Abstract

Small molecular weight probes that can show a fluorescence signaling response upon binding to RNAs are promising for RNA imaging in living cells. Live-cell RNA imaging probes that can achieve a large light-up ability (>100-fold) and high Φbound value for RNA (>0.50) have been rarely reported to date. Here, benzo[c,d]indole-oxazolopyridine (BIOP), an unsymmetrical monomethine cyanine analogue, was newly developed as a bright and large light-up probe for imaging of nucleolar RNA in living cells. BIOP served as a yellow-emissive probe (λem = 570 nm) and exhibited a significant light-up response upon RNA binding (770-fold) with a high Φbound value (0.52). We demonstrated the advantages of BIOP over a commercially available RNA-staining probe, SYTO RNA select, for robust and sensitive RNA sensing by a systematic comparison of fluorescent properties for RNA. In addition, BIOP was found to possess high membrane permeability and low cytotoxicity in living cells. The examination of live-cell imaging revealed that BIOP exhibited emission in the nucleolus upon binding to nucleolar RNA much stronger than that of SYTO RNA select. Furthermore, BIOP facilitated the highly sensitive imaging of nucleolar RNA, in which 50 nM BIOP can stain nucleolar RNA in living cells with a 20 min incubation.
Introduction
With increasing knowledge about the diverse roles of RNA within cells, much attention has been paid to RNA-binding fluorescent probes as powerful and versatile tools for the study of RNA functions.1 In comparison to protein-based2 and oligonucleotide-based probes,3 small molecular weight probes for RNA analysis have been less developed despite their versatility and easy-to-use properties.4 This is mainly because this class of probes usually shows the binding preference for DNA over RNA.5 SYTO RNA select is one of the commercially available probes with RNA selectivity. While SYTO RNA is almost nonfluorescent in solution, its emission is greatly enhanced by more than 100-fold upon binding to RNA. Such a light-up ability renders this probe useful as a sensitive RNA-sensing probe.6 On the other hand, the usefulness of SYTO RNA select as an RNA-imaging probe in cells has been severely limited. This probe has low photostability4,7 and a relatively short emission wavelength (λem = 530 nm) that suffers interference from cellular autofluorescence. Moreover, the low cellular permeability of SYTO RNA select is also problematic, resulting in less applicability toward the RNA imaging in living cells.8
Considerable work has been done in order to overcome these drawbacks of SYTO RNA select, resulting in the realization of improved photostability, long emission wavelength, and compatibility with live-cell imaging.9 Several probes were shown to be applicable to the staining of nucleolar RNA in living cells, offering useful tools for the analysis of nucleolar RNA dynamics.9a9b On the other hand, less attention has been paid to the brightness and light-up property of this class of probes despite the fact that these factors are crucial for achieving sensitive imaging. Indeed, live-cell RNA-imaging probes to meet the large light-up ability (>100-fold) and high Φbound value for RNA (>0.50) have been rarely reported to date. In this context, Wang et al. recently developed a new styryl dye-based green-emissive probe (λem = 511 nm), PI2, that exhibited a large light-up response and high Φbound for RNA (Φfree = 0.0039, Φbound = 0.577).10 However, a high concentration of PI2 (5.0 μM) was required for live-cell imaging experiments, presumably due to its low permeability in living cells. It was reported that the viability of 3T3 cells was decreased to 70–75% with an incubation concentration of 5–20 μM P12 for 24 h.10 Our group designed unsymmetrical monomethine cyanine-based probes with a distinct light-up switch for RNA.11 Benzo[c,d]indole-quinoline (BIQ) showed a significant enhancement of the emission by more than 100-fold upon binding to RNA,11a whose fluorogenic “off–on” ability in deep-red region (λem > 650 nm) is outstanding among small probes for live-cell RNA imaging. We also demonstrated nucleolar RNA staining in living MCF7 cells even with 500 nM of the BIQ derivative with improved binding affinity.11b However, BIQ analogues have a relatively moderate fluorescence quantum yield in the bound state with RNA (Φbound < 0.02), resulting in low brightness of the probe in live-cell imaging.
In this work, we report on a new benzo[c,d]indole-containing monomethine cyanine with improved brightness and light-up properties for RNA sensing. We focused on the oxazolopyridine heterocycle for the probe design because cyanine dyes containing this heterocycle exhibited a large fluorescence quantum yield in the bound state with nucleic acids.12 Here, the oxazolo[5,4-c]pyridine heterocycle was conjugated with a benzo[c,d]indole unit (Scheme 1), yielding the benzo[c,d]indole-oxazolopyridine (BIOP) probe. BIOP works as a yellow-emissive probe (λem = 570 nm) and exhibits a significant light-up response upon RNA binding (770-fold) with a high Φbound value (0.52). Based on such promising signaling functions, we demonstrate the potential of BIOP for nucleolar RNA imaging in living cells.
Scheme 1. Synthesis of Benzo[c,d]indole-oxazolo[5,4-c]pyridine (BIOP).
Reagents and conditions: (a) potassium ethyl xanthate, EtOH, reflux, 93%; (b) iodomethane, DMF, 61%; (c) methyl p-toluenesulfonate, 130 °C, 28%; (d) 1,2-dimethylbenzo[c,d]indol-1-ium iodide (3), NEt3, MeCN, 40 °C, 14%.
Results and Discussion
Synthesis of BIOP
For the synthesis of BIOP (Scheme 1), 5-methyl-2-(methylthio)oxazolo[5,4-c]pyridin-5-ium tosylate (2) was first prepared in three steps from the commercially available 4-aminopyridin-3-ol (1) (Scheme S1).13,14 Compound 2 was then combined with the benzo[c,d]indole unit (3)11,15 by a condensation reaction, which affords BIOP (purity >95%) as characterized by 1H NMR and ESI-MS (Figures S1 and S2). Further details of the synthesis are shown in the Supporting Information.
Fluorescence Response of BIOP for RNA
First, we investigated the fluorescence response of BIOP (1.0 μM) to calf thymus DNA orEscherichia coli (E. coli) total RNA, as these nucleic acids from biological species were utilized in many studies for RNA-targeting fluorescent probes.9 Measurements were done at 25 °C in a 10 mM sodium phosphate buffer (pH 7.0). As shown in Figure 1, we observed negligible emission in the absence of nucleic acid (fluorescence quantum yield (Φfree) = 0.00038). This can be explained by the nonradiative energy loss through free rotation around the monomethine linker between corresponding heterocycles, as typically observed for various monomethine cyanine dyes.16 The addition of E. coli total RNA (1.0 mM per nucleotide) resulted in a significant fluorescence enhancement of BIOP in the yellow spectral range (λem = 570 nm). We observed a hyperchromicity and a red shift of the absorption band of BIOP when bound to RNA (Figure S3), which suggests the intercalative binding mode of BIOP for RNA. Thus, the observed light-up response would arise from the suppressed rotation upon intercalation into the base pairs in the RNA.16,17 The light-up factor (I/I0, where I and I0 denote the fluorescence intensities in the presence and absence of a target, respectively) of BIOP is determined to be 770-fold. Such a large light-up property is well reflected in the remarkable increase in the fluorescence quantum yield upon binding to RNA. Significantly, the fluorescence quantum yield of BIOP in the bound state (Φbound) was determined to be 0.52. This value is 1 order of magnitude larger compared to that of BIQ derivatives that were previously developed by us.11 The brightness, defined as the product of the fluorescence quantum yield and molecular extinction coefficient, reaches 1.1 × 104 for BIOP bound to RNA (Table 1). Apparently, the integration of the oxazolopyridine unit into the monomethine cyanine scaffold leads to large Φbound value for RNA binding. In addition, the response for RNA is larger than that for DNA (Figure 1), which indicates the selective response of BIOP toward RNA like BIQ derivatives.11
Figure 1.
Fluorescence spectra of BIOP (1.0 μM) in the absence (a) and presence of (b) 1.0 mM E. coli total RNA or (c) 1.0 mM ctDNA. Measurements were done in 10 mM sodium phosphate buffer solution (pH 7.0) containing 100 mM NaCl and 1.0 mM EDTA. Excitation: 530 nm. Temperature: 25 °C.
Table 1. Photophysical Properties and Fluorescence Signaling Ability for RNA Sensing.
| BIOP | SYTO RNA select | |
|---|---|---|
| emission wavelengtha | 570 nm | 530 nm |
| light-up factorb | 770 | 500 |
| Φboundb | 0.52 | 0.36 |
| εabs,maxa | 2.2 × 104 cm–1 M–1 | 3.9 × 104 cm–1 M–1 |
| brightness (εabs, max × Φbound) | 1.1 × 104 | 1.4 × 104 |
| photostabilityb | high | low |
Values obtained in 10 mM sodium phosphate buffer solution (pH 7.0) containing 100 mM NaCl and 1.0 mM EDTA.
[Probe] = 1.0 μM, [E. coli total RNA] = 1.0 mM.
In order to gain further insights into binding of BIOP to RNA, we examined the fluorescence response for synthetic double-stranded (ds) and single-stranded (ss) RNA (Figure S4). The fluorescence intensity was much enhanced for rG/rC and rA/rU compared to that of rC, rA, and rU. It is thus highly likely that BIOP binding arises from the intercalation into the base pairs in the RNA.16,17 The comparison with synthetic DNA reveals the selective response for RNA, which is consistent with the results for biological nucleic acids (cf. Figure 1). We note that BIOP displayed a strong light-up response for both rG and dG, presumably due to the possible binding to G-quadruplex structures similar to various monomethine cyanines.11,18 We then estimate the binding affinity of BIOP for the rG/rC duplex by fluorescence titration experiments (Figure S5). The apparent dissociation constant (Kd) was determined to be 8.1 ± 1.6 μM (N = 3). The binding affinity of BIOP is stronger than BIQ having a quinoline counterpart (Kd > 22 μM),11a indicating that the use of oxazolo[5,4-c]pyridine is also advantageous for the enhanced binding affinity.
Comparison with SYTO RNA Select
Here, the response properties of BIOP were directly compared with those of commercially available SYTO RNA select for E. coli total RNA under the identical conditions (cf. Figure 1). We found several advantages of BIOP over SYTO RNA select (Figure S6 and Table 1). Taken together with the longer emission wavelength compared to that of SYTO RNA select, both light-up properties and Φbound values for BIOP are superior by more than 1.4-fold compared to those of SYTO RNA select (I/I0 = 500-fold and Φbound = 0.36). These results indicate more favorable fluorescence signaling ability of BIOP over SYTO RNA select for RNA sensing. In addition, BIOP possesses a stronger resistance to photobleaching than that of SYTO RNA select (Figure S7). The fluorescence intensity of SYTO RNA select bound to RNA decreased by as much as 75% after 160 min continuous irradiation, whereas the decrease in the fluorescence intensity of BIOP was very small (7%). While the clear selectivity for RNA over DNA is a promising function of SYTO RNA select in solutions (Figure S6), a distinct advantage of BIOP can be seen when applied to live-cell imaging.
Fluorescence Imaging of Living Cells
Figure 2A shows the fluorescence image of living MCF7 cells after incubation of 0.5 μM BIOP for 20 min using TRITC filter sets in the fluorescence microscopy. We observed strong emission in the yellow spectral region in the nucleolus and cytoplasm. This indicates good cell plasma and nuclear membrane permeability of BIOP in living cells. Considering the abundance of RNA, such as rRNA, in the nucleolus,4,7−11 it is highly likely that the observed signal in the nucleolus originates from the light-up response of BIOP for nucleolar RNA. Meanwhile, the emission in the cytoplasm was found to be due to the accumulation of BIOP in the mitochondria by examination of a series of organelle-selective staining probes (Figure S8, Pearson’s correlation coefficient: 0.78). This would be due to possible interaction of cationic BIOP with a negative mitochondria membrane, as observed for various cyanine analogues.19 In order to confirm the origin of the emission in the nucleolus, deoxyribonuclease (DNase) and ribonuclease (RNase) digestion experiments were done in fixed and permeabilized MCF7 cells (Figure 2B). BIOP exhibited strong emission in the nucleolus in fixed and permeabilized cells, similar to the case of living cells. The fluorescence signal of BIOP in the nucleolus dramatically diminished in the case of RNase, whereas the signal remained almost unaffected after the DNase treatment. These results showed that BIOP exhibited strong emission by binding to nucleolar RNA. We also noticed the emission disappearance in the mitochonria after the DNase treatment. This result suggests that the emission observed in the mitochondria originates from the binding of BIOP to mitochondrial DNA.20
Figure 2.

(A) Fluorescence image of living MCF7 cells incubated with 0.5 μM BIOP for 20 min. Fluorescence intensity profile along the white line is also shown, where the fluorescence in the nucleolus is highlighted. (B) Fluorescence images of fixed-permeabilized MCF7 cells stained with 0.5 μM BIOP before (control) and after treatment of RNase or DNase. Fluorescence intensity profiles along the white line are also shown. (C) Fluorescence image of living MCF7 cells incubated with 0.5 μM SYTO RNA select for 20 min. Fluorescence intensity profile along the white line is also shown. Scale bar: 15 μm.
The live-cell imaging performance of BIOP for MCF7 cells was then compared with that of SYTO RNA select under the identical measurement conditions ([probe] = 0.5 μM, 20 min incubation). In sharp contrast to BIOP, SYTO RNA select displayed negligible emission in the nucleolus, whereas we observed weak emission in the cytoplasm (Figure 2C). The observed imaging pattern of SYTO RNA select is consistent with the previous reports.8b9c Clearly, BIOP can serve as a more useful probe for the imaging of nucleolar RNA in living cells compared to SYTO RNA select.
Significantly, the excellent imaging properties of BIOP enable the staining of nucleolar RNA even at a concentration as low as 50 nM (Figure 3). The emission in the nucleolus is clearly observed and can be distinguishable from that in the nucleus. Several fluorescent probes have been so far shown to possess an imaging performance for nucleolar RNA in living cells better than that with SYTO RNA select;7−11,21 however, to the best of our knowledge, nucleolar RNA imaging has never been accomplished at a 50 nM probe concentration. Thus, BIOP can work as the most sensitive imaging probe for nucleolar RNA in living cells. The high RNA-sensing ability of BIOP would contribute to this improved imaging sensitivity of nucleolar RNA. In addition, it is highly likely that the high membrane permeability of BIOP also provides a better imaging performance. Low probe concentration should be beneficial as it can greatly reduce the damage to the biological systems inside the cells.8a,22
Figure 3.
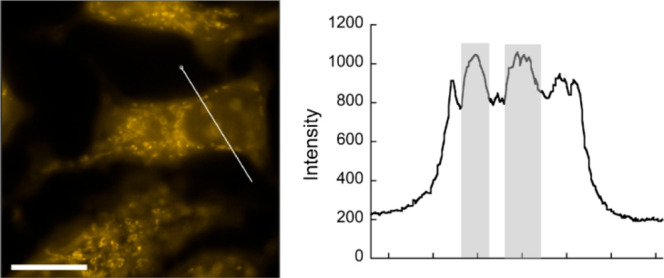
Fluorescence image of living MCF7 cells incubated with 50 nM BIOP for 20 min. Fluorescence intensity profile along the white line is also shown, where the fluorescence in the nucleolus is highlighted. Scale bar: 15 μm.
Cytotoxicity of live-cell imaging probes is also an important factor for the practical use toward accurate analysis of living cells. We assessed the cytotoxicity of BIOP after 20 min incubation in living MCF7 cells with the Alamar blue assay (Figure S9A). Negligible cytotoxicity was found for the BIOP concentration below 5.0 μM, which shows that the nucleolar RNA staining with 20 min incubation of BIOP has no influence on the cell viability. In addition, more than 90% cell viability was observed in the case of 24 h incubation of 1.0 μM BIOP (Figure S9B). It is consistent with the observation that BIOP clearly stained the nucleolus without any morphological change after 24 h incubation (Figure S10). BIOP thus holds great potential for the long-term monitoring of nucleolar RNA in living cells, which offers a useful tool for the analysis of nucleolar RNA dynamics during mitosis and apotosis induced by anticancer drugs.9a
It should be also noted that BIOP has a good counterstaining compatibility with the nuclear selective Hoechst 33342, a blue-emissive DNA-staining probe (Figure S11, Pearson’s correlation coefficient: 0.12). As the excitation and emission profiles of BIOP are orthogonal to Hoechst 33342, the combination of these probes allows the simultaneous visualization of DNA and RNA distribution in living cells.7a,9a,9c We also demonstrated the applicability of BIOP to living HeLa cells (Figure S12). As observed for MCF7 cells, BIOP was able to stain the nucleolar RNA under the above condition ([BIOP] = 1.0 μM, 20 min incubation). This shows the versatility of BIOP as a useful imaging probe for nucleolar RNA in living cells.
Conclusions
In summary, benzo[c,d]indole-oxazolopyridine, BIOP, was newly developed as a useful probe for RNA sensing and nucleolar RNA imaging in living cells. BIOP has brighter and larger light-up response for RNA as well as higher photostability compared to that of SYTO RNA select. Highly sensitive imaging of nucleolar RNA in living cells was achieved by virtue of the excellent RNA-sensing properties and high membrane permeability. This sensitivity of BIOP is superior compared to the previously developed probes with better imaging performance for nucleolar RNA in living cells over SYTO RNA select. Moreover, BIOP possesses low cytotoxicity in living cells. While there is some room for improvement of the probe function, such as the Stokes shift9a and nucleolus-targeting ability, we believe that BIOP can be the most useful candidate for practical use toward spatiotemporal analysis of nucleolar RNA in living cells.
Acknowledgments
This work was supported by Grant-in-Aid for Challenging Research (Pioneering) (No. 21K18207) and for Young Scientists (A) (No. 17H04881) from the Japan Society for the Promotion of Science (JSPS).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c02408.
Experimental details, absorption spectra, fluorescence response, photostability, live-cell imaging, cytotoxicity of BIOP (PDF)
Author Contributions
K.H. and Y.S. conceived the study. K.H. and M.S. designed and performed all experiments with help from N.T. and Y.Y. K.H., Y.S., and S.N. wrote the manuscript. Y.S. and S.N. supervised the research.
The authors declare no competing financial interest.
Supplementary Material
References
- Suseela Y. V.; Narayanaswamy N.; Pratihar S.; Govindaraju T. Far-red fluorescent probes for canonical and no-canonical nucleic acid structures: current progress and future implications. Chem. Soc. Rev. 2018, 47, 1098–1131. 10.1039/C7CS00774D. [DOI] [PubMed] [Google Scholar]
- Rath A. K.; Rentmeister A. Genetically encoded tools for RNA imaging in living cells. Curr. Opin. Biotechnol 2015, 31, 42–49. 10.1016/j.copbio.2014.07.012. [DOI] [PubMed] [Google Scholar]
- Tomoike F.; Abe H. RNA imaging by chemical probes. Adv. Drug Delivery Rev. 2019, 147, 44–58. 10.1016/j.addr.2019.08.001. [DOI] [PubMed] [Google Scholar]
- Song G.; Sun Y.; Liu Y.; Wang X.; Chen M.; Miao F.; Zhang W.; Yu X.; Jin J. Low molecular weight fluorescent probes with good photostability for imaging RNA-rich nucleolus and RNA in cytoplasm in living cells. Biomaterials 2014, 35, 2103–2112. 10.1016/j.biomaterials.2013.11.052. [DOI] [PubMed] [Google Scholar]
- a Wilson W. D.; Ratmeyer L.; Zhao M.; Strekowski L.; Boykin D. The search for structure-specific nucleic acid-interactive drugs: Effects of compound structure on RNA versus DNA interaction strength. Biochemistry 1993, 32, 4098–4104. 10.1021/bi00066a035. [DOI] [PubMed] [Google Scholar]; b Sato Y.; Ichihashi T.; Nishizawa S.; Teramae N. Strong and selective binding of amiloride to an abasic site in RNA duplexes: Thermodynamic characterization and microRNA detection. Angew. Chem. Int. Ed 2012, 51, 6369–6372. 10.1002/anie.201201790. [DOI] [PubMed] [Google Scholar]
- a Wu Y.; Liu Y.; Lu C.; Lei S.; Li J.; Du G. Quantification of RNA by a fluorometric method using the SYTO RNA Select stain. Anal. Biochem. 2020, 606, 113857. 10.1016/j.ab.2020.113857. [DOI] [PubMed] [Google Scholar]; b de Voogt W. S.; Tanenbaum M. E.; Vader P. Illuminating RNA trafficking and functional delivery by extracellular vesicles. Adv. Drug Delivery Rev. 2021, 174, 250–264. 10.1016/j.addr.2021.04.017. [DOI] [PubMed] [Google Scholar]
- a Lu Y.-J.; Deng Q.; Hu D.-P.; Wang Z.-Y.; Huang B.-H.; Du Z.-Y.; Fang Y.-X.; Wong W.-L.; Zhang K.; Chow C.-F. A molecular fluorescent dye for specific staining and imaging of RNA in live cells: A novel ligand integration from classical thiazole orange and styryl compounds. Chem. Commun. 2015, 51, 15241–15244. 10.1039/C5CC05551B. [DOI] [PubMed] [Google Scholar]; b Liu J.; Zhang S.; Zhang C.; Dong J.; Shen C.; Zhu; Xu H.; Fu M.; Yang G.; Zhang X. A water-soluble two-photon ratiometric triarylboron probe with nucleolar targeting by preferential RNA binding. Chem. Commun. 2017, 53, 11476–11479. 10.1039/C7CC06045A. [DOI] [PubMed] [Google Scholar]
- a Li Q.; Kim Y.; Namm J.; Kulkarni A.; Rosania G. R.; Ahn Y.-H.; Chang Y.-T. RNA-selective, live cell imaging probes for studying nuclear structure and function. Chem. Biol. 2006, 13, 615–623. 10.1016/j.chembiol.2006.04.007. [DOI] [PubMed] [Google Scholar]; b Cao C.; Wei P.; Li R.; Zhong Y.; Li X.; Xue F.; Shi Y.; Yi T. Ribosomal RNA-selective light-up fluorescent probe for rapidly imaging the nucleolus in live cells. ACS Sens 2019, 4, 1409–1416. 10.1021/acssensors.9b00464. [DOI] [PubMed] [Google Scholar]
- a Zhou B.; Liu W.; Zhang H.; Wu J.; Liu S.; Xu H.; Wang P. Imaging nucleolar RNA in living cells using a highly photostable deep-red fluorescent probe. Biosens. Bioelectron 2015, 68, 189–196. 10.1016/j.bios.2014.12.055. [DOI] [PubMed] [Google Scholar]; b Yao Q.; Li H.; Xian L.; Xu F.; Xia J.; Fan J.; Du J.; Wang J.; Peng X. Differentiating RNA from DNA by a molecular fluorescent probe based on the “door-bolt” mechanism biomaterials. Biomaterials 2018, 177, 78–87. 10.1016/j.biomaterials.2018.05.050. [DOI] [PubMed] [Google Scholar]; c Guo L.; Chan M. S.; Xu D.; Tam D. Y.; Bolze F.; Lo P. K.; Wong M. S. Indole-based cyanine as a nuclear RNA-selective two-photon fluorescent probe for live cell imaging. ACS Chem. Biol. 2015, 10, 1171–1175. 10.1021/cb500927r. [DOI] [PubMed] [Google Scholar]
- Wang C.; Lu Y.-J.; Cai S.-Y.; Long W.; Zheng Y.-Y.; Lin J.-W.; Yan Y.; Huang X.-H.; Wong W.-L.; Zhang K.; Chow C. F. Advancing small ligands targeting RNA for better binding affinity and specificity: A study of structural influence through molecular design approach. Sens. Actuators B 2018, 262, 386–394. 10.1016/j.snb.2018.02.004. [DOI] [Google Scholar]
- a Yoshino Y.; Sato Y.; Nishizawa S. Deep-red light-up signaling of benzo[c,d]indole-quinoline monomethine cyanine for imaging of nucleolar RNA in living cells and for sequence-selective RNA analysis. Anal. Chem. 2019, 91, 14254–14260. 10.1021/acs.analchem.9b01997. [DOI] [PubMed] [Google Scholar]; b Sato Y.; Igarashi Y.; Suzuki M.; Higuchi K.; Nishizawa S. Deep-red fluorogenic cyanine dyes carrying an amino-group-terminated side chain for improved RNA detection and nucleolar RNA imaging. RSC Adv. 2021, 11, 35436–35439. 10.1039/D1RA05872J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Kovalska V. B.; Tokar V. P.; Losytskyy M. Y.; Deligeorgiev T.; Vassilev A.; Gadjev N.; Drexhage K. H.; Yarmoluk S. M. Studies of monomeric and homodimeric oxazolo[4,5-b]pyridinium cyanine dyes as fluorescent probes for nucleic acids visualization. J. Biochem. Biophys. Methods 2006, 68, 155–165. 10.1016/j.jbbm.2006.04.006. [DOI] [PubMed] [Google Scholar]; b Hövelmann F.; Gaspar I.; Ephrussi A.; Seitz O. Brightness enhanced DNA FIT-probes for wash-free RNA imaging in tissues. J. Am. Chem. Soc. 2013, 135, 19025–19032. 10.1021/ja410674h. [DOI] [PubMed] [Google Scholar]
- Walczynski K.; Zuiderveld O. P.; Timmerman H. Non-imidazole histamine H3 ligands. Part III. New 4-n-propylpiperazines as non-imidazole histamine H3-antagonists. Eur. J. Med. Chem. 2005, 40, 15–23. 10.1016/j.ejmech.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Zhang S.; Fan J.; Li Z.; Hao N.; Cao J.; Wu T.; Wang J.; Peng X. A bright red fluorescent cyanine dye for live-cell nucleic acid imaging, with high photostability and a large Stokes shift. J. Mater. Chem. B 2014, 2, 2688–2693. 10.1039/C3TB21844A. [DOI] [PubMed] [Google Scholar]
- Deligeorgiev T. G.; Gadjev N. I.; Drexhage K. H. Styryl dyes containing the benz[c,d]indolium hetrocycles. Dyes Pigm 1991, 15, 215–223. 10.1016/0143-7208(91)80006-U. [DOI] [Google Scholar]
- Nygren J.; Svanvik N.; Kubista M. The interactions between the fluorescent dye thiazole orange and DNA. Biopolymers 1998, 46, 39–51. . [DOI] [PubMed] [Google Scholar]
- Yarmoluk S. M.; Lukashov S. S.; Losytskyy M. Y.; Akerman B.; Kornyushyna O. S. Interactions of cyanine dyes with nucleic acids: XXVI. Intercalation of the trimethine cyanine dye cyan 2 into double-stranded DNA: study by spectral luminescence methods. Spectrochim. Acta, Part A 2002, 58, 3223–3232. 10.1016/S1386-1425(02)00100-2. [DOI] [PubMed] [Google Scholar]
- Lubitz I.; Zikich D.; Kotlyar A. Specific high-affinity binding of thiazole orange to triplex and G-quadruplex DNA. Biochemistry 2010, 49, 3567–3574. 10.1021/bi1000849. [DOI] [PubMed] [Google Scholar]
- a Carreon J. R.; Stewart K. M.; Mahon K. P. Jr.; Shin S.; Kelley S. O. Cyanine dye conjugates as probes for live cell imaging. Bioorg. Med. Chem. Lett. 2007, 17, 5182–5185. 10.1016/j.bmcl.2007.06.097. [DOI] [PubMed] [Google Scholar]; b Kurutos A.; Orehovec I.; Saftic D.; Horvat L.; Crnolatac I.; Piantanida I.; Deligeorgiev T. Cell penetrating, mitochondria targeting multiply charged DABCO-cyanine dyes. Dyes Pigm 2018, 158, 517–525. 10.1016/j.dyepig.2018.05.035. [DOI] [Google Scholar]
- a Shen Y.; Shao T.; Fang B.; Du W.; Zhang M.; Liu J.; Liu T.; Tian X.; Zhang Q.; Wang A.; Yang J.; Wu J.; Tian Y. Visualization of mitochondrial DNA in living cells with super-resolution microscopy using thiophene-based terpyridine Zn (II) complexes. Chem. Commun. 2018, 54, 11288–11291. 10.1039/C8CC06276E. [DOI] [PubMed] [Google Scholar]; b Chen Y.; Wei X.-R.; Sun R.; Xu Y.-J.; Ge J.-F. The application of azonia-cyanine dyes for nucleic acids imaging in mitochondria. Sens. Actuators B 2019, 281, 499–506. 10.1016/j.snb.2018.10.146. [DOI] [Google Scholar]
- a Du W.; Wang H.; Zhu Y.; Tian X.; Zhang M.; Zhang Q.; De Souza S. C.; Wang A.; Zhou H.; Zhang Z.; Wu J.; Tian Y. Highly hydrophilic, two-photon fluorescent terpyridine derivatives containing quaternary ammonium for specific recognizing ribosome RNA in living cells. ACS Appl. Mater. Interfaces 2017, 9, 31424–31432. 10.1021/acsami.7b08068. [DOI] [PubMed] [Google Scholar]; b Wang L.; Xia Q.; Liu R.; Qu J. A red fluorescent probe for ribonucleic acid (RNA) detection, cancer cell tracing and tumor growth monitoring. Sens. Actuators B 2018, 273, 935–943. 10.1016/j.snb.2018.07.018. [DOI] [Google Scholar]
- Sun W.; Fan J.; Hu C.; Cao J.; Zhang H.; Xiong X.; Wang J.; Cui S.; Sun S.; Peng X. A two-photon fluorescent probe with near-infrared emission for hydrogen sulfide imaging in biosystems. Chem. Commun. 2013, 49, 3890–3892. 10.1039/c3cc41244j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




