Abstract
SARS-CoV-2 infection has been reported to be associated with a positive direct antiglobulin test (DAT). In this study, an analysis of 40 consecutive coronavirus disease 2019 (COVID-19) cases from December 2020 to September 2021 in Japan revealed that patients of 70 years and over were predisposed to a positive DAT. DAT positivity was related to a decrease in the hemoglobin level. Anemia in DAT-positive COVID-19 patients was attributed to hemolysis, which was corroborated by high reticulocyte counts and an increase in the red blood cell distribution width. Human leukocyte antigen (HLA)-DRB1*12:01 and DRB1*12:02 were exclusively found in DAT-positive COVID-19 patients. In silico assays for the Spike protein of SARS-CoV-2 predicted several common core peptides that met the criteria for a B cell epitope and strong binding to both HLA-DRB1*12:01 and DRB1*12:02. Among these peptides, the amino acids sequence TSNFR, which is found within the S1 subunit of SARS-CoV-2 Spike protein, is shared by human blood group antigen Rhesus (Rh) CE polypeptides. In vitro analysis showed that the expression of HLA-DR in CD4+ T cells and CD8+ T cells from a DAT-positive patient was increased after pulsation with TSNFR-sequence-containing peptides. In summary, positive DAT is related to enhanced anemia and to HLA-DR12 in the Japanese population. A peptide sequence within SARS-CoV-2 Spike protein may act as an epitope for IgG binding to RBCs in DAT-positive COVID-19 patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00277-022-04921-9.
Keywords: Red blood cell, COVID-19, Direct antiglobulin test, Human leukocyte antigen, Epitope
Introduction
Red blood cell (RBC) sensitization by immunoglobulins (Ig) and complement during infections with various pathogens is well known. A correlation between coronavirus disease 2019 (COVID-19) and positive direct antiglobulin test (DAT) has been reported [1–4]. The frequency of positive DAT was reported to be 80% in 20 UK COVID-19 patients and 40% in 113 Italian COVID-19 patients [5, 6]. These cases were mostly IgG-specific DAT, although various phenotypes of autoimmune hemolytic anemia (AIHA), such as warm type, cold type, or mixed type, have been reported [2, 3, 7].
Regardless of DAT results, a lower hemoglobin level is observed among COVID-19 patients and these low levels are associated with a poor prognosis [8, 9]. Clinical manifestations of AIHA vary [2, 5, 6]; i.e., laboratory data may indicate the presence [10] or absence [6] of hemolysis, and onset of symptoms can occur in infants [11] or adults [2, 10]. Recently, a large study revealed that the severity of anemia was significantly higher in DAT-positive patients than in DAT-negative patients, and that the severity correlated with a high requirement of RBC transfusion and high red blood cell distribution width (RDW) values [6].
The immunological mechanisms underlying positive DAT in COVID-19 patients are not understood. Possible mechanisms include enhanced binding of IgG and/or modification of the RBC membrane by cytokine storm-induced hyper-inflammation [6, 12]. Another possibility is molecular mimicry between SARS-CoV-2 protein and proteins in human cells, including RBCs [13]. In this study, we analyzed the significance of positive DAT for anemia and inflammation in Japanese COVID-19 patients. We also performed In silico and in vitro analyses to predict the epitopes recognized by an RBC-bound antibody.
Materials and methods
Subjects
Forty COVID-19 patients, who provided written informed consent, were enrolled. All patients had positive SARS-CoV-2 RT-PCR test results of nasopharyngeal swabs. They had been hospitalized and discharged alive after treatment for COVID-19 infection. Peripheral blood samples were collected from the patients both on admission and on discharge between December 2020 and March 2021 (period A), and between April 2021 and September 2021 (period B). The analysis was performed on laboratory data from the first 14 days after the onset of COVID-19-related symptoms. The study was approved by the ethical committee of Fujita Health University (HM20-177).
RBC-bound antibody test
RBC-bound antibodies were determined by DAT using bead column agglutination technology (CAT) (Ortho BioVue System DAT/IDAT cassettes, Ortho Clinical Diagnostics, Tokyo, Japan). A 5% RBC suspension (10 µL) was added to the reaction chamber of each cassette tube, and the cassettes were centrifuged at 55 × g for 2 min and 199 × g for 3 min. DAT reactivity was evaluated in accordance with the manufacturer’s instructions. COVID-19 patients were considered DAT-positive when the reactivity of samples on admission and/or on discharge was positive. When peripheral blood samples were available, flow cytometric analysis was performed. RBC suspensions were stained with fluorescent dye-conjugated antibodies for 20 min at room temperature. The antibodies used were as follows: fluorescein isothiocyanate (FITC)-conjugated F(ab′)2 goat anti-human IgG Fc (Invitrogen/Thermo Fisher Scientific, Waltham, MA, polyclonal), phycoerythrin (PE)-conjugated mouse anti-C3b/iC3b (BioLegend, San Diego, CA, clone 3E7/C3b), allophycocyanin (APC)-conjugated mouse anti-human CD235a (Invitrogen/Thermo Fisher Scientific, clone HIR2). For peptide pulse experiments, peripheral blood mononuclear cells (PBMCs) obtained from a DAT-positive patient were pulsed with NH2-TSNFRVQPT-COOH (Eurofins Genomics, Bayern, Germany) at final concentrations of 100 nM and 1 μM for 3 days. Then, the cells were stained for 20 min at 4°C with APC-conjugated mouse anti-human CD3 (BD Pharmingen, Franklin Lakes, NJ, clone UCHT1), Pacific orange (PO)-conjugated mouse anti-human CD4 (Invitrogen/Thermo Fisher Scientific, clone RPA-T4), Pacific Blue (PB)-conjugated mouse anti-human CD8 (BD Horizon, Franklin Lakes, NJ, clone PRA-T8), and PE-conjugated mouse anti-human HLA-DR (Invitrogen/Thermo Fisher Scientific, clone L243). After washing, the data were acquired using a Gallios or CytoFLEX (Beckman Coulter Inc., Brea, CA) and analyzed using FlowJo software (Tree Star, Ashland, OR).
Scanning electron microscope imaging
For scanning electron microscope (SEM) imaging, RBCs were first fixed overnight at 4°C in 1/2 Karnovsky fixative and washed with 0.1 M phosphate buffer and then fixed a second time with 1% osmium tetroxide for 1 h. The dehydration step was performed with an ascending series of ethanol concentrations. After drying, the fixed samples were immersed in t-butyl alcohol, lyophilized, and then coated with platinum. The samples were observed through a JEOL JSM-7610Fplus scanning electron microscope (JEOL Ltd., Tokyo, Japan) (voltage, 5.0 kV; magnification, 3000 ×).
Human leukocyte antigen analysis
Genomic DNA was prepared from peripheral blood using a QIAGEN DNA extraction kit, in accordance with the manufacturer’s instructions (QIAGEN K.K., Tokyo, Japan). The HLA-DRB1 genotyping of COVID-19 patients was performed by a method combining polymerase chain reaction amplification and sequence-specific oligonucleotide probes and Luminex flow cytometry [14, 15].
In silico epitope analyses
In silico assays for peptide binding to HLA-DRB1 were performed using NetMHCIIpan version 4.0 [16]. Strong binding and weak binding levels were defined as %Rank values below 2% and 10%, respectively. A B cell epitope analysis was performed using a BepiPred Linear Epitope Prediction 2.0 [17]. Peptide searches was executed using UniProtKB analysis [18].
Statistical analysis
The Student’s t-test or Chi-squared test was used for the analysis. Statistical significances were expressed as *P < 0.05 and **P < 0.01.
Results
DAT positivity in COVID-19 patients
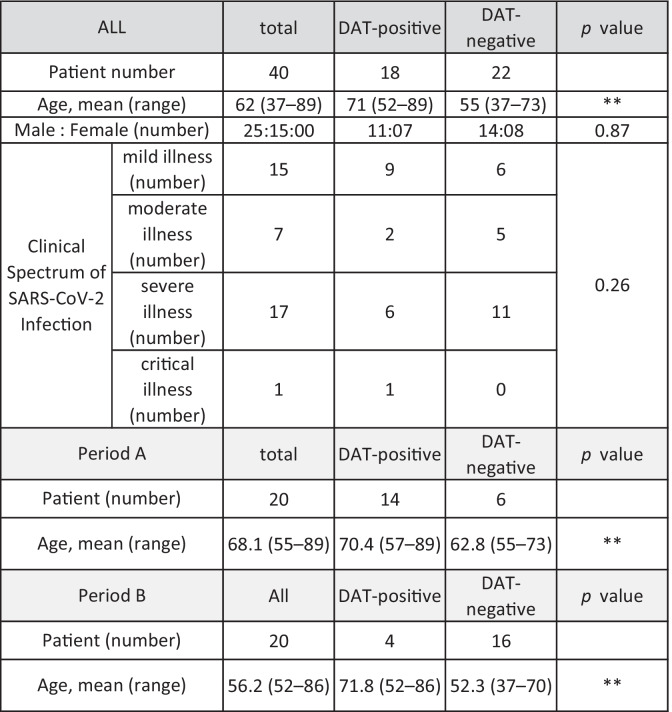
A positive DAT result was found in 18 out of 40 (45%) COVID-19 patients using CAT (Table 1). Among these patients, 11 patients (61%) were positive only for IgG, seven patients (39%) were positive for both IgG and C3d/C3b, and none were positive only for C3d/C3b (Table S1). The sex and clinical spectrum of SARS-CoV-2 infection were not different between DAT-positive patients and DAT-negative patients, whereas the average age of DAT-positive patients was significantly higher than that of DAT-negative patients (Table 1). This was true in both periods; the average age of DAT-positive patients was high, at around 70 years old (70.4 years old in period A, 71.8 years old in period B), compared with that of DAT-negative patients (62.8 years old in period A, 52.3 years old in period B). The frequency of DAT positivity was 70% in period A and 20% in period B. The DAT-positive cases determined by CAT were confirmed to be positive for IgG and complement (Fig. 1). Twenty-one patients were given steroid therapy based on the severity of COVID-19 (Table S1). Among these patients, six were DAT-positive patients and of these the status of DAT results was unchanged in four cases after steroid therapy.
Table 1.
DAT results in COVID-19 patients. The clinical spectrum of SARS-CoV-2 Infection was evaluated by the guideline defined by NIH (https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/). DAT, direct antiglobulin test. **p < 0.01
Fig. 1.
IgG antibody binds to RBCs in COVID-19 patients. a–c The pattern of DAT results in COVID-19 patients using CAT. Representative images of IgG-positive/complement C3-positive (a), IgG-positive/C3 negative (b), and IgG-negative/C3-negative (c) cases are shown. d–i The pattern of IgG and complement binding to CD235a+ RBCs in COVID-19 patients as revealed by flow cytometric analysis. Representative dot plots of IgG-positive/C3b-positive (Q2) (d, g), IgG-positive/C3b-negative (Q2) (e, h), and IgG-negative/C3b-negative (Q2) (f, i) are shown
Hemoglobin levels are decreased in DAT-positive COVID-19 patients
We next analyzed the relationship between DAT results and hemoglobin levels in the COVID-19 patients. The DAT-positive patients showed a significantly lower hemoglobin level (11.2 ± 2.0 g/dL) than that of DAT-negative patients (13.5 ± 1.4 g/dL) (Fig. 2a). The RBC distribution width-standard deviation (RDW-SD) was significantly higher in the DAT-positive patients than in DAT-negative patients (Fig. 2b). A lower hemoglobin level and a higher RDW-SD value in DAT-positive patients were observed in both period A (Fig. 2c, d) and period B (Fig. 2e, f). The mean corpuscular volume indicated a normal average RBC size in both groups, with average values of 88.8 fL in DAT-positive patients and 88.5 fL in DAT-negative patients (Fig. 2g). The reticulocyte level was significantly higher in DAT-positive patients than in DAT-negative patients (Fig. 2h). Collectively, these results suggest the occurrence of hemolysis-related anemia in DAT-positive COVID-19 patients.
Fig. 2.
RBC parameters in COVID-19 patients. a–f Hemoglobin (Hb) and RDW-SD in COVID-19 patients with ( +) or without (–) positive DAT in total period (a, b), in period A (c, d), and in period B (e, f). g, h. The values of mean corpuscular volume (MCV) (g) and reticulocytes (h) in COVID-19 patients with ( +) or without (–) positive DAT. Data are shown as box plots. *p < 0.1; **p < 0.01
IgG-bound RBCs are impaired in COVID-19 patients
The average level of indirect bilirubin in the DAT-positive COVID-19 patients was 0.45 mg/dL, 0.48 mg/dL, and 0.42 mg/dL in the total period, period A, and period B, respectively. In the DAT-negative COVID-19 patients, it was slightly lower, at 0.40 mg/dL in all periods; however, this difference was not significant (Fig. 3a–c). The serum ferritin level was not higher in DAT-positive COVID-19 patients (Fig. 3d). Enhanced inflammation in the DAT-positive COVID-19 patients was suggested by high serum interleukin (IL)-6 levels (Fig. 3e), which was corroborated by a significantly higher level of serum C-reactive protein (CRP) (Fig. 3f). The D-dimer value was significantly higher in the DAT-positive COVID-19 patients (Fig. 3g–i). SEM of RBCs from the peripheral blood samples revealed more irregular membranes and spikes in RBCs from DAT-positive COVID-19 patients than in those from DAT-negative COVID-19 patients (Fig. 3j–l). These results suggested that DAT positivity may be associated with higher levels of hemolysis in the setting of increased inflammation and immune activation compared to DAT-negative patients, but this is not definitive with conflicting laboratory values.
Fig. 3.
Hemolysis-related parameters in COVID-19 patients. a–c The values of serum indirect bilirubin (I-bil) in COVID-19 patients with ( +) or without (–) positive DAT in total period (a), in period A (b), and in period B (c). d–f The values of ferritin (d), IL-6 (e), and CRP (f) in COVID-19 patients with ( +) or without (–) positive DAT. g–i D-dimer values of COVID-19 patients with ( +) or without (–) positive DAT in total period (g), in period A (h), and in period B (i). Data are shown as box plots. *p < 0.05; **p < 0.01. Morphology of peripheral blood RBCs from patients with DAT-positive (j) and DAT-negative (k) COVID-19 patients and from a healthy control (l). Images were obtained by SEM. Accelerating voltage, 5.0 kV; original magnification, 3000 × ; working distance, 8.1 mm; white bars, 1 μm
HLA-DR12 is associated with positive DAT in COVID-19 patients
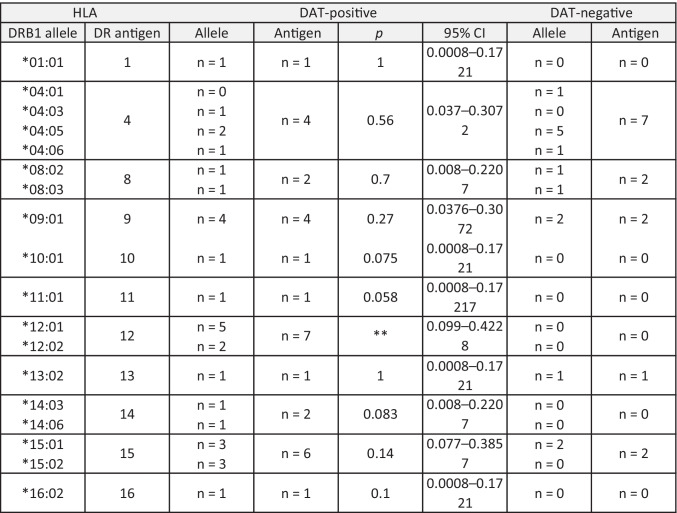
We analyzed HLA-DRB1 alleles in the COVID-19 patients. Among the 18 alleles identified in this study, HLA-DRB1*12:01 and HLA-DRB1*12:02 were found exclusively in the DAT-positive COVID-19 patients (Table 2). Taking the HLA-DR allele frequency in the Japanese population into consideration, HLA-DR12 was the only antigen that was significantly associated with the DAT-positive results in COVID-19 patients (Table 2, S2).
Table 2.
HLA-DRB1 typing of COVID-19 patients. The number of cases that had the corresponding HLA-DRB1 allele and antigen is shown. DAT direct antiglobulin test, CI confidence interval. n = 22. **p < 0.01
We conducted an in silico peptide binding assay to explore whether Spike protein of SARS-CoV-2 interacted with HLA-DRB1*12:01/DRB1*12:02 (Supplemental data 1). From 1259 peptide sequences, 26 strong binding peptides and 128 weak binding peptides were identified for HLA-DRB1*12:01, and 18 strong binding peptides and 115 weak binding peptides were identified for HLA-DRB1*12:02 (Fig. 4). The strong binding peptides shared 11 and 10 types of core peptides (nine amino acids) for HLA-DRB1*12:01 and HLA-DRB1*12:02, respectively, and seven core peptides were common for both HLA-DRB1*12:01 and HLA-DRB1*12:02 (Figs. 4, 5b). We also performed B cell epitope prediction analysis for SARS-CoV-2 Spike protein and found 34 peptides with lengths ranging from 1 to 62 amino acids (Fig. 4, Supplemental data 2). Two out of seven common core peptides that showed strong binding to both HLA-DRB1*12:01 and HLA-DRB1*12:02 in NetMHCIIpan analysis did not overlap with the 34 peptides predicted by B cell epitope prediction analysis (Figs. 4, 5c). Among the remaining five common core peptides, two were in the S1 subunit and three were in the S2 subunit of SARS-CoV-2 Spike protein (Figs. 4, 5c, d). UniProt peptide search analysis of the five common core peptides identified the TSNFR amino acid sequence, which is found within the IYQTSNFRV sequence, a common core peptide with strong binding to both HLA-DRB1*12:01 and HLA-DRB1:12:02, within the Rhesus (Rh) CE polypeptide of the Rh blood group system (Figs. 4, 5c, d, and Supplemental data 2). We further performed an in vitro assay and found that the expression of HLA-DR in CD4+ T cells and CD8+ T cells from a DAT-positive patients increased after pulsation with TSNFR-sequence containing peptides (Fig. 5e, f). These results suggested that the peptide sequence that was predicted to have strong binding to HLA-DR12 and to meet the criteria for a B cell epitope and that was shared by the RhCE polypeptide and SARS-CoV-2 Spike protein, is a possible epitope for the IgG that binds to RBCs in DAT-positive COVID-19 patients.
Fig. 4.
Algorithm for in silico analyses. The amino acid sequence of SARS-CoV-2 Spike protein was searched for using NetMHCIIpan version 4.0 and BepiPred Linear Epitope Prediction 2.0. The predicted peptides were further filtered through a UniProt peptide search to identify a target peptide sequence
Fig. 5.
In silico and in vitro peptide analyses. a, b Venn diagrams of the number of a core peptide sequences that showed weak binding (a) and strong binding (b) to HLA-DRB1*12:01 and HLA-DRB1*12:02. The data were obtained from NetMHCIIpan version 4.0. c In silico analysis for seven common core peptides obtained from (b). The predicted B cell epitope amino acid positions were obtained by a BepiPred Linear Epitope Prediction 2.0. NA, not applicable; ND, not detected. d Schematic diagram of domain structures of SARS-CoV-2 Spike protein. NTD, N-terminal domain; RBD, receptor-binding domain; RBM, receptor-binding motif, SD, subdomain; S2′, S2′ protease cleavage site; FP, fusion peptide; HR, heptad repeat; CH, central helix; CD, connector domain; TM, transmembrane domain; CT, cytoplasmic tail. Numbers indicate the positions of the amino acid sequence. Triangles indicate the locations of common core peptides, the colors of which are black (strong binding to HLA-DR12, predicted B cell epitope, shared sequence with RBC membrane protein), gray (strong binding to HLA-DR12 and predicted B cell epitope), or white (strong binding to HLA-DR12 but not a predicted B cell epitope). e, f Flow cytometric analysis showing the expression of HLA-DR on CD4+ T cells (e) and CD8+ T cells (f) that were pulsed with or without a TSNFR-containing peptide at the indicated concentrations
Discussion
At the beginning of the COVID-19 outbreak, we observed an increasing frequency of DAT positivity in routine cross-matching tests. In a consecutive series of analyses of COVID-19 patients, the frequency of DAT positivity was 70% in period A (December 2020 to March 2021, corresponding to the period of B.1.1.214 strain predominance in Japan), and 45% in period B (April 2021 to September 2021, corresponding to the period of B.1.1.7 strain predominance in Japan). The average age of DAT-positive COVID-19 patients was similar, at around 70 years old in both periods. By contrast, the average age of total COVID-19 patients was lower in period B than in period A. Vaccination against SARS-CoV-2 had not yet been provided to the community in period A. It began at the beginning of period B, from April 2021, during which priority was given to people aged 70 years and over (Fig. S1). This vaccination program was probably the cause of the reduction in the average age of total COVID-19 patients and the reduction of DAT positivity frequency in period B. The fact that old age was a significant factor related to DAT positivity is likely because age-induced alterations of innate and adaptive immunity result in immunosenescense, which induces a pro-inflammatory traits and increases susceptibility to autoimmune diseases [19, 20].
In this study, multiple RBC-related parameters including a higher value of RDW-SD, a marker of RBC destruction and/or shortened RBC lifespan, higher reticulocytes, and the obvious alteration of RBC morphology, demonstrated the presence of enhanced hemolysis in DAT-positive COVID-19 patients [21, 22]. AIHA is manifested by the destruction or increased clearance of RBCs due to RBC-bound antibodies [23]. IgG-bound RBCs are typically phagocytosed or destroyed in the reticuloendothelial system [24]. Our data did not support enhanced extravascular hemolysis in DAT-positive COVID-19 patients because significantly increased serum ferritin levels were not observed. Because there were increases in the serum levels of CRP, IL-6 and d-dimers, it is conceivable that inflammation caused endothelial damage in the DAT-positive COVID-19 patients. The immune reaction mediated by IgG-binding to RBCs and/or the endothelial damage presumably contributed to the hemolysis and anemia in the DAT-positive COVID-19 patients.
The immunological mechanisms underlying positive DAT in COVID-19 patients are not well understood [6, 9, 13]. Since the HLA system orchestrates immune regulation, it has been speculated that certain HLA alleles are associated with susceptibility to positive DAT in COVID-19. Our data suggests for the first time an association between HLA-DR12 and DAT positivity in COVID-19 patients in the Japanese population. Differences in HLA allele distribution in COVID-19 patients were reported in China, where the frequency of HLA-DRB1*04:06 was higher, and that of HLA-DRB1*12:02 was lower, in COVID-19 patients than in a control population [25]. By contrast, a genome-wide association study in Italy and Spain showed no significant associations between HLA allele distribution and susceptibility to COVID-19 [26]. Overall, the data showing altered HLA allele distributions in COVID-19 patients are still inconsistent. In the previous SARS-CoV outbreak, HLA-DR12 was more frequent in patients in the Vietnamese population [27]. The SARS-CoV-2 sequence displays considerable homology with that of SARS-CoV, but the two viruses have distinct amino acid substitution patterns [28]. Thus, the incorporation of HLA allele data from SARS-infected patients when analyzing COVID-19 outcomes needs to be done with care. With regard to the association between disease severity and certain HLA-DR alleles in COVID-19, a correlation for HLA-DRB1*15:01 was reported in Italian patients [25].
Most recently, rapid extracellular antigen profiling has demonstrated that COVID-19 patients produce diverse functional autoantibodies and exhibit a high prevalence of antibodies against immunomodulatory proteins, reactivities of which are associated with disease severity [29]. Furthermore, the insightful theory that anti-idiotype antibodies play roles in “long-COVID” syndrome may explain complex effects on multiple organ systems and the production of diverse antibodies after SARS-CoV-2 infection [30]. Further research is needed to understand the pathology of diverse immunological and hematological manifestations in COVID-19 patients.
The aim of this study was to investigate the production of RBC-bound IgG in COVID-19 patients. AIHA occurs in the early phase of SARS-CoV-2 infection and seems to correlate with cytokine storms [4]. Immune reactions, including the T cell response and antibody production against SARS-CoV-2 infection, are different between convalescent patients who achieve virus clearance and recovery, and those who die [31]. Therefore, this study was performed using blood samples and laboratory data from the first 14 days after the onset of COVID-19-related symptoms and was performed only for patients who were discharged alive.
In summary, positive DAT is related to enhanced anemia and to HLA-DR12. A peptide sequence within SARS-CoV-2 Spike protein, which met the criteria for both strong binding to HLA-DR12 and being a B cell epitope, may be an epitope for IgG binding to RBCs in DAT-positive COVID-19 patients.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 Table S1 (PPTX 57 KB)
Supplementary file2 Table S2 (PPTX 46 KB)
Supplementary file3 Figure S1 (PPTX 65 KB)
Supplementary file4 Supplemental data 1 (XLSX 240 KB)
Supplementary file5 Supplemental data 2 (XLSX 11 KB)
Supplementary file6 Supplemental data 3 (XLSX 55 KB)
Acknowledgements
The authors thank Ms. Michiko Osawa, Ms. Yuki Mizutani, Dr. Yoko Toyama, Prof. Kazuyoshi Imaizumi, and Prof. Kuniaki Saito for their contributions to this work.
Author contribution
HM, SF, YS, HA, and YMi: conception and study design, collection of study materials, data collection, data analysis and interpretation, financial support, and manuscript writing. YMa: conception and study design, data analysis and interpretation, and manuscript writing. All authors approved the final version of the manuscript and agreed to all aspects of the work.
Funding
This work was supported in part by Grants-in-Aid from JSPS KAKENHI (Grant Number 21K15655 for HM, 19K17856 for SF and 18K08323, 22K07485 for YM).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lazarian G, Quinquenel A, Bellal M, Siavellis J, Jacquy C, Re D, Merabet F, Mekinian A, Braun T, Damaj G, Delmer A, Cymbalista F. Autoimmune haemolytic anaemia associated with COVID-19 infection. Br J Haematol. 2020;190:29–31. doi: 10.1111/bjh.16794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lopez C, Kim J, Pandey A, Huang T, DeLoughery TG. Simultaneous onset of COVID-19 and autoimmune haemolytic anaemia. Br J Haematol. 2020;190:31–32. doi: 10.1111/bjh.16786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huscenot T, Galland J, Ouvrat M, Rossignol M, Mouly S, Sène D, APHP Lariboisière COVID Group SARS-CoV-2-associated cold agglutinin disease: a report of two cases. Ann Hematol. 2020;99:1943–1944. doi: 10.1007/s00277-020-04129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vega Hernández P, Borges Rivas Y, Ortega Sánchez E, Marqués Cabrero A, Remedios Mateo L, Silvera Roig P, Infante Quintanar A, Díaz-Delgado Peñas R, Sánchez Escudero V, García-García ML. Autoimmune hemolytic anemia in a pediatric patient with severe acute respiratory syndrome coronavirus 2 infection. Pediatr Infect Dis J. 2020;39:e288. doi: 10.1097/INF.0000000000002809. [DOI] [PubMed] [Google Scholar]
- 5.Platton S, Mendes N, Booth C, Lancut J, Lee K, Regan F, Green L. Positive direct antiglobulin tests in patients with COVID-19. Transfusion. 2021;61:333–334. doi: 10.1111/trf.16156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berzuini A, Bianco C, Paccapelo C, Bertolini F, Gregato G, Cattaneo A, Erba E, Bandera A, Gori A, Lamorte G, Manunta M, Porretti L, Revelli N, Truglio F, Grasselli G, Zanella A, Villa S, Valenti L, Prati D. Red cell-bound antibodies and transfusion requirements in hospitalized patients with COVID-19. Blood. 2020;136:766–768. doi: 10.1182/blood.2020006695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zagorski E, Pawar T, Rahimian S, Forman D. Cold agglutinin autoimmune haemolytic anaemia associated with novel coronavirus (COVID-19) Br J Haematol. 2020;190:e183–e184. doi: 10.1111/bjh.16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan BE, Chong VCL, Chan SSW, Lim GH, Lim KGE, Tan GB, Mucheli SS, Kuperan P, Ong KH. Hematologic parameters in patients with COVID-19 infection. Am J Hematol. 2020;95:E131–E134. doi: 10.1002/ajh.25774. [DOI] [PubMed] [Google Scholar]
- 9.Taneri PE, Gómez-Ochoa SA, Llanaj E, Raguindin PF, Rojas LZ, Roa-Díaz ZM, Salvador D, Jr, Groothof D, Minder B, Kopp-Heim D, Hautz WE, Eisenga MF, Franco OH, Glisic M, Muka T. Anemia and iron metabolism in COVID-19: a systematic review and meta-analysis. Eur J Epidemiol. 2020;35:763–773. doi: 10.1007/s10654-020-00678-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capes A, Bailly S, Hantson P, Gerard L, Laterre PF. COVID-19 infection associated with autoimmune hemolytic anemia. Ann Hematol. 2020;99:1679–1680. doi: 10.1007/s00277-020-04137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenzweig JD, McThenia SS, Kaicker S. SARS-CoV-2 infection in two pediatric patients with immune cytopenias: a single institution experience during the pandemic. Pediatr Blood Cancer. 2020;67:e28503. doi: 10.1002/pbc.28503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berentsen S. New insights in the pathogenesis and therapy of cold agglutinin-mediated autoimmune hemolytic anemia. Front Immunol. 2020;11:590. doi: 10.3389/fimmu.2020.00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lucchese G, Flöel A. Molecular mimicry between SARS-CoV-2 and respiratory pacemaker neurons. Autoimmun Rev. 2020;19:102556. doi: 10.1016/j.autrev.2020.102556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh Y, Mizuki N, Shimada T, Azuma F, Itakura M, Kashiwase K, Kikkawa E, Kulski JK, Satake M, Inoko H. High-throughput DNA typing of HLA-A, -B, -C, and -DRB1 loci by a PCR-SSOP-Luminex method in the Japanese population. Immunogenetics. 2005;57:717–729. doi: 10.1007/s00251-005-0048-3. [DOI] [PubMed] [Google Scholar]
- 15.Matsuno T, Matsuura H, Fujii S, Suzuki R, Sugiura Y, Miura Y. Anti-Fya-mediated delayed hemolytic transfusion reaction following emergency-release red blood cell transfusion: possible involvement of HLA-DRB1*04:03 in the Japanese population. Int J Hematol. 2022;115:440–445. doi: 10.1007/s12185-021-03242-3. [DOI] [PubMed] [Google Scholar]
- 16.Reynisson B, Barra C, Kaabinejadian S, Hildebrand WH, Peters B, Nielsen M. Improved prediction of MHC II antigen presentation through integration and motif deconvolution of mass spectrometry MHC eluted ligand data. J Proteome Res. 2020;19:2304–2315. doi: 10.1021/acs.jproteome.9b00874. [DOI] [PubMed] [Google Scholar]
- 17.Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linton PJ, Dorshkind K. Age-related changes in lymphocyte development and function. Nat Immunol. 2004;5:133–139. doi: 10.1038/ni1033. [DOI] [PubMed] [Google Scholar]
- 20.Lindstrom TM, Robinson WH. Rheumatoid arthritis: a role for immunosenescence? J Am Geriatr Soc. 2010;58:1565–1575. doi: 10.1111/j.1532-5415.2010.02965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foy BH, Carlson JCT, Reinertsen E, Padros I, Valls R, Pallares Lopez R, Palanques-Tost E, Mow C, Westover MB, Aguirre AD, Higgins JM. Association of red blood cell distribution width with mortality risk in hospitalized adults with SARS-CoV-2 infection. JAMA Netw Open. 2020;3:e2022058. doi: 10.1001/jamanetworkopen.2020.22058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel HH, Patel HR, Higgins JM. Modulation of red blood cell population dynamics is a fundamental homeostatic response to disease. Am J Hematol. 2015;90:422–428. doi: 10.1002/ajh.23982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Go RS, Winters JL, Kay NE. How I treat autoimmune hemolytic anemia. Blood. 2017;129:2971–2979. doi: 10.1182/blood-2016-11-693689. [DOI] [PubMed] [Google Scholar]
- 24.Brodsky RA. Warm autoimmune hemolytic anemia. N Engl J Med. 2019;381:647–654. doi: 10.1056/NEJMcp1900554. [DOI] [PubMed] [Google Scholar]
- 25.Wang W, Zhang W, Zhang J, He J, Zhu F. Distribution of HLA allele frequencies in 82 Chinese individuals with coronavirus disease-2019 (COVID-19) HLA. 2020;96:194–196. doi: 10.1111/tan.13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Severe Covid-19 GWAS Group, Ellinghaus D, Degenhardt F et al (2020) Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med 383:1522–1534. 10.1056/NEJMoa2020283 [DOI] [PMC free article] [PubMed]
- 27.Keicho N, Itoyama S, Kashiwase K, Phi NC, Long HT, Ha LD, Ban VV, Hoa BK, Hang NT, Hijikata M, Sakurada S, Satake M, Tokunaga K, Sasazuki T, Quy T. Association of human leukocyte antigen class II alleles with severe acute respiratory syndrome in the Vietnamese population. Hum Immunol. 2009;70:527–531. doi: 10.1016/j.humimm.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, Zhao S, Teng T, Abdalla AE, Zhu W, Xie L, Wang Y, Guo X. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12:244. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang EY, Mao T, Klein J, Dai Y, Huck JD, Jaycox JR, Liu F, Zhou T, Israelow B, Wong P, Coppi A, Lucas C, Silva J, Oh JE, Song E, Perotti ES, Zheng NS, Fischer S, Campbell M, Fournier JB, Wyllie AL, Vogels CBF, Ott IM, Kalinich CC, Petrone ME, Watkins AE; Yale IMPACT Team, Dela Cruz C, Farhadian SF, Schulz WL, Ma S, Grubaugh ND, Ko AI, Iwasaki A, Ring AM (2021) Diverse functional autoantibodies in patients with COVID-19. Nature 595:283–288. 10.1038/s41586-021-03631-y [DOI] [PubMed]
- 30.Murphy WJ, Longo DL. A possible role for anti-idiotype antibodies in SARS-CoV-2 infection and vaccination. N Engl J Med. 2022;386:394–396. doi: 10.1056/NEJMcibr2113694. [DOI] [PubMed] [Google Scholar]
- 31.Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 Table S1 (PPTX 57 KB)
Supplementary file2 Table S2 (PPTX 46 KB)
Supplementary file3 Figure S1 (PPTX 65 KB)
Supplementary file4 Supplemental data 1 (XLSX 240 KB)
Supplementary file5 Supplemental data 2 (XLSX 11 KB)
Supplementary file6 Supplemental data 3 (XLSX 55 KB)