Abstract
The bacterial diversity assessed from clone libraries prepared from rRNA (two libraries) and ribosomal DNA (rDNA) (one library) from polychlorinated biphenyl (PCB)-polluted soil has been analyzed. A good correspondence of the community composition found in the two types of library was observed. Nearly 29% of the cloned sequences in the rDNA library were identical to sequences in the rRNA libraries. More than 60% of the total cloned sequence types analyzed were grouped in phylogenetic groups (a clone group with sequence similarity higher than 97% [98% for Burkholderia and Pseudomonas-type clones]) represented in both types of libraries. Some of those phylogenetic groups, mostly represented by a single (or pair) of cloned sequence type(s), were observed in only one of the types of library. An important difference between the libraries was the lack of clones representative of the Actinobacteria in the rDNA library. The PCB-polluted soil exhibited a high bacterial diversity which included representatives of two novel lineages. The apparent abundance of bacteria affiliated to the beta-subclass of the Proteobacteria, and to the genus Burkholderia in particular, was confirmed by fluorescence in situ hybridization analysis. The possible influence on apparent diversity of low template concentrations was assessed by dilution of the RNA template prior to amplification by reverse transcription-PCR. Although differences in the composition of the two rRNA libraries obtained from high and low RNA concentrations were observed, the main components of the bacterial community were represented in both libraries, and therefore their detection was not compromised by the lower concentrations of template used in this study.
Investigations of microbial composition and diversity in natural and anthropogenically impacted or created habitats is important in the characterization of such habitats, since microbes are key players in many environmental processes. Over the last few years, cultivation-independent methodologies, particularly the sequence analysis of cloned 16S ribosomal RNA genes (16S rDNA), have proven to be powerful tools for investigating the microbial diversity of environmental samples (10). At least as important is the specific identification of the metabolically active microorganisms, since these are responsible for the microbially driven environmental processes. For example, knowledge of the active microorganisms in polluted habitats is relevant to the development of optimal in situ bioremediation strategies, as well as contributing to the identification of yet-undescribed (i.e., not yet-cultured) bacteria which may play important, albeit unknown, roles in pollutant degradation or other community processes.
Since metabolically active cells usually contain higher numbers of ribosomes than quiescent cells (23), a 16S rRNA library generated from total extracted rRNA may be considered to reflect predominantly the diversity of the metabolically active members of the community. Several reports on the analysis of bacterial communities using 16S rRNA have been published (7, 20, 22, 36, 37). However, it is not currently known whether rRNA and rDNA libraries will be significantly different, since it is not known which proportion of microbial community is quiescent. A comparison of results obtained from rRNA and rDNA libraries has been attempted by Miskin et al. (20) in a study of an anoxic sediment sample. These authors observed a few identical sequences in the two types of library and concluded that the libraries did not have a degree of coverage of the diversity in the sample high enough to enable valid comparisons.
We have undertaken such a comparison with a degree of diversity coverage that should permit conclusions. In the present study we describe a 16S rRNA gene clone library, obtained by PCR amplification from total DNA extracted from a polychlorinated biphenyl (PCB)-polluted soil, and compare it with a previously described 16S rRNA library obtained by reverse transcription-PCR (RT-PCR) (22) and an unreported rRNA library generated from a 1:500 dilution of the original template RNA. A high species diversity was found in both types of library, though it was clear from rarefaction plots that, even though some 404 clones were analyzed, not all of the bacterial diversity in that habitat had been revealed. A considerable percentage of rDNA clones were also represented in the rRNA libraries and, in general, there was a qualitative correspondence of clone frequency in the two types of libraries, with representatives of the alpha and beta subdivisions of Proteobacteria and the Acidobacterium phylum dominating.
MATERIALS AND METHODS
Total DNA and RNA extraction.
The sample used for total nucleic acid (DNA and RNA) extraction was taken from the upper few centimeters of the surface of a soil in an area near Wittenberg, Germany, where high concentrations of PCB were detected (22), weighed, and frozen at −70°C until processing. Total nucleic acids were extracted from the soil using a protocol described previously (22). The extracted nucleic acids were pelleted and washed with 70% ethanol, dried, and resuspended in 300 μl of deionized water. An aliquot of the sample was digested with 30 U of RNase-free DNase I (Roche Diagnostics, GmbH, Mannheim, Germany) at 37°C for 2 h in 10 mM sodium acetate–0.5 mM MgSO4 (pH 5.0). Both total RNA and total DNA were purified using Microcon microconcentrators 100 (Millipore GmbH, Eschborn, Germany), according to the manufacturer's instructions. Aliquots of purified and nonpurified total RNA and total DNA were analyzed by electrophoresis on a 1% (wt/vol) agarose gel and staining with ethidium bromide.
RT-PCR amplification of 16S rRNA, PCR amplification of 16S rRNA genes, and cloning of the amplification products.
The region of the 16S rRNA between nucleotide positions 27 and 518 (Escherichia coli 16S rRNA gene sequence numbering), corresponding to approximately one-third of the entire 16S rRNA, was targetted for reverse transcription-PCR (RT-PCR) amplification from the extracted template RNA. RT-PCR analyses were performed with ca. 230 ng and 460 pg (dilution, 1:500) of the total RNA, using rTth DNA polymerase (Applied Biosystems, Weiterstadt, Germany) as described previously (22). Nearly the entire 16S rRNA gene, between positions 27 and 1492 (E. coli 16S rRNA gene sequence numbering), was amplified by PCR. PCR mixtures contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM concentrations of deoxynucleoside triphosphates, 0.5 μM concentrations of primers, approximately 80 ng of DNA, and 2.5 U of AmpliTaq DNA polymerase (Applied Biosystems). PCRs were performed in a GeneAmp 9600 thermocycler (Applied Biosystems) with the following conditions: an initial denaturation step at 94°C for 2 min, followed by 30 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C, and a final extension step of 10 min at 72°C.
The PCR and RT-PCRs were carried out in triplicate, and the resulting products were pooled before gel purification and cloning. The cloning procedure has been detailed previously (22). Three clone libraries were generated: one with the PCR amplification products from total DNA and two with the RT-PCR amplification products from undiluted and 1:500-diluted RNA, respectively.
Sequencing of cloned RT-PCR products.
The nucleotide sequences of the cloned products were determined from plasmid DNA preparations (obtained using Qiawell 8 or QiaSpin plasmid extraction kits [Qiagen GmbH, Hilden, Germany]) using the ABI PRISM dRhodamine and BigDye Terminator Cycle Sequencing kits and ABI373 and ABI377 Sequencers (Applied Biosystems) according to the manufacturer's instructions. Vector primers T3 and T7 were used for the sequencing reactions.
Assignment of cloned sequences to established phylogenetic divisions.
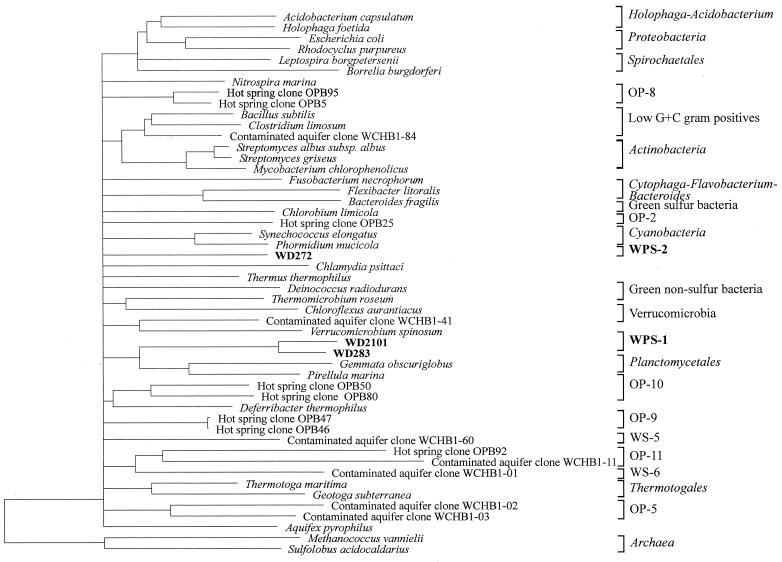
Cloned 16S rRNA sequences were compared initially with reference sequences contained in the EMBL Nucleotide Sequence Database (2) using the FASTA program (25) and subsequently aligned with 16S rRNA reference sequences in the ARB package (http://www.mikro.biologie.tu-muenchen.de) (32). Ambiguous positions were excluded from similarity calculations. Evolutionary distances, derived from sequence-pair dissimilarities using the Jukes and Cantor algorithm (12), were calculated using the DNADIST program from the Phylogeny Inference Package (PHYLIP) included in the ARB package. For the calculation of the dendrogram shown in Fig. 2, cloned sequences were aligned with 16S rRNA sequences representative of the main bacterial divisions. Dendrograms were calculated using neighbor joining; the least-squares algorithm of Fitch-Margoliash of the FITCH program; parsimony (DNAPARS), and maximum-likelihood (DNA_ML) algorithms of the PHYLIP package included in the ARB software. Hypervariable regions in the 16S rRNA molecule were excluded from the calculation as described elsewhere (14). Branches whose phylogenetic position in the dendrogram changed depending on the method of analysis used were collapsed back to the previous consistent node by introducing multifurcations.
FIG. 2.
Dendrogram of the novel bacterial lineages WPS-1 and WPS-2 and sequences representative of different bacterial divisions. Evolutionary distances and the phylogenetic relationships were calculated using distance, parsimony, and maximum-likelihood methods. Only nearly-complete sequences from the 16S rDNA clones were included in the calculation. Branches whose position varied with the treeing method used were collapsed back to the previous consistent node. The hypervariable regions in the 16S rRNA molecule (14) were excluded from the calculation. The archaeal sequences were used as outgroup.
Rarefaction analyses and diversity indexes.
Rarefaction calculations were done using the software Analytic Rarefaction (version 1.2; Stratigraphy Laboratory, University of Georgia [www.uga.edu/∼strata/Software.html]). Shannon diversity index (H) and equitability (J) values were calculated as previously described (3).
In situ hybridization.
The same soil samples from the PCB-polluted soil used for the elaboration of the libraries were fixed at 4°C for 16 h in 4% paraformaldehyde–phosphate-buffered saline (PBS), composed of 0.13 M NaCl, 7 mM Na2HPO4, and 3 mM NaH2PO4 (pH 7.2). After fixation, the samples were washed in PBS three times and stored in ethanol-PBS (1:1 [vol/vol]) at −20°C. The soil slurry was vortexed for 1 min and diluted in PBS. Hybridizations were carried out on 0.22 μm-pore-size polycarbonate filters (Millipore) after filtration of the diluted soil slurries. Oligonucleotide probes were synthesized with Cy3 fluorochrome at the 5′ end (Interactiva Biotechnologie GmbH, Ulm, Germany). The probes used were EUB338 for the domain Bacteria (1), ALF1b for the alpha subclass of Proteobacteria, BET42a for the beta subclass of Proteobacteria (used with competitor), GAM42a for the gamma subclass of Proteobacteria (used with competitor) (19), PLA886 for planctomycetes (used with competitor) (21), HGC69a for Actinobacteria (formerly gram-positive bacteria with a high G+C content) (28), SUBU1237 for Burkholderia and Suterella spp. (31), and the antisense probe NON338 (1). Hybridizations and microscopy counts of hybridized and DAPI (4′,6-diamidino-2-phenylindole)-stained cells were performed as previously described (29), except that an additional prehybridization step using 1% (for EUB338) or 2% (for the other probes) of blocking reagent (Roche) was introduced in order to reduce unspecific binding of the probes to soil particles. The slides were examined with an Axiophot II microscope (Zeiss, Jena, Germany).
Nucleotide sequence data.
The sequence data of the cloned 16S rRNA obtained by RT-PCR with undiluted RNA template was deposited in the EMBL database under the accession numbers AJ233467 to AJ233589. The new sequence data reported in this study have been deposited under accession numbers AJ292571 to AJ292689 for the sequence data corresponding to cloned 16S rDNA and AJ292771 to AJ292925 for cloned 16S rRNA obtained with the diluted RNA template.
RESULTS
Comparison of the composition of the 16S rDNA library and the 16S rRNA libraries.
The bacterial diversity in an acidic PCB-polluted soil near Wittenberg (Germany) was analyzed by amplification of 16S rDNA from total DNA extracted from the soil and compared with the diversity observed in two clone libraries generated from extracted RNA, which should be more representative of the metabolically active bacteria in the soil. The 16S rDNA cloned sequences determined were designated with a number preceded by the letters “W” (for Wittenberg) and “D” (from DNA) to differentiate them from the cloned sequences WR (from RNA).
The predominant bacterial divisions present in the 16S rDNA library were also the most numerous in both libraries obtained from 16S rRNA, i.e., cloned sequence types affiliated to the alpha, beta, and gamma subdivisions of the Proteobacteria (30) and to the Holophaga-Acidobacterium phylum (16).
The 5′-partial sequences of 34 clones from the 16S rDNA clone library (28.6% of the total number of 16S rDNA clones analyzed) were identical to those of clones from the 16S rRNA libraries and belonged to the four predominant divisions mentioned above: 21, 6, and 4 cloned sequences clustered within the beta, alpha, and gamma subdivisions of the Proteobacteria, respectively, and three cloned sequences within the Holophaga-Acidobacterium phylum.
In order to simplify the comparison of the sequences obtained in the analysis of the three libraries from Wittenberg, cloned sequence types with >97% similarity were considered to constitute a phylogenetic group, except for the collection of cloned sequence types related to Pseudomonas and Burkholderia spp., for which a higher similarity threshold (ca. 98%) was set.
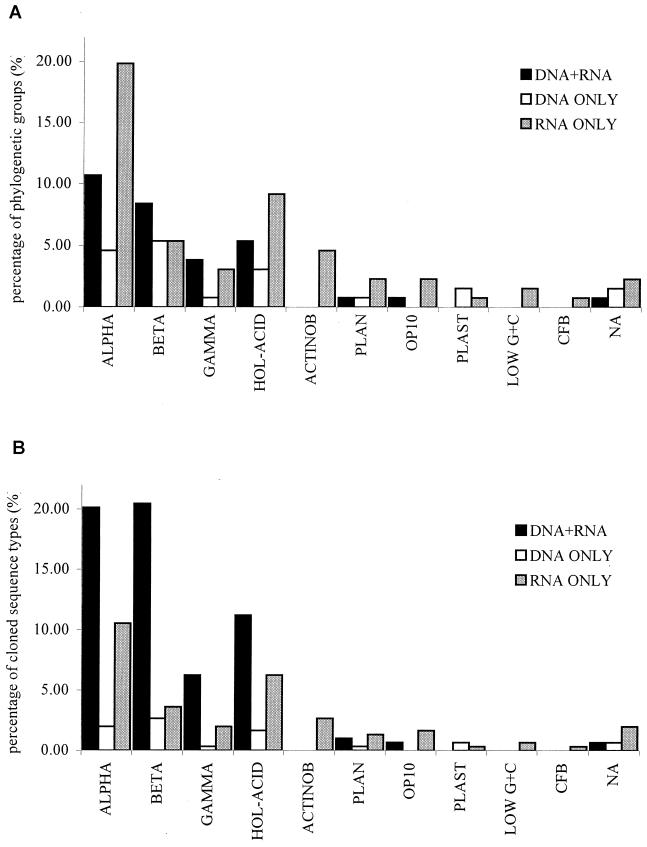
The number of phylogenetic groups, cloned sequences and cloned sequence types for the different bacterial divisions are shown in Table 1. A total of 29.3% of the phylogenetic groups, which included most of the sequence types observed (183 cloned sequence types, representing 60.4% of the total sequence types; Fig. 1), were found in both types of libraries. Table 2 shows the affiliation of rDNA cloned sequence types representative of each of the phylogenetic groups observed in rDNA and rRNA libraries. The identification of the clones present in both types of libraries agreed with the results derived from previous analyses on the microbial community from Wittenberg PCB-contaminated soil (22), i.e., (i) a diverse set of cloned sequence types affiliated with the alpha subclass of Proteobacteria, with a high proportion of cloned sequence types related to 16S rRNA sequences of Sphingomonas and acidiphilic genera within this subdivision, (ii) a large number of cloned sequence types related to Burkholderia and Variovorax-related 16S rRNA sequences in the beta subclass of the Proteobacteria, (iii) a prevalence of cloned sequence types affiliated with Nevskia ramosa within the gamma subclass of the Proteobacteria, (iv) a high diversity within the cloned sequence types affiliated with the Holophaga-Acidobacterium phylum, and (v) the presence of cloned sequence types related to 16S rRNA sequences of Isosphaera spp. within the Planctomycetales.
TABLE 1.
Number of phylogenetic groups, clones, and cloned sequence types from PCB-polluted soil for each of the bacterial divisions observed in either the 16S rDNA library, the two 16S rRNA libraries, or both types of library simultaneously
| Bacterial division | No. of phylogenetic groupsa observed in PCB-polluted soil 16S rDNA and 16S rRNA libraries
|
|||||
|---|---|---|---|---|---|---|
| Total | DNA library | RNA libraries | DNA plus RNA libraries | DNA library only | RNA libraries only | |
| Proteobacteria | ||||||
| Alpha subdivision | 46 (128/99) | 20 | 40 | 14 (88/61) | 6 (6/6) | 26 (34/32) |
| Beta subdivision | 25 (124/81) | 18 | 18 | 11 (105/62) | 7 (8/8) | 7 (11/11) |
| Gamma subdivision | 10 (42/26) | 6 | 9 | 5 (35/19) | 1 (1/1) | 4 (6/6) |
| Holophaga-Acidobacterium | 23 (68/58) | 11 | 19 | 7 (41/34) | 4 (5/5) | 12 (22/19) |
| Actinobacteria | 6 (9/8) | 0 | 6 | 0 | 0 | 6 (9/8) |
| Planctomycetales | 5 (9/8) | 2 | 4 | 1 (3/3) | 1 (1/1) | 3 (5/4) |
| Candidate division OP10 | 4 (7/7) | 1 | 4 | 1 (2/2) | 0 | 3 (5/5) |
| Plastids | 3 (3/3) | 2 | 1 | 0 | 2 (2/2) | 1 (1/1) |
| Low G+C gram positives | 2 (2/2) | 0 | 2 | 0 | 0 | 2 (2/2) |
| Cytophaga-Flavobacterium-Bacteroides | 1 (1/1) | 0 | 1 | 0 | 0 | 1 (1/1) |
| Not affiliated (WPS-1 and WPS-2) | 6 (11/10) | 3 | 4 | 1 (2/2) | 2 (2/2) | 3 (7/6) |
| Total | 131 (404/303) | 63 | 108 | 40 (276/183) | 23 (25/25) | 68 (103/95) |
Phylogenetic group: a clone group with sequence similarity higher than 97% (98% for Burkholderia and Pseudomonas-type clones). Values in parentheses indicate the number of clones/cloned sequence types.
FIG. 1.
Comparison of the representation of different bacterial divisions in the 16S rDNA library, 16S rRNA libraries, or both. (A) Phylogenetic groups. (B) Cloned sequence types. ALPHA, BETA, and GAMMA, the alpha, beta, and gamma subdivisions of the Proteobacteria; HOL-ACID, Holophaga-Acidobacterium; ACTINOB, Actinobacteria; PLAN, Planctomycetales; OP10, candidate division OP10; PLAST, plastids; LOW G+C, low-G+C-content gram positives; CFB, Cytophaga-Flavobacterium-Bacteroides; NA, not affiliated (lineages WPS-1 and WPS-2).
TABLE 2.
Identification of representative cloned sequence types present in phylogenetic groups found in both the 16S rDNA and the 16S rRNA clone libraries from PCB-polluted soil
| Bacterial division | Phylogenetic group | No. of sequences/ no. of sequence types | Representative clone | Closest relative in database (EMBL accession no.) | Length (nt)a | % Identity |
|---|---|---|---|---|---|---|
| Alpha-Proteobacteria | A-1 | 6/5 | WD208 | Sphingomonas asaccharolytica IFO 10564 (Y09639) | 1,410 | 95.32 |
| A-2 | 7/4 | WD225 | Phenylobacterium immobile (Y18216) | 1,404 | 97.08 | |
| A-3 | 5/3 | WD229 | Magnetospirillum sp. strain MSM-4 (Y17390) | 1,409 | 93.75 | |
| A-4 | 4/4 | WD2103 | Azospirillum sp. strain ASP-1 (X92464) | 1,408 | 90.13 | |
| A-7 | 2/2 | WD238 | Rhodopila globiformis DSM 161 (D86513) | 1,428 | 92.58 | |
| A-8 | 10/6 | WD248 | Acidosphaera rubrifaciens HS-AP3 (D86512) | 1,414 | 94.70 | |
| A-9 | 16/9 | WD249 | Sphingomonas subartica KF1 (X94102) | 1,411 | 96.17 | |
| A-10 | 5/4 | WD252 | Sphingomonas pruni IFO 15498 (Y09637) | 1,409 | 98.58 | |
| A-11 | 4/4 | WD267 | Afipia genosp. 13 strain G8991 (U87784) | 1,411 | 95.46 | |
| A-12 | 2/1 | WD271 | Gluconacetobacter liquefaciens LMG 1382 (X75617) | 1,413 | 92.43 | |
| A-13 | 11/5 | WD275 | Bradyrhizobium sp. strain Pe4 (AF159437) | 1,410 | 99.01 | |
| A-15 | 4/4 | WD295 | Gluconacetobacter sacchari IF2-6 (AF127412) | 1,413 | 94.55 | |
| A-16 | 6/6 | WD297 | Caulobacter vibriodes ATCC 11764 (AJ227755) | 1,408 | 97.23 | |
| A-19 | 6/4 | WD2108 | Gluconacetobacter sacchari IF2-6 (AF127412) | 1,417 | 96.47 | |
| Beta-Proteobacteria | B-2 | 11/3 | WD291 | Xylophilus ampelinus ATCC 33914 (AF078758) | 1,455 | 98.14 |
| B-3 | 10/4 | WD2115 | Variovorax sp. strain WFF52 (AB003627) | 1,457 | 99.18 | |
| B-5 | 8/5 | WD2102 | Rubrivivax gelatinosus (D16213) | 1,451 | 96.35 | |
| B-13 | 6/6 | WD221 | Burkholderia sp. strain LB400 (U86373) | 739 | 97.29 | |
| B-14 | 8/8 | WD202 | Burkholderia sp. strain N3P2 (U37344) | 1,453 | 98.97 | |
| B-15 | 5/4 | WD263 | Burkholderia sp. strain NF100 (AB025790) | 1,456 | 97.25 | |
| B-16 | 5/5 | WD206 | Burkholderia sp. strain NF100 (AB025790) | 1,455 | 97.80 | |
| B-17 | 5/3 | WD237 | Burkholderia sp. strain N2P5 (U37342) | 748 | 98.80 | |
| B-18 | 38/17 | WD227 | Burkholderia sp. strain Dha-54 (AJ011508) | 1,455 | 98.28 | |
| B-19 | 3/3 | WD268 | Burkholderia sp. strain N2P5 (U37342) | 1,451 | 97.80 | |
| B-21 | 6/4 | WD2116 | Burkholderia glathei ATCC 29195 (AB021374) | 1,455 | 97.25 | |
| Gamma-Proteobacteria | G-1 | 2/1 | WD259 | Pseudomonas sp. strain PsK (AF105389) | 1,459 | 99.32 |
| G-2 | 3/2 | WD260 | Ectothiorhodospira sp. strain Bogoria Red (AF084511) | 1,462 | 87.41 | |
| G-3 | 13/4 | WD280 | Nevskia ramosa (AJ001343) | 1,450 | 96.14 | |
| G-4 | 15/10 | WD284 | Nevskia ramosa (AJ001343) | 1,450 | 96.69 | |
| G-5 | 2/2 | WD2124 | Ectothiorhodospira sp. strain Bogoria Red (AF084511) | 1,463 | 86.74 | |
| Holophaga-Acidobacterium | H-2 | 7/5 | WD207 | Acidobacterium capsulatum (D26171) | 1,424 | 95.37 |
| H-4 | 7/7 | WD247 | Clone UA1 (AF200696) | 1,422 | 96.06 | |
| H-5 | 6/4 | WD217 | Clone UA3 (AF200699) | 1,426 | 92.50 | |
| Holophaga-Acidobacterium | H-6 | 5/3 | WD228 | Clone TRB82 (AF047646) | 1,398 | 97.14 |
| H-7 | 5/3 | WD243 | Clone TRB82 (AF047646) | 1,402 | 96.36 | |
| H-10 | 4/4 | WD261 | Clone DA052 (Y07646) | 1,461 | 93.29 | |
| H-11 | 7/7 | WD277 | Clone TRB82 (AF047646) | 1,400 | 95.14 | |
| Planctomycetales | P-4 | 3/3 | WD2112 | Isosphaera sp. strain Schlesner 640 (X81959) | 1,444 | 92.45 |
| Candidate division OP10 | O-1 | 2/2 | WD294 | Clone SJA-22 (AJ009456) | 1,439 | 86.17 |
| Not affiliated (WS-I) | N-3 | 2/2 | WD2101 | Planctomycete strain 394 (AJ231192) | 1,395 | 75.41 |
nt, nucleotides.
An interesting set of cloned sequence types represented in rRNA and rDNA libraries appeared to be distantly related to clones retrieved from a trichlorobenzene-transforming consortium which were reported to be members of the candidate division OP10 (34). Note that four of these cloned sequence types had, on the basis of partial sequences, been previously assigned as members of the class Actinobacteria since they appeared to be related to Acidimicrobium ferrooxidans (22).
About 52 and 18% of the phylogenetic groups were present exclusively in the rRNA libraries or in the rDNA library, respectively (Fig. 1), although the majority of these were represented by a single or two cloned sequence types (a few contained more [up to five]). Most of the phylogenetic groups unique to the rDNA library were closely related to groups found in both rRNA and rDNA libraries. However, clone sequence types affiliated with three bacterial divisions not observed in the 16S rDNA library were observed exclusively in the rRNA libraries, namely, cloned sequence types clustering within the Actinobacteria (constituting a diverse set related to the 16S rRNA sequences of genera such as Gordonia, Curtobacterium, Geodermatophilus, and Terrabacter and the soil cloned sequence type TM146, a member of the group I TM clones [27]), the low-G+C-content gram positives (Clostridium-like sequence types), and the Cytophaga-Flavobacterium-Bacteroides phylum, with sequence types related to 16S rRNA sequences of Sphingobacterium.
New bacterial lineages.
A total of 10 cloned sequence types retrieved from Wittenberg soil were not affiliated with any described bacterial divisions and are proposed here as representatives of two new bacterial lineages, which we designated WPS-1 for “Wittenberg polluted soil” (nine cloned sequence types) and WPS-2 (one sequence type). The dendrogram in Fig. 2 shows the phylogenetic positions among the Bacteria for WPS-1 and WPS-2.
The WPS-1 sequence types formed a diverse collection of related sequences with similarities between them ranging from 82.7 to 99.8% and included cloned sequence types from the rDNA library analyzed (two sequence types) and both rRNA libraries analyzed (seven sequence types). Comparison of the almost-complete 16S rDNA sequence of the clones from the rDNA library with sequences available in databases suggests that WPS-1 might be a deeply branching lineage, distantly related to the Planctomycetales (similarities of <76% to the closest relatives). The phylogenetic relationship between WPS-1 and the Planctomycetales was supported by all the treeing methods employed in the analysis.
The WPS-2 lineage was represented by a single cloned sequence type, WD272, which was only observed in the 16S rDNA library. Analysis of this cloned sequence with the program CHIMERA_CHECK (version 2.7) (18) and careful checking of its base pairing (with the aid of the ARB package) ruled out the possibility that this sequence was a chimeric product. The sequence similarity of clone WD272 to cloned sequence types in the WPS-1 lineage ranged between 63.2 and 67.8%. The phylogenetic position of this cloned sequence type could not be determined consistently by the different treeing methods used in the present analysis. Three treeing methods (neighbor joining, parsimony, and maximum likelihood) indicated the proximity of WPS-2 to the cyanobacteria. While for the first two methods WPS-2 branched from the radiation to the cyanobacteria, it appeared to branch outside the cyanobacterial lineage in the maximum parsimony tree. In the tree calculated using FITCH, WPS-2 branched from the radiation to the deinococci. By using FASTA searches, the closest 16S rDNA sequences to that of WD272 were the sequences of clones SJA-5, SJA-22, and WCHB1-84 retrieved from chlorinated hydrocarbon-degrading communities (5, 34).
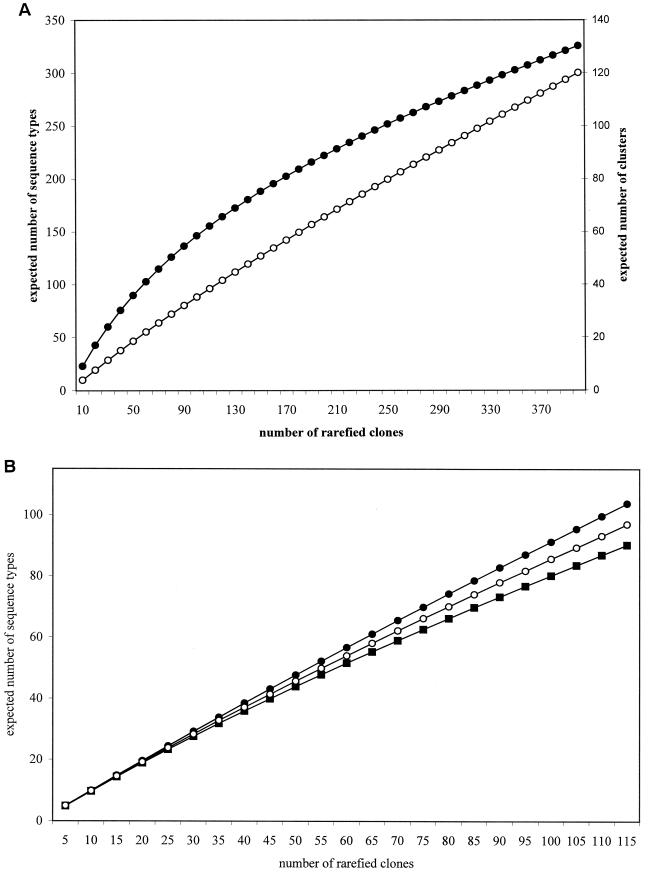
Rarefaction analysis and diversity indexes.
The cloned sequences represented in the different libraries from Wittenberg were subjected to rarefaction analysis. Two sets of data were rarefied: one comprising the cloned sequence types and another comprising the phylogenetic groups established. Despite the fact that only 362 of 404 cloned sequences (65%) in the three libraries were unique, the rarefaction analysis suggests that the number of clones screened is insufficient to circumscribe the bacterial diversity in the PCB-polluted soil (Fig. 3A). The data from the cloned sequence types in each one of the three libraries generated in this study were also rarefied. According to the rarefaction plots, the expected number of sequence types in the 16S rDNA library was lower than that for the 16S rRNA libraries, suggesting a lower diversity within the 16S rDNA library (Fig. 3B). This result is consistent with the higher percentage of redundant cloned sequences observed in the 16S rDNA library. The expected number of sequence types in the library obtained with diluted RNA was lower than in the one generated from undiluted RNA, as expected. Shannon diversity indices (calculated for the three libraries from cloned sequence types) were higher and very similar for the libraries generated from RNA (H = 4.71 and 4.72 for the library obtained from undiluted and diluted RNAs, respectively), as opposed to the library obtained from DNA (H = 4.41). Equitability was higher for the library obtained from undiluted RNA (J = 0.97), slightly lower for the library obtained from diluted RNA (J = 0.94) and, finally, lower for the library generated from DNA (J = 0.92). These results agree with the rarefaction plots and show a higher diversity of the libraries generated from RNA.
FIG. 3.
Rarefaction analysis. (A) Expected number of phylogenetic groups (clusters) (●) and cloned sequence types (○) after rarefaction of the total sequence data from Wittenberg PCB-polluted soil. (B) Expected number of cloned sequence types for each one of the clone libraries analyzed (DNA, ■; undiluted RNA, ●; 1:500-diluted RNA, ○).
Effect of template dilution prior to RT-PCR on the composition of the 16S rRNA libraries.
One general concern in the analysis of bacterial diversity by PCR amplification of 16S rRNA genes, particularly with samples from soil environments, is the frequent necessity to dilute the extracted DNA to overcome inhibition of the PCR and the effect that dilution might have on the compositions of derived clone libraries. This effect has been explored with sediment samples in which the PCRs were carried out with very low template DNA concentrations, in the order of picograms, and clone libraries were screened by restriction fragment length polymorphism analysis (4). On the other hand, although the abundance of a certain 16S rRNA (or rDNA) sequence type in a clone library cannot be directly correlated with the abundance in the environment of the bacterium represented by that sequence, the presence of cloned sequence types in libraries obtained with highly diluted template might be considered to be indicative of their predominance in that particular sample. We have studied the effect of RNA template dilution concentrations (in nanograms) most frequently obtained with soil samples by comparing the compositions of two libraries obtained by RT-PCR amplification of 16S rRNA from total RNA (undiluted and at a 1:500 dilution) and cloning of the resulting products. As expected, a higher percentage of sequence redundancy characterized the cloned sequences obtained from diluted RNA (27% sequence redundancy and J = 0.94 compared with 17% redundancy and J = 0.97 in the library obtained with undiluted RNA). Eighty-one clones with identical sequences were obtained from both libraries.
Table 3 shows the number of phylogenetic groups, clones, and cloned sequence types which were observed in the rRNA libraries. Nearly two-thirds of the cloned sequence types were observed in both rRNA libraries and were precisely the cloned sequence types within the predominant phylogenetic groups observed in both libraries, including cloned sequence types similar to the 16S rRNA sequence of species of (i) Sphingomonas, Caulobacter, Bradyrhizobium, Phenylobacterium, Magnetospirillum, Acidosphaera, and Gluconoacetobacter, within the alpha subclass of the Proteobacteria, (ii) Burkholderia, Xylophilus ampelinus, and Rubrivivax gelatinosus in the beta subclass, (iii) Nevskia ramosa within the gamma subclass of the Proteobacteria, and (iv) Acidobacterium capsulatum and the cloned sequences UA1 and UA3 (26), TRB82 (6), and HRS-56 (24) in the Holophaga-Acidobacterium phylum.
TABLE 3.
Number of phylogenetic groups, clones, and cloned sequence types from PCB-polluted soil for each of the bacterial divisions observed in either the library generated from undiluted RNA, the one generated from diluted RNA, and in both types of libraries simultaneously
| Bacterial division | No. of phylogenetic groupsa observed in 16S rRNA libraries obtained with undiluted and diluted RNA
|
|||
|---|---|---|---|---|
| Total | Both rRNA libraries | Undiluted RNA only | 1:500-diluted RNA only | |
| Proteobacteria | ||||
| Alpha subdivision | 40 (97/78) | 13 (67/50) | 17 (19/18) | 10 (11/10) |
| Beta subdivision | 18 (74/57) | 10 (59/44) | 8 (15/13) | 0 |
| Gamma subdivision | 9 (33/22) | 3 (25/15) | 3 (3/3) | 3 (4/4) |
| Holophaga-Acidobacterium | 19 (48/42) | 8 (37/31) | 3 (3/3) | 8 (8/8) |
| Actinobacteria | 6 (9/8) | 1 (2/2) | 3 (3/3) | 2 (4/3) |
| Planctomycetales | 4 (7/6) | 2 (5/4) | 1 (1/1) | 1 (1/1) |
| Candidate division OP10 | 4 (6/6) | 0 | 3 (4/4) | 1 (2/2) |
| Plastids | 1 (1/1) | 0 | 0 | 1 (1/1) |
| Low G+C gram positives | 2 (2/2) | 0 | 0 | 2 (2/2) |
| Cytophagales | 1 (1/1) | 0 | 0 | 1 (1/1) |
| Not affiliated (WPS-1) | 4 (8/7) | 1 (5/4) | 0 | 3 (3/3) |
| Total | 108 (285/230) | 38 (200/150) | 38 (48/45) | 32 (37/35) |
Phylogenetic group: a clone group with sequence similarity higher than 97% (98% for Burkholderia and Pseudomonas-type clones). Values in parentheses indicate the number of clones/cloned sequence types.
Certain cloned sequence types, almost exclusively represented by a single sequence type, were only found in the library prepared from diluted RNA, including clone sequence types affiliated with the low-G+C-content gram-positive bacteria, plastids, and the Cytophaga-Flavobacterium-Bacteroides. Dilution of the template RNA also had an effect on the abundance of certain sequence types in the resulting libraries. For example, sequence types related to the 16S rDNA sequence of Nevskia ramosa were represented by 5 clones in the library from undiluted RNA and by 18 clones in the library from diluted RNA.
Analysis of the community composition by FISH.
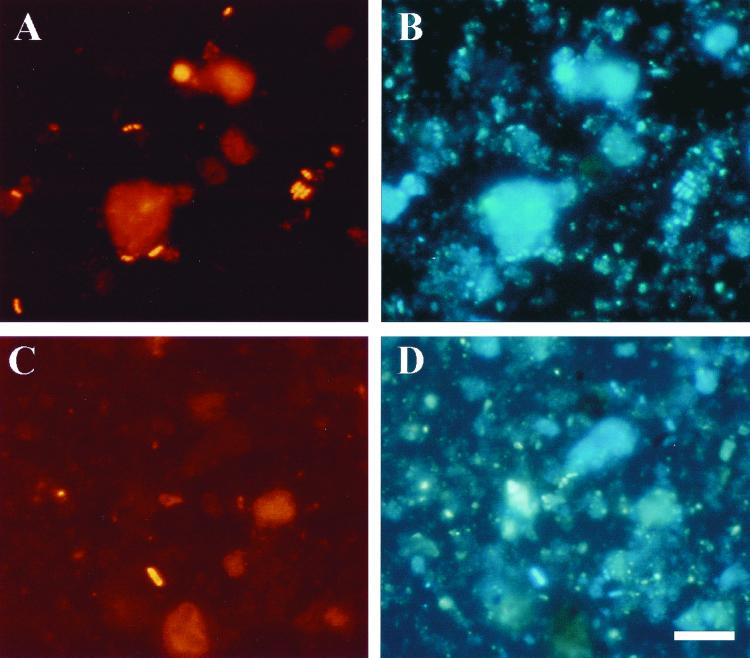
In order to confirm the high abundance of bacteria related to the beta subclass of the Proteobacteria, and Burkholderia spp. in particular, in the bacterial community of the Wittenberg PCB-polluted soil, fluorescence in situ hybridization (FISH) analysis with specific oligonucleotide probes was carried out. The same soil sample employed for the construction of the clone libraries was used for the FISH experiments. High background fluorescence was observed with Wittenberg soil samples due to nonspecific binding of the fluorescently labeled probes. The problem was partially solved by introducing a prehybridization treatment with blocking reagent (see Materials and Methods for details) prior to the addition of the fluorescently labeled probe, which resulted in lower background levels, though this made the counting of DAPI-stained cells more difficult. Total cell counts, determined by DAPI staining of the soil sample, were 1.3 × 109 cells g of soil−1, values which were in accordance with what has been reported previously for Wittenberg soil (35). About 66% of the DAPI-stained cells hybridized with the EUB338 probe for Bacteria and most exhibited strong fluorescent signals (Fig. 4). The abundance of members of the alpha subclass of the Proteobacteria was comparable to that of the beta subclass, with 4.2 and 5.3% of the total DAPI-stained cells hybridizing with probes ALF1b and BET42a, respectively. Some 3.4% of the DAPI-stained cells hybridized with the probe SUBU1237 for Burkholderia and Suterella spp. (31), which represented almost 64% of the bacteria belonging to the beta subclass of the Proteobacteria detected (Fig. 4). These proportions are consistent with those obtained from the clone libraries, which indicated that bacteria affiliated to the genus Burkholderia are abundant and most probably metabolically active in this soil. The PLA886 probe hybridized with spherical cells, whose morphology is that of Isosphaera pallida (9), the closest cultivated relative of the planctomycete cloned sequence types found in Wittenberg soil libraries.
FIG. 4.
FISH of a PCB-polluted soil sample from Wittenberg. (A) Hybridization with probe EUB338, specific for Bacteria. (B) Identical microscopic field for DAPI staining. (C and D) Identical microscopic fields showing results of hybridization with probe SUBU1237 specific for Burkholderia and Suterella spp. (C) and of staining with DAPI (D). Bar, 10 μm (applies to all panels).
DISCUSSION
A knowledge of the diversity of microbial communities inhabiting polluted environments is useful since it provides clues about the type of bacteria able to adapt to and to exploit such habitats. In this study, the bacterial community in a soil highly polluted with PCBs has been analyzed by cloning and sequencing 16S rDNA amplified from total DNA extracted from the soil, and the results obtained have been compared with those of two 16S rRNA clone libraries, which are assumed to better reflect the metabolically active bacteria. Despite the inherent methodological differences in the generation of the PCR and RT-PCR amplification products used for cloning, a good correspondence was observed, with both types of libraries exhibiting similar bacterial community compositions, in terms of the major constituents. A considerable number of identical cloned sequences were observed in all libraries. Phylogenetic groups represented in both types of libraries also contained the majority of the cloned sequence types observed. The results confirmed the abundance of sequence types related to the beta subclass of the Proteobacteria, and the predominance of sequence types related to Burkholderia, Variovorax paradoxus, Xylophilus ampelinus, Nevskia ramosa, Sphingomonas sp., members of the Rhodopila globiformis phylogenetic branch, and Acidobacterium capsulatum. These results support the potential functional importance of these bacterial groups in PCB-polluted Wittenberg soil. There were also some differences in the composition of the rDNA and rRNA libraries. For example, some phylogenetic groups were found only in one or the other of the rDNA or rRNA clone libraries (although they were mostly represented by one or two cloned sequence types), and cloned sequence types affiliated with the Actinobacteria, CFB, and low-G+C-content gram positives were observed only in the rRNA libraries.
In a study carried out with freshwater sediment samples, Miskin et al. (20) were unable to compare the results of the rDNA and rRNA libraries because of the low number of identical sequences observed and the low degree of coverage of the diversity present in the sediment that was reflected in the clone libraries. Although, as shown by the rarefaction plots, the diversity in Wittenberg soil was considerable, the coverage values for Wittenberg PCB-polluted soil libraries were high (33.6% for rDNA and 29% for rRNA, calculated as described previously [20]) in contrast to values of 4 and 6% in the freshwater sediment (20). This might be due to the high abundance of certain sequence types in Wittenberg clone libraries (such as Burkholderia) and to the higher number of clones analyzed in our study.
It is important to point out that the results presented here are merely indicative and not definitive. Determination of the significance of the observed similarities and differences between the rDNA and the rRNA libraries from this PCB-polluted soil will require more-comprehensive studies involving statistical analysis. Some of the differences observed in rDNA and rRNA libraries might be explained with regard to biology and/or methodology, however. The presence of single sequence types exclusively in the rDNA library could, theoretically, be indicative of quiescent bacteria present in the sample. The contrary situation, namely, the presence of rare cloned sequence types exclusively in the 16S rRNA library, is unexpected since the cells (active or not) contain both RNA and DNA. Several methodological factors might have contributed to these differences between the rDNA and rRNA libraries. First, clone selection from a complex collection is a random event, which may result in the selection of a rare clone and distort comparisons of different clone libraries. Therefore, comparisons should not be based on rare clones that appear only once or twice in the libraries. This aspect emphasizes once more the frequently discussed issue of how representative are clone libraries of the high bacterial diversity in soils and how many clones should be screened in order to obtain a representative picture of the composition of the bacterial soil community. Second, there was a slight difference in the specificity of the reverse primers used for the PCR and the RT-PCR (primer 1492R has a narrower specificity than primer 518R [14]). The rationale behind the use of primer 518R for the RT-PCRs instead of primer 1492R was to minimize bias caused by early termination of the RT at modified bases in some 16S rRNA molecules (a potential bias for RT-PCR amplification from rRNA), as was shown by Weller et al. (36, 37). Finally, a third factor might be different starting concentrations of the different templates in the amplification reactions (since this will depend on the abundances in the environment of each of the different bacterial populations, the number of rRNA genes, and the ribosomal content per cell), which would result in bias in the proportion of different PCR amplicons (33).
Despite these uncertainities in comparing the libraries, the combined use of rDNA and rRNA to analyze the bacterial community in this PCB-polluted soil has resulted in an expanded view of the bacterial diversity in this soil. The results from the rRNA libraries allowed us to identify bacteria which were presumably metabolically active and therefore responsible for the functionality of the community in this polluted soil. On the other hand, results from the rDNA library have allowed us to identify other members of the community, as well as enabling more precise phylogenetic assignment of the cloned sequences, which in the case of rRNA clones is limited by the short length of the amplified product (17).
Data obtained from the PCB-polluted Wittenberg soil confirmed the presence of sequence types affiliated with the proposed candidate division OP10 (11), which appear to be related to sequence types retrieved from a trichlorobenzene-transforming consortium (34). Since these sequence types were observed in both rRNA and rDNA libraries, there is strong evidence for the metabolic activity of the bacteria represented by these sequences in the PCB-polluted soil. However, because no cultured representatives of candidate division OP10 have been described, the metabolic capabilities of bacteria within this division remain unknown. Our results also show the presence of two putative new bacterial lineages in Wittenberg soil, one of which, WPS-1, appeared to be phylogenetically related to the Planctomycetales and was represented by several cloned sequence types in both rRNA and rDNA libraries. The phylogenetic position of the second novel bacterial lineage, WPS-2, remains unclear (the analysis had to be based in a unique cloned sequence since no other representatives of this phylogenetic group were found in the clone libraries).
The main feature of the bacterial community in this highly PCB-polluted soil, compared with other soil communities, was the abundance of a low-diversity set of sequence types affiliated to the beta subclass of the Proteobacteria, mostly related to the genus Burkholderia. FISH analysis indicated abundances of approximately 4.3 × 107 cells hybridizing with the probe SUBU1237 per g of soil, and a strong fluorescent signal was observed for the cells hybridizing with this probe (as shown in Fig. 4). FISH results confirmed the abundance and thus presumably the high activity of bacteria of the genus Burkholderia in the PCB-polluted soil. Also consistent with this conclusion is the high number of Burkholderia isolates obtained from this Wittenberg site samples (W. R. Abraham et al., unpublished data). Although representatives of this genus are frequently found in the rhizosphere, they are not usually abundant in bulk soil (8, 13). The only publication reporting abundant Burkholderia-related (and Janthinobacterium-related) sequences in soil is one of a clone library obtained from an acidic thermal soil in New Zealand (15). In that case, the authors speculated on the possibility that these cloned sequences might represent either inactive or nonviable cells, since these bacteria are considered to be mesophilic and characteristic of neutral pH environments. The detection here of abundant, presumably metabolically active, Burkholderia-type cells in a low-pH PCB-polluted soil attests to their ability to flourish in acidic soils. A high abundance of Burkholderia has also been reported in a bacterial community degrading aromatic hydrocarbons in a trickle-bed bioreactor (31).
Finally, comparison of the two rRNA libraries obtained from undiluted template and 1:500-diluted template revealed discrepancies previously observed (4) of cloned sequence types appearing only in the library obtained from diluted RNA. These differences may be attributed to PCR kinetics bias, which results in the early inhibition of amplification of abundant templates, while less-abundant templates continue to be amplified (33). The effects of template dilution seemed to be complex, affecting both the occurrence and the abundance of sequence types, and biased according to sequence types (i.e., sequence types affiliated with the gamma subclass of the Proteobacteria and with the Holophaga-Acidobacterium phylum were characteristically more numerous in the library from diluted RNA). However, as comparison of the rDNA and rRNA libraries revealed, both rRNA libraries reflected the major phylogenetic groups and sequence types representing the bacterial community in the PCB-polluted soil. Therefore, dilution of the template prior to amplification by PCR, which is frequently necessary after nucleic acid extraction from soil environments, does not seem to compromise assessment of the bacterial diversity in the sample (at least for not very low template concentrations). On the other hand, the abundance in both rRNA libraries of related 16S rRNA sequence groups is consistent with the predominance of certain bacterial populations in this polluted soil, bacteria related to Sphingomonas, Burkholderia, Nevskia, and Acidobacterium spp., among others.
ACKNOWLEDGMENTS
This work was supported by a grant from the German Ministry for Education and Research (project no. 0319433C). During part of the work, B.N. was the recipient of a postdoctoral fellowship from the Spanish Ministry for Education and Culture. K.N.T. acknowledges the support of the Fonds der Chemischen Industrie.
We thank Annette Krüger and Carsten Strömpl for their excellent sequencing work. B.N. acknowledges A. M. Osborn for many late-hour fruitful discussions and Frank-Oliver Glöckner for kindly sharing his expertise in the use of the ARB package. We also thank Steve M. Holland at the University of Georgia for the rarefaction program Analytic Rarefaction 1.2.
REFERENCES
- 1.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker W, van den Broek A, Camon E, Hingamp P, Sterk P, Stoesser G, Tuli M A. The EMBL nucleotide sequence database. Nucleic Acids Res. 2000;28:19–23. doi: 10.1093/nar/28.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begon M, Harper J L, Townsend C R. Ecology: individuals, population and community. Oxford, England: Blackwell Scientific Publications; 1986. The nature of the community; pp. 591–628. [Google Scholar]
- 4.Chandler D P, Fredrickson J K, Brockman F J. Effect of the PCR template concentration on the composition and distribution of total community 16S rDNA clone libraries. Mol Ecol. 1997;6:475–482. doi: 10.1046/j.1365-294x.1997.00205.x. [DOI] [PubMed] [Google Scholar]
- 5.Dojka M A, Hugenholtz P, Haak S K, Pace N R. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards K J, Goebel B M, Rodgers T M, Schrenk M O, Gihring T M, Cardona M M, Hu B, McGuire M M, Hamers R J, Pace N R, Banfield J F. Geomicrobiology of pyrite (FeS2) dissolution: case study at Iron Mountain, California. Geomicrobiol J. 1999;16:155–179. [Google Scholar]
- 7.Felske A, Rheims H, Wolterink A, Stackebrandt E, Akkermans A D L. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology. 1997;143:2983–2989. doi: 10.1099/00221287-143-9-2983. [DOI] [PubMed] [Google Scholar]
- 8.Felske A, Wolterink A, van Lis R, Akkermans A D L. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands) Appl Environ Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giovannoni S J, Schabtach E, Castenholz R W. Isosphaera pallida, gen. and comb. nov., a gliding, budding eubacterium from hot springs. Arch Microbiol. 1987;147:276–284. doi: 10.1128/jb.169.6.2702-2707.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Head I, Saunders J R, Pickup R W. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microbiol Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 11.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H M, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 13.Kuske C R, Barns S M, Busch J D. Diverse uncultivated bacterial groups from soils of the arid Southwestern United States that are present in many geographic regions. Appl Environ Microbiol. 1997;63:3614–3621. doi: 10.1128/aem.63.9.3614-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley & Sons, Ltd.; 1991. pp. 115–175. [Google Scholar]
- 15.Liesack W, Janssen P H, Rainey F A, Ward-Rainey N, Stackebrandt E. Microbial diversity in soil: the need for a combined approach using molecular and cultivation techniques. In: van Elsas J D, Trevors J T, Wellington E M H, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker, Inc; 1997. pp. 375–439. [Google Scholar]
- 16.Ludwig W, Bauer S H, Bauer M, Held I, Kirchof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer K-H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 17.Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M, Schleifer K-H. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- 18.Maidak B L, Cole J R, Lilburn T G, Parker C T, Jr, Saxman P R, Stredwick J M, Garrity G M, Li B, Olsen G J, Pramanik S, Schmidt T M, Tiedje J M. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 2000;28:173–174. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 20.Miskin I P, Farrimond P, Head I M. Identification of novel lineages as active members of microbial populations in a freshwater sediment using a rapid RNA extraction procedure and RT-PCR. Microbiology. 1999;145:1977–1987. doi: 10.1099/13500872-145-8-1977. [DOI] [PubMed] [Google Scholar]
- 21.Neef A, Amann R, Schlesner H, Schleifer K-H. Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology. 1998;144:3257–3266. doi: 10.1099/00221287-144-12-3257. [DOI] [PubMed] [Google Scholar]
- 22.Nogales B, Moore E R B, Abraham W-R, Timmis K N. Identification of the metabolically-active members of a bacterial community in a polychlorinated biphenyl-polluted moorland soil. Environ Microbiol. 1999;1:199–212. doi: 10.1046/j.1462-2920.1999.00024.x. [DOI] [PubMed] [Google Scholar]
- 23.Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 24.Nüsslein K, Tiedje J M. Soil bacterial community shift correlated with change from forest to pasture vegetation in a tropical soil. Appl Environ Microbiol. 1999;65:3622–3626. doi: 10.1128/aem.65.8.3622-3626.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radajewski S, Ineson P, Parekh N R, Murrell J C. Stable-isotope probing as a tool in microbial ecology. Nature. 2000;403:646–649. doi: 10.1038/35001054. [DOI] [PubMed] [Google Scholar]
- 27.Rheims H, Spröer C, Rainey F A, Stackebrandt E. Molecular biological evidence for the occurrence of uncultured members of the actinomycete line of descent in different environments and geographical locations. Microbiology. 1996;142:2863–2870. doi: 10.1099/13500872-142-10-2863. [DOI] [PubMed] [Google Scholar]
- 28.Roller C, Wagner M, Amann R I, Ludwig W, Schleifer K-H. In situ probing of gram positive bacteria with high DNA G+C content using 23S rRNA-targeted oliogonucleotides. Microbiology. 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- 29.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K-H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stackebrandt E, Murray R G E, Trüper H G. Proteobacteria classis nov., a name for the phylogenetic taxon that include the ‘purple bacteria and their relatives’. Int J Syst Bacteriol. 1988;38:321–325. [Google Scholar]
- 31.Stoffels M, Amann R, Ludwig W, Hekmat D, Schleifer K-H. Bacterial community dynamics during start-up of a trickle-bed bioreactor degrading aromatic compounds. Appl Environ Microbiol. 1998;64:930–939. doi: 10.1128/aem.64.3.930-939.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strunk O, Gross O, Reichel B, May M, Hermann S, Stuckmann N, Nonhoff B, Lenke M, Ginhart A, Vilbig A, Ludwig T, Bode A, Schleifer K-H, Ludwig W. ARB: a software environment for sequence data. Munich, Germany: Department of Microbiology, Technische Universität München; 1998. [Google Scholar]
- 33.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Wintzingerode F, Selent B, Hegeman W, Göbel U B. Phylogenetic analysis of an anareobic, trichlorobenzene-transforming microbial consortium. Appl Environ Microbiol. 1999;65:283–286. doi: 10.1128/aem.65.1.283-286.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinbauer M G, Beckmann C, Höfle M G. Utility of green fluorescent nucleic acid dyes and aluminum oxide membrane filters for rapid epifluorescence enumeration of soil and sediment bacteria. Appl Environ Microbiol. 1998;64:5000–5003. doi: 10.1128/aem.64.12.5000-5003.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weller R, Ward D M. Selective recovery of 16S rRNA sequences from natural microbial communities in the form of cDNA. Appl Environ Microbiol. 1989;55:1818–1822. doi: 10.1128/aem.55.7.1818-1822.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weller R, Weller J W, Ward D M. 16S rRNA sequences of uncultivated hot spring cyanobacterial mat inhabitants retrieved as randomly primed cDNA. Appl Environ Microbiol. 1991;57:1146–1151. doi: 10.1128/aem.57.4.1146-1151.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]