Abstract
Membrane fusion requires tethers, SNAREs of R, Qa, Qb, and Qc families, and chaperones of the SM, Sec17/SNAP, and Sec18/NSF families. SNAREs have N-domains, SNARE domains that zipper into 4-helical RQaQbQc coiled coils, a short juxtamembrane (Jx) domain, and (often) a C-terminal transmembrane anchor. We reconstitute fusion with purified components from yeast vacuoles, where the HOPS protein combines tethering and SM functions. The vacuolar Rab, lipids, and R-SNARE activate HOPS to bind Q-SNAREs and catalyze trans-SNARE associations. With SNAREs initially disassembled, as they are on the organelle, we now report that R- and Qa-SNAREs require their physiological juxtamembrane (Jx) regions for fusion. Swap of the Jx domain between the R- and Qa-SNAREs blocks fusion after SNARE association in trans. This block is bypassed by either Sec17, which drives fusion without requiring complete SNARE zippering, or transmembrane-anchored Qb-SNARE in complex with Qa. The abundance of the trans-SNARE complex is not the sole fusion determinant, as it is unaltered by Sec17, Jx swap, or the Qb-transmembrane anchor. The sensitivity of fusion to Jx swap in the absence of a Qb transmembrane anchor is inherent to the SNAREs, because it remains when a synthetic tether replaces HOPS.
INTRODUCTION
The highly conserved pathway of intracellular membrane fusion is essential for eukaryotic subcellular protein delivery, cell growth, secretion of hormones and other proteins, endocytosis, autophagy, and neurotransmission (Wickner and Rizo, 2017). Membrane fusion is mediated by conserved protein families. Rab GTPases mark each organelle and bind effectors to mediate tethering, a prerequisite for fusion. SNARE proteins anchored to each tethered membrane are catalyzed to engage in trans (between membranes) to form tetravalent trans-SNARE complexes. SNAREs have N-domains, followed by characteristic SNARE domains of approximately 60 amino acyl residues with heptad-repeat apolar residues, a short juxtamembrane (Jx) region, and (often) a C-terminal trans-membrane (TM) anchor. Individual SNARE domains are random coil, but as they zipper together in the N- to C-direction (Sorensen et al., 2006), they form α-helices that assemble in a left-handed 4-SNARE complex. Heptad-repeat apolar residues form the core of the 4-SNARE complex, but in the center of the 4-helical coiled coil, each SNARE has an inward-oriented glutaminyl (Q) or arginyl (R) residue (Sutton et al., 1998). SNAREs are in four conserved families, termed R, Qa, Qb, and Qc, and SNARE complexes are of RQaQbQc composition (Fasshauer et al., 1998). Because high levels of SNAREs alone can mediate fusion (Weber et al., 1998), the rate of fusion has been thought to simply reflect the abundance of trans-complexes between SNARE domains. SNARE assembly is catalyzed by Sec1/Munc18 (SM) family proteins, alone or in oligomeric complex (Fiebig et val., 1999; Sorensen et al., 2006; Baker et al., 2015; Orr et al., 2017; Jiao et al., 2018), while SNARE complex disassembly is mediated by Sec17/α-SNAP and the AAA-family ATPase Sec18/NSF (Söllner et al., 1993). Sec17 and Sec18 also assemble onto a platform of partially zippered SNAREs to directly contribute to the fusion event per se (Schwartz and Merz, 2009; Schwartz et al., 2017; Zick et al., 2015; Song et al, 2021). Together, these proteins induce a striking restructuring of the lipids in the two closely apposed bilayers to form a single continuous fused membrane. This fusion joins the aqueous lumenal compartments that each membrane had enclosed, all without release of contents into the cytosol as would be seen with membrane joining through lysis and reannealing.
We study membrane fusion with yeast vacuoles, which undergo both fission and homotypic fusion. Wada et al. (1992) identified VAM (vacuole morphology) genes for membrane fusion, encoding the vacuolar Rab Ypt7, two SNAREs, and each of the six subunits of a large homotypic fusion and vacuole protein sorting multisubunit protein termed HOPS (Seals et al., 2000). Many of these genes were also identified in the vps (vacuole protein sorting) screens of the Emr (Scott Emr, Cornell University) and Stevens labs (Thomas Stevens, University of Oregon). Vacuolar fusion requires Nyv1, Vam3, Vti1, and Vam7, the R, Qa, Qb, and Qc SNAREs of the organelle, respectively; these are referred to hereafter as simply R, Qa, Qb, and Qc. Fusion of purified yeast vacuoles (Haas et al., 1994) requires Ypt7, each SNARE, Sec17, Sec18, and HOPS. HOPS is a hexameric complex of Vps 11, 16, 18, 33, 39, and 41. Both Vps39 and Vps41 have direct affinity for vacuole-anchored Ypt7 (Brett et al., 2008; Bröcker et al., 2012), explaining how HOPS catalyzes the tethering of the organelle (Stroupe et al., 2006) or even of liposomes bearing only this Rab (Hickey and Wickner, 2010). Vps33 is the vacuolar SM protein that directly binds the R and Qa SNARE domains, in parallel and in register, an essential step in the catalysis of SNARE assembly (Baker et al., 2015). HOPS also has direct affinity for the Qb and Qc SNAREs (Stroupe et al., 2006; Song et al., 2020). HOPS is activated to catalyze SNARE complex assembly by binding to Ypt7 and vacuolar lipids (Torng et al., 2020; Torng and Wickner, 2021). HOPS can bind Sec17 (Song et al., 2021) and Sec17 stimulates HOPS-dependent fusion, both in an in vitro reconstituted fusion reaction with vacuolar lipids, Rab, SNAREs, HOPS, and Sec18 (Zick et al., 2015; Song et al., 2017) and in vivo (Schwartz et al., 2017).
While SNARE domains have received exhaustive study and apolar TM domains may simply serve as membrane anchors, there has been less examination of the short Jx domains that connect them. Genetic studies (Van Komen et al., 2005; DeMill et al., 2014; Singer-Lahat et al., 2018) have shown the importance of the Jx domain. The functions of the JxR and JxQa regions have been evaluated with SNARE proteoliposomes, either alone or with accessory proteins such as Munc13 or Munc18-1. Substantially lengthening the Jx domain can block fusion (McNew et al., 1999), especially if it increases the distance from the SNARE domain to two tryptophans in the R-SNARE Jx domain (Hu et al., 2021). Addition of helix-disrupting prolyl residues into the Jx domain can also inhibit fusion (Hu et al., 2021), suggesting a need for α-helical structure. The R-SNARE Jx domain is surface active in that it partitions to a water:membrane interface, and the substitution of another surface-active peptide for this Jx domain still allows fusion (Rathore et al., 2019). Both basic and hydrophobic residues of the Qa Jx domain are needed (Rathore et al., 2019). These findings suggest two possible roles for Jx domains, one to connect the SNARE and membrane anchors, perhaps propagating a continuous α-helical structure between them, and the other to poise a bilayer-disrupting peptide sequence immediately adjacent to the two apposed membranes (Hu et al., 2021). A simple swap of the R- and Qa-Jx domains might well preserve either or both of these functions.
Yeast vacuole R and Qa SNAREs have short, basic Jx domains of similar sizes, and thus their swap would be expected to have little effect on fusion. However, we now report that the specific association of the basic, polar, and short Jx domains of the vacuolar Qa- and R-SNAREs with their physiological SNARE domain is crucial for fusion. Either Sec17 or a synthetic preassembly of the Q-SNAREs during proteoliposome preparation, providing a second TM anchor to the Q-SNARE complex, can bypass the requirement for both R- and Qa-Jx domains to be present and for each Jx domain to associate with its physiological SNARE. Fusion with entirely wild-type SNARE domains requires either the correct match between the Jx and SNARE domains, Sec17, or the transmembrane anchor on the Qb SNARE. None of these alters the level of trans-SNARE complex.
RESULTS
The vacuolar R, Qa, and Qb SNAREs have 8–10 amino acyl residue basic juxtamembrane (Jx) regions linking their SNARE domains and transmembrane (TM) domains (Figure 1A). Earlier studies suggested that such small length differences would have little effect on fusion (McNew et al., 1999). The Jx domains lie near the apposed membranes in the SNARE-zippered prefusion structure (Figure 1B). To explore their functions, we systematically replaced the juxtamembrane plus transmembrane regions (JxTM) of R and Qa with the JxTM of either R, Qa, or Qb. The purified R and Qa recombinant proteins were incorporated along with the Rab Ypt7 into proteoliposomes bearing lumenal reporter proteins, either Cy5-streptavidin or biotinylated phycoerythrin (Figure 1Ci). In mixtures of these proteoliposomes, the fluorophores are separated by at least the thickness of two lipid bilayers, too far for FRET (fluorescence resonance energy transfer). Upon fusion, the mixing of lumenal contents allows tight binding of biotin to streptavidin, bringing the two fluorophores into intimate contact and yielding a strong FRET signal (Zucchi and Zick, 2011). Fusion reactions also have HOPS, sQb (the soluble form of Qb without its membrane anchor), and Qc as well as excess nonfluorescent streptavidin to quench any FRET signal from proteoliposome lysis.
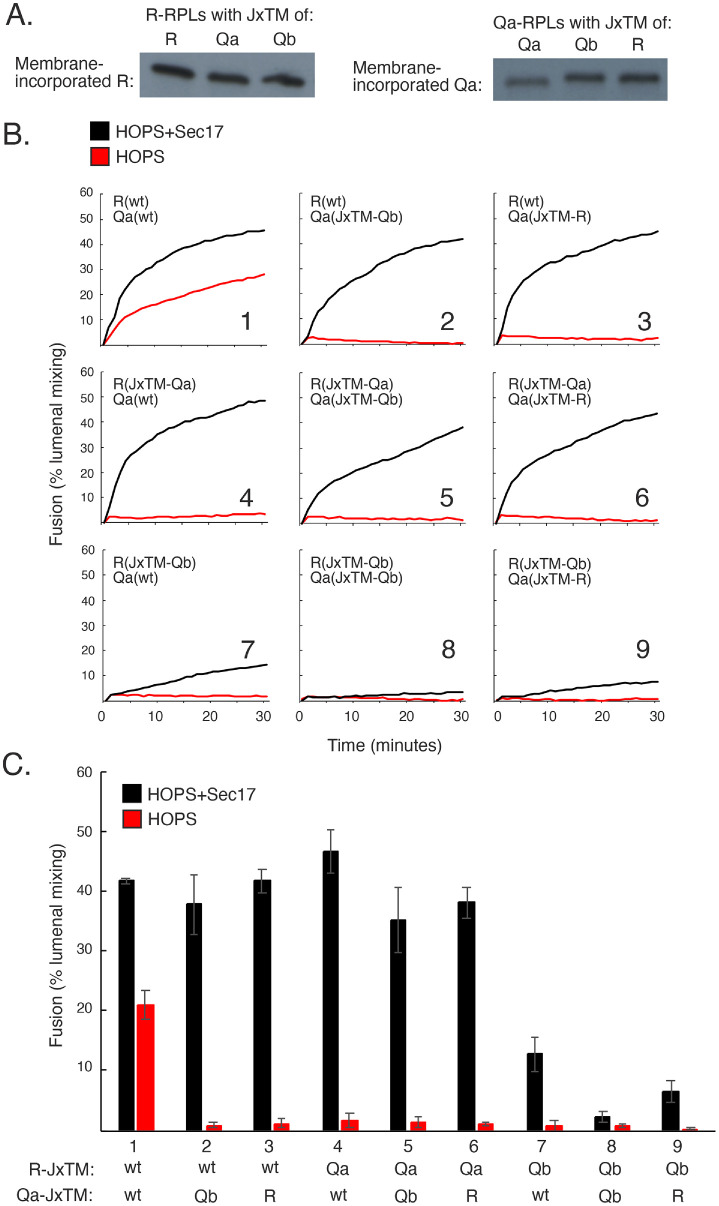
FIGURE 1:
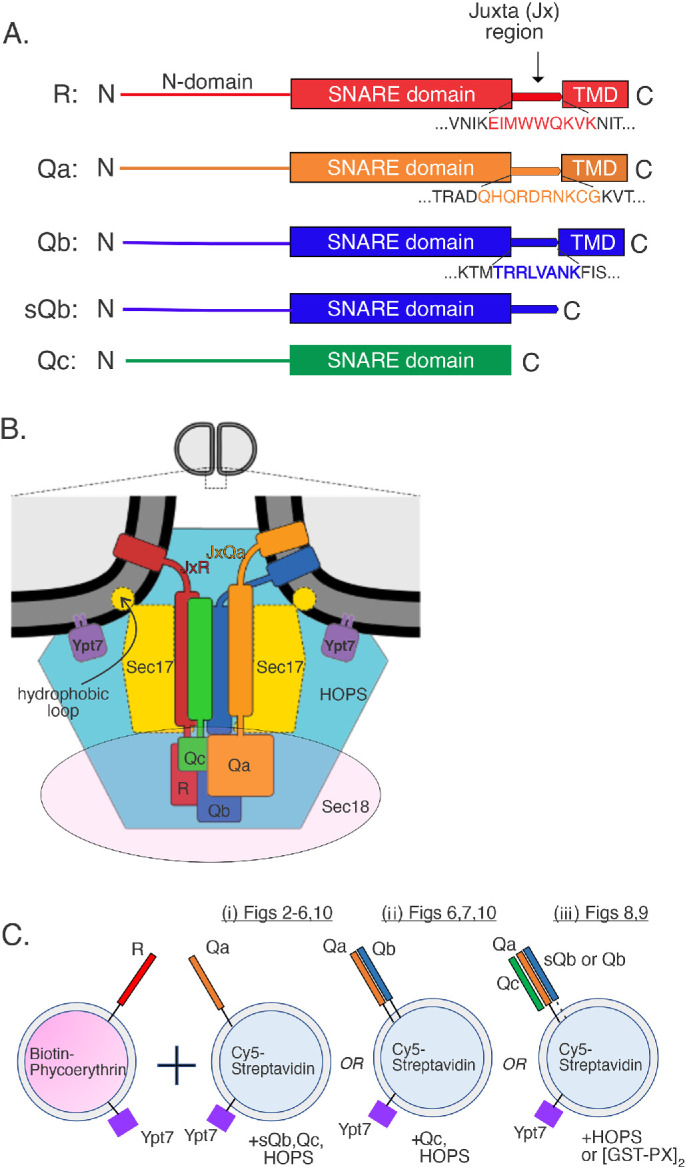
Schematics. (A) Juxtamembrane (Jx) regions of three vacuolar SNAREs. (B) Working model of the spatial and binding relationships among the four vacuolar SNAREs, the Rab GTPase Ypt7, Sec17, Sec18, and HOPS. (C) FRET assays measure the FRET between Cy5-streptavidin and biotinylated phycoerythrin, initially trapped in the lumens of fusion partner proteoliposomes but then mixed and bound together after fusion. Fusion is between R proteoliposomes and either (i) Qa, (ii) QaQb, or (iii) QaQbQc proteoliposomes, where the Qb is either soluble or membrane bound.
Proteoliposomes with Ypt7 and R, bearing either its own JxTM or those of Qa or Qb, were incubated with proteoliposomes bearing Ypt7 and Qa with its own JxTM or those of R or Qb. We deliberately chose to begin fusion incubations with separate, uncomplexed SNAREs, because the ubiquitous Sec17/Sec18 chaperones keep SNAREs disassembled in cells except when undergoing HOPS-mediated trans-SNARE assembly; very little of the Qb SNARE on isolated vacuoles is in complex with Qa (Collins et al., 2005), and HOPS protects trans-SNARE complexes, but not cis-SNARE complexes, from Sec17/Sec18-mediated disassembly (Xu et al., 2010). The wild-type and mutant R and Qa SNAREs incorporated into proteoliposomes to the same levels (Figure 2A). Upon addition of a mixture of HOPS, soluble Qb without membrane anchor, and Qc, the proteoliposomes with SNAREs bearing their own JxTM domains underwent fusion (Figure 2B, panel 1, red curve) with modest stimulation by Sec17 (black curve), as reported (Song et al., 2020). In contrast, fusion with HOPS was lost when either the R or Qa SNARE had a nonnative JxTM region (Figure 2, B, panels 2–9, red curves, and C). Many fusion reactions were restored by 500 nM Sec17 (Figure 2, B, panels 2–6, black curves, and C), including some with Qa(JxTM-Qb) (panels 2 and 5), but there was minimal restoration of fusion by Sec17 with R(JxTM-Qb) proteoliposomes as one of the fusion partners (panels 7–9).
FIGURE 2:
Sec17 is required for fusion when the JxTM of even one membrane-integrated SNARE is substituted for another. (A) Comparable levels of JxTM-substituted or wild-type SNAREs incorporate into membranes. R and Qa proteoliposomes were prepared with Ypt7 and their own JxTMs or substituted JxTMs and analyzed by Western blot for SNARE incorporation. (B) R and Qa require their own JxTMs for fusion to occur without Sec17. Fusion between R and Qa proteoliposomes, where the R and Qa had their own JxTM or a substituted JxTM, was performed as described in Materials and Methods. Fusion competency was measured by FRET, with 100 nM soluble Qb and Qc, 50 nM HOPS, and with or without 600 nM Sec17. (C) Averages and SDs of fusion in the first 20 min of incubation are shown.
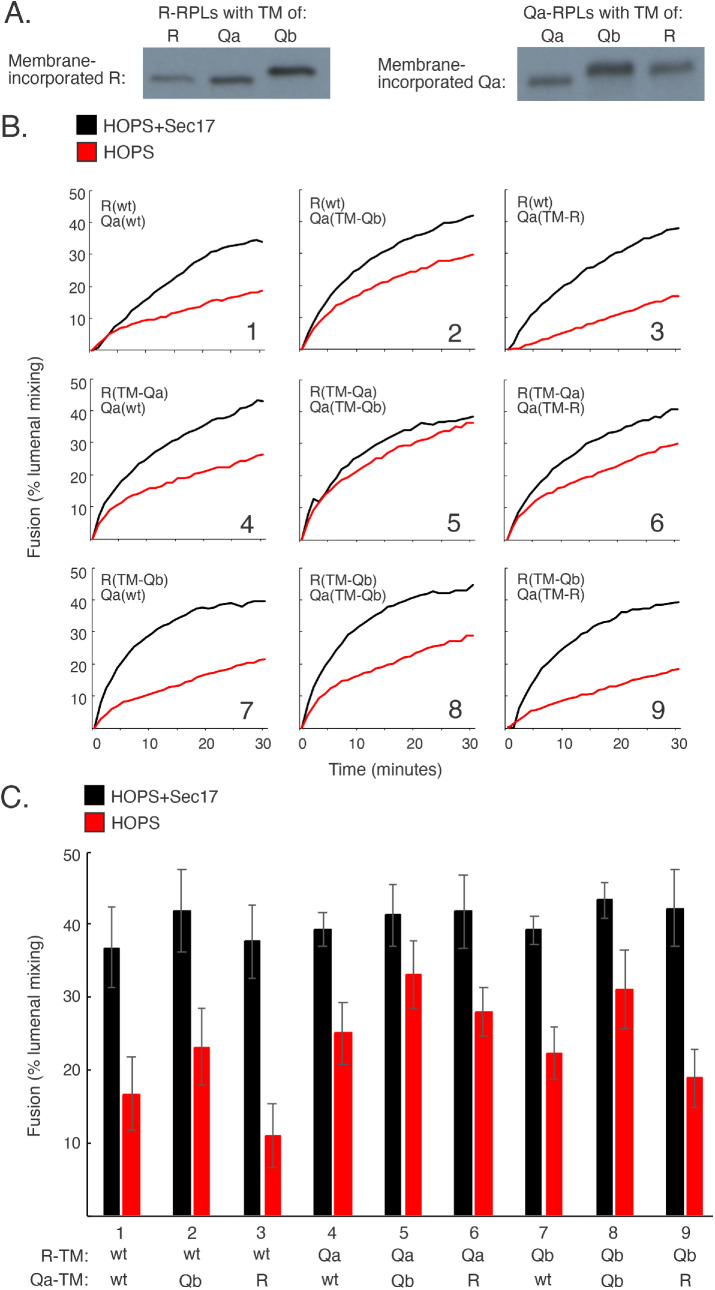
Interchangeable TM domains, SNARE-specific Jx domains
To test the functional specificity of the TM domain for the R and Qa SNAREs, each was prepared with its own Jx domain but with the R, Qa, and Qb TM domain. There was little effect of these apparently interchangeable TM domains on fusion with HOPS alone (Figure 3B, panels 1–9, red) or with Sec17 stimulation (black). The TM domains appear to fulfill only a simple membrane-anchor function, suggesting that the stark differences seen with varied JxTM domains are due to a specificity of each SNARE for its Jx region. To confirm this, we prepared R(JxQa) and Qa(JxR), each with its own TM region, and then assembled proteoliposomes with Ypt7 and comparable amounts of either wild-type R and Qa or these SNAREs with exchanged Jx regions, R(JxQa) or Qa(JxR) (Figure 4A). The fusion without Sec17 between proteoliposomes with wild-type R and Qa (Figure 4, B, panel 1, and C, red) was lost when either or both SNARE had a nonnative Jx region (panels 2–4, red). Fusion was restored by 500 nM Sec17 (black) or by lower Sec17 plus Sec18/ATPγS (green). Earlier studies showed that fusion blocked by arrested zippering due to SNARE-domain truncation is restored by either moderate levels of Sec17 or lower levels of Sec17 with Sec18 (Zick et al., 2015; Schwartz et al., 2017; Song et al., 2021); here we see the same pattern of fusion loss and restoration with all wild-type, full-length SNARE domains but swapped Jx domains. Because both of the Jx regions that had been swapped are basic, have similar lengths, and would be closely apposed to the bilayers after zippering is complete, it is particularly noteworthy that there is no fusion without Sec17 between R(JxTM-Qa) and Qa(JxTM-R) proteoliposomes (Figure 2B, panel 6) or between R(JxQa) and Qa(JxR) proteoliposomes (Figure 4, B, panel 4, and C).
FIGURE 3:
Transmembrane (TM) domain exchange among the SNAREs has little effect on fusion. (A) Comparable levels of SNAREs with substituted TMs incorporate into proteoliposomes. R and Qa RPLs were prepared as described with Ypt7 and either their own TMs or substituted TMs. Levels of incorporation were analyzed by immunoblot. (B) R and Qa proteoliposomes with substituted TMs remain fusion competent without Sec17. Fusion assays, as described in Materials and Methods, contained 100 nM sQb, 100 nM Qc, 50 nM HOPS, and 600 nM Sec 17 where present. (C) Means and SDs are shown for fusion at 20 min from three repeat experiments.
FIGURE 4:
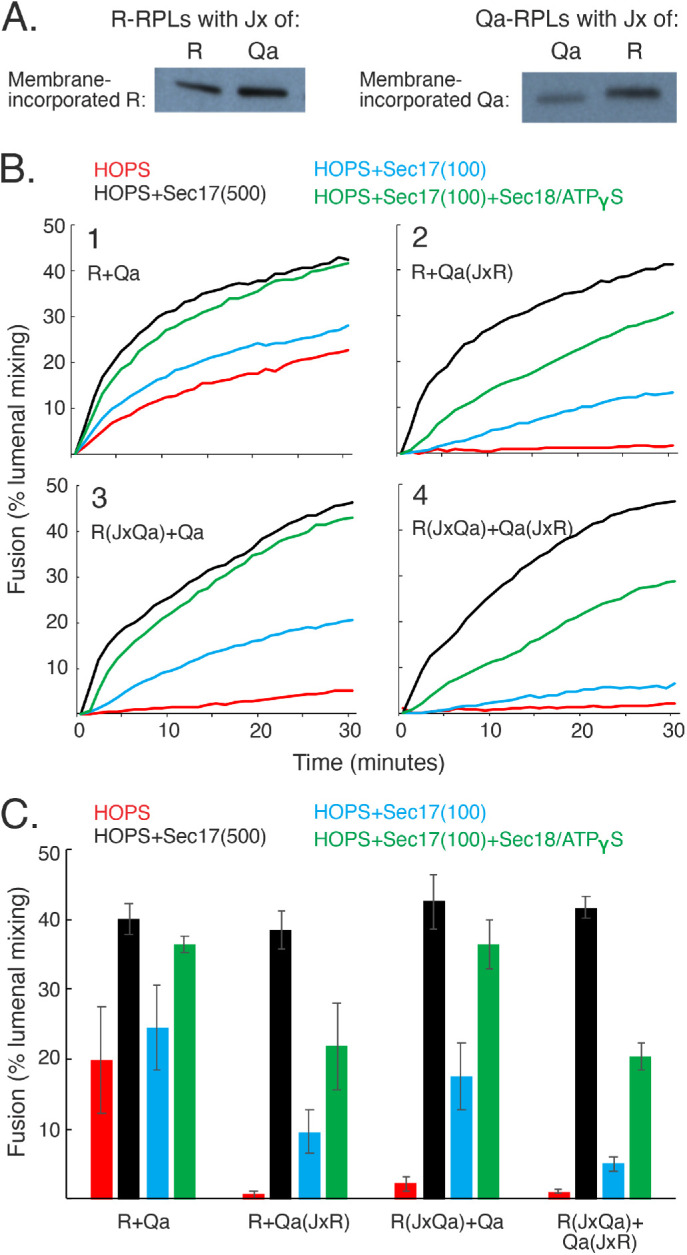
Substitution or swap of the Jx regions of R and/or Qa strongly inhibits R+Qa fusion without Sec17. Moderate levels of Sec17 (500 nM) are sufficient for full restoration of fusion, and lower Sec17 levels (100 nM) are stimulated by 300 nM Sec18 plus MgATPγS. (A) SNAREs with exchanged Jx regions incorporate into proteoliposomes as well as wild-type SNAREs. R and Qa proteoliposomes were prepared as described with Ypt7 and either wild-type or exchanged Jx regions. SNARE incorporation was analyzed by immunoblot. (B) R and Qa fusion reactions, where one or both membrane-bound SNARE has an exchanged Jx region, require Sec17. Sec17 at moderate levels or low Sec17 with Sec18/ATPgS relieves the block imposed by Jx exchange. Fusion assays were as described in Materials and Methods, with 50 nM HOPS, 100 or 500 nM Sec17, and 300 nM Sec18 where added. All reactions contained 1 mM MgATPγS. (C) Means and SDs are shown for fusion at 20 min from three repeat experiments
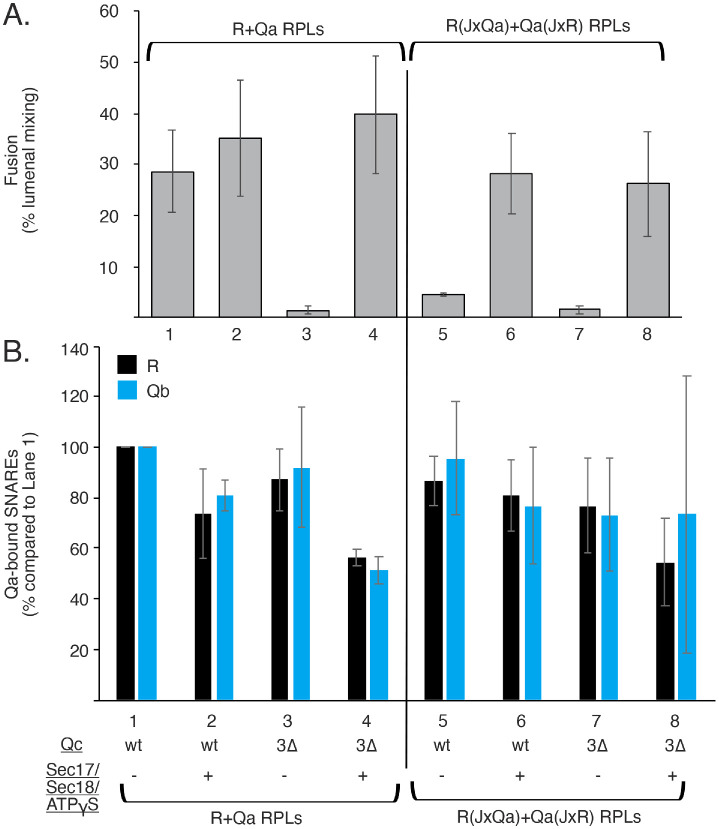
Jx function when zippering is arrested
SNARE domains are believed to be random coil before zippering (Fasshauer et al., 1997; Sutton et al., 1998) and to zipper in an N- to C-terminal direction (Fiebig et al., 1999; Sorensen et al., 2006). SNARE zippering is arrested by deletion of the C-terminal four heptads of Qc, the Qc3Δ mutation, and fusion can be restored by Sec17, in vitro (Schwartz and Merz, 2009) or in vivo (Schwartz et al., 2017), aided by Sec18 (Zick et al., 2015; Song et al., 2021). Zippering should concentrate the Jx domains at the zone of membrane apposition for fusion (Figure 1B). It was unclear whether fusion loss due to Jx exchange would show a synthetic defect with loss due to SNARE-domain C-terminal truncation, suggesting distinct contributions to fusion, or whether when combined they would still be bypassed by Sec17/Sec18/ATPγS. We therefore examined whether Sec17/Sec18/ATPγS will rescue fusion with Jx-swapped R and Qa SNAREs when zippering is arrested by Qc3Δ. HOPS-catalyzed fusion with wild-type SNAREs (Figure 5A, lane 1, and Supplemental Figure S1) is blocked by the Qc3Δ mutation (lane 3), but this block is relieved by Sec17/Sec18/ATPγS (lane 4), as reported (Zick et al., 2015; Schwartz et al., 2017; Song et al., 2017, 2021). When the R and Qa Jx domains are swapped (lanes 5–8), fusion with either full-length Qc (lane 5) or Qc3Δ (lane 7) is blocked. In either case, Sec17/Sec18/ATPγS fully restores fusion (lanes 6 and 8). The proteoliposomes from each of these incubations were solubilized, the Qa was isolated by immunoprecipitation, and Qa-bound R and Qb SNAREs were assayed by immunoblot. The striking effects on fusion of SNARE truncation, Jx swap, and Sec17/Sec18/ATPγS did not reflect corresponding changes in the levels of trans-SNARE complex, assayed by R:Qa association (Figure 5B, black bars) or in HOPS-mediated cis-SNARE assembly, assayed as Qa:Qb association (blue bars). SNARE-specific Jx function may be needed only after zippering brings them into an initial SNARE assembly near the fusion site (Figure 1B), and a Sec17/Sec18/ATPγS bypass of the need for complete zippering also bypasses the need for SNARE-specific Jx function.
FIGURE 5:
Even with arrested SNARE zippering, Sec17/Sec18/ATPgS restores fusion to proteoliposomes bearing SNAREs with swapped Jx domains, whereas neither Jx swap, zippering arrest, nor Sec17/Sec18/ATPgS affects trans-SNARE complex assembly. Ypt7/R and Ypt7/Qa proteoliposomes were prepared as described in Materials and Methods, with either their wild-type Jx domains or swapped Jx domains. Fusion reactions and immunoprecipitations were conducted as in Materials and Methods, with 50 nM HOPS and 100 nM sQb, but higher levels (2 µM) of wild-type Qc or Qc3∆. The indicated reactions had 500 nM Sec17 and 250 nM Sec18. All reactions contained 1 mM MgATPγS. (A) The means and SDs of fusion after 20 min of three replicate incubations are shown. (B) UN-SCAN-IT software (Silk Scientific, Orem, UT) was used to compare pixel intensities of Western blot bands to that of lane 1 with wild-type SNAREs. Means and SDs of three such analyses are shown. See Supplemental Figure S1 for typical kinetic data and a representative Western blot image.
SNARE associations, in cis and in trans, and the ensuing fusion
Sec17, Sec18, and ATP disassemble cis-SNARE complexes left from prior rounds of fusion (Mayer et al., 1996) to allow the liberated SNAREs to engage in the next round of trans-associations, catalyzed by HOPS (Torng et al., 2020). Continuous cis-SNARE disassembly is efficient, as only 10% of the Qb is bound to Qa on isolated vacuoles (Collins et al., 2005), and HOPS is needed to protect trans-SNARE complexes from Sec17/Sec18-mediated disassembly (Xu et al., 2010). In model reconstitution studies, Qa and Qb SNAREs in detergent mixed micellar solution assemble during the extensive dialysis used for QaQb proteoliposome preparation, bypassing HOPS assembly functions (Baker et al., 2015; Orr et al., 2017; Jiao et al., 2018; Song et al., 2020). We therefore explored the effects of HOPS, Sec17, and the Jx domain:SNARE match on SNARE associations and on fusion. Ypt7/R proteoliposomes were incubated with Ypt7/Qa or Ypt7/QaQb proteoliposomes (Figure 1C, i or ii) plus HOPS, Qc, and Sec17 where indicated (Figure 6 and Supplemental Figure S2). sQb was included where Qb was not integrally bound to proteoliposomes. Fusion was measured as the FRET due to protected lumenal compartment mixing (Figure 6A). Aliquots from these incubations were solubilized, the Qa-SNARE immunoprecipitated by bead-bound antibodies, and the Qa-bound proteins released with SDS and analyzed by SDS–PAGE and immunoblot (Figure 6B). Some incubations had proteoliposomes with swapped Jx regions, that is, with R(JxQa) and Qa(JxR).
FIGURE 6:
The block to fusion from swapped Jx domains is relieved by Sec17 or by having wild-type Qb anchored to the membrane by its TM domain. Proteoliposomes with Ypt7 and SNAREs were prepared as described in Materials and Methods with R (wild-type) or R having the Jx region of Qa, with Qa (wild-type) or Qa having the Jx region of R, or with QaQb with wild-type Qa or with Qa having the Jx region of R. Fusion reactions and immunoprecipitations were conducted as described in Materials and Methods, with 100 nM Qc, 100 nM sQb (where Qb was not present on the proteoliposomes), 50 nM HOPS, and with or without 500 nM Sec17, as indicated. (A) Average FRETs with SDs are shown at 20 min for three replicates. (B) Samples from each incubation were solubilized in RIPA buffer and analyzed for Qa-bound Qb and R as described in Materials and Methods, and bands were analyzed with UN-SCAN-IT software for pixel intensity as compared with lane 8 with all wild-type SNAREs. Two y-axis scales are shown to display differences of bound sQb in lanes 1–6. Averages and SDs of the three replicates are shown. See Supplemental Figure S2 for typical kinetic data and a representative Western blot image.
Without HOPS, there was almost no fusion (Figure 6A, lanes 1 and 2) and only background Qa-association with R- or sQb-SNAREs (Figure 6B, lanes 1 and 2, black and blue bars). R proteoliposomes were also essential for HOPS-dependent association of sQb with Qa (Figure 6B, lane 3 vs. lane 13, black bars), in accord with earlier studies (Torng et al., 2020). With R and Qa proteoliposomes, HOPS-dependent fusion was accompanied by Qa association with both R and sQb (lane 3), with only modest effects of Sec17 on fusion and on the association of R and sQb with Qa (lane 4). The swap of Jx regions between R and Qa caused fusion to become dependent on Sec17 (lanes 5 and 6), yet neither the Jx swap nor Sec17 altered the HOPS-dependent Qa association with Qb or R (lanes 3–6). Sec17 alone, without HOPS, supports neither fusion nor these SNARE associations (lane 2). The Sec17 restoration of fusion when the Jx domains are swapped (lane 6) cannot be ascribed to enhanced SNARE associations (lanes 3–6). The preassembly of Qa with Qb bypassed the requirement that Qa have its normal Jx sequence for fusion (contrast lanes 3 and 5 with sQb to lanes 8 and 11 with membrane-bound Qb).
The capacity of preassembled QaQb to bypass the fusion block from Jx swap between R and Qa might have been due either to the higher levels of Qa:Qb complex that had assembled during proteoliposome preparation in comparison to HOPS-assembled Qa:sQb complex (Figure 6B, lanes 3–6 vs. lanes 7–12, blue bars), to wild-type Qb being bound to the membrane in complex with Qa rather than the initial localization of sQb in solution, or to Qb having a TM domain. We therefore tested each of these possibilities.
Effects of the levels of Qa:Qb complex on fusion with Jx swap
Qa and Qb alone form a stable complex (Song et al., 2020), and HOPS enables Ypt7/QaQb proteoliposomes to engage with Ypt7/R proteoliposomes to form a productive fusion intermediate (Harner and Wickner, 2018; Song et al., 2020). To mimic the low levels of HOPS-mediated sQb in complex with Qa (Figure 6B, lanes 3–6), we prepared R and QaQb, as well as R(JxQa) and Qa(JxR)Qb proteoliposomes with both Qa or Qa(JxR) and Qb, at 1:16,000 molar ratios to lipid or with the Qb-SNARE reduced by serial twofold levels to reduce the level of Qa:Qb or Qa(JxR):Qb complex. All reconstituted proteoliposomes (RPLs) were prepared with Ypt7 at 1:8000 molar ratios to lipid. Fusion was smoothly diminished by lowered levels of Qb, but at each level the fusion was comparable when the R and Qa had wild-type or swapped Jx domains (Figure 7 and Supplemental Figure S3). The relative sensitivity of fusion with Qa to a Jx-domain swap (Figure 6, lane 3 vs. lane 5) in comparison to fusion with QaQb (Figure 6, lane 8 vs. lane 11) was therefore not simply due to the low levels of sQb:Qa complex in comparison to Qa:Qb complex (Figure 6, lanes 3–6 vs. lanes 7–12).
FIGURE 7:
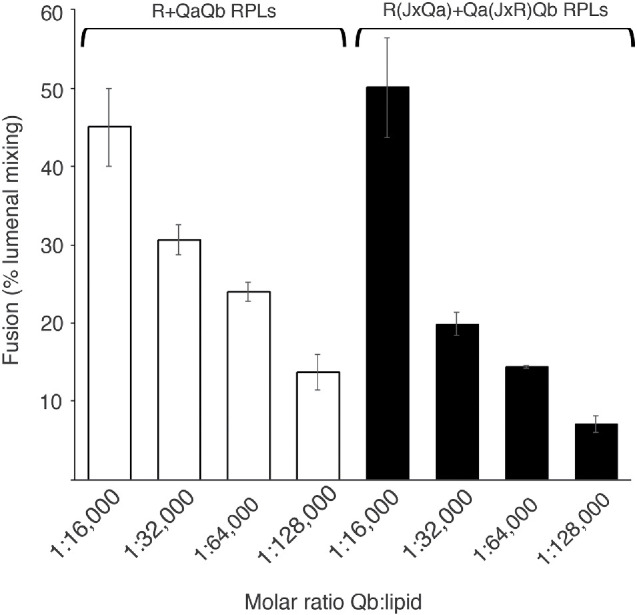
When wild-type (anchored) Qb levels are reduced to limit QaQb complex, fusion remains insensitive to the Jx-domain swap. R(JxQa) + Qa(JxR)Qb proteoliposomes and R + QaQb proteoliposomes give comparable fusion, even as the level of anchored Qb is steadily reduced. R and R(JxQa) proteoliposomes as well as QaQb and Qa(JxR)Qb proteoliposomes were prepared with Ypt7 as described in Materials and Methods. The molar ratio of Qb:lipid was varied from 1:16,000 to 1:128,000 by successive twofold dilutions of the initial stock of purified Qb. Fusion was assayed as described in Materials and Methods except that the liposome pairs were nucleotide exchanged together in a 10 µl volume, and the remaining soluble components were prepared as one mix, also contributing 10 µl of volume to the total 20 µl reaction. Concentrations of each component in 20 µl remain as described in Materials and Methods, with 50 nM HOPS and 100 nM Qc but no Sec17 or Sec18. Proteoliposomes had wild-type R and Qa (open bars) or swapped JxR and JxQa (black). Mean and SD values of fusion after 20 min are presented for triplicate experiments. See Supplemental Figure S3 for typical kinetic data.
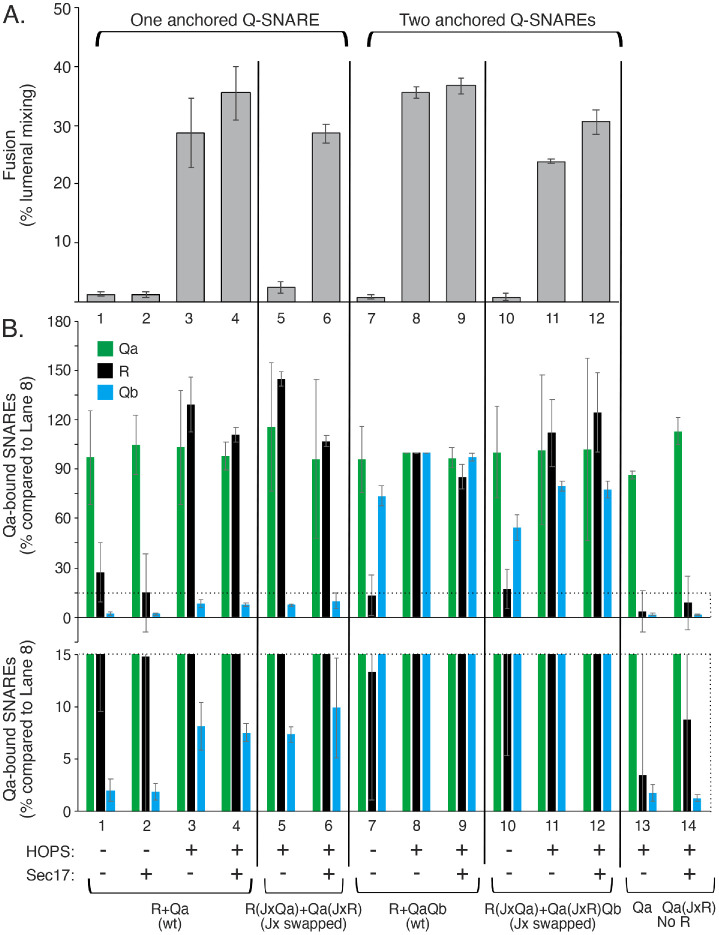
Jx swap when sQb is preassembled with Qa on a membrane
To examine the possible role of preassembly of the Qb SNARE with Qa, we prepared Ypt7/3Q-SNARE proteoliposomes bearing sQb, which lacks a TM domain, instead of full-length Qb, which has a TM domain. QasQbQc and Qa(JxR)sQbQc (Figure 1Ciii) were made by the same method of detergent dialysis from protein/lipid/detergent mixed micelles. The Qc SNARE was included in proteoliposome preparation because the QaQb complex is less stable than the QaQbQc complex (Song et al., 2020; see also Figure 8A, lanes 2 and 4 vs. lanes 1 and 3). sQb incorporated into proteoliposomes at levels that were comparable to those of proteoliposomes with membrane-anchored Qb at a 1:128,000 molar ratio to lipids and at somewhat higher levels when Qc was included in the reconstitution (Figure 8A). These QasQbQc proteoliposomes fused well (Figure 8B, lane 9, and Supplemental Figure S4), even better than with wild-type Qb in 3Q-SNARE complex (lanes 1 and 5). Strikingly, while fusion with membrane-anchored (wild-type) Qb is little affected by swap of the R and Qa Jx domains (Figure 8B, lane 1 vs. lane 4 and lane 5 vs. lane 8), the R and Qa Jx-domain swap with Qa-bound sQb strongly suppressed fusion (Figure 8B, lane 9 vs. lane 12). Fusion was just as fully suppressed when only one Jx domain was altered, that is, with R(wild-type) + Qa(JxR)sQbQc proteoliposomes (lane 10) or R(JxQa) + Qa(wild-type)sQbQc proteoliposomes (lane 11), as when both of these domains were swapped, that is, R(JxQa) + Qa(JxR)sQbQc proteoliposomes (lane 12). The sensitivity of fusion to unphysiological Jx assignment is thus bypassed by Qa-associated full-length Qb with its TM anchor, but not by Qa-associated sQb, which lacks the TM anchor. The difference is not initial membrane localization, but rather the presence or absence of the TM domain on Qb versus sQb.
FIGURE 8:
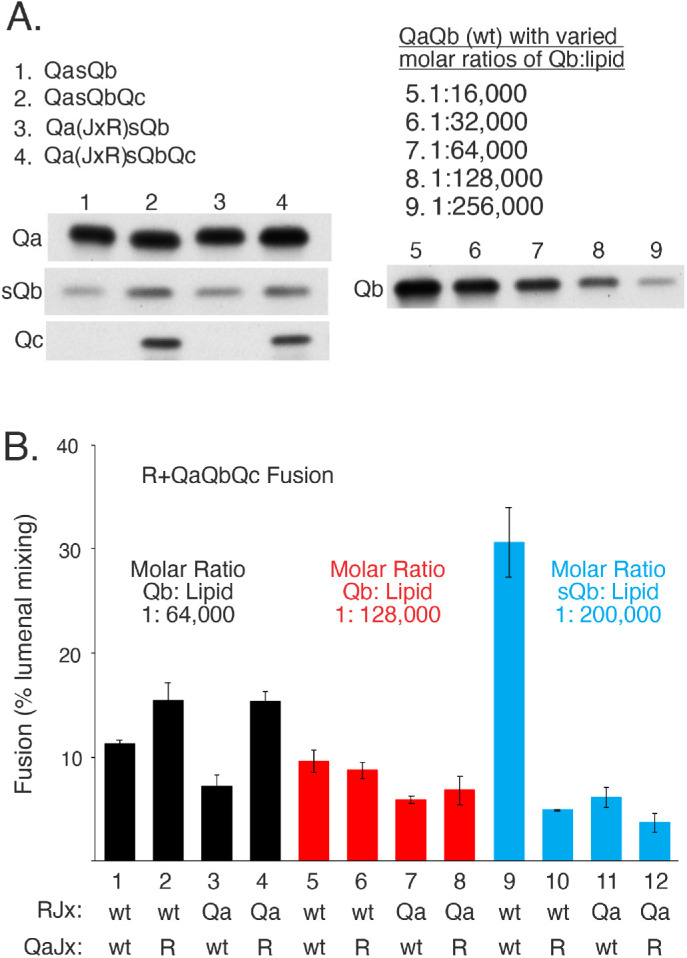
R and QaQbQc proteoliposomes fuse with little sensitivity to Jx swap, but R and QasQbQc proteoliposome fusion is strongly inhibited by Jx-domain swap between R and Qa. (A) Proteoliposome preparation. R and R(JxQa) proteoliposomes and QasQbQc and Qa(JxR)sQbQc proteoliposomes were prepared with Ypt7 as described in Materials and Methods, but with 5× the molar concentrations of sQb and Qc (where present) as Qa in the initial mixed micellar solution to drive SNARE complex assembly. Unincorporated SNAREs are removed during proteoliposome flotation. QaQb proteoliposomes were also prepared as described, with a 1:16,000 molar ratio of Qa:lipid, but with the indicated molar ratios of wild-type Qb to lipid, serving as standards for the level of sQb assembly into proteoliposomes. After isolation and adjusting each to 2 mM lipid, equivalent volumes of the indicated proteoliposomes were analyzed by SDS gel and immunoblot for their content of Qa, Qc, and sQb or Qb. sQb and Qb samples were analyzed on the same gel and blot. (B) Fusion of R and 3Q proteoliposomes. R and Qa were at 1:16,000 molar ratios to lipid and were either wild-type or R(JxQa) or Qa(JxR). Wild-type Qb was incorporated at 1:64,000 and 1:128,000 molar ratios to lipid to approach the lower levels of sQb incorporated. Fusion was assayed as described in Figure 7, but without any soluble SNARE additions, Sec17, or Sec18. Mean and SDs of fusion for triplicate reactions after 20 min are shown. See Supplemental Figure S4 for typical kinetic data.
Jx specificity is inherent to SNAREs rather than HOPS
To test whether HOPS governs the need for Jx:SNARE-domain match and the capacity of membrane-anchored Qb to restore fusion after Jx-domain swap, we employed a synthetic tether, dimeric glutathione-S-transferase fused to a PX domain, [GST-PX]2, which tethers by binding to PI3P in each membrane (Song and Wickner, 2019). When all three Q-SNAREs are preassembled on one fusion partner, [GST-PX]2 supports fusion as well as HOPS (Song and Wickner, 2019). Ypt7/R proteoliposomes were incubated with Ypt7/QasQbQc proteoliposomes, and fusion was initiated by addition of either HOPS (Figure 9, lane 1) or [GST-PX]2 (lane 5). In each case, fusion was inhibited by the Jx swap (lanes 2 and 6). In contrast, when wild-type, TM-anchored Qb was assembled into Ypt7/3Q proteoliposomes instead of sQb, fusion with either HOPS or [GST-PX]2 was not inhibited by the Jx-domain swap (lane 3 vs. lane 4 and lane 7 vs. lane 8). The specificity of fusion for Jx association with its natural SNARE domain and its bypass by a second Q-SNARE membrane anchor is not limited to fusion reactions with a particular physiological tether such as HOPS or with an SM protein but is apparently inherent to SNARE function.
FIGURE 9:
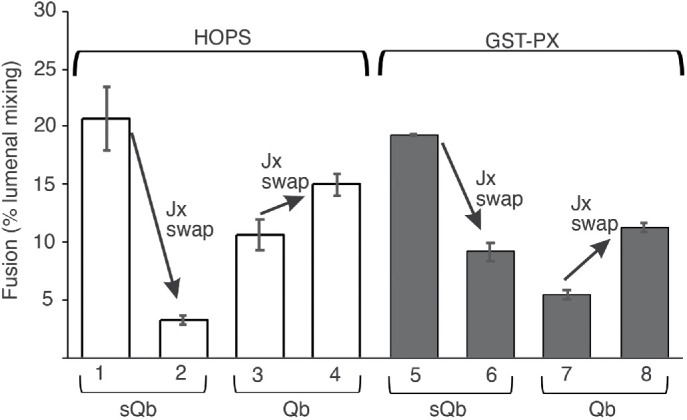
Jx domain specificity, and its bypass by anchoring Qb through its TM domain, is specific to SNAREs and not to a specific tether or SM protein. Proteoliposomes with R, QasQbQc, and QaQb(1:64,000 molar ratio to lipid)Qc, as well as the Jx-domain swapped R and Qa versions of each, were prepared as in Figure 8. Each R- and 3Q-SNARE pairing was assayed for fusion as described in Materials and Methods, but without Sec17 or Sec18, and with either 43 nM HOPS (open bars) or 1 mM GST-PX (filled bars) added as tethering agent at t = 0, as described (Song and Wickner, 2019). Average and SDs are shown of fusion at 20 min of incubation. Typical kinetic data can be found in Supplemental Figure S5.
Fusion in the presence of Sec18 and ATP
To avoid any possible complication from trans-SNARE complex disassembly (Starai et al., 2008; Xu et al., 2010), the foregoing studies did not have Sec18 and hydrolyzable ATP. Does Jx swap still inhibit fusion, and is this inhibition still relieved by Sec17 or by anchored Qb, when Sec18 and ATP are present? We examined the fusion of Ypt7/R and Ypt7/Qa proteoliposomes with Sec18 and ATP as well as HOPS, sQb, and Qc (Figure 10 and Supplemental Figure S6). Fusion with Ypt7 and wild-type R and Qa proteoliposomes (lane 1) is inhibited by swap of the R and Qa Jx domains (lane 2), but fusion is restored by the addition of Sec17 (lane 4). This restoration relies on the apolarity of the Sec17 N-terminal loop, because the Sec17-F21SM22S mutations, which abolish the N-loop apolarity (Zick et al., 2015; Schwartz et al., 2017; Song et al., 2017), prevent fusion restoration (lane 6). With wild-type, anchored Qb (lanes 7–12), Jx swap has a smaller effect on fusion (lanes 7 and 8), with little effect of Sec17 (lane 10) and independent of the Sec17 apolar N-loop (lane 12). Fusion loss from R and Qa Jx swap and its bypass by either Sec17 or membrane-anchored and Qa-associated Qb is thus seen in either the presence or the absence of Sec18 and ATP.
FIGURE 10:
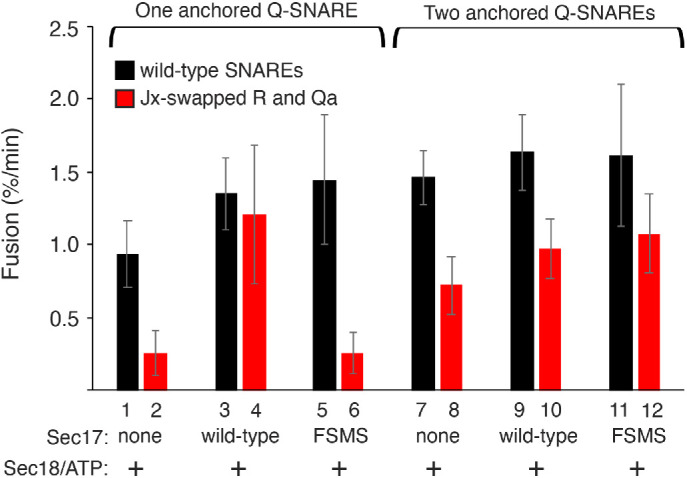
In the presence of Sec18 and MgATP, fusion of R+Qa proteoliposomes remains sensitive to the Jx swap, and reversal of this block by Sec17 requires its hydrophobic loop. With membrane-anchored Qb, there is less fusion inhibition by the Jx swap, with little effect of Sec17 or its hydrophobic loop. Proteoliposomes were prepared as described in Figure 6, with either one or two anchored Q-SNAREs. Fusion reactions were conducted as described in Materials and Methods with 50 nM HOPS, 100 nM Qc, 100 nM sQb (where Qa was the only anchored SNARE), 500 nM Sec17 or Sec17 FSMS where added, and with 250 nM Sec18 and 1 mM hydrolyzable MgATP. Averages and SDs of fusion rates from minutes 0–20 are shown from three replicate experiments. Typical kinetic data are shown in Supplemental Figure S6.
DISCUSSION
Even with all wild-type SNARE domains, and with invariant levels of trans-SNARE complex, fusion still relies on other SNARE domains and SNARE-binding factors. Fusion between R and Qa proteoliposomes is blocked when the Jx region of R is changed to that of Qa, the Jx region of Qa is changed to that of R, or both are changed, termed the “swap.” These assays contain HOPS, sQb, and Qc. While changing either R or Qa’s Jx region to the other, or the swap, blocks fusion, there is little or no effect on the level of physical association between R and Qa in trans (Figure 6). Each of these blocks to fusion, all with entirely wild-type SNARE domains, can be relieved by either addition of Sec17 alone, addition of lower concentrations of Sec17 with Sec18, or inclusion of membrane-anchored Qb (but not sQb) in cis to Qa during proteoliposome preparation. In cells, the vacuolar SNAREs are kept in a disassembled state by the continuous action of the ubiquitous Sec17 and Sec18 chaperones, and little of the Qb is found in complex with Qa on isolated vacuoles (Collins et al., 2005). The SNAREs assemble only during catalysis by HOPS (Baker et al., 2015; Orr et al., 2017; Jiao et al., 2018; Torng et al., 2020), which protects them from Sec17/Sec18 disassembly (Xu et al., 2010).
To date there are three models for reconstitution of vacuolar Qb function: 1) adding recombinant sQb without a membrane anchor to Qa proteoliposomes (Zick and Wickner, 2013), 2) incorporating full-length Qa and Qb into the same proteoliposomes by dialysis from detergent/lipid/protein mixed micelles (Fukuda et al., 2000), and 3) preparing proteoliposomes with anchored Qa and sQb by the same method (Figure 8). Each model has its limitations. While wild-type Qb has a membrane anchor, fusion reconstituted with disassembled sQb mimics the largely disassembled state of the SNAREs found on vacuoles. Soluble Qb lacks a TM anchor that can otherwise compensate for altered R or Qa Jx domain function. Coincorporation of TM-anchored Qa and Qb in the same proteoliposomes during dialysis allows their stable association in complex with each other (Figure 6, lanes 7–12), an association that may bypass important SNARE assembly functions of HOPS (Song et al., 2020).
These studies contribute to our current model of fusion. Vacuoles, and vacuolar proteoliposomes, are tethered by HOPS and Ypt7 (Stroupe et al., 2006; Hickey and Wickner, 2010). HOPS is activated by association with Ypt7, R, phosphatidylinositol, and phosphatidylinositol-3P to catalyze SNARE complex assembly (Torng et al., 2020; Torng and Wickner, 2021). SNAREs zipper N to C through the four SNARE domains (Sorensen et al., 2006). During zippering, Sec18 and several Sec17s associate with the zippering SNARE platform. Sec17 apolar loops, as well as the completion of zippering, promote fusion (Schwartz and Merz, 2009; Zick et al., 2015; Schwartz et al., 2017; Song et al., 2021). While Sec17 and Sec18 enhance the rate of fusion (Figures 2–4 and Song et al., 2020), there are three known conditions where Sec17 and Sec18 are essential: with SNAREs that are C-terminally truncated to block zippering (Schwartz and Merz, 2009; Song et al., 2021), with suboptimal lipid compositions (Zick et al., 2015), and with exchanged Jx domains (Figures 2, 4, 6, and 10). Fusion loss upon Jx-domain swap is particularly striking because, with two membranes poised to fuse, there is no known difference between the forces acting on the membrane bearing the R-SNARE and the membrane with the Q-SNAREs. The Jx regions of R and Qa are of similar lengths and sizes, but R has dual tryptophans, seen in the Jx region of some other R-SNAREs such as Snc2. Jx domains may be needed to connect SNARE and TM domains in continuous α-helices and to provide membrane disruption by tryptophan membrane insertion, but these functions alone might have been impervious to a swap of Jx domains. The positional stringency for JxR and JxQa is consistent with a need for physiological aminoacyl side chain interactions between these Jx domains with other neighbors in the four-helical bundle. In any case, this seems to reflect specific interactions among SNAREs rather than with, for example, HOPS, as it is still seen when the HOPS tether is replaced by an artificial tether such as dimeric GST-PX (Figure 9).
We note that there is more fusion in proportion to the Qb:Qa complex with sQb than with wild-type, membrane-anchored Qb (Figures 6 and 8). Where the sQb is recruited to association with Qa by HOPS and the fusion was comparable to that with membrane bound Qb (Figure 6), there was far less sQb bound to Qa than when Qb and Qa were both initially bound to the same membranes. When sQb was initially incorporated into membranes as part of the QasQbQc ternary complex, there was more fusion than seen with QaQbQc complex with similar amounts of bound sQb or Qb (Figure 8). The basis of the high fusion activity of sQb is unknown. One working model is that completion of zippering is impeded by the need to move the Qb membrane anchor around that of Qa as zippering proceeds, and sQb might relieve this constraint, but this model is untested.
Many components have a vital role in fusion: Sec17, Sec18, a tether and an SM SNARE assembly catalyst (which are combined in HOPS), the SNAREs through their SNARE, Jx, and TM domains, and key lipids such as sterol, acidic lipids, small headgroup lipids, and their fluid fatty acyl chains. Genetic ablation or selective omission from reconstituted proteoliposome fusion reactions can effectively illuminate the individual roles of components. Thus fusion can occur either without completion of SNARE zippering (through the apolar N-loop of Sec17), without Sec17 or Sec18 (through complete zippering), without key lipids (through unphysiologically elevated SNARE levels), or, as shown here, without correct associations between Jx and SNARE domains (through the apolar N-domain of Sec17 or the TM anchor of Qb). Physiological functions of each component are revealed as others that could compensate for their absence are, for analysis, stripped away.
Jx domains connect SNARE and trans-membrane domains and are proposed to form a continuous α-helix structure between them (McNew et al., 1999; Stein et al., 2009). These cannot be the only Jx functions, as they would be preserved during the swap of the Jx domain between R and Qa. The tryptophans in the JxR domain may serve as a membrane-disrupting wedge, yet fusion is strongly impaired in R (wild-type) plus Qa(JxR) or R(JxQa) plus Qa(JxR) proteoliposome pairings. The full fusion seen upon Sec17 addition or Qb membrane anchoring shows that the Jx-altered SNAREs are not simply denatured or inert. Sec17 acts without SNARE zippering, while the Jx domains are believed to be random coil until zippering is complete; the Sec17 bypass of all the Jx duplications or swap is therefore not surprising. The bypass of Jx swap by preassembly of Qa and Qb suggests that fusion can be supported through the bound Qb via its TM domain. This bypass is not limited to fusion with HOPS, as fusion with a synthetic tether also needs the correct Jx:SNARE-domain match (Figure 9). As a working model, we suggest that each Jx domain conveys its own SNARE-domain zippering energy to the membrane, but that Jx swap can be bypassed through full-length Qb once it is in a zippered 4-SNARE complex, conveying the signal through the Qb transmembrane domain. SNARE domains are not the only determinants of fusion.
MATERIALS AND METHODS
Reagents
Lipids were purchased from Avanti Polar Lipids (Alabaster, AL) with the exceptions of PI3P (p hosphatidylinositol 3-phosphate diC16) from Echelon Biosciences (Salt Lake City, UT) and ergosterol from Fluka (St. Louis, MO). Octylglucoside (n-octyl-ß-d-glucopyranoside) was from Anatrace (Maumee, OH), Cy5-labeled streptavidin from LGC Clinical Diagnostics (Milford, MA), biotin-conjugated R-phycoerythrin from Life Technologies Corporation (Eugene, OR), and Histodenz from Sigma-Aldrich (St. Louis, MO). Dialysis tubing was from Repligen Corporation (Waltham, MA) and Bio-Beads SM-2 Resin from Bio-Rad Laboratories (Hercules, CA). Underivatized streptavidin and Pierce Protein-A Magnetic Beads were from Thermo Scientific (Waltham, MA).
Recombinant gene construction
For the Qa and R-SNAREs with exchanged juxta-transmembrane (JxTMs) or transmembrane (TM) domains, the Qa and R SNARE genes without those regions were generated by PCR using Phusion HiFi DNA polymerase from NEB. The JxTMs and the TMs of Qa, R, and Qb were amplified using the same technology. For each mutant protein an In-fusion HD Cloning Kit (Takara Bio USA, Mountain View, CA) was used to simultaneously clone the two relevant inserts into an MBP-pParallel1 vector that had been previously digested with BamHI and Sal1.
Forward (F) and Reverse (R) oligonucleotides:
For MBP-R without its JxTM (Nyv1 1-220):
F: AGGGCGCCATGGATCCGATGAAACGCTTTAATGTAAG
R: TATATTGACCGCTTTGCGCC
For MBP-Qa without its JxTM (Vam3 1-252):
F: AGGGCGCCATGGATCCGATGTCCTTTTTCGACATCGA
R: TGCTCTGGTTAATTGTTTGT
For MBP-R without its TM (Nyv1 1-230):
F: AGGGCGCCATGGATCCGATGAAACGCTTTAATGTAAG
R: TTTGACCTTCTGCCACCA
For MBP-Qa without its TM (Vam3 1-263):
F: AGGGCGCCATGGATCCGATGTCCTTTTTCGACATCGA
R: ACCGCATTTGTTACGGTC
Qa with the JxTM (221-253) of the R-SNARE:
F: CAATTAACCAGAGCAAAAGAAATAATGTGGTGGCA
R: AGTTGAGCTCGTCGATTACCACAGATAGAAAAACAT
Qa with the JxTM (187-217) of the Qb-SNARE:
F: CAATTAACCAGAGCAACTAGAAGGCTAGTTGCTAA
R: AGTTGAGCTCGTCGATTATTTAAACTTTGAGAA
R with the JxTM (253-283) of the Qa-SNARE:
F: AAAGCGGTCAATATAGACCAGCATCAGAGGGACCG
R: AGTTGAGCTCGTCGACTAACTTAATACAGCAAGCA
R with the JxTM (187-217) of the Qb-SNARE:
F: AAAGCGGTCAATATAACTAGAAGGCTAGTTGCTAA
R: AGTTGAGCTCGTCGATTATTTAAACTTTGAGAA
Qa with the TM (231-253) of the R-SNARE:
F: GACCGTAACAAATGCGGTAATATTACGTTATTAACT
R: AGTTGAGCTCGTCGATTACCACAGATAGAAAAACAT
Qa with the TM (195-217) of the Qb-SNARE:
F: GACCGTAACAAATGCGGTTTCATAAGCTATGCCATT
R: AGTTGAGCTCGTCGATTATTTAAACTTTGAGAA
R with the TM (264-283) of the Qa-SNARE:
F: TGGTGGCAGAAGGTCAAAAAGGTCACCCTAATCATT
R: AGTTGAGCTCGTCGACTAACTTAATACAGCAAGCA
R with the TM (195-217) of the Qb-SNARE:
F: TGGTGGCAGAAGGTCAAATTCATAAGCTATGCC
R: AGTTGAGCTCGTCGATTATTTAAACTTTGAGAA
To create the Jx swapped R and Qa SNAREs, where each keeps its own TMs, MBP-pParallel-Vam3 and MBP-pParallel-Nyv1 were amplified without their Jx regions using NEB’s Phusion HiFi DNA polymerase and the following oligos:
pParallel1_Nyv1 R: TTTTATATTGACCGCTTTGC
pParallel1_Nyv1 F: AATATTACGTTATTAACTTT
pParallel1_Vam3 R: GTCTGCTCTGGTTAATTGTT
pParallel1_Vam3 F: AAGGTCACCCTAATCATTAT
Complementary oligos encompassing the entire Jx regions of the Qa and R SNAREs were annealed and then cloned into the PCR-amplified Jx-deficient MBP-pParallel-Qa or R plasmids using an In-fusion HD Cloning Kit from Takara Bio USA (Mountain View, CA).
Nyv1_Vam3 Jx(254-263)_ Nyv1TM
Upper:GCGGTCAATATAAAACAGCATCAGAGGGACCGTAACAAATGCGGTAATATTACGTTATTA
Lower:TAATAACGTAATATTACCGCATTTGTTACGGTCCCTCTGATGCTGTTTTATATTGACCGC
Vam3_Nyv1 Jx(222-230)_Vam3TM
Upper:TTAACCAGAGCAGACGAAATAATGTGGTGGCAGAAGGTCAAAAAGGTCACCCTAATC
Lower:GATTAGGGTGACCTTTTTGACCTTCTGCCACCACATTATTTCGTCTGCTCTGGTTAA
Protein purification
MBP-tagged mutant SNAREs were purified as described for MBP-Ypt7-TM (Song et al., 2020). The GST-tagged Nyv1 used as a competitor of distinct molecular weight in the immunoprecipitations was purified as described in Song et al. (2020). Vam7, GST-tagged Vam3, and untagged full-length Vti1 and Nyv1 (Mima et al., 2008; Schwartz and Merz, 2009; Zucchi and Zick, 2011) were purified as described, and Vti1 and Nyv1 were buffer exchanged (Zucchi and Zick, 2011). The tags were removed from GST-Vam3 and MBP-Ypt7-TM by the addition of TEV to the mixed micellar detergent/lipid/protein solution before dialysis. Sec17 and Vam7∆3 (Schwartz and Merz, 2009), Sec18 (Mayer et al., 1996), and HOPS and MBP-sVti1 (Zick and Wickner, 2013) were purified as described. MBP-sVti1 was cleaved of its tag with TEV before use in fusion reactions or proteoliposome preparation. GST-PX was purified as described (Fratti et al., 2007).
Proteoliposome preparation
Vacuolar mimic lipid (VML) proteoliposomes were prepared with SNAREs and Ypt7-TM as described (Song and Wickner, 2019). In brief, lipids in chloroform were mixed with ß-octylglucoside, dried for 30 min at room temperature under a stream of nitrogen and then for 3 h in a speedvac, and solubilized in 5× concentrated Rb150+Mg (100 mM HEPES/NaOH, pH 7.4, 750 mM NaCl, 50% glycerol, 5 mM MgCl2). SNARE proteins were added at a 1:16,000 molar ratio of protein to lipid unless otherwise indicated, and Ypt7-TM was added at a 1:8000 molar ratio to lipid. Biotin-phycoerythrin was added to lipid–detergent mixes containing the R-SNARE, and Cy5-streptavidin was added to mixes with one or more of the Q-SNAREs. His6-TEV (1 mM) was added to all mixtures. Lipid–protein mixes in detergent were dialyzed against Rb150+Mg (20 mM HEPES/NaOH, pH 7.4, 150 mM NaCl, 10% glycerol, 1 mM MgCl2) in 25K MWCO dialysis tubing (12 mm flat width) for at least 16 h at 4°C in the dark and then floated through Histodenz gradients and harvested. Concentrations were determined by lipid phosphate analysis, and proteoliposomes were snap-frozen at 2 mM lipid phosphate concentration.
Fusion assay
Each complete fusion reaction was in 20 µl. Unless otherwise indicated, R and Q proteoliposomes, all of which contain Ypt7 and each in a volume of 5 µl, were separately nucleotide exchanged as follows: proteoliposomes (R or Q) (2.5 µl) with 2 mM lipid were mixed with 9.6 µM underivatized streptavidin, 40 µM GTP, and 2 mM EDTA, incubated for 10 min at 27°C and then supplemented with 4 mM MgCl2. Final concentrations of these components in a 20 µl reaction are 0.5 mM total lipid, 4.8 µM streptavidin, 20 µM GTP, 1 mM EDTA, and 2 mM MgCl2. Mixes of 1) soluble SNAREs, 2) HOPS, and 3) Sec17±Sec18/ATPγS were prepared separately; their combined volumes were 10 µl per reaction, and final concentrations were 100 nM each soluble SNARE, 50 nM HOPS, 100, 500, or 600 nM Sec17 as indicated, 250 or 300 nM Sec18 as indicated, and 1 mM ATPγS where added, mixed with an additional 1 mM MgCl2. Each separate mix was incubated in a 384-well plate at 27°C for 10 min. Reactions were started by transferring 5 µl of R-SNARE mix to a new set of wells, followed swiftly by 5 µl of Q-SNARE(s) mix, then the soluble SNARE mix, the HOPS mix, and finally the Sec17±Sec18/MgATPγS mix for a total volume of 20 µl. FRET between the Cy5-streptavidin and biotin-phycoerythrin (Ex: 565, Em: 670, CO: 630) was recorded in a Molecular Devices SpectraMax Gemini XPS plate reader (Sunnyvale, CA).
Immunoprecipitations
In addition to the 20 µl fusion reactions in microplates described above, 40 µl reactions were simultaneously prepared, components separately incubated in PCR strips in a 27°C water bath, and fusion reactions started in 0.5 ml Eppendorf tubes in a 27°C water bath. After 20 min, the microplate reactions were removed from the plate reader and 18 µl of the reactions in the wells was added to the complementary reactions in the Eppendorf tubes. A portion of each (48 µl) was added to a 1.5 ml tube containing 6 µl of Pierce Protein A Magnetic Beads, 6 µg affinity-purified αVam3, and 5 µM GST-3C-Nyv1 in RIPA buffer (20 mM HEPES/NaOH, pH 7.4, 150 mM NaCl, 0.2% bovine serum albumin, 1% Triton-X 100, 1% sodium cholate, 0.1% SDS) in a total volume of 150 µl. Reactions were nutated end-over-end at room temperature for 2 h, beads were washed three times in 1 ml RIPA buffer, and antibody-bound proteins were eluted by adding 48 µl 1× SDS buffer with ß-mercaptoethanol and boiling for 2 min at 95°C. Samples were cooled, harvested from the magnetic beads, and analyzed by immunoblot.
Supplementary Material
Acknowledgments
This work was supported by grant R35GM118037 from the National Institute of General Medical Sciences. We thank Michael Zick, Gustav Lienhard, Christopher Shoemaker, Jose Rizo, and Henry Higgs for helpful comments.
Abbreviations used:
- Jx
the region of each SNARE between its SNARE domain and its apolar membrane anchor
- RPL
reconstituted proteoliposome
- TEV
tobacco etch virus protease
- TM
trans-membrane anchor.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-11-0583) on February 16, 2022.
REFERENCES
- Baker RW, Jeffrey PD, Zick M, Phillips BP, Wickner WT, Hughson FM (2015). A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science 349, 1111–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett CL, Plemel RL, Lobinger BT, Vignali M, Fields S, Merz AJ (2008). Efficient termination of vacuolar Rab GTPase signaling requires coordinated action by a GAP and a protein kinase. J Cell Biol 182, 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröcker C, Kuhlee A, Gatsogiannis C, Balderhaar HJK, Hönscher C, Engelbrecht-Vadre S, Ungermann C, Raunser S (2012). Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS). tethering complex. Proc Natl Acad Sci USA 109, 1991–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins KM, Thorngren NL, Fratti RA, Wickner WT (2005). Sec17p and HOPS, in distinct SNARE complexes, mediate SNARE complex disruption or assembly for fusion. EMBO J 24, 1775–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMill CM, Xiu X, Kisiel M, Bolotta A, Stewart BA (2014). Investigation of the juxtamembrane region of neuronal-Synaptobrevin in synaptic transmission at the Drosophila neuromuscular juction. J Neurophysiol 112, 1356–1366. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Bruns D, Shen B, Jahn R, Brünger AT (1997). A structural change occurs upon binding of syntaxin to SNAP-25. J Biol Chem 272, 4582–4590. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Sutton RBB, Brunger AT, Jahn R (1998). Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA 95, 15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig KM, Rice LM, Pollock E, Brünger AT (1999). Folding intermediates of SNARE complex assembly. Nat Struct Biol 6, 117–123. [DOI] [PubMed] [Google Scholar]
- Fratti R, Collins KM, Hickey CM, Wickner W (2007). Stringent 3Q:1R composition of the SNARE 0-layer can be bypassed for fusion by compensatory SNARE mutation or by lipid bilayer modification. J Biol Chem 282, 14861–14867. [DOI] [PubMed] [Google Scholar]
- Fukuda R, McNew JA, Weber T, Parlati F, Engel T, Nickel W, Rothman JE, Söllner T (2000). Functional architecture of an intracellular membrane t-SNARE. Nature 407, 198–202. [DOI] [PubMed] [Google Scholar]
- Haas A, Conradt B, Wickner W (1994). G-protein ligands inhibit in vitro reactions of vacuole inheritance. J Cell Biol 126, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner M, Wickner W (2018). Assembly of intermediates for rapid membrane fusion. J Biol Chem 293, 1346–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey CM, Wickner W (2010). HOPS initiates vacuole docking by tethering membranes before trans-SNARE complex assembly. Mol Biol Cell 21, 2297–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zhu L, Ma C (2021). Structural roles for the juxtamembrane linker region and transmembrane region of synaptobrevin 2 in membrane fusion. Front Cell Dev Biol 8, 609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J, He M, Port SA, Baker RW, Xu Y, Qu H, Xiong Y, Wang Y, Jin H, Eisemann TJ, et al. (2018). Munc18-1 catalyzes neuronal SNARE assembly by templating SNARE association. eLife 7, e41771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Wickner W, Haas A (1996). Sec18p (NSF)-driven release of Sec17p (α-SNAP) can precede docking and fusion of yeast vacuoles. Cell 85, 83–94. [DOI] [PubMed] [Google Scholar]
- Mima J, Hickey C, Xu H, Jun Y, Wickner W (2008). Reconstituted membrane fusion requires regulatory lipids, SNAREs and synergistic SNARE chaperones. EMBO J 27, 2031–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew JA, Weber T, Engelman DM, Söllner TH, Rothman JE (1999). The length of the flexible SNAREpin juxtamembrane region is a critical determinant of SNARE-dependent fusion. Mol Cell 4, 415–421. [DOI] [PubMed] [Google Scholar]
- Orr A, Song H, Rusin SF, Kettenbach AN, Wickner W (2017). HOPS catalyzes the interdependent assembly of each vacuolar SNARE into a SNARE complex. Mol Biol Cell 28, 975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore SS, Liu Y, Hu H, Wan C, Lee M, Yin Q, Stowell MHB, Shen J (2019). Intracellular vesicle fusion requires a membrane-destabilizing peptide located at the juxtamembrane region of the v-SNARE. Cell Rep 29, 4583–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz ML, Merz AJ (2009). Capture and release of partially zipped trans-SNARE complexes on intact organelles. J Cell Biol 185, 535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz ML, Nickerson DP, Lobingier BT, Plemel RL, Duan M, Angers CG, Zick M, Merz A (2017). Sec17 (α-SNAP) and an SM-tethering complex regulate the outcome of SNARE zippering in vitro and in vivo. eLife 6, e27396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals D, Eitzen G, Margolis N, Wickner WT, Price A (2000). A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc Natl Acad Sci USA 97, 9402–9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Lahat D, Barak-Broner N, Sheinin A, Greitzer-Antes D, Michaelevski I, Lotan I (2018). The dual function of the polybasic juxtamembrane region of syntaxin 1A in clamping spontaneous release and stimulating Ca2+-triggered release in neuroendocrine cells. J Neurosci 3, 220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE (1993). A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell 75, 409–418. [DOI] [PubMed] [Google Scholar]
- Song H, Orr A, Duan M, Merz AJ, Wickner W (2017). Sec17/Sec18 act twice, enhancing membrane fusion and then disassembling cis-SNARE complexes. eLife 6, e26646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Orr AS, Lee M, Harner ME, Wickner WT (2020). HOPS recognizes each SNARE, assembling ternary trans-complexes for rapid fusion upon engagement with the 4th SNARE. eLife 9, e53559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Torng TL, Orr AS, Brunger AT, Wickner WT (2021). Sec17/Sec18 can support membrane fusion without help from completion of SNARE zippering. eLife 10, e67578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Wickner W (2019). Tethering guides fusion-competent trans-SNARE assembly. Proc Natl Acad Sci USA 116, 13952–13957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen JB, Wiederhold K, Müller EM, Milosevic I, Nagy G, de Groot B, Grubmüller H, Fasshauer D (2006). Sequential N to C-terminal SNARE complex assembly drives priming and fusion of secretory vesicles. EMBO J 25, 955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai VJ, Hickey CM, Wickner W (2008). HOPS proofreads the trans-SNARE complex for yeast vacuole fusion. Mol Biol Cell 19, 2500–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A, Weber G, Wahl MC, Jahn R (2009). Helical extension of the neuronal SNARE complex into the membrane. Nature 460, 525–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroupe C, Collins KM, Fratti RA, Wickner W (2006). Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J 25, 1579–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT (1998). Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353. [DOI] [PubMed] [Google Scholar]
- Torng T, Song H, Wickner W (2020). Asymmetric Rab activation of vacuolar HOPS to catalyze SNARE complex assembly. Mol Biol Cell 31, 1060–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torng T, Wickner W (2021). Phosphatidylinositol and phosphatidylinositol-3-phosphate activate HOPS to catalyze SNARE assembly, allowing small headgroup lipids to support the terminal steps of membrane fusion. Mol Biol Cell 32, ar19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Komen JS, Bai X, Rodkey TL, Schaub J, McNew JA (2005). The polybasic juxtamembrane region of Sso1p is required for SNARE function in vivo. Eukaryotic Cell 4, 2017–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Ohsumi Y, Anraku Y (1992). Genes for directing vacuole morphogenesis in Saccharomyces cerevisiae. I. Isolation and characterization of two classes of vam mutants. J Biol Chem 267, 18665–18670. [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachi M, Parlati F, Söllner TH, Rothman JE (1998). SNAREpins: minimal machinery for membrane fusion. Cell 92, 759–772. [DOI] [PubMed] [Google Scholar]
- Wickner W, Rizo J (2017). A cascade of multiple proteins and lipids catalyzes membrane fusion. Mol Biol Cell 28, 707–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Jun Y, Thompson J, Yates J, Wickner W (2010). HOPS prevents the disassembly of trans-SNARE complexes by Sec17p/Sec18p during membrane fusion. EMBO J 29, 1948–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick M, Orr A, Schwartz ML, Merz AJ, Wickner WT (2015). Sec17 can trigger fusion of trans-SNARE paired membranes without Sec18. Proc Natl Acad Sci USA 112, E2290–E2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick M, Wickner W (2013). The tethering complex HOPS catalyzes assembly of the soluble SNARE Vam7 into fusogenic trans-SNARE complexes. Mol Biol Cell 24, 3746–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchi PC, Zick M (2011). Membrane fusion catalyzed by a Rab, SNAREs, and SNARE chaperones is accompanied by enhanced permeability to small molecules and by lysis. Mol Biol Cell 22, 4635–4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.