Abstract
Ultraviolet (UV) light can inactivate SARS-CoV-2. However, the practicality of UV light is limited by the carcinogenic potential of mercury vapor-based UV lamps. Recent advances in the development of krypton chlorine (KrCl) excimer lamps hold promise, as these emit a shorter peak wavelength (222 nm), which is highly absorbed by the skin's stratum corneum and can filter out higher wavelengths. In this sense, UV 222 nm irradiation for the inactivation of virus particles in the air and surfaces is a potentially safer option as a germicidal technology. However, these same physical properties make it harder to reach microbes present in complex solutions, such as saliva, a critical source of SARS-CoV-2 transmission. We provide the first evaluation for using a commercial filtered KrCl excimer light source to inactivate SARS-CoV-2 in saliva spread on a surface. A conventional germicidal lamp (UV 254 nm) was also evaluated under the same condition. Using plaque-forming units (PFU) and Median Tissue Culture Infectious Dose (TCID50) per milliliter we found that 99.99% viral clearance (LD99.99) was obtained with 106.3 mJ/cm2 of UV 222 nm for virus in DMEM and 2417 mJ/cm2 for virus in saliva. Additionally, our results showed that the UV 254 nm had a greater capacity to inactivate the virus in both vehicles. Effective (after discounting light absorption) LD99.99 of UV 222 nm on the virus in saliva was ∼30 times higher than the value obtained with virus in saline solution (PBS), we speculated that saliva might be protecting the virus from surface irradiation in ways other than just by intensity attenuation of UV 222 nm. Due to differences between UV 222/254 nm capacities to interact and be absorbed by molecules in complex solutions, a higher dose of 222 nm will be necessary to reduce viral load in surfaces with contaminated saliva.
Keywords: SARS-CoV-2, Disinfection, UV-C, 222 nm, Krypton chlorine lamp
1. Introduction
SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) is a respiratory virus responsible for the coronavirus disease 2019 (COVID-19) pandemic [1,2]. SARS-CoV-2 infects humans mainly through the contact of a healthy person's mucous membranes (e.g., lungs, eyes, mouth, and nose) with virus-containing droplets of saliva and sputum circulating in the air from a contaminated individual [3]. As COVID-19 evolves, the body's inflammatory response to the virus can dangerously harm the neurological, cardiac, gastrointestinal, and respiratory systems [4], with a wide variety of symptoms, such as fever, cough, dyspnea, nausea, and anosmia [4]. By the time this article was prepared, almost half a billion people were infected by the virus worldwide, with more than six million deaths associated with COVID-19 [5].
Although there has been significant progress in the development of vaccines and small molecule inhibitors, there is no cure for the disease, and there is much to be learned about the virus and COVID-19 [[6], [7], [8], [9]]. Many antivirals, antibodies, and other drugs have been tested via clinical trials, but only a few drugs have been approved by the FDA [10,11]. Moreover, due to the inadequate global distribution of approved drugs and vaccines, variants of concern can frequently appear [12,13]. In this sense, the continuous development of effective preventive methods, such as UV irradiation for the inactivation of virus particles in the air and surfaces, becomes vital.
UV radiation represents electromagnetic radiation with a wavelength from 100 to 400 nm. The UV spectra are commonly subdivided into UV-A (315-400 nm), UV-B (280-315 nm), and UV-C (100-280 nm); further technical subcategories of UV-C exist. Radiation-based disinfection methods rely on the already well-known germicide effect of UV-C light, as it can be applied to the inactivation of bacteria, bacterial spores, fungi, viruses (including coronavirus), and protists from the skin, object surfaces, water, and air [[14], [15], [16]]. UV inactivation is controlled by wavelength and total exposure (commonly referred to as UV dose). Germicidal efficacy varies with UV-C wavelength and depends on the targeted microbe or virus. Although the germicidal action varies even among microorganism species, a common feature is a local peak in the 260–270 nm region, where UV-C induces nucleic acid breakdown, resulting in the inactivation of the pathogen. The discovery of this process, coupled with readily available mercury vapor-based UV lamps, led to the popularity of upper-room ultraviolet germicidal irradiation (UVGI) systems (emitting predominantly at 254 nm), which can be found in heating and ventilation systems and air conditioning equipment, all of which are often used in hospitals and other public spaces [[17], [18], [19], [20]]. Although widespread, human exposure to mercury vapor-based UVGI systems should be restricted since they harm the corneal cells of the eyes and the outermost layer of skin cells, potentially causing cancer.
Wavelengths below 230 nm are generally highly effective for inactivation due to the absorbance of photons by both nucleic acids and proteins. Interestingly, however, due to its physical properties, UV light below 230 nm barely penetrates human tissues at a level similar to that of 254 nm wavelength, and it is often recommended as a potentially safer option as a germicidal technology [21]. Filtered KrCl excimer lamps emitting primarily at 222 nm are commercially available and have been reported to display comparable effective dose to 254 nm lamps regarding the inactivation of SARS-CoV-2 when either surface-dried or diluted in liquid [[22], [23], [24]]. However, while target pathogens can readily absorb this shorter UV-C wavelength, surrounding media can also readily attenuate this radiation, resulting in a higher irradiation dose. It is important to note that airborne viral pathogens are often contained within saliva and aerosol droplets, which also contain relatively high concentrations of unrelated proteins and other macromolecules. These compounds may limit the penetration of far UV-C radiation into human fluids relevant to SARS-CoV-2 transmission and may influence the delivery of photons to the target pathogens [25]. The effects of absorbance by natural human saliva proteins (and other constituents) are understood qualitatively. However, the direct comparison between the dose received by a solution with the target element and the effective dose (the dose able to inactivate the target after light absorbance through the liquid column) of UV 222 nm and 254 nm when SARS-CoV-2 is diluted in this vehicle remains elusive. We demonstrate that human saliva significantly attenuates the effects of UV 222 nm light, and a much higher dose of 222 nm light than of 254 nm light is required to achieve the same LD percentage. These results raise concerns about the practical use of this type of radiation for environmental disinfection.
2. Materials and methods
2.1. UV-C devices
Two test apparatuses were designed, optimized, fabricated, and calibrated to enable accurate and controlled UV-C (at 254 nm or 222 nm) treatment of test samples. The top chamber contained the UV-C light source, the electronic driver, and a chronometer to control the exposure times of the samples to the light. Samples were treated in the bottom chamber using UV-C light generated with a low-pressure mercury lamp 8 W HNS (Osram), generating a peak wavelength at 254 nm, or a 15 W Care222 B1 illuminator KrCl excimer lamp with a filtering system for wavelengths > 240 nm (Ushio). The irradiance levels for the lamps inside the treatment chamber were measured using calibrated UV-C sensor systems, HD2302.0 Delta OHM with detector LP 471 UV-C (260 nm) or UIT2400 Handheld Light Meter 222 nm radiometer, which provided irradiance patterns and levels from which optimal treatment locations could be derived. Additionally, the irradiation spectrum of the KrCl excimer lamp was measured using the spectrometer QEPro (Ocean Insights) from 200–950 nm with a 400 µm optical fiber XSR-UV–Vis (Ocean Insights). Data were acquired using Ocean View software in triplicate and are presented as the average of the three spectra (with standard deviation) collected by analysis.
2.2. Virus irradiance assays
The viral strain HIAE-02 SARS-CoV-2/SP02/human/2020/BRA (GenBank accession number: MT126808.1) was initially isolated from the first COVID-19 patient from Brazil and was kindly provided by Dr. Edison Durigon from USP-São Paulo. Viruses were propagated in Vero CCL81 cells cultivated in Dulbecco's modified Eagle's medium (DMEM; Cultilab, Campinas, Brazil) for 36-48 h and clarified by centrifugation. Virus titer was defined by PFU/mL in a biosafety level 3 laboratory (BSL3) laboratory. The confirmation of the identity of the virus and copy number determination were performed using RT–qPCR as described below. For the UV absorption studies utilizing human saliva, we collected and pooled 1 mL of saliva samples from 8 volunteers according to the principles of the Declaration of Helsinki previously approved by the ethics committees Centro Universitário de Paulínia - UNIFACP (CAAE: 46338521.3.0000.0123). DMEM and dermal cell basal medium (ATCC) were also used.
The virus stock had a concentration of 2×106 PFU/mL or 4×106 PFU/mL and was diluted in 1 mL of PBS 1X, DMEM, or natural human saliva from donors to a final concentration of 2×105 PFU/mL. The assays were performed in a 35 mm dishes. The human saliva was previously centrifuged at 805 x g for 10 min at 4°C before dilution to decrease viscosity. All irradiation experiments with SARS-CoV-2 were performed in a BSL3.
2.3. TCID50/mL determination assay
Vero CCL18 cells were plated in DMEM containing 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin in 96-well plates at 104 cells per well. After incubation for 24 h at 37°C and 5% CO2 to promote adhesion of the cells, the plate was taken to the BSL3 laboratory for infection with inactivated SARS-CoV-2 or was left untreated followed by treatment with UV-C 222 nm or 254 nm at different doses. Infection was performed in triplicate, starting with an initial MOI of 1, followed by three serial dilutions with a dilution factor of 10. Next, the cells were incubated with the virus (or only medium as a negative control) in DMEM without serum and antibiotics for 1 h at 37°C. After this period, the supernatant was removed, and fresh DMEM containing serum and antibiotics was added, followed by incubation for 48 h at 37°C and 5% CO2. Next, the cells were stained with 2 μM Hoechst-33342 (Invitrogen), followed by 4% methanol-free formaldehyde fixation. Data were collected with an Operetta plate reader microscope (PerkinElmer), and the analysis was performed with Columbus 2.4 software (PerkinElmer). To quantify the cytopathic effect (CPE) induced by SARS-CoV-2, images of all fields in the well were acquired, and the number and area of nuclei stained with Hoechst-33342 per well were determined. CPE was defined as the number of dead cells (total number of cells in nonirradiated control subtracted from the total number of cells in the irradiated condition) added to the number of sick cells (cells with condensed nuclei). To define the viral titer, the Sun method [26], modified by Kärber by incorporating the Bliss weighting method [27], was used to calculate the median lethal dose. This methodology gives the 99% infection dose (ID) value as:
The method used provides values identical to those of Spearman-Kärber but with the presentation of a 95% confidence interval (CI) through the calculation of the standard error (SE), as follows:
where p is the infection rate at a given dilution and n is the number of test units per dilution. TCID50/mL is the reciprocal value of ID50. A spreadsheet provided by Lei et al. [28] was used in this work. By applying the Poisson distribution, the TCID50/mL was multiplied by 0.7 to predict the PFU/ml.
2.4. PFU/mL determination assay
Vero CCL18 cells were plated in DMEM containing 10 % FBS and 1 % penicillin and streptomycin in 24-well plates at 104 cells per well. After 24 h at 37°C and 5 % CO2, the plate was taken to the BSL3 laboratory for infection with inactivated SARS-CoV-2 or lack of treatment followed by UV-C 222 nm or 254 nm treatment as described above. Infection was performed in triplicate, starting with an initial MOI of 1, followed by five serial dilutions with a dilution factor of 10. Cells were incubated with the virus (or only medium as a negative control) in serum-free DMEM and without antibiotics for 1 h at 37°C. After this period, the supernatant was removed, and 1 % CMC semisolid medium (1 % carboxymethylcellulose in DMEM) containing serum and antibiotics was added. Cells were incubated for 72 h at 37°C and 5 % CO2 for viral replication. The medium was discarded, and the cells were fixed in 8% methanol-free formaldehyde and stained with 1% methylene blue. Lysis plaques formed by the virus were counted, corrected for the dilution factor, and expressed as PFU (plaque-forming units) per mL, defined by the following formula:
VT = virus titer
PN = plaque number (at a chosen dilution)
VD = virus dilution chosen to count the plaques
V= inoculated volume
2.5. Lethal dose (LD) calculation
All assays were performed in two or three independent experiments, each performed in three technical replicates. First, the surviving fraction (surviving fraction, S) of the virus was calculated by dividing the titer number at each UV dose (TiterUVdose) by the titer at dose zero (TiterControl), namely, S = TiterUVdose/TiterControl. Then, a 1-phase exponential fitting using the [S] value and UV doses (D, mJ/cm2) was performed using GraphPad Prism. Using this approach, virus survival [S] was described by the equation:
Y = Y0e(-k*X) where Y = S, X = dose, Y0 is the value when X = 0, and k is the decay factor (inactivation rate) of the curve. The adjustment was performed considering that the curve plateaued at 0 (curve touches the X-axis for S = 0). The LD99.99 or UV dose that inactivated 99.99 % of the virus was calculated as:
LD99.99 = (1/k) x (ln(Y0/1–0.999))
The following equation was used to calculate the effective dose (De):
De = D x (1-e−(Abs 1cm x 2.303 x h) / Abs1cm x 2.303 x h) x DF x PDF where Abs1cm is the absorption value at a 1 cm path length, h is the liquid column in the assay, DF is the divergence factor as in [29], and PDF is the Petri dish factor as in a previous study [29].
2.6. RT–qPCR
An RNA/DNA shield (Zymo) was added to protect the viral RNA, and then lysis buffer (Zymo) was added to lyse cells and viral particles, releasing the genetic material of the virus. Using the manufacturer's recommendations, RNA extraction was performed with the Quick-RNA Viral Kit (Zymo). Quantification of SARS-CoV-2 RNA was performed according to the one-step RT–qPCR protocol of a previous report [30] using primers and probes for the virus gene E (forward primer: 5′- ACA GGT ACG TTA ATA GTT AAT AGC GT-3′, reverse primer: 5′-ATA TTG CAG CAG TAC GCA CAC A-3′, probe: 5′-6FAM-ACA CTA GCC ATC CTT ACT GCG CTT CG- QSY-3′). The reactions were performed in a final volume of 12 μL, with 3 μL of TaqMan fast virus 1-step master mix (Applied Biosystems), 800 nM of the primers (Exxtend), 400 nm of the probe (Thermo Fischer), and 6 μL of the sample diluted 100-fold in RNAse-free ultrapure water. The reaction cycle was performed using StepOne 7500 RT–PCR equipment (Applied Biosystems) and consisted of 1 cycle at 25°C for 10 min and 1 cycle at 95°C for 2 min followed by 45 cycles at 95°C for 5 s and 60°C for 30 s. Samples were analyzed in duplicate, and the wells containing water in place of samples were assessed as negative reaction controls. The copy number was obtained from a standard curve performed with different known concentrations of the SARS-CoV-2 E gene cloned into the pGEM vector and corrected for the dilution factor. The original E gene template was kindly provided by Dr. Marcio Chaim Bajgelman (CNPEM, Brazil) (data not published).
3. Results
3.1. UV-C irradiation device
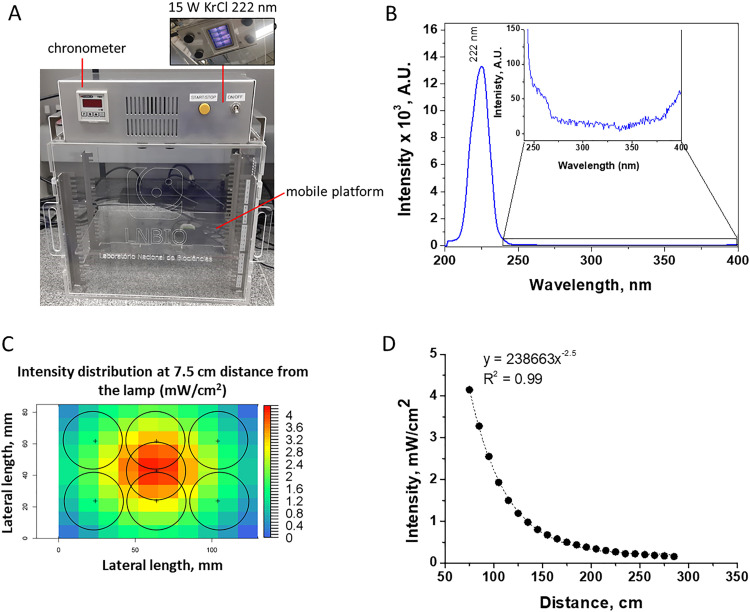
We built a UV-nontransparent acrylic box with the Care222 Filtered Far UV-C Excimer Lamp 12 W B1 module (Ushio) (Fig. 1 A) to perform the virus irradiation experiment. We also built a second similar device to perform control assays with a regular UV 254 nm low-pressure mercury lamp (data not shown). The box contained a mobile platform adjusted for the sample to receive the appropriate irradiation intensity (mW/cm2). In addition, a chronometer was connected to the device to control the irradiation time to obtain the planned irradiation dose (mJ/cm2). First, we characterized the irradiation spectrum of both lamps by using a luminometer, confirming the emission peak at the desired wavelengths (222 nm in Fig. 1B, data not shown for UV 254 nm). Notably, it was confirmed that Care222 did not have significant emission above 222 nm, precisely above 240 nm up to 400 nm, with the peak at ∼222 nm corresponding to 98% of the total emission in this range.
Fig. 1.
UV device built for the assays. (A) UV device built for 222 nm irradiation assays with the features described. (B) Light emission spectra of the KrCl excimer lamp; inset showing the intensity between 240–400 nm. (C) Irradiation intensity distribution along the seven 35 mm petri dish positions used for the assay, at 7.5 cm from the bottom of the lamp; each circle delineates the circumference of the dish. (D) Irradiation intensity variation as a function of the distance from the lamp at the central position is shown in (C).
Additionally, we used suitable radiometers to measure the irradiance intensity at marked areas on the platform. The diameter of the probes used in each radiometer was equivalent to that of the petri dish (35 mm) used in the assays, so the obtained value was an integration of the equivalent area (222 nm in Fig. 1C, data not shown for UV 254 nm). The measured irradiation intensity at different distances from the lamp was fitted in a first-order exponential equation (Fig. 1D).
3.2. Lethal dose of UV-C radiation at 222/254 nm for 99.99 % SARS-CoV-2 inactivation in different liquid vehicles
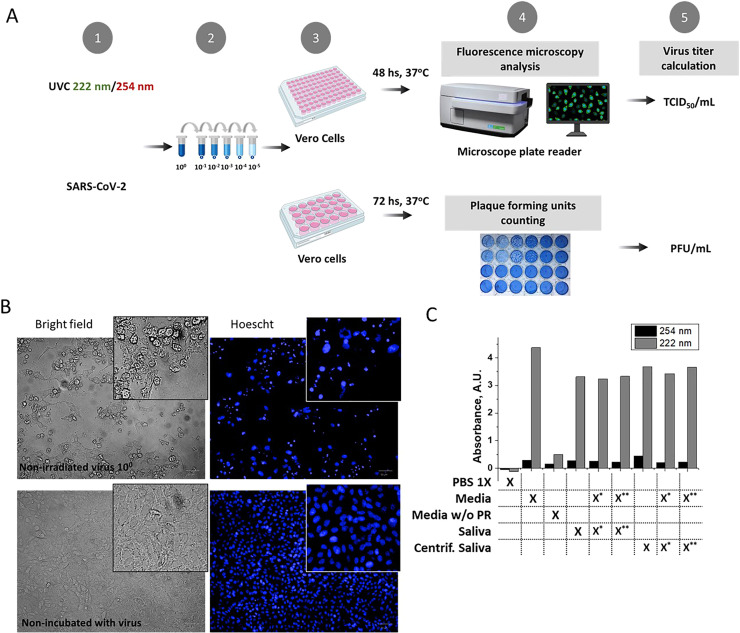
To obtain the lethal dose of UV-C at 222 nm and 254 nm required to inactivate 99.99% of SARS-CoV-2 in solution (LD99.99), the viral stock was serially diluted in cell culture media (DMEM), phosphate buffered saline (PBS), or pooled donor-derived human saliva and irradiated at the desired UV doses. The nonirradiated virus was used as the negative control. The virucide effect of UV-C was evaluated by two methodologies (Fig. 2 A), plaque-forming unit (PFU) and median tissue culture infectious dose (TCID50), and the viral titer was given as PFU/mL and TCID50/mL. The virus incubation induced the death of Vero cells and morphological alterations in the nuclei, mainly condensation (Fig. 2 B). This information was used to measure the cytopathic effect. Viral titers were used to calculate the surviving fraction (S), defined as the irradiated virus's viral titer divided by the nonirradiated virus's viral titer (NI). S values were plotted against the irradiation dose values, and a 1-phase exponential adjustment (setting the plateau at 0) was performed. The exponential equation obtained was used to calculate the LD99.99 values of UV radiation at 254 nm and 222 nm based on the values of k (inactivation rate) and Y0 (S when X (dose) = 0) obtained. The decay rate k and LD99.99 for each condition are displayed in Table 1 . LD99.99 values are presented in Table 2 . To account for UV absorption by the vehicle used in the assay, we measured this value for the different solutions (Fig. 2 C), and we used it to calculate the effective irradiation dose.
Fig. 2.
Scheme of the experimental approach used and vehicle UV absorption. (A) In step 1, the viral solution distributed in a 35 mm petri dish was irradiated with the UV 222 or 254 nm lamp or was untreated for the appropriate times. In step 2, the irradiated solution was serially diluted. Then, serial dilutions were inoculated into a 96-well (up to 10−3) or 24-well (up to 10−5) plate in Vero cells for one hour. After this process, viral solutions were removed, and the cells were incubated for the indicated times (step 3). Cells were evaluated by microscopy or visually inspected (step 4) for detection and calculation of the cytopathic effect or the ability to form plaques at different dilutions, respectively (step 5). (B). Representative bright-field images and Hoechst staining showing cells incubated with virus or untreated. (C) 222 nm and 254 nm light absorption values from a 1 mm path length (liquid column equivalent to the one obtained with 1 ml in a 35 mm petri dish) were measured for the different vehicles used for virus dilution: PBS 1X, DMEM (Media), dermal media (Media without – w/o – phenol red), saliva and centrifuged saliva. Virus stock (in DMEM) was mixed with saliva at different proportions (* saliva/media 10/1 v/v; ** saliva/media 20/1 v/v).
Table 1.
Values of k (inactivation rate) and Y0 were obtained with irradiation with 222 nm or 254 nm lamps and two different methodologies used to calculate virus titer. The results represent fitted values ± standard deviation (SD) from 2-4 biological replicates.
| Inactivation rate, k (cm2/mJ) |
|||||
|---|---|---|---|---|---|
|
PFU/ml |
TCID50/ml |
||||
| λ | Vehicle | k | SD | k | SD |
| 254 nm | DMEM | 1.0 | 0.1 | 0.91 | 0.07 |
| Saliva | 0.88 | 0.07 | 0.72 | 0.07 | |
| 222 nm | DMEM | 0.087 | 0.009 | 0.05 | 0.01 |
| Saliva | 0.004 | 0.002 | 0.0004 | 0.0002 | |
| PBS 1X | 1.6 | 0.4 | 1.4 | 0.2 | |
| Y0 | |||||
| PFU/ml | TCID50/ml | ||||
| λ | Vehicle | Y0 | SD | Y0 | SD |
| 254 nm | DMEM | 1.00 | 0.04 | 1.00 | 0.03 |
| Saliva | 1.000 | 0.003 | 1.000 | 0.007 | |
| 222 nm | DMEM | 1.00 | 0.03 | 1.0 | 0.1 |
| Saliva | 1.0 | 0.2 | 1.0 | 0.2 | |
| PBS 1X | 1.0 | 0.1 | 0.99 | 0.08 | |
Table 2.
LD99.99 was obtained with the two different methodologies used for virus titer calculation. The results are presented as the calculated values with 95% confidence interval (CI) values from 2-4 biological replicates. Importantly, the effective dose calculations considered the 222 nm and 254 nm light absorption values.
| LD99,99 (CI 95%), mJ/cm2 |
Effective LD99,99 (CI 95%), mJ/cm2 |
||||
|---|---|---|---|---|---|
| λ | Vehicle | PFU/mL, 95% CI | TCID50-PFU/mL, 95% CI | PFU/mL, 95% CI | TCID50-PFU/mL, 95% CI |
| 254 nm | DMEM | 8.8 (10.4 - 7.3) | 10.2 (11.2 - 9.1) | 5.6 (6.5 – 4.6) | 6.4 (7.0 – 5.8) |
| Saliva | 10.4 (11.4 - 9.0) | 12.8 (14.2 - 11.0) | 7.0 (7.7 – 6.1) | 8.7 (9.6 – 7.4) | |
| 222 nm | DMEM | 106.3 (120.0 - 92.1) | 176.8 (259.8 - 117.6) | 9.4 (10.6 – 8.1) | 15.6 (22.9 – 10.4) |
| Saliva | 2417 (6070 - 1127) | 20658 (38310 - 13183) | 277 (695 - 129) | 2368 (4391 – 1511) | |
| PBS 1X | 5.9 (8.2 - 4.0) | 6.7 (8.3 - 5.3) | 5.8 (8.1 – 3.9) | 6.5 (8.1 – 5.2) | |
4. Discussion
In this study, we used two methodologies (TCID50/mL and PFU/mL) for calculating the infective viral titer of SARS-CoV-2 to determine the radiation exposure dose (mJ/cm2) of UV 254 nm and UV 222 nm, which led to a 99.99% loss of infective capacity of virus diluted in liquid culture medium, in PBS, and human saliva. Using the PFU/mL values as a reference, we discovered that the LD99.99 values for UV 254 nm and UV 222 nm were 8.8 mJ/cm2 and 106.3 mJ/cm2, respectively, for SARS-CoV-2 diluted in DMEM. However, when the virus was diluted in human saliva, a dose of 2417.7 mJ/cm2 at 222 nm was necessary for 99.99% inactivation of the virus, while only 10.4 mJ/cm2 of UV 254 nm was required for the same effect. Curiously, it was recently published that saliva inactivate viruses from the Coronaviridae family (as HCoV-OC43) [31]. Applying absorption correction factors for the culture medium and saliva [32,33], the calculated effective LD99.99 of 222 nm was 9.3 mJ/cm2 and 318.4 mJ/cm2 SARS-CoV-2 when diluted in DMEM or saliva, respectively. Accordingly, in PBS, which has negligible absorption at 222 nm, only 5.9 mJ/cm2 UV 222 nm was required for 99.99 % viral clearance. Since the effective LD99.99 of UV 222 nm on virus diluted in saliva was approximately thirty times higher than the values obtained for the virus diluted in PBS, we speculated that components of the saliva might be protecting the virus from irradiation, probably in ways other than just by attenuating the UV 222 nm intensity. A recent work using artificial saliva and the Feline Infectious Peritonitis Virus showed no interference of the vehicle with UV 222 nm action, which may be explained by the composition of the artificial solution [34].
It was reported that the low concentrations observed in low contamination environments (multiplicity of infection, MOI = 0.05), the mean concentration found in sputum from COVID-19 patients (MOI = 5), and the concentration found in the sputum of terminally ill COVID-19 patients (MOI = 1000) require different doses of 254 nm UV radiant exposure for viral inactivation [35]. As an example of the effect of viral concentration on the demanded lethal dose, UV254 nm doses lower than 4 mJ/cm2 lead to complete virus inactivation at MOI = 0.05. In contrast, doses ≥ 16.7 mJ/cm2 are required to inactivate the virus at MOI = 1000 [36]. Furthermore, it is known that the sputum of patients with COVID-19 has, on average, 7×106 genomic copies/mL, with a maximum of 2.35×109 copies/mL [37]. Since our assays were carried out in viral solution with 2×105 PFU/mL (∼1.7×109 genomic copies/mL), our studies reveal irradiation doses for 99.99% viral annihilation at physiologically relevant viral concentrations.
Based on these results and the conditions of our experimental setup (15 cm away from a straight irradiation direction from a 15 W lamp in a UV nontransparent chamber), we can state that exposing a viral solution of 2×105/mL diluted in saliva would require 6 minutes of UV 254 nm irradiation and 25 minutes of 222 nm irradiation for 99 % viral inactivation. We provide the lethal dose equation for other calculations and the k and Y0 constant values (Table 1). In addition, we provide the equation that explains the intensity irradiation as a function of the lamp distance (Fig. 2C-D) measured approximately 90 degrees from the center of the lamp.
Recent work showed that doses of 1.7 and 1.2 mJ/cm2 UV 222 nm inactivated 99.9% of aerosolized alpha coronavirus HCoV-229E and beta HCoV-OC43, respectively [38]. Another work revealed that 1 and 3 mJ/cm2 222 nm UV irradiation resulted in 88.5 and 99.7 % reductions in viable SARS-CoV-2 (TCID50-based). In this study, the viral solution was allowed to dry on a surface and then was irradiated, although information about the initial vehicle of the virus was not provided [39]. Plaque infectivity assays demonstrated an effective DMEM-diluted SARS-CoV-2 LD90 of 1.6 mJ/cm2 of UV 222 nm [33]. Curiously, an inactivation of up to 99.99 % of SARS-CoV-2 was achieved with a dose of less than 8 mJ/cm2 with the virus diluted in DMEM [40]. The value found by Jung et al. [40] is very close to what we obtained when the effective dose was calculated (after discounting media light absorption) or when the virus was diluted in PBS.
In this study, we explored the effect of irradiation on SARS-CoV-2 diluted in human saliva. To the best of our knowledge, this is the first time that the effect of natural human saliva as a vehicle fluid was evaluated for UV 222 nm irradiation of SARS-CoV-2. Saliva comprises a variety of electrolytes, including sodium, potassium, calcium, magnesium, bicarbonate, and phosphates. In addition, proteins, such as enzymes, immunoglobulins, and mucins, and nitrogenous products, such as urea and ammonia, are also found in saliva [41]. Mucins serve to clean, aggregate or even bind to oral microorganisms. In this sense, saliva components highly absorb far-UV-C light due to specific chemical groups found amply in the components cited [42]. Of all vehicles tested in this project, however, DMEM was the vehicle with the highest UV absorption at 222 nm.
Respiratory fluids are expected to contain salts, proteins (mucins), and lung surfactants, and a study of the enveloped virus ϕ6 labeled with fluorescent lipids and mucin showed that viruses in a droplet do not associate with mucins [43]. Thus, the interaction of the SARS-CoV-2 virus with components of saliva and airborne respiratory fluids may be different, and the viral inactivation capacity of 222 nm light will vary significantly between these two vehicles. Furthermore, respiratory droplets have microscopic dimensions (< 1 µm) and, therefore, a very small optical path of UV light absorbing medium, which may potentiate the action of the light [44,45].
It is known that people become infected with SARS-CoV-2 through exposure to respiratory fluids carrying the infectious virus [4,46]. Exposure occurs through the inhalation of very fine respiratory droplets and aerosol particles; the deposition of respiratory droplets and particles on exposed mucous membranes in the mouth, nose, or eyes by direct splashes and sprays; and by touching mucous membranes with contaminated hands. In this sense, we believe that it is essential to evaluate the ability of lamps to inactivate the virus in its primary means of contamination: aerosol drops in a formulation of respiratory fluids. To this end, given the dangerousness of SARS-CoV-2, studies should be carried out with murine coronavirus variants, which are not dangerous to humans, from the beta coronavirus genus MHV, a virus recently shown to be a suitable study model to mimic SARS-CoV-2 [47]. In fact, factors such as the presence of encapsulation and the size of genetic material (and whether it is single or double-stranded) are the factors that will influence the dose required for viral inactivation.
5. Conclusions
-
•
Our studies of UV 222 nm inactivation of SARS-CoV-2 diluted in different vehicles showed the following:
-
•
A dose of 2417 mJ/cm2 UV 222 nm is required for 99.99% virus inactivation when viruses are diluted in natural human saliva, while a dose of only 10.4 mJ/cm2 of UV 254 nm is necessary to reach the same effectiveness.
-
•
After applying correction factors to account for the saliva UV 222 nm absorption and other light interferences, the effective LD99.99 of 222 nm is 318.4 mJ/cm2.
-
•
These results highlight that, due to the differences in the energy of both wavelengths and the consequent differences in their capacities to interact with molecules of the solution, a higher dose of UV 222 nm will be necessary to achieve the same effectiveness as that of UV-C 254 nm when viruses are present in complex solutions, such as saliva.
-
•
Since the effective LD99.99 of UV 222 nm on virus diluted in human saliva was still almost 30 times higher than the values obtained for the virus diluted in PBS, we speculated that components of the human saliva might protect the virus from irradiation, probably in ways other than just by attenuating the UV 222 nm intensity.
-
•
Our findings indicate that UV 222 nm is not the best option for sterilizing surfaces containing saliva contaminated with SARS-CoV-2, compared to UV 254 nm. Although UV 222 nm is significantly attenuated in virus diluted in saliva (with a liquid column of ∼1 mm), it will be worth studying the effect of this irradiation on a surrogate model of SARS-CoV-2 aerosolized in respiratory fluids to evaluate the effectiveness of this wavelength in such conditions.
Appendix A. Supplementary data
There is no appendix or supplementary data in this manuscript.
Authors contribution
RSC, CVZN and SMGD co-wrote the first draft which was revised by the entire group of authors. WC, TJS, AB, VCT built and characterized the devices. RSC, JFS, AN, RSNP, DA, MAF, AG, MC, and ALBA, performed the assays and/or analyzed data. SMGD and REM co-supervised the work. FG and JLPM provided access and supported activities at the BSL3 laboratory.
CRediT authorship contribution statement
Renata Sesti-Costa: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Cyro von Zuben Negrão: Writing – review & editing, Writing – original draft, Data curation. Jacqueline Farinha Shimizu: Writing – review & editing, Methodology, Investigation. Alice Nagai: Writing – review & editing, Methodology, Investigation. Renata Spagolla Napoleão Tavares: Writing – review & editing. Douglas Adamoski: Writing – review & editing, Methodology, Data curation. Wanderley Costa: Writing – review & editing, Methodology. Marina Alves Fontoura: Writing – review & editing, Methodology. Thiago Jasso da Silva: Writing – review & editing, Methodology. Adriano de Barros: Writing – review & editing, Methodology. Alessandra Girasole: Writing – review & editing, Methodology. Murilo de Carvalho: Writing – review & editing, Methodology, Data curation. Veronica de Carvalho Teixeira: Writing – review & editing, Methodology. Andre Luis Berteli Ambrosio: Writing – review & editing, Conceptualization. Fabiana Granja: Writing – review & editing, Resources, Methodology. José Luiz Proença-Módena: . Rafael Elias Marques: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Sandra Martha Gomes Dias: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Acknowledgements
We would like to acknowledge the financial support of Instituto Tecnológico Vale/Vale S.A., Embrapii, CNPEM and CIBFar (FAPESP CEPID 2013/07600-3).
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krishnan A., Hamilton J.P., Alqahtani S.A., A.Woreta T. A narrative review of coronavirus disease 2019 (COVID-19): clinical, epidemiological characteristics, and systemic manifestations. Internal Emergency Med. 2021;16:815–830. doi: 10.1007/s11739-020-02616-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samet J.M., Prather K., Benjamin G., Lakdawala S., Lowe J.M., Reingold A., Volckens J., Marr L.C. Airborne transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): what we know. Clin. Infect. Dis. 2021;73:1924–1926. doi: 10.1093/cid/ciab039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krishnan A., Hamilton J.P., Alqahtani S.A., A.Woreta T. A narrative review of coronavirus disease 2019 (COVID-19): clinical, epidemiological characteristics, and systemic manifestations. Internal Emergency Med. 2021;16:815–830. doi: 10.1007/s11739-020-02616-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO Coronavirus (COVID-19) Dashboard, Avaible from Https://Covid19.Who.Int/. Website Accessed in 01rs, April, 2022. (n.d.).
- 6.Schneider S.A., Hennig A., Martino D. Relationship between COVID-19 and movement disorders: a narrative review. Eur. J. Neurol. 2022;29:1243–1253. doi: 10.1111/ENE.15217. [DOI] [PubMed] [Google Scholar]
- 7.Chandra A., Johri A. A peek into pandora's box: COVID-19 and neurodegeneration. Brain Sci. 2022;12 doi: 10.3390/BRAINSCI12020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scapaticci S., Neri C.R., Marseglia G.L., Staiano A., Chiarelli F., Verduci E. The impact of the COVID-19 pandemic on lifestyle behaviors in children and adolescents: an international overview. Ital. J. Pediatr. 2022;48:22. doi: 10.1186/S13052-022-01211-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdi A., AlOtaiby S., Al Badarin F., Khraibi A., Hamdan H., Nader M. Interaction of SARS-CoV-2 with cardiomyocytes: insight into the underlying molecular mechanisms of cardiac injury and pharmacotherapy. Biomed. Pharmacother. 2022;146 doi: 10.1016/j.biopha.2021.112518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akinbolade S., Coughlan D., Fairbairn R., McConkey G., Powell H., Ogunbayo D., Craig D. Combination therapies for COVID-19: an overview of the clinical trials landscape. Br. J. Clin. Pharmacol. 2022;88:1590–1597. doi: 10.1111/bcp.15089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basu D., Chavda V.P., Mehta A.A. Therapeutics for COVID-19 and post COVID-19 complications: an update. Current Res. Pharmacol. Drug Discovery. 2022;3 doi: 10.1016/j.crphar.2022.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmad J., Ahmed S., Mir A., Shinde M., Bender O. The SARS-CoV-2 mutations versus vaccine effectiveness: new opportunities to new challenges. J. Infect. Public Health. 2022;15:228–240. doi: 10.1016/j.jiph.2021.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel R., Kaki M., Potluri V.S., Kahar P., Khanna D. A comprehensive review of SARS-CoV-2 vaccines: Pfizer, Moderna & Johnson & Johnson. Hum. Vaccin. Immunother. 2022;18:1–12. doi: 10.1080/21645515.2021.2002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rattanakul S., Oguma K. Inactivation kinetics and efficiencies of UV-LEDs against Pseudomonas aeruginosa, Legionella pneumophila, and surrogate microorganisms. Water Res. 2018;130:31–37. doi: 10.1016/j.watres.2017.11.047. [DOI] [PubMed] [Google Scholar]
- 15.Cheng Y., Chen H., Sánchez Basurto L.A., Protasenko V.V., Bharadwaj S., Islam M., Moraru C.I. Inactivation of listeria and E. coli by Deep-UV LED: effect of substrate conditions on inactivation kinetics. Sci. Rep. 2020;10:3411. doi: 10.1038/s41598-020-60459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D., Kang D. UVC LED irradiation effectively inactivates aerosolized viruses. Appl. Environ. Microbiol. 2018;84:1–11. doi: 10.1128/AEM.00944-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Orazio A., D'Alessandro D. Air bio-contamination control in hospital environment by UV-C rays and HEPA filters in HVAC systems. Annali DiIgiene. 2020;32:449–461. doi: 10.7416/AI.2020.2369. [DOI] [PubMed] [Google Scholar]
- 18.Song L., Li W., He J., Li L., Li T., Gu D., Tang H. Development of a pulsed xenon ultraviolet disinfection device for real-time air disinfection in ambulances. J. Healthc. Eng. 2020:2020. doi: 10.1155/2020/6053065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller S.L., Linnes J., Luongo J. Ultraviolet germicidal irradiation: future directions for air disinfection and building applications. Photochem. Photobiol. 2013;89:777–781. doi: 10.1111/PHP.12080. [DOI] [PubMed] [Google Scholar]
- 20.Kim D., Kang D. UVC LED irradiation effectively inactivates aerosolized viruses. Appl. Environ. Microbiol. 2018;84:1–11. doi: 10.1128/AEM.00944-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hessling M., Haag R., Sieber N., Vatter P. The impact of far-UVC radiation (200-230 nm) on pathogens, cells, skin, and eyes - a collection and analysis of a hundred years of data. GMS Hyg. Infect. Control. 2021;16:Doc07. doi: 10.3205/dgkh000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R.T. Robinson, N. Mahfooz, O. Rosas-Mejia, Y. Liu, N.M. Hull, SARS-CoV-2 disinfection in aqueous solution by UV<sub>222</sub> from a krypton chlorine excilamp, MedRxiv. (2021) 2021.02.19.21252101.
- 23.Kitagawa H., Nomura T., Nazmul T., Omori K., Shigemoto N., Sakaguchi T., Ohge H. Effectiveness of 222-nm ultraviolet light on disinfecting SARS-CoV-2 surface contamination. Am. J. Infect. Control. 2021;49:299–301. doi: 10.1016/j.ajic.2020.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung W.K., Park K.T., Lyoo K.S., Park S.J., Park Y.H. Demonstration of antiviral activity of far-uvc microplasma lamp irradiation against sars-cov-2. Clin. Lab. 2021;67:1955–1958. doi: 10.7754/Clin.Lab.2020.201140. [DOI] [PubMed] [Google Scholar]
- 25.Hessling M., Haag R., Sieber N., Vatter P. The impact of far-UVC radiation (200-230 nm) on pathogens, cells, skin, and eyes - a collection and analysis of a hundred years of data. GMS Hyg. Infect. Control. 2021;16 doi: 10.3205/dgkh000378. Doc07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.RY C.S. A practical combined method for computing the median lethal dose (LD50) Acta Pharm. Sin. 1963;10:65–74. [Google Scholar]
- 27.CI B. The determination of the dose-the proportion responding curve from small numbers. Q. J. Pharma Pharmacol. 1938;11:192–216. [Google Scholar]
- 28.Lei C., Yang J., Hu J., Sun X. On the calculation of TCID50 for quantitation of virus infectivity. Virol. Sin. 2021;36:141–144. doi: 10.1007/s12250-020-00230-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson R.T., Mahfooz N., Rosas-Mejia O., Liu Y., Hull N.M. SARS-CoV-2 disinfection in aqueous solution by UV222 from a krypton chlorine excilamp. MedRxiv. 2021 doi: 10.1126/science.372.6548.1301-a. [DOI] [Google Scholar]
- 30.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., Van Der Veer B., Van Den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enwemeka C.S., Bumah V.V., Castel J.C., Suess S.L. Pulsed blue light, saliva and curcumin significantly inactivate human coronavirus. J. Photochem. Photobiol. B. 2022;227 doi: 10.1016/j.jphotobiol.2021.112378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linden K.G., Darby J.L. Estimating effective germicidal dose from medium pressure UV lamps. J. Environ. Eng. 1997;123:1142–1149. doi: 10.1061/(ASCE)0733-9372(1997)123:11(1142). [DOI] [Google Scholar]
- 33.R.T. Robinson, N. Mahfooz, O. Rosas-Mejia, Y. Liu, N.M. Hull, SARS-CoV-2 disinfection in aqueous solution by UV222 from a krypton chlorine excilamp, MedRxiv. (2021). 10.1126/science.372.6548.1301-a.
- 34.Gardner A., Ghosh S., Dunowska M., Brightwell G. Virucidal efficacy of blue LED and far-UVC light disinfection against feline infectious peritonitis virus as a model for SARS-CoV-2. Viruses. 2021;13:1436. doi: 10.3390/v13081436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biasin M., Bianco A., Pareschi G., Cavalleri A., Cavatorta C., Fenizia C., Galli P., Lessio L., Lualdi M., Tombetti E., Ambrosi A., Redaelli E.M.A., Saulle I., Trabattoni D., Zanutta A., Clerici M. UV-C irradiation is highly effective in inactivating SARS-CoV-2 replication. Sci. Rep. 2021;11:1–7. doi: 10.1038/s41598-021-85425-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biasin M., Bianco A., Pareschi G., Cavalleri A., Cavatorta C., Fenizia C., Galli P., Lessio L., Lualdi M., Tombetti E., Ambrosi A., Redaelli E.M.A., Saulle I., Trabattoni D., Zanutta A., Clerici M. UV-C irradiation is highly effective in inactivating SARS-CoV-2 replication. Sci. Rep. 2021;11:1–7. doi: 10.1038/s41598-021-85425-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 38.Buonanno M., Welch D., Shuryak I., Brenner D.J. Far-UVC light (222 nm) efficiently and safely inactivates airborne human coronaviruses. Sci. Rep. 2020;10:1–8. doi: 10.1038/s41598-020-67211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kitagawa H., Nomura T., Nazmul T., Omori K., Shigemoto N., Sakaguchi T., Ohge H. Effectiveness of 222-nm ultraviolet light on disinfecting SARS-CoV-2 surface contamination. Am. J. Infect. Control. 2021;49:299–301. doi: 10.1016/j.ajic.2020.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jung W.K., Park K.T., Lyoo K.S., Park S.J., Park Y.H. Demonstration of antiviral activity of far-uvc microplasma lamp irradiation against sars-cov-2. Clin. Lab. 2021;67:1955–1958. doi: 10.7754/Clin.Lab.2020.201140. [DOI] [PubMed] [Google Scholar]
- 41.Barancheshme F., Philibert J., Noam-Amar N., Gerchman Y., Barbeau B. Assessment of saliva interference with UV-based disinfection technologies. J. Photochem. Photobiol. B. 2021;217 doi: 10.1016/j.jphotobiol.2021.112168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.E. Pretsch, P. (Philippe) Bühlmann, M. Badertscher, Structure determination of organic compounds : tables of spectral data, (2009) 433.
- 43.Vejerano E.P., Marr L.C. Physico-chemical characteristics of evaporating respiratory fluid droplets. J. R. Soc., Interface. 2018;15:1–10. doi: 10.1098/rsif.2017.0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papineni R.S., Rosenthal F.S. The size distribution of droplets in the exhaled breath of healthy human subjects. J. Aerosol Med. 1997;10:105–116. doi: 10.1089/jam.1997.10.105. [DOI] [PubMed] [Google Scholar]
- 45.Welch D., Buonanno M., Grilj V., Shuryak I., Crickmore C., Bigelow A.W., Randers-Pehrson G., Johnson G.W., Brenner D.J. Far-UVC light: a new tool to control the spread of airborne-mediated microbial diseases. Sci. Rep. 2018;8:1–7. doi: 10.1038/s41598-018-21058-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Basu D., Chavda V.P., Mehta A.A. Therapeutics for COVID-19 and post COVID-19 complications: an update. Current Res. Pharmacol. Drug Discovery. 2022;3 doi: 10.1016/j.crphar.2022.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pendyala B., Patras A., Pokharel B., D'Souza D. Genomic modeling as an approach to identify surrogates for use in experimental validation of SARS-CoV-2 and HuNoV inactivation by UV-C treatment. Front. Microbiol. 2020;11:1–9. doi: 10.3389/fmicb.2020.572331. [DOI] [PMC free article] [PubMed] [Google Scholar]