Abstract
Steep vertical gradients of oxidants (O2 and NO3−) in Puget Sound and Washington continental margin sediments indicate that aerobic respiration and denitrification occur within the top few millimeters to centimeters. To systematically explore the underlying communities of denitrifiers, Bacteria, and Archaea along redox gradients at distant geographic locations, nitrite reductase (nirS) genes and bacterial and archaeal 16S rRNA genes (rDNAs) were PCR amplified and analyzed by terminal restriction fragment length polymorphism (T-RFLP) analysis. The suitablility of T-RFLP analysis for investigating communities of nirS-containing denitrifiers was established by the correspondence of dominant terminal restriction fragments (T-RFs) of nirS to computer-simulated T-RFs of nirS clones. These clones belonged to clusters II, III, and IV from the same cores and were analyzed in a previous study (G. Braker, J. Zhou, L. Wu, A. H. Devol, and J. M. Tiedje, Appl. Environ. Microbiol. 66:2096–2104, 2000). T-RFLP analysis of nirS and bacterial rDNA revealed a high level of functional and phylogenetic diversity, whereas the level of diversity of Archaea was lower. A comparison of T-RFLPs based on the presence or absence of T-RFs and correspondence analysis based on the frequencies and heights of T-RFs allowed us to group sediment samples according to the sampling location and thus clearly distinguish Puget Sound and the Washington margin populations. However, changes in community structure within sediment core sections during the transition from aerobic to anaerobic conditions were minor. Thus, within the top layers of marine sediments, redox gradients seem to result from the differential metabolic activities of populations of similar communities, probably through mixing by marine invertebrates rather than from the development of distinct communities.
In the ocean, the nitrogen budget is largely unbalanced, as the removal of inorganic nitrogen exceeds the total input up to fourfold, primarily due to sedimentary denitrification (10, 23). Denitrification is a dissimilatory microbial redox process where nitrogen oxides (NO3−, NO2−) are reduced stepwise to gaseous end products (NO, N2O, N2) which are concurrently released into the environment. The dominant sites of nitrogen loss are continental margin sediments, although they constitute only about 10% of the total marine sediment surface area (9, 13, 35). Marine sedimentary respiration is characterized by a stratification due to redox reactions consuming oxidants in the order oxygen, nitrate and manganese, iron, and sulfate (15), where the steepness of gradients is a function of reducible carbon input (5). Although the impact of microbial community structure on the development of these gradients and thus on the denitrification process in marine sediments is critical, our understanding of the underlying microbial populations is still limited.
Insights into community structures in environmental samples were achieved by the use of molecular tools such as 16S rRNA genes (rDNAs), which avoid the limitations of culturability (1, 36). Communities of Bacteria and Archaea have been successfully explored using terminal restriction fragment length polymorphism (T-RFLP) analysis of amplified total community 16S rDNA (8, 20, 22, 33). However, a group-specific 16S rDNA approach is not suitable for community analysis of denitrifying bacteria, as this functional group is widely distributed over the phylogenetic tree (31, 38). The genetic diversity of denitrifiers in marine sediments was explored by cloning nirK and nirS genes, which encode copper- and cytochrome cd1-containing nitrite reductases, respectively, key enzymes in the denitrification process (4). The PCR method to detect nir genes was highly specific for nirS and evaluated novel and diverse denitrifier communities in selected marine sediment samples (4). To more systematically explore bacterial communities on a functional level, PCR-amplified functional genes such as nirS genes can be used with subsequent T-RFLP analysis. This has been demonstrated for mercury resistance (mer) genes (6), ammonia monooxygenase (amoA) genes (19), and nosZ, which encodes nitrous oxide reductase (30).
In this study, we present a polyphasic DNA-based approach to investigate whether communities of denitrifying bacteria, Bacteria, and Archaea reflect redox gradients within marine sediment cores from different geographic locations by PCR amplification of nirS and 16S rDNAs and subsequent T-RFLP community analysis.
MATERIALS AND METHODS
Sediment sampling.
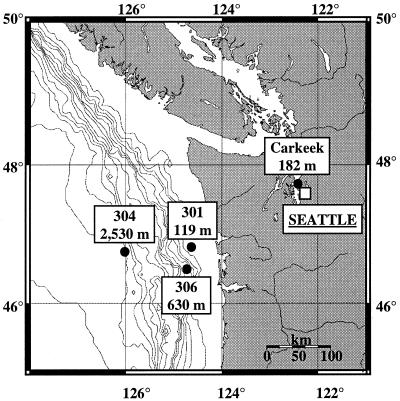
Sections from three sediment cores along a transect at the Washington margin (station 301, 119 m; station 306, 630 m; station 304, 2,530 m [water depth]) and from one sediment core from Puget Sound (Carkeek; water depth 182 m) were investigated (Fig. 1). Sediment cores (depth, 10 cm) from the Washington margin were collected in November 1997, subsampled vertically in 0.5- or 1.0-cm-thick sections, and stored in sterile polypropylene bags at −70°C until DNA was extracted in Michigan. The Puget Sound sediment core was collected in March 1998. Sections from depths of 1.0 to 1.5, 1.5 to 2.0, and 6.0 to 6.5 cm were subsampled and placed in sterile polypropylene bags. Samples were immediately stored on ice, shipped to Michigan on dry ice, and stored at −20°C until DNA was extracted. The sediment cores were sampled, and pore water oxygen, nitrate, and ammonia profiles (Fig. 2) were determined as described by Devol and Christensen (13).
FIG. 1.
Locations and water depths of the sampling stations at Puget Sound and the Washington margin.
FIG. 2.
Profiles of the oxidants oxygen, nitrate, nitrite, and ammonium within sediment cores from Puget Sound (Carkeek) and the Washington margin (cores 301, 306, and 304).
DNA extraction.
DNAs from 1-g sediment subsamples (station 301, sections from depths of 0.0 to 0.5 cm [301/1], 0.5 to 1.0 cm [301/2], 1.0 to 1.5 cm [301/3], 1.5 to 2.0 cm [301/4], 4.0 to 5.0 cm [301/5], and 9.0 to 10.0 cm [301/6]; station 306, 0.0 to 0.5 cm [306/1], 0.5 to 1.0 cm [306/2], 1.0 to 1.5 cm [306/3], 1.5 to 2.0 cm [306/4], and 6.0 to 7.0 cm [306/5]; station 304, 2.0 to 3.0 cm [304/1], 3.0 to 4.0 cm [304/2], and 4.0 to 5.0 cm [304/3]; and Puget Sound core sections, 1.0 to 1.5 cm [C/1], 1.5 to 2.0 cm [C/2], and 6.0 to 6.5 cm [C/3]) were extracted by the freeze-thaw procedure according to the method of van Elsas and Smalla (34) with an additional proteinase K treatment (50 μl of a 20-mg ml−1 solution) after incubation with sodium dodecyl sulfate. The DNAs were quantified and analyzed spectrophotometrically by taking point measurements at 230, 260, and 280 nm. The concentrations of all DNA extracts were adjusted to 100 ng μl−1.
PCR conditions.
PCR amplifications of nirS genes and bacterial and archaeal 16S rDNAs from total environmental DNA extracts were performed with a total volume of 50 μl in a model 9600 thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.). Fragments of nirS genes (approximately 890 bp) were amplified from 100 ng of total environmental DNA extracts using the primer pair nirS1F-nirS6R and the PCR method developed by Braker et al. (3) with modifications (4). The forward primer nirS1F was 5′-end labeled with 6-carboxyfluorescein (Operon Inc., Alameda, Calif.). Bacterial and archaeal 16S rDNAs from 100 ng of total environmental DNA extract were amplified in reaction mixtures containing 10 pmol of each primer, 200 μM each deoxyribonucleoside triphosphate, 400 ng of bovine serum albumin (Roche Molecular Biochemicals, Indianapolis, Ind.) μl−1, 150 mM MgCl2 (Gibco BRL, Gaithersburg, Md.), 0.5 U of Taq polymerase (Gibco BRL), and 1/10 volume of a 10× PCR buffer provided with the enzyme. After a denaturation step of 5 min at 95°C, amplification reactions were performed with 30 cycles of denaturation (1 min, 95°C), primer annealing (1 min, 57°C), and primer extension (3 min, 72°C) and a final extension step of 7 min at 72°C. Primers used for amplification of eubacterial 16S rDNAs (8-27F, 5′-AGAGTTTGATCMTGGCTCAG-3′, with M for A or C; 1392-1407R, 5′-ACGGGCGGTGTGTACA-3′) were described by Amann et al. (1) and modified by C. L. Moyer (unpublished results). Archaeal 16S rDNA primers were described by Moyer et al. (25). Forward primers for amplification of both bacterial and archaeal 16S rDNAs were 5′-end labeled with 5-hexachlorofluorescein (Operon Inc.).
Products of three replicate PCRs each for bacterial and archaeal 16S rDNA and those of five PCRs for nirS were combined. Aliquots (5 and 10 μl) of 16S rDNA and nirS PCR products were analyzed by electrophoresis on 0.8 and 2% (wt/vol) agarose gels (Gibco BRL), followed by 15 min of staining with ethidium bromide (0.5 mg liter−1). Bands were visualized by UV excitation. nirS PCR products were concentrated by ethanol precipitation (2). PCR products except for the archaeal 16S rDNA PCR products were loaded onto an analytical gel from which bands were eluted in 35 μl of sterile filtered destilled water using a QIAquick gel extraction kit (Qiagen, Chatsworth, Calif.). The archaeal 16S rDNA products were purified with a QIAquick PCR purification kit (Qiagen). The eluted or purified products were again separated on agarose gels to confirm purity and similar concentrations of the purified PCR products.
16S rDNA and nirS T-RFLPs.
Aliquots (5 μl) were cleaved for 2 h in a water bath at 37 and 65°C (TaqI) with 5 U of restriction endonuclease in the manufacturer's recommended reaction buffers. Hydrolysis was performed with three different restriction endonucleases in digestions with a single tetrameric enzyme each (for bacterial 16S rDNAs, HaeIII [GG′CC] [where the prime shows the site of cleavage], HhaI [GCG′C], and MspI [C′CGG]; for archaeal 16S rDNAs, HaeIII, HhaI, and RsaI [GT′CA]; and for nirS genes, HhaI, MspI, and TaqI [T′GCA]; Gibco BRL). To prevent the internal standard from cleavage, the restriction endonucleases were deactivated by heating the reaction mixture to 65°C (80°C for TaqI) for 10 min after the reaction was completed. Aliquots (2 μl) of the digest were mixed with 2 μl of deionized formamide, 0.5 μl of loading buffer (Applied Biosystems Instruments [ABI], Foster City, Calif.), and 0.5 μl of a DNA fragment length standard (TAMRA GS 2500; ABI). After denaturing of the DNA at 94°C for 5 min and immediate chilling on ice, aliquots (2.5 μl) were loaded onto a 36-cm-long 6% denaturing polyacrylamide gel of an automated DNA sequencer (373 ABI Stretch). Electrophoresis was run for 14 h with limits of 1,680 V and 40 mA. After electrophoresis, the lengths of fluorescently labeled terminal restriction fragments (T-RFs) were analyzed by comparison with the internal standard using GeneScan 3.1 software (ABI).
Analysis of T-RFLPs.
For each sample, peaks over a threshold of 50 units above background fluorescence were analyzed by manually aligning fragments to the size standard. To avoid detection of primers and uncertainties of size determination, terminal fragments smaller than 35 bp and larger than 825 bp were excluded from the analysis. Reproducibility of patterns was confirmed for repeated T-RFLP analysis of nirS using the same DNA extracts from two samples. Communities were characterized by the numbers of peaks and the heights of the peaks. The relative abundance of T-RFs within the sections was determined by calculating the ratio between the peak height of each peak and the total peak height of all peaks within one sample. Ratios were converted to percentages, and the results are displayed as histograms. Gene-specific T-RFLPs from sections within and between cores were compared by correspondence analysis (32) of combined results from three different cleavages using the procedure CORRESPONDENCE from the SAS statistical package (version 6.12; SAS Institute, Cary, N.C.) by considering numbers of peaks and peak heights. Additionally, T-RFLPs were analyzed by the presence or absence of T-RFs by calculating dendrograms based on a 1/0 matrix (1, presence; 0, absence of a given T-RF) with the CLIQUE or RESTML function for restriction sites from the PHYLIP, version 3.5c, program package (14).
A computer-simulated hydrolysis of 34 marine nirS sequences (EMBL accession numbers AJ248401 to AJ248427, AJ248429 to AJ248432, and AJ248435 to AJ248437) from the Puget Sound sediment samples and one sample from the Washington margin described by Braker et al. (4) was performed for the restriction endonucleases used in this study with the MAP program of the Genetics Computer Group program package (16). T-RFs obtained from the computer simulation were compared to fragments occurring within T-RFLPs from sediment samples. Lengths of predominant bacterial and archaeal 16S rDNA T-RFs were theroretically compared to those of aligned sequences using the TAP T-RFLP function of the Ribosomal Database Project program Beta2, release 7.1 (http://www.rdp.cme.edu).
RESULTS
Diversity of nirS genes and 16S rDNAs in marine sediments.
Three restriction endonucleases determined to yield the highest numbers and most even size distribution of T-RFs were chosen out of 10 tetrameric enzymes to cut amplified nirS genes and 16S rDNAs from one sediment sample each from Puget Sound and the Washington margin. The highest numbers of different T-RFs in all samples were detected from amplified nirS genes (T-RF = 173, HhaI; T-RF = 139, MspI; T-RF = 131, TaqI). Cleavage of amplified bacterial 16S rDNAs yielded a total of 73 (HaeIII), 70 (MspI), and 59 (HhaI) different T-RFs. The numbers of total fragments obtained from archaeal 16S rDNAs were significantly lower (T-RF = 40, HaeIII; T-RF = 28, RsaI; T-RF = 22, HhaI). These enzymes were used for subsequent experiments.
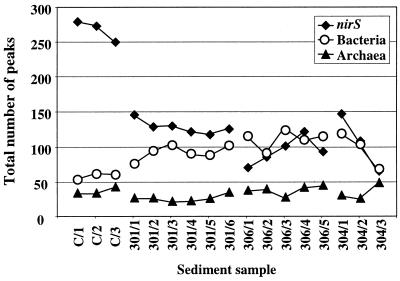
Results of individual cleavages of each gene showed the same trends for all samples. Thus, they were combined for each gene and sample to yield the total number of T-RFs, although the levels of resolution for individual cleavages were different in terms of fragment numbers (Fig. 3). For denitrification genes, extremely high numbers of T-RFs were observed for the Puget Sound samples, whereas diversity was reduced twofold to a level similar to that of the bacterial 16S rDNAs at the Washington margin. Diversity decreased slightly with depth within cores except for the 306 core, where it doubled within the top 2 cm and then decreased for the 6.0- to 7.0-cm sample (306/5). For bacterial 16S rDNAs, the number of T-RFs was twofold higher at the Washington margin than at Puget Sound, showing a slight increase in diversity with increasing distance from shore. The numbers of different T-RFs remained low for archaeal 16S rDNAs within cores from both geographic locations. Levels of diversity within cores for bacterial and archaeal 16S rDNA slightly increased with depth within cores except for bacterial 16S rDNAs within core 304. However, the most dramatic changes in the diversity of the three genes with depth were observed within the most offshore core (304). nirS and bacterial diversity was reduced by more than twofold; in contrast, archaeal diversity increased by a factor of 2 (Fig. 3).
FIG. 3.
Total numbers of T-RFs derived from amplified nirS genes and 16S rDNAs (Bacteria and Archaea) within sediment samples from Puget Sound (C/1 to C/3) and the Washington margin (301/1 to 301/6; 306/1 to 306/5; 304/1 to 304/3). Total numbers were calculated from cleavages with three restriction endonucleases.
Evaluation of the denitrifier, bacterial, and archaeal communities.
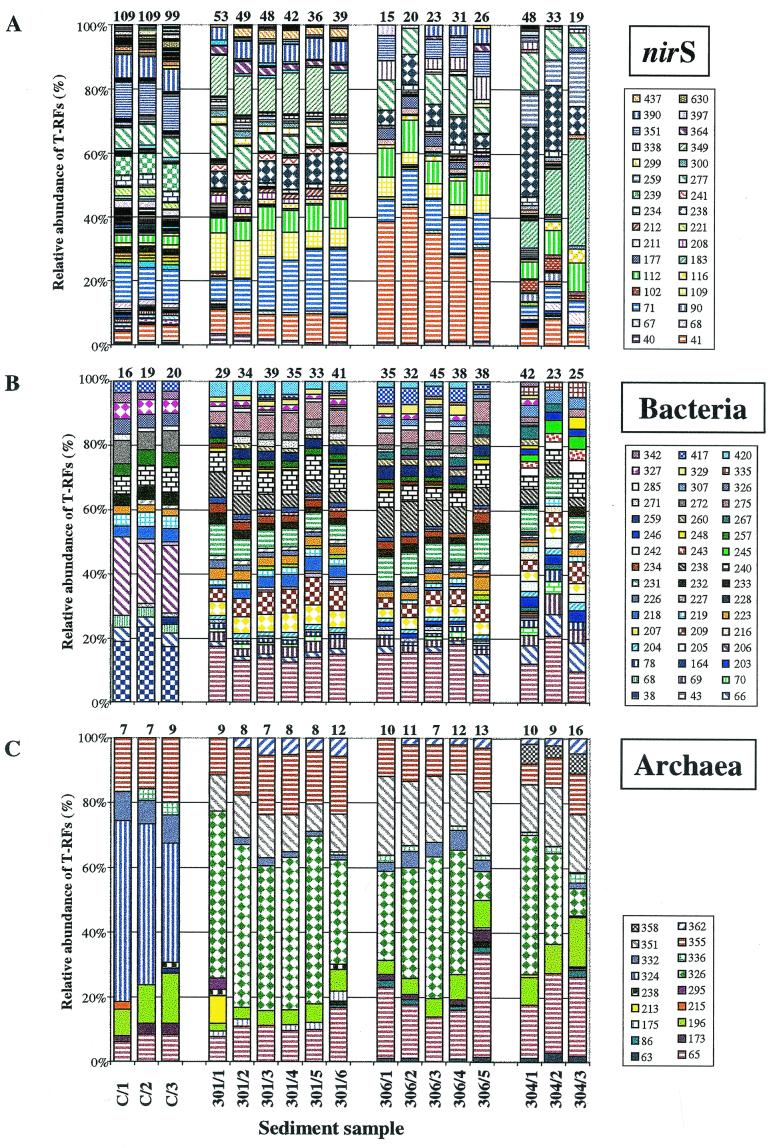
T-RFLPs were compared by calculating the relative abundances of individual T-RFs within samples (Fig. 4). Histograms are displayed after cleavage with HhaI for nirS and with HaeIII for bacterial 16S rDNA, which yielded the highest number of T-RFs and thus represented the highest level of resolution. However, results were similar for these two genes with the other two endonucleases. For Archaea, results after cleavage with HhaI are shown because cleavage with HaeIII did not reveal results similar to those with HhaI and RsaI, although they yielded the highest number of T-RFs.
FIG. 4.
Relative abundances of T-RF of amplified nirS genes (A) and 16S rDNAs of Bacteria (B) and Archaea (C) within sediment samples from Puget Sound (C/1 to C/3) and the Washington margin (301/1 to 301/6; 306/1 to 306/5; 304/1 to 304/3). Diagrams show results after cleavage with HhaI (nirS), HaeIII (Bacteria), and HhaI (Archaea). Numbers on top of the columns indicate the numbers of detectable T-RFs for individual samples. Numbers in the keys indicate the lengths of the T-RFs in base pairs for fragments with a relative abundance of more than 2%.
Puget Sound sediment and Washington margin sediment communities were clearly separated by comparison based on the relative abundances of T-RFs derived from the three genes under consideration. Only a few T-RFs were found to be shared between these different geographic locations. For nirS after cleavage with HhaI, four of the predominant T-RFs (41 bp [3.4 to 42.6%], 71 bp [2.3 to 20.3%], 112 bp [1.4 to 9.0%], and 277 bp [4.6 to 12.1%]) occurred within samples of both locations. Only one fragment (240 bp [3.9 to 7.7%]) was common within all bacterial 16S rDNA profiles after cleavage with HaeIII, whereas three were found for the archaeal genes (65 bp [6.0 to 32.3%], 196 bp [2.3 to 15.7%], and 355 bp [6.4 to 19.9%]) after cleavage with HhaI. On the other hand, some fragments were unique to one sampling location. A nirS fragment of 239 bp (6.0 to 8.6%) was restricted to Puget Sound, whereas a fragment of 238 bp (5.2 to 21.8%) occurred exclusively at the Washington margin. Other nirS fragments were even restricted to one core, such as the 349-bp (10.4 to 13.9%) and 437-bp (0.9 to 2.9%) fragments to core 301, the 177-bp (1.3 to 3.6%) fragment to core 306, and the 259-bp (8.1 to 16.9%) fragment to core 304. T-RFLPs based on bacterial 16S rDNAs from Puget Sound were very different from those found at the Washington margin. Therefore, a number of fragments were found to be restricted to this location, such as fragments of 38 (17.1 to 23.4%), 68 (2.8 to 3.7%), 206 (21.1 to 28.6%), and 232 (3.6 to 3.7%) bp. At the Washington margin, samples were more similar at sampling locations 301 and 306 than at station 304. This is indicated by common fragments of 234 (1.7 to 3.3%), 259 (2.5 to 4.4%), and 420 (0.8 to 5.2%) bp occurring only at stations 301 and 306 and fragments of 205 (2.6 to 3.0%), 242 (2.3 to 3.7%), 243 (2.1 to 3.7%), 245 (2.2 to 4.1%), and 335 (2.7 to 5.0%) bp being restricted to core 304. More T-RFs (3 out of 22) were shared in all samples from both geographic locations (Puget Sound and the Washington margin) for the archaeal 16S rDNAs, whereas one fragment (324 bp [36.9 to 56%]) was found only in the Puget Sound samples. Three T-RFs (326 [8.5 to 51.5%], 351 [8.6 to 24.3%], and 362 [0.0 to 5.7%] bp) were restricted to the Washington margin, one T-RF was found exclusively in core 301 (175 [0.0 to 3.0%]), and one was found exclusively in core 304 (358 [3.7 to 6.3%]). However, within the same sampling location changes in communities seemed rather restricted to the relative abundance of T-RFs than to differences in the presence or absence of T-RFs. Again, the most obvious changes were observed within the 304 core, while communities within the other cores remained rather stable with increasing depth. For example, the nirS fragment of 183 bp increased about fourfold and the fragment of 238 bp increased about threefold with depth. A decrease of fivefold was observed for the archaeal 16S rDNA fragment of 326 bp within core 304 but also in core 306, whereas bacterial communities seemed to remain more stable as no significant changes within cores were observed.
Comparison of the denitrifier, bacterial, and archaeal communities.
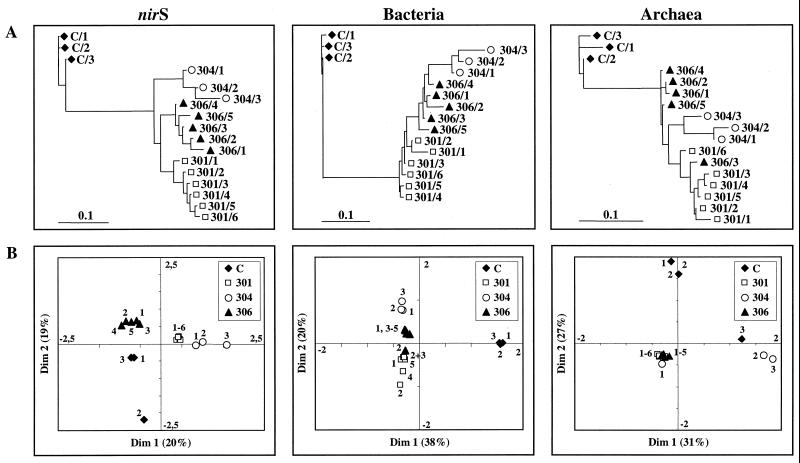
T-RFLPs were compared on the basis of the presence or absence of T-RFs and by correspondence analysis. Results from the comparison of T-RFLPs based on the presence or absence of T-RFs derived from combined cleavages with three enzymes were analyzed by cluster analysis and are displayed as dendrograms (Fig. 5A). Dendrograms of T-RFLPs from Puget Sound show a clear distinction from those obtained from the Washington margin. T-RFLPs were grouped together according to the sampling station except for the archaeal 16S rDNA patterns, of which one pattern from core 306 (306/3) was grouped with those from core 301. Generally, patterns obtained from within cores were grouped closely together. Again, the most obvious differences were detected within core 304.
FIG. 5.
Relationship of T-RFLPs of amplified nirS genes and 16S rDNAs (Bacteria and Archaea) within sediment samples from Puget Sound (C/1 to C/3) and the Washington margin (301/1 to 301/6; 306/1 to 306/5; 304/1 to 304/3). Dendrograms calculated from the presence and absence of peaks (A) and correspondence analysis using combined results from three individual cleavages (B) are shown. Dim, dimension.
To compare communities by considering two parameters, the number of terminal fragments present (richness) and their relative abundance (eveness), T-RFLPs were analyzed by ordination using correspondence analysis (Fig. 5B). Correspondence analysis summarizes multivariate data in scatter diagrams and assumes an underlying structure to the data. The occurrence of T-RFs is determined by a few unknown environmental variables, which correspondence analysis tries to recover and with which it tries to arrange T-RFLPs in a two-dimensional diagram. Thus, the closer the scattered data points are to each other, the more similar communities are in their responses to these variables in species composition and abundance. For T-RFLPs derived from nirS genes and bacterial 16S rDNAs, results were very consistent with those obtained from analyses of relative abundance and the presence or absence of T-RFs. Communities from different sampling locations were clearly separated, and patterns from within sampling locations clustered tightly together with two exceptions, the C/2 sample for nirS and the 306/2 sample for bacterial 16S rDNA. This clustering pattern was not found for archaeal 16S rDNAs. One tight archaeal cluster was found from all Washington margin samples except for samples 304/2 and 304/3. The Puget Sound archaeal profiles were clearly separated, although one sample (C/3) did not cluster with the other two from this location.
Assignment of nirS clones and archaeal species to T-RFs.
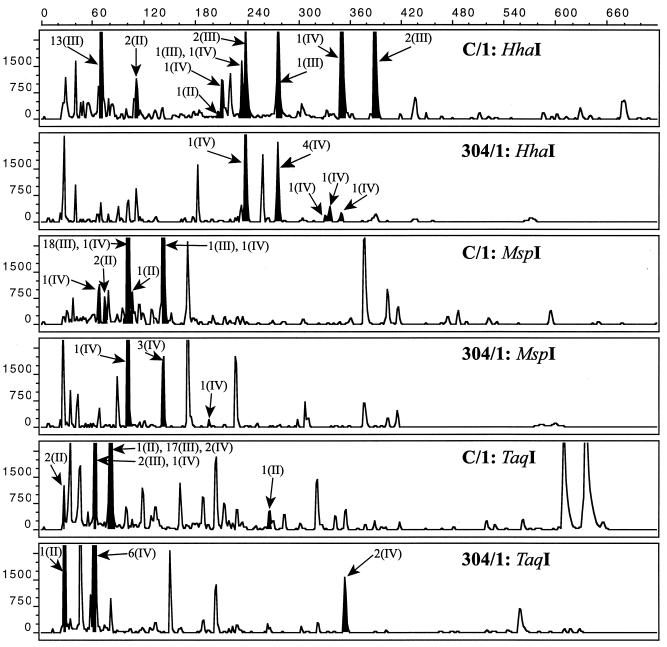
In a computer simulation, nirS sequences from 34 marine sediment clones obtained in a previous study (4) were cleaved with the chosen restriction endonucleases for nirS. The lengths of these theoretical T-RFs were calculated, and clones were assigned to peaks found in the chromatograms from two sediment samples (C/1 and 304/1) (Fig. 6). For the Puget Sound profile, at least one clone could be assigned to most of the dominant peaks and clones corresponded with few exceptions to fragments which represented the dominant T-RFs within the community. Due to the lower number of sequenced clones from the Washington margin, clones could be assigned only to the majority of the dominant T-RFs. Generally, clones that could be assigned to a certain T-RF all belonged to the same cluster of nirS sequences (II, III, and IV) that were found in Pacific Northwest marine sediments. For the T-RFLPs from Puget Sound, some clones from different clusters were represented by the same T-RF, especially for the samples cleaved with TaqI. However, these clusters were resolved by cleavage with the other enzymes (data not shown), but then other clones from different clusters were represented by the same T-RF.
FIG. 6.
Comparison of T-RFs of amplified nirS genes to nirS clone fragments from in silico digestion of sequences from marine sediment samples. T-RFs were detected in sediment samples from Puget Sound (C/1) and the Washington margin (304/1); cloned nirS fragments were obtained from environmental DNA extracts from the same core. Shaded peaks and arrows indicate clones corresponding to T-RFs. x axis, size (base pairs); y axis, relative fluorescence units.
It was not possible to assign bacterial species to T-RFs due to the high level of diversity found within T-RFLPs from amplified bacterial 16S rDNAs using the TAP T-RFLP tool of the Ribosomal Database Project. However, due to lower levels of diversity, archaeal species were found to correspond to T-RFs (Table 1). Considering that a difference of ±2 bp in the sizes of the T-RFs is likely to occur due to the nature of the gel separation, we found T-RFs corresponding to methanogenic species as well as to the cluster of marine crenarchaeota from cold habitats (Sta. Barbara bacterioplankton clone SBAR 12 [11]; Woods Hole bacterioplankton clone WHARQ [11]; Pele's Vent archaeal DNA clones PVA2, −3, and −4 [25]; and clone Antarctic 12 [12]. T-RFs corresponding to almost all known methanogen species (Methanococcus sp., Methanosaeta sp., Methanoculleus sp., Methanohalophilus sp., Methanolobus sp., and Methanocorpusculum sp.) were found within samples from all sampling stations. Marine crenarchaeota from cold habitats were indicated by two out of three cleavages, and they occurred in high relative abundance in the Puget Sound samples; an example is the 324-bp fragment of the HhaI cleavage (Fig. 4).
TABLE 1.
Observed and predicted T-RFs of archaeal 16S rDNAs from marine communities
| Size of observed T-RFs (bp) with:
|
Sizes of predicted T-RFs (bp) with:
|
Archaeal species or clone | ||
|---|---|---|---|---|
| HaeIII | HhaI | RsaI | HaeIII, HhaI, RsaI | |
| 71 | 196 | 608 | 69, 194, 606 | Methanococcus sp. |
| 71, 196, 608 | Methanococcus sp. | |||
| 71 | 326 | 80 | 70, 327, 80 | Methanoculleus sp. |
| 241 | 326 | 263 | 242, 327, 263 | Methanococcus sp. |
| 215 | 326 | 263 | 216, 328, 264 | Methanofollis sp. |
| 215 | 326 | 244 | 217, 329, 244 | Methanohalophilus sp. |
| 215 | 196 | 80 | 214, 195, 79 | Methanoculleus sp. |
| 217, 198, 82 | Methanolobus sp. | |||
| 215 | 196 | 242 | 214, 195, 241 | Methanosaeta sp. |
| 215 | 324 | 263 | 213, 325, 261 | Sta. Barbara Channel bacterioplankton clone SBAR 12 |
| 213, 324, 261 | Woods Hole bacterioplankton clone WHARQ | |||
| 214, 326, 262 | Pele's Vents archaeal DNA clones PVA2, PVA3, and PVA4 | |||
| 214, 325, 262 | Clone Antarctic 12 | |||
| NDa | 213 and 215 | 263 | 22, 214, 263 | Methanococcus sp. |
| ND | 326 | 263 | 22, 327, 263 | Methanocorpusculum sp. |
| ND | 86 | 272 | 22, 86, 272 | Methanococcus sp. |
ND, not detected.
DISCUSSION
In a previous study, high diversity and novel clusters of denitrifiers were found by cloning of nitrite reductase genes (nirK and nirS), screening, and sequencing of clones obtained from a limited number of marine sediment samples from the Pacific Northwest (4). To more systematically investigate marine sediment communities of denitrifiers, Bacteria, and Archaea along the redox gradients in Puget Sound and continental margin sediments (Fig. 2), T-RFLP analysis was applied as a fingerprint method. T-RFLP analysis is independent of cloning and thus applicable to higher numbers of samples. It has been shown to be semiquantitative (20) and generates fingerprints of precise sizes that are reproducible in their T-RF patterns and peak heights from replicate samples (28, 30) with a level of resolution similar to that of denaturing gradient gel electrophoresis (24). When we analyzed the nirS T-RFLP reproducibility of two samples, patterns derived from different DNA extracts, PCR amplifications, and restriction hydrolysis were virtually identical, in agreement with published results (28, 30). Peak height as a meaningful measure of gene abundance is supported by a positive correlation of relative T-RF abundance and quantitative dot blot hybridization of Roseobacter sp.-specific 16S rDNAs in a marine algal bloom (17). Furthermore, relative abundances of T-RFs were demonstrated to be independent of the number of PCR cycles, varying within a range of 5 to 10% until PCR cycle 27, and thereafter remained stable for the majority of peaks (29).
The molecular method to detect cytochrome cd1 containing denitrifiers was highly specific and evaluated a high level of diversity and novel marine nirS genes in Puget Sound and Washington margin sediments (4). In the previous study, 29 out of 37 nirS sequences were obtained from clones from the same Puget Sound samples investigated in this study and from one sample from the 304 core (section, 0.0 to 0.5 cm). However, some nirS sequences, although obtained from a different Washington margin core (1936 m; section, 0.5 to 1.0 cm), showed RFLPs identical to those of clones from the 304 core, suggesting identical or very similar sequences, and were included in our analysis; thus, 34 nirS sequences were analyzed. Computer-simulated restriction hydrolysis of these 34 clones and comparison of the calculated T-RF lengths to T-RFLPs derived from marine sediment samples revealed high congruence of simulated T-RFs and T-RFs occurring in natural samples, especially for Puget Sound T-RFLPs. Since T-RFLP analysis clusters sequences according to restriction sites and since differences in patterns within sediment cores were minor (Fig. 4), simulated T-RFs derived from clones from the entire core (Puget Sound) or even a different sample (core 304, 0.0 to 0.5 cm) could be assigned to environmental patterns from individual samples (Fig. 6). Dominant T-RFs within the environmental profiles were generally represented by clones found in the cloning experiment even though clones and T-RFLP patterns were derived from different batches of DNA extracts (Puget Sound samples) or from DNA extracts from spatially close samples from the same core (304 core). Furthermore, the selected restriction endonucleases (HhaI, MspI, and TaqI), with few exceptions, grouped the clones according to the novel and habitat-specific clusters II, III, and IV of marine nirS sequences found by the cloning approach (4), suggesting an appropriate level of resolution of the T-RFLP method for nirS (Fig. 6). These results on one hand provide information about the dominant groups of sequences found within these sediment samples and indicate on the other hand that cloned and sequenced nirS genes were indeed derived from dominant groups of nirS genes. The level of resolution was determined by the restriction endonuclease. Enzymes cleaving at GC-rich regions of the nirS genes (HhaI and MspI) resolved sequences according to marine clusters II, III, and IV unlike with TaqI (T′GCA), suggesting cleavage of a more conserved restriction site.
Besides nirS genes, diversity and shifts in sediment communities were also explored for bacterial and archaeal 16S rDNAs. Diversity expressed as total numbers of different T-RFs ranked nirS above bacterial 16S rDNAs and bacterial rDNAs above archaeal 16S rDNAs for all restriction endonucleases. This ranking corresponded to expected levels of diversity due to more conserved 16S rDNAs and faster evolutionary rates for functional genes, especially inducible genes such as nirS. The very high level of diversity of nirS genes within Puget Sound sediment samples (Fig. 3) was in good agreement with results from rarefaction analysis of nirS clones from Puget Sound (4). Intermediate levels of diversity were detected for bacterial 16S rDNAs, whereas diversity was constantly low for archaeal 16S rDNAs. Within cores, the most obvious changes in diversity were observed for nirS genes and 16S rDNAs within the 304 core, where the number of T-RFs decreased three- and twofold with depth for nirS and Bacteria, respectively, but slightly increased for Archaea.
Monitoring community structure by relative abundances of T-RFs (Fig. 4) differentiated microbial communities according to geographic location, although some groups were abundant at both Puget Sound and the Washington margin. For the Washington margin, the most obvious differences between cores were generally observed for nirS genes, especially for the 304 core, in accordance with its much greater ocean depth (Fig. 1) and lesser carbon input (26). Patterns derived from the other genes were characterized by minor changes, indicating that on the 16S rDNA level, nirS-containing denitrifiers probably did not comprise a major fraction of the total bacterial or archaeal community. Within cores, differences seem to be restricted to the presence or absence of T-RFs rather than to relative abundance, indicating only minor differences in the compositions of communities along the redox gradients.
To statistically evaluate differences within and between locations and cores, we used analyses based on the presence or absence of T-RFs and correspondence analysis based on three cleavages. Both methods were in agreement in grouping T-RFLPs of the three genes into two distinct clusters from Puget Sound and the Washington margin (Fig. 5A). Within the Washington margin cluster, the T-RFLPs from cores were grouped into separate subclusters, with the most dissimilar profiles being found within the 304 core and thus confirming the observations from the histograms (Fig. 4). However, for the archaeal 16S rDNAs, correspondence analysis (Fig. 5B) showed discrepancies from clustering by the presence or absence of T-RFs. Presumably due to restriction sites in more conserved regions of the 16S rDNA, individual cleavages with HaeIII did not distinguish Puget Sound and Washington margin archaeal communities, despite yielding the highest number of peaks, and RsaI separated communities from these geographic locations but not within cores. Cleavage with HhaI reflected an intermediate level of resolution and could distinguish communities between locations and within cores, although it yielded the lowest number of T-RFs.
T-RFs derived from the archaeal 16S rDNAs could be assigned to methanogens and branches of crenarchaeaota clones from cold marine habitats (Table 1). Both groups are common inhabitants of marine environments (11, 12, 37). However, fragments were also found to be specific for Archaea currently known to be halophilic and alkalophilic (27); an abundance of these types in those ocean habitats is unlikely. These organisms might share T-RFs with other organisms not yet included in the database but express a different phenotype. In addition, the occurrence of both halophiles and alkalophiles was supported only by two T-RFs, with the predicted third T-RF being smaller (22 bp) than peaks analyzed.
Our results indicate that different populations of organisms containing nirS and bacterial and archaeal 16S rDNAs develop under the selection of differing environmental conditions at distant geographic locations rather than due to the steep vertical redox gradients. Scala and Kerkhof (30) found a major influence of geographic distance (i.e., meters to kilometers) on the structures of marine denitrifying communities as determined by T-RFLP analysis of nosZ. Besides large distances on the microbial scale, one major factor that might influence the selection of different communities is the differing amounts of carbon resources at our sites. Puget Sound sediment has more carbon, which is also less degraded than is Washington margin sediment carbon (18). Furthermore, among the Washington margin sites, the amount of carbon decreases with greater distance from shore (26). The lack of community difference over the dramatic changes in redox gradients was initially surprising. A study of Wadden Sea sediment cores using in situ hybridization found most of the major phylogenetic groups at similar relative abundances throughout the redox gradient (21), which is in agreement with our results (Fig. 1). We therefore hypothesize that mixing events by marine invertebrates cause few apparent vertical differences in the microbial community structure despite the strong redox gradients. Mixing data for conserved markers at the same depth zone of the Washington margin (7) are consistent with this explanation.
ACKNOWLEDGMENTS
We thank Carl Ramm for advice on correspondence analysis, Michael Thomm for hosting G. Braker in his group at Kiel University, and Jizhong Zhou for his contributions to this project.
This work was supported by DOE grant DE-FG02-98ER62535 and NSF grant DEB 9120006 to the Center for Microbial Ecology.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kinston E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1991. [Google Scholar]
- 3.Braker G, Fesefeldt A, Witzel K-P. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Environ Microbiol. 1998;64:3769–3775. doi: 10.1128/aem.64.10.3769-3775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braker G, Zhou J, Wu L, Devol A H, Tiedje J M. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl Environ Microbiol. 2000;66:2096–2104. doi: 10.1128/aem.66.5.2096-2104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandes J A, Devol A H. Simultaneous nitrate and oxygen respiration in coastal sediments: evidence for discrete diagenesis. J Mar Res. 1995;52:771–797. [Google Scholar]
- 6.Bruce K D. Analysis of mer gene subclasses within bacterial communities in soils and sediments resolved by fluorescent-PCR–restriction fragment length polymorphism profiling. Appl Environ Microbiol. 1997;63:4914–4919. doi: 10.1128/aem.63.12.4914-4919.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpenter R, Peterson M L, Bennett J T. 210Pb-derived sediment accumulation and mixing rates for the Washington continental slope. Mar Chem. 1982;48:135–164. [Google Scholar]
- 8.Chin K-J, Lukow T, Conrad R. Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field. Appl Environ Microbiol. 1999;65:2341–2349. doi: 10.1128/aem.65.6.2341-2349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen J P, Murray J W, Devol A H, Codispoti L A. Denitrification in continental shelf sediments has major impact on the oceanic nitrogen budget. Global Biogeochem Cycles. 1987;1:97–116. [Google Scholar]
- 10.Codispoti L A. Is the ocean loosing nitrate? Nature. 1995;376:724. [Google Scholar]
- 11.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLong E F, Wu K Y, Prézelin B B, Jovine R V M. High abundance of archaea in antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 13.Devol A H, Christensen J P. Benthic fluxes and nitrogen cycling in sediments of the continental margin of the eastern North Pacific. J Mar Res. 1993;51:345–372. [Google Scholar]
- 14.Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- 15.Froehlich P N, Klinkhammer G P, Bender M L, Luedtke N A, Heath G R, Cullen D, Dauphin P, Hammond D, Hartman B, Maynard V. Early oxidation of organic matter in pelagic sediments of the equatorial Atlantic, suboxic diagenesis Geochim. Cosmochim Acta. 1979;43:1075–1090. [Google Scholar]
- 16.Genetics Computer Group. GCG program manual. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 17.Gonzáles J M, Simó R, Massana R, Covert J S, Casamayor E O, Pedrós-Alió C, Moran M A. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl Environ Microbiol. 2000;66:4237–4246. doi: 10.1128/aem.66.10.4237-4246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedges J I, Hu F S, Devol A H, Hartnett H E, Tsamakis E, Keil R G. Sedimentary organic matter preservation: a test for selective degradation under oxic conditions. Am J Sci. 1999;299:529–555. [Google Scholar]
- 19.Horz H-P, Rotthauwe J-H, Luckow T, Liesack W. Identification of the major subgroups of ammonia-oxidizing bacteria in environmental samples by T-RFLP analysis of amoA PCR products. J Microb Methods. 2000;39:197–204. doi: 10.1016/s0167-7012(99)00119-0. [DOI] [PubMed] [Google Scholar]
- 20.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llobet-Brossa E, Rosselló-Mora R, Amann R I. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl Environ Microbiol. 1998;64:2691–2696. doi: 10.1128/aem.64.7.2691-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lüdemann H, Arth I, Liesack W. Spatial changes in the bacterial community structure along a vertical oxygen gradient in flooded paddy soil cores. Appl Environ Microbiol. 2000;66:754–762. doi: 10.1128/aem.66.2.754-762.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Middelburg J J, Soetaert K, Herman P M J, Heip C H R. Denitrification in marine sediments: a model study. Global Biogeochem Cycles. 1996;10:661–673. [Google Scholar]
- 24.Moeseneder M, Arrieta J M, Muyzer G, Winter C, Herndl G. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:3518–3525. doi: 10.1128/aem.65.8.3518-3525.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moyer C L, Tiedje J M, Dobbs F C, Karl D M. Diversity of deep-sea hydrothermal vent Archaea from Loihi Seamount, Hawaii. Deep-Sea Res. 1998;45:303–317. [Google Scholar]
- 26.Murray J W, Kuivila K M. Organic matter diagenesis in the northeast Pacific: transition from aerobic red clay to suboxic hemipelagic sediments. Deep-Sea Res. 1990;37:59–80. [Google Scholar]
- 27.Oren A. Life at high salt concentrations. In: Dworkin M, editor. The prokaryotes. Springer-Verlag, New York, N.Y. ( http://rizzo.springer-ny.com:6336/contents/.) 1999. [Google Scholar]
- 28.Osborn A M, Moore E R B, Timmis K N. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ Microbiol. 2000;2:39–50. doi: 10.1046/j.1462-2920.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- 29.Ramakrishnan B, Lueders T, Conrad R, Friedrich M. Effect of soil aggregate size on methanogenesis and archaeal community structure in anoxic rice field soil. FEMS Microbiol Ecol. 2000;32:261–270. doi: 10.1111/j.1574-6941.2000.tb00719.x. [DOI] [PubMed] [Google Scholar]
- 30.Scala D J, Kerkhof L J. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl Environ Microbiol. 2000;66:1980–1986. doi: 10.1128/aem.66.5.1980-1986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shapleigh J P. The denitrifying prokarotes. In: Dworkin M, editor. The prokaryotes. Springer-Verlag, New York, N.Y. ( http://rizzo.springer-ny.com:6336/contents/.) 1999. [Google Scholar]
- 32.ter Braak C J F. Ordination. In: Jongman R H G, ter Braak C J F, van Tongeren O F R, editors. Data analysis in community and landscape ecology. Cambridge, United Kingdom: Cambridge University Press; 1995. pp. 91–105. [Google Scholar]
- 33.van der Maarel M J E C, Artz R R D, Haanstra R, Forney L J. Association of marine archaea with the digestive tracts of two marine fish species. Appl Environ Microbiol. 1998;64:2894–2898. doi: 10.1128/aem.64.8.2894-2898.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Elsas J D, Smalla K. Extraction of microbial community DNA from soils. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1.3.3/1–1.3.3/11. [Google Scholar]
- 35.Walsh J J. Importance of continental margins in the marine biogeochemical cycling of carbon and nitrogen. Nature. 1991;350:53–55. [Google Scholar]
- 36.Ward D M, Weller R, Bateson M M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 37.Whitman W B, Bowen T L, Boone D R. The methanogenic bacteria. In: Dworkin M, editor. The prokaryotes. Springer-Verlag, New York, N.Y. ( http://rizzo.springer-ny.com:6336/contents/.) 1999. [Google Scholar]
- 38.Zumft W G. The denitrifying prokaryotes. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. New York, N.Y: Springer Verlag; 1992. pp. 554–582. [Google Scholar]