Abstract
Background
Height is an important anthropometric measurement and is associated with many health-related outcomes. Genome-wide association studies (GWASs) have identified hundreds of genetic loci associated with height, mainly in individuals of European ancestry.
Methods
We performed genome-wide association analyses and replicated previously reported GWAS-determined single nucleotide polymorphisms (SNPs) in the Taiwanese Han population (Taiwan Biobank; n = 67,452). A genetic instrument composed of 251 SNPs was selected from our GWAS, based on height and replication results as the best-fit polygenic risk score (PRS), in accordance with the clumping and p-value threshold method. We also examined the association between genetically determined height (PRS251) and measured height (phenotype). We performed observational (phenotype) and genetic PRS251 association analyses of height and health-related outcomes.
Results
GWAS identified 6843 SNPs in 89 genomic regions with genome-wide significance, including 18 novel loci. These were the most strongly associated genetic loci (EFEMP1, DIS3L2, ZBTB38, LCORL, HMGA1, CS, and GDF5) previously reported to play a role in height. There was a positive association between PRS251 and measured height (p < 0.001). Of the 14 traits and 49 diseases analyzed, we observed significant associations of measured and genetically determined height with only eight traits (p < 0.05/[14 + 49]). Height was positively associated with body weight, waist circumference, and hip circumference but negatively associated with body mass index, waist-hip ratio, body fat, total cholesterol, and low-density lipoprotein cholesterol (p < 0.05/[14 + 49]).
Conclusions
This study contributes to the understanding of the genetic features of height and health-related outcomes in individuals of Han Chinese ancestry in Taiwan.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12916-022-02450-w.
Keywords: Height, Genome-wide association studies, Genetic single nucleotide polymorphisms, Polygenic risk score, Health-related outcomes
Background
Height is the growth phenotype during the entire developmental period from infancy to adulthood and becomes relatively stable in adulthood [1–3]. Previous studies have reported that social and environmental factors can influence height. Some of these factors include educational attainment, smoking, alcohol consumption, and regular exercise [3–9]. Higher levels of education [4, 5] and regular exercise [9] are associated with increased growth and height. In contrast, exposure to smoking or drinking may cause bone mass loss, reduced growth, and reduced height [10–12]. Height is determined by polygenic inheritance under complex and multi-locus genetic regulation [13–15]. Genome-wide association studies (GWAS) of height have identified hundreds of genetic loci (or single nucleotide polymorphisms [SNPs]) with genome-wide significance, especially in individuals of European ancestry [15–26]. These identified genetic loci are associated with proteins of the tyrosine phosphatase family, insulin-like growth factors, proteins involved in skeletal development and mitosis, fibroblast growth factors, the Wnt/β-catenin pathway, Hedgehog signaling, and cancer-associated pathways. These findings highlight the polygenic, complex, and multilocus genetic regulation of height.
Height is associated with several health-related outcomes later in life [27–34]. For instance, taller people tend to have a higher risk of cancer [28] and cancer-related mortality [27] but have a reduced risk of CVD [27, 29], CVD-related mortality [27, 29], type 2 diabetes [34], better retention of cognitive function [30–32], and healthy aging [33]. Height can be measured as a genetic component using a polygenic risk score (PRS). PRS is the sum of the weighted risk alleles from a combination of independent SNPs, usually with genome-wide significance, derived from GWAS results [13, 35, 36]. PRS serves as a genetic instrument variable and can be used to assess associations with health-related outcomes without confounding [37, 38]. Genetically determined taller height (in those with European ancestry) is also associated with an increased risk of cancers [39–44] and cancer-related mortality [45, 46] but a reduced risk of CVD [42, 47–50]. The precise shared genetic loci between height and health-related outcomes are yet to be elucidated, especially in individuals of Han Chinese ancestry. In addition, the mechanisms on how shared genetic loci contribute to both height and health-related outcomes remain unclear.
Therefore, this study aimed to identify the genetic architecture for height in individuals from the Taiwan Biobank—a community-based biobank in Taiwan. We also performed observational and genetic PRS analyses of height and health-related outcomes.
Methods
Taiwan Biobank
The Taiwan Biobank is a database for phenotypic and genomic measurements of the Taiwanese population that was established in 2012. The study recruited volunteers aged 30–70 years with no history of malignancy at enrollment (Twbiobank; https://www.twbiobank.org.tw/new_web/) [51, 52]. All volunteers were residents of Taiwan and provided informed consent. Participants completed questionnaires and underwent interviews, anthropometric measurements, and blood and urine tests to collect demographic, lifestyle, and genomic data.
Taiwan Biobank phenotypes
Anthropometric measurements, including height, body weight, waist circumference, hip circumference, and body fat percentage, were obtained from participants in the Taiwan Biobank (Additional file 1: Table S1). Body mass index (BMI) was calculated as BMI = body weight/body height2. The waist-hip ratio (WHR) was calculated as WHR = waist circumference/ hip circumference. Anthropometric measurements were stratified by sex and analyzed using the mean and standard deviation (SD), where data were normalized to one SD before further analysis.
Blood pressure and lipid and glucose levels were quantitatively measured in participants in the Taiwan Biobank. Systolic blood pressure (SBP), diastolic blood pressure (DBP), total cholesterol (TC), triglyceride (TG), low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), fasting glucose, and hemoglobin (Hb) A1c levels were obtained (Additional file 1: Table S1).
Participants were asked to report their health status using questionnaires and interviews. According to participants’ self-reported health status (comorbidities) in the Taiwan Biobank, 10 broad categories of 49 diseases were investigated in our study as follows (Additional file 1: Table S1): (1) orthopedic or joint disorders: osteoporosis, arthritis, rheumatoid arthritis, osteoarthritis, and gout; (2) lung and respiratory diseases: asthma and emphysema or chronic bronchitis; (3) cardiovascular diseases: valvular heart disease, coronary artery disease, heart arrhythmia, cardiomyopathy, congenital heart defect, other type of heart disease, hyperlipidemia, hypertension, and stroke; (4) diabetes: type 1 diabetes and type 2 diabetes; (5) digestive diseases: peptic ulcer disease, gastroesophageal reflux disease, and irritable bowel syndrome; (6) Mental or emotional disorders: depression, bipolar disorder, postpartum depression, obsessive-compulsive disorder, alcohol addiction or drug abuse, and schizophrenia; (7) nervous system disorders: epilepsy, migraine, multiple sclerosis, Parkinson’s disorder, and dementia; (8) other types of disease: gallstones, kidney stones, kidney failure, and vertigo; (9) eye diseases: cataract, glaucoma, dry eye syndrome, retinal detachment, floaters, blindness, color blindness, and others; and (10) female diseases: severe menstrual cramps, uterine fibroids, ovarian cysts, endometriosis, and uterine/cervical polyps.
Study population
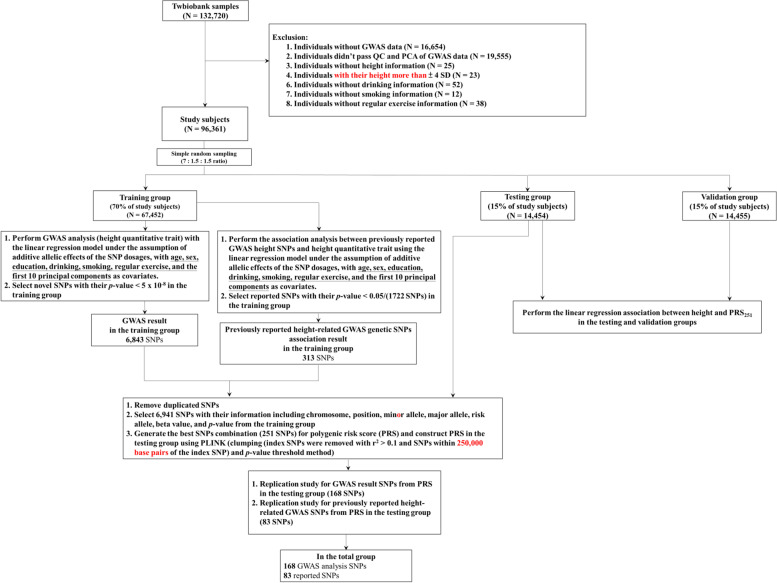
A total of 132,720 participants were selected from the Taiwan Biobank (Fig. 1). The exclusion criteria were as follows: (1) individuals who did not have GWAS data (N = 16,654), (2) individuals who did not pass the quality control (QC) and principal component analysis (PCA) of GWAS data (N = 19,555), (3) individuals who did not have height information (N = 25), (4) individuals with their height more than ±4 SD (N = 23), (5) individuals without drinking information (N = 52), (6) individuals without smoking information (N = 12), and (7) individuals without regular exercise information (N = 38). The criteria for drinking included current drinkers for at least 6 months; smoking criteria included current smokers for at least 6 months; finally, regular exercise criteria included participants performing regular exercise currently, for at least 6 months.
Fig. 1.
Flowchart of the study design and analysis process
Finally, 96,361 participants of Han Chinese ancestry were included in this study (Fig. 1) and assigned to the training, testing, and validation groups using a simple random sampling method (7:1.5:1.5 ratio). The training group (N = 67,452 participants) comprised 70% of the total study population and underwent GWAS based on height (Additional file 2: Table S2; Figs. 2 and 3). Before the height GWAS analysis, the measured height (phenotype) was stratified by sex and subsequently mean-centered and normalized to one SD. GWAS for height was then performed using a linear regression model with the assumption of additive allelic effects of SNP dosages, with adjusted covariates including age, sex, education, drinking, smoking, regular exercise, and the first 10 PCAs (Additional file 4: Fig. S2), using the PLINK software (version 1.9, 2.0) [3–9, 20, 53–56]. A genome-wide significance value was used (p < 5.00E−8 for the additive test).
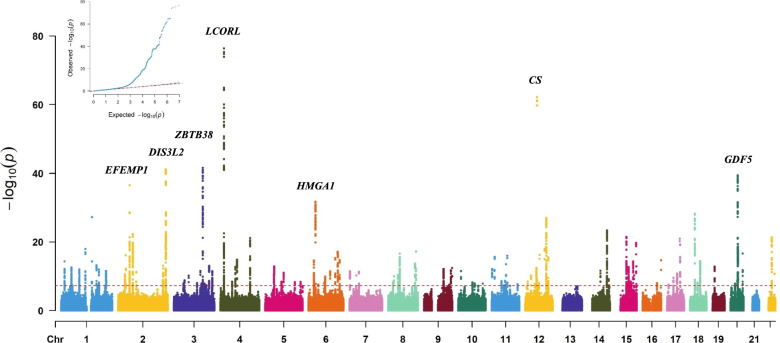
Fig. 2.
Manhattan plot for height with Han Chinese ancestry
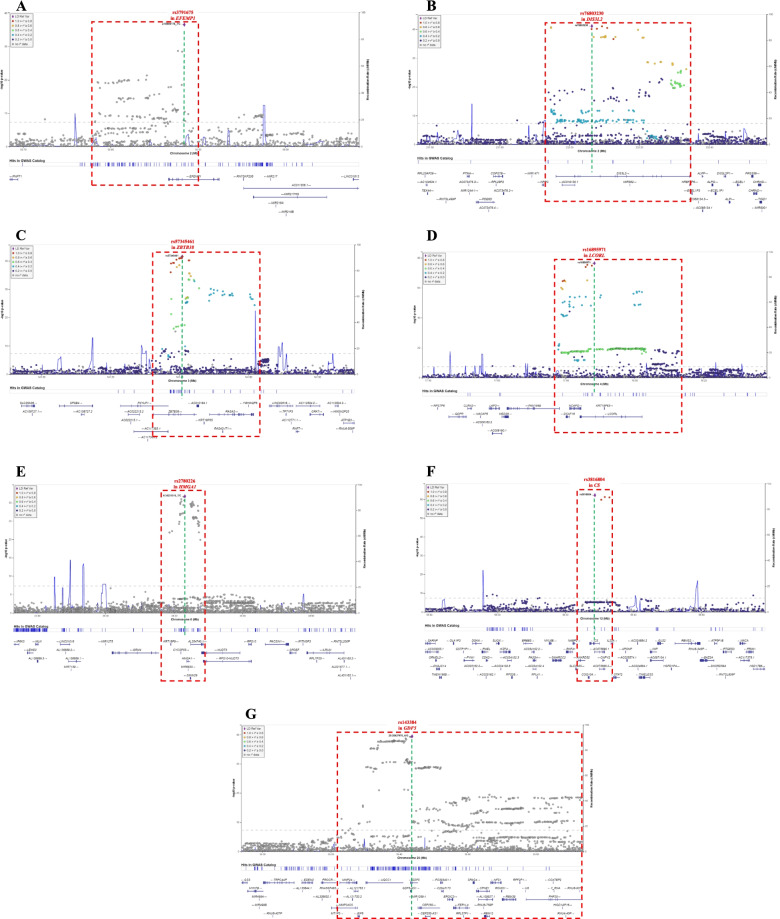
Fig. 3.
Regional plots for the independent signals at seven genetic loci for height in individuals with Han Chinese ancestry. A rs3791675 in EGF containing fibulin extracellular matrix protein 1 (EFEMP1). B rs76803230 in DIS3 like 3′-5′ exoribonuclease 2 (DIS3L2). C rs57345461 in zinc finger and BTB domain containing 38 (ZBTB38). D rs16895971 in ligand dependent nuclear receptor corepressor like (LCORL). E rs2780226 in high mobility group AT-hook 1 (HMGA1). F rs3816804 in citrate synthase (CS). G rs143384 in growth differentiation factor 5 (GDF5). Each plot shows the –log10 p-value on the y-axis for each SNP and the SNP position in the genome region on the x-axis. The top significant SNP is shown by a purple diamond; genes in its proximity are shown below each plot. LD with nearby SNPs is measured using R2 values, according to the 1000 Genomes Project Phase 3 East Asia Summit data, and is indicated by the color of each circle
We also ensured that our GWAS findings of the training group replicated previously identified height-associated genetic variants (https://www.ebi.ac.uk/gwas/efotraits/EFO_0004339). The reported GWAS height-associated genetic variants were mainly from individuals of European ancestry (excluding the genetic variants of infant or child height traits) and were downloaded from the GWAS catalog website. After removal of the repeated SNPs, 1722 reported GWAS body height-related SNPs were obtained from the GWAS catalog (Additional file 3: Table S3). These SNPs were replicated in our cohort, and we further identified 313 GWAS SNPs associated with height in our cohort (p < 0.05/1722 SNPs) (Additional file 3: Table S3).
The testing group (N = 14,454 participants) comprised 15% of the total study population and was used to select the best-fit PRS, to investigate the association between genetically determined height (PRS251) and measured height (phenotype) using linear regression analysis (Fig. 4). The validation group (n = 14,455 participants) comprised 15% of the total population and was used to determine the association between genetically determined height (PRS251) and measured height (phenotype) using linear regression analysis (Fig. 4). This study was approved by the Human Studies Committee of the China Medical University Hospital, Taichung, Taiwan (approval number: CMUH107-REC3-074).
Fig. 4.
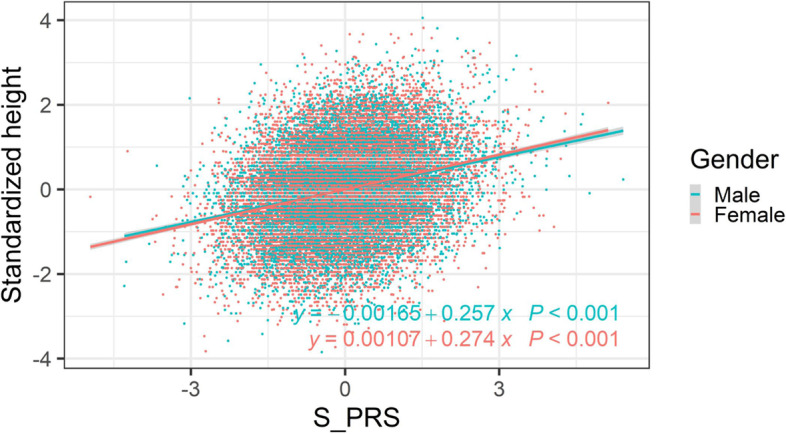
Association between genetically determined height (PRS251) and measured height (phenotype). Measured height (cm) and calculated polygenic risk score (PRS) for height in the testing and validation groups were stratified by sex, mean-centered, and normalized to one standard deviation (SD), respectively (males, N = 10,919; females, N = 17,990). The normalized measured height is shown on the y-axis and normalized genetically determined height (PRS251) is shown on the x-axis
QC of the original data
Genomic DNA from the Taiwan Biobank was genotyped using Axiom genome-wide TWB1 (653,291 SNPs) or TWB2 (752,921 SNPs) array plates based on the Axiom genome-wide array plate system, according to the manufacturer’s instructions (Affymetrix Inc., Santa Clara, CA, USA). Genotyping was performed at the National Genotyping Center of Academia Sinica, Taipei, Taiwan (http://ncgm.sinica.edu.tw/affymetrix_tech_01.html) (https://www.biobank.org.tw/fd.php).
Genotypic data were then subjected to QC procedures (individual QC and SNP QC) in the Taiwan Biobank (https://www.biobank.org.tw/fd.php). The exclusion criteria for individual QC were as follows: (1) individuals with a missing call rate of > 5%, (2) a heterozygosity rate of >±5 SD, (3) individual identity by descent (IBD) score of ≥ 0.125, and (4) individuals who did not fit the East Asia Summit ancestry PCA. Similar to the results of a previous study [51], most individuals from the Taiwan Biobank were of Han Chinese ancestry. The exclusion criteria for SNP QC were as follows: (1) SNPs with a missing call rate of >5%, (2) SNPs with Hardy–Weinberg equilibrium (HWE) p-value of <1 × 10−5, and (3) SNPs with a minor allele frequency (MAF) of <5%.
Imputation
Qualified genotype data were then subjected to an imputation procedure to maximize the number of SNPs in the Taiwan Biobank (https://www.biobank.org.tw/fd.php). First, the SNPs of the qualified genotype data were excluded based on the following criteria: (1) SNPs with MAF of <1%, (2) SNPs with a HWE p-value of <1 × 10−5, and (3) SNP with a missing call rate > 5% using the PLINK software (versions 1.9 and 2.0, http://zzz.bwh.harvard.edu/plink/). SHAPEIT2 (v2.r790) was used to phase the genotypes into full haplotypes (https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html). Third, imputation was performed using IMPUTE2 (v2.3.1, https://mathgen.stats.ox.ac.uk/impute/impute_v2.html), according to the pooled reference panel [Taiwan Biobank (TWB) + East Asian (EAS)]. The pooled reference panel comprised 973 phased individuals with the TWB panel from the Taiwan Biobank [57] and 504 phased individuals from the EAS panel [57, 58] (The Phase 3 1000 Genomes Project reference panel; The 1000 Genomes Project Consortium, 2010). The pooled reference panel with TWB and EAS ancestry groups was used to improve imputation accuracy [57, 58]. The following imputed SNPs were excluded: (1) SNP with a missing call rate of > 5%, (2) SNPs with MAF < 0.01%, and (3) SNPs with IMPUTE2 information score of < 0.3.
QC for this study
Imputed GWAS data were obtained from the Taiwan Biobank. In our study, SNP QC and individual QC procedures were applied before GWAS of height (Fig. 1). SNPs were excluded from the SNP QC based on the following criteria: (1) SNP with a missing call rate of > 5%, (2) SNPs with HWE p-value of < 1 × 10−6; and (3) SNPs with MAF < 0.01%. After SNP QC, the remaining SNPs were used to perform ancestry PCA for the population structure analysis. The exclusion criteria for individual QC were as follows: (1) individual with a missing call rate > 5%, (2) heterozygosity rate > ±5 SD, (3) individual IBD score ≥0.125, and (4) individuals who did not fit the East Asia Summit ancestry PCA. Participants of non-Chinese ancestry, with evidence of relatedness, or with DNA contamination were excluded.
PRS calculation
The PRS was calculated in the testing group using PLINK software (versions 1.9 and 2.0) [35, 53, 59], based on the statistical results of the 6,941 SNPs in the training group (Fig. 1). The 6941 SNPs comprised SNPs with genome-wide significance (p < 5 × 10−8) and SNPs that were replicated from previously reported body-height GWAS SNPs (p < 0.05/1722 SNPs; Fig. 1).
The 6941 SNPs were then subjected to the clumping procedure (within the range of 250,000 base pairs of the index SNP, where SNPs were removed when r2 > 0.1), according to the estimated linkage disequilibrium (LD) among the SNPs in the testing group (Additional file 4: Fig. S1A). After clumping, 251 SNPs were obtained. These 251 SNPs were used to select the “best-fit” PRS according to a series of cutoff values for height-associated p-value thresholds (including 5 × 10−15, 5 × 10−14, 5 × 10−13, 5 × 10−12, 5 × 10−11, 5 × 10−10, 5 × 10−9, 5 × 10−8, 5 × 10−7, 5 × 10−6, and 5 × 10−5) in the testing group. The p-value cutoff (5 × 10−5) was adopted by the “best-fit” PRS with the largest explicable phenotype r2 using only the PRS (PRS r2 = 0.0712, SNP number = 251; Additional file 4: Fig. S1A). In total, 251 SNPs were obtained for the best-fit PRS calculations for all participants. For each participant, the genetically determined height (PRS value) was calculated [35, 53, 59] using 251 SNPs obtained after the clumping protocol. Data centering and standardization were also performed for the PRS height data.
Statistical analyses
Genotype and imputed genotype data were used for GWAS analysis, as previously described. The HWE for the SNPs in the controls was evaluated using chi-square (χ2) tests. Lewontin’s D and R2 values were used to evaluate the inter-marker coefficient of LD for haplotype block analysis [60]. The confidence interval (CI) for LD was used to construct haplotype blocks by resampling [61, 62]. LocusZoom was used to plot the resulting significant locus [63].
Measured height (phenotype) served as the exposure variables. Sixty-three health-related outcomes, including 14 traits and 49 diseases, were used as outcome variables. A multivariate linear regression model was made for continuous outcome variables (14 traits), with adjustments for age, sex, education, drinking, smoking, regular exercise, and 10 PCAs [3–9, 20]. Multivariate logistic regression analysis was performed for binary outcome variables (49 diseases), with adjustments for age, sex, education, drinking, smoking, regular exercise, and 10 PCAs [3–9, 20].
The genetically determined height (PRS251) also served as the exposure variable. Sixty-three health-related outcomes, including 14 traits and 49 diseases, were used as outcome variables. A multivariate linear regression model was performed for continuous outcome variables (14 traits), with adjustments for age, sex, education, drinking, smoking, regular exercise, and 10 PCAs [3–9, 20]. Multivariate logistic regression analysis was performed for binary outcome variables (49 diseases), with adjustments for age, sex, education, drinking, smoking, regular exercise, and 10 PCAs [3–9, 20]. PLINK software (versions 1.9 and 2.0) and R packages for Windows were used for all statistical analyses.
Results
GWAS of the quantitative trait of height in Han Chinese in Taiwan
The Manhattan and QQ plots for the adult-height GWAS results are shown in Fig. 2. GWAS association analysis identified 6843 SNPs in 89 genomic regions with genome-wide significance (p < 5.00E−08 [5 × 10−8], not shown). The top lead SNPs were selected in 89 genomic regions with significant associations (p < 5 × 10−8) using an LD of < 0.2 (Additional file 2: Table S2). Among these, 18 novel lead SNPs within 18 novel regions, 48 novel lead SNPs within 48 reported regions, and 23 lead SNPs within 23 reported regions were found (Additional file 2: Table S2). Moreover, among these 89 genomic regions, the seven lead SNPs were located within seven genetic loci (Fig. 2). These seven genetic loci were located near the following genes: EGF-containing fibulin extracellular matrix protein 1 (EFEMP1), DIS3 like 3′-5′ exoribonuclease 2 (DIS3L2), zinc finger and BTB domain containing 38 (ZBTB38), ligand-dependent nuclear receptor corepressor like (LCORL), high-mobility group AT-hook 1 (HMGA1), citrate synthase (CS), and growth differentiation factor 5 (GDF5). The seven lead SNPs from these seven genetic loci are shown in Additional file 2: Table S2.
Regional plots of the lead SNPs and their neighboring SNPs from these seven genetic loci are shown in Fig. 3. Among them, two lead SNPs were novel (Fig. 3B, C; chromosome 2, rs76803230 in DIS3L2; chromosome 3, rs57345461 in ZBTB38), whereas the remaining five lead SNPs were previously reported (Fig. 3A, D–G). On chromosome 2, the lead SNP rs76803230 was located in the intronic region of DIS3L2 (risk allele: T, beta = 0.0681, [95% CI:0.0583–0.0780], p = 7.47E−42 [7.47 × 10−42]; Additional file 2: Table S2; Fig. 3B). On chromosome 3, the lead SNP rs57345461 was located in the intronic region of ZBTB38 (risk allele: T, beta = 0.0723, [95% CI: 0.0619–0.0827], p = 2.54E−42 [2.54 × 10−42]; Additional file 2: Table S2; Fig. 3C).
In the previously reported SNPs on chromosome 2, only a handful reached genome-wide significance associated with height, where the lead SNP rs3791675 was located in the intronic region of EFEMP1 (risk allele: C; training group: beta = 0.0753, [95% CI: 0.0638–0.0869], p = 2.78E-37 [2.78 × 10−37]; Additional file 2: Table S2; Fig. 3A). This SNP has been associated with body height, BMI-adjusted waist circumference, pelvic organ prolapse, and BMI-adjusted WHR [23, 64–66]. On chromosome 4, the lead SNP rs16895971 was located in the 3-untranslated region (3UTR) of LCORL (risk allele: T, beta = 0.1018, [95% CI: 0.0911–0.1125], p = 3.69E−77 [3.69 × 10−77]; Additional file 2: Table S2; Fig. 3D). This SNP has been associated with body height in East Asians [67]. On chromosome 6, the lead SNP rs2780226 was located in the 5 untranslated region (UTR) of HMGA1 (risk allele: C, beta = 0.0948, [95% CI: 0.0791–0.1104], p = 1.75E−32 [1.75 × 10−32]; Additional file 2: Table S2; Fig. 3E). This SNP has been associated with body height, BMI-adjusted waist circumference, and birth weight [15, 68, 69]. On chromosome 12, the lead SNP rs3816804 was located in the intronic region of CS (risk allele: C, beta = 0.1124, [95% CI: 0.0993–0.1255], p = 6.35E−63 [6.35 × 10−63]; Additional file 2: Table S2; Fig. 3F). This SNP has also been associated with body height in East Asians [70]. On chromosome 20, the lead SNP rs143384 was located in the 5-UTR of GDF5 (risk allele: G, beta = 0.0738, [95% CI: 0.0629–0.0847], p = 3.61E−40 [3.61 × 10−40]; Additional file 2: Table S2; Fig. 3G). This SNP has been associated with body height, BMI-adjusted hip circumference, BMI-adjusted WHR, and body fat [15, 64, 71, 72].
Replication of previously reported GWAS-determined SNPs in the Taiwan Han population
The previously reported GWAS-determined SNPs for height were obtained from the GWAS catalog (https://www.ebi.ac.uk/gwas/efotraits/EFO_0004339) and used to replicate the reported SNPs in the training group using the linear regression model, as described previously. In this study, an association analysis identified 313 SNPs that were significantly associated with height (p < 0.05/1722 SNPs; Additional file 3: Table S3).
In this study, GWAS-identified 6843 SNPs, and 313 of the reported SNPs were combined. After removing duplicate SNPs, 6941 SNPs were associated with height (Fig. 1). These 6941 SNPs were then applied to exclude SNPs with strong LD and to select the best SNP combination for the best-fit PRS calculation in the testing group, using PLINK software (versions 1.9 and 2.0) [53]. This resulted in the identification of independent genetic signals for the best-fit PRS with 251 SNPs. These 251 SNPs included 168 GWAS-identified SNPs (Table 1) and 83 previously reported GWAS-determined SNPs (Table 2). These results show that 168 novel GWAS-identified SNPs and 83 reported SNPs were associated with height in individuals of Han Chinese ancestry in Taiwan.
Table 1.
Newly identified SNPs associated with height in Taiwan
| No. | rs ID | Nearest Gene | Chr. | Position | Minor allele | Major allele | Risk allele | Training group (N = 67,452) (p < 5 ×10−8) | Testing group (N = 14,454) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | 95% CI | P-value | Beta | 95% CI | P-value | ||||||||||
| 1 | rs56265117 | MFAP2 | 1 | 16980428 | C | T | C | 0.040 | 0.030 | 0.050 | 4.66E−15 | 0.042 | 0.021 | 0.064 | 1.31E−04 |
| 2 | rs76910682 | 1 | 50408548 | A | G | A | 0.037 | 0.025 | 0.050 | 7.45E−09 | 0.010 | −0.018 | 0.037 | 4.97E−01 | |
| 3 | rs12098132 | FAF1 | 1 | 50661852 | A | C | A | 0.060 | 0.044 | 0.077 | 8.05E−13 | 0.049 | 0.013 | 0.086 | 8.04E−03 |
| 4 | rs61115731 | FAF1 | 1 | 50932429 | C | G | C | 0.061 | 0.044 | 0.077 | 2.48E−13 | 0.054 | 0.018 | 0.090 | 2.96E−03 |
| 5 | rs140830175 | LINC01562 | 1 | 51197120 | T | C | T | 0.058 | 0.041 | 0.075 | 3.85E−11 | 0.054 | 0.016 | 0.092 | 5.26E−03 |
| 6 | rs4926705 | 1 | 55953858 | A | G | A | 0.039 | 0.025 | 0.053 | 4.55E−08 | 0.045 | 0.015 | 0.075 | 3.12E−03 | |
| 7 | rs3806340 | PKN2-AS1 | 1 | 88683110 | G | T | T | 0.036 | 0.026 | 0.045 | 9.75E−13 | 0.015 | −0.006 | 0.037 | 1.59E−01 |
| 8 | rs7530513 | KYAT3 | 1 | 88944228 | A | G | G | 0.033 | 0.023 | 0.044 | 7.58E−11 | 0.027 | 0.005 | 0.049 | 1.64E−02 |
| 9 | rs7513580 | 1 | 118307286 | A | G | G | 0.046 | 0.036 | 0.057 | 1.01E−18 | 0.041 | 0.019 | 0.064 | 2.98E−04 | |
| 10 | rs10489289 | DNM3 | 1 | 172254949 | C | T | C | 0.043 | 0.032 | 0.054 | 6.19E−14 | 0.028 | 0.003 | 0.052 | 2.68E−02 |
| 11 | rs12047271 | 1 | 184044357 | C | T | C | 0.036 | 0.026 | 0.046 | 8.73E−13 | 0.052 | 0.030 | 0.073 | 2.52E−06 | |
| 12 | rs1046017 | TGFB2, TGFB2-OT1 | 1 | 218443793 | C | G | G | 0.039 | 0.028 | 0.050 | 2.75E−12 | 0.023 | −0.001 | 0.047 | 5.71E−02 |
| 13 | rs7538503 | 1 | 219615188 | G | A | G | 0.037 | 0.025 | 0.049 | 1.13E−09 | 0.029 | 0.003 | 0.054 | 2.92E−02 | |
| 14 | rs2367623 | LTBP1 | 2 | 33202983 | A | C | A | 0.035 | 0.024 | 0.045 | 2.80E−11 | 0.021 | −0.001 | 0.043 | 6.47E−02 |
| 15 | rs143098957 | LTBP1 | 2 | 33263857 | T | G | G | 0.043 | 0.030 | 0.056 | 1.02E−10 | 0.039 | 0.011 | 0.068 | 6.66E−03 |
| 16 | rs17019115 | FEZ2 | 2 | 36575874 | C | G | G | 0.029 | 0.019 | 0.039 | 2.49E−08 | 0.009 | −0.013 | 0.031 | 4.11E−01 |
| 17 | rs4670703 | 2 | 37385957 | C | A | C | 0.036 | 0.026 | 0.046 | 6.39E−13 | 0.012 | −0.010 | 0.033 | 2.84E−01 | |
| 18 | rs79121675 | 2 | 55724807 | A | C | A | 0.085 | 0.056 | 0.114 | 6.55E−09 | 0.077 | 0.016 | 0.139 | 1.41E−02 | |
| 19 | rs146446706 | LOC112268416, EFEMP1 | 2 | 55870880 | T | C | T | 0.151 | 0.111 | 0.191 | 2.34E−13 | 0.030 | −0.059 | 0.119 | 5.04E−01 |
| 20 | rs1824305 | 2 | 71179325 | T | C | C | 0.051 | 0.041 | 0.061 | 4.96E−23 | 0.047 | 0.025 | 0.068 | 2.55E−05 | |
| 21 | rs57092473 | 2 | 71440419 | A | G | G | 0.044 | 0.034 | 0.054 | 4.47E−18 | 0.045 | 0.024 | 0.067 | 4.20E−05 | |
| 22 | rs1913671 | EIF2AK3 | 2 | 88600365 | T | C | C | 0.035 | 0.025 | 0.044 | 5.04E−12 | 0.027 | 0.006 | 0.048 | 1.22E−02 |
| 23 | rs1118150 | DIRC3 | 2 | 217415545 | A | C | A | 0.034 | 0.022 | 0.045 | 5.54E−09 | 0.013 | −0.011 | 0.038 | 2.90E−01 |
| 24 | rs484085 | USP37 | 2 | 218531961 | C | T | T | 0.036 | 0.024 | 0.047 | 1.19E−09 | 0.020 | −0.005 | 0.045 | 1.10E−01 |
| 25 | rs422702 | CFAP65 | 2 | 219037931 | C | T | C | 0.036 | 0.026 | 0.046 | 5.30E−13 | 0.045 | 0.023 | 0.066 | 4.45E−05 |
| 26 | rs374935766 | 2 | 231850100 | C | G | G | 0.175 | 0.117 | 0.234 | 4.07E−09 | 0.102 | −0.019 | 0.222 | 9.82E−02 | |
| 27 | rs33994242 | 2 | 231915948 | G | A | A | 0.043 | 0.028 | 0.058 | 2.01E−08 | 0.059 | 0.027 | 0.091 | 3.61E−04 | |
| 28 | rs76803230 | DIS3L2 | 2 | 232063990 | G | T | T | 0.068 | 0.058 | 0.078 | 7.47E−42 | 0.078 | 0.057 | 0.099 | 7.90E−13 |
| 29 | rs146229392 | DIS3L2 | 2 | 232064573 | C | G | G | 0.092 | 0.061 | 0.123 | 6.79E−09 | 0.079 | 0.013 | 0.144 | 1.84E−02 |
| 30 | rs3748967 | DIS3L2 | 2 | 232333663 | A | G | G | 0.055 | 0.045 | 0.065 | 6.35E−27 | 0.061 | 0.040 | 0.083 | 2.85E−08 |
| 31 | rs894857163 | GIGYF2 | 2 | 232726985 | T | C | C | 0.280 | 0.191 | 0.369 | 6.79E−10 | 0.248 | 0.056 | 0.440 | 1.15E−02 |
| 32 | rs11130111 | CCDC12 | 3 | 46968315 | C | T | C | 0.028 | 0.018 | 0.038 | 2.58E−08 | 0.019 | −0.002 | 0.040 | 7.92E−02 |
| 33 | rs12495173 | KIF9-AS1, KIF9 | 3 | 47241409 | T | C | T | 0.040 | 0.026 | 0.054 | 1.12E−08 | 0.042 | 0.012 | 0.072 | 6.08E−03 |
| 34 | rs1209842003 | 3 | 52186981 | T | G | G | 0.263 | 0.177 | 0.350 | 2.53E−09 | 0.387 | 0.194 | 0.580 | 8.58E−05 | |
| 35 | rs754871503 | NT5DC2 | 3 | 52525012 | C | T | T | 0.268 | 0.181 | 0.355 | 1.38E−09 | 0.464 | 0.269 | 0.660 | 3.22E−06 |
| 36 | rs1328122506 | SFMBT1 | 3 | 52913981 | A | C | C | 0.260 | 0.173 | 0.346 | 3.59E−09 | 0.471 | 0.280 | 0.662 | 1.34E−06 |
| 37 | rs13086339 | RYBP | 3 | 72428668 | C | A | C | 0.033 | 0.023 | 0.043 | 6.55E−11 | 0.013 | −0.008 | 0.035 | 2.22E−01 |
| 38 | rs11710894 | BOC | 3 | 113273112 | T | C | C | 0.029 | 0.019 | 0.039 | 9.04E−09 | 0.012 | −0.010 | 0.033 | 2.98E−01 |
| 39 | rs4073154 | H1FX-AS1 | 3 | 129316642 | A | G | G | 0.038 | 0.027 | 0.049 | 2.71E−12 | 0.028 | 0.005 | 0.051 | 1.56E−02 |
| 40 | rs7632556 | 3 | 134087102 | A | G | G | 0.030 | 0.020 | 0.041 | 1.37E−08 | 0.047 | 0.024 | 0.070 | 5.71E−05 | |
| 41 | rs57345461 | ZBTB38 | 3 | 141407983 | T | A | T | 0.072 | 0.062 | 0.083 | 2.54E−42 | 0.070 | 0.047 | 0.092 | 1.04E−09 |
| 42 | rs1104288 | RSRC1 | 3 | 158249730 | A | C | C | 0.029 | 0.019 | 0.039 | 4.04E−08 | 0.008 | −0.014 | 0.031 | 4.67E−01 |
| 43 | rs12639337 | FNDC3B | 3 | 172279149 | G | C | C | 0.038 | 0.028 | 0.048 | 8.28E−14 | 0.038 | 0.016 | 0.060 | 5.45E−04 |
| 44 | rs9790124 | RTP2, LOC100131635 | 3 | 187712899 | G | A | G | 0.041 | 0.029 | 0.052 | 3.34E−12 | 0.023 | −0.002 | 0.048 | 7.31E−02 |
| 45 | rs116972792 | FAM184B | 4 | 17648794 | A | G | G | 0.080 | 0.052 | 0.108 | 1.53E−08 | 0.036 | −0.027 | 0.099 | 2.62E−01 |
| 46 | rs16895971 | LCORL | 4 | 17883363 | C | T | T | 0.102 | 0.091 | 0.113 | 3.69E−77 | 0.080 | 0.057 | 0.104 | 1.42E−11 |
| 47 | rs16896140 | LCORL | 4 | 17957655 | C | T | C | 0.085 | 0.060 | 0.111 | 6.25E−11 | 0.063 | 0.008 | 0.118 | 2.41E−02 |
| 48 | rs2724485 | LCORL | 4 | 17968075 | C | T | T | 0.028 | 0.018 | 0.038 | 3.41E−08 | 0.035 | 0.013 | 0.056 | 1.64E−03 |
| 49 | rs148309730 | 4 | 18085973 | G | A | A | 0.093 | 0.064 | 0.122 | 3.64E−10 | 0.047 | −0.014 | 0.108 | 1.31E−01 | |
| 50 | rs76924442 | 4 | 18118006 | A | G | G | 0.072 | 0.053 | 0.090 | 2.18E−14 | 0.031 | −0.009 | 0.071 | 1.32E−01 | |
| 51 | rs4698216 | 4 | 18128100 | T | C | T | 0.045 | 0.035 | 0.055 | 2.43E−18 | 0.034 | 0.012 | 0.055 | 2.62E−03 | |
| 52 | rs56281640 | 4 | 56899650 | A | G | A | 0.030 | 0.020 | 0.040 | 3.41E−09 | 0.036 | 0.014 | 0.058 | 1.20E−03 | |
| 53 | rs10027494 | ADAMTS3 | 4 | 72541929 | A | T | A | 0.033 | 0.022 | 0.045 | 1.94E−08 | 0.009 | −0.016 | 0.035 | 4.72E−01 |
| 54 | rs1662840 | 4 | 81235255 | T | C | T | 0.052 | 0.039 | 0.065 | 1.40E−15 | 0.060 | 0.032 | 0.088 | 2.06E−05 | |
| 55 | rs117072351 | HHIP | 4 | 144676679 | T | C | T | 0.091 | 0.060 | 0.121 | 4.51E−09 | 0.028 | −0.038 | 0.095 | 4.04E−01 |
| 56 | rs12654242 | 5 | 42365278 | G | A | G | 0.051 | 0.035 | 0.067 | 4.54E−10 | 0.062 | 0.027 | 0.097 | 4.64E−04 | |
| 57 | rs4273617 | GHR | 5 | 42695369 | G | A | G | 0.051 | 0.038 | 0.065 | 1.23E−13 | 0.035 | 0.006 | 0.064 | 1.80E−02 |
| 58 | rs6453386 | SCAMP1 | 5 | 78408512 | C | G | C | 0.033 | 0.022 | 0.044 | 3.80E−09 | 0.004 | −0.020 | 0.028 | 7.24E−01 |
| 59 | rs985296 | MEF2C-AS1 | 5 | 89081827 | A | G | G | 0.034 | 0.024 | 0.044 | 8.81E−12 | 0.036 | 0.014 | 0.057 | 1.09E−03 |
| 60 | rs184923695 | ARHGAP26 | 5 | 143097754 | A | G | G | 0.054 | 0.036 | 0.072 | 4.62E−09 | 0.071 | 0.033 | 0.110 | 2.97E−04 |
| 61 | rs186405009 | SLC17A1 | 6 | 25823049 | A | G | A | 0.180 | 0.124 | 0.236 | 3.59E−10 | 0.099 | −0.030 | 0.228 | 1.31E−01 |
| 62 | rs811041 | 6 | 26225804 | C | G | C | 0.047 | 0.035 | 0.059 | 2.28E−15 | 0.050 | 0.025 | 0.075 | 1.05E−04 | |
| 63 | rs181680390 | BTN3A3 | 6 | 26452046 | A | T | A | 0.126 | 0.081 | 0.171 | 4.06E−08 | 0.099 | −0.003 | 0.201 | 5.67E−02 |
| 64 | rs185780403 | 6 | 26745946 | A | C | A | 0.132 | 0.086 | 0.177 | 1.26E−08 | 0.100 | −0.003 | 0.202 | 5.62E−02 | |
| 65 | rs192632187 | 6 | 27113326 | C | T | C | 0.137 | 0.091 | 0.182 | 3.75E−09 | 0.116 | 0.015 | 0.217 | 2.50E−02 | |
| 66 | rs183108303 | ZNF204P | 6 | 27370053 | T | G | T | 0.126 | 0.082 | 0.170 | 2.53E−08 | 0.117 | 0.017 | 0.218 | 2.15E−02 |
| 67 | rs182819650 | 6 | 27823957 | C | T | C | 0.129 | 0.085 | 0.174 | 1.31E−08 | 0.108 | 0.008 | 0.208 | 3.48E−02 | |
| 68 | rs182706663 | 6 | 28116420 | T | C | T | 0.127 | 0.083 | 0.172 | 2.34E−08 | 0.094 | −0.006 | 0.193 | 6.47E−02 | |
| 69 | rs2299870 | PPARD | 6 | 35417160 | G | C | G | 0.068 | 0.044 | 0.091 | 1.66E−08 | 0.033 | −0.019 | 0.084 | 2.17E−01 |
| 70 | rs1564926 | CD2AP | 6 | 47502410 | C | T | C | 0.030 | 0.020 | 0.041 | 2.70E−08 | 0.034 | 0.011 | 0.058 | 4.20E−03 |
| 71 | rs145101575 | BCKDHB | 6 | 80344040 | A | G | A | 0.062 | 0.042 | 0.082 | 1.18E−09 | 0.053 | 0.009 | 0.096 | 1.79E−02 |
| 72 | rs62424499 | 6 | 80678133 | T | A | A | 0.035 | 0.024 | 0.046 | 3.66E−10 | 0.042 | 0.018 | 0.065 | 5.78E−04 | |
| 73 | rs1145861 | 6 | 80940193 | C | G | G | 0.037 | 0.026 | 0.048 | 1.15E−10 | 0.041 | 0.016 | 0.065 | 1.15E−03 | |
| 74 | rs13197753 | 6 | 104924810 | G | C | G | 0.037 | 0.026 | 0.047 | 3.82E−12 | 0.047 | 0.024 | 0.069 | 4.91E−05 | |
| 75 | rs78638402 | 6 | 129995451 | G | C | C | 0.069 | 0.048 | 0.089 | 6.24E−11 | 0.029 | −0.016 | 0.074 | 2.09E−01 | |
| 76 | rs6926186 | L3MBTL3 | 6 | 130029149 | G | A | G | 0.053 | 0.034 | 0.072 | 2.11E−08 | 0.029 | −0.011 | 0.070 | 1.51E−01 |
| 77 | rs1040525 | ADGRG6 | 6 | 142382532 | T | C | C | 0.044 | 0.034 | 0.054 | 7.17E−18 | 0.046 | 0.024 | 0.067 | 4.12E−05 |
| 78 | rs73780873 | ESR1 | 6 | 151829789 | A | G | A | 0.045 | 0.034 | 0.056 | 1.87E−15 | 0.018 | −0.006 | 0.042 | 1.36E−01 |
| 79 | rs1182176 | GNA12 | 7 | 2834967 | G | A | A | 0.046 | 0.033 | 0.059 | 3.16E−12 | 0.063 | 0.035 | 0.090 | 1.18E−05 |
| 80 | rs185053690 | KBTBD2 | 7 | 32868716 | T | G | G | 0.158 | 0.111 | 0.206 | 6.20E−11 | 0.205 | 0.104 | 0.307 | 7.61E−05 |
| 81 | rs2960429 | LOC102723446, LOC105375264 | 7 | 46008459 | G | C | C | 0.035 | 0.025 | 0.045 | 4.87E−12 | 0.020 | −0.001 | 0.042 | 6.40E−02 |
| 82 | rs62452707 | 7 | 46530573 | T | A | T | 0.029 | 0.019 | 0.038 | 1.25E−08 | 0.027 | 0.006 | 0.049 | 1.20E−02 | |
| 83 | rs6557667 | LOXL2 | 8 | 23390501 | C | T | C | 0.035 | 0.023 | 0.046 | 5.71E−09 | 0.013 | −0.012 | 0.039 | 3.14E−01 |
| 84 | rs74476179 | EXTL3 | 8 | 28740152 | A | G | G | 0.111 | 0.078 | 0.145 | 9.56E−11 | 0.134 | 0.063 | 0.205 | 2.17E−04 |
| 85 | rs10957084 | LOC105375821 | 8 | 48444248 | G | A | G | 0.038 | 0.027 | 0.049 | 3.92E−11 | 0.022 | −0.002 | 0.046 | 7.47E−02 |
| 86 | rs181231559 | PLAG1 | 8 | 56188663 | C | T | T | 0.185 | 0.123 | 0.246 | 3.55E−09 | 0.152 | 0.023 | 0.282 | 2.10E−02 |
| 87 | rs6984782 | 8 | 56223330 | C | T | T | 0.081 | 0.062 | 0.099 | 2.42E−17 | 0.071 | 0.030 | 0.111 | 6.13E−04 | |
| 88 | rs112083368 | 8 | 56438778 | G | C | C | 0.061 | 0.042 | 0.081 | 7.16E−10 | 0.024 | −0.018 | 0.066 | 2.68E−01 | |
| 89 | rs3886938 | GSDMC | 8 | 129725300 | T | G | T | 0.043 | 0.032 | 0.054 | 2.01E−14 | 0.046 | 0.022 | 0.070 | 1.33E−04 |
| 90 | rs566313810 | 8 | 134043298 | T | C | T | 0.272 | 0.191 | 0.352 | 4.62E−11 | 0.305 | 0.114 | 0.496 | 1.73E−03 | |
| 91 | rs1213791479 | 8 | 134354346 | C | T | C | 0.283 | 0.206 | 0.359 | 5.41E−13 | 0.245 | 0.063 | 0.427 | 8.42E−03 | |
| 92 | rs368372931 | ZFAT | 8 | 134609000 | G | A | G | 0.139 | 0.094 | 0.184 | 1.51E−09 | 0.146 | 0.043 | 0.250 | 5.36E−03 |
| 93 | rs1246647183 | ZFAT | 8 | 134627377 | A | T | A | 0.345 | 0.267 | 0.423 | 6.19E−18 | 0.307 | 0.119 | 0.494 | 1.38E−03 |
| 94 | rs12541381 | ZFAT | 8 | 134637605 | A | G | G | 0.034 | 0.023 | 0.045 | 4.95E−10 | 0.059 | 0.036 | 0.082 | 6.14E−07 |
| 95 | rs56119276 | 9 | 95518374 | C | T | C | 0.037 | 0.027 | 0.047 | 1.05E−12 | 0.023 | 0.001 | 0.045 | 4.50E−02 | |
| 96 | rs4743291 | 9 | 95758524 | T | A | A | 0.036 | 0.024 | 0.048 | 6.27E−09 | 0.024 | −0.003 | 0.050 | 7.86E−02 | |
| 97 | rs10985794 | 9 | 122844338 | C | T | T | 0.035 | 0.023 | 0.048 | 2.68E−08 | 0.024 | −0.003 | 0.051 | 8.51E−02 | |
| 98 | rs10901208 | FUBP3 | 9 | 130587253 | T | C | T | 0.035 | 0.025 | 0.045 | 1.91E−12 | 0.031 | 0.010 | 0.052 | 4.40E−03 |
| 99 | rs35859988 | CCDC3 | 10 | 12902646 | T | C | C | 0.046 | 0.033 | 0.059 | 2.66E−12 | 0.033 | 0.005 | 0.061 | 2.28E−02 |
| 100 | rs10998375 | TET1 | 10 | 68665362 | A | G | G | 0.044 | 0.029 | 0.058 | 5.85E−09 | 0.042 | 0.010 | 0.073 | 9.99E−03 |
| 101 | rs4979861 | 10 | 79371839 | A | G | A | 0.030 | 0.020 | 0.040 | 1.18E−08 | 0.011 | −0.011 | 0.034 | 3.33E−01 | |
| 102 | rs291979 | GRK5 | 10 | 119370285 | A | G | A | 0.035 | 0.023 | 0.047 | 1.43E−08 | 0.012 | −0.014 | 0.039 | 3.66E−01 |
| 103 | rs1003484 | IGF2, INS-IGF2 | 11 | 2146388 | G | A | G | 0.036 | 0.026 | 0.046 | 7.41E−13 | 0.058 | 0.037 | 0.080 | 1.00E−07 |
| 104 | rs78899385 | PSMA1 | 11 | 14538479 | T | C | C | 0.081 | 0.062 | 0.101 | 2.17E−16 | 0.067 | 0.025 | 0.109 | 1.60E−03 |
| 105 | rs76778262 | PDE3B | 11 | 14815707 | C | A | A | 0.083 | 0.057 | 0.108 | 1.67E−10 | 0.074 | 0.019 | 0.130 | 8.79E−03 |
| 106 | rs4752839 | CELF1 | 11 | 47473650 | A | G | A | 0.033 | 0.022 | 0.044 | 2.43E−09 | 0.058 | 0.034 | 0.081 | 1.72E−06 |
| 107 | rs763648441 | LTBP3 | 11 | 65546523 | C | A | C | 0.365 | 0.245 | 0.484 | 2.16E−09 | 0.299 | 0.051 | 0.548 | 1.82E−02 |
| 108 | rs594318 | FOXRED1 | 11 | 126277818 | C | G | C | 0.030 | 0.019 | 0.040 | 2.13E−08 | 0.028 | 0.006 | 0.051 | 1.37E−02 |
| 109 | rs4763719 | ETV6 | 12 | 11724486 | G | A | A | 0.030 | 0.020 | 0.040 | 3.28E−09 | 0.021 | −0.001 | 0.042 | 5.78E−02 |
| 110 | rs57454081 | 12 | 27950661 | T | C | T | 0.048 | 0.033 | 0.062 | 6.68E−11 | 0.027 | −0.005 | 0.058 | 9.50E−02 | |
| 111 | rs76467375 | 12 | 46655632 | T | C | T | 0.040 | 0.026 | 0.054 | 1.43E−08 | 0.024 | −0.006 | 0.054 | 1.20E−01 | |
| 112 | rs10444558 | 12 | 53661701 | T | A | T | 0.042 | 0.031 | 0.054 | 3.36E−13 | 0.054 | 0.029 | 0.078 | 1.84E−05 | |
| 113 | rs139121417 | RAB5B | 12 | 55986276 | T | C | C | 0.045 | 0.031 | 0.059 | 5.68E−10 | 0.055 | 0.024 | 0.086 | 4.71E−04 |
| 114 | rs11834895 | HMGA2, HMGA2-AS1 | 12 | 65853230 | G | C | C | 0.033 | 0.021 | 0.044 | 4.84E−08 | 0.018 | −0.007 | 0.044 | 1.58E−01 |
| 115 | rs151174669 | 12 | 65980466 | T | C | C | 0.069 | 0.046 | 0.092 | 7.72E−09 | −0.004 | −0.056 | 0.048 | 8.76E−01 | |
| 116 | rs7971647 | SOCS2 | 12 | 93590078 | C | T | C | 0.046 | 0.036 | 0.056 | 1.10E−18 | 0.041 | 0.019 | 0.063 | 3.33E−04 |
| 117 | rs80328976 | WASHC3 | 12 | 102022590 | C | G | G | 0.058 | 0.047 | 0.069 | 3.78E−26 | 0.075 | 0.052 | 0.098 | 1.98E−10 |
| 118 | rs1986854 | WASHC3 | 12 | 102023697 | C | T | C | 0.043 | 0.027 | 0.058 | 3.00E−08 | 0.050 | 0.018 | 0.082 | 2.45E−03 |
| 119 | rs12424129 | LINC02456 | 12 | 102281461 | C | T | T | 0.060 | 0.049 | 0.070 | 9.80E−28 | 0.072 | 0.049 | 0.095 | 8.79E−10 |
| 120 | rs12228148 | LINC02456 | 12 | 102313697 | A | G | A | 0.050 | 0.037 | 0.063 | 5.00E−14 | 0.059 | 0.030 | 0.087 | 4.74E−05 |
| 121 | rs17032833 | LOC105369944 | 12 | 102561342 | C | T | T | 0.039 | 0.029 | 0.049 | 1.40E−14 | 0.025 | 0.003 | 0.046 | 2.42E−02 |
| 122 | rs117988169 | HVCN1 | 12 | 110672222 | T | G | G | 0.059 | 0.041 | 0.078 | 2.96E−10 | 0.011 | −0.029 | 0.051 | 5.90E−01 |
| 123 | rs3782886 | BRAP | 12 | 111672685 | C | T | T | 0.045 | 0.034 | 0.056 | 4.75E−16 | 0.019 | −0.005 | 0.043 | 1.21E−01 |
| 124 | rs116873087 | NAA25 | 12 | 112074109 | C | G | G | 0.046 | 0.035 | 0.057 | 2.94E−16 | 0.016 | −0.008 | 0.040 | 2.00E−01 |
| 125 | rs11066359 | RPH3A | 12 | 112607850 | T | C | C | 0.029 | 0.019 | 0.039 | 1.39E−08 | 0.015 | −0.007 | 0.037 | 1.70E−01 |
| 126 | rs2072134 | OAS3 | 12 | 112971371 | A | G | G | 0.039 | 0.026 | 0.052 | 2.44E−09 | 0.012 | −0.016 | 0.040 | 4.01E−01 |
| 127 | rs12590263 | TC2N | 14 | 91852154 | A | G | A | 0.033 | 0.023 | 0.042 | 7.92E−11 | 0.012 | −0.010 | 0.033 | 2.80E−01 |
| 128 | rs7143616 | ATXN3 | 14 | 92065114 | C | A | A | 0.053 | 0.043 | 0.063 | 4.03E−24 | 0.041 | 0.019 | 0.063 | 3.09E−04 |
| 129 | rs3759556 | 14 | 100725962 | G | A | A | 0.071 | 0.049 | 0.094 | 6.89E−10 | 0.073 | 0.024 | 0.123 | 3.66E−03 | |
| 130 | rs35443927 | 14 | 103383378 | T | C | C | 0.029 | 0.019 | 0.039 | 4.28E−08 | 0.026 | 0.004 | 0.049 | 2.07E−02 | |
| 131 | rs2663534 | 15 | 50883261 | C | T | T | 0.030 | 0.020 | 0.040 | 4.54E−09 | 0.027 | 0.005 | 0.049 | 1.41E−02 | |
| 132 | rs8041967 | MIR4713HG, CYP19A1 | 15 | 51252127 | A | G | A | 0.039 | 0.029 | 0.049 | 1.75E−14 | 0.040 | 0.019 | 0.062 | 2.43E−04 |
| 133 | rs28723025 | CYP19A1 | 15 | 51304114 | A | C | C | 0.054 | 0.043 | 0.064 | 3.03E−22 | 0.054 | 0.031 | 0.078 | 5.60E−06 |
| 134 | rs2162062 | VPS13C | 15 | 61987738 | A | G | G | 0.038 | 0.028 | 0.049 | 1.95E−13 | 0.014 | −0.008 | 0.037 | 2.09E−01 |
| 135 | rs2415130 | MYO9A | 15 | 71950213 | A | G | A | 0.028 | 0.018 | 0.038 | 2.08E−08 | 0.020 | −0.002 | 0.041 | 7.45E−02 |
| 136 | rs55763892 | PARP6 | 15 | 72247839 | T | C | C | 0.032 | 0.021 | 0.043 | 3.00E−09 | 0.029 | 0.006 | 0.052 | 1.40E−02 |
| 137 | rs6495171 | SIN3A | 15 | 75373282 | A | G | A | 0.031 | 0.020 | 0.042 | 2.70E−08 | 0.024 | 0.000 | 0.048 | 4.67E−02 |
| 138 | rs1526080 | ADAMTSL3 | 15 | 83921168 | G | A | A | 0.045 | 0.033 | 0.056 | 4.54E−15 | 0.024 | −0.001 | 0.048 | 5.58E−02 |
| 139 | rs938608 | ACAN | 15 | 88855374 | T | G | G | 0.041 | 0.029 | 0.053 | 5.26E−11 | 0.064 | 0.038 | 0.091 | 1.83E−06 |
| 140 | rs138351276 | ACAN | 15 | 88859943 | G | A | A | 0.102 | 0.069 | 0.134 | 6.62E−10 | 0.169 | 0.096 | 0.241 | 5.54E−06 |
| 141 | rs28456063 | 15 | 98637993 | T | C | C | 0.078 | 0.062 | 0.095 | 2.07E−20 | 0.102 | 0.066 | 0.138 | 3.09E−08 | |
| 142 | rs897377828 | IGF1R, IRAIN | 15 | 98649099 | A | G | G | 0.260 | 0.185 | 0.335 | 1.20E−11 | 0.157 | 0.001 | 0.313 | 4.80E−02 |
| 143 | rs2573650 | ADAMTS17 | 15 | 99973892 | G | A | A | 0.037 | 0.027 | 0.047 | 1.38E−13 | 0.047 | 0.025 | 0.068 | 1.87E−05 |
| 144 | rs4619391 | WWP2 | 16 | 69780860 | T | A | T | 0.030 | 0.019 | 0.040 | 9.78E−09 | 0.023 | 0.001 | 0.045 | 3.75E−02 |
| 145 | rs114509338 | SF3B3 | 16 | 70574733 | C | T | T | 0.033 | 0.021 | 0.044 | 1.72E−08 | 0.000 | −0.024 | 0.025 | 9.74E−01 |
| 146 | rs116560331 | POLR2A | 17 | 7488366 | A | G | G | 0.062 | 0.040 | 0.084 | 2.80E−08 | 0.049 | 0.002 | 0.095 | 4.04E−02 |
| 147 | rs113934718 | ATAD5 | 17 | 30887862 | A | C | C | 0.048 | 0.033 | 0.063 | 2.75E−10 | 0.024 | −0.008 | 0.057 | 1.44E−01 |
| 148 | rs67474242 | LRRC37A2, WNT3 | 17 | 46777685 | A | G | G | 0.031 | 0.021 | 0.041 | 1.36E−09 | 0.000 | −0.022 | 0.021 | 9.86E−01 |
| 149 | rs2411374 | 17 | 48945636 | C | T | C | 0.037 | 0.026 | 0.047 | 1.49E−11 | 0.021 | −0.002 | 0.044 | 7.40E−02 | |
| 150 | rs6504608 | ZNF652 | 17 | 49347319 | A | C | C | 0.030 | 0.020 | 0.041 | 1.41E−08 | 0.014 | −0.009 | 0.037 | 2.30E−01 |
| 151 | rs9905385 | 17 | 61420889 | A | G | A | 0.053 | 0.042 | 0.064 | 1.08E−21 | 0.046 | 0.022 | 0.069 | 1.67E−04 | |
| 152 | rs2320125 | CD79B | 17 | 63930958 | C | T | C | 0.046 | 0.036 | 0.056 | 4.63E−20 | 0.040 | 0.019 | 0.061 | 2.11E−04 |
| 153 | rs11651289 | 17 | 63936114 | T | C | T | 0.070 | 0.047 | 0.094 | 3.45E−09 | 0.073 | 0.024 | 0.121 | 3.41E−03 | |
| 154 | rs4239437 | CABLES1 | 18 | 23152260 | T | C | C | 0.074 | 0.061 | 0.087 | 5.92E−29 | 0.053 | 0.025 | 0.082 | 2.24E−04 |
| 155 | rs9807648 | TMEM241 | 18 | 23344958 | G | A | G | 0.033 | 0.022 | 0.045 | 3.28E−08 | 0.030 | 0.004 | 0.056 | 2.16E−02 |
| 156 | rs4349223 | FHOD3 | 18 | 36541412 | A | C | C | 0.029 | 0.019 | 0.039 | 6.93E−09 | 0.021 | −0.001 | 0.042 | 5.91E−02 |
| 157 | rs12606199 | DYM, LOC100129878 | 18 | 49045546 | A | G | G | 0.046 | 0.034 | 0.057 | 3.49E−15 | 0.023 | −0.002 | 0.048 | 7.04E−02 |
| 158 | rs201707253 | DYM | 18 | 49342419 | T | G | G | 0.042 | 0.031 | 0.052 | 6.17E−14 | 0.017 | −0.006 | 0.041 | 1.53E−01 |
| 159 | rs3843750 | SLC44A2 | 19 | 10637397 | C | G | C | 0.039 | 0.029 | 0.050 | 1.55E−13 | 0.039 | 0.016 | 0.061 | 8.68E−04 |
| 160 | rs754332 | KIZ, KIZ-AS1 | 20 | 21197259 | A | G | A | 0.029 | 0.019 | 0.039 | 1.28E−08 | 0.018 | −0.004 | 0.040 | 1.04E−01 |
| 161 | rs61016611 | 20 | 35624229 | A | G | A | 0.062 | 0.043 | 0.081 | 1.23E−10 | 0.058 | 0.017 | 0.099 | 5.31E−03 | |
| 162 | rs3827030 | PHF20 | 20 | 35887025 | G | A | G | 0.060 | 0.047 | 0.074 | 1.87E−19 | 0.042 | 0.014 | 0.071 | 3.47E−03 |
| 163 | rs8183892 | 20 | 36980155 | T | C | T | 0.041 | 0.031 | 0.051 | 2.79E−16 | 0.028 | 0.006 | 0.049 | 1.18E−02 | |
| 164 | rs4608 | RPN2 | 20 | 37236651 | T | C | T | 0.044 | 0.034 | 0.054 | 2.23E−18 | 0.026 | 0.005 | 0.048 | 1.61E−02 |
| 165 | rs8121252 | GNAS | 20 | 58901754 | T | C | C | 0.048 | 0.037 | 0.059 | 2.30E−17 | 0.053 | 0.029 | 0.077 | 1.42E−05 |
| 166 | rs5754190 | SYN3, LOC105373002 | 22 | 32654480 | C | T | C | 0.052 | 0.042 | 0.063 | 4.32E−22 | 0.050 | 0.027 | 0.073 | 1.90E−05 |
| 167 | rs4821086 | SYN3 | 22 | 32687867 | A | C | A | 0.077 | 0.052 | 0.102 | 2.10E−09 | 0.039 | −0.015 | 0.093 | 1.59E−01 |
| 168 | rs7290267 | MIRLET7BHG | 22 | 46088855 | G | A | G | 0.052 | 0.037 | 0.068 | 1.75E−11 | 0.062 | 0.029 | 0.095 | 2.30E−04 |
SNP, single nucleotide polymorphism; No., number; Chr., chromosome; 95% CI, 95% confidence interval; GWAS, genome-wide association study
This analysis was performed under the additive inheritance model. These SNPs were ordered by chromosome and position. Positions were based on the NCBI GRCh38 version. Genes were identified based on the gene containing the SNP or the nearest gene (within 100 kb up- or downstream) to the SNP
The measured (phenotypic) heights (cm) were stratified by sex, mean-centered, and normalized to one standard deviation (SD) before height GWAS analysis
Beta-value calculation was conducted according to the defined risk alleles
Table 2.
Association of previously reported GWAS height SNPs with height in Taiwan
| No. | rs ID | Gene | Chr. | Position | Minor allele | Major allele | Risk allele | Training group (N = 67,452) (p < 0.05/1722) | Testing group (N = 14,454) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | 95% CI | P-value | Beta | 95% CI | P-value | ||||||||||
| 1 | rs2300092 | MTOR | 1 | 11206407 | T | C | T | 0.027 | 0.014 | 0.039 | 2.62E−05 | 0.024 | −0.003 | 0.050 | 8.51E−02 |
| 2 | rs3014240 | CCDC17 | 1 | 45623553 | C | G | C | 0.025 | 0.015 | 0.036 | 9.06E−07 | 0.032 | 0.010 | 0.054 | 4.17E−03 |
| 3 | rs2666504 | 1 | 62169409 | C | T | C | 0.024 | 0.013 | 0.034 | 5.80E−06 | 0.015 | −0.007 | 0.037 | 1.84E−01 | |
| 4 | rs11205303 | MTMR11 | 1 | 149934520 | C | T | C | 0.057 | 0.047 | 0.067 | 5.69E−28 | 0.033 | 0.011 | 0.055 | 3.26E−03 |
| 5 | rs6587515 | 1 | 150636412 | A | G | A | 0.026 | 0.014 | 0.037 | 2.27E−05 | 0.018 | −0.008 | 0.043 | 1.71E−01 | |
| 6 | rs1325596 | PAPPA2 | 1 | 176824930 | G | A | A | 0.032 | 0.021 | 0.044 | 6.10E−08 | 0.043 | 0.018 | 0.068 | 9.05E−04 |
| 7 | rs10911212 | LAMC1 | 1 | 183055334 | C | T | C | 0.024 | 0.014 | 0.034 | 2.83E−06 | 0.007 | −0.015 | 0.029 | 5.30E−01 |
| 8 | rs4472734 | PTPN14 | 1 | 214444842 | C | T | C | 0.028 | 0.018 | 0.037 | 4.96E−08 | 0.013 | −0.009 | 0.034 | 2.48E−01 |
| 9 | rs10165255 | CYS1 | 2 | 10059474 | A | G | A | 0.030 | 0.016 | 0.044 | 1.70E−05 | 0.037 | 0.008 | 0.066 | 1.30E−02 |
| 10 | rs6735681 | 2 | 15983051 | T | C | T | 0.023 | 0.013 | 0.033 | 5.34E−06 | 0.014 | −0.007 | 0.036 | 1.98E−01 | |
| 11 | rs780094 | GCKR | 2 | 27518370 | T | C | C | 0.022 | 0.012 | 0.031 | 1.69E−05 | 0.025 | 0.004 | 0.046 | 2.22E−02 |
| 12 | rs3755206 | CRIM1 | 2 | 36456285 | G | T | T | 0.054 | 0.042 | 0.067 | 7.94E−17 | 0.054 | 0.026 | 0.081 | 1.35E−04 |
| 13 | rs6544743 | LOC102723904 | 2 | 44163230 | T | G | T | 0.024 | 0.013 | 0.035 | 1.52E−05 | 0.005 | −0.018 | 0.029 | 6.55E−01 |
| 14 | rs3791675 | EFEMP1 | 2 | 55884174 | C | T | C | 0.075 | 0.064 | 0.087 | 2.78E−37 | 0.072 | 0.047 | 0.097 | 2.06E−08 |
| 15 | rs1432559 | LOC105374690 | 2 | 55962483 | G | T | G | 0.055 | 0.030 | 0.080 | 1.32E−05 | 0.073 | 0.019 | 0.126 | 8.21E−03 |
| 16 | rs4241349 | ANTXR1 | 2 | 69103152 | G | A | G | 0.026 | 0.014 | 0.037 | 9.55E−06 | 0.039 | 0.015 | 0.064 | 1.78E−03 |
| 17 | rs76709099 | IHH | 2 | 219055182 | A | C | C | 0.064 | 0.047 | 0.080 | 8.72E−14 | 0.061 | 0.025 | 0.097 | 9.16E−04 |
| 18 | rs2564923 | 3 | 53069246 | A | G | A | 0.027 | 0.016 | 0.038 | 1.49E−06 | 0.034 | 0.011 | 0.058 | 4.25E−03 | |
| 19 | rs9841212 | 3 | 134473096 | C | T | T | 0.024 | 0.013 | 0.036 | 2.79E−05 | 0.003 | −0.021 | 0.027 | 8.10E−01 | |
| 20 | rs1055153 | WWTR1 | 3 | 149657086 | T | G | G | 0.046 | 0.030 | 0.062 | 1.33E−08 | 0.013 | −0.022 | 0.047 | 4.69E−01 |
| 21 | rs6774762 | GHSR | 3 | 172447200 | G | A | A | 0.037 | 0.024 | 0.050 | 1.80E−08 | 0.032 | 0.005 | 0.060 | 2.16E−02 |
| 22 | rs7697556 | 4 | 72649596 | C | T | T | 0.037 | 0.027 | 0.047 | 1.77E−13 | 0.030 | 0.009 | 0.051 | 5.37E−03 | |
| 23 | rs17017911 | GUSBP5 | 4 | 143559481 | G | A | A | 0.022 | 0.012 | 0.032 | 2.19E−05 | 0.016 | −0.005 | 0.038 | 1.40E−01 |
| 24 | rs6845999 | HHIP-AS1 | 4 | 144644674 | T | C | T | 0.058 | 0.046 | 0.070 | 7.29E−22 | 0.062 | 0.037 | 0.088 | 1.52E−06 |
| 25 | rs4240326 | 4 | 144918112 | A | G | A | 0.028 | 0.017 | 0.039 | 7.17E−07 | 0.018 | −0.006 | 0.042 | 1.50E−01 | |
| 26 | rs301901 | NIPBL | 5 | 37046524 | A | G | A | 0.024 | 0.014 | 0.034 | 2.04E−06 | 0.016 | −0.005 | 0.038 | 1.32E−01 |
| 27 | rs4865956 | 5 | 55586677 | T | A | T | 0.024 | 0.014 | 0.035 | 5.07E−06 | 0.018 | −0.005 | 0.040 | 1.22E−01 | |
| 28 | rs7706662 | CEP120 | 5 | 123419868 | T | C | C | 0.023 | 0.013 | 0.032 | 7.16E−06 | −0.014 | −0.035 | 0.008 | 2.09E−01 |
| 29 | rs2908532 | 5 | 142242319 | A | C | A | 0.027 | 0.016 | 0.037 | 1.23E−06 | 0.010 | −0.013 | 0.033 | 3.93E−01 | |
| 30 | rs2974438 | SLIT3 | 5 | 168823898 | A | G | G | 0.030 | 0.017 | 0.043 | 3.53E−06 | 0.020 | −0.008 | 0.047 | 1.61E−01 |
| 31 | rs12153391 | SMIM23 | 5 | 171776434 | A | C | C | 0.029 | 0.019 | 0.039 | 1.22E−08 | 0.016 | −0.006 | 0.038 | 1.46E−01 |
| 32 | rs4868126 | 5 | 171856465 | T | G | G | 0.031 | 0.020 | 0.041 | 4.13E−09 | 0.041 | 0.019 | 0.063 | 2.39E−04 | |
| 33 | rs722585 | GMDS, HCG17 | 6 | 1775629 | A | G | G | 0.023 | 0.013 | 0.034 | 1.61E−05 | 0.060 | 0.037 | 0.083 | 3.26E−07 |
| 34 | rs78566116 | 6 | 32428369 | T | G | G | 0.040 | 0.022 | 0.057 | 5.70E−06 | 0.039 | 0.002 | 0.075 | 3.87E−02 | |
| 35 | rs2780226 | 6 | 34231315 | C | T | C | 0.095 | 0.079 | 0.110 | 1.75E−32 | 0.072 | 0.038 | 0.106 | 2.87E−05 | |
| 36 | rs12209223 | FILIP1, LOC101928540 | 6 | 75454873 | A | C | A | 0.050 | 0.033 | 0.067 | 1.02E−08 | 0.057 | 0.020 | 0.095 | 2.76E−03 |
| 37 | rs648831 | BCKDHB | 6 | 80246491 | C | T | T | 0.024 | 0.014 | 0.034 | 1.53E−06 | 0.017 | −0.004 | 0.038 | 1.22E−01 |
| 38 | rs3805859 | BCKDHB | 6 | 80339229 | C | A | C | 0.025 | 0.015 | 0.035 | 5.62E−07 | 0.017 | −0.005 | 0.038 | 1.23E−01 |
| 39 | rs113898003 | L3MBTL3 | 6 | 130020090 | C | T | T | 0.042 | 0.032 | 0.052 | 4.72E−16 | 0.045 | 0.023 | 0.068 | 5.45E−05 |
| 40 | rs7765757 | EPB41L2 | 6 | 131050608 | C | T | T | 0.038 | 0.022 | 0.053 | 1.29E−06 | 0.038 | 0.006 | 0.070 | 2.12E−02 |
| 41 | rs3020359 | ESR1 | 6 | 152044128 | T | C | C | 0.023 | 0.013 | 0.033 | 1.11E−05 | 0.035 | 0.013 | 0.057 | 1.78E−03 |
| 42 | rs73029259 | LOC107986666 | 6 | 163690316 | A | T | A | 0.037 | 0.020 | 0.054 | 1.99E−05 | 0.033 | −0.004 | 0.069 | 7.95E−02 |
| 43 | rs57246313 | 7 | 25850077 | A | G | A | 0.024 | 0.014 | 0.034 | 3.01E−06 | 0.026 | 0.004 | 0.048 | 1.93E−02 | |
| 44 | rs1007358 | 7 | 46161757 | G | A | G | 0.036 | 0.023 | 0.049 | 1.12E−07 | 0.041 | 0.012 | 0.069 | 5.43E−03 | |
| 45 | rs42377 | CDK6 | 7 | 92614358 | A | G | A | 0.035 | 0.019 | 0.052 | 2.44E−05 | 0.007 | −0.029 | 0.042 | 7.15E−01 |
| 46 | rs445 | CDK6 | 7 | 92779056 | T | C | C | 0.025 | 0.015 | 0.035 | 1.86E−06 | 0.029 | 0.007 | 0.050 | 1.08E−02 |
| 47 | rs76364830 | DLC1 | 8 | 13514611 | A | G | G | 0.041 | 0.023 | 0.059 | 9.41E−06 | 0.059 | 0.019 | 0.098 | 3.93E−03 |
| 48 | rs10958476 | PLAG1 | 8 | 56183249 | C | T | C | 0.030 | 0.017 | 0.042 | 2.60E−06 | 0.014 | −0.013 | 0.041 | 3.04E−01 |
| 49 | rs7842996 | 8 | 77194904 | A | T | A | 0.031 | 0.019 | 0.043 | 6.50E−07 | 0.033 | 0.007 | 0.060 | 1.47E−02 | |
| 50 | rs7817087 | 8 | 116552698 | A | G | G | 0.026 | 0.016 | 0.035 | 4.15E−07 | 0.010 | −0.012 | 0.031 | 3.81E−01 | |
| 51 | rs6992491 | 8 | 128185657 | G | C | G | 0.022 | 0.012 | 0.032 | 1.68E−05 | 0.011 | −0.011 | 0.033 | 3.22E−01 | |
| 52 | rs10120219 | LOC105376158 | 9 | 95602265 | C | T | T | 0.036 | 0.026 | 0.046 | 7.29E−13 | 0.040 | 0.018 | 0.061 | 2.77E−04 |
| 53 | rs34575265 | 9 | 106181520 | T | C | C | 0.022 | 0.012 | 0.032 | 2.66E−05 | 0.011 | −0.011 | 0.033 | 3.16E−01 | |
| 54 | rs7858562 | ZNF483, PTGR1 | 9 | 111562668 | G | A | A | 0.025 | 0.014 | 0.037 | 2.33E−05 | 0.035 | 0.009 | 0.060 | 7.19E−03 |
| 55 | rs12344818 | 9 | 115728289 | T | C | C | 0.031 | 0.019 | 0.043 | 1.73E−07 | −0.006 | −0.031 | 0.020 | 6.65E−01 | |
| 56 | rs3789280 | PAPPA | 9 | 116191093 | A | T | A | 0.036 | 0.020 | 0.053 | 1.23E−05 | 0.018 | −0.018 | 0.053 | 3.30E−01 |
| 57 | rs12338076 | QSOX2 | 9 | 136229894 | C | A | C | 0.040 | 0.029 | 0.050 | 3.63E−13 | 0.045 | 0.022 | 0.068 | 1.50E−04 |
| 58 | rs779933 | ZMIZ1 | 10 | 79158760 | A | G | G | 0.028 | 0.017 | 0.039 | 8.37E−07 | 0.038 | 0.014 | 0.062 | 2.22E−03 |
| 59 | rs2648725 | PCGF5 | 10 | 91255322 | A | T | A | 0.044 | 0.024 | 0.064 | 1.61E−05 | 0.052 | 0.009 | 0.095 | 1.77E−02 |
| 60 | rs1938679 | 11 | 69457328 | T | C | C | 0.038 | 0.029 | 0.048 | 2.98E−14 | 0.032 | 0.010 | 0.053 | 3.84E−03 | |
| 61 | rs645935 | SERPINH1 | 11 | 75568245 | C | T | T | 0.042 | 0.032 | 0.051 | 9.90E−17 | 0.034 | 0.012 | 0.055 | 2.03E−03 |
| 62 | rs59917308 | 12 | 56264924 | T | C | T | 0.069 | 0.039 | 0.099 | 5.37E−06 | −0.005 | −0.069 | 0.059 | 8.81E−01 | |
| 63 | rs3816804 | CS | 12 | 56286961 | T | C | C | 0.112 | 0.099 | 0.126 | 6.35E−63 | 0.114 | 0.085 | 0.142 | 3.14E−15 |
| 64 | rs2277339 | PRIM1 | 12 | 56752285 | G | T | T | 0.037 | 0.025 | 0.049 | 1.81E−09 | 0.040 | 0.014 | 0.066 | 2.91E−03 |
| 65 | rs10747784 | 12 | 57857579 | G | A | G | 0.027 | 0.015 | 0.038 | 4.71E−06 | 0.019 | −0.005 | 0.044 | 1.21E−01 | |
| 66 | rs10878984 | 12 | 69434754 | C | T | T | 0.044 | 0.034 | 0.054 | 5.99E−17 | 0.025 | 0.003 | 0.047 | 2.84E−02 | |
| 67 | rs3847787 | CRADD | 12 | 93813756 | G | A | G | 0.022 | 0.012 | 0.033 | 1.91E−05 | 0.031 | 0.009 | 0.053 | 6.20E−03 |
| 68 | rs2093210 | C14orf39 | 14 | 60490561 | T | C | C | 0.043 | 0.031 | 0.055 | 2.54E−12 | 0.027 | 0.001 | 0.053 | 4.52E−02 |
| 69 | rs910316 | TMED10 | 14 | 75159339 | A | C | A | 0.029 | 0.016 | 0.042 | 1.20E−05 | 0.030 | 0.002 | 0.058 | 3.67E−02 |
| 70 | rs7156335 | ITPK1 | 14 | 92939887 | C | T | C | 0.049 | 0.028 | 0.070 | 4.46E−06 | 0.033 | −0.013 | 0.080 | 1.56E−01 |
| 71 | rs12592845 | 15 | 48392761 | T | C | C | 0.028 | 0.015 | 0.041 | 1.99E−05 | 0.065 | 0.037 | 0.093 | 5.11E−06 | |
| 72 | rs975210 | TLE3 | 15 | 70072013 | A | G | A | 0.033 | 0.019 | 0.048 | 7.70E−06 | 0.004 | −0.028 | 0.036 | 8.25E−01 |
| 73 | rs750460 | LOXL1 | 15 | 73949165 | A | G | G | 0.047 | 0.031 | 0.063 | 1.16E−08 | 0.048 | 0.012 | 0.083 | 8.87E−03 |
| 74 | rs8025068 | ARID3B | 15 | 74577704 | G | T | G | 0.023 | 0.013 | 0.032 | 6.66E−06 | 0.028 | 0.006 | 0.049 | 1.10E−02 |
| 75 | rs4467054 | ADAMTS17 | 15 | 100255167 | G | T | G | 0.029 | 0.018 | 0.039 | 9.87E−08 | 0.038 | 0.014 | 0.061 | 1.45E−03 |
| 76 | rs258324 | CDK10 | 16 | 89687847 | T | G | T | 0.043 | 0.032 | 0.054 | 1.97E−15 | 0.046 | 0.023 | 0.069 | 1.13E−04 |
| 77 | rs74494415 | GALR1 | 18 | 77260182 | T | C | C | 0.040 | 0.022 | 0.058 | 8.97E−06 | 0.026 | −0.013 | 0.064 | 1.88E−01 |
| 78 | rs1741344 | 20 | 4121153 | C | T | C | 0.032 | 0.019 | 0.044 | 3.37E−07 | 0.040 | 0.014 | 0.067 | 2.99E−03 | |
| 79 | rs967417 | 20 | 6640246 | G | A | G | 0.033 | 0.020 | 0.046 | 1.10E−06 | 0.040 | 0.011 | 0.068 | 5.95E−03 | |
| 80 | rs3213180 | E2F1 | 20 | 33675818 | C | G | G | 0.032 | 0.022 | 0.043 | 1.83E−09 | 0.043 | 0.020 | 0.066 | 2.71E−04 |
| 81 | rs143384 | GDF5 | 20 | 35437976 | G | A | G | 0.074 | 0.063 | 0.085 | 3.61E−40 | 0.056 | 0.032 | 0.079 | 4.19E−06 |
| 82 | rs2235363 | ZHX3 | 20 | 41179129 | G | A | G | 0.023 | 0.013 | 0.032 | 7.22E−06 | 0.011 | −0.010 | 0.032 | 3.10E−01 |
| 83 | rs11537645 | UBE2C | 20 | 45812764 | G | C | C | 0.044 | 0.026 | 0.061 | 1.45E−06 | 0.008 | −0.031 | 0.046 | 7.01E−01 |
SNP, single nucleotide polymorphism; GWAS, genome-wide association study; No., number; Chr., chromosome; 95% CI, 95% confidence interval
The measured (phenotypic) heights (cm) were stratified by sex, mean-centered, and normalized to one standard deviation (SD) before height GWAS analysis
Beta-value calculation was performed in agreement with the defined risk alleles
Association between the genetically determined height (PRS251) and the measured height (phenotype)
The association between genetically determined height (PRS251) and measured height (phenotype) was investigated in the testing and validation groups, where height was stratified by sex (male: N = 10,919; female: N = 17,990; Fig. 4). For males, the regression line indicated that a 1-SD increase in PRS251 was associated with a 0.257-SD increase in normalized measured height (slope = 0.257; p < 0.001; green line). For females, the regression line indicated that a 1-SD increase in PRS251 was associated with a 0.274-SD increase in normalized measured height (slope = 0.274; p < 0.001; red line).
Furthermore, to assess the validity of our findings, we replicated the association between genetically determined height (PRS237) and measured heights (phenotype) in another cohort, kindly provided by the Big Data Center in China Medical University Hospital (CMUH), Taichung, Taiwan (Additional file 4: Fig. S3). As shown, only 237 of the 251 SNPs were available from the independent cohort of the Big Data Center at CMUH (Additional file 5: Table S4). The measured height (phenotype) and genetically determined height (PRS237) were normalized (standardized) by sex. For males, the regression line indicated that a 1-SD increase in PRS237 was associated with a 0.0972-SD increase in normalized measured height (slope = 0.0972; p < 0.001; green line; Additional file 4: Fig. S3). For females, the regression line indicated that a 1-SD increase in (PRS237) was associated with a 0.104-SD increase in normalized measured height (slope = 0.104; p < 0.001; red line; Additional file 4: Fig. S3).
Associations of height with health-related outcomes
In this study, we performed both observational (phenotype) and genetic PRS association analyses of height with 63 health-related outcomes using the Taiwan Biobank (Fig. 5). We examined the association between observational (phenotype) height with 63 health-related outcomes, including 14 traits and 49 diseases, in 67,452 individuals of Han Chinese ancestry (Fig. 5). Similar analyses of the association between genetic PRS of height and height were performed (Fig. 5). The genetically determined height of PRS251 (251 SNPs) applied in this analysis was calculated from our GWAS results, consisting of 168 GWAS-identified SNPs (Table 1) and 83 previously reported GWAS-determined SNPs (Table 2). The estimated beta values (95% CI) for the 14 traits are shown in Fig. 5A–C. The estimated odds ratios (95% CI) for the 49 diseases are also shown in Fig. 5D–M. After adjusting for age, sex, education, drinking, smoking, regular exercise, and 10 PCA results, our analyses showed that observational (phenotype) height was associated with eight of the 14 traits (p < 0.05/[14 + 49]; Table 3). Further analyses confirmed that genetic (PRS251) height was also associated with these eight traits (Table 3). No significant associations were observed between the measured and genetic PRS height with the 49 diseases (p > 0.05/[14 + 49]; Fig. 5D–M). Among anthropometric traits, observational height was positively associated with body weight, waist circumference, and hip circumference but negatively associated with BMI, WHR, and body fat (Table 3). Genetic PRS height was associated with increased body weight (beta = 1.2182, 95% CI = 1.1405–1.2959), waist circumference (beta = 0.4462, 95% CI = 0.3754–0.5171), and hip circumference (beta = 0.6006, 95% CI = 0.5488–0.6523), and a decreased BMI (beta = −0.0837, 95% CI = (−0.1110)–(−0.0563)), WHR (beta = −0.0008, 95% CI = (−0.0012)–(−0.0003)), and body fat (beta = −0.1401, 95% CI = (−0.1856)–(−0.0946)).
Fig. 5.
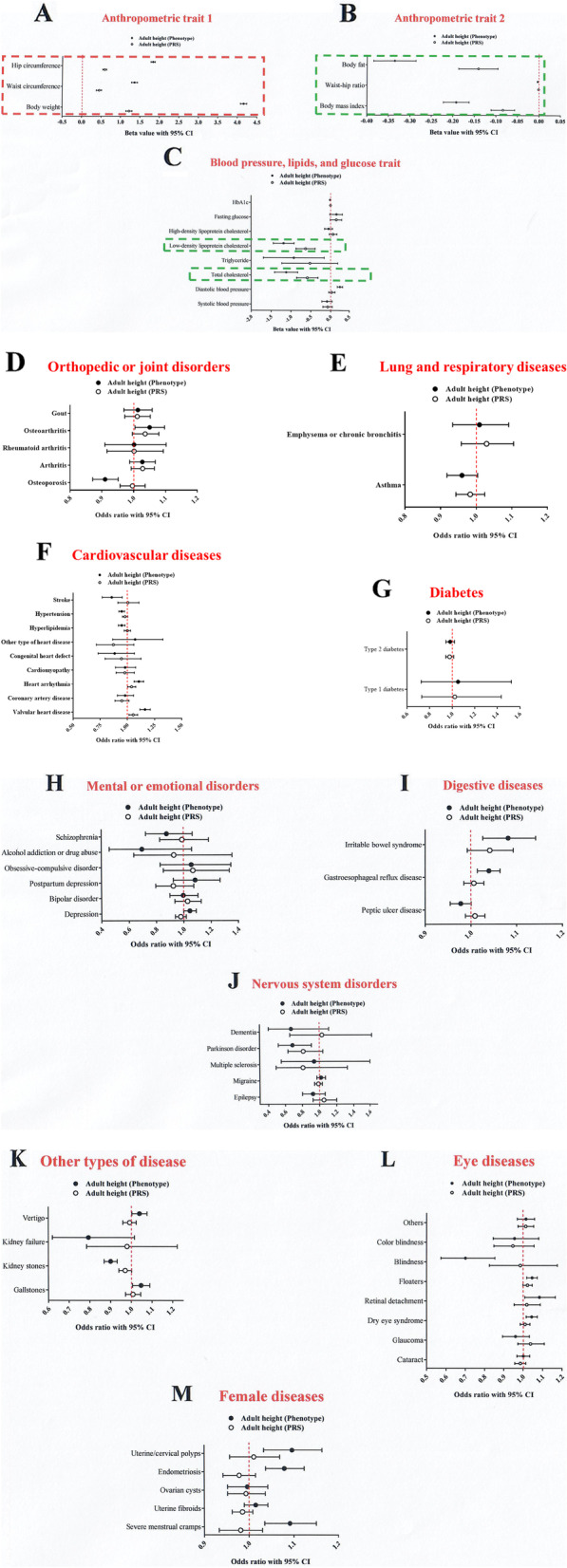
Observational (phenotype) and genetic PRS251 associations of height with 63 health-related outcomes. Beta value and 95% confidence interval (CI) per standard deviation (SD) increase in height are shown for A anthropometric trait 1 (hip circumference, waist circumference, and body weight), B anthropometric trait 2 (body fat, waist-hip ratio, and body mass index), and C blood pressure, blood lipid level, and blood glucose level (including systolic blood pressure (SBP), diastolic blood pressure (DBP), total cholesterol (TC), triglyceride (TG), low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), fasting glucose, and HbA1c). Odds ratio and 95% CI per SD increase in height are shown for D orthopedic or joint disorders (osteoporosis, arthritis, rheumatoid arthritis, osteoarthritis, and gout), E lung and respiratory diseases (asthma and emphysema or chronic bronchitis), F cardiovascular diseases (valvular heart disease, coronary artery disease, heart arrhythmia, cardiomyopathy, congenital heart defect, other type of heart disease, hyperlipidemia, hypertension, and stroke), G diabetes (type 1 diabetes and type 2 diabetes), H mental or emotional disorders (depression, bipolar disorder, postpartum depression, obsessive-compulsive disorder, alcohol addiction or drug abuse, and schizophrenia), I digestive diseases (peptic ulcer disease, gastroesophageal reflux disease, and irritable bowel syndrome), J nervous system disorders (epilepsy, migraine, multiple sclerosis, Parkinson’s disorder, and dementia), K other types of disease (gallstones, kidney stones, kidney failure, and vertigo), L eye diseases (cataract, glaucoma, dry eye syndrome, retinal detachment, floaters, blindness, color blindness, and others), and M female diseases (severe menstrual cramps, uterine fibroids, ovarian cysts, endometriosis, and Uterine/cervical polyps)
Table 3.
Significant association between phenotypic and genetically determined height with eight traits
| Height (exposure) | 8 traits (outcome) | Beta | 95% confidence interval | P-value | |
|---|---|---|---|---|---|
| Anthropometric trait 1 | |||||
| Height (Phenotype) | Body weight (Phenotype) | 4.162 | 4.083 | 4.241 | 0.00E+00 |
| Height (PRS) | Body weight (Phenotype) | 1.218 | 1.141 | 1.296 | 7.81E−206 |
| Height (Phenotype) | Waist circumference (Phenotype) | 1.363 | 1.287 | 1.440 | 1.56E−264 |
| Height (PRS) | Waist circumference (Phenotype) | 0.446 | 0.375 | 0.517 | 5.81E−35 |
| Height (Phenotype) | Hip circumference (Phenotype) | 1.845 | 1.790 | 1.900 | 0.00E+00 |
| Height (PRS) | Hip circumference (Phenotype) | 0.601 | 0.549 | 0.652 | 4.53E−114 |
| Anthropometric trait 2 | |||||
| Height (Phenotype) | Body mass index (Phenotype) | −0.192 | −0.222 | −0.162 | 1.36E−36 |
| Height (PRS) | Body mass index (Phenotype) | −0.084 | −0.111 | -0.056 | 2.01E−09 |
| Height (Phenotype) | Waist-hip ratio (Phenotype) | −0.003 | −0.003 | −0.002 | 1.75E−24 |
| Height (PRS) | Waist-hip ratio (Phenotype) | −0.001 | −0.001 | 0.000 | 7.18E−04 |
| Height (Phenotype) | Body fat (Phenotype) | −0.335 | −0.385 | −0.286 | 3.82E−40 |
| Height (PRS) | Body fat (Phenotype) | −0.140 | −0.186 | −0.095 | 1.58E−09 |
| Blood pressure, lipids, and glucose trait | |||||
| Height (Phenotype) | Total cholesterol (Phenotype) | −1.117 | −1.407 | −0.828 | 4.13E−14 |
| Height (PRS) | Total cholesterol (Phenotype) | −0.587 | −0.853 | −0.321 | 1.55E−05 |
| Height (Phenotype) | Low-density lipoprotein cholesterol (Phenotype) | −1.180 | −1.439 | −0.921 | 4.68E−19 |
| Height (PRS) | Low-density lipoprotein cholesterol (Phenotype) | −0.629 | −0.867 | −0.391 | 2.25E−07 |
PRS polygenic risk score
Height phenotype indicates the measured height (normalized using Z-scores)
Height polygenic risk score (PRS) indicates the height calculated from 251 SNPs (Tables 1 and 2)
Multivariate linear regression analysis was used with adjustment factors (age, sex, educational attainment, drinking, smoking, regular exercise, and 10 principal components analysis data). P-value (p < 0.05/(14+49)) was highlighted in bold italic. P-value less than 2.23E−308 were expressed as 0.00E+00
Regarding blood pressure, and lipid and glucose levels, observational height was negatively associated with TC and LDL-C (Table 3). Genetic PRS height was associated with decreased TC (beta = −0.5869, 95% CI = (−0.8530)–(−0.3207)) and LDL-C levels (beta = −0.6291, 95% CI = (−0.8672)–(−0.3910)).
Discussion
We reported a genetic profile for height in the Han Chinese population using genome-wide SNP analysis and a replication study in the Taiwan Biobank—a community-based database in Taiwan. This is the first large-scale finding on the genetic basis for height and health-related outcomes in individuals of Han Chinese ancestry in Taiwan. Our study results are consistent with the genetic profile of height observed mainly in individuals of European ancestry [15–26]. Accordingly, our findings support the validity of height in observational (phenotype) studies and are consistent with the health-related outcomes of this phenotype [73–81].
In this study, we identified 6843 SNPs with genome-wide significance in 89 genomic regions, including 18 novel loci. Among these, we identified seven independent lead SNPs at seven genetic loci (two of these lead SNPs were novel: chromosome 2, rs76803230 in DIS3L2; chromosome 3, rs57345461 in ZBTB38) with genome-wide significance. DIS3L2 encodes one of the subunits of the RNA exosome, and its genetic variants are associated with height in individuals of European ancestry [21, 82] and East Asian ancestry [83, 84]. ZBTB38 is a zinc finger transcriptional activator that binds methylated DNA and is associated with apoptosis. ZBTB38 genetic variants are associated with height in individuals of European ancestry [17, 18] and East Asian ancestry [20, 23]. The remaining five lead SNPs were reported previously [15, 23, 64–72]. The lead SNP rs3791675 in EFEMP1 encodes an extracellular matrix glycoprotein of the fibulin family and has been associated with body height, BMI-adjusted waist circumference, pelvic organ prolapse, and BMI-adjusted WHR [23, 64–66]. The lead SNP rs16895971 in LCORL, a transcription factor involved in spermatogenesis, has been associated with height in East Asians [67]. The lead SNP rs2780226 in HMGA1 encodes a chromatin-associated protein that regulates gene transcription and metastatic progression of cancer cells and has been associated with body height, BMI-adjusted waist circumference, and birth weight [15, 68, 69]. The lead SNP rs3816804 in CS has also been associated with height in East Asians [70]. The lead SNP rs143384 in GDF5, which encodes a secreted ligand of the transforming growth factor-beta superfamily, regulates the development of numerous tissue and cell types and has been associated with body height, BMI-adjusted hip circumference, BMI-adjusted WHR, and body fat [15, 64, 71, 72]. Our observations report novel lead SNPs in the Han Chinese population that share an overlapping genetic architecture for height, mainly discovered in individuals of European ancestry.
Our observational (phenotype) analyses showed that height was associated with eight traits. Furthermore, our PRS251 analyses confirmed that genetic height was also associated with these eight traits. Taller height was associated with decreased BMI, WHR, body fat, TC, and LDL-C, but with increased body weight, waist circumference, and hip circumference. For anthropometric traits, we observed that both observational (phenotype) and genetically determined height (PRS251) were associated with BMI, WHR, body fat, body weight, waist circumference, and hip circumference. Taller height was associated with decreased BMI, WHR, and body fat, but increased body weight, waist circumference, and hip circumference. Our findings are consistent with previous observational (phenotype) studies that reported an inverse association between height and obesity-related traits, including BMI, WHR, and body fat [73, 74]. As expected, taller adults had lower rates of obesity [73, 74]. Our findings also support previous observational (phenotype) studies that reported positive associations between height and body weight [73–75], waist circumference [74, 75], and hip circumference [75]. Furthermore, taller adults had increased body weight and waist and hip circumferences. The results of our genetic PRS of height association were also in agreement with previous genetic correlation studies, mainly conducted in individuals of European ancestry [78–81]. There was a negative correlation between genetically determined height and BMI [78–80], and positive correlations between genetically determined height with waist and hip circumference [80, 81]. Our findings may reflect a partial genetic overlap between height and anthropometric traits including BMI, WHR, body fat, body weight, waist circumference, and hip circumference. However, genetic correlations in individuals of Han Chinese ancestry remain to be elucidated.
Regarding blood pressure and lipids and glucose levels, both observational (phenotype) and genetically determined height (PRS251) were inversely associated with TC and LDL-C. Our findings are consistent with those of previous observational (phenotype) studies that reported an inverse association between height and blood lipid levels [76, 77, 85]. Taller adults have lower levels of TC and LDL-C [76, 77, 85]. The results of our genetic PRS of height are also in agreement with previous studies, mainly conducted in individuals of European ancestry [50, 80, 81]. Negative genetic correlations between height and TC were found [80, 81]. A taller genetic PRS was associated with lower LDL-C levels in individuals of European ancestry [50]. Our results also suggest that genetically taller individuals of Han Chinese ancestry have lower levels of TC and LDL-C.
Conclusions
This large-scale assessment of the genetic architecture of height in the Han Chinese population of Taiwan quantified the extent of the shared genetic basis with individuals of European ancestry. Our observational and genetic study supports the relevance of height to the etiology of various health-related outcomes, especially those regarding anthropometric traits and blood lipids.
Supplementary Information
Additional file 1: Table S1. Basic characteristics of the study participants at enrollment.
Additional file 2: Table S2. The top lead SNPs in 89 genomic regions that were significantly associated with height (p < 5 × 10−8), using LD (r2 < 0.2), in individuals of Han Chinese ancestry.
Additional file 3: Table S3. Replication of a previous GWAS of body height in the SNPs of the training group (313 of 1722 SNPs).
Additional file 4: Figure S1. The clumping and p-value threshold method identifies the “best-fit” SNP number for the polygenic risk score (PRS) calculation, according to the largest explainable phenotype correlation r2 using only PRS (PRS r2 and SNP number). The x-axis shows the p-value thresholds from the height of GWAS results. The y-axis represents the explainable phenotypic correlation r2 using only the PRS (PRS r2). The p-values above the bars show the statistical significance of the associations between genetically determined height (PRS) and measured height (phenotype). (A) 251 SNPs were obtained from 6,941 SNPs (novel and reported SNPs; PRS r2 = 0.0712, SNP number = 251). (B) 194 SNPs were obtained from 6,843 SNPs (novel SNPs; PRS r2 = 0.0622, SNP number = 194). (C) 154 SNPs were obtained from 313 SNPs (reported SNPs; PRS r2 = 0.0706, SNP number = 154). Figure S2. Scree plot identifying the number of principal component analyses (PCA) needed for the correction of population structure in the height GWAS study, using pcadapt (an R package used to determine the number of principal components). Figure S3. Association between genetically determined height (PRS237) and measured height (phenotype) in an independent cohort of the Big Data Center in China Medical University Hospital in Taiwan. The measured height (cm) and calculated polygenic risk score (PRS) for height were stratified by sex, mean-centered, and normalized to one standard deviation (SD; males, N = 46,310; females, N = 54,728). The normalized measured height is represented on the y-axis, and normalized genetically determined height (PRS237) is represented on the x-axis.
Additional file 5: Table S4. Characteristics of 237 out of the 251 SNPs associated with height in the independent cohort of the Big Data Center at China Medical University Hospital in Taiwan.
Acknowledgements
We are grateful to the Health Data Science Center at the China Medical University Hospital, the iHi Clinical Research and iHi Genomics Platform from the Big Data Center of China Medical University Hospital, Taiwan Biobank (TWBR10812-06), and the National Center for Genome Medicine of the National Core Facility Program for Biotechnology for data exploration; administrative, technical, and statistical analysis; and funding support.
Abbreviations
- BMI
Body mass index
- CI
Confidence interval
- CS
Citrate synthase
- CVD
Cardiovascular disease
- DBP
Diastolic blood pressure
- DIS3L2
DIS3 like 3′-5′ exoribonuclease 2
- EFEMP1
EGF-containing fibulin extracellular matrix protein 1
- GDF5
Growth differentiation factor 5
- GWAS
Genome-wide association study
- HDL-C
High-density lipoprotein cholesterol
- HMGA1
High-mobility group AT-hook 1
- IBD
Individual identity by descent
- LCORL
Ligand-dependent nuclear receptor corepressor like
- LD
Linkage disequilibrium
- LDL-C
Low-density lipoprotein cholesterol
- LM
Linear regression model
- MAF
Minor allele frequency
- PCA
Principal component analysis
- PRS
Polygenic risk score
- QC
Quality control
- SBP
Systolic blood pressure
- SD
Standard deviation
- SNP
Single nucleotide polymorphism
- TC
Total cholesterol
- TG
Triglyceride
- WHR
Waist-hip ratio
- ZBTB38
Zinc finger and BTB domain containing 38
Authors’ contributions
YJL and FJT conceptualized this study. FJT supervised the study. JSC, CFC, WML, CHC, and CHW collected data for this study. JSC, CFC, and CHC analyzed and computed the genetic risk scores. JSC performed the statistical analyses. YJL drafted the manuscript in consultation with FJT and intellectual inputs from WDL, MLC, WCC, CWL, THL, CCL, SMH, and CHT. All authors contributed to critical revision of the manuscript. YJL and FJT were primarily responsible for the final content. All the authors have read and approved the final manuscript.
Funding
We thank the China Medical University, Taiwan (CMU-110-S-17, CMU-110-S-24, CMU110-MF-115, and CMU110-MF-49); the China Medical University Hospital, Taiwan (DMR-111-062 and DMR-111-153); and the Ministry of Science and Technology, Taiwan (MOST 108-2314-B-039-044-MY3, MOST 109-2320-B-039-035-MY3, MOST 109-2410-H-039-002, and MOST 110-2410-H-039-002) for supporting this publication. The funding entities had no role in the study design, data collection, data analysis, interpretation, or authorship of the manuscript.
Availability of data and materials
Individual-level Taiwan Biobank data are available upon application to the Taiwan Biobank (https://www.twbiobank.org.tw/new_web/).
Declarations
Ethics approval and consent to participate
The study procedures were approved by the Human Studies Committee of the China Medical University Hospital, Taichung, Taiwan (approval number: CMUH107-REC3-074). All recruited patients provided written informed consent upon enrollment.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ying-Ju Lin, Email: yjlin.kath@gmail.com.
Fuu-Jen Tsai, Email: d0704@mail.cmuh.org.tw.
References
- 1.Dubois L, Ohm Kyvik K, Girard M, Tatone-Tokuda F, Perusse D, Hjelmborg J, Skytthe A, Rasmussen F, Wright MJ, Lichtenstein P, et al. Genetic and environmental contributions to weight, height, and BMI from birth to 19 years of age: an international study of over 12,000 twin pairs. PLoS One. 2012;7(2):e30153. doi: 10.1371/journal.pone.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jelenkovic A, Hur YM, Sund R, Yokoyama Y, Siribaddana SH, Hotopf M, et al. Genetic and environmental influences on adult human height across birth cohorts from 1886 to 1994. Elife. 2016;5:e20320. [DOI] [PMC free article] [PubMed]
- 3.Jelenkovic A, Sund R, Hur YM, Yokoyama Y, Hjelmborg JV, Moller S, Honda C, Magnusson PK, Pedersen NL, Ooki S, et al. Genetic and environmental influences on height from infancy to early adulthood: an individual-based pooled analysis of 45 twin cohorts. Sci Rep. 2016;6:28496. doi: 10.1038/srep28496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyrrell J, Jones SE, Beaumont R, Astley CM, Lovell R, Yaghootkar H, Tuke M, Ruth KS, Freathy RM, Hirschhorn JN, et al. Height, body mass index, and socioeconomic status: mendelian randomisation study in UK Biobank. BMJ. 2016;352:i582. doi: 10.1136/bmj.i582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magnusson PK, Rasmussen F, Gyllensten UB. Height at age 18 years is a strong predictor of attained education later in life: cohort study of over 950,000 Swedish men. Int J Epidemiol. 2006;35(3):658–663. doi: 10.1093/ije/dyl011. [DOI] [PubMed] [Google Scholar]
- 6.German A, Mesch G, Hochberg Z. People are taller in countries with better environmental conditions. Front Endocrinol (Lausanne) 2020;11:106. doi: 10.3389/fendo.2020.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravnborg TL, Jensen TK, Andersson AM, Toppari J, Skakkebaek NE, Jorgensen N. Prenatal and adult exposures to smoking are associated with adverse effects on reproductive hormones, semen quality, final height and body mass index. Hum Reprod. 2011;26(5):1000–1011. doi: 10.1093/humrep/der011. [DOI] [PubMed] [Google Scholar]
- 8.Carter RC, Jacobson JL, Sokol RJ, Avison MJ, Jacobson SW. Fetal alcohol-related growth restriction from birth through young adulthood and moderating effects of maternal prepregnancy weight. Alcohol Clin Exp Res. 2013;37(3):452–462. doi: 10.1111/j.1530-0277.2012.01940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alves JGB, Alves GV. Effects of physical activity on children's growth. J Pediatr (Rio J) 2019;95(Suppl 1):72–78. doi: 10.1016/j.jped.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Lucas R, Fraga S, Ramos E, Barros H. Early initiation of smoking and alcohol drinking as a predictor of lower forearm bone mineral density in late adolescence: a cohort study in girls. PLoS One. 2012;7(10):e46940. doi: 10.1371/journal.pone.0046940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Loughlin J, Karp I, Henderson M, Gray-Donald K. Does cigarette use influence adiposity or height in adolescence? Ann Epidemiol. 2008;18(5):395–402. doi: 10.1016/j.annepidem.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Huang W, Lei X, Ridder G, Strauss J, Zhao Y. Health, height, height shrinkage, and SES at older ages: evidence from China. Am Econ J Appl Econ. 2013;5(2):86–121. doi: 10.1257/app.5.2.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crouch DJM, Bodmer WF. Polygenic inheritance, GWAS, polygenic risk scores, and the search for functional variants. Proc Natl Acad Sci U S A. 2020;117(32):18924–18933. doi: 10.1073/pnas.2005634117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan Y, Holmen OL, Dauber A, Vatten L, Havulinna AS, Skorpen F, Kvaloy K, Silander K, Nguyen TT, Willer C, et al. Common variants show predicted polygenic effects on height in the tails of the distribution, except in extremely short individuals. PLoS Genet. 2011;7(12):e1002439. doi: 10.1371/journal.pgen.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan J, Kutalik Z, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46(11):1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, Eyheramendy S, Voight BF, Butler JL, Guiducci C, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40(5):584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40(5):575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JJ, Lee HI, Park T, Kim K, Lee JE, Cho NH, Shin C, Cho YS, Lee JY, Han BG, et al. Identification of 15 loci influencing height in a Korean population. J Hum Genet. 2010;55(1):27–31. doi: 10.1038/jhg.2009.116. [DOI] [PubMed] [Google Scholar]
- 20.Okada Y, Kamatani Y, Takahashi A, Matsuda K, Hosono N, Ohmiya H, Daigo Y, Yamamoto K, Kubo M, Nakamura Y, et al. A genome-wide association study in 19 633 Japanese subjects identified LHX3-QSOX2 and IGF1 as adult height loci. Hum Mol Genet. 2010;19(11):2303–2312. doi: 10.1093/hmg/ddq091. [DOI] [PubMed] [Google Scholar]
- 21.Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, Sulem P, Thorlacius S, Gylfason A, Steinberg S, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40(5):609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Ferreira T, Morris AP, Medland SE, Genetic Investigation of ATC, Replication DIG, Meta-analysis C. Madden PA, Heath AC, Martin NG, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44(4):369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, Yoon D, Lee MH, Kim DJ, Park M, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41(5):527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 24.Chan Y, Salem RM, Hsu YH, McMahon G, Pers TH, Vedantam S, Esko T, Guo MH, Lim ET, Consortium G, et al. Genome-wide analysis of body proportion classifies height-associated variants by mechanism of action and implicates genes important for skeletal development. Am J Hum Genet. 2015;96(5):695–708. doi: 10.1016/j.ajhg.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soranzo N, Rivadeneira F, Chinappen-Horsley U, Malkina I, Richards JB, Hammond N, Stolk L, Nica A, Inouye M, Hofman A, et al. Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5(4):e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin YJ, Cheng CF, Wang CH, Liang WM, Tang CH, Tsai LP, et al. Genetic architecture associated with familial short stature. J Clin Endocrinol Metab. 2020;105(6):dgaa131. [DOI] [PubMed]
- 27.Emerging Risk Factors C Adult height and the risk of cause-specific death and vascular morbidity in 1 million people: individual participant meta-analysis. Int J Epidemiol. 2012;41(5):1419–1433. doi: 10.1093/ije/dys086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green J, Cairns BJ, Casabonne D, Wright FL, Reeves G, Beral V, Million Women Study c Height and cancer incidence in the Million Women Study: prospective cohort, and meta-analysis of prospective studies of height and total cancer risk. Lancet Oncol. 2011;12(8):785–794. doi: 10.1016/S1470-2045(11)70154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paajanen TA, Oksala NK, Kuukasjarvi P, Karhunen PJ. Short stature is associated with coronary heart disease: a systematic review of the literature and a meta-analysis. Eur Heart J. 2010;31(14):1802–1809. doi: 10.1093/eurheartj/ehq155. [DOI] [PubMed] [Google Scholar]
- 30.Case A, Paxson C. Height, health, and cognitive function at older ages. Am Econ Rev. 2008;98(2):463–467. doi: 10.1257/aer.98.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guven C, Lee WS. Height and cognitive function at older ages: is height a useful summary measure of early childhood experiences? Health Econ. 2013;22(2):224–233. doi: 10.1002/hec.1827. [DOI] [PubMed] [Google Scholar]
- 32.Guven C, Lee WS. Height, aging and cognitive abilities across Europe. Econ Hum Biol. 2015;16:16–29. doi: 10.1016/j.ehb.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Ma W, Hagan KA, Heianza Y, Sun Q, Rimm EB, Qi L. Adult height, dietary patterns, and healthy aging. Am J Clin Nutr. 2017;106(2):589–596. doi: 10.3945/ajcn.116.147256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shrestha S, Rasmussen SH, Pottegard A, Angquist LH, Jess T, Allin KH, Bjerregaard LG, Baker JL. Associations between adult height and type 2 diabetes mellitus: a systematic review and meta-analysis of observational studies. J Epidemiol Community Health. 2019;73(7):681–688. doi: 10.1136/jech-2018-211567. [DOI] [PubMed] [Google Scholar]
- 35.Choi SW, Mak TS, O'Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc. 2020;15(9):2759–2772. doi: 10.1038/s41596-020-0353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janssens A. Validity of polygenic risk scores: are we measuring what we think we are? Hum Mol Genet. 2019;28(R2):R143–R150. doi: 10.1093/hmg/ddz205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye J, Wu C, Chu X, Wen Y, Li P, Cheng B, Cheng S, Liu L, Zhang L, Ma M, et al. Evaluating the effect of birth weight on brain volumes and depression: an observational and genetic study using UK Biobank cohort. Eur Psychiatry. 2020;63(1):e73. doi: 10.1192/j.eurpsy.2020.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teumer A. Common methods for performing Mendelian randomization. Front Cardiovasc Med. 2018;5:51. doi: 10.3389/fcvm.2018.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thrift AP, Gong J, Peters U, Chang-Claude J, Rudolph A, Slattery ML, Chan AT, Esko T, Wood AR, Yang J, et al. Mendelian randomization study of height and risk of colorectal cancer. Int J Epidemiol. 2015;44(2):662–672. doi: 10.1093/ije/dyv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cornish AJ, Law PJ, Timofeeva M, Palin K, Farrington SM, Palles C, Jenkins MA, Casey G, Brenner H, Chang-Claude J, et al. Modifiable pathways for colorectal cancer: a mendelian randomisation analysis. Lancet Gastroenterol Hepatol. 2020;5(1):55–62. doi: 10.1016/S2468-1253(19)30294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang B, Shu XO, Delahanty RJ, Zeng C, Michailidou K, Bolla MK, et al. Height and breast cancer risk: evidence from prospective studies and Mendelian randomization. J Natl Cancer Inst. 2015;107(11):djv219. [DOI] [PMC free article] [PubMed]
- 42.Lai FY, Nath M, Hamby SE, Thompson JR, Nelson CP, Samani NJ. Adult height and risk of 50 diseases: a combined epidemiological and genetic analysis. BMC Med. 2018;16(1):187. doi: 10.1186/s12916-018-1175-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dixon-Suen SC, Nagle CM, Thrift AP, Pharoah PDP, Ewing A, Pearce CL, Zheng W, Australian Ovarian Cancer Study G, Chenevix-Trench G. Fasching PA, et al. Adult height is associated with increased risk of ovarian cancer: a Mendelian randomisation study. Br J Cancer. 2018;118(8):1123–1129. doi: 10.1038/s41416-018-0011-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dusingize JC, Olsen CM, An J, Pandeya N, Law MH, Thompson BS, Goldstein AM, Iles MM, Webb PM, Neale RE, et al. Body mass index and height and risk of cutaneous melanoma: Mendelian randomization analyses. Int J Epidemiol. 2020;49(4):1236–1245. doi: 10.1093/ije/dyaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davies NM, Gaunt TR, Lewis SJ, Holly J, Donovan JL, Hamdy FC, Kemp JP, Eeles R, Easton D, Kote-Jarai Z, et al. The effects of height and BMI on prostate cancer incidence and mortality: a Mendelian randomization study in 20,848 cases and 20,214 controls from the PRACTICAL consortium. Cancer Causes Control. 2015;26(11):1603–1616. doi: 10.1007/s10552-015-0654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ong JS, An J, Law MH, Whiteman DC, Neale RE, Gharahkhani P, MacGregor S. Height and overall cancer risk and mortality: evidence from a Mendelian randomisation study on 310,000 UK Biobank participants. Br J Cancer. 2018;118(9):1262–1267. doi: 10.1038/s41416-018-0063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nuesch E, Dale C, Palmer TM, White J, Keating BJ, van Iperen EP, Goel A, Padmanabhan S, Asselbergs FW, Investigators EP-N et al. Adult height, coronary heart disease and stroke: a multi-locus Mendelian randomization meta-analysis. Int J Epidemiol. 2016;45(6):1927–1937. doi: 10.1093/ije/dyv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stefan N, Haring HU, Hu FB, Schulze MB. Divergent associations of height with cardiometabolic disease and cancer: epidemiology, pathophysiology, and global implications. Lancet Diabetes Endocrinol. 2016;4(5):457–467. doi: 10.1016/S2213-8587(15)00474-X. [DOI] [PubMed] [Google Scholar]
- 49.Marouli E, Del Greco MF, Astley CM, Yang J, Ahmad S, Berndt SI, Caulfield MJ, Evangelou E, McKnight B, Medina-Gomez C, et al. Mendelian randomisation analyses find pulmonary factors mediate the effect of height on coronary artery disease. Commun Biol. 2019;2:119. doi: 10.1038/s42003-019-0361-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelson CP, Hamby SE, Saleheen D, Hopewell JC, Zeng L, Assimes TL, Kanoni S, Willenborg C, Burgess S, Amouyel P, et al. Genetically determined height and coronary artery disease. N Engl J Med. 2015;372(17):1608–1618. doi: 10.1056/NEJMoa1404881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen CH, Yang JH, Chiang CWK, Hsiung CN, Wu PE, Chang LC, Chu HW, Chang J, Song IW, Yang SL, et al. Population structure of Han Chinese in the modern Taiwanese population based on 10,000 participants in the Taiwan Biobank project. Hum Mol Genet. 2016;25(24):5321–5331. doi: 10.1093/hmg/ddw346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin WY, Chan CC, Liu YL, Yang AC, Tsai SJ, Kuo PH. Performing different kinds of physical exercise differentially attenuates the genetic effects on obesity measures: Evidence from 18,424 Taiwan Biobank participants. PLoS Genet. 2019;15(8):e1008277. doi: 10.1371/journal.pgen.1008277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marees AT, de Kluiver H, Stringer S, Vorspan F, Curis E, Marie-Claire C, Derks EM. A tutorial on conducting genome-wide association studies: quality control and statistical analysis. Int J Methods Psychiatr Res. 2018;27(2):e1608. doi: 10.1002/mpr.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsiao YJ, Chuang HK, Chi SC, Wang YY, Chiang PH, Teng PC, et al. Genome-wide polygenic risk score for predicting high risk glaucoma individuals of Han Chinese ancestry. J Pers Med. 2021;11(11):1169. [DOI] [PMC free article] [PubMed]
- 56.Lin YJ, Ho TJ, Lin TH, Hsu WY, Huang SM, Liao CC, Lai CH, Liu X, Tsang H, Lai CC, et al. P-coumaric acid regulates exon 12 splicing of the ATP7B gene by modulating hnRNP A1 protein expressions. Biomedicine (Taipei) 2015;5(2):10. doi: 10.7603/s40681-015-0010-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei CY, Yang JH, Yeh EC, Tsai MF, Kao HJ, Lo CZ, Chang LP, Lin WJ, Hsieh FJ, Belsare S, et al. Genetic profiles of 103,106 individuals in the Taiwan Biobank provide insights into the health and history of Han Chinese. NPJ Genom Med. 2021;6(1):10. doi: 10.1038/s41525-021-00178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi S, Yuan N, Yang M, Du Z, Wang J, Sheng X, Wu J, Xiao J. Comprehensive Assessment of Genotype Imputation Performance. Hum Hered. 2018;83(3):107–116. doi: 10.1159/000489758. [DOI] [PubMed] [Google Scholar]
- 59.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 61.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 62.Lin YJ, Wan L, Wu JY, Sheu JJ, Lin CW, Lan YC, Lai CH, Hung CH, Tsai Y, Tsai CH, et al. HLA-E gene polymorphism associated with susceptibility to Kawasaki disease and formation of coronary artery aneurysms. Arthritis Rheum. 2009;60(2):604–610. doi: 10.1002/art.24261. [DOI] [PubMed] [Google Scholar]
- 63.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tachmazidou I, Suveges D, Min JL, Ritchie GRS, Steinberg J, Walter K, Iotchkova V, Schwartzentruber J, Huang J, Memari Y, et al. Whole-genome sequencing coupled to imputation discovers genetic signals for anthropometric traits. Am J Hum Genet. 2017;100(6):865–884. doi: 10.1016/j.ajhg.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Olafsdottir T, Thorleifsson G, Sulem P, Stefansson OA, Medek H, Olafsson K, Ingthorsson O, Gudmundsson V, Jonsdottir I, Halldorsson GH, et al. Genome-wide association identifies seven loci for pelvic organ prolapse in Iceland and the UK Biobank. Commun Biol. 2020;3(1):129. doi: 10.1038/s42003-020-0857-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Christakoudi S, Evangelou E, Riboli E, Tsilidis KK. GWAS of allometric body-shape indices in UK Biobank identifies loci suggesting associations with morphogenesis, organogenesis, adrenal cell renewal and cancer. Sci Rep. 2021;11(1):10688. doi: 10.1038/s41598-021-89176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sakaue S, Kanai M, Tanigawa Y, Karjalainen J, Kurki M, Koshiba S, Narita A, Konuma T, Yamamoto K, Akiyama M, et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat Genet. 2021;53(10):1415–1424. doi: 10.1038/s41588-021-00931-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Plotnikov D, Williams C, Guggenheim JA. Association between birth weight and refractive error in adulthood: a Mendelian randomisation study. Br J Ophthalmol. 2020;104(2):214–219. doi: 10.1136/bjophthalmol-2018-313640. [DOI] [PubMed] [Google Scholar]
- 69.Graff M, Scott RA, Justice AE, Young KL, Feitosa MF, Barata L, Winkler TW, Chu AY, Mahajan A, Hadley D, et al. Genome-wide physical activity interactions in adiposity - a meta-analysis of 200,452 adults. PLoS Genet. 2017;13(4):e1006528. doi: 10.1371/journal.pgen.1006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hao Y, Liu X, Lu X, Yang X, Wang L, Chen S, Li H, Li J, Cao J, Chen J, et al. Genome-wide association study in Han Chinese identifies three novel loci for human height. Hum Genet. 2013;132(6):681–689. doi: 10.1007/s00439-013-1280-9. [DOI] [PubMed] [Google Scholar]
- 71.Rask-Andersen M, Karlsson T, Ek WE, Johansson A. Genome-wide association study of body fat distribution identifies adiposity loci and sex-specific genetic effects. Nat Commun. 2019;10(1):339. doi: 10.1038/s41467-018-08000-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wojcik GL, Graff M, Nishimura KK, Tao R, Haessler J, Gignoux CR, Highland HM, Patel YM, Sorokin EP, Avery CL, et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570(7762):514–518. doi: 10.1038/s41586-019-1310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bosy-Westphal A, Plachta-Danielzik S, Dorhofer RP, Muller MJ. Short stature and obesity: positive association in adults but inverse association in children and adolescents. Br J Nutr. 2009;102(3):453–461. doi: 10.1017/S0007114508190304. [DOI] [PubMed] [Google Scholar]
- 74.Han TS, Seidell JC, Currall JE, Morrison CE, Deurenberg P, Lean ME. The influences of height and age on waist circumference as an index of adiposity in adults. Int J Obes Relat Metab Disord. 1997;21(1):83–89. doi: 10.1038/sj.ijo.0800371. [DOI] [PubMed] [Google Scholar]
- 75.Heymsfield SB, Heo M, Pietrobelli A. Are adult body circumferences associated with height? Relevance to normative ranges and circumferential indexes. Am J Clin Nutr. 2011;93(2):302–307. doi: 10.3945/ajcn.110.005132. [DOI] [PubMed] [Google Scholar]
- 76.Lee MY, Nam GE, Han K, Kim DH, Kim YH, Cho KH, Park YG. Association between height and hypercholesterolemia in adults: a nationwide population-based study in Korea. Lipids Health Dis. 2019;18(1):198. doi: 10.1186/s12944-019-1148-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oh NK, Song YM, Kim SH, Park MJ. Short stature is associated with increased risk of dyslipidemia in Korean adolescents and adults. Sci Rep. 2019;9(1):14090. doi: 10.1038/s41598-019-50524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vogelezang S, Bradfield JP, Ahluwalia TS, Curtin JA, Lakka TA, Grarup N, Scholz M, van der Most PJ, Monnereau C, Stergiakouli E, et al. Novel loci for childhood body mass index and shared heritability with adult cardiometabolic traits. PLoS Genet. 2020;16(10):e1008718. doi: 10.1371/journal.pgen.1008718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, ReproGen C, Psychiatric Genomics C, Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control C. Duncan L, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi H, Mancuso N, Spendlove S, Pasaniuc B. Local genetic correlation gives insights into the shared genetic architecture of complex traits. Am J Hum Genet. 2017;101(5):737–751. doi: 10.1016/j.ajhg.2017.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo MH, Hirschhorn JN, Dauber A. Insights and implications of genome-wide association studies of height. J Clin Endocrinol Metab. 2018;103(9):3155–3168. doi: 10.1210/jc.2018-01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Estrada K, Krawczak M, Schreiber S, van Duijn K, Stolk L, van Meurs JB, Liu F, Penninx BW, Smit JH, Vogelzangs N, et al. A genome-wide association study of northwestern Europeans involves the C-type natriuretic peptide signaling pathway in the etiology of human height variation. Hum Mol Genet. 2009;18(18):3516–3524. doi: 10.1093/hmg/ddp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.He M, Xu M, Zhang B, Liang J, Chen P, Lee JY, Johnson TA, Li H, Yang X, Dai J, et al. Meta-analysis of genome-wide association studies of adult height in East Asians identifies 17 novel loci. Hum Mol Genet. 2015;24(6):1791–1800. doi: 10.1093/hmg/ddu583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Akiyama M, Ishigaki K, Sakaue S, Momozawa Y, Horikoshi M, Hirata M, Matsuda K, Ikegawa S, Takahashi A, Kanai M, et al. Characterizing rare and low-frequency height-associated variants in the Japanese population. Nat Commun. 2019;10(1):4393. doi: 10.1038/s41467-019-12276-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Henriksson KM, Lindblad U, Agren B, Nilsson-Ehle P, Rastam L. Associations between body height, body composition and cholesterol levels in middle-aged men. the coronary risk factor study in southern Sweden (CRISS) Eur J Epidemiol. 2001;17(6):521–526. doi: 10.1023/A:1014508422504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Basic characteristics of the study participants at enrollment.
Additional file 2: Table S2. The top lead SNPs in 89 genomic regions that were significantly associated with height (p < 5 × 10−8), using LD (r2 < 0.2), in individuals of Han Chinese ancestry.
Additional file 3: Table S3. Replication of a previous GWAS of body height in the SNPs of the training group (313 of 1722 SNPs).
Additional file 4: Figure S1. The clumping and p-value threshold method identifies the “best-fit” SNP number for the polygenic risk score (PRS) calculation, according to the largest explainable phenotype correlation r2 using only PRS (PRS r2 and SNP number). The x-axis shows the p-value thresholds from the height of GWAS results. The y-axis represents the explainable phenotypic correlation r2 using only the PRS (PRS r2). The p-values above the bars show the statistical significance of the associations between genetically determined height (PRS) and measured height (phenotype). (A) 251 SNPs were obtained from 6,941 SNPs (novel and reported SNPs; PRS r2 = 0.0712, SNP number = 251). (B) 194 SNPs were obtained from 6,843 SNPs (novel SNPs; PRS r2 = 0.0622, SNP number = 194). (C) 154 SNPs were obtained from 313 SNPs (reported SNPs; PRS r2 = 0.0706, SNP number = 154). Figure S2. Scree plot identifying the number of principal component analyses (PCA) needed for the correction of population structure in the height GWAS study, using pcadapt (an R package used to determine the number of principal components). Figure S3. Association between genetically determined height (PRS237) and measured height (phenotype) in an independent cohort of the Big Data Center in China Medical University Hospital in Taiwan. The measured height (cm) and calculated polygenic risk score (PRS) for height were stratified by sex, mean-centered, and normalized to one standard deviation (SD; males, N = 46,310; females, N = 54,728). The normalized measured height is represented on the y-axis, and normalized genetically determined height (PRS237) is represented on the x-axis.
Additional file 5: Table S4. Characteristics of 237 out of the 251 SNPs associated with height in the independent cohort of the Big Data Center at China Medical University Hospital in Taiwan.
Data Availability Statement
Individual-level Taiwan Biobank data are available upon application to the Taiwan Biobank (https://www.twbiobank.org.tw/new_web/).