV·z/DoF) values in Figure 3d, pink). The HMGA2·DNA22 1:1 complex started unfolding at ~190 meV (9+) while the DNA-free HMGA2 already presented unfolding events at ~90 meV (9+), suggesting that the DNA substrate stabilizes the HMGA2·DNA complex and restricts the HMGA2 unfolding. In addition, CIU profiles for the HMGA2·DNA22 1:2 complex did not reveal any transition as compared to HMGA2·DNA22 1:1, further supporting the hypothesis that the DNA substrate stabilizes the HMGA2 protein in the complex. Finally, complementary CID experiments were used to evaluate the HMGA2·DNA22 1:1 and HMGA2·DNA22·HOE 1:1:1 binding affinity (Figure 3e). Note that the CID experiments for HMGA2·DNA22 1:2 and HMGA2·DNA22·HOE 1:1:2 are not provided due to low signal intensity. As the collisional activation increases, a decrease in the molecular ion signal was observed, arising from the dissociation of the complex. Comparison between the CID profiles of the HMGA2·DNA22 1:1 and HMGA2·DNA22·HOE 1:1:1 showed that the HMGA2·DNA22·HOE complex dissociates faster as compared to the HMGA2·DNA22 complex. This suggests that the binding affinity becomes weaker when HMGA2 binds to the DNA major groove rather than the case when binding to the DNA minor groove.
3.3. Characterization of HMGA2·DNA50 complex using nESI-(CIU)−TIMS–CID-MS and solution fluorescence anisotropy.
Native nESI-MS analysis of HMGA2 in the presence of a longer DNA stem loop (DNA50) in an equimolar ratio resulted in the observation of the HMGA2·DNA50 complexes (26.9 kDa) with a 1:1 binding stoichiometry over a narrow charge-state distribution (7+ to 9+, blue squares in Figure 4a). The presence of a lower charge-state distribution as compared to the HMGA2·DNA22 complex probably derived from the longer DNA50 stem loop which has two AT-rich binding regions (Figure 1b) that may interact with multiple AT-hook motifs, thus accounting for the maintenance of more compact conformations. This suggests that upon DNA50 binding the structural heterogeneity of HMGA2 is significantly reduced, supporting a transition from disordered to ordered. The TIMS profiles showed similar TIMSCCSN2 values (~2175 Å2) for all three charge states (blue square traces in Figure 4b). Note that results similar to those observed for HMGA2·DNA22 were obtained when HMGA2·DNA50 was compared to the DNA-free HMGA2 protein. In addition, the mobility band widths of HMGA2·DNA50 were found to be like the one observed for the HMGA2·DNA22 1:1 complex (green square traces in Figure 4b), despite having a larger DNA. This feature suggests that the structural heterogeneity of HMGA2 is even more restricted upon DNA50 binding, which is also consistent with relatively close TIMSCCSN2 values and ion mobility width when comparing HMGA2·DNA22 1:2 (green triangle traces in Figure 4b) with HMGA2·DNA50. In the case of the HMGA2· DNA50 complex, the DNA50 substrate, containing two AT-rich binding regions, can interact with multiple AT-hook regions.These findings suggest multiple HMGA2·DNA50 binding scenarios: (i) AT-hook 1,2 and DNA50 while CTMP interacts with AT-hook 3, (ii) AT-hook 1,3 and DNA50 while CTMP interacts with AT-hook 2, and (iii) AT-hook 2,3 and DNA50 while CTMP interacts with AT-hook 1, as illustrated in Figure 4c.
Figure 4.
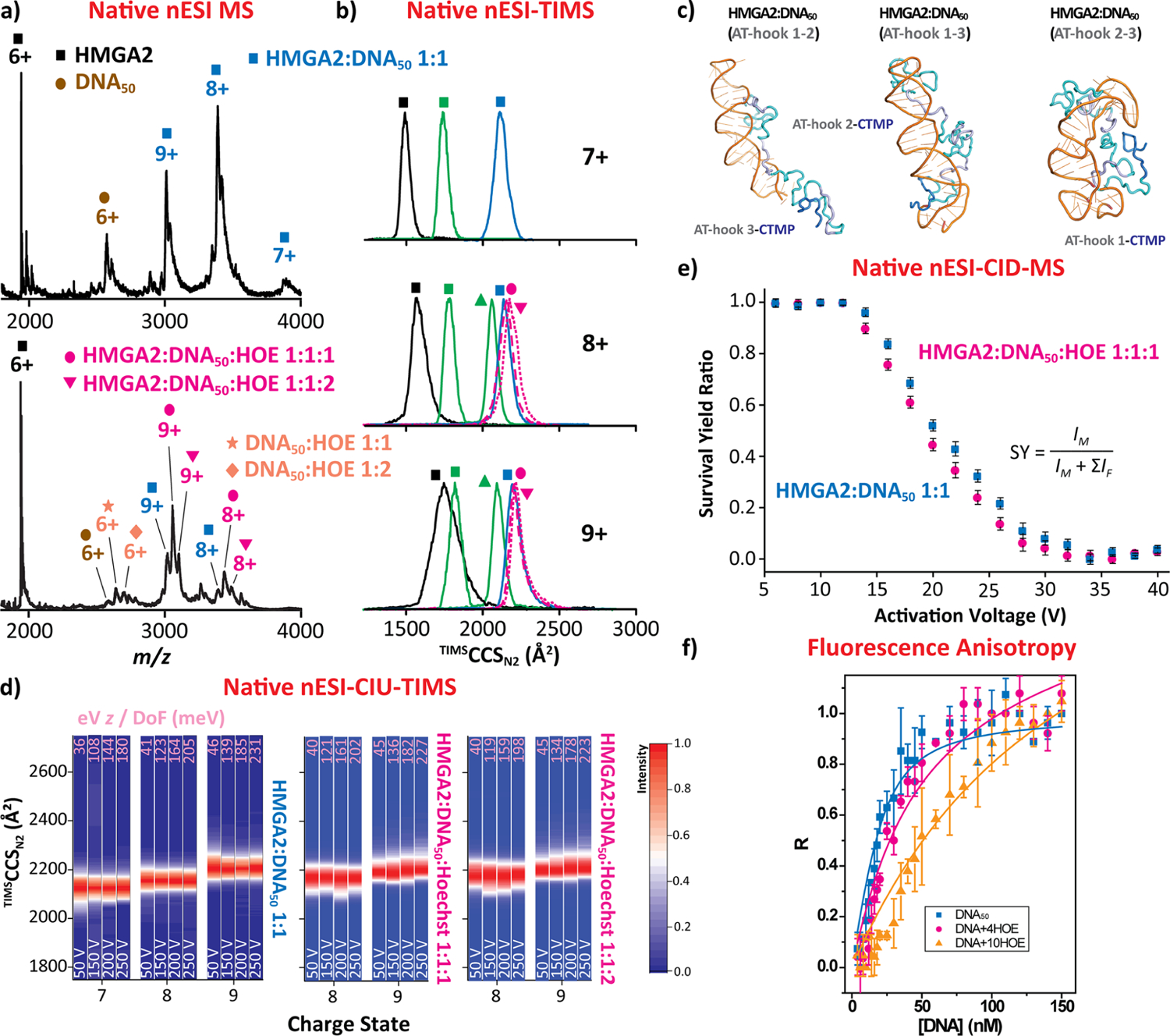
HMGA2·DNA50 (blue) and HMGA2·DNA50·HOE (magenta) native nESI generated mass spectra (a), native ion mobility distributions (b), representative intra- and intermolecular interactions, for which DNA, AT-Hook segments, and CTMP are colored in orange, gray, and blue, respectively (c), CIU fingerprints (d), collision-induced dissociations curves (e), and solution fluorescence anisotropy (f). Ion mobility distributions of HMGA2·DNA22 1:1 (green squares) and 1:2 (green triangles) are shown for direct comparison with the HMGA2·DNA50 1:1 complex. The voltage difference (ΔV) of the CIU is denoted in white, and the eV z/DoF (in meV) values ae denoted in pink for each CIU profile. Note that individual TIMS spectra for each CIU condition are shown in Figure S5. The CID and anisotropy data represent the mean value ± standard deviation (SD) of three independent experiments.
TIMS–MS experiments inserting the minor groove binder HOE to the DNA50 assessed the potential of HMGA2 to bind to AT-rich DNA major grooves (as previously, HOE was added in excess, ×4). Native nESI-MS analysis revealed that HMGA2 binds to the DNA50·HOE complex, resulting in a 1:1:1 and 1:1:2 binding stoichiometry (magenta circles and inverted triangles in Figure 4a), for which both HMGA2· DNA50·HOE complexes displayed a narrow charge-state distribution (8+ and 9+). The presence of HMGA2·DNA50·HOE 1:1:2 suggests that HMGA2 is able to bind to the two major grooves of the long stem loop DNA. The TIMS spectra for both HMGA2·DNA50·HOE complexes showed TIMSCCSN2 values (~2200 Å2) and ion mobility band widths comparable to those of HMGA2·DNA50 (magenta circles and inverted triangle traces in Figure 4b). As showcased for HMGA2· DNA22, these features suggest that the conformational and binding dynamics are not disturbed when DNA50·HOE is added to the HMGA2 protein and will probably not alter the nuclear activities when bound to the major groove of DNA oligomers.
The conformational changes and stability of the HMGA2·DNA50 complexes were investigated using CIU and CID (Figures 4d,e and S5). The CIU range accessible (up to 230 meV, pink values) in this platform did not reveal any unfolding transitions for all HMGA2·DNA50 complexes, which also supports the hypothesis that the larger DNA substrate further stabilizes the HMGA2 protein in the complex in agreement with the CIU results for the HMGA2·DNA22 1:2 complex. This increase in stability upon binding to DNA50 as compared to DNA22 was reflected through their CIU profiles. In fact, the HMGA2·DNA22 1:1 complex started unfolding at ~190 meV (9+), while HMGA2·DNA50 did not show any unfolding events (Figures 3d and 4d). Moreover, CID experiments were used to assess the HMGA2·DNA50 1:1 and HMGA2·DNA50·HOE 1:1:1 binding affinity (Figure 4e). Note that the CID experiments for HMGA2·DNA50·HOE 1:1:2 are not provided due to low signal intensity. Comparison between the two CID profiles showed that the HMGA2·DNA50·HOE complex dissociates at slightly lower collisional activation as compared to the HMGA2·DNA50 complex. This result suggests that the binding affinity becomes weaker when HMGA2 binds to the major groove of the large stem loop DNA in agreement with previous observation on HMGA2·DNA22.
Complementary fluorescence anisotropy solution experiments were performed on the HMGA2·DNA50 in the absence and presence of HOE (Figure 4f). DNA50 and DNA50·HOE solutions were titrated into 20 nM of TMR labeled HMGA2 in 10 mM Tris–HCl (pH 8.0) containing 200 mM NaCl. Note that the His-tag does not affect the binding of HMGA2 to AT-rich DNA sequences, as previously reported.49 Fitting of HMGA2 binding to DNA50 yielded a Kd of 7.3 ± 1.1 nM (blue squares in Figure 4f), which was consistent with our previously published results.39 However, results from the titration of DNA50·HOE at a molar ratio of 1:4 could not be fitted to a single binding site model. These results were then fitted to a model of two independent binding sites, yielding two dissociation constants: 23.6 ± 5.6 nM and 391 ± 143 nM (magenta circles in Figure 4f). This result suggests that two independent binding reactions occurred in this titration experiment: one for HMGA2 binding to DNA50 with a Kd of 23.6 ± 5.6 nM and another one for HMGA2 binding to the DNA50·HOE complex with a much higher Kd of 391 ± 143 nM. The high Kd of the HMGA2·DNA50·HOE complex stems from the fact that the minor groove of the DNA50 AT-rich regions was occupied by the minor groove binder HOE. As a result, HMGA2 binds to the major groove, resulting in a much higher Kd in agreement with the gas-phase experiments.18 If this hypothesis is correct, a further increase of HOE concentration in the binding reaction mixture should shift the binding curve to the right. At a point where the minor groove of the AT rich region of the DNA50 oligomer is fully occupied by HOE, HMGA2 can only bind to the major groove of the DNA50·HOE complex with a higher Kd. Indeed, increasing the molar ratio of HOE to 10 (DNA50·HOE 1:10) significantly shifts the binding curve to the right (orange triangles in Figure 4f). Fitting this binding data to a single binding site model yields a Kd of 137 ± 3 nM, indicating that HMGA2 binds to the major groove of DNA50.
4. CONCLUSIONS
Native ion mobility-mass spectrometry analysis of HMGA2 indicated a conformational heterogeneity dependence with the charge state and significantly more conformational broadening in the DNA-free from when compared to the HMGA2·DNA complexes. CIU experiments showed higher energetic stability of HMGA2 upon DNA binding, and the larger the DNA the greater the stability.
Gas-phase and solution experimental evidence demonstrated that HMGA2 binds to the minor and/or major (when the minor groove is occupied by HOE) grooves of the AT-rich DNA oligomers. CID profiles showed that the HMGA2·DNA·HOE complexes dissociate at lower collisional activation when compared to the HMGA2·DNA complexes, in good agreement with the higher dissociation constants (Kd) observed in solution in the presence of HOE.
Native (CIU)−TIMS–CID-MS prove to be a powerful and complementary biophysical tool capable of providing structural insights from disordered proteins and their complexes that are inaccessible using traditional methods. These discoveries of HMGA2 binding to the DNA major groove have great biological consequences since a variety of DNA-binding proteins carry the AT-hook core motifs. That is, HMGA2-like proteins can effectively bind to DNA minor and/or major grooves and still participate in nuclear activities, such as transcription, chromatin remodeling, and DNA repair.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the financial support from the National Science Foundation Division of Chemistry, under CAREER award CHE-1654274, with cofunding from the Division of Molecular and Cellular Biosciences to F.F.-L. Funding from the National Institutes of Health (R35GM139658) and the Robert A. Welch Foundation (F-1155) is acknowledged by J.S.B. This work is also supported by NIH grant 1R21AI125973 to F. L.
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/jasms.2c00101
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jasms.2c00101.
Figures that illustrate the TIMS–MS instrument showing the convex electrode TIMS geometry and TIMS operation, TIMS profiles as a function of the collision activation for the multiply protonated species of the free HMGA2 and HMGA2·DNA and HMGA2·DNA·HOE complexes, and nESI MS profile of the HMGA2·DNA22 complexes with 1:3 excess DNA22 in solution (PDF)
The authors declare no competing financial interest.
Contributor Information
Kevin Jeanne Dit Fouque, Department of Chemistry and Biochemistry, Florida International University, Miami, Florida 33199, United States.
Sarah N. Sipe, Department of Chemistry, University of Texas, Austin, Texas 78712, United States.
Alyssa Garabedian, Department of Chemistry and Biochemistry, Florida International University, Miami, Florida 33199, United States.
German Mejia, Department of Chemistry and Biochemistry, Florida International University, Miami, Florida 33199, United States.
Linjia Su, Department of Chemistry and Biochemistry, Florida International University, Miami, Florida 33199, United States.
Md Lokman Hossen, Department of Physics, Florida International University, Miami, Florida 33199, United States.
Prem P. Chapagain, Department of Physics and Biomolecular Sciences Institute, Florida International University, Miami, Florida 33199, United States.
Fenfei Leng, Department of Chemistry and Biochemistry and Biomolecular Sciences Institute, Florida International University, Miami, Florida 33199, United States.
Jennifer S. Brodbelt, Department of Chemistry, University of Texas, Austin, Texas 78712, United States.
Francisco Fernandez-Lima, Department of Chemistry and Biochemistry and Biomolecular Sciences Institute, Florida International University, Miami, Florida 33199, United States.
REFERENCES
- (1).Navarra A; Musto A; Gargiulo A; Petrosino G; Pierantoni GM; Fusco A; Russo T; Parisi S Hmga2 is necessary for Otx2-dependent exit of embryonic stem cells from the pluripotent ground state. BMC Biol. 2016, 14, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Zhou X; Benson KF; Ashar HR; Chada K Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 1995, 376 (6543), 771–774. [DOI] [PubMed] [Google Scholar]
- (3).Chung J; Zhang X; Collins B; Sper RB; Gleason K; Simpson S; Koh S; Sommer J; Flowers WL; Petters RM; Piedrahita JA High mobility group A2 (HMGA2) deficiency in pigs leads to dwarfism, abnormal fetal resource allocation, and cryptorchidism. Proc. Natl. Acad. Sci. U.S.A 2018, 115 (21), 5420–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Weedon MN; Lettre G; Freathy RM; Lindgren CM; Voight BF; Perry JR; Elliott KS; Hackett R; Guiducci C; Shields B; Zeggini E; Lango H; Lyssenko V; Timpson NJ; Burtt NP; Rayner NW; Saxena R; Ardlie K; Tobias JH; Ness AR; Ring SM; Palmer CN; Morris AD; Peltonen L; Salomaa V; Diabetes Genetics; Davey Smith G; Wellcome Trust Case Control; Groop, L. C.; Hattersley AT; McCarthy MI; Hirschhorn JN; Frayling TM A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat. Genet 2007, 39 (10), 1245–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Yang TL; Guo Y; Zhang LS; Tian Q; Yan H; Guo YF; Deng HW HMGA2 is confirmed to be associated with human adult height. Ann. Hum. Genet 2010, 74 (1), 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Stein JL; Medland SE; Vasquez AA; Hibar DP; Senstad RE; Winkler AM; Toro R; Appel K; Bartecek R; Bergmann Ør.; Bernard M; Brown AA; Cannon DM; Chakravarty MM; Christoforou A; Domin M; Grimm O; Hollinshead M; Holmes AJ; Homuth G; Hottenga J-J; Langan C; Lopez LM; Hansell NK; Hwang KS; Kim S; Laje G; Lee PH; Liu X; Loth E; Lourdusamy A; Mattingsdal M; Mohnke S; Maniega SM; Nho K; Nugent AC; O’Brien C; Papmeyer M; Putz B; Ramasamy A; Rasmussen J; Rijpkema M; Risacher SL; Roddey JC; Rose EJ; Ryten M; Shen L; Sprooten E; Strengman E; Teumer A; Trabzuni D; Turner J; van Eijk K; van Erp TGM; van Tol M-J; Wittfeld K; Wolf C; Woudstra S; Aleman A; Alhusaini S; Almasy L; Binder EB; Brohawn DG; Cantor RM; Carless MA; Corvin A; Czisch M; Curran JE; Davies G; de Almeida MAA; Delanty N; Depondt C; Duggirala R; Dyer TD; Erk S; Fagerness J; Fox PT; Freimer NB; Gill M; Goring HHH; Hagler DJ; Hoehn D; Holsboer F; Hoogman M; Hosten N; Jahanshad N; Johnson MP; Kasperaviciute D; Kent JW; Kochunov P; Lancaster JL; Lawrie SM; Liewald DC; Mandl R; Matarin M; Mattheisen M; Meisenzahl E; Melle I; Moses EK; Muhleisen TW; Nauck M; Nothen MM; Olvera RL; Pandolfo M; Pike GB; Puls R; Reinvang I; Renteria ME; Rietschel M; Roffman JL; Royle NA; Rujescu D; Savitz J; Schnack HG; Schnell K; Seiferth N; Smith C; Steen VM; Valdes Hernandez MC; Van den Heuvel M; van der Wee NJ; Van Haren NEM; Veltman JA; Volzke H; Walker R; Westlye LT; Whelan CD; Agartz I; Boomsma DI; Cavalleri GL; Dale AM; Djurovic S; Drevets WC; Hagoort P; Hall J; Heinz A; Jack CR; Foroud TM; Le Hellard S; Macciardi F; Montgomery GW; Poline JB; Porteous DJ; Sisodiya SM; Starr JM; Sussmann J; Toga AW; Veltman DJ; Walter H; Weiner MW; Alzheimer’s Disease Neuroimaging; Bis JC; Ikram MA; Smith AV; Saguenay Youth Study; Gudnason V; Tzourio C; Vernooij MW; Launer LJ; DeCarli C; Seshadri S; Andreassen OA; Apostolova LG; Bastin ME; Blangero J; Brunner HG; Aging Research in Genomic Epidemiology; Buckner RL; Cichon S; Coppola G; de Zubicaray GI; Deary IJ; Donohoe G; de Geus EJC; Espeseth T; Fernandez G; Glahn DC; Grabe HJ; Hardy J; Hulshoff Pol HE; Jenkinson M; Kahn RS; McDonald C; McIntosh AM; McMahon FJ; McMahon KL; Meyer-Lindenberg A; Morris DW; Muller-Myhsok B; Nichols TE; Ophoff RA; Paus T; Pausova Z; Penninx BW; Potkin SG; Samann PG; Saykin AJ; Schumann G; Smoller JW; Wardlaw JM; Weale ME; Martin NG; Franke B; Wright MJ; Thompson PM Identification of common variants associated with human hippocampal and intracranial volumes. Nat. Genet 2012, 44 (5), 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Ashar HR; Chouinard RA Jr.; Dokur M; Chada K In vivo modulation of HMGA2 expression. Biochim. Biophys. Acta 2010, 1799 (1–2), 55–61. [DOI] [PubMed] [Google Scholar]
- (8).Hauke S; Leopold S; Schlueter C; Flohr AM; Murua Escobar H; Rogalla P; Bullerdiek J Extensive expression studies revealed a complex alternative splicing pattern of the HMGA2 gene. Biochim. Biophys. Acta 2005, 1729 (1), 24–31. [DOI] [PubMed] [Google Scholar]
- (9).Fedele M; Battista S; Manfioletti G; Croce CM; Giancotti V; Fusco A Role of the high mobility group A proteins in human lipomas. Carcinogenesis 2001, 22 (10), 1583–1591. [DOI] [PubMed] [Google Scholar]
- (10).Kumar MS; Armenteros-Monterroso E; East P; Chakravorty P; Matthews N; Winslow MM; Downward J HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature 2014, 505 (7482), 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- (11).Sun M; Song CX; Huang H; Frankenberger CA; Sankarasharma D; Gomes S; Chen P; Chen J; Chada KK; He C; Rosner MR HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc. Natl. Acad. Sci. U.S.A 2013, 110 (24), 9920–9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Anand A; Chada K In vivo modulation of Hmgic reduces obesity. Nat. Genet 2000, 24 (4), 377–380. [DOI] [PubMed] [Google Scholar]
- (13).Sun T; Fu M; Bookout AL; Kliewer SA; Mangelsdorf DJ MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol. Endocrinol 2009, 23 (6), 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Fusco A; Fedele M Roles of HMGA proteins in cancer. Nat. Rev. Cancer 2007, 7 (12), 899–910. [DOI] [PubMed] [Google Scholar]
- (15).Alonso N; Guillen R; Chambers JW; Leng F A rapid and sensitive high-throughput screening method to identify compounds targeting protein-nucleic acids interactions. Nucleic Acids Res. 2015, 43 (8), e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Chen B; Young J; Leng F DNA bending by the mammalian high-mobility group protein AT hook 2. Biochemistry 2010, 49 (8), 1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Reeves R; Nissen MS The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem 1990, 265 (15), 8573–8582. [PubMed] [Google Scholar]
- (18).Garabedian A; Jeanne Dit Fouque K; Chapagain PP; Leng F; Fernandez-Lima F AT-hook peptides bind the major and minor groove of AT-rich DNA duplexes. Nucleic Acids Res. 2022, 50 (5), 2431–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Frost L; Baez MA; Harrilal C; Garabedian A; Fernandez-Lima F; Leng F The Dimerization State of the Mammalian High Mobility Group Protein AT-Hook 2 (HMGA2). PloS One 2015, 10 (6), e0130478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Reeves R HMGA proteins: flexibility finds a nuclear niche? Biochem. Cell Biol 2003, 81 (3), 185–195. [DOI] [PubMed] [Google Scholar]
- (21).Reeves R Molecular biology of HMGA proteins: hubs of nuclear function. Gene 2001, 277 (1–2), 63–81. [DOI] [PubMed] [Google Scholar]
- (22).Reeves R High mobility group (HMG) proteins: Modulators of chromatin structure and DNA repair in mammalian cells. DNA Repair 2015, 36, 122–136. [DOI] [PubMed] [Google Scholar]
- (23).Huth JR; Bewley CA; Nissen MS; Evans JN; Reeves R; Gronenborn AM; Clore GM The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol 1997, 4 (8), 657–665. [DOI] [PubMed] [Google Scholar]
- (24).Fonfria-Subiros E; Acosta-Reyes F; Saperas N; Pous J; Subirana JA; Campos JL Crystal structure of a complex of DNA with one AT-hook of HMGA1. PloS One 2012, 7 (5), e37120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Pierson NA; Chen L; Valentine SJ; Russell DH; Clemmer DE Number of solution states of bradykinin from ion mobility and mass spectrometry measurements. J. Am. Chem. Soc 2011, 133 (35), 13810–13813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Beveridge R; Chappuis Q; Macphee C; Barran P Mass spectrometry methods for intrinsically disordered proteins. Analyst 2013, 138 (1), 32–42. [DOI] [PubMed] [Google Scholar]
- (27).Ruotolo BT; Hyung SJ; Robinson PM; Giles K; Bateman RH; Robinson CV Ion mobility-mass spectrometry reveals long-lived, unfolded intermediates in the dissociation of protein complexes. Angew. Chem., Int. Ed 2007, 46 (42), 8001–8004. [DOI] [PubMed] [Google Scholar]
- (28).Knapman TW; Morton VL; Stonehouse NJ; Stockley PG; Ashcroft AE Determining the topology of virus assembly intermediates using ion mobility spectrometry-mass spectrometry. Rapid Commun. Mass Spectrom 2010, 24 (20), 3033–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Dixit SM; Polasky DA; Ruotolo BT Collision induced unfolding of isolated proteins in the gas phase: past, present, and future. Curr. Opin. Chem. Biol 2018, 42, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Fernandez-Lima FA; Kaplan DA; Suetering J; Park MA Gas-phase separation using a Trapped Ion Mobility Spectrometer. Int. J. Ion Mobil. Spectrom 2011, 14 (2–3), 93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ridgeway ME; Bleiholder C; Mann M; Park MA Trends in trapped ion mobility – Mass spectrometry instrumentation. Trends Anal. Chem 2019, 116, 324–331. [Google Scholar]
- (32).Jeanne Dit Fouque K; Fernandez-Lima F Recent advances in biological separations using trapped ion mobility spectrometry – mass spectrometry. Trends Anal. Chem 2019, 116, 308–315. [Google Scholar]
- (33).Liu FC; Ridgeway ME; Park MA; Bleiholder C Tandem trapped ion mobility spectrometry. Analyst 2018, 143 (10), 2249–2258. [DOI] [PubMed] [Google Scholar]
- (34).Jeanne Dit Fouque K; Garabedian A; Leng F; Tse-Dinh YC; Ridgeway ME; Park MA; Fernandez-Lima F Trapped Ion Mobility Spectrometry of Native Macromolecular Assemblies. Anal. Chem 2021, 93 (5), 2933–2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Garabedian A; Bolufer A; Leng F; Fernandez-Lima F Peptide Sequence Influence on the Conformational Dynamics and DNA binding of the Intrinsically Disordered AT-Hook 3 Peptide. Sci. Rep 2018, 8 (1), 10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Jeanne Dit Fouque K; Garabedian A; Leng F; Tse-Dinh YC; Fernandez-Lima F Microheterogeneity of Topoisomerase IA/IB and Their DNA-Bound States. ACS Omega 2019, 4 (2), 3619–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Cui T; Wei S; Brew K; Leng F Energetics of binding the mammalian high mobility group protein HMGA2 to poly(dA-dT)2 and poly(dA)-poly(dT). J. Mol. Biol 2005, 352 (3), 629–645. [DOI] [PubMed] [Google Scholar]
- (38).Lakowicz JR Fluorescence Anisotropy. In Principles of Fluorescence Spectroscopy; Springer US: Boston, M.,Ed.; 2006; pp 353–382. [Google Scholar]
- (39).Cui T; Leng F Specific recognition of AT-rich DNA sequences by the mammalian high mobility group protein AT-hook 2: a SELEX study. Biochemistry 2007, 46 (45), 13059–13066. [DOI] [PubMed] [Google Scholar]
- (40).Fernandez-Lima FA; Kaplan DA; Park MA Note: Integration of trapped ion mobility spectrometry with mass spectrometry. Rev. Sci. Instrum 2011, 82 (12), 126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Hernandez DR; Debord JD; Ridgeway ME; Kaplan DA; Park MA; Fernandez-Lima F Ion dynamics in a trapped ion mobility spectrometer. Analyst 2014, 139 (8), 1913–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Ridgeway ME; Lubeck M; Jordens J; Mann M; Park MA Trapped ion mobility spectrometry: A short review. Int. J. Mass Spectrom 2018, 425, 22–35. [Google Scholar]
- (43).Michelmann K; Silveira JA; Ridgeway ME; Park MA Fundamentals of trapped ion mobility spectrometry. J. Am. Soc. Mass Spectrom 2015, 26 (1), 14–24. [DOI] [PubMed] [Google Scholar]
- (44).Silveira JA; Michelmann K; Ridgeway ME; Park MA Fundamentals of Trapped Ion Mobility Spectrometry Part II: Fluid Dynamics. J. Am. Soc. Mass Spectrom 2016, 27 (4), 585–595. [DOI] [PubMed] [Google Scholar]
- (45).Hanwell MD; Curtis DE; Lonie DC; Vandermeersch T; Zurek E; Hutchison GR Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform 2012, 4 (1), 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Yan Y; Tao H; He J; Huang SY The HDOCK server for integrated protein-protein docking. Nat. Protoc 2020, 15 (5), 1829–1852. [DOI] [PubMed] [Google Scholar]
- (47).Zhong Y; Han L; Ruotolo BT Collisional and Coulombic unfolding of gas-phase proteins: high correlation to their domain structures in solution. Angew. Chem., Int. Ed 2014, 53 (35), 9209–9212. [DOI] [PubMed] [Google Scholar]
- (48).Haq I; Ladbury JE; Chowdhry BZ; Jenkins TC; Chaires JB Specific binding of hoechst 33258 to the d-(CGCAAATTTGCG)2 duplex: calorimetric and spectroscopic studies. J. Mol. Biol 1997, 271 (2), 244–257. [DOI] [PubMed] [Google Scholar]
- (49).Su L; Bryan N; Battista S; Freitas J; Garabedian A; D’Alessio F; Romano M; Falanga F; Fusco A; Kos L; Chambers J; Fernandez-Lima F; Chapagain PP; Vasile S; Smith L; Leng F Identification of HMGA2 inhibitors by AlphaScreen-based ultra-high-throughput screening assays. Sci. Rep 2020, 10 (1), 18850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


