Abstract
During cell cycle progression in metazoans, the kinetochore is assembled at mitotic onset and disassembled during mitotic exit. Once assembled, the kinetochore complex attached to centromeres interacts directly with the spindle microtubules, the vehicle of chromosome segregation. This reassembly program is assumed to be absent in budding and fission yeast, because most kinetochore proteins are stably maintained at the centromeres throughout the entire cell cycle. Here, we show that the reassembly program of the outer kinetochore at mitotic onset is unexpectedly conserved in the fission yeast Schizosaccharomyces pombe. We identified this behavior by removing the Rabl chromosome configuration, in which centromeres are permanently associated with the nuclear envelope beneath the spindle pole body during interphase. In addition to having evolutionary implications for kinetochore reassembly, our results aid the understanding of the molecular processes responsible for kinetochore disassembly and assembly during mitotic entry.
INTRODUCTION
The three-dimensional architecture of the yeast genome is characterized by the evolutionarily conserved Rabl chromosome configuration, which is defined by the stable association of centromeres and telomeres with the nuclear envelope (NE; Jin et al., 1998, 2000; Gerlich and Ellenberg, 2003; Berger et al., 2008; Duan et al., 2010; Taddei and Gasser, 2012; Mizuguchi et al., 2014). The NE comprises the inner nuclear membrane (INM) and the outer nuclear membrane (ONM). INM proteins play key roles in the interaction of the NE with chromatin (Czapiewski et al., 2016; Fernandez-Alvarez and Cooper, 2017a). In particular, in fission yeast, centromeres are clustered together in a kinetochore-dependent manner at the INM, beneath the spindle pole body (SPB, the centrosome equivalent in yeast) and opposite the nucleolus (Funabiki et al., 1993; Ding et al., 1997). The linkage between centromeres and the INM occurs via the SPB-associated linker of nucleoskeleton and cytoskeleton (LINC) complex, which comprises the KASH-domain ONM proteins (Kms1 and Kms2) and the SUN-domain INM protein (Sad1; Hagan and Yanagida, 1995; Shimanuki et al., 1997; Hiraoka and Dernburg, 2009; Unruh et al., 2018). Sad1 plays an essential role in supporting the associations between centromeres and the SPB. These associations are also strengthened by the protein Csi1, which bridges Sad1 and outer kinetochore proteins, and by the conserved LEM-domain INM protein Lem2, which localizes at the nuclear periphery and the SPB (Hou et al., 2012; Barrales et al., 2016; Fernandez-Alvarez and Cooper, 2017a). The Rabl configuration is thought to reflect the positioning of the chromosomes during their segregation from the preceding mitosis; in metazoans, the Rabl configuration is dismantled at the mitotic exit, but in yeast, it is maintained throughout the subsequent interphase (Cremer and Cremer, 2001; Taddei and Gasser, 2012; Mizuguchi et al., 2014, 2015). Why the Rabl configuration in yeast is not disassembled at mitotic exit but is maintained throughout interphase is not known. Recently, it has been observed in fission yeast that the interaction of at least one centromere with the SPB during interphase is required to trigger SPB insertion into the NE, a crucial event for nucleating the spindle microtubules. Hence, disassembly of the Rabl configuration abolishes SPB insertion and spindle formation, leading to cellular lethality (Fernandez-Alvarez et al., 2016).
Attachment of centromeres to the SPB, and thus the maintenance of the Rabl configuration, is supported by the kinetochore, which is built on the centromeres (Cheeseman, 2014). This large complex comprises around 80 proteins identified in humans, and its major components are conserved throughout eukaryotes. The kinetochore can be subdivided into two distinct regions: the inner kinetochore, which interacts with chromatin, and the outer kinetochore, which constitutes the platform for interacting with spindle microtubules. Therefore, the kinetochore establishes the chromosomal attachment place for spindle microtubules, the motors that drive chromosome distribution to daughter cells (Cheeseman and Desai, 2008).
Kinetochore composition is dynamically regulated during the cell cycle in metazoans (Hara and Fukagawa, 2018). Some kinetochore proteins are constitutively present at centromeres, establishing the centromere-associated network (CCAN), but most are recruited to the kinetochore during late G2, prophase, or mitosis. In this manner, proteins of the outer kinetochore, such as Mis12 and Ndc80, are recruited precisely to the centromeres in late interphase and prophase, respectively. Once the chromosomes have segregated, Ndc80 and Mis12 are depleted in an orderly way following the onset of anaphase and the end of mitosis, respectively (Cheeseman and Desai, 2008; Nagpal and Fukagawa, 2016; Dhatchinamoorthy et al., 2018). This well-regulated recruitment of kinetochore components to the centromeres is assumed to be absent in Schizosaccharomyces pombe. In the fission yeast, most of the outer kinetochore components, such as Ndc80 and Nuf2, are constitutively present at centromeric regions throughout the cell cycle (Saitoh et al., 1997; Nabetani et al., 2001; Wigge and Kilmartin, 2001; Hayashi et al., 2004; Biggins, 2013), and only the components of the DASH complex, an essential element of the kinetochore that is required for the biorientation of sister chromatids, are recruited during mitosis (Cheeseman et al., 2001; Janke et al., 2002; Liu et al., 2005).
Two hypotheses aim to explain the absence of the outer kinetochore disassembly/assembly program at mitotic onset in fission yeast. The first hypothesis posits that the disassembly/assembly program is coordinated with NE breakdown in metazoans (Guttinger et al., 2009; Smoyer and Jaspersen, 2014; Hattersley et al., 2016). Thus, the fact that the NE is not disassembled before mitosis in S. pombe suggests that a kinetochore disassembly/assembly program might not be an efficient mechanism of outer kinetochore formation, as it involves the active transit of proteins from the cytoplasm to the nucleus. The second hypothesis suggests that the preservation of the outer kinetochore structure during interphase might be justified by its crucial role in maintaining the Rabl configuration (Takahashi et al., 2000; Asakawa et al., 2005). Thus, in this case, the absence of an assembly program in fission yeast mitosis would be, in principle, independent of the absence of proper NE breakdown.
It is challenging to determine which hypothesis explains the absence of the outer kinetochore disassembly/assembly program in fission yeast. This challenge could be addressed by studying the behavior of the kinetochore in cells without the interphase Rabl configuration. However, it has been difficult to generate Rabl configuration–deficient cells without compromising either kinetochore structure or cell viability. For instance, mutations to Nuf2 or Ndc80 partially remove the Rabl configuration but also alter kinetochore structure (Nabetani et al., 2001; Asakawa et al., 2005; Hsu and Toda, 2011); in contrast, the presence of a thermosensitive allele of sad1 (sad1.2) at a restrictive growth temperature (36°C) abolishes all centromere–SPB associations but immediately leads to cell death (Fernandez-Alvarez et al., 2016). Hence, the identification of a new genetic background in which the intact kinetochore is completely disconnected from the SPB at 32°C (the standard growth temperature of fission yeast) without dramatically impairing cell viability would help to reveal the behavior of kinetochore proteins in the absence of the Rabl configuration during yeast interphase. With that goal in mind, we found that the combination of the sad1.2 allele and the deletion of csi1 at the semipermissive temperature of 32°C generates severe centromere dissociation defects. However, most of the cells are still viable due to occasional centromere interactions with the SPB, which are sufficient to trigger spindle formation. Thus, sad1.2 csi1∆ represents a new scenario in which it is possible to characterize the behavior of kinetochores dissociated from the SPB independently of their essential function in maintaining the Rabl configuration.
Here, we show that key elements of the outer kinetochore structure are lost in interphase sad1.2 csi1∆ cells. More unexpectedly, we show that similar to the situation in metazoans, the outer kinetochore in yeast is reassembled in late G2. These results suggest that the outer kinetochore assembly program at mitotic onset is conserved in fission yeast but has not been observed thus far, because it is masked by the Rabl configuration. Our observations establish S. pombe as a model organism for studying the mechanisms behind kinetochore assembly, which is highly conserved in metazoans and has enormous relevance to faithful chromosome segregation during cell cycle progression.
RESULTS AND DISCUSSION
The Rabl configuration in fission yeast is not supported by the cytoskeleton
We explored different approaches to removing interphase centromere-SPB associations at 32°C without altering the stability of either the kinetochore or the SPB. In budding yeast, centromere–SPB associations require nuclear microtubules (Jin et al., 2000; Bystricky et al., 2004), but studies in fission yeast have shown by electron microscopy an apparent absence of microtubules in the nuclear microenvironment between centromeres and the SPB (Ding et al., 1997; Appelgren et al., 2003). To rule out a role for microtubules in supporting centromere–SPB associations, we evaluated the state of these associations on addition of the microtubule-depolymerizing drug carbendazim (also known as methyl-2-benzimidazole carbamate, or MBC). These experiments confirmed that, in contrast to the situation in Saccharomyces cerevisiae, the maintenance of the Rabl configuration in S. pombe is independent of nuclear microtubules (Supplemental Figure 1, A–D).
We also considered whether actin is involved in maintaining the Rabl configuration, as actin has a well characterized role in promoting telomere positioning at the NE during budding yeast meiotic prophase (Trelles-Sticken et al., 2005). We disrupted actin by adding latrunculin A (LatA), an actin polymerization inhibitor (Riedl et al., 2008; Huang et al., 2012). Although LatA treatment led to major structural defects in actin, centromere–SPB associations persisted (Supplemental Figure 1, B–E). Hence, we discount a major role for cytoskeleton motors in maintaining the Rabl configuration.
Phosphomutants and phosphomimetics of the sad1.2 allele do not show defects in centromere dissociation from the spindle pole body
Another approach to addressing the complete disruption of the Rabl configuration in S. pombe is to use the thermosensitive Sad1 allele sad1.2. The Sad1.2 protein harbors two single substitutions, Thr-3-Ser and Ser-52-Pro, the latter of which disrupts Ser-52, a validated phosphorylation site for the cyclin-dependent protein kinase Cdc2/CDK-1 (Carpy et al., 2014; Fernandez-Alvarez et al., 2016; Swaffer et al., 2016). All three centromeres dissociate from the SPB when sad1.2 cells are grown at 36°C (Supplemental Figure 1F). However, this scenario leads to cell lethality as a result of failures in SPB insertion into the NE and in spindle formation (Fernandez-Alvarez et al., 2016). In contrast, the growth of sad1.2 cells at a semipermissive temperature (32°C) produces only partial dissociation of the centromeres from the SPB and does not completely disrupt the Rabl configuration (Fernandez-Alvarez and Cooper, 2017b).
To improve the penetrance of centromere declustering in sad1.2 cells at 32°C, we tested whether any other phosphomutants or phosphomimetics at Thr-3 and Ser-52 increases the percentage of cells showing centromere declustering during interphase. However, this analysis showed that only the combination of Thr-3-Ser and Ser-52-Pro leads to centromere declustering from SPB; no other combinations produce centromere dissociation defects (Supplemental Figure 1G). In addition, analysis of cellular growth on MBC-containing media showed that hypersensitivity to MBC was higher for sad1.2 cells than for the other sad1 mutant allele combinations (Supplemental Figure 1H). Previous studies have shown that hypersensitivity to microtubule-depolymerizing drugs (MBC or TBZ) in mutants showing centromere–SPB dissociation might be linked to problems in chromosome recapture for the spindle during mitosis; the fact that centromeres are dissociated and located far from the SPB, the major microtubule nucleator center, would complicate their capture (Hou et al., 2012). These observations argue against the possibility that centromere–SPB associations are regulated only by phosphorylation of the Sad1 residues Thr-3 and Ser-52. Therefore, the association of centromeres with Sad1 must be controlled by other complementary mechanisms, which are probably altered by the Thr-3-Ser and Ser-52-Pro substitutions but not by cytoskeleton motors.
Loss of Csi1 in sad1.2 cells leads to a higher rate of total centromere–spindle pole body dissociation
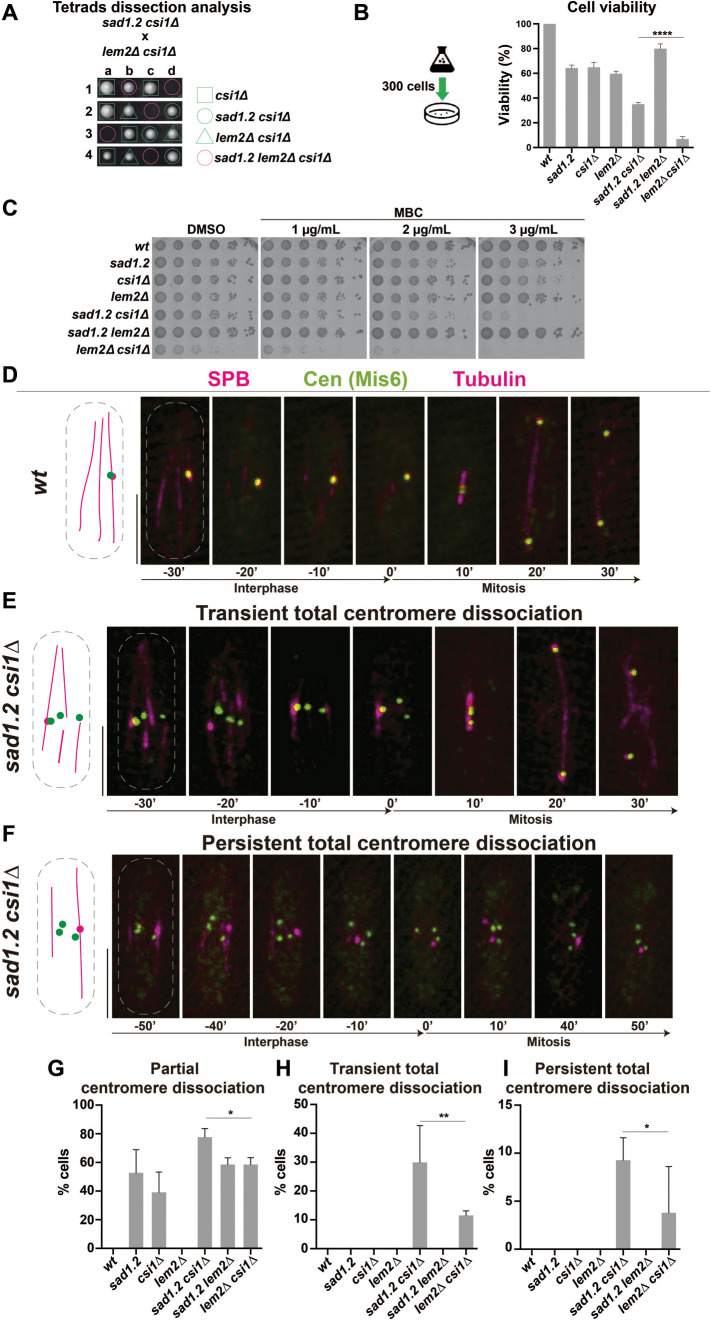
The aforementioned observations indicate that the Rabl configuration in S. pombe is independent of microtubules and actin. Our analysis of the sad1.2 phosphomutant and phosphomimetic alleles also suggests that only the combination of Thr-3-Ser and Ser-52-Pro at sad1 leads to centromere dissociation from the SPB. Hence, to increase the penetrance of centromere–SPB dissociations in sad1.2 cells, we constructed strains combining mutations in sad1, csi1, and lem2, since the Sad1-interacting factor Csi1 and the LEM-domain INM protein Lem2 could be the only major regulators in maintaining the centromere–SPB associations (Hou et al., 2012; Barrales et al., 2016; Fernandez-Alvarez and Cooper, 2017b). We found a strong negative interaction in the triple mutant sad1.2 lem2∆ csi1∆ (Figure 1A): in 69.3% of asci dissected (n = 62), spores were unable to germinate or produced very small colonies. The remaining sad1.2 lem2∆ csi1∆ spores were able to generate colonies. This rate of suppression might be explained by the compensatory increase in lnp1 gene expression, which frequently suppresses the loss of lem2, as has been observed previously (Tange et al., 2016). Due to these severe viability defects, we ceased working with the triple mutant. Analysis of the behavior of all possible double mutant combinations showed that double loss of Csi1 and Lem2 also severely hindered cell viability (Figure 1B). These defects have been associated with defective pericentromeric heterochromatin identity, which impairs kinetochore proteins’ association with centromeres. This leads to chromosome loss and subsequent growing defects on MBC-containing media, as has been reported previously (Hou et al., 2012; Barrales et al., 2016; Figure 1C). The other double mutant showing cellular growth defects was sad1.2 csi1∆, although these defects were weaker than those in lem2∆ csi1∆; sad1.2 csi1∆ also showed increased sensitivity to MBC (Figure 1, B and C).
FIGURE 1:
Loss of Csi1 in sad1.2 cells leads to a higher rate of Rabl configuration-deficient cells. (A) Tetrad analysis of h- sad1.2 csi1∆ crossed with h+ lem2∆ csi1∆ shows a strong negative genetic interaction between sad1.2, lem2∆, and csi1∆ when spores harbor the three mutations. Spores were grown at 32°C for 5 d. (B) Cell viability relative to wt cells was evaluated by colony formation assays. Cells were cultured in liquid medium to 107 cells/mL, 300 cells spotted onto YE4S plates, and incubated at 32°C for 5 d (n > 500 colonies per genotype were scored in four independent experiments). Data were subjected to Fisher’s exact test; ****, p-value < 0.0001. (C) Serial dilutions (fivefold) of log-phase cultures were spotted and grown on rich media with DMSO (control) and rich media containing MBC. Plates were incubated at 32°C for 48 h. (D–F) Frames from films of proliferating cells carrying Sid4-mCherry (endogenously tagged; SPB), Mis6-GFP (endogenously tagged; centromeres), and ectopically expressed mCherry-Atb2 (tubulin). Scale bar represents 5 µm. (G–I) Centromere–SPB association patterns. >30 cells were scored for each genotype in at least three independent experiments. p-values were determined by Fisher’s exact test; **, 0.001< p < 0.01; *, 0.01< p < 0.05.
Due to the correlation between centromere dissociation from the SPB and MBC sensitivity, these experiments pointed to the double mutants sad1.2 csi1∆ and lem2∆ csi1∆ as possibly having greater centromere–SPB dissociation than the single mutants csi1∆ and sad1.2. Around 10–15% of lem2∆ csi1∆ cells show all centromeres transiently disconnected from the SPB (Barrales et al., 2016; Fernandez-Alvarez and Cooper, 2017b), but no information has been obtained yet about sad1.2 csi1∆ cells. For this reason, we investigated centromere–SPB associations in sad1.2 csi1∆ cells. For comparative purposes, we included all single and double mutant combinations and allocated centromere dissociation phenotypes into two categories: 1) partial centromere–SPB dissociation, where at least one centromere is detached from the SPB during the analysis (example in –30′ frame in Figure 1E); and 2) total centromere–SPB dissociation, where all three centromeres are dissociated from the SPB. In the latter category, we established two subtypes: transient, where at least one frame in interphase showed total centromere–SPB dissociation during our time-lapse analysis (example in –20′ frame in Figure 1E); or persistent, where centromeres did not interact at all with the SPB at any time during the analysis in interphase (Figure 1F). In the case of transient total centromere–SPB dissociation, cells are still able to divide because one centromere–SPB interaction is sufficient to trigger SPB insertion into the NE, which allows spindle formation (Fernandez-Alvarez et al., 2016; Figure 1E). In contrast, in persistent total centromere–SPB dissociation, SPB insertion and thus spindle formation are abolished (Fernandez-Alvarez et al., 2016; Figure 1F). Notably, we found the highest penetrance of centromere–SPB dissociation in sad1.2 csi1Δ cells: around 80% of sad1.2 csi1Δ cells showed centromere clustering defects (Figure 1G). Most importantly, ∼25% of these mutant cells showed transient total centromere–SPB dissociation, which was never seen in the csi1Δ, lem2Δ or sad1.2 single mutants (Figure 1H). In contrast, ∼9% of sad1.2 csi1Δ cells displayed persistent total centromere–SPB dissociation (Figure 1I). This population of cells might explain the slight reduction in cellular viability, because, as described above, persistent total centromere–SPB dissociation leads to cell death. Intriguingly, although the cell growth defects and MBC sensitivity of the lem2∆ csi1∆ strain are more severe than those of the sad1.2 csi1Δ genotype (Figure 1C), the penetrance of centromere–SPB dissociation is lower in lem2∆ csi1∆ than in sad1.2 csi1Δ (Figures 1, G–I). This suggests that some of the growth defects could be unrelated to the loss of centromere–SPB contacts. For instance, the role of Lem2 in the maintenance of centromeric heterochromatin and nuclear envelope integrity might be behind the strong growth defects in lem2∆ csi1Δ (Barrales et al., 2016; Tange et al., 2016; Kume et al., 2019). In conclusion, we identified sad1.2 csi1Δ cells as an optimal strain for exploring the behavior of the kinetochore in Rabl configuration-deficient cells, because they present higher penetrance of total centromere–SPB dissociation and weaker defects in cell viability than lem2Δ csi1Δ cells.
Inner kinetochore components are stably maintained at the centromeres during interphase in sad1.2 csi1∆ cells
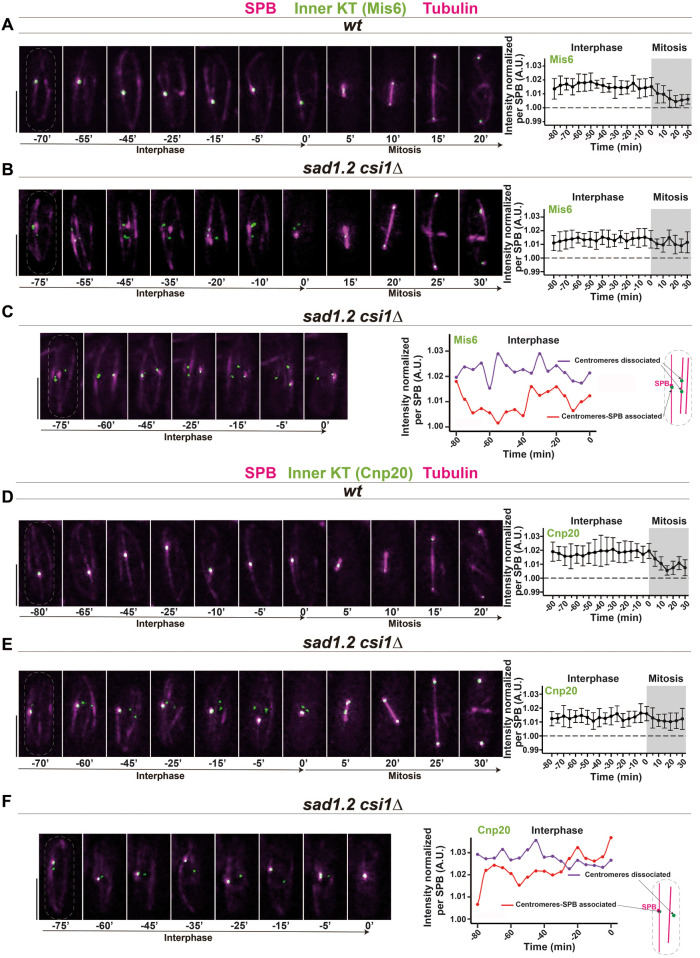
To address the behavior of kinetochore proteins during cell cycle progression in fission yeast with (wt) and without the Rabl configuration (sad1.2 csi1∆), we followed the focal intensity of endogenously GFP-tagged outer and inner kinetochore proteins on exponentially growing cells by live fluorescence microscopy. We analyzed Mis6 and Cnp20 (CENP-I and CENP-T orthologues, respectively) as representative members of the inner kinetochore (Takahashi et al., 2000; Hou et al., 2012). Protein intensity levels of all images acquired from living cells were quantified (see Methods for details), and we delineated the focal intensity of both kinetochore proteins normalized per SPB signal (visualized via Sid4-mCherry). A comparison of the sum of Mis6-GFP signals in wt and sad1.2 csi1∆ cells confirmed the presence of a stable Mis6-GFP signal throughout interphase in both strains (Figure 2, A and B). By analyzing the sum of the centromeres dissociated from the SPB and the sum of the centromeres associated with the SPB independently, we found that both centromere locations show a stable presence of Mis6-GFP during mitotic interphase (Figure 2C). Consistent with the previous results, our analysis of the inner kinetochore protein Cnp20 showed that it has behavior similar to that of Mis6-GFP: wt and sad1.2 csi1∆ cells showed a stable signal in interphase independent of whether or not centromeres are associated with the SPB (Figure 2, D–F). Together, these observations indicate that the inner kinetochore, at least Mis6 and Cnp20, is stably associated with the centromeres in Rabl configuration-deficient cells.
FIGURE 2:
Mis6 and Cnp20 are stably associated with the centromeres in Rabl configuration-deficient cells. (A–F) Frames from films of mitotic cells carrying Sid4-mCherry (SPB), ectopically expressed mCherry-Atb2 (tubulin), and endogenously tagged Mis6-GFP, A–C, or Cnp20-GFP, D–F. Bars, 5 µm. Mean of total Mis6-GFP, A, B, and Cnp20-GFP, D, E intensities through interphase and mitosis were quantified. Ten cells during more than three independent experiments were monitored for focal intensity of Mis6-GFP and Cnp20-GFP. Error bars represent standard deviations; t = 0 min is just before SPB separation. (C, F) Quantification of Mis6-GFP and Cnp20-GFP focus intensity separately in centromeres with and without SPB association in sad1.2 csi1∆ cells. (A, D) In wt cells, all Mis6-GFP and Cnp20-GFP signals localize to the SPB during interphase. (B, C, E, F) sad1.2 csi1∆ cells showing centromere dissociation from the SPB also show stable signals through interphase.
We also explored the behavior of Mis12, a component of the NMS (Ndc80-MIND-Spc7) complex (Obuse et al., 2004; see Supplemental Figure 2A for a schematic representation of the main elements of the fission yeast outer kinetochore). Mis12 is also assembled and disassembled at mitotic onset and exit, respectively, during the metazoan cell cycle, but is constantly attached to centromeres in yeast (Biggins, 2013). Analysis of Mis12-GFP in wt and Rabl configuration-deficient cells (sad1.2 csi1∆) showed a stable signal across interphase (Supplemental Figure 2, B and C), similar to the behavior of the inner kinetochore proteins Mis6 and Cnp20. However, the endogenous GFP-tagging of Mis12 greatly reduced the penetrance of centromere–SPB dissociation defects in sad1.2 csi1∆ cells (Supplemental Figure 2D). Hence, the tagging of Mis12 is able to stabilize the interaction between the kinetochore and Sad1, reducing the impact of sad1.2 and csi1 mutations on centromere dissociation from the SPB.
Ndc80 and Nuf2 are dissociated from centromeres during interphase in the Rabl configuration-deficient cells sad1.2 csi1∆
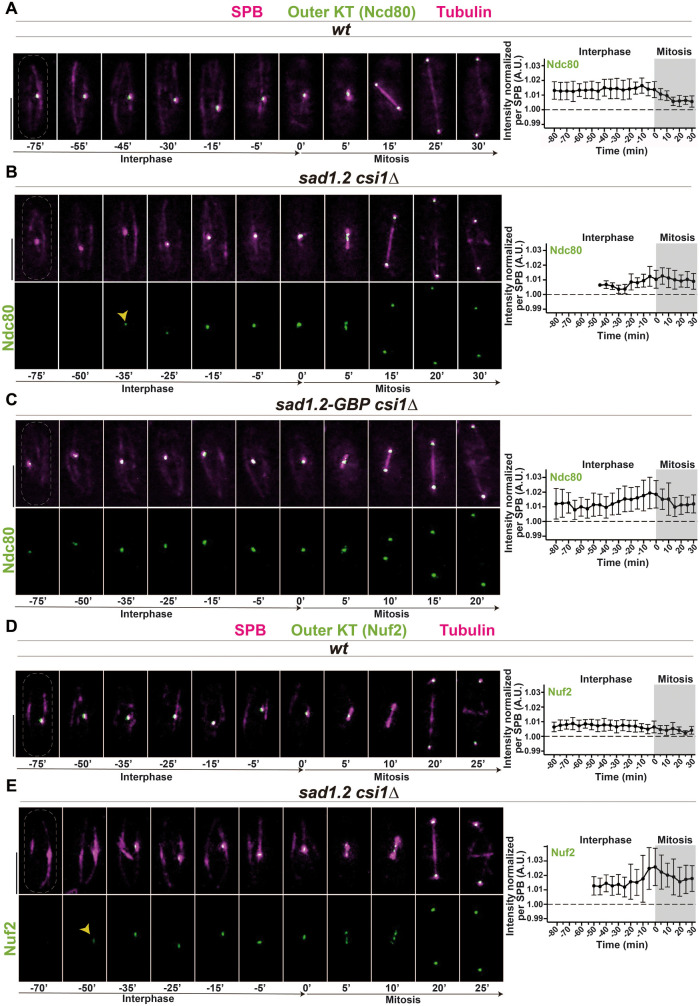
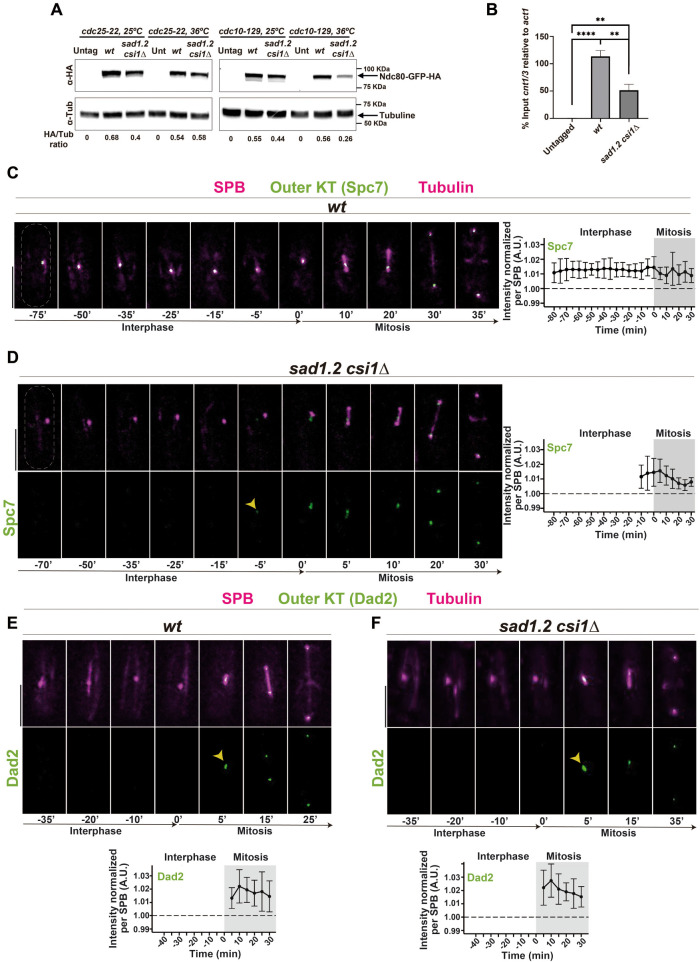
We studied two representative members of the outer kinetochore, Ndc80 and Nuf2 (part of the Ndc80 complex; see Supplemental Figure 2A), which are absent in interphase in metazoans but localize at centromeres throughout the cell cycle in budding and fission yeast (Liu et al., 2005; Biggins, 2013). Contrary to our observations for the inner kinetochore, we could not detect a clear signal for Ndc80-GFP at the centromeres in the vast majority of sad1.2 csi1∆ interphase cells (Figure 3, A–B; Supplemental Figure 3A). This loss of signal at the centromeres during interphase was also observed for Nuf2, which indicates that the absence of a normal signal is a general defect in the Ndc80 complex (Figure 3, D–E). To confirm this observation, we analyzed the inner and outer kinetochore together using Mis6-mCherry and Ndc80-GFP, respectively. In agreement with our previous observations, we found that the deletion of Csi1 in sad1.2 cells leads to the loss of the Ndc80-GFP signal but not of Mis6-mCherry in interphase, and in cases where sad1.2 csi1∆ cells displayed the Ndc80-GFP signal (about 20% of cells; Supplemental Figure 3A), most of these centromeres (>90%) colocalized with the SPB (Supplemental Figure 3B). This suggests that the loss of Csi1 or/and Sad1.2 might destabilize the outer kinetochore, Ndc80 and Nuf2, and thus debilitate the centromere–SPB association, which has been observed using thermosensitive alleles of Ndc80 and Nuf2 (Asakawa et al., 2005). Western blot analysis of cultures enriched in G1, using the thermosensitive allele cdc10-129 (Tormos-Perez et al., 2016), indicated that Ndc80 protein levels are reduced in sad1.2 csi1∆ cells from those in wt settings (Figure 4A). Furthermore, ChIP-qPCR analysis showed lower enrichment of Ndc80 at sad1.2 csi1∆ centromeres (1 and 3) than in wt (Figure 4B).
FIGURE 3:
Ndc80 and Nuf2 are dissociated from centromeres during interphase and reassembled at mitotic onset in sad1.2 csi1∆ cells. (A–E) Frames from films of mitotic cells carrying SPB and tubulin markers as in Figure 2, and endogenously tagged Ndc80-GFP, A–C, or Nuf2-GFP, D, E. Bars, 5 µm. Means of total Ndc80-GFP and Nuf2-GFP intensities were quantified as in Figure 2. Ten cells during more than three independent experiments were monitored. Error bars represent standard deviations; t = 0 min is just before SPB separation. (C) The GBP-GFP system was used to force centromere–SPB interactions (see Supplemental Figure 3C). Association with centromeres and levels of Ndc80 protein in interphase are recovered in sad1.2-GBP csi1∆ settings compared with wt settings.
FIGURE 4:
Rabl configuration-deficient cells efficiently complete outer kinetochore assembly. (A) Western blot analysis of protein samples from cells synchronized in G2/M (left) or G1 (right). Labels to the left indicate the antibodies used to probe the blots. The data shown are from a single representative experiment out of two repeats and quantifications from below are the averages of the HA signal relative to the tubulin control from two independent experiments. (B) ChIP analysis of Ndc80 levels at centromeres 1 and 3 in cells synchronized in G1. Error bars represent standard deviations of three biological replicates. p-values were determined by t test, **** p < 0.0001, ** p < 0.005. (C–F) Frames from films of mitotic cells carrying SPB and tubulin markers as in Figure 2, and endogenously tagged Spc7-GFP, C, D, or Dad2-GFP, E, F. Bars, 5 µm. Means of total Spc7-GFP and Dad2-GFP intensities were quantified as in Figure 2. Ten cells during more than three independent experiments were quantified. Error bars represent standard deviations; t = 0 min is just before the separation of the SPBs.
To test the hypothesis that the absence of the normal Ndc80 signal is the cause, and not the consequence, of the centromere–SPB dissociation in sad1.2 csi1∆ cells, we ectopically targeted Ndc80-GFP to Sad1.2 using the GFP binding protein GBP to recruit GFP-tagged proteins (Rothbauer et al., 2006; Fernandez-Alvarez et al., 2016; Supplemental Figure 3C). Using this system, we confirmed that the Ndc80-GFP signal is constant throughout interphase in all sad1.2 csi1∆ cells analyzed (Figure 3C). Moreover, we found that these cells did not show centromere–SPB dissociation and consequently restored the sad1.2 csi1∆ growth defects on TBZ-containing media (Supplemental Figure 3D). Together, these results indicate that the phenotype of centromere–SPB dissociation in sad1.2, csi1∆, and sad1.2 csi1∆ cells might be explained by the loss of Ndc80 at centromeres, which would weaken the interaction of the kinetochore with the SPB. Outer kinetochore instability and, consequently, centromere–SPB dissociation could also be caused by loss of proximity of the centromeres to the NE region below the SPB, which could provide the necessary nuclear microenvironment to maintain kinetochore and centromere identity, as has recently been suggested by the Allshire lab (Wu et al., 2021).
Although the outer kinetochore is crucial for maintaining the interaction of the centromeres with the SPB in fission yeast (Nabetani et al., 2001; Asakawa et al., 2005), interactions in the absence of the outer kinetochore have been characterized. For instance, during meiotic prophase, the outer kinetochore is dissembled and reassembled to prepare the kinetochore for the segregation of homologous chromosomes in meiosis I (Hayashi et al., 2006). At the beginning of meiotic prophase, the centromeres are dissociated from the SPB while the telomeres are moved by cytoskeleton motors to the SPB to form the telomere bouquet (Yoshida et al., 2013), the prophase-specific chromosomal configuration where the telomeres cluster together at the SPB (Chikashige et al., 1994; Chikashige et al., 2006). In the case of telomere bouquet mutants, such as bqt1∆, the absence of a telomere–SPB interaction allows sporadic and short-lived centromere–SPB interactions (lasting around 30 min) during meiotic prophase, even in the absence of Ndc80 and Nuf2 at the centromeres (Fennell et al., 2015). Transient total centromere–SPB dissociation, one of the most common centromere dissociation phenotypes in sad1.2 csi1∆ cells, might reflect a situation similar to meiotic prophase in bouquet-deficient meiocytes, in which centromeres are able to interact with the SPB for short periods of times when little of Ndc80 is present. Current studies aim to decipher the molecular basis of centromere anchoring to the SPB.
The outer kinetochore is reassembled at mitotic onset
Although we did not observe normal Ndc80 and Nuf2 signals in interphase sad1.2 csi1∆ cells, these proteins are clearly visible at the centromeres in mitotic cells. In fact, western blot analysis confirmed that Ndc80 protein levels were similar in G2/M phase–cultured cells with and without the Rabl configuration (Figure 4A). This observation indicates that the outer kinetochore can be rebuilt at mitotic onset to prepare cells for chromosome segregation. To establish the dynamics of outer kinetochore reassembly, we quantified the Ndc80-GFP and Nuf2-GFP signals using the same methodology used for Mis6, Cnp20, and Mis12. We found that Ndc80-GFP and Nuf2-GFP signals accumulate during late interphase, around 40–20 min before separation of the duplicated SPBs (Figure 3, B and E). This accumulation of Ndc80 at the centromeres is never seen in a wt setting, and it precedes the increment of the protein levels observed during anaphase (Dhatchinamoorthy et al., 2017). Hence, the active accumulation of the outer kinetochore, or at least its core proteins Ndc80 and Nuf2, at mitotic onset in fission yeast is similar in timing to that seen in metazoans.
Next, to understand the behavior of other kinetochore proteins, we analyzed Spc7 (KNL1 ortholog in humans), a member of the NMS complex (Supplemental Figure 2A). Analysis of endogenously GFP-tagged Spc7 showed that while the signal is stably maintained throughout interphase in wt settings, it is dramatically reduced at the centromeres in interphase and recovered at mitotic onset in sad1.2 csi1∆ cells, similarly to the behavior observed for Ndc80 and Nuf2 (Figure 4, C and D). In other words, the stability of Spc7 at the centromeres is compromised in sad1.2 csi1∆ cells, and cells with single mutations to Sad1 and Csi1 did not show any clear loss of Spc7 signal (100% of 50 cells analyzed). However, unlike Ndc80 and Nuf2, which are recruited around 40–20 min before mitosis, Spc7 is recruited around 10–5 min before mitosis (Figure 4D). Hence, our data indicate that Ndc80 and Nuf2 are recruited to the centromere before Spc7, which suggests that centromere dissociation from the SPB in sad1.2 csi1∆ cells is due to the loss of Ndc80 and/or Nuf2 rather than the loss of Spc7.
To confirm the ability of Rabl configuration–deficient cells (sad1.2 csi1∆) to complete outer kinetochore assembly efficiently, we studied the behavior of the recruitment of Dad2, a member of the DASH complex in S. pombe. Wt and sad1.2 csi1∆ cells showed similar recruitment profiles at the beginning of mitosis (Figure 4, E and F), confirming that one of the final stages of the outer kinetochore assembly is performed correctly in sad1.2 csi1∆ cells, which ensures chromosome segregation. Our results indicate the existence of a program in fission yeast to actively recruit the outer kinetochore that is triggered at the beginning of mitosis in a manner reminiscent of the assembly program in metazoans.
The high conservation of the outer kinetochore disassembly/assembly program during cell cycle progression appears to have a remarkable exception in yeast, where this program is assumed to be absent. Here, we present evidence that the program might actually be present in fission yeast mitosis but masked by the Rabl configuration. A plausible explanation is that the maintenance of the Rabl configuration during interphase appears in evolution later than the outer kinetochore disassembly/assembly program. According to this hypothesis, the function of the Rabl configuration in controlling SPB insertion into the NE, a yeast-specific mechanism, favors Ndc80 and Nuf2 remaining stable at the centromeres to preserve centromere–SPB interactions. Using the double mutant sad1.2 csi1∆, we found that Ndc80 and Nuf2 are lost from the centromere during interphase, which likely destroys the Rabl configuration. Unexpectedly, we found that Ndc80 and Nuf2 are recruited to the centromeres with a timing reminiscent of that seen in metazoans. We also found that Spc7 is recruited to the centromeres before mitosis but slightly later than Ndc80 and Nuf2 recruitment. Hence, we establish four groups of kinetochore proteins based on their pattern of location across interphase in Rabl configuration-deficient cells (sad1.2 csi1∆): 1) persistent signal throughout interphase: Mis6 and Cnp20; 2) early recruitment in the transition from G2 to M: Ndc80 and Nuf2; 3) late recruitment in the transition from G2 to M: Spc7; 4) assembly at mitosis onset: Dad2. In contrast, we identified only two groups in wt settings: 1) persistent signal throughout interphase: Mis6, Cnp20, Ndc80, Nuf2, and Spc7; 2) assembly at mitosis onset: Dad2 (Figure 5). This program of kinetochore reassembly follows stages similar to those occurring during fission yeast meiotic prophase, in which there are two stages of outer kinetochore reconstruction, the first one formed by the Ndc80 complex and the second one by the NMS complex (Spc7), followed by the assembly of the DASH complex (Dad2). Hence, S. pombe cells might harbor the kinetochore reassembly program in mitosis and meiosis but use it only in meiosis, because the maintenance of the Rabl configuration by the outer kinetochore, a process that might have appeared later in evolution, overlaps the outer kinetochore disassembly/assembly program. However, in contrast to the scenario in meiosis, our results regarding Ndc80 protein levels in sad1.2 csi1∆ indicate that the outer kinetochore proteins are degraded, not just relocated, as has been observed during meiotic prophase (Asakawa et al., 2005; Hayashi et al., 2006). Unexpectedly, our results suggest that the mechanism controlling the disassembly and reassembly of the outer kinetochore might also be conserved in fission yeast. Discovering the existence of this mechanism in S. pombe opens up the possibility of future studies using this yeast model to explore the metazoan outer kinetochore reconstruction program in mitosis.
FIGURE 5:
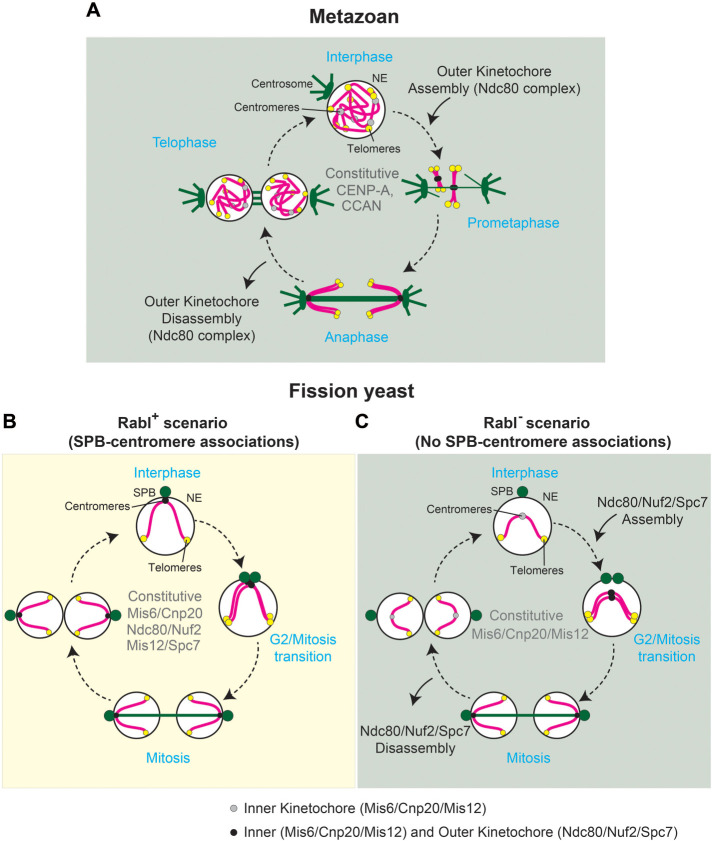
Pattern of location of kinetochore proteins across interphase in Rabl configuration-deficient cells (sad1.2 csi1∆). (A) In metazoans, CENP-A and CCAN (constitutive centromere-associated network) are constitutively associated with centromeres during the cell cycle. Ndc80 complex is assembled and accumulated at centromeres in prophase, and it is delocalized during late anaphase–telophase. (B) Fission yeast cells show persistent signal of inner and outer kinetochore proteins throughout interphase. (C) Outer kinetochore components are recruited to the centromeres in Rabl configuration-deficient cells with a timing reminiscent of that seen in metazoans.
METHODS
Request a protocol through Bio-protocol.
Strains and growth conditions
Strains used are listed in Supplementary Table 1. Growth conditions and molecular biology approaches were used as described previously (Moreno et al., 1991). Gene deletion and C-terminal tagging were performed as described by Bahler et al. (1998) and Fennell et al. (2015). Insertions of mCherry-Atb2 at the aur1 locus (Hashida-Okado et al., 1998) utilized pYC19-mCherryAtb2 (Nakamura et al., 2011) provided by T. Toda (Hiroshima University). Haploid cells were usually grown at 32°C in YE4S or EMM2 medium. Final concentrations of aureobasidin A (0.5 µg/ml), nourseothricin (100 µg/ml clonNAT), G418 (150 µg/ml geneticin), and hygromycin B (300 µg/ml) were added for selection purpose.
MBC and TBZ sensitivity test
Strains were grown to exponential phase (1 × 107–1.4 × 107 cells/mL) at 32°C and normalized to 107 cells/mL, and fivefold serial dilutions were spotted onto YE4S plates containing different concentrations of MBC (carbendazim, Sigma-Aldrich) or TBZ (tiabendazole, Sigma-Aldrich). The plates were incubated at 32°C for 48–72 h.
Colony formation assays
Strains were grown to exponential phase (1 × 107–1.4 × 107 cells/mL) at 32°C and normalized to 107 cells/mL, and cell viability was determined by plating 300 cells in triplicate onto YE4S plates and counting colony-forming units after five days incubation at 32°C. Percentage of viable cells from each genotype is normalized respect to the colony number of wt cells.
Fluorescence microscopy, live analysis, and quantification
Fluorescence microscopy images were generated using the DeltaVision microscope system (Applied Precision, Seattle, WA). Cells were adhered to 35 mm glass culture dishes (MatTek) using 0.2 mg/ml soybean lectin (Sigma-Aldrich) and immersed in EMM (with required supplements). Time-lapse imaging was carried out at 32°C in an Environmental Chamber with a DeltaVision Spectris (Applied Precision) comprising an Olympus IX70 widefield inverted epifluorescence microscope, an Olympus UPlanSapo 100× NA 1.4 oil immersion objective, and a Photometrics CCD CoolSnap HQ camera. Images were acquired over 15 focal planes at a 0.4-μm step size. For the quantification of protein fluorescence intensity, sum-projected raw microscopy data were used. Foci intensity time series were obtained after detection with a Laplacian of Gaussian filter and tracking with the LAP algorithm (TrackMate plugin in ImageJ). Tracks were time aligned according to the SPB duplication events; time zero was set as the last frame before the SPB duplication. Intensities were normalized to background mean intensity. Background was measured by taking the mean of four regions of interest (circle area of 18 pixels each), in this case fluorescence intensity of the cell, excluding the SPBs.
Spot selection of the SPB-associated/dissociated centromeres was performed in a semiautomated manner with a custom-written ImageJ macro and the Trackmate plugin in ImageJ. The correspondence of the SPB-associated centromere is established by colocalization with the SPB signal. Satellite foci are all those outside the SPBs colocalization area. Under our image conditions, SPBs have a size of 3 × 3 pixels, so we designate the signal as the mean of the nine pixels with the most intensity inside the designated area, and nine more pixels for each satellite focus. The mean intensity is set as the signal and is normalized with respect to background mean intensity. Image processing for representation was performed by deconvolving and combining each color channel into a 2D image using the maximum-intensity projection setting in softWoRx (Applied Precision) from raw microscopy data. Combined maximum Z-projection images were treated using ImageJ and Adobe Photoshop CS5 Extended.
Carbendazim and latrunculin treatments
For carbendazim treatment, a working solution of YE4S +MBC (15 µg/ml) was used. Strains were grown to OD (600 nm) = 0.3–0.4 in YE4S. Lectin (50 µl; 0.2 µg/ml; Sigma-Aldrich, L1395) was used for cell immobilization on a µ-Slide 8-well uncoated (Ibidi GmbH). YE4S+MBC (experiment) or YE4S (control) medium was used for filming cells. On the other hand, for latrunculin A treatment, exponentially growing cells were incubated for 10 min in 3 ml of YE4S rich medium with a total concentration of latrunculin A of 5 µM (15 µl of a 1 mM stock). After incubation, cells were immobilized with lectin as in MBC treatment for image acquisition. Images were taken with 100 ms and 50 ms exposure time for fluorescent and brightfield channels, respectively, and 13 focal planes with a 0.5-µm step size, using a spinning disk confocal microscopy system (Photometrics Evolve camera; Olympus 100 × 1.4 NA oil immersion objective; Roper Scientific). For the colocalization analysis, maximum Z-projection images of interphase cells, those with one single Sid4-mCherry dot (SPB), were subjected to colocalization analysis. For each cell, an axis containing the centers of both Sid4-mCherry and Mis6-GFP (centromeres) dots was drawn, and the intensity of the pixels from both channels was measured, normalized, and plotted along this axis. The final intensity profiles were used to measure the distance between the dots, defined as the micrometers between the x-coordinates of the maximum intensity of both profiles, considered to correspond with the center of the dots.
Cell cycle synchronizations in G1 and G2/M
Cell enrichment in G1 or G2/M phases, using strains harboring the cdc10-129 or cdc25-22 thermosensitive allele, respectively, was performed following a protocol from the Moreno lab (Tormos-Perez et al., 2016). Cells were grown in rich medium at 25°C to 107 cells/ml and transferred at 36°C for 4 h to arrest. G1 or G2/M enrichment was checked by quantifying the percentage of binucleated cells and septum formation.
Western blot analysis
Samples of cdc10-129 or cdc25-22 cells were obtained from 10 ml of exponentially growing cultures (107 cells/mL) at 25°C (asynchronous) or grown at 36°C for 4 h (enriched in G1 or G2/M phases, respectively). Cells were collected by centrifugation at 4°C, and cell pellets were flash-frozen in liquid nitrogen and stored at –80°C until use.
Protein extracts were prepared from trichloroacetic acid-treated cells as described (Grallert and Hagan, 2017). Briefly, cell pellets corresponding to 5 × 107 cells per condition and strain were removed from storage at –80°C and mixed with ice-cold 20% TCA and cold acid-washed glass beads (Sigma-Aldrich). Cell integrity was disrupted by FastPrep-24 (MP Biomedicals) for four cycles of 20 s at 4 m/s. Then ice-cold 5% TCA was added, and cell lysate was recovered. Next, samples were centrifuged, and supernatant was discarded. The protein pellet was washed with ice-cold 100% acetone and centrifuged. All previous manipulations were performed at 4°C. Supernatant was discarded and the protein pellet was resuspended at room temperature in SDS loading buffer and 1 M Tris-HCl pH 8 to raise the pH of the sample to neutral. Samples were denatured by heating, centrifuged, and loaded in a 10% SDS–PAGE gel (Biorad) to separate proteins. HA-tagged proteins were detected with an anti-HA antibody (Biolegend) and tubulin, with an anti-tubulin antibody (Sigma-Aldrich). The secondary antibody was horseradish peroxidase (HRP)-coupled anti-mouse IgG (Sigma-Aldrich). Visualization was performed using the SuperSignal WestFemto Maximum Sensitivity Substrate (ThermoFisher) in a Chemidoc MP imaging system (Biorad). Image processing for representation was performed using Adobe Photoshop CS5 Extended and signal quantifications were performed in ImageJ using the gel analysis tool.
ChIP-qPCR experiments
Based on protocols from Cam and Whitehall (2016) and Migeot and Hermand (2018), we performed ChIP-qPCR experiments as follows: samples of 2 × 109 cdc10-129 synchronized cells at 36°C for 4 h in YE4S were cross-linked with 1% formaldehyde (Sigma-Aldrich). Cross-linking was quenched by adding 150 mM glycin. All the next manipulations were performed at 4°C. Cell pellets were obtained by centrifugation, washed two times with prechilled PBS 1X, flash-frozen in liquid nitrogen, and stored at –80°C until use. The frozen cell pellets were thawed in ice, resuspended in ice-cold ChIP buffer I (50 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA pH 8, 0.1% Na deoxycholate, 0.1% SDS, 1 mM PMSF, proteinase inhibitor cocktail [Roche cOmplete, EDTA-free]) and mixed with cold acid-washed glass beads (Sigma-Aldrich) and cell integrity was disrupted by FastPrep-24 (MP Biomedicals) for six cycles of 20 s at 6 m/s. After cell lysis was checked under the microscope, cell lysate was recovered. Next, samples were centrifuged and the cross-linked chromatin appeared as a transparent layer around the pellet of cell debris. This pellet was resuspended with a Pasteur pipet in ice-cold ChIP buffer I and incubated on a rocking platform for 1 h. Next, samples were centrifuged and the pellet was resuspended in ice-cold ChIP buffer I before sonication in milliTUBE 1 ml AFA Fiber (Covaris) using a M220 Focused ultrasonicator (Covaris; 20% duty, PIP75w, 200 cycles/burst for 30 min) to obtain an average DNA fragment size of ∼200–400 bp. The sonicated samples were incubated in a rotating wheel for 30 min and centrifuged to clarify chromatin supernatant. This solution was precleared with pan mouse IgG protein G magnetic dynabeads (ThermoFisher) for 3 h in a rocking platform. Then the chromatin solution was incubated overnight with anti-HA antibody (Biolegend) in a rocking platform. The next day, the solution was centrifuged and the supernatant was incubated with protein G magnetic beads for 3 h in a rocking platform. Beads were separated from chromatin solution and washed three times with ice-cold ChIP buffer I, two times with ice-cold ChIP buffer II (50 mM HEPES-KOH, pH 7.5, 500 mM NaCl, 1% Triton X-100, 1 mM EDTA pH 8, 0.1% Na deoxycholate, 0.1% SDS), and two times with ice-cold ChIP buffer III (10 mM Tris HCl, pH 8, 250 mM LiCL, 0.5% NP40, 0.5% Na deoxycholate, 1 mM EDTA pH 8). Then beads were washed with TE buffer and DNA was eluted with elution buffer (50 mM Tris HCl, pH 8, 10 mM EDTA pH 8, 1% SDS) at 65°C for 20 min. Both input and ChIP samples were treated with proteinase K to de–crosslink proteins from DNA. The next day, samples were treated with RNase A and DNA was purified using a QIAquick PCR purification kit (Qiagen).
Quantitative PCRs were performed as Migeot and Hermand (2018) described and the percentage of input from cnt1/3 fragments was calculated for each sample relative to act1 as housekeeping control, using the ΔΔCt method. Oligonucleotides sequences used in this study are shown in Supplemental Table 2.
Supplementary Material
Acknowledgments
We thank all laboratory members for critical comments on the manuscript; Alejandra Cano for technical support; and the CABD microscopy facility technician Katherina García. We would like to thank the Genetics Department and Springboard lab for their useful discussion and comments, especially Víctor Carranco for technical support. We thank Sergio Moreno and Juan Jiménez for the cdc10-129 and cdc25-22 strains, respectively. This work was supported by the Spanish government, Plan Nacional project PGC2018-098118-A-I00, Ramon y Cajal program, RyC-2016-19659 to A.F.-A. and the program “Escalera de Excelencia” of the Junta de Castilla y León Ref. CLU-2017-03, co-funded by the P.O. FEDER of Castilla y León 14-20 and by the Pablo de Olavide University “Ayuda Puente Predoctoral” fellowship (PPI1803) to A.P.-S., and by the Spanish Education and Professional Formation Ministry, Research Collaboration Grant to D.L.-P. The CABD is an institution funded by Pablo de Olavide University, the Consejo Superior de Investigaciones Científicas (CSIC), and the Junta de Andalucía.
Abbreviations used:
- CCAN
centromere-associated network
- GBP
GFP-binding protein
- INM
inner nuclear membrane
- LatA
latrunculin A
- LINC
linker of nucleoskeleton and cytoskeleton complex
- MBC
methyl-2-benzimidazole carbamate or carbendazim
- NE
nuclear envelope
- ONM
outer nuclear membrane
- SPB
spindle pole body
- TBZ
tiabendazole.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-09-0600) on March 11, 2022.
REFERENCES
- Appelgren H, Kniola B, Ekwall K (2003). Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J Cell Sci 116, 4035–4042. [DOI] [PubMed] [Google Scholar]
- Asakawa H, Hayashi A, Haraguchi T, Hiraoka Y (2005). Dissociation of the Nuf2–Ndc80 complex releases centromeres from the spindle-pole body during meiotic prophase in fission yeast. Mol Biol Cell 16, 2325–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Barrales RR, Forn M, Georgescu PR, Sarkadi Z, Braun S (2016). Control of heterochromatin localization and silencing by the nuclear membrane protein Lem2. Genes Dev 30, 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AB, Cabal GG, Fabre E, Duong T, Buc H, Nehrbass U, Olivo-Marin JC, Gadal O, Zimmer C (2008). High-resolution statistical mapping reveals gene territories in live yeast. Nat Methods 5, 1031–1037. [DOI] [PubMed] [Google Scholar]
- Biggins S (2013). The composition, functions, and regulation of the budding yeast kinetochore. Genetics 194, 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K, Heun P, Gehlen L, Langowski J, Gasser SM (2004). Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc Natl Acad Sci USA 101, 16, 495–16, 500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Whitehall S (2016). Chromatin Immunoprecipitation (ChIP) in Schizosaccharomyces pombe. Cold Spring Harb Protoc, 10.1101/pdb.prot091546. [DOI] [PubMed] [Google Scholar]
- Carpy A, Krug K, Graf S, Koch A, Popic S, Hauf S, Macek B (2014). Absolute proteome and phosphoproteome dynamics during the cell cycle of Schizosaccharomyces pombe (Fission Yeast). Mol Cell Proteomics. 13, 1925–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM (2014). The kinetochore. Cold Spring Harb Perspect Biol 6, a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Brew C, Wolyniak M, Desai A, Anderson S, Muster N, Yates JR, Huffaker TC, Drubin DG, Barnes G (2001). Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J Cell Biol 155, 1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A (2008). Molecular architecture of the kinetochore–microtubule interface. Nat Rev Mol Cell Biol 9, 33–46. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y (1994). Telomere-led premeiotic chromosome movement in fission yeast. Science 264, 270–273. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y (2006). Meiotic proteins Bqt1 and Bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 125, 59–69. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C (2001). Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2, 292–301. [DOI] [PubMed] [Google Scholar]
- Czapiewski R, Robson MI, Schirmer EC (2016). Anchoring a leviathan: how the nuclear membrane tethers the genome. Front Genet 7, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhatchinamoorthy K, Mattingly M, Gerton JL (2018). Regulation of kinetochore configuration during mitosis. Curr Genet 64, 1197–1203. [DOI] [PubMed] [Google Scholar]
- Dhatchinamoorthy K, Shivaraju M, Lange JJ, Rubinstein B, Unruh JR, Slaughter BD, Gerton JL (2017). Structural plasticity of the living kinetochore. J Cell Biol 216, 3551–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, West RR, Morphew M, Oakley BR, McIntosh JR (1997). The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol Biol Cell 8, 1461–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS (2010). A three-dimensional model of the yeast genome. Nature 465, 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennell A, Fernández-álvarez A, Tomita K, Cooper JP (2015). Telomeres and centromeres have interchangeable roles in promoting meiotic spindle formation. J Cell Biol 208, 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Alvarez A, Bez C, O’Toole ET, Morphew M, Cooper JP (2016). Mitotic nuclear envelope breakdown and spindle nucleation are controlled by interphase contacts between centromeres and the nuclear envelope. Dev Cell 39, 544–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Alvarez A, Cooper JP (2017a). Chromosomes orchestrate their own liberation: nuclear envelope disassembly. Trends Cell Biol 27, 255–265. [DOI] [PubMed] [Google Scholar]
- Fernandez-Alvarez A, Cooper JP (2017b). The functionally elusive RabI chromosome configuration directly regulates nuclear membrane remodeling at mitotic onset. Cell Cycle 16, 1392–-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M (1993). Cell cycle–dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol 121, 961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Ellenberg J (2003). Dynamics of chromosome positioning during the cell cycle. Curr Opin Cell Biol 15, 664–671. [DOI] [PubMed] [Google Scholar]
- Grallert A, Hagan IM (2017). Preparation of protein extracts from Schizosaccharomyces pombe using trichloroacetic acid precipitation. Cold Spring Harb Protoc (2017). [DOI] [PubMed] [Google Scholar]
- Guttinger S, Laurell E, Kutay U (2009). Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol 10, 178–191. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M (1995). The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol 129, 1033–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M, Fukagawa T (2018). Kinetochore assembly and disassembly during mitotic entry and exit. Curr Opin Cell Biol 52, 73–81. [DOI] [PubMed] [Google Scholar]
- Hashida-Okado T, Yasumoto R, Endo M, Takesako K, Kato I (1998). Isolation and characterization of the aureobasidin A-resistant gene, aur1R, on Schizosaccharomyces pombe: roles of Aur1p+ in cell morphogenesis. Curr Genet 33, 38–45. [DOI] [PubMed] [Google Scholar]
- Hattersley N, Cheerambathur D, Moyle M, Stefanutti M, Richardson A, Lee KY, Dumont J, Oegema K, Desai A (2016). A nucleoporin docks protein phosphatase 1 to direct meiotic chromosome segregation and nuclear assembly. Dev Cell 38, 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi A, Asakawa H, Haraguchi T, Hiraoka Y (2006). Reconstruction of the kinetochore during meiosis in fission yeast Schizosaccharomyces pombe. Mol Biol Cell 17, 5173–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M (2004). Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118, 715–729. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Dernburg AF (2009). The SUN rises on meiotic chromosome dynamics. Dev Cell 17, 598–605. [DOI] [PubMed] [Google Scholar]
- Hou H, Zhou Z, Wang Y, Wang J, Kallgren SP, Kurchuk T, Miller EA, Chang F, Jia S (2012). Csi1 links centromeres to the nuclear envelope for centromere clustering. J Cell Biol 199, 735–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu KS, Toda T (2011). Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Curr Biol 21, 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Huang Y, Yu H, Subramanian D, Padmanabhan A, Thadani R, Tao Y, Tang X, Wedlich-Soldner R, Balasubramanian MK (2012). Nonmedially assembled F-actin cables incorporate into the actomyosin ring in fission yeast. J Cell Biol 199, 831–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Tanaka TU, Lechner J, Schiebel E (2002). Four new subunits of the Dam1–Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J 21, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, Trelles-Sticken E, Scherthan H, Loidl J (1998). Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J Cell Biol 141, 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin QW, Fuchs J, Loidl J (2000). Centromere clustering is a major determinant of yeast interphase nuclear organization. J Cell Sci 113(Pt 11), 1903-–1912. [DOI] [PubMed] [Google Scholar]
- Kume K, Cantwell H, Burrell A, Nurse P (2019). Nuclear membrane protein Lem2 regulates nuclear size through membrane flow. Nat Commun 10, 1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, McLeod I, Anderson S, Yates JR, 3rd, He X (2005). Molecular analysis of kinetochore architecture in fission yeast. EMBO J 24, 2919–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeot V, Hermand D (2018). Chromatin immunoprecipitation-polymerase chain reaction (ChIP-PCR) detects methylation, acetylation, and ubiquitylation in S. pombe. Methods Mol Biol 1721, 25–34. [DOI] [PubMed] [Google Scholar]
- Mizuguchi T, Barrowman J, Grewal SI (2015). Chromosome domain architecture and dynamic organization of the fission yeast genome. FEBS Lett 589, 2975–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi T, Fudenberg G, Mehta S, Belton JM, Taneja N, Folco HD, FitzGerald P, Dekker J, Mirny L, Barrowman J, Grewal SIS (2014). Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature 516, 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Nabetani A, Koujin T, Tsutsumi C, Haraguchi T, Hiraoka Y (2001). A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma 110, 322–334. [DOI] [PubMed] [Google Scholar]
- Nagpal H, Fukagawa T (2016). Kinetochore assembly and function through the cell cycle. Chromosoma 125, 645–659. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Arai A, Takebe Y, Masuda M (2011). A chemical compound for controlled expression of nmt1-driven gene in the fission yeast Schizosaccharomyces pombe. Anal Biochem 412, 159–164. [DOI] [PubMed] [Google Scholar]
- Obuse C, Iwasaki O, Kiyomitsu T, Goshima G, Toyoda Y, Yanagida M (2004). A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat Cell Biol 6, 1135–1141. [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nat Methods 5, 605–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Tillib S, Nowak D, Schermelleh L, Gahl A, Backmann N, Conrath K, Muyldermans S, Cardoso MC, Leonhardt H (2006). Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat Methods 3, 887–889. [DOI] [PubMed] [Google Scholar]
- Saitoh S, Takahashi K, Yanagida M (1997). Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell 90, 131–143. [DOI] [PubMed] [Google Scholar]
- Shimanuki M, Miki F, Ding DQ, Chikashige Y, Hiraoka Y, Horio T, Niwa O (1997). A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol Gen Genet 254, 238–249. [DOI] [PubMed] [Google Scholar]
- Smoyer CJ, Jaspersen SL (2014). Breaking down the wall: the nuclear envelope during mitosis. Curr Opin Cell Biol 26, 1–9. [DOI] [PubMed] [Google Scholar]
- Swaffer MP, Jones AW, Flynn HR, Snijders AP, Nurse P (2016). CDK substrate phosphorylation and ordering the cell cycle. Cell 167, 1750–1761 e1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Gasser SM (2012). Structure and function in the budding yeast nucleus. Genetics 192, 107–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M (2000). Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288, 2215–2219. [DOI] [PubMed] [Google Scholar]
- Tange Y, Chikashige Y, Takahata S, Kawakami K, Higashi M, Mori C, Kojidani T, Hirano Y, Asakawa H, Murakami Y, et al. (2016). Inner nuclear membrane protein Lem2 augments heterochromatin formation in response to nutritional conditions. Genes Cells 21, 812–832. [DOI] [PubMed] [Google Scholar]
- Tormos-Perez M, Perez-Hidalgo L, Moreno S (2016). Fission yeast cell cycle synchronization methods. Methods Mol Biol 1369, 293–308. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken E, Adelfalk C, Loidl J, Scherthan H (2005). Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J Cell Biol 170, 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unruh JR, Slaughter BD, Jaspersen SL (2018). Functional analysis of the yeast LINC complex using fluctuation spectroscopy and super-resolution imaging. Methods Mol Biol 1840, 137–161. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kilmartin JV (2001). The Ndc80p complex from Saccharomycescerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol 152, 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WMT, Kelly DA, Pidoux AL, Allshire RC (2021). Centromere identity is dependent on nuclear spatial organization. bioRxiv. 2021.12.16.473016. [DOI] [PMC free article] [PubMed]
- Yoshida M, Katsuyama S, Tateho K, Nakamura H, Miyoshi J, Ohba T, Matsuhara H, Miki F, Okazaki K, Haraguchi T, et al. (2013). Microtubule-organizing center formation at telomeres induces meiotic telomere clustering. J Cell Biol 200, 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.