Abstract
Context
Biomarkers that can accurately predict risk of type 1 diabetes (T1D) in genetically predisposed children can facilitate interventions to delay or prevent the disease.
Objective
This work aimed to determine if a combination of genetic, immunologic, and metabolic features, measured at infancy, can be used to predict the likelihood that a child will develop T1D by age 6 years.
Methods
Newborns with human leukocyte antigen (HLA) typing were enrolled in the prospective birth cohort of The Environmental Determinants of Diabetes in the Young (TEDDY). TEDDY ascertained children in Finland, Germany, Sweden, and the United States. TEDDY children were either from the general population or from families with T1D with an HLA genotype associated with T1D specific to TEDDY eligibility criteria. From the TEDDY cohort there were 702 children will all data sources measured at ages 3, 6, and 9 months, 11.4% of whom progressed to T1D by age 6 years. The main outcome measure was a diagnosis of T1D as diagnosed by American Diabetes Association criteria.
Results
Machine learning–based feature selection yielded classifiers based on disparate demographic, immunologic, genetic, and metabolite features. The accuracy of the model using all available data evaluated by the area under a receiver operating characteristic curve is 0.84. Reducing to only 3- and 9-month measurements did not reduce the area under the curve significantly. Metabolomics had the largest value when evaluating the accuracy at a low false-positive rate.
Conclusion
The metabolite features identified as important for progression to T1D by age 6 years point to altered sugar metabolism in infancy. Integrating this information with classic risk factors improves prediction of the progression to T1D in early childhood.
Keywords: type 1 diabetes, prediction, integration, machine learning
The development of type 1 diabetes (T1D) is driven by an interaction between genetic and environmental factors. The relationships and roles of human leukocyte antigen (HLA) and other genes as they affect development of islet autoimmunity and subsequent progression to T1D continues to be refined (1-4). Environmental and biomarker discovery research, as well as examination of the interplay between potential risk factors and gene variants, has provided insights into T1D risk and potential pathogenic mechanisms (5-11).
The Environmental Determinants of Diabetes in the Young (TEDDY) study has followed thousands of children who are at increased genetic risk of T1D and has collected diverse data, such as infant characteristics, family history, diet, genetics, islet autoantibodies (IAAbs), and metabolomics. Previous analyses have generated a considerable list of risk factors associated with T1D, such as high-risk genotypes, genetic risk scores (GRS) computed from T1D-associated single-nucleotide variations (SNVs; formerly single-nucleotide polymorphisms [SNPs]), prebiotic or probiotic exposure, and timing of gluten exposure. A key goal of TEDDY, as well as other large cohort studies, is to develop models to predict onset of T1D. Diagnosing children as early as possible has the potential to reduce the risk of diabetic ketoacidosis at onset as well as reduce the risk of subsequent complications by blunting initial hyperglycemia and reducing subsequent glucose excursions through improved glycemic control (12-15). One approach to address the challenge of prediction is machine learning, which can build mathematical models to discriminate individuals into groups based on multiple risk factors and observed data for defined outcomes of interest (16-22).
Machine learning predictive models are at the center of precision medicine because they can predict the future outcome of an individual as a probability estimate based on the current input data for that person. Within individual binary risk factors, such as HLA genotype, all patients are essentially given a binary probability, the same probability for all individuals if they have the at-risk HLA genotype. By extending this concept to quantitative variables, such as GRS (23), more refined probabilities can be assigned using classical statistical machine learning methods, such as logistic regression (4). Adding additional risk factors to the machine learning model can continue to refine these individual-based predictions and, depending on the discriminatory power of the feature, can either increase or decrease the overall accuracy of the model. Thus, these models can be interrogated to identify the specific risk factors that work well together, as a multivariate panel, to enable segregation of the class of interest. In the context of T1D research, predictive modeling, to date, has mostly focused on the separation of controls from those with T1D based on genetics (4, 23). However, more recently, prognostic evaluations using other measures have become common as they have clear applicability to screening in high-risk children (6).
Prior work in T1D birth cohorts have demonstrated associations between genetic background and specific environmental exposures with T1D-related outcomes of interest, specifically for children younger than 6 years, based on genetic screening and the prospective follow-up information collected on the TEDDY enrolled children beginning at birth (6). Herein, we use machine learning to evaluate the potential of multiple sources of information on each TEDDY participant collected during infancy, specifically before age 9 months, to predict the likelihood they will develop T1D by age 6 years. To make the predictions we explore a combination of demographic data, such as sex, family history, and dietary information, in combination with genetic and untargeted plasma metabolomic profiles. Although the statistical association of the risk factors used have been evaluated via multiple studies, this is the first attempt to integrate all these factors with metabolomics into a single machine learning model from which the probability of development of T1D by age 6 years can be used to evaluate accuracy. We then evaluate prediction models at each time point to determine the benefit of screening at 1, 2, or 3 time points, as well as the age at which the screening is performed. The data-driven feature selection approach allows a collection of specific demographics, dietary, genetic, and metabolomic features to be identified and used to accurately classify children at age 9 months into their 6-year T1D risk outcome.
Materials and Methods
Study Design and Measurements
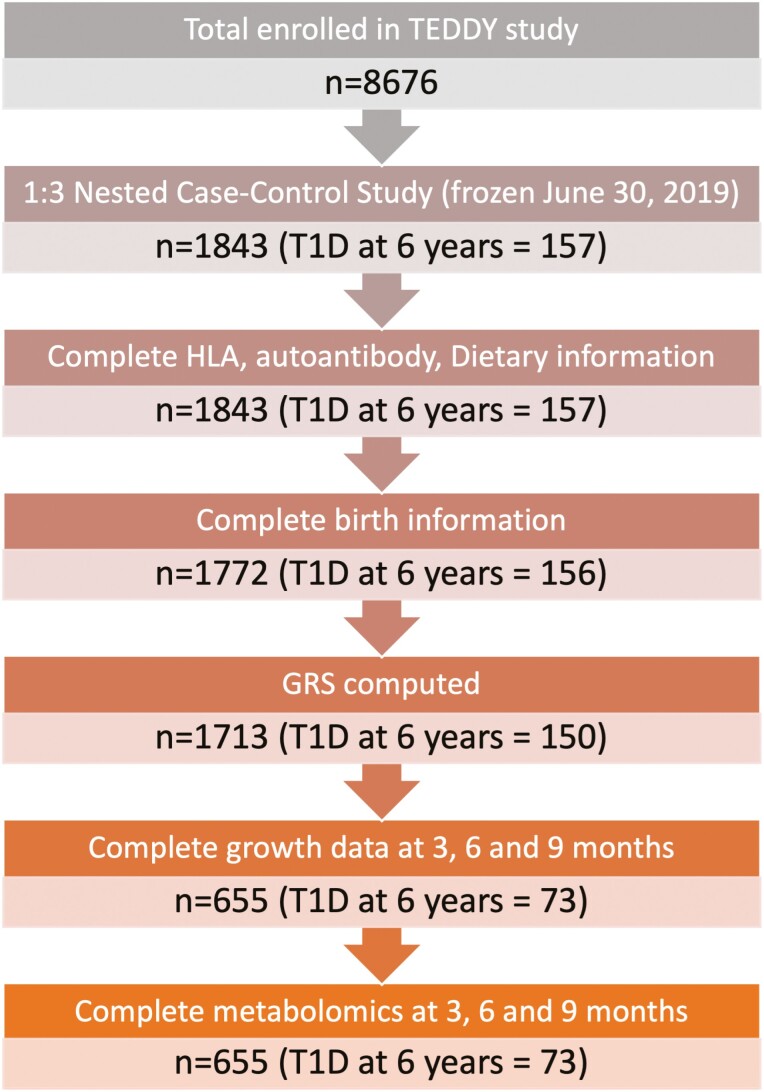
TEDDY is a prospective cohort study following children recruited from the general population at birth, based on having increased risk of T1D, identified through T1D-associated high-risk HLA genotypes or a relative with T1D. TEDDY participants were recruited before age 4.5 months at 6 study sites in 4 countries: the United States, Germany, Sweden, and Finland (24). The ethics committee or institutional review board approved the TEDDY study as applicable to each country. Written informed consents were obtained from a parent or primary caregiver for all participants for genetic screening and the prospective follow-up separately. Participants were evaluated for the development of islet autoimmunity every 3 months thereafter until either the development of T1D or age 4 years. After age 4 years, those with autoantibody seroconversion continued visits every 3 months and the remainder were evaluated every 6 months. We used features that were collected before or at age 9 months, including those participants with clinic visits at approximately age 3, 6, and 9 months. As seen in Fig. 1, there are 8676 children enrolled in TEDDY. This analysis focused on the 1843 distinct children selected for the 1:3 matched, nested, case-control study used for omics analyses (25). Of these 1843 children, 655 had complete data available from the TEDDY Data Coordinating Center, including demographic data (eg, infant diet, family history), birth measurements, GRS, HLA genotypes, IAAb status at age 9 months, and metabolomics data at age 3, 6, and 9 months. T1D was diagnosed using American Diabetes Association criteria (25, 26).
Figure 1.
Flowchart of selection of children from The Environmental Determinants of Diabetes in the Young (TEDDY) cohort.
General infant attributes
There were 24 factors that do not require an assay-based test that are considered as the general infant attributes (GIA) data set. This included an initial set of 4 infant birth data (sex, gestational age, birth length, birth weight), 12 growth measures (height and weight at each of the 3 time points, as well as the change from ages 3 to 6, 3 to 9, and 6 to 9 months), 3 family history data points (any first-degree relative with T1D [FDR-T1D], father with T1D, mother with T1D), and 5 dietary variables (exposure to formula with cow milk by either age 28 days or 6 months, exposure to prebiotics or probiotics by either age 28 days or 6 months, gluten exposure by age 6 months). We tested if the distributions of samples from the 6 clinical centers (Colorado, Georgia, and Washington, USA; and Finland, Germany, and Sweden) were different between our progressors and nonprogressors to T1D by age 6 years. There was not a significant difference (P value = ~0.58; χ2 test of independence) between the fraction from each center that progressed to T1D by age 6 years and those that did not.
Islet autoantibody measurements
Radiobinding assays in 2 laboratories (Barbara Davis Center, Aurora, Colorado, USA, and the University of Bristol Laboratory, Bristol, UK) were used to measure islet autoantibodies; insulin autoantibody (IAA), glutamic acid decarboxylase autoantibody (GADA), and insulinoma-associated antigen 2 autoantibody (IA-2A) as previously described (27, 28). In the 2020 Islet Autoantibody Standardization Workshop, the sensitivities were at minimum 99% for all assays and the specificities were 62%, 78%, and 72% for IAA, GADA, and IA-2A, respectively (29). Children who had 2 or more consecutive confirmed positive samples were defined as persistently IAAb positive unless it was determined to be due to maternal transfer or they developed T1D before the next sample collection. This data set is defined with a categorical variable as either IAAb positive or negative as defined earlier for IAA, GADA, or IA-2A at their 9-month visit. The 3- and 6-month time points were excluded because of small counts (< 5 participants).
Genetic risk score and human leukocyte antigen
When the children were aged 9 to 12 months, the HLA-DR and HLA-DQ genotypes were confirmed by reverse blot hybridization at the central HLA Reference Laboratory at Roche Molecular Systems as previously described (30). For this study these genotypes were translated to 5 categorical variables (DR3/4 [HLA-DR3/HLA-DR4], DR4/4, DR3/3, DR4/8, or other) as previously described (5). The T1D GRS as a single quantitative variable for each TEDDY participant was computed as a weighted sum across 41 T1D-associated non-HLA region SNVs (effect size as weights for the SNV) as previously described (23).
Metabolomics
Untargeted plasma metabolomics were carried out across multiple time points for TEDDY participants based on an existing nested, case-control design (25). This analysis profiled 10 522 plasma samples in which primary metabolites were quantified from citrated plasma using gas chromatography–time-of-flight mass spectrometry (GC-TOF MS) at the NIH West Coast Metabolomics Center at the University of California, Davis. The GC-TOF MS metabolomics data acquisition followed previously described protocols (31) followed by data processing and compound identification using the BinBase algorithm (32) and normalization using Random Forest normalization (33). From these data we specifically extracted the children who had complete quantified metabolomics data collected at the 3-, 6-, and 9-month visits, which yielded a total of 139 known metabolites. All metabolomics data were analyzed as relative abundance on the log2 scale.
Statistical Analysis
Statistical analyses were performed for initial feature filtering (34, 35). For features that were normally distributed, which included the T1D GRS, a standard 2-sample t test was performed. For all categorical features, a Fisher exact test was performed. For those features that did not have a normal distribution (gestational age, growth data, log2-transformed metabolomics data,), a Wilcoxon rank sum test was performed. All statistics were completed using MatLab (R2019a) software.
Machine Learning
Machine learning was performed with a naive Bayes classifier in MatLab (R2019a). Results for each participant were evaluated in the context of the predicted probabilities from 5-fold cross-validation (CV). Models were assessed based on a receiver operating characteristic (ROC) curve, for which the area under the ROC curve (AUC) serves as an overall metric of performance at all possible classification thresholds. A random classifier returns an AUC of 0.5 and a perfect classifier yields an AUC of 1.0. Feature-importance matrices were generated using Repeated Optimization for Feature Interpretation (ROFI) (20, 36). ROFI uses statistical sampling and optimization to identify subsets of features that have a multivariate representation that increases the ability to predict the TEDDY participants who develop T1D from those that do not by age 6 years. The repeated analyses component of ROFI yields importance metrics for each feature that describes the likelihood that a sample will be included as important to the machine learning model. ROFI was implemented using 20 repetitions of 5-fold CV of naive Bayes within a simulated annealing optimization routine (v = 1E-3), convergence criteria of 1E-5, AUC as the optimization metric, and 100 repetitions of the model output. The final subset of features was selected based on frequency of inclusion in the optimized model as previously described with code available in the peppuR package to implement ROFI (https://github.com/pmartR/peppuR) (22).
Results
Summary cohort characteristics of the 31 nonmetabolite features for the 655 TEDDY participants included in this study are given in Table 1. Of the 655 children, 73 (11.2%) developed T1D by age 6 years. Before the machine learning analysis, the 139 metabolites at each of the 3 time points and the 31 features in Table 1 were subjected to a statistics filter to remove features that do not show any statistical association with diagnosis of T1D by age 6 years (34, 35). Since the goal was to evaluate the utility of multiple risk factors in the context of machine learning, an uncorrected and liberal threshold (P < .1) was used for filtering to retain those that may have a weak statistical association but may be important in the context of a multivariate signature. There were 23 time-based metabolite abundances also selected at this threshold; 8 metabolites at age 3 months, 11 metabolites at age 6 months, and 4 metabolites at age 9 months. Five of the 24 GIA features were retained: FDR-T1D, FDR-T1D (father), gestational age, weight at age 9 months, and exposure to formula with cow milk by age 6 months. IAAb status at age 9 months was positively associated with progression to T1D by age 6 years, as well as the T1D GRS and 2 of the HLA genotypes (DR3/4 and DR4/4). The largest Pearson correlation between any 2 quantitative features was approximately 0.5, suggesting that duplicative features should not dramatically affect model generation and feature selection. Within the qualitative features, only the family history metrics (FDR-T1D and FDR-T1D [father]) had high similarity as FDR-T1D (father) is a subset of FDR-T1D.
Table 1.
Characteristics of The Environmental Determinants of Diabetes in the Young (TEDDY) participants categorized for machine learning based on type 1 diabetes outcome at age 6 years
| Data | Feature | Positive T1D at age 6 y | Negative T1D at age 6 y | P |
|---|---|---|---|---|
| No. of participants | 73 | 582 | ||
| GIA | Female | 40 (54.8%) | 263 (45.2%) | .136 |
| GIA | T1D first-degree relative | 24 (32.9%) | 124 (21.3%) | .037 |
| GIA | T1D first-degree relative is mother | 5 (6.8%) | 41 (7.0%) | ≥ .999 |
| GIA | T1D first-degree relative is father | 14 (19.2%) | 64 (11.0%) | .054 |
| GIA | Gestational Age, wk | 39.57 | 40.00 | .006 |
| GIA | Length at birth, cm | 50.80 | 51.00 | .991 |
| GIA | Length at age 3 mo | 62.60 | 62.50 | .641 |
| GIA | Length at age 6 mo | 68.60 | 68.40 | .500 |
| GIA | Length at age 9 mo | 72.60 | 72.50 | .458 |
| GIA | Growth age 3 to 6 mo | 5.60 | 50.50 | .876 |
| GIA | Growth age 3 to 9 mo | 10.0 | 10.0 | .861 |
| GIA | Growth age 6 to 9 mo | 4.50 | 4.40 | .861 |
| GIA | Weight at birth, kg | 3.60 | 3.53 | .214 |
| GIA | Weight at age 3 mo | 6.57 | 6.51 | .228 |
| GIA | Weight at age 6 mo | 8.13 | 8.03 | .128 |
| GIA | Weight at age 9 mo | 9.24 | 9.16 | .084 |
| GIA | Weight gain at age 3 to 6 mo | 1.52 | 1.48 | .246 |
| GIA | Weight gain at age 3 to 9 mo | 2.64 | 2.52 | .250 |
| GIA | Weight gain at age 6 to 9 mo | 1.06 | 1.05 | .477 |
| GIA | Formula (cow milk) before 28 d | 33 (42.2%) | 272 (46.7%) | .901 |
| GIA | Formula (cow milk) before 6 mo | 48 (65.8%) | 451 (77.5%) | .040 |
| GIA | Formula (prebiotic or probiotic) before age 28 d | 16 (21.9%) | 120 (20.6%) | .761 |
| GIA | Formula (prebiotic or probiotic) before age 6 mo | 25 (34.2%) | 203 (34.9%) | ≥ .999 |
| GIA | Gluten before age 6 mo | 30 (41.1%) | 265 (45.5%) | .533 |
| GRS | GRS | 10.5 | 10.2 | .002 |
| HLA | DR3/4 | 43 (58.9%) | 217 (37.3%) | 4.37E-16 |
| HLA | DR4/4 | 8 (11.0%) | 117 (20.1%) | .081 |
| HLA | DR3/3 | 8 (11.0%) | 90 (15.5%) | .385 |
| HLA | DR4/8 | 8 (11.0%) | 106 (18.2%) | .142 |
| HLA | Other | 6 (8.2%) | 52 (8.9%) | ≥ .999 |
| IAAb | Persistent islet autoantibody positive at age 9 mo | 23 (31.5%) | 11 (1.9%) | .001 |
Numbers for qualitative features are percentages as evaluated by Fisher exact test and for qualitative features are the median as evaluated by Wilcoxon rank sum test with the exception of the GRS, which is the mean and evaluated via 2-sample t test. Features in bold italics are used in machine learning.
Abbreviations: GIA, general infant attributes; GRS, genetic risk score; HLA, human leukocyte antigen; T1D, type 1 diabetes.
Feature Selection
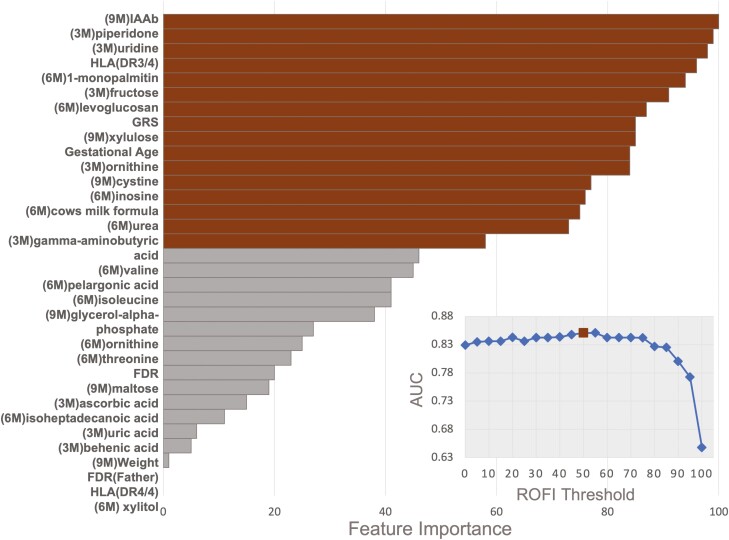
We used ROFI to acquire feature-importance metrics for each of the 32 features included in the initial modeling. Fig. 2 gives the importance metric of each feature, which is the likelihood of inclusion in the model when optimizing AUC within a naive Bayes classifier. A common threshold used in this approach is 50%, which did provide a higher overall accuracy with respect to the AUC (inset to Fig. 2) in this setting. At a ROFI feature-importance threshold of 50%, there were 16 total features. As seen in the inset to Fig. 2, the AUC did not increase when including additional features after these 16 features (red triangle). The final set of 16 features reduced the feature set to 3 genetic and the immunologic features: DR3/4, T1D GRS, and IAAb positivity at age 9 months, 1 infant characteristic (gestational age), 1 dietary marker (cow milk formula < age 6 months), and 11 time-specific metabolite measurements.
Figure 2.
Importance of each feature to the predictive model for all 32 features where brown indicates above the 50% threshold (16 features). Of the 16 features there were 5 nonmetabolite features (islet autoantibody [IAAb] at age 9 months, DR3/4, genetic risk score [GRS], gestational age, and exposure to cow’s milk before age 6 months), 3 metabolites measured at age 3 months, 5 metabolites measured at age 6 months, and 3 metabolites measured at age 9 months. Inset is the area under the curve (AUC) value achieved by feature reduction using ROFI-based thresholds where the x-axis on the left indicates the Repeated Optimization for Feature Interpretation (ROFI) feature importance score and the y-axis on the right indicates the average AUC of the model based on the threshold. The ROFI-based threshold of 50%, brown square, is a common threshold in this is case also relates to a near optimal AUC.
Screening Age and Sampling Time for Metabolite Measurements
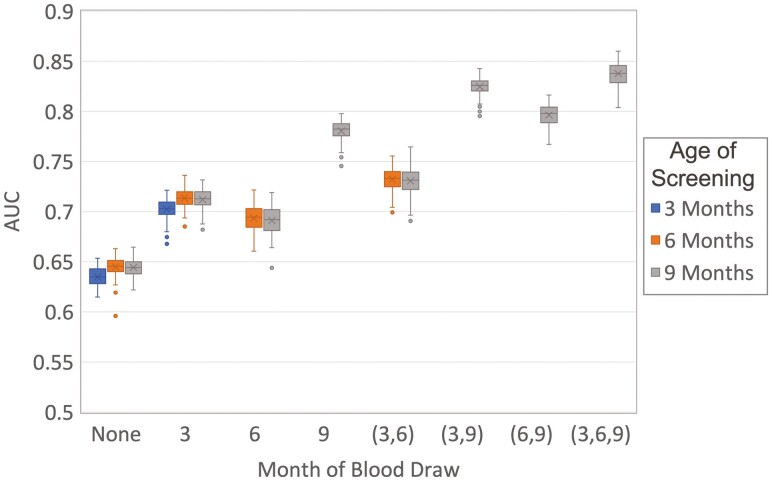
The feature selection in Fig. 2 is a combination of markers across 3 time points, which in practice would require 3 sequential blood samples from a child. To evaluate how well the prediction can be made with less sampling, ROFI was used to evaluate the models based on screening age, as well as the time point(s) for which samples would be drawn as constraints. Each model was optimized in the same fashion as for the full data set, and the final evaluation was based on a 50% feature-importance threshold capturing variability in the CV process through the 100 repetitions of the optimization process. This resulted in 14 distinct models, each with 100 estimates of the AUC, based on the screening age and time points at which blood samples would be available at that screening age. As seen in Fig. 3, if no samples are collected screening was based only on the demographics and yielded a low overall AUC. Adding in metabolomics, HLA and GRS increased the AUC at ages 3 and 6 months; however, there was a dramatic increase in accuracy adding the 9 months as both the sample collection and the primary screening age. This is due to the importance of IAAb as a feature measured at age 9 months in terms of prediction. In adding more blood draws, the increase remained highest when the 9-month sampling was included for the same reason regarding the strong predictive power of the IAAb data set. A Kruskal-Wallis test with a Tukey post hoc adjustment indicated that screening at age 9 months and including all 3 time points does not yield a statistically larger AUC than screening at age 9 months and including only metabolites from ages 3 and 9 months (P = .997). The model including only metabolomics from ages 3 and 9 months returns 13 features, of which the top 16 overlap completely for those specific time points with the addition of glycerol-alpha-phosphate. If only a single time point for a blood draw is selected, then the 9-month time point AUC is significantly larger than all the remaining sampling and screening age combinations. It includes 7 features, which completely overlaps with the top 16 in the full model excluding the 3-and 6-month metabolites.
Figure 3.
Box plot of the area under the curve (AUC) values from 100 repetitions of 5-fold cross-validation based on the number and timing of blood samples drawn from children, where each box represents the month(s) at which a blood draw is taken. Statistical comparisons between AUC values for each sampling/screening combination was performed with a Kruskal-Wallis test with a Tukey post hoc adjustment.
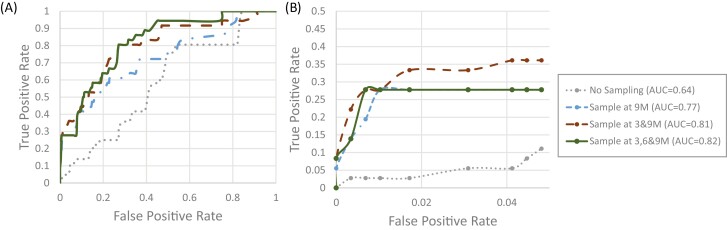
Fig. 4A gives representative global ROC curves associated with the largest AUC at no, 1, 2, or 3 sampling time points assuming a screening age of 9 months. Based on the prior Kruskal-Wallis analysis associated with Fig. 3, the ROC curves visually show the same pattern in which the 3- and 9-month sampling was not statistically different from all 3 time points. The single time point of age 9 months was significantly smaller overall; however, when focusing on a small range of predictions within a false-positive rate (FPR) of less than 0.05, the 3 were more similar, although the 2-sample model of ages 3 and 9 months is highest at the lowest FPRs (Fig. 4B). Specifically, if the percentage of false positives was set to 5% (~29 of the 582 negative participants), then the true-positive rate (TPR) dramatically increased from approximately 7.6% (~5 of the 73 T1D participants) with no sampling to approximately 38% (~28 of the 73 T1D participants) by adding the single time point of age 9 months and adding the second sampling increased the TPR by another 2%.
Figure 4.
Representative receiver operating characteristic curve based on data available at different sampling time point(s) when screening at age 9 months evaluating A, the entire range of false positive, and B, truncated range to 5% false positives. The dotted line represents no blood draws (ie, prediction is based solely on demographic data), the dash-dot line represents a single blood draw at age 9 months, the dashed line blood draws at ages 3 and 9 months, and the solid line includes data from blood draws at all 3 ages.
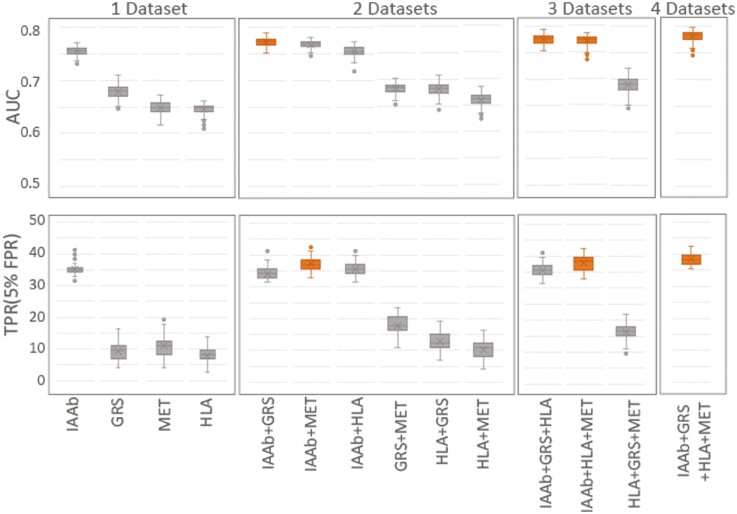
Assay Evaluation
An advantage of machine learning is the inclusion of the multivariate nature in which the various assay types work together in combination with infant measurement, demographic, and dietary information to make a prediction. The full evaluations in Figs. 2 to 4 assume that all 4 distinct assays (IAAb, HLA, GRS, and metabolomics) were included, which may not be necessary in practice. Selecting the 9-month sampling and screening time point as the best single evaluation point, each assay was evaluated individually in addition to the infant measurement, demographic, and dietary information and both the AUC and TPR at a set 5% FPR were used for evaluation (Fig. 5). The addition of the IAAb measurement yielded the largest increase both in the AUC and TPR. When evaluating 2 data sets, adding GRS to the IAAb data set yielded the largest AUC; however, adding the metabolomics to the IAAb data set was the largest gain for the TPR. With 3 data sets, adding the GRS or metabolomics to the IAAb with the HLA data had nearly the same AUC as each other and using all data sets. The maximum TPR with 3 data sets was with IAAb, HLA, and metabolomics. The AUC and TPR values for the 14 models were compared with a Kruskal-Wallis test with a Tukey post hoc test adjustment, which found that using only a 2-data set model of IAAb and GRS returned AUC values that were not statistically different from the full model. Based on the TPR, the best 2-data set model was IAAb and metabolomics, which again was not statistically different from the full model.
Figure 5.
Change in A, area under the curve (AUC), and B, true positive rate (TPR), as collections of assays are included in the model under the constraint that data is collected and evaluated at age 9 months. Statistical comparisons between AUC and TPR values for each data set combination was performed with a Kruskal-Wallis test with a Tukey post hoc adjustment.
Discussion
Here we demonstrated that an overall good prediction of T1D outcomes at age 6 years can be achieved by evaluating children for a small profile of features at age 9 months and integrating this information with their HLA typing, a T1D GRS, and IAAb status at age 9 months. Improvements in prediction can be achieved by adding an additional metabolite screening at age 3 months. It is important to recognize that features are selected as a multivariate model and, thus, univariate interpretations have limited utility as applied to complex biological networks. The machine learning framework allows assignment of individualized probabilities of progressing to T1D before age 6 years in children at a moderately high genetic risk at a 9-month screening, providing opportunities for monitoring, prevention of diabetic ketoacidosis at onset, or enrollment in immune intervention trials.
The top nonmetabolite features identified has having high feature importance (see Fig. 2) are established risk factors, such as IAAb, HLA, GRS, and gestational age (3, 6, 10, 23, 37). We further identified the 9-month age to be the optimal single screening and sampling time point; adding the 3-month as a secondary blood sampling time point increased the AUC from 0.77 to 0.81. We further evaluated the individual assays in the context of the 9-month screening age, noting IAAb as the most predictive data set, and adding either the GRS or metabolomics data to the IAAb gave the largest increase in accuracy, dependent on the metric of evaluation. Likely for clinical utility, a machine learning model would be tailored to a predefined TPR or FPR, and in the context of this study the most robust data types are IAAb and metabolomics, the combination of these two data types having both high AUC and TPR (see Fig. 5).
A limitation of this study is the size and imbalance of the cohort available for the machine learning: 73 children who progressed to T1D vs 582 who were not diagnosed with T1D at age 6 years. In addition, TEDDY is focused on high-risk children and as such the initial case-control design of the metabolomics study has a large proportion of children who are autoantibody positive by age 6 years (130), which likely makes these results specific to children of highest risk. Further, some features, such as gestational age, have a small change on the order of days, which may limit its utility in practice. A second limitation to this study is the use of global metabolomics measurements, and thus it is essential these findings be further validated through targeted metabolomic assay development and evaluation on independent studies, especially since metabolite measurements can change over time.
Our analysis of TEDDY plasma, untargeted metabolomic data, found that approximately 62% of the top metabolites in terms of feature importance, greater than 50, overlap with identified significant longitudinal metabolome profiles associated with the appearance of a first IAAb of multiple autoantibodies (38). It is also observed that several metabolites may be dietary based, such as piperidone and ascorbic acid (39). We note that the observed metabolites in the machine learning model suggest that children who develop T1D before age 6 years may have different sugar metabolite profiles as infants, as the selection of fructose, levoglucosan, glycerol-alpha-phosphate, and xylulose suggest that the carbohydrate metabolism is altered. In addition to altered carbohydrate metabolism, we observed the pentose phosphate pathway and purine degradation metabolism as potential targets of interest in the pathophysiology of T1D. Uridine and inosine are intermediates of the purine degradation pathway, including adenosine 5′-triphosphate, that has uric acid as the end product. Uric acid has been shown to be associated with insulin resistance and type 2 diabetes (40) but its association with T1D is not known. There are also metabolites of the pentose phosphate pathway, such as xylulose, observed in samples taken months preceding the onset of T1D. The alteration of carbohydrate levels, pentose phosphate pathway, and adenosine 5′-triphosphate could be due to a dysfunction of β cells and improper insulin secretion, as these processes are regulated by insulin (41, 42). In fact, β-cell dysfunction has been observed up to 5 years before the onset of T1D (43). Our results suggest alterations in sugar metabolism may start or develop early in infancy in individuals who develop T1D as younger children, but further focused studies are needed to evaluate these hypotheses.
Acknowledgments
The TEDDY Study Group
Colorado Clinical Center: Marian Rewers, MD, PhD, principal investigator (PI),1,4,6,9,10 Aaron Barbour, Kimberly Bautista,11 Judith Baxter,8,911 Daniel Felipe-Morales, Brigitte I Frohnert, MD,2,13 Marisa Stahl, MD,12 Patricia Gesualdo,2,6,11,13 Rachel Haley, Michelle Hoffman,11,12,13 Rachel Karban,11 Edwin Liu, MD,12 Alondra Munoz, Jill Norris, PhD,2,3,11 Stesha Peacock, Hanan Shorrosh, Andrea Steck, MD,3,13 Megan Stern11, Kathleen Waugh.6,7,11 University of Colorado, Anschutz Medical Campus, Barbara Davis Center for Childhood Diabetes, Aurora, Colorado, USA.
Finland Clinical Center: Jorma Toppari, MD, PhD, PI,¥^1,4,10,13 Olli G Simell, MD, PhD, Annika Adamsson, PhD,^11 Sanna-Mari Aaltonen,^ Suvi Ahonen,*±§ Mari Åkerlund,*±§ Leena Hakola,*± Anne Hekkala, MD,µ¤ Henna Holappa,µ¤ Heikki Hyöty, MD, PhD,*±6 Anni Ikonen,µ¤ Jorma Ilonen, MD, PhD,¥¶3 Sanna Jokipuu,^ Leena Karlsson,^ Jukka Kero, MD, PhD,¥^3,13 Miia Kähönen,µ¤11,13 Mikael Knip, MD, PhD,*± Minna-Liisa Koivikko,µ¤ Katja Kokkonen,*± Merja Koskinen,*± Mirva Koreasalo,*±§2 Kalle Kurppa, MD, PhD,*±12 Salla Kuusela, MD,*± Jarita Kytölä,*± Sinikka Lahtinen,*± Jutta Laiho, PhD,*6 Tiina Latva-aho,µ¤ Laura Leppänen,^ Katri Lindfors, PhD,*12 Maria Lönnrot, MD, PhD,*±6 Elina Mäntymäki,^ Markus Mattila,*± Maija Miettinen,§2 Katja Multasuo,µ¤ Teija Mykkänen,µ¤ Tiina Niininen,±*11 Sari Niinistö,§2 Mia Nyblom,*± Sami Oikarinen, PhD,*±6 Paula Ollikainen,µ¤ Zhian Othmani,¥ Sirpa Pohjola,µ¤ Jenna Rautanen,±§ Anne Riikonen,*±§2 Minna Romo,^ Satu Simell, MD, PhD,¥12 Aino Stenius,µ¤11 Päivi Tossavainen, MD,µ¤ Mari Vähä-Mäkilä,¥ Eeva Varjonen,^11 Riitta Veijola, MD, PhD,µ¤13 Irene Viinikangas,µ¤ Suvi M Virtanen, MD, PhD,*±§2 ¥University of Turku, Turku, Finland,*Tampere University, Tampere, Finland,µUniversity of Oulu, Oulu, Finland,^Turku University Hospital, Hospital District of Southwest Finland, Turku, Finland,±Tampere University Hospital, Tampere, Finland,¤Oulu University Hospital, Oulu, Finland, §Finnish Institute for Health and Welfare, Helsinki, Finland.
Georgia/Florida Clinical Center: Richard McIndoe, PhD, PI,4,10 Jin-Xiong She, PhD, Desmond Schatz, MD,*4,7,8 Diane Hopkins,11 Leigh Steed,11,12,13 Jennifer Bryant,11 Katherine Silvis,2 Michael Haller, MD,*13 Melissa Gardiner,11 Ashok Sharma, Laura Jacobsen, MD,*13 John Marks, DHSc,*11,13 P D Towe.* Center for Biotechnology and Genomic Medicine, Augusta University, Augusta, Georgia, USA.*University of Florida, Pediatric Endocrinology, Gainesville, Florida, USA.
Germany Clinical Center: Anette G Ziegler, MD, PI,1,3,4,10 Ezio Bonifacio, PhD,* Cigdem Gezginci, Anja Heublein, Eva Hohoff,¥2 Sandra Hummel, PhD,2 Annette Knopff7, Charlotte Koch, Sibylle Koletzko, MD,¶12 Claudia Ramminger,11 Roswith Roth, PhD8, Jennifer Schmidt, Marlon Scholz, Joanna Stock,8,11,13 Katharina Warncke, MD,13 Lorena Wendel, Christiane Winkler, PhD.2,11 Forschergruppe Diabetes e.V. and Institute of Diabetes Research, Helmholtz Zentrum München, Forschergruppe Diabetes, and Klinikum rechts der Isar, Technische Universität München, Neuherberg, Germany.*Center for Regenerative Therapies, TU Dresden, Dresden, Germany,¶Dr. von Hauner Children’s Hospital, Department of Gastroenterology, Ludwig Maximillians University Munich, Munich, Germany;¥University of Bonn, Department of Nutritional Epidemiology, Bonn, Germany.
Sweden Clinical Center: Åke Lernmark, PhD, PI,1,3,4,5,6,8,9,10 Daniel Agardh, MD, PhD,6,12 Carin Andrén Aronsson, PhD,2,11,12 Maria Ask, Rasmus Bennet, Corrado Cilio, PhD, MD,6 Susanne Dahlberg, Malin Goldman Tsubarah, Emelie Ericson-Hallström, Annika Björne Fors, Lina Fransson, Thomas Gard, Monika Hansen, Susanne Hyberg, Berglind Jonsdottir, MD, PhD,11 Helena Elding Larsson, MD, PhD,6,13 Marielle Lindström, Markus Lundgren, MD, PhD,13 Marlena Maziarz, PhD, Maria Månsson Martinez, Jessica Melin,11 Zeliha Mestan, Caroline Nilsson, Yohanna Nordh, Kobra Rahmati, Anita Ramelius, Falastin Salami, Anette Sjöberg, Carina Törn, PhD,3 Ulrika Ulvenhag, Terese Wiktorsson, Åsa Wimar.13 Lund University, Lund, Sweden.
Washington Clinical Center: William A Hagopian, MD, PhD, PI,1,3,4,6,7,10,12,13 Michael Killian,6,7,11,12 Claire Cowen Crouch,11,13 Jennifer Skidmore,2 Christian Chamberlain, Brelon Fairman, Arlene Meyer, Jocelyn Meyer, Denise Mulenga,11 Nole Powell, Jared Radtke, Shreya Roy, Davey Schmitt, Sarah Zink . Pacific Northwest Research Institute, Seattle, Washington, USA.
Pennsylvania Satellite Center: Dorothy Becker, MD, Margaret Franciscus, MaryEllen Dalmagro-Elias Smith,2 Ashi Daftary, MD, Mary Beth Klein, Chrystal Yates . Children’s Hospital of Pittsburgh of UPMC, Pittsburgh, Pennsylvania, USA.
Data Coordinating Center: Jeffrey P Krischer, PhD, PI,1,4,5,9,10 Rajesh Adusumali, Sarah Austin-Gonzalez, Maryouri Avendano, Sandra Baethke, Brant Burkhardt, PhD,6 Martha Butterworth,2 Nicholas Cadigan, Joanna Clasen, Kevin Counts, Christopher Eberhard, Steven Fiske,8 Laura Gandolfo, Jennifer Garmeson, Veena Gowda, Belinda Hsiao, Christina Karges, Qian Li, PhD,2,3 Shu Liu, Xiang Liu, PhD,2,3,8,13 Kristian Lynch, PhD,6,8 Jamie Malloy, Cristina McCarthy,11 Jose Moreno, Hemang M Parikh, PhD,3,8 Cassandra Remedios, Chris Shaffer, Susan Smith,11 Noah Sulman, PhD, Roy Tamura, PhD,1,2,11,12,13 Dena Tewey, Michael Toth, Ulla Uusitalo, PhD,2 Kendra Vehik, PhD,4,5,6,8,13 Ponni Vijayakandipan, Melissa Wroble, Jimin Yang, PhD, RD,2 Kenneth Young, PhD. (Past staff: Michael Abbondondolo, Lori Ballard, Rasheedah Brown, David Cuthbertson, Stephen Dankyi, David Hadley, PhD, Kathleen Heyman, Francisco Perez Laras, Hye-Seung Lee, PhD, Colleen Maguire, Wendy McLeod, Aubrie Merrell, Steven Meulemans, Ryan Quigley, Laura Smith, PhD.8,11) University of South Florida, Tampa, Florida, USA.
Project Scientist: Beena Akolkar, PhD.1,3,4,5,6,7,9,10 National Institutes of Diabetes and Digestive and Kidney Diseases, Bethesda, Maryland, USA.
Autoantibody Reference Laboratories: Liping Yu, MD,^5 Dongmei Miao, MD,^ Kathleen Gillespie,*5 Alistair Williams,* Kyla Chandler,* Ilana Kelland,* Yassin Ben Khoud,* Matthew Randell.*^Barbara Davis Center for Childhood Diabetes, University of Colorado Denver, Aurora, Colorado, USA;*Bristol Medical School, University of Bristol, Bristol, UK.
Cortisol Laboratory: Elisabeth Aardal Eriksson, MD, PhD, Ing-Marie Lundgren, Ewa Lönn Karlsson, Dzeneta Nezirevic Dernroth, PhD. Department of Clinical Chemistry, Linköping University Hospital, Linköping, Sweden.
Dietary Biomarkers Laboratory: Iris Erlund, PhD2, Irma Salminen, Jouko Sundvall, Nina Kangas, Petra Arohonka . Finnish Institute for Health and Welfare, Helsinki, Finland.
HbA 1c Laboratory: Randie R Little, PhD, Curt Rohlfing. Diabetes Diagnostic Laboratory, Department of Pathology, University of Missouri School of Medicine, Columbia, Missouri, USA.
HLA Reference Laboratory: William Hagopian, MD, PhD,3 Christian Chamberlain, Jared Radtke, Sarah Zink . Pacific Northwest Research Institute, Seattle, Washington, USA (previously Henry Erlich, PhD,3 Steven J. Mack, PhD, Anna Lisa Fear. Center for Genetics, Children’s Hospital Oakland Research Institute, Oakland, California, USA.)
Autoantibody Reference Laboratories: Liping Yu, MD,^5 Dongmei Miao, MD,^ Kathleen Gillespie,*5 Alistair Williams,* Kyla Chandler,* Ilana Kelland,* Yassin Ben Khoud,* Matthew Randell.*^Barbara Davis Center for Childhood Diabetes, University of Colorado Denver, Aurora, Colorado, USA;*Bristol Medical School, University of Bristol, Bristol, UK.
HLA Reference Laboratory: William Hagopian, MD, PhD,3 Christian Chamberlain, Jared Radtke, Sarah Zink . Pacific Northwest Research Institute, Seattle, Washington, USA (previously Henry Erlich, PhD,3 Steven J. Mack, PhD, Anna Lisa Fear. Center for Genetics, Children’s Hospital Oakland Research Institute, Oakland, California, USA.)
Metabolomics Laboratory: Oliver Fiehn, PhD, Bill Wikoff, PhD, Brian Defelice, Dmitry Grapov, PhD, Tobias Kind, PhD, Mine Palazoglu, Luis Valdiviez, Benjamin Wancewicz, Gert Wohlgemuth, Joyce Wong . UC Davis Metabolomics Center, Davis, California, USA.
SNP Laboratory: Stephen S Rich, PhD3, Wei-Min Chen, PhD3, Suna Onengut-Gumuscu, PhD,3 Emily Farber, Rebecca Roche Pickin, PhD, Jonathan Davis, Jordan Davis, Dan Gallo, Jessica Bonnie, Paul Campolieto . Center for Public Health Genomics, University of Virginia, Charlottesville, Virginia, USA.
Repository: Sandra Ke, Niveen Mulholland, PhD. NIDDK Biosample Repository at Fisher BioServices.
Other Contributors: Thomas Briese, PhD,6 Columbia University. Todd Brusko, PhD,5 University of Florida, Gainesville, Florida, USA. Suzanne Bennett Johnson, PhD,8,11 Florida State University, Tallahassee, Florida, USA. Eoin McKinney, PhD,5 University of Cambridge. Tomi Pastinen, MD, PhD,5 The Children’s Mercy Hospital. Eric Triplett, PhD,6 University of Florida, Gainesville, Florida, USA.
Committees:
1Ancillary Studies, 2Diet, 3Genetics, 4Human Subjects/Publicity/Publications, 5Immune Markers, 6Infectious Agents, 7Laboratory Implementation, 8Psychosocial, 9Quality Assurance, 10Steering, 11Study Coordinators, 12Celiac Disease, 13Clinical Implementation.
Glossary
Abbreviations
- AUC
area under the receiver operating characteristic curve
- CV
cross-validation
- FDR-T1D
first-degree relative with T1D
- FPR
false-positive rate
- GADA
glutamic acid decarboxylase antibody
- GC-TOF MS
gas chromatography–time-of-flight mass spectrometry
- GIA
general infant attributes
- GRS
genetic risk scores
- HLA
human leukocyte antigen
- IA-2A
insulinoma-associated antigen 2 autoantibody
- IAA
insulin autoantibody
- IAAb
islet autoantibody
- ROC
receiver operating characteristic curve
- ROFI
Repeated Optimization for Feature Interpretation
- SNV
single-nucleotide variation
- T1D
type 1 diabetes
- TEDDY
The Environmental Determinants of Diabetes in the Young
- TPR
true-positive rate
Contributor Information
Bobbie-Jo M Webb-Robertson, Biological Sciences Division, Pacific Northwest National Laboratory, Richland, Washington 99352, USA; Colorado School of Public Health, University of Colorado Anschutz Medical Campus, Aurora, Colorado 80045, USA.
Ernesto S Nakayasu, Biological Sciences Division, Pacific Northwest National Laboratory, Richland, Washington 99352, USA.
Brigitte I Frohnert, Barbara Davis Center for Childhood Diabetes, School of Medicine, University of Colorado Anschutz Medical Campus, Aurora, Colorado 80045, USA.
Lisa M Bramer, Biological Sciences Division, Pacific Northwest National Laboratory, Richland, Washington 99352, USA.
Sarah M Akers, Computing & Analytics Division, Pacific Northwest National Laboratory, Richland, Washington 99352, USA.
Jill M Norris, Colorado School of Public Health, University of Colorado Anschutz Medical Campus, Aurora, Colorado 80045, USA.
Kendra Vehik, Health Informatics Institute, Morsani College of Medicine, University of South Florida, Tampa, Florida 33612, USA.
Anette-G Ziegler, Institute of Diabetes Research, Helmholtz Zentrum München, 85764 Neuherberg, Germany; Kilinikum rechts der Isar, Technische Universität München, 80333 Munich, Germany; Forschergruppe Diabetes e.V., 85764 Neuherberg, Germany.
Thomas O Metz, Biological Sciences Division, Pacific Northwest National Laboratory, Richland, Washington 99352, USA.
Stephen S Rich, Center for Public Health Genomics, University of Virginia, Charlottesville, Virginia 22908, USA.
Marian J Rewers, Barbara Davis Center for Childhood Diabetes, School of Medicine, University of Colorado Anschutz Medical Campus, Aurora, Colorado 80045, USA.
Financial Support
This research was performed by an external analytic partner under the auspices of the TEDDY Study Group, which was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), National Institute of Environmental Health Sciences (NIEHS), Centers for Disease Control and Prevention (CDC), and JDRF (grant Nos. U01 DK63829, U01 DK63861, U01 DK63821, U01 DK63865, U01 DK63863, U01 DK63836, U01 DK63790, UC4 DK63829, UC4 DK63861, UC4 DK63821, UC4 DK63865, UC4 DK63863, UC4 DK63836, UC4 DK95300, UC4 DK100238, UC4 DK106955, UC4 DK112243, UC4 DK117483, U01 DK124166, and contract No. HHSN267200700014C). TEDDY is supported in part by the National Institutes of Health/National Center for Advancing Translational Sciences Clinical and Translational Science Awards to the University of Florida (grant No. UL1 TR000064) and the University of Colorado (grant No. UL1 TR002535).
Disclosures
The authors have nothing to disclose.
Data Availability
The data sets generated and analyzed during the present study will be made available in the NIDDK Central Repository at https://repository.niddk.nih.gov/studies/teddy.
References
- 1. Regnell SE, Lernmark Å. Early prediction of autoimmune (type 1) diabetes. Diabetologia. 2017;60(8):1370-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rich SS, Concannon P. Role of type 1 diabetes-associated SNPs on autoantibody positivity in the Type 1 Diabetes Genetics Consortium: overview. Diabetes Care. 2015;38(Suppl 2):S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Törn C, Hadley D, Lee HS, et al. . TEDDY Study Group. Role of type 1 diabetes-associated SNPs on risk of autoantibody positivity in the TEDDY study. Diabetes. 2015;64(5):1818-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Winkler C, Krumsiek J, Buettner F, et al. . Feature ranking of type 1 diabetes susceptibility genes improves prediction of type 1 diabetes. Diabetologia. 2014;57(12):2521-2529. [DOI] [PubMed] [Google Scholar]
- 5. Krischer JP, Liu X, Lernmark Å, et al. . TEDDY Study Group. The influence of type 1 diabetes genetic susceptibility regions, age, sex, and family history on the progression from multiple autoantibodies to type 1 diabetes: a TEDDY study report. Diabetes. 2017;66(12):3122-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krischer JP, Lynch KF, Lernmark Å, et al. . TEDDY Study Group. Genetic and environmental interactions modify the risk of diabetes-related autoimmunity by 6 years of age: the TEDDY Study. Diabetes Care. 2017;40(9):1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frederiksen BN, Kroehl M, Fingerlin TE, et al. . Association between vitamin D metabolism gene polymorphisms and risk of islet autoimmunity and progression to type 1 diabetes: the Diabetes Autoimmunity Study in the Young (DAISY). J Clin Endocrinol Metab. 2013;98(11):E1845-E1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mishra SP, Wang S, Nagpal R, et al. . Probiotics and prebiotics for the amelioration of type 1 diabetes: present and future perspectives. Microorganisms. 2019;7(3):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Uusitalo U, Lee HS, Aronsson CA, et al. . TEDDY Study Group. Early infant diet and islet autoimmunity in the TEDDY Study. Diabetes Care. 2018;41(3):522-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Steck AK, Vehik K, Bonifacio E, et al. . TEDDY Study Group. Predictors of progression from the appearance of islet autoantibodies to early childhood diabetes: The Environmental Determinants of Diabetes in the Young (TEDDY). Diabetes Care. 2015;38(5):808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu X, Vehik K, Huang Y, et al. . TEDDY Study Group. Distinct growth phases in early life associated with the risk of type 1 diabetes: the TEDDY Study. Diabetes Care. 2020;43(3):556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bowden SA, Duck MM, Hoffman RP. Young children (< 5 yr) and adolescents (> 12 yr) with type 1 diabetes mellitus have low rate of partial remission: diabetic ketoacidosis is an important risk factor. Pediatr Diabetes. 2008;9(3 Pt 1):197-201. [DOI] [PubMed] [Google Scholar]
- 13. Larsson HE, Vehik K, Gesualdo P, et al. . TEDDY Study Group. Children followed in the TEDDY study are diagnosed with type 1 diabetes at an early stage of disease. Pediatr Diabetes. 2014;15(2):118-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ludvigsson J. Immune intervention at diagnosis—should we treat children to preserve beta-cell function? Pediatr Diabetes. 2007;8(Suppl 6):34-39. [DOI] [PubMed] [Google Scholar]
- 15. Steck AK, Larsson HE, Liu X, et al. . TEDDY Study Group. Residual beta-cell function in diabetes children followed and diagnosed in the TEDDY Study compared to community controls. Pediatr Diabetes. 2017;18(8):794-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marshall SM. Precision diabetes: a realistic outlook on a promising approach. Diabetologia. 2017;60(5):766-768. [DOI] [PubMed] [Google Scholar]
- 17. Mohan V, Radha V. Precision diabetes is slowly becoming a reality. Med Princ Pract. 2019;28(1):1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Udler MS, McCarthy MI, Florez JC, Mahajan A. Genetic risk scores for diabetes diagnosis and precision medicine. Endocr Rev. 2019;40(6):1500-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kosorok MR, Laber EB. Precision medicine. Annu Rev Stat Appl. 2019;6:263-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frohnert BI, Webb-Robertson BJM, Bramer LM, et al. . Predictive modeling of type 1 diabetes stages using disparate data sources. Diabetes. 2020;69(2):238-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rosenfeld A, Graham DG, Jevons S, et al. . Development and validation of a risk prediction model to diagnose Barrett’s oesophagus (MARK-BE): a case-control machine learning approach. Lancet Digit Health. 2020;2(1):E37-E48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Webb-Robertson BJM, Bramer LM, Stanfill BA, et al. . Prediction of the development of islet autoantibodies through integration of environmental, genetic, and metabolic markers. J Diabetes. 2021;13(2):143-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bonifacio E, Beyerlein A, Hippich M, et al. . TEDDY Study Group. Genetic scores to stratify risk of developing multiple islet autoantibodies and type 1 diabetes: a prospective study in children. PLoS Med. 2018;15(4):e1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rewers M, Hyöty H, Lernmark Å, et al. . TEDDY Study Group. The Environmental Determinants of Diabetes in the Young (TEDDY) Study: 2018 update. Curr Diab Rep. 2018;18(12):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee HS, Burkhardt BR, McLeod W, et al. . TEDDY Study Group. Biomarker discovery study design for type 1 diabetes in The Environmental Determinants of Diabetes in the Young (TEDDY) Study. Diabetes Metab Res Rev. 2014;30(5):424-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Puavilai G, Chanprasertyotin S, Sriphrapradaeng A. Diagnostic criteria for diabetes mellitus and other categories of glucose intolerance: 1997 criteria by the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus (ADA), 1998 WHO consultation criteria, and 1985 WHO criteria. World Health Organization. Diabetes Res Clin Pract. 1999;44(1):21-26. [DOI] [PubMed] [Google Scholar]
- 27. Yu L, Rewers M, Gianni R, et al. . Antiislet autoantibodies usually develop sequentially rather than simultaneously. J Clin Endocrinol Metab. 1996;81(12):4264-4267. [DOI] [PubMed] [Google Scholar]
- 28. Bonifacio E, Yu L, Williams AK, et al. . Harmonization of glutamic acid decarboxylase and islet antigen-2 autoantibody assays for National Institute of Diabetes and Digestive and Kidney Diseases consortia. J Clin Endocrinol Metab. 2010;95(7):3360-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lampasona V, Pittman DL, Williams AJ, et al. . Islet Autoantibody Standardization Program 2018 workshop: interlaboratory comparison of glutamic acid decarboxylase autoantibody assay performance. Clin Chem. 2019;65(9):1141-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hagopian WA, Erlich H, Lernmark Å, et al. . TEDDY Study Group. The Environmental Determinants of Diabetes in the Young (TEDDY): genetic criteria and international diabetes risk screening of 421 000 infants. Pediatr Diabetes. 2011;12(8):733-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fiehn O. Metabolomics by gas chromatography-mass spectrometry: combined targeted and untargeted profiling. Curr Protoc Mol Biol. 2016;114:30.4.1-30.4.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kind T, Wohlgemuth G, Lee DY, et al. . FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009;81(24):10038-10048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fan S, Kind T, Cajka T, et al. . Systematic error removal using random forest for normalizing large-scale untargeted lipidomics data. Anal Chem. 2019;91(5):3590-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ditzler G, Polikar R, Rosen G. A sequential learning approach for scaling up filter-based feature subset selection. IEEE Trans Neural Netw Learn Syst. 2018;29(6):2530-2544. [DOI] [PubMed] [Google Scholar]
- 35. Wasserman L, Roeder K. High dimensional variable selection. Ann Stat. 2009;37(5A):2178-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Webb-Robertson BJM, Bramer LM, Reehl SM, et al. . ROFI—the use of repeated optimization for feature interpretation. International Conference on Computational Science and Computational Intelligence (CSCI), Las Vegas, NV, USA. IEEE; 2016:29-33.
- 37. Khashan AS, Kenny LC, Lundholm C, et al. . Gestational age and birth weight and the risk of childhood type 1 diabetes: a population-based cohort and sibling design study. Diabetes Care. 2015;38(12):2308-2315. [DOI] [PubMed] [Google Scholar]
- 38. Li Q, Parikh H, Butterworth MD, et al. . TEDDY Study Group. Longitudinal metabolome-wide signals prior to the appearance of a first islet autoantibody in children participating in the TEDDY Study. Diabetes. 2020;69(3):465-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mattila M, Erlund I, Lee HS, et al. . TEDDY Study Group. Plasma ascorbic acid and the risk of islet autoimmunity and type 1 diabetes: the TEDDY Study. Diabetologia. 2020;63(2):278-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Katsiki N, Papanas N, Fonseca VA, Maltezos E, Mikhailidis DP. Uric acid and diabetes: is there a link? Curr Pharm Des. 2013;19(27):4930-4937. [DOI] [PubMed] [Google Scholar]
- 41. Titchenell PM, Lazar MA, Birnbaum MJ. Unraveling the regulation of hepatic metabolism by insulin. Trends Endocrinol Metab. 2017;28(7):497-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wagle A, Jivraj S, Garlock GL, Stapleton SR. Insulin regulation of glucose-6-phosphate dehydrogenase gene expression is rapamycin-sensitive and requires phosphatidylinositol 3-kinase. J Biol Chem. 1998;273(24):14968-14974. [DOI] [PubMed] [Google Scholar]
- 43. Evans-Molina C, Sims EK, DiMeglio LA, et al. . Type 1 Diabetes TrialNet Study Group. β Cell dysfunction exists more than 5 years before type 1 diabetes diagnosis. JCI Insight. 2018;3(15):e120877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated and analyzed during the present study will be made available in the NIDDK Central Repository at https://repository.niddk.nih.gov/studies/teddy.