Abstract
In an effort to efficiently discover genes in the diazotrophic endophyte of maize, Klebsiella pneumoniae 342, DNA from strain 342 was hybridized to a microarray containing 96% (n = 4,098) of the annotated open reading frames from Escherichia coli K-12. Using a criterion of 55% identity or greater, 3,000 (70%) of the E. coli K-12 open reading frames were also found to be present in strain 342. Approximately 24% (n = 1,030) of the E. coli K-12 open reading frames are absent in strain 342. For 1.6% (n = 68) of the open reading frames, the signal was too low to make a determination regarding the presence or absence of the gene. Genes with high identity between the two organisms are those involved in energy metabolism, amino acid metabolism, fatty acid metabolism, cofactor synthesis, cell division, DNA replication, transcription, translation, transport, and regulatory proteins. Functions that were less highly conserved included carbon compound metabolism, membrane proteins, structural proteins, putative transport proteins, cell processes such as adaptation and protection, and central intermediary metabolism. Open reading frames of E. coli K-12 with little or no identity in strain 342 included putative regulatory proteins, putative chaperones, surface structure proteins, mobility proteins, putative enzymes, hypothetical proteins, and proteins of unknown function, as well as genes presumed to have been acquired by lateral transfer from sources such as phage, plasmids, or transposons. The results were in agreement with the physiological properties of the two strains. Whole genome comparisons by genomic interspecies microarray hybridization are shown to rapidly identify thousands of genes in a previously uncharacterized bacterial genome provided that the genome of a close relative has been fully sequenced. This approach will become increasingly more useful as more full genome sequences become available.
Klebsiella spp. are common endophytes of maize, and several independent isolations of this organism from maize have been reported to date (3a, 4, 5, 6, 11). By labeling them with green fluorescent protein, these strains have been shown to easily reenter maize after isolation (4, 5). Palus et al. (12) showed that these Klebsiella endophytes are diazotrophic. Chelius and Triplett (5) showed that one of these strains produces dinitrogenase reductase protein within roots, provided that a carbon source is added to the maize seedlings.
Klebsiella pneumoniae and Escherichia coli are closely related enteric bacteria. Strain K-12 of Escherichia coli was fully sequenced by Blattner et al. (3). The availability of that sequence allowed Richmond et al. (13) to construct a microarray containing 96% of the open reading frames (ORFs) from the K-12 genome. These arrays have been used to assess gene expression in E. coli following heat shock (13). In that work, the number of genes known to respond to heat shock increased from 23 to 96. Tao et al. (17) examined gene expression in E. coli using these arrays following culture in minimal and rich media. Genes involved in the synthesis of building blocks and those under RpoS regulation were induced with cells cultured in minimal medium.
Here the use of these microarrays is presented for rapid gene discovery in a close relative of E. coli. Others have done comparisons of whole genomes in silico using the complete or nearly complete genome sequences available in the databases (1, 2, 15, 16). The primary advantage of the microarray approach is that it allows the identification of thousands of genes in an organism without any need for sequencing, provided that an ORF microarray for a fully sequenced close relative is available. The disadvantage of this approach is that it indicates only the genes in common between the fully sequenced relative and the strain of interest; genes unique to a Klebsiella isolate from maize compared to E. coli remain unknown. However, identification of the unique genes by subtraction and sequence analysis in conjunction with the microarray analysis described here provides an efficient means for whole genome characterization.
MATERIALS AND METHODS
DNA isolation, nebulization, and labeling.
DNA isolation from strains K-12 and 342 was done by a standard sodium dodecyl sulfate lysis technique (14). Nebulization, fluorescent labeling, and microarray hybridization were done as described by Richmond et al. (13). DNA from K-12 and 342 were labeled by primer extension using Cy3 and Cy5 labels, respectively. The hybridization temperature was 60°C. Other hybridization temperatures were attempted with unsatisfactory results. At a hybridization temperature of 55°C, the background was too high on the microarray. At 65°C, the hybridization signals from the positive controls were too low.
The E. coli ORF microarray.
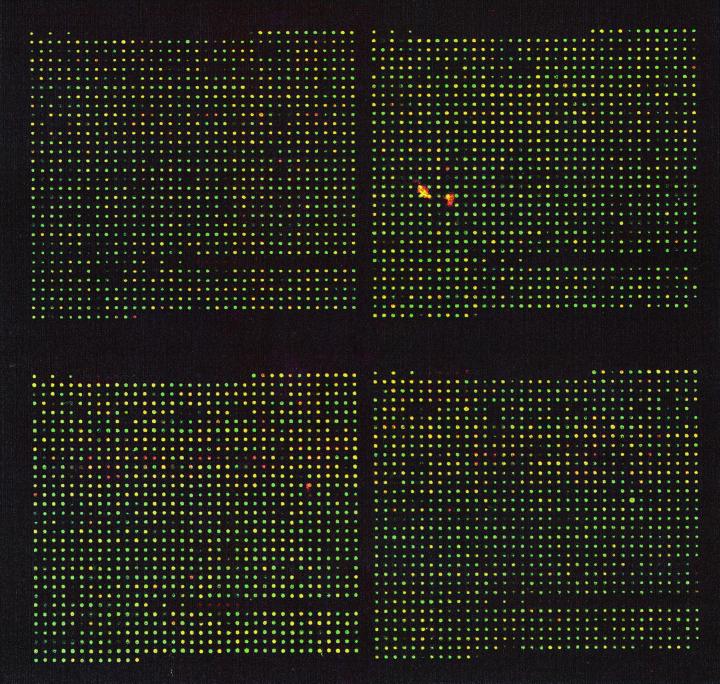
The high-density microarray of E. coli ORFs were prepared as described by Richmond et al. (13). The microarray is in four blocks, each containing 1,152 spots (32 rows by 36 columns). The array includes negative controls such as salmon sperm DNA and yeast tRNA genes. Data from the negative controls were averaged and used to subtract the background fluorescence. Internal controls are included in the experiment by the simultaneous hybridization of K-12 DNA to the microarray with the Klebsiella DNA. Of the 4,290 ORFs in E. coli K-12, 192 ORFs (4.5%) were not on the microarray because of the inability to amplify these products. The results presented here represent a compilation of eight separate experiments. A typical microarray from these experiments is shown in Fig. 1.
FIG. 1.
A typical microarray showing the hybridization K. pneumoniae 342 DNA with the ORF chip of E. coli K-12. DNA from E. coli K-12 is also hybridized as an internal control. Where spots appear green, only E. coli DNA has bound to the ORFs on the chip. Where the spots appear green, red, or yellow, the ORFs have been bound with K-12 DNA only, 342 DNA only, or DNA from both strains, respectively. Negative controls include salmon sperm DNA and yeast tRNA genes. Background fluorescence obtained from the negative controls is averaged and subtracted from the fluorescence values of all other spots.
Data acquisition, analysis, and graphical representation.
Scanning the microarray after hybridization was done using a ScanArray 5000 confocal laser scanner (GSI-Lumonics, Inc.) as described by Richmond et al. (13). The signal intensity of the two fluors was determined using ScanAlyze software (http://rana.stanford.edu/software/) and Quantarray. Microarray data were analyzed using Quantarray (GSI Lumonics, Inc.) and an E. coli database designed using Microsoft Access (J. D. Glasner and F. R. Blattner, unpublished results). The assessment of the metabolic functions present or absent in strain 342 was done using this database as well as Eco Cyc (7). Genome level graphics depicted here were prepared using GeneScene from DNAStar.
Criteria chosen for the presence or absence of an E. coli ORF in K. pneumoniae 342.
Using the Quantarray analysis software, a normalized fluorescence ratio of each spot was obtained by dividing the fluorescence value obtained from 342-labeled DNA by the fluorescence value obtained by K-12-labeled DNA. The geometric mean of the eight replicates of each spot was determined for 98.8% of 4,030 spots for which the coefficient of variance was <0.4. For the remaining 48 spots, the hybridization images were visually inspected and a geometric mean of the fluorescence ratio was recalculated for spots that contained replicates that were unreliable due to high background or weak signal intensity.
The geometric mean fluorescence ratio, coefficient of variance, and standard error of the 4,030 spots were 1.0402, 0.2000, and 0.0761, respectively. The 4,030 spots is 260 less than the total number (4,290) of annotated ORFs in the E. coli K-12 genome because it excludes the 192 ORFs not on the chip and the 68 spots for which the signal was too low to make a judgment regarding the presence or absence in K. pneumoniae 342.
A set of 83 genes known to be in common between E. coli and K. pneumoniae was used to place the fluorescence ratios into three categories as follows. Genes with an average fluorescence ratio of >0.8696 corresponded to genes with >75% identity between the two organisms at the nucleotide level. Where the average ratio was between 0.3534 and 0.8696, the genes are expected to have between 55 and 75% identity. At average ratios of <0.3534, little or no identity should be expected.
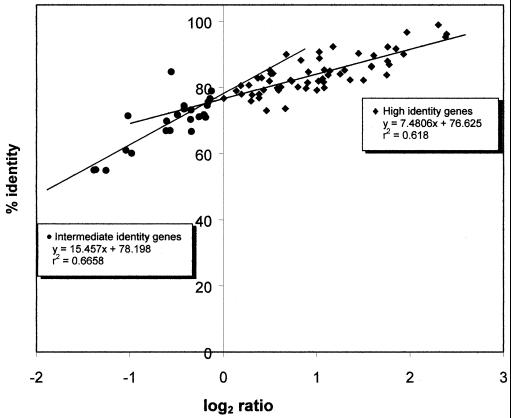
The fluorescence value cutoff values of 0.8696 and 0.3534 were determined as follows. A standard curve was prepared to determine the level of identity between E. coli K-12 and K. pneumoniae 342 genes (Fig. 2). Eighty-three genes of known identity between E. coli and K. pneumoniae were obtained from the GenBank database. Following eight microarray hybridizations between genomic DNA of K. pneumoniae 342 and E. coli K-12, the geometric mean of the fluorescence ratio for each of these genes was determined. The log2 of these geometric means was then plotted against the known percent identity of these genes. A biphasic plot was observed. The junction of the regression lines of these two phases was defined as the cutoff between high and intermediate levels of identity. This cutoff was found to be 75% identity. Genes with an identity of >75% are considered to be of high identity. Intermediate identity was defined as those genes falling between 55 and 75% identity. The lower cutoff value of 55% identity is based on the observation that none of the genes found in the GenBank database in common between these two species have an identity that is <55%. The regression lines of these two phases were used to determine the percent identity level of each of the E. coli K-12 genes that have homologs in K. pneumoniae 342.
FIG. 2.
Standard curve used to determine the level identity between E. coli K-12 and K. pneumoniae 342 genes. Eighty-three genes of known homology between E. coli and K. pneumoniae were obtained from the GenBank database. Following eight microarray hybridizations, the geometric mean of the fluorescence ratio for each of these genes was determined. The log2 of these geometric means was then plotted against the known percent identity of these genes. A biphasic plot was observed, and the regression lines for each phase, including their equations and r2 values, are shown within the plot. The two phases contain negative and positive log2 ratio values, respectively. The junction of the regression lines of these two phases is defined as the cutoff between high and intermediate levels of identity. This cutoff was found to be 75%. Genes with an identity of >75% are considered to be of high identity. Intermediate identity was defined as those genes falling between 55 and 75% identity. This lower cutoff value is based on the observation that none of the genes found in the GenBank database that are in common between these two species have an identity that is <55%. The regression lines of these two phases were used to determine the percent identity level of each of the E. coli K-12 genes that have homologs in K. pneumoniae 342.
Spotfire analysis software was used to categorize sets of genes according to their functions when known (7). For each set of genes within a known function, a weight value was calculated. The weight values were used to determine whether each functional category was highly conserved, conserved, or of low identity. The weight value was calculated as follows: weight value = percent genes with >75% identity + [(percent genes with 55 to 75% identity) × (0.55/0.75)]. Functions were categorized as highly conserved when the weight value was >75% and the percentage of genes with >75% identity was >50%. A function was also categorized as highly conserved between the two organisms when the percentage of genes with >75% identity within that function was ≥60%. Where the weight value is <60 and the percentage of genes with >75% homology within a given function is <40%, that function is defined as having little of no identity between the two organisms. Functions falling between these two categories were defined as being of intermediate identity.
RESULTS
Overview of results.
Of the 4,098 ORFs from E. coli K-12 present on the microarry, 1,894 genes were ≥75% identical with K. pneumoniae DNA. Another 1,106 ORFs were identical at the 55 to 75% level. Thus, a total of 3,000 ORFs of E. coli K-12 hybridized to DNA from K. pneumoniae 342. This represents 70% of the ORFs in K-12. About 24% of the K-12 ORFs, i.e., 1,030, showed little or no identity with DNA from K. pneumoniae 342. For 68 ORFs, the signal was too low from the positive control (K-12 DNA hybridization) to determine the extent of hybridization with 342 DNA. Thus, no estimate of conservation can be made with these ORFs. The complete list of K-12 genes that are present or absent in strain 342 can be found online (http://agronomy.wisc.edu/∼triplett/index.html).
The K-12 genes were subdivided into 23 broad functional categories (Table 1). Within each category, the number of genes that are ≥75% identical, 55 to 75% identical, of low identity, or of uncertain identity, as well as the number of genes that are absent on the microarray, are listed. The categories of these functions were determined by calculating a weight value for all of the genes in each category that takes into account the proportion of genes that have high, intermediate, or low identity between the two strains. Table 1, function group A (Table 1A), lists the functions where the weight value is >71 where >60% of the genes of a given function are >75% identical. When the weight value is between 57 and 71, i.e., between 40 and 60% of the genes of a given function are >75% identical, that function is said to have intermediate identity between the two organisms (Table 1B). Where <40% of the genes involved in a function are >75% identical and the weight value is <57, that function is defined as having little or no identity between the two organisms (Table 1C).
TABLE 1.
Broad functional categories where K. pneumoniae 342 genes are mostly of high identity (group A), mostly of intermediate identity (group B), or have little or no identity (group C) with genes from E. coli K-12a
| Group | Function category | No. of genes at level of DNA identity
|
% Total >75% | Wt (%) | |||||
|---|---|---|---|---|---|---|---|---|---|
| >75% | 55–75% | <55% | Uncertain | Absent | Total | ||||
| A | Functions with high identity | ||||||||
| Amino acid biosynthesis and metabolism | 104 | 17 | 4 | 1 | 5 | 131 | 79 | 89 | |
| Nucleotide biosynthesis and metabolism | 47 | 6 | 1 | 1 | 3 | 58 | 81 | 89 | |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 71 | 26 | 2 | 3 | 2 | 103 | 69 | 87 | |
| Fatty acid and phospholipid metabolism | 30 | 14 | 2 | 0 | 2 | 48 | 63 | 84 | |
| Translation and posttranslational modification | 138 | 15 | 15 | 5 | 9 | 182 | 76 | 82 | |
| Regulatory function | 28 | 11 | 3 | 1 | 2 | 45 | 62 | 80 | |
| Transport and binding proteins | 178 | 54 | 39 | 5 | 5 | 281 | 63 | 77 | |
| DNA replication, recombination, modification, and repair | 71 | 24 | 14 | 3 | 3 | 115 | 62 | 77 | |
| Energy metabolism | 145 | 49 | 41 | 1 | 7 | 243 | 60 | 74 | |
| Transcription, RNA processing, and degradation | 32 | 10 | 7 | 0 | 6 | 55 | 58 | 72 | |
| Subtotals for each category | 844 | 225 | 128 | 20 | 44 | 1,261 | 67 | 80 | |
| B | Functions with intermediate identity | ||||||||
| Other known genes | 9 | 13 | 2 | 1 | 1 | 26 | 35 | 71 | |
| Carbon compound catabolism | 64 | 38 | 23 | 0 | 5 | 130 | 49 | 71 | |
| Central intermediary metabolism | 96 | 48 | 36 | 1 | 8 | 188 | 51 | 70 | |
| Membrane proteins | 4 | 6 | 2 | 0 | 1 | 13 | 31 | 65 | |
| Putative transport proteins | 60 | 38 | 40 | 0 | 8 | 146 | 41 | 60 | |
| Structural proteins | 15 | 14 | 9 | 1 | 3 | 42 | 36 | 60 | |
| Cell processes (includes adaptation and protection) | 76 | 46 | 54 | 3 | 9 | 188 | 40 | 58 | |
| Subtotals for each category | 324 | 203 | 165 | 6 | 35 | 733 | 44 | 65 | |
| C | Functions with little identity | ||||||||
| Hypothetical, unclassified, unknown | 541 | 510 | 477 | 27 | 79 | 1,634 | 33 | 56 | |
| Cell structure | 64 | 42 | 66 | 1 | 9 | 182 | 35 | 52 | |
| Putative enzymes | 77 | 72 | 83 | 3 | 16 | 251 | 31 | 52 | |
| Putative regulatory proteins | 32 | 42 | 47 | 5 | 7 | 133 | 24 | 47 | |
| Phage, transposon, or plasmid | 11 | 11 | 58 | 6 | 1 | 87 | 13 | 22 | |
| Putative chaperones | 1 | 1 | 6 | 0 | 1 | 9 | 11 | 19 | |
| Subtotals for each category | 726 | 678 | 737 | 42 | 113 | 2,296 | 32 | 53 | |
| Totals across all functions | 1,894 | 1,106 | 1,030 | 68 | 192 | 4,290 | |||
The “uncertain” category refers to genes for which the fluorescence signal from the positive control E. coli DNA was too low to obtain a reasonable fluorescence ratio. The “absent” category refers to E. coli genes that were not on the microarray. In the far right column, note that all data are sorted by a weight factor that takes into account the number of genes of intermediate as well as high identity, calculated as follows: weight = (% genes with >75% identity) + [(% genes with 55 to 75% identity) × (0.55/0.75)].
Genes and functions with >75% identity between the two organisms.
Functions that are highly conserved between 342 and K-12 include genes that code for RNA and/or proteins involved in replication, transcription, translation, regulation, transport, protein or nucleic acid binding, amino acid metabolism, energy metabolism, fatty acid and phospholipid metabolism, the synthesis of prosthetic groups, cofactors, and carriers, and nucleotide metabolism (Table 1A). These 10 broad functional categories are further divided into 34 more specific functions (Table 2A). For example, energy metabolism includes more-specific categories such as the tricarboxylic acid cycle, glycolysis, ATP–proton-motive-force interconversion, and aerobic respiration. The transport function in Table 1A is further divided into several specific mechanisms of small molecule transport in Table 2A, including anions, cations, amino acids and amines, nucleosides, purines, and pyrimidines.
TABLE 2.
Specific functional categories where K. pneumoniae 342 genes are mostly of high identity (group A), mostly of intermediate identity (group B), or have little or no identity (group C) with genes from E. coli K-12a
| Group | Specific function category | No. of genes at level of DNA identity
|
% Total >75% | Wt (%) | |||||
|---|---|---|---|---|---|---|---|---|---|
| >75% | 55–75% | <55% | Uncertain | Absent | Total | ||||
| A | Specific functions with high identity | ||||||||
| Pyruvate dehydrogenase | 5 | 0 | 0 | 0 | 0 | 5 | 100 | 100 | |
| Entner-Douderoff | 3 | 0 | 0 | 0 | 0 | 3 | 100 | 100 | |
| Miscellaneous glucose metabolism | 3 | 0 | 0 | 0 | 0 | 3 | 100 | 100 | |
| ATP–proton-motive-force interconversion | 9 | 0 | 0 | 0 | 0 | 9 | 100 | 100 | |
| Plasmid-related functions | 1 | 0 | 0 | 0 | 0 | 1 | 100 | 100 | |
| Pyrimidine ribonucleotide biosynthesis | 9 | 1 | 0 | 0 | 0 | 10 | 90 | 97 | |
| 2′-Deoxyribonucleotide metabolism | 8 | 1 | 0 | 0 | 0 | 9 | 89 | 97 | |
| Glycolysis | 15 | 2 | 0 | 0 | 0 | 17 | 88 | 97 | |
| Ribosomal protein synthesis and modification | 53 | 1 | 0 | 1 | 1 | 56 | 95 | 96 | |
| Colicin-related functions | 4 | 1 | 0 | 0 | 0 | 5 | 80 | 95 | |
| Fatty acid and phosphatidic acid biosynthesis | 18 | 5 | 0 | 0 | 0 | 23 | 78 | 94 | |
| Amino acid biosynthesis | 93 | 11 | 1 | 1 | 3 | 109 | 85 | 93 | |
| Transport of amino acids and amines | 46 | 7 | 3 | 0 | 0 | 56 | 82 | 91 | |
| Degradation of amines | 8 | 0 | 1 | 0 | 0 | 9 | 89 | 89 | |
| Osmotic adaptation | 8 | 6 | 0 | 0 | 0 | 14 | 57 | 89 | |
| Phospholipid synthesis | 9 | 1 | 0 | 0 | 1 | 11 | 82 | 88 | |
| Fermentation | 14 | 5 | 0 | 0 | 1 | 20 | 70 | 88 | |
| Proteins: translation and modification | 29 | 1 | 2 | 1 | 1 | 34 | 85 | 87 | |
| Sulfur metabolism | 8 | 1 | 1 | 0 | 0 | 10 | 80 | 87 | |
| Amino acyl tRNA synthesis; tRNA modification | 31 | 5 | 0 | 1 | 3 | 40 | 78 | 87 | |
| Tricarboxylic acid cycle | 14 | 1 | 0 | 0 | 2 | 17 | 82 | 87 | |
| Phosphorus compounds | 11 | 5 | 0 | 0 | 1 | 17 | 65 | 86 | |
| Detoxification | 8 | 2 | 0 | 1 | 0 | 11 | 73 | 86 | |
| RNA synthesis modification and DNA transcription | 15 | 5 | 0 | 0 | 2 | 22 | 68 | 85 | |
| Salvage of nucleosides and nucleotides | 13 | 3 | 1 | 1 | 0 | 18 | 72 | 84 | |
| Biosynthesis of cofactors, carriers | 74 | 29 | 5 | 3 | 2 | 113 | 65 | 84 | |
| Polyamine biosynthesis | 6 | 1 | 0 | 0 | 1 | 8 | 75 | 84 | |
| DNA replication, repair, restriction/modification | 56 | 18 | 6 | 1 | 2 | 83 | 67 | 83 | |
| Transport of nucleosides, purines, and pyrimidines | 5 | 0 | 1 | 0 | 0 | 6 | 83 | 83 | |
| Aerobic respiration | 25 | 2 | 5 | 0 | 0 | 32 | 78 | 83 | |
| Global regulatory functions | 33 | 12 | 3 | 1 | 2 | 51 | 65 | 82 | |
| Chaperone folding and ushering | 5 | 1 | 0 | 0 | 1 | 7 | 71 | 82 | |
| Transport of anions | 12 | 8 | 2 | 0 | 0 | 22 | 55 | 81 | |
| Purine ribonucleotide biosynthesis | 17 | 1 | 1 | 0 | 3 | 22 | 77 | 81 | |
| Transport of cations | 30 | 15 | 5 | 1 | 0 | 51 | 59 | 80 | |
| Glyoxylate bypass | 4 | 0 | 1 | 0 | 0 | 5 | 80 | 80 | |
| Degradation of proteins, peptides, and glycogen | 21 | 3 | 3 | 0 | 2 | 29 | 72 | 80 | |
| Murein sacculus and peptidoglycan | 22 | 6 | 2 | 0 | 3 | 33 | 67 | 80 | |
| Adaptations, atypical conditions | 9 | 5 | 2 | 0 | 0 | 16 | 56 | 79 | |
| Polysaccharides (cytoplasmic) | 4 | 1 | 1 | 0 | 0 | 6 | 67 | 79 | |
| Transport of protein or peptide secretion | 16 | 4 | 4 | 0 | 0 | 24 | 67 | 79 | |
| Basic proteins: synthesis and modification | 4 | 2 | 0 | 1 | 0 | 7 | 57 | 78 | |
| Nucleotide interconversions | 7 | 3 | 2 | 0 | 0 | 12 | 58 | 77 | |
| Transport of other | 7 | 3 | 0 | 0 | 2 | 12 | 58 | 77 | |
| Nonoxidative branch, pentose pathway | 6 | 0 | 1 | 0 | 1 | 8 | 75 | 75 | |
| Gluconeogenesis | 3 | 0 | 0 | 0 | 1 | 4 | 75 | 75 | |
| Transport of carbohydrates, organic acids, and alcohols | 53 | 13 | 16 | 0 | 2 | 84 | 63 | 75 | |
| Cell division | 20 | 5 | 4 | 1 | 2 | 32 | 63 | 75 | |
| Subtotals for each category | 874 | 196 | 73 | 14 | 39 | 1,196 | 73 | 75 | |
| B | Specific functions with intermediate identity | ||||||||
| Undefined in central intermediary metabolism | 1 | 2 | 0 | 0 | 0 | 3 | 33 | 82 | |
| Lipoprotein | 4 | 5 | 0 | 1 | 0 | 10 | 40 | 77 | |
| Nucleotide hydrolysis | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 73 | |
| Anaerobic respiration | 44 | 19 | 15 | 1 | 1 | 80 | 55 | 72 | |
| Sugar-nucleotide biosynthesis or conversions | 7 | 8 | 2 | 0 | 1 | 18 | 39 | 71 | |
| Degradation of amino acids | 9 | 5 | 3 | 0 | 1 | 18 | 50 | 70 | |
| Degradation of carbon compounds | 53 | 35 | 25 | 0 | 0 | 113 | 47 | 70 | |
| Drug or analog sensitivity | 19 | 11 | 5 | 0 | 5 | 40 | 48 | 68 | |
| Degradation of DNA | 13 | 5 | 6 | 0 | 1 | 25 | 52 | 67 | |
| Degradation of RNA | 5 | 2 | 0 | 1 | 2 | 10 | 50 | 65 | |
| Pool, multipurpose conversions of intermediate metabolism | 30 | 12 | 17 | 0 | 4 | 63 | 48 | 62 | |
| Subtotal for each category | 185 | 106 | 73 | 3 | 15 | 382 | 48 | 69 | |
| C | Specific functions with little identity | ||||||||
| Degradation of polysaccharides | 1 | 1 | 0 | 0 | 1 | 3 | 33 | 58 | |
| Cell killing | 1 | 1 | 0 | 1 | 0 | 3 | 33 | 58 | |
| Amino sugars | 5 | 1 | 3 | 0 | 1 | 10 | 50 | 57 | |
| Outer membrane constituents | 4 | 7 | 5 | 0 | 0 | 16 | 25 | 57 | |
| Unknown function | 445 | 450 | 382 | 23 | 63 | 1,363 | 33 | 57 | |
| Ribosomes: maturation and modification | 2 | 1 | 1 | 0 | 1 | 5 | 40 | 55 | |
| Electron transport | 7 | 9 | 9 | 0 | 0 | 25 | 28 | 54 | |
| Not classified | 345 | 301 | 352 | 21 | 62 | 1,081 | 32 | 52 | |
| Surface polysaccharides and antigens | 8 | 4 | 5 | 0 | 4 | 21 | 38 | 52 | |
| Oxidative branch, pentose pathway | 1 | 0 | 0 | 0 | 1 | 2 | 50 | 50 | |
| Lipopolysaccharide | 3 | 3 | 7 | 0 | 0 | 13 | 23 | 40 | |
| Degradation of fatty acids | 3 | 1 | 5 | 0 | 1 | 10 | 30 | 37 | |
| Inner membrane | 0 | 2 | 1 | 0 | 1 | 4 | 0 | 37 | |
| Phage-related functions and prophages | 5 | 7 | 11 | 6 | 0 | 29 | 17 | 35 | |
| Surface structures | 3 | 10 | 44 | 0 | 0 | 57 | 5 | 18 | |
| Chemotaxis and mobility | 0 | 2 | 10 | 0 | 0 | 12 | 0 | 12 | |
| Transposon-related functions | 2 | 4 | 49 | 0 | 3 | 58 | 3 | 9 | |
| Subtotals for each category | 835 | 804 | 884 | 51 | 138 | 2,712 | 31 | 53 | |
| Total across all functions | 1,894 | 1,106 | 1,030 | 68 | 192 | 4,290 | |||
The “uncertain” category refers to genes for which the fluorescence signal from the positive control E. coli DNA was too low to obtain a reasonable fluorescence ratio. The “absent” category refers to E. coli genes that were not on the microarray. In the far right column, note that all data are sorted by a weight factor that takes into account the number of genes of intermediate as well as high identity, calculated as follows: weight = (% genes with >75% identity) + [(% genes with 55 to 75% identity) × (0.55/0.75)].
Genes and functions of intermediate identity (55 to 75%) between the two organisms.
Functions with an intermediate level of identity included carbon compound metabolism, membrane proteins, structural proteins, putative transport proteins, cell processes such as adaptation and protection, and central intermediary metabolism (Table 1B). The specific functions of intermediate identity include the degradation of amino acids, carbon compounds, DNA, and RNA (Table 2B). Genes involved in various aspects of nucleotide metabolism, including nucleotide hydrolysis, nucleotide interconversions, sugar-nucleotide biosynthesis or interconversions, are also of intermediate conservation (Table 2B). Other processes, such as anaerobic respiration, ribosome maturation and modification, and lipoprotein synthesis, are also of intermediate identity (Table 2B).
Genes and functions of little or no identity between the two organisms.
Broad categories of genes with little or no identity between the two organisms include those that are hypothetical, unclassified, or of unknown function and many other genes whose roles are only putative whether they are enzymes, chaperones, or regulatory proteins (Table 1C). Also, proteins involved in cell structure are not highly conserved as are those genes that were acquired by E. coli through lateral transfer either by phage, by transposons, or by plasmids (Table 1C). More specific functions of low homology include electron transport, cell killing, degradation of polysaccharides, surface structures, outer and inner membrane proteins, and mobility functions (Table 2C).
Physiological confirmation of the microarray results.
The physiology of the two strains is consistent with the microarray results. Phenotypes were examined that are present in E. coli K-12 but missing in K. pneumoniae 342. The genes for such phenotypes can be examined in the microarray analysis. Since this analysis shows only genes that are in common between the two organisms and genes that are found only in K-12, phenotypes that are unique to strain 342 could not be assessed. According to Bergey's manual (9) and as confirmed by a physiological analysis of strains K-12 and 342 (data not shown), two phenotypes are described for E. coli that are not present in K. pneumoniae. First, strains of E. coli are capable of producing indole from tryptophan, whereas strains of K. pneumoniae are not. In agreement with that observation, this microarray analysis shows that strain 342 lacks a homolog of the E. coli tryptophanase gene that is responsible for indole formation. A second phenotype found in E. coli but lacking in klebsiellae is motility. In agreement with the physiology, most of the E. coli genes necessary for flagellum biosynthesis and function are missing in strain 342.
When we consider functions that are held in common by both strains, of the genes necessary for the uptake and metabolism of specific carbon sources, including glycerol, arabinose, ribose, d-xylose, galactose, glucose, fructose, mannose, rhamnose, mannitol, sorbitol, N-acetylglucosamine, maltose, lactose, melibiose, trehalose, l-fucose, and gluconate, 68% are >75% identical and another 24% are 55 to 75% identical between strains 342 and K-12 based on the criteria defined here. Of the remainder, 4% have no match between the two organisms and another 4% were not available on the microarray.
Organization of the E. coli genes in common with strain 342 as well as those that are lacking in strain 342.
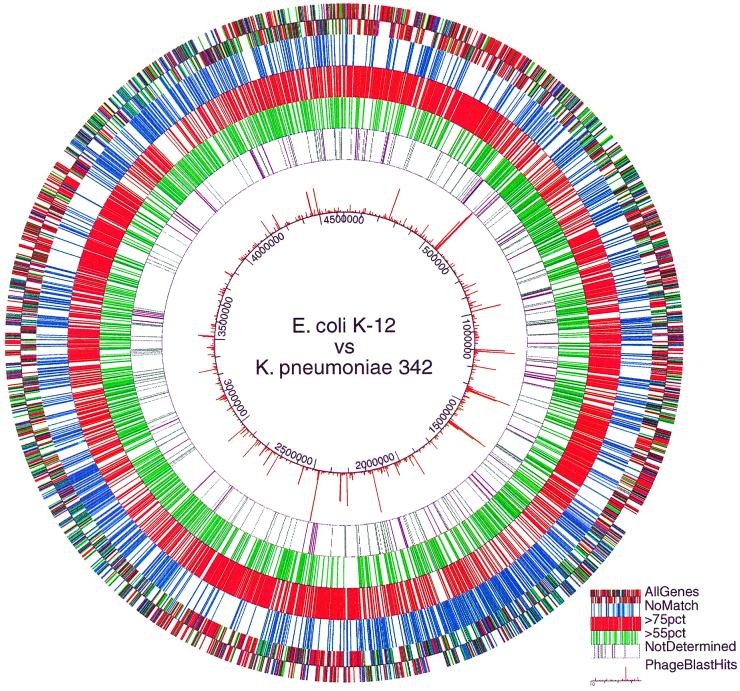
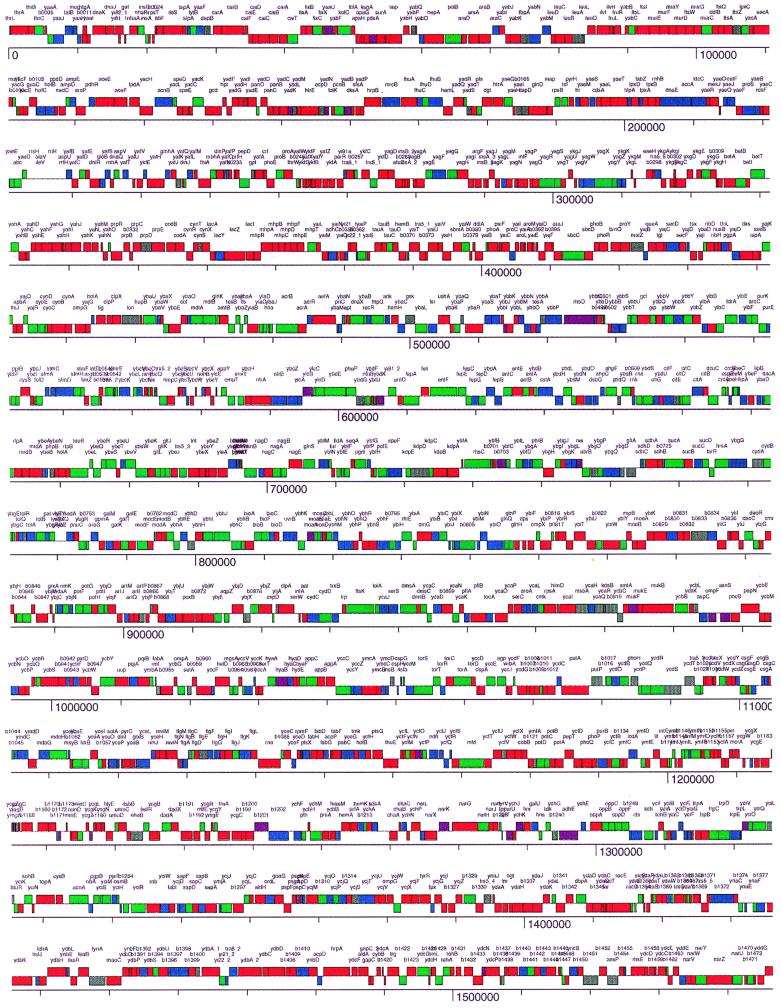
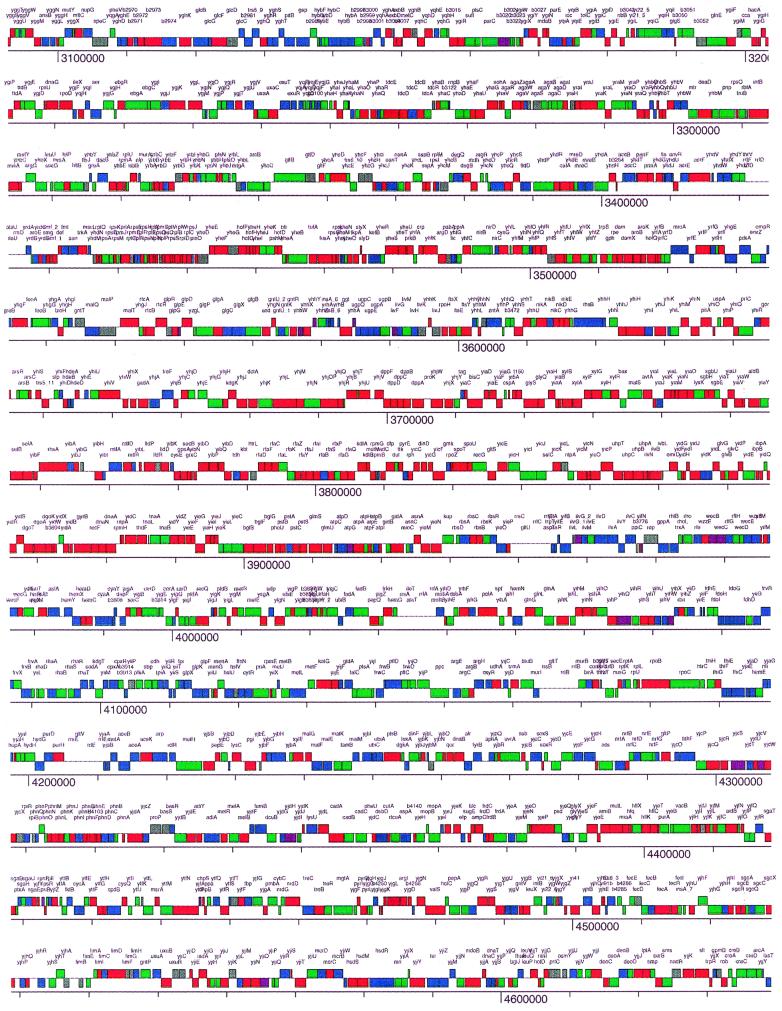
An assessment was made of the organization of the genes in common between these two organisms (Fig. 3 and 4). This assessment allowed us to examine the organization of the genes missing in strain 342 that are present in K-12 (Fig. 3 and 4). The intention here was to discover whether the genes in common between the two organisms were randomly dispersed throughout the K-12 genome or whether they were in large, contiguous regions. The same questions were of interest with regard to those K-12 genes with no homologs in strain 342. In Fig. 3, the genes in common between K-12 and 342 are shown to be dispersed throughout the K-12 genome, but they are not randomly dispersed. The same can be said with regard to the organization of the set of K-12 genes that are missing in the 342 genome. There are large clusters of genes in the K-12 genome that are present in both organisms, and these clusters are dispersed throughout the genome. These clusters vary in size from 100 to 400 kb. Similarly sized clusters of K-12 genes not found in the 342 genome are also observed.
FIG. 3.
Circular genome level representation of the distribution of those genes in common between strains K. pneumoniae 342 and E. coli K-12, as well as those missing in 342 but present in K-12. The outer two concentric rings represent the strand of transcription of the genes. Genes represented in the five outermost rings that are >75% identical, 55 to 75% identical, and of low homology are shown in red, green, and blue, respectively. Genes represented in gray on the sixth concentric ring from the outside are the K-12 genes that are not on the ORF array used in these experiments. Also found in the sixth ring are genes represented in purple for which an identity placement category could not be reliably made.
FIG. 4.
Genome level, high-resolution representation of the homology of each gene in common between K. pneumoniae 342 and E. coli K-12. Genes that are ≥75% identical, 55 to 75% identical, and of little or no identity are shown in red, green, and blue, respectively. In gray are those ORFs that were not present on the microarray. ORFs in purple had a very low fluorescence from the K-12 positive control DNA, preventing the assignment of an identity level. The colored bars are on two levels, which correspond to the strand being transcribed.
DISCUSSION
An efficient means to discover many of the genes and much of the metabolism of an organism without full genome sequencing is presented here. This was made possible by the availability of a microarray that contains nearly all of the ORFs in E. coli K-12. This has allowed the discovery of 3,000 genes that are in common between the two organisms. Success in this work was possible because of the very close phylogenetic relationship between E. coli and K. pneumoniae. For example, the homology of the 16S rRNA genes of these two organisms is 97%. Similar experiments are now in progress to determine whether two less closely related organisms can be compared using this approach. Because this approach depends on high identity at the nucleotide level, there are circumstances where this approach would not be useful. For example, it is not useful for two organisms that possess a large number of proteins in common that are very similar at the amino acid level but are not highly conserved at the nucleotide level.
To determine whether this approach can be useful for a microorganism of interest, microarrays from the closest fully sequenced relative must be available. If ORF microarrays are available for that organism, a preliminary examination of the nucleotide identity between known genes from both organisms should be assessed. Such an assessment should provide useful identity cutoff values to allow the investigator to determine whether the stringency conditions can be adjusted sufficiently to permit the acquisition of useful data for two organisms.
The use of oligonucleotide microarrays, such as those made by Affymetrix, Inc., for this purpose was also considered. Oligonucleotide microarrays require perfect matches for sets of 20-mer oligonucleotides. Just below each set is another set of 20-mer oligonucleotides that each contains a one-base mismatch compared to the oligonucleotide above it. A positive match between E. coli and K. pneumoniae with such a chip would require that many of the oligonucleotides are identical within each gene's set of oligonucleotides. Since the average genes in common between E. coli and K. pneumoniae may be only 85% homologous at the nucleotide level, it is unlikely that several stretches of 20-base perfect matches exist for most genes. Thus, an oligonucleotide microarray may give many false negatives compared to an ORF microarray that does not depend on such perfect matches. Although oligonucleotide microarrays are ideal for expression analysis, they are less than ideal for comparing the genomes of two organisms as described here.
Confirmation of the usefulness of ORF microarrays for whole genome comparisons requires that the results agree with what is known about the physiology of the two organisms. As described above, the microarray results described here agree with the physiology of the two organisms. In addition, less conservation for these functions should be expected as they could tolerate a high rate of mutation. For example, structural proteins in the inner and outer membranes can be expected to remain embedded in membranes provided that transmembrane domains exist. Since the specific sequence of the hydrophobic amino acids is not crucial, there can be significant changes in this nucleotide sequence of the genes coding for these amino acids. This would lead to a lower identity for certain structural functions. These microarray results confirmed these expectations. Genes involved in cellular structure were generally of intermediate homology.
In contrast, some processes essential for metabolism would be expected to be highly conserved. For example, it is not surprising that these results show that genes coding for pyruvate dehydrogenase, glycolysis, ribosomal proteins, the tricarboxylic acid cycle, and amino acid biosynthesis are all highly conserved between these two organisms.
The list of gene categories that are of little or no identity also are as predicted. Genes obtained by K-12 by lateral transfer are usually absent in strain 342. Genes from K-12 of unknown function or with a putative function are also not common in strain 342. For example, known regulatory genes have high identity between the two organisms, while putative regulatory elements are usually absent in strain 342.
The long-term objective of this approach is to learn as much about an organism of interest as quickly and efficiently as possible. With these data in hand, the next step is to clone the DNA that is unique to the strain of interest compared with the fully sequenced relative. An example of such a subtraction protocol is that described by Lisitsyn et al. (10). This protocol has had many uses, such as the isolation and analysis of strain-specific DNA from Neisseria sp. (8, 18). The strain-specific DNA can then be sequenced partially with a onefold random shotgun sequence coverage followed by sequence to closure of those regions of interest. The genome size of strain 342 is 4.8 megabases (Herlache and E. W. Triplett, unpublished results). Assuming that the 3,000 genes of strain 342 discovered here represent about 3.3 Mb of the genome (based on 1.1 kb/gene), about 1.5 Mb of the genome of strain 342 remains to be characterized. Perhaps 200 kb of this genome is of primary interest to us with regard to its ability to interact with maize. Regions of interest can then be fully sequenced and characterized by mutagenesis.
Characterizing a genome by these steps is approximately 10-fold less expensive than fully sequencing a genome. The approach described here is not nearly as informative as the full DNA sequence of an organism. However, where resources are limited, this approach can be a valuable, inexpensive alternative for the discovery of thousands of genes in a prokaryotic organism.
ACKNOWLEDGMENTS
This work was supported by the Consortium for Plant Biotechnology Research and the College of Agricultural and Life Sciences, University of Wisconsin—Madison.
We thank Craig Richmond, Hongfan Jin, and Sandra Splinter for their help and discussions during the microarray experiments.
REFERENCES
- 1.Andrade M A, Ouzounis C, Sander C, Tamames J, Valencia A. Functional classes in the three domains of life. J Mol Evol. 1999;49:551–557. doi: 10.1007/pl00006576. [DOI] [PubMed] [Google Scholar]
- 2.Bansa A K. An automated comparative analysis of 17 complete microbial genomes. Bioinformatics. 1999;15:900–908. doi: 10.1093/bioinformatics/15.11.900. [DOI] [PubMed] [Google Scholar]
- 3.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3a.Chelius, M. K., and E. W. Triplett. The diversity of Archaea and Bacteria in the roots of Zea mays L. Microb. Ecol., in press. [DOI] [PubMed]
- 4.Chelius M K, Triplett E W. Diazotrophic endophytes associated with maize. In: Triplett E W, editor. Prokaryotic nitrogen fixation: a model system for the analysis of a biological process. Norfolk, United Kingdom: Horizon Scientific Press; 2000. pp. 779–792. [Google Scholar]
- 5.Chelius M K, Triplett E W. Immunolocalization of dinitrogenase reductase produced by Klebsiella pneumoniae in association with Zea mays L. Appl Environ Microbiol. 2000;66:783–787. doi: 10.1128/aem.66.2.783-787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher P J, Petrini O, Scott H M L. The distribution of some fungal and bacterial endophytes in maize (Zea mays L.) New Phytol. 1992;122:299–305. doi: 10.1111/j.1469-8137.1992.tb04234.x. [DOI] [PubMed] [Google Scholar]
- 7.Karp P D, Riley M, Paley S M, Pellegrini-Toole A, Krummenacker M. Eco Cyc: encyclopedia of Escherichia coli genes and metabolism. Nucleic Acids Res. 1999;27:55–58. doi: 10.1093/nar/27.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klee S R, Nassif X, Kusecek B, Merker P, Beretti J-L, Achtman M, Tinsley C R. Molecular and biological analysis of eight genetic islands that distinguish Neisseria meningitides from the closely related pathogen Neisseria gonorrhoeae. Infect Immun. 2000;68:2082–2095. doi: 10.1128/iai.68.4.2082-2095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. [Google Scholar]
- 10.Lisitsyn N, Lisitsyn N, Wigler M. Cloning the differences between two complex genomes. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 11.McInroy J A, Kloepper J W. Survey of indigenous bacterial endophytes from cotton and sweet corn. Plant Soil. 1995;173:337–342. [Google Scholar]
- 12.Palus J A, Borneman J, Ludden P W, Triplett E W. Isolation and characterization of endophytic diazotrophs from Zea mays L. and Zea luxurians Iltis and Doebley. Plant Soil. 1996;186:135–142. [Google Scholar]
- 13.Richmond C S, Glasner J D, Mau R, Jin H F, Blattner F R. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T, editors. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 15.Suckow J M, Suzuki M. Genomic DNA shuffling in archaebacteria—comparison of the genomes of two Pyrococcus species. Proc Jpn Acad Ser B Phys Biol Sci. 1999;75:10–15. [Google Scholar]
- 16.Tamames J, Casari G, Ouzounis C, Valencia A. Conserved clusters of functionally related genes in two bacterial genomes. J Mol Evol. 1997;44:66–73. doi: 10.1007/pl00006122. [DOI] [PubMed] [Google Scholar]
- 17.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tinsley C R, Nassif X. Analysis of the genetic differences between Neisseria meningitides and Neisseria gonorrhoeae: two closely related bacteria expressing two different pathogenicities. Proc Natl Acad Sci USA. 1996;93:11109–11114. doi: 10.1073/pnas.93.20.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]