Abstract
The mammalian target of rapamycin (mTOR) is a large protein kinase that assembles into two multisubunit protein complexes, mTORC1 and mTORC2, to regulate cell growth in eukaryotic cells. While significant progress has been made in our understanding of the composition and structure of these complexes, important questions remain regarding the role of specific sequences within mTOR important for complex formation and activity. To address these issues, we have used a molecular genetic approach to explore TOR complex assembly in budding yeast, where two closely related TOR paralogues, TOR1 and TOR2, partition preferentially into TORC1 versus TORC2, respectively. We previously identified an ∼500-amino-acid segment within the N-terminal half of each protein, termed the major assembly specificity (MAS) domain, which can govern specificity in formation of each complex. In this study, we have extended the use of chimeric TOR1-TOR2 genes as a “sensitized” genetic system to identify specific subdomains rendered essential for TORC2 function, using synthetic lethal interaction analyses. Our findings reveal important design principles underlying the dimeric assembly of TORC2 as well as identifying specific segments within the MAS domain critical for TORC2 function, to a level approaching single-amino-acid resolution. Together these findings highlight the complex and cooperative nature of TOR complex assembly and function.
INTRODUCTION
The mammalian target of rapamycin (mTOR) network is a conserved regulator of eukaryotic cell growth and cellular homeostasis, contributes to aging, and is dysregulated in many human diseases, including cancer (Wullschleger et al., 2006; Saxton and Sabatini, 2017; Mossmann et al., 2018; Magaway et al., 2019). The central component in this network is the large (∼280 kDa) mTOR protein, a Ser/Thr protein kinase and member of the conserved PIKK-related family of protein kinases (Bosotti et al., 2000; Wullschleger et al., 2006). In addition to a conserved C-terminal kinase domain and adjacent FRB domain that interacts with rapamycin, mTOR is composed almost entirely of helical repeats, including N-terminal and middle HEAT (N-HEAT and M-HEAT, respectively) and FAT/TPR repeat domains (Andrade and Bork, 1995; Andrade et al., 2001; Knutson, 2010; Tafur et al., 2020). mTOR functions as part of two distinct protein complexes, mTORC1 and mTORC2, where mTORC1 is uniquely inhibited by the macrolide antibiotic rapamycin (Kim et al., 2002; Loewith et al., 2002; Jacinto et al., 2004; Sarbassov et al., 2004; Wullschleger et al., 2006; Eltschinger and Loewith, 2016; Tafur et al., 2020). In addition to mTOR, mTORC1 includes as core components Raptor and mLST8/GβL, while mTORC2 includes Rictor, SIN1, and mLST8/GβL (Hara et al., 2002; Kim et al., 2002, 2003; Sarbassov et al., 2004; Eltschinger and Loewith, 2016). Association with complex-specific partners and regulators is proposed to control their intracellular localization as well as governing substrate selection and regulating mTOR kinase activity (Eltschinger and Loewith, 2016; Yang et al., 2017).
Our understanding of the structure and function of mTORC1 and mTORC2 has advanced considerably in recent years, aided by parallel studies in budding yeast, Saccharomyces cerevisiae, where two TOR paralogues, TOR1 and TOR2, assemble with a conserved core set of orthologous proteins to form TORC1 and TORC2 (Loewith et al., 2002; Wedaman et al., 2003; Reinke et al., 2004; Tafur et al., 2020). Specifically, TORC1 contains either TOR1 or TOR2, as well as KOG1 (orthologue of Raptor), whereas TORC2 contains TOR2, AVO1 (orthologue of SIN1), and AVO3 (orthologue of Rictor). Both complexes contain LST8 (orthologue of mLST8/GβL) as a common subunit, as well as a number of nonconserved, complex-specific proteins, including AVO2 in TORC2 (Loewith et al., 2002; Wedaman et al., 2003; Reinke et al., 2004; Tafur et al., 2020). Models derived from biochemical and ultrastructural studies of both yeast and mammalian complexes indicate that they likely function as dimers and possess an overall rhomboid-like, symmetrical shape, determined primarily by the superhelical topology of the large N-terminal domain of TOR/mTOR (Wullschleger et al., 2005; Yang et al., 2013, 2016, 2017; Gaubitz et al., 2015; Aylett et al., 2016; BaretiL et al., 2016; Karuppasamy et al., 2017; Chen et al., 2018; Stuttfeld et al., 2018; Scaiola et al., 2020). High-resolution (∼3.0 Å) Cryogenic electron microscopy (Cryo-EM) models have emerged recently for both mTORC1 and mTORC2 (Yang et al., 2017; Scaiola et al., 2020), revealing key architectural details as well as insights into their regulation, for example evidence for conformation changes within the mTORC1 active site following interaction with the upstream activator Rheb (Yang et al., 2017). Despite these advances, many important structural features within these models remain unresolved, particularly for mTORC2 (Scaiola et al., 2020). Moreover, the role of specific amino acids within TOR/mTOR for complex assembly and function remains almost completely unknown. Finally, while structural models provide important information about macromolecular organization, to be useful they must be paired with other methods to examine structure–function relationships involved in complex assembly and activity, as well as test predictions regarding the significance of observed protein–protein contacts.
To address these gaps and to complement ongoing structural studies, we have employed a molecular genetic approach to interrogate the importance of specific elements within TOR1 and TOR2, taking advantage of their unique behavior in yeast, where TOR1 assembles exclusively into TORC1 and TOR2 assembles preferentially into TORC2 (Loewith et al., 2002; Wedaman et al., 2003; Reinke et al., 2004). By constructing a set of TOR1-TOR2 chimeras, we previously identified an ∼500-amino-acid domain corresponding to the N-HEAT domain, termed the major assembly specificity (MAS) domain, that can direct assembly of both TOR proteins into TORC1 versus TORC2 (Hill et al., 2018) (Figure 1A). In our present study, we have extended this approach to probe the importance of predicted quaternary interactions involving specific TORC2 partners as well as exploring the role of specific features within the TOR2 MAS domain for TORC2 function. Our findings reveal unanticipated complexity for TORC2 assembly as well as identifying specific subdomains within the TOR2 MAS critical for TORC2 activity.
FIGURE 1:
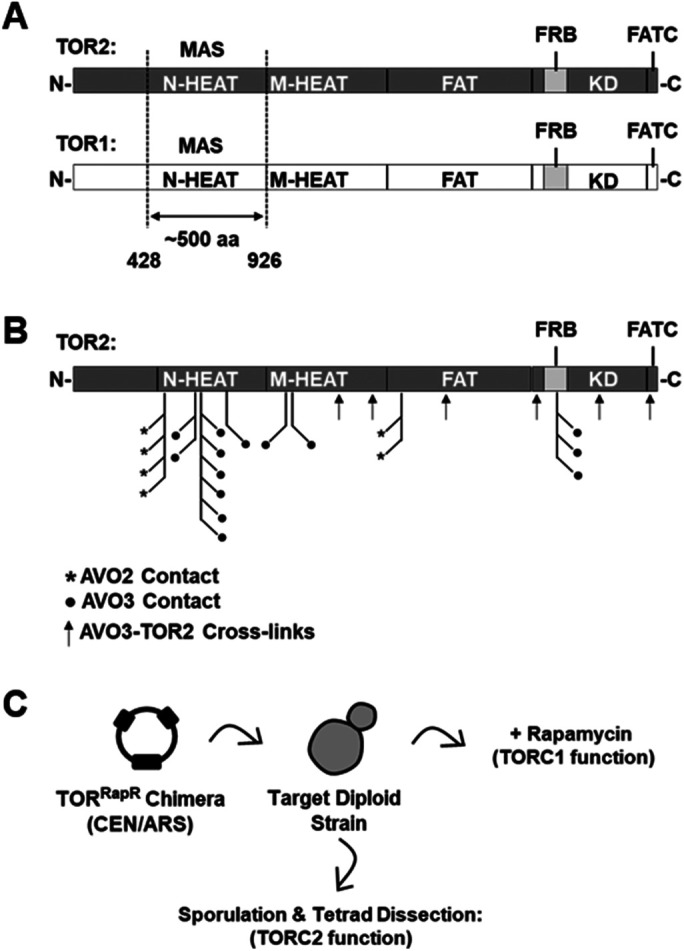
Overview and experimental approach. (A) Schematics showing the domain architecture of TOR1 and TOR2. The major assembly specificity (MAS) domain, which corresponds to the N-HEAT region of both TOR1 and TOR2 (Hill et al., 2018), is depicted. (B) Predicted amino acid contacts between TOR2 and TORC2 components AVO2 (asterisks) and AVO3 (filled circles) within a model of TORC2 determined by Cryo-EM (Karuppasamy et al., 2017). Contacts were identified using the structure analysis software tool UCSF Chimera (Pettersen et al., 2004). Identified protein cross-links (Gaubitz et al., 2015) between TOR2 and AVO3 (arrows) are also indicated. (C) Schematic of genetic approach to test chimeric TOR1-TOR2 genes for TORC1 and TORC2 function. See the text for details.
RESULTS
Strategy and experimental approach
Our prior study revealed that distinct regions of TOR2, in addition to the MAS/N-HEAT domain (for simplicity referred to here as TOR2 MAS), are sufficient but not necessary to confer TORC2 function (Hill et al., 2018). These findings suggest that significant sequence flexibility exists within TOR2 to govern TORC2 assembly. In this context, we observed that many predicted sites of interaction between TOR2 and its TORC2-specific partners AVO2 and AVO3, based on a medium-resolution Cryo-EM structural model and protein–protein cross-linking data, map to discontinuous segments of TOR2 within several TOR1-TOR2 chimeras (Gaubitz et al., 2015; Karuppasamy et al., 2017; Hill et al., 2018) (Figure 1B). We therefore sought to use these chimeras to interrogate the importance of predicted quaternary interactions for TORC2 assembly and function, using a synthetic sick/lethal (SSL) genetic interaction approach (Kaiser and Schekman, 1990; Guarente, 1993). This approach takes advantage of the fact that two mutations in interacting gene products, including within multiprotein complexes, often yield exacerbated phenotypes compared with either single mutation alone. Moreover, using genetic approaches, including synthetical lethality, to explore the function of paralogous gene products can yield information about both the architecture and the evolution of protein complexes (Mirny and Gelfand, 2002; Varga et al., 2021). Our general strategy, adapted from our prior study (Hill et al., 2018), was to introduce chimeric TOR genes into target heterozygous diploid strains, followed by sporulation, tetrad dissection, and phenotypic analysis to assess TOR2 and, by extension, TORC2 function (Figure 1C). Unless stated otherwise, each chimera also harbored the dominant rapamycin resistance (RapR) allele (Heitman et al., 1991) to allow for qualitative assessment of TORC1 activity (Figure 1C).
Evidence for essential quaternary interactions within TORC2
On the basis of our prior findings (Hill et al., 2018), we reconstructed representative chimeras that span the range of predicted TOR2 interactions with AVO2 and AVO3 (Figures 1B and 2A). Because the essential TORC2-specific subunit AVO1 (orthologue of mammalian SIN1) is predicted to interact exclusively with the C-terminal region of TOR2 (Gaubitz et al., 2015; Karuppasamy et al., 2017), we did not address its role in TORC2 assembly in this study. Chimeras were constructed using synthetic gene synthesis and seamless gene cloning methods, where each construct contained three copies of the HA epitope at the N-terminus (see Materials and Methods). Western blot analysis demonstrated that each chimera produced normal levels of TOR protein (Figure 2D).
FIGURE 2:
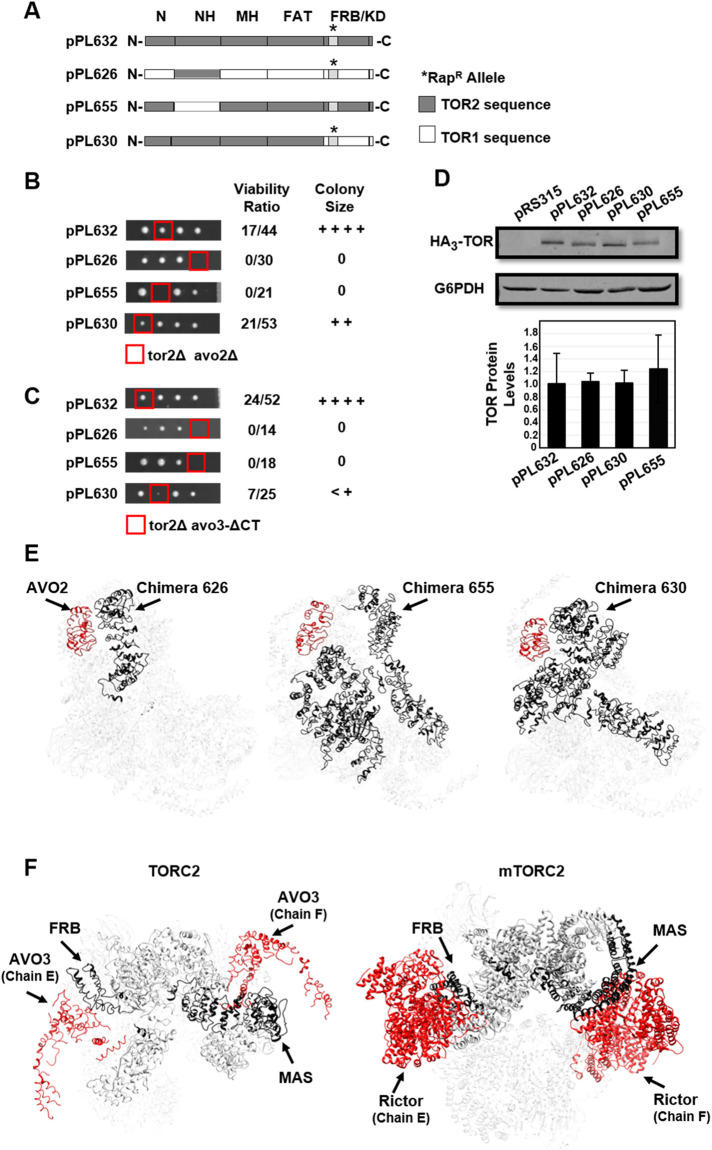
Synthetic genetic interactions involving TORC2 components. (A) Chimeric TOR1-TOR2 genes constructed for this study. Plasmid pPL632 expresses full-length TOR2. Plasmid pPL626 expresses the TOR2 MAS domain within the context of TOR1. The reciprocal chimera, pPL655, expresses the TOR1 MAS domain within the context of TOR2. Plasmid pPL630 expresses the complete N-terminal region of TOR2, including the FAT domain, but contains the TOR1 FRB and kinase domains. Where indicated, constructs harbor the rapamycin resistance mutation (RapR); this mutation corresponds to the TOR1-1 allele (S1972R) for sequences corresponding to TOR1 and the TOR2-1 allele (S1975R) for sequences corresponding to TOR2. (B) Chimeras described in A were introduced into a double heterozygous TOR2/tor2Δ AVO2/avo2Δ strain, followed by sporulation and tetrad dissection. Viable haploid progeny were genotyped by growth on selective media (see Materials and Methods). Viability was determined based on the number of tor2Δ avo2Δ haploid progeny that carried a plasmid compared with total tor2Δ progeny that carried a plasmid. The relative colony size was assessed following incubation at 30°C for 2 d on YPD solid media. ++++ corresponds to wild-type growth, and 0 corresponds to no growth. (C) Chimeras described in A were introduced into a double heterozygous TOR2/tor2Δ AVO3/avo3-Δ CT strain, followed by sporulation and tetrad dissection. Viable haploid progeny were genotyped by growth on selective media (see Materials and Methods). Viability was determined based on the number of tor2Δ avo3-Δ CT haploid progeny that carried a plasmid compared with total tor2Δ progeny that carried a plasmid. The relative colony size was assessed as in B. (D) Western blot analysis of TOR1-TOR2 chimeras. The indicated plasmids were introduced into a TOR2/tor2Δ heterozygous diploid strain (lanes 2–5). Cells were grown to mid–log phase in selective media, and protein extracts were prepared and analyzed by SDS–PAGE and immunoblotting. Blots were probed with anti-HA to detect plasmid-expressed TOR chimeras or anti-G6PDH (Zwf1) as a loading control. Plasmid pRS315 is an empty control vector. TOR protein levels were quantified following normalization to the G6PDH signal and represent averages of three independent experiments (± SD). (E) Modeling TOR2-specific regions in the TOR2 MAS domain (pPL626) (left panel) or TOR1 MAS domain (pPL655) (right panel) chimeras, respectively. TOR2-specific regions are highlighted in gray in the schematic diagrams and are shown in black in the Cryo-EM model for TORC2 (Karuppasamy et al., 2017). AVO2 is shown in red. Protein chains are labeled according to nomenclature described in Karuppasamy et al. (2017). (F) Left panel: TOR2 MAS and FRB domains within TOR2 (chain A) are shown in black and both copies of AVO3 are shown in red within the Cryo-EM model for TORC2 (Karuppasamy et al., 2017). Proximities between the FRB domain and one copy of AVO3 (chain E) and between the MAS domain and the second copy of AVO3 (chain F) are apparent. Protein chains are labeled according to nomenclature described in Karuppasamy et al. (2017). Right panel: mTOR MAS and FRB domains within mTOR (chain A) are shown in black and both copies of Rictor are shown in red within the Cryo-EM model for mTORC2 (Scaiola et al., 2020). Proximities between the FRB domain and one copy of Rictor (chain E) and between the MAS domain and the second copy of Rictor (chain F) are apparent. Protein chains are labeled according to nomenclature described in Scaiola et al. (2020).
We first tested the ability of these chimeras to replace endogenous TOR2 within TORC2 in the absence of the nonessential TORC2 component AVO2. Plasmids expressing these chimeras were introduced into a double heterozygous TOR2/tor2Δ AVO2/avo2Δ strain, followed by sporulation and tetrad dissection. For comparison, we also tested a plasmid that expressed full-length TOR2 (pPL632). As expected and consistent with our previous findings (Hill et al., 2018), cells singly deleted for either tor2Δ or avo2Δ were viable in the presence of each of these chimeras. We next scored the frequency of isolating haploid strains that harbored both tor2Δ and avo2Δ mutations and a chimera plasmid (described as percent viability) as well as scoring relative colony sizes of resulting viable haploid strains. We observed that full-length TOR2 and a chimera containing the entire N-terminus of TOR2 (pPL630) (Helliwell et al., 1994; Hill et al., 2018) were each viable in tor2Δ avo2Δ cells (Figure 2B). By contrast, chimeras containing either the TOR2 MAS domain (pPL626) or the reciprocal chimera containing the TOR1 MAS domain (pPL655) failed to support the growth of tor2Δ avo2Δ cells (Figure 2B). We conclude from these results that TOR2-specific sequences throughout the N-terminal region of TOR2 are required for proper TORC2 function in the absence of AVO2.
To extend these results, we next tested the ability of these chimeras to provide TOR2 function in a strain that harbored a truncated allele of AVO3 lacking 147 amino acids at its C-terminus (termed avo3-ΔCT). Previous studies demonstrated that this allele is functional yet causes TORC2 to become sensitive to rapamycin, where it is proposed to increase access of the FRBP-rapamycin complex to the FRB domain (Gaubitz et al., 2015). Given that AVO3 is encoded by an essential gene and, therefore, we could not analyze a null allele, we used avo3-ΔCT as a possible hypomorphic mutant. As expected, full-length TOR2 (pPL632) supported viability in tor2Δ avo3-ΔCT cells (Figure 2C). By contrast, as was observed in avo2Δ cells, both the TOR2 MAS domain (pPL626) and TOR1 MAS domain (pPL655) chimeras displayed synthetic lethal interactions with the avo3-ΔCT allele (Figure 2C). Interestingly, we observed that the chimera containing the entire N-terminus of TOR2 (pPL630) conferred a severe synthetic phenotype in combination with the avo3-ΔCT allele (Figure 2C). We conclude from these results that TOR2-specific determinants throughout the length of the protein, including the FRB and kinase domains, are required for proper TORC2 function in cells harboring this truncated allele of AVO3.
A recent structural model for yeast TORC2, based on Cryo-EM analysis, includes only limited assigned densities corresponding to AVO2 and AVO3 (Karuppasamy et al., 2017). Despite these limitations, we observed important correlations between chimera-specific phenotypes, described above, and the proximity of TOR2-specific sequences to both proteins. For AVO2, the TOR1 MAS (pPL655) and TOR2 MAS (pPL626) are each missing TOR2-unique sequences adjacent to AVO2, whereas these sequences are present in the complete N-terminal TOR2 chimera (pPL630) (Figure 2E). We note that each copy of AVO2 interacts with TOR2-specific sequences within a single TOR2 chain, suggesting that loss of AVO2 disrupts the assembly of individual TORC2 monomers (Figure 2E).
By contrast, only full-length TOR2 is functional in combination with the avo3-ΔCT allele, consistent with the fact that AVO3 makes extensive contacts throughout TOR2. Intriguingly, contacts with AVO3 are partitioned with respect to elements in a single TOR2 protein. Thus, the N-terminal MAS domain is predicted to interact with one AVO3 monomer, whereas the C-terminus, including the FRB domain, is in proximity to a second AVO3 monomer (Figure 2F, left panel). On the basis of our phenotypic analyses, we suggest that assembly and/or stability of TORC2 is likely to require interactions between TORC2 monomers that are anchored, in part, by interactions involving AVO3. Significantly, this arrangement of interactions is conserved in mTORC2, where in a recent high-resolution structural model (Scaiola et al., 2020), the mTOR MAS domain contacts one Rictor monomer and the FRB domain interacts with a second copy of Rictor (Figure 2F, right panel).
The molecular basis for the observed synthetic interactions between TOR1-TOR2 chimeras and the avo3-ΔCT allele is unknown, as amino acids corresponding to this deletion are not resolved in the published model for TORC2 (Gaubitz et al., 2015; Karuppasamy et al., 2017). However, one possibility is that loss of C-terminal sequences in AVO3 disrupts interactions between monomers that make TORC2 activity dependent on the presence of TOR2-specific sequences adjacent to the FRB and/or kinase domain(s).
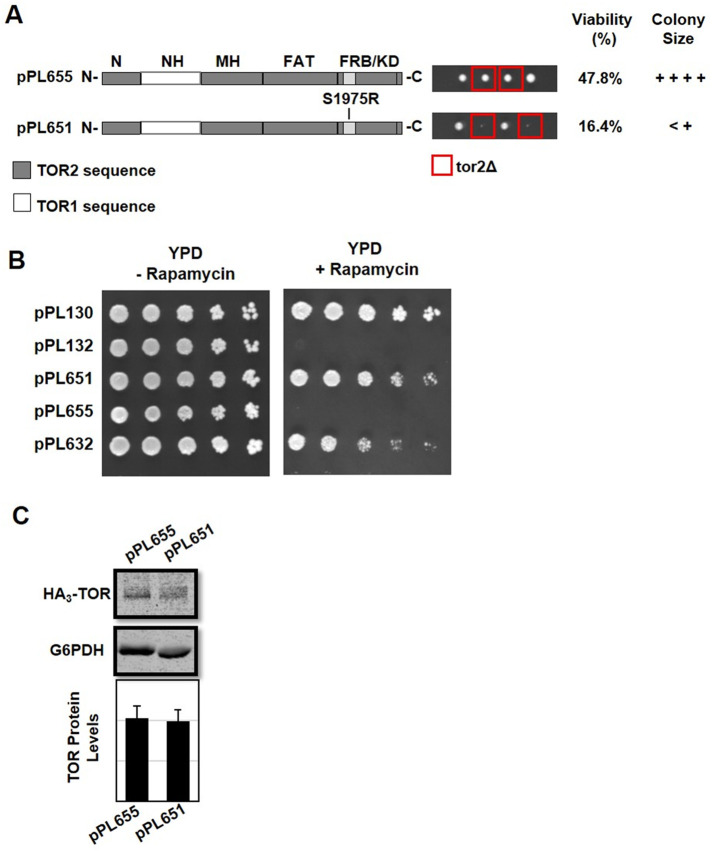
In the above experiments, except for the TOR1 MAS domain chimera (pPL655), all constructs contained the RapR allele and, as expected, conferred robust rapamycin resistance as well as rescuing the lethality of a tor2Δ strain (Figure 3). By contrast, when the RapR allele was introduced into the TOR1 MAS domain chimera (pPL655), the resulting construct (pPL651) conferred strong rapamycin resistance yet provided only weak rescue of the lethality of a tor2Δ strain (Figure 3). Western blot analysis demonstrated that chimeras 651 and 655 produced equivalent levels of protein, suggesting that deficiencies in steady state levels of TOR do not explain these phenotypic differences (Figure 3). While we do not understand the molecular basis for this apparent synthetic genetic interaction, it is consistent with our conclusion, described above, that functional interactions between the MAS and FRB domains are required for proper TORC2 function.
FIGURE 3:
Introducing the TOR2-1 allele into the TOR1 MAS domain chimera is deleterious to TORC2 but not TORC1 function. (A) The TOR1 MAS domain chimera (pPL655) and the TOR1 MAS domain chimera containing the S1975R (RapR) TOR2-1 allele (pPL651) were introduced into heterozygous TOR2/tor2Δ cells and analyzed by tetrad dissection. Viability was determined based on the number of viable tor2Δ haploid progeny that carried a plasmid compared with all viable haploid progeny that carried a plasmid. The relative colony size was assessed following incubation at 30°C for 2 d on YPD solid media. (B) TOR2/tor2Δ cells carrying the indicated plasmids were tested for rapamycin resistance. Cells were grown in selective media to mid–log phase, serially diluted, and plated onto solid agar plates containing SCD minus leucine or SCD minus leucine and 0.2 μg/ml rapamycin. Plates were incubated at 30°C for 2 d and then photographed. As controls, plasmid pPL130 expresses the rapamycin resistance TOR1-1 allele and plasmid pPL132 expresses wild-type TOR1. (C) Western blot analysis of TOR1-TOR2 chimeras. TOR2/tor2Δ cells carrying the indicated plasmids were grown in selective media to mid–log phase, and protein extracts were prepared and analyzed as described in the legend to Figure 2D.
A sensitized genetic system to identify TOR2 elements essential for TORC2 function
Our above findings revealed novel genetic interactions between distinct TOR1-TOR2 chimeras and TORC2-specific partners. We reasoned that we could employ a similar approach to identify synthetic interactions within TOR2 (i.e., intragenic interactions), as an approach to refine our understanding of structural elements within TOR2 important for TORC2 function. We focused on the TOR2 MAS domain, where we showed previously that approximately 500 amino acids of TOR2 are sufficient to confer TORC2 function within the context of an otherwise full-length TOR1 protein (Hill et al., 2018). We constructed several new chimeras using the TOR2 MAS domain construct (pPL626) as a starting platform (see Materials and Methods). TOR2 activity was assessed by transformation of a TOR2/tor2Δ diploid strain, followed by sporulation and tetrad dissection. Here we scored both the frequency of finding haploid strains that harbored both a tor2Δ deletion and a chimera plasmid (described as percent viability) and their relative colony size. In parallel, we assessed the rapamycin resistance of each strain as a measure of TORC1 function as well as monitoring steady state levels of TOR protein by Western blot analysis.
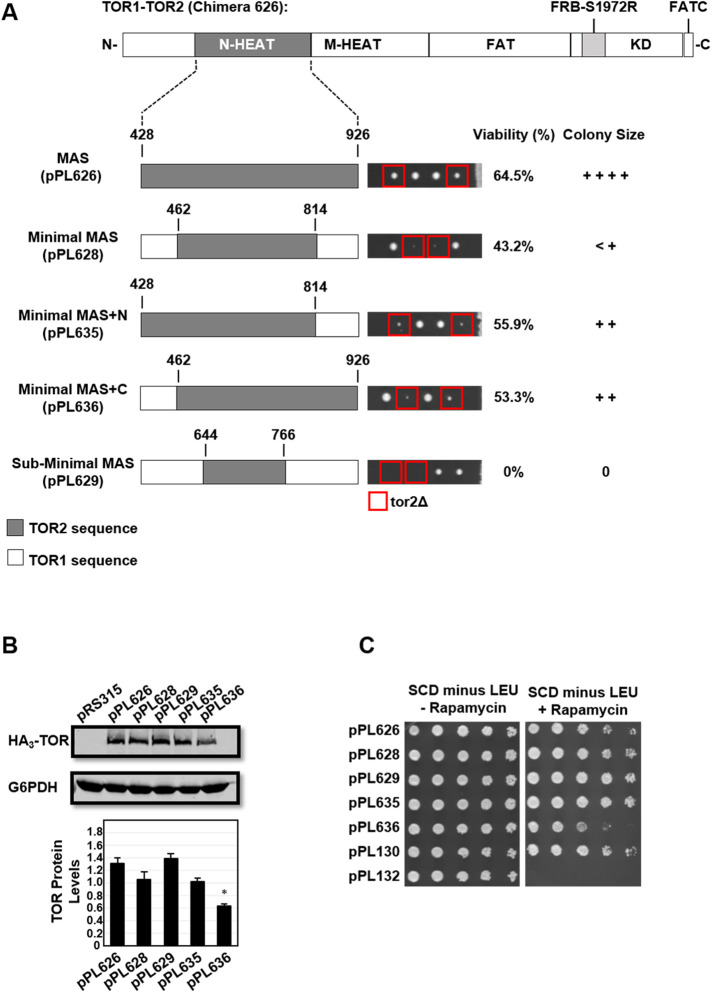
In a first approach, we constructed “Minimal” (pPL628) and “Sub-Minimal” (pPL629) TOR2 MAS domain chimeras (Figure 4A). The TOR1/2 boundaries of these chimeras encompassed a central core of predicted TOR2 contacts for AVO2 and AVO3 (pPL628) or, alternatively, for AVO3 alone (pPL629) (Figure 1B) (Gaubitz et al., 2015; Hill et al., 2018). Surprisingly, the Minimal TOR2 MAS domain (pPL628) afforded only very limited TOR2 function, and the Sub-Minimal TOR2 MAS domain (pPL629) was completely nonfunctional as TOR2 (Figure 4A). Both chimeras produced normal levels of protein and conferred rapamycin resistance, consistent with an observed defect in TORC2 activity alone (Figure 4, B and C). To test whether the impaired phenotype of the Minimal TOR2 MAS domain was due to loss of TOR2-specific sequences at either end of this domain, we constructed two additional chimeras that restored TOR2 sequences at either the N-terminal side (Minimal MAS+N; pPL635) or the C-terminal side (Minimal MAS+C; pPL636), respectively. Each construct displayed only minor improvement in TOR2 phenotypes (Figure 4A). We conclude from this analysis that sequences outside the central core of the TOR2 MAS domain are required for proper TORC2 function. Surprisingly, we observed that Minimal MAS+C (pPL636) chimera also resulted in reduced rapamycin resistance and a significant reduction in the steady state level of TOR protein (Figure 4, B and C).
FIGURE 4:
Testing the functionality of the central core of the TOR2 MAS domain. (A) TOR2 MAS domain plasmid pPL626 containing the S1972R (RapR) TOR1-1 allele was modified to create the indicated TOR1-TOR2 chimeras. Amino acid numbering refers to TOR2 protein sequence. “N” and “C” refer to N- and C-termini, respectively. Plasmids were introduced into heterozygous TOR2/tor2Δ cells and analyzed by tetrad analysis as described in the legend to Figure 3A. (B) Western blot analysis of TOR1-TOR2 chimeras. TOR2/tor2Δ cells carrying the indicated plasmids were grown in selective media to mid–log phase, and protein extracts were prepared and analyzed by SDS–PAGE and immunoblotting, as described in the legend to Figure 2D. (C) TOR2/tor2Δ cells carrying the indicated plasmids were tested for rapamycin resistance as described in the legend to Figure 3B.
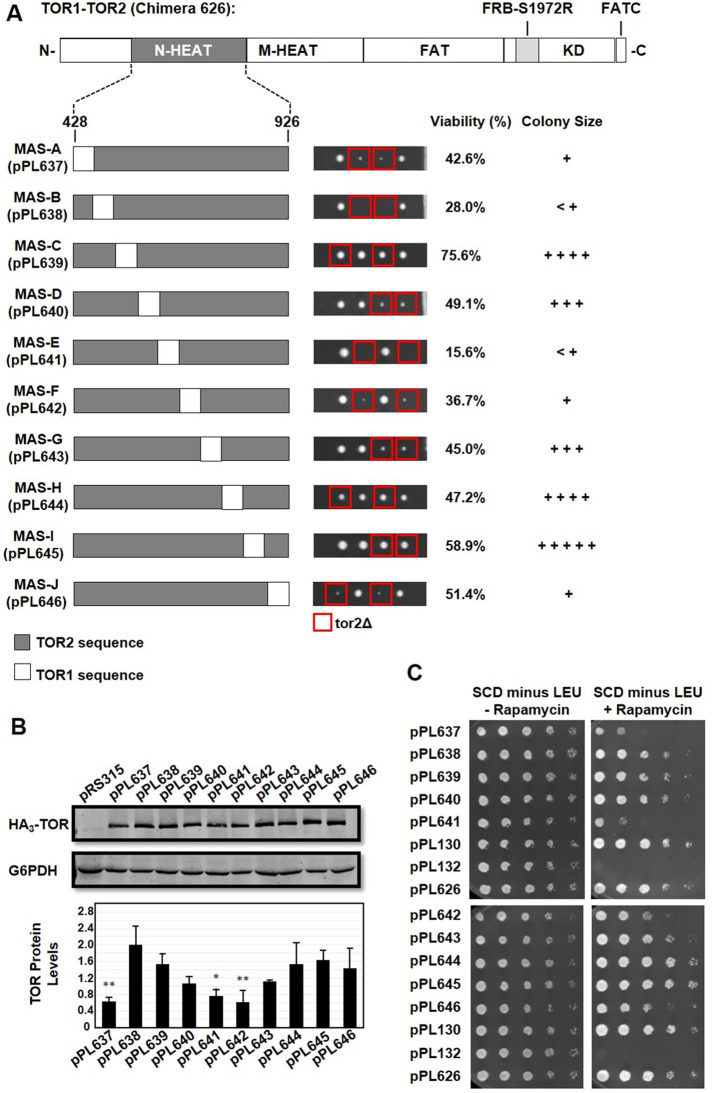
In a second approach, we tested whether the TOR2 MAS domain would tolerate the introduction of short stretches of TOR1 sequence interspersed throughout its length and retain TOR2 function. Accordingly, we constructed 10 chimeras (TOR2 MAS-A through TOR2 MAS-J), each representing a contiguous series of ∼50-amino-acid segments of TOR1-specific sequences that replaced corresponding TOR2 sequences (Figure 5A) (see Materials and Methods). To maximize the likelihood of proper protein folding, chimeras were designed to maximize two criteria where possible: 1) TOR1-TOR2 boundaries (“switch points”) were placed where TOR1 and TOR2 sequences were identical; and 2) each construct encompassed complete predicted α-helical repeat units (Perry and Kleckner, 2003; Knutson, 2010; Karuppasamy et al., 2017). Chimeras were introduced into a TOR2/tor2Δ strain and analyzed as above. Here we observed surprising phenotypic diversity with respect to the level of rescue of tor2Δ lethality (Figure 5A). The most deleterious phenotypes were observed for chimeras that swapped sequences within the central region (chimeras E and F) of the MAS domain as well as at the N- and C-termini (chimeras A, B, and J) (Figure 5A). In particular, chimeras MAS-B and MAS-E were most impaired in their ability to rescue the lethality of a tor2Δ strain (Figure 5A). Together these results both confirm that outer sequences of the TOR2 MAS domain are important for TORC2 function and highlight the importance of the central core of this domain.
FIGURE 5:
Systematic functional dissection of the TOR2 MAS domain. (A) TOR2 MAS domain plasmid pPL626containing the S1972R (RapR) TOR1-1 allele was modified to create the indicated TOR1-TOR2 chimeras. Each construct, MAS-A through MAS-J, contains approximately 50 amino acids of TOR1 sequence dispersed along the length of the TOR2 MAS domain. Plasmids were introduced into heterozygous TOR2/tor2Δ cells and analyzed by tetrad analysis as described in the legend to Figure 3A. (B) Western blot analysis of TOR1-TOR2 chimeras. TOR2/tor2Δ cells carrying the indicated plasmids were prepared and analyzed as described in the legend to Figure 2D. (C) TOR2/tor2Δ cells carrying the indicated plasmids were tested for rapamycin resistance as described in the legend to Figure 3B.
All chimeras tested in this second approach produced full-length TOR protein and conferred resistance to rapamycin, an indication that they all assemble and function within TORC1 (Figure 5, B and C). However, three chimeras, MAS-A (pPL637), MAS-E (pPL641), and MAS-F (pPL642), resulted in significantly reduced steady state levels of protein and conferred weaker rapamycin resistance (Figure 5, B and C). We note that chimeras MAS-A (pPL637) and Minimal MAS+C (pPL636) share overlapping TOR1-specific sequences at the N-terminus of the TOR2 MAS domain, highlighting the sensitivity of this region to changes in TOR2-specific sequences (compare Figures 4 and 5). In addition, our findings that chimeras MAS-E and MAS-F result in reduced levels of TOR protein and impaired rapamycin resistance indicate that the central core of the TOR2 MAS domain is also sensitive to the precise composition of TOR1 versus TOR2 sequences and impact TOR complex assembly and/or TOR protein stability.
Fine dissection of TOR2 elements crucial for TORC2 function
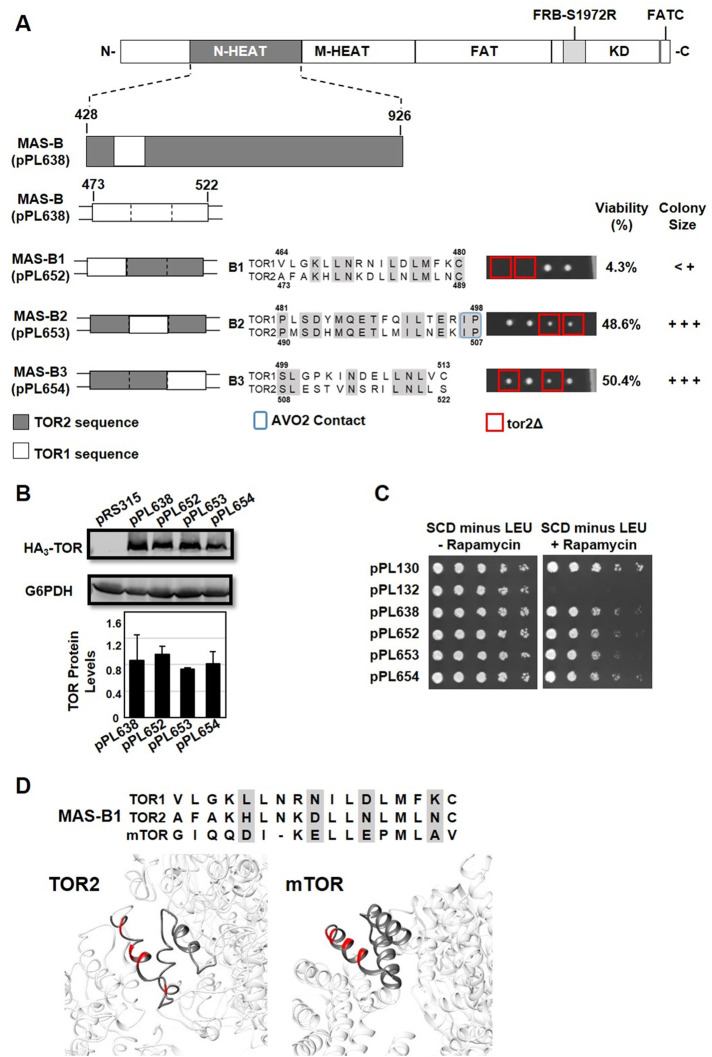
To begin to understand, at the single-amino-acid level, the importance of elements within the TOR2 MAS domain, we focused on chimera TOR2 MAS-B, as introduction of TOR1-specific sequences into this relatively discrete region severely disrupted TORC2 function without affecting overall protein levels or TORC1 activity. We subdivided TOR2 MAS-B into three separate chimeras, MAS-B1 through MAS-B3, each representing approximately 6–10 amino acids of the TOR1 sequence within the context of an otherwise complete TOR2 MAS domain (Figure 6A). Each construct was introduced into a TOR2/tor2Δ strain and analyzed as above.
FIGURE 6:
Identification of functional elements within the TOR2 MAS-B chimera. (A) MAS-B plasmid pPL638 containing the S1972R (RapR) TOR1-1 allele was modified to create the indicated TOR1-TOR2 chimeras. Each construct, B1 through B3, contains approximately 6–10 amino acids of TOR1 sequence within an otherwise TOR2 MAS domain, as depicted in the indicated sequence alignments for TOR1 and TOR2. Predicted amino acids contacts between TOR2 and AVO2 are boxed in blue in construct TOR2 MAS B-2. Plasmids were introduced into heterozygous TOR2/tor2Δ cells and analyzed by tetrad dissection as described in the legend to Figure 3A. (B) Western blot analysis of TOR1-TOR2 chimeras. TOR2/tor2Δ cells carrying the indicated plasmids were prepared and analyzed as described in the legend to Figure 2D. (C) TOR2/tor2Δ cells carrying the indicated plasmids were tested for rapamycin resistance as described in the legend to Figure 3B. (D) Sequence alignment of MAS-B1 in TOR1, TOR2, and mTOR, highlighting four nonconservative amino acid changes between TOR1 and TOR2 (shaded). Structures depict the MAS-B1 element within Cryo-EM structures of TORC2 (left) (Karuppasamy et al., 2017) and mTORC2 (right) (Scaiola et al., 2020). The four amino acids shaded in the alignment are highlighted in red in both structures.
We observed that chimera MAS-B1 (pPL652) was severely impaired in its ability to rescue the lethality of a tor2Δ strain (Figure 6A). By contrast, chimeras MAS-B2 (pPL653) and MAS-B3 (pPL654) both provided near-wild-type rescue (Figure 6A). All three constructs produced normal levels of TOR protein and conferred strong rapamycin resistance (Figure 6, B and C). Remarkably, chimera MAS-B1 contains only 10 amino acids that differ between TOR1 and TOR2, with respect to the 498 amino acids that constitute the TOR2 MAS domain, affirming the utility of this approach to reveal amino acids crucial for TORC2 function (Figure 6A). We observed that chimera MAS-B2 includes two predicted amino acid contacts for AVO2 within MAS-B (Figure 6A). However, these amino acids are identical in TOR1 and TOR2; thus, it is unlikely that they contribute directly to complex specificity, a conclusion consistent with our findings that this chimera supports full TOR2 activity.
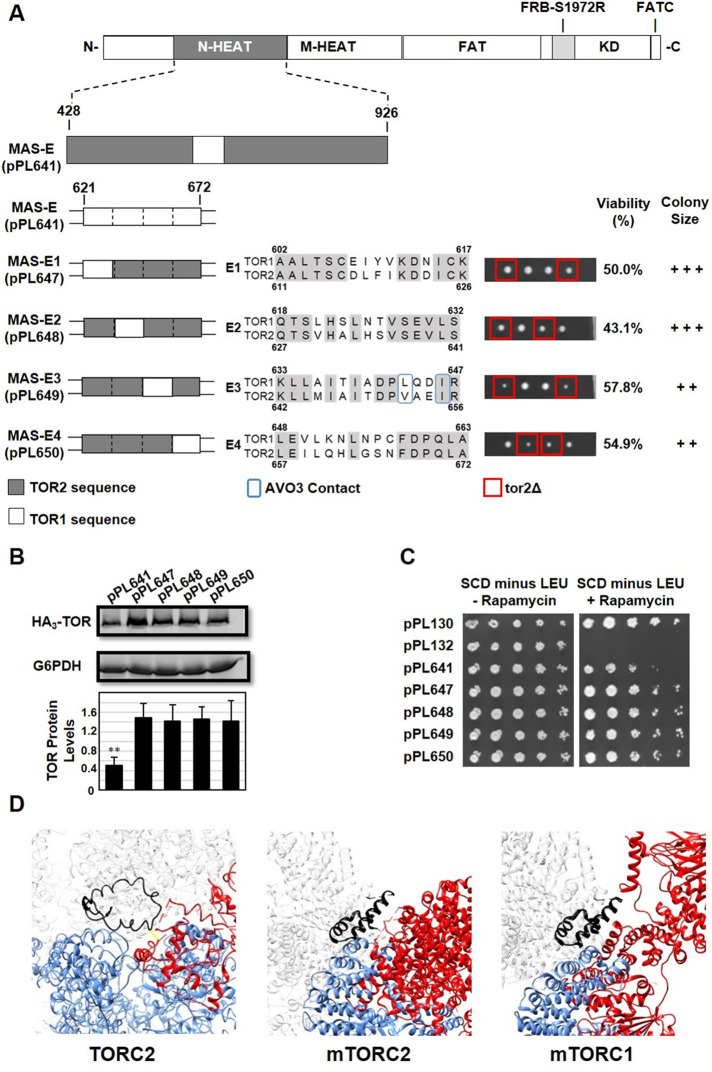
We used a similar approach to dissect chimera MAS-E, which is significantly impaired in TORC2 activity but showed defects in TORC1 as well (Figure 5). We subdivided chimera MAS-E into four separate constructs, MAS-E1 through MAS-E4, where each chimera possessed only four- to six-amino-acid differences between TOR1 and TOR2 with respect to the TOR2 MAS domain (Figure 7A). Interestingly, in contrast to the complete MAS-E chimera, all four constructs rescued a tor2Δ allele, produced normal levels of TOR protein, and provided strong rapamycin resistance (Figure 7, A–C). We note that two constructs, MAS-E3 (pPL649) and MAS-E4 (pPL650), displayed modest growth defects in tor2Δ cells, where MAS-E3 is the location of predicted sites of interaction between TOR2 and AVO3 (Figure 7A). We conclude from these findings that the TOR2 MAS domain can tolerate smaller clusters of TOR1 sequences within its central core and maintain adequate TORC2 activity, produce stable TOR protein, and function within TORC1.
FIGURE 7:
Identification of functional elements within the TOR2 MAS-E chimera. (A) MAS-E plasmid pPL641 containing the S1972R (RapR) TOR1-1 allele was modified to create the indicated TOR1-TOR2 chimeras. Each construct, E1 through E4, contains approximately 6–10 amino acids of TOR1 sequence within an otherwise TOR2 MAS domain, as depicted in the indicated sequence alignments for TOR1 and TOR2. We note that chimera E1 includes four additional TOR1-specific amino acids that are not present in MAS-E. Predicted amino acids contacts between TOR2 and AVO3 are boxed in blue in E3. Plasmids were introduced into heterozygous TOR2/tor2Δ cells and analyzed as described in the legend to Figure 3A. (B) Western blot analysis of TOR1-TOR2 chimeras. TOR2/tor2Δ cells carrying the indicated plasmids were prepared and analyzed as described in the legend to Figure 2D. (C) TOR2/tor2Δ cells carrying the indicated plasmids were tested for rapamycin resistance as described in the legend to Figure 3B. (D) Sequences corresponding to MAS-E (black) are highlighted in Cryo-EM structures for TORC2, mTORC2, and mTORC1, as indicated. Also shown are the indicated partners within each complex (red) as well as portions of TOR2 or mTOR (blue) corresponding to the other protein chain within each structure. Regions of AVO3 or Rictor absent within these structures are indicated by dashed red lines in each model. Protein chains are labeled according to nomenclature described in Karuppasamy et al. (2017).
DISCUSSION
In this study, we have used chimeric TOR1-TOR2 proteins to extend our understanding of specific determinants within TOR2 that are critical for TORC2 assembly and function. For example, the pattern of synthetic genetic interactions between TOR chimeras and mutant alleles of TORC2-specific components AVO2 and AVO3, examined within the context of a structural model for TORC2, suggests important design features for assembly of TORC2. In particular, our observation that proper TORC2 activity requires interactions between TOR2-specific elements within the context of the same TOR2 chain and both copies of AVO3 suggests that stable assembly of TORC2 requires complex quaternary interactions across the dimer interface. Because this arrangement of physical interactions is conserved between mTOR and Rictor, we predict that interactions between monomers will also turn out to be crucial for mTORC2 assembly and/or stability. Interestingly, an inspection of the structure of mTORC1 reveals that Raptor also makes contacts with both copies of mTOR within the mTORC1 dimer (Yang et al., 2017), raising the interesting possibility that both mTORC1 and mTORC2 assemble as obligate dimers.
We previously identified the TOR2 MAS domain as a contiguous ∼500-amino-acid segment within the N-terminus of TOR2 that is sufficient to confer TORC2 activity in the context of a TOR1-TOR2 chimera. We were surprised that we were unable to narrow this domain further, even though most predicted interactions between TOR2 and both AVO2 and AVO3 are located within a central ∼200-amino-acid core of this domain. Instead, we systematically exchanged discrete segments within the TOR2 MAS domain, by introducing a “moving window” of TOR1 sequence, to identify specific clusters of essential TOR2-specific sequences. This approach employed a TOR2 MAS domain chimera as a “sensitized” genetic background to reveal the importance of specific TOR2 sequences. Remarkably, for one segment, TOR2 MAS-B1, we were able to resolve this element to near-amino-acid resolution.
At a structural level, chimera MAS-B1 corresponds to the N-terminal helix of HEAT repeat 10, where it is positioned at the exterior of the MAS domain (Karuppasamy et al., 2017) (Figure 6D). Within this segment are four nonconservative amino acid differences between TOR1 and TOR2, all predicted to lie on an external-facing edge of the helix (Figure 6D). At present, there are no predicted interactions involving these residues. Thus, one possibility is that this helix interacts with portions of AVO2 or AVO3 that have yet to be identified at the ultrastructural level or, alternatively, with a different component essential for TORC2 assembly and/or function, for example, the TTT-R2TP cochaperone complex required for proper (m)TOR folding (Takai et al., 2007, 2010; Kim et al., 2013). Examination of a structural model for mTORC2 reveals that this same helix within mTOR is similarly positioned at the outer surface of mTORC2 (Figure 6D) (Scaiola et al., 2020). In this context, we note that, as for AVO3, a substantial portion of Rictor (approximately 35%) remains unidentified within the mTORC2 structure (Scaiola et al., 2020).
The segment of TOR2 corresponding to MAS-E is predicted to be involved in interactions with both AVO3 and the other TOR2 chain within the TORC2 dimer interface (Karuppasamy et al., 2017; Hill et al., 2018) (Figure 7D). This arrangement of interactions is conserved in mTORC2, where corresponding sequences in mTOR are located adjacent to Rictor as well as the other mTOR chain (Scaiola et al., 2020) (Figure 7D). Interestingly, for both AVO3 and Rictor, in the present structural models significant portions of both proteins are missing directly adjacent to MAS-E, suggesting that there may be additional associations with this region of TOR (Karuppasamy et al., 2017; Scaiola et al., 2020). Moreover, in a recent structural model for mTORC1, Raptor displays extensive interactions with sequences corresponding to MAS-E (Yang et al., 2017) (Figure 7D), providing a possible rationale for why changes in this region affect both TORC1 and TORC2 activity. Alternatively, this region includes intermolecular contacts between the two mTOR proteins within the dimeric structures of mTORC1 and mTORC2 (Yang et al., 2017; Scaiola et al., 2020), as well as for TOR2 within TORC2 (Karuppasamy et al., 2017).
We were surprised to find that several chimeras we constructed resulted in reduced steady state levels of protein, likely contributing to impaired TORC1 as well as TORC2 behavior. One possibility is that the introduction of shorter TOR1-specific sequences within these regions, for example in chimeras MAS-A (pPL637) and Minimal MAS+C (pPL636), impairs localized folding, despite these sequences functioning normally within the context of a complete TOR1 protein. A more interesting possibility is that such a chimera may juxtapose sequences specific for TORC1 versus TORC2 assembly, therefore causing localized disruptions and/or competition for crucial quaternary interactions for complex-specific partners or chaperones, resulting in destabilized chimeras susceptible to degradation. As described above, consistent with the latter possibility is the observation that chimera TORC2 MAS-E involves sequences predicted to be involved in multiple protein–protein interactions within both complexes. The MAS domain is also predicted to interact with the TTT-R2TP cochaperone complex (Takai et al., 2007, 2010).
Our functional studies highlight the cooperative nature of TOR complex assembly, where distinct domains distributed throughout TOR2 are normally sufficient for the formation of TORC2. By employing TOR1-TOR2 chimeras, we have been able to identify specific regions by making them essential for TORC2 identity. These findings now point the way for more detailed analyses of their role in TORC2 assembly and function. Our approach underscores the utility of analyzing paralogues to understand the molecular basis of assembly, as well as evolution, of large protein complexes (Mirny and Gelfand, 2002; Capra et al., 2012; de Juan et al., 2013; Schick et al., 2019; Varga et al., 2021). Owing to the conservation of several important elements within mTOR, including the MAS domain, our findings point to the likely importance of these regions for both mTORC1 and mTORC2 assembly. We anticipate that applying a similar structure–function approach to mTOR will be a successful avenue for testing the importance of amino acid contacts identified in both mammalian complexes. This includes testing the importance of defined interactions between mTOR and complex-specific partners during dimer assembly as well as determining whether essential elements within the MAS domain interact with currently unidentified interacting components essential for mTOR complex assembly and signaling.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Strains, media, and general methods
Strains and plasmids used in this study are listed in Tables 1 and 2, respectively. Cells were cultured in YPD (2% yeast extract, 1% peptone, and 2% dextrose) or synthetic complete dextrose (SCD) medium (0.8% yeast nitrogen base without amino acids, pH 5.5, 2% dextrose) supplemented with amino acids as described (Sherman, 1991). Rapamycin (Sigma-Aldrich, St. Louis, MO) was dissolved in dimethyl sulfoxide (DMSO) and added to SCD medium and agar plates to a final concentration of 0.2 µg/ml. Hygromycin B (Sigma-Aldrich, St. Louis, MO) was dissolved in water and added to agar plates at a final concentration of 200 µM. Yeast transformations were performed using a lithium acetate procedure (Gietz and Woods, 2002).
TABLE 1:
Saccharomyces cerevisiae strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| MPR1 | TB50a avo3-dCT(-157aa) [HpH] | Gaubitz et al., 2015 |
| PLY 362 | avo2::TRP W303a | Reinke et al., 2004 |
| PLY 1760 | W303a/α tor2::HIS/TOR2 [pPL632] | This study |
| PLY 1761 | W303a/α tor2::HIS/TOR2 [pPL655] | This study |
| PLY 1762 | W303a/α tor2::HIS/TOR2 [pPL630] | This study |
| PLY 1763 | W303a/α tor2::HIS/TOR2 [pPL626] | This study |
| PLY 1764 | TOR2/tor2::HIS AVO2/avo2::TRP [pPL632] | This study |
| PLY 1765 | TOR2/tor2::HIS AVO2/avo2::TRP [pPL626] | This study |
| PLY 1766 | TOR2/tor2::HIS AVO2/avo2::TRP [pPL655] | This study |
| PLY 1767 | TOR2/tor2::HIS AVO2/avo2::TRP [pPL630] | This study |
| PLY 1768 | TOR2/tor2::HIS AVO3/avo3-dCT::HpH [pPL632] | This study |
| PLY 1769 | TOR2/tor2::HIS AVO3/avo3-dCT::HpH [pPL626] | This study |
| PLY 1770 | TOR2/tor2::HIS AVO3/avo3-dCT::HpH [pPL655] | This study |
| PLY 1771 | TOR2/tor2::HIS AVO3/avo3-dCT::HpH [pPL630] | This study |
| PLY 1772 | W303a/α tor2::HIS/TOR2 [pPL628] | This study |
| PLY 1773 | W303a/α tor2::HIS/TOR2 [pPL629] | This study |
| PLY 1774 | W303a/α tor2::HIS/TOR2 [pPL635] | This study |
| PLY 1775 | W303a/α tor2::HIS/TOR2 [pPL636] | This study |
| PLY 1776 | W303a/α tor2::HIS/TOR2 [pPL637] | This study |
| PLY 1777 | W303a/α tor2::HIS/TOR2 [pPL638] | This study |
| PLY 1778 | W303a/α tor2::HIS/TOR2 [pPL639] | This study |
| PLY 1779 | W303a/α tor2::HIS/TOR2 [pPL640] | This study |
| PLY 1780 | W303a/α tor2::HIS/TOR2 [pPL641] | This study |
| PLY 1781 | W303a/α tor2::HIS/TOR2 [pPL642] | This study |
| PLY 1782 | W303a/α tor2::HIS/TOR2 [pPL643] | This study |
| PLY 1783 | W303a/α tor2::HIS/TOR2 [pPL644] | This study |
| PLY 1784 | W303a/α tor2::HIS/TOR2 [pPL645] | This study |
| PLY 1785 | W303a/α tor2::HIS/TOR2 [pPL646] | This study |
| PLY 1786 | W303a/α tor2::HIS/TOR2 [pPL652] | This study |
| PLY 1787 | W303a/α tor2::HIS/TOR2 [pPL653] | This study |
| PLY 1788 | W303a/α tor2::HIS/TOR2 [pPL654] | This study |
| PLY 1789 | W303a/α tor2::HIS/TOR2 [pPL647] | This study |
| PLY 1790 | W303a/α tor2::HIS/TOR2 [pPL648] | This study |
| PLY 1791 | W303a/α tor2::HIS/TOR2 [pPL649] | This study |
| PLY 1792 | W303a/α tor2::HIS/TOR2 [pPL650] | This study |
TABLE 2: .
Plasmids used in this study.
| Plasmid | Description | Chimera name | Source |
|---|---|---|---|
| pRS315 | LEU2 CEN/ARS | Sikorski and Hieter, 1989 | |
| pPL130 | LEU2 CEN/ARS TOR1-1 | Reinke et al., 2004 | |
| pPL132 | LEU2 CEN/ARS TOR1 | Reinke et al., 2004 | |
| pPL172 | pPL132, TOR2 114-1770 | Hill et al., 2018 | |
| pPL273 | pPL130, TOR2 428-946 | Hill et al., 2018 | |
| pPL321 | pPL130, TOR2 | Hill et al., 2018 | |
| pPL630 | pPL172, with S1972R | N-term TOR2 | This study |
| pPL632 | pPL321, with S195R | Full-length TOR2 | This study |
| pPL651 | pPL632, TOR1 428-946, with S1975R | TOR1 MAS S1975R | This study |
| pPL655 | pPL632, TOR1 428-946 | TOR1 MAS | This study |
| pPL626 | pPL273 | TOR2 MAS | This study |
| pPL628 | pPL626, TOR2 462-814 | Minimal MAS | This study |
| pPL629 | pPL626, TOR2 644-766 | Sub-Minimal MAS | This study |
| pPL635 | pPL626, TOR2 462-926 | Minimal MAS + N | This study |
| pPL636 | pPL626, TOR2 428-814 | Minimal MAS + C | This study |
| pPL637 | pPL626, TOR1 423-472 | TOR2 MAS-A | This study |
| pPL638 | pPL626, TOR1 473-522 | TOR2 MAS-B | This study |
| pPL639 | pPL626, TOR1 523-572 | TOR2 MAS-C | This study |
| pPL640 | pPL626, TOR1 573-622 | TOR2 MAS-D | This study |
| pPL641 | pPL626, TOR1 623-672 | TOR2 MAS-E | This study |
| pPL642 | pPL626, TOR1 673-722 | TOR2 MAS-F | This study |
| pPL643 | pPL626, TOR1 723-772 | TOR2 MAS-G | This study |
| pPL644 | pPL626, TOR1 773-823 | TOR2 MAS-H | This study |
| pPL645 | pPL626, TOR1 824-873 | TOR2 MAS-I | This study |
| pPL646 | pPL626, TOR1 874-946 | TOR2 MAS-J | This study |
| pPL652 | pPL626, TOR1 473-489 | TOR2 MAS-B1 | This study |
| pPL653 | pPL626, TOR1 490-507 | TOR2 MAS-B2 | This study |
| pPL654 | pPL626, TOR1 508-522 | TOR2 MAS-B3 | This study |
| pPL647 | pPL626, TOR1 611-626 | TOR2 MAS-E1 | This study |
| pPL648 | pPL626, TOR1 627-641 | TOR2 MAS-E2 | This study |
| pPL649 | pPL626, TOR1 642-656 | TOR2 MAS-E3 | This study |
| pPL650 | pPL626, TOR1 657-672 | TOR2 MAS-E4 | This study |
Construction and analysis of yeast strains for synthetic genetic interactions
Double heterozygous TOR2/tor2Δ AVO2/avo2Δ diploid strains carrying individual control or chimera plasmids were constructed by mating tor2Δ haploid strains carrying a plasmid to an avo2Δ haploid strain. Diploids were selected by growth on SCD minus leucine and tryptophan media. Following sporulation, haploid progeny were analyzed by scoring the presence or absence of markers for tor2Δ (growth on SCD minus histidine media), avo2Δ (growth on SCD minus tryptophan), and the relevant plasmid (growth on SCD minus leucine media). Double heterozygous TOR2/tor2Δ AVO3/avo3-ΔCT diploid strains carrying individual control or chimera plasmids were constructed by mating tor2Δ haploid strains carrying a plasmid to an avo3-ΔCT haploid strain. Diploids were identified following growth in SCD minus leucine media by their ability to form spores in sporulation media. Following sporulation, haploid progeny were analyzed by scoring the presence or absence of markers for tor2Δ (growth on SCD minus histidine media), avo3-ΔCT (growth in the presence of hygromycin), and the relevant plasmid (growth on SCD minus leucine media).
Plasmid construction
Plasmids used in this study are listed in Table 2. Plasmid construction was carried out in conjunction with Genscript (Piscataway, NJ). Plasmids pPL626, pPL630, and pPL632 were constructed using previously constructed parental plasmids (Hill et al., 2018) and used as templates for construction of new chimeras, as indicated in Table 2. The precise positions of switch points and specific chimeric sequences for each plasmid are listed in Supplemental Table S1, and all sequences are provided in Supplemental Table S2. Synthetic DNA sequences were subcloned into expression vectors by fragment exchange using two unique restriction sites or by modified Gibson Assembly (Casini et al., 2015) (see Supplemental Table S1). Plasmids were sequenced in their entirety to confirm the accuracy of construction.
Whole cell extraction, Western blot analysis, and quantification
Cells were grown overnight to mid–logarithmic phase (A600 = 2.0) in SCD minus leucine media at 30°C. Protein extracts were prepared using the NaOH cell lysis method (Dilova et al., 2004). Equivalent amounts of extract were loaded onto SDS–PAGE gels and then transferred to nitrocellulose membranes. Membranes were probed with α-HA (12CA5; 1:5000 dilution; Covance) and α-G6PDH (1:100,000 dilution; Sigma-Aldrich) primary antibodies. Secondary antibodies conjugated to IRDye (1:5000 dilution; LI-COR Biosciences) were used, and blots were imaged using either an Odyssey Infrared Imaging System (LI-COR Biosciences) or an Azure Sapphire Biomolecular Imager. Images were quantified using LI-COR Image Studio Lite.
Averages of three independent biological replicates are presented with means ± SD. The p values were calculated using Student’s t test: *p between 0.05 and 0.01 and **p ≤ 0.01.
Molecular modeling
Molecular graphics and analyses were performed with the UCSF Chimera package (Pettersen et al., 2004). The following Cryo-EM models were used for analysis: S. cerevisiae TORC2 (PDB accession number 6EMK) (Karuppasamy et al., 2017); human mTORC2 (PDB accession number 6ZWM) (Scaiola et al., 2020); and human mTORC1 (PDB accession number 6BCX) (Yang et al., 2017).
Supplementary Material
Acknowledgments
We thank A. Cuyegkeng for assistance with sequence alignments and modeling using UCSF Chimera, A. Cuyegkeng and M. Cadden for help with early designs of TOR2 MAS domain chimeras, and R. Loewith for the avo3-ΔCT strain. We thank E. Baldwin, K. Kaplan, and J. Nunnari for discussions and E. Baldwin, C. Fraser, M. Hall, R. Loewith, and K. Shiozaki for comments on the manuscript. This work was supported by research funds provided by the Department of Molecular and Cellular Biology and the College of Biological Sciences (to T. P.). Chimera was developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institute of General Medical Sciences P41-GM103311).
Abbreviations used:
- TOR
target of rapamycin
- TORC1
TOR complex 1
- TORC2
TOR complex 2.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-12-0611) on March 16, 2022.
REFERENCES
- Andrade MA, Bork P (1995). HEAT repeats in the Huntington’s disease protein. Nat Genet 11, 115–116. [DOI] [PubMed] [Google Scholar]
- Andrade MA, Petosa C, O’Donoghue SI, Müller CW, Bork P (2001). Comparison of ARM and HEAT protein repeats. J Mol Biol 309, 1–18. [DOI] [PubMed] [Google Scholar]
- Aylett CHS, Sauer E, Imseng S, Boehringer D, Hall MN, Ban N, Maier T (2016). Architecture of human mTOR complex 1. Science 351, 48–52. [DOI] [PubMed] [Google Scholar]
- Baretić D, Berndt A, Ohashi Y, Johnson CM, Williams RL (2016). Tor forms a dimer through an N-terminal helical solenoid with a complex topology. Nat Commun 7, 11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosotti R, Isacchi A, Sonnhammer EL (2000). FAT: a novel domain in PIK-related kinases. Trends Biochem Sci 25, 225–227. [DOI] [PubMed] [Google Scholar]
- Capra EJ, Perchuk BS, Skerker JM, Laub MT (2012). Adaptive mutations that prevent crosstalk enable the expansion of paralogous signaling protein families. Cell 150, 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casini A, Storch M, Baldwin GS, Ellis T (2015). Bricks and blueprints: methods and standards for DNA assembly. Nat Rev Mol Cell Biol 16, 568–576. [DOI] [PubMed] [Google Scholar]
- Chen X, Liu M, Tian Y, Li J, Qi Y, Zhao D, Wu Z, Huang M, Wong CCL, Wang HW, et al. (2018). Cryo-EM structure of human mTOR complex 2. Cell Res 28, 518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Juan D, Pazos F, Valencia A (2013). Emerging methods in protein co-evolution. Nat Rev Genet 14, 249–261. [DOI] [PubMed] [Google Scholar]
- Dilova I, Aronova S, Chen JC-Y, Powers T (2004). Tor signaling and nutrient-based signals converge on Mks1p phosphorylation to regulate expression of Rtg1.Rtg3p-dependent target genes. J Biol Chem 279, 46527–46535. [DOI] [PubMed] [Google Scholar]
- Eltschinger S, Loewith R (2016). TOR complexes and the maintenance of cellular homeostasis. Trends Cell Biol 26, 148–159. [DOI] [PubMed] [Google Scholar]
- Gaubitz C, Oliveira TM, Prouteau M, Leitner A, Karuppasamy M, Konstantinidou G, Rispal D, Eltschinger S, Robinson GC, Thore S, et al. (2015). Molecular basis of the rapamycin insensitivity of target of rapamycin complex 2. Mol Cell 58, 977–988. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350, 87–96. [DOI] [PubMed] [Google Scholar]
- Guarente L (1993). Synthetic enhancement in gene interaction: a genetic tool come of age. Trends Genet 9, 362–366. [DOI] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K (2002). Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189. [DOI] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN (1991). Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253, 905–909. [DOI] [PubMed] [Google Scholar]
- Helliwell SB, Wagner P, Kunz J, Deuter-Reinhard M, Henriquez R, Hall MN (1994). TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol Biol Cell 5, 105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A, Niles B, Cuyegkeng A, Powers T (2018). Redesigning TOR kinase to explore the structural basis for TORC1 and TORC2 assembly. Biomolecules 8, E36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Rüegg MA, Hall A, Hall MN (2004). Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 6, 1122–1128. [DOI] [PubMed] [Google Scholar]
- Kaiser CA, Schekman R (1990). Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 61, 723–733. [DOI] [PubMed] [Google Scholar]
- Karuppasamy M, Kusmider B, Oliveira TM, Gaubitz C, Prouteau M, Loewith R, Schaffitzel C (2017). Cryo-EM structure of Saccharomyces cerevisiae target of rapamycin complex 2. Nat Commun 8, 1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-H, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (2002). mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175. [DOI] [PubMed] [Google Scholar]
- Kim D-H, Sarbassov DD, Ali SM, Latek RR, Guntur KVP, Erdjument-Bromage H, Tempst P, Sabatini DM (2003). GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell 11, 895–904. [DOI] [PubMed] [Google Scholar]
- Kim SG, Hoffman GR, Poulogiannis G, Buel GR, Jang YJ, Lee KW, Kim BY, Erikson RL, Cantley LC, Choo AY, Blenis J (2013). Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol Cell 49, 172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson BA (2010). Insights into the domain and repeat architecture of target of rapamycin. J Struct Biol 170, 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN (2002). Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 10, 457–468. [DOI] [PubMed] [Google Scholar]
- Magaway C, Kim E, Jacinto E (2019). Targeting mTOR and metabolism in cancer: lessons and innovations. Cells 8, E1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA, Gelfand MS (2002). Using orthologous and paralogous proteins to identify specificity-determining residues in bacterial transcription factors. J Mol Biol 321, 7–20. [DOI] [PubMed] [Google Scholar]
- Mossmann D, Park S, Hall MN (2018). mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat Rev Cancer 18, 744–757. [DOI] [PubMed] [Google Scholar]
- Perry J, Kleckner N (2003). The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell 112, 151–155. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Reinke A, Anderson S, McCaffery JM, Yates J, Aronova S, Chu S, Fairclough S, Iverson C, Wedaman KP, Powers T (2004). TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem 279, 14752–14762. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim D-H, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (2004). Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 14, 1296–1302. [DOI] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM (2017). mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaiola A, Mangia F, Imseng S, Boehringer D, Berneiser K, Shimobayashi M, Stuttfeld E, Hall MN, Ban N, Maier T (2020). The 3.2-Å resolution structure of human mTORC2. Sci Adv 6, eabc1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick S, Rendeiro AF, Runggatscher K, Ringler A, Boidol B, Hinkel M, Májek P, Vulliard L, Penz T, Parapatics K, et al. (2019). Systematic characterization of BAF mutations provides insights into intracomplex synthetic lethalities in human cancers. Nat Genet 51, 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F (1991). Getting started with yeast. Methods Enzymol 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuttfeld E, Aylett CH, Imseng S, Boehringer D, Scaiola A, Sauer E, Hall MN, Maier T, Ban N (2018). Architecture of the human mTORC2 core complex. eLife 7, e33101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafur L, Kefauver J, Loewith R (2020). Structural Insights into TOR Signaling. Genes (Basel) 11, E885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Wang RC, Takai KK, Yang H, de Lange T (2007). Tel2 regulates the stability of PI3K-related protein kinases. Cell 131, 1248–1259. [DOI] [PubMed] [Google Scholar]
- Takai H, Xie Y, de Lange T, Pavletich NP (2010). Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev 24, 2019–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J, Kube M, Luck K, Schick S (2021). The BAF chromatin remodeling complexes: structure, function, and synthetic lethalities. Biochem Soc Trans 49, 1489–1503. [DOI] [PubMed] [Google Scholar]
- Wedaman KP, Reinke A, Anderson S, Yates J, McCaffery JM, Powers T (2003). Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol Biol Cell 14, 1204–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN (2006). TOR signaling in growth and metabolism. Cell 124, 471–484. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Oppliger W, Hall MN (2005). Molecular organization of target of rapamycin complex 2. J Biol Chem 280, 30697–30704. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang J, Liu M, Chen X, Huang M, Tan D, Dong MQ, Wong CC, Wang J, Xu Y, Wang HW, et al. (2016). 4.4 Å resolution cryo-EM structure of human mTOR complex 1. Protein Cell 7, 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Jiang X, Li B, Yang HJ, Miller M, Yang A, Dhar A, Pavletich NP (2017). Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature 552, 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP (2013). mTOR kinase structure, mechanism and regulation. Nature 497, 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.