Abstract
Plasmid profiling and amplified fragment length polymorphism (AFLP) analysis were used to genotype 50 Escherichia coli strains from poultry carcasses. Thirty different plasmid profiles were evident, and clustering of the AFLP data showed that they were a distinctly heterogeneous group of strains. Susceptibility testing against five antimicrobial agents used in the South African poultry industry showed all strains to be susceptible to danofloxacin and colistin, while the majority (96%) were resistant to two tetracyclines.
Escherichia coli forms part of the bacterial population of the chicken gastrointestinal tract. In poultry processing, E. coli is regarded as an indicator of fecal contamination (19). Levels of E. coli associated with poultry carcasses can increase or decrease during processing depending on factors such as levels of fecal contamination on live birds, length of time and temperature of scalding, efficiency of evisceration, bacterial load and temperature of the immersion chiller water, and hygienic practices in the abattoir (23). E. coli is also regarded as a major pathogen of worldwide importance in commercially produced poultry and can result in significant economic losses (20). Poultry-associated diseases caused by pathogenic E. coli strains include colibacilliosis and airsacculitis, which can cause high morbidity and mortality in poultry (20). To control and prevent poultry diseases, breeders are known to administer subtherapeutic and therapeutic levels of antimicrobial agents to chickens via feed and water (7). This practice also improves feed efficiency and accelerates weight gain (7). The administration of antimicrobial agents to poultry, however, has provided a selection pressure for antimicrobial resistance genes, and as a result, many bacteria associated with chickens and poultry meat are now resistant to antimicrobial agents (32, 36).
Several molecular typing techniques, including plasmid profiling, random amplified polymorphic DNA analysis, pulsed-field gel electrophoresis, and ribotyping have been used to characterize and determine epidemiological relationships of E. coli strains (1, 17, 26, 30, 34). Amplified fragment length polymorphism (AFLP) analysis, based on the principles of restriction fragment length polymorphism analysis and PCR amplification (25, 37), is a high-resolution typing method which has been used to differentiate between strains of Campylobacter jejuni and Campylobacter coli (9); E. coli O157:H7 (24), Helicobacter pylori (16), Streptococcus pyogenes (8), Pseudomonas fluorescens, and Pseudomonas putida (15); and Lactobacillus plantarum and Leuconostoc mesenteroides (27).
In this study, E. coli strains from poultry carcasses were analyzed to determine their susceptibilities to antimicrobial agents used in the South African poultry industry, and genetic relationships based on plasmid profiling and AFLP analysis.
Bacterial strains.
The 50 strains examined (Table 1) were obtained from a microbiological survey of a poultry abattoir where bacterial counts and populations associated with the neck skins of carcasses at six processing stages were determined (13, 14). The API 20E system (bioMérieux, Marcy l'Etoile, France) was used to confirm the identity of the E. coli strains. O- and K-antigen serogrouping of the strains was performed by the Onderstepoort Veterinary Institute of the Agricultural Research Council (Onderstepoort, South Africa). Standard E. coli antisera were used, excluding the antisera against antigens K21, K64, K65, K77, K92, and K100 through K102 (31). All 50 strains were O rough and K minus.
TABLE 1.
Summary of profiles of 50 E. coli strains from poultry carcasses
| Processing stage | Strain | Antimicrobial resistancea | Plasmid profile | AFLP profile |
|---|---|---|---|---|
| After defeathering (nb = 9) | 181 | CT, OT | P5 | A15 |
| 183 | CT, OT | P9 | A12 | |
| 184 | CT, OT | P2 | A9 | |
| 185 | CT, OT | P21 | A10 | |
| 186 | NM | P28 | A30 | |
| 187 | CT, OT | P17 | A8 | |
| 188 | CT, OT | P2 | A7 | |
| 189 | NM, CT, OT | P16 | A13 | |
| 192 | CT, OT | P19 | A13 | |
| Before evisceration (n = 9) | 193 | CT, OT | P14 | A13 |
| 195 | CT, OT | P3 | A13 | |
| 196 | CT, OT | P27 | A34 | |
| 197 | CT, OT | P15 | A4 | |
| 198 | CT, OT | P9 | A5 | |
| 199 | NM, CT, OT | P7 | A6 | |
| 201 | CT, OT | P26 | A14 | |
| 202 | NM, CT, OT | P29 | A3 | |
| 204 | CT, OT | P2 | A18 | |
| After evisceration (n = 12) | 205 | CT, OT | P2 | A18 |
| 206 | CT, OT | P14 | A17 | |
| 207 | CT, OT | P2 | A38 | |
| 208 | NM, CT, OT | P23 | A24 | |
| 209 | CT, OT | P19 | A9 | |
| 210 | NM | P30 | A32 | |
| 211 | CT, OT | P2 | A1 | |
| 212 | CT, OT | P19 | A2 | |
| 213 | CT, OT | P19 | A2 | |
| 214 | NM, CT, OT | P6 | A19 | |
| 215 | CT, OT | P18 | A16 | |
| 216 | CT, OT | P19 | A20 | |
| After spray washing (n = 11) | 217 | Susceptible | P27 | A31 |
| 218 | CT, OT | P13 | A21 | |
| 219 | CT, OT | P13 | A21 | |
| 220 | CT, OT | P24 | A36 | |
| 221 | NM | P1 | A37 | |
| 222 | CT, OT | P10 | A11 | |
| 224 | NM, CT, OT | P22 | A11 | |
| 225 | CT, OT | P12 | A22 | |
| 226 | CT, OT | P4 | A29 | |
| 227 | CT, OT | P4 | A33 | |
| 228 | CT, OT | P12 | A41 | |
| After immersion chilling (n = 8) | 231 | CT, OT | P2 | A40 |
| 232 | Susceptible | P25 | A35 | |
| 233 | NM, CT, OT | P2 | A27 | |
| 235 | CT, OT | P20 | A39 | |
| 236 | CT, OT | P7 | A28 | |
| 238 | CT, OT | P11 | A23 | |
| 239 | CT, OT | P2 | A15 | |
| 240 | CT, OT | P11 | A25 | |
| After packaging (n = 1) | 248 | CT, OT | P8 | A26 |
Abbreviations: CT, chlortetracycline; OT, oxytetracycline; NM, neomycin.
n, number of strains.
Strains were stored at −70°C in tryptone soya broth (TSB) (Oxoid, Basingstoke, United Kingdom) supplemented with 15% (vol/vol) glycerol.
Antimicrobial susceptibility testing.
MICs for the E. coli strains of the five antimicrobial agents used in the South African poultry industry were determined by the microdilution method according to the guidelines of the National Committee for Clinical Laboratory Standards (NCCLS) (29). Reference powders were kindly provided by Pfizer (Groton, Conn.) (danofloxacin) and Logos AgVet (Midrand, South Africa) (colistin sulfate, neomycin sulfate, chlortetracycline hydrochloride, and oxytetracycline base). Mueller-Hinton broth (Oxoid) was supplemented with cations, and concentrations of test strains were standardized to 5 × 105 CFU/ml (29). MICs were read after 18 h of incubation at 37°C. The MIC was interpreted as the lowest concentration that visibly inhibited growth. E. coli ATCC 25922 was used as the quality control reference strain (29).
MIC ranges and MICs at which 50 and 90% of the strains tested are inhibited (MIC50s and MIC90s, respectively) are shown in Table 2. MIC breakpoints for resistance and susceptibility have not been established by the NCCLS for any of the antimicrobial agents tested here. For purposes of this study, therefore, MIC breakpoints were assigned to each of the antimicrobial agents that were based on breakpoints established by the NCCLS for related antibiotics (29). The strains analyzed here were thus considered resistant when MICs were ≥4 μg/ml for danofloxacin and ≥16 μg/ml for neomycin, chlortetracycline, and oxytetracycline. An arbitrary MIC breakpoint for resistance to colistin of ≥16 μg/ml was used, since NCCLS interpretative standards have not been established for the polymyxin class of antibiotics, to which colistin belongs. Using these breakpoints, all but two of the strains (strains 217 and 232 [Table 1] were resistant to at least one and at most three of the antimicrobial agents. The majority (76%) of the strains were resistant to the two tetracyclines only, while 14% were resistant to the tetracyclines as well as neomycin. The remaining three isolates were resistant to neomycin only (Table 1). All the strains were susceptible to danofloxacin and colistin, with MIC90s of ≤0.125 and 1 μg/ml, respectively (Table 2). Similarly, Watts et al. (38) reported the MIC90 of danofloxacin for E. coli isolates of veterinary origin to be ≤0.015 μg/ml. Danofloxacin belongs to the new fluoroquinolone class of antimicrobials, which are highly effective against gram-negative bacilli (6, 12). Their use in the poultry industry, however, is thought to be inappropriate due to cross-resistance with fluoroquinolones used to treat important human enteric infections (10, 11). Fluoroquinolone resistance has been reported for Salmonella serotypes (21, 28), Campylobacter jejuni (10), and E. coli (11). The susceptibilities of the strains in this study to colistin were in agreement with those reported in a Spanish study where 468 E. coli strains of avian origin were susceptible to this antimicrobial agent (6). Resistance to colistin reportedly does not commonly develop in bacteria originally susceptible to this antimicrobial agent (22), which could possibly explain the narrow range and low MICs obtained for the E. coli strains in this study. Neomycin is an aminoglycoside and is primarily active against Escherichia spp., but it is also effective against other genera of the Enterobacteriaceae (22). In our study, 20% of the E. coli strains tested were resistant to this antimicrobial agent. Conversely, 90% of the strains were resistant to the two tetracyclines, chlortetracycline, and oxytetracycline (MIC90s of 128 and >512 μg/ml, respectively) (Table 2). This high level of resistance is of concern due to possible cross-resistance with antibiotics used in human medicine. Recent studies have suggested a link between the use of antimicrobial agents in poultry and other food-producing animals, and the emergence of human pathogens with decreased susceptibilities or complete resistance to antibiotics used for treatment of human infections (4, 5, 28).
TABLE 2.
MICs of five antimicrobial agents for 50 E. coli strains associated with poultry carcasses
| Antimicrobial agent | MIC (μg/ml)
|
||
|---|---|---|---|
| Range | 50%a | 90%b | |
| Danofloxacin | ≤0.125–1 | ≤0.125 | ≤0.125 |
| Colistin | 0.5–1 | 1 | 1 |
| Neomycin | 2–>512 | 8 | 16 |
| Chlortetracycline | 4–128 | 64 | 128 |
| Oxytetracycline | 4–>512 | >512 | >512 |
50%, MIC50.
90%, MIC90.
Antimicrobial resistance typing was a poor tool for differentiating between strains in this study since the majority (76%) shared the same profile, that is, resistance to the two tetracyclines (Table 1).
Plasmid profiles.
Plasmid DNA was extracted by the alkaline lysis method from overnight cultures grown in TSB at 37°C (18). Plasmids were separated on 0.8% agarose gels, viewed under UV transillumination, and photographed. Lactococcus lactis subsp. lactis DSM 4645 plasmids were used as molecular size markers (3).
All but one of the strains contained between one and six plasmids, with sizes ranging from 1.5 to 89 kb. One, two, or four plasmids were harbored by almost equal proportions of the strains (24, 28, and 24%, respectively). Overall, however, plasmid profiles obtained for all the strains were diverse, with 30 profiles emanating from the 50 isolates (Table 1). Twenty of these profiles were unique, while the remaining 10 were shared by at least two and at most nine strains. These nine strains contained a single 89-kb plasmid (profile P2 [Table 1]) which was also present in 86% of strains containing more than one plasmid. Profile P19 was shared by five strains, while profiles P4, P7, P9, P11 through P14, and P27 were shared by two strains each (Table 1).
Seven of the nine strains isolated from carcasses after the defeathering stage had different plasmid profiles, while the profiles of all the strains originating from carcasses before evisceration were different (Table 1). Conversely, 3 and 4 of the 12 strains from carcasses after evisceration shared profiles P2 and P19, respectively, while two strains each from carcasses after spray washing shared profiles P4, P12, and P13. Furthermore, three and two strains from carcasses after the immersion chilling stage displayed profiles P2 and P11, respectively (Table 1).
No apparent correlation was found between the plasmid profiles of the strains and their resistance patterns to the antimicrobial agents (Table 1).
AFLP analysis.
The NucleoSpin C & T kit (Macherey-Nagel, Düren, Germany) was used to extract genomic DNA from 1-ml cultures grown in TSB at 37°C for 18 h. DNA concentrations were estimated by agarose gel electrophoresis with diluted samples of λ DNA (Boehringer Mannheim GmbH, Mannheim, Germany). The AFLP ligation and preselective amplification kit (Perkin-Elmer, Foster City, Calif.) was used for AFLP reactions, which were each performed on 250 to 500 ng of DNA as described previously (15). Amplified fragments were separated on denaturing 4% polyacrylamide sequencing gels, which were run on a model S2 sequencing gel apparatus (Gibco, BRL Life Technologies, Gaithersburg, Md.) at 50 W with 1× Tris-borate-EDTA (TBE) buffer in the upper compartment and 1× TBE supplemented with 0.5 M sodium acetate in the lower compartment (2). AFLP fingerprints were detected by the modified silver staining method described previously (15). Gels were air dried overnight and then scanned with a Hewlett-Packard ScanJet IIcx scanner. AFLP patterns were analyzed with the GelCompar software (version 4.0; Applied Maths, Kortrijk, Belgium). Gels were normalized by including the 1-kb Plus ladder (Gibco) at four-lane intervals on every gel as a standard. After conversion, normalization, and background subtraction, levels of similarity between the AFLP fingerprints were calculated by using the Pearson product-moment correlation coefficient (r). Strains were clustered by using the unweighted pair group method with arithmetic averages (UPGMA) (33).
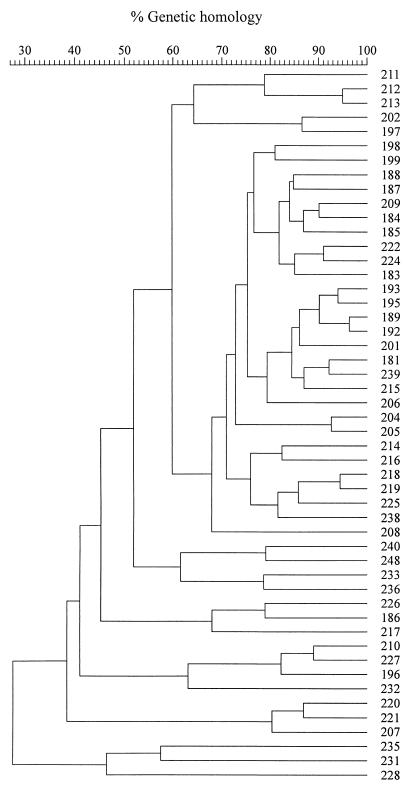
AFLP reactions generated between 26 and 44 detectable bands per strain. The AFLP fingerprints of one E. coli strain from carcasses after evisceration and three strains from carcasses after spray washing are shown in Fig. 1 as typical examples. The dendrogram generated by clustering of the AFLP data by UPGMA is shown in Fig. 2. Clearly, the AFLP fingerprints of the 50 strains analyzed were highly heterogeneous, with the highest level of homology observed between the strains being 96% and the lowest level of homology being 27%. Thus, none of the AFLP fingerprints were shown by the software to be 100% homologous, even though they appeared to be identical by visual inspection. Slight variations in band width and mobility as well as background intensities could explain this discrepancy. Similarity levels obtained here are thus not absolute but were nevertheless useful for determining relationships among the E. coli strains. For purposes of this study, therefore, strains whose AFLP patterns were >90% similar were assumed to be closely related genetically. At a delineation level of 90%, therefore, 41 different AFLP fingerprints were generated for the 50 E. coli strains (Table 1). Strains with homology levels of >90% included strains 212 and 213 from carcasses after evisceration, strains 218 and 219, as well as strains 222 and 224 from carcasses after spray washing (Fig. 2; Table 1). Strains isolated from carcasses at different stages of processing were also found to be genetically related. These included strains 181 and 239 from carcasses after defeathering and immersion chilling, respectively; strains 184 and 209 from carcasses after defeathering and evisceration, respectively; and strains 204 and 205 from carcasses before and after evisceration, respectively (Fig. 2; Table 1). Finally, strains 189, 192, 193, and 195 formed the largest cluster of related strains, with two of the strains originating from carcasses after defeathering and two strains from carcasses before evisceration (Fig. 2; Table 1). The heterogeneous nature of the AFLP fingerprints of the strains possibly indicates a large number of contamination sources of carcasses with E. coli. Sources could include the farm and processing environments as well as the processing equipment.
FIG. 1.
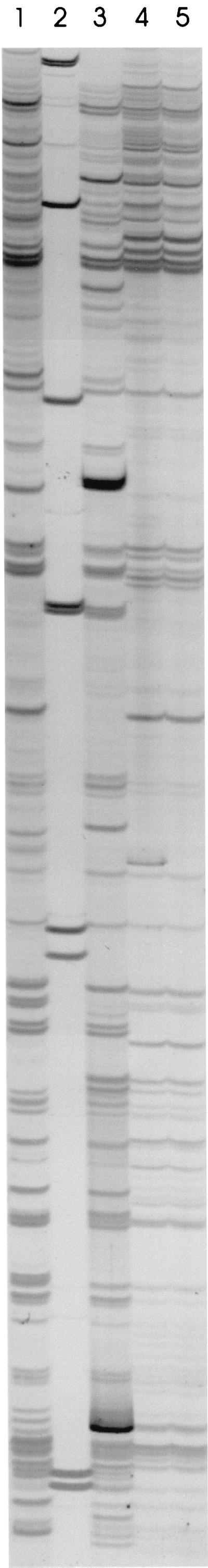
AFLP patterns of one E. coli strain from carcasses after evisceration (strain 216) and three strains from carcasses after spray washing (strains 217, 218, and 219). Lane 1, strain 216; lane 2, 1-kb Plus ladder; lane 3, strain 217; lane 4, strain 218; lane 5, strain 219.
FIG. 2.
Dendrogram based on AFLP fingerprints of 50 E. coli strains from poultry carcasses. The dendrogram was constructed by using UPGMA. Levels of similarity between AFLP fingerprints were calculated using the Pearson product-moment correlation coefficient.
Comparison of the plasmid and AFLP profiles obtained for each of the strains showed that in almost all cases, strains that shared plasmid profiles did not also share the same AFLP profiles (Table 1). Plasmid profiling has previously been shown to be of limited value as a genotyping method compared to other molecular typing techniques, mainly due to the instability of plasmids, poor reproducibility due to the variable presence of extra bands from open and linear forms of the plasmids, the presence of plasmids that appear to be similar, or simply the absence of plasmids in some of the strains (1, 35). In the present study, some of the strains that were regarded as genetically closely related by AFLP analysis also shared plasmid profiles, that is, strains 204 and 205 (level of homology, 93%), strains 212 and 213 (level of homology, 95%), and strains 218 and 219 (level of homology, 95%) (Fig. 2; Table 1). The remainder of the related strains, however, displayed different plasmid profiles. For instance, strains 189 and 192, whose AFLP fingerprints were 96% similar, differed not only in their plasmid profiles but also in their antimicrobial resistance profiles (Fig. 2; Table 1). In order for the AFLP technique to be sensitive enough to discern these polymorphisms in small genomes, such as those in bacteria, it may have to be modified to cover more alleles. This could be achieved by using a restriction endonuclease that cuts more frequently than the ones used in the present study.
In conclusion, the high-resolution genotyping method of AFLP analysis showed that the strains isolated from poultry carcasses during processing were genetically diverse. This suggests multiple sources of contamination of carcasses with E. coli. To pinpoint these sources, a study including isolates from the environment and equipment, as well as intestinal contents of carcasses and workers' hands, would have to be conducted.
Acknowledgments
Leigh Morgan is acknowledged for technical assistance with the AFLP analyses.
The National Research Foundation is acknowledged for financial support.
REFERENCES
- 1.Aarestrup F M, Jorsal S E, Ahrens P, Jensen N E, Meyling A. Molecular characterization of Escherichia coli strains isolated from pigs with edema disease. J Clin Microbiol. 1997;35:20–24. doi: 10.1128/jcm.35.1.20-24.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aarts H J M, van Lith L A J T, Keijer J. High-resolution genotyping of Salmonella strains by AFLP-fingerprinting. Lett Appl Microbiol. 1998;26:131–135. doi: 10.1046/j.1472-765x.1998.00302.x. [DOI] [PubMed] [Google Scholar]
- 3.Anderson D G, McKay L L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bager F, Madsen M, Christensen J, Aarestrup F M. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms. Prev Vet Med. 1997;31:95–112. doi: 10.1016/s0167-5877(96)01119-1. [DOI] [PubMed] [Google Scholar]
- 5.Bates J, Jordens J Z, Griffiths D T. Farm animals as a putative reservoir for vancomycin-resistant enterococcal infection in man. J Antimicrob Chemother. 1994;34:507–516. doi: 10.1093/jac/34.4.507. [DOI] [PubMed] [Google Scholar]
- 6.Blanco J E, Blanco M, Mora A, Blanco J. Prevalence of bacterial resistance to quinolones and other antimicrobials among avian Escherichia coli strains isolated from septicemic and healthy chickens in Spain. J Clin Microbiol. 1997;35:2184–2185. doi: 10.1128/jcm.35.8.2184-2185.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bower C K, Daeschel M A. Resistance responses of microorganisms in food environments. Int J Food Microbiol. 1999;50:33–44. doi: 10.1016/s0168-1605(99)00075-6. [DOI] [PubMed] [Google Scholar]
- 8.Desai M, Efstratiou A, George R, Stanley J. High-resolution genotyping of Streptococcus pyogenes serotype M1 isolates by fluorescent amplified-fragment length polymorphism analysis. J Clin Microbiol. 1999;37:1948–1952. doi: 10.1128/jcm.37.6.1948-1952.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duim B, Wassenaar T M, Rigter A, Wagenaar J. High-resolution genotyping of Campylobacter strains isolated from poultry and humans with amplified fragment length polymorphism fingerprinting. Appl Environ Microbiol. 1999;65:2369–2375. doi: 10.1128/aem.65.6.2369-2375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Endtz H P, Ruijs G J, van Klingeren B, Jansen W H, van der Reyden T, Mouton R P. Quinolone resistance in Campylobacter isolated from man and poultry following the introduction of fluoroquinolones in veterinary medicine. J Antimicrob Chemother. 1991;27:199–208. doi: 10.1093/jac/27.2.199. [DOI] [PubMed] [Google Scholar]
- 11.Garau J, Xercavins M, Rodríguez-Carballeira M, Gómez-Vera J R, Coll I, Vidal D, Llovet T, Ruíz-Bremón A. Emergence and dissemination of quinolone-resistant Escherichia coli in the community. Antimicrob Agents Chemother. 1999;43:2736–2741. doi: 10.1128/aac.43.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Rodríguez J A, Fresnadillo M J, García M I, García-Sánchez E, García-Sánchez J E, Trujillano I the Spanish Study Group on Quinolone Resistance. Multicenter Spanish study of ciprofloxacin susceptibility in gram-negative bacteria. Eur J Clin Microbiol Infect Dis. 1995;14:456–459. doi: 10.1007/BF02114906. [DOI] [PubMed] [Google Scholar]
- 13.Geornaras I, de Jesus A, van Zyl E, von Holy A. Microbiological survey of a South African poultry processing plant. J Basic Microbiol. 1995;35:73–82. doi: 10.1002/jobm.3620350204. [DOI] [PubMed] [Google Scholar]
- 14.Geornaras I, de Jesus A E, van Zyl E, von Holy A. Bacterial populations associated with poultry processing in a South African abattoir. Food Microbiol. 1996;13:457–465. [Google Scholar]
- 15.Geornaras I, Kunene N F, von Holy A, Hastings J W. Amplified fragment length polymorphism fingerprinting of Pseudomonas strains from a poultry processing plant. Appl Environ Microbiol. 1999;65:3828–3833. doi: 10.1128/aem.65.9.3828-3833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson J R, Slater E, Xerry J, Tompkins D S, Owen R J. Use of an amplified-fragment length polymorphism technique to fingerprint and differentiate isolates of Helicobacter pylori. J Clin Microbiol. 1998;36:2580–2585. doi: 10.1128/jcm.36.9.2580-2585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordillo M E, Reeve G R, Pappas J, Mathewson J J, DuPont H L, Murray B E. Molecular characterization of strains of enteroinvasive Escherichia coli O143, including isolates from a large outbreak in Houston, Texas. J Clin Microbiol. 1992;30:889–893. doi: 10.1128/jcm.30.4.889-893.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grattard F, Pozzetto B, Berthelot P, Rayet I, Ros A, Lauras B, Gaudin O G. Arbitrarily primed PCR, ribotyping, and plasmid pattern analysis applied to investigation of a nosocomial outbreak due to Enterobacter cloacae in a neonatal intensive care unit. J Clin Microbiol. 1994;32:596–602. doi: 10.1128/jcm.32.3.596-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grau F H. Microbial ecology of meat and poultry. In: Pearson A M, Dutson T R, editors. Advances in meat research. 2. Meat and poultry microbiology. Westport, Conn: AVI Publishing Company, Inc.; 1986. pp. 1–47. [Google Scholar]
- 20.Gross W G. Diseases due to Escherichia coli in poultry. In: Gyles C L, editor. Escherichia coli in domestic animals and humans. Wallingford, United Kingdom: CAB International; 1994. pp. 237–259. [Google Scholar]
- 21.Hakanen A, Siitonen A, Kotilainen P, Huovinen P. Increasing fluoroquinolone resistance in Salmonella serotypes in Finland during 1995–1997. J Antimicrob Chemother. 1999;43:145–148. doi: 10.1093/jac/43.1.145. [DOI] [PubMed] [Google Scholar]
- 22.Huber W G. Aminoglycosides, macrolides, lincosamides, polymyxins, chloramphenicol, and other antibacterial drugs. In: Booth N H, McDonald L E, editors. Veterinary pharmacology. 6th ed. Ames: Iowa State University Press; 1988. pp. 822–848. [Google Scholar]
- 23.International Commission on Microbiological Specifications for Foods. Microorganisms in foods. 6. Microbial ecology of food commodities. London, United Kingdom: Blackie Academic & Professional; 1998. [Google Scholar]
- 24.Iyoda S, Wada A, Weller J, Flood S J, Schreiber E, Tucker B, Watanabe H. Evaluation of AFLP, a high-resolution DNA fingerprinting method, as a tool for molecular subtyping of enterohemorrhagic Escherichia coli O157:H7 isolates. Microbiol Immunol. 1999;43:803–806. doi: 10.1111/j.1348-0421.1999.tb02473.x. [DOI] [PubMed] [Google Scholar]
- 25.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 26.Kariuki S, Gilks C, Kimari J, Obanda A, Muyodi J, Waiyaki P, Hart C A. Genotype analysis of Escherichia coli strains isolated from children and chickens living in close contact. Appl Environ Microbiol. 1999;65:472–476. doi: 10.1128/aem.65.2.472-476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunene N F, Geornaras I, von Holy A, Hastings J W. Characterization and determination of origin of lactic acid bacteria from a sorghum-based fermented weaning food by analysis of soluble proteins and amplified fragment length polymorphism fingerprinting. Appl Environ Microbiol. 2000;66:1084–1092. doi: 10.1128/aem.66.3.1084-1092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malorny B, Schroeter A, Helmuth R. Incidence of quinolone resistance over the period 1986 to 1998 in veterinary Salmonella isolates from Germany. Antimicrob Agents Chemother. 1999;43:2278–2282. doi: 10.1128/aac.43.9.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7–A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 30.Oethinger M, Conrad S, Kaifel K, Cometta A, Bille J, Klotz G, Glauser M P, Marre R, Kern W V The International Antimicrobial Therapy Cooperative Group of the European Organization for Research and Treatment of Cancer. Molecular epidemiology of fluoroquinolone-resistant Escherichia coli bloodstream isolates from patients admitted to European cancer centers. Antimicrob Agents Chemother. 1996;40:387–392. doi: 10.1128/aac.40.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ørskov F, Ørskov I. Serotyping of Escherichia coli. Methods Microbiol. 1984;14:43–112. [Google Scholar]
- 32.Quednau M, Ahrné S, Petersson A C, Molin G. Antibiotic-resistant strains of Enterococcus isolated from Swedish and Danish retailed chicken and pork. J Appl Microbiol. 1998;84:1163–1170. doi: 10.1046/j.1365-2672.1998.00463.x. [DOI] [PubMed] [Google Scholar]
- 33.Sneath P H A, Sokal R R. Numerical taxonomy: the principles and practice of numerical classification. W. H. San Francisco, Calif: Freeman; 1973. [Google Scholar]
- 34.Tarkka E, Åhman H, Siitonen A. Ribotyping as an epidemiological tool for Escherichia coli. Epidemiol Infect. 1994;112:263–274. doi: 10.1017/s0950268800057678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsen H-Y, Lin J S, Hu H H, Liu P R, Wang T K. Use of pulsed field gel electrophoresis as an epidemiological tool for analysis of sporadic associated strains of Salmonella typhi isolated in Taiwan. J Appl Microbiol. 1999;86:761–768. doi: 10.1046/j.1365-2672.1999.00720.x. [DOI] [PubMed] [Google Scholar]
- 36.Turtura G C, Massa S, Ghazvinizadeh H. Antibiotic resistance among coliform bacteria isolated from carcasses of commercially slaughtered chickens. Int J Food Microbiol. 1990;11:351–354. doi: 10.1016/0168-1605(90)90029-5. [DOI] [PubMed] [Google Scholar]
- 37.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watts J L, Salmon S A, Sanchez M S, Yancey R J., Jr In vitro activity of premafloxacin, a new extended-spectrum fluoroquinolone, against pathogens of veterinary importance. Antimicrob Agents Chemother. 1997;41:1190–1192. doi: 10.1128/aac.41.5.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]