Abstract
Context
Approximately 70% of infertile men are diagnosed with idiopathic (abnormal semen parameters) or unexplained (normozoospermia) infertility, with the common feature of lacking etiologic factors. Follicle-stimulating hormone (FSH) is essential for initiation and maintenance of spermatogenesis. Certain single-nucleotide variations (SNVs; formerly single-nucleotide polymorphisms [SNPs]) (ie, FSHB c.–211G > T, FSHR c.2039A > G) are associated with FSH, testicular volume, and spermatogenesis. It is unknown to what extent other variants are associated with FSH levels and therewith resemble causative factors for infertility.
Objective
We aimed to identify further genetic determinants modulating FSH levels in a cohort of men presenting with idiopathic or unexplained infertility.
Methods
We retrospectively (2010-2018) selected 1900 men with idiopathic/unexplained infertility. In the discovery study (n = 760), a genome-wide association study (GWAS) was performed (Infinium PsychArrays) in association with FSH values (Illumina GenomeStudio, v2.0). Minor allele frequencies (MAFs) were analyzed for the discovery and an independent normozoospermic cohort. In the validation study (n = 1140), TaqMan SNV polymerase chain reaction was conducted for rs11031005 and rs10835638 in association with andrological parameters.
Results
Imputation revealed 9 SNVs in high linkage disequilibrium, with genome-wide significance (P < 4.28e-07) at the FSHB locus 11p.14.1 being associated with FSH. The 9 SNVs accounted for up to a 4.65% variance in FSH level. In the oligozoospermic subgroup, this was increased up to 6.95% and the MAF was enhanced compared to an independent cohort of normozoospermic men. By validation, a significant association for rs11031005/rs10835638 with FSH (P = 4.71e-06/5.55e-07) and FSH/luteinizing hormone ratio (P = 2.08e-12/6.4e-12) was evident.
Conclusions
This GWAS delineates the polymorphic FSHB genomic region as the main determinant of FSH levels in men with unexplained or idiopathic infertility. Given the essential role of FSH, molecular detection of one of the identified SNVs that causes lowered FSH and therewith decreases spermatogenesis could resolve the idiopathic/unexplained origin by this etiologic factor.
Keywords: genome-wide association study (GWAS), single-nucleotide variation (SNV), follicle-stimulating hormone (FSH), idiopathic male infertility
In Western countries, infertility affects at least 15% of couples during reproductive age, and in 50% of all cases a male factor contributes essentially (1). Impaired spermatogenesis can be attributed to oncologic or genetic causes in 4% to 10% of infertile men, rendering about 72% of infertile men with no or only mild etiologic factors, insufficient to explain their impaired fertility status (2). In infertile men with unknown etiology, 2 conditions are to be distinguished: unexplained and idiopathic male infertility. By definition, unexplained infertility comprises men in whom female factors and other etiologic factors are missing; these men show normozoospermia in their semen analyses. Different from idiopathic infertile men, who also lack etiologic and female factors, but who display abnormal semen parameters (3-5). In this very heterogeneous condition, there is an urgent need to improve the diagnostic workup by identifying genetic and endocrine factors influencing spermatogenesis.
The pituitary follicle-stimulating hormone (FSH) is essential for initiation and maintenance of spermatogenesis in humans. During the prenatal and prepubertal stages, FSH stimulates Sertoli cell (SC) proliferation and by this determines their final number and the potential for spermatogenic output. In the adult testis, FSH is mandatory for qualitative and quantitative spermatogenesis. Together with testosterone, signals for germ cell maturation are evoked via the SCs (6-9). In addition, signaling for antiapoptotic survival factors, and for adhesion complexes between SCs and germ cells, is initiated (10).
Compared with luteinizing hormone (LH), which is stored in the gonadotropic cells and released on gonadotropin-releasing hormone stimulus, FSH is directly secreted on stimulus into the bloodstream (11, 12). Thus, the transcription of FSHB messenger RNA is not only the rate-limiting step, it also determines FSH serum level.
Genetic variants in the promoter region of the FSHB genes are therefore capable of changing FSH serum levels by modulating FSHB transcription. A single-nucleotide variation (SNV) in the promoter of the FSHB gene c.-211G > T (rs10835638) was found to prominently regulate transcription and thereby biosynthesis of the FSHB subunit. The reduced transcriptional activity of the gene by 50% presumably is due to an impaired binding of the transcription factor LHX3 in the presence of a T-allele (13).
The SNVs in the FSHR gene (rs1394205, rs6165, and rs6166) are known to affect the sensitivity of the receptor by altering signaling pathways and its timing of expression and activation (14-16).
Clinically, an SNV in the promoter of the FSHB gene c.-211G > T was shown to be associated with reduction in testicular volume, reduced sperm counts, and serum FSH levels in patients with nonazoospermic infertility of unknown origin, carrying 1 or 2 T-alleles compared to GG homozygous men. Patients carrying the T-allele do not adequately upregulate FSH serum levels to achieve full spermatogenesis (17).
However, there is a surprising lack of data to which extent other SNVs might affect FSH levels/action on spermatogenesis.
We therefore aimed to identify further genetic determinants modulating FSH levels in a cohort of men presenting with idiopathic or unexplained infertility (oligozoospermia or normozoospermia). To this end we performed a genome-wide association study (GWAS) and identified (discovery study) and confirmed (validation study) several FSH-associated SNVs in a genomic region covering the FSHB gene. The observed effect was found to be even more pronounced in oligozoospermic men, being a subgroup of the initial discovery cohort.
Materials and Methods
Study Population
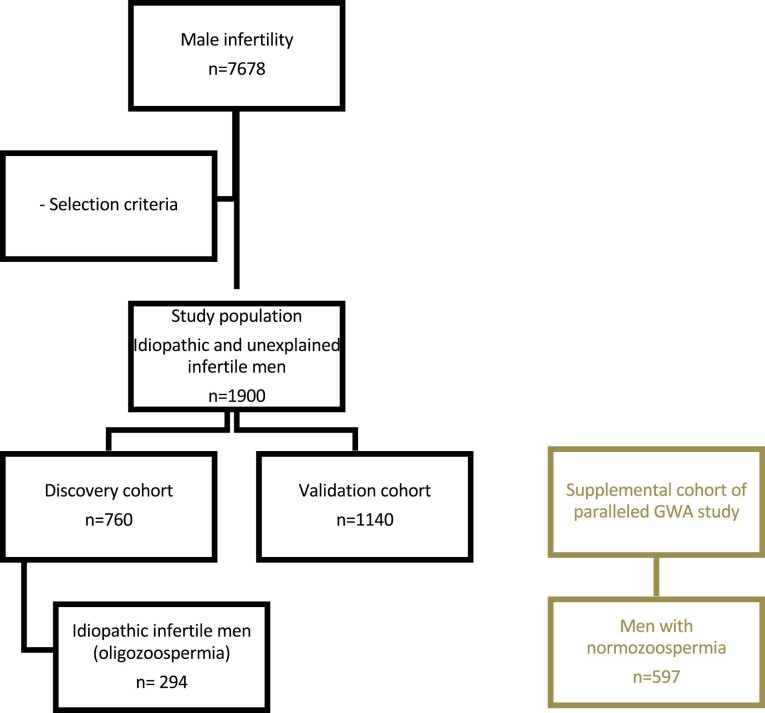
The study population included men with idiopathic or unexplained male infertility and comprised 2 cohorts: i) a discovery cohort (n = 760) for GWAS to identify FSH-associated SNVs, and ii) a validation cohort (n = 1140) for TaqMan polymerase chain reaction (PCR) assay for validation of 2 SNVs (Fig. 1). By analyzing more than 1800 patients, statistical power reached more than 80% in the respective analyses (see power analysis later in this work).
Figure 1.
Selection of study population. In a retrospective query of our database with an 8-year time range, approximately 7678 patients infertile men were selected. After applying strict selection criteria, a study population of 1900 men with idiopathic or unexplained male infertility were identified. The discovery cohort, to perform the genome-wide association (GWA) analysis, comprised 760 men according to our power analysis. The validation cohort, to perform the TaqMan single-nucleotide variation polymerase chain reaction, comprised 1140 men. The supplemental cohort for minor allele frequency comparison consists of men with normozoospermia.
Discovery and Validation Cohort
White patients of Caucasian origin were diagnosed with idiopathic or unexplained male infertility, meaning no major factors contributing to impaired fertility had been identified, leading to a unique and highly selected study cohort (unexplained: normozoospermia, idiopathic: abnormal semen parameters). The patients had visited our center from 2010 through 2018 for an infertility workup. To select for male infertility without etiologic factors, we applied the following selection criteria: inclusion of men with FSH greater than or equal to 1 IU/L, total testosterone greater than or equal to 8 nmol/L, bitesticular volume greater than or equal to 20 nmol/L, and total sperm count (TSC) greater than or equal to 1 Mill. sperm/ejaculate. Exclusion criteria included men with karyotype anomalies, azoospermia factor deletions, current or former oncological diseases and/or gonadotoxic treatments, single testis, and female partner infertility. By applying the combination of these criteria, we excluded men with hypogonadotropic/hypergonadotropic hypogonadism (18). The threshold in TSC of 1 Mill sperm was chosen because of the increasing risk of underlying, undiscovered genetic causes in more severe phenotypes of infertility (19). For all patients that we included in the study and whose sperm concentration was less than 5 mill/mL, we performed additional genetic testing (karyotype, azoospermia factor deletions) to rule out commonly known genetic factors.
These clinical selection criteria were supplemented by procedural parameters like availability of sufficient DNA (for GWAS or TaqMan PCR, respectively). Selected men for the discovery and validation study were chosen chronologically by their appearance/visit at the Centre of Reproductive Medicine and Andrology (CeRA).
All men provided written informed consent for the evaluation of their clinical data and genetic analysis of the donated DNA samples. The study was carried out in accordance with the protocols approved by the ethics committee of the medical faculty and the state medical board (Az. 2017-139-f-S and 2013-255-f-S).
Clinical Workup and Laboratory Analyses
Routine clinical workup included physical examination, ultrasound of the testes, and hormone and semen analysis. Hormone analysis included measurement of FSH, LH, and testosterone. These procedures have been described previously (20).
Ejaculates were obtained by masturbation after sexual abstinence of 3 to 7 days, at 2 time points, and semen analysis was carried out according to the World Health Organization guidelines (21).
Genomic DNA Isolation
DNA was isolated from EDTA-preserved peripheral blood samples using the FlexiGene DNA Kit from Qiagen (Qiagen) according to the manufacturer’s specifications. The concentration and quality of samples were measured with the FLUOstar Omega spectrometer (BMG Labtech).
Genome-wide Genotyping and Quality Control
A total of 760 idiopathic infertile men (discovery cohort) were genotyped using the PsychArray-24 v1.3 DNA Analysis Bead Chip from Illumina, including 595 427 SNVs. This array contains proven tag SNVs, which are informative SNVs in a region of the genome with high linkage disequilibrium for distinct haplotypes. Genotyping was performed at the Life & Brain Research Center (Bonn, Germany). Data were analyzed using PLINK v1.90b4; quality control of the genotype data was performed at the SNV and individual level. Exclusion of SNVs was due to call rates of less than 0.95, P values from Hardy-Weinberg equilibrium test of less than .001, and minor allele frequencies (MAFs) of less than 0.01. Exclusion of patient samples was performed by genotype missing per individual greater than 5% and population outliers as determined by multidimensional scaling (MDS).
Genotyping (Validation Cohort and Internal Control)
The SNVs rs11031005 and rs10835638 were analyzed by TaqMan PCR assays and allelic discrimination (Genotyping Assay C__32036788_10 for rs11031005 and C__27829553_10 for rs10835638) using the StepOnePlus detection system (Applied Biosystems) as described elsewhere (20).
Stratification Analysis
Two-dimension reduction routines provided by PLINK v1.90b4 were performed to assess population structure. Top principal components were used as covariates in association analysis regressions to correct for population structure. Two-dimensional MDS was used to identify clustering of samples and select population outliers. The quantiles of the observed P values for the GWAS experiment were compared with the quantiles of a uniform distribution using a quantile-quantile plot (Q-Q plot) in the R-project software 3.6.
Linkage Disequilibrium Analysis
Pairwise linkage disequilibrium (LD) was calculated with PLINK v1.90b4 (using parameter-indep-pairwise 50 5 0.2). LD blocks were computed using PLINK’s-blocks command, which is based on the algorithm described by Gabriel et al (22).
Genotype Imputation
Genome-wide imputation was conducted after QC filtering (SNV and sample genotyping rates > 0.95, Hardy-Weinberg P value < .001). The reference panel was based on the 1000 Genomes Project (phase 3). First, haplotypes were phased using SHAPEIT2 v2.r837. We used IMPUTE2 v2.3.2 to impute genotypes using a confidence threshold of 0.95 and a block size of 5 Mbp. After imputation and QC filtering, 7 242 305 markers for 742 samples remained for analysis of the discovery cohort.
As an internal control for imputation quality, we compared imputed genotypes of SNVs rs11031005 and rs10835638 with genotypes as acquired by assay (for discovery cohort). Imputation data and genotyping data of rs11031005 and rs10835638 showed an overlap of 95.15% and 97.44%, respectively, thus indicating high concordance between imputed and actual genotypes.
Statistical Analysis
Individual reproductive parameters were normalized using a Box-Cox transformation before association analysis. Quantitative association analysis was conducted using PLINK v1.90b4. SNVs with MAFs less than 1% were excluded from the analysis to increase statistical robustness. P values were computed using PLINK’s linear regression model (--assoc –linear) with the top 5 principal components and patient age as covariates to stratify for population and age effects. Explained variance was computed as coefficient of determination (r², as computed by PLINK’s --assoc command). PLINK –perm was used to generate empirical P values for 100 000 permutations.
Plain Bonferroni correction is too conservative for GWAS of smaller sample size as it assumes independence between different SNVs. As a result, we corrected for the number of LD blocks instead. This results in a genome-wide significance threshold of 4.28e-7 for the infertile discovery cohort and a threshold of 4.52e-7 with inclusion of the fertile cohort (0.05/number of LD blocks). We also report SNVs reaching the suggestive significance threshold of 8.56e-6 for the infertile discovery cohort and 9.03e-6 with inclusion of the fertile cohort (1/number of LD blocks) for further research and replication.
Power Analysis
We used QUANTO to compute the sample size required to associate FSH levels with SNV genotypes with a statistical power of π = 0.80 (23, 24).
For the GWAS power analysis, we assumed a minimum MAF of 5%, a coefficient of determination (r²) of 0.05, and a target P value of 1e-7 using 2-sided testing. For the validation cohort we assumed a minimum MAF of 5%, a β coefficient of –0.3, and a target P value of .05 using one-sided testing.
Results
Reproductive Parameters of Discovery Cohort
Using our in-house database Androbase (25), we retrospectively selected from more than 7678 infertile patients 1900 men with idiopathic or unexplained male infertility, fulfilling the selection criteria and presenting with a complete data set and available DNA samples. A total of 760 men were part of the discovery cohort, whereas 1140 men were selected for the validation cohort (see Fig. 1). The median age of these patients was 36 years. The majority of patients (79.4%) had an FSH serum level within the normal range (1-7 IU/L) with a mean of 5.3 IU/l (±3.9) (Table 1). Mean bitesticular volume was 39.6 mL (±14.2 mL). Ejaculate analysis, according to World Health Organization criteria, revealed a mean TSC of 106.3 million sperm/ejaculate (±135.1) (see Table 1).
Table 1.
Reproductive parameters of the discovery cohort, oligozoospermic subgroup, and validation cohort
| Discovery cohort (n = 742) | Oligozoospermic subgroup of discovery cohort (n = 294) | Validation cohort (n = 1123) | |
|---|---|---|---|
| Age, y | 36 ± 5.7 | 35.9 ± 5.7 | 35 ± 6.2 |
| 35 (28-46) | 35 (28-46) | 35 (26-45) | |
| FSH, IU/L (1-7] | 5.3 ± 3.9 | 7.3 ± 4.8 | 5.4 ± 4.5 |
| 4.2 (1.7-13.2) | 5.9 (2.3-16.1) | 4.2 (1.7-13.1) | |
| n FSH 1-7 IU/L, % | 79, 40% | 61, 90% | 79, 30% |
| n FSH > 7 IU/L, % | 20, 60% | 38, 10% | 20, 70% |
| LH, IU/L (2-10) | 3.3 ± 1.7 | 4 ± 2 | 3.6 ± 2 |
| 2.9 (1.4-6.6) | 3.7 (1.6-7.5) | 3.2 (1.5-6.8) | |
| FSH/LH | 1.7 ± 0.9 | 1.9 ± 1 | 1.6 ± 1.1 |
| 1.5 (0.6-3.4) | 1.8 (0.7-3.9) | 1.4 (0.5-3.3) | |
| Testosterone, nmol/L (≥ 12) | 17.1 ± 6.1 | 17.2 ± 6.4 | 16.4 ± 6.4 |
| 16.4 (9.2-28.5) | 16.1 (9-28.2) | 15.3 (9.1-27.3) | |
| Bitestic. volume, mL | 39.6 ± 14.2 | 34.6 ± 12.1 | 41.2 ± 14 |
| 37 (21.1-63) | 33 (20-58) | 40 (21-67) | |
| Ejaculate volume, mL | 4 ± 1.7 | 3.7 ± 1.6 | 3.9 ± 1.6 |
| 3.7 (1.8-7) | 3.5 (1.7-6.6) | 3.5 (1.8-7) | |
| Total sperm count, Mill/ejac | 106.3 ± 135.1 | 14.5 ± 11.1 | 109.5 ± 158.9 |
| 58.7 (2.1-380.9) | 11.8 (1.3-35) | 57.2 (2.1-370.7) | |
| n ≥ 39 Mill/ejac, % | 60,40% | 0,00% | 57,30% |
| n < 39 Mill/ejac, % | 39,60% | 100,00% | 42,70% |
| Sperm conc, Mill/mL | 28.4 ± 34.4 | 4.6 ± 4.3 | 30.1 ± 41.5 |
| 16.4 (0.6-104.4) | 3.5 (0.4-12.7) | 16.4 (0.6-104.7) | |
| Sperm ab-motility, % | 43.8 ± 12.4 | 37.1 ± 13.6 | 43.2 ± 13.2 |
| 46 (18.9-58.1) | 38 (14-55) | 46 (17-58) | |
| Sperm morphology | 3.8 ± 2 | 2.9 ± 1.6 | 4.5 ± 2.6 |
| 4 (1-7) | 3 (1-6) | 4 (1-9) |
Data are presented as mean ± SD and median (5th-95th percentile). Reference range of hormones is indicated in parentheses.
Abbreviations: Bitestic, bitesticular; ejac, ejaculation; FSH, follicle-stimulating hormone; LH, luteinizing hormone.
Genome-wide Association Study Delineates the Genomic Locus 11p.14.1 as the Major Genetic Determinant Associated With Follicle-Stimulating Hormone Levels
Of the 760 selected well-characterized men, 742 remained for analysis after quality control of the GWAS data. MDS showed that samples are clustered tightly (26). Plotting observed P values against the expected (.1) uniform distribution in a quantile-quantile plot and revealed similar distribution of observed and expected P values. There was an overrepresentation of small P values that correspond to the markers of interest for FSH association (26).
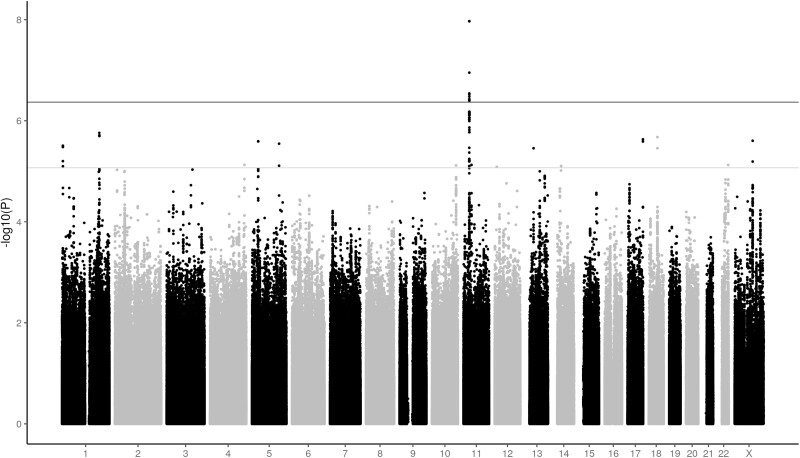
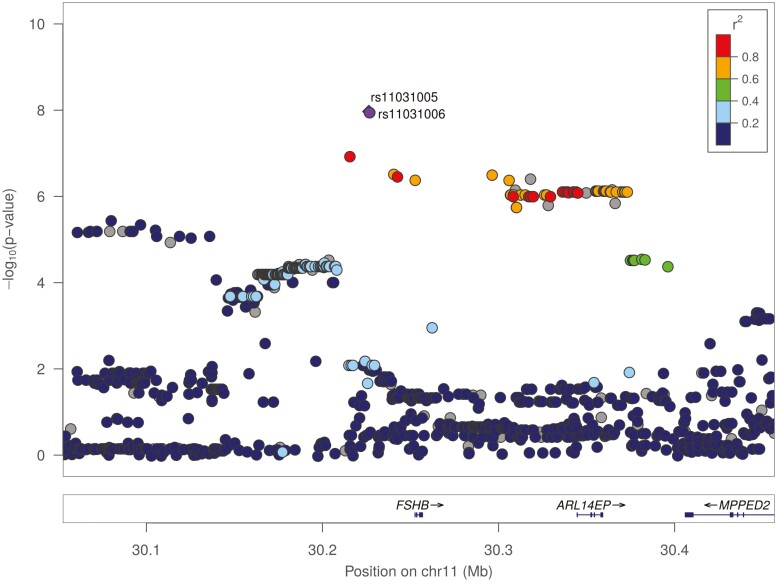
Imputation of the GWAS data was performed to account for nongenotyped polymorphisms. Nine SNVs, all located on chromosome 11p.14.1 and including rs10835638, reached genome-wide significance (P < 4.28e-07) (Table 2, Fig. 2). Six of the 9 SNVs are located upstream and 3 are located downstream of the FSHB gene (Fig. 3). All 9 SNVs are in strong LD (26). In addition, 94 SNVs reached suggestive P value (P < 8.56e-6), with 69 located on chromosome 11 (26).
Table 2.
The genomic region on chromosome 11p14.1 containing the FSHB gene and corresponding single-nucleotide variations
| Chr | Position | ID | β | P | % variance explained, % | % variance explained in patients with total sperm count < 39 Mill/ejac (n = 294), % | MAF | Distance to FSHB | Publicationsa |
|---|---|---|---|---|---|---|---|---|---|
| 11 | 30226356 | rs11031005 | –0.3984 | 1.07E-08 | 4.65 | 6.95 | 0.163 | 26 207 bp (upstream) | (27-33) |
| 11 | 30226528 | rs11031006 | –0.3984 | 1.07E-08 | 4.65 | 6.95 | 0.163 | 26 035 bp (upstream) | (27, 28, 30, 31, 34-46) |
| 11 | 30215261 | rs11031002 | –0.3933 | 1.12E-07 | 3.98 | 6.61 | 0.139 | 37 302 bp (upstream) | (29, 31-33, 47) |
| 11 | 30240178 | rs11031010 | –0.3549 | 2.89E-07 | 3.67 | 5.18 | 0.169 | 12 385 bp (upstream) | (36, 48-52) |
| 11 | 30296055 | rs12364889 | –0.3554 | 3.01E-07 | 3.66 | 5.18 | 0.168 | 39 231 bp (downstream) | – |
| 11 | 30242287 | rs74485684 | –0.334 | 3.32E-07 | 3.69 | 5.63 | 0.19 | 10 276 bp (upstream) | (29) |
| 11 | 30317839 | rs202057396 | –0.3353 | 3.73E-07 | 3.72 | 5.92 | 0.191 | 61 015 bp (downstream) | – |
| 11 | 30252352 | rs10835638 | –0.351 | 3.97E-07 | 3.60 | 5.12 | 0.17 | 211 bp (upstream) | (16-18, 21, 31-33, 39, 53-71) |
| 11 | 30305675 | rs11031033 | –0.3542 | 3.99E-07 | 3.61 | 4.89 | 0.168 | 48 851 bp (downstream) | – |
After imputation analyses 9 SNVs, all located on chr.11p14.1, are significantly associated with FSH levels. An additive linear model was applied, including the top 5 principal component and patient age as covariates. All coordinates are reported according to hg19 reference genome. β Values correspond to each SNV’s linear regression coefficient. The effect of the respective SNVs on FSH serum level is indicated in column % variance explained. Relevant literature for respective SNVs are presented.
Abbreviations: bp, base pair; Chr, chromosome; FSH, follicle-stimulating hormone; ID, identification; MAF, minor allele frequency; SNV, single-nucleotide variation.
a Publications from Ensembl (https://www.ensembl.org/index.html).
Figure 2.
Discovery cohort: association of imputed single-nucleotide variations (SNVs) with follicle-stimulating hormone (FSH) levels. Manhattan plot of imputed genome-wide association study data depicts SNVs associated with FSH. X axis: genomic coordinates of tested SNVs on respective chromosomes. Y axis: significance level on a –log10 scale. The suggestive significance threshold is indicated by the gray horizontal line (P = 8.56e-6). The genome-significance threshold is indicated by the black horizontal line (P = 4.28e-7).
Figure 3.
Genomic region of chromosome 11p14.1. Regional association plot of 11p14.1 visualized by LocusZoom. Dots present individual single-nucleotide variations (SNVs). Colors indicate pair-wise r² values between each SNV and rs11031005, describing patterns of linkage disequilibrium around FSHB (for color figure refer online version).
The effect of the 9 significant SNVs on FSH serum level was calculated for each single SNV as a coefficient of determination (see Table 2). The SNVs rs11031005 and rs11031006 each explained 4.65% of FSH serum level variance, which reveals a stronger effect on FSH serum level, when compared to rs10835638 (FSHB c.-211 G > T), with 3.6% variance.
Thus, we identified by GWAS the genomic locus 11p.14.1 hosting 9 SNVs that are significantly associated with FSH levels in men with idiopathic or unexplained infertility.
Minor Allele Frequencies of Identified Single-Nucleotide Variations (Discovery Cohort vs. Independent Normozoospermic Genome-wide Association Study Cohort)
We decided to further focus on the MAFs of the 9 SNVs in the genomic region of the FSHB gene in distinct patient populations. From a parallel GWA study performed in 597 men with normozoospermia, we obtained data on the MAFs and compared this to the MAFs of the discovery cohort. When combining all MAFs of the identified 9 SNVs in 1 model, no significant difference (P = .48) was observed between the discovery cohort and the normozoospermic men. However, when comparing MAFs between the 297 oligozoospermic men of the discovery cohort (18.40% MAF) and the cohort of 597 normozoospermic men (16.54% MAF), significant differences were observed (P = .004).
Reproductive Parameters of Patients from the Independent Validation Cohort
The same selection process as for the discovery cohort was applied to the validation cohort. The median age of the analyzed men in the validation cohort was 35 years; median FSH level was 5.2 IU/L. The majority of patients (79.3%) had an FSH serum level within regular range (see Table 1). The mean bitesticular volume was 41.2 mL (±14 mL). Ejaculate analysis revealed a mean TSC of 109.5 Mill. sperm/ejaculate (±158.9) (see Table 1).
Both cohorts, the discovery and validation cohort, were homogenous in terms of age, sperm, and reproductive parameters (see Table 1).
Validation Study Confirms Results of Genome-wide Association Study Analysis
The validation by specific TaqMan PCR assay was performed for the SNVs rs11031005 and rs10835638. The latter was chosen because its effect on distinct andrological parameters was already shown by us and others, while the SNV rs11031005 displayed the highest β value compared to the other SNVs. Association analyses of FSH and other reproductive parameters was performed.
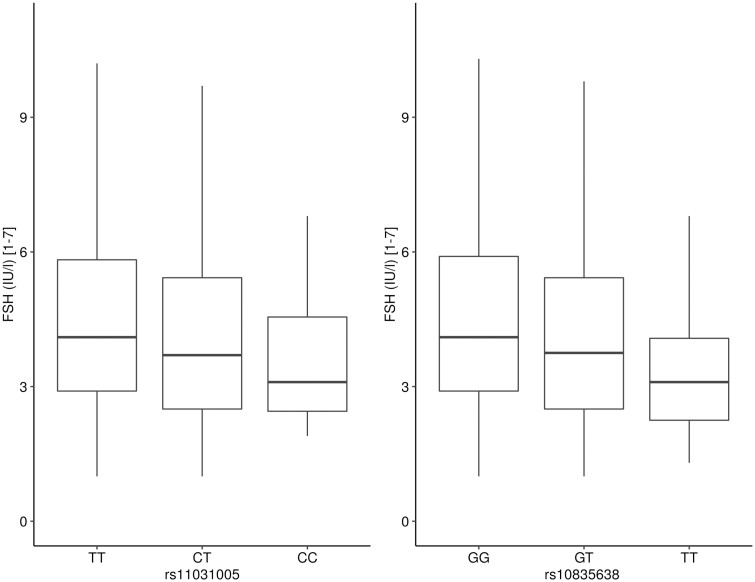
Using an additive model, which assumes a codominant effect of alleles in which the heterozygotes should generally exhibit intermediate levels, the SNV rs11031005 was significantly associated with FSH serum levels (P = 4.71e-06) and FSH/LH ratio (P = 2.08e-12) (see Table 3). In comparison, the previously described FSHB c.-211 SNV (rs10835638) revealed a significant association with FSH levels (P = 5.55e-07) and FSH/LH ratio (P = 6.4e-12). However, an association with bitesticular volume was not evident after Bonferroni correction (P = 4.93e-03) (see Table 3). The validity of an additive model for SNV analysis becomes evident when analyzing the different FSHB genotypes and their effect on FSH levels (Fig. 4).
Table 3.
Validation cohort: comparative analysis of reproductive parameters corresponding to single-nucleotide variations rs11031005 and rs10835638
| Androl. Parameters | rs10835638 | rs11031005 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Additive linear model | Genotype | Additive linear model | |||||||
| GG (n = 803) | GT (n = 294) | TT (n = 26) | P | beta | TT (n = 831) | CT (n = 269) | CC (n = 23) | P | beta | |
| FSH, IU/l (1-7) | 5.8 ± 4.9 | 4.6 ± 3.2 | 3.6 ± 2.1 | 5.55e-07** | -0.29 (0.06) | 5.7 ± 4.8 | 4.6 ± 3.3 | 4 ± 2.3 | 4.71e-06** | -0.27 (0.06) |
| 4.4 (1.8-14.2) | 3.9 (1.5-10) | 3.1 (1.6-8) | 4.4 (1.8-14.1) | 3.8 (1.5-10.1) | 3.1 (1.9-8.7) | |||||
| LH, IU/l (2-10) | 3.6 ± 1.8 | 3.8 ± 2.3 | 3.9 ± 1.4 | 1.63e-01 | 0.08 (0.06) | 3.6 ± 1.8 | 3.8 ± 2.4 | 4 ± 1.4 | 3.64e-02* | 0.13 (0.06) |
| 3.2 (1.5-6.6) | 3.2 (1.5-7) | 3.5 (2.2-6.4) | 3.2 (1.5-6.6) | 3.2 (1.5-7.2) | 3.8 (2.2-6.6) | |||||
| FSH/LH | 1.7 ± 1.2 | 1.3 ± 0.7 | 1 ± 0.8 | 6.4e-12** | -0.4 (0.06) | 1.7 ± 1.2 | 1.3 ± 0.6 | 1.1 ± 0.9 | 2.08e-12** | -0.42 (0.06) |
| 1.4 (0.6-3.6) | 1.2 (0.5-2.7) | 0.8 (0.5-1.6) | 1.4 (0.6-3.6) | 1.2 (0.4-2.5) | 0.8 (0.5-1.7) | |||||
| Testosterone, nmol/l (≥ 12) | 16.4 ± 6.4 | 16.2 ± 6.2 | 18.7 ± 7 | 8.75e-01 | 0.01 (0.06) | 16.5 ± 6.4 | 16 ± 6.1 | 18.2 ± 6.9 | 7.36e-01 | -0.02 (0.06) |
| 15.3 (9.1-27.3) | 14.9 (9.2-25.7) | 15.6 (11.5-29.1) | 15.3 (9.1-27.6) | 14.9 (8.9-25) | 15.5 (11.4-28.4) | |||||
| Bitestic. Volume, ml | 41.8 ± 14.2 | 40 ± 13.3 | 35 ± 13.3 | 4.93e-03* | -0.16 (0.06) | 41.8 ± 14.2 | 39.8 ± 13.2 | 35 ± 13.7 | 5.42e-03* | -0.17 (0.06) |
| 40 (22-67) | 39 (21-64.3) | 34 (19.8-52) | 40 (22-67) | 39 (21-64.6) | 34 (19.4-52.6) | |||||
| Ejaculate volume, ml | 3.9 ± 1.6 | 3.7 ± 1.6 | 4 ± 1.4 | 3.04e-01 | -0.06 (0.06) | 3.9 ± 1.6 | 3.8 ± 1.6 | 4.1 ± 1.5 | 7.9e-01 | -0.02 (0.06) |
| 3.6 (1.8-7) | 3.4 (1.8-7) | 3.9 (1.8-6.8) | 3.5 (1.8-7) | 3.5 (1.8-7.1) | 4 (1.8-6.9) | |||||
| Total sperm count, Mill/ejac | 113.5 ± 169.8 | 98.6 ± 127.2 | 108.9 ± 130.8 | 9.25e-01 | -0.01 (0.06) | 112.4 ± 168.4 | 101.2 ± 128.5 | 103.6 ± 129.1 | 8.93e-01 | 0.01 (0.06) |
| 59 (1.9-371.3) | 52.5 (3.1-370.3) | 51.7 (2.9-365.6) | 58.3 (1.9-371.4) | 55 (3.1-363) | 37.4 (2.9-367.3) | |||||
| Sperm conc., Mill/ml | 30.6 ± 43 | 28.9 ± 37 | 32.1 ± 42.9 | 7.4e-01 | 0.02 (0.06) | 30.5 ± 42.7 | 29.3 ± 37.6 | 29.1 ± 40.2 | 7.36e-01 | 0.02 (0.06) |
| 17 (0.6-103.1) | 15.1 (0.8-108.4) | 14.2 (0.7-121.5) | 16.9 (0.6-103.4) | 15.4 (0.8-109.9) | 13 (0.6-95) | |||||
| Sperm ab-motility, % | 43.5 ± 13.2 | 42.6 ± 13.4 | 40.5 ± 13.8 | 1.61e-01 | -0.08 (0.06) | 43.5 ± 13.2 | 42.5 ± 13.2 | 40.2 ± 14 | 1.31e-01 | -0.09 (0.06) |
| 47 (17-59) | 46 (17.6-58) | 42 (17-57) | 47 (17-60) | 45 (17-58) | 40 (16.4-57) | |||||
| Sperm morphology | 4.4 ± 2.5 | 4.6 ± 2.7 | 4.6 ± 2.2 | 5.09e-01 | 0.04 (0.06) | 4.4 ± 2.5 | 4.7 ± 2.8 | 4.5 ± 2.3 | 3.85e-01 | 0.06 (0.07) |
| 4 (1-9) | 4 (1-10.8) | 5 (1-7) | 4 (1-9) | 4 (1-11) | 5 (1-7.3) | |||||
Two of the 9 identified single-nucleotide variations (discovery study) were analyzed via reverse transcriptase–polymerase chain reaction in an independent validation cohort (n = 1140 men with idiopathic infertility). Data are presented as mean ± SD and median (5th-95th percentile). Genotype-trait associations are calculated using an additive linear model with age added as a covariate. For sperm parameters, abstinence time was also included as a covariate.
Trends are marked in italics (*P < .05).
Statistically significant associations are marked in bold (**P < 2.50e-03).
Abbreviations: Androl., andrological; Bitestic, bitesticular; conc., concentration; ejac, ejaculation; FSH, follicle-stimulating hormone; LH, luteinizing hormone.
Figure 4.
Association between single-nucleotide variations (SNVs) rs11031005 and rs10835638 and follicle-stimulating hormone (FSH) serum levels using an additive model. Box plot depicting FSH serum levels for genotypes in the validation cohort (n = 1140). Medians are drawn as straight lines. Upper and lower hinges correspond to the 25th and 75th percentiles, respectively. Upper and lower whiskers extend to 1.5 interquartile ranges. Data beyond the end of the whiskers are not shown.
Applying a recessive model, where strong effects are expected only in minor allele homozygotes, and therefore, pooled wild-type and heterozygotes are compared with homozygotes (GG, GT, vs TT), a significant association of the analyzed SNVs with FSH/LH could be observed (26).
The results were further corroborated when combining the discovery and validation cohort: the SNVs rs11031005 and rs10835638 again were significantly associated with FSH serum level and FSH/LH ratio. In addition, when combining both cohorts the previously observed trend of an association to bitesticular volume is significant for both SNVs (rs10835638 P = 2.36e-04 and rs11031005 P = 5.17e-04) (26).
Thus, the SNV rs11031005 identified by GWAS in the frame of the discovery study was validated in an independent cohort of idiopathic and unexplained infertile men and a significant association with FSH levels and FSH/LH ratios was shown.
Effect of Identified Single-Nucleotide Variations on Follicle-Stimulating Hormone Serum Level Is Pronounced in Oligozoospermic Men
To further determine the effect of the identified SNVs, we performed a subgroup analysis in oligozoospermic patients (idiopathic infertility) of the discovery cohort (TSC < 39 Mill/ejac. [n = 294]).
We noticed a markedly increase in explained variances for all significant SNVs (see Table 2). For example, the SNV rs11031005 explains 6.95% of the variance in the oligozoospermic subgroup as compared to 4.65% for all patients in the discovery cohort (oligozoospermic and normozoospermic men). Again, the effect of the percentage variance explained in the described cohort was stronger in rs11031005 compared to rs10835638, which was previously known as the major genetic determinant to affect FSH action.
We identified and validated a genomic locus at chromosome 11p.14.1, containing the FSHB, to be the main determinant for genetically regulating FSH levels. The identified 9 SNVs, which are in high LD, include the previously described rs10835638. This SNV was outperformed, with respect to the effect on FSH serum level, by the rs11031005 identified in this study. The effects of the identified SNVs were even more enhanced in men with a more severe phenotype (oligozoospermia—discovery cohort) when compared to the entire discovery cohort.
Discussion
Male infertility is a highly complex disease, and for the majority of men no leading factor for infertility can be identified, resulting in the diagnosis of idiopathic or unexplained infertility. Intensive emphasis has been given to the effect of genetic variants on male infertility. GWASs have been performed in infertile men to identify SNVs associated with azoospermia and/or oligozoospermia, but with conflicting results (58, 72). Those may be due to the heterogeneous causes of impaired spermatogenesis, different ethnicities with different SNV compositions, or a lack of proper patient selection criteria.
Given the essential role of FSH for the initiation and maintenance of full spermatogenesis, we designed and compiled a GWAS (in the so far biggest Caucasian cohort of idiopathic and unexplained infertile men) in which we aimed to identify further genetic variants modifying FSH action. Our study is the first to delineate a genomic region on chromosome 11p.14.1 including the FSHB gene as of crucial importance in the regulation of FSH serum levels in infertile men with unknown origin. In this region we identified 9 SNVs (all in strong LD) located upstream and downstream from the FSHB gene that are significantly associated with FSH levels. In the validation of the SNVs rs11031005 and rs10835638 in an independent cohort, significant associations with FSH levels and FSH/LH ratio were evident.
Functional Impact of Identified Single-Nucleotide Variations
Functional studies have been exclusively performed on SNV FSHB c.-211G > T (rs10835638) located in the promoter of the FSHB gene. The reduced transcriptional activity is explained by an impaired binding of the transcription factor LHX3 (13). These studies neglect the presence of several LHX3 binding sites and the fact that the SNV is only adjacent to an LHX3 binding site. Therefore, it is tempting to speculate that the complete genomic FSHB gene region acts as a functional entity. Ruf-Zamojski et al (73) performed ATAC-Seq (assay for transposase-accessible chromatin with high-throughput sequencing) and identified an open chromatin region –40 to –90 kb upstream from the FSHB gene, which might represent novel upstream enhancers. A recent study by Bohaczuk et al (74) identified a region bearing a putative SF1 binding site, which could possibly act as such an enhancer of FSHB expression. This was further corroborated by Ruf-Zamojski and colleagues (75), who deleted a genomic region containing the SNV rs11031006 upstream from the FSHB gene which lead to an increased in basal and stimulated transcription of the FSHB gene. This further supports our hypothesis that the FSHB genomic region contains several enhancer and other regulatory elements that are modulated by the presence of several SNVs, and thereby regulate FSHB transcription. It is noteworthy that the rs11031005 described and investigated in this study is located in this region. Further studies have to reveal the functional implications of the SNVs in the enhancer regions in conjunction with the already described promoter effect.
Effect of Single-Nucleotide Variations on Follicle-stimulating Hormone Serum Level Variance and Clinical Relevance
The explained variance, which is the coefficient of determination, depicts the effect of individual SNVs on the variance of the FSH serum level. The 9 significant SNVs each explained 3.6% to 4.65% variance in FSH serum level in the analyzed cohort. This is a more powerful effect than that of the 3 SNVs (rs10835638, rs1394205, and rs6166) demonstrated by Grigorova and colleagues (66) who presented a variance of 2.3% on FSH serum level.
The effect is even more pronounced when analyzed in an oligozoospermic subgroup; here, the rs11031005 explains up to 6.95% of FSH serum level variance and even outperforms the rs10835638, which was so far considered the SNV with the highest effect on FSH serum level.
There is seemingly a sex difference concerning genotypes of distinct SNVs and their opposed effect on FSH serum level (54, 76). However, large GWASs performed in (menopausal/infertile) women identified a significant association of SNVs that we identified too, as rs11031005 and/or rs11031006 on FSH, LH, and sex hormone levels (30-32, 46). These results further strengthen our finding that the region 11.p14.1 is crucial for FSH levels.
The increased effect of the SNVs on FSH serum level in oligozoospermic men is in line with the increased MAF that we found in the oligozoospermic subgroup of the discovery cohort compared to an independent normozoospermic cohort. This again demonstrates the diverse effect of the SNVs on distinct phenotypes of infertility.
The different effect of the respective SNVs on the variance in FSH levels can be explained by the high but not complete LD among the SNVs. In case of rs11031005 and rs10835638, the LD is 74%. The frequency of association of their different alleles is high, but not 100%, which explains their slightly different effect on FSH serum level variance, and also explains that rs11031005 is a seemingly better predictor of FSH serum levels than the previously identified rs10835638 SNV. The magnitude of variance calculated for the SNVs in this study is in line with other described SNVs, with a strong effect on certain clinical parameters (77, 78).
The identified polymorphic genomic region outperforms other already identified and studied SNVs such as the SNVs of the FSH receptor by nearly a magnitude (65, 67). These results are in line with a cluster analysis that we performed in a large cohort of idiopathic and unexplained infertile men, in which we tried to delineate subgroups in this heterogeneous patient cohort via an unbiased cluster approach. We identified subgroups, interestingly across all phenotypes of infertility, with the strongest segregation markers in which the 2 subgroups differing significantly were FSHB c.-211 G > T, FSH, and bitesticular volume (79). This strongly supports the notion of a contributing factor in this genetic region.
In another study we found that azoospermic patients, lacking major genetic causes and carrying a T-allele in rs10835638, had significantly reduced chances for positive sperm retrieval rate via testicular sperm extraction compared to wild-type carriers (20). In a consecutive project we found that the SC number is not affected by the T-allele (80). This is in line with our recent findings; in case of an unfavorable SNV carrier/combination this obviously does not lead to a reduced SC number in the prepubertal phase, but during adulthood to the clinically well-described phenotype of reduced spermatogenesis.
The fact that the FSHB genomic region and its corresponding SNVs play a pivotal role in regulating FSH serum levels, which is supported by the calculated variance explained (see Table 2), has immediate clinical implications on a diagnostic level. The genotyping of the identified SNVs tags patients that have reduced FSH upregulation. We suggest including the genotyping of rs11031005 and/or rs10835638 after standardized andrological infertility workup, particularly in men with infertility of unknown origin. Therewith the amount of idiopathic and unexplained infertile men could be reduced tremendously.
Limitations
While we were able to identify 9 significant markers close to FSHB, our GWAS analysis should nevertheless be treated with caution because of sample size as observed effect sizes were smaller than our assumed r² value of 0.05. As a result, we would inherently miss genetic markers with only small effects on FSH levels. Nevertheless, our data might serve a further purpose in meta-analyses in future related GWAS analyses with higher sample sizes.
Decreased FSH serum level was an exclusion criterion; however, we did not include an upper limit for FSH serum level, nor subgroup analyses based on distinct FSH values. This was mainly because of the lack normal distribution via Box-Cox normalization.
Conclusion
Our unbiased approach to identifying genetic variants affecting FSH revealed that a genomic region on chromosome 11p14.1 bearing the FSHB gene seems to be the main genetic determinant affecting FSH serum levels and thereby FSH action. Other previously studied genetic variants such as in the FSHR gene seem to be less important in modifying FSH action and are outperformed by the strong effect of the identified FSHB SNVs. Consequently, SNVs in this region can be used for diagnostic purposes to identify a contributing etiologic factor in men with infertility of so far unknown origin.
Acknowledgments
We thank all participating patients for donating a blood sample for scientific use, and the clinicians who took care of the patients. We are grateful to the technical staff of the Centre of Reproductive Medicine and Andrology, Nicole Terwort, Sabine Forsthoff, Elisabeth Lahrmann, and Reinhild Sandhowe-Klaverkamp, for hormone and molecular analysis. In addition, we are indebted to Daniela Hanke, Jolanta Körber, Sabine Strüwing, and Raphaele Kürten for performing semen analysis. We thank Alexander Busch for valuable discussion on results of the study.
Glossary
Abbreviations
- FSH
follicle-stimulating hormone
- GWAS
genome-wide association study
- LD
linkage disequilibrium
- LH
luteinizing hormone
- MAF
minor allele frequency
- MDS
multidimensional scaling
- PCR
polymerase chain reaction
- SC
Sertoli cell
- SNV
single-nucleotide variation
- TSC
total sperm count
Contributor Information
Maria Schubert, Department of Clinical and Surgical Andrology, Centre of Reproductive Medicine and Andrology, University of Münster, Münster, North Rhine-Westphalia 48149, Germany.
Lina Pérez Lanuza, Department of Pediatric Hematology and Oncology, University Children’s Hospital Münster, Münster, North Rhine-Westphalia 48149, Germany.
Marius Wöste, Institute of Medical Informatics, University of Münster, Münster, North Rhine-Westphalia 48149, Germany.
Martin Dugas, Institute of Medical Informatics, University of Münster, Münster, North Rhine-Westphalia 48149, Germany; Institute of Medical Informatics, Heidelberg University Hospital, D-69120 Heidelberg, Germany.
F David Carmona, Department of Genetics and Institute of Biotechnology, University of Granada, Granada, Andalusia 18016, Spain; Instituto de Investigación Biosanitaria ibs.GRANADA, Granada, Andalusia 18012, Spain.
Rogelio J Palomino-Morales, Instituto de Investigación Biosanitaria ibs.GRANADA, Granada, Andalusia 18012, Spain; Department of Biochemistry and Molecular Biology I, Faculty of Sciences, University of Granada, Granada, Andalusia 18071, Spain.
Yousif Rassam, Department of Clinical and Surgical Andrology, Centre of Reproductive Medicine and Andrology, University of Münster, Münster, North Rhine-Westphalia 48149, Germany.
Stefanie Heilmann-Heimbach, Institute of Human Genetics, University of Bonn, School of Medicine & University Hospital, Bonn, North Rhine-Westphalia 53127, Germany.
Frank Tüttelmann, Institute of Reproductive Genetics, University of Münster, Münster, North Rhine-Westphalia 48149, Germany.
Sabine Kliesch, Department of Clinical and Surgical Andrology, Centre of Reproductive Medicine and Andrology, University of Münster, Münster, North Rhine-Westphalia 48149, Germany.
Jörg Gromoll, Institute of Reproductive and Regenerative Biology, Centre of Reproductive Medicine and Andrology, University of Münster, Münster, North Rhine-Westphalia 48149, Germany.
Author Contributions
L.P.L., M.S., M.W., and J.G. designed the study, compiled the available literature, and wrote the manuscript; L.P.L. performed the experimental work; S.K. was responsible for patient phenotyping; M.S. and Y.R. were responsible for the patient cohort selection; S.H.H. performed genotyping; D.C. and R.P. initiated and contributed the GWAS data on the normozoospermic group; M.W. performed all bioinformatic and statistical analysis; L.P.L., M.S., M.W., and J.G. designed the figures; and F.T., M.D., and S.K. gave valuable comments prior to the design of the study and during discussions of the outcome.
Financial Support
This work was supported by grants from the German Research Foundation of the CRU326 (Clinical Research Unit Male Germ Cells: From Genes to Function), and GR 1547/24-2.
Disclosures
The authors have nothing to disclose.
Data Availability
The data sets generated and/or analyzed during the present study are not publicly available but are available from the corresponding author on reasonable request.
References
- 1. Kliesch S. Diagnosis of male infertility: diagnostic work-up of the infertile man. Eur Urol Suppl. 2014;13:73-82. [Google Scholar]
- 2. Tüttelmann F, Ruckert C, Röpke A. Disorders of spermatogenesis: perspectives for novel genetic diagnostics after 20 years of unchanged routine. Med Genet. 2018;30(1):12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schlegel PN, Sigman M, Collura B, et al. Diagnosis and treatment of infertility in men: AUA/ASRM guideline 2021. J Urol. 2021;205(1):36-43. [DOI] [PubMed] [Google Scholar]
- 4. Hamada A, Esteves SC, Nizza M, Agarwal A. Unexplained male infertility: diagnosis and management. Int Braz J Urol. 2012;38(5):576-594. [DOI] [PubMed] [Google Scholar]
- 5. Sigman M, Lipshultz LI, Howards SS. Office evaluation of the subfertile male. In: Lipshultz LI, Howards SS, Niederberger CS, eds. Infertility in the Male. Cambridge University Press; 2009:176. [Google Scholar]
- 6. Sharpe RM, McKinnell C, Kivlin C, Fisher JS. Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood. Reproduction. 2003;125(6):769-784. [DOI] [PubMed] [Google Scholar]
- 7. Allan CM, Haywood M, Swaraj S, et al. A novel transgenic model to characterize the specific effects of follicle-stimulating hormone on gonadal physiology in the absence of luteinizing hormone actions. Endocrinology. 2001;142(6):2213-2220. [DOI] [PubMed] [Google Scholar]
- 8. Allan CM, Garcia A, Spaliviero J, et al. Complete Sertoli cell proliferation induced by follicle-stimulating hormone (FSH) independently of luteinizing hormone activity: evidence from genetic models of isolated FSH action. Endocrinology. 2004;145(4):1587-1593. [DOI] [PubMed] [Google Scholar]
- 9. Oduwole OO, Peltoketo H, Huhtaniemi IT. Role of follicle-stimulating hormone in spermatogenesis. Front Endocrinol (Lausanne). 2018;9:763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ruwanpura SM, McLachlan RI, Meachem SJ. Hormonal regulation of male germ cell development. J Endocrinol. 2010;205(2):117-131. [DOI] [PubMed] [Google Scholar]
- 11. McNeilly AS. The control of FSH secretion. Acta Endocrinol Suppl (Copenh). 1988;288:31-40. [PubMed] [Google Scholar]
- 12. Das N, Kumar TR. Molecular regulation of follicle-stimulating hormone synthesis, secretion and action. J Mol Endocrinol. 2018;60(3):R131-R155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benson CA, Kurz TL, Thackray VG. A human FSHB promoter SNP associated with low FSH levels in men impairs LHX3 binding and basal FSHB transcription. Endocrinology. 2013;154(9):3016-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nordhoff V, Sonntag B, von Tils D, et al. Effects of the FSH receptor gene polymorphism p.N680S on cAMP and steroid production in cultured primary human granulosa cells. Reprod Biomed Online. 2011;23(2):196-203. [DOI] [PubMed] [Google Scholar]
- 15. Simoni M, Casarini L. Mechanisms in endocrinology: genetics of FSH action: a 2014-and-beyond view. Eur J Endocrinol. 2014;170(3):R91-R107. [DOI] [PubMed] [Google Scholar]
- 16. Gromoll J, Simoni M. Genetic complexity of FSH receptor function. Trends Endocrinol Metab. 2005;16(8):368-373. [DOI] [PubMed] [Google Scholar]
- 17. Schubert M, Pérez Lanuza L, Gromoll J. Pharmacogenetics of FSH action in the male. Front Endocrinol (Lausanne). 2019; 10:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jungwirth A, Diemer T, Kopa Z, Krausz C, Minhas S, Tournaye H. EAU Guidelines on male infertility. Presented at: EAU Annual Congress; 2019; Barcelona, Spain.
- 19. Punab M, Poolamets O, Paju P, et al. Causes of male infertility: a 9-year prospective monocentre study on 1737 patients with reduced total sperm counts. Hum Reprod. 2017;32(1):18-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Busch AS, Tüttelmann F, Cremers JF, et al. FSHB –211 G > T polymorphism as predictor for TESE success in patients with unexplained azoospermia. J Clin Endocrinol Metab. 2019;104(6):2315-2324. [DOI] [PubMed] [Google Scholar]
- 21. Cooper TG, Noonan E, von Eckardstein S, et al. World Health Organization reference values for human semen characteristics. Hum Reprod Update. 2010;16(3):231-245. [DOI] [PubMed] [Google Scholar]
- 22. Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225-2229. [DOI] [PubMed] [Google Scholar]
- 23. Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002;155(5):478-484. [DOI] [PubMed] [Google Scholar]
- 24. Gauderman WJ. Sample size requirements for matched case-control studies of gene-environment interaction. Stat Med. 2002;21(1):35-50. [DOI] [PubMed] [Google Scholar]
- 25. Tüttelmann F, Luetjens CM, Nieschlag E. Optimising workflow in andrology: a new electronic patient record and database. Asian J Androl. 2006;8(2):235-241. [DOI] [PubMed] [Google Scholar]
- 26. Schubert M, Pérez Lanuza L, Wöste M, et al. Supplementary data for “A GWAS in Idiopathic/Unexplained Infertile Men Detects a Genomic Region Determining Follicle-Stimulating Hormone Levels.” figshare. Online deposition date: 11 March 2022. 10.6084/m9.figshare.19329824 [DOI] [PMC free article] [PubMed]
- 27. Gallagher CS, Mäkinen N, Harris HR, et al. Genome-wide association and epidemiological analyses reveal common genetic origins between uterine leiomyomata and endometriosis. Nat Commun. 2019;10:4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kichaev G, Bhatia G, Loh PR, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet. 2019;104(1):65-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sapkota Y, Steinthorsdottir V, Morris AP, et al. iPSYCH-SSI-Broad Group. Meta-analysis identifies five novel loci associated with endometriosis highlighting key genes involved in hormone metabolism. Nat Commun. 2017;8:15539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pau CT, Mosbruger T, Saxena R, Welt CK. Phenotype and tissue expression as a function of genetic risk in polycystic ovary syndrome. PLoS One. 2017;12(1):e0168870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mbarek H, Steinberg S, Nyholt DR, et al. Identification of common genetic variants influencing spontaneous dizygotic twinning and female fertility. Am J Hum Genet. 2016;98(5):898-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ruth KS, Campbell PJ, Chew S, et al. Genome-wide association study with 1000 Genomes imputation identifies signals for nine sex hormone-related phenotypes. Eur J Hum Genet. 2016;24(2):284-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ruth KS, Beaumont RN, Tyrrell J, et al. Genetic evidence that lower circulating FSH levels lengthen menstrual cycle, increase age at menopause and impact female reproductive health. Hum Reprod. 2016;31(2):473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cardoso-Dos-Santos AC, Tagliani-Ribeiro A, Matte U, Schuler-Faccini L. Genetic variants linked to folliculogenesis and successful pregnancy are not associated with twin births in a twins’ town. J Matern Fetal Neonatal Med. 2020;33(20):3431-3438. [DOI] [PubMed] [Google Scholar]
- 35. Papadakis G, Kandaraki EA, Tseniklidi E, Papalou O, Diamanti-Kandarakis E. Polycystic ovary syndrome and NC-CAH: distinct characteristics and common findings. a systematic review. Front Endocrinol (Lausanne). 2019;10:388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shaaban Z, Khoradmehr A, Jafarzadeh Shirazi MR, Tamadon A. Pathophysiological mechanisms of gonadotropins- and steroid hormones-related genes in etiology of polycystic ovary syndrome. Iran J Basic Med Sci. 2019;22(1):3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mortlock S, Restuadi R, Levien R, et al. Genetic regulation of methylation in human endometrium and blood and gene targets for reproductive diseases. Clin Epigenet. 2019;11:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang XZ, Pang YL, Wang X, Li YH. Computational characterization and identification of human polycystic ovary syndrome genes. Sci Rep. 2018;8:12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laisk T, Kukuškina V, Palmer D, et al. Large-scale meta-analysis highlights the hypothalamic-pituitary-gonadal axis in the genetic regulation of menstrual cycle length. Hum Mol Genet. 2018;27(24):4323-4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Matalliotakis M, Zervou MI, Matalliotaki C, et al. The role of gene polymorphisms in endometriosis. Mol Med Rep. 2017;16(5):5881-5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim JJ, Choi YM, Hong MA, et al. FSH receptor gene p. Thr307Ala and p. Asn680Ser polymorphisms are associated with the risk of polycystic ovary syndrome. J Assist Reprod Genet. 2017;34(8):1087-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mbarek H, Dolan CV, Boomsma DI. Two SNPs associated with spontaneous dizygotic twinning: effect sizes and how we communicate them. Twin Res Hum Genet. 2016;19(5):418-421. [DOI] [PubMed] [Google Scholar]
- 43. Rosenfield RL, Ehrmann DA. The pathogenesis of polycystic ovary syndrome (PCOS): the hypothesis of PCOS as functional ovarian hyperandrogenism revisited. Endocr Rev. 2016;37(5):467-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hayes MG, Urbanek M, Ehrmann DA, et al. Genome-wide association of polycystic ovary syndrome implicates alterations in gonadotropin secretion in European ancestry populations. Nat Commun. 2015;6:7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dumesic DA, Oberfield SE, Stener-Victorin E, Marshall JC, Laven JS, Legro RS. Scientific statement on the diagnostic criteria, epidemiology, pathophysiology, and molecular genetics of polycystic ovary syndrome. Endocr Rev. 2015;36(5):487-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Day FR, Hinds DA, Tung JY, et al. Causal mechanisms and balancing selection inferred from genetic associations with polycystic ovary syndrome. Nat Commun. 2015;6:8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Perry JRB, Hsu YH, Chasman DI, et al. kConFab Investigators; ReproGen Consortium. DNA mismatch repair gene MSH6 implicated in determining age at natural menopause. Hum Mol Genet. 2014;23(9):2490-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Laven JSE. Follicle stimulating hormone receptor (FSHR) polymorphisms and polycystic ovary syndrome (PCOS). Front Endocrinol (Lausanne). 2019;10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Branavan U, Muneeswaran K, Wijesundera S, Jayakody S, Chandrasekharan V, Wijeyaratne C. Identification of selected genetic polymorphisms in polycystic ovary syndrome in Sri Lankan women using low cost genotyping techniques. PLoS One. 2018;13(12):e0209830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wood MA, Rajkovic A. Genomic markers of ovarian reserve. Semin Reprod Med. 2013;31(6):399-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Qin Y, Sun M, You L, et al. ESR1, HK3 and BRSK1 gene variants are associated with both age at natural menopause and premature ovarian failure. Orphanet J Rare Dis. 2012;7:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. He C, Kraft P, Chasman DI, et al. A large-scale candidate gene association study of age at menarche and age at natural menopause. Hum Genet. 2010;128(5):515-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Trevisan CM, de Oliveira R, Christofolini DM, Barbosa CP, Bianco B. Effects of a polymorphism in the promoter region of the follicle-stimulating hormone subunit beta (FSHB) gene on female reproductive outcomes. Genet Test Mol Biomarkers. 2019;23(1):39-44. [DOI] [PubMed] [Google Scholar]
- 54. Conforti A, Esteves SC, Cimadomo D, et al. Management of women with an unexpected low ovarian response to gonadotropin. Front Endocrinol (Lausanne). 2019;10:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Batiha O, Alahmad NA, Sindiani A, Bodoor K, Shaaban S, Al-Smadi M. Genetics of female infertility: molecular study of newborn ovary homeobox gene in poor ovarian responders. J Hum Reprod Sci. 2019;12(2):85-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Busch AS, Hagen CP, Assens M, Main KM, Almstrup K, Juul A. Differential impact of genetic loci on age at thelarche and menarche in healthy girls. J Clin Endocrinol Metab. 2018;103(1):228-234. [DOI] [PubMed] [Google Scholar]
- 57. Rull K, Grigorova M, Ehrenberg A, et al. FSHB –211 G > T is a major genetic modulator of reproductive physiology and health in childbearing age women. Hum Reprod. 2018;33(5):954-966. [DOI] [PubMed] [Google Scholar]
- 58. Liu SY, Zhang CJ, Peng HY, et al. Strong association of SLC1A1 and DPF3 gene variants with idiopathic male infertility in Han Chinese. Asian J Androl. 2017;19(4):486-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Grigorova M, Punab M, Poolamets O, Adler M, Vihljajev V, Laan M. Genetics of sex hormone-binding globulin and testosterone levels in fertile and infertile men of reproductive age. J Endocr Soc. 2017;1(6):560-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu Q, Zhang J, Zhu P, et al. The susceptibility of FSHB –211G > T and FSHR G-29A, 919A > G, 2039A > G polymorphisms to men infertility: an association study and meta-analysis. BMC Med Genet. 2017;18:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tamburino L, La Vignera S, Tomaselli V, Condorelli RA, Mongioì LM, Calogero AE. Impact of the FSHB gene –211G/T polymorphism on male gonadal function. J Assist Reprod Genet. 2017;34(5):671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Simoni M, Santi D, Negri L, et al. Treatment with human, recombinant FSH improves sperm DNA fragmentation in idiopathic infertile men depending on the FSH receptor polymorphism p.N680S: a pharmacogenetic study. Hum Reprod. 2016;31(9):1960-1969. [DOI] [PubMed] [Google Scholar]
- 63. Busch AS, Tüttelmann F, Zitzmann M, Kliesch S, Gromoll J. The FSHB –211G > T variant attenuates serum FSH levels in the supraphysiological gonadotropin setting of Klinefelter syndrome. Eur J Hum Genet. 2015;23(5):700-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Punab AM, Grigorova M, Punab M, et al. Carriers of variant luteinizing hormone (V-LH) among 1593 Baltic men have significantly higher serum LH. Andrology. 2015;3(3):512-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Grigorova M, Punab M, Poolamets O, et al. Study in 1790 Baltic men: FSHR Asn680Ser polymorphism affects total testes volume. Andrology. 2013;1(2):293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Grigorova M, Punab M, Punab AM, et al. Reproductive physiology in young men is cumulatively affected by FSH-action modulating genetic variants: FSHR –29G/A and c.2039 A/G, FSHB –211G/T. PLoS One. 2014;9(4):e94244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tüttelmann F, Laan M, Grigorova M, Punab M, Sõber S, Gromoll J. Combined effects of the variants FSHB –211G > T and FSHR 2039A > G on male reproductive parameters. J Clin Endocrinol Metab. 2012;97(10):3639-3647. [DOI] [PubMed] [Google Scholar]
- 68. Laan M, Grigorova M, Huhtaniemi IT. Pharmacogenetics of follicle-stimulating hormone action. Curr Opin Endocrinol Diabetes Obes. 2012;19(3):220-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Grigorova M, Punab M, Zilaitienė B, et al. Genetically determined dosage of follicle-stimulating hormone (FSH) affects male reproductive parameters. J Clin Endocrinol Metab. 2011;96(9):E1534-E1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Grigorova M, Punab M, Poolamets O, et al. Increased prevalance [sic] of the –211 T allele of follicle stimulating hormone (FSH) beta subunit promoter polymorphism and lower serum FSH in infertile men. J Clin Endocrinol Metab. 2010;95(1): 100-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Grigorova M, Punab M, Ausmees K, Laan M. FSHB promoter polymorphism within evolutionary conserved element is associated with serum FSH level in men. Hum Reprod. 2008;23(9):2160-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen Z, Tao S, Gao Y, et al. Genome-wide association study of sex hormones, gonadotropins and sex hormone-binding protein in Chinese men. J Med Genet. 2013;50(12):794-801. [DOI] [PubMed] [Google Scholar]
- 73. Ruf-Zamojski F, Fribourg M, Ge Y, et al. Regulatory architecture of the LβT2 gonadotrope cell underlying the response to gonadotropin-releasing hormone. Front Endocrinol (Lausanne). 2018;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bohaczuk SC, Thackray VG, Shen J, Skowronska-Krawczyk D, Mellon PL. FSHB transcription is regulated by a novel 5′ distal enhancer with a fertility-associated single nucleotide polymorphism. Endocrinology. 2021;162(1):bqaa181. doi: 10.1210/endocr/bqaa181. PMID: 33009549; PMCID: PMC7846141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ruf-Zamojski F, Zhang Z, Zamojski M, et al. Single nucleus multi-omics regulatory landscape of the murine pituitary. Nat Commun. 2021;12(1):2677. doi: 10.1038/s41467-021-22859-w. PMID: 33976139; PMCID: PMC8113460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Schüring AN, Busch AS, Bogdanova N, Gromoll J, Tüttelmann F. Effects of the FSH-β-subunit promoter polymorphism –211G->T on the hypothalamic-pituitary-ovarian axis in normally cycling women indicate a gender-specific regulation of gonadotropin secretion. J Clin Endocrinol Metab. 2013;98(1):E82-E86. [DOI] [PubMed] [Google Scholar]
- 77. Perry JRB, McMahon G, Day FR, Ring SM, Nelson SM, Lawlor DA. Genome-wide association study identifies common and low-frequency variants at the AMH gene locus that strongly predict serum AMH levels in males. Hum Mol Genet. 2016;25(2):382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li Z, Chen J, Yu H, et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet. 2017;49(11):1576-1583. [DOI] [PubMed] [Google Scholar]
- 79. Krenz H, Sansone A, Kliesch S, Gromoll J, Schubert M. FSHB genotype identified as a relevant diagnostic parameter revealed by cluster analysis of men with idiopathic infertility. Front Endocrinol (Lausanne). 2021;12:780403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Schubert M, Kaldewey S, Pérez Lanuza L, et al. Does the FSHB c.–211G > T polymorphism impact Sertoli cell number and the spermatogenic potential in infertile patients? Andrology. 2020;8(5):1030-1037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated and/or analyzed during the present study are not publicly available but are available from the corresponding author on reasonable request.