Abstract
The oxidation of methane in anoxic marine sediments is thought to be mediated by a consortium of methane-consuming archaea and sulfate-reducing bacteria. In this study, we compared results of rRNA gene (rDNA) surveys and lipid analyses of archaea and bacteria associated with methane seep sediments from several different sites on the Californian continental margin. Two distinct archaeal lineages (ANME-1 and ANME-2), peripherally related to the order Methanosarcinales, were consistently associated with methane seep marine sediments. The same sediments contained abundant 13C-depleted archaeal lipids, indicating that one or both of these archaeal groups are members of anaerobic methane-oxidizing consortia. 13C-depleted lipids and the signature 16S rDNAs for these archaeal groups were absent in nearby control sediments. Concurrent surveys of bacterial rDNAs revealed a predominance of δ-proteobacteria, in particular, close relatives of Desulfosarcina variabilis. Biomarker analyses of the same sediments showed bacterial fatty acids with strong 13C depletion that are likely products of these sulfate-reducing bacteria. Consistent with these observations, whole-cell fluorescent in situ hybridization revealed aggregations of ANME-2 archaea and sulfate-reducing Desulfosarcina and Desulfococcus species. Additionally, the presence of abundant 13C-depleted ether lipids, presumed to be of bacterial origin but unrelated to ether lipids of members of the order Desulfosarcinales, suggests the participation of additional bacterial groups in the methane-oxidizing process. Although the Desulfosarcinales and ANME-2 consortia appear to participate in the anaerobic oxidation of methane in marine sediments, our data suggest that other bacteria and archaea are also involved in methane oxidation in these environments.
Microbially mediated oxidation of methane in anoxic marine systems is a globally significant process, with up to 90% of the oceanic methane production recycled in anaerobic marine sediments (35). Anaerobic consumption of methane is geochemically and biologically important, since it significantly decreases the flux of methane from marine sediments to the atmosphere. The process transforms terminally reduced carbon into forms that are more readily accessible to a larger group of microorganisms in anoxic sediments. Localized chemosynthetic communities benefit from large quantities of hydrogen sulfide (2), generated as a by-product of the anaerobic oxidation of methane. Geochemical evidence supporting anaerobic oxidation of methane (AOM) is well documented in the literature and is based on stable isotopic signatures (7), pore water chemical profiles (5, 23), inhibitor studies (17, 21), and sample incubations with radiotracers (21, 23). The results of these studies led to the hypothesis that AOM is mediated by a consortium consisting of a methanogen operating in reverse (producing hydrogen and carbon dioxide from methane) and a hydrogen-scavenging, sulfate-reducing partner (21). Despite the indirect evidence supporting microbially mediated AOM, identifying the individual consortium members and the actual mechanism involved has been difficult.
The recent discoveries of methane-derived, isotopically light archaeal lipids in seep-associated sediments and carbonates provided compelling chemotaxonomic evidence for the direct involvement of archaea in anaerobic methane utilization (11, 18, 19, 30, 42). Hinrichs et al. (18) identified isotopially depleted lipid biomarkers and archaeal 16S rRNA genes (rDNAs) occurring together in cold seep sediment samples from the Eel River Basin, where AOM is thought to actively occur. Results of this study corroborated the involvement of methanogenic lineages in AOM, identifying two potential archaeal groups related to the aceticlastic Methanosarcinales (ANME-1 and ANME-2) as likely candidates for the methane-oxidizing archaea in anoxic marine sediments.
Further studies of Eel River Basin seep sediments and additional seep sites in Santa Barbara Basin confirmed the presence of extremely depleted archaeal lipids, in addition to identifying isotopically depleted bacterial fatty acids and glycerol ethers, most likely originating from the AOM syntrophic partners (19). Similar 13C-depleted microbial lipids were recently observed in hydrate-associated sediments from the Cascadia Margin (4, 11) and Mediterranean mud volcanoes (30), as well as in surface sediments and seep carbonates from the Black Sea (41). The observation of both archaeal and bacterial lipids that are highly 13C depleted suggests a close coupling of and a transfer of carbon between these two groups, providing additional evidence for a syntrophic association of archaea and bacteria (19).
In this study, we conducted cultivation-independent 16S rDNA surveys on a variety of samples from different seep environments, in which the activities of anaerobic methanotrophic microbes are indicated by the presence of 13C-depleted biomarkers. We surveyed and compared bacterial and archaeal groups present at geographically distant methane seep sites, as well as in control sediments. Whole-cell fluorescent in situ hybridization experiments were also conducted to confirm the identities of AOM consortium members at these sites, extending preliminary observations of a previous study (4).
MATERIALS AND METHODS
Site description and sampling.
Sediment samples were obtained from the Eel River Basin and Santa Barbara Basin at a water depth of approximately 500 m by means of the remotely operated vehicle Ventana. Samples were collected with push cores or hydraulic piston cores and subsequently stored in a nitrogen atmosphere at 4°C until processed on shore ∼0.5 to 6 h after collection. Sample, location, area description, and SO42− and H2S levels are summarized in Table 1. Sediment samples from push cores were extruded upwards in 3-cm-thick sections under a nitrogen atmosphere. Samples were then subdivided for in situ hybridization (0.5 g [wet weight] of sediment plus 1 ml of 50% ethanol–2% NaCl [1:1]), lipid and nucleic acid analyses, (approximately 15 g of sediment frozen in liquid nitrogen), and pore water chemistry (remaining sediment sample).
TABLE 1.
Summary of samples used in the construction of SSU rDNA libraries and their physical and chemical descriptions
| Samplea | Depth (cm) | Latitude, longitude (°) | Physical or chemical description | SO4 (mM) | H2S (mM) | rDNA library designation | PCR primers | No. of clones screened |
|---|---|---|---|---|---|---|---|---|
| Eel-Hpc4 | 0–4 | 40.47, −124.35 | Bacterial mat | NAe | NA | Eel-TE | B27f-U1492r | 52 |
| Eel-Hpc4 | 20–22 | 40.47, −124.35 | Bacterial mat | NA | NA | Eel-BE | B27f-U1492r | 121 |
| Eel-pc36 | 4–7 | 40.48, −124.36 | Clam patch, high CH4 | 20 | +d | Eel-36e | B27f-U1492r | 18 |
| Eel-pc36 | 4–7 | 40.48, −124.36 | Clam patch, high CH4 | 20 | + | Eel-36a | A20f-U1492r | 121 |
| SB-pc24 | 14–16 | 34.10, −119.99 | Calyptogena clam patch | 5.9b | 11c | SB-24e | B27f-U1492r | 91 |
| SB-pc24 | 14–16 | 34.10, −119.99 | Calyptogena clam patch | 5.9 | 11 | SB-24a | A20f-U1492r | 90 |
| SB-pc17 | 6–8 | 34.25, −119.97 | Bacterial mat | 11.5 | 10 | SB-17a | A20f-U1492r | 77 |
| SB-pc7 | 12–14 | 34.27, −119.96 | Reference core, background | 28 | 0 | SB-7a | A20f-U1492r | 25 |
SB, Santa Barbara Basin; Eel, Eel River Basin. Water depth ranged between 519 and 556 m for both the Santa Barbara and Eel River basin sites.
SO42− values for Santa Barbara Basin samples were provided by Jon Martin (personal communication).
H2S values for Santa Barbara Basin samples were provided by Jim Barry (personal communication).
+, H2S detected but not quantified.
NA, not available.
Pore water chemistry.
Pore waters were squeezed from 3-cm-thick core intervals using a pressurized gas sediment squeezer (34) and collected into attached air-tight 60-ml disposable syringes (Becton Dickinson, Mountain View, Calif.). Sulfate concentrations in pore water were measured using a DIONEX DX-120 ion chromatograph equipped with an AS-9HC column. Sulfate was eluted using 0.028 M Na2CO3 at a flow rate of 1.0 ml min−1. Samples were diluted 1:100 (vol/vol) in deionized water prior to analysis. Measured values were standardized against IAPSO standard seawater (28.9 mM SO42−). Dissolved methane was extracted from pore waters by adding an equal volume of nitrogen gas to the syringe and then shaking for 2 min prior to measurement with a gas chromatograph (GC) equipped with a flame ionization detector. Pore waters used for analysis of dissolved hydrogen sulfide were obtained by centrifugation under oxygen-free conditions (2) and measured using a colorimetric assay (8).
Lipid analysis.
Sediment samples (∼10 g [wet weight]) were sterilized by ultrasonication (ultrasonic bath) in 20 to 40 ml of a mixture of dichloromethane (DCM) and methane (9:1, vol/vol) for ∼30 min. After evaporation of the residual solvent, the sediments were oven dried at ∼50°C. Experiments have shown that oven drying largely suppresses degradation of analytes by microbial activities (K.-U. Hinrichs, unpublished data). Free lipids were extracted from dried and homogenized sediments using a DIONEX Accelerated Solvent Extraction 200 system at 100°C and 1,000 lb/in2, with DCM-methanol (90:1, vol/vol) as the solvent. Extracts were separated into four fractions of increasing polarities using SUPELCO LC-NH2 glass cartridges (500 mg of sorbent) and a sequence of solvent mixtures of increasing polarities (hydrocarbons −4 ml of n-hexane, ketones and esters −6 ml of n-hexane–DCM [3:1], alcohols −5 ml of DCM-acetone [9:1], and carboxylic acids −2% formic acid in DCM). Individual compounds were quantified and identified using a Hewlett-Packard (HP) model 6890 GC equipped with a J&W model DB-5 (60-m length, 0.32-mm inner diameter, and 0.25-μm film thickness) capillary column and coupled to an HP model 5973 mass-selective detector. Stable carbon isotopic compositions of individual compounds were determined using a Finnigan Delta Plus mass spectrometer coupled to an HP model 6890 GC and equipped with a column identical to that described above. Column temperatures of both GC systems were programmed from 40°C (1 min under isothermal conditions) to 130°C at a rate of 20°/min and then to 320°C (60 min under isothermal conditions) at 4°C/min. Alcohols were analyzed as their trimethylsilyl ethers after reaction with N,O-bis(trimethylsilyl)trifluoroacetamide reagent (plus 1% trimethylchlorosilane) in DCM. Reported δ-13C values are means of results of at least two analyses and have been corrected for the presence of carbon atoms added during derivatization. Differences between individual analyses were generally less than 1‰.
Nucleic acid extraction.
For 16S rDNA analysis, total nucleic acids were extracted from sediment samples, with each sample consisting of a 3-cm depth interval. Cell lysis and DNA extraction from 0.5 g (wet weight) of sediment were conducted using a Bio 101 (Vista, Calif.) Fastprep beadbeating machine (Bio 101) and a Fast soil prep kit (MoBio Inc., San Diego, Calif). The protocol for the MoBio kit was modified by initially beadbeating the sample using the Fastprep machine (speed 4.5 for 20 s), followed by two 5-min incubations at 70°C. The remainder of the extraction procedure was carried out according to the manufacturer's instructions. This procedure typically produced large-molecular-size DNA (>20 kb). Nucleic acids from three independent extractions were pooled and purified using a small-scale CsCl density gradient as previously described (9).
16S rDNA library construction and screening.
Small-subunit (SSU) rDNAs were amplified by PCR with purified DNA samples from Santa Barbara and Eel River basin cold seep sediments. PCR mixtures (50 μl) contained a 0.2 μM concentration of either bacterium-specific (27f and 1492r) or archaeon-specific (20f and 1492r) primers. Reaction mixtures also contained 5 μl of PCR buffer (containing 2 mM MgCl2), 2.5 mM each deoxynucleotide triphosphate, and 0.025 U of Taq polymerase (Promega, Madison, Wis.).
PCR conditions for archaeal libraries (Eel-36a, SB-24a, SB-17a, and SB-7a).
Archaeal 16S rDNAs from the CsCl-purified DNAs were amplified for 30 cycles (1.5 min of denaturation at 94°C, 30 s of annealing at 55°C, and 7 min of elongation at 72°C) using archaeon-specific primers (A20f, 5′-TTCCGGTTGATCCYGCCRG-3′; U1492r, 5′-GGTTACCTTGTTACGACTT-3′). The exception in this procedure was archaeal library Eel-36a, which was constructed under similar PCR conditions but amplified for only 20 cycles to minimize bias associated with high cycle numbers (38).
PCR conditions for bacterial libraries (Eel-36e, Eel-TE, Eel-BE, and SB-24e).
Bacterial 16S rDNAs were PCR amplified for either 30 cycles (Eel-TE, Eel-BE, and SB-PC24) or 20 cycles (Eel-PC36) using a bacterium-specific forward primer (B27f, 5′-AGAGTTTGATCCTGGCTCAG-3′) and a universal reverse primer (U1492r, 5′-GGTTACCTTGTTACGACTT-3′). PCR conditions were the same as stated above for the archaeal library construction.
Cloning and sequencing.
Amplicons were pooled from three reactions and cleaned using a Qiaquick PCR purification kit (Qiagen, Valencia, Calif.) for all 16S rDNA libraries. Cleaned products were then cloned with a TA cloning vector kit according to the instructions of the manufacturer (Invitrogen, Carlsbad, Calif.). Screening for the libraries was conducted by restriction fragment length polymorphism analysis on M13F- and M13R-amplified products using either HaeIII or RsaI (Promega). M13-amplified PCR products were initially diluted 1:20 in PCR buffer. Five microliters of the diluted product was then used in the restriction digest containing 0.5 μl of enzyme and 2 μl of buffer in a 20-μl total volume, according to the manufacturer's instructions. Unique clones were identified and plasmids were purified either with a Wizard genomic DNA purification kit (Promega) or by electroelution by an automated miniprep protocol (McConnell, La Jolla, Calif.). Cleaned plasmid preparations were quantified and sequenced using a Thermo Sequenase Fluorescent Labeled Primer Cycle Sequencing kit (Amersham, Braunschweig, Germany) and an automated model 4000L or 4200 DNA sequencer (LI-COR BioTech, Lincoln, Nebr.). Double-stranded sequencing was completed using a suite of primers targeting 16S rDNA.
Phylogenetic analysis.
Sequences from clones were submitted to GenBank for preliminary analysis using the BLAST program of the Ribosomal Database Project to identify putative close phylogenetic relatives (27). Sequences were aligned to their nearest neighbor with the automated alignment tool of the ARB program package (O. Strunk and W. Ludwig [ed.], ARB: a software environment for sequence data. 1999. [http://www.mikro.biologie.tu-muenchen.de]). Phylogenetic trees were generated using the SEQBOOT, DNADIST, and NEIGHBOR programs of the PHYLIP version 3.5 software (12). The Kimura two-parameter model was used to estimate evolutionary distance, and 1,000 bootstrap analyses were performed to assign confidence levels to the nodes in the trees.
Fluorescent in situ hybridization (FISH).
Selected Eel River Basin sediments, displaying chemical pore water profiles diagnostic of active AOM (Fig. 1) and containing 13C-depleted biomarkers, were screened for the presence of archaeal-bacterial aggregations. Sediment samples (0.5 cm3) stored in 2% NaCl–ethanol (1:1) were diluted (1:10) in phosphate-buffered saline and treated with 15 s of mild sonication at 32 A (Sonics and Materials Inc., Danbury, Conn.). (Prior fixation with formaldehyde was not absolutely required for successful hybridization with the oligonucleotide probes.) Diluted samples were centrifuged briefly for 5 s at 5,000 rpm to pellet large sediment particles, and 50 to 70 μl of the supernatant was filtered onto a 0.2-μm-pore-size GTTP polycarbonate filter. Filters were then treated for 2 min with a phosphate-buffered saline–ethanol (1:1) solution and dried before hybridization. Hybridization and wash buffers were synthesized according to the method of Glöckner et al. (15) using 30% formamide in the hybridization buffer and 80 mM NaCl in the wash solution. Oligonucleotide probes were labeled with either Cy3 or fluorescein isothiocyanate fluorochromes (Genset S.A., Paris, France). Oligonucleotide probes targeting the bacterial Desulfosarcinales-Desulfococcus group (DSS658, TCCACTTCCCTCTCCCAT) and the seep-specific archaeal ANME-2 group (EelMSMX932, AGCTCCACCCGTTGTAGT) have been previously reported (4, 32). General archaeal and bacterial probes, AR915 and EUB338, were used as previously described (1). Hybridization was conducted at 46°C for 2 h and followed by a wash at 48°C for 15 min. Washed filters were stained with a dilute 4′6′-diamidino-2-phenylindole (DAPI) solution (5 μg/ml) for 1 min and examined under epifluorescence microscopy with an Axiophot 2 microscope using a 100× PlanAPO objective (Zeiss, Thornwood, N.Y.). Archaeal-bacterial dually stained aggregates were photographed using a Spot SP100 cooled digital color charge-coupled-device camera (Diagnostic Instruments, Inc., Sterling Heights, Mich.). Captured images were overlaid in Adobe Photoshop 4.0.
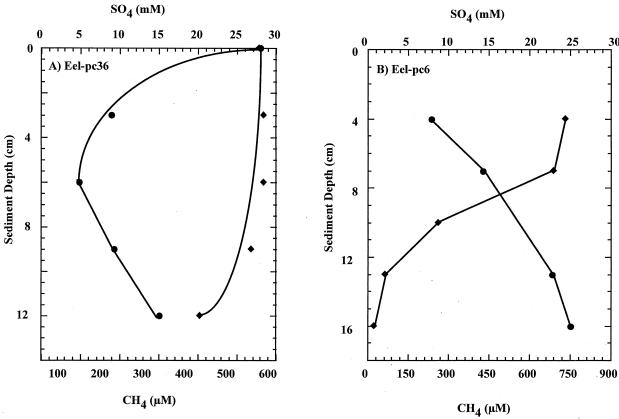
FIG. 1.
Pore water profiles of SO42− (⧫) and CH4 (●) for select Eel River Basin seep samples. (A) Profile for a seep core used in the generation of the clone libraries Eel-36a and Eel-36e; (B) chemical profile for a seep core in which ANME-2 and Desulfosarcinales aggregates were detected by FISH (depth, 3 to 6 cm) (Fig. 5).
Nucleotide sequence accession numbers.
The rDNA sequences were submitted to GenBank and have been assigned the following accession numbers: AF354126 to AF354167.
RESULTS
Chemical profiling of cold seep sediments.
Two active methane seep sites within the Eel River Basin were targeted for extensive chemical and microbial analysis in August 1999. Depth profiles for 14 push cores with an average length of 15 cm were analyzed for concentrations of SO42−, CH4, and CO2. Nine of the 14 seep-associated cores had profiles characteristic of active anaerobic methane oxidization, with shallow sulfate depletion depths that reached undetectable levels of SO42− at depths of <15 cm. Pore water chemistry features, typical for continental margin sediments with abundant methane (i.e., the intersection of methane and sulfate minima in a so-called transition zone) (23, 28), were rarely observed in seep sediments in the Eel River Basin. Instead, high concentrations of methane and sulfate often occurred together in the upper 10 cm (Fig. 1). Some seep cores contained high concentrations of methane (up to 6.6 mM), but little to no sulfate depletion was observed. This is most likely reflective of non-steady-state conditions caused by dynamic fluid and gas transport in active seep zones (43). Independent of the pore water sulfate concentration, the majority of the seep cores profiled (seven out of nine) also contained characteristic archaeal and bacterial lipid biomarkers indicative of AOM consortia (18, 19, 30).
Santa Barbara Basin seep sediments, including seep cores SB-pc24 and SB-pc17 used in 16S rDNA library construction, displayed high hydrogen sulfide concentrations (up to 11.5 mM) and low sulfate concentrations (<5 mM) at 10 cm of sediment depth. Nonseep reference cores from both environments showed minimal sulfate depletion throughout the core and undetectable levels of methane, hydrogen sulfide, and isotopically depleted lipid biomarkers (Table 1; data not shown).
Chemotaxonomic analysis of archaeal diversity.
Chemotaxonomic analysis of seep-associated sedimentary lipids from the Eel River and Santa Barbara basins revealed high concentrations of diverse, extremely δ-13C-depleted archaeal lipids, the most prominent being archaeol, sn-2-hydroxyarchaeol, saturated and unsaturated 2,6,10,15,19-pentamethylicosane (PMI), and crocetane. Depth profiles of sedimentary lipid biomarkers in Eel River Basin seep core Eel-pc36 revealed abundant, 13C-depleted archaeal and bacterial lipids throughout the core (Table 2). The archaeal lipids archaeol and sn-2-hydroxyarchaeol displayed relatively constant δ-13C values of less than −100‰ at all depths, suggestive of constant fractionation by archaeal groups under conditions of excess methane availability. In contrast to the isotopic values of the archaeal lipid fraction, profiles of bacterial lipids (fatty acids and sn-1-monoalkylglycerolethers [sn-1-MAGE]) in the core were more variable, showing decreasing δ-13C values with increasing depth, likely due to the decreasing δ-13C of pore water substrates. Evidence for an isotopically depleted carbon pool at depth is also reflected in the isotopic values of authigenic carbonates extracted from the same core, with δ-13C values being significantly lower in more deeply buried samples (Table 2).
TABLE 2.
Concentrations and δ-13C levels of archaeal and bacterial lipids and δ-13C levels of total organic carbon and carbonate of Eel-pc36
| Depth interval (cm) | Archaeol
|
sn-2-Hydroxyarchaeol
|
Crocetane
|
MAGE cis-ω7-16:1
|
δ-13C (‰) of total organic carbon | δ-13C (‰) of CO32− | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| δ-13C (‰) | Concn (μg/g) | δ-13C (‰) | Concn (μg/g) | δ-13C (‰) | Concn (μg/g) | δ-13C (‰) | Concn (μg/g) | |||
| 0–3 | −101.1 | 1.3 | −105.2 | 3.8 | NDa | ND | −76.7 | 1.00 | −28.8 | −18.0 |
| 3–6 | −100.6 | 1.3 | −105.8 | 2.7 | −91.6 | 0.3 | −77.6 | 1.16 | −28.9 | −24.0 |
| 6–9 | −102.6 | 1.5 | −105.5 | 2.3 | ND | ND | −80.8 | 0.95 | −29.7 | −32.5 |
| 9–12 | −102.1 | 1.1 | −105.7 | 1.0 | ND | ND | −86.5 | 1.03 | −28.5 | −29.9 |
ND, not determined.
Phylogenetic diversity of archaea.
Culture-independent analysis of archaeal assemblages within seep sediments containing isotopically depleted archaeal lipid biomarkers revealed five distinct archaeal phylogenetic clades affiliated with both the Euryarchaeota and Crenarchaeota (Table 3). Of these, two groups peripherally related to the Methanosarcinales, ANME-2 and ANME-1, represented a significant proportion of the total rDNA clones in methane-laden sediments.
TABLE 3.
Major archaeal lineages represented in 16S rDNA librariesa
| Archaea | Presence in or absence from indicated sample (% of total organisms) in 16S rDNA library:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Eel-TAb | Eel-BAb | Eel-36a | SB-24a | SB-17a | SB-7a | pISA (39) | GBc | MT/C (29) | APA (46) | JTB (26) | |
| Eel River Basin
|
Santa Barbara Basin
|
||||||||||
| Eel-Hpc4 (0–4 cm) | Eel-Hpc4d (20–22 cm) | Eel-pc36d (4–7 cm) | SB-pc24d (13–16 cm) | SB-pc17 (4–7 cm) | SB-pc7 (4–7 cm) | Hydrothermal vent sediments from Japan | Seep sediments from Guaymas Basin |
Coastal salt marsh, Essex, United Kingdom |
Deep-sea sediments from the Atlantic | Cold seep sediments from the Japan Trench | |
| ANME-1 | + (55) | + (84) | + (7.5) | − | + (18) | − | + | + | − | − | − |
| ANME-2 | + | + (16) | + (71) | + (67) | + (22) | − | − | + | + | − | − |
| Methanosarcinales | − | − | + (6) | + (9) | − | − | − | − | + | − | − |
| Crenarchaeota | − | − | + (2) | + (2) | + (4) | − | + | − | − | + | + |
| Thermoplasmales | + | − | + (2) | + (3) | + (41) | + | + | − | + | + | + |
+, sequence type detected; −, not detected. Numbers in parentheses beside 16S rDNA library designations (pISA, GB, MT/C, APA, and JTB) are reference numbers.
Data were previously reported in reference 18.
Data are from Andreas Teske (personal communication).
δ-13C-depleted archaeal lipid biomarkers detected in methane seep sediment samples included archaeol, sn-2-hydroxyarchaeol, dihydrophytol, unsaturated PMI, and crocetane. Archaeal biomarkers were not detected in sample SB-17a or in nonseep control sample SB-7a. Crocetane was not detected in sample Eel-BA. δ-13C values of these lipids ranged from −81 to −129‰.
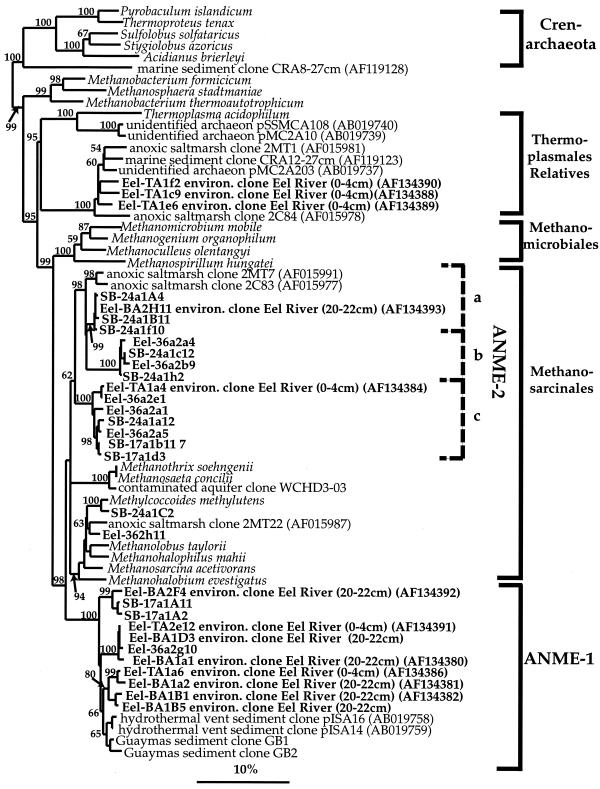
16S rDNAs from the ANME-2 clade (85 to 89% similar to Methylococcoides methylutens) were most frequently recovered, representing 21 to 71% of the total clones in two of the three archaeal libraries prepared from Eel River and Santa Barbara basin sites (Table 3). Phylotypes grouping within ANME-2 from the Eel River and Santa Barbara basins formed three distinct clusters of highly related sequences (>97% similarity) (designated subgroups a, b, and c), with maximum overall sequence similarity of 87% for this group (Fig. 2). The diversity of ANME-2 phylotypes detected within single samples was high, with representative sequences falling into two or more subgroups. The exception to this was library SB-17a, which contained phylotypes clustering only within subgroup c. The SB-17a rDNA clone library was atypical compared to other seep libraries overall, with the majority of phylotypes being associated with relatives of the Thermoplasmales (Table 3).
FIG. 2.
Phylogenetic tree showing relationships of 16S rDNA archaeal clone sequences from Santa Barbara Basin and Eel River Basin seep sites (in boldface) to selected cultured and environmental euryarchaeotal sequences in the database. Boldface Eel River Basin clones containing accession numbers were previously reported in reference 18. Environmental sequences included 2MT and 2C from anoxic salt marsh sediments (29), CRA from deep sea sediments (46), pISA from hydrothermal vent sediments (39). The tree was generated by neighbor-joining analysis and corrected with a mask that included only 60% of the conserved regions. One thousand bootstrap analyses were performed, and percentages greater than 50% are reported. The bar represents a 10% estimated sequence divergence. environ., environmental.
The putative methane-oxidizing archaeal group ANME-1, recently described from Eel River Basin methane seep sediments, was repeatedly detected at methane seep sites in the Eel River and Santa Barbara basins. ANME-1-related phylotypes represented 7.5 and 18% of the total archaeal clones screened in the 16S rDNA libraries Eel-36a and SB-17a, respectively. Like the ANME-2 group, the ANME-1 group contains a significant amount of intragroup diversity, with the most distant sequences in this clade sharing 90% similarity to each other. However, in contrast to the greater ANME-1 diversity previously reported from a single Eel River Basin cold seep, fewer ANME-1-related sequences were recovered in this study and there was less diversity within single samples (Fig. 2). The ANME-1 phylotypes recently reported from methane-rich, hydrothermally active settings formed a separate clade (39) and were more closely related to each other than to the ANME-1 cold seep sequences (>96%) (Fig. 2).
Two other methanogen-like sequences, closely related to cultured members of the Methanosarcinales, made up less than 10% of the SB-24a and EEL-36a libraries (Table 3). Phylotype SB-24a1C2, representing 9% of the clones from the Santa Barbara Basin library, is highly similar (98.8% similarity) to the obligate methylotroph Methanococcoides burtonii, previously isolated from an antarctic lake (13). Eel River Basin sequence types, represented by Eel-36a2H11, were also closely related to the methylotrophic Methanococcoides spp. (>95% similarity) and made up 6% of the Eel-36a library.
Remaining euryarchaeotal sequences were similar to rDNAs previously recovered from marine sediments (29) that are distantly related to the Thermoplasmales (Fig. 2). Relatives of the Thermoplasmales were a minor proportion of the total clones, representing <3% of organisms in all seep libraries except the Santa Barbara Basin library SB-17a. 13C-depleted archaeon-specific biomarkers were below detection levels in SB-pc17, which in addition to the high proportion of Thermoplasmales phylotypes may indicate low methane flux at this site. Sequences related to the Thermoplasmales represented 100% of the clones analyzed in library SB-7a, constructed from sediments from a control site in Santa Barbara Basin, where neither ANME-2 or ANME-1 clones, sulfate depletion, high sulfide levels, nor 13C-depleted lipid biomarkers were observed.
Like the Thermoplasmales-related phylotypes, crenarchaeotal sequences were present in both Eel River and Santa Barbara basin sites and comprised a relatively low percentage (<4%) of clones in the methane seep libraries (Table 3). Crenarchaeotal sequences from both sites were highly similar to each other (98.9%) and were most closely related to rDNAs recovered from other marine sedimentary environments (46, 47).
Lipid chemotaxonomy of bacteria.
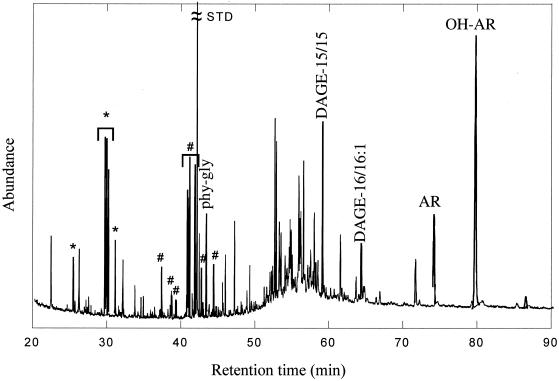
Two classes of lipids with significant contributions from bacterial, syntrophic partners participating in the AOM consortium were generally present at seep sites in the Eel River and Santa Barbara basins. These compounds are fatty acids ranging from C14 to C18 in carbon number and monoalkylglycerolethers (MAGE) and dialkylglycerolethers (DAGE) with nonisoprenoidal alkyl moieties and chain lengths from C14 to C18. The participation of organisms that produce MAGE and DAGE in AOM is indicated by δ-13C values that require more or less exclusive utilization of methane-derived carbon for biosynthesis (approximately −100‰ for most MAGE and selected fatty acids [19]). In addition to the MAGE and fatty acids, the sedimentary alcohols showed very similar structural and isotopic features, suggesting that certain syntrophic bacterial members of the AOM consortium synthesize them as well. Fatty alcohols are known products of some bacteria, but no systematic surveys, i.e., comparable to those of fatty acids, have been undertaken on alcohol contents in microorganisms. Very similar relative amounts of MAGE and fatty alcohols with carbon isotopic compositions ranging from −100 to −60‰ occur commonly in sediments with active AOM (e.g., a representative chromatogram is illustrated in Fig. 3).
FIG. 3.
Reconstructed ion chromatogram of the alcohol fraction from sample Eel-pc 36 (6 to 9 cm). Labeled peaks designate compounds with isotopic compositions indicating a partial or exclusive derivation from anaerobic methanotrophic microbes. ∗, C14 to C17 acyclic alcohols from bacteria; #, sn-1-MAGE with ether-linked C14 to C18 acyclic alcohol moieties from bacteria. The numbers beside “DAGE” designate carbon numbers of ether-linked alkyl moieties expressed as numbers, e.g., DAGE-15/15 is diether with two C15-alkyl moieties. phy-gly, sn-1-monophytanylglycerolether (archaea); AR, archaeol (archaea); OH-AR, sn-2-hydroxyarchaeol (archaea); STD, standard.
Phylogenetic diversity of δ-proteobacteria.
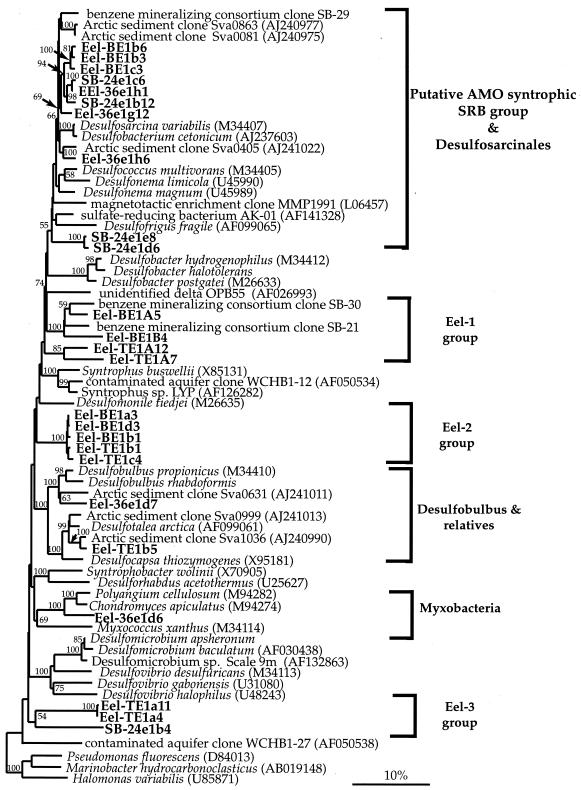
Table 4 shows the major δ-proteobacterial lineages detected in four cold seep sediment samples that contain 13C-depleted bacterial lipids. Analysis of 16S rDNA sequences recovered from the same sediment samples revealed a predominance of δ-proteobacterial phylotypes, represented by six distinct lineages. The majority of the seep libraries contained representatives from two or three major groups within the δ-proteobacteria (Table 4). These sequence clusters were each related by >81% sequence similarity and grouped among both cultured sulfate reducers (i.e., Desulfosarcinales and Desulfobulbus spp.) and novel clusters so far represented only by environmental sequences originating from similar environments (groups Eel-1, Eel-2, and Eel-3) (Fig. 4).
TABLE 4.
Major 16S rDNA phylotypes associated with sulfate-reducing bacteria
| δ-Proteobacteria | Presence in or absence from indicated sample in 16S rDNA library (reference):
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Eel-TE | Eel-BE | Eel-36ea | SB-24ea | ODPB (3) | GCAb | JTB (26) | TAYNAYA (6) | sva (47) | |
| Eel-Hpc4 (0–4 cm) | Eel-Hpc4 (22 cm) | Eel-pc36 (4–7 cm) | SB-pc24 (13–16 cm) | Gas hydrate sediment from the Cascadia Margin |
Hydrocarbon seep sediments | Japan Trench cold seep sediments | Antarctic fjord sediments | Shelf sediment from the Arctic Ocean | |
| Desulfosarcina | + | + | + | + | + | + | + | + | + |
| Seep Eel-1 | + | + | − | − | − | + | − | − | − |
| Desulfobulbus | + | − | + | − | + | − | − | − | + |
| Seep Eel-2 | + | + | − | − | + | − | − | − | − |
| Seep Eel-3 | + | − | − | + | − | − | − | − | + |
| Myxobacterium | − | − | + | − | − | − | + | − | + |
Fatty acids associated with sulfate-reducing bacteria (10, 40) detected in methane seep sediment samples included n-C14:0, n-C16:1 (ω:5), and 10Me-C16:0. Levels of δ-13C in sedimentary fatty acids ranged from −52 to −103‰. Note that δ-13C levels in photosynthetically derived lipids are approximately −25‰.
FIG. 4.
Phylogenetic tree showing relationships of 16S rDNA δ-proteobacterial clone sequences from Santa Barbara Basin and Eel River Basin seeps (in boldface) to selected cultured and environmental proteobacterial sequences in the database. Environmental sequences included sva from arctic sediment (47) and SB from a benzene-mineralizing enrichment culture (31). The tree was generated by neighbor-joining analysis and corrected with a mask that included only 60% of the conserved regions. One thousand bootstrap analyses were preformed, and percentages greater than 50% are reported. The bar represents a 10% estimated sequence divergence.
Sequences related to the Desulfosarcinales (91% similarity to Desulfosarcina variabilis) were the most common phylotype recovered, being detected in all four 16S rDNA libraries and representing up to 17.6% of total clones in library SB-24e. These ubiquitous rDNA sequences formed a distinct clade, more closely related to each other than to known sequences in the database, with greater than 94% similarity over a 1,300-bp region. Although recovered from geographically distant sites, some phylotypes within this group from both Eel River Basin (Eel-36e1h1) and Santa Barbara Basin (SB-24e1c6) were nearly identical, with only a 0.8% sequence difference detected (Fig. 4).
In addition to phylotypes related to the Desulfosarcinales, 16S rDNAs affiliated with the propionate-utilizing Desulfobulbus were also recovered from two sites in Eel River Basin. Desulfobulbus-related clones were closely related to environmental clones recovered from arctic sediments (92% similar to environmental clone sva0631) as well as methane hydrate-associated sediments from Cascadia Margin (Fig. 4) (3, 33). Additionally, phylotypes clustering within the Eel-2 clade were also peripherally related to Desulfobulbus members (∼88% similarity to Desulfobulbus propionicus). Eel-2 phylotypes comprised a significant percentage of clones recovered from Eel River Basin seep sample Eel-Hpc4 (17%) and were detected in both shallow (4- to 7-cm) and deep (20- to 22-cm) sediment sections (Table 4). Similar to those within Eel-2 clones, phylotypes within the Eel-1 clade were also recovered from both depth intervals from seep core Eel-Hpc4. Eel-1 phylotypes represented 12% of the total clones screened from library Eel-BE (constructed from the 20- to 22-cm sediment interval) and are closely affiliated with environmental clones recovered from a sulfate-reducing benzene-mineralizing enrichment (∼92% similarity) (Fig. 4) (31). In addition, Eel-3 phylotypes, detected in both the Eel River and Santa Barbara basins were not closely related to known δ-proteobacteria, with only 76% similarity to Desulfovibrio gabonensis and 80% similarity to an environmental clone recovered from cold seeps in Japan (26). Eel-3 phylotypes were highly similar to each other (99%) and comprised 5 of the 52 clones screened from library Eel-TE.
Comparison of bacterial 16S rDNA diversity between Santa Barbara and Eel River basin sites.
Four other major lineages of the domain Bacteria were represented in 16S rDNA libraries from both Santa Barbara Basin (SB-24e) and Eel River Basin (Eel-BE) cold seep libraries, including γ-proteobacteria, the Flexibacter-Bacteroides-Cytophaga division, candidate division OP-9, and the Acidobacterium-Holophaga group (Table 5). In both the SB-24e and Eel-BE libraries, δ-proteobacteria related to sulfate-reducing bacteria were the most abundant phylotypes recovered, representing 25 and 36.4% of total clones screened, respectively. In addition to sulfate-reducing phylotypes, library SB-24e also contained a large percentage of ɛ-proteobacterial phylotypes (26%) related to other sulfide-oxidizing chemoautotrophic microorganisms (93.5% similar to Thiomicrospira denitrificans). Related ɛ-proteobacterial phylotypes were also recovered in one Eel River library, Eel-36e (∼92% similarity to SB-24e phylotypes), similar to those found in other seep environments, including methane-rich anoxic sediments from the Cascadia Margin, the Japan Trench, and Sagami Bay (3, 26, 44) (Table 5).
TABLE 5.
Major bacterial lineages represented in 16S rRNA gene libraries from methane seeps
| Organisms | Closest relative | Presence in or absence from indicated sample (% of total organisms) in 16S rDNA librarya:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| SB-24e | Eel-BE | Eel-TEc | Eel-36ec | JTB (26) | GCAb | TAY-NAYA (6) | sva (47) | ||
| Santa Barbara Basin SB-pc24 (13–16 cm) | Eel River Basin Eel-Hpc4 (20–22 cm) | Eel River Basin Eel-pc36 (4–7 cm) |
Santa Barbara Basin SB-pc24 (13–16 cm) |
Cold seep sediment from Japan |
Oil seep sediments | Antarctic fjord sediment from Taynaya Bay | Shelf sediments from the Arctic Ocean | ||
| δ-Proteobacteria | + (25) | + (36.4) | + | + | + | + | + | + | |
| ɛ-Proteobacteria | S-oxidizing symbionts | + (26) | − | − | + | + | + | − | − |
| γ-Proteobacteria | (S-oxidizing symbionts + others) | + (4.3) | + (3.3) | + | + | + | + | + | + |
| α-Proteobacteria | Rhizobium spp. | − | − | − | + | + | − | + | + |
| Planctomyces | Pirellula spp. | + (3.3) | + (1) | + | − | − | − | − | + |
| FBCd | Cytophaga spp. | + (3.3) | + (6.6) | + | − | + | − | + | + |
| Graciliutes | Haloanaerobium spp. | − | + (2.5) | − | − | − | + | − | − |
| Gram-positive bacteria | Propionigenium spp. | − | + (4.1) | + | − | + | + | + | + |
| Green nonsulfur organisms | + (5.5) | − | − | − | − | + | − | + | |
| Holophaga | Acidobacterium spp. | + (3.3) | + (2.5) | + | − | − | + | + | + |
| Candidate division OP-9 | + (3.3) | + (21) | − | − | − | + | + | − | |
| Candidate division OP-11 | − | + (4.1) | − | − | − | − | − | − | |
| Spirochetes | + (5.5) | − | − | − | − | − | − | − | |
Numbers in parentheses beside library designations are reference numbers.
Hydrocarbon seep. Data are from K. R. O'Neill (GenBank database accession no. AF154080 to AF154106).
Based on a partial library screening of Eel-TE (52 clones) and Eel-36e (18 clones).
FBC, Flexibacter-Bacteroides-Cytophaga division.
Diverse phylotypes related to the γ-proteobacteria were detected in all four bacterial libraries, with many phylotypes being related to sequences recovered from arctic sediments (85 to 90% similarity) (33). Similar to the ɛ-proteobacterial representatives, a number of γ-proteobacteria phylotypes recovered from Eel River and Santa Barbara basin sites were related to both free-living and symbiotic sulfide oxidizers (91% similarity), as well as other environmental sequences recovered from similar seep habitats (26). Additional groups represented within the γ-proteobacteria included SB-24e clones highly similar to Halomonas variabilis (95.3% similarity) and Eel-TE clones related to the aerobic methane-oxidizing Methylobacter luteus (95.7% similarity) and the psychrophile Colwellia psychroerythrus (97.7% similarity).
Relatives of candidate division OP-9, originally described from hot spring environments, comprised a significant proportion of the total clones screened in library Eel-BE (21%). Sequences highly related to the Eel River OP-9 phylotypes (96.7% similarity) were also detected in the Santa Barbara Basin library but comprised only a relatively small percentage of the total clones (∼3%) (Table 5). Included in the seep-associated OP-9 cluster were environmental clones from cold seeps in the Japan Trench (26), hydrocarbon-loaded seep sediments (GenBank accession no. AF154106), and a benzene-mineralizing consortium clone SB-15 (31). Sequences related to the recently described Acidobacterium-Holophaga division were also detected but in low abundance (>4%).
Planctomyces spp. and Cytophagales spp.-related phylotypes were common in Santa Barbara and Eel River basin libraries, representing 3.3 and 6.6% of the clones, respectively. Environmental sequences related to the Cytophagales appear to have a widespread distribution in marine sediments, detected in both seep-associated and other marine sedimentary environments, including the Arctic Ocean, an antarctic fjord, and the Japan Trench (6, 26, 33). Planctomyces-related sequences were less frequently detected in marine sediment diversity surveys than Cytophagales-related sequences. Related phylotypes were previously reported in arctic and Puget Sound sediments (16, 33). Although frequently reported in molecular surveys of marine sediments, including those from seep environments, sequences clustering within the gram-positive bacteria comprised only a small percentage (4%) of the total clones screened in this study. Gram-positive bacterial phylotypes were detected only in 16S rDNA libraries from the Eel River Basin.
In situ whole-cell hybridization (FISH).
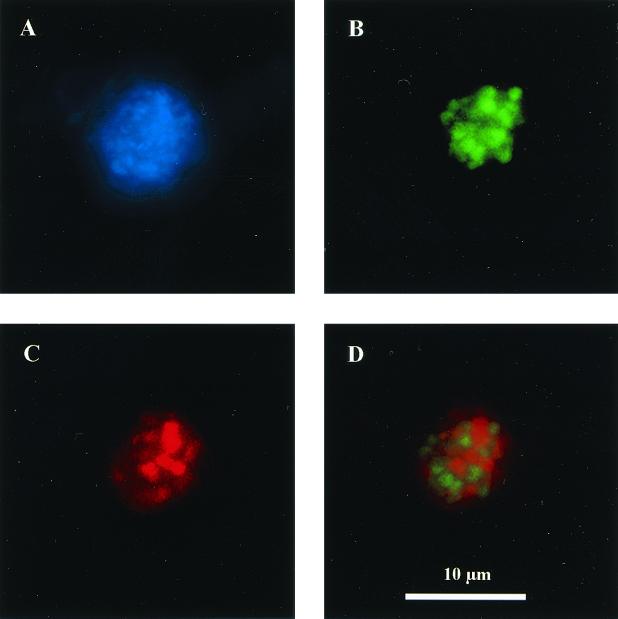
We used previously designed (4) 16S rDNA-targeted oligonucleotide probes to visualize microbial groups that are thought to be involved in the anaerobic oxidation of methane. Oligonucleotide probes specific for members of the Desulfosarcinales and Desulfococcoides groups (DSS658) and the newly described archaeal ANME-2 group (EelMSMX932), as well as general archaeal and bacterial probes (AR915 and EUB338), were hybridized with cells from methane seep sediment samples (4, 32). A total of four cores (Eel-pc6 [4 to 7 cm], Eel-pc36 [4 to 7 cm], Eel-pc21 [1 to 4 cm], and Eel-pc54 [3 to 4 cm]) from Eel River Basin seeps contained cells hybridizing with archaeal ANME-2 and sulfate-reducing DSS658 probes. Archaeal and bacterial cells hybridizing with the EelMSMX932 and DSS658 probes occurred together as structured aggregates, comprised of irregular coccoid cells 1 to 2 μm in diameter (Fig. 5). Archaeal and sulfate-reducing bacterial aggregates were highly varied in size, ranging between 5 and 30 μm in diameter, and displayed an irregular staining pattern with DAPI.
FIG. 5.
Whole-cell FISH of putative methane-oxidizing consortia in methane seep sediments. (A) DAPI stain of aggregates from a 3- to 6-cm sediment depth from Eel River Basin cold seeps. Sediment preperations were simultaneously hybridized with a fluoroscein-labeled probe for Desulfosarcina-Desulfococcus members (DSS658) (B) and a Cy-3 labeled archaeal ANME-2-group specific probe (EelMSMX932) (C). (D) Overlay of both Cy-3 ANME-2 group (red) and fluoroscein Dulsulfosarcinales (green) results to show the architecture of the aggregate. Scale bar = 10 μm.
DISCUSSION
One of the more important biogeochemical processes influencing carbon turnover in continental margin environments is the anaerobic oxidation of methane. Although there is convincing biogeochemical evidence for archaeon and sulfate reducer cooperative involvement in AOM, identification of the potential organisms involved has been reported only very recently (4, 18). Combining environmental 16S rDNA surveys and whole-cell in situ hybridization with chemotaxonomic surveys of 13C-depleted sedimentary lipids from geographically distant methane seep sites, we confirm and extend previous observations of AOM consortia in marine sediments. Our data indicate the widespread occurrence of specific groups of methane-consuming archaea and sulfate-reducing bacteria involved in AOM. Our data also support preliminary phylogenetic evidence (4, 18) identifying some of the key microbial groups mediating this specialized process in the methane-rich, anaerobic environments.
Archaeal diversity within methane seeps.
Levels of total seep-related archaeal SSU rDNA diversity detected from both Eel River Basin and Santa Barbara Basin were very similar. Five major phylogenetic groups were represented and included sequences highly related to cultured Methanosarcinales, the newly described ANME-2 clade, phylotypes more distantly related to cultured members within the Methanosarcinales (ANME-1 group), and low-temperature relatives of the Thermoplasmales and Crenarchaeota. Comparisons to other 16S rDNA phylogenetic surveys of sedimentary archaea recovered from both seep and nonseep marine habitats suggest that the ANME-1 and ANME-2 groups are specific to anoxic seep-associated environments but that phylotypes related to the Thermoplasmales and low-temperature Crenarchaeota are more broadly distributed in marine sediments.
Coinciding with the archaeal genotypes recovered, sedimentary biomarkers related to the archaea were also abundant, comprised of at least four distinct lipids with extremely low levels of δ-13C (down to −129‰). Two of the most prominent biomarkers, sn-2-hydroxyarchaeol and PMI, are specifically abundant in members of the Methanosarcinales and were correlated with the recovery of Methanosarcinales-related ANME-2 rDNA clones from seep samples within Santa Barbara Basin (SB-pc24) and Eel River Basin (Eel-pc36). These findings strongly support the hypotheses that ANME-2 members are one source of the isotopically light archaeal lipids and that they actively consume methane within a variety of methane-rich anaerobic marine environments. Boetius et al. (4) came to similar conclusions based on FISH results in methane-rich sediments at Hydrate Ridge that also contain the same, isotopically light archaeal lipids. In addition, the uniform isotopic values and lipid concentrations in sedimentary vertical profiles suggest that the composition of methane-consuming archaea does not change significantly with depth. Similar 13C-depleted diphytanylglycerolethers (archaeol and sn-2-hydroxyarchaeol) have been previously detected in other methane-laden sedimentary environments, including seep sediments in the Mediterranean and at the Cascadia Margin, suggesting that Methanosarcinales relatives and, more specifically, members of the ANME-2 group may be active methane consumers in high-flux methane-rich marine environments worldwide (4, 19, 30). Furthermore, ANME-2-related phylotypes were previously recovered from highly reduced, methane-rich salt marsh sediments, suggesting that this group's involvement in AOM might extend to shallow, warmer water coastal flats as well (29). In addition to containing archaeol derivatives, selected samples from both the Santa Barbara and Eel River basins contained 13C-depleted crocetane, previously attributed to anaerobic methane-utilizing microorganisms (11, 42). Although microbial sources of crocetane are unknown, its structure with the tail-to-tail linkage of isoprene units suggests an archaeal origin, different from that of the producers of hydroxyarchaeol, which appear to exclusively synthesize regularly linked C20 isoprenoid chains (19, 42). The presence of both 13C-depleted glycerolethers and crocetane in methane seep sediments suggests that the anaerobic methanotrophic community is comprised of at least two different archaeal groups. Further investigation is necessary to determine whether crocetane is derived from members of the seep-associated archaeal group ANME-1 or from another archaeal source.
The total archaeal chemotaxonomic and SSU phylogenetic diversity found in this study was also in good agreement with earlier findings from an Eel River Basin methane seep site (18). In the previous study, however, the concentrations of the signature lipids were lower and the relative clone recovery of the putative methane-oxidizing groups, namely, ANME-1 and ANME-2 Methanosarcinales relatives, proved to be markedly different. In contrast to our findings where ANME-2-related clones made up a large majority of the 16S rDNA libraries, Hinrichs et al. (18) reported a predominance of clones (up to 84%) affiliated with the ANME-1 group, with a lower proportion of phylotypes clustering within the ANME-2 clade. This discrepancy in ANME-1 or ANME-2 representation may be due to differences in anaerobic methane-oxidizing activities or sedimentary methane or fluid flux or may represent methodological artifacts, for example, differential nucleic acid recoveries during extraction and purification, or biases in PCR-generated clone libraries (38). Although methodological biases or artifacts are possibilities, geochemical, chemotaxonomic and isotopic evidence also point to a highly active anaerobic methane-oxidizing community in samples where the ANME-2 phylotypes were predominant.
Putative sulfate-reducing syntrophic partners.
Substantial indirect evidence for the involvement of sulfate-reducing bacteria in AOM has been reported and is based on maximum sulfate reduction rates that coincide with maximum methane oxidation rates (4), high hydrogen sulfide levels (2), inhibition experiments (17, 21), linear or concave pore water sulfate profiles (5, 23), and increased numbers of cultivable sulfate-reducing bacteria in sediments where AOM occurs (48). An additional line of evidence is based on the detection of 13C-depleted fatty acids associated with sulfate-reducing bacteria, as well as extremely 13C-depleted archaeon-specific biomarkers, strongly suggesting that anaerobic methane oxidation is mediated by archaeal and sulfate reducer consortia (4, 19, 30). In all seep samples examined, methanotrophic archaeal biomarkers were generally more depleted in 13C than bacterial lipids, with both groups being significantly more depleted than photosynthetic products. Similar patterns of 13C depletion in archaeal and bacterial biomarkers were recently reported from Mediterranean mud volcanoes where AOM is presumed to occur (30). The presence of 13C-depleted bacterial lipids suggests that syntrophic sulfate-reducing partners likely assimilate a carbon intermediate, perhaps acetate or carbon dioxide produced by the methanotrophic archaea, in addition to participating in interspecies hydrogen transfer (19).
Chemotaxonomic, isotopic, and geochemical evidence strongly supports the involvement of sulfate-reducing bacteria in the anaerobic oxidation of methane as a functional group. The question still remains, however, as to whether all AOM consortia contain a highly specialized sulfate-reducing bacterial partner or whether the role of H2 (and/or acetate)-scouring syntrophic partners can be filled by a diversity of different sulfate-reducing bacteria. On a broad scale, the high abundance of clones related to sulfate-reducing δ-proteobacteria recovered from all seep-associated clone libraries was in good agreement with the geochemical and chemotaxonomic evidence. Much of the diversity associated with the δ-proteobacteria varied between sampling sites, although many phylotypes were related to environmental clones recovered from both seep-associated and nonseep marine sedimentary environments. Of the δ-proteobacterial phylotypes affiliated with the sulfate-reducing bacteria, phylotypes related to the Desulfosarcinales were predominant in both sites and formed a distinct cluster of highly related sequences that were exclusively recovered from seep environments. Included in the Desulfosarcinales cluster were sequences recovered from Japan Trench cold seep sediments and gas hydrate-bearing sediments from the Cascadia Margin (3, 26). In addition to being a common component of seep-associated bacterial communities, members of the Desulfosarcinales have also been shown to be dominant in other marine environments, comprising up to 73% of the total sulfate-reducing bacteria detected in shallow arctic sediments (32). In contrast to the novel archaeal groups ANME-1 and ANME-2 detected exclusively in methane-rich, anoxic habitats, seep-associated sulfate-reducing bacterial phylotypes are related to groups with a more widespread distribution in marine sediments.
Unique δ-proteobacterial groups (Eel-1, Eel-2, and Eel-3) were all recovered from a single seep sample (Eel-Hpc4) from the Eel River Basin. Eel-1 and Eel-2 were related to environmental sequences recovered from similar environments, including hydrocarbon seep and Guaymas Basin sediments (A. Teske, personal communication) and may directly or indirectly be associated with the anaerobic oxidation of methane. Alternatively, the Eel-1 group, whose closest relative was recovered from an anaerobic benzene-mineralizing enrichment, may possibly be involved in sulfate-mediated anaerobic hydrocarbon degradation (31). Although these groups comprised a significant fraction of the total bacterial diversity recovered in a particular sample (29% of the total clones), the limited detection of these groups overall makes it difficult to assess how significant they are in an AOM-based community.
Chemotaxonomic evidence for additional AOM bacterial groups.
Extremely 13C-depleted sedimentary bacterial lipids recovered from methane seeps at both locations were structurally diverse and likely originated from multiple bacterial sources. Whereas the fatty acid distribution in seep environments studied here is consistent with that of products from cultured sulfate-reducing bacteria (10, 24, 40), the alkylglycerolethers MAGE and DAGE are not known products from mesophilic sulfate reducers. The only reports of these types of ether lipids have been made for the most deeply branching thermophilic bacteria, Thermodesulfotobacterium commune (25) and Aquifex pyrophilus (22), suggesting that ether lipids in the bacterial kingdom may be limited to bacteria located close to the root of the phylogenetic tree (14). The absence of MAGE and DAGE in δ-proteobacteria studied to date implies that other microbes likely play an important role in AOM as well.
Implications of whole-cell in situ hybridization techniques.
Recently, Boetius et al. (4) using fluorescent whole-cell hybridization, described a close association between members of the Desulfosarcinales and archaea affiliated with the ANME-2 group in methane hydrate-containing sediments from Cascadia Margin. We also were able to demonstrate similar aggregations of Desulfosarcinales relatives and Methanosarcinales-related ANME-2 members in multiple samples from the Eel River Basin, where chemical, chemotaxonomic, and/or phylogenetic evidence of active anaerobic oxidation of methane was present (Fig. 1 and 5). The presence of aggregations of these two groups is not surprising since members of both the Desulfosarcinales and Methanosarcinales have aggregating qualities, often existing as sarcina-like cell clusters both in culture and in situ (32, 37). Coccoid cells hybridizing with the same Desulfosarcina-Desulfococcus-specific probe as that used in this study were previously detected in sulfidogenic sludge granules in association with methanogenic archaea (36). The physical association of sulfate-reducing Desulfosarcinales relatives and ANME-2 archaeal types suggests syntrophic cooperation between these two microbial groups and strongly supports the hypothesis that the mediation of methane oxidation in anaerobic methane seep sediments is controlled by a sulfate-reducing-bacterial–archaeal consortium. The fact that Desulfosarcina relatives have been shown to aggregate with the ANME-2 group in four separate seep locations—three sites in the Eel River Basin (this study) and one site from the Cascadia Margin (4)—implies a tight coupling between these two microbial types. Consistent with this association, 16S rDNA sequence types affiliated with these groups were relatively proportional in the bacterial and archaeal libraries. Samples dominated by ANME-2 phylotypes in the archaeal SSU clone library were also found to contain abundant Desulfosarcinales relatives in the bacterial SSU clone library (e.g., SB-24e), and in samples with low ANME-2 group recovery, the corresponding Desulfosarcinales-related phylotypes were less abundant (e.g., Eel-BE).
Possible mechanisms involved in AOM.
There is still much uncertainty regarding the specifics of the archaeal and sulfate-reducing-bacterial association in AOM and the mechanisms controlling this process (18, 21, 30, 45). In the context of our findings, members of the archaeal ANME-2 group fall within the Methanosarcinales, which are known utilizers of acetate and methylated compounds. Furthermore, members of the Methanosarcinales have been shown to produce minor amounts of acetate and methanol from methane oxidation in anaerobic culture-based experiments (49). This suggests the possibility that the ANME-2 group may be oxidizing methane to acetate and hydrogen (45) instead of the previously postulated CO2-H2 production (20). Such methanotrophic archaeal products may be efficiently channeled to the sulfate-reducing partner. According to Valentine and Reeburgh (45), the anaerobic oxidation of two molecules of methane to acetate and hydrogen under physical and chemical conditions typical of methane seep sites generates 35 kJ per mol of CH4, compared to 25 kJ per mol of CH4 with H2 and CO2 as end products. Cultured members of the Desulfosarcinales are capable of utilizing acetate and hydrogen, and uncultured relatives of Desulfosarcinales in multispecies sulfidogenic sludge granules were shown to actively consume acetate and hydrogen (36). If the seep-associated Desulfosarcinales relatives actively metabolize both acetate and hydrogen produced by methanotrophic archaea, this would explain the isotopically depleted δ-13C signature detected in bacterial lipids detected in the cold seep sites.
Conclusions.
Our culture-independent surveys of microbial assemblages within methane-rich sediments from geographically distant sites along the California Continental Margin have provided further insight into microbial diversity and population structure in these specialized anoxic habitats. Combining phylogenetic information and whole-cell in situ hybridization with chemotaxonomic and isotopic surveys of sedimentary lipids has allowed us to identify specific microorganisms associated with the anaerobic oxidization of methane. Independent lines of evidence obtained in this study strongly support the involvement of a syntrophic consortium, consisting of methanotrophic archaea related to the Methanosarcinales, and sulfate-reducing bacteria phylogenetically similar to the Desulfosarcinales in the mediation of methane oxidation in anaerobic methane-rich, marine sediments in a diversity of settings. Although ANME-2–Desulfosarcinales consortia appear to be widespread in shallow methane-rich marine systems, this association is probably only one element in the larger AOM puzzle; chemotaxonomic and isotopic evidence indicates that additional, as yet unidentified microbial groups are also involved in this process (19). To date, there has been no straightforward assignment of 16S rDNAs to producers of the 13C-depleted archaeal lipid crocetane or producers of the quantitatively significant bacterial ether lipids and related fatty alcohols and acids. However, phylogenetic 16S rDNA surveys from this study suggest that lineages such as the archaeal ANME-1 group and bacterial phylotypes related to candidate division OP-9 and δ-proteobacterial Eel-2 are specific to methane seeps and may also play a role in methane cycling in anoxic marine sediments. Further research needs to be conducted to determine the involvement of these microorganisms in AOM within shallow methane seep sites as well as in more steady-state deep subsurface systems. Preliminary work that builds on this study by combining FISH and ion microprobe mass spectrometry to obtain stable isotope readings of individual cell aggregates is now providing evidence for the involvement of specific phylogenetic groups in AOM (V. J. Orphan and C. H. House, unpublished data).
ACKNOWLEDGMENTS
Funding for this project was provided by the David and Lucile Packard Foundation and a NASA isotopic biogeochemistry grant, NAG5-9422, to J.M.H. K.-U.H. thanks the Hanse Institute of Advanced Study in Delmenhorst, Germany, for a fellowship, during which the manuscript was completed.
We thank Andreas Teske, Jon Martin, Jim Barry, and Thomas Naehr for graciously supplying data used in this study. We also thank Shana Goffredi for helpful comments on the manuscript; Josh Plant, Christopher Lovera, and the crew of the R.V. Point Lobos for their invaluable assistance in sample collection and processing; and A. Boetius and D. Valentine for sharing their unpublished manuscripts with us.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry J P, Kochevar R E, Baxter C H. The influence of pore-water chemistry and physiology on the distribution of vesicomyid clams at cold seeps in Monterey Bay: implications for patterns of chemosynthetic community organization. Limnol Oceanogr. 1997;42:318–328. [Google Scholar]
- 3.Bidle K, Kastner M, Bartlett D H. A phylogenetic analysis of microbial communities associated with methane hydrate containing marine fluids and sediments in the Cascadia margin (ODP site 892B) FEMS Microbiol Lett. 1999;177:101–108. doi: 10.1111/j.1574-6968.1999.tb13719.x. [DOI] [PubMed] [Google Scholar]
- 4.Boetius A, Ravenschlag K, Schubert C J, Rickert D, Widdel F, Gieseke A, Amann R, Jørgensen B B, Witte U, Pfannkuche O. Microscopic identification of a microbial consortium apparently mediating anaerobic methane oxidation above marine gas hydrate. Nature (London) 2000;407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 5.Borowski W, Paull C K, Ussler W. Global and local variations of interstitial sulfate gradients in deep-water, continental margin sediments: sensitivity to underlying methane and gas hydrates. Mar Geol. 1999;159:131–154. [Google Scholar]
- 6.Bowman J, Rea S M, McCammon S A, McMeekin T A. Diversity and community structure within anoxic sediment from marine salinity meromictic lakes and a coastal meromictic marine basin, Vestfold Hills, Eastern Antarctica. Environ Microbiol. 2000;2:227–237. doi: 10.1046/j.1462-2920.2000.00097.x. [DOI] [PubMed] [Google Scholar]
- 7.Burns S J. Carbon isotopic evidence for coupled sulfate reduction-methane oxidation in Amazon Fan sediments. Geochim Cosmochim Acta. 1998;62:797–804. [Google Scholar]
- 8.Cline J. Spectrophotometric determination of hydrogen sulphide in natural waters. Limnol Oceanogr. 1969;14:454–458. [Google Scholar]
- 9.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dowling N J E, Widdel F, White D C. Phospholipid ester-linked fatty acid biomarkers of acetate-oxidizing sulphate-reducers and other sulphide-forming bacteria. J Gen Microbiol. 1986;132:1815–1825. [Google Scholar]
- 11.Elvert M, Suess E, Whiticar M J. Anaerobic methane oxidation associated with marine gas hydrates: superlight C-isotopes from saturated and unsaturated C20 and C25 irregular isoprenoids. Naturwissenschaften. 1999;86:295–300. [Google Scholar]
- 12.Felsenstein J. PHYLIP-Phylogeny inference package (version 3.5) Cladistics. 1993;5:164–166. [Google Scholar]
- 13.Franzmann P D, Springer N, Ludwig W, de Macario C, Rohde M. A methanogenic bacterium from ACE Lake, Antarctica: Methanococcoides burtonii sp. nov. Syst Appl Microbiol. 1992;15:573–582. [Google Scholar]
- 14.Gambacorta A, Trincone A, Nicolaus B, Lama L. Unique features of lipids in Archaea. Syst Appl Microbiol. 1994;16:518–527. [Google Scholar]
- 15.Glöckner F, Fuchs B, Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray J P, Herwig R P. Phylogenetic analysis of bacterial communities in marine sediments. Appl Environ Microbiol. 1996;62:4049–4059. doi: 10.1128/aem.62.11.4049-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen L B, Finster K, Fossing H, Iversen N. Anaerobic methane oxidation in sulfate depleted sediments: effects of sulfate and molybdate additions. Aquat Microb Ecol. 1998;14:195–204. [Google Scholar]
- 18.Hinrichs K-U, Hayes J M, Sylva S P, Brewer P G, DeLong E F. Methane-consuming archaebacteria in marine sediments. Nature (London) 1999;398:802–805. doi: 10.1038/19751. [DOI] [PubMed] [Google Scholar]
- 19.Hinrichs K-U, Summons R E, Orphan V J, Sylva S P, Hayes J M. Molecular and isotopic analysis of anaerobic methane-oxidizing communities in marine sediments. Org Geochem. 2000;31:1685–1701. [Google Scholar]
- 20.Hoehler T M, Alperin M J. Anaerobic methane oxidation by a methangen-sulfate reducer consortium: geochemical evidence and biochemical considerations. In: Lidstrom M E, Tabita F R, editors. Microbial growth on C1 compounds. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. [Google Scholar]
- 21.Hoehler T M, Alperin M J, Albert D B, Martens C S. Field and laboratory studies of methane oxidation in an anoxic marine sediment: evidence for a methanogen-sulfate reducer consortium. Global Biogeochem Cycles. 1994;8:451–463. [Google Scholar]
- 22.Huber R, Wilharm T, Huber D, Trincone A, Burggraf S, Rachel R, Rockinger I, Fricke H, Stetter K O. Aquifex pyrophilus gen. nov., sp. nov., represents a novel group of marine hyperthermophilic hydrogen-oxidizing bacteria. Syst Appl Microbiol. 1992;15:340–351. [Google Scholar]
- 23.Iversen N, Jørgensen B B. Anaerobic methane oxidation rates at the sulfate-methane transition in marine sediments from Kattegat and Skagerrak (Denmark) Limnol Oceanogr. 1985;30:944–955. [Google Scholar]
- 24.Kaneda T. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol Rev. 1991;55:288–302. doi: 10.1128/mr.55.2.288-302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langworthy T A, Holzer G, Zeikus J G, Tornabene T G. Iso- and anteiso-branched glycerol diethers of thermophilic anaerobe Thermodesulfotobacterium commune. Syst Appl Microbiol. 1983;4:1–17. doi: 10.1016/S0723-2020(83)80029-0. [DOI] [PubMed] [Google Scholar]
- 26.Li L, Kato C, Horikoshi K. Microbial diversity in sediments collected from the deepest cold-seep area, the Japan Trench. Mar Biotechnol. 1999;1:391–400. doi: 10.1007/pl00011793. [DOI] [PubMed] [Google Scholar]
- 27.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acid Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martens C S, Berner R A. Interstitial water chemistry of anoxic Long Island Sound sediments. 1. Dissolved gases. Limnol Oceanogr. 1977;22:10–25. [Google Scholar]
- 29.Munson M A, Nedwell D B, Embley T M. Phylogenetic diversity of archaea in sediment samples from a coastal salt marsh. Appl Environ Microbiol. 1997;63:4729–4733. doi: 10.1128/aem.63.12.4729-4733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pancost R D, Sinninghe Damste J S, de Lint S, van der Maarel M J E C, Gottschal J C The Medinaut Shipboard Scientific Party. Biomarker evidence for widespread anaerobic methane oxidation in Mediterranean sediments by a consortium of methanogenic archaea and bacteria. Appl Environ Microbiol. 2000;66:1126–1132. doi: 10.1128/aem.66.3.1126-1132.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phelps C, Kerkhof L, Young L. Molecular characterization of a sulfate-reducing consortium which mineralizes benzene. FEMS Microb Ecol. 1998;27:269–279. [Google Scholar]
- 32.Ravenschlag K, Sahm K, Knoblauch C, Jørgensen B, Amann R. Community structure, cellular rRNA content, and activity of sulfate-reducing bacteria in marine Arctic sediments. Appl Environ Microbiol. 2000;66:3592–3602. doi: 10.1128/aem.66.8.3592-3602.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravenschlag K, Sahm K, Pernthaler J, Amann R. High bacterial diversity in permanently cold marine sediments. Appl Environ Microbiol. 1999;65:3982–3989. doi: 10.1128/aem.65.9.3982-3989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reeburgh W S. An improved interstitial water sampler. Limnol Oceanogr. 1967;12:163–165. [Google Scholar]
- 35.Reeburgh W S, Whalen S C, Alperin M J. The role of methylotrophy in the global methane budget. In: Murrell J C, Kelly D P, editors. Microbial growth on C1 compounds. Proceedings of the 7th International Symposium. Washington, D.C.: American Society for Microbiology; 1993. pp. 1–14. [Google Scholar]
- 36.Santegoeds C M, Damgaard L R, Hesselink G, Zopfi J, Lens P, Muyzer G, de Beer D. Distribution of sulfate-reducing and methanogenic bacteria in anaerobic aggregates determined by microsensor and molecular analyses. Appl Environ Microbiol. 1999;65:4618–4629. doi: 10.1128/aem.65.10.4618-4629.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sekiguchi Y, Kamagata Y, Nakamura K, Ohashi A, Harada H. Fluorescence in situ hybridization using 16S rRNA-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules. Appl Environ Microbiol. 1999;65:1280–1288. doi: 10.1128/aem.65.3.1280-1288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki M, Giovannoni S. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takai K, Horikoshi K. Genetic diversity of Archaea in deep-sea hydrothermal vent environments. Genetics. 1999;152:1285–1297. doi: 10.1093/genetics/152.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor J, Parkes R J. The cellular fatty acids of the sulphate-reducing bacteria, Desulfobacter sp., Desulfobulbus sp., and Desulvovibrio desulfuricans. J Gen Microbiol. 1983;129:3303–3309. [Google Scholar]
- 41.Thiel V, Peckmann J, Richnow H H, Luth U, Reitner J, Michaelis W. Molecular signals for anaerobic methane oxidation in Black Sea seep carbonates and a microbial mat. Mar Chem. 2001;73:97–112. [Google Scholar]
- 42.Thiel V, Peckmann J, Seifert R, Wehrung P, Reitner J, Michaelis W. Highly isotopically depleted isoprenoids: molecular markers for ancient methane venting. Geochim Cosmochim Acta. 1999;63:3959–3966. [Google Scholar]
- 43.Tyron M D, Brown K M, Torres M E, Trehu A M, McManus J, Collier R W. Measurement of transience and downward fluid flow near episodic methane gas vents, Hydrate Ridge, Cascadia. Geology. 1999;27:1075–1078. [Google Scholar]
- 44.Urakawa H, Tsukamoto K K, Ohwada K. Microbial diversity in marine sediments from Sagami Bay and Tokyo Bay, Japan, as determined by 16S rRNA gene analysis. Microbiology. 1999;145:3305–3315. doi: 10.1099/00221287-145-11-3305. [DOI] [PubMed] [Google Scholar]
- 45.Valentine D L, Reeburgh W S. New perspectives on anaerobic methane oxidation. Environ Microbiol. 2000;2:477–484. doi: 10.1046/j.1462-2920.2000.00135.x. [DOI] [PubMed] [Google Scholar]
- 46.Vetriani C, Jannasch H W, MacGregor B J, Stahl D A, Reysenbach A-L. Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl Environ Microbiol. 1999;65:4375–4384. doi: 10.1128/aem.65.10.4375-4384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vetriani C, Reysenbach A-L, Dore J. Recovery and phylogenetic analysis of archaeal rRNA sequences from continental shelf sediments. FEMS Microbiol Lett. 1998;161:83–88. doi: 10.1111/j.1574-6968.1998.tb12932.x. [DOI] [PubMed] [Google Scholar]
- 48.Wellsbury P, Goodman K, Cragg B, Parkes R. The geomicrobiology of deep marine sediments from Blake Ridge containing methane hydrate (sites 994, 995, and 997) Proc Ocean Drill Prog Sci Results. 2000;164:379–391. [Google Scholar]
- 49.Zehnder A J B, Brock T D. Methane formation and oxidation by methanogenic bacteria. J Bacteriol. 1979;137:420–432. doi: 10.1128/jb.137.1.420-432.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]