INTRODUCTION
To control their size, budding yeast couple cell growth to progression through the cell cycle transition known as Start in the G1 phase of the cell cycle (Johnston et al., 1977; Di Talia et al., 2007; Doncic et al., 2011). While the pre-Start G1 duration is highly variable, on average, smaller-born cells spend more time in pre-Start G1 before entering their first cell cycle.
One mechanism through which cell growth drives the Start transition is the dilution of Whi5, an inhibitor of the SBF transcription factor crucial for cell cycle–dependent gene expression (Schmoller et al., 2015). However, since we first reported the Whi5 dilution mechanism, two recent studies (Dorsey et al., 2018; Litsios et al., 2019) directly contradicted our claim that cell growth during G1 dilutes Whi5 to lower its concentration. Instead, Litsios et al. propose that there is a two- to threefold increase in the total protein synthesis rate before Start, which drives a strong increase in the concentration of the G1 cyclin Cln3 (Litsios et al., 2019). Given the importance of accurate protein concentration measurements for a mechanistic understanding of G1 cell cycle control, we revisited the question of whether Whi5 is diluted in pre-Start G1 using independent strains, experimental setups, and analysis pipelines in multiple laboratories. We present data from five independent laboratories confirming Whi5 dilution and find no evidence for a dramatic increase of global protein synthesis directly before Start. Taken together, this analysis firmly establishes Whi5 dilution dynamics as one of likely several mechanisms that link cell growth to cell cycle progression through the G1 phase of the cell division cycle.
The Whi5 dilution model requires that the concentration of Whi5 in newborn cells decreases with their cell size at birth, and that dilution by cell growth causes a continuous decrease of Whi5 concentration during G1. While the size-dependent Whi5 concentration at birth is based on an unusual cell size–independent transcriptional mechanism and chromatin-based partitioning (Schmoller et al., 2015; Swaffer et al., 2021), the latter is explained by two key observations. First, Whi5 is a stable protein (Schmoller et al., 2015; Gomar-Alba et al., 2017). Second, transcription of WHI5 mRNA is strongly cell cycle–dependent and peaks in S/G2 (Spellman et al., 1998; Pramila et al., 2006; Granovskaia et al., 2010). Indeed, it has long been known that WHI5 is a cell cycle–dependent transcript as it ranks among the top 300 most periodically expressed budding yeast genes in the Cyclebase database (Santos et al., 2015). This cell cycle dependence was recently confirmed using single molecule FISH (Qu et al., 2019; Swaffer et al., 2021). Thus, unless translational control perfectly compensates for the transcriptional oscillation, cell growth during G1—the period in which Whi5 mRNA is only weakly expressed—will inevitably decrease Whi5 protein concentration. Importantly, dilution of Whi5 does not imply that there is absolutely no synthesis of Whi5 protein during G1, just that its synthesis rate in G1 is significantly lower than in S/G2/M.
Whi5 dilution during pre-Start G1 was observed using live-cell wide-field fluorescence microscopy of Whi5 in asynchronously cycling single cells (Schmoller et al., 2015). This analysis required careful subtraction of background and cell autofluorescence before the total fluorescence intensity of tagged Whi5 can be extracted as a proxy for the total amount of Whi5 protein. Simultaneous estimation of cell volume based on cell segmentations obtained from phase contrast images can then be used to calculate the relative change of Whi5 concentration over time. Using this approach, we found that Whi5 protein concentrations directly reflect its transcriptional regulation. In G1, Whi5 is weakly expressed, which leads to its dilution in this phase of the cell cycle. This is followed by a strong increase in Whi5 synthesis in the budded phase of the cell cycle (Schmoller et al., 2015).
Since our initial report, dilution of Whi5 has been confirmed using live-cell microscopy by the Tang and Murray labs (Supplemental Figure S1; Qu et al., 2019; Barber et al., 2020), and by the Schneider and Aldea labs (Supplemental Figures S2 and S3). In addition, dilution of Whi5 in G1 was verified using immunoblots as shown in Schmoller et al. for G1 arrested cells (Schmoller et al., 2015). Lucena et al. shows an elutriation experiment with normally sized cells where the ratio of Whi5 and Cln3 protein amounts decreases as cells grow through G1 phase after release from poor into rich media (see Supplemental Figure S4 for quantitation of this experiment from Lucena et al., 2018). The latter was confirmed recently by Sommer et al., who conclude that “Whi5 concentration decreased by ∼30% in rich carbon and 20% in poor carbon, consistent with the fact that Whi5 protein levels do not change substantially” [in G1] (Sommer et al., 2021). We note that the ambiguous term “levels” in Sommer et al. refers to protein amounts. Importantly, these verifications of Whi5 dilution were made using different strains, microscopes, and analysis methods. However, in contrast to the growing consensus that Whi5 concentration decreases in pre-Start G1, two recent studies, Litsios et al. (2019) and Dorsey et al. (2018), reported constant Whi5 concentrations during G1. In an effort to understand this discrepancy, we carefully reinvestigated the claims of these studies.
RESULTS
Whi5 is diluted in G1 in Litsios et al.’s own fluorescence microscopy measurements
Litsios et al. use live-cell fluorescence microscopy to measure the concentration of Whi5 in asynchronously cycling cells, which is similar to the experiments performed by us and others described above. However, Litsios et al. concludes that they saw no or very little dilution of Whi5: “While it was recently proposed that the timing of Start is determined by the dilution of Whi5 (Schmoller et al., 2015), accumulating evidence from more recent studies contradicts this model. In accordance with our findings, Dorsey et al. (2018) did not observe any dilution of Whi5 under different genetic backgrounds and nutrient conditions, attributing reported changes in the Whi5 concentration to photobleaching.” We note here that we performed control experiments that excluded the possibility that photobleaching causes the observed decrease of Whi5 concentration during G1 in our original study (cf. Figure S10 of Schmoller et al., 2015). A more than threefold change in the fluorescence exposure had no effect on the measured Whi5 concentration dynamics (Schmoller et al., 2015). If photobleaching were causing the dilution effect, then increasing fluorescence exposure would lead to more rapid “dilution.” This is clearly not the case. Moreover, using the same imaging conditions and the same mCitrine fluorophore, we found that all other regulators of Start we examined, including Swi4, Bck2, and Whi3, did not show similar dilution (Schmoller et al., 2015). Again, if photobleaching of mCitrine were causing apparent “dilution,” then one would expect to have seen a similar “dilution” effect in all these proteins examined in the same imaging conditions. Finally, the quantitative imaging approach used to identify Whi5 dilution can be used to detect the well-known cell cycle dependence of budding yeast histones, which—similar to Whi5—are not produced during pre-Start G1 and show a peak of expression during S phase (Claude et al., 2021).
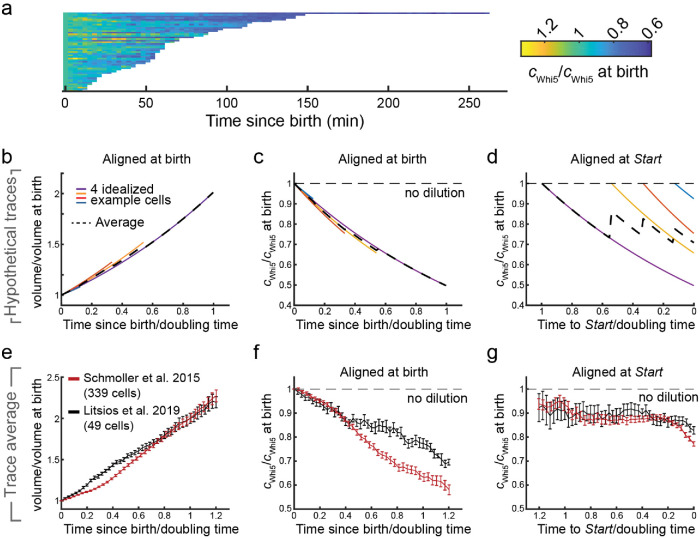
Having again excluded the possibility that Whi5 dilution is an artifact of photobleaching, we turn our attention to the discrepancy between our observation and Litsios et al.’s conclusion that Whi5 shows little or no dilution. We sought to examine their data more carefully, which indicated that the concentration of Whi5 was constant during G1 (cf. Figure 5a in Litsios et al., 2019). Upon request of the raw data, we received 49 single-cell traces of concentration and cell volume corresponding to “Whi5-mCherry WF-2” published in Litsios et al. Upon examination of these individual Whi5-mCherry concentration traces, we were surprised to see that Whi5 concentration decreased in 44 of 49 cells from birth to Start (Figure 1A). To draw the conclusion that Whi5 concentration was constant from their microscopy data, Litsios et al. performed an unusual normalization and averaging procedure. They normalized the Whi5 concentration of each individual cell to the time of cell birth, and then aligned all single-cell traces at Start to calculate the time evolution of the average shown in their Figure 5a that shows only a modest decrease in Whi5 concentration. However, given the large variability of pre-Start-G1 durations, aligning cell traces at Start while simultaneously normalizing to the concentration at birth masks the underlying dynamics and is inadequate to determine whether or not Whi5 is diluted by cell growth in single cells. To make this point absolutely clear, consider four cells that all grow with a similar exponential growth rate, but show varying durations of the period from birth to Start (Figure 1B). Assuming ideal dilution of a stable pool of Whi5, the Whi5 concentration in each cell will then decrease in inverse proportion to cell volume as cells grow during G1. If normalized to the initial concentration at birth, each cell will then show a similar decrease over time. Thus, even though each single-cell trace will have a different length, the mean of the traces aligned at birth will accurately represent the average dynamics of Whi5 dilution (Figure 1C). If we instead align all normalized traces at Start, as Litsios et al. did, it becomes obvious that the resulting mean does not reflect the dynamics of dilution, but instead strongly depends on the distribution of pre-Start-G1 durations (Figure 1D).
FIGURE 1:
Whi5 is diluted during pre-Start G1. (A) Single-cell traces of Whi5-mCherry concentration reported by Litsios et al. are shown over time from cell birth to Start. Each trace is normalized on the initial value at cell birth. n = 49 cells. (B–D) Illustration of the effect of single-cell normalization and alignment on the observed apparent dynamics of Whi5 dilution. We assume four idealized cells with different pre-Start G1 duration, each growing exponentially with similar growth rate (B, volume doubling time is estimated from the mean behavior) and diluting a stable pool of Whi5. The mean accurately reflects the single-cell dilution if concentration traces are normalized and aligned at cell birth (C). If single-cell traces are normalized at birth and aligned at Start instead, the mean does not reflect the single-cell dynamics and strongly depends on the distribution of pre-Start-G1 duration (D). (E–G) Data by Litsios et al. on Whi5-mCherrry (Litsios et al., 2019) and Schmoller et al. on Whi5-mCitrine (Schmoller et al., 2015) show similar Whi5 dilution if plotted accurately. Because the data sets were obtained using different growth media, we first determined the volume doubling time from the mean relative volume growth over time (E). If normalized and aligned at birth, both data sets show dilution of Whi5 as cells progress through pre-Start G1 (F). If normalized at birth and aligned at Start, dilution is largely masked in the mean behavior (G).
Given these problems associated with normalizing single-cell traces at one time point, such as cell birth, and aligning them at another, such as Start, we wondered whether the fact that Litsios et al. did not observe Whi5 dilution may be simply an artifact due to their unusual alignment and normalization procedure. We therefore replotted the raw data of Whi5-mCherry concentration shown in Litsios et al., and found that Whi5 is clearly diluted during G1, with dynamics that are comparable to those we previously observed (Figure 1, E and F; cf. Figure 1f in Schmoller et al., 2015). We note that we have rescaled time with the amount of time it takes a cell to double in volume in the two different media conditions (based on the averages shown in Figure 1E). In contrast, if we align our data at Start while normalizing at birth, Whi5-dilution is masked due to this inadequate averaging procedure (Figure 1G).
In summary, the microscopy data shown in Litsios et al. show clear dilution of Whi5 and are qualitatively consistent with the data of Schmoller et al. and several other groups. Whether the smaller quantitative difference in the dilution behavior observed in the two data sets reflects a strain- or condition-dependent difference or is due to different image analysis protocols is unclear. To clarify this, we requested raw image files from the authors of Litsios et al., but did not receive them.
Mass-spectrometry data from Litsios et al. are inconclusive
As an approach orthogonal to fluorescence microscopy, Litsios et al. employed mass spectrometry to determine relative changes in Cln3 and Whi5 protein concentrations as cells that were initially in G1 progressed through the cell cycle (Figure 4e and f, and extended data, Figure 5b, in Litsios et al., 2019). They used centrifugal elutriation to enrich for pre-Start cells and monitored changes in the relative abundance of Whi5 and Cln3 peptides as these semisynchronous cultures progressed through the cell cycle. Likely because the abundance of Whi5 and Cln3 peptides are too low to detect using conventional data-dependent acquisition (DDA) proteomics, the authors used parallel reaction monitoring (PRM) to quantify the relative abundance of two Whi5 and two Cln3 peptides.
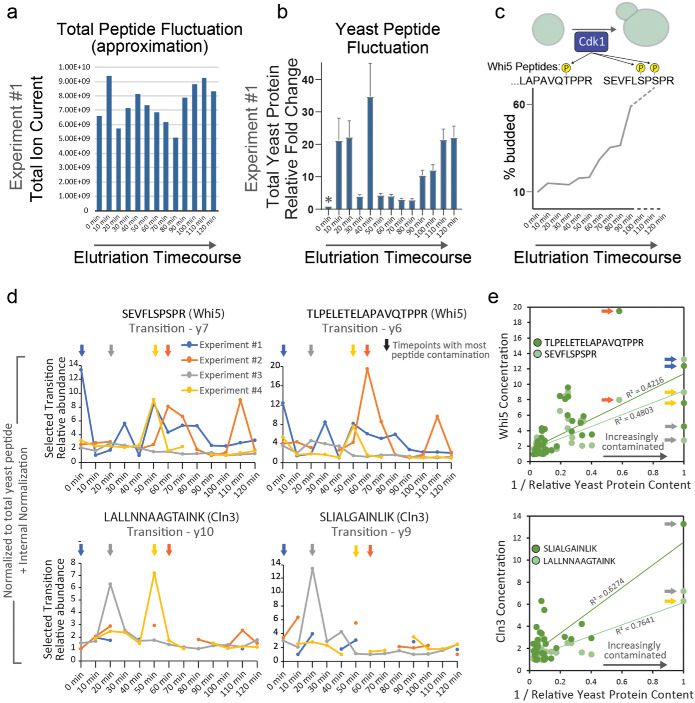
The authors performed four independent elutriation time courses and claim that their quantification of Whi5 and Cln3 peptides via targeted proteomics supports their measurements of Whi5 and Cln3 made using microscopy. However, we reviewed their proteomic analysis (see Materials and Methods, Supplemental Figure S5, and Supplemental Table S1 for a complete description of our reanalysis) and found that it is complicated by a striking inconsistency in the content of their protein samples. Despite a relatively consistent injection of bulk peptide material into their liquid chromatography–mass spectrometry (LC-MS) (Figure 2A, as inferred by the approximate total ion current [TIC] from Litsios et al.’s raw data), the peptide peak areas of several common yeast housekeeping proteins vary significantly from run to run (one example is shown in Figure 2B; similar plots for all experiments are shown in Supplemental Figure S5). This suggests that the proportion of peptides originating from yeast proteins varies greatly from sample to sample, and that, in some cases, nearly all the injected mixture is composed of foreign peptides due to contamination. Though not acknowledged in their article, the authors were aware of this contamination (personal correspondence). They suspected that the yeast pellets obtained from the elutriation time course were insufficiently washed and that the source of the contaminating peptides was remnant peptone from the YPD yeast media. Because proteins in peptone are animal in origin, the authors attempted to correct for the suppressive presence of animal peptides by applying a normalization to a set of yeast housekeeping proteins (like those depicted in Figure 2B).
FIGURE 2:
Mass-spectrometry data is inconclusive. (A) Total peptide load estimated for each time point in the elutriation time course based on the measurement of total ion current (TIC). Experiment no. 1 is shown as an example (see Supplemental Figure S5 for all replicate experiments). (B) Total yeast peptide amount based on the measurement of six yeast housekeeping peptides. Data from Experiment #1 is shown as an example. MS1 peak areas of peptides from Enolase, GAPDH, and ADH1 were used to estimate changes to the total amount of yeast protein. Yeast peptide fold change was calculated relative to the time point with the least amount of yeast peptide signal (denoted by asterisk). These fold changes were used to normalize the PRM measurements of Whi5 and Cln3 peptides. Error bars represent SEM. (C) Top, schematic of Whi5 peptides indicating known phosphorylation sites of Cdk1. Bottom, bud index from Litsios et al. indicating the degree of cell cycle synchrony and progression through the cell cycle. Whi5 is highly phosphorylated at the G1/S transition (see Supplemental Figure S6). (D) PRM measurements (normalized to the amount of total yeast protein) for Cln3 and Whi5 peptides throughout the elutriation time course. All four experiments are plotted. Litsios et al. used the single, most intense transition to quantify the elution curves for Whi5 and Cln3 peptides. We used the same ions in our reanalysis. Missing data points represent PRM measurements that did not pass a quality metric designated by the authors (Qvalue > 0.01). Arrows denote the time points in each of the four replicate time courses that contained the largest amount of foreign peptide contamination (i.e., the least amount of yeast peptide). Supplemental Table S1 contains a step-by-step derivation of the plotted data points. (E) The concentration calculated for each Whi5 and Cln3 peptide throughout all experimental time courses (i.e., the data points from D) plotted against the extent of peptide contamination in each of those time points. The extent of foreign peptide contamination is inferred from the relative amount of yeast peptides in each sample (plotted in B and Supplemental Figure S5), which was also used in the determination of relative concentration. Arrows correspond to the time points in D. The most contaminated sample in Experiment #2 was manually excluded by Litsios et al., thus the orange arrow points to the second most contaminated time point. The arrows that are absent in the Cln3 plot correspond to time points where the PRM measurement of Cln3 did not pass the required quality metric and were excluded.
We think the following points demonstrate this proteomic data set is not suited to measure the concentration changes of Cln3 and Whi5 through the cell cycle. 1) The extreme extent of the MS1-level normalization means that the Whi5 and Cln3 PRM signals are regularly adjusted by more than 20-fold, even between adjacent time points (Figure 2B and Supplemental Figure S5). 2) Both Whi5 peptides monitored by the PRM analysis contain phosphosites targeted by the cyclin-dependent kinase Cdk1 at Start, which lead to Whi5’s nuclear export (Costanzo et al., 2004; De Bruin et al., 2004; Wagner et al., 2009; Doncic et al., 2011). The stoichiometry of Whi5 phosphorylation after Start is high and can be inferred from its phospho-dependent shift in a phostag gel (Supplemental Figure S6). Thus, the concentration of nonphosphorylated forms of these peptides, which is what the PRM analysis measures, should decrease significantly when cells proceed through Start and are in S phase (see peptide schematic in Figure 2C showing Cdk sites and bud index throughout elutriation time course). That Litsios et al. do not observe a decrease in the concentration of unphosphorylated Whi5 peptides through the cell cycle suggests that their measurements do not reflect in vivo changes in Whi5 concentration. 3) Though the authors reported that four biological replicates were used to calculate the changes in Cln3 concentration, many of the data points surrounding Start (the period in which they see a “pulse”) were excluded based on a PRM quality control metric (Figure 2D and Supplemental Table S1). 4) When the experiments are plotted separately, the noisy nature of the data is more evident, and it is clear that the Cln3 “pulse” reported at t = 20 min is primarily driven by experiment no. 3 and is less or not apparent in the other experiments. We also note that the Whi5 time courses contain many nonreproducible single data point “pulses” in concentration. The authors of Litsios et al. confirmed the correspondence of our reanalysis with the data they used to generate the plots in Figure 4e of their article. 5) When considering all their experiments together, nearly every peak in concentration corresponds to the time points within each time course that contain the greatest extent of peptide contamination (see arrows in Figure 2D), which are therefore subjected to the most adjustment during normalization. In fact, when all the time points from all time courses are plotted together, the derived Whi5 concentrations correlate with the extent of peptide contamination (Figure 2E). Cln3 exhibits a similar, though less visually dramatic correlation (Figure 2E), which could be explained by the fact that many of the most contaminated Cln3 time points are excluded by the Qvalue (some of these time points have no detectable Cln3 PRM signal). This observation indicates that the “normalization” used to derive Cln3 and Whi5 concentrations by Litsios et al. is significantly biased by the contamination in each of their peptide samples.
Given this analysis, we do not think that the presented mass-spectrometry data support the claim that Cln3 concentration sharply increases in G1 before Start nor the claim that Whi5 is not diluted. Importantly, because the microscopy data presented by Litsios et al. show a clear dilution of Whi5 when aligned correctly (Figure 1), the mass-spectrometry data is not consistent with the microscopy data presented in the same study. Moreover, similar elutriation experiments were performed independently by two other research groups, both of which found that Whi5 was diluted during G1 (Lucena et al., 2018; Chen et al., 2020).
There is no dramatic increase in protein synthesis rates leading up to Start
A central part of the model suggested by Litsios et al. is that protein synthesis rates increase two- to threefold in the leadup to Start. This is important because an increase in the protein synthesis rate would drive concentration changes for highly unstable proteins, such as the cell cycle activator Cln3. This is because the change in the amount of an unstable protein, p, synthesized at a constant rate, s, and degraded in a first-order reaction at a rate d, is given by dp/dt = s − dp. Because we are interested in rapidly degraded proteins, the concentrations will reach steady state so that p = s/d, that is, the amount of the unstable protein is directly proportional to the protein synthesis rate. Then, the concentration of the unstable protein in a cell of volume v is [p] = s/(dv) (we note that the nuclear volume, where these proteins reside, is approximately proportional to the cell volume; Jorgensen et al., 2007; Walters et al., 2014).
To support the claim that Cln3 amounts increase severalfold leading up to Start, Litsios et al. present fluorescence microscopy data of cells expressing CLN3-2A-sfGFP. sfGFP is separated from Cln3 upon translation through the viral self-cleaving 2A sequence, which enables imaging despite the fact that Cln3 is very unstable. The total amount of sfGFP in each cell at each time point is then calculated by multiplying the mean pixel intensity by the estimated area. Then, the Cln3 synthesis rate is calculated by taking the first derivative of a Gaussian process smoothing fit of the total GFP curve. Because the Cln3 synthesis rate is proportional to Cln3 abundance, they estimate an approximately twofold increase in Cln3 abundance (not concentration) around Start (Figure 5c in Litsios et al., 2019).
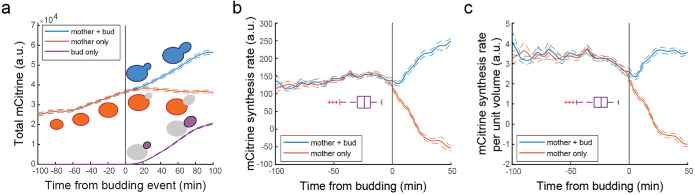
A strong prediction of the model proposed by Litsios et al. is that the increase in Cln3 synthesis reflects a global increase in protein synthesis rates and should therefore be visible in the production rates of most proteins. Supporting this view, Litsios et al. also examined sfGFP expressed from a constitutive TEF1 promoter and found an increase in protein synthesis similar to that of their Cln3 reporter. To independently test the claim that there is a two- to threefold increase in global protein synthesis rate before Start, we decided to reanalyze some of our own data on cells expressing mCitrine fluorescent protein from an ACT1 promoter (data from Chandler-Brown et al., 2017, Figures 1–4). We observe that the total cellular fluorescence in the mother body increases smoothly up until bud emergence. After budding, cell growth mostly serves to increase bud volume rather than the mother cell body (Figure 3A and Supplemental Figure S7). We can then use these fluorescence traces to estimate the protein synthesis rate (Figure 3B) and then divide by the cell volume to estimate the concentration changes of unstable proteins including Cln3 (Figure 3C). Note that to do this, it is important to analyze protein synthesis on a single-cell level. This is because the dynamics of the mean fluorescence intensity shown in Figure 3A will depend on the distribution of G1 durations of the individual cells. Because larger cells with more fluorescence tend to have shorter G1 durations, more of them will be included at the time points closer to Start leading to an increase in the mean total fluorescence.
FIGURE 3:
Protein synthesis rates do not dramatically increase before Start. (A) Mean cellular mCitrine dynamics expressed from an integrated ACT1pr-mCitrine allele. Data are shown for the whole yeast, mother cell body only, and for the bud only. The fluorescence microscopy data of 163 ACT1pr-mCitrine cells was from Chandler-Brown et al. (2017). Each trace was aligned to the time of budding (black vertical line). The solid lines represent the mean value and the dashed lines represent the standard error at each time point. (B) Mean cellular mCitrine synthesis rate. Single-cell traces associated with the data shown in A were smoothed using a Gaussian process regression function using a similar approach to that used by Litsios et al. Then, the rate was calculated by subtracting the adjacent time point and dividing by the 3 min time between movie frames. Each trace was aligned to the time of the budding (black vertical line). The solid lines represent the mean value and the dashed lines represent the standard error at each time point. The boxplot shows the distribution of time points at which each cell passes Start (Whi5 nuclear exit). (C) The concentration of unstable proteins such as Cln3 was estimated as the rate of mCitrine synthesis per unit volume in ACT1pr-mCitrine cells (see text). Data for individual mCitrine synthesis rates from B were divided by the volume at each time point. Each trace was aligned to the time of budding (black vertical line). The solid lines represent the mean value and the dashed lines represent the standard error at each time point. The boxplot shows the distribution of time points at which each cell passes Start (Whi5 nuclear exit).
In contrast to Litsios et al., we estimate a much more constant concentration of unstable proteins, like Cln3, before Start. Following Start and around bud emergence, we see a slight decrease in the protein synthesis rate, which is followed by an increase to a similar protein synthesis rate per unit mass as before Start. We see no evidence of a two- to threefold increase in the global protein synthesis rate. These results are consistent with our previous results examining a stabilized Cln3 protein expressed from the endogenous CLN3 locus (Schmoller et al., 2015). Because the increase in protein synthesis rate proposed by Litsios et al. is rather dramatic, it should be observable using bulk populations of synchronized cells. However, bulk radiolabeling experiments looking at total protein synthesis rates in cells synchronized by elutriation did not find any changes in global protein synthesis rate during the cell cycle (Elliott and McLaughlin, 1978). Thus, while there may be some specific proteins whose abundance increases through G1, this is unlikely to reflect changes in the global protein synthesis rate before Start.
Live-cell microscopy presented by Dorsey et al. results in extreme photobleaching and a severe reduction in cell growth
Dorsey et al. (2018) used two different fluorescence microscopy approaches to measure the cell-size dependence of Whi5. First, they use a complex confocal-based sN&B imaging approach to measure local protein concentrations at single time points. Second, they use a live-cell imaging approach to measure the dynamics of Whi5 concentration in single cells progressing through the cell cycle.
These live-cell imaging experiments presented by Dorsey et al. are directly comparable to our experiments demonstrating Whi5 dilution. However, in contrast to our measurements (Schmoller et al., 2015), measurements made by others (Qu et al., 2019; Barber et al., 2020), and our reanalysis of data from Litsios et al., Dorsey et al. did not observe dilution of Whi5 during G1 (cf. Figure 3a–c in Dorsey et al., 2018). It is, however, noteworthy that Dorsey et al. draw this conclusion based on the analysis of only four cells, which is far below the typical number of cells analyzed in most single-cell studies. Moreover, the imaging protocol of Dorsey et al. results in dramatic photobleaching, leading to an ∼3.7-fold decrease of the fluorescence intensity after 2 h despite an exceptionally low frame rate of 20 min (cf. Figure 3a of Dorsey et al.). Instead of reducing the light intensity or exposure time, Dorsey et al. proceed with their analysis by correcting for this strong photobleaching using a GFP-only control analyzed in three cells. In our experience, such strong photobleaching is correlated with a level of phototoxicity that either kills cells, or results in strongly reduced cell growth. Indeed, Dorsey et al. note in their Methods section that “Cells grew more slowly in the microfluidics device than in free cultures.” Accordingly, for the three cells in their Figure 3a, Dorsey et al. report cell growth of only ∼20% in 2 h, which is far less than the expected mass doubling of budding yeast on glucose media. In summary, Dorsey et al. support their claim that Whi5 is not diluted during G1 on the analysis of four cells, which are exposed to extreme photobleaching and are therefore hardly growing or dying. Thus, the fact that they do not observe Whi5 dilution by growth can likely be attributed to the large experimental error and the fact that the cells are barely growing due to phototoxicity.
In addition to these live-cell imaging experiments, Dorsey et al. use an sN&B imaging approach to measure Whi5 concentrations in cells at single time points. In contrast to our results, they do not observe a decrease of Whi5 concentration with cell size. Because we do not have any expertise with the sN&B imaging approach, we are not in a position to identify the origin of this discrepancy. However, we note that the results presented by Dorsey et al. show some simple inconsistencies. For example, in Figure 3g–j of their article, they find, using conventional fluorescence microscopy, that Whi5 concentrations are ∼2.5–4-fold higher in glycerol/ethanol compared with glucose media. In contrast, based on their sN&B analysis, Whi5 concentrations are independent of the carbon source (Dorsey et al., 2018). Neither result is consistent with Qu et al., which showed that Whi5 concentration is higher in poor compared with rich media (Qu et al., 2019). Finally, we noticed that many of the cells analyzed by Dorsey et al. appear to be surprisingly small (including cells grown on glucose media that are smaller than 5 fL as shown in their Figures 2b and 3f), which indicates large experimental errors from their cell segmentation.
While we have difficulties assessing the sN&B experiments by Dorsey et al., their live-cell experiments that are directly comparable to our own suffer serious experimental problems that render them uninterpretable. At this point, we are aware of at least nine independent research groups (including the authors) that have corroborated Whi5 dilution. These studies used both immunoblots and fluorescence microscopy, and were done using different strain backgrounds, microscopes, and analysis methods. This leaves the conclusions drawn by Dorsey et al. based on their sN&B experiments quite isolated.
DISCUSSION
We previously identified the dilution of Whi5 as one molecular mechanism through which cell growth drives progression through Start in budding yeast (Schmoller et al., 2015). Since our initial study, two publications questioned whether Whi5 was diluted during G1 by cell growth (Dorsey et al., 2018; Litsios et al., 2019). Here, we reanalyze this work, and present new data from independent laboratories verifying Whi5 dilution. Combined with the published results of other independent research groups, this leads us to arrive at a consensus that Whi5 is indeed diluted during G1. Importantly, our reanalysis here of Litsios et al.’s fluorescence microscopy data shows that Whi5 is diluted during pre-Start G1 in their data as well. Thus, the mass-spectrometry data presented by Litsios et al. is not only inconsistent with our findings, but also with their own microscopy data. Our analysis suggests that the mass-spectrometry data are not reliable, possibly due to contamination. This is supported by the fact that they do not observe a decrease in the concentration of the two monitored Whi5 peptides, which both contain Cdk sites that are phosphorylated at Start and therefore should hardly be detected after Start in their unphosphorylated form.
On average, Litsios et al.’s data show that Whi5 concentration decreases by 20% before Start, which is consistent with the average amount of cell growth during G1 in these conditions. As discussed in a recent paper by Qu et al., the relative amount of Whi5 dilution increases as cell growth rates decrease and G1 durations increase in poorer nutrient conditions (Qu et al., 2019). That cells grown on rich media exhibit shorter pre-Start G1 phases and therefore dilute Whi5 less is fully consistent with weaker G1/S cell-size control that has been observed in these conditions (Di Talia et al., 2007). Nevertheless, even on glucose media, deletion of Whi5 results in significantly weaker size control at Start (Garmendia-Torres et al., 2018). In any case, we note the model proposed by Litsios et al. does not provide any mechanism for G1/S size control as no experiments were performed that isolated cell size rather than time in G1 as a variable.
In addition to claiming Whi5 is not diluted in G1, Litsios et al. present a revised model of Start that requires a two- to threefold increase in protein synthesis rates before Start that causes a pulse of Cln3 synthesis. We find this claim to be highly unlikely based on both the analysis of fluorescent reporters we present here, but also because of the fact that such a large cell cycle–dependent change in the protein synthesis rate should have been detected in the classic bulk radiolabeling studies, but was not (Elliott and McLaughlin, 1978).
Finally, we emphasize our claim that Whi5 dilution is one mechanism through which cell growth in G1 promotes cell cycle progression. We never claimed Whi5 dilution is the only such mechanism. In fact, we anticipate the discovery of additional mechanisms based on the similar principle of size-dependent concentration changes as a recent study showed that cell growth triggers relative concentration changes of a broader group of cell cycle regulators (Chen et al., 2020). Indeed, Cln3 may well contribute in some way to cell-size control. The live-cell imaging data presented by Litsios et al. is consistent with a moderate increase of Cln3 concentration during G1 (Supplemental Figure S8), which is similar in magnitude to the increase of ∼25% in the first 12 min of G1 that we observed using a stabilized Cln3 allele (Figure 1b in Schmoller et al., 2015). A stronger increase of Cln3 concentration during G1 of small cells released from elutriation has been observed before using Western blots (see also Supplemental Figure S4D; Thorburn et al., 2013; Zapata et al., 2014; Lucena et al., 2018; Sommer et al., 2021). However, in these experiments, elutriation release was often combined with a media switch from poor to rich nutrients. Because the concentration of the highly unstable Cln3 is tightly linked to the biosynthetic capacity of the cell, it is hard to assess to what extent this increase in Cln3 concentration is a consequence of cell cycle progression or cell-size increase rather than simply due to the media switch or recovery from the stress induced by centrifugal elutriation. Indeed, a recent study found that the increase of Cln3 concentration after centrifugal elutriation of cells grown on a poor carbon source is far less dramatic if cells are released into poor rather than rich media (Sommer et al., 2021). Future studies will be needed to clarify this question and, in any case, we do not dispute the importance of Cln3, whose concentration is clearly important for G1 duration (Cross, 1988; Nash et al., 1988; Tyers et al., 1993; Liu et al., 2015; Blank et al., 2018).
The observation of Whi5 dilution in budding yeast inspired a similar study in human cells, where it was found that cell growth in G1 dilutes the retinoblastoma protein Rb, a functional orthologue of Whi5, cell cycle inhibitor, and tumor suppressor (Zatulovskiy et al., 2020). While both yeast and human cells use inhibitor dilution mechanisms to couple cell growth to cell division, Whi5 and Rb share no sequence similarity and have different evolutionary histories (Medina et al., 2016). That both serve as diluted cell-size sensors suggests inhibitor dilution mechanisms are frequently employed to control cell size across eukaryotes. However, getting to the molecular basis of these biosynthetic mechanisms controlling cell size first requires building a consensus on how different protein concentrations are impacted by cell growth as we here provide.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Schneider lab live-cell microscopy
Nikon Eclipse setup with custom microfluidics.
Live-cell time-lapse microscopy was performed in a custom-made microfluidic device made of polydimethylsiloxane (PDMS) and a glass coverslip that allows trapping of cells in a dedicated region of interest, limiting colony growth in the XY plane. Cells were grown on synthetic complete media with 2% glycerol, 1% ethanol as a carbon source (SCGE) overnight to exponential phase before the experiment. During the experiment, SCGE was supplied at a constant flow of 20 µl/min, enabling imaging of colony growth over several generations.
A Nikon Eclipse Ti-E with SPECTRA X light engine illumination and an Andor iXon Ultra 888 camera were used for epifluorescence microscopy. A plan-apo λ 60×/1.4 numerical aperture (NA) Ph3 oil immersion objective was used to take phase contrast and fluorescence images with a 3-min frame rate. For automated focusing, the built-in Nikon perfect focus system was used during the experiment. mCitrine fluorescence was imaged by exposure for 400 ms, illuminating with the SPECTRA X light engine at 504 nm and ∼16 mW (25%) power.
Temperature control was achieved by setting both a custom-made heatable insertion and an objective heater to 30°C.
Identical settings were used for each of the experiments.
Image analysis
Cell segmentation.
Cells were automatically segmented based on phase contrast images using the Matlab-based Phylocell software developed in the Gilles Charvin lab (Goulev et al., 2017). Segmentation results were visually inspected and manually corrected if necessary.
Calculation of cell volume.
To calculate cell volume based on 2D phase contrast images, we first aligned the segmented cell area along its major axis. Next, we divided the cell into slices perpendicular to the major axis, each 1 pixel in width. To approximate cell volume, we then assumed rotational symmetry of each slice around its middle axis parallel to the cell’s major axis. We then calculated for each slice the volume of the resulting cylinder with 1 pixel in height and diameter the width of the respective slice. Finally, we summed the volumes of each cylinder to obtain total cell volume.
Protein amount calculation.
To calculate changes in Whi5 amount over time, we used a strain where Whi5 was endogenously tagged with mCitrine. In this case, the total fluorescence signal produced by mCitrine should directly correlate with Whi5 amount. To measure the mCitrine signal, we first estimated the background fluorescence using two areas of 200 × 200 pixels containing no cells and subtracted its median value from all pixels. Then, we calculated the total fluorescence for each cell separately as a sum of intensities of all pixels within a cell counter.
Cell cycle analysis.
We analyzed 100 cells for the Whi5-mCitrine strain and 59 cells for the wild-type control (KSY108-1 and MMY116-2C, respectively; Schmoller et al., 2015) during their first cell cycle. Next, we estimated the median G1 beginning (birth time) as 69 min and the median time of cytokinesis as 96 min from the bud emergence. For cell cycle analyses shown in Supplemental Figure S2B, we divided each total fluorescence value of each cell by the corresponding cell volume to account for differences in the cell volume over time. For Supplemental Figure S2B, we corrected for the wild-type autofluorescence by subtracting its mean value for each time point from values of Whi5-mCitrine strain for the same time points. Then, we additionally normalized each fluorescence value of each cell by the fluorescence value at birth. Finally, we pooled all data for Whi5-mCitrine, and plotted the mean fluorescence from the median G1 beginning (birth time) to bud emergence. For Supplemental Figure S2A, we pooled all data for the Whi5-mCitrine strain without any correction or normalization and plotted the mean fluorescence from the median G1 beginning (birth time) to the median time of cytokinesis. For both subpanels of Supplemental Figure S2, 95% confidence intervals were determined from 50,000 bootstrap samples.
Reanalysis of mass-spectrometry data
Our reanalysis of the mass-spectrometry experiments performed by Litsios et al. utilized the raw MS acquisition files and SpectroDive output files that were uploaded to the PRIDE repository (PXD015327). We calculated the approximate TIC for each experimental time point from the corresponding raw data file (see “TIC” tab of SupplementalTable S1). To quantify changes in the amount of yeast peptide from time point to time point, we used a corresponding DDA spectral library and the Skyline analysis software to extract the chromatograms for several hundred yeast peptides in each experimental time point (DDA acquisition files were provided upon request from the authors of Litsios et al.). Similar to what was performed by Litsios et al., we selected a set of six peptides from three different yeast housekeeping proteins (Enolase, ADH1, and GAPDH) and used the average fluctuation in their peptide peak areas to approximate changes in the total amount of yeast protein within each sample (see “normalization” tab of Supplemental Table S1). These peptides behaved similarly to many other yeast housekeeping genes and their peaks consistently traced to a similar retention time (see “normalization” tab of Supplemental Table S1).
The supplemental file related to the PRM analysis that was directly uploaded with the Litsios et al. article (Supplemental Table 4 from Litsios et al.) contains the top seven Q3 transitions for each Cln3 and Whi5 peptide but no quantitative information. A set of text files uploaded to PRIDE (“PRIDE_experiment1_spectrodive” and “PRIDE_experiment2_spectrodive”) contained the Cln3 and Whi5 PRM quantitation acquired from a set of four to six Q3 transitions. To recreate the analysis from Litsios et al., we normalized their published PRM measurements of the Cln3 and Whi5 peptides to the MS1 peptide peak areas of three yeast housekeeping proteins we extracted using Skyline. Litsios et al. used a single Q3 transition (the most abundant one) to quantify Whi5 and Cln3 abundance. A step-by-step demonstration of our calculations is laid out in Supplemental Table S1. The authors of Litsios et al. confirmed the correspondence of our reanalysis with the data they used to generate the plots in their article.
Methods Aldea Lab
Time-lapse microscopy and measurements of volume growth rate in G1 were as described by Ferrezuelo et al. (2012). Photobleaching during acquisition was negligible (<0.1% per time point) and background autofluorescence was always subtracted. The cellular concentration of fluorescent fusion proteins was obtained by dividing the integrated fluorescence signal within the projected area of the cell by its volume.
Methods Lucena et al.
Cell size was measured using a Coulter counter (Channelizer Z2; Beckman Coulter) as previously described in Lucena et al. The percentage of budded cells was measured by counting the number of small unbudded cells over a total of more than 200 cells using a Zeiss Axioskop 2 (Carl Zeiss) and an AxioCam HRm camera with a 63×/1.4 NA objective. Densitometric quantification of Whi5 and Cln3 Western blot signals was performed using ImageJ (Schneider et al., 2012) normalizing over the loading control band.
Supplementary Material
Acknowledgments
This work was generously supported by an NIH MIRA award R35 GM-134858, by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) through project 431480687, and by the Human Frontier Science Program (Career Development Award to K.M.S.). We thank Matthias Heinemann, Andreas Milias-Argeitis, and Alexander Schmidt for discussing their work with us.
Abbreviations used:
- DDA
data-dependent acquisition
- LC-MS
liquid chromatography–mass spectrometry
- PRM
parallel reaction monitoring
- TIC
total ion current.
Footnotes
REFERENCES
- Barber F, Amir A, Murray AW (2020). Cell size regulation in budding yeast does not depend on linear accumulation of Whi5. PNAS 117, 14243–14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank HM, Callahan M, Pistikopoulos IPE, Polymenis AO, Polymenis M (2018). Scaling of G1 duration with population doubling time by a cyclin in Saccharomyces cerevisiae. Genetics 210, 895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler-Brown D, Schmoller KM, Winetraub Y, Skotheim JM (2017). The adder phenomenon emerges from independent control of pre- and post-Start phases of the budding yeast cell cycle. Curr Biol 27, 2774–2783.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhao G, Zahumensky J, Honey S, Futcher B (2020). Differential scaling of gene expression with cell size may explain size control in budding yeast. Mol Cell 78, 359–370.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claude K-L, Bureik D, Adarska P, Singh A, Schmoller KM (2021). Transcription coordinates histone amounts and genome content. Nat Comm 12, 4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M (2004). CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117, 899–913. [DOI] [PubMed] [Google Scholar]
- Cross FR (1988). DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol 8, 4675–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bruin RAM, McDonald WH, Kalashnikova TI, Yates J, Wittenberg C (2004). Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117, 887–898. [DOI] [PubMed] [Google Scholar]
- Doncic A, Falleur-Fettig M, Skotheim JM (2011). Distinct interactions select and maintain a specific cell fate. Mol Cell 43, 528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR (2007). The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature 448, 947–951. [DOI] [PubMed] [Google Scholar]
- Dorsey S, Tollis S, Cheng J, Black L, Notley S, Tyers M, Royer CA (2018). G1/S transcription factor copy number is a growth-dependent determinant of cell cycle commitment in yeast. Cell Syst 6, 539–554.e11. [DOI] [PubMed] [Google Scholar]
- Elliott SG, McLaughlin CS (1978). Rate of macromolecular synthesis through the cell cycle of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA 75, 4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrezuelo F, Colomina N, Palmisano A, Garí E, Gallego C, Csikász-Nagy A, Aldea M (2012). The critical size is set at a single-cell level by growth rate to attain homeostasis and adaptation. Nat Commun 3, 1012. [DOI] [PubMed] [Google Scholar]
- Garmendia-Torres C, Tassy O, Matifas A, Molina N, Charvin G (2018). Multiple inputs ensure yeast cell size homeostasis during cell cycle progression. Elife 7, e34025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomar-Alba M, Méndez E, Quilis I, Bañó MC, Igual JC (2017). Whi7 is an unstable cell-cycle repressor of the Start transcriptional program. Nat Commun 8, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulev Y, Morlot S, Matifas A, Huang B, Molin M, Toledano MB, Charvin G (2017). Nonlinear feedback drives homeostatic plasticity in H2O2 stress response. Elife 6, e23971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granovskaia MV, Jensen LJ, Ritchie ME, Toedling J, Ning Y, Bork P, Huber W, Steinmetz LM (2010). High-resolution transcription atlas of the mitotic cell cycle in budding yeast. Genome Biol 11, R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston GC, Pringle JR, Hartwell LH (1977). Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res 105, 79–98. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B (2007). The size of the nucleus increases as yeast cells grow. Mol Biol Cell 18, 3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsios A, Huberts DHEW, Terpstra HM, Guerra P, Schmidt A, Buczak K, Papagiannakis A, Rovetta M, Hekelaar J, Hubmann G, et al. (2019). Differential scaling between G1 protein production and cell size dynamics promotes commitment to the cell division cycle in budding yeast. Nat Cell Biol 21, 1382–1392. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang X, Yang X, Liu S, Jiang L, Qu Y, Hu L, Ouyang Q, Tang C (2015). Reliable cell cycle commitment in budding yeast is ensured by signal integration. Elife 4, e03977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena R, Alcaide-Gavilán M, Schubert K, He M, Domnauer MG, Marquer C, Klose C, Surma MA, Kellogg DR (2018). Cell size and growth rate are modulated by TORC2-dependent signals. Curr Biol 28, 196–210.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina EM, Turner JJ, Gordân R, Skotheim JM, Buchler NE (2016). Punctuated evolution and transitional hybrid network in an ancestral cell cycle of fungi. Elife 5, e09492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash R, Tokiwa G, Anand S, Erickson K, Futcher AB (1988). The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J 7, 4335–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramila T, Wu W, Miles S, Noble WS, Breeden LL (2006). The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev 20, 2266–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Jiang J, Liu X, Wei P, Yang X, Tang C (2019). Cell cycle inhibitor Whi5 records environmental information to coordinate growth and division in yeast. Cell Rep 29, 987–994.e5. [DOI] [PubMed] [Google Scholar]
- Santos A, Wernersson R, Jensen LJ (2015). Cyclebase 3.0: A multi-organism database on cell-cycle regulation and phenotypes. Nucleic Acids Res 43, D1140–D1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoller KM, Turner JJ, Kõivomägi M, Skotheim JM (2015). Dilution of the cell cycle inhibitor Whi5 controls budding yeast cell size. Nature 526, 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer RA, DeWitt JT, Tan R, Kellogg DR (2021). Growth-dependent signals drive an increase in early G1 cyclin concentration to link cell cycle entry with cell growth. eLife 10, e64364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B (1998). Comprehensive identification of cell cycle–regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaffer MP, Chandler-Brown D, Langhinrichs M, Marinov G, Kundaje A, Schmoller KM, Skotheim JM (2021). Size-independent mRNA synthesis and chromatin-based partitioning mechanisms generate and maintain constant amounts of protein per cell. Mol Cell, 81, 4861–4875.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn RR, Gonzalez C, Brar GA, Christen S, Carlile TM, Ingolia NT, Sauer U, Weissman JS, Amon A (2013). Aneuploid yeast strains exhibit defects in cell growth and passage through START. Mol Biol Cell 24, 1274–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Futcher B (1993). Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J 12, 1955–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner MV, Smolka MB, de Bruin RAM, Zhou H, Wittenberg C, Dowdy SF (2009). Whi5 regulation by site specific CDK-phosphorylation in Saccharomyces cerevisiae. PLoS One 4, e4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters AD, May CK, Dauster ES, Cinquin BP, Smith EA, Robellet X, D’Amours D, Larabell CA, Cohen-Fix O (2014). The yeast polo kinase Cdc5 regulates the shape of the mitotic nucleus. Curr Biol 24, 2861–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata J, Dephoure N, MacDonough T, Yu Y, Parnell EJ, Mooring M, Gygi SP, Stillman DJ, Kellogg DR (2014). PP2ARts1 is a master regulator of pathways that control cell size. J Cell Biol 204, 359–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatulovskiy E, Zhang S, Berenson DF, Topacio BR, Skotheim JM (2020). Cell growth dilutes the cell cycle inhibitor Rb to trigger cell division. Science 369, 466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.