Abstract
We developed a novel approach for improving the production of antibiotic from Streptomyces coelicolor A3(2) by inducing combined drug-resistant mutations. Mutants with enhanced (1.6- to 3-fold-higher) actinorhodin production were detected at a high frequency (5 to 10%) among isolates resistant to streptomycin (Strr), gentamicin (Genr), or rifampin (Rifr), which developed spontaneously on agar plates which contained one of the three drugs. Construction of double mutants (str gen and str rif) by introducing gentamicin or rifampin resistance into an str mutant resulted in further increased (1.7- to 2.5-fold-higher) actinorhodin productivity. Likewise, triple mutants (str gen rif) thus constructed were found to have an even greater ability for producing the antibiotic, eventually generating a mutant able to produce 48 times more actinorhodin than the wild-type strain. Analysis of str mutants revealed that a point mutation occurred within the rpsL gene, which encodes the ribosomal protein S12. rif mutants were found to have a point mutation in the rpoB gene, which encodes the β-subunit of RNA polymerase. Mutation points in gen mutants still remain unknown. These single, double, and triple mutants displayed in hierarchical order a remarkable increase in the production of ActII-ORF4, a pathway-specific regulatory protein, as determined by Western blotting analysis. This reflects the same hierarchical order observed for the increase in actinorhodin production. The superior ability of the triple mutants was demonstrated by physiological analyses under various cultural conditions. We conclude that by inducing combined drug-resistant mutations we can continuously increase the production of antibiotic in a stepwise manner. This new breeding approach could be especially effective for initially improving the production of antibiotics from wild-type strains.
Members of the genus Streptomyces produce a wide variety of secondary metabolites that include about half of the known microbial antibiotics. Many of these compounds have been applied in both human medicine and agriculture. Streptomyces coelicolor A3(2), the most fully genetically characterized streptomycete, is an appropriate strain for studying the regulation of antibiotic production (11, 29, 30). It produces at least four antibiotics, including the blue-pigmented polyketide antibiotic actinorhodin that is normally produced in stationary-phase cultures (9, 17). Much progress has been made in elucidating the organization of antibiotic biosynthesis gene clusters in several Streptomyces species, and a number of pathway-specific regulatory genes have been identified that are required for the activation of their cognate biosynthetic genes (18, 21, 34, 46, 53). In the actinorhodin biosynthetic pathway, actII-ORF4 plays such a pathway-specific regulatory role. The production of actinorhodin is mediated at the transcriptional level through activation of the actII-ORF4 promoter (22). In addition to pathway-specific regulatory genes, S. coelicolor possesses several genes with pleiotropic effects on antibiotic production. These genes fall into two classes: those that affect only antibiotic production (absA, absB, afsB, afsR, and abaA) (1, 8, 19, 31, 32) and those that affect both antibiotic production and morphological differentiation (bldA, bldB, bldC, bldD, bldG, and bldH) (28). Many other factors influence directly or indirectly the production of actinorhodin in S. coelicolor A3(2). It has been stressed that ppGpp (guanosine 3′-diphosphate, 5′-diphosphate), which is responsible for the so-called stringent response, plays a role in triggering the onset of antibiotic production, including the production of actinorhodin in S. coelicolor (7, 37, 41, 48, 49, 51).
We reported previously that certain mutations conferring streptomycin resistance give rise to secondary metabolite production (by an unknown mechanism) without the requirement for ppGpp in Streptomyces lividans and S. coelicolor A3(2) (25, 51, 55). Later, we demonstrated that the introduction of a specific str mutation into other bacterial genera gave rise to a marked increase in antibiotic productivity, thus further elucidating the mechanism in S. coelicolor A3(2) (33). Recently, we found that acquisition of resistance to other aminoglycoside antibiotics, such as gentamicin and Geneticin, also confers the ability to produce actinorhodin in S. lividans 66, which normally does not produce actinorhodin (H. Hu and K. Ochi, unpublished). Furthermore, by inducing mutations conferring resistance to rifampin, an ansamycin antibiotic, we were able to restore the impaired actinorhodin production in the relA and relC mutants of S. coelicolor A3(2) (Y. Tozawa, J. Xu, and K. Ochi, unpublished data). These results offer some available strategies for improving the productivity of antibiotics. Since the development of methods to improve the production of antibiotics is of considerable industrial and economic importance, we attempted in the present study to develop novel approaches for improving antibiotic-producing strains, especially focusing on methods to induce combined drug-resistant mutations. Some physiological aspects of the mutant strains are also described.
MATERIALS AND METHODS
Bacterial strains and preparation of mutants.
The wild-type strain (1147) of S. coelicolor A3(2) and its mutants used in this study are listed in Table 1. Spontaneous streptomycin-resistant (Strr), gentamicin-resistant (Genr), Geneticin-resistant (Gner ), or rifampin-resistant (Rifr) mutants were obtained from colonies that grew within 7 days after spores (or cells) were spread on GYM agar containing various concentrations of the above drugs (see Table 2).
TABLE 1.
Strains of S. coelicolor A3(2) used in this study
| Straina | Relevant genotype | Source (reference)b |
|---|---|---|
| 1147 | Prototrophic wild type | D. A. Hopwood (30) |
| S-1 | str-11 | Streptomycin-resistant mutant from 1147 |
| S-2 | str-12 | Streptomycin-resistant mutant from 1147 |
| S-3 | str-13 | Streptomycin-resistant mutant from 1147 |
| G-1 | gen-1 | Gentamicin-resistant mutant from 1147 |
| G-2 | gen-2 | Gentamicin-resistant mutant from 1147 |
| G-3 | gen-3 | Gentamicin-resistant mutant from 1147 |
| R-1 | rif-1 | Rifampin-resistant mutant from 1147 |
| R-2 | rif-2 | Rifampin-resistant mutant from 1147 |
| R-3 | rif-3 | Rifampin-resistant mutant from 1147 |
| SGe-1 | str-11 gne-1 | Geneticin-resistant mutant from S-1 |
| SGe-2 | str-11 gne-2 | Geneticin-resistant mutant from S-1 |
| SGe-3 | str-11 gne-3 | Geneticin-resistant mutant from S-1 |
| SG-1 | str-11 gen-4 | Gentamicin-resistant mutant from S-1 |
| SG-2 | str-11 gen-5 | Gentamicin-resistant mutant from S-1 |
| SG-3 | str-11 gen-6 | Gentamicin-resistant mutant from S-1 |
| SR-1 | str-11 rif-4 | Rifampin-resistant mutant from S-1 |
| SR-2 | str-11 rif-5 | Rifampin-resistant mutant from S-1 |
| SR-3 | str-11 rif-6 | Rifampin-resistant mutant from S-1 |
| SR-4 | str-11 rif-7 | Rifampin-resistant mutant from S-1 |
| SR-5 | str-11 rif-8 | Rifampin-resistant mutant from S-1 |
| SR-6 | str-11 rif-9 | Rifampin-resistant mutant from S-1 |
| SR-7 | str-11 rif-10 | Rifampin-resistant mutant from S-1 |
| SGR-1 | str-11 gen-4 rif-11 | Rifampin-resistant mutant from SG-1 |
| SGR-2 | str-11 gen-4 rif-12 | Rifampin-resistant mutant from SG-1 |
| SGR-3 | str-11 gen-4 rif-13 | Rifampin-resistant mutant from SG-1 |
S, G, and R, str, gen, and rif mutants, respectively; SGe, SG, and SR, str gne, str gen, and str rif double mutants, respectively; SGR, str gen rif triple mutant.
All mutant strains isolated in this study were spontaneous antibiotic-resistant mutants.
TABLE 2.
Screening and antibiotic productivity of drug-resistant mutants
| Strain | Actinorhodin productivity (OD633)a | MIC (μg/ml)b of: | Concn of antibiotic used for screening (μg/ml) | Frequency (%) of mutants producing increased actinorhodinc | Mean productivity detected (OD633 ± SE)d | Highest productivity detected (OD633) |
|---|---|---|---|---|---|---|
| S. coelicolor 1147 (wild type) | 0.77 | Gentamicin (0.1) | 1.0 | 5 (4/80) | 1.06 ± 0.20 | 1.25 |
| Streptomycin (1.0) | 5 | 6 (7/120) | 1.17 ± 0.11 | 1.39 | ||
| Rifampin (10) | 200 | 10 (15/150) | 1.85 ± 0.21 | 2.32 | ||
| S-1 | 1.28 | Geneticin (0.2) | 2.5 | 13 (15/112) | 1.87 ± 0.17 | 2.22 |
| Gentamicin (0.1) | 2.5 | 14 (14/104) | 2.35 ± 0.35 | 2.80 | ||
| Rifampin (10) | 200 | 18 (21/116) | 2.47 ± 0.32 | 3.14 | ||
| SG-1 | 2.02 | Rifampin (10) | 200 | 10 (8/80) | 6.00 ± 0.73 | 6.88 |
,d The optical density of supernatant at 633 nm (OD633) was determined after 6 days of cultivation at 30°C, using a 25-ml test tube containing 5 ml of R4 medium. All measurements were done in triplicate, and the mean value is presented. Mean productivity is the mean value of the productivity of mutants producing increased actinorhodin. SE, standard error.
Determined after 2 days of incubation on GYM agar.
Mutants producing more antibiotic than starting strain. Numbers in parentheses show the number of mutants producing more antibiotic divided by the number of mutants tested.
Media and growth conditions.
GYM, R3, and R4 media were described previously (48, 55). All strains were stored as spore suspensions at −20°C. For use in each experiment, spore suspensions were spread onto GYM agar plates and incubated for 7 to 10 days at 30°C to allow for sporulation. Sterile distilled water (5 ml) was added to each plate, and the surface was gently scraped to release the spores. Suspensions were collected by centrifugation and washed twice with distilled water. Before being used for inoculation, the spores were dispersed for 10 min in a sonic bath. The concentrations of spores were about 2 × 109 spores per ml. A total of 0.5 ml of spore suspension was inoculated into 150 ml of media. Cultivation of cells was carried out with 500-ml flasks containing 150 ml of media and incubated on a rotary shaker (200 rpm) at 30°C. In some experiments, cultivation was performed by using 25-ml test tubes each containing 5 ml of media and incubated on a reciprocal shaker (350 rpm) at 30°C.
Assay for actinorhodin.
Culture samples (1 ml) were taken at each time point and adjusted to pH 8.0. After centrifugation was carried out at 1,100 × g for 5 min, the amount of the blue-colored antibiotic actinorhodin was determined by measuring the optical density of supernatants at 633 nm. Measurements were always taken from duplicate or triplicate cultures, and the mean values are presented in Table 2 or legends to Fig. 2, 3, and 4.
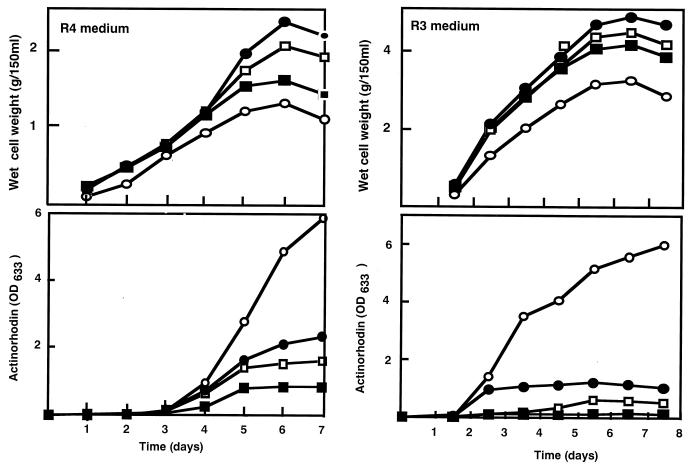
FIG. 2.
Comparison of actinorhodin production between media R3 (□) and R4 (▪). Actinorhodin production was determined after 6 days of incubation.
FIG. 3.
Growth and actinorhodin production in media R4 and R3. Symbols: ▪, 1147 (wild type); □, S-1 (str); ●, SG-1 (str gen); ○, SGR-1 (str gen rif).
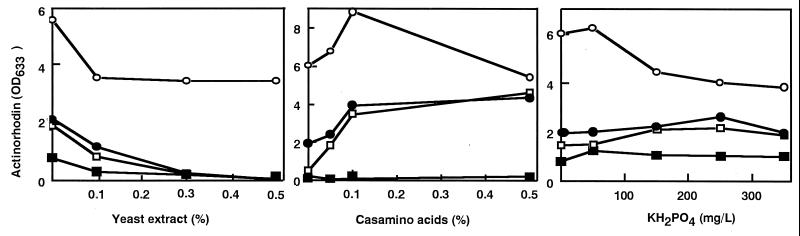
FIG. 4.
The effect of yeast extract, Casamino Acids, and KH2PO4 on the production of actinorhodin. Strains were grown in R4 medium supplemented with various concentrations of yeast extract, Casamino Acids, or KH2PO4 for 6 days. Symbols: ▪, 1147 (wild type); □, S-1 (str); ●, SG-1 (str gen) ; ○, SGR-1 (str gen rif).
Determination of MIC and resistance.
The lowest concentration of an antibiotic that totally inhibited growth through a 48-h incubation period at 30°C on GYM agar was defined as the MIC. The resistance levels were determined similarly to the MIC.
Mutation analyses of the rpsL and rpoB genes.
The rpsL gene fragment of the streptomycin-resistant mutant (S-1, S-2, or S-3) was obtained by PCR using the mutants' genomic DNA as a template and the synthetic oligonucleotide primers P1 (forward: 5′-ATTCGGCACACAGAAAC-3′) and P2 (reverse: 5′-AGAGGAGAACCGTAGAC-3′), which were designed from the sequence for S. lividans (DDBJ accession no. D83746). ExTaq (Takara) was used to perform PCR according to the manufacturer's instructions. A Perkin-Elmer Cetus thermal cycler was used at the following conditions: 5 min of incubation at 96°C; 30 cycles of 96°C for 18 s, 55°C for 12 s, and 72°C for 30 s; and a final step at 72°C for 10 min. PCR products were directly sequenced by the dideoxy chain termination procedure (54) using the BigDye Terminator Cycle Sequencing kit (Perkin-Elmer Applied Biosystems, Foster City, Calif.). The sequence data were analyzed with the GENETIX program (Software Development Co., Tokyo, Japan). The partial rpoB gene fragment (nucleotides 600 to 1300) of the rifampin-resistant mutants (R-1, R-2, R-3, etc.) was obtained by PCR using the mutants' genomic DNA as a template and the synthetic oligonucleotide primers P3 (forward: 5′-GGCCGCTACAAGGTGAACAAGAAG-3′) and P4 (reverse: 5′-CGATGACGAAGCGGTCCTCC-3′), which were designed from the sequence for S. coelicolor M145. PCR and DNA sequencing were performed under the same conditions as those for the rpsL gene.
Western blotting analysis.
Cultures were grown on GYM agar plates covered with cellophane sheets at 30°C for 36 to 72 h. Cells were scraped off the cellophane sheet; suspended in 20 mM Tris-HCl (pH 7.0) containing 1 mM EDTA, 1 mM dithiothreitol, 10% (vol/vol) glycerol, and 0.5 mM phenylmethylsulfonyl fluoride; and disrupted by sonication as described previously by Gramajo et al. (23). Protein concentrations were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Western blots were developed with the enhanced chemiluminescence Western blotting detection system for chemiluminescent detection as specified by the manufacturer. Polyclonal antiserum against the ActII-ORF4 protein was prepared in rabbits and used as a primary antibody at a dilution of 1:3,000.
RESULTS
Construction of combined resistant mutants.
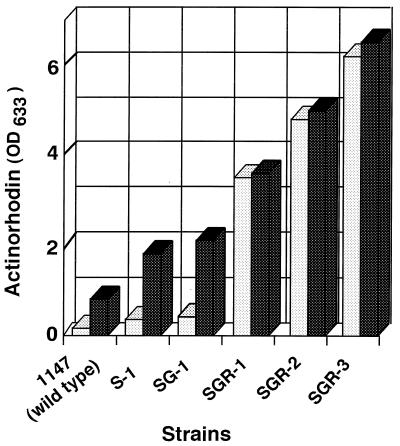
First, we introduced a single drug-resistant mutation into S. coelicolor wild-type strain 1147; the results are summarized in Table 2. The mutants with enhanced actinorhodin production (we tested 80 to 150 resistant isolates per antibiotic) were detected at a high frequency (5 to 10%) among streptomycin-resistant, gentamicin-resistant, or rifampin-resistant isolates. The highest productivity detected for each mutant strain ranged from 1.6 to 3 times the wild-type production level (Table 2). It is notable that the gen mutants with enhanced actinorhodin productivity all demonstrated low levels of resistance (a threefold-higher MIC) to gentamicin, whereas the str or rif mutants revealed either a low or high level of resistance (5- to 100-fold-higher MIC of streptomycin; 5- to 40-fold-higher MIC of rifampin). Several representative strains are listed in Tables 1 and 3.
TABLE 3.
Summary of mutations on the S. coelicolor rpsL or rpoB gene resulting in amino acid exchange
| Strain | Position in rpsL genea | Amino acid position (exchange) | Position in rpoB geneb | Amino acid position (exchange) | Resistance level (μg/ml)c to:
|
|||
|---|---|---|---|---|---|---|---|---|
| STRd | GEN | RIF | GNE | |||||
| 1147 | —e | 1 | 0.1 | 10 | 0.2 | |||
| S-1 | 262A→G | 88 (Lys→Glu) | 100 | 0.1 | 10 | 0.2 | ||
| S-2 | NDf | 5 | 0.2 | 10 | 0.2 | |||
| S-3 | ND | 10 | 0.1 | 5 | 0.1 | |||
| G-1 | ND | 1 | 0.3 | 10 | 0.2 | |||
| G-2 | ND | 1 | 0.3 | 10 | 0.2 | |||
| G-3 | ND | 1 | 0.3 | 10 | 0.2 | |||
| R-1 | 1049G→A | 350 (Arg→His) | 1 | 0.1 | 400 | 0.1 | ||
| R-2 | 1040A→G | 347 (His→Arg) | 1 | 0.1 | 400 | 0.2 | ||
| R-3 | 1049G→T | 350 (Arg→Phe) | 1 | 0.1 | 400 | 0.2 | ||
| SGe-1 | 262A→G | 88 (Lys→Glu) | 100 | 0.5 | ||||
| SGe-2 | 262A→G | 100 | 1 | |||||
| SGe-3 | 262A→G | 100 | 1 | |||||
| SG-1 | 262A→G | 100 | 0.3 | |||||
| SG-2 | 262A→G | 100 | 0.3 | |||||
| SG-3 | 262A→G | 100 | 0.3 | |||||
| SR-1 | 262A→G | 995T→C | 332 (Leu→Arg) | 50 | 50 | |||
| SR-2 | 262A→G | 1154C→T | 385 (Pro→Leu) | 50 | 150 | |||
| SR-3 | 262A→G | 1179C→G | 393 (Ile→Met) | 50 | 400 | |||
| SR-4 | 262A→G | 1011C→A | 337 (Asp→Glu) | 100 | 400 | |||
| SR-5 | 262A→G | 1010A→G | 337 (Asp→Gly) | 100 | 400 | |||
| SR-6 | 262A→G | 1028C→T | 343 (Ser→Leu) | 50 | 400 | |||
| SR-7 | 262A→G | 1048C→T | 350 (Arg→Cys) | 50 | 400 | |||
| SGR-1 | 262A→G | 1039C→T | 347 (His→Tyr) | 100 | 0.2 | 400 | ||
| SGR-2 | 262A→G | 1041C→A | 347 (His→Gln) | 100 | 0.2 | 400 | ||
| SGR-3 | 262A→G | ND | 50 | 0.3 | 300 | |||
,b Numbering originates from the start codon (GTG or ATG) of the open reading frame.
Determined after 4 days of cultivation on GYM agar.
STR, GEN, RIF, and GNE, streptomycin, gentamicin, rifampin, and geneticin, respectively.
—, wild-type rpsL gene.
Mutations were not detected (ND) within the rpsL or rpoB gene.
Next, we constructed double mutants by generating spontaneous Genr , Rifr, or Gner mutants from the str mutant S-1, which was used as the starting strain (Table 2). The frequency of double mutants producing a greater amount of actinorhodin was as high as 13 to 18%, and the highest productivity detected ranged from 1.7 to 2.5 times that of strain S-1. These results indicate that all of the combinations of each single-resistance mutation resulting in the generation of double mutants used here (str gne, str gen, and str rif) are effective for increasing the production of actinorhodin. Representative strains are listed in Tables 1 and 3.
Finally, triple mutants were constructed by generating spontaneous rif mutants from an str gen double mutant (SG-1) as the starting strain (Table 2). The frequency of triple mutants producing a greater amount of actinorhodin was as high as 10%, and the highest productivity detected was 3.5 times higher than that of the starting str gen double-mutant strain. Thus, the third mutation (rif) was effective for increasing actinorhodin productivity in strains containing double mutations. Consistent with these results, no cross-resistance was detected among mutants resistant to streptomycin, gentamicin, or rifampin (Table 3).
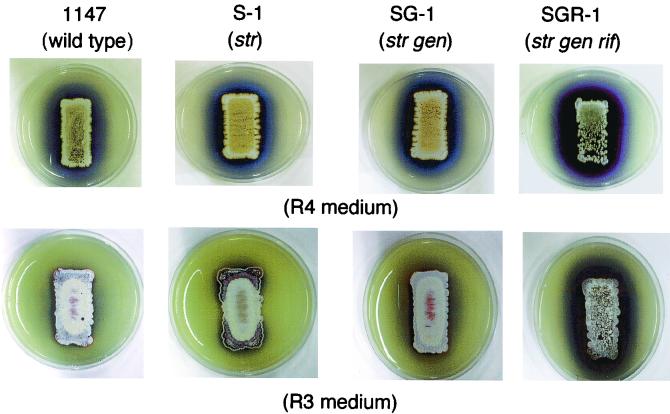
As examined on R4 and R3 agar plate cultures, single and double mutants (strains S-1 and SG-1) grew as well as the wild-type strain and produced abundant aerial mycelia and spores. However, the triple mutant (SGR-1) grew somewhat more slowly and produced a smaller amount of aerial mycelia and spores (Fig. 1). It is evident from the results shown in Fig. 1 that also parallel those from liquid cultures (see below) that actinorhodin productivity increased in the following hierarchical order: single, double, and triple mutants.
FIG. 1.
Ability to produce aerial mycelia and actinorhodin in S. coelicolor wild-type strain 1147 and mutant strains. Spores were inoculated on R4 or R3 agar plates and incubated at 30°C for 6 days. A blue color represents the pigmented antibiotic actinorhodin.
Mutational analyses of the mutants.
There is strong evidence that streptomycin resistance frequently results from a mutation in the rpsL gene, which encodes the ribosomal protein S12 (33, 55), while rifampin resistance results from a mutation in the rpoB gene, which encodes the β-subunit of RNA polymerase (24, 35, 59). We therefore sequenced and compared the rpsL gene and rpoB gene from the mutants to the wild-type strain. As summarized in Table 3, the str mutant with high resistance to streptomycin (strain S-1) contained a mutation within the rpsL gene, where the altered nucleotide (from A to G) was found at position 262, resulting in an alteration of Lys-88 to Glu. str mutants with low levels of streptomycin resistance (S-2 and S-3) showed no mutation in the rpsL gene. Although certain mutations in the16S rRNAs or the rpsD gene, which encodes the ribosomal S4 protein, have been known to confer streptomycin resistance (4, 43), no mutation was found in either.
Although gentamicin and Geneticin, as well as streptomycin, are classified as aminoglycoside antibiotics, none of the gen and gne mutants (single or double) examined gave rise to a mutation in the rpsL gene (Table 3). It is known that methylation of a specific 16S rRNA site (G-1405 or A-1408) elicits high-level resistance to gentamicin, as demonstrated by the gentamicin-producing strain Micromonospora purpurea (3). Also, mutations in the rplF gene, which codes for the ribosomal L6 protein, are reported to give rise to a low level of resistance to gentamicin in Escherichia coli (5, 14). However, as examined in the three gen mutants (G-1, G-2, and G-3), no mutation was detected in either the 16 S rRNAs (rrnA, rrnB, rrnC, rrnD, rrnE, and rrnF) or the rplF gene (data not shown).
When sequencing the rpoB gene in the rif mutants, we focused on a specified region (nucleotides 600 to 1300) which includes the so-called rif domain as detected previously in E. coli (35). The sequencing data revealed that most of the rif mutants possess a point mutation in this region. However, only in strain SGR-3 did we not detect a mutation in this region, suggesting that there may be a mutation in a part of the rpoB gene not sequenced (Table 3). The rif alleles detected could be divided into two clusters; cluster I covers amino acids 331 to 352, while cluster II covers amino acids 385 to 393. Most of the rif alleles conferred a high level of resistance (>300 μg/ml) against rifampin, except for two alleles (Lys-332 to Arg and Pro-385 to Leu) which conferred less resistance (Table 3).
Physiological characterization of the mutants.
Production of actinorhodin by wild-type and mutant strains was medium dependent. This was especially pronounced in the wild-type, single-mutant, and double-mutant strains (Fig. 2). In contrast, triple (str gen rif) mutants produced a high level of actinorhodin, irrespective of the medium used for cultivation, indicating the superior ability of these triple mutants to produce the antibiotic. Although triple mutants ultimately produced a high level of actinorhodin, production commenced at the same time as for the wild-type strain when examined with R4 and R3 media (Fig. 3). The triple mutants (i.e., SGR-1) (Fig. 3) revealed a somewhat reduced growth rate. It should be noted that the actinorhodin production by the triple mutant continued for a longer period of time (4 days) than by the single or double mutants (2 days).
Next, we studied how varying the nutritional source can effect actinorhodin production, using R4 medium as a basal medium. As summarized in Fig. 4, supplementation of yeast extract resulted in the severe impairment of actinorhodin productivity. This result was less pronounced in the triple mutant. Unlike yeast extract, Casamino Acids were effective for enhancing actinorhodin production in the single, double, or triple mutants but not the wild-type strain, demonstrating the efficacy of those drug-resistant mutations. KH2PO4 had virtually no effect on actinorhodin productivity (Fig. 4).
Finally, we compared the actinorhodin productivity of wild-type and mutant strains using GYM, R3, and R4 media. Multiple mutations were always effective for increasing actinorhodin productivity in any of the media used, giving rise to 48-, 40-, and 9-fold increases in antibiotic production in GYM, R3, and R4 media, respectively. Thus, we conclude that by inducing combined drug-resistant mutations we can continuously increase the productivity of actinorhodin in a stepwise manner.
Genetic characterization of the mutants.
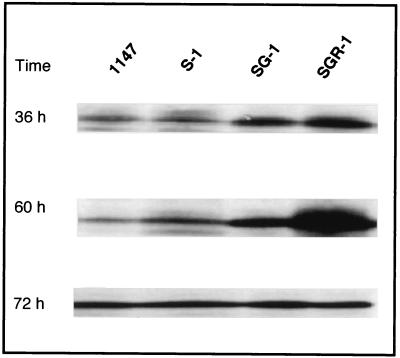
The ActII-ORF4 protein has been characterized as a DNA binding protein that positively regulates the transcription of the actinorhodin biosynthesis genes (2). Transcription of actII-ORF4 is growth-phase dependent in liquid culture, reaching a maximum output during the transition from exponential to stationary phase (22). We therefore analyzed the expression level of the ActII-ORF4 protein by Western analysis in the wild-type and mutant strains (Fig. 5). Cells were grown on GYM agar covered with a cellophane sheet, scraped off, disrupted by sonication, and then subjected to Western analysis (see Materials and Methods). Wild-type strain 1147 produced a basal level of ActII-ORF4 during the middle (36 h) and late (60 h) growth phases that increased when cells entered into the stationary phase (72 h). It should be stressed that the amount of ActII-ORF4 increased (especially at transition phase) in the following hierarchical order: single (S-1), double (SG-1), and triple (SGR-1) mutants (Fig. 5). The increase in ActII-ORF4 was especially pronounced in the triple mutant (SGR-1) at 60 h, reflecting the superior ability (see above) of this strain. The dramatically increased expression of ActII-ORF4 is transient, as the expression of the triple mutant decreased later (at 72 h) to the same level as that of the wild-type strain (Fig. 5).
FIG. 5.
Western blotting analysis of the ActII-ORF4 protein, a pathway-specific positive regulator in the actinorhodin biosynthesis pathway. Cultures were grown at 30°C for the denoted time on a GYM agar plate covered with a cellophane sheet (see Materials and Methods). Each lane contained 20 μg of total proteins.
DISCUSSION
In the present study, we successfully developed a new approach for improving antibiotic producers by inducing combined drug-resistant mutations. This method not only results in a remarkable increase in antibiotic productivity (48-fold higher in GYM medium) but also makes it possible to generate positive mutations at a high frequency (5 to 15%). Although much progress has been made in improving antibiotic producers (10, 39, 40), our method is characterized by the host cell's amenability (generation of spontaneous drug-resistant mutation) and the method's applicability to a number of microorganisms (if not all) as demonstrated previously with other bacteria (33, 55). It should also be emphasized that combined resistant mutations (triple mutations) demonstrated no significant impairment in growth or sporulation under the conditions tested. However, since the triple mutants (e.g., SGR-1) had a reduced growth rate (Fig. 3), it is highly likely that the effect of the mutations is actually to alter growth rates and the timing of entrance into secondary metabolism.
A high level of resistance to streptomycin has been previously shown to result from a point mutation in the rpsL gene, which encodes the ribosomal protein S12 (20, 25, 27, 33, 42, 55). In agreement with this, our str mutant (S-1), which showed a high level of resistance, contained a mutation (Lys-88 to Glu) in the rpsL gene. However, those strains with low levels of resistance (S-2 and S-3) showed no mutation in the rpsL gene. It has been reported that mutations in the 16S rRNAs (530 loop region) or the rpsD gene, which encodes the ribosomal S4 protein, can confer streptomycin resistance (4, 43). Gentamicin resistance is known to result from a mutation in the rplF gene, which codes for the ribosomal L6 protein (5, 38). Mutations occur within their putative RNA-binding sites (14). Nevertheless, we could not detect this mutation in our mutant strains, which had a low level of resistance to streptomycin or gentamicin, although we analyzed every 16S rRNA, rpsD, and rplF sequence. However, it is possible that our mutants have a mutation in certain ribosomal components other than the 16S rRNAs, S4, and L6 proteins.
Aminoglycosides are the best characterized as a class of antibiotics that bind directly to rRNA, cause a decrease in translational accuracy, and inhibit translocation of the ribosome (15, 16). These antibiotics bind to a conserved sequence in the rRNA that is near the site of codon-anticodon recognition in the aminoacyl-tRNA site (A site) of the 30S subunit. Aminoglycoside binding stabilizes the tRNA-mRNA interaction in the A site by decreasing tRNA dissociation rates; this decrease interferes with proofreading steps that ensure translational fidelity (36). The action of streptomycin on bacterial ribosomes has been studied in great detail (reviewed by Wallace et al. [58] and Cundliffe [13]), and among the numerous effects attributed to this drug, the misreading of the mRNA codons is the best known. Gentamicin (and also Geneticin) belongs to the kanamycin class of aminoglycosides but is structurally different from streptomycin when classified on the basis of structural characteristics. The L6 mutations are drastic because they result in large deletions of an RNA-binding region; thus, they may indirectly affect proofreading by locally distorting the EF-Tu–GTP–aminoacyl tRNA-binding site on the large subunit (14, 60). It is well known that S12 mutations, which can confer streptomycin resistance, can in general increase the accuracy of protein synthesis. More recently, defined regions of the 16S rRNA have also come to be associated with the ribosome's accuracy function (43, 45, 52). It is notable that translational accuracy can also be affected by two components of the 50S subunit, the ribosomal protein L6 and the 2660 loop region of the 23S rRNA. Mutations have been identified in these components that result in a decreased rate of translation, greater accuracy in protein synthesis, and increased resistance to many of the misreading-inducing aminoglycoside antibiotics, in particular gentamicin (5, 38, 44, 57). Failure in identifying these mutations in our mutant strains may be due to the fact that, unlike the previously identified mutations as described above, we are dealing with mutations which confer only a low level of (or even slight) resistance to streptomycin or gentamicin.
Previous studies dealing with various bacteria have indicated that mutations in the rpoB gene, which codes for the β-subunit of RNA polymerase, are responsible for the acquisition of resistance to rifampin (24, 35, 56, 59). Almost all the mutations found are located on a specified conserved region of the rpoB gene that can be divided into three clusters: I, II, and III. In the present study, most of the rif mutants exhibited point mutations in either cluster I or II, resulting in an amino acid alteration in one of eight sites. All of these sites correspond to previously known positions conferring rifampin resistance, although some new substitution types have been found. These results are apparently related to previous findings that show that the guanine nucleotide ppGpp is a pivotal signal molecule for the onset of antibiotic production (7, 41, 47, 51), since ppGpp has been proven to bind to the β-subunit of E. coli RNA polymerase (12). ppGpp [and (p)ppGpp] is believed to be responsible for the stringent response, which causes an immediate cessation of RNA synthesis and other cellular reactions (for reviews, see Cashel et al. [6]). Strains with mutated relA (which codes for ppGpp synthetase) or relC = rplK (which codes for the ribosomal L11 protein) fail to synthesize normal levels of ppGpp. Although the relA and relC mutants of various Streptomyces spp. exhibit a severely impaired ability to produce antibiotics due to the failure to synthesize ppGpp (7, 37, 48, 50, 51), we have found that the acquisition of certain rif mutations by S. coelicolor relA and relC mutants restores the antibiotic productivity lost in these mutants (Y. Tozawa, J. Xu, and K. Ochi, unpublished data). These rif mutants, like the rif mutants used in the present study, have mutations in the β-subunit of RNA polymerase. The dependence of S. coelicolor on ppGpp to initiate antibiotic production is therefore apparently bypassed by certain mutations in RNA polymerase. It is possible that the remarkable enhancement of ActII-ORF4 expression which accompanies the overproduction of actinorhodin (Fig. 5) is based on the independence of cells on ppGpp in initiating the secondary metabolism. The mutant RNA polymerase may function by mimicking the ppGpp bound form.
Despite the lack of detail on the molecular level for the effects of these mutations, overexpression of the ActII-ORF4 protein by introducing str, gen, and rif mutations accounts well for the observed hierarchical increase in actinorhodin productivity, since ActII-ORF4 plays a crucial role in activating the genes necessary for actinorhodin biosynthesis (2). Antibiotic production is in general subjected to the suppressive effects caused by an excess of nutrients such as carbon, nitrogen, and phosphate sources (26). In particular, ammonium and phosphate both appear to be major regulators of antibiotic production in S. coelicolor A3(2) and their control systems may be interrelated in some way (26). Consistent with this notion, our results revealed that actinorhodin production in wild-type and mutant strains is more or less medium dependent, displaying more production in R4 medium (containing less yeast extract and phosphate) than in R3 medium (Fig. 3). It is important to point out that the triple (str gen rif) mutants constructed in the present study all revealed less sensitivity to such suppressive effects (Fig. 2 through 4). Our new breeding approach could be effective especially for initially improving the production of antibiotics from wild-type strains and may be effective not only for antibiotic production but also for certain enzyme production linked with secondary metabolism.
ACKNOWLEDGMENTS
This work was supported by a grant from the Organized Research Combination System (ORCS) of the Science and Technology Agency of Japan. Haifeng Hu is a recipient of a fellowship from the Science and Technology Agency of Japan (STA fellowship).
We are grateful to Alexander Lezhava for his help in performing Western blotting and Yuzuru Tozawa for his comments about rifampin-resistant mutations.
REFERENCES
- 1.Adamidis T, Riggle P, Champness W. Mutations in a new Streptomyces coelicolor locus which globally block antibiotic biosynthesis but not sporulation. J Bacteriol. 1990;172:2962–2969. doi: 10.1128/jb.172.6.2962-2969.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias P, Fernández-Moreno M A, Malpartida F. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J Bacteriol. 1999;181:6958–6968. doi: 10.1128/jb.181.22.6958-6968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beauclerk A A D, Cundliffe E. Sites of action of two ribosomal RNA methylases responsible for resistance to aminoglycosides. J Mol Biol. 1987;193:661–671. doi: 10.1016/0022-2836(87)90349-4. [DOI] [PubMed] [Google Scholar]
- 4.Bjorkman J, Samuelsson P, Andersson D I, Hughes D. Novel ribosomal mutations affecting translational accuracy, antibiotic resistance and virulence of Salmonella typhimurium. Mol Microbiol. 1999;31:53–58. doi: 10.1046/j.1365-2958.1999.01142.x. [DOI] [PubMed] [Google Scholar]
- 5.Buckel P, Buchberger A, Böck A, Wittmann H-G. Alteration of ribosomal protein L6 in mutants of Escherichia coli resistant to gentamicin. Mol Gen Genet. 1977;158:47–54. doi: 10.1007/BF00455118. [DOI] [PubMed] [Google Scholar]
- 6.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 7.Chakraburtty R, White J, Takano E, Bibb M. Cloning, characterization and disruption of a (p)ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) Mol Microbiol. 1996;19:357–368. doi: 10.1046/j.1365-2958.1996.390919.x. [DOI] [PubMed] [Google Scholar]
- 8.Champness W, Riggle P, Adamidis T. Loci involved in regulation of antibiotic synthesis. J Cell Biochem. 1990;14A:88. [Google Scholar]
- 9.Chater K F. Aspects of multicellular differentiation in Streptomyces coelicolor A3(2) In: Hershberger C L, Queener S W, Hegeman G, editors. Genetics and molecular biology of industrial microorganisms. Washington, D.C.: American Society for Microbiology; 1989. pp. 99–107. [Google Scholar]
- 10.Chater K F. The improving prospects for yield increase by genetic engineering in antibiotic-producing streptomycetes. Bio/Technology. 1990;8:115–121. doi: 10.1038/nbt0290-115. [DOI] [PubMed] [Google Scholar]
- 11.Chater K F, Hopwood D A. Streptomyces. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 83–89. [Google Scholar]
- 12.Chatterji D, Fujita N, Ishihama A. The mediator for stringent control, ppGpp, binds to the β-subunit of Escherichia coli RNA polymerase. Genes Cells. 1998;3:279–287. doi: 10.1046/j.1365-2443.1998.00190.x. [DOI] [PubMed] [Google Scholar]
- 13.Cundliffe E. Recognition sites for antibiotics within rRNA. In: Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function, and evolution. Washington, D.C.: American Society for Microbiology; 1990. pp. 479–490. [Google Scholar]
- 14.Davies C, Bussiere D E, Golden B L, Porter S J, Ramakrishnan V, White S W. Ribosomal proteins S5 and L6: high-resolution crystal structures and roles in protein synthesis and antibiotic resistance. J Mol Biol. 1998;279:873–888. doi: 10.1006/jmbi.1998.1780. [DOI] [PubMed] [Google Scholar]
- 15.Davies J, Gorini L, Davis B D. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol Pharmacol. 1965;1:93–106. [PubMed] [Google Scholar]
- 16.Davies J, Davies B D. Misreading of ribonucleic acid code words induced by aminoglycoside antibiotics. J Biol Chem. 1968;243:3312–3316. [PubMed] [Google Scholar]
- 17.Demain A L, Aharoowitz Y, Martin J F. Metabolic control of secondary biosynthetic pathways. In: Vining L C, editor. Biochemistry and genetic regulation of commercially important antibiotics. London, England: Addison-Wesley; 1983. pp. 49–72. [Google Scholar]
- 18.Distler J, Ebert A, Mansouri K, Pissowotzki P, Stockmann M, Piepersberg W. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: nucleotide sequence of three genes and analysis of transcriptional activity. Nucleic Acids Res. 1987;15:8041–8056. doi: 10.1093/nar/15.19.8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernández-Moreno M A, Martín-Triana A J, Martínez E, Niemi J, Kieser H M, Hopwood D A, Malpartida F. abaA, a new pleiotropic regulatory locus for antibiotic production in Streptomyces coelicolor. J Bacteriol. 1992;174:2958–2967. doi: 10.1128/jb.174.9.2958-2967.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finken M, Kirschner P, Meier A, Wrede A, Böttger E C. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol Microbiol. 1993;9:1239–1246. doi: 10.1111/j.1365-2958.1993.tb01253.x. [DOI] [PubMed] [Google Scholar]
- 21.Geistlich M, Losick R, Turner J R, Rao R N. Characterization of a novel regulatory gene governing the expression of a polyketide synthase gene in Streptomyces ambofaciens. Mol Microbiol. 1992;6:2019–2029. doi: 10.1111/j.1365-2958.1992.tb01374.x. [DOI] [PubMed] [Google Scholar]
- 22.Gramajo H C, Takano E, Bibb M J. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol Microbiol. 1993;7:837–845. doi: 10.1111/j.1365-2958.1993.tb01174.x. [DOI] [PubMed] [Google Scholar]
- 23.Gramajo H C, White J, Hutchinson C R, Bibb M J. Overproduction and localization of components of the polyketide synthase of Streptomyces glaucescens involved in the production of the antibiotic tetracenomycin C. J Bacteriol. 1991;173:6475–6483. doi: 10.1128/jb.173.20.6475-6483.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heep M, Beck D, Bayerdorffer E, Lehn N. Rifampin and rifabutin resistance mechanism in Helicobacter pylori. Antimicrob Agents Chemother. 1999;43:1497–1499. doi: 10.1128/aac.43.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hesketh A, Ochi K. A novel method for improving Streptomyces coelicolor A3(2) for production of actinorhodin by introduction of rpsL (encoding ribosomal protein S12) mutations conferring resistance to streptomycin. J Antibiot. 1997;50:532–535. doi: 10.7164/antibiotics.50.532. [DOI] [PubMed] [Google Scholar]
- 26.Hobbs G, Frazer C M, Gardner D C J, Flett F, Oliver S G. Pigmented antibiotic production by Streptomyces coelicolor A3(2): kinetics and the influence of nutrients. J Gen Microbiol. 1990;136:2291–2296. [Google Scholar]
- 27.Honore N, Cole S T. Streptomycin resistance in mycobacteria. Antimicrob Agents Chemother. 1994;38:238–242. doi: 10.1128/aac.38.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hopwood D A. Towards an understanding of gene switching in Streptomyces, the basis of sporulation and antibiotic production. Proc R Soc Lond B Biol Sci. 1988;235:121–138. doi: 10.1098/rspb.1988.0067. [DOI] [PubMed] [Google Scholar]
- 29.Hopwood D A, Chater K F, Bibb M J. Genetics of antibiotic production in Streptomyces coelicolor A3(2) In: Vining L C, Stuttard C, editors. Genetics and biochemistry of antibiotic production. Newton, Mass: Butterworth-Heinemann; 1995. pp. 65–102. [DOI] [PubMed] [Google Scholar]
- 30.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, England: John Innes Foundation; 1985. [Google Scholar]
- 31.Horinouchi S, Kito M, Nishiyama M, Furuya K, Hong S K, Miyake K, Beppu T. Primary structure of AfsR, a global regulatory protein for secondary metabolite formation in Streptomyces coelicolor A3(2) Gene. 1990;95:49–56. doi: 10.1016/0378-1119(90)90412-k. [DOI] [PubMed] [Google Scholar]
- 32.Horinouchi S, Hara O, Beppu T. Cloning of a pleiotropic gene that positively controls biosynthesis of A-factor, actinorhodin, and prodigiosin in Streptomyces coelicolor A3(2) and Streptomyces lividans. J Bacteriol. 1983;155:1238–1248. doi: 10.1128/jb.155.3.1238-1248.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hosoya Y, Okamoto S, Muramatsu H, Ochi K. Acquisition of certain streptomycin-resistant (str) mutations enhances antibiotic production in bacteria. Antimicro Agents Chemother. 1998;42:2041–2047. doi: 10.1128/aac.42.8.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunter I S, Baumberg S. Molecular genetics of antibiotic formation. In: Baumberg S, Hunter I S, Rhodes P M, editors. Microbial products: new approaches. Cambridge, England: Cambridge University Press; 1989. pp. 121–162. [Google Scholar]
- 35.Jin D J, Gross C A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 36.Karimi R, Ehrenberg M. Dissociation rate of cognate peptidyl-tRNA from the A-site of hyper-accurate and error-prone ribosomes. Eur J Biochem. 1994;226:355–360. doi: 10.1111/j.1432-1033.1994.tb20059.x. [DOI] [PubMed] [Google Scholar]
- 37.Kawamoto S, Zhang D, Ochi K. Molecular analysis of the ribosomal L11 protein gene (rplK = relC) of Streptomyces griseus and identification of a deletion allele. Mol Gen Genet. 1997;255:549–560. doi: 10.1007/s004380050528. [DOI] [PubMed] [Google Scholar]
- 38.Kuhberger R, Piepersberg W, Petzet A, Buckel P, Böck A. Alteration of ribosomal protein L6 in gentamicin-resistant strains of Escherichia coli: effects on fidelity of protein synthesis. Biochemistry. 1979;18:187–193. doi: 10.1021/bi00568a028. [DOI] [PubMed] [Google Scholar]
- 39.Lai R, Khanna R, Kaur H, Khanna M, Dhingra N, Lai S, Gartemann K H, Eichenlaub R, Ghosh P K. Engineering antibiotic producers to overcome the limitations of classical strain improvement programs. Crit Rev Microbiol. 1996;22:201–255. doi: 10.3109/10408419609105481. [DOI] [PubMed] [Google Scholar]
- 40.Lee S H, Rho Y T. Improvement of tylosin fermentation by mutation and medium optimization. Lett Appl Microbiol. 1999;28:142–144. doi: 10.1046/j.1365-2672.1999.00478.x. [DOI] [PubMed] [Google Scholar]
- 41.Martinez-Costa O H, Arias P, Romero N M, Parro V, Mellado R P, Malpartida F. A relA/spoT homologous gene from Streptomyces coelicolor A3(2) controls antibiotic biosynthetic genes. J Biol Chem. 1996;271:10627–10634. doi: 10.1074/jbc.271.18.10627. [DOI] [PubMed] [Google Scholar]
- 42.Meier A, Kirschner P, Bange F-C, Vogel U, Böttger E C. Genetic alterations in streptomycin-resistant Mycobacterium tuberculosis: mapping of mutations conferring resistance. Antimicrob Agents Chemother. 1994;38:228–233. doi: 10.1128/aac.38.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melancon P, Lemieux C, Brakier-Gingras L. A mutation in the 530 loop of Escherichia coli 16S ribosomal RNA causes resistance to streptomycin. Nucleic Acids Res. 1988;16:9631–9639. doi: 10.1093/nar/16.20.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melançon P, Tapprich W E, Brakier-Gingras L. Single-base mutations at position 2661 of Escherichia coli 23S rRNA increase efficiency of translational proofreading. J Bacteriol. 1992;174:7896–7901. doi: 10.1128/jb.174.24.7896-7901.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montandon P E, Wagner R, Stutz E. E. coli ribosomes with a C912 to U base change in the 16S rRNA are streptomycin resistant. EMBO J. 1986;5:3705–3708. doi: 10.1002/j.1460-2075.1986.tb04703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narva K E, Feitelson J S. Nucleotide sequence and transcriptional analysis of the redD locus of Streptomyces coelicolor A3(2) J Bacteriol. 1990;172:326–333. doi: 10.1128/jb.172.1.326-333.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ochi K. Occurrence of the stringent response in Streptomyces sp. and its significance for the initiation of morphological and physiological differentiation. J Gen Microbiol. 1986;132:2621–2631. doi: 10.1099/00221287-132-9-2621. [DOI] [PubMed] [Google Scholar]
- 48.Ochi K. Metabolic initiation of differentiation and secondary metabolism by Streptomyces griseus: significance of the stringent response (ppGpp) and GTP content in relation to A factor. J Bacteriol. 1987;169:3608–3616. doi: 10.1128/jb.169.8.3608-3616.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ochi K. A relaxed (rel) mutant of Streptomyces coelicolor A3(2) with a missing ribosomal protein lacks the ability to accumulate ppGpp, A-factor and prodigiosin. J Gen Microbiol. 1990;136:2405–2412. doi: 10.1099/00221287-136-12-2405. [DOI] [PubMed] [Google Scholar]
- 50.Ochi K. Streptomyces relC mutants with an altered ribosomal protein ST-L11 and genetic analysis of a Streptomyces griseus relC mutant. J Bacteriol. 1990;172:4008–4016. doi: 10.1128/jb.172.7.4008-4016.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ochi K, Zhang D, Kawamoto S, Hesketh A. Molecular and functional analysis of the ribosomal L11 and S12 protein genes (rplK and rpsL) of Streptomyces coelicolor A3(2) Mol Gen Genet. 1997;256:488–498. doi: 10.1007/pl00008614. [DOI] [PubMed] [Google Scholar]
- 52.Powers T, Noller H F. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10:2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raibaud A, Zalacain M, Holt T G, Tizard R, Thompson C J. Nucleotide sequence analysis reveals linked N-acetyl hydrolase, thioesterase, transport, and regulatory genes encoded by the bialaphos biosynthetic gene cluster of Streptomyces hygroscopicus. J Bacteriol. 1991;173:4454–4463. doi: 10.1128/jb.173.14.4454-4463.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shima J, Hesketh A, Okamoto S, Kawamoto S, Ochi K. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2) J Bacteriol. 1996;178:7276–7284. doi: 10.1128/jb.178.24.7276-7284.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singer M, Jin D J, Walter W A, Cross C A. Genetic evidence for the interaction between cluster I and cluster III rifampicin resistant mutations. J Mol Biol. 1993;231:1–5. doi: 10.1006/jmbi.1993.1251. [DOI] [PubMed] [Google Scholar]
- 57.Tapprich W E, Dahlberg A E. A single-base mutation at position 2661 in Escherichia coli 23S ribosomal RNA affects the binding of ternary complexes to the ribosome. EMBO J. 1990;9:2649–2655. doi: 10.1002/j.1460-2075.1990.tb07447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wallace B J, Tai P-C, Davies B D. Streptomycin and related antibiotics. In: Hahn F E, editor. Antibiotics V—mechanism of action of antibacterial agents. New York, N.Y: Springer-Verlag; 1979. pp. 272–303. [Google Scholar]
- 59.Wichelhaus T A, Schäfer V, Brade V, Böddinghaus B. Molecular characterization of rpoB mutations conferring cross-resistance to rifamycins on methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:2813–2816. doi: 10.1128/aac.43.11.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshizawa S, Fourmy D, Puglisi J D. Structural origins of gentamicin antibiotic action. EMBO J. 1998;17:6437–6448. doi: 10.1093/emboj/17.22.6437. [DOI] [PMC free article] [PubMed] [Google Scholar]