Abstract
Germ granules are cytoplasmic assemblies of RNA-binding proteins (RBPs) required for germ cell development and fertility. During the first four cell divisions of the Caenorhabditis elegans zygote, regulated assembly of germ (P) granules leads to their selective segregation to the future germ cell. Here we investigate the role of DLC-1, a hub protein implicated in stabilization and function of diverse protein complexes, in maintaining P granule integrity. We find that DLC-1 directly interacts with several core P granule proteins, predominantly during embryogenesis. The loss of dlc-1 disrupts assembly of P granule components into phase-separated organelles in the embryos, regardless of whether or not DLC-1 directly interacts with these proteins. Finally, we infer that P granule dispersal in the absence of dlc-1 is likely independent of DLC-1’s function as a subunit of the dynein motor and does not result from a loss of cell polarity.
INTRODUCTION
RNA/protein granules are membraneless organelles serving as hubs for RNA metabolism and implicated in development and disease (Shin and Brangwynne, 2017; Gomes and Shorter, 2019). Assembly of protein and RNA constituents of the granules leads to their demixing from the surrounding cytoplasm through liquid–liquid phase separation (LLPS; Banani et al., 2017; Shin and Brangwynne, 2017; Mittag and Parker, 2018). Multivalent protein–protein and protein–RNA interactions mediated by intrinsically disordered and structured domains of RNA-binding proteins (RBPs) can drive LLPS in vitro (Banani et al., 2017; Shin and Brangwynne, 2017; Mittag and Parker, 2018). Beyond these modes of interaction, tuning phase separation properties of RNA granule components through association with additional protein partners remains relatively unexplored. Here, we report that the hub protein DLC-1 (dynein light chain 1) plays an essential role in promoting in vivo assembly of Caenorhabditis elegans germ granules.
Germ (P) granules of C. elegans are a well-studied model of cytoplasmic RNA granules (Strome and Wood, 1982; Voronina, 2013; Marnik and Updike, 2019). Dynamic LLPS of embryonic P granules ensures their segregation to the posterior blastomere in the first cell division and eventually to the cells of the germline lineage (Figure 1A; Strome and Wood, 1982; Brangwynne et al., 2009). The core components of embryonic P granules include RGG domain proteins PGL-1 and its paralogue PGL-3 (Kawasaki et al., 1998, 2004), a family of germline DEAD-box RNA helicases, GLH-1, -2, -3, and -4 (Gruidl et al., 1996; Kuznicki et al., 2000), and paralogous intrinsically disordered proteins MEG-3 and MEG-4 (Wang et al., 2014). Asymmetric assembly of embryonic P granules requires the presence of multiple core components: phase separation of PGL proteins is disrupted in meg-3/meg-4 double mutants (Wang et al., 2014; Smith et al., 2016) and also in the glh-1 mutant (Kawasaki et al., 1998, 2000; Spike et al., 2008; Hanazawa et al., 2011). Conversely, pgl-1/pgl-3 double mutant embryos display defective assembly of GLH-1, MEG-3, and MEG-4 into granules (Hanazawa et al., 2011; Wang et al., 2014; Smith et al., 2016). In addition, pgl-1/-3 knockdown also led to a reduction in localization of other proteins to P granules, including IFE-1 (Amiri et al., 2001), MEX-3 (Hanazawa et al., 2011), and POS-1 (Hanazawa et al., 2011), suggesting that PGLs are important for recruiting other P granule components. The model of embryonic P granule assembly proposes asymmetric phase separation of MEG and PGL protein driven by an anterior–posterior gradient in RNA association linked to embryonic polarity regulators (Saha et al., 2016; Smith et al., 2016).
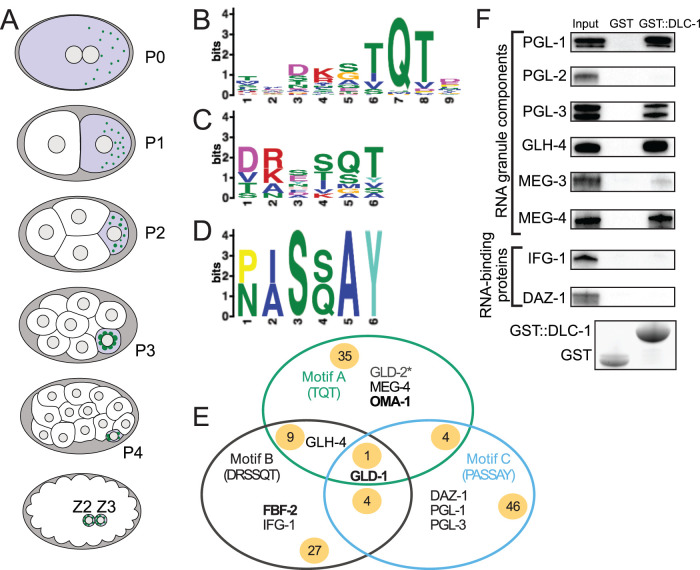
FIGURE 1:
Motif scans identify C. elegans DLC-1–interacting proteins. (A) Schematics of early C. elegans embryos from the one-cell stage to the ∼100-cell stage. Germ cells are highlighted in purple, and P granules are depicted as green puncta. (B) MEME analysis of published LC8-binding sites generates the canonical binding motif A (“TQT”) similar to the one reported previously (Rapali et al., 2011b). (C) MEME analysis of divergent LC8-binding sites including the sites from GLD-1 (Ellenbecker et al., 2019) and FBF-2 (Wang et al., 2016) generates an alternative motif B (“DRRSQT”). (D) MEME analysis of divergent LC8-binding sites including the site from GLD-1 and additional atypical LC8-binding sites from Rodríguez-Crespo et al. (2001) generates an alternative motif C (“PASSAY”). (E) Scanning the C. elegans proteome with motifs A–C returns 126 proteins previously identified as RBPs (Tamburino et al., 2013; Matia-González et al., 2015). The green, black, and blue circles represent the results from each respective motif scan. Examples of RBPs found exclusively by each motif scan are listed in each circle. Examples of RBPs found by more than one scan are shown in regions where circles overlap. The numbers on yellow background indicate how many RBPs were identified in each specific group or overlap between groups. The RBPs listed have been tested for interaction with DLC-1 in this report or previously. Bold text indicates RBPs previously found to interact with DLC-1, while an asterisk indicates no interaction with DLC-1 in a previous study. (F) GST pull downs confirm several interactions between GST::DLC-1 and RBPs identified by the proteome scan. All RBPs tested were 6xHis tagged with the exception of MEG-3, MEG-4, and DAZ-1, which were MBP tagged. RBP constructs were detected by Western blotting with anti-6xHis or anti-MBP antibodies, and GST alone or GST::DLC-1 was detected by stain-free chemistry. PGL-2 and MEG-3 were included for comparison against their paralogues PGL-1, PGL-3, and MEG-4, which were recovered with the motif scan.
Although several core P granule components can undergo LLPS as purified proteins in vitro (Elbaum-Garfinkle et al., 2015; Saha et al., 2016; Smith et al., 2016; Zhang et al., 2018; Putnam et al., 2019), genetic approaches identified a number of additional factors required for in vivo P granule assembly (Updike and Strome, 2009; Voronina and Seydoux, 2010). The genes promoting P granule formation are involved in diverse cellular processes spanning cell cycle, protein degradation, RNA splicing, translational control, and nucleocytoplasmic transport. These findings suggest that P granule assembly is not simply driven by the presence of the core components and accessible RNA but are fine-tuned by other genes and cellular processes. Here, we report the contribution of the bimolecular scaffold DLC-1 to embryonic P granule assembly in C. elegans embryos.
C. elegans DLC-1 is an LC8 family protein originally described as a subunit of the axonemal dynein motor (Pfister et al., 1982) and later found in cytoplasmic dynein motor complexes (King and Patel-King, 1995; Wilson et al., 2001). LC8 genes are highly conserved across eukaryotes (Wickstead and Gull, 2007), and are essential in C. elegans (Kamath et al., 2003), Drosophila melanogaster (Dick et al., 1996), and mice (King et al., 2019). LC8 proteins bind a large number of protein interaction partners beyond the dynein motor components (Rapali et al., 2011b) and display a broader intracellular distribution than the dynein motor (King et al., 1996; Stelter et al., 2007; Wang et al., 2016; Jespersen et al., 2019). Therefore, LC8 is thought to be a hub that facilitates the assembly of many protein complexes important for a wide variety of processes, such as transcriptional regulation, tumor suppression, and apoptosis (Barbar, 2008; Rapali et al., 2011b; Jespersen and Barbar, 2020). LC8 forms homodimers with two symmetrical grooves where its partners bind (Liang et al., 1999). By engaging both binding sites, LC8 can promote homodimerization of its interaction partners and facilitate changes in their conformation, mediating enhanced affinity for additional ligands or changes in function (Rapali et al., 2011b; Clark et al., 2015). Such conformational changes upon LC8 binding were shown for several interaction partners, including Swallow, where LC8 association with a disordered monomeric segment favors formation of a dimeric coiled-coil fold (Wang et al., 2004; Kidane et al., 2013). LC8-binding sites in partner proteins are typically short linear peptides that form a beta sheet upon binding to LC8 (Barbar, 2008). While the LC8-binding consensus motif has been proposed (Rapali et al., 2011a), not every peptide conforming to the motif can bind LC8 and some LC8-binding peptides are widely divergent (Clark et al., 2015). Despite the ever-growing list of known LC8-binding proteins, the field is far from capturing the diversity of LC8 partners and therefore the full range of functions that LC8 provides to the cells is still unknown.
We previously identified C. elegans LC8 protein DLC-1 as a cofactor for the germline RBPs FBF-2 (Wang et al., 2016) and GLD-1 (Ellenbecker et al., 2019). DLC-1 promoted the function of these RBPs and the localization of FBF-2 to P granules. Through RIP-seq, we found that DLC-1 associates with more than 2700 mRNAs in C. elegans (Day et al., 2018), suggesting that DLC-1 might interact with many RBPs. In this report, we use an in silico interaction motif scanning approach to predict additional RBPs interacting with DLC-1. We find that DLC-1 directly interacts with several P granule components and is important for P granule assembly in embryos. Our findings highlight the importance of cofactors in regulating in vivo phase separation of germ granules.
RESULTS
In silico and in vitro identification of RBPs that bind DLC-1
Given the high number of DLC-1–associated mRNAs identified using RIP-seq (Day et al., 2018), we hypothesized that DLC-1 interacts with multiple C. elegans RBPs in addition to the previously described association with FBF-2 (Wang et al., 2016) and GLD-1 (Ellenbecker et al., 2019). To predict additional DLC-1–interacting RBPs, we used a bioinformatic approach to identify RBPs containing LC8 interaction motifs with the online MEME tool (Bailey, 2011; details in Materials and Methods). Attempting to represent the diversity of DLC-interacting peptides, we used different combinations of LC8-interacting peptides to generate and test three possible DLC-1–interacting motifs (Figure 1, B–D; details in Materials and Methods). Motif A contains the conserved “TQT” residues and resembles the canonical LC8 interaction motif (Rapali et al., 2011b) (Figure 1B). This motif best represents the peptides containing “TQT” residues present in more than half of input sequences but minimally reflects divergent DLC-1–interacting peptides. Motifs B and C deemphasize the overrepresented “TQT” residues of motif A and reflect greater diversity of LC8 interaction sites.
Motifs A–C were then used to scan the C. elegans proteome to identify proteins that contained at least one instance of one of these interaction motifs. To identify RBPs, the output from these proteome scans was compared against comprehensive lists of C. elegans RBPs (Tamburino et al., 2013; Matia-González et al., 2015). Together, scans with these three motifs yielded a total of 108 RBPs predicted to interact with DLC-1 by a single motif scan each and an additional 18 RBPs that were identified by more than one motif scan (Figure 1E; Supplemental Table 2). Motif scans recovered known DLC-1 RBP interactors GLD-1 (Ellenbecker et al., 2019) and FBF-2 (Wang et al., 2016). In addition, several DLC-1 interactors predicted by the motif scan approach have been previously recovered by high-throughput yeast two-hybrid screens (Li et al., 2004; Simonis et al., 2009). We conclude that motif scans are capable of retrieving DLC-1 interaction partners. Surprisingly, all three motif scans identified multiple putative DLC-1 interactors that are also components of P granules, including GLD-1 (Jones et al., 1996), PGL-1 (Kawasaki et al., 1998), PGL-3 (Kawasaki et al., 2004), GLH-4 (Kuznicki et al., 2000), MEG-4 (Wang et al., 2014), and GLD-2 (Wang et al., 2002). P granules are non–membrane bound cytoplasmic assemblies of RNAs and RBPs required for fertility and posttranscriptional mRNA regulation; however, the mechanism of their assembly is not fully understood. The emerging role of DLC-1 as a hub for protein complex assembly led us to test whether it directly interacts with P granule RBPs, which might indicate its role in P granule assembly.
To determine whether DLC-1 might directly interact with predicted RBP partners, DLC-1 fused to GST and six putative interacting RBPs tagged with 6xHis or maltose-binding protein (MBP) were expressed in bacteria and tested for direct interaction using in vitro GST pull downs. Tested RBPs included PGL-1, PGL-3, GLH-4, MEG-4, IFG-1, and DAZ-1. Four of these (PGL-1, PGL-3, GLH-4, MEG-4) directly interacted with DLC-1 (Figure 1F). Interaction between DLC-1 and PGL-3 was previously observed in a high-throughput yeast two-hybrid screen (Li et al., 2004). DAZ-1 did not directly interact with DLC-1, suggesting that a reported interaction between the mouse orthologues Dazl and LC8 (Lee et al., 2006) is not conserved, and neither did IFG-1. PGL and MEG proteins are members of protein families containing several paralogous proteins each, so we investigated whether the paralogues not recovered by motif scans were able to interact with DLC-1. MEG-3, a paralogue of MEG-4 with 71% identity (Wang et al., 2014), weakly interacted with DLC-1 compared to MEG-4 (Figure 1F). By contrast, PGL-2 did not interact with DLC-1 at all (Figure 1F). In our previous attempts to identify DLC-1–interacting RBPs based on the overlap of mRNA targets, GST pulldowns confirmed 37% of predicted binding partners (Day et al., 2018; N. Day, unpublished). By contrast, in vitro interaction testing of RBPs with a putative DLC-1–binding motif validated 67% of the candidates (Figure 1F). We conclude that bioinformatic analysis has successfully enriched DLC-1–interacting proteins within the list of putative binding partners.
DLC-1 is incorporated into PGL-1–containing complexes in the adult germline
To test whether DLC-1-RBP interactions identified in vitro are observed in vivo, we used an in situ approach to observe and quantify these interactions. For these experiments, we focused on core P granule components PGL-1 and PGL-3 (Kawasaki et al., 1998, 2004). We first confirmed that DLC-1 is coexpressed with PGL-1 and PGL-3 in the adult germline using coimmunostaining (Supplemental Figure S1). 3xFLAG::DLC-1 was uniformly expressed throughout the germline as previously reported (Dorsett and Schedl, 2009; Wang et al., 2016; Day et al., 2020) (Supplemental Figure S1, Aii, Bii, and Cii). GFP alone used as a control was uniformly distributed throughout the germline (Supplemental Figure S1Aiii). Expression patterns of GFP-tagged PGL-1 and PGL-3 in the germline (Supplemental Figure S1, Biii and Ciii) showed both proteins localized to P granules in germ cells and oocytes, with PGL-1 expressed throughout the germline and PGL-3 expressed from the midpachytene region to the oocytes, as previously observed (Kawasaki et al., 2004). Both PGL-1 and PGL-3 are coexpressed with DLC-1, suggesting that they could interact in the germline.
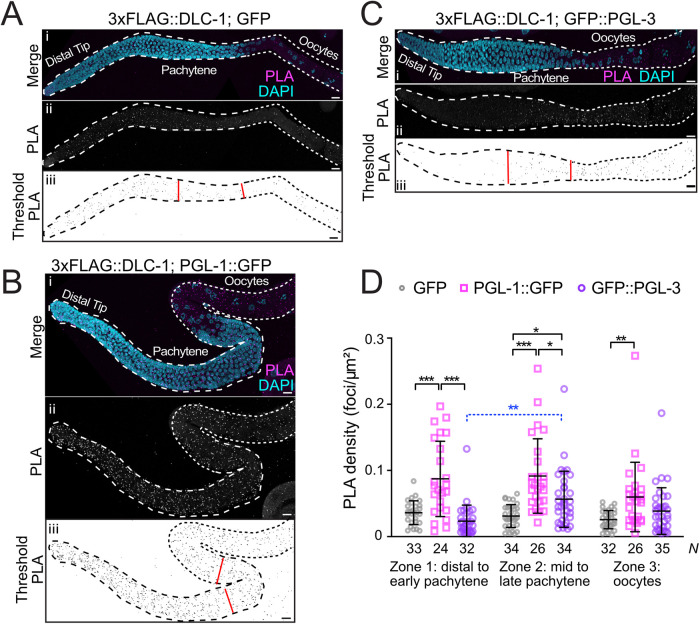
To visualize and quantify interactions between DLC-1 and PGL-1 or PGL-3 in vivo, we carried out a Duolink in situ proximity ligation assay (PLA), a procedure that produces fluorescent signal when the target epitopes are closer than 40 nm in situ (Bagchi et al., 2015). We performed PLA in strains coexpressing 3xFLAG::DLC-1 with GFP-tagged PGL-1 or PGL-3 using the same anti-FLAG and anti-GFP primary antibodies that were used for immunofluorescence and quantitatively analyzed the signal following the previously described workflow (Day et al., 2020) (Figure 2). As a negative control, we used the animals coexpressing 3xFLAG::DLC-1 with GFP, as DLC-1 and GFP are not expected to interact and any PLA signal produced in this strain would be due to spurious proximity between the epitopes. For quantification each germline was separated into three zones, where zone 1 comprised the distal end to early pachytene, zone 2 spanned mid to late pachytene, and zone 3 included all oocytes (demarcated by red lines in subpanels iii of Figure 2). Among the three protein pairs tested, 3xFLAG::DLC-1; PGL-1::GFP germlines had the highest mean PLA density in zone 1 (P < 0.0001; Figure 2D). By contrast, the PLA density in 3xFLAG::DLC-1; GFP::PGL-3 germlines was at the background level in zone 1 but showed a moderate yet statistically significant increase in zone 2 (P < 0.0167; Figure 2D). This is likely explained by the expression pattern of PGL-3, which starts in zone 2 at the midpachytene (Supplemental Figure S1Ciii). PGL-1’s PLA density in zone 2 was still significantly higher compared to PGL-3’s (P < 0.0167), suggesting that DLC-1 predominantly interacts with PGL-1 in this zone. PLA density was also quantified in the oocytes (zone 3, Figure 2D), where P granules change their localization from perinuclear to cytoplasmic during oogenesis and oocyte maturation (Strome and Wood, 1982; Pitt et al., 2000). While the mean PLA densities for 3xFLAG::DLC-1; PGL-1::GFP were similar in zones 1 and 2, we observed a more than 30% decrease in oocytes of zone 3 (Figure 2D). 3xFLAG::DLC-1; GFP::PGL-3 also exhibited a 30% reduction in mean PLA density for zone 3 compared to zone 2, resulting in PLA density close to background. On the basis of these data, we conclude that DLC-1 preferentially interacts with PGL-1 over PGL-3 in the germline. Further, the differences in PLA density between coexpressed protein pairs and the decrease in PLA density in the oocytes suggest that coexpression of partner proteins is not the sole determinant of DLC-1/PGL interactions in the germline.
FIGURE 2:
Proximity ligation detects formation of DLC-1/PGL complexes in the adult germline. (A–C) Extruded adult germlines of 3xFLAG::DLC-1; GFP (A), 3xFLAG::DLC-1; PGL-1::GFP (B), and 3xFLAG::DLC-1; GFP::PGL-3 (C) with PLA in magenta and DNA labeled with DAPI (4′,6-diamidino-2-phenylindole; cyan). The individual PLA channels (Aii, Bii, Cii) are also shown in grayscale for better contrast. For quantification, the PLA foci in the grayscale PLA channels were subjected to the particle thresholding procedure (Aiii, Biii, Ciii). Red lines separate the three zones used for quantitative analysis in D. In each image, the stem cells and meiotic pachytene are outlined with dashed lines, while the oocytes are outlined with dotted lines. Images were acquired with a confocal microscope. Scale bars are 10 µm. (D) The PLA density (number of PLA foci per μm2) was measured for germlines of each genetic background segmented into three zones (see Materials and Methods) as denoted by the red lines (Aiii, Biii, Ciii). Average and SD are indicated for each column. The number of germlines analyzed (N) for each strain in each zone is shown below the graph. Differences in PLA density for each protein pair and each zone were evaluated by one-way analysis of variance (ANOVA) followed by Sidak’s multiple comparison test. The sole significant cross-zone difference in PLA density is shown with a blue dashed bracket. Asterisks denote statistical significance (***, P < 0.001; **, P < 0.01; *, P < 0.05). Data are representative of three biological replicates.
DLC-1 is incorporated into PGL-containing RNPs in embryos
We further investigated potential interactions between DLC-1 and PGL-1 or PGL-3 in developing embryos. We first established that 3xFLAG::DLC-1 is coexpressed with GFP alone or PGL-1::GFP and GFP::PGL-3 throughout embryonic development (Supplemental Figure S2). Interestingly, anti-FLAG immunostaining in embryos at the ∼100-cell stage shows that 3xFLAG::DLC-1 is enriched in primordial germ cells Z2 and Z3 (yellow asterisk, Supplemental Figure S2, D, H, and L), also recognized by the localization of PGL-1 or PGL-3. The overlapping patterns of expression of DLC-1 in somatic and germ cells and PGL-1 and PGL-3 in germ cells suggest that DLC-1 could interact with these RBPs in developing embryos.
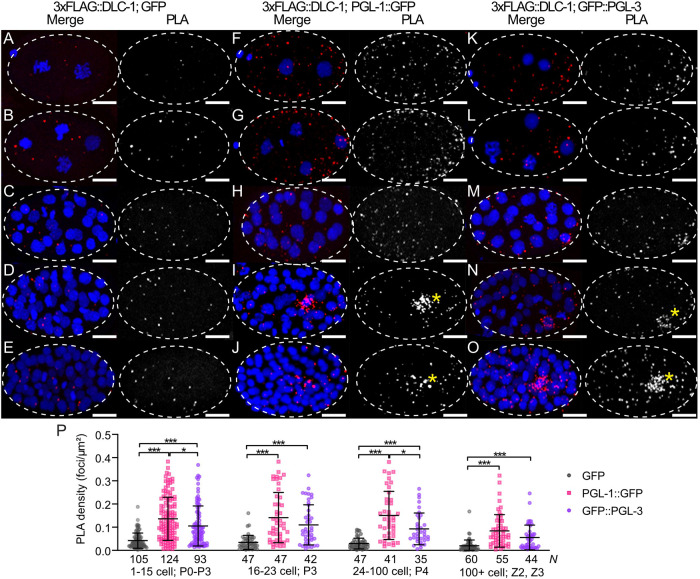
Using PLA, we quantified interactions between DLC-1 and PGL-1 or PGL-3 in embryos across several different stages of development grouped based on cytoplasmic or perinuclear P granule localization and the germ cell identity (Figure 3). At every stage, the average PLA densities observed in 3xFLAG::DLC-1; PGL-1::GFP and 3xFLAG::DLC-1; GFP::PGL-3 embryos were significantly higher than in the negative control (P < 0.001; Figure 3P). Meanwhile, we observed only minor differences between the PLA densities for DLC-1/PGL-1 and DLC-1/PGL-3 complexes. The average PLA density values for both PGL-1 and PGL-3 in the embryo were higher than those observed in the adult germlines (Figure 2D), even though we quantified PLA density across the whole embryo rather than just in the germ cell. This suggests that incorporation of DLC-1 into PGL-1 or PGL-3 RNPs is more prevalent in embryos than in the germlines or oocytes. Surprisingly, PLA foci for 3xFLAG::DLC-1 with PGL-1::GFP or GFP::PGL-3 were present throughout the early embryos, including both somatic and germ cells (Figure 3, F–O) in contrast to the dramatic accumulation of PGL-1 and PGL-3 in P granules in embryonic germ cells (Supplemental Figure S2, column iii). We concluded that DLC-1 might be a part of PGL-1 and PGL-3 RNP complexes not only in germ cells but also in somatic cells (Hird et al., 1996; Zhang et al., 2009). Interestingly, in later-stage embryos DLC/PGL PLA foci appeared enriched in the germ cells (yellow asterisks, Figure 3, I, J, N, and O), which we quantitatively analyzed next.
FIGURE 3:
DLC-1 forms complexes with PGL-1 and PGL-3 in early embryos. (A–O) PLA (red) in 3xFLAG::DLC-1; GFP (A–E), 3xFLAG::DLC-1; PGL-1::GFP (F-J), and 3xFLAG::DLC-1; GFP::PGL-3 (K–O) embryos. Rows 1 (A, F, and K) and 2 (B, G, and L): two- and four-cell-stage embryos, where cytoplasmic P granules segregate into the germ cell P1 or P2. Row 3 (C, H, and M): 16–23-cell embryos, where P granules transition to the perinuclear location in the late P3 germ cell. Row 4 (D, I, and N): 24–100-cell embryos, where P granules are located at the nucleus of the P4 germ cell. Row 5 (E, J, and O): embryos at ∼100-cell stage, where autophagy clears PGL proteins from somatic cells, with P granules remaining in Z2 and Z3 primordial germ cells. DNA is labeled with DAPI (blue in the merged images). The PLA channels are also shown in grayscale for better contrast. Images were acquired using a confocal microscope. Yellow asterisks in I, J, N, and O indicate PLA foci enriched in germ cells. Embryos in this and the following figures are oriented with anterior to the left. Scale bars = 10 µm. (P) PLA density (PLA foci per μm2) was measured within each whole embryo coexpressing 3xFLAG::DLC-1 with either of GFP (negative control), PGL-1::GFP, or GFP::PGL-3. The values were subsequently binned into groups based on developmental stage of the embryo (as indicated along the X-axis). Average and SD are indicated for each column. N values for each strain and developmental stage are shown under each column. Differences in PLA density for each protein pair were evaluated by one-way ANOVA followed by Sidak’s multiple comparison test. Asterisks denote statistical significance (***, P < 0.001; *, P < 0.05). Data are representative of four biological replicates.
DLC-1/PGL RNPs are enriched in germ cells
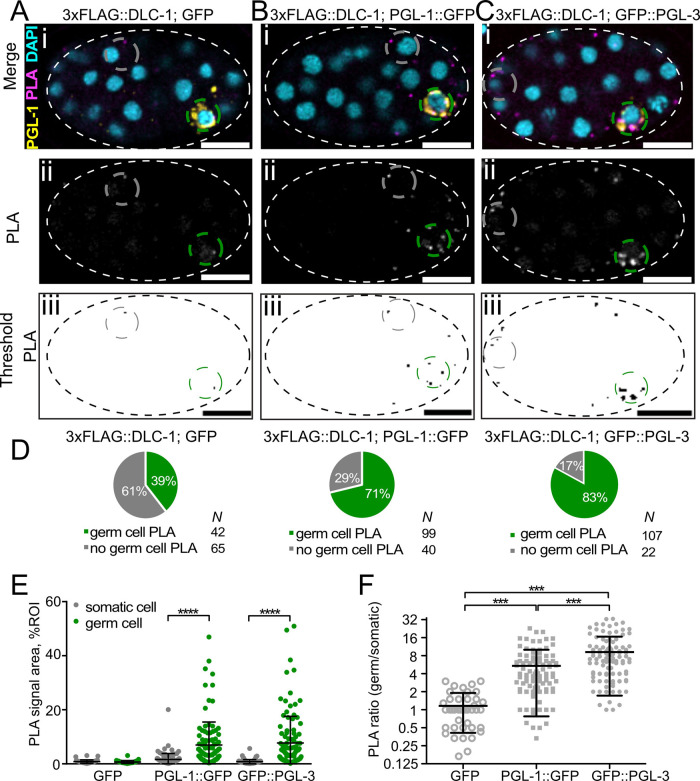
PGL proteins that partition to somatic cells during early embryonic cell divisions are gradually cleared by autophagy (Zhang et al., 2009). To evaluate whether there is a preference for DLC-1/PGL complex formation in the germ cells, we evaluated distribution of PLA foci in the P4-stage embryos, before substantial clearance of PGL proteins in the somatic cells. Using PLA in tandem with immunostaining against PGL-1 to mark P granules (Figure 4, A–C), we first scored how often PLA foci were observed in the germ cells (Figure 4D). Both 3xFLAG::DLC-1; GFP::PGL-3 and 3xFLAG::DLC-1; PGL-1::GFP embryos had a high prevalence of PLA foci in germ cells (83% and 71% of embryos, respectively). By contrast, the prevalence of germ cell PLA foci was lower in 3xFLAG::DLC-1; GFP embryos (39%; Figure 4D) and significant differences in the proportion of embryos with germ cell PLA foci were revealed by a Chi-square test (P < 10–19 for PGL-1::GFP, P < 10–29 for GFP::PGL-3 vs. GFP alone). We conclude that PGL-3/DLC-1 and PGL-1/DLC-1 complexes are both present in germ cells above background level. To evaluate whether either of the PGL/DLC complexes was enriched in germ cells to a greater extent, we quantitatively compared the germ cell PLA signal to that observed in the somatic cells.
FIGURE 4:
DLC-1/PGL complexes are enriched in P4 and Z2/Z3 germ cells. (A–C) PLA foci (magenta) in 3xFLAG::DLC-1; GFP (A), 3xFLAG::DLC-1; PGL-1::GFP (B), and 3xFLAG::DLC-1; GFP::PGL-3 (C) single confocal planes of embryos at approximately the 40-cell stage. DNA is labeled with DAPI (cyan). P granules (yellow) are immunostained by anti–PGL-1. The PLA channels are also shown in grayscale (Aii, Bii, Cii) for better contrast. For quantification, the PLA foci in the grayscale PLA channels of each embryo image were subject to particle thresholding (Aiii, Biii, Ciii). The green dashed circle delineates the germ cell ROI, while the gray dashed circle delineates the somatic cell. Scale bars = 10 µm. (D–F) Quantitative analysis of PLA. The PLA signal in test and control strains was evaluated by three different metrics using the same embryo images for all analyses. PGL-1::GFP data were collected across five biological replicates, GFP::PGL-3 data in four biological replicates, and GFP data in three biological replicates. (D) DLC-1/PGL complexes are present in germ cells. Pie charts represent proportions of embryos that had PLA foci present in germ cells. The number of observations (N) for each group is shown next to each pie chart legend. (E) The relative area occupied by the PLA signal of DLC-1/PGL complexes is greater in the germ cells than in the somatic cells. The relative PLA area in somatic and germ cells (gray and green circles) is plotted for embryos of each strain. The embryos with no signal in the germ cells were excluded from this data set. Average and SD are shown for each column. Differences in relative PLA areas were evaluated by one-way ANOVA followed by comparisons of somatic vs. germ cells for each strain with Sidak’s multiple comparison test (****, P < 0.0001). (F) The germ cell enrichment of DLC-1/PGL-3 complex is greater than that of the DLC-1/PGL-1 complex. The ratios of relative PLA areas in germ cell over somatic cell of the same embryo are plotted for each strain (the Y-axis is log-scale). Average and SD are indicated for each column. Differences in germ/somatic cell PLA area ratio for each target protein were evaluated by one-way ANOVA followed by Tukey multiple comparison test. Asterisks denote statistical significance (***, P < 0.001).
To quantify germ cell PLA signal, we measured the relative area that PLA foci occupy in germ and somatic cells of the same embryo (green vs. gray dashed outlines in Figure 4, A–C). This metric was more informative than PLA density as it reflected the differences in the size of PLA foci and was robust to merging of PLA foci. These measurements were performed only on embryos where PLA signal was present in the germ cells (Figure 4D). The average relative areas occupied by PLA foci in somatic and germ cells were not significantly different in the GFP control (Figure 4E). By contrast, for both PGL-1::GFP and GFP::PGL-3, the relative PLA area in germ cells was significantly greater than in somatic cells (P < 0.0001; Figure 4E). We concluded that DLC-1/PGL–containing RNPs are enriched in the germ cells despite the substantial presence of PGL proteins in the somatic cells at this stage, confirming qualitative observations. To determine whether one of the DLC-1/PGL complexes might be enriched in germ cells to a greater extent, we calculated the ratios between relative PLA areas in the germ and somatic cells (Figure 4F) for each individual embryo plotted in Figure 4E. We found that 3xFLAG::DLC-1; GFP::PGL-3 had significantly higher germ cell/soma PLA enrichment compared to the 3xFLAG::DLC-1; PGL-1::GFP, while both DLC-1/PGL complexes were enriched in the germ cells more than the DLC-1/GFP control (P < 0.001; Figure 4F). We conclude that both DLC-1/PGL complexes are significantly enriched in the germ cells and the DLC-1/PGL-3 RNP complex might be more selective for the germ cells compared to the somatic cells.
Loss of DLC-1 disrupts P granule assembly in the embryo
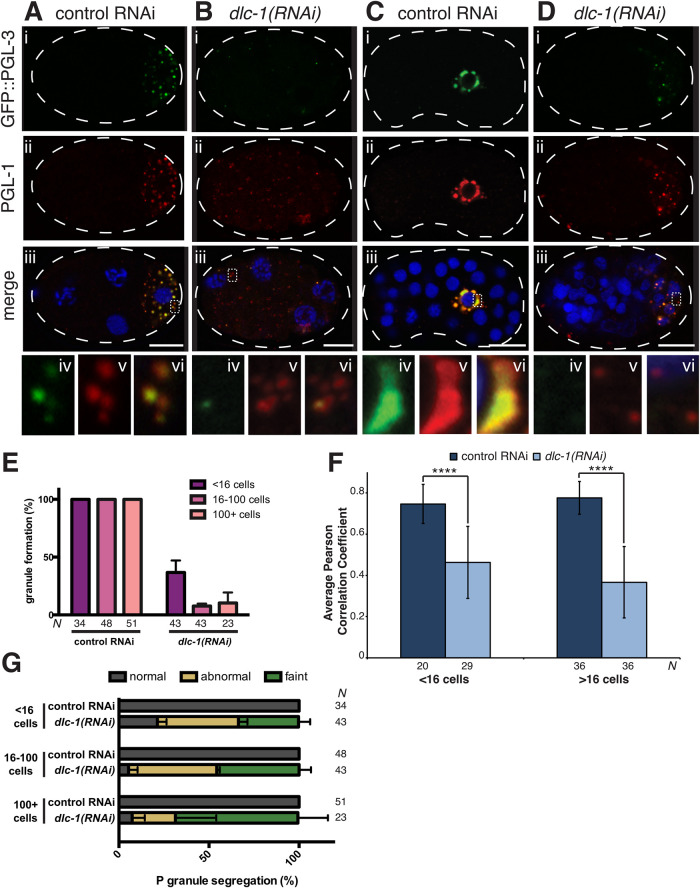
Because DLC-1 interacts with multiple core P granule components in vitro (Figure 1F) and is incorporated into RNP complexes with PGL-1 and PGL-3 in vivo (Figures 2–4), we hypothesized that loss of DLC-1 might disrupt the assembly of PGLs into P granules. To test this hypothesis, we documented the effect of dlc-1 knockdown on PGL-1 and PGL-3. RNAi-mediated dlc-1 knockdown at 20°C results in 100% embryonic lethality. We found that PGL protein assembly into granules is severely disrupted in both early and later-stage embryos following dlc-1(RNAi) (Figure 5, B and D). Many embryos had completely dispersed faint PGL signal, and in cases where PGL granules assembled, they were smaller than in the control (compare Figure 5, Cvi and Dvi) and in some instances contained only one of the PGL proteins (Figure 5, Bvi and Dvi). Scoring P granule formation in the dlc-1(RNAi) embryos revealed significant disruption of P granule assembly across developmental stages (Figure 5E). Quantitative evaluation of PGL-1/PGL-3 colocalization by Pearson’s correlation coefficient analysis based on Costes’ automatic threshold (Costes et al., 2004) revealed that the PGL-1/PGL-3 correlation coefficient was significantly reduced in both early and late dlc-1(RNAi) embryos compared to the control (P < 0.0001; Figure 5F). This reduction in PGL-1/PGL-3 colocalization upon the loss of dlc-1 was intriguing, given that PGL-1 and PGL-3 directly interact in vitro (Kawasaki et al., 2004). Additionally, we observed frequent missegregation of PGL proteins into multiple embryonic cells (Figure 5, D and G). By contrast, PGL protein assembly into perinuclear granules in the adult germline was not affected by dlc-1(RNAi), as reported previously (Supplemental Figure S3) (Wang et al., 2016). We concluded that DLC-1 promotes PGL-1/3 assembly into P granules in the embryo.
FIGURE 5:
dlc-1 is required for PGL-1 and PGL-3 assembly into embryonic P granules. (A–D) P granule components PGL-1 (red) and GFP::PGL-3 (green) detected by immunostaining of control or dlc-1(RNAi) embryos. (A, B) Four-cell embryos. (C, D) Forty-cell embryos. DNA is labeled with DAPI (blue). Panels iv–vi are zoomed-in regions (dotted outlines) of panel iii to highlight differences in P granule assembly and size between control RNAi and dlc-1(RNAi)–treated embryos. Images were acquired using a confocal microscope. Scale bars in i–iii are 10 µm. (E) P granules are disrupted in a large fraction of dlc-1(RNAi) embryos. Plotted are the average percents of embryos with distinct cytoplasmic PGL-1 foci following control or dlc-1(RNAi). Error bars represent the SD, and the number of embryos observed (N) for each condition is indicated below each bar. Data were collected in three biological replicates. (F) Pearson’s correlation analysis quantifying colocalization between PGL-1 and GFP::PGL-3 in wild-type and dlc-1(RNAi) embryos. Plotted values are means ± SD. The difference between the control and dlc-1(RNAi) was significant in both early and later-stage embryos. The P values were determined using a two-tailed/equal variance t test where **** = P < 0.0001. The number of embryos observed (N) in each RNAi experiment is noted under each bar. Data were collected in three biological replicates. (G) Defects in P granule segregation are frequently observed in dlc-1(RNAi) embryos. P granule segregation was classified as normal for each developmental stage (gray) or abnormal with P granules found in somatic cells (dark olive). A fraction of dlc-1(RNAi) embryos had very diffuse PGL-1 signal, where it was hard to ascertain the fidelity of segregation into specific cells (faint, light olive color). Plotted values are means ± SD. The number of embryos observed (N) in each RNAi condition is indicated next to each bar. The same embryos were scored to generate panels E and G.
To test whether dlc-1(RNAi) leads to a reduction in the levels of PGL proteins, we documented the levels of endogenous PGL-1 and transgenic GFP::PGL-3 in the control and dlc-1(RNAi) embryos. We find that the levels of PGL-1 and GFP::PGL-3 in the embryo lysates are not affected by dlc-1(RNAi) (Figure 6), suggesting that the observed effect of dlc-1 depletion on PGL protein assembly is independent of any change in protein abundance.
FIGURE 6:
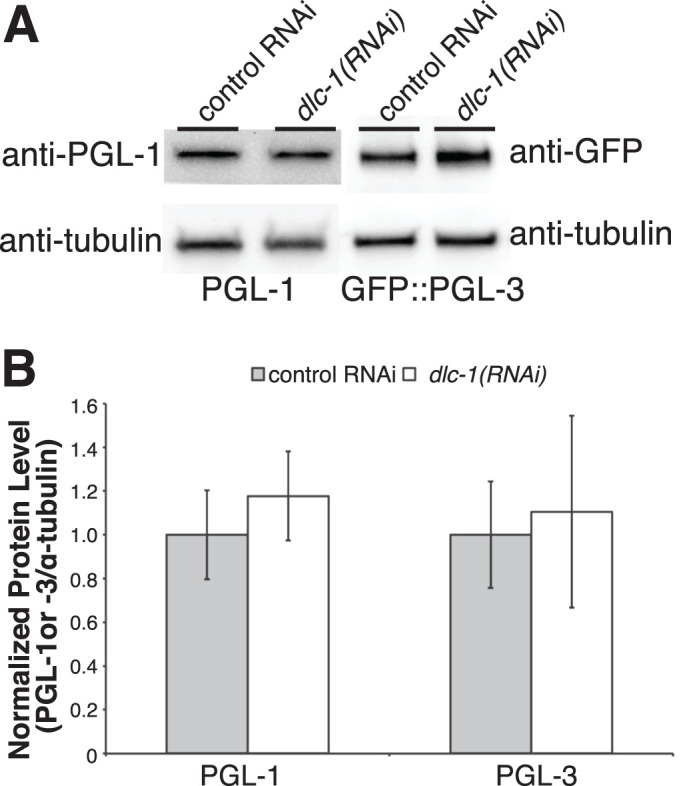
dlc-1(RNAi) does not affect the levels of PGL-1 and PGL-3 in embryos. (A) Western blot of embryo lysates following control or dlc-1(RNAi). PGL-1 and GFP::PGL-3 abundance does not decrease following dlc-1(RNAi). Tubulin is used as a loading control. (B) Average PGL-1 and GFP::PGL-3 protein levels normalized to tubulin over three biological replicates. Error bars represent the SD from the mean. No significant differences were observed between the control and dlc-1(RNAi) (PGL-1: P > 0.3; PGL-3: P > 0.7), calculated by Student’s unpaired t test.
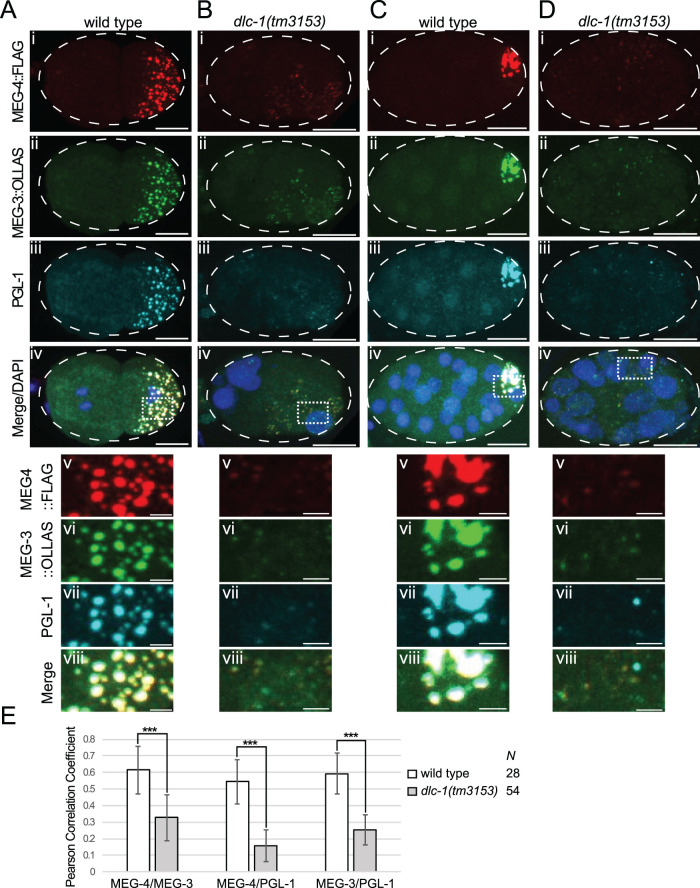
To assess whether defects in PGL-1/3 assembly into P granules upon dlc-1(RNAi) reflect more general disruption of P granule structure, we assessed the effects of dlc-1 mutation on CRISPR-tagged MEG-3::OLLAS and MEG-4::3xFLAG. By GST pull down, MEG-3 only weakly interacted with DLC-1 (Figure 1F); therefore it was not clear whether its localization and assembly into P granules might be disrupted by a loss of DLC-1. Both MEG proteins along with endogenous PGL-1 were examined in wild type or dlc-1 loss of function mutant embryos at 20°C (embryos produced by the homozygous dlc-1-mutant mothers). In dlc-1 mutants, segregation and perinuclear localization of P granules containing MEG-4, MEG-3, and PGL-1 were disrupted in both early and later-stage embryos (Figure 7, B and D). While large P granules in the wild-type embryos contain all three core P granule components (row viii, Figure 7, A and C), P granules that form in dlc-1 mutants appear smaller in size and occasionally lack some components (rows v–viii, Figure 7, B and D), suggesting that P granule proteins fail to assemble into the normal complex. For each pair of P granule proteins tested, we observed a significant decrease in the colocalization coefficient between the wild type and dlc-1 mutant (P < 0.0005; Figure 7E), in agreement with the qualitative observations. Interestingly, the dlc-1 mutation affected colocalization of MEG-3 and MEG-4 with PGL-1, even though both MEGs directly interact with PGL-1 in vitro (Wang et al., 2014). Overall, we conclude that DLC-1 promotes both assembly and localization of multiple P granule components in developing embryos, including those that do not directly interact with DLC-1.
FIGURE 7:
dlc-1 is required for embryonic P granule integrity. (A–D) Maximum-intensity projections of wild-type or dlc-1 mutant embryos coimmunostained for P granule components MEG-4::3xFLAG (red), MEG-3::OLLAS (green), and PGL-1 (cyan), respectively. DNA was labeled with DAPI (blue in the merged images). (A, B) Two- to four-cell-stage embryos. (C, D) Twenty-three- to twenty-four-cell-stage embryos. Rows v–viii are zoomed-in regions (dotted outlines in panel iv) split into single channels for clarity. Images were acquired using a confocal microscope. Scale bars: 10 µm (i–iv); 2 µm (v–viii). (E) Average colocalization coefficients (Pearson correlation based on Costes’ automatic threshold) for each indicated pair of P granule proteins in wild-type and dlc-1 mutant embryos. For each pair of P granule proteins examined, the difference between the wild type and mutant was significantly different. The P values were determined using a two-tailed, equal variance t test, *** = P < 0.0005. Error bars represent SD from the mean. The numbers of wild-type or dlc-1 mutant embryos observed (N) are denoted in the bar plot legend. The data were collected over three biological replicates.
DLC-1 function in embryonic P granule assembly likely does not reflect dynein motor activity
DLC-1 is a multifunctional protein that contributes to a variety of cellular protein complexes including the dynein motor (Rapali et al., 2011b). RNAi depletion of dynein motor components dhc-1 (the heavy chain) and dyci-1 (the intermediate chain) was previously reported to lead to P granule dispersal (Updike and Strome, 2009). Therefore, we considered whether the requirement for dlc-1 in embryonic P granule assembly was linked to its dynein motor function. However, RNA interference (RNAi) depletion of maternal dhc-1 or dyci-1 resulted in loss of oocyte production in the majority of treated animals as previously reported (Gönczy et al., 1999; Schmidt et al., 2005; unpublished data). The few dhc-1(RNAi) or dyci-1(RNAi) embryos that we were able to analyze failed cytokinesis and proceeded to massively endoreplicate DNA. At the early one-cell stage, these embryos were able to assemble cytoplasmic P granules that never enlarged with the progression of endoreplicative nuclear cycles (unpublished data). It is possible that P granule “dispersal” in these backgrounds resulted from a failure to partition the negative regulators of P granule assembly, which is normally achieved by cytokinesis. We were able to circumvent the requirement for maternal dhc-1 by disrupting dynein motor function in the embryos using a temperature-sensitive mutant allele, dhc-1(or195ts), that is inactivated within ∼1 min of an upshift to the restrictive temperature (Schmidt et al., 2005). We documented P granule phenotypes in the embryos produced by the mutant adult mothers shifted to the restrictive temperature of 26°C for 2 h by immunostaining for endogenous PGL-1 (Figure 8). Inactivation of dhc-1(or195ts) at the elevated temperature resulted in variable phenotypes along the timeline of embryonic development. The cytoplasmic P granules in the early embryos (germ cell stage P1 to early P3) were often missegregated, despite forming foci of a size similar to those observed in the control (Figure 8, Aiii, D, and E). The P granules in the 16–100-cell embryos, which are perinuclear in the late P3 or P4 control germ cell (Figure 8Bi), failed to segregate to a single embryonic blastomere and were often dispersed at the restrictive temperature in dhc-1(or195ts) (Figure 8, Biii, D, and E). By contrast, once Z2 and Z3 primordial germ cells formed in ∼100-cell embryos, presumably before the shift to a restrictive temperature, P granules were no longer affected by inactivation of dhc-1 (Figure 8, Ciii, D, and E). Conversely, P granules appeared normal in the dhc-1(or195ts) strain at the permissive temperature (Figure 8, Ai, Bi, and Ci) and in the wild-type strain at both temperatures (Supplemental Figure S4). We conclude that dynein motor is required for normal P granule segregation during embryonic cell divisions likely through its contribution to mitotic spindle positioning and embryonic polarity (Gönczy et al., 1999; Rose and Gönczy, 2014). Despite their missegregation in the early embryos, P granules maintain their integrity upon dynein motor inactivation. Furthermore, in the nondividing Z2 and Z3 primordial germ cells, P granule integrity and perinuclear localization are not affected by the disruption of the dynein motor. Overall, this suggests that dynein motor activity likely does not contribute to stabilization of embryonic P granules. We speculate that disruption of P granules in rapidly dividing 16–100-cell embryos upon inactivation of dhc-1 is secondary to their missegregation.
FIGURE 8:
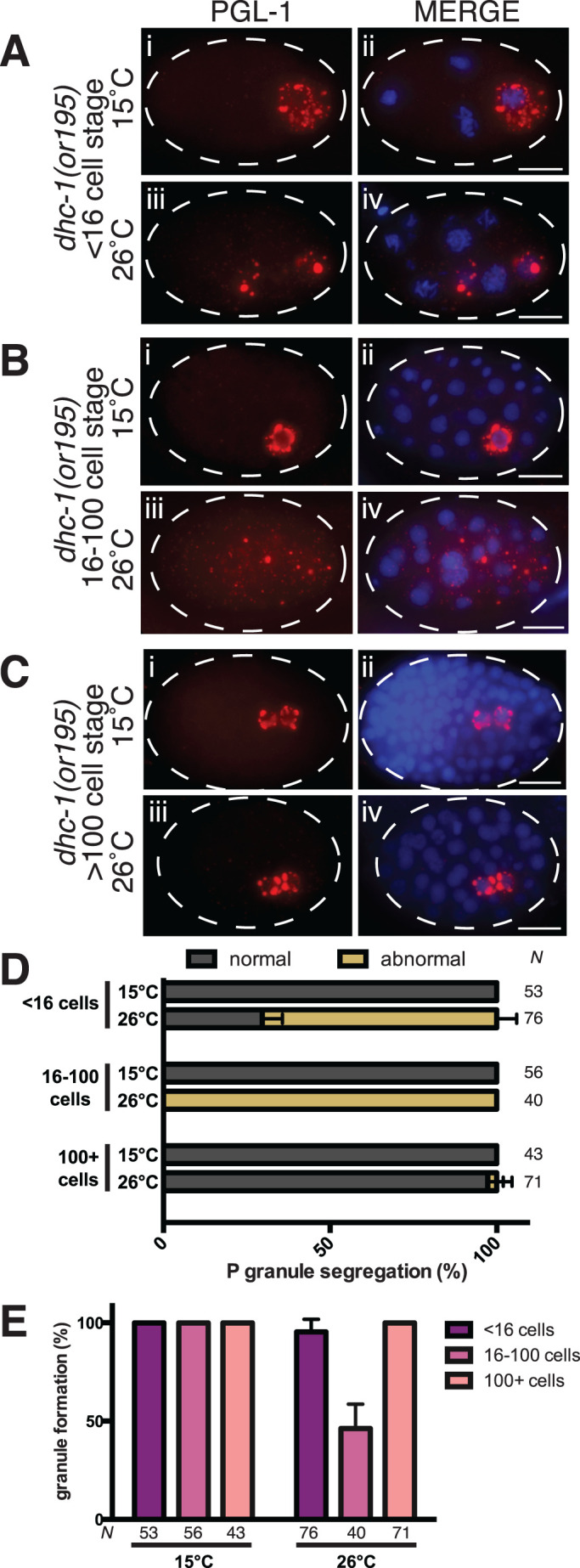
P granule integrity is likely independent of dynein motor function. (A–C) PGL-1 (red) detected by immunostaining of dhc-1(or195) embryos at the permissive (15°C, subpanels i and ii) or restrictive (26°C, subpanels iii and iv) temperature. (A) Four-cell embryos. (B) Approximately 40–cell embryos. (C) Approximately 100-cell embryos. DNA is labeled with DAPI (blue). Images were acquired using an epifluorescence microscope. Scale bars: 10 µm. (D) Inactivation of dynein motor causes defects in P granule segregation in the early embryos. P granule segregation was classified as normal for each developmental stage (gray) or abnormal where P granules were localized to somatic cells (tan). The number of embryos scored (N) in each experiment is indicated next to each bar. Data were collected in three biological replicates. (E) Perinuclear P granules are disrupted in 16- to 100-cell embryos upon inactivation of dynein motor. Plotted are the average percent of embryos with PGL-1 granules of a normal size following incubation at either permissive (15°C) or restrictive (26°C) temperature. Error bars represent the SD, and the number of embryos observed (N) in each experiment is below each bar. The same embryos were scored to generate panels D and E.
DLC-1 requirement for P granule assembly is independent of cell polarity
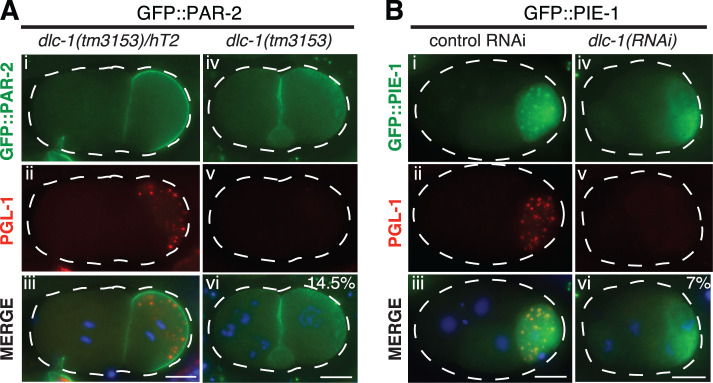
P granule segregation and integrity during embryonic asymmetric cell divisions depend on the establishment of cortical asymmetry of conserved cell polarity regulators, the PAR proteins (Lang and Munro, 2017). Polarized distribution of PARs then drives asymmetric localization of cytoplasmic components such as RBP PIE-1 and P granules (Mello et al., 1996; Griffin, 2015). To assess whether disruption of P granules following dlc-1(RNAi) is secondary to a failure of polarity establishment, we assessed whether GFP::PAR-2 achieved its normal posterior localization in dlc-1 mutant embryos. If P granule disruption depends on the loss of PAR polarity, we expect to observe P granule assembly defects only in the embryos that have lost PAR-2 localization. By contrast, we observed that in all 2–4-cell embryos where P granules were most severely dispersed (reflecting ∼15% of scored embryos), localization of GFP::PAR-2 was normal (Figure 9A, iv–vi). To determine whether P granule dispersal correlated with a failure to generate cytoplasmic asymmetries, we assessed localization of GFP::PIE-1 following dlc-1(RNAi). We found that in all 2–4-cell dlc-1(RNAi) embryos where P granules were severely dispersed (corresponding to 7% of scored embryos), GFP::PIE-1 formed a cytoplasmic gradient (Figure 9B, iv–vi), suggesting that P granule assembly defects were not associated with a general loss of cytoplasmic asymmetries. We conclude that P granule dispersal upon the loss of dlc-1 does not reflect a general disruption of cell polarity, supporting a specific role for DLC-1 in P granule assembly.
FIGURE 9:
Normal polarity in two- to four-cell embryos with dispersed P granules. GFP fusion proteins (green) and PGL-1 (red) detected by immunostaining in two- to four-cell embryos. (A) GFP::PAR-2 transgene in the progeny of dlc-1 heterozygous (i–iii) or homozygous mutant (iv–vi) mothers. (B) GFP::PIE-1 transgene in the progeny of hermaphrodites treated with control (i–iii) or dlc-1(RNAi) (iv–vi). DNA is labeled with DAPI (blue). Percent of scored embryos with severely dispersed P granules is shown in merge subpanels (vi). Early embryos were scored in three biological replicates, and N = 48 (GFP::PAR-2) and 55 (GFP::PIE-1). Images were acquired using an epifluorescence microscope. Scale bars: 10 µm.
DISCUSSION
In this study, we report the in vitro and in vivo interactions between DLC-1 and several P granule component proteins. We find that in vivo interactions between DLC-1 and PGL proteins are more pronounced in the embryo than in the adult germline and dlc-1 knockdown or null mutant interferes with embryonic P granule formation, likely independent of DLC-1’s contribution to the dynein motor. Together, our results support a model that DLC-1 is an essential facilitator of embryonic P granule assembly in C. elegans (Figure 10).
FIGURE 10:
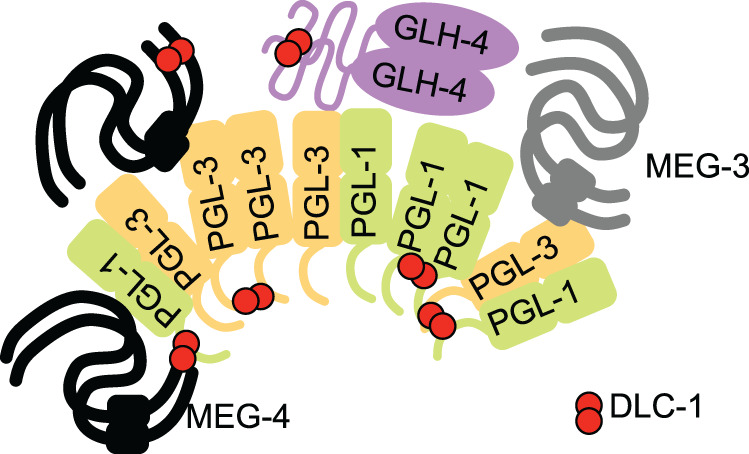
Model of DLC-1 contribution to P granule assembly in C. elegans embryo. P granules assemble through multivalent interactions of their constituent proteins. Structured domains are pictured as ovals or rectangles, and presumed disordered proteins or domains are indicated with lines. PGL-1 and PGL-3 form homo/heterodimers and likely oligomeric assemblies. Both PGLs and MEGs interact with RNAs, which is omitted for clarity. We speculate that binding of DLC-1 dimer (red) provides additional multivalent contacts that stabilize protein complexes and might additionally facilitate conformational changes promoting assembly of P granule components in the embryo.
Bioinformatics identifies new DLC-1–binding partners
The interaction motif scan identified a number of RBPs as candidate interactors of DLC-1, including several P granule components. Using GST pull downs, we confirmed direct interaction between DLC-1 and PGL-1, PGL-3, GLH-4, and MEG-4 (Figure 1F). These binding partners were recovered using a combination of three different interaction motifs to scan the proteome, suggesting that there is diversity in the protein sequences recognized by DLC-1 that cannot be captured in a single degenerate motif. Diversity of DLC-1 interaction sites outside of the canonical TQT interaction motif is important to consider in future bioinformatic searches for DLC-1 partners. Further work is needed to test whether the predicted binding sites in fact mediate the interaction between DLC-1 and its P granule partners.
Dynamic interaction of DLC-1 with PGL-1 and PGL-3
While DLC-1 is coexpressed with PGL-1 and PGL-3 both in the adult germline and in embryos, PLA revealed that DLC-1/PGL interactions are dynamic and change with development (Figures 2D and 3P). In the adult germline, DLC-1/PGL-1 complexes are more abundant than DLC-1/PGL-3 complexes (Figure 2D). In the distal region of the germline (zone 1), this difference reflects the lack of PGL-3 expression (Supplemental Figure S1C). The differences in DLC-1/PGLs interactions through zones 2 and 3 might similarly stem from unequal expression levels of PGL-1 and PGL-3. However, it is also possible that the interaction between PGLs and DLC-1 is regulated by varying accessibility of binding sites that might be due to posttranslational modifications or interactions with additional binding partners. Both DLC-1/PGL complexes decreased in abundance in the oocytes compared to the mid to late pachytene (Figure 2D). This might reflect P granule remodeling at the time when they lose their perinuclear localization and become cytoplasmic. We find that DLC-1/PGL interactions are most pronounced during embryonic development and are detected in both somatic and germ cells (Figure 3, F–O). As development proceeds and P granules become perinuclear, the DLC-1/PGL interactions become significantly enriched in the germ cells, with DLC-1/PGL-3 complex enrichment being more pronounced (Figure 4, E and F). Elucidation of the mechanisms behind temporal and spatial regulation of DLC-1/PGL interactions remains the subject of future research.
DLC-1 facilitates embryonic P granule assembly
Many P granule components including LAF-1 (Elbaum-Garfinkle et al., 2015), MEG-3 (Smith et al., 2016; Putnam et al., 2019), PGL-1 (Zhang et al., 2018), and PGL-3 (Saha et al., 2016; Zhang et al., 2018) can phase separate on their own and form condensates similar to P granules in vitro. Moreover, ectopic expression of PGL-3 or PGL-1 in mammalian tissue culture cells and somatic C. elegans cells results in granule formation, while GLH-1 is unable to form ectopic granules (Updike and Strome, 2009; Hanazawa et al., 2011). However, phase separation of P granule components in embryonic germ cells is interdependent on each other’s presence. For example, PGL-1 localization to P granules and asymmetric segregation depend on glh-1 (Kawasaki et al., 1998; Spike et al., 2008; Hanazawa et al., 2011) as well as on meg-3 and meg-4 (Wang et al., 2014; Smith et al., 2016). Conversely, pgl-3 and pgl-1 together are required for assembly of GLH-1, MEG-3, and MEG-4 into granules (Hanazawa et al., 2011; Wang et al., 2014; Smith et al., 2016). The complex regulation of P granule assembly in vivo is not fully understood and likely involves contributions from additional factors.
We found that dlc-1 is required for embryonic P granule assembly (Figures 5 and 7), in agreement with a previous report that localization of transgenic GFP-tagged PGL-1 in embryos was disrupted by dlc-1(RNAi) (Updike and Strome, 2009). We extended the previous findings by documenting defects in localization of CRISPR-tagged or endogenous MEG-3/4 and PGL-1 proteins, as well as transgenic GFP::PGL-3 upon dlc-1(RNAi) and in dlc-1 mutant embryos. We find that these defects in PGL-1/3 localization are not linked to changes in PGL-1/3 abundance (Figure 6). Our quantitative analysis indicated that colocalization between all four P granule components significantly decreased upon the loss of dlc-1 (Figures 5F and 7E). The loss of dlc-1 affected colocalization of MEG-3 with other P granule proteins, even though MEG-3 and DLC-1 do not directly interact. Together, these results suggested complete failure of P granule assembly and phase separation rather than selective loss of DLC-1–interacting components from a stable resilient structure. Interestingly, in vitro experiments have found direct interaction between PGL-1 and PGL-3 (Kawasaki et al., 2004), MEG-3 or MEG-4 and PGL-1 (Wang et al., 2014), and MEG-4 and PGL-3 (N.D., unpublished). While many P granule proteins can interact without intermediaries in vitro, our data imply that P granule assembly in vivo requires additional factors such as dlc-1 and is not driven by association of core components alone.
We further considered whether the effect of dlc-1 depletion on P granules is indirect and mediated either by DLC-1’s activity in the dynein motor complex or through affecting cell polarity of early embryonic cell divisions. Although the dynein motor was previously hypothesized to promote P granule integrity (Updike and Strome, 2009), we find that P granule dispersal in conditions where the dynein motor is disrupted is secondary to failing cell divisions (Figure 8 and unpublished data). By contrast, dlc-1 knockdown embryos continue cell divisions until the ∼100-cell stage, suggesting that the dynein motor function is not fully eliminated and P granule assembly is compromised despite the maintenance of residual dynein motor activity. As P granule segregation depends on faithful execution of asymmetric cell divisions, we have tested whether P granule dispersal correlated with defective localization of an upstream polarity regulator PAR-2 or with disruption of cytoplasmic polarity assessed by localization of RBP PIE-1. We find that the early embryos with dispersed P granules displayed normal localization of both GFP::PAR-2 and GFP::PIE-1 (Figure 9), suggesting that this dispersal is not a result of the general loss of cytoplasmic asymmetries.
We hypothesize that DLC-1 promotes P granule assembly by functioning as a bimolecular hub stabilizing multimeric protein complexes (Barbar, 2008; Clark et al., 2015) (Figure 10). Multivalent interactions are key to forming phase-separating molecular assemblies (Banani et al., 2017). PGL proteins homo- and heterodimerize (Kawasaki et al., 1998, 2004) and possess at least two dimerization interfaces (Hanazawa et al., 2011; Aoki et al., 2018, 2021). Multivalency of PGL-1 self-interactions is important because mutagenesis of a single dimerization interface disrupts P granule assembly in the adult germline (Aoki et al., 2021). Because LC8 family proteins have been implicated in stabilizing homodimeric complexes (Clark et al., 2015), it is possible that DLC-1 stabilizes PGL-1 or PGL-3 dimers. Additionally, because a number of DLC-1–binding partners in P granules interact among themselves (Kawasaki et al., 2004; Wang et al., 2014), it is conceivable that DLC-1 might similarly stabilize heterodimeric protein complexes. We speculate that cross-linking of core P granule components by DLC-1 strengthens their associations and contributes to efficient phase separation in embryos. Testing this hypothesis requires mapping the DLC-1–binding sites in its P granule component partners and remains the subject for future studies.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Nematode strains and culture
C. elegans strains (Supplemental Table 3) were cultured as per standard protocols (Brenner, 1974) at 20°C or 24°C (if containing a GFP-tagged transgene or edited gene). The 3xFLAG::dlc-1(mntSi13); gfp::pgl-3(mntIs9) strain (UMT420) was generated by crossing UMT282 males with JH2469. The 3xFLAG::dlc-1(mntSi13); pgl-1::gfp(ax3122) strain (UMT432) was generated by crossing UMT282 males with JH3269. The dlc-1(tm3153)/hT2; meg-3::ollas (ax3051) meg-4::3xFLAG(ax2080) strain (UMT398) was generated by crossing UMT351 males with JH3374.
Bioinformatics
Published biochemically verified LC8-binding sites (Supplemental Table 1) were analyzed by the online motif discovery tool Multiple EM for Motif Elicitation https://meme-suite.org/meme/ (MEME; Bailey and Elkan, 1994; accessed 07/2016). To generate consensus motifs, the classic motif discovery mode was selected with the site distribution set to zero or one occurrence per sequence and the advanced settings set at the default parameters (i.e., background model: 0-order model of sequences, minimum width: 6, maximum width: 50). For each group of peptide sequences (Supplemental Table 1) submitted to online MEME, the motif with the lowest E value was chosen as the consensus motif. Motif A was derived using all 53 previously reported LC8-interacting peptides (Rapali et al., 2011b; dominated by TQT-containing peptides) together with DLC-1 interaction sites on FBF-2 (Wang et al., 2016) and GLD-1 (Ellenbecker et al., 2019) identified by our lab (Supplemental Table 1). Motif B was generated from 10 divergent interaction sequences in the initial data set including the DLC-1 interaction sites on FBF-2 (Wang et al., 2016) and GLD-1 (Ellenbecker et al., 2019). Motif C was produced with the 10-peptide data set where the low-complexity and serine-rich FBF-2 interaction sites were replaced with sequences containing the “QVD” residues reported in Rodríguez-Crespo et al. (2001). Each motif was then submitted to the Find Individual Motif Occurrences (FIMO; Grant et al., 2011) module of the online MEME suite to scan the C. elegans proteome (Ensembl Release 96, WBcel235) with default parameters. Proteins that contained at least one instance of the motif and met the threshold for a significant match of P < 0.0001 were sorted to the output. To identify RBPs, the output was compared against comprehensive lists of C. elegans RBPs (Tamburino et al., 2013; Matia-González et al., 2015); the full list of identified RBPs is provided in Supplemental Table 2.
In vitro pull downs
Full-length proteins were amplified from Bristol N2 cDNA and cloned into pDEST17 (Thermo Fisher Scientific) to generate 6xHis-tagged proteins or into pMALc2 to generate MBP-tagged proteins to improve solubility. All constructs were confirmed by sequencing and expressed in BL21(DE3) Escherichia coli cultures grown to a 600 nm optical density of 0.6. Expression of MBP- or 6xHis- tagged proteins was then induced with 0.1 mM IPTG (Isopropyl β-d-1-thiogalactopyranoside) at 15°C for 16–18 h. Expression of GST alone or GST::DLC-1 was induced with 1 mM IPTG at 37°C for 4 h. Bacterial pellets were frozen overnight at –80°C and lysed for 1 h at 4°C with a lysis buffer (20 mM Tris, pH 7.5, 250 mM NaCl, 10% glycerol, 1 mM MgCl2, 0.1% Triton X-100, 10 mM 2-mercaptoethanol, 1 mM PMSF (phenylmethylsulfonyl fluoride), Roche protease inhibitor cocktail, 0.5 mg/ml lysozyme, 6 µg/ml DNase 1) at 1/15 volume of the initial culture. Pull downs were performed by directly adding precleared extracts to GST alone or GST::DLC-1–bound glutathione sepharose beads (Sigma-Aldrich) in binding buffer (20 mM Tris, pH 7.5, 250 mM NaCl, 10% glycerol, 0.1% Triton X-100, 10 mM 2-mercaptoethanol, 1 mM PMSF, Roche protease inhibitor cocktail, and 0.5 mg/ml bovine serum albumin [BSA]). The binding reactions were incubated for 3 h at 15°C and washed four times with wash buffer (10 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.1% NP-40, and 0.5 mg/ml BSA) before elution with SDS sample buffer with 10 mM dithiothreitol at 90°C for 10 min.
Western blotting
Western blotting of in vitro pull downs was performed as described (Day et al., 2018). The primary antibodies used were anti-6xHis (Sigma-Aldrich; Catalogue #H1029) at 1:2000 and anti-MBP (Developmental Studies Hybridoma Bank; Catalogue #DSHB-MBP-3D7) (Park et al., 2016) at 1:800.
C. elegans embryo samples were harvested by hypochlorite treatment of synchronous young adult worms reared on dlc-1 or control RNAi, frozen in liquid nitrogen, and stored at –80°C. Frozen embryos were lysed in 50 mM HEPES, pH 7.4, 1 mM EGTA (Ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid tetrasodium salt), 3 mM MgCl2, 150 mM KCl, 10% glycerol, 0.05% NP-40, Roche protease inhibitor cocktail, 1 mM PMSF lysis buffer, ultrasonicated (Qsonica), and clarified by centrifugation. Three biological replicates of embryos were analyzed by Western blotting with primary antibodies recognizing endogenous PGL-1 (Developmental Studies Hybridoma Bank; Catalogue #K76) at 1:500 (1.3 μg/ml), GFP (Invitrogen; Catalogue #G10362) at 1:1000, and α-tubulin (Sigma-Aldrich; DM1a) at 1:1000.
RNAi
RNAi was performed as previously described (Day et al., 2018), with the exception of growing nematodes at 20°C. This temperature was still permissible for GFP::PGL-3 expression and also ensured that dlc-1(RNAi) worms were able to produce embryos.
Nematode dissection, immunostaining, and imaging
The procedure for dissection, immunostaining, and imaging of germlines was as described in Day et al. (2018). For immunostaining embryos, worms were dissected at the vulva to release embryos. The primary and secondary antibodies and their dilution factors used for immunostaining 3xFLAG-, GFP- and OLLAS-tagged proteins, and endogenous P granule proteins are described in Supplemental Table 4. The images were acquired with a Leica DFC300G camera attached to a Leica DM5500B microscope or with a Zeiss LSM 880 confocal microscope, stitched together using FIJI/ImageJ (if needed), and cropped using Adobe Photoshop CS3. Colocalization analysis was performed on confocal images in FIJI/ImageJ using the Just Another Colocalization Plugin (JACoP; Bolte and Cordelières, 2006). In Figure 5, P granules were imaged with the same laser power/gain for panels A, B, and D (early and dispersed P granules) but with a lower laser power for panel C (wild-type late-embryo P granules) to avoid overexposure.
PLA, imaging, and quantification
PLA of germlines and embryos was performed as previously described (Day et al., 2020) using Duolink In situ reagents (Sigma-Aldrich) as per the manufacturer’s protocol. Antibody dilutions are reported in Supplemental Table 4. Briefly, the PLA procedure is performed on dissected young adult nematodes. Following fixation, blocking, and incubation with the primary antibodies to label the proteins of interest, the tissues are incubated with PLA probes (secondary antibodies linked to DNA oligos). The PLA probes are linked through a ligation reaction, amplified by a PCR, and labeled by red fluorophores. Labeled adult germlines and embryos were imaged using a Zeiss 880 confocal microscope. PLA focus densities were quantified using the previously described FIJI-based workflow (Day et al., 2020), where PLA foci were identified by thresholding and the number of foci per μm2 in each region of interest (ROI) was determined. PLA density in the germline was analyzed separately in three standardized ROIs corresponding to distinct developmental stages (see Figure 2, Aiv, Biv, and Civ). Zone 1 encompassed the distal tip region through the early pachytene. Zone 2 is composed of the proximal half of the pachytene, where the midpachytene region was defined as starting at the 16th cell row distal to the first oocyte in diplotene, based on the pattern of expression for GFP::PGL-3 in the germline. Finally, zone 3 encompassed all oocytes. PLA density in embryos was determined with the whole embryo as the ROI. The anti–PGL-1 antibody was used to coimmunostain PGL-1 to mark P granules in the germ cell during embryo PLA experiments. The antibody was incorporated into the primary antibody solution along with the anti-FLAG and anti-GFP antibodies and incubated for the same time and temperature (overnight, 4°C). When the PLA probes were added to the sample the next day, the anti-mouse immunoglobulin M Alexa 488 was also included in the mix using the same dilution as with the immunostaining of embryos and incubated at the same time and temperature (1 h, 37°C). For quantification of the relative area of PLA in germ cells, a single focal plane with the best PGL-1 immunostaining signal was chosen. An ROI that encompassed the germ cell and P granules was drawn using the ellipse tool. This germ cell ROI was duplicated and placed on a somatic cell (ideally on a cell on the opposite side of the embryo) containing PLA foci, to ensure that the same area of measurement is used. The image was then subjected to the same particle threshold as was used to quantify whole embryo PLA. Quantification of relative area of PLA within each ROI was obtained using the same workflow as above. For embryos where PLA was observed only in the germ cell but not in the somatic cell in the same focal plane, the relative area for the somatic cell was substituted with the minimum value observed among somatic cells in the data set.
Supplementary Material
Acknowledgments
We thank the members of the Voronina laboratory for helpful discussions. All research was performed at the University of Montana. Some nematode strains used in this study were provided by the Caenorhabditis Genetics Center funded by the National Institutes of Health (NIH) (P40OD010440). Confocal microscopy was performed in the University of Montana BioSpectroscopy Core Research Laboratory operating with support from NIH awards P20GM103546 and S10OD021806. This work was supported by the NIH grant GM109053 to E.V.
Abbreviations used:
- LLPS
liquid–liquid phase separation
- PLA
proximity ligation assay
- RBP
RNA-binding protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-05-0275) on March 11, 2022.
REFERENCES
- Amiri A, Keiper BD, Kawasaki I, Fan Y, Kohara Y, Rhoads RE, Strome S (2001). An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development 128, 3899–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki ST, Lynch TR, Crittenden SL, Bingman CA, Wickens M, Kimble J (2021). C. elegans germ granules require both assembly and localized regulators for mRNA repression. Nat Commun 12, 996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki ST, Porter DF, Prasad A, Wickens M, Bingman CA, Kimble J (2018). An RNA-binding multimer specifies nematode sperm fate. Cell Rep 23, 3769–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi S, Fredriksson R, Wallén-Mackenzie Å (2015). In situ proximity ligation assay (PLA). Methods Mol Biol 1318, 149–159. [DOI] [PubMed] [Google Scholar]
- Bailey TL (2011). Dreme: motif discovery in transcription factor chip-seq data. Bioinformatics 27, 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2, 28–36. [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbar E (2008). Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry 47, 503–508. [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelières FP (2006). A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224(Pt 3), 213–232. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, Hyman AA (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732. [DOI] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SA, Jespersen N, Woodward C, Barbar E (2015). Multivalent IDP assemblies: unique properties of LC8-associated, IDP duplex scaffolds. FEBS Lett 589(Pt A), 2543–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S (2004). Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J 86, 3993–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day NJ, Ellenbecker M, Voronina E (2018). Caenorhabditis elegans DLC-1 associates with ribonucleoprotein complexes to promote mRNA regulation. FEBS Lett 592, 3683–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day NJ, Wang X, Voronina E (2020). In situ detection of ribonucleoprotein complex assembly in the C. elegans germline using proximity ligation assay. J Vis Exp 2020, DOI: 10.3791/60982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick T, Ray K, Salz HK, Chia W (1996). Cytoplasmic dynein (dDlc1) mutations cause morphogenetic defects and apoptotic cell death in Drosophila melanogaster. Mol Cell Biol 16, 1966–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett M, Schedl T (2009). A role for dynein in the inhibition of germ cell proliferative fate. Mol Cell Biol 29, 6128–6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP (2015). The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci USA 112, 7189–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenbecker M, Osterli E, Wang X, Day NJ, Baumgarten E, Hickey B, Voronina E (2019). Dynein light chain DLC-1 facilitates the function of the germline cell fate regulator GLD-1 in Caenorhabditis elegans. Genetics 211, 665–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E, Shorter J (2019). The molecular language of membraneless organelles. J Biol Chem 294, 7115–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P, Pichler S, Kirkham M, Hyman AA (1999). Cytoplasmic dynein is required for distinct aspects of mtoc positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol 147, 135–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin EE (2015). Cytoplasmic localization and asymmetric division in the early embryo of Caenorhabditis elegans. Wiley Interdiscip Rev Dev Biol 4, 267–282. [DOI] [PubMed] [Google Scholar]
- Gruidl ME, Smith PA, Kuznicki KA, McCrone JS, Kirchner J, Roussell DL, Strome S, Bennett KL (1996). Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proc Natl Acad Sci USA 93, 13837–13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanazawa M, Yonetani M, Sugimoto A (2011). PGL proteins self associate and bind RNPs to mediate germ granule assembly in C. elegans. J Cell Biol 192, 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird SN, Paulsen JE, Strome S (1996). Segregation of germ granules in living Caenorhabditis elegans embryos: cell-type-specific mechanisms for cytoplasmic localisation. Development 122, 1303–1312. [DOI] [PubMed] [Google Scholar]
- Jespersen N, Barbar E (2020). Emerging features of linear motif-binding hub proteins. Trends Biochem Sci 45, 375–384. [DOI] [PubMed] [Google Scholar]
- Jespersen N, Estelle A, Waugh N, Davey NE, Blikstad C, Ammon YC, Akhmanova A, Ivarsson Y, Hendrix DA, Barbar E (2019). Systematic identification of recognition motifs for the hub protein LC8. Life Sci Alliance 2, e201900366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Francis R, Schedl T (1996). GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev Biol 180, 165–183. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. (2003). Systematic functional analysis of the caenorhabditis elegans genome using RNAi. Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- Kawasaki I, Amiri A, Fan Y, Meyer N, Dunkelbarger S, Motohashi T, Karashima T, Bossinger O, Strome S (2004). The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics 167, 645–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I, Shim YH, Kirchner J, Kaminker J, Wood WB, Strome S (1998). PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94, 635–645. [DOI] [PubMed] [Google Scholar]
- Kidane AI, Song Y, Nyarko A, Hall J, Hare M, Löhr F, Barbar E (2013). Structural features of LC8-induced self-association of Swallow. Biochemistry 52, 6011–6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A, Hoch NC, McGregor NE, Sims NA, Smyth IM, Heierhorst J (2019). Dynll1 is essential for development and promotes endochondral bone formation by regulating intraflagellar dynein function in primary cilia. Hum Mol Genet 28, 2573–2588. [DOI] [PubMed]
- King SM, Barbarese E, Dillman JF, Patel-King RS, Carson JH, Pfister KK (1996). Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved Mr 8,000 light chain. J Biol Chem 271, 19358–19366. [DOI] [PubMed] [Google Scholar]
- King SM, Patel-King RS (1995). The M(r) = 8,000 and 11,000 outer arm dynein light chains from Chlamydomonas flagella have cytoplasmic homologues. J Biol Chem 270, 11445–11452. [DOI] [PubMed] [Google Scholar]
- Kuznicki KA, Smith PA, Leung-Chiu WM, Estevez AO, Scott HC, Bennett KL (2000). Combinatorial RNA interference indicates GLH-4 can compensate for GLH-1; these two P granule components are critical for fertility in C. elegans. Development 127, 2907–2916. [DOI] [PubMed] [Google Scholar]
- Lang CF, Munro E (2017). The PAR proteins: from molecular circuits to dynamic self-stabilizing cell polarity. Development 144, 3405–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Lee S, Kim B, Chang S, Kim SW, Paick JS, Rhee K (2006). Dazl can bind to dynein motor complex and may play a role in transport of specific mRNAs. EMBO J 25, 4263–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. (2004). A map of the interactome network of the metazoan C. elegans. Science 303, 540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Jaffrey SR, Guo W, Snyder SH, Clardy J (1999). Structure of the Pin/LC8 dimer with a bound peptide. Nat Struct Biol 6, 735–740. [DOI] [PubMed] [Google Scholar]
- Marnik EA, Updike DL (2019). Membraneless organelles: P granules in Caenorhabditis elegans. Traffic 20, 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matia-González AM, Laing EE, Gerber AP (2015). Conserved mRNA-binding proteomes in eukaryotic organisms. Nat Struct Mol Biol 22, 1027–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Schubert C, Draper B, Zhang W, Lobel R, Priess JR (1996). The PIE-1 protein and germline specification in C. elegans embryos. Nature 382, 710–712. [DOI] [PubMed] [Google Scholar]
- Mittag T, Parker R (2018). Multiple modes of protein-protein interactions promote RNP granule assembly. J Mol Biol 430, 4636–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YN, Glover RA, Daniels KJ, Soll DR (2016). Generation and validation of monoclonal antibodies against the maltose binding protein. Monoclon Antib Immunodiagn Immunother 35, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister KK, Fay RB, Witman GB (1982). Purification and polypeptide composition of dynein ATPases from Chlamydomonas flagella. Cell Motil 2, 525–547. [DOI] [PubMed] [Google Scholar]
- Pitt JN, Schisa JA, Priess JR (2000). P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev Biol 219, 315–333. [DOI] [PubMed] [Google Scholar]
- Putnam A, Cassani M, Smith J, Seydoux G (2019). A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat Struct Mol Biol 26, 220–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapali P, Radnai L, Süveges D, Harmat V, Tölgyesi F, Wahlgren WY, Katona G, Nyitray L, Pál G (2011a). Directed evolution reveals the binding motif preference of the LC8/DYNLL hub protein and predicts large numbers of novel binders in the human proteome. PLoS One 6, e18818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapali P, Szenes Á, Radnai L, Bakos A, Pál G, Nyitray L (2011b). DYNLL/LC8: A light chain subunit of the dynein motor complex and beyond. FEBS J 278, 2980–2996. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Crespo I, Yélamos B, Roncal F, Albar JP, Ortiz de Montellano PR, Gavilanes F (2001). Identification of novel cellular proteins that bind to the LC8 dynein light chain using a pepscan technique. FEBS Lett 503, 135–141. [DOI] [PubMed] [Google Scholar]
- Rose L, Gönczy P (2014). Polarity establishment, asymmetric division and segregation of fate determinants in early C. elegans embryos. WormBook. 1–43. [DOI] [PubMed]
- Saha S, Weber CA, Nousch M, Adame-Arana O, Hoege C, Hein MY, Osborne-Nishimura E, Mahamid J, Jahnel M, Jawerth L, et al. (2016). Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell 166, 1572–1584.e1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt DJ, Rose DJ, Saxton WM, Strome S (2005). Functional analysis of cytoplasmic dynein heavy chain in Caenorhabditis elegans with fast-acting temperature-sensitive mutations. Mol Biol Cell 16, 1200–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382. [DOI] [PubMed] [Google Scholar]
- Simonis N, Rual JF, Carvunis AR, Tasan M, Lemmens I, Hirozane-Kishikawa T, Hao T, Sahalie JM, Venkatesan K, Gebreab F, et al. (2009). Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Methods 6, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Calidas D, Schmidt H, Lu T, Rasoloson D, Seydoux G (2016). Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. eLife 5, e21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C, Meyer N, Racen E, Orsborn A, Kirchner J, Kuznicki K, Yee C, Bennett K, Strome S (2008). Genetic analysis of the Caenorhabditis elegans GLH family of P-granule proteins. Genetics 178, 1973–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Kunze R, Flemming D, Höpfner D, Diepholz M, Philippsen P, Böttcher B, Hurt E (2007). Molecular basis for the functional interaction of dynein light chain with the nuclear-pore complex. Nat Cell Biol 9, 788–796. [DOI] [PubMed] [Google Scholar]
- Strome S, Wood WB (1982). Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans . Proc Natl Acad Sci USA 79, 1558–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburino AM, Ryder SP, Walhout AJ (2013). A compendium of Caenorhabditis elegans RNA binding proteins predicts extensive regulation at multiple levels. G3 (Bethesda) 3, 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike DL, Strome S (2009). A genomewide RNAi screen for genes that affect the stability, distribution and function of P granules in Caenorhabditis elegans. Genetics 183, 1397–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E (2013). The diverse functions of germline P granules in Caenorhabditis elegans. Mol Reprod Dev 80, 624–631. [DOI] [PubMed] [Google Scholar]
- Voronina E, Seydoux G (2010). The C. elegans homolog of nucleoporin Nup98 is required for the integrity and function of germline P granules. Development 137, 1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Smith J, Chen BC, Schmidt H, Rasoloson D, Paix A, Lambrus BG, Calidas D, Betzig E, Seydoux G (2014). Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans . eLife 3, e04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J (2002). A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419, 312–316. [DOI] [PubMed] [Google Scholar]
- Wang L, Hare M, Hays TS, Barbar E (2004). Dynein light chain LC8 promotes assembly of the coiled-coil domain of Swallow protein. Biochemistry 43, 4611–4620. [DOI] [PubMed] [Google Scholar]
- Wang X, Olson JR, Rasoloson D, Ellenbecker M, Bailey J, Voronina E (2016). Dynein light chain DLC-1 promotes localization and function of the PUF protein FBF-2 in germline progenitor cells. Development 143, 4643–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K (2007). Dyneins across eukaryotes: A comparative genomic analysis. Traffic 8, 1708–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MJ, Salata MW, Susalka SJ, Pfister KK (2001). Light chains of mammalian cytoplasmic dynein: Identification and characterization of a family of LC8 light chains. Cell Motil Cytoskeleton 49, 229–240. [DOI] [PubMed] [Google Scholar]
- Zhang G, Wang Z, Du Z, Zhang H (2018). MTOR regulates phase separation of PGL granules to modulate their autophagic degradation. Cell 174, 1492–1506.e1422. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, Zhao Y, Li Z, Song B, Han J, et al. (2009). SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell 136, 308–321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.