Abstract
How nuclear pore complexes (NPCs) assemble in the intact nuclear envelope (NE) is only rudimentarily understood. Nucleoporins (Nups) accumulate at the inner nuclear membrane (INM) and deform this membrane toward the outer nuclear membrane (ONM), and eventually INM and ONM fuse by an unclear mechanism. In budding yeast, the integral membrane protein Brl1 that transiently associates with NPC assembly intermediates is involved in INM/ONM fusion during NPC assembly but leaving the molecular mechanism open. AlphaFold predictions indicate that Brl1-like proteins carry as common motifs an α-helix with amphipathic features (AαH) and a disulfide-stabilized, anti-parallel helix bundle (DAH) in the perinuclear space. Mutants with defective AαH (brl1F391E, brl1F391P, brl1L402E) impair the essential function of BRL1. Overexpression of brl1F391E promotes the formation of INM and ONM enclosed petal-like structures that carry Nups at their base, suggesting that they are derived from an NPC assembly attempt with failed INM/ONM fusion. Accordingly, brl1F391E expression triggers mislocalization of Nup159 and Nup42 and to a lesser extent Nsp1, which localize on the cytoplasmic face of the NPC. The DAH also contributes to the function of Brl1, and AαH has functions independent of DAH. We propose that AαH and DAH in Brl1 promote INM/ONM fusion during NPC assembly.
INTRODUCTION
Nuclear pore complexes (NPCs) are conserved, macromolecular complexes of eukaryotic cells that facilitate transport of proteins and ribonucleoproteins from the nucleoplasm into the cytoplasm and the import of proteins into the nucleus. NPCs have an octagonal symmetry consisting of ∼30 NPC components called nucleoporins (Nups). Nups assemble into stable subcomplexes known in yeast as Nup82, channel, inner ring, Y, and Nup116-Gle2 complexes (Beck and Hurt, 2017). These subcomplexes function as the modular building blocks of NPCs. The central inner ring complex is flanked on the cytoplasmic and nuclear sides by the Y-complex. The Nup82 complex that interacts with the Y-complex is located on the cytoplasmic side of the NPC. The nuclear basket is attached to NPCs via the Y-complex on the nuclear side (Alber et al., 2007; Allegretti et al., 2020). Some Nups contain repeats of the amino acids phenylalanine-glycine (FG) that localize in the central transport channel but also extend into the nucleoplasm and cytoplasm. FG repeats function as a selective permeability barrier for the transport of macromolecules between the nucleus and cytoplasm (Frey and Gorlich, 2007).
In vertebrates NPCs assemble in two cell cycle phases by distinct mechanisms (Otsuka and Ellenberg, 2018). With mitotic exit, new NPCs assemble in small membrane openings of chromatin-associated endoplasmic reticulum (ER) sheets (Otsuka et al., 2018). Subsequently, NPCs enlarge during the assembly of the inner ring complex and finally are embedded in the reforming nuclear envelope (NE). This suggests that the postmitotic NPC assembly pathway is not dependent on the fusion of the INM with the ONM. Interphase NPC assembly starts by the deposition of Nups at the INM of the intact NE, followed by INM deformation and fusion of the INM and ONM that is an essential step for interphase NPC assembly (Doucet et al., 2010; Otsuka et al., 2016).
Budding yeast with its closed mitosis uses only the interphase pathway for NPC assembly. In contrast to human cells, NPC intermediates are not observed in wild-type (WT) yeast cells probably because the assembly process is relatively rapid (Winey et al., 1997). Interestingly, however, mutations in genes coding for Nups can lead to the accumulation of deformations of the INM, so-called herniations, that extend into the perinuclear space (Wente and Blobel, 1993; Aitchison et al., 1995; Murphy et al., 1996; Rampello et al., 2020). At least some of these herniations arise from a defect in NPC assembly, for example the failure of the fusion of the INM with the ONM (Onischenko et al., 2017; Zhang et al., 2018). The morphology of herniations suggests that in budding yeast NPC assembly also starts at the INM followed by the subsequent fusion of the INM with the ONM and the embedding of the NPC into INM/ONM fusion sites (Zhang et al., 2018).
In yeast, the paralogous Brl1 and Brr6 together with the interacting protein Apq12 form a module that functions in NPC assembly as indicated by the association of the three proteins with NPC assembly intermediates and the accumulation of herniations in conditional lethal BRL1 and BRR6 cells and cells with APQ12 deletion, without being components of fully assembled NPCs (de Bruyn Kops and Guthrie, 2001; Saitoh et al., 2005; Hodge et al., 2010; Tamm et al., 2011; Lone et al., 2015; Zhang et al., 2018, 2021). All components of the Apq12, Brl1 and Brr6 module are integral membrane proteins of the INM, ONM, and ER that carry two transmembrane regions. These are connected by amino acid stretches in the perinuclear space, while N- and C-termini of the three proteins either localize in the nucleoplasm or cytoplasm, dependent on their INM or ONM localization (Zhang et al., 2018, 2021). Brl1 and Brr6 are special because they harbor two conserved disulfide bridges in the perinuclear space domain, of which at least one is essential for the function of Brl1 (Tamm et al., 2011; Zhang et al., 2018).
BRL1 and APQ12 genetically and functionally interact with NUP116 coding for an FG Nup that has an additional role in NPC assembly, probably by functioning as a scaffold during interphase NPC assembly (Scarcelli et al., 2007; Onischenko et al., 2017; Zhang et al., 2018). Interestingly, overexpression of BRL1 but not of BRR6 suppressed the growth and herniation phenotype of nup116∆ cells at 37°C, indicating that BRL1 functions in some way in the fusion of the INM/ONM during NPC assembly (Zhang et al., 2018).
Recently, we described that Apq12 carries a short amphipathic alpha helix that connects the two transmembrane regions (Zhang et al., 2021). Mutations disrupting the amphipathic nature of this helix in Apq12 affect NPC assembly similar to APQ12 deletion. Here we analyzed Brl1 and Brr6-like proteins by the AlphaFold Protein Structure Database (Jumper et al., 2021) that predicts an α-helix with amphipathic features (AαH) and a disulfide-stabilized, anti-parallel helix bundle (DAH) as common perinuclear space characteristics of Brl1/Brr6-like proteins. We show that brl1F391E, brl1F391P, and brl1L402E mutations in the predicted AαH impair the essential function of budding yeast BRL1. Overexpression of brl1F391E is highly toxic for cells because it induces an abortive process of NPC assembly that probably fails because of a defect in INM/ONM fusion. This promotes mislocalization of the cytoplasmic nucleoporins Nup159, Nsp1, and Nup42, while other Nups such as Nup82 show normal NE localization. The DAH also contributes to the function of Brl1, and brl1F391E-overexpression toxicity is not affected by mutations in the DAH, suggesting that AαH can function independent of a fully intact DAH. We suggest that the AαH and DAH are the functional elements of Brl1 that promote INM/ONM fusion during NPC assembly.
RESULTS
Brl1 carries an AαH and DAH in the perinuclear space
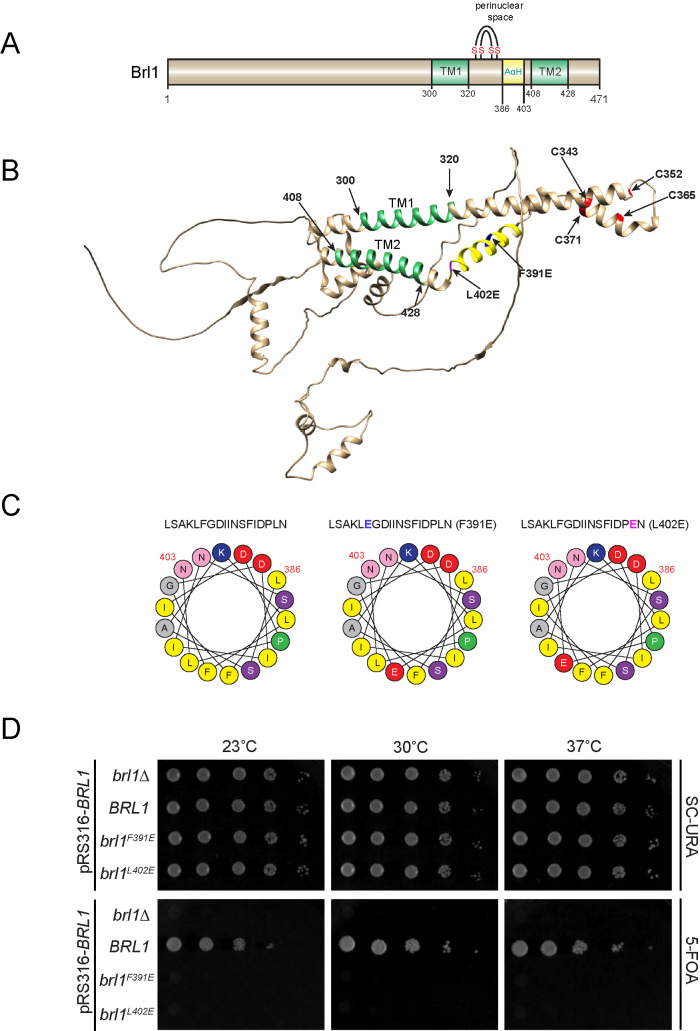
Brl1 and Brr6 are conserved proteins of organisms with closed mitosis where they function in processes related to INM/ONM fusion (Tamm et al., 2011; Zhang et al., 2018). The perinuclear space region of these proteins is probably important for this function. We used the AlphaFold Protein Structure Database (Jumper et al., 2021) to compare the perinuclear regions of Brl1/Brr6-like proteins from Saccharomyces cerevisiae, Schizosaccharomyces pombe, Candida albicans, and Plasmodium falciparum (Figure 1, A and B, and Supplemental Figure S1, A–D) in order to identify conserved motifs. This analysis indicated two common structural elements in the perinuclear space. As outlined for S. cerevisiae Brl1, Brl1 is predicted to contain an α-helix with amphipathic features (AαH, amino acids 386–403 in Brl1) with charged residues on the hydrophilic side and hydrophobic amino acid residues on the opposite side (Figure 1C) and an anti-parallel helix bundle (DAH, amino acids 321–372 in Brl1) stabilized by two disulfide bridges (Figure 1, A and B, Cys marked in red, Cys343-Cys371 and Cys352-Cys365). The inner two cysteines of the DAH (Cys352 and Cys365) are essential for the function of Brl1 (Zhang et al., 2018). The transmembrane region 1 (TM1, amino acids 300–320) of Brl1 is followed by the DAH and the AαH in the perinuclear space that connects to the TM2 (Figure 1, A and B, green; 408–428).
FIGURE 1:
Brl1 carries an AαH that is essential for its function. (A) Domain organization of Brl1. The two transmembrane (TM) domains and an amphipathic helix (AαH) are shown. The two disulfide bridges in Brl1 are indicated. (B) Brl1 prediction from the AlphaFold Protein Structure Database. The two TM regions are colored green. The AαH is shown in yellow. The four critical cysteine residues are shown in red. (C) Heliquest predictions (Gautier et al., 2008) of the AαH, AαHF391E, and AαHL402E helices. The amino acids marked in blue and purple (top row) indicate amino acid changes introduced in the AαH in order to disrupt its amphipathic nature. (D) Growth test of BRL1, brl1F391E, and brl1L402E using a plasmid shuffle approach. Tenfold serial dilutions were spotted onto SC-Ura and 5-FOA plates that were incubated at the indicated temperatures.
We first tested the importance of the predicted AαH in S. cerevisiae Brl1 by changing single amino acids in the hydrophobic part of the helix to glutamic acid (F391E, L402E) or to the helix-deforming proline (F391P) (Figure 1C and Supplemental Figure S2B). We analyzed the function of the brl1F391E, brl1F391P, and brl1L402E mutant alleles by a plasmid shuffle approach. The shuffle strain (brl1∆ pRS316-BRL1) was unable to grow on 5-fluoroorotic acid (5-FOA), which is toxic for cells expressing the URA3 gene consistent with the essential function of BRL1 (Figure 1D and Supplemental Figure S2B) (Saitoh et al., 2005). A chromosomally integrated BRL1 (Figure 1D and Supplemental Figure S2B) allowed growth of the shuffle strain on 5-FOA because the URA3-based BRL1 plasmid was lost in some cells and the essential function of the URA3-BRL1 gene was taken over by the chromosomally inserted BRL1. In contrast, chromosomally integrated brl1F391E, brl1F391P, and brl1L402E did not allow growth of brl1∆ pRS316-BRL1 cells on 5-FOA at 23°C, 30°C, and 37°C (Figure 1D and Supplemental Figure S2B), indicating that these mutations impaired the essential function of BRL1. In contrast, brl1∆ pRS316-BRL1 cells with brl1F391E, brl1F391P, and brl1L402E grew equally well on plates without 5-FOA (Figure 1D and Supplemental Figure S2B).
In a similar manner, we analyzed the function of mutations in the DAH (Supplemental Figure S2A; conserved amino acid residues are enlarged in blue [Gardner et al., 2021]). Brl1C343Y supported the growth of cells at 30°C but they were unable to grow at 37°C, indicating a conditional lethal phenotype (Supplemental Figure S2B). brl1C365S C371S strongly affected the growth of cells at 30°C and 37°C. Other mutations in the DAH did not affect the growth of cells at 30°C and 37°C, suggesting that these single-amino-acid changes did not impair the function of Brl1.
Overexpression of brl1 F391E, brl1 F391P, and brl1 L402E leads to growth defects and petal-like NE deformations
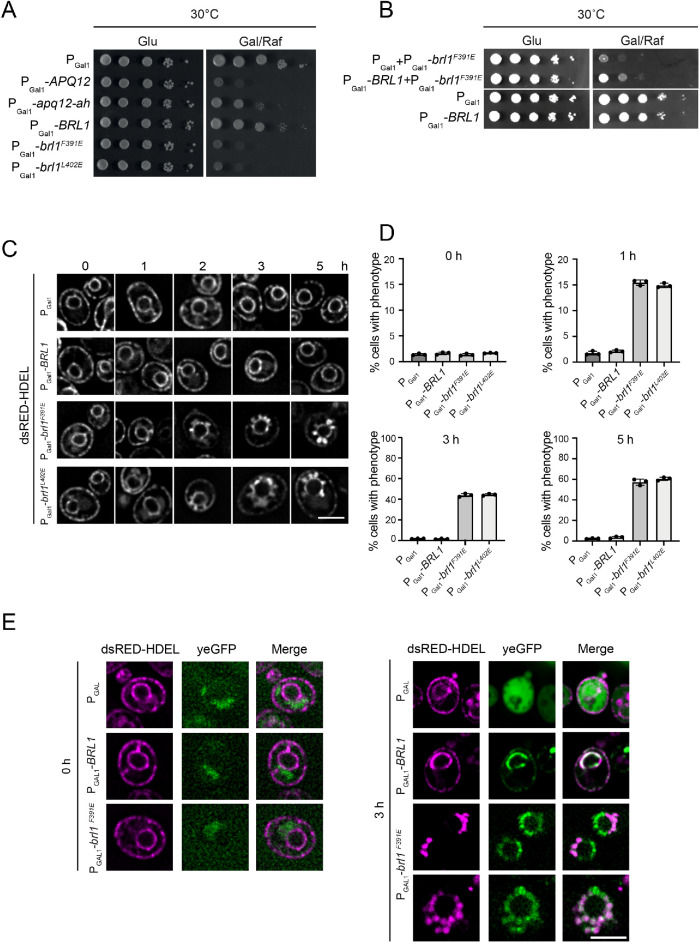
We tested whether expression of brl1F391E, brl1F391P, and brl1L402E is dominant lethal in BRL1 WT cells. Overexpression of APQ12, which is toxic for cells, and of apq12-ah, which carries mutations in the amphipathic helix relieving this toxicity, were used as controls (Figure 2A) (Zhang et al., 2021). Cells with the empty plasmid pGal1 or pGal1-BRL1 grew well on glucose and galactose plates. In contrast, cells harboring the pGal1-brl1F391E, pGal1-brl1F391P, or pGal1-brl1L402E plasmids showed severe growth defects on plates with the inducing galactose, while they were able to form colonies on the repressing glucose plates (Figure 2A and Supplemental Figure S2C). We tested whether the competition between Brl1 and Brl1F391E could be responsible for the growth defects. If this is the case, elevated Brl1 levels should reverse the pGal1-brl1F391E induced defect. Indeed, BRL1 co-overexpression partially rescued the growth defect caused by pGal1-brl1F391E (Figure 2B). This was not observed when the pGal1 and pGal1-brl1F391E plasmids were combined. In addition, we established that toxicity of pGal1-brl1F391E overexpression was not dependent on the functions of APQ12, BRR6, or NUP116, although overexpression of WT BRL1 suppressed the growth defect of nup116∆ cells at 37°C (Supplemental Figure S3, A–C) (Lone et al., 2015; Zhang et al., 2018). Taking the results together, Brl1F391E causes toxicity at least partly in competition with Brl1 but independent of APQ12, NUP116, and BRR6.
FIGURE 2:
Overexpression of BRL1 mutants with defective AαH causes formation of petal-like structures at the NE. (A) Overexpression of brl1F391E and brl1L402E is toxic for cells. WT cells with the control plasmid PGal1 or PGal1-BRL1, PGal1-brl1F391E, PGal1-brl1L402E were spotted in 10-fold serial dilutions onto glucose (Glu) and galactose/raffinose (Gal/Raf) plates at 30°C. PGal1-APQ12 and PGAL1-apq12-ah were used as controls for overexpression toxicity and growth on galactose/raffinose (Gal/Raf) plates (Zhang et al., 2021). (B) BRL1 overexpression slightly suppresses PGal1-brl1F391E toxicity. WT yeast cells containing the indicated plasmids were grown on glucose or galactose/raffinose plates at 30°C. (C) Overexpression of brl1F391E and brl1L402E triggers the formation of petal-like structures along the NE. BRL1, brl1F391E, and brl1L402E were overexpressed under the galactose-inducible promoter PGal1 in cells carrying the dsRED-HDEL NE/ER marker for 0, 1, 2, 3, and 5 h. Cells were imaged by microscopy after galactose induction. Scale bar: 3 µm. (D) Graph showing the percentage of cells with NE petals upon BRL1, brl1F391E, brl1L402E overexpression over time. n = 3. Shown is the mean with SD. (E) brl1F391E localizes to sites of membrane overproliferation. Fluorescence image analysis of PGal1-yeGFP, PGal1-BRL1-yeGFP, and PGal1-brl1F391E-yeGFP cells carrying the dsRED-HDEL marker with (3 h) and without (0 h) promoter induction. Scale bar: 5 µm.
We analyzed the morphology of the NE using the dsRed-HDEL reporter, which labels the NE and the ER. Overexpression of pGal1-brl1F391E, pGal1-brl1F391P, and pGal1-brl1L402E triggered the formation of petal-like extensions of the NE (Figure 2C and Supplemental Figure S2D). This phenotype was observed as early as 1 h after brl1F391E and brl1L402 expression with rising numbers of affected cells over the induction time (Figure 2D). Cells carrying the PGal1 control plasmid or PGal1-BRL1 did not develop these petals (Figure 2C and Supplemental Figure S2D).
To detect the localization of the Brl1F391E, we overexpressed yeGFP-tagged BRL1 in dsRED-HDEL–expressing cells. pGal1-yeGFP and pGal1-BRL1-yeGFP were used as controls. Without overexpression of the constructs, a weak background signal was detected in the yeGFP channel (Figure 2E; 0 h). Overexpressed yeGFP accumulated in the cytoplasm, as expected. Brl1-yeGFP localized at the NE and more weakly at the cortical ER as indicated by its colocalization with dsRED-HDEL at both locations (Figure 2E; 3 h) (Zhang et al., 2018). Brl1F391E-yeGFP was associated with the petal-like extensions at the NE that were marked by dsRED-HDEL (Figure 2E). Taking the results together, overexpression of mutants that affect the AαH in Brl1 deforms the NE into petal-like structures.
Integrity of the DAH in Brl1 is not important for brl1 F391E-overexpression phenotypes
The DAH is a prominent feature of the perinuclear domain of Brl1 (Figure 1B). Here, by combining the F391E mutation in the AαH of Brl1 with mutations in the DAH, we asked whether these functional regions cooperate in the formation of the NE petals seen upon brl1F391E overexpression. For this purpose, we first tested the overexpression phenotypes of brl1C343Y and brl1C365S C371S affecting the disulfide bonds in the DAH of Brl1 and brl1Y347H, brl1N353D, brl1T355A, brl1P356G, brl1A360D, or brl1W368Y in the loop connecting the two anti-parallel helices in the DAH (Supplemental Figure S2A). Importantly, C343, Y347, A360, C365, W368, and C371 are conserved residues of Brl1/Brr6-like proteins (Gardner et al., 2021). In repeated experiments, PGal1 overexpression of brl1C343Y and brl1C365S C371S was mildly toxic for cells compared with the BRL1 control while the other brl1DAH mutants grew as the BRL1 control (Supplemental Figure S2C). We next combined the loop mutants with the F391E mutation in AαH in order to see whether the toxic impact of brl1F391E overexpression requires an intact DAH loop. PGal1 overexpression of brl1F391E C343Y, brl1F391E C365S C371S, brl1F391E Y347H, brl1F391E A360D, brl1F391E W368Y, brl1F391E N353D, brl1F391E T355A, or brl1F391E P356G was still toxic as in the case of brl1F391E (Supplemental Figure S2C). In addition, all BRL1 constructs with F391E still induced petal-like structures irrespectively of the DAH mutations (Supplemental Figure S2D), suggesting that the toxicity of overexpressed brl1F391E and the formation of the NE petals do not require a fully intact DAH (especially valid for brl1C365S C371S, which impairs the function of BRL1; Supplemental Figure S2B). Interestingly, brl1C343Y overexpression also had the ability to induce petal-like structures, which were not observed upon overexpression of brl1C365S C371S, brl1Y347H, brl1A360D, brl1W368Y, brl1N353D, brl1T355A, or brl1P356G, indicating a specific function of C343 in Brl1. Why brl1C343Y overexpression is less toxic than overexpressed brl1F391E is unclear (Supplemental Figure S2C); however, we noticed before that not all NE deformations, such as the formation of herniations in apq12∆ cells, affect cell growth (Zhang et al., 2021).
brl1 F391E overexpression promotes growth of the INM and ONM toward the cytoplasm
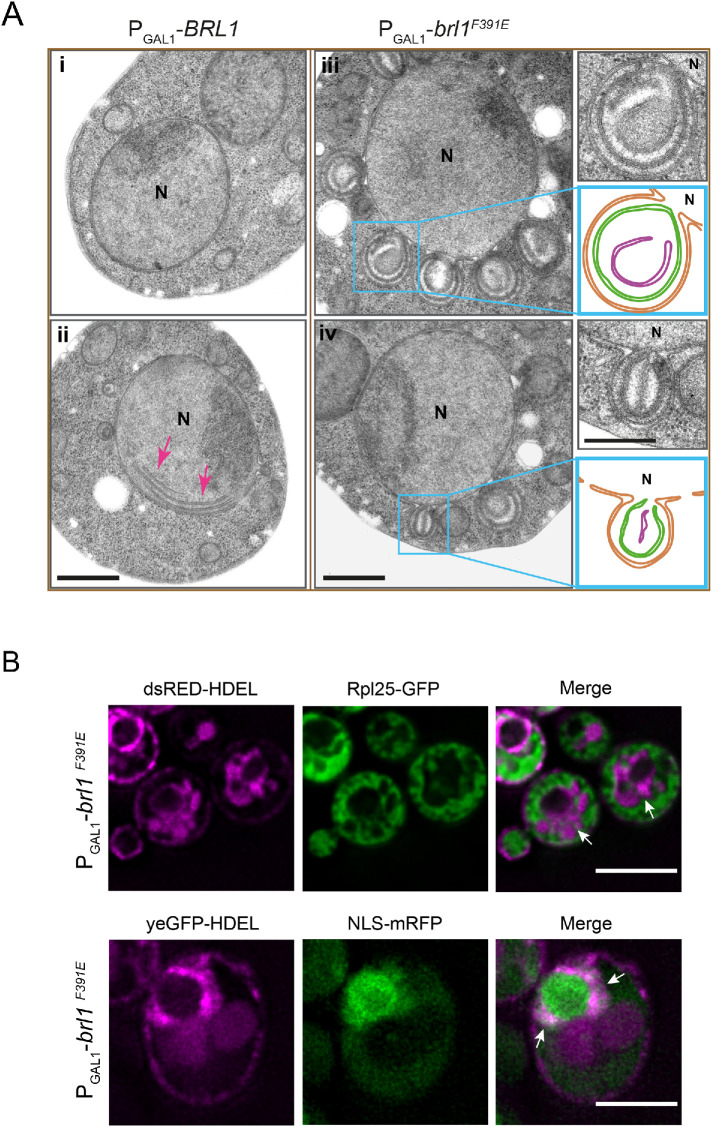
To gain an understanding of the nature of the petal-like structures, we analyzed the PGal1-brl1F391E phenotype by thin-section electron microscopy (EM). Similar structures were observed after 1–5 h of brl1F391E overexpression (Figure 3A). EM analysis identified the NE petals as large cytoplasm-directed ONM/INM deformations that were open toward the nucleus (Figure 3A, iii and iv, and Supplemental Figure S2E). The NE petals often contained horseshoe and ring-like double-membrane assemblies. We also observed that the petals detached from the nucleus and were then localized in the cytoplasm (Supplemental Figure S2F). In contrast, 1–3 h of PGal1-BRL1 overexpression hardly affected NE integrity (Figure 3Ai). However, after 5 h of overexpression, we noticed membrane sheets in the nucleus that were devoid of NPCs (Figure 3Aii, red arrows), consistent with published data (Zhang et al., 2018).
FIGURE 3:
Overexpression of brl1F391E induces formation of petal-like structures at the NE. (A) Electron micrographs of WT cells expressing PGal1-BRL1 or PGal1-brl1F391E 5 h after addition of galactose. The images on the right show enlargements of the boxed brl1F391E-induced petals. Abbreviation: N, nucleus. Scale bars: 500 nm. (B) Petals originating from overexpression of brl1F391E do not contain cytoplasmic material. brl1F391E was overexpressed in cells carrying the membrane marker dsRED-HDEL and the cytoplasmic Rpl25-yeGFP (top). Cells were imaged after 3 h of galactose induction. Arrows indicate NE petals that were marked by dsRED-HDEL but lacked the cytoplasmic Rpl25-yeGFP. In addition, PGal1-brl1F391E cells with yeGFP-HDEL and NLS-mRFP were imaged after 3 h of PGal1 induction (bottom). The arrows indicate yeGFP-HDEL–labeled petals that carried a weak NLS-mRFP signal. Scale bars: 5 µm.
We analyzed the content of these NE-derived petal-like structures that arise after PGal1-brl1F391E overexpression. We used the ribosomal Rpl25-yeGFP as cytoplasmic and NLS-mRFP as nuclear markers. Overexpression of PGal1-brl1F391E in dsRED-HDEL RPL25-yeGFP cells and analysis of the petals by fluorescence microscopy identified that the dsRED-HDEL marked NE extensions as being devoid of a Rpl25-yeGFP signal (Figure 3B, top), demonstrating that the NE petals do not contain cytoplasmic proteins. PGal1-brl1F391E expression in yeGFP-HDEL NLS-mRFP cells revealed that GFP-HDEL labeled petals that overlapped with a weak but clearly detectable NLS-mRFP signal (Figure 3B, bottom), indicating that the petals are connected with the nucleus. Taking the results together, the INM/ONM extensions that arise by brl1F391E overexpression are connected with the nucleus and do not contain cytoplasmic material.
brl1 F391E expression affects localization of the cytoplasmic nucleoporins Nup159, Nsp1, and Nup42
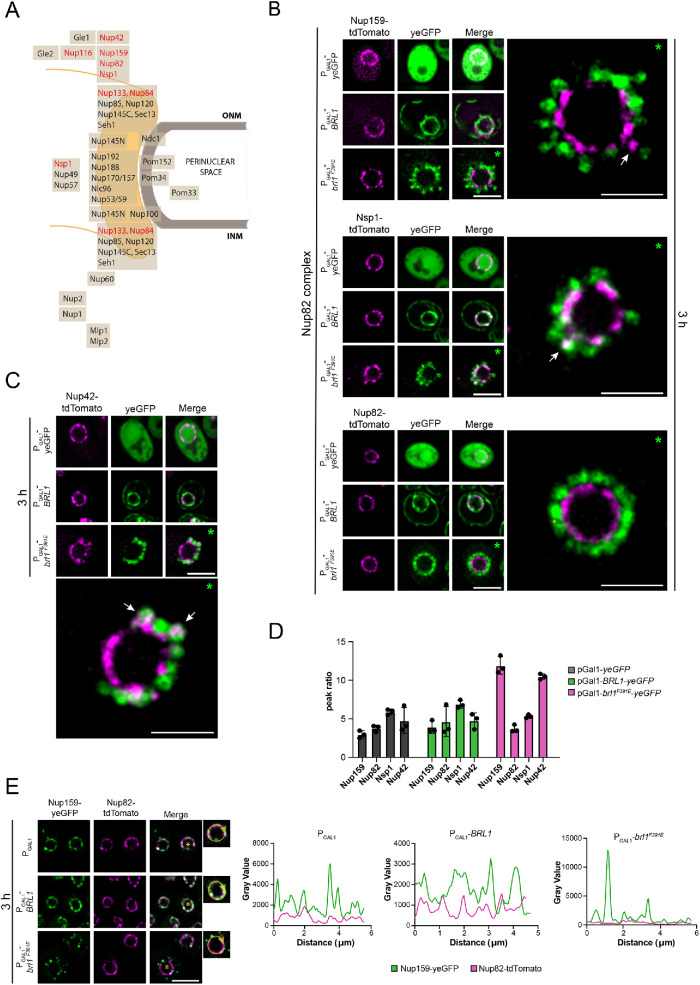
The deformation of the NE in response to brl1F391E overexpression (Figure 3A) prompted us to analyze how PGal1 expressed yeGFP, BRL1-yeGFP, and brl1F391E-yeGFP impact localization of tdTomato-tagged Nups. Specifically, we tested the Nup82 complex components Nup159, Nup82, and Nsp1, the cytoplasmic nucleoporins Nup116 and Nup42, and the Y-complex proteins Nup133 and Nup84 (Figure 4A; Supplemental Figures S4 and S5). Without the induction of the PGal1 promoter all tested Nups showed smooth NE localization (Supplemental Figures S4A and S5A). Overexpression of yeGFP or BRL1-yeGFP did not affect NE localization of any of the Nups (Figure 4, B and C, and Supplemental Figure S5A). Similarly, overexpression of brl1F391E-yeGFP had no detectable impact on the NE distribution of Nup82, Nup84, Nup116, and Nup133 (Figure 4B and Supplemental Figures S4B and S5A). However, we noticed that brl1F391E-yeGFP overexpression affected NE localization of the cytoplasmic nucleoporins Nup159, Nup42, and Nsp1 (Figure 4, B and C, arrows, and Supplemental Figure S4B). Nup159, Nsp1, and Nup42 were no longer uniformly distributed along the NE and also colocalize with some brl1F391E-yeGFP–labeled petals in 43%, 35%, and 74% of the cells, respectively (Figure 4, B and C, arrows). Mislocalization of Nups became further apparent by calculation of the tdTomato peak ratios of the NE line scans. Upon PGal1 induction of yeGFP, BRL1-yeGFP, and brl1F391E-yeGFP, the peak ratio was between 3 and 7 for Nup159, Nup82, Nsp1, and Nup42 with the exception of brl1F391E-yeGFP for NUP159-tdTomato and NUP42-tdTomato, where we measured a peak ratio of 13 as a reflection of the Nup159-tdTomato and Nup42-tdTomato clustering (Figure 4D). This increase was not observed in brl1F391E-yeGFP NSP1-tdTomato cells probably because of the lower penetrance of the phenotype (only 35%) and a less strong clustering phenotype.
FIGURE 4:
Overexpression of brl1F391E affects NE localization of Nup159, Nsp1, and Nup42. (A) Schematic view of the NPC showing the distribution of Nups relative to the ONM and INM. (B) Overexpression of brl1F391E-yeGFP affects localization of Nup159 and Nsp1. brl1F391E was overexpressed for 3 h in cells carrying different tdTomato-tagged components of the Nup82 subcomplex. Nup159-tdTomato (43%) and to a lesser extent Nsp1-tdTomato (35%) showed clustering upon brl1F391E overexpression. Nup82-tdTomato showed uniform localization along the NE even when brl1F391E was overexpressed. The nucleus on the right shows a fivefold enlargement of the nucleus from PGal1-brl1F391E cells on the left. The white arrows on the enlarged nucleus indicate mislocalization of Nup119 and Nsp1. The line scans of the NE signals from NUP159-tdTomato, NUP82-tdTomato, and NSP1-tdTomato cells after 3 h of pGal1 induction are shown in Supplemental Figure S4B. Scale bars: 5 µm; enlargements: 2 µm. (C) PGal1-brl1F391E overexpression affects NE localization of Nup42-tdTomato in 74% of the cells. Nup42-tdTomato was analyzed by fluorescence microscopy in cells expressing yeGFP, BRL1-yeGFP, and brl1F391E-yeGFP for 3 h at 30°C. The nucleus on the right shows a fivefold enlargement of the nucleus from PGal1-brl1F391E-yeGFP cells on the left. The white arrow on the enlarged nucleus indicates mislocalization of Nup42. The line scan of the NE signal from NUP42-tdTomato cells is shown in Supplemental Figure S4B. Scale bars: 5 µm; enlargements: 2 µm. (D) The peak max:min ratio of tdTomato-tagged Nup159, Nup82, Nsp1, and Nup42 cells carrying the indicated PGal1 plasmids after 3 h of PGal1 induction. NE line scans of the tdTomato signal (Supplemental Figure S4B) were used to calculate the peak max:min ratios. (E) PGal1-brl1F391E overexpression in NUP159-yeGFP NUP82-tdTomato cells affects NE localization of Nup159 but not of Nup82. Line scans of the merged signals are shown on the right. Scale bar: 5 µm. (B, C) Green asterisks (*) indicate the nuclei that were used for the enlargement. (E) Yellow asterisks (*) in the nucleus correspond to the enlarged cell on the right used for the scan analysis of the NE signal shown on the right.
We used superresolution structured illumination microscopy (SIM) to confirm the distribution of the Nup82 complex components and Nup42 upon brl1F391E overexpression. After PGal1-BRL1 induction the NPCs were smoothly distributed along the NE, while the NPC distribution in PGal1-brl1F391E–overexpressing cells was affected for Nup159, Nup42, and Nsp1 but not for Nup82 (Supplemental Figure S6), thus confirming the data from conventional fluorescence microscopy.
We directly compared NE localization of Nup159-yeGFP with Nup82-tdTomato and Nup116-tdTomato in the same cell. The Nup159-yeGFP localization was affected upon overexpression of brl1F391E, while Nup82-tdTomato (Figure 4E and Supplemental Figure S4C) and Nup116-tdTomato (Supplemental Figure S5B) still showed a smooth NE distribution. The peak trend of the scan analysis reflected the clustering behavior of Nup159 after 3 h of PGal1-brl1F391E overexpression compared with the PGal1 and PGal1-BRL1 controls (Figure 4E and Supplemental Figure S5B). Thus, a subset of cytoplasmic Nups is affected by brl1F391E overexpression.
We next asked whether brl1F391E overexpression blocks assembly of NPC cores. This was addressed using the recombination-induced tag exchange (RITE) system (Terweij et al., 2013), which allows switching of tags via Cre-induced recombination from NUP188-mCherry to NUP188-yeGFP. Specifically, we tested whether brl1F391E overexpression blocked incorporation of the newly synthesized inner ring complex protein NUP188-yeGFP into the NE, which is expected when Brl1F391E completely blocks NPC assembly. However, we observed that upon switching expression of NUP188-mCherry to NUP188-yeGFP in PGal1-brl1F391E overexpressing cells, Nup188-yeGFP was still incorporated into the NE as indicated by the green fluorescent NE signal similar to PGal1 and PGal1-BRL1 cells (Supplemental Figure S7, A and B). Thus, PGal1-brl1F391E expression does not block the assembly of core NPC structures.
The brl1 F391E-induced petals carry NPC proteins at their NE base
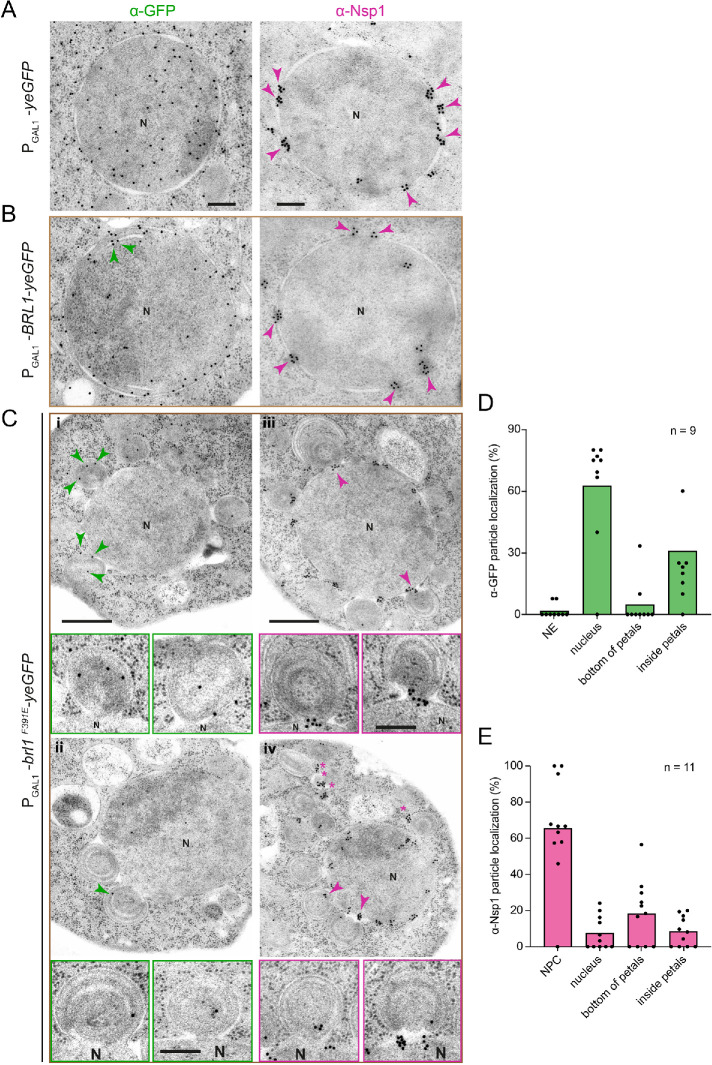
The brl1F391E-induced petals could result from an attempt of NPC assembly that failed because of a defect in the fusion between the INM and ONM. Therefore, the nuclear membranes continuously expand and a restart of this process could explain the dome-like double membranes inside the petals. If this model is correct, we would expect that Nups are associated with the petal base that connects it with the NE. The analysis of the localization of Nsp1 (anti-Nsp1 antibodies followed by protein A-gold) and Brl1-yeGFP (anti-GFP antibodies combined with protein A-gold) by immuno-EM tested this notion. WT control cells stained for GFP did not show gold particles associated with any cellular structure, indicating that background binding of the antibodies and protein A-gold was very low (Supplemental Figure S8A). In yeGFP-expressing cells, the yeGFP signal localized throughout the cells (Figure 5A) and the Nsp1 signal (15 nm gold particles) was detected clustered at NPCs (Figure 5A, magenta arrowheads). Overproduced Brl1-yeGFP was mainly detected along the NE (Figure 5B). In some cells, membrane sheets were detected inside the NE that showed the Brl1-yeGFP label (Figure 5B, green arrowheads). The Nsp1 staining was at NPCs (Figure 5B, magenta arrowheads). Brl1F391E-yeGFP associated with the NE-associated petals (Figure 5Ci, green arrowheads) and localized inside the nucleus (Figure 5, Cii and D). Strikingly, in PGal1-brl1F391E-yeGFP cells the Nps1 signal was at NPCs, inside petal structures (Figure 5Civ and enlargements) and at the base of the petals (Figure 5C, iii and iv, magenta arrowheads and enlargements, and 5E).
FIGURE 5:
Nsp1 localizes at the base of the PGal1-brl1F391E-induced petals. (A–E) Cells expressing (A) yeGFP, (B) BRL1-yeGFP, and (C) brl1F391E-yeGFP for 3 h at 30°C were analyzed for the localization of yeGFP or the yeGFP-tagged Brl1 and Nsp1 by immuno-EM. (A) Magenta arrowheads point toward Nsp1 at NPCs. (B) Green arrowheads point toward Brl1-yeGFP at membrane sheets in the nucleus, and magenta arrowheads point toward Nsp1 at NPCs. (C) Green arrowheads highlight Brl1F391E in NE attached petals (i and ii). Nsp1 is found beside NPCs on the base of the NE-associated petals (iii and iv; magenta arrowheads). Enlargements from cells i–iv show NE-associated petals with Brl1 (green) or Nsp1 signal (magenta) at the base of the petals. (A, B) Scale bars: 200 nm. (C) Scale bar: 500; enlargements: 200 nm. (D, E) Quantification of gold particles reflecting the localization of brl1F391E-yeGFP (D) or Nsp1 (E) from 9 and 11 cells, respectively. Abbreviation: N, nucleus.
To verify the Nsp1 immuno-EM result, we analyzed the localization of Nups using the monoclonal antibody MAB414 that recognizes a subset of yeast Nups (Aris and Blobel, 1989). Immuno-EM of brl1F391E-yeGFP cells using MAB414 detected Nups at the base of the petal structure similar to the Nsp1 antibodies, confirming the result of the Nsp1 staining (Supplemental Figure S8B).
DISCUSSION
How NPCs assemble into the intact NE of interphase human cells or the NE of budding yeast with its closed mitosis is still largely unknown (Beck and Hurt, 2017; Otsuka and Ellenberg, 2018). Previously, in search for functional elements that could be involved in membrane fusion, we identified an amphipathic α-helix in Apq12 that resides in the perinuclear space and connects the two transmembrane regions (Zhang et al., 2018, 2021). We showed that the amphipathic α-helix of Apq12, most likely by inducing local phosphatidic acid (PA) accumulation at the NE, promotes NPC assembly. AlphaFold Protein Structure Database predictions of Brl1/Brr6-like proteins identified an α-helix with amphipathic features in the perinuclear space close to TM2 (Figure 1B; Supplemental Figure S1). An additional prominent feature of the perinuclear space of Brl1/Brr6-like proteins is the DAH linked to TM1. The DAH is stabilized by two disulfide bonds (Figure 1B). We previously showed that the inner disulfide bond of Brl1 (Cys352-Cys365) is essential for the function of the protein (Zhang et al., 2018), while Cys371 (Cys371Arg or Cys371Ser) mutations were reported to cause a conditional lethal growth defect (Saitoh et al., 2005; Stirling et al., 2011; Zhang et al., 2018). brl1Y347H also causes temperature-sensitive growth defect (Supplemental Figure S2B). In conclusion, this emphasizes the importance of this stabilized DAH for the function of Brl1.
Mutations in the predicted AαH (brl1F391E, brl1L402E) that affect its amphipathic nature or deform the helix (brl1F391P) disrupt the essential function of BRL1 (Figure 1C and Supplemental Figure S2B). However, brl1F391E, brl1F391, or brl1L402E overexpression shows that it is not a simple loss of function because it leads to a severe growth defect (Figure 2) and a phenotype coined by a novel type of herniation, petal-like structures, that similar to “conventional” herniations carry NPC proteins on an INM/ONM opening close to the base at the NE (Thaller and Lusk, 2018; Zhang et al., 2018). This opening connects the petal with the nucleus as suggested by EM analysis and the observation that nuclear yeGFP diffuses inside the petals (Figure 3B). The size of the petals is much larger than conventional herniations (Wente and Blobel, 1993; Thaller and Lusk, 2018), suggesting that brl1F391E overexpression promotes membrane growth or bypasses a control mechanism that normally stops membrane expansion during NPC assembly. Another unusual feature in brl1F391E cells is horseshoe-like structures and closed double-membrane rings inside the petals. At present, we do not understand how these membranous structures form. Fusion between the INM and ONM at the base of the petals could give rise to these double-membrane enclosures.
We observed that brl1F391E overexpression does not block NPC assembly per se, as judged based on the proper NE localization of core Nups and the incorporation of newly synthesized Nup188 after PGal1-brl1F391E induction (Supplemental Figure S7), but affects proper incorporation of the cytoplasmic Nsp1, Nup42, and Nup159 into the NPC. These observations are consistent with an NPC assembly attempt that fails because of an INM/ONM fusion defect and therefore failure in the recruitment of cytoplasmic Nups to the NPC core that is still shielded by the NE from the cytoplasm. It is therefore likely that some cytoplasmic Nups are incorporated from the cytoplasmic side after the NPC core assembled in the NE from the nuclear side and fusion of both NE membranes. Such a delayed recruitment of cytoplasmic Nups compared with core Nups was also observed during the interphase assembly of human NPC where the cytoplasmic filament protein NUP358 becomes incorporated later than the Y-complex component NUP107 (Otsuka et al., 2016).
An attractive model is that the AαH, which is connected to the TM2 and therefore close to the INM, embeds its hydrophobic phase into the INM on the perinuclear side. During the NPC assembly process, the AαH could become repositioned, either released from the INM or transferred to the ONM, as soon as the INM/ONM come close together because of the deformation of the INM during NPC assembly. In our model, Brl1F391E competes with WT Brl1 on NPC assembly sites as suggested by the partial suppression of the brl1F391E growth defect by overexpressed BRL1 (Figure 2B). Brl1F391E could be specifically defective in INM/ONM fusion while leaving other properties of Brl1, such as binding to NPC assembly intermediates, unaffected. It is also possible that the F391E mutation partially activates Brl1 because the affected AαH can no longer be inserted into the NE and then promote expansion of the INM and ONM without allowing proper fusion of the INM/ONM.
The DAH in Brl1/Brr6-like proteins proteins is a conserved feature that is essential for the function of these proteins as is the AαH (Lo Presti et al., 2007; Tamm et al., 2011; Zhang et al., 2018). The close proximity of DAH and AαH in the perinuclear space raises the possibility of a cooperation of both functional elements of Brl1. However, our data do not support a functional link between the AαH and the DAH at least for the overexpressed brl1F391E, because brl1F391E DAH double mutations did not affect the strong toxic effect of brl1F391E overexpression or the formation of NE petals (Supplemental Figure S2, C and D).
How do the two paralogues, Brl1 and Brr6, which both have essential functions (de Bruyn Kops and Guthrie, 2001; Hodge et al., 2010; Lone et al., 2015; Zhang et al., 2018), cooperate together in NPC assembly? AlphaFold also predicts a DAH in Brr6. This DAH is connected to an α-helix, similar to what we observed for Brl1 (Supplemental Figure S1A). Because Brl1 is predominately associated with the INM while Brr6 is enriched at the ONM (Zhang et al., 2018), both proteins could have similar functions from distinct sites of the NE. Consistent with this notion is the observation that BRL1 can rescue the cold sensitivity of the brr6-1 allele, indicating a partial functional overlap between the two genes in S. cerevisiae (Saitoh et al., 2005). Interestingly, S. pombe encodes only BRR6 (SpBRL1) but not BRL1 (Tamm et al., 2011) and SpBRL1 is able to rescue the essential functions of S. cerevisiae BRL1 and BRR6 (Saitoh et al., 2005). Perhaps SpBRL1 localizes on the INM and ONM in S. pombe or interacts with binding partners that in S. cerevisiae are divided between Brl1 and Brr6.
Brl1 and Brr6 are conserved proteins of organisms with closed mitosis (Tamm et al., 2011), raising the question about the proteins that take over their function in vertebrates. Knowing the principals and structural elements in the model organism, yeast, will help in identifying proteins in vertebrates that promote NE fusion during interphase NPC assembly.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Yeast strains and plasmids
All strains and plasmids used in this study are listed in Supplemental Table S1. The yeast strains used are derived from ESM-356-1 (MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1). PCR-based methods were used for endogenous gene tagging and gene deletions (Knop et al., 1999; Janke et al., 2004). Yeast strains were grown in YPD (yeast extract, peptone, and glucose), SC (synthetic complete) medium, or SC-selection medium lacking a base or amino acid (Rose and Fink, 1987) at 23°C, 30°C, or 37°C. To induce the expression of proteins under the PGal1 promoter, the raffinose-based SC media was supplemented with galactose added to a final concentration of 2%. Alkaline lysis and trichloroacetic acid precipitations were used to prepare yeast extracts to allow analysis of protein levels by immunoblotting (Janke et al., 2004). The growth defect test was performed, growing cells overnight in selective medium and adjusting the cell density to OD600 = 1. Afterward the cell suspension was 10-fold serial diluted onto selection plates that were incubated at the indicated temperatures.
Electron microscopy
High-pressure frozen yeast samples were freeze-substituted, sectioned, and labeled for EM as described: cells were collected onto a 0.45 μm polycarbonate filter (Millipore) using vacuum filtration and then high-pressure frozen with a HPM010 (Abra-Fluid, Switzerland) freezing machine. Cells were freeze-substituted using the EM-AFS2 device (Leica Microsystems, Vienna, Austria; freeze substitution solution: 0.1% glutaraldehyde, 0.2% uranyl acetate, 1% water—dissolved in anhydrous acetone) and stepwise infiltrated with Lowicryl HM20 resin (Polysciences, Warrington, PA), started by a low temperature of –90°C. For polymerization the samples were finally exposed to UV light for 48 h at –45°C and were gradually warmed up to 20°C. Resin blocks with embedded yeast cells were serially sectioned using a Reichert Ultracut S Microtome (Leica Instruments, Vienna, Austria) to a thickness of 80 nm. Poststaining with 3% uranyl acetate and lead citrate was performed. Sections were imaged on a Jeol JE-1400 (Jeol Ltd., Tokyo, Japan) operating at 80 kV equipped with a 4k x 4k digital camera (F416; TVIPS, Gauting, Germany). Micrographs were adjusted in brightness and contrast using ImageJ (National Institutes of Health, Bethesda, MD).
For immunolabeling on sections of yeast cells, primary antibodies were used against green fluorescent protein (GFP), Nsp1, or MAB414. Samples were prepared similarly as described above with the exception that the glutaraldehyde was omitted from the freeze substitution solution. Sections on slot grids were treated with blocking buffer (1.5% bovine serum albumin, 0.1% fish skin gelatin in phosphate-buffered saline [PBS]) and then incubated with the primary antibody (rabbit anti-GFP [homemade], mouse anti-Nsp1, and mouse MAB414) followed by PBS washes and incubation with the linker protein rabbit anti-mouse and finally marked with protein A-gold conjugates (15 nm; Utrecht University, Utrecht, Netherlands). As a control, thin sections of WT cells not expressing GFP were incubated with anti-GFP antibodies followed by protein A-gold or antigen-containing cells were incubated only with protein A-gold. Poststaining as usual was performed with 3% uranyl acetate and lead citrate.
Fluorescence microscopy
A DeltaVision RT system (Olympus IX71 based; Applied Precision Ltd.) equipped with a Photometrics CoolSnap HQ camera (Roper Scientific), a 100×/1.4-NA Super-Plan Apochromat oil objective (Olympus), a four-color Standard Insight SSI module light source, a workstation with a CentOS operating system, and softWoRx software (Applied Precision Ltd.) was used for cell imaging. Imaging was done at 30°C or 37°C with GFP or mCherry channels. The imaging was conducted with the same exposure and illumination setting to allow direct comparison of the results, and one single stack was used for the analysis. Images were deconvolved with softWoRx software (Applied Precision) and processed with ImageJ (National Institutes of Health, Bethesda, MD). Imaging experiments and quantifications were performed three times and analyzed with GraphPad Prism software.
SIM
For SIM analysis, cells were fixed on a glass coverslip for 30 min with 4% paraformaldehyde and 2% sucrose in PBS buffer. After several wash steps with PBS, the coverslips were mounted on glass slides with Prolong Glass mounting medium. A Nikon N-SIM microscope system (equipped with total internal reflection fluorescence Apochromat 100 × 1.49 NA oil immersion objective and a single photon-detection, electron-multiplying, charge-coupled device camera [iXon3 DU-897E; Andor Technology]) was used to image the samples. tdTomato-tagged proteins were imaged using a 561 nm laser combined with emission bandpass filter 610/60. A single stack of nucleus cross-sections was imaged, and images were reconstructed using NIS imaging and image analysis software (Nikon).
RITE
We used the RITE system published by Terweij et al. (2013). Estradiol (1 µM [final concentration]) was added to the culture to induce recombination. At the same time, expression from the Gal1 promoter was induced by adding 2% galactose. Cells were incubated for 2 h at 30°C with shaking and then imaged.
Antibodies
The antibodies of this study were as follows: rabbit anti-GFP (immuno-EM, 1:5; gift from M. Seedorf, Zentrum für Molekulare Biologie, Heidelberg, Germany), mouse anti-Nsp1 (immuno-EM, 1:100; ab4641; Abcam), and mouse anti-MAB 414 (immuno-EM, 1:100; ab24609; Abcam).
Supplementary Material
Acknowledgments
We acknowledge the electron microscopy and Nikon imaging facilities of Heidelberg University for their support. This work is supported by a grant from the Deutschen Forschungsgemeinschaft (DFG) to E. S. (DFG Schi 295/5-3).
Abbreviations used:
- AαH
amphipathic alpha helix
- DAH
disulfide-stabilized. Anti-parallel helix bundle
- ER
endoplasmic reticulum
- FG
phenylalanine glycine
- 5-FOA
5-fluoroorotic acid
- INM
inner nuclear membrane
- NE
nuclear envelope
- NPC
nuclear pore complex
- Nup
nucleoporin
- ONM
outer nuclear membrane
- PA
phosphatidic acid
- SIM
structured illumination microscopy
- TM
transmembrane.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-12-0616) on March 16, 2022.
REFERENCES
- Aitchison JD, Blobel G, Rout MP (1995). Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J Cell Biol 131, 1659–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang WH, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. (2007). The molecular architecture of the nuclear pore complex. Nature 450, 695–701. [DOI] [PubMed] [Google Scholar]
- Allegretti M, Zimmerli CE, Rantos V, Wilfling F, Ronchi P, Fung HKH, Lee CW, Hagen W, Turonova B, Karius K, et al. (2020). In-cell architecture of the nuclear pore and snapshots of its turnover. Nature 586, 796–800. [DOI] [PubMed] [Google Scholar]
- Aris JP, Blobel G (1989). Yeast nuclear-envelope proteins cross react with an antibody against mammalian pore complex proteins. J Cell Biol 108, 2059–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Hurt E (2017). The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol 18, 73–89. [DOI] [PubMed] [Google Scholar]
- de Bruyn Kops A, Guthrie C (2001). An essential nuclear envelope integral membrane protein, Brr6p, required for nuclear transport. EMBO J 20, 4183–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet CM, Talamas JA, Hetzer MW (2010). Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell 141, 1030–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Gorlich D (2007). A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130, 512–523. [DOI] [PubMed] [Google Scholar]
- Gardner JM, O’Toole E, Jaspersen SL (2021). A mutation in budding yeast BRR6 affecting nuclear envelope insertion of the spindle pole body. MicroPubl Biol 2021, 10.17912/micropub.biology.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier R, Douguet D, Antonny B, Drin G (2008). HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics 24, 2101–2102. [DOI] [PubMed] [Google Scholar]
- Hodge CA, Choudhary V, Wolyniak MJ, Scarcelli JJ, Schneiter R, Cole CN (2010). Integral membrane proteins Brr6 and Apq12 link assembly of the nuclear pore complex to lipid homeostasis in the endoplasmic reticulum. J Cell Sci 123, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M (2004). A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962. [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E (1999). Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Lone MA, Atkinson AE, Hodge CA, Cottier S, Martinez-Montanes F, Maithel S, Mene-Saffrane L, Cole CN, Schneiter R (2015). Yeast integral membrane proteins Apq12, Brl1, and Brr6 form a complex important for regulation of membrane homeostasis and nuclear pore complex biogenesis. Eukaryot Cell 14, 1217–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti L, Cockell M, Cerutti L, Simanis V, Hauser PM (2007). Functional characterization of Pneumocystis carinii brl1 by transspecies complementation analysis. Eukaryot Cell 6, 2448–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R, Watkins JL, Wente SR (1996). GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol Biol Cell 7, 1921–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E, Tang JH, Andersen KR, Knockenhauer KE, Vallotton P, Derrer CP, Kralt A, Mugler CF, Chan LY, Schwartz TU, Weis K (2017). Natively unfolded FG repeats stabilize the structure of the nuclear pore complex. Cell 171, 904–917.e919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka S, Bui KH, Schorb M, Hossain MJ, Politi AZ, Koch B, Eltsov M, Beck M, Ellenberg J (2016). Nuclear pore assembly proceeds by an inside-out extrusion of the nuclear envelope. eLife 5, e19071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka S, Ellenberg J (2018). Mechanisms of nuclear pore complex assembly—two different ways of building one molecular machine. FEBS Lett 592, 475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka S, Steyer AM, Schorb M, Heriche JK, Hossain MJ, Sethi S, Kueblbeck M, Schwab Y, Beck M, Ellenberg J (2018). Postmitotic nuclear pore assembly proceeds by radial dilation of small membrane openings. Nat Struct Mol Biol 25, 21–28. [DOI] [PubMed] [Google Scholar]
- Rampello AJ, Laudermilch E, Vishnoi N, Prophet SM, Shao L, Zhao CG, Lusk CP, Schlieker C (2020). Torsin ATPase deficiency leads to defects in nuclear pore biogenesis and sequestration of MLF2. J Cell Biol 219, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Fink GR (1987). KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell 48, 1047–1060. [DOI] [PubMed] [Google Scholar]
- Saitoh Y, Ogawa K, Nishimoto T (2005). Brl1p—a novel nuclear envelope protein required for nuclear transport. Traffic 6, 502–517. [DOI] [PubMed] [Google Scholar]
- Scarcelli JJ, Hodge CA, Cole CN (2007). The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J Cell Biol 178, 799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling PC, Bloom MS, Solanki-Patil T, Smith S, Sipahimalani P, Li ZJ, Kofoed M, Ben-Aroya S, Myung K, Hieter P (2011). The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet 7, e1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm T, Grallert A, Grossman EP, Alvarez-Tabares I, Stevens FE, Hagan IM (2011). Brr6 drives the Schizosaccharomyces pombe spindle pole body nuclear envelope insertion/extrusion cycle. J Cell Biol 195, 467–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terweij M, van Welsem T, van Deventer S, Verzijlbergen KF, Menendez-Benito V, Ontoso D, San-Segundo P, Neefjes J, van Leeuwen F (2013). Recombination-induced tag exchange (RITE) cassette series to monitor protein dynamics in Saccharomyces cerevisiae. G3 (Bethesda, Md.) 3, 1261–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaller DJ, Lusk CP (2018). Fantastic nuclear envelope herniations and where to find them. Biochem Soc Trans 46, 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Blobel G (1993). A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J Cell Biol 123, 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Yarar D, Giddings TH Jr, Mastronarde DN (1997). Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol Biol Cell 8, 2119–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Khan A, Neuner A, Vitale J, Rink K, Lüchtenborg C, Brügger B, Söllner TH, Schiebel E (2021). A short perinuclear amphipathic α-helix in Apq12 promotes nuclear pore complex biogenesis. Open Biol, doi: 10.1098/rsob.210250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Neuner A, Rüthnick D, Sachsenheimer T, Lüchtenborg C, Brügger B, Schiebel E (2018). Brr6 and Brl1 locate to nuclear pore complex assembly sites to promote their biogenesis. J Cell Biol 217, 877–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.