Abstract
Synthetic peptides are commonly used in biomedical science for many applications in basic and translational research. While peptide synthesis is generally easy and reliable, the chemical nature of some amino acids as well as the many steps and chemical compounds involved can render the synthesis of some peptide sequences difficult. Identification of these problematic sequences and mitigation of issues they may present can be important for the reliable use of peptide reagents in several contexts. Here, we assembled a large dataset of peptides that were synthesized using standard Fmoc chemistry and whose identity was validated using mass spectrometry. We analyzed the mass spectra to identify errors in peptide syntheses and sought to develop a computational tool to predict the likelihood that any given peptide sequence would be synthesized accurately. Our model, named Peptide Synthesis Score (PepSySco), is able to predict the likelihood that a peptide will be successfully synthesized based on its amino acid sequence.
Introduction
Synthetic peptides are commonly used in biomedical science, including basic biology studies of epitope immunogenicity, protein–protein interactions, and substrate specificity of enzymes.1 Peptides are also used as therapeutics, such as in personalized cancer vaccines.2 The broad use of synthetic peptides makes it important to understand what peptides can be synthesized with ease versus those that cannot.
Peptide synthesis is a complex process with multiple steps: (1) deprotection of the N-terminal of the growing peptide chain, (2) activation of the incoming amino acid C-terminal by a coupling agent, and (3) coupling of the C-terminal of the incoming amino acid chain with the N-terminal of the growing peptide chain.3 While this process was traditionally performed manually, today, peptide synthesizers allow for automation and high-throughput production of peptides.4 Due to the many steps and chemical compounds involved and the chemical nature of specific amino acids, the synthesis of some sequences can be problematic and present challenges. For example, longer peptide chains are susceptible to incomplete deprotection and coupling reactions.5
As a means to assess the accuracy of peptide synthesis and to validate the identity of the synthesized peptides, mass spectrometry (MS) is often used. A typical tandem MS experiment results in two types of spectra: (i) MS1 spectra, wherein each peak depicts the mass-to-charge ratio (m/z) of the measured peptides that is proportional to the ion’s molecular weight and, hence, allows the derivation of the measured peptide mass, and (ii) MS2 spectra, which depict the m/z of peptide fragment ions upon fragmentation. The latter type of spectra allows one to derive the precise peptide sequence in addition to its molecular weight. Here, we make use of a large set of individually recorded MS1 spectra per ordered peptide to determine the success of synthesis and to train our model Peptide Synthesis Score (PepSySco). We validate our model using an independent set of MS1 and MS2 spectra recorded from 9604 peptides measured in 8 batches.
While not always true, in general, the highest peak in a MS1 spectrum is the ion with the greatest relative abundance and is referred to as the base peak; for comparative purposes, the base peak is assigned a relative intensity of 100% (Figure 1).6 Additional peaks, if present, can be compared to the base peak and relative abundance values determined.
Figure 1.
Examples of MS1 spectra of failed and successful peptide syntheses. (A) MS1 spectrum of a successful synthesis. A single peak at 100% relative abundance at a molecular weight of 1644.6 kDa which matches the molecular weight of the expected synthesized peptide. (B) MS1 spectrum of a lower quality synthesis. The peak at 100% relative abundance at a molecular weight of 1264.9 kDa matches the molecular weight of the expected synthesized peptide. There are, however, additional peaks at other molecular weights with lower relative abundance values. (C) MS1 spectrum of a problematic synthesis. The molecular weight of the expected synthesized peptide is 1566.1 kDa, which corresponds to the right-most peak. This peak is only fourth in relative abundance, and there are several other peaks at higher and lower relative abundance values. Those peaks are associated with lower molecular weights indicating the presence of shorter peptides.
We use the MS1 results to examine the relative abundance values associated with each m/z value and compare them with the expected molecular weight of the peptide of interest. While MS1 spectra of high-quality syntheses typically include one prominent peak with a mass weight corresponding to the desired peptide (Figure 1A), MS1 spectra of lower quality syntheses generally include more than one peak, representative of peptides or fragments other than the desired peptide (Figure 1B). Additional peaks are also frequently present due to common biochemical events such as the reduction or oxidation of residues or the presence of additional ions such as sodium. Peptide syntheses for which the MS1 spectrum does not include any signal for the desired peptide would generally be considered as failed (Figure 1C).
In the last 2 decades, our groups at the La Jolla Institute for Immunology (LJI) have ordered tens of thousands of synthesized peptides. To evaluate the quality of the synthetic peptides, samples are routinely randomly selected for quality control by MS1. The primary purpose of these spot-checks is to provide a fast and cost-efficient quality check (QC) to determine what particular set of peptides may have systematic issues, necessitating re-synthesis of the entire set.
In this study, we analyzed a large set of such peptide synthesis MS1-based QC analyses. We retrieved the information from the MS1 results and compared them to the expected peptide sequences. About 3.6% of the peptide syntheses analyzed showed no or minor signal for the desired peptide, which would be considered a complete synthesis failure. Furthermore, in about 14% of the cases, the ordered peptide was not the majority of the peptide mass in solution, suggesting a potentially problematic sequence for synthesis.
Due to the importance of peptide synthesis in biomedical research, we sought to develop a computational tool to predict the likelihood that any given peptide sequence would be synthesized accurately. To do so, we analyzed 1917 MS1 spectra of the synthesized peptides measured using a PE SCIEX150 mass spectrometer and performed statistical analyses on different aspects of the peptide sequences and the biochemical properties of the contained amino acids. Using this data, we trained a machine learning (ML) model that is able to predict the likelihood that a peptide will be successfully synthesized based on its amino acid sequence. We verified our new model named PepSySco using a large independent peptide dataset (9604 peptides) measured via an Orbitrap Fusion Lumos mass spectrometer. PepSySco predicted successful peptide syntheses with >80% overall accuracy in the validation peptide dataset.
Results
Vast Majority of Peptides Were Synthesized Correctly
We analyzed 1917 MS1 spectra measured via a PE SCIEX150 mass spectrometer corresponding to 1771 unique synthetic peptides that were picked randomly for QC from over 23,000 synthesized peptides (Supporting Information Table 1). Out of the 1917 analyzed MS1 spectra, 28.8% had only one peak, 30.3% had two peaks, 22.3% had three peaks, 11.2% had four peaks, 4.2% had five peaks, and 4.2% had six peaks or more. For 92% of the analyzed peptides, the peak at the peptide’s molecular weight was the highest peak (defined as 100% relative intensity). 2% of all peptides had their correct m/z peak as the 2nd highest intensity and only 1% at the 3rd or worse position. 5% of the analyzed MS1 spectra had no representation of the desired peptide at all.
For the purpose of further analyses, we wanted to classify peptide syntheses into two categories: (1) successful and (2) problematic or, more stringently for our present purpose, failed. We defined a synthesis to be successful when the majority of the measured molecular weight peaks were related to the expected molecular weight of the desired peptide. We calculated the “abundance ratio” as the sum of the relative abundance values of the m/z peaks matching the ordered peptide divided by the sum of the relative abundance values of all m/z peaks observed in the MS1 spectrum. We considered MS1 spectra with an abundance ratio <50% as indicative of problematic (or failed) syntheses and MS1 spectra with an abundance ratio >50% as successful (Figure 2). The MS1 spectra for 87% of the analyzed peptides met the abundance ratio threshold of >50% and were considered as successful syntheses.
Figure 2.
Abundance ratio histogram. We calculated the “abundance ratio” as the sum of the relative abundance values of the m/z peaks matching the ordered peptide divided by the sum of the relative abundance values of all m/z peaks observed in the MS1 spectrum. We considered MS1 spectra with an abundance ratio <50% (red line) as indicative of problematic (or failed) syntheses and MS1 spectra with an abundance ratio >50% as successful. The MS1 spectra for 87% of the analyzed peptides met the abundance ratio threshold of >50% and were considered as successful syntheses.
Our dataset contained peptides of 8–25 amino acid length. As expected, for longer peptides (i.e., longer than 12 amino acids), synthesis was problematic significantly more often than for shorter peptides: out of the 1408 unique longer peptides, synthesis failed for 218 peptides (15%), while out of the unique 363 shorter peptides, only 13 (4%) failed (p-value = 2.468 × 10–11, Fisher’s exact test).
Discrepancies between Expected and Measured Peptide Molecular Weight
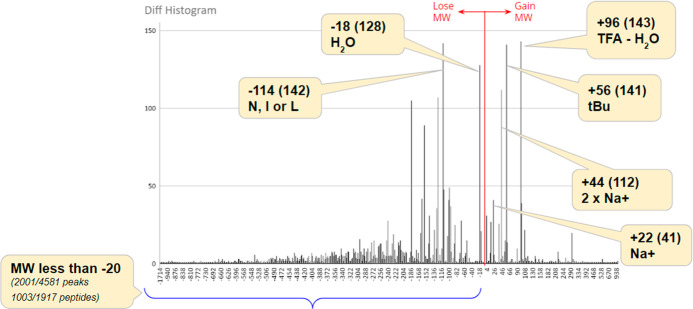
Next, we compared the measured molecular weight in the peptide MS1 results to the expected calculated molecular weight and analyzed whether discrepancies in molecular weight can be explained by artifacts left over from the synthesis or MS preparation processes. We considered all m/z 4581 peaks in the analyzed 1917 MS1 spectra. We found 153 cases where the molecular weight difference indicates a gain of a single or double sodium ion (NA+), a major component of the buffers used in MS preparation. 141 peaks had a molecular weight difference of +56 kDa, which can be explained by a tert-butyl (C4H9) residue left over from the synthesis process. 143 peaks had a difference of +96 kDa, which matches the weight of TFA minus water. TFA is used in the synthesis and MS sample preparation processes. 128 peaks matched exactly the expected molecular weight when the molecular weight of a water molecule was subtracted. None of these variances were associated with a failed synthesis.
2001 of the 4581 MS1 peaks analyzed had a molecular weight smaller than expected by more than 20 kDa, which may indicate an incomplete synthesis. These differences might be explained by the synthesized peptide missing specific amino acids. For example, a molecular weight difference of −114 kDa might be explained by the peptide missing asparagine (N), isoleucine (I), or leucine (L). 1003 out of the 1917 MS1 spectra (52%) had at least one peak with more than 20 kDa molecular weight difference, which could result from a faulty addition of two amino acids rather than one in one of the synthesis steps. Both kinds of synthesis errors could impinge upon the analysis of post-translationally spliced peptides, which are efficiently produced by proteasomes and other proteases.7 Pipelines have been developed to avoid identifying putative spliced peptide products by MS, which, in reality, were produced by peptide hydrolysis of synthesis error polypeptides.8,9
An overview of observed discrepancies between expected and measured peptide molecular weights is shown by way of example in Figure 3. A molecular weight difference of −18 occurred 128 times in our dataset and most likely represents dehydration. Some impurities are formed during TFA treatment in the peptide synthesis process which would explain some of the commonly observed molecular weight differences in our dataset: the difference of +44 occurred 112 times and could represent the addition of Trp(COOH) or the addition of two sodium ions (NA+). The weight difference of +96 occurred 143 times and could represent trifluoroacetylation or addition of TFA–H2O. The weight difference of +56 occurred 141 times and could represent the addition of tBu.
Figure 3.
Histogram of discrepancies between expected and measured peptide molecular weight. The chart is a histogram highlighting differences in the molecular weight of various peaks present in a representative spectrum compared to the desired peptide’s molecular weight. The values are rounded to the nearest 2. A difference of 0, which indicates an exact match, was removed from this chart for visualization purposes. Difference values higher than 0 indicate the molecular weight gained by the peptide. Difference values lower than 0 indicate that molecular weight dropped from the peptide. Possible explanations for various gains and losses are provided.
Success of Peptide Synthesis Can Be Predicted Using Machine Learning
We next wanted to utilize the analyzed datasets to train a ML method that can predict the likelihood of a successful synthesis for a given peptide. We defined a set of 22 features to describe the peptide sequences, as detailed in the Methods section. Some of the features are as simple as the peptide length or counting the occurrence of each amino acid in the peptide, while other features are, for example, based on the biochemical properties of individual amino acids.
We used all of these features (X1–X22) and trained multiple classification algorithms to predict the likelihood of successful peptide synthesis (Figure 4): naive Bayes/Gaussian,10 logistic regression,11 K-nearest neighbor,12 support vector machine,13 decision tree,14 and random forest.15 For each, we performed 10-fold cross-validation, repeated 10 times.16,17 We performed a receiver operating characteristics (ROC) analysis and calculated the area under the ROC curve (AUC) to assess the performance of each model. We found that the best performance was consistently achieved with the naive Bayes algorithm.
Figure 4.
(A) ML process. Shows the process of finding the best features and the best ML algorithm and validating the trained model against other data sources. (B) 10-fold cross-validation of different predictors. This set of runs shows that naive Bayes consistently provided performed best. (C) ROC of the final prediction model. Best performance was achieved using a naive Bayes classifier considering the peptide length and amino acid properties according to the Janin index.
We next performed feature reduction as a lower number of features can improve performance and can also reduce the computational effort and complexity of the model.18 In order to perform feature reduction, we tested all possible feature permutations and utilized 2-fold cross-validation and repeated 10 times. We assessed the performance of each feature combination by calculating AUC and found that the best results were obtained by combining the features “peptide length” (X1) and the “Janin index” (X15) with an AUC of 0.773 ± 0.015, while using only peptide length that achieved an AUC of 0.668 ± 0.007 and using only Janin index that achieved an AUC of 0.725 ± 0.020 (Supporting Information Table 2). Using the complete set of features achieved an AUC of 0.759 ± 0.021 (Supporting Information Table 2).
At this point, we performed a complete re-train on the entire dataset using a naïve Bayes classifier with the two selected features and achieved an AUC of 0.773 on the training dataset. Our final model, PepSySco, provides a score from 0 to 1, with a higher score indicating more likely success at synthesis.
Validation of the Prediction Model on an Independent Dataset
To validate the PepSySco prediction model, we measured a larger and independent dataset of 9604 synthesized peptides using a more sensitive mass spectrometer, i.e., an Orbitrap Fusion Lumos (see the Methods section). The MS method used for the measurement of this dataset recapitulated what has been used for the identification of peptides bound to major histocompatibility molecules class I (MHC-I). Peptides were grouped in 8 library batches, with each measured at two concentrations (100 and 500 fmol of each peptide were loaded in the mass spectrometer). These libraries contained 9, 10, or 15 amino acid long peptides related to CD4+ and CD8+ T cell response to dengue and varicella zoster (VZV) viruses. With this validation dataset, we adopted an alternative strategy to estimate the proportion of synthesis errors in the peptide library. We analyzed the MS2 spectra, which are the result of the fragmentation of peptide precursors via higher-energy collisional dissociation (HCD). MS2 spectra are commonly used for the identification of peptide sequences in complex samples such as MHC-I bound peptides and can allow the identification of sequences that slightly differ in their sequence.19−25 For each original peptide sequence theoretically contained in the peptide library, we computed all amino acid combination resulted from the missed insertion of one or more amino acid as well as the faulty addition of one or more amino acid during the peptide synthesis. To this end, we used a method originally developed to identify post-translationally spliced peptides.8,26 By doing so, a database was generated that was used for the identification of MS2 spectra matching the original peptide sequences and the cognate synthesis errors. The success ratio R was defined as the ratio of the number of MS2 spectra matching the original peptide divided by the number of MS2 spectra matching the associated synthesis error sequences. A success ratio of R = 1 would indicate a perfect synthesis with decreasing R values indicating decreasing synthesis qualities. In the measured peptide library, 2,35,441 MS2 spectra matching the original peptide sequences and 46,842 cognate synthesis errors were identified. Table 1 shows the summary of different peptide lengths passing different success rate thresholds and clearly highlights that peptide syntheses for longer sequences tend to be more problematic: while 98% of the analyzed 9-mer peptides pass the success ratio of 0.5, only 70% of the analyzed 15-mer peptides do.
Table 1. Validation Dataset, Success Rate Threshold per Peptide Lengtha.
| success rate threshold | 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | 0.9 | 1 |
|---|---|---|---|---|---|---|---|---|---|---|
| 9 | 100.0% | 99.9% | 99.6% | 99.4% | 98.4% | 97.2% | 94.6% | 89.0% | 79.8% | 70.9% |
| 10 | 99.6% | 99.2% | 98.8% | 97.7% | 95.4% | 91.9% | 85.8% | 76.0% | 65.2% | 53.1% |
| 15 | 96.3% | 91.3% | 85.0% | 77.0% | 69.9% | 62.5% | 55.2% | 47.7% | 41.1% | 33.9% |
| total | 99.2% | 98.3% | 97.0% | 95.1% | 92.6% | 89.5% | 84.7% | 77.1% | 67.6% | 57.7% |
For each peptide length and each success rate threshold, the fraction of peptides passing the threshold is summarized.
We used PepSySco to also predict the likelihood of a successful synthesis for all peptides in the validation dataset and performed an ROC analysis considering the different success ratio thresholds. Our model consistently achieved AUC values of >0.8 for success rate thresholds >0.5, indicating that our method was able to accurately predict synthesis success on this independent validation dataset (Figure 5).
Figure 5.
ROC analysis of peptide synthesis success predictions on the independent MS2-based dataset. We used PepSySco to predict the likelihood of a successful synthesis for all peptides in the validation dataset and performed an ROC analysis considering the different MS2 success rate thresholds (shown here in different colors).
Comparing Prediction Performance of PepSySco to the ThermoFisher Scientific Analysis Tool
The peptide synthesis and proteotypic peptide analyzing tool provided by ThermoFisher Scientific (https://www.thermofisher.com/us/en/home/life-science/protein-biology/peptides-proteins/custom-peptide-synthesis-services/peptide-analyzing-tool.html) predicts the ease of peptide synthesis and purification. The tool grades the input peptides on a scale from A to C, with A predicting no anticipated issues and C predicting a challenging synthesis. Peptides are additionally graded on a scale from 1 to 5 to predict compatibility with quantitative MS workflows based on hydrophobicity, with 1 indicating a very hydrophilic peptide and 5 a very hydrophobic one.
We used the ThermoFisher Scientific tool to predict ease of synthesis for our list of 1,771 peptides and performed an ROC analysis. With an AUC of 0.651, this tool performed worse than our model PepSysCo (Figure 6A).
Figure 6.
ROC analysis for PepSysCo and the ThermoFisher Scientific tool for (A) our training dataset and (B) the independent MS2-based validation dataset.
We also used the ThermoFisher tool to predict ease of synthesis of the 9604 synthetic peptides in our validation peptide dataset. When considering peptide syntheses passing the success rate threshold of 0.5 as successful, and syntheses that do not pass this threshold as failed, PepSySco achieved an AUC of 0.806, while the ThermoFisher Scientific tool only achieved an AUC of 0.567 (Figure 6B). PepSySco also outperformed the ThermoFisher Scientific tool when higher success rate thresholds were considered (Supporting Information Figure 1).
Discussion
Peptides, which are short amino acid polymers, are a crucial reagent for research projects designed to study biological phenomena such as immune system recognition of a wide variety of perturbations, from infectious disease to autoimmunity and cancer. For example, generating peptides to recruit tumor-specific T cells is essential for developing personalized cancer vaccines, or characterizing peptide targets of T cells is critical for understanding the immune response to pathogens, such as SARS-CoV-2. As such, the capacity to identify and synthesize peptide sets for use in various platforms is essential.27−30
Peptides are composed of various combinations of the 20 different naturally occurring amino acids and represent complex molecules from a synthesis standpoint. For many studies in an immunological context, peptides typically vary in length from about 8 to 25 amino acids. Out of about 23,000 synthesized peptides at the La Jolla Institute of Immunology, we analyzed over 1700 peptides with MS for QC. We found that for 1540 of the 1771 (87%) unique peptides we examined by MS1, the most abundant MS1 peaks represented the expected peptide. In 13% of the MS1 spectra, the majority of the observed peaks did not match the mass of the expected peptide, and these problematic syntheses were defined operationally as failed.
We defined 22 different features based on various properties of the peptide amino acid sequences and trained a ML model to predict the likelihood of successful peptide synthesis. The most predictive model was achieved when the features “peptide length” and “Janin index”31 were combined in a naïve Bayes model. We expected the peptide length to play an important role because, with a growing amino acid chain, the likelihood of peptide fragmentation or missed residues also grows. Indeed, our analyses showed that longer peptides are more prone to failed synthesis than shorter ones.
The Janin index being more predictive than other hydrophobicity scales we considered was more surprising. The Janin index was determined by examining proteins with known 3D structures and defining the hydrophobicity of a residue based on its localization in the structure, that is, whether the residue is accessible on the protein surface or buried inside a globular structure.31 In contrast, the Kyte–Doolittle scale, for example, which is the most widely known hydrophobicity scale, was determined by directly inspecting the amino acid structures and assessing the physicochemical properties of the side chains.32 It is possible that the way the Janin index is determined captures unique contexts that are specifically relevant during the peptide synthesis process.
It is important to note that the peptides being analyzed in the present study have been produced using high throughput synthesis methods that are expected to produce crude materials of at least 70–75% purity, on average. This synthesis approach is the most cost-effective way to generate the large peptide sets necessary for probing reactivity to large proteomes of various viruses, bacteria, or cancer antigens. Other methods are available that are more likely to produce high-quality peptides without issues. Still, these methods require additional steps of HPLC purification, which are costly and not feasible for large-scale studies, but also do not guarantee successful synthesis.
The peptides analyzed in this study were all synthesized using standard Fmoc chemistry.33 The conditions of Fmoc chemistry are milder as compared to the more senior Boc chemistry, which led to a shift in the field in the late 1990s, and Fmoc became the chemistry of choice in a majority of peptide laboratories.3 However, the solvents commonly used in Fmoc chemistry, such as DMF, NMP, and dichloromethane (DCM), are known to be hazardous and considerable research has been done in recent years to identify less toxic solvents as alternatives.34−36 It is currently unclear how PepSySco will perform on peptides produced with synthesis chemistries other than the standard Fmoc chemistry, and the effectiveness of PepSySco might be limited to peptides synthesized with this specific methodology. We plan to evaluate PepSySco on peptide sets that were synthesized using other methods when they become available to us in the future.
In this study, we have highlighted commonly occurring problems during peptide synthesis. Importantly, many problems can be avoided by expert evaluation of each peptide and optimizing the synthesis procedure accordingly. For instance, several impurities are formed during TFA treatment and could be avoided by optimizing the deprotection procedure. The goal of our tool is, however, to quickly evaluate thousands of peptides and pick peptides that are likely easy to synthesize without the need for expert intervention. It is also important to note that our current evaluation of different peaks observed in the MS spectra is simplistic. For instance, we assumed that a difference of −18 between the expected and measured mass weight represents the loss of a water molecule. A mass weight difference of −18 could however also represent an aspartimide formation at aspartic acid residues. We plan to further improve such evaluations in the future when we have more data available for analysis.
We believe PepSySco will be useful for researchers when assembling sets of peptides for synthesis by identifying peptides that are very likely to have a successful synthesis and identifying potentially problematic sequences. For instance, PepSySco could be used for the critical planning of in vitro digestions of synthetic polypeptides by proteases and the downstream identification and characterization of MHC-I-restricted epitope candidates.37−44 Users can use different PepSySco thresholds depending on their needs. For example, a PepSySco threshold of 0.85 covered about 51% of the peptides in our dataset and predicted successful synthesis with an accuracy of 95%. In contrast, a PepSySco threshold of 0.99 covered only 12% of the peptides in our dataset but provided 98% accuracy.
PepSySco is freely available to the scientific community at http://tools.iedb.org/pepsysco.
Methods
Peptide Synthesis and Mass Spectrometry
All peptides analyzed in this study were synthesized as crude material on a 1 mg scale using the SYRO II peptide synthesizer and using standard Fmoc chemistry. Wang resins were utilized for all amino acids except Cys and Pro, for which preloaded chloro-trityl resins were used. The loading capacity of the resins used was between 0.3–0.4 mmol/g, and the equivalent of the coupling reagents used was 5. Coupling was done for 40 min at room temperature with O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate (TBTU) and 4-methylmorpholine as reagents. Dimethylformamide (DMF), ACS certified at >99% purity, was utilized as the solvent and 40% piperidine in DMF for Fmoc-deprotection. This step was repeated twice, for 6 min each time, and DMF washes were performed after Fmoc-deprotection and coupling steps, six and four times respectively. All peptides were synthesized identically and subject to QC analysis by MS. The same vendor was used for all peptides.
MS measurement of the first peptide library (training dataset) was performed on the ion-spray PE SCIEX150 mass spectrometer. Briefly, following lyophilization, randomly selected samples were taken using a pipet tip and dissolved in methanol. Samples were then infused onto a mass spectrometer at 100 μL/min, with the instrument mass range set to 500–2000. Because concentration can vary, enough events were captured until a clear visual signal was obtained. As the obtained data tends to reflect doubly charged ions, the captured data are reprocessed to represent molecular weights associated with singly charged ions. Final spectra were saved as pdf files for subsequent analysis.
The synthetic peptide library for the validation dataset consisted of 9,604 synthetic peptides, which were subdivided into 8 batches, with each measured at 2 concentrations (100 and 500 fmol of each peptide were loaded in the mass spectrometer). We obtained 32 MS RAW files (8 synthetic peptide library batches × 2 concentrations × 2 technical replicates). The RAW MS data were first converted with ms-convert 4.1.12 from ProteoWizard45 to the open mzML format, which is readable by most MS software platforms. MzML files were analyzed by both Mascot and PEAKS DB. Two databases in FASTA format were generated: (i) a target database containing all synthetic peptide sequences included in the peptide library and (ii) a synthesis error database. Both databases were used to search the MS1/MS2 spectra of the validation dataset. As we were only interested in assigning the exact peptide sequence as found in the target database or the synthesis error database, enzyme specificities were set to “no processing” in PEAKS DB. Precursor mass tolerances were set to 5 ppm, and fragment ion mass tolerances were set to 0.02 Da for measurements on Fusion Lumos. MS2 spectra assigned by PEAKS, after filtering peptides for 1% FDR, were extracted.
MS measurement of these peptide samples was performed using an Orbitrap Fusion Lumos mass spectrometer coupled to an Ultimate 3000 RSLC nano pump (both from ThermoFisher Scientific). Briefly, peptides were loaded and separated by a nanoflow HPLC (RSLC Ultimate 3000) on an Easy-spray C18 nano column (50 cm length, 75 mm internal diameter; ThermoFisher Scientific), coupled on-line to a nano-electrospray ionization Orbitrap Fusion Lumos mass spectrometer (ThermoFisher Scientific). Peptides were eluted with a linear gradient of 5–45% buffer B (80% ACN, 0.1% formic acid) at a flow rate of 300 nL/min over 90 min at 50 °C. The instrument was programmed in Xcalibur 4.1 to acquire MS data using a “Universal” method by defining a 3s cycle time between a full MS scan and MS/MS fragmentation. This method takes advantage of multiple analyzers in the Orbitrap Fusion Lumos and drives the system to use all available parallelizable time, resulting in a decreased dependence on method parameters (such as Data Dependent Acquisition; DDA). We acquired one full-scan MS spectrum at a resolution of 1,20,000 at 200 m/z with an automatic gain control (AGC) target value of 2 × 105 ions and a scan range of 350–1550 m/z. The MS2 fragmentation was conducted using HCD collision energy (30%) with an Orbitrap resolution of 30,000 at 200 m/z. The AGC target value was set up as 5 × 104 with a max injection time of 120 ms. A dynamic exclusion of 30 s and 1–4 included charged states defined within this method.
Peptide Library Datasets
The MS graph batches for the training dataset were taken from data from the laboratory of Prof. Sette. The analyzed sets included peptides derived from sequences from various antigens of Cytomegalovirus, Mycobacterium tuberculosis, Zika virus, chikungunya virus, SARS-CoV-2, pertussis, tetanus, yellow fever virus, rhinovirus, metapneumovirus, influenza, Plasmodium falciparum, and other sources. The sets included peptides predicted for class I and II binding studies and CD4 and CD8 T cell recognition assays, and sets of overlapping peptides spanning entire protein antigens. There were 30 peptide synthesis batches of 23,279 peptides and 1917 MS1 graphs, out of which 1771 were unique peptide sequences. We collected the tables detailing the target peptide sequence for each peptide synthesis batch. We manually retrieved the relative intensity and molecular weight for each MS result and each MS1 peak within the result.
The validation peptide library contained 9, 10, or 15 amino acid long peptides related to CD4+ and CD8+ T cell response to dengue and VZV viruses. The dengue and VZV synthetic peptides utilized in this study were selected for analysis because they were already available in-house and synthesized for separate epitope identification studies. The selection and characterization of these peptides were described previously.46−53 Each of the peptides in synthetic peptide libraries was derived from respective dengue and VZV proteomes. Peptides were originally selected for other studies based on bioinformatic analyses of predicted capacity to bind various common MHC-I and -II alleles in the general worldwide population. The set of dengue protein sequences of provenance represent all four dengue serotypes and several different variant isolates. The VZV peptides were primarily derived from the attenuated varicella vaccine strain vOka and a few variant isolates.
Databases, Datasets, and Algorithm Availability
The MS proteomics data we generated have been deposited to the ProteomeXchange consortium via the PRIDE54 partner repository with the dataset identifier PXD025346.
The developed tool PepSySco is freely available at http://tools.iedb.org/pepsysco.
Tools
We used custom JavaScript and Python scripts to aggregate data and perform calculations. We used Python packages scikit learn and pandas to train ML models. MS2 post-analysis was performed using custom scripts in R.
Features Considered for Machine Learning
We considered a total of 22 peptide features for training our prediction model:
X1 Length of Peptide
The lengths of peptides in our dataset ranged between 8 and 25 amino acids, with the majority (58%) being 15 residues. The vast majority of failed peptide synthesis (98%) appeared at a peptide length of 15.
X2 Number of Aliphatic-Hydrophobic Residues in the Peptide
Counting the number of peptides that are aliphatic-hydrophobic (I, L, M, and V).
X3 Length of the Longest Aliphatic-Hydrophobic Stretch within the Peptide
Counting the highest consecutive sequence of contiguous residues that are aliphatic-hydrophobic (I, L, M, and V).
X4 N-Terminal Amino Acid—Vectorized
Since we identified in the MS1 spectra analyses that particular amino acids tend to be more often dropped at the end of the peptide synthesis, at the N-terminal side of the peptide, we used the vectorized left amino acid as a feature.
X5 All Amino Acids—Vectorized
Vectorization, or “bag of letters”, measuring the number of each amino acid in the peptide, is represented as a series of 20 numbers.
X6 N-Terminal Amino Acid Drop
Many spectra indicated the loss, or dropping, of a single amino acid residue. Often, in the dataset analyzed, this occurred at the N-terminus. Here, the spectra are analyzed with this consideration.
X7 Amino Acid Drop at Any Position
As with X6, this feature considers amino acid drops at any position within the sequence.
X8 Broken Amino Acid Bonds at Any Position
Here, instead of considering the dropped amino acid itself, the bond to the right of the dropped amino acid is considered. There are 20 amino acids; hence, there are 20 × 20 = 400 possible amino acid bonds.
X9 C-Terminal Amino Acid Drop
As with X6, here, amino acid drops on the C-terminus were considered.
X10 Kyte–Doolittle Amino Acid Hydrophobicity Index
Kyle–Doolittle amino acid index conversion was performed,32 averaging the conversion of all amino acids in the peptide and dividing by the peptide length. This index is a common measure for peptide hydrophobicity.
X11 Hopp–Woods Amino Acid Hydrophobicity Index
Hopp–Woods amino acid index conversion was performed,55 averaging the conversion of all amino acids in the peptide and dividing by the peptide length. This index is a hydrophilicity scale based on the individual amino acid water solubility.
X12 Cornette Amino Acid Hydrophobicity Index
Cornette amino acid index conversion was performed,56 averaging the conversion of all amino acids in the peptide and dividing by the peptide length. This index is a hydrophobicity scale based on 28 other published scales computed for optimality.
X13 Eisenberg Amino Acid Hydrophobicity Index
Eisenberg amino acid index conversion was performed,57 averaging the conversion of all amino acids in the peptide and dividing by the peptide length. This index is based on the calculation of hydrophobic dipole moments of areas within a polypeptide chain and of the energy needed to move the residue from the inside of the protein to its surface.
X14 Rose Amino Acid Hydrophobicity Index
Rose amino acid index conversion was performed,58 averaging the conversion of all amino acids in the peptide and dividing by the peptide length. This hydrophobicity scale is correlated to the average area of buried amino acids in globular proteins.
X15 Janin Amino Acid Hydrophobicity Index
Janin amino acid index conversion was performed,31 averaging the conversion of all amino acids in the peptide and dividing by the peptide length. The Janin scale provides an indication for the surface accessibility of the amino acid residues of globular proteins.
X16 Engelman GES Amino Acid Hydrophobicity Index
Engelman GES amino acid index conversion was performed,59 averaging the conversion of all amino acids in the peptide and dividing by the peptide length. This scale is based on the energy required for separating amino chains in aqueous solutions and membranes.
X17 Number of Acidic Residues in the Peptide
The number of acidic residues was counted (D and E).
X18 Number of Small Polar Residues in the Peptide
The number of residues that are small and polar was counted (C, S, and T).
X19 Number of Small Residues in the Peptide
The number of residues that are small was counted (A, G, and P).
X20 Number of Large Polar Residues in the Peptide
The number of residues that are large and polar was counted (N and Q).
X21 Number of Basic Residues in the Peptide
The number of basic residues was counted (H, K, and R).
X22 Number of Aromatic Residues in the Peptide
The number of aromatic residues was counted (F, W, and Y).
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award number U24CA248138 and by the National Institute of Allergy and Infectious Diseases (NIAID) under award number 75N93019C00001. M.M. and J.L. were in part supported by (i) Cancer Research UK [C67500; A29686] and the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London and/or the NIHR Clinical Research Facility to M.M. and (ii) the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no 945528) to J.L. We thank S. Lyham and X. Yang (KCL) for technical assistance.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c02425.
Training dataset of synthesized peptides; and ROC analysis for the ThermoFisher Scientific tool on the independent MS2-based validation dataset using different success rate thresholds (PDF)
Performance of different features and pairwise combination of features for predicting the success of peptide synthesis (XLSX)
The authors declare no competing financial interest.
Supplementary Material
References
- Szymczak L. C.; Kuo H.-Y.; Mrksich M. Peptide Arrays: Development and Application. Anal. Chem. 2018, 90, 266–282. 10.1021/acs.analchem.7b04380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. S.; Mardis E. R. Applications of Immunogenomics to Cancer. Cell 2017, 168, 600–612. 10.1016/j.cell.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawikowski M.; Fields G. B.. Introduction to Peptide Synthesis. Current Protocols in Protein Science, 2012. chapter, unit-18.1. [DOI] [PubMed] [Google Scholar]
- Mijalis A. J.; Thomas D. A.; Simon M. D.; Adamo A.; Beaumont R.; Jensen K. F.; Pentelute B. L. A fully automated flow-based approach for accelerated peptide synthesis. Nat. Chem. Biol. 2017, 13, 464–466. 10.1038/nchembio.2318. [DOI] [PubMed] [Google Scholar]
- Al-Warhi T. I.; Al-Hazimi H. M. A.; El-Faham A. Recent development in peptide coupling reagents. J. Saudi Chem. Soc. 2012, 16, 97–116. 10.1016/j.jscs.2010.12.006. [DOI] [Google Scholar]
- Downard K.Mass spectrometry: a foundation course; Royal Society of Chemistry, 2007. [Google Scholar]
- Liepe J.; Ovaa H.; Mishto M. Why do proteases mess up with antigen presentation by re-shuffling antigen sequences?. Curr. Opin. Immunol. 2018, 52, 81–86. 10.1016/j.coi.2018.04.016. [DOI] [PubMed] [Google Scholar]
- Specht G.; Roetschke H. P.; Mansurkhodzhaev A.; Henklein P.; Textoris-Taube K.; Urlaub H.; Mishto M.; Liepe J. Large database for the analysis and prediction of spliced and non-spliced peptide generation by proteasomes. Sci. Data 2020, 7, 146. 10.1038/s41597-020-0487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishto M. Commentary: Are there indeed spliced peptides in the immunopeptidome?. Mol. Cell. Proteomics 2021, 20, 100158. 10.1016/j.mcpro.2021.100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.The Optimality of Naive Bayes, 2004; Vol. 6. [Google Scholar]
- Bishop C. M.Pattern Recognition and Machine Learning, 2008; Vol. 34, p 9. [Google Scholar]
- Bentley J. L. Multidimensional binary search trees used for associative searching. Commun. ACM 1975, 18, 509–517. 10.1145/361002.361007. [DOI] [Google Scholar]
- Wu T.-F.; Lin C.-J.; Weng R. C. Probability Estimates for Multi-class Classification by Pairwise Coupling. J. Mach. Learn. Res. 2004, 5, 975–1005. [Google Scholar]
- Dumont M.; Marée R.; Wehenkel L.; Geurts P.. Fast Multi-class Image Annotation with Random Subwindows and Multiple Output Randomized Trees, 2009, pp 196–203.
- Geurts P.; Ernst D.; Wehenkel L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. 10.1007/s10994-006-6226-1. [DOI] [Google Scholar]
- Breiman L.; Spector P. Submodel Selection and Evaluation in Regression. The X-Random Case. Int. Stat. Rev. 1992, 60, 291–319. 10.2307/1403680. [DOI] [Google Scholar]
- Rao R. B.; Fung G.; Rosales R.. On the Dangers of Cross-Validation. An Experimental Evaluation, Proceedings of the 2008 SIAM International Conference on Data Mining, 2008; pp 588–596.
- Islam M. T.; Xing L. A data-driven dimensionality-reduction algorithm for the exploration of patterns in biomedical data. Nat. Biomed. Eng. 2021, 5, 624–635. 10.1038/s41551-020-00635-3. [DOI] [PubMed] [Google Scholar]
- Liepe J.; Marino F.; Sidney J.; Jeko A.; Bunting D. E.; Sette A.; Kloetzel P. M.; Stumpf M. P. H.; Heck A. J. R.; Mishto M. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 2016, 354, 354–358. 10.1126/science.aaf4384. [DOI] [PubMed] [Google Scholar]
- Liepe J.; Sidney J.; Lorenz F. K. M.; Sette A.; Mishto M. Mapping the MHC Class I-Spliced Immunopeptidome of Cancer Cells. Cancer Immunol Res 2019, 7, 62–76. 10.1158/2326-6066.cir-18-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faridi P.; Woods K.; Ostrouska S.; Deceneux C.; Aranha R.; Duscharla D.; Wong S. Q.; Chen W.; Ramarathinam S. H.; Lim Kam Sian T. C. C.; Croft N. P.; Li C.; Ayala R.; Cebon J. S.; Purcell A. W.; Schittenhelm R. B.; Behren A. Spliced Peptides and Cytokine-Driven Changes in the Immunopeptidome of Melanoma. Cancer Immunol Res 2020, 8, 1322–1334. 10.1158/2326-6066.cir-19-0894. [DOI] [PubMed] [Google Scholar]
- Sarkizova S.; Klaeger S.; Le P. M.; Li L. W.; Oliveira G.; Keshishian H.; Hartigan C. R.; Zhang W.; Braun D. A.; Ligon K. L.; Bachireddy P.; Zervantonakis I. K.; Rosenbluth J. M.; Ouspenskaia T.; Law T.; Justesen S.; Stevens J.; Lane W. J.; Eisenhaure T.; Lan Zhang G.; Clauser K. R.; Hacohen N.; Carr S. A.; Wu C. J.; Keskin D. B. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat. Biotechnol. 2020, 38, 199–209. 10.1038/s41587-019-0322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastri A.; Liao H.; Muller J.; Purcell A. W.; Ternette N. The Choice of HLA-Associated Peptide Enrichment and Purification Strategy Affects Peptide Yields and Creates a Bias in Detected Sequence Repertoire. Proteomics 2020, 20, e2070175 10.1002/pmic.202070175. [DOI] [PubMed] [Google Scholar]
- Wilhelm M.; Zolg D. P.; Graber M.; Gessulat S.; Schmidt T.; Schnatbaum K.; Schwencke-Westphal C.; Seifert P.; de Andrade Krätzig N.; Zerweck J.; Knaute T.; Bräunlein E.; Samaras P.; Lautenbacher L.; Klaeger S.; Wenschuh H.; Rad R.; Delanghe B.; Huhmer A.; Carr S. A.; Clauser K. R.; Krackhardt A. M.; Reimer U.; Kuster B. Deep learning boosts sensitivity of mass spectrometry-based immunopeptidomics. Nat. Commun. 2021, 12, 3346. 10.1038/s41467-021-23713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz Cuevas M. V.; Hardy M.-P.; Hollý J.; Bonneil É.; Durette C.; Courcelles M.; Lanoix J.; Côté C.; Staudt L. M.; Lemieux S.; Thibault P.; Perreault C.; Yewdell J. W. Most non-canonical proteins uniquely populate the proteome or immunopeptidome. Cell Rep. 2021, 34, 108815. 10.1016/j.celrep.2021.108815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepe J.; Mishto M.; Textoris-Taube K.; Janek K.; Keller C.; Henklein P.; Kloetzel P. M.; Zaikin A. The 20S Proteasome Splicing Activity Discovered by SpliceMet. PLoS Comput. Biol. 2010, 6, e1000830 10.1371/journal.pcbi.1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale A. S.; Satyanarayanajois S. Peptides and peptidomimetics as immunomodulators. Immunotherapy 2014, 6, 755–774. 10.2217/imt.14.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower D. R. Designing immunogenic peptides. Nat. Chem. Biol. 2013, 9, 749–753. 10.1038/nchembio.1383. [DOI] [PubMed] [Google Scholar]
- Ng C. X.; Lee S. H. The Potential Use of Anticancer Peptides (ACPs) in the Treatment of Hepatocellular Carcinoma. Curr. Cancer Drug Targets 2020, 20, 187–196. 10.2174/1568009619666191111141032. [DOI] [PubMed] [Google Scholar]
- Mobilizing peptides in immunity. Nat. Chem. Biol. 2013, 9(), 747. 10.1038/nchembio.1409 [DOI] [PubMed] [Google Scholar]
- Janin J. Surface and inside volumes in globular proteins. Nature 1979, 277, 491–492. 10.1038/277491a0. [DOI] [PubMed] [Google Scholar]
- Kyte J.; Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Hansen P. R.; Oddo A. Fmoc Solid-Phase Peptide Synthesis. Methods Mol. Biol. 2015, 1348, 33–50. 10.1007/978-1-4939-2999-3_5. [DOI] [PubMed] [Google Scholar]
- Egelund P. H. G.; Jadhav S.; Martin V.; Johansson Castro H.; Richner F.; Le Quement S. T.; Dettner F.; Lechner C.; Schoenleber R.; Sejer Pedersen D. Fmoc-Removal with Pyrrolidine Expands the Available Solvent Space in Green Solid-Phase Peptide Synthesis. ACS Sustainable Chem. Eng. 2021, 9, 14202–14215. 10.1021/acssuschemeng.1c04770. [DOI] [Google Scholar]
- Martelli G.; Cantelmi P.; Tolomelli A.; Corbisiero D.; Mattellone A.; Ricci A.; Fantoni T.; Cabri W.; Vacondio F.; Ferlenghi F.; Mor M.; Ferrazzano L. Steps towards sustainable solid phase peptide synthesis: use and recovery of N-octyl pyrrolidone. Green Chem. 2021, 11, 4095–4106. 10.1039/d1gc00910a. [DOI] [Google Scholar]
- Martin V.; Egelund P. H. G.; Johansson H.; Thordal Le Quement S.; Wojcik F.; Sejer Pedersen D. Greening the synthesis of peptide therapeutics: an industrial perspective. RSC Adv. 2020, 10, 42457–42492. 10.1039/d0ra07204d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebstein F.; Textoris-Taube K.; Keller C.; Golnik R.; Vigneron N.; Van den Eynde B. J.; Schuler-Thurner B.; Schadendorf D.; Lorenz F. K. M.; Uckert W.; Urban S.; Lehmann A.; Albrecht-Koepke N.; Janek K.; Henklein P.; Niewienda A.; Kloetzel P. M.; Mishto M. Proteasomes generate spliced epitopes by two different mechanisms and as efficiently as non-spliced epitopes. Sci. Rep. 2016, 6, 24032. 10.1038/srep24032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishto M.; Mansurkhodzhaev A.; Ying G.; Bitra A.; Cordfunke R. A.; Henze S.; Paul D.; Sidney J.; Urlaub H.; Neefjes J.; Sette A.; Zajonc D. M.; Liepe J. An in silico-in vitro Pipeline Identifying an HLA-A*02:01+ KRAS G12V+ Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients. Front. Immunol. 2019, 10, 2572. 10.3389/fimmu.2019.02572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishto M.; Rodriguez-Hernandez G.; Neefjes J.; Urlaub H.; Liepe J. Response: Commentary: An In Silico-In Vitro Pipeline Identifying an HLA-A*02:01+ KRAS G12V+ Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients. Front. Immunol. 2021, 12, 679836. 10.3389/fimmu.2021.679836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkers C. R.; de Jong A.; Schuurman K. G.; Linnemann C.; Geenevasen J. A. J.; Schumacher T. N. M.; Rodenko B.; Ovaa H. Peptide Splicing in the Proteasome Creates a Novel Type of Antigen with an Isopeptide Linkage. J. Immunol. 2015, 195, 4075–4084. 10.4049/jimmunol.1402454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalet A.; Robbins P. F.; Stroobant V.; Vigneron N.; Li Y. F.; El-Gamil M.; Hanada K.; Yang J. C.; Rosenberg S. A.; Van den Eynde B. J. An antigenic peptide produced by reverse splicing and double asparagine deamidation. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, E323–E331. 10.1073/pnas.1101892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron N.; Stroobant V.; Chapiro J.; Ooms A.; Degiovanni G.; Morel S.; van der Bruggen P.; Boon T.; Van den Eynde B. J. An antigenic peptide produced by peptide splicing in the proteasome. Science 2004, 304, 587–590. 10.1126/science.1095522. [DOI] [PubMed] [Google Scholar]
- Textoris-Taube K.; Cammann C.; Henklein P.; Topfstedt E.; Ebstein F.; Henze S.; Liepe J.; Zhao F.; Schadendorf D.; Dahlmann B.; Uckert W.; Paschen A.; Mishto M.; Seifert U. ER-aminopeptidase 1 determines the processing and presentation of an immunotherapy-relevant melanoma epitope. Eur. J. Immunol. 2020, 50, 270–283. 10.1002/eji.201948116. [DOI] [PubMed] [Google Scholar]
- Textoris-Taube K.; Keller C.; Liepe J.; Henklein P.; Sidney J.; Sette A.; Kloetzel P. M.; Mishto M. The T210M Substitution in the HLA-a*02:01 gp100 Epitope Strongly Affects Overall Proteasomal Cleavage Site Usage and Antigen Processing. J. Biol. Chem. 2015, 290, 30417–30428. 10.1074/jbc.m115.695189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers M. C.; Maclean B.; Burke R.; Amodei D.; Ruderman D. L.; Neumann S.; Gatto L.; Fischer B.; Pratt B.; Egertson J.; Hoff K.; Kessner D.; Tasman N.; Shulman N.; Frewen B.; Baker T. A.; Brusniak M.-Y.; Paulse C.; Creasy D.; Flashner L.; Kani K.; Moulding C.; Seymour S. L.; Nuwaysir L. M.; Lefebvre B.; Kuhlmann F.; Roark J.; Rainer P.; Detlev S.; Hemenway T.; Huhmer A.; Langridge J.; Connolly B.; Chadick T.; Holly K.; Eckels J.; Deutsch E. W.; Moritz R. L.; Katz J. E.; Agus D. B.; MacCoss M.; Tabb D. L.; Mallick P. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Sullivan N. L.; Rouphael N.; Yu T.; Banton S.; Maddur M. S.; McCausland M.; Chiu C.; Canniff J.; Dubey S.; Liu K.; Tran V.; Hagan T.; Duraisingham S.; Wieland A.; Mehta A. K.; Whitaker J. A.; Subramaniam S.; Jones D. P.; Sette A.; Vora K.; Weinberg A.; Mulligan M. J.; Nakaya H. I.; Levin M.; Ahmed R.; Pulendran B. Metabolic Phenotypes of Response to Vaccination in Humans. Cell 2017, 169, 862–877. 10.1016/j.cell.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.; McCausland M.; Sidney J.; Duh F.-M.; Rouphael N.; Mehta A.; Mulligan M.; Carrington M.; Wieland A.; Sullivan N. L.; Weinberg A.; Levin M. J.; Pulendran B.; Peters B.; Sette A.; Ahmed R. Broadly reactive human CD8 T cells that recognize an epitope conserved between VZV, HSV and EBV. PLoS Pathog. 2014, 10, e1004008 10.1371/journal.ppat.1004008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D.; Angelo M. A.; Grifoni A.; O’Rourke P. H.; Sidney J.; Paul S.; De Silva A. D.; Phillips E.; Mallal S.; Premawansa S.; Premawansa G.; Wijewickrama A.; Peters B.; Sette A. HLA-DRB1 Alleles Are Associated With Different Magnitudes of Dengue Virus-Specific CD4+T-Cell Responses. J. Infect. Dis. 2016, 214, 1117–1124. 10.1093/infdis/jiw309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D.; Angelo M. A.; de Azeredo E. L.; Sidney J.; Greenbaum J. A.; Fernando A. N.; Broadwater A.; Kolla R. V.; De Silva A. D.; de Silva A. M.; Mattia K. A.; Doranz B. J.; Grey H. M.; Shresta S.; Peters B.; Sette A. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, E2046–E2053. 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D.; Angelo M. A.; Bangs D. J.; Sidney J.; Paul S.; Peters B.; de Silva A. D.; Lindow J. C.; Diehl S. A.; Whitehead S.; Durbin A.; Kirkpatrick B.; Sette A. The Human CD8 + T Cell Responses Induced by a Live Attenuated Tetravalent Dengue Vaccine Are Directed against Highly Conserved Epitopes. J. Virol. 2015, 89, 120–128. 10.1128/jvi.02129-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D.; Cerpas C.; Angelo M. A.; Bangs D. J.; Sidney J.; Paul S.; Peters B.; Sanches F. P.; Silvera C. G. T.; Costa P. R.; Kallas E. G.; Gresh L.; de Silva A. D.; Balmaseda A.; Harris E.; Sette A. Human CD8+T-Cell Responses Against the 4 Dengue Virus Serotypes Are Associated With Distinct Patterns of Protein Targets. J. Infect. Dis. 2015, 212, 1743–1751. 10.1093/infdis/jiv289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D.; Bangs D. J.; Sidney J.; Kolla R. V.; De Silva A. D.; de Silva A. M.; Crotty S.; Peters B.; Sette A. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, E4256–E4263. 10.1073/pnas.1505956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D.; Angelo M. A.; Sidney J.; Peters B.; Shresta S.; Sette A. Immunodominance changes as a function of the infecting dengue virus serotype and primary versus secondary infection. J. Virol. 2014, 88, 11383–11394. 10.1128/jvi.01108-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y.; Csordas A.; Bai J.; Bernal-Llinares M.; Hewapathirana S.; Kundu D. J.; Inuganti A.; Griss J.; Mayer G.; Eisenacher M.; Pérez E.; Uszkoreit J.; Pfeuffer J.; Sachsenberg T.; Yılmaz Ş.; Tiwary S.; Cox J.; Audain E.; Walzer M.; Jarnuczak A. F.; Ternent T.; Brazma A.; Vizcaíno J. A. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopp T. P.; Woods K. R. A computer program for predicting protein antigenic determinants. Mol. Immunol. 1983, 20, 483–489. 10.1016/0161-5890(83)90029-9. [DOI] [PubMed] [Google Scholar]
- Cornette J. L.; Cease K. B.; Margalit H.; Spouge J. L.; Berzofsky J. A.; DeLisi C. Hydrophobicity scales and computational techniques for detecting amphipathic structures in proteins. J. Mol. Biol. 1987, 195, 659–685. 10.1016/0022-2836(87)90189-6. [DOI] [PubMed] [Google Scholar]
- Eisenberg D.; Schwarz E.; Komaromy M.; Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 1984, 179, 125–142. 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Rose G. D.; Geselowitz A. R.; Lesser G. J.; Lee R. H.; Zehfus M. H. Hydrophobicity of amino acid residues in globular proteins. Science 1985, 229, 834–838. 10.1126/science.4023714. [DOI] [PubMed] [Google Scholar]
- Engelman D. M.; Steitz T. A.; Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 1986, 15, 321–353. 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.