Abstract
Aberrant expression of adenosine triphosphate-binding cassette subfamily C (ABCC), one of the largest superfamilies and transporter gene families of membrane proteins, is associated with various tumors. However, its relationship with liver hepatocellular carcinoma (LIHC) remains unclear.
We used the Oncomine, UALCAN, Human Protein Atlas, GeneMANIA, GO, Kyoto Encyclopedia of Genes and Genomes (KEGG), TIMER, and Kaplan–Meier Plotter databases. On May 20, 2021, we searched these databases for the terms ABCC1, ABCC2, ABCC3, ABCC4, ABCC5, ABCC6, ABCC7, ABCC8, ABCC9, ABCC10, ABCC11, ABCC12, ABCC13, and “liver cancer.” The exposure group comprised LIHC patients, and the control group comprised normal patients (those with noncancerous liver tissue). All patients shown in the retrieval language search were included. We compared the mRNA expression of these proteins in LIHC and control patients to examine the potential role of ABCC1–13 in LIHC.
Relative to the normal liver tissue, mRNA expression of ABCC1/2/3/4/5/6/10 was significantly upregulated (P < .001), and that of ABCC9/11 significantly downregulated (both P < .001), in LIHC. ABCC mRNA expression varied with gender (P < .05), except for ABCC11–13; with tumor grade (P < 0.05), except for ABCC7/12/13; with tumor stage (P < .05), except for ABCC11–13; and with lymph node metastasis status (P < .05), except for ABCC7/8/11/12/13. Based on KEGG enrichment analysis, these genes were associated with the following pathways: ABC transporters, Bile secretion, Antifolate resistance, and Peroxisome (P < .05). Except for ABCC12/13, the ABCCs were significantly associated with B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil, and dendritic cell infiltration (P < .05). High mRNA expression of ABCC1/4/5/8 (P < .05) and low expression of ABCC6/7/9/12/13 (P < .05) indicated poor prognosis. Prognostic significance was indicated for ABCC2/13 for both men and women (P < .05); for ABCC1/6/12/13 for tumor grades 1–3 (P < .05); for ABCC5/11/12/13 for all tumor stages (P < .05); for ABCC1/11/12/13 for American Joint Committee on Cancer T stages 1–3 (P < .05); and for ABCC1/5/6/13 for vascular invasion. None showed prognostic significance for microvascular invasion (P < .05).
We identified ABCC1/2/3/4/5/6/9/10/11 as potential diagnostic markers, and ABCC1/4/5/6/7/8/9/12/13 as prognostic markers, of LIHC. Our future work will promote the use of ABCCs in the diagnosis and treatment of LIHC.
Keywords: ABCC family, bioinformatics, diagnostic marker, liver hepatocellular carcinoma, prognostic marker
1. Introduction
Liver hepatocellular carcinoma (LIHC), the fourth leading cause of cancer-related deaths worldwide, accounts for 90% of primary liver cancers. In 2018, it caused ca. 625,000 deaths worldwide.[1,2] LIHC is caused by factors such as hepatitis B, hepatitis C virus infection, and aflatoxin exposure. Its onset is insidious, with no obvious symptoms; it is usually discovered in the middle and late stages, when the best opportunity for surgical resection, the most frequently recommend treatment, has already passed. Early detection is therefore important. Other current typical treatments include antiviral therapy, liver transplantation, and chemotherapy. In spite of the available treatments, prognosis remains poor,[3] and screening of novel LIHC biomarkers is required to improve early diagnosis and prognosis.[4] AFP performs poorly as a marker for LIHC detection, increasing the rate of missed diagnoses. Molecular targeted therapy and immunotherapy for LIHC have emerged as research hotspots. For example, therapies targeting Bak1, TAA, and other proteins have therapeutic effects on LIHC, but need to be improved.[5,6]
The adenosine triphosphate-binding cassette subfamily C (ABCC) superfamily is another promising target, Its aberrant expression is associated with various tumors.[10–14] It consists of 48 ABCC transporters, being one of the largest superfamilies of membrane proteins in prokaryotes and eukaryotes.[7] It is also the largest transporter gene family: the members bind with ATP and use this energy to drive the transport of sugars, metal ions, compounds, and other molecules.[8] There are 7 sub-families, ABCA, ABCB, ABCC (ABCC1–13),[9] ABCD, ABCE, ABCF, and ABCG. ABCC1 (also known as MRP1) can promote the excretion of heterogeneous and endogenous organic anions, and confer multidrug resistance via the efflux of active drugs, thus protecting human organs and tissues from cytotoxicity. ABCC1 is associated with progression and drug resistance in various cancers, including LIHC, prostate cancer, and colon cancer.[15–18]
ABCC2 (MRP2) plays an important role in the transportation of endogenous and exogenous substances, as well as drug absorption, distribution, and excretion, and is associated with colorectal cancer, renal cell carcinoma, multiple myeloma, and other tumors.[19–22] ABCC3 (MRP3), which is responsible for binding, hydrolysis, and ATP release during molecular transport, is vital in the transport and regulation of different organic and toxic compounds. It has great potential for improving cancer treatment and survival.[23] ABCC4 (MRP4), which can transport various organic anionic compounds out of cells, is widely used as a drug transporter in tumors, and is associated with colon cancer, pancreatic cancer, and esophageal squamous cell carcinoma.[24–26] ABCC5 (MRP5), is an organic anion transporter with excellent ability to transport nucleotides and nucleotide analogs, and is associated with breast cancer bone metastasis and prostate cancer progression.[27–29] ABCC6 is an ATP-dependent transmembrane transporter, mainly expressed in the liver and kidney, and is a potential target for tumor treatment.[30,31] ABCC7 (CFTR), mainly expressed in colon tissue and skin, regulates ion and liquid transport in epithelial tissues.[32] ABCC8 and ABCC9 are indispensable to the KATP channel, and are closely associated with neonatal diabetes, pulmonary hypertension, and other diseases.[33–35] ABCC10 (MRP7), which plays a role in drug resistance, transports various chemotherapeutic drugs, including taxanes, epothilone B, and vinca alkaloids.[36] ABCC11 (MRP8) is associated with the risk of breast cancer.[37] A harmful mutation of ABCC12 (MRP9) can cause cholestasis.[38] ABCC13 (MRP10), a pseudogene, is highly expressed in human fetal liver.[39,40]
Nonetheless, little is known about the role of ABCCs in LIHC. We therefore examined their expression and prognostic value in LIHC patients, via a retrospective bioinformatics-driven approach.
2. Materials and methods
2.1. Oncomine database
We used the online Oncomine database (http://www.oncomine.org), a cancer microarray database for genome-wide expression analysis, to analyze ABCC mRNA expression in various cancers. Oncomine includes 715 datasets and 86733 samples, covering 35 cancer types.[41] We determined statistical difference via the Student t test, and determined differences in mRNA expression based on P < .0001, fold change = 1.5, and gene grade = 10%.
2.2. UALCAN database
UALCAN (http://ualcan.path.uab.edu) is an online database that uses data from The Cancer Genome Atlas database, and contains RNA-seq data for 31 cancer types.[42] We performed a genome-wide analysis of ABCC expression in LIHC, using UALCAN data for 371 LIHC patients and 50 normal controls (patients with noncancerous liver tissue), accounting for gender, tumor grade, tumor stage, and lymph node metastasis. UALCAN provides all statistically significant results (P < .05). We excluded records with transcripts per million (TPM) < 1.
2.3. Human protein atlas
From the Human Protein Atlas (HPA) (http://www.proteinatlas.org), which collects representative immunohistochemistry-based protein expression data for nearly 20 highly common cancers,[43] we obtained immunohistochemical images of ABCC protein expression in clinical specimens from patients with LIHC and normal tissues. We selected HPA records with P < .05.
2.4. GeneMANIA database
Using GeneMANIA (http://www.genemania.org), an online tool that uses available genomics and proteomics data to generate hypotheses involving gene function,[44] we analyzed the functional association network between ABCC family members and their related genes. The advanced statistics option is a maximum synthetic attribute of 10 and a maximum synthetic gene of 20. GeneMANIA considers P < .05 statistically significant.
2.5. Gene ontology (GO) and kyoto encyclopedia of genes and genomes pathway enrichment
The GO database (http://geneontology.org) comprehensively describes the attributes of genes and gene products in an organism in terms of the molecular function of the genes, the function of cell components, and the biological processes involved.[45] KEGG (http://www.kegg.jp) integrates information on genome, chemistry, and system function.[46] We used the Bioconductor plugin in R for GO and KEGG enrichment analysis, and considered P < .05 statistically significant.
2.6. TIMER database
The TIMER (https://cistrome.shinyapps.io/timer) database uses systematic analysis of microarray expression data to detect immune-cell penetration in tumor tissues, and to determine its association with various cancers, or with gene expression. We quantitatively analyzed the penetration ratios of 6 types of immune cells (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells).[47] We used TIMER to evaluate the immune infiltration of ABCC family members in LIHC, and analyzed the Spearman correlation between these 6 types of immune cells and ABCC mRNA expression. Statistical significance was set at P < .05.
2.7. Kaplan-meier plotter
Kaplan–Meier plotter (http://kmplot.com/analysis) is an online database for prognostic analysis of various types of cancer. It is based on the data sets of 3 major medical centers, in Berlin, Bethesda, and Melbourne.[48–51] For 364 LIHC patients, we evaluated overall survival (OS), determined the prognostic significance, and accounted for gender, tumor grade, tumor stage, American Joint Committee on Cancer (AJCC) T stage, and vascular invasion, with 95% confidence intervals and logarithmic P values. We used an OS chart to compare OS in the high- and low-expression groups. P < .05 was considered statistically significant. The probe numbers used to study ABCC1–13 were, respectively, 202804-at, 206155-at, 214979-at, 203196-at, 22636363-at, 214033-at, 205043-at, 210245-at, 208561-at, 213485-s-at, 224146-s-at, 1552590-at and 1552582-at.
2.8. Ethical approval
These analyses were based on online open-access databases, hence this article does not contain any research conducted by any author on human participants or animals, nor can it be followed up and updated.
2.9. Statistical analysis
SPSS 25.0 (IBM, Armonk, NY) was used for statistical analysis. Results were considered significant at P < .05. For Cox proportional hazard regression analysis, the 95% confidence interval (CI) and hazard ratio (HR) were used for risk assessment.
2.10. Data management and collection
We obtained records from the Oncomine, UALCAN, HPA, TIMER, and Kaplan–Meier plotter databases on May 20, 2021. Records were obtained by searching these databases using the terms ABCC1, ABCC2, ABCC3, ABCC4, ABCC5, ABCC6, ABCC7, ABCC8, ABCC9, ABCC10, ABCC11, ABCC12, ABCC13 and “liver cancer”. The exposure group comprised patients with LIHC, and the control group comprised normal (noncancerous) tissue samples. All patients included in the search language search were included. The search was not restricted based on race, country, gender, or language. Two researchers (SD and MXT) independently reviewed the eligibility of the data, and XZ resolved any discrepancies. Disagreements over eligibility were resolved via discussion. The research selection process conformed to the STROBE guidelines. To ensure the validity and reliability of the results, SD and WS independently conducted statistical analysis. LYY reviewed the data to detect potential bias that could arise during subgroup analysis.
3. Results
3.1. mRNA expression of ABCCs in LIHC patients
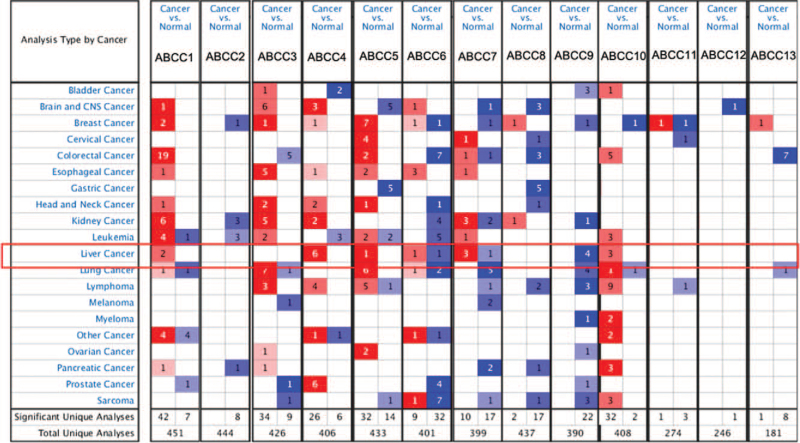
Using the Oncomine database, we compared ABCC transcription in 20 cancers and normal tissues: mRNA expression of members ABCC1/4/5/6/7/10 was significantly higher, whereas that of ABCC9 was significantly lower, in LIHC tissue (P < .05). In the Roessler Liver 2 dataset,[52] the mRNA expression of ABCC6/7 was lower in LIHC tissue than in normal tissue (Fig. 1, Table 1).
Figure 1.
Transcriptional expression of different ABCCs family members in 20 types of cancer. The data was compared by t-test. The cut-off P value and the fold change were as follows: P value <.0001, fold change = 1.5, gene grade = 10%. Red means overexpression, blue means overexpression.
Table 1.
Transcriptional expression of ABCCs family members between LIHC and normal liver tissue (Oncomine).
| Types of LIHC vs liver | Fold change | P value | t-test | Ref | |
| ABCC1 | Cirrhosis | 1.899 | 3.86E-5 | 4.923 | Wurmbach Liver[25] |
| Cirrhosis | 1.770 | 5.02E-12 | 9.502 | Mas Liver[26] | |
| ABCC4 | Hepatocellular Carcinoma | 2.186 | 2.45E-10 | 8.359 | Mas Liver[26] |
| Cirrhosis | 2.324 | 6.25E-11 | 9.954 | Mas Liver[26] | |
| Cirrhosis | 2.272 | 3.89E-6 | 6.409 | Wurmbach Liver[25] | |
| Hepatocellular Carcinoma | 2.321 | 1.13E-5 | 4.853 | Wurmbach Liver[25] | |
| Hepatocellular Carcinoma | 1.605 | 7.22E-8 | 5.495 | Chen Liver[27] | |
| Hepatocellular Carcinoma | 2.074 | 3.38E-34 | 14.058 | Roessler Liver 2[28] | |
| ABCC5 | Hepatocellular Carcinoma | 2.304 | 5.23E-9 | 7.701 | Wurmbach Liver[25] |
| ABCC6 | Hepatocellular Carcinoma | 1.808 | 6.12E-11 | 6.841 | Chen Liver[27] |
| Hepatocellular Carcinoma | −2.256 | 2.43E-31 | −12.883 | Roessler Liver 2[28] | |
| ABCC7 | Cirrhosis | 9.813 | 2.02E-8 | 8.400 | Wurmbach Liver[25] |
| Cirrhosis | 3.519 | 7.59E-27 | 16.473 | Wurmbach Liver[25] | |
| Hepatocellular Carcinoma | 1.800 | 7.45E-7 | 5.541 | Mas Liver26 | |
| Hepatocellular Carcinoma | −2.019 | 9.57E-26 | −11.217 | Roessler Liver 2[28] | |
| ABCC9 | Hepatocellular Carcinoma | −5.857 | 5.48E-13 | −9.922 | Wurmbach Liver[25] |
| Cirrhosis | −7.525 | 5.11E-13 | −15.057 | Wurmbach Liver[25] | |
| Liver Cell Dysplasia | −3.146 | 5.49E-6 | −5.632 | Wurmbach Liver[25] | |
| Hepatocellular Carcinoma | −1.630 | 1.02E-10 | −8.376 | Roessler Liver[28] | |
| ABCC10 | Hepatocellular Carcinoma | 2.553 | 9.77E-8 | 7.557 | Wurmbach Liver[25] |
| Hepatocellular Carcinoma | 1.839 | 4.47E-47 | 16.858 | Roessler Liver 2[28] | |
| Hepatocellular Carcinoma | 1.561 | 1.16E-5 | 5.232 | Roessler Liver[28] |
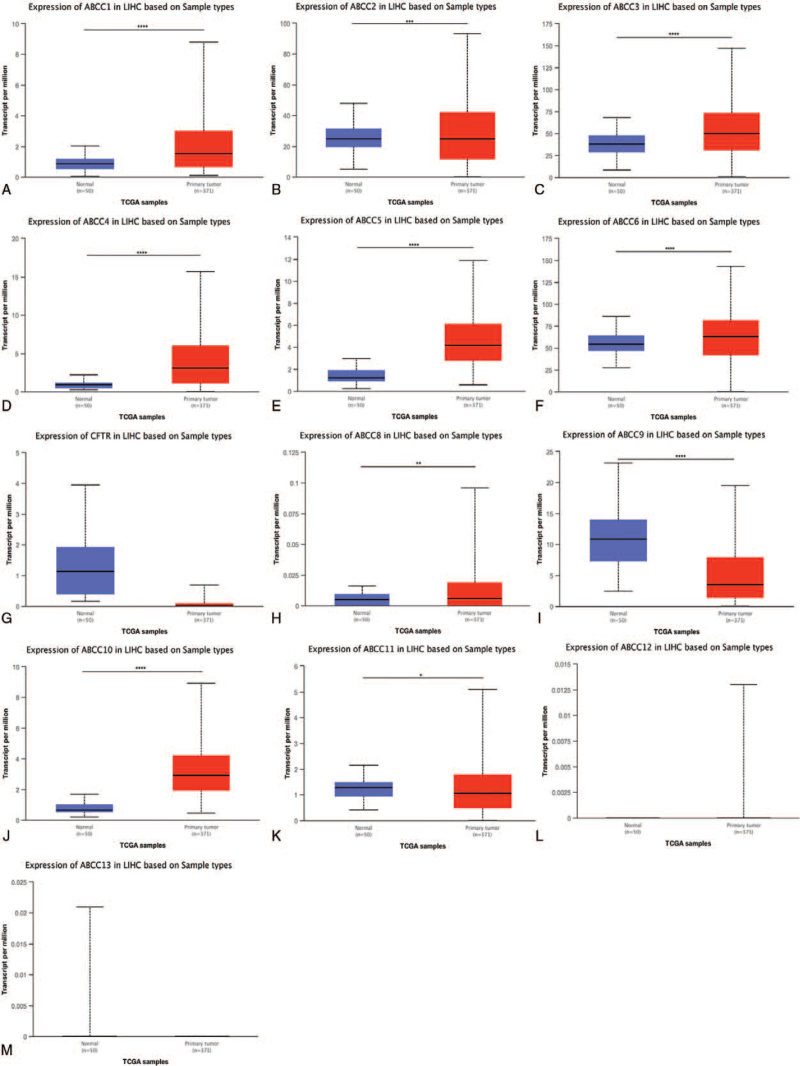
We then verified these results using the UALCAN database. Relative to normal liver tissue, mRNA expression was upregulated for ABCC1/2/3/4/5/10 (P < .0001) and ABCC6 (P < .001) (Fig. 2), and downregulated in ABCC9/11 (P < 0.0001). ABCC8/12/13 were excluded from all analyses, because they had TPM < 1.
Figure 2.
mRNA expression of different ABCCs family members in LIHC patients and normal liver tissues. The mRNA expression of different ABCCs family members in LIHC patients from the TCGA database (A–M). ∗ P < .05, ∗∗ P < .01, ∗∗∗ P < .001, ∗∗∗∗ P < .0001.
ABCC protein expression in LIHC was evaluated using the HPA database: that of ABCC2 and ABCC12 was downregulated, and that of ABCC3/4/8/9 was upregulated (Fig. 3).
Figure 3.
Representative immunohistochemical images of different ABCCs family members in LIHC tissues and normal liver tissues (HPA database). The expression of ABCC3, ABCC4, ABCC8 and ABCC9 increased, and the expression of ABCC2 and ABCC12 decreased.
3.1.1. ABCC mRNA expression in LIHC by gender
We compared 50 patients with normal (noncancerous) liver tissue, 245 male LIHC patients, and 117 female LIHC patients: except for ABCC2 and ABCC7, ABCC mRNA expression differed significantly between men and women (Fig. 4). Relative to normal liver tissue, mRNA expression in LIHC was significantly upregulated for ABCC1/3/4/5/6/10 in both men and women (Fig. 4A, C, D, E, F, J; P < .0001); that of ABCC9 was significantly downregulated in men (P < .05) and women (P < .01) (Fig. 4I); that of ABCC2 was upregulated in men (Fig. 4B, P < .0001); and that of ABCC7 was downregulated in women (Fig. 4G, P < .05).
Figure 4.
The relationship between the mRNA expression of ABCCs family members and the sex of LIHC patients. Box plots showed the mRNA expression (A–M) of family members of ABCCs in normal individuals and LIHC patients of different genders. ∗ P < .05; ∗∗ P < .01; ∗∗∗ P < .001, ∗∗∗∗ P < .0001.
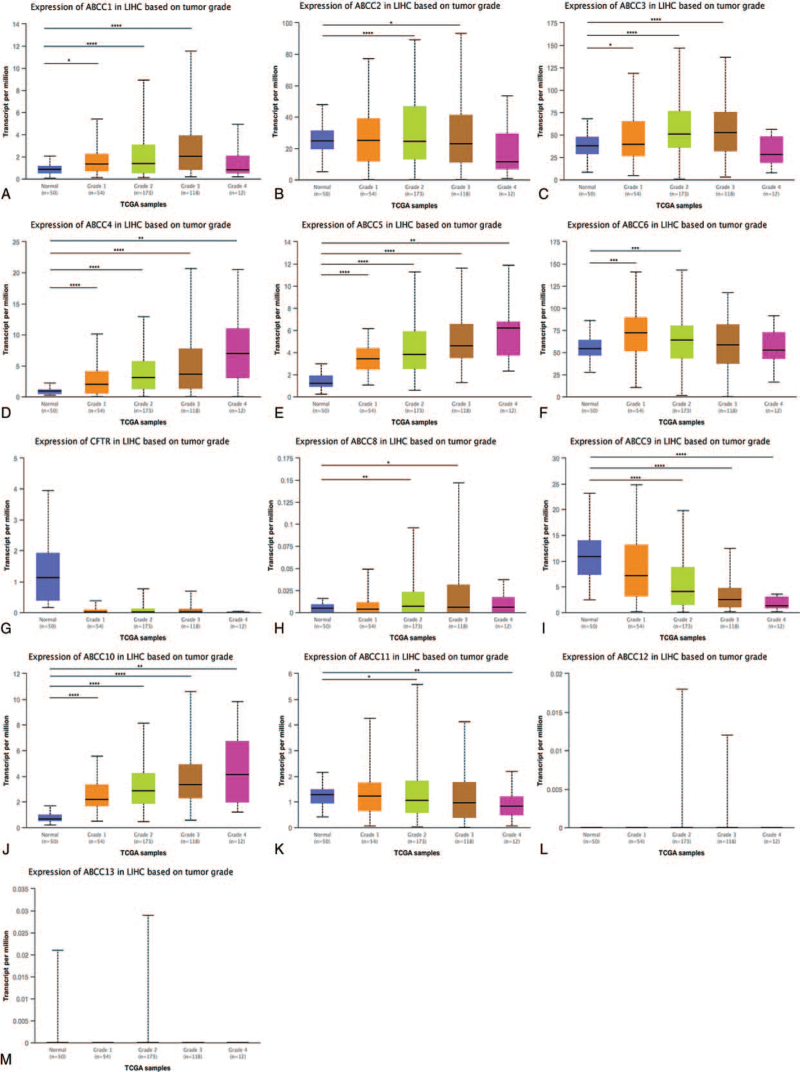
3.1.2. ABCC mRNA expression in LIHC by tumor grade
We compared mRNA expression in 50 patients with normal (noncancerous) liver tissue, 54 grade 1 LIHC patients, 173 grade 2 patients, 118 grade 3 patients, and 12 grade 4 patients: members ABCC4/5/10 were highly upregulated in all grades (Fig. 5D, E, J; P < .01). For the other ABCC members (excluding ABCC7, Fig. 5G), mRNA expression did not differ significantly between the control and LIHC samples. LIHC tumor grade was significantly correlated with ABCC, for all ABCC members (Fig. 5A–F, H–K; P < .05).
Figure 5.
The mRNA expression of ABCCs family members is correlated with the tumor grade of LIHC. The box plot showed the normal individuals or LIHC patients in Grade 1: Well differentiated (low grade), Grade 2: Moderately differentiated (intermediate grade), Grade 3: Poly differentiated (high grade) or Grade 4: Undifferentiated (high grade) (A–M) mRNA expression of ABCCs family members. ∗ P < .05; ∗∗ P < .01; ∗∗∗ P < .001, ∗∗∗∗ P < .0001.
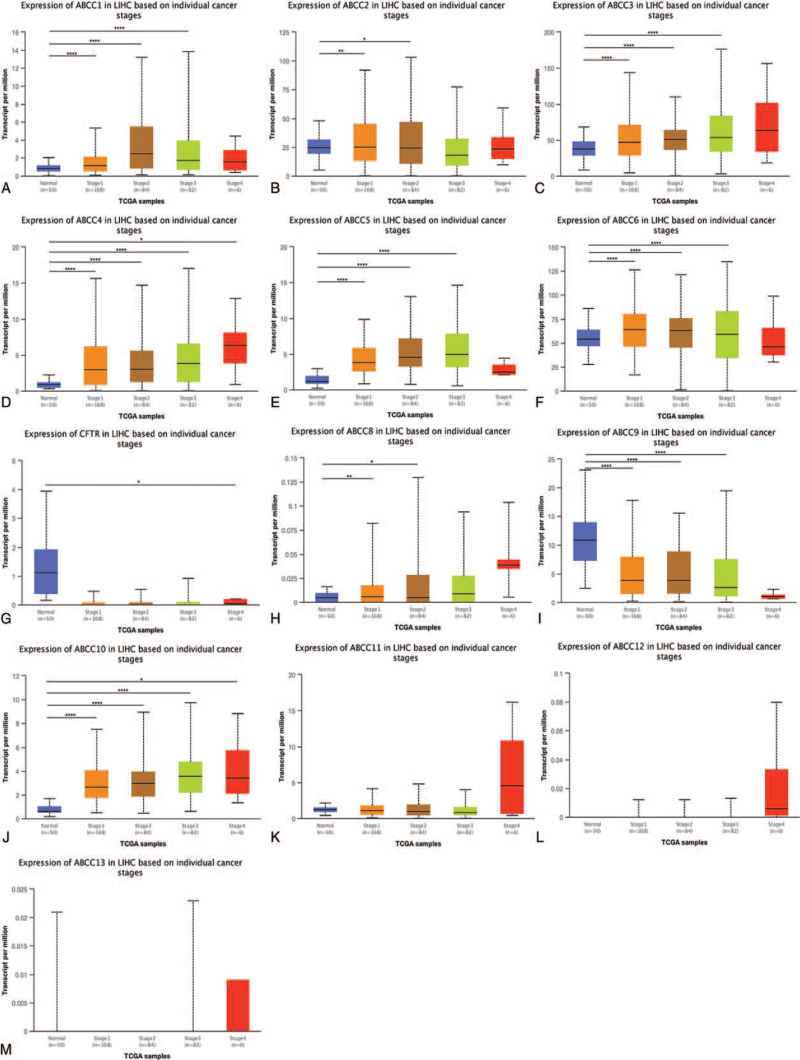
3.1.3. ABCC mRNA expression in LIHC patients by tumor stage
We compared 50 patients with normal liver tissue with 168 stage 1, 84 stage 2, 82 stage 3, and 6 stage 4 LIHC patients: for all stages, mRNA expression was upregulated for ABCC4 (P < .05, Fig. 6D), but was not significantly different for ABCC8/11/12/13 (Fig. 6H, K, L, M). For the remaining ABCC members, mRNA expression was correlated with stage (Fig. 6A-G, I, J; P < .05).
Figure 6.
The mRNA expression of ABCCs family members is correlated with the tumor stage of LIHC patients. The box plot shows the mRNA expression of ABCCs family members in normal individuals and LIHC patients in stage 1, stage 2, stage 3 and stage 4 (A–M). ∗ P < .05; ∗∗ P < .01; ∗∗∗ P < .001, ∗∗∗∗ P < .0001.
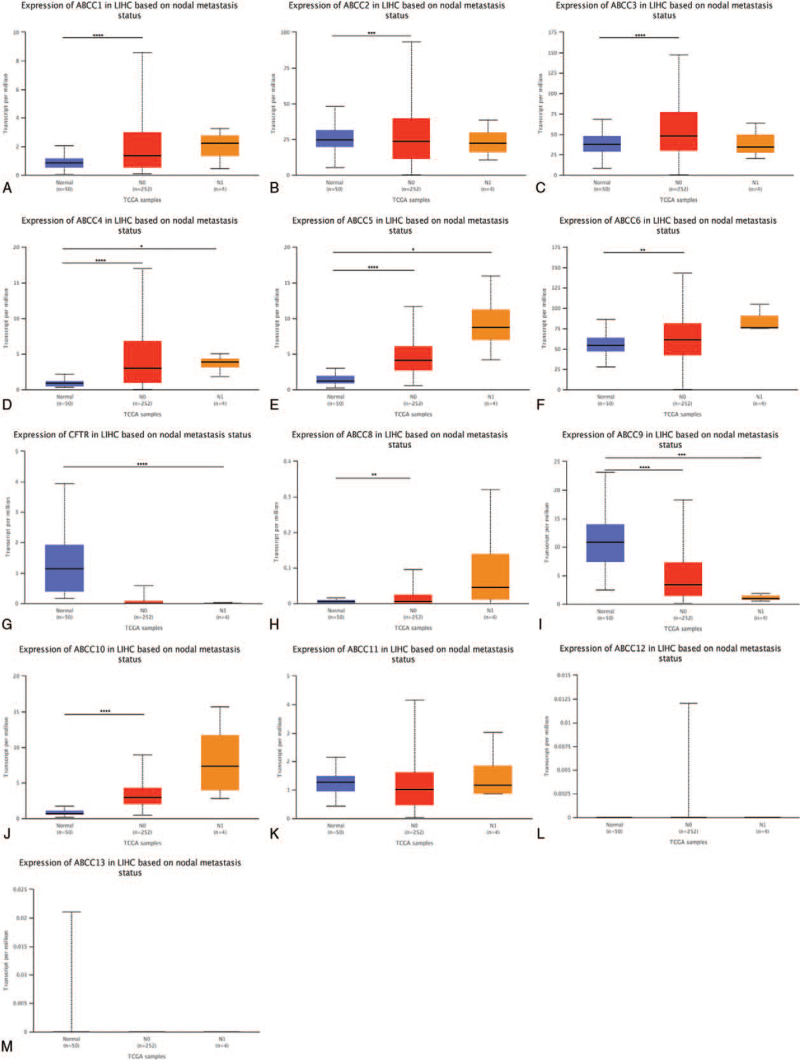
3.1.4. ABCC mRNA expression in LIHC by lymph node metastasis status
We compared 50 patients with normal liver tissue, with 252 lymph node metastasis status N0 and 4 N1 status LIHC patients (Fig. 7). mRNA expression was highly upregulated for ABCC4 and ABCC5 for both N0 and N1 (Fig. 7D, E; P < .05); that of ABCC9 was significantly downregulated (Fig. 7I, P < .001); that of ABCC7/11 was not associated with lymphatic node metastasis status (Fig. 7G, K; P ≥ .05); and that of ABCC1/2/3/6/10 was associated with lymph node metastasis (Fig. 7A, B, C, F, J; P < .05).
Figure 7.
The mRNA expression of ABCCs family members is correlated with the status of lymph node metastasis in patients with LIHC. The box plot showed the mRNA expression of ABCCs family members in normal individuals or lymph node metastasis states N0 or N1 (A–M). ∗ P < .05; ∗∗ P < .01; ∗∗∗ P < .001, ∗∗∗∗ P < .0001.
3.2. Functional enrichment of ABCCs in LIHC
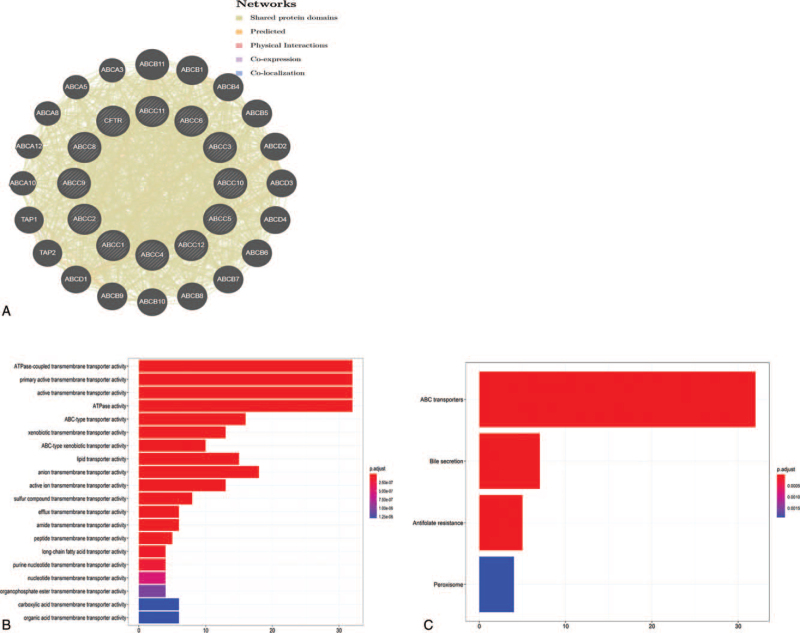
We constructed a network of ABCCs and their 20 related genes using GeneMANIA (Fig. 8A). ABCC members interacted with the following proteins: ABCB11, ABCB1, ABCB4, ABCB5, ABCD2, ABCD3, ABCD4, ABCB6, ABCB7, ABCB8, ABCB10, ABCB9, ABCD1, TAP2, TAP1, ABCA10, ABCA12, ABCA8, ABCA5, and ABCA3.
Figure 8.
Functional enrichment of members of the ABCCs family in LIHC. A. Analyze the network of ABCCs family members and their 20 related genes by GeneMANIA. B. GO enrichment analysis ranked the top 20 pathways; C. KEGG pathway analysis.
We analyzed the GO functions and pathways of ABCCs and their 20 related genes, via the Bioconductor plugin in R. The top 10 functions and pathways were GO:0042626 (ATPase-coupled transmembrane transporter activity), GO:0015399 (primary active transmembrane transporter activity), GO:0022804 (active transmembrane transporter activity), GO:0016887 (ATPase activity), GO:0140359 (ABC-type transporter activity), GO:0042910 (xenobiotic transmembrane transporter activity), GO:0008559 (ABC-type xenobiotic transporter activity), GO:0005319 (lipid transporter activity), GO:0008509 (anion transmembrane transporter activity), and GO:0022853 (active ion transmembrane transporter activity) (Fig. 8B, P < .05). The primary enriched KEGG pathways were as follows: ABC transporters, Bile secretion, Antifolate resistance, and Peroxisome (Fig. 8C; P < .05).
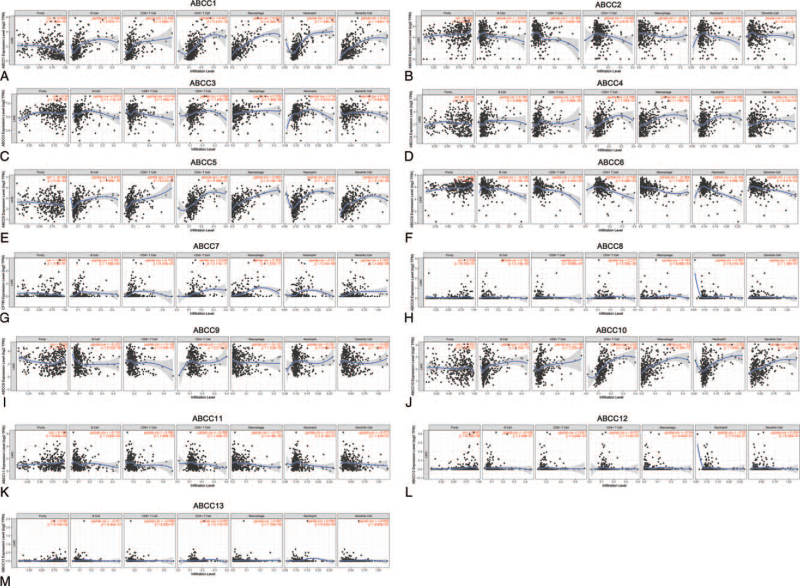
3.3. Correlation between ABCC mRNA expression and LIHC immune infiltration
We used the TIMER database to determine the correlation between ABCC mRNA expression and the level of immune infiltration in LIHC (Fig. 9). The mRNA expression of members ABCC1/4/5/10 was positively correlated with B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil, and dendritic cell infiltration (Fig. 9A, D, E, J; P < .05). ABCC6/7 was negatively correlated with infiltration by these cells (Fig. 9F, G; all P < .01). mRNA expression of ABCC2 was negatively correlated with CD8+ T cell infiltration (Fig. 9B; P < .001); that of ABCC3 was positively correlated with CD4+ T cell, macrophage, and neutrophil infiltration (Fig. 9C; P < .001); that of ABCC8 was positively correlated with CD4+ T cell and macrophage infiltration (Fig. 9H; P < .001); that of ABCC9 was negatively correlated with B cell and macrophage infiltration (Fig. 9I; P < .001); and that of ABCC11 was negatively correlated with B cell infiltration (Fig. 9K; P < .05). There were no correlations between ABCC12/13 mRNA expression and immune-cell infiltration (Fig. 9M, L; P ≥ .05). In summary, for most of the ABCC members, mRNA expression was correlated with immune-cell infiltration in LIHC.
Figure 9.
The relationship between the mRNA expression of ABCCs family members and the level of immune infiltration in LIHC. The mRNA expression of ABCCs family members were significantly correlated with the level of immune infiltration in LIHC (A–M).
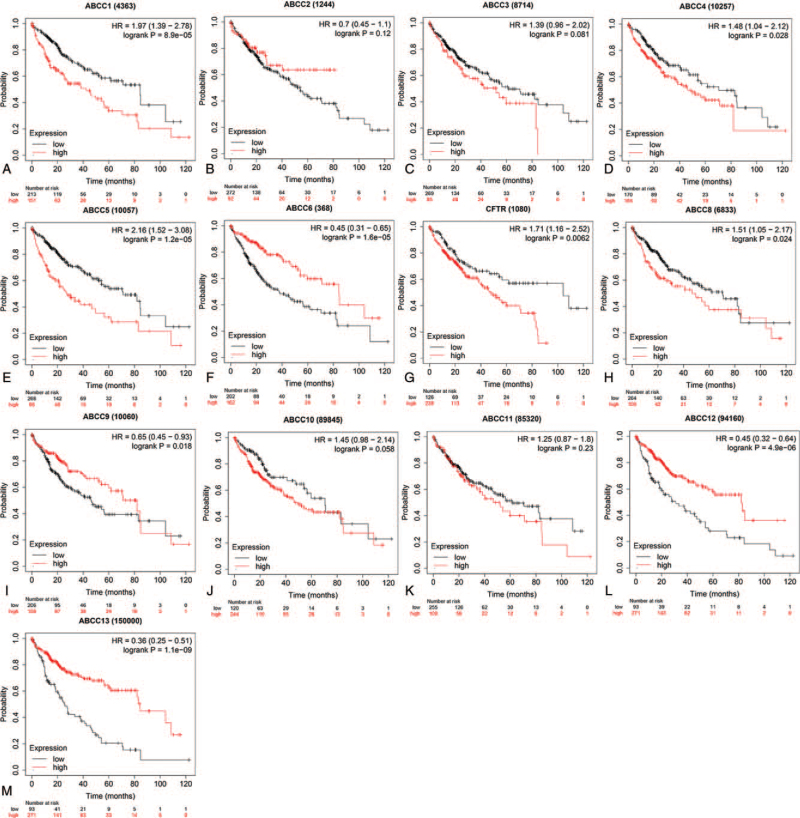
3.4. Correlation between ABCC mRNA expression and OS
ABCC mRNA expression was associated with OS in LIHC patients. Poor prognosis was associated with high mRNA expression of members ABCC1/4/5/8 (Fig. 10A, D, E, H; P < .05) and low mRNA expression of members ABCC6/7/9/12/13 (Fig. 10F, G, I, L, M; P < .05).
Figure 10.
The prognostic value of mRNA expression of ABBCs family members in LIHC patients. Compare the survival curves of high and low expression of ABBCs family members of LIHC patients in Kaplan–Meier plotter.
ABCC2 and ABCC13 showed prognostic significance in both men and women (Table 2; all P < .05). ABCC1/5–9/12 showed prognostic significance in men (ABCC7, P < .05; the others, P < .01), and ABCC2/13 showed prognostic significance in women (P < .05).
Table 2.
Correlation analysis between ABCCs and gender.
| Gene | Gender | Cases | HR | 95% CI | P value |
| ABCC1 | Male | 246 | 2.71 | 1.74–4.24 | .0000051 |
| Female | 118 | 0.71 | 0.39–1.27 | .2449 | |
| ABCC2 | Male | 246 | 0.57 | 0.34–0.96 | .0337 |
| Female | 118 | 1.97 | 1.1–3.5 | .0193 | |
| ABCC3 | Male | 246 | 1.37 | 0.86–2.18 | .1857 |
| Female | 118 | 1.45 | 0.73–2.86 | .2868 | |
| ABCC4 | Male | 246 | 1.52 | 0.97–2.38 | .0643 |
| Female | 118 | 1.8 | 0.98–3.45 | .075 | |
| ABCC5 | Male | 246 | 2.61 | 1.67–4.09 | .000012 |
| Female | 118 | 1.86 | 0.93–3.72 | .0757 | |
| ABCC6 | Male | 246 | 0.35 | 0.23–0.55 | .000002 |
| Female | 118 | 0.53 | 0.27–1.03 | .0579 | |
| ABCC7 | Male | 246 | 1.98 | 1.16–3.35 | .0101 |
| Female | 118 | 1.63 | 0.88–3.02 | .1195 | |
| ABCC8 | Male | 246 | 1.88 | 1.18–2.98 | .0067 |
| Female | 118 | 0.75 | 0.42–1.34 | .3235 | |
| ABCC9 | Male | 246 | 0.42 | 0.25–0.72 | .001 |
| Female | 118 | 1.48 | 0.82–2.67 | .1906 | |
| ABCC10 | Male | 246 | 1.41 | 0.88–2.26 | .1464 |
| Female | 118 | 1.72 | 0.91–3.25 | .0925 | |
| ABCC11 | Male | 246 | 1.34 | 0.85–2.12 | .1991 |
| Female | 118 | 1.72 | 0.93–3.17 | .0819 | |
| ABCC12 | Male | 246 | 0.31 | 0.2–0.48 | .000000034 |
| Female | 118 | 1.57 | 0.87–2.84 | .1294 | |
| ABCC13 | Male | 246 | 0.27 | 0.17–0.42 | 7.1E-10 |
| Female | 118 | 0.46 | 0.26–0.83 | .0083 |
ABCC1/6/12/13 had prognostic significance for tumor grades 1 to 3 (P < .05), ABCC5/7 for grades 2/3 (P < .05), and ABCC3/4/8/11 for grade 2 (P < .05). Tumor grade 4 was excluded because of its small sample size (n = 12) (Table 3).
Table 3.
Correlation analysis between ABCCs and staging.
| GENE | Grade | Cases | HR | 95% CI | P value |
| ABCC1 | 1 | 55 | 0.31 | 0.1–0.94 | .0286 |
| 2 | 174 | 2.59 | 1.53–4.38 | .00025 | |
| 3 | 118 | 1.97 | 1.01–3.83 | .0429 | |
| 4 | 12 | – | – | – | |
| ABCC2 | 1 | 55 | 1.65 | 0.61–4.45 | .3199 |
| 2 | 174 | 0.56 | 0.29–1.08 | .0804 | |
| 3 | 118 | 1.34 | 0.73–2.44 | .3449 | |
| 4 | 12 | – | – | – | |
| ABCC3 | 1 | 55 | 2.51 | 0.88–7.11 | .0742 |
| 2 | 174 | 1.84 | 1.07–3.14 | .0241 | |
| 3 | 118 | 0.58 | 0.31–1.1 | .0895 | |
| 4 | 12 | – | – | – | |
| ABCC4 | 1 | 55 | 2.36 | 0.93–5.99 | .0629 |
| 2 | 174 | 1.79 | 1.03–3.11 | .0352 | |
| 3 | 118 | 1.29 | 0.66–2.52 | .4495 | |
| 4 | 12 | – | – | – | |
| ABCC5 | 1 | 55 | 1.75 | 0.66–4.63 | .2534 |
| 2 | 174 | 2.39 | 1.44–3.97 | .00055 | |
| 3 | 118 | 2.76 | 1.51–5.06 | .00058 | |
| 4 | 12 | – | – | – | |
| ABCC6 | 1 | 55 | 0.18 | 0.07–0.47 | 8.90E-05 |
| 2 | 174 | 0.56 | 0.33–0.95 | .0286 | |
| 3 | 118 | 0.32 | 0.14–0.72 | .0039 | |
| 4 | 12 | – | – | – | |
| ABCC7 | 1 | 55 | 2.14 | 0.84–5.48 | .105 |
| 2 | 174 | 2.24 | 1.2–4.18 | .009 | |
| 3 | 118 | 0.45 | 0.21–0.93 | .0273 | |
| 4 | 12 | – | – | – | |
| ABCC8 | 1 | 55 | 0.51 | 0.19–1.35 | .1696 |
| 2 | 174 | 1.81 | 1.08–3.04 | .0233 | |
| 3 | 118 | 1.65 | 0.85–3.18 | .1333 | |
| 4 | 12 | – | – | – | |
| ABCC9 | 1 | 55 | 0.37 | 0.13–1.08 | .0578 |
| 2 | 174 | 0.65 | 0.39–1.1 | .1069 | |
| 3 | 118 | 0.51 | 0.23–1.15 | .1001 | |
| 4 | 12 | – | – | – | |
| ABCC10 | 1 | 55 | 2.57 | 0.97–6.83 | .0505 |
| 2 | 174 | 1.49 | 0.89–2.49 | .1248 | |
| 3 | 118 | 1.38 | 0.74–2.57 | .3051 | |
| 4 | 12 | – | – | – | |
| ABCC11 | 1 | 55 | 0.46 | 0.18–1.21 | .1057 |
| 2 | 174 | 1.98 | 1.18–3.33 | .0086 | |
| 3 | 118 | 0.65 | 0.35–1.21 | .1711 | |
| 4 | 12 | – | – | – | |
| ABCC12 | 1 | 55 | 0.39 | 0.15–1 | .0436 |
| 2 | 174 | 0.43 | 0.25–0.72 | .0011 | |
| 3 | 118 | 0.46 | 0.25–0.84 | .0102 | |
| 4 | 12 | – | – | – | |
| ABCC13 | 1 | 55 | 0.27 | 0.1–0.69 | .0033 |
| 2 | 174 | 0.31 | 0.19–0.52 | 3.00E-06 | |
| 3 | 118 | 0.33 | 0.18–0.61 | .00017 | |
| 4 | 12 | – | – | – |
We combined tumor stages 3 and 4, because of the small sample size of stage 4 (n = 4). ABCC5/11/12/13 showed prognostic significance for all stages (P < .05), ABCC1/8 for stage 1 (P < .01), ABCC6/7 for stage 2 (P < .05), and ABCC4/6/9/10 for the combined stage 3 + 4 (P < .05) (Table 4).
Table 4.
Correlation analysis between ABCCs and tumor grade.
| GENE | Stage | Cases | HR | 95% CI | P value |
| ABCC1 | 1 | 170 | 2.4 | 1.29–4.47 | .0045 |
| 2 | 83 | 2.23 | 0.96–5.17 | .0559 | |
| 3 + 4 | 87 | 1.71 | 0.91–3.21 | .0899 | |
| ABCC2 | 1 | 170 | 1.84 | 0.98–3.47 | .0552 |
| 2 | 83 | 1.81 | 0.81–4.07 | .1443 | |
| 3 + 4 | 87 | 0.67 | 0.35–1.28 | .2236 | |
| ABCC3 | 1 | 170 | 1.49 | 0.78–2.83 | .2261 |
| 2 | 83 | 0.6 | 0.27–1.33 | .2079 | |
| 3 + 4 | 87 | 0.57 | 0.32–1.04 | .0634 | |
| ABCC4 | 1 | 170 | 1.44 | 0.77–2.71 | .2489 |
| 2 | 83 | 1.94 | 0.86–4.35 | .1019 | |
| 3 + 4 | 87 | 2.07 | 1.02–4.19 | .0396 | |
| ABCC5 | 1 | 170 | 3.35 | 1.41–7.95 | .0036 |
| 2 | 83 | 4.03 | 1.83–8.85 | .00018 | |
| 3 + 4 | 87 | 2.36 | 1.3–4.3 | .0039 | |
| ABCC6 | 1 | 170 | 0.59 | 0.32–1.08 | .0831 |
| 2 | 83 | 0.31 | 0.12–0.78 | .0086 | |
| 3 + 4 | 87 | 0.39 | 0.21–0.75 | .0036 | |
| ABCC7 | 1 | 170 | 1.66 | 0.87–3.16 | .1174 |
| 2 | 83 | 2.84 | 1.11–7.3 | .0242 | |
| 3 + 4 | 87 | 0.63 | 0.33–1.22 | .1682 | |
| ABCC8 | 1 | 170 | 3.32 | 1.39–7.95 | .0043 |
| 2 | 83 | 0.59 | 0.26–1.35 | .2094 | |
| 3 + 4 | 87 | 0.62 | 0.32–1.19 | .1485 | |
| ABCC9 | 1 | 170 | 0.78 | 0.42–1.43 | .4186 |
| 2 | 83 | 2.09 | 0.71–6.1 | .1693 | |
| 3 + 4 | 87 | 0.34 | 0.16–0.71 | .0027 | |
| ABCC10 | 1 | 170 | 2.19 | 0.97–4.93 | .0525 |
| 2 | 83 | 0.67 | 0.3–1.5 | .3293 | |
| 3 + 4 | 87 | 2.17 | 1.1–4.27 | .0219 | |
| ABCC11 | 1 | 170 | 2.09 | 1–4.39 | .0458 |
| 2 | 83 | 2.25 | 1.02–4.97 | .0393 | |
| 3 + 4 | 87 | 0.5 | 0.27–0.92 | .0233 | |
| ABCC12 | 1 | 170 | 0.51 | 0.27–0.95 | .0312 |
| 2 | 83 | 0.37 | 0.17–0.81 | .0101 | |
| 3 + 4 | 87 | 0.43 | 0.24–0.78 | .0041 | |
| ABCC13 | 1 | 170 | 0.41 | 0.22–0.75 | .0029 |
| 2 | 83 | 0.37 | 0.17–0.81 | .0092 | |
| 3 + 4 | 87 | 0.4 | 0.22–0.72 | .0018 |
ABCC1/11/12/13 showed prognostic significance for AJCC T stages 1–3 (P < .05), ABCC5/ABCC6 for AJCC T 2 and 3 (P < .01), ABCC10 for AJCC T 1 (P < .05), ABCC7 for AJCC T 2 (P < .05), and ABCC4/9/10 for AJCC T 3 (P < .05) (Table 5). We excluded AJCC T 4 because of its small sample size (n = 13).
Table 5.
Correlation analysis between ABCCs and AJCC T classification.
| GENE | AJCC_T | Cases | HR | 95% CI | P-value |
| ABCC1 | 1 | 180 | 2.1 | 1.15–3.82 | .0129 |
| 2 | 90 | 2.52 | 1.12–5.7 | .0214 | |
| 3 | 78 | 1.97 | 1.04–3.73 | .034 | |
| 4 | 13 | – | – | – | |
| ABCC2 | 1 | 180 | 1.71 | 0.94–3.11 | .078 |
| 2 | 90 | 2.02 | 0.96–4.29 | .0609 | |
| 3 | 78 | 0.66 | 0.35–1.25 | .2009 | |
| 4 | 13 | – | – | – | |
| ABCC3 | 1 | 180 | 1.37 | 0.73–2.59 | .32 |
| 2 | 90 | 1.7 | 0.64–4.5 | .2817 | |
| 3 | 78 | 0.6 | 0.32–1.12 | .1048 | |
| 4 | 13 | – | – | – | |
| ABCC4 | 1 | 180 | 1.46 | 0.8–2.67 | .2154 |
| 2 | 90 | 2.05 | 0.97–4.31 | .0543 | |
| 3 | 78 | 2.04 | 1–4.18 | .0461 | |
| 4 | 13 | – | – | – | |
| ABCC5 | 1 | 180 | 2.42 | 1.17–5.02 | .0143 |
| 2 | 90 | 3.62 | 1.75–7.47 | .0002 | |
| 3 | 78 | 2.89 | 1.53–5.43 | .00063 | |
| 4 | 13 | – | – | – | |
| ABCC6 | 1 | 180 | 0.61 | 0.34–1.09 | .0933 |
| 2 | 90 | 0.28 | 0.11–0.69 | .0031 | |
| 3 | 78 | 0.39 | 0.2–0.77 | .0048 | |
| 4 | 13 | – | – | – | |
| ABCC7 | 1 | 180 | 1.74 | 0.86–3.53 | .1216 |
| 2 | 90 | 3.02 | 1.2–7.63 | .0144 | |
| 3 | 78 | 1.64 | 0.84–3.22 | .15 | |
| 4 | 13 | – | – | – | |
| ABCC8 | 1 | 180 | 3.17 | 1.41–7.15 | 3.17 |
| 2 | 90 | 0.61 | 0.28–1.31 | .1989 | |
| 3 | 78 | 1.72 | 0.92–3.21 | .0877 | |
| 4 | 13 | – | – | – | |
| ABCC9 | 1 | 180 | 0.71 | 0.4–1.27 | .2509 |
| 2 | 90 | 1.8 | 0.73–4.42 | .1948 | |
| 3 | 78 | 0.31 | 0.14–0.68 | .0021 | |
| 4 | 13 | – | – | – | |
| ABCC10 | 1 | 180 | 2.2 | 0.98–4.91 | .0496 |
| 2 | 90 | 1.29 | 0.61–2.73 | .5053 | |
| 3 | 78 | 2.94 | 1.37–6.31 | .0041 | |
| 4 | 13 | – | – | – | |
| ABCC11 | 1 | 180 | 2.28 | 1.09–4.74 | .0237 |
| 2 | 90 | 2.18 | 1.04–4.57 | .0347 | |
| 3 | 78 | 0.52 | 0.27–0.99 | .0432 | |
| 4 | 13 | – | – | – | |
| ABCC12 | 1 | 180 | 0.5 | 0.27–0.91 | .0202 |
| 2 | 90 | 0.44 | 0.21–0.91 | .024 | |
| 3 | 78 | 0.46 | 0.25–0.84 | .0104 | |
| 4 | 13 | – | – | – | |
| ABCC13 | 1 | 180 | 0.43 | 0.24–0.77 | .00238 |
| 2 | 90 | 0.37 | 0.18–0.76 | .0048 | |
| 3 | 78 | 0.37 | 0.19–0.7 | .0016 | |
| 4 | 13 | – | – | – |
ABCC1/5/6/13 showed prognostic significance for vascular and microvascular invasion (P < .05), ABCC4/12 for vascular invasion (P < .01), and ABCC3/8/9/11 for microvascular invasion (P < .05) (Table 6). We did not analyse macrovascular invasion because of its small sample size (n = 16).
Table 6.
Correlation analysis between ABCCs and Vascular invasion.
| GENE | Vascular invasion | Cases | HR | 95% CI | P value |
| ABCC1 | None | 203 | 1.92 | 1.13–3.28 | .0149 |
| Micro | 90 | 2.24 | 1.02–4.9 | .038 | |
| Macro | 16 | – | – | – | |
| ABCC2 | None | 203 | 1.42 | 0.83–2.42 | .198 |
| Micro | 90 | 0.56 | 0.25–1.23 | .1397 | |
| Macro | 16 | – | – | – | |
| ABCC3 | None | 203 | 1.39 | 0.8–2.39 | .2395 |
| Micro | 90 | 2.35 | 1.08–5.09 | .0261 | |
| Macro | 16 | – | – | – | |
| ABCC4 | None | 203 | 1.79 | 1.04–3.08 | .035 |
| Micro | 90 | 1.37 | 0.62–3.04 | .4307 | |
| Macro | 16 | – | – | – | |
| ABCC5 | None | 203 | 1.72 | 1.03–2.87 | .0366 |
| Micro | 90 | 2.92 | 1.35–6.31 | .0043 | |
| Macro | 16 | – | – | – | |
| ABCC6 | None | 203 | 0.48 | 0.28–0.83 | .0067 |
| Micro | 90 | 0.32 | 0.11–0.93 | .027 | |
| Macro | 16 | – | – | – | |
| ABCC7 | None | 203 | 1.64 | 0.89–3.02 | .1073 |
| Micro | 90 | 0.41 | 0.14–1.18 | .088 | |
| Macro | 16 | – | – | – | |
| ABCC8 | None | 203 | 1.35 | 0.78–2.34 | .2849 |
| Micro | 90 | 3.01 | 1.4–6.45 | .003 | |
| Macro | 16 | – | – | – | |
| ABCC9 | None | 203 | 0.77 | 0.45–1.31 | .3293 |
| Micro | 90 | 0.43 | 0.18–1.01 | .0451 | |
| Macro | 16 | – | – | – | |
| ABCC10 | None | 203 | 1.77 | 0.97–3.23 | .0599 |
| Micro | 90 | 0.52 | 0.24–1.11 | .0868 | |
| Macro | 16 | – | – | – | |
| ABCC11 | None | 203 | 1.65 | 0.91–2.97 | .0932 |
| Micro | 90 | 2.29 | 1.05–5 | .0319 | |
| Macro | 16 | – | – | – | |
| ABCC12 | None | 203 | 0.35 | 0.21–0.6 | 4.60E-05 |
| Micro | 90 | 1.85 | 0.86–4 | .1116 | |
| Macro | 16 | – | – | – | |
| ABCC13 | None | 203 | 0.33 | 0.19–0.54 | 7.00E-06 |
| Micro | 90 | 0.35 | 0.16–0.74 | .0044 | |
| Macro | 16 | – | – | – |
4. Discussion
Novel diagnostic and prognostic markers are urgently required in LIHC, and prior work has suggested ABCCs as promising candidates. The objective of this study was to describe the roles and mechanisms of action of ABCCs in LIHC. We established the diagnostic value of ABCCs in LIHC by comparing their mRNA expression in LIHC and normal (noncancerous) liver tissue: ABCC1/2/3/4/5/6/10 were upregulated, and ABCC9/11 downregulated, in LIHC. ABCC mRNA expression was associated with gender, grade, stage, and lymph node metastasis status. ABCC1–9/10/11 therefore provide potential diagnostic markers for LIHC. We found that ABCCs interact mainly with ABCB11, ABCB1, ABCB4, ABCB5, and other ABCCs, and function by, for instance, participating in ATPase-coupled transmembrane transporter activity and interacting with ABC transporters.
Our findings show that ABCCs are potential targets for LIHC immunotherapy: ABCC mRNA expression was correlated with B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil, and dendritic cell infiltration. This indicates that ABCCs play a key role in LIHC, possibly by regulating the immune response. Further, our findings reveal the prognostic value of ABCCs in LIHC: poor prognosis was associated with high mRNA expression of ABCC1/4/5/8 and low expression of ABCC6/7/9/12/13. Upregulated mRNA expression was observed for ABCC2/13 in both men and women; for ABCC1/6/12/13 in tumor grades 1–3; for ABCC5/11/12/13 in all tumor stages; and for ABCC1/11/12/13 in AJCC T stages 1–3. ABCC1/5/6/13 showed prognostic significance in vascular and microvascular invasion.
Our finding that ABCC expression is disrupted in LIHC, and that this family has prognostic value, is consistent with prior findings. ABCC1 is overexpressed in non-small cell lung cancer tissue, serum, and cells;[53] further, it is significantly highly expressed in breast cancer.[54] ABCC2 is significantly highly expressed in ovarian cancer.[55] ABCC3 is upregulated in the malignant ascites of ovarian cancer, possibly due to the growth of ovarian cancer spheroids.[56] ABCC3 and ABCC6 expression is higher in high-grade than in low-grade serous carcinoma.[57] ABCC4 is overexpressed in colorectal cancer, in which it may be associated with phenotypic transition, which regulates cell migration in a cyclic nucleotide-dependent manner.[58] ABCC5 is significantly overexpressed in prostate cancer, in which its expression is positively correlated with cell proliferation, migration, and invasion.[29] In esophageal squamous cell carcinoma, ABCC7 overexpression can activate the p38 signaling pathway; this activation is associated with a good prognosis.[59] ABCC8 mRNA expression is a new independent prognostic indicator of glioma: high expression is associated with longer survival.[60] ABCC9 is downregulated in prostate cancer.[61] In colorectal cancer, ABCC10 downregulation reduces survival, and low ABCC11 protein expression increases the risk of cancer recurrence.[62] ABCC12 may become a useful target for breast cancer immunotherapy: although it is not expressed in normal (noncancerous) breast tissue, it is highly expressed in breast cancer.[63] Little is known about ABCC13 expression in relation to tumors; however, it is highly expressed in human fetal liver.[64]
To the best of our knowledge, we are the first to determine that ABCCs can be used as markers for the diagnosis, treatment, and prognosis of LIHC, providing new ideas and targets to this end. Prior work has revealed that ABCC1/2/3 are associated with LIHC diagnosis and prognosis,[65,66] and that ABCCs may be upregulated in untreated LIHC tissue, mediated by cellular microRNAs.[67] However, these studies examined a limited number of ABCC members and associations, without addressing mRNA or protein expression, molecular function, immune infiltration, or prognosis. Because they were based on animal and human experiments, these studies had research biases. Further, their samples were too small to adequately describe the diagnostic and prognostic value of ABCCs.
Our study has different limitations. First, the analysis was database-driven and retrospective. Second, for some ABCC members, there was insufficient transcription and expression data. Our selection of subgroups and of the study sample may have introduced biases into the analysis. Our future experimental and clinical prospective research will address these limitations. To verify these findings, studies using animal experiments and larger cohorts are needed.
5. Conclusion
To the best of our knowledge, this is the first study to identify ABCCs as potential markers for LIHC diagnosis, treatment, and prognosis. We identified ABCC1/2/3/4/5/6/9/10/11 as potential diagnostic markers, and ABCC1/4/5/6/7/8/9/1/13 as prognostic markers for LIHC. Although much remains to be discovered about the roles of ABCCs in LIHC, this work provides insight and potential targets for the diagnosis and treatment of LIHC. Our future work will promote the use of ABCCs in the diagnosis and treatment of LIHC.
Acknowledgments
The authors are grateful to the various departments of Changchun University of Chinese Medicine for their support. We would like to thank Editage (www.editage.com) for English language editing.
Author contributions
Conceptualization: Shen Dong, Tiejun Liu.
Data curation: Shen Dong, Liu Yangyang, Song Wang.
Formal analysis: Liu Yangyang.
Funding acquisition: Xiong Zhuang.
Investigation: Shen Dong.
Methodology: Shen Dong, Tiejun Liu.
Software: Xiangtong Meng.
Supervision: Tiejun Liu.
Validation: Xiaohao Xu, Tiejun Liu.
Writing – original draft: Shen Dong.
Writing – review & editing: Xiong Zhuang.
Footnotes
Abbreviations: ABCC = adenosine triphosphate-binding cassette subfamily C, AJCC = American Joint Committee on Cancer, GO = gene ontology database, HPA = human protein atlas, KEGG = kyoto encyclopedia of genes and genomes, LIHC = liver hepatocellular carcinoma, OS = overall survival.
How to cite this article: Meng X, Dong S, Yangyang L, Wang S, Xu X, Liu T, Zhuang X. Adenosine triphosphate-binding cassette subfamily C members in liver hepatocellular carcinoma: bioinformatics-driven prognostic value. Medicine. 2022;101:7(e28869).
SD and MX contributed equally to this work.
This research was funded by the National Natural Science Foundation of China (81804007), Natural Science Foundation of Jilin Province (20200201590JC), and Jilin Province Education Department “Thirteenth Five-Year Plan” Science and Technology Project (JJKH20200885KJ).
The authors have no conflicts of interests to disclose.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
References
- [1].Kanwal F, Singal AG. Surveillance for hepatocellular carcinoma: current best practice and future direction. Gastroenterology 2019;157:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gordan JD, Kennedy EB, Abou-Alfa GK, et al. Systemic therapy for advanced hepatocellular carcinoma: ASCO guideline. J Clin Oncol 2020;38:4317–45. [DOI] [PubMed] [Google Scholar]
- [3].Chen LT, Martinelli E, Cheng AL, et al. Pan-Asian adapted ESMO Clinical Practice Guidelines for the management of patients with intermediate and advanced/relapsed hepatocellular carcinoma: a TOS-ESMO initiative endorsed by CSCO, ISMPO, JSMO, KSMO, MOS and SSO. Ann Oncol 2020;31:334–51. [DOI] [PubMed] [Google Scholar]
- [4].Wan L, Guo L, Hu Y, et al. Comparing the diagnostic value of serum oligosaccharide chain (G-test) and alpha-fetoprotein for hepatitis B virus-related liver cancer. Clin Biochem 2021;89:44–50. (2.573). [DOI] [PubMed] [Google Scholar]
- [5].Zhu J, Tang B, Lv X, et al. Identifying apoptosis-related transcriptomic aberrations and revealing clinical relevance as diagnostic and prognostic biomarker in hepatocellular carcinoma. Front Oncol 2020;10:519180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dal BM, De ME, Baboci L, et al. New insights into the pharmacological, immunological, and CAR-T-cell approaches in the treatment of hepatocellular carcinoma. Drug Resist Updat 2020;51:100702. [DOI] [PubMed] [Google Scholar]
- [7].Xu HQ, Ma M, Ma YP, et al. Identification and expression characterization of ATP-binding cassette (ABC) transporter genes in melon fly. Insects 2021;12:undefined.(2.22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu X. ABC Family Transporters. Adv Exp Med Biol 2019;1141:13–00. (2.45). [DOI] [PubMed] [Google Scholar]
- [9].Lee Jyh-Yeuan. Rosenbaum daniel M. Transporters revealed. Cell 2017;168:951–3. [DOI] [PubMed] [Google Scholar]
- [10].Brodeur GM. Knowing your ABCCs: novel functions of ABCC transporters. J Natl Cancer Inst 2011;103:1207–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mao X, He Z, Zhou F, et al. Prognostic significance and molecular mechanisms of adenosine triphosphate-binding cassette subfamily C members in gastric cancer. Medicine (Baltimore) 2019;98:e18347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang L, Huang P, Huang C, et al. Varied clinical significance of ATP-binding cassette C sub-family members for lung adenocarcinoma. Medicine (Baltimore) 2021;100:e25246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Robey RW, Pluchino KM, Hall MD, et al. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat Rev Cancer 2018;18:452–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hoffmann K, Xiao Z, Franz C, et al. Involvement of the epidermal growth factor receptor in the modulation of multidrug resistance in human hepatocellular carcinoma cells in vitro. Cancer Cell Int 2011;11:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Conseil G, Cole SPC. The first cytoplasmic loop in the core structure of the ABCC1 (Multidrug Resistance Protein 1; MRP1) transporter contains multiple amino acids essential for its expression. Int J Mol Sci 2021;22:9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Emmanouilidi A, Casari I, Gokcen AB, et al. Inhibition of the lysophosphatidylinositol transporter ABCC1 reduces prostate cancer cell growth and sensitizes to chemotherapy. Cancers (Basel) 2020;12:2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gao Q, Li X, Xu Y, et al. IRE1α-targeting downregulates ABC transporters and overcomes drug resistance of colon cancer cells. Cancer Lett 2020;476:67–74. [DOI] [PubMed] [Google Scholar]
- [18].Satyananda V, Oshi M, Tokumaru Y, et al. Sphingosine 1-phosphate (S1P) produced by sphingosine kinase 1 (SphK1) and exported via ABCC1 is associated with hepatocellular carcinoma (HCC) progression. Am J Cancer Res 2021;11:4394–407. [PMC free article] [PubMed] [Google Scholar]
- [19].Le MT, Phan TV, Tran-Nguyen VK, et al. Prediction model of human ABCC2/MRP2 efflux pump inhibitors: a QSAR study. Mol Divers 2021;25:741–51. [DOI] [PubMed] [Google Scholar]
- [20].Zhu Y, Huang S, Chen S, et al. SOX2 promotes chemoresistance, cancer stem cells properties, and epithelial-mesenchymal transition by β-catenin and Beclin1/autophagy signaling in colorectal cancer. Cell Death Dis 2021;12:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Saleeb RM, Farag M, Lichner Z, et al. Modulating ATP binding cassette transporters in papillary renal cell carcinoma type 2 enhances its response to targeted molecular therapy. Mol Oncol 2018;12:1673–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Macauda A, Castelli E, Buda G, et al. Inherited variation in the xenobiotic transporter pathway and survival of multiple myeloma patients. Br J Haematol 2018;183:375–84. [DOI] [PubMed] [Google Scholar]
- [23].Ramírez-Cosmes A, Reyes-Jiménez E, Zertuche-Martínez C, et al. The implications of ABCC3 in cancer drug resistance: can we use it as a therapeutic target? Am J Cancer Res 2021;11:4127–40. [PMC free article] [PubMed] [Google Scholar]
- [24].Kryczka J, Sochacka E, Papiewska-Pająk I, et al. Implications of ABCC4-Mediated cAMP Eflux for CRC Migration. Cancers (Basel) 2020;12:3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Carozzo A, Yaneff A, Gómez N, et al. Identification of MRP4/ABCC4 as a Target for Reducing the Proliferation of Pancreatic Ductal Adenocarcinoma Cells by Modulating the cAMP Efflux. Mol Pharmacol 2019;96:13–25. [DOI] [PubMed] [Google Scholar]
- [26].Sun Y, Shi N, Lu H, et al. ABCC4 copy number variation is associated with susceptibility to esophageal squamous cell carcinoma. Carcinogenesis 2014;35:1941–50. [DOI] [PubMed] [Google Scholar]
- [27].Ritter CA, Jedlitschky G, Meyer H, et al. Cellular export of drugs and signaling molecules by the ATP-binding cassette transporters MRP4 (ABCC4) and MRP5 (ABCC5). Drug Metab Rev 2005;37:253–78. [DOI] [PubMed] [Google Scholar]
- [28].Mourskaia AA, Amir E, Dong Z, et al. ABCC5 supports osteoclast formation and promotes breast cancer metastasis to bone. Breast Cancer Res 2012;14:R149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ji G, He S, Huang C, et al. Upregulation of ATP binding cassette subfamily C member 5 facilitates prostate cancer progression and enzalutamide resistance via the CDK1-mediated AR Ser81 phosphorylation pathway. Int J Biol Sci 2021;17:1613–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bisaccia F, Koshal P, Abruzzese V, et al. Structural and functional characterization of the ABCC6 transporter in hepatic cells: role on PXE, cancer therapy and drug resistance. Int J Mol Sci 2021;22:2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ostuni A, Carmosino M, Miglionico R, et al. Inhibition of ABCC6 transporter modifies cytoskeleton and reduces motility of HepG2 cells via purinergic pathway. Cells 2020;9:1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].He L, Aleksandrov AA, An J, et al. Restoration of NBD1 thermal stability is necessary and sufficient to correct (F508 CFTR folding and assembly. J Mol Biol 2015;427:106–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pipatpolkai T, Usher S, Stansfeld PJ, et al. New insights into K channel gene mutations and neonatal diabetes mellitus. Nat Rev Endocrinol 2020;16:378–93. [DOI] [PubMed] [Google Scholar]
- [34].Southgate L, Machado RD, Gräf Stefan, et al. Molecular genetic framework underlying pulmonary arterial hypertension. Nat Rev Cardiol 2020;17:85–95. [DOI] [PubMed] [Google Scholar]
- [35].Zhang R, Li SW, Liu L, et al. TRIM11 facilitates chemoresistance in nasopharyngeal carcinoma by activating the β-catenin/ABCC9 axis via p62-selective autophagic degradation of Daple. Oncogenesis 2020;9:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kathawala RJ, Wang YJ, Ashby CR, et al. Recent advances regarding the role of ABC subfamily C member 10 (ABCC10) in the efflux of antitumor drugs. Chin J Cancer 2014;33:223–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ishiguro J, Ito H, Tsukamoto M, et al. A functional single nucleotide polymorphism in ABCC11, rs17822931, is associated with the risk of breast cancer in Japanese. Carcinogenesis 2019;40:537–43. [DOI] [PubMed] [Google Scholar]
- [38].Pham DH, Kudira R, Xu L, et al. Deleterious variants in ABCC12 are detected in idiopathic chronic cholestasis and cause intrahepatic bile duct loss in model organisms. Gastroenterology 2021;161: 287-300.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhou SF, Wang LL, Di YM, et al. Substrates and inhibitors of human multidrug resistance associated proteins and the implications in drug development. Curr Med Chem 2008;15:1981–2039. [DOI] [PubMed] [Google Scholar]
- [40].Li D, Zhao W, Zhang X, et al. NEFM DNA methylation correlates with immune infiltration and survival in breast cancer. Clin Epigenetics 2021;13:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Rhodes D, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data mining platform. Neoplasia (New York NY) 2004;6:01–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017;19:649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Thul PJ, Lindskog C. The human protein atlas: a spatial map of the human proteome. Protein Sci 2018;27:233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38: W214-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Carbon S, Ireland A, Mungall CJ, et al. Amigo: online access to ontology and annotation data. Bioinformatics 2009;25:288–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kanehisa M, Furumichi M, Sato Y, et al. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res 2021;49:D545–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nagy A, Munkárcsy G, Győrffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep 2021;11:6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wurmbach E, Chen Y, Khitrov G, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 2007;45:938–47. [DOI] [PubMed] [Google Scholar]
- [50].Mas VR, Maluf DG, Archer KJ, et al. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med 2009;15:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chen X, Cheung ST, So S, et al. Gene expression patterns in human liver cancers. Mol Biol Cell 2002;13:1929–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Roessler S, Jia HL, Budhu A, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res 2010;70:10202–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Cole Susan PC. Targeting multidrug resistance protein 1 (MRP1, ABCC1): past, present, and future. Annu Rev Pharmacol Toxicol 2014;54:95–117. [DOI] [PubMed] [Google Scholar]
- [54].Vulsteke C, Lambrechts D, Dieudonné A, et al. Genetic variability in the multidrug resistance associated protein-1 (ABCC1/MRP1) predicts hematological toxicity in breast cancer patients receiving (neo-)adjuvant chemotherapy with 5-fluorouracil, epirubicin and cyclophosphamide (FEC). Ann Oncol 2013;24:1513–25. [DOI] [PubMed] [Google Scholar]
- [55].Suzuki H, Sug Iyama Y. Single nucleotide polymorphism s inmultidrug resistance associated protein 2 (MRP2/ABCC2): its impact on drug d isposition. Adv Drug Deliv Rev 2002;54:1311–31. [DOI] [PubMed] [Google Scholar]
- [56].Mitra A, Yoshida-Court K, Solley TN, et al. Extracellular vesicles derived from ascitic fluid enhance growth and migration of ovarian cancer cells. Sci Rep 2021;11:9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Elsnerova K, Mohelnikova-Duchonova B, Cerovska E, et al. Gene expression of membrane transporters: importance for prognosis and progression of ovarian carcinoma. Oncol Rep 2016;35:2159–70. [DOI] [PubMed] [Google Scholar]
- [58].Kryczka J, Sochacka E, Papiewska-Pająk I, et al. Implications of ABCC4-mediated cAMP eflux for CRC migration. Cancers (Basel) 2020;12:undefined. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Matsumoto Y, Shiozaki A, Kosuga T, et al. Expression and role of CFTR in human esophageal squamous cell carcinoma. Ann Surg Oncol 2021;undefined:undefined. [DOI] [PubMed] [Google Scholar]
- [60].Zhou K, Liu Y, Zhao Z, et al. ABCC8 mRNA expression is an independent prognostic factor for glioma and can predict chemosensitivity. Sci Rep 2020;10:12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Demidenko R, Razanauskas D, Daniunaite K, et al. Frequent down-regulation of ABC transporter genes in prostate cancer. BMC Cancer 2015;15:683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Krizkova V, Dubova M, Susova S, et al. Protein expression of ATP-binding cassette transporters ABCC10 and ABCC11 associates with survival of colorectal cancer patients. Cancer Chemother Pharmacol 2016;78:595–603. [DOI] [PubMed] [Google Scholar]
- [63].Bera TK, Iavarone C, Kumar V, et al. MRP9, an unusual truncated member of the ABC transporter superfamily, is highly expressed in breast cancer. Proc Natl Acad Sci U S A 2002;99:6997–7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zhou Shu-Feng, Wang Lin-Lin, Di Yuan Ming, et al. Substrates and inhibitors of human multidrug resistance associated proteins and the implications in drug development. Curr Med Chem 2008;15:1981–2039. [DOI] [PubMed] [Google Scholar]
- [65].Zhang H, You Y, Zhu Z. The human RNA surveillance factor Up-frameshift 1 inhibits hepatic cancer progression by targeting MRP2/ABCC2. Biomed Pharmacother 2017;92:365–72. [DOI] [PubMed] [Google Scholar]
- [66].Nies AT, König J, Pfannschmidt M, et al. Expression of the multidrug resistance proteins MRP2 and MRP3 in human hepatocellular carcinoma. Int J Cancer 2001;94:492–9. [DOI] [PubMed] [Google Scholar]
- [67].Borel F, Han R, Visser A, et al. Adenosine triphosphate-binding cassette transporter genes up-regulation in untreated hepatocellular carcinoma is mediated by cellular microRNAs. Hepatology 2012;55:821–32. [DOI] [PubMed] [Google Scholar]