Abstract
16S ribosomal DNA terminal restriction fragment patterns from rat fecal samples were analyzed to track the dynamics of Lactobacillus acidophilus NCFM and discern bacterial populations that changed during feeding with NCFM. Lactobacillus johnsonii and Ruminococcus flavefaciens were tentatively identified as such bacterial populations. The presence of L. johnsonii was confirmed by isolation from feces.
Many efforts to study microbial communities in the gastrointestinal tract have focused on the large bowel and consequently on fecal samples (9, 10, 17, 22, 23, 24). Several methods for following the complex communities in the large bowel have been employed. However, because of the abundance and diversity of bacteria in feces, this has proved a difficult task. One method well suited for studying complex bacterial communities is the analysis of terminal restriction fragment (TRF) patterns (also known as terminal restriction fragment length polymorphism analysis), which can provide a rapid and reproducible means observing bacterial population dynamics. The goal of this report was to explore a method for analyzing TRF patterns that can be used to track the dynamics of specific populations of bacteria in complex communities such as those in feces.
TRF patterns are created by endonuclease digestion of DNA from a PCR using one fluorescently labeled primer. Only the terminal restriction fragments are visualized after electrophoretic separation on a DNA sequencing machine. When the target of PCR is 16S ribosomal DNA (rDNA), then the TRF pattern reflects the taxonomic diversity of the bacteria in the sample (7, 11). This method was used to compare bacterial communities from different environments with clear success (7, 10, 11, 12, 13, 15, 20). Comparing TRF patterns taken at different times can also monitor temporal changes in bacterial community structure.
While TRF pattern analysis allows rapid monitoring of environments over time and space, it does have drawbacks. The ability of TRF patterns to accurately describe complex bacterial communities is complicated by variations in the conserved 16S rDNA sequences commonly used as PCR priming sites. Primer selection can dramatically alter the picture that is presented in a TRF pattern, because only a fraction of the bacterial 16S rDNA sequences in a sample will be amplified. In this study we used primers shown to hybridize well with 90% (46f) and 99% (536r) of the ∼1,500 bacterial 16S rDNA sequences tested in a study by Brunk et al. (5). In addition to the coverage provided by PCR primers, there is the concern of TRF length overlap. Phylogenetically distant bacteria might produce TRFs of different lengths when digested with one endonuclease but result in TRFs of the same length when digested with a different enzyme. Thus, a more complete picture of the bacterial community is provided by an analysis of TRF patterns derived from multiple-enzyme digests. The use of several enzyme-derived TRF patterns can also provide data that allow the identification of bacteria involved in population shifts during a study (2, 3, 16).
The samples for this study came from an experiment reported by Rao et al. on colon carcinogenesis in rats fed L. acidophilus NCFM (18). Rao et al. showed that dietary NCFM suppressed the formation of precancerous lesions in the colons of rats injected with azoxymethane (18). The rats were injected with azoxymethane at 7 weeks and sacrificed after reaching 16 weeks of age. We received combined fecal samples collected at 7 and 16 weeks of age from three groups of rats (one cage per group). Each sample was stored at −70°C and consisted of pellets collected over a 24-h period from each cage (three rats per cage). Each group was fed the same diet except that the amount of NCFM was varied. Table 1 shows the sample names, diets, and ages of the rats these samples came from. This study used TRF pattern analysis to follow NCFM content in the feces and monitor changes in the fecal bacterial communities.
TABLE 1.
Rat fecal samples with respective diets as received from the American Health Foundation for use in this study
| Fecal sample | Age (wk) | Probiotics in the diet |
|---|---|---|
| YR 0% | 7 | Control |
| YR 2% | 7 | 2% L. acidophilus (4.2 × 109 CFU/g) |
| YR 4% | 7 | 4% L. acidophilus (8.4 × 109 CFU/g) |
| OR 0% | 16 | Control |
| OR 2% | 16 | 2% L. acidophilus (4.2 × 109 CFU/g) |
| OR 4% | 16 | 4% L. acidophilus (8.4 × 109 CFU/g) |
Creating TRF patterns for analysis.
Each sample was homogenized by pulverization under liquid nitrogen. DNA was extracted from 100 mg of sample using a MoBio (Solano Beach, Calif.) Ultraclean Soil DNA Kit by the manufacturer's protocol. Amplification of the template DNA was performed by using a 5′-FAM-labeled primer, 46f (5′-FAM-GCYTAACACATGCAAGTCGA; Applied Biosystems Inc., Fremont, Calif.), and unlabeled primer 536r (5′-GTATTACCGCGGCTGCTGG). Reactions were carried out in triplicate with the following reagents in 50-μl reaction mixtures: template DNA, 10 ng; 1× buffer (Promega); deoxynucleoside triphosphates, 0.6 mM; bovine serum albumin, 0.8 μg/liter; MgCl2, 3.5 mM; 46f, 0.2 μM; 536r, 0.2μM; and Taq DNA polymerase (Promega), 2 U. Reaction temperatures and cycling for fecal samples were as follows: 94°C for 2 min; 35 cycles of 94°C for 2 min, 48.5°C for 1 min, and 72°C for 1 min; and one cycle of 72°C for 10 min. Primers were removed and amplicons were concentrated using the MoBio PCR Clean-Up kit according to the manufacturer's protocol. Fluorescently labeled DNA (200 ng) was cut with one restriction endonuclease enzyme—MspI, DpnII, or HaeIII (2.0 to 4.0 U; New England Biolabs, Beverly, Mass.)—in the manufacturer's recommended reaction buffers. Reaction mixtures were incubated for 5 h at 37°C and then immersed in a 65°C water bath for 20 min. After ethanol precipitation the DNA was dissolved in 18 μl of formamide (Bio-Rad, Benecia, Calif.), with 1 μl each of Genescan Rox 500 (Applied Biosystems) and Rox 550-700 (BioVentures, Murfreesboro, Tenn.) size standards. The DNA was denatured at 95°C for 5 min and snap-cooled on ice. Samples were run on an ABI Prism 310 Genetic Analyzer. Genescan 3.1 software with a 50-U detection threshold, Local Southern size matching, and heavy smoothing were used to quantify the electropherogram output.
Sample data consisted of the size (base pairs) and peak area for each TRF peak in a pattern. Because the amount of DNA loaded on the capillary cannot be accurately controlled, the sum of all TRF peak areas in a pattern (total peak area) varied between TRF patterns. To compensate for this variation, it was necessary to normalize peak detection thresholds and peak areas. The peak detection threshold was normalized by creating artificial detection thresholds for each sample. The new threshold value was created by multiplying a pattern's relative DNA ratio (the ratio of total peak area in the pattern to the total peak area in the sample with the smallest total peak area) by 580 area units (the area of a peak at the 50-U detection threshold). TRF peaks with areas less than the new threshold value for a sample were removed from the data set (Table 2). Peak areas were then normalized by converting the value of each remaining peak area to parts per million of the new total area.
TABLE 2.
Example of truncation procedure used to determine smallest observable peak for each sample in a data set
| Sample | Total area (area units) | Ratio of total peak areas | Threshold value (area units) |
|---|---|---|---|
| Smallest | 200,000 | 1:1 | 580a |
| Big | 400,000 | 2:1 | 1,160 |
| Bigger | 2,000,000 | 10:1 | 5,800 |
Minimum detectable peak area with Genescan software.
Monitoring Lactobacillus acidophilus NCFM in feces.
All the TRF patterns for rats fed NCFM included a large TRF peak 2 to 3 bp smaller than that predicted for NCFM by rDNA sequence analysis (Table 3). A pure culture of NCFM produced the same size TRF peak as the one seen in the fecal patterns. This difference between predicted and observed TRF length has been described previously, though not fully explained (4, 7, 11, 13).
TABLE 3.
TRF peak matches to organisms from the RDP database
| TRF peak seta and organism | TRF matching length for:
|
||
|---|---|---|---|
| MspI | DpnII | HaeIII | |
| TRF peaks in the L. acidophilus set | 141 | 156 | 207 |
| Lactobacillus acidophilus NCFM | 144 | 158 | 209 |
| Lactobacillus crispatus | 143 | 157 | 208 |
| Lactobacillus amylovorus | 141 | 155 | 206 |
| TRF peaks in the L. johnsonii set | 149 | 287 | 293 |
| Lactobacillus acidophilus ssp. johnsonii | 151 | 288 | 294 |
| Lactobacillus gasseri | 151 | 288 | 294 |
| Lactobacillus johnsonii | 151 | 288 | 294 |
| TRF peaks in the R. flavefaciens set | 243 | 233 | 264 |
| Ruminococcus flavefaciens (5 strains) | 244 | 234 | 264 |
| Unidentified rumen bacterium (3 clones) | 244 | 234 | 264 |
| Ruminococcus flavefaciens (1 strain) | 246 | 236 | 266 |
| Unidentified rumen bacterium (1 clone) | 246 | 236 | 266 |
TRF peak sets (data in boldface type) were seen in TRF patterns (Fig. 3), while TRF lengths ascribed to organisms were predicted from 16S rDNA sequences in the RDP database.
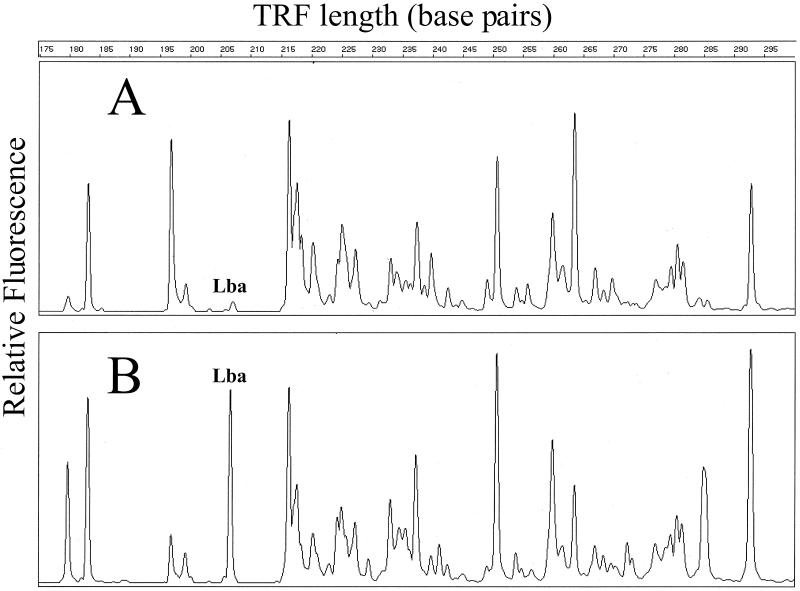
Visual inspection of the TRF patterns shows a large NCFM-associated peak in feces from rats fed NCFM, in contrast to a very small peak in those not fed NCFM (Fig. 1). Fortunately, there appeared to be very few bacteria with the same TRF peaks as NCFM in the feces of rats not fed NCFM (Table 4). Regardless of the enzyme used to create a TRF pattern, TRF peak areas increased with dietary NCFM content. In fact, the relative amount of peak area attributable to NCFM doubled in rats fed twice as much NCFM (Table 4). This does not necessarily imply that the feces harbored twice as many live cells of NCFM because PCR was performed on DNA extracted directly from the feces and could have been isolated from nonviable cells.
FIG. 1.
Close-up of HaeIII-derived TRF patterns from young rats in the study. The TRF patterns from feces of rats not fed NCFM (A) and from feces from rats fed 2% NCFM (B) are shown. Peaks of interest in this study are labeled for ease of discussion (Lba, L. acidophilus NCFM).
TABLE 4.
Peak areas from each fecal sample for each TRF peak set
| Fecal sample | Mean (SD) peak area (% of total area) when indicated enzyme was used
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
L. acidophilus TRF peak set
|
L. johnsonii TRF peak set
|
R. flavefaciens TRF peak set
|
|||||||
| Msp141 | Dpn156 | Hae207 | Msp149 | Dpn287 | Hae293 | Msp243 | Dpn233 | Hae264 | |
| YR 0% | 0.1 (0.0) | 0.0 (0.0) | 0.4 (0.4) | 1.8 (1.0) | 2.1 (0.4) | 3.0 (0.4) | 5.1 (1.5) | 4.3 (0.7) | 4.4 (1.4) |
| YR 2% | 9.5 (1.2) | 9.7 (4.1) | 6.1 (2.6) | 6.2 (1.9) | 6.7 (0.1) | 6.4 (0.3) | 1.8 (0.9) | 2.9 (0.9) | 2.9 (1.0) |
| YR 4% | 13.5 (3.3) | 15.3 (1) | 15.2 (10.8) | 1.8 (0.8) | 3.2 (0.6) | 2.5 (0.3) | 0.5 (0.2) | 1.2 (0.1) | 0.8 (0.4) |
| OR 0% | 0.0 (0.0) | 0.0 (0.0) | 0.2 (0.2) | 0.9 (0.1) | 1.7 (0.4) | 2.0 (0.7) | 4.3 (0.4) | 7.2 (0.4) | 9.6 (0.4) |
| OR 2% | 4.6 (1.3) | 8.5 (3.9) | 6.1 (1.3) | 1.1 (0.2) | 2.2 (0.4) | 1.9 (0.5) | 3.4 (0.4) | 6.1 (0.7) | 6.5 (1.7) |
| OR 4% | 8.7 (4.1) | 11.6 (1) | 9.6 (2.7) | 0.3 (.01) | 1.9 (1.8) | 0.5 (0.2) | 3.6 (2.1) | 7.1 (1.0) | 7.0 (0.5) |
The ability of TRF patterns to detect the relative amount of NCFM DNA in the feces suggests that TRF patterns may be used for the relative quantitation of some bacteria in complex communities. In fact, Clement and Kitts reported a one-to-one correspondence between the proportion of NCFM cells added to a fecal sample and the percentage of the total TRF peak area present as L. acidophilus TRF peaks (6). This correspondence may be influenced by both PCR primer homology and 16S rDNA copy number in L. acidophilus. Previous papers have postulated that an average bacterial community has 3.8 16S rDNA copies per genome (8) and L. acidophilus has four copies (19).
Monitoring changes in fecal communities.
While the number of samples collected in the study of Rao et al. was too small to produce statistically significant data (18), we were interested in testing methods for analyzing TRF patterns to detect specific organisms that contribute to differences in bacterial communities. A combination of principal component analysis (PCA) and database matching was investigated.
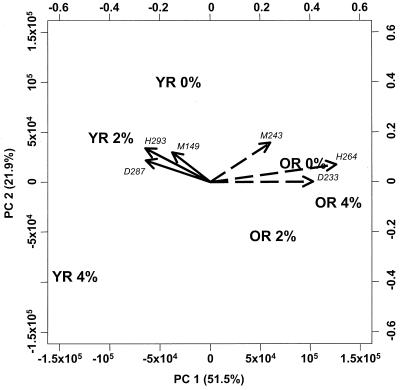
Covariance PCA (21) was performed on normalized data sets consisting of TRF peak areas combined from all three enzyme-derived TRF patterns. Samples from rats with NCFM in their diet (samples YR 2%, YR 4%, OR 2%, and OR 4% [Table 1]) were analyzed after removing peaks specific to NCFM to prevent PCA from separating the samples based solely on the dominant NCFM peaks. PCA produces a collection of loadings for each principal component that describes the relative contribution of each variable to the principal component score for a sample (21). This allows an investigator to focus on those variables (TRF peaks in this case) that contribute to differences in principal component scores between samples. The TRF peaks with large loadings can then be matched to database-predicted TRF lengths for presumptive identification of significant bacterial populations. The first principal component (PC1) appeared to separate samples based on the age of rats (sample group YR versus OR [Fig. 2]). The second principal component (PC2) appeared to separate samples by NCFM content (0% versus 2 and 4% [Fig. 2]). This suggested that some bacteria differed in levels of abundance in the rat feces depending on age and/or diet.
FIG. 2.
PCA of three combined TRF patterns for each fecal sample. Data for each sample consist of TRF peak areas from all patterns made after digestion with each of the three enzymes. Arrows represent principal component loadings for the TRF peak sets listed in Table 3: arrows with solid lines, L. johnsonii set; arrows with dashed lines, R. flavifaciens set. The TRF lengths (base pairs) and enzymes (M, MspI; D, DpnII; H, HaeIII) are indicated. Percent variation covered by each principal component is indicated in parentheses in the axis titles below and to the left, along with principal component scores. The right and top axes show values for principal component loadings.
For each sample, TRF pattern data from the three enzyme digests were used to look for matches to database-predicted TRF lengths. 16S rDNA sequences from the Ribosomal Database Project (RDP) 7.1 (14) were used to create a new database containing the calculated TRF peak sizes for 6,358 organisms after PCR with primers 46f and 536r. To identify bacteria in a fecal sample, each organism from the database was compared to the TRF peaks from three different enzyme digests using a Microsoft Excel macro. Differences are commonly reported between observed TRF lengths and those predicted by sequence analysis (5, 6, 11, 13). To compensate for this discrepancy, an observed TRF (from a sample) was allowed to be within +1 to −4 of the predicted TRF length (in the database).
TRF peaks with large, congruent loadings for PC1 were then used to search through database matches to identify presumptive bacterial species whose abundance differed with the age of the rats. Two sets of TRF peaks clearly fit these criteria and were thus chosen for closer investigation (Fig. 2). The TRF peak sets were named for bacteria in the database that matched the set (Table 3). We could now monitor two TRF peak sets that showed different dynamics during the study of Rao et al (18).
Lactobacillus johnsonii TRF peak set.
Peak areas from the L. johnsonii set were more abundant (1.8 to 6.7%) in young rats than in old rats (0.3 to 2.2%) as visualized in the PCA graph (Fig. 2). Peak areas decreased as the rats aged, regardless of dietary NCFM (Table 4). A similar decrease in NCFM peak areas was seen as the rats aged. Since both species of lactobacillus decreased in relative abundance, perhaps something changed in the older rat's cecal environment that made it less hospitable to lactobacillus species in general.
To confirm the presence of L. johnsonii in the feces, three different bacteria were isolated at random from two fecal samples (YR 0% and YR 4%). Rat fecal samples were diluted and plated on MRS agar (Difco, Sparks, Md.). Colonies were picked and streaked twice for purity. DNA from each isolate was then processed for TRF analysis as described above. All three bacteria produced TRF peaks identical to the L. johnsonii TRF set. Extracted DNA samples were amplified for sequencing by PCR as described above except that the forward primer was replaced with unlabeled 8df (5′-AGAGTTTGTTCMTGGCTCAG). Sequencing reaction mixtures (10 μl) contained DNA, 4 ng; primer, 1 μM; ABI Big Dye (Perkin-Elmer), 4 μl; and PCR water, 0.4 μl. Samples were run on an ABI 377 DNA sequencer, and the resulting sequences were analyzed in Autoassembler (Perkin-Elmer). A BLAST search of the sequences gave L. johnsonii as a 99% match. This indicates not only that L. johnsonii was actually present in the feces but also that it was numerically abundant enough to be easily isolated after the freezing and processing steps the feces were taken through.
Ruminococcus flavefaciens TRF peak set.
In seven-week-old rats the R. flavefaciens set represented an average of 5% of the total peak area in control rats, while those fed NCFM showed an average of 1.7%. By age 16 weeks, the average peak area of the R. flavefaciens set had increased to 6% in the NCFM-fed rats and 7% in the controls (Table 4). In contrast with L. johnsonii, this suggests that R. flavefaciens was adversely effected by NCFM at an early stage but eventually reached levels similar to those of the controls in spite of the continuing presence of NCFM. Perhaps NCFM altered the cecal environment in such a way that R. flavefaciens growth was inhibited in young rats. The increase in R. flavefaciens TRF peak areas to levels similar to those seen in control rats by 16 weeks of age suggests that this effect was only temporary. Perhaps the inhibitory effect against R. flavefaciens in young rats was diminished as the levels of NCFM dropped in the older rats. Rao et al. found that feeding NCFM correlated with a significant decrease in fecal β-glucuronidase activity (18). It is not unreasonable, therefore, to expect to find altered TRF peak areas corresponding to bacteria known to produce β-glucuronidase enzymes. Intriguingly, Ruminococcus species have been recently reported to express β-glucuronidase (1).
Conclusions.
Although no obvious mechanism behind the beneficial health effects seen in the Rao et al. study could be ascertained here, TRF patterns proved to be a useful tool for monitoring the effects of probiotic dietary supplements on bacterial community structure. TRF pattern analysis clearly has the ability to detect changes in bacterial communities due to the introduction of probiotic supplements. Unfortunately, the statistical significance of the observed differences in TRF patterns could not be accurately assessed in this case because the feces of three rats per treatment were combined during sampling. However, TRF patterns were able to identify organisms involved in the dynamics of bacterial community structure, a confirmation of suggestions by researchers using this tool (2, 3, 10, 16). The isolation of fecal L. johnsonii strains producing the exact TRF peaks seen in fecal TRF patterns after a database search predicted the presence of this bacterium confirmed the accuracy of this method. The ability to associate TRF peaks with organisms allowed a deeper understanding of bacterial community dynamics with some unforeseen results. The addition of L. acidophilus NCFM to the young rat's diet appeared to inhibit the growth of an R. flavifaciens strain and decrease β-glucuronidase activity (18). Why this happened, whether it is a reproducible effect, and what the significance of this interaction might be are now topics for investigation in a new study designed specifically to explore these findings.
Acknowledgments
Thanks are due to the Environmental Biotechnology Institute at Cal Poly San Luis Obispo for funding this study and to the Unocal Corporation for their support of TRF studies that has provided the foundation for this study at the EBI.
We also thank the staff at EBI that helped with the research: Raul Cano, Brian Clement, Tobe Cox, and Alice Hamrick.
REFERENCES
- 1.Akao T. Influence of various bile acids on the metabolism of glycyrrhizin and glycyrrhetic acid by Ruminococcus sp. PO1–3 of human intestinal bacteria. Biol Pharm Bull. 1999;22:787–793. doi: 10.1248/bpb.22.787. [DOI] [PubMed] [Google Scholar]
- 2.Avaniss-Aghajani E, Jones K, Chapman D, Brunk C. A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences. BioTechniques. 1994;17:144–149. [PubMed] [Google Scholar]
- 3.Avaniss-Aghajani E, Jones K, Holtzman A, Aronson T, Glover N, Boian M, Froman S, Brunk C F. Molecular technique for rapid identification of mycobacteria. J Clin Microbiol. 1996;34:98–102. doi: 10.1128/jcm.34.1.98-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhard A E, Field K G. Identification of non-point sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl Environ Microbiol. 2000;66:1587–1594. doi: 10.1128/aem.66.4.1587-1594.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunk C F, Avaniss-Aghajani E, Brunk C A. A computer analysis of primer and probe hybridization potential with bacterial small-subunit rRNA sequences. Appl Environ Microbiol. 1996;62:872–879. doi: 10.1128/aem.62.3.872-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clement B, Kitts C L. Isolating PCR quality DNA from human feces with a soil DNA kit. BioTechniques. 2000;28:640–646. doi: 10.2144/00284bm06. [DOI] [PubMed] [Google Scholar]
- 7.Clement B G, Kehl L E, DeBord K L, Kitts C L. Terminal restriction fragment patterns (TRFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J Microbiol Methods. 1998;31:135–142. [Google Scholar]
- 8.Fogel G B, Collins C R, Li J, Brunk C F. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb Ecol. 1999;38:93–113. doi: 10.1007/s002489900162. [DOI] [PubMed] [Google Scholar]
- 9.Franks A H, Harmsin H J M, Raangs G C, Jansen G J, Schut F, Welling G W. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-trageted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–3345. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leser T D, Lindecrona R H, Jensen T K, Jensen B B, Møller K. Changes in bacterial community structure in the colon of pigs fed different experimental diets and after infection with Brachyspira hyodysenteriae. Appl Environ Microbiol. 2000;66:3290–3296. doi: 10.1128/aem.66.8.3290-3296.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu W, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W, Marsh T L, Forney L J. Determination of the microbial diversity of anaerobic-aerobic activated sludge by a novel molecular biological technique. Water Sci Technol. 1998;37:417–422. [Google Scholar]
- 13.Lüdemann H, Arth I, Liesack W. Spatial changes in the bacterial community structure along a vertical oxygen gradient in flooded paddy soil cores. Appl Environ Microbiol. 2000;66:754–762. doi: 10.1128/aem.66.2.754-762.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maidak B L, Cole J R, Lilburn T G, Parker C T, Jr, Saxman P R, Stredwick J M, Garrity G M, Li B, Olsen G J, Pramanik S, Schmidt T M, Tiedje J M. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 2000;28:173–174. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsh T L, Liu W T, Forney L J, Cheng H. Beginning a molecular analysis of the eukaryal community in activated sludge. Water Sci Technol. 1998;37:455–460. [Google Scholar]
- 16.Marsh T L, Saxman P, Cole J, Tiedje J. Terminal restriction fragment length polymorphism analysis program, a Web-based research tool for microbial community analysis. Appl Environ Microbiol. 2000;66:3616–3620. doi: 10.1128/aem.66.8.3616-3620.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCartney A, Wenzhi W, Tannock G. Molecular analysis of the composition of the bifidobacterial and Lactobacillus microflora of humans. Appl Environ Microbiol. 1996;62:4608–4613. doi: 10.1128/aem.62.12.4608-4613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao C V, Sanders M E, Indranie C, Simi B, Reddy B S. Prevention of colonic preneoplastic lesions by the probiotic Lactobacillus acidophilus NCFMTM in F344 rats. Int J Oncol. 1999;14:939–944. doi: 10.3892/ijo.14.5.939. [DOI] [PubMed] [Google Scholar]
- 19.Roussel Y, Colmin C, Simonet J M, Decaris B. Strain characterization, genome size and plasmid content in the Lactobacillus acidophilus group hansen and mocquot. J Appl Bacteriol. 1993;74:549–556. [PubMed] [Google Scholar]
- 20.Scala D J, Kerkhof L J. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl Environ Microbiol. 2000;66:1980–1986. doi: 10.1128/aem.66.5.1980-1986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma S. Applied multivariate techniques. New York, N.Y: John Wiley and Sons; 1996. [Google Scholar]
- 22.Tannock G W, Munro K, Harmsen H J M, Welling G W, Smart J, Gopal P K. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl Environ Microbiol. 2000;66:2578–2588. doi: 10.1128/aem.66.6.2578-2588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van der Maarel M J E C, Artz R R E, Haanstra R, Forney L J. Association of marine Archaea with the digestive tracts of two marine fish species. Appl Environ Microbiol. 1998;64:2894–2898. doi: 10.1128/aem.64.8.2894-2898.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson K H, Blitchington R B. Human colonic biota studied by ribosomal DNA sequence analysis. Appl Environ Microbiol. 1996;62:2273–2278. doi: 10.1128/aem.62.7.2273-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]