Significance
Cancers initiate through the effects of oncogenic mutations on susceptible cell-of-origin states. To explore the interplay between initiating mutations, cell-of-origin states, and cancer cell behaviors, we examined retinal cell responses to N-Myc overexpression, which induces a form of retinoblastoma distinct from the RB1-mutant subtype. Whereas pRB loss induces proliferation and tumorigenesis in maturing cone precursors that retain differentiation markers and progress through an indolent premalignant stage, N-Myc induced proliferation in immature cone precursors, deconstrained cone precursor identity, and enabled more rapid oncogenic transformation dependent on a gene (SOX9) that is normally expressed in retinal progenitor cells. These findings illustrate that distinct cone precursor states are sensitive to different cancer-initiating mutations and follow markedly different paths toward phenotypically distinct retinoblastoma tumors.
Keywords: retinoblastoma, cone precursor, MYCN, cancer cell of origin, dedifferentiation
Abstract
Most retinoblastomas develop from maturing cone precursors in response to biallelic RB1 loss and are dependent on cone maturation-related signaling. Additionally, ∼2% lack RB1 mutations but have MYCN amplification (MYCNA), N-Myc protein overexpression, and more rapid and invasive growth, yet the MYCNA retinoblastoma cell of origin and basis for its responses to deregulated N-Myc are unknown. Here, using explanted cultured retinae, we show that ectopic N-Myc induces cell cycle entry in cells expressing markers of several retinal types yet induces continuous proliferation and tumorigenesis only in cone precursors. Unlike the response to RB1 loss, both immature cone arrestin-negative (ARR3–) and maturing ARR3+ cone precursors proliferate, and maturing cone precursors rapidly dedifferentiate, losing ARR3 as well as L/M-opsin expression. N-Myc–overexpressing retinal cells also lose cell lineage constraints, occasionally coexpressing the cone-specific RXRγ with the rod-specific NRL or amacrine-specific AP2α and widely coexpressing RXRγ with the progenitor and Müller cell–specific SOX9 and retinal ganglion cell-specific BRN3 and GAP43. Mechanistically, N-Myc induced Cyclin D2 and CDK4 overexpression, pRB phosphorylation, and SOX9-dependent proliferation without a retinoma-like stage that characterizes pRB-deficient retinoblastoma, despite continuous p16INK4A expression. Orthotopic xenografts of N-Myc–overexpressing retinal cells formed tumors with retinal cell marker expression similar to those in MYCN-transduced retinae and MYCNA retinoblastomas in patients. These findings demonstrate the MYCNA retinoblastoma origin from immature and lineage-deconstrained cone precursors, reveal their opportunistic use of an undifferentiated retinal progenitor cell feature, and illustrate that different cancer-initiating mutations cooperate with distinct developmental stage–specific cell signaling circuitries to drive retinoblastoma tumorigenesis.
Cancers develop in response to diverse oncogenic changes that converge on selected cell signaling pathways. However, even for a specific cancer type, different driver mutations may engender distinct tumor behaviors and therapeutic sensitivities. For most cancers, it is unclear whether different driver mutations generate distinct phenotypes solely because of their different signaling effects or due to the different cell-of-origin states on which they operate (1, 2). Retinoblastoma is a childhood retinal tumor that may reveal relationships between initiating oncogenic mutations, cell-of-origin states, and tumor behavior, as the tumors initiate either in response to biallelic RB1 inactivation, in ∼98% of cases, or in response to MYCN amplification (MYCNA), in ∼2%, and exhibit distinct subtype-specific behaviors (3, 4). Thus, retinoblastoma provides an opportunity to assess how different retinal cell states respond to pRB loss versus N-Myc overexpression to enable tumorigenesis.
pRB-deficient retinoblastomas are thought to arise from maturing long/medium wavelength (L/M) cone precursors based on their predominant L/M cone-like protein expression pattern and the pRB-deficient cone precursors’ ability to proliferate and form retinoblastoma-like masses both in explanted fetal retina and in stem cell–derived retinal organoids (5–8). In explanted retina, only maturing cone arrestin+ (ARR3+) pRB-depleted cone precursors enter the cell cycle, reflecting roles for a maturation-associated increase in N-Myc, MDM2, and other proliferation-related proteins (5, 6, 8). Maturing pRB-deficient cone precursors enter the cell cycle and begin to proliferate within 1 week of pRB loss, form retinoblastoma-specific histologic structures called Flexner–Wintersteiner rosettes within 1 month, largely exit the cell cycle to produce indolent retinoma-like lesions within several months, and subsequently emerge as retinoblastoma-like masses (8), recapitulating the stages of retinoblastoma genesis inferred from analyses of tumor-adjacent tissue (9). Cone precursor–derived retinoblastoma-like masses first become evident at a postfertilization tissue age of ∼8–9 mo in long-term cultured retinae, in orthotopic xenografts, and in genetically predisposed (RB1+/−) children (6, 8, 10, 11), consistent with the existence of a temporally constrained oncogenic program. Finally, pRB-deficient retinoblastomas progress to more aggressive, less differentiated forms with highly recurrent somatic copy number alterations (SCNAs) including 1q+, 2p+, 6p+, and 16q–, and less recurrent nucleotide variants (12–15).
Less is known of MYCNA retinoblastoma origins. MYCNA tumors usually arise earlier (median age 4.5 mo vs. 24 mo), grow faster, respond less well to therapy, and are more metastatic (3, 16, 17). While most MYCNA retinoblastomas have fewer copy number alterations and lack highly recurrent SCNAs, later-diagnosed cases can have high aneuploidy, as in RB1−/− retinoblastomas (3, 16, 18). Histologically, MYCNA retinoblastomas resemble MYCNA neuroblastoma more than RB1−/− retinoblastoma, with high nucleus-to-cytoplasm ratios, prominent nucleoli, a lack of Flexner–Wintersteiner rosettes, and pRB inactivated by phosphorylation rather than mutation (3, 4). While MYCNA retinoblastomas express markers of cones as well as rods (3, 19) and transcriptomically resemble a subset of RB1−/− retinoblastomas (15), their cell of origin and its responses to N-Myc overexpression are unknown.
To address the MYCNA retinoblastoma origin, oncogenic responses, and relationship to RB1−/− retinoblastoma, we examined effects of N-Myc overexpression in explanted developing human retinae. We report that N-Myc induces retinal cell type–specific yet developmental stage–independent cell proliferation, deconstrains cell lineage–related protein expression, enables reexpression of a critical cell survival–related retinal progenitor cell transcription factor, and recapitulates the distinctive behavior of the MYCNA retinoblastoma subtype.
Results
MYCN Transduction and Overexpression.
To define cell type–specific effects of N-Myc overexpression, we infected cultured fetal retinae with BE-MYCN lentivirus expressing N-Myc and green fluorescent protein (GFP) or with the BE-GFP vector control (Fig. 1 A and B) and assessed N-Myc expression and cell cycle entry in transduced (GFP+) cells in each retinal cell type. The BE-GFP bidirectional promoter uses an EF1α enhancer-promoter paired with a minimal CMV promoter to codrive MYCN and GFP expression (20). Explanted retinae ranged from 16 to 18 weeks gestation, when there is a peripheral-to-central differentiation gradient with retinal progenitor cells and early born retinal neurons in the peripheral retina and postmitotic retinal cells in the central retina (Fig. 1A). The cultured retinae largely retain tissue architecture yet with loss of most retinal ganglion cells by 1–2 wk after explant and optic nerve transection (8).
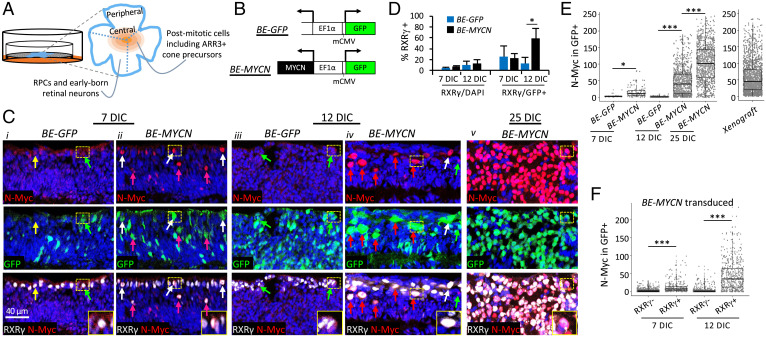
Fig. 1.
Lentivirus-mediated N-Myc overexpression in explanted retinae. (A) Explanted retinae were transduced with lentivirus, cut into sectors containing central and peripheral regions (dashed lines), and cultured semidry for 7–12 d (BE-GFP) or 7–25 d (BE-MYCN). The peripheral to central maturation gradient is represented by concentric shading centered on the fovea. (B) Bidirectional lentiviral vectors used to overexpress N-Myc and GFP from EF1α promoter-enhancer and minimal CMV promoter (mCMV). (C) N-Myc and GFP immunostaining of different sectors of the same retinae at 7, 12, and 25 DIC. Yellow arrows, nontransduced GFP–,RXRγ+ cones with low N-Myc; green arrows, transduced GFP+,RXRγ+ with low N-Myc; white arrows, transduced GFP+,RXRγ+ with high N-Myc; pink arrows, transduced GFP+,RXRγ+ with high N-Myc and displaced away from outer nuclear layer. Retinal sectors were fixed and embedded with identical protocols and immunostained and imaged in parallel with identical settings. Inset, enlarged view of GFP+,RXRγ+ cells with high N-Myc. Scale bar, 40 μm. (D) Percentage of total (DAPI+) cells and transduced (GFP+) cells identified as cones (RXRγ+) at 7 and 12 d after transduction with BE-GFP vector or BE-MYCN. 13,306–16,670 cells from four to eight retinal regions examined for each timepoint and condition. (E) N-Myc expression in GFP+ cells in retinal explants transduced with BE-GFP or BE-MYCN at 7, 12, and 25 DIC or in xenografts stained and imaged with identical conditions. (F) N-Myc expression in RXRγ+ and RXRγ– cells at 7 or 12 d after BE-MYCN infection. Box plots show interquartile range and median. Significance assessed by t test. *P < 0.01; ***P < 0.0001. Data are representative of two BE-GFP– and two BE-MYCN–transduced retinae.
Retinae were infected on the day of explant and sectors examined for GFP and N-Myc at 7, 12, and 25 d in culture (DIC) (Fig. 1C). At 7 DIC, GFP was distributed across all retinal layers in both the BE-GFP and BE-MYCN groups. In control BE-GFP–infected retinae, endogenous N-Myc was detected in GFP– and GFP+ cone precursors (Fig. 1Ci), identified by their outer nuclear layer (ONL) position and RXRγ expression, as in uncultured retinae (5). In contrast, BE-MYCN–infected retinae had higher N-Myc in GFP+,RXRγ+ cone precursors both in the ONL and displaced toward the inner retina (Fig. 1Cii). 12-DIC BE-GFP–infected retinae resembled those at 7 DIC but with a greater proportion of GFP+ cells, whereas 12-DIC BE-MYCN–infected retinae had higher GFP and N-Myc expression in RXRγ+ cells (Fig. 1Ciii, iv) and a higher proportion of GFP+,RXRγ+ cells (Fig. 1D). N-Myc was also far higher after MYCN transduction than after pRB depletion (SI Appendix, Fig. S1). The transduced RXRγ+,N-Myc+ cells often formed foci in peripheral and central retinae at 12 DIC and formed larger foci that disrupted retinal architecture at 25 DIC (Fig. 1Civ, v and SI Appendix, Figs. S1 and S2). Cells in these foci also expressed the L/M cone marker TRβ2 (SI Appendix, Fig. S2B). Quantitative image analyses using Weka machine learning–based segmentation (21) across entire retina sections (SI Appendix, Fig. S3) confirmed higher N-Myc in BE-MYCN versus control retinae at 7 and 12 DIC and still higher N-Myc at 25 DIC (Fig. 1E). Moreover, N-Myc levels were 5 times higher in MYCN-transduced RXRγ+ cone precursors than in RXRγ– noncones at 7 DIC and were 12 times higher at 12 DIC (Fig. 1F). Thus, cone precursors expressed ectopic N-Myc at higher levels than other retinal cell types.
N-Myc–Induced Cone Precursor Proliferation and SOX9 Misexpression.
We next examined N-Myc effects on cell cycle entry in each surviving retinal cell type by costaining for GFP, cell type markers, and Ki67 at 12, 25, and 56 DIC (Fig. 2 A–C and SI Appendix, Fig. S4). Markers included RXRγ (cones), NRL (rods), AP2α (amacrine cells), PROX1 (horizontal, amacrine, and G2/M phase progenitor cells (22)), VSX2 (bipolar, progenitor, and Müller cells), and SOX9 (progenitor and Müller cells). Automated segmentation was used for unbiased detection of each marker in hundreds to thousands of transduced (GFP+) cells in multiple images tiled across explanted retina sections.
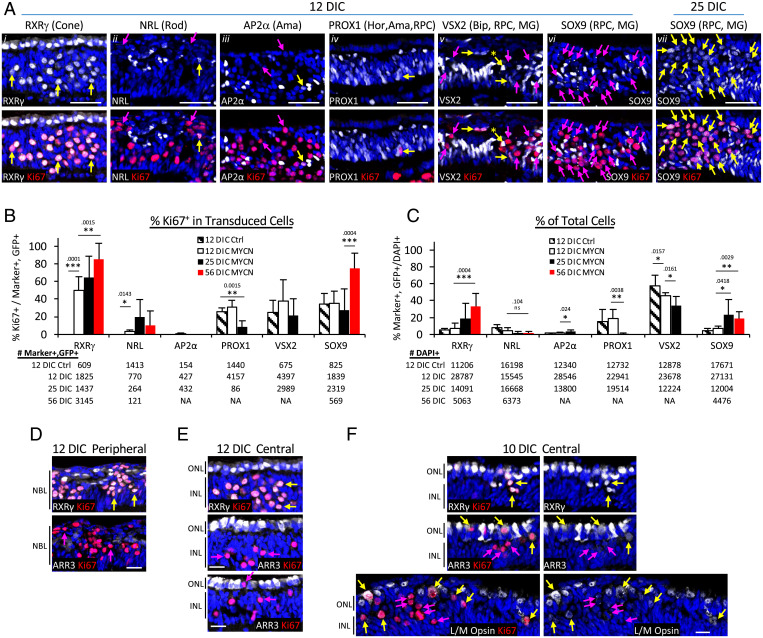
Fig. 2.
MYCN overexpression induces cone precursor cell cycle entry and SOX9 misexpression. (A) Immunofluorescence costaining of Ki67 with retinal cell markers RXRγ, NRL, AP2α, PROX1, VSX2, and SOX9 in retinal explants 12 or 25 DIC after BE-MYCN infection. Yellow arrows, transduced GFP+,marker+,Ki67+. Pink arrows, transduced GFP+,marker–,Ki67+. Scale bars, 40 μm. GFP staining, enlarged views of marker+, Ki67+ cells, and additional 25 DIC data are in SI Appendix, Fig. S4. (B and C) Quantitation of Ki67 coexpression with each marker in transduced (GFP+) cells (B) and overall proportion of transduced marker-positive cells (C) in 12-DIC BE-GFP transduced explants and 12, 25, and 56 DIC BE-MYCN–transduced explants. The numbers of marker+,GFP+ cells (A) or DAPI+ cells (B) are indicated below each panel. Values and error bars represent means and SDs of seven or eight separately imaged regions across entire retina sections. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant. Results are representative of immunostaining in two BE-MYCN-transduced retinae. (D–F) Cone precursor maturation-independent cell cycle entry. Peripheral and central regions of explant retinae at 10 or 12 d after BE-MYCN transduction coimmunostained with pan cone marker (RXRγ) or cone maturation markers (ARR3 or L/M-opsin) and with Ki67. Each group shows similar region in adjacent or near-adjacent section. Arrows colored as in (A). NBL, neuroblastic layer. Scale bars, 20 µm.
First focusing on cone precursors, at 12 DIC, all MYCN-transduced cells in RXRγ+ foci were Ki67+ (Fig. 2Ai and SI Appendix, Fig. S4A). When cells outside these foci were included, ∼50% of MYCN-transduced cones but no control vector-infected cones were Ki67+ at 12 DIC (P < 0.0001), rising to 86% at 56 DIC (P < 0.01, Fig. 2B). Concurrently, the proportion of RXRγ+,GFP+ cells in cultured retina increased from 5 to 33% (P < 0.001), indicative of continuous cone precursor proliferation (Fig. 2C). Moreover, RXRγ+,Ki67+ cells costained for photoreceptor marker CRX but not retinal ganglion cell marker BRN3 at 7 and 12 DIC (SI Appendix, Fig. S5), consistent with cone precursor identity at the time of initial cell cycle entry.
In contrast, only ∼3% of MYCN-transduced (and no control vector-transduced) NRL+ cells were Ki67+ at 12 DIC, similar proportions were Ki67+ at 25 and 56 DIC (Fig. 2Aii and B and SI Appendix, Fig. S4), and the proportion of GFP+,NRL+ cells trended to lower levels from 12 DIC to 56 DIC (P = 0.10, Fig. 2C). About 2% of GFP+,NRL+ cells coexpressed RXRγ at 12 and 25 DIC, and >80% coexpressed RXRγ at 56 DIC (SI Appendix, Fig. S6), indicative of NRL misexpression in the expanding RXRγ+ cone precursor population. Concordantly, at early timepoints, GFP+,NRL+,Ki67+ cells were often detected in RXRγ+,Ki67+ foci (Fig. 2Aii). Similarly, MYCN failed to induce robust cell cycle entry in amacrine cells, as only 0.3% of MYCN-transduced GFP+,AP2α+ cells were Ki67+ at 12 DIC and no AP2α+,Ki67+ cells were detected at later timepoints (Fig. 2 Aiii and B, and SI Appendix, Fig. S4). Although 12 DIC GFP+,AP2α+,Ki67+ cells were detected in RXRγ+, Ki67+ foci (Fig. 2Aiii), only rare (<1%) GFP+,AP2α+ cells coexpressed RXRγ (SI Appendix, Fig. S6). These findings are consistent with rare N-Myc–induced cell cycle entry in AP2α+ amacrine cells or rare AP2α misexpression in cone precursor foci.
In contrast to the MYCN-dependent expression of Ki67 in RXRγ+, NRL+, or AP2α+ cells, Ki67 was detected in similar proportions of PROX1+ or VSX2+ cells in MYCN-transduced and control retinae (Fig. 2Aiv–v and B). Most transduced PROX1+,Ki67+ and VSX2+, Ki67+cells were in the peripheral retina, as expected for retinal progenitor cells that proliferate independent of ectopic N-Myc. However, after MYCN transduction, rare PROX1+,Ki67+ and VSX2+,Ki67+ cells were detected in central retina inner nuclear layer (INL) positions consistent with Müller glia (Fig. 2Aiv), and some VSX2+, Ki67+ cells had INL positions and high VSX2 characteristic of bipolar cells (Fig. 2Av). Still, the GFP+,PROX1+ and GFP+,VSX2+ populations declined between 12 and 25 DIC (Fig. 2C), showing no evidence of N-Myc–induced proliferation.
Last, we examined cells expressing the retinal progenitor and Müller cell marker SOX9 (23). At 12 DIC, ∼35% of transduced SOX9+ cells were Ki67+ in BE-MYCN–infected and control BE-GFP–infected retinae (Fig. 2B), with most SOX9+, Ki67+ cells in peripheral regions consistent with proliferating progenitors. However, in contrast to progenitor markers PROX1 and VSX2, the GFP+,SOX9+ population increased from 12 to 25 DIC and GFP+,SOX9+,Ki67+ cells increased at 56 DIC (Fig. 2 B and C). Moreover, although SOX9 was not detected in Ki67+ foci at 12 DIC, it was detected throughout such foci at 25 and 56 DIC (Fig. 2Avi–vii and SI Appendix, Fig. S4 B and C), where it coexpressed with RXRγ (SI Appendix, Fig. S6).
In sum, N-Myc induced cell cycle entry and proliferating foci in explanted fetal retina. The vast majority of the N-Myc–induced Ki67+ cells expressed the cone-specific RXRγ and TRβ2, and the expanding RXRγ+ cells adopted SOX9 expression and comprised >30% of explanted retinal cells by 56 DIC. Ki67 was rarely detected in MYCN-transduced NRL+ or AP2α+ cells, and at least a subset of such cells coexpressed RXRγ, indicative of NRL or AP2α misexpression in cone precursors, yet these populations declined over time. Ki67 was less often detected in central retina PROX1+ or VSX2+ cells, whose proportions also declined. Thus, N-Myc induced continuous proliferation and, subsequently, SOX9 misexpression solely in cone precursor populations.
N-Myc–Induced Proliferation Is Maturation Independent and Associated with Dedifferentiation and pRB Phosphorylation.
As pRB loss enables cell cycle entry solely in maturing ARR3+ cone precursors (8), we examined whether N-Myc also induces cell cycle entry in a specific cone maturation state. At 12 DIC, N-Myc induced production of RXRγ+,Ki67+ foci both in the retinal periphery, which harbors immature ARR3– cone precursors, and in the central retina, which has maturing ARR3+ cone precursors (Fig. 2 D and E and SI Appendix, Fig. S2). However, in the central retina, most Ki67+ cells within RXRγ+ foci were ARR3–, in contrast to the ARR3+,Ki67– cone precursors in the nearby ONL (Fig. 2E). We then analyzed a MYCN-transduced central retina at 10 DIC, when smaller RXRγ+,Ki67+ foci extended from the ONL into the INL (Fig. 2F). In this setting, Ki67+ cells in the ONL had detectable but diminished ARR3 relative to their Ki67– neighbors, whereas those in the INL had little or no ARR3 (Fig. 2F). Similarly, L/M-opsin was detected in Ki67– and Ki67+ cone precursors in the ONL and was weaker or undetectable in Ki67+ cells displaced from the ONL (Fig. 2F). Thus, N-Myc induced cell cycle entry in immature and maturing cone precursors and reduced expression of the cone maturation-related ARR3 and L/M-opsin proteins.
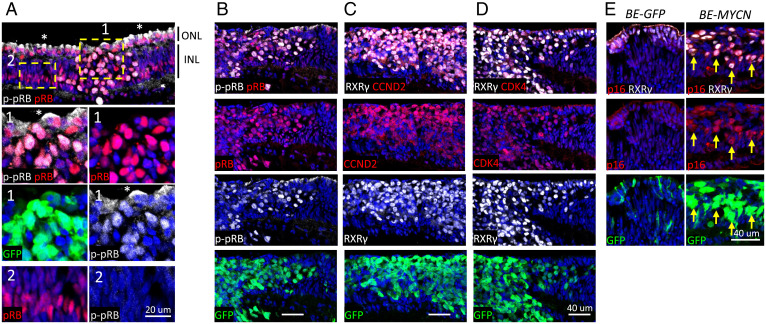
To assess how N-Myc induces cone precursor proliferation while retaining wild type RB1, we evaluated pRB Ser-807,811 phosphorylation and expression of pRB kinases. At 12 DIC, phosphorylated (p-) pRB(S807, S811) coimmunostained with total pRB throughout proliferating cone precursor foci (Fig. 3A, region 1) but was not detected in pRB+ Müller cells (Fig. 3A, region 2) identified by their characteristic nuclear positions and high pRB expression (24). The p-pRB(S807, S811)+ cone precursors highly coexpressed Cyclin D2 and CDK4 (Fig. 3 B–D), contrasting with undetectable Cyclin D1 and weak Cyclin D3 and CDK6, and consistent with Myc-induced Cyclin D2 expression (25). As in pRB-depleted cone precursors (8), N-Myc induced p16INK4a at 12 DIC. Moreover, p16 was sustained in highly proliferating cone precursor masses at 25 and 56 DIC (Fig. 3E and SI Appendix, Fig. S7), implying that p16 levels were insufficient to inhibit pRB phosphorylation and proliferation.
Fig. 3.
pRB phosphorylation, Cyclin D2/CDK4 induction, and p16INK4A expression in MYCN-induced cone precursor foci. (A) MYCN-transduced retina coimmunostained for total pRB and p-pRB(S807, S811) at 12 DIC. Insets show proliferating cone precursor focus (region 1) and Müller glia nuclei (region 2). p-pRB(S807, S811) signal in cone precursor cytoplasm (*) is nonspecific since it was not diminished by pRB knockdown. (B–D) MYCN-transduced cone precursor focus costained for pRB and p-pRB(S807, S811) (B), RXRγ and Cyclin D2 (C), or RXRγ and CDK4 (D) at 25 DIC. (E) Control (BE-GFP) and MYCN-transduced 12 DIC retina costained for RXRγ and p16INK4A. Arrows, cells coexpressing GFP, RXRγ, and p16. Scale bars, 40 µm. Results are representative of immunostaining in two BE-MYCN–infected retinae.
MYCN-Transduced Retinae Yield Tumors Resembling MYCNA Retinoblastoma.
To determine whether MYCN-transduced cone precursors can form tumors, we FACS isolated cone-enriched populations by using cone surface markers (6) or a transduced cone-specific (OPN1LW-CFP) fluorescent reporter, followed by BE-MYCN infection, culture for 1 wk, and either further culture or subretinal xenograft in NSG mice. However, with both cone enrichment strategies, all MYCN-transduced cells died within 2 wk in vitro and failed to form tumors in vivo, in contrast to the behavior of shRB1-transduced cells (6). The propensity for MYCN-induced cell death was also evident in RXRγ+,Ki67+ foci (SI Appendix, Fig. S4D).
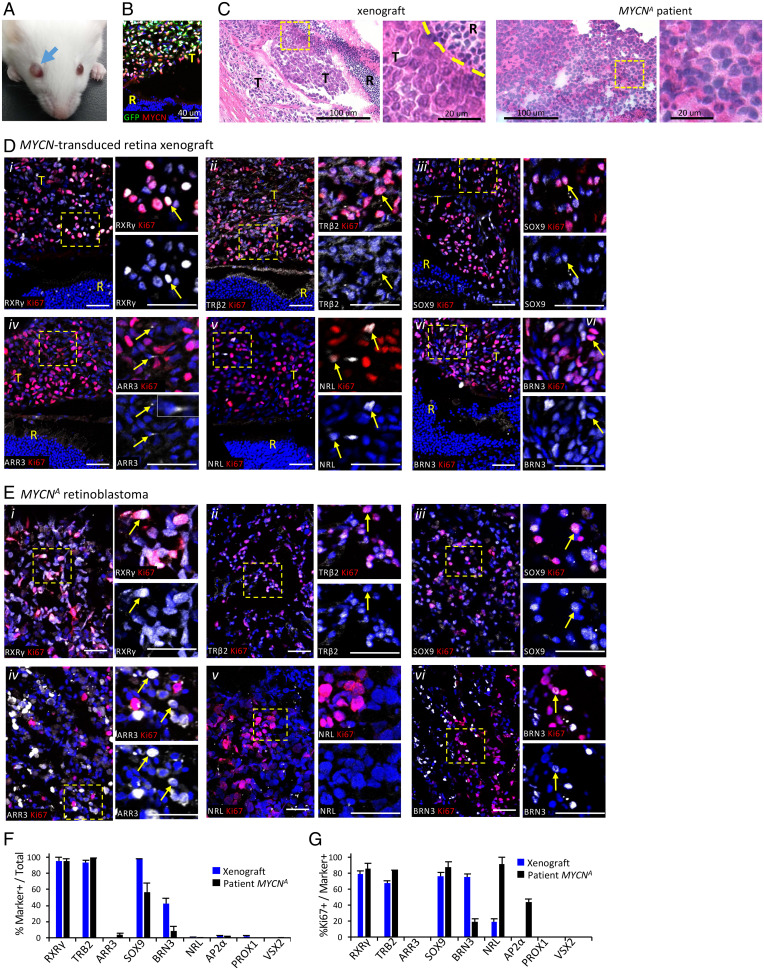
As an alternative strategy, subretinal xenografts were performed with a mixed population of 105 cells from 14-DIC BE-MYCN or control BE-GFP transduced retinae. After 47 d, tumors were seen in three of four mice engrafted with MYCN-transduced cells but in none of four engrafted with control cells (Fig. 4A). The tumors appeared more rapidly than engraftments with pRB-depleted retinal cells (mean 325 d) (6). Detailed analysis of one xenograft revealed tumor cells with high N-Myc expression as in RXRγ+ foci in explanted retina (Fig. 1E and 4B) and histology similar to that of a MYCNA patient tumor, with high nuclear:cytoplasmic ratio and prominent nucleoli (Fig. 4C). Most tumor cells costained for pRB and p-pRB(S807, S811), as in cultured retinae and MYCNA tumors (SI Appendix, Fig. S8 A and B) (4).
Fig. 4.
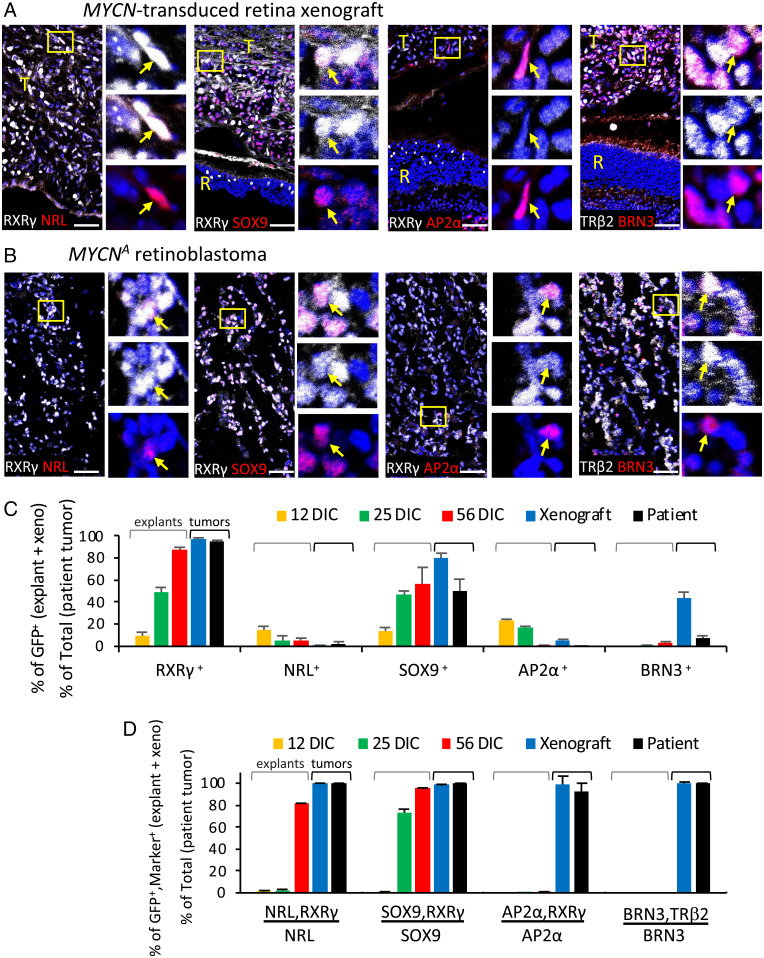
Xenografts of MYCN-transduced retinal explants mimic MYCNA retinoblastoma. (A) Intraocular tumor at 47 d postxenograft of 14-DIC MYCN-transduced retina. (B) N-Myc expression in xenograft GFP+ cells; quantitation in Fig. 1E. (C) Hematoxylin and eosin staining of xenograft and patient MYCNA retinoblastoma. Scale bars, 100 µm (Left); 20 µm (Right). (D and E) Immunofluorescence costaining of Ki67 with cell type markers RXRγ, TRβ2, SOX9, ARR3, NRL, BRN3 in xenograft (D) and MYCNA retinoblastoma (E). RXRγ, TRβ2, and SOX9 are detected in cones and Müller cells in mouse retina (R) as well as in xenograft tumor (T), whereas ARR3 and NRL were not detected in mouse retina with human-specific antibodies. Dashed boxes, regions enlarged at right. Arrows, cells coexpressing each marker. Scale bar, 40 μm. (F and G) Proportions of xenograft and MYCNA patient tumor cells expressing each marker (F) and proportion of marker+ cells also Ki67+ (G). One xenograft tumor and one MYCNA tumor analyzed; 1,130–8,277 GFP+ xenograft cells and 2,067–38,146 MYCNA tumor cells examined in two to nine separate stainings per marker.
We next examined whether the expression of retinal cell markers in MYCN-transduced retinae is recapitulated in the xenograft and in a patient-derived MYCNA retinoblastoma by costaining for cell type markers, Ki67, and GFP, followed by Weka-based quantitation. As all xenograft cells expressing the human-specific HuNu antigen were GFP+, only GFP+ cells were examined in the xenograft, whereas all cells were examined in the MYCNA tumor. Similar to cone precursor foci in 25 and 56 DIC retinae (Fig. 2), >90% of the xenograft and MYCNA tumor cells expressed cone markers RXRγ and TRβ2, and most xenograft and patient tumor cells coexpressed SOX9 (Fig. 4 Di–iii, Ei–iii, and F). More than 80% of RXRγ+ and SOX9+ cells were Ki67+ (Fig. 4G), revealing that these cells made up the main proliferating population. ARR3 was detected only rarely, at low levels, and in subcellular structures in Ki67+ cells of xenograft and MYCNA tumors but was detected at higher levels and throughout the cytoplasm in ∼3% of MYCNA tumor cells, mostly in localized regions and never coexpressing Ki67 (Fig. 4 Div, Eiv, F, and G). Thus, the xenograft and MYCNA retinoblastoma cells coexpressed cone factors RXRγ and TRβ2 with the progenitor factor SOX9, and rare nonproliferating MYCNA tumor cells coexpressed ARR3.
Similar to MYCN-transduced cultured retinae, only rare cells in the xenograft and MYCNA tumor expressed NRL, AP2α, or PROX1, and occasional NRL+ and AP2α+ cells were Ki67+ (Fig. 4 Dv, Ev, F, and G). However, unlike cultured retinae, many xenograft and MYCNA tumor cells expressed BRN3, and among those, most BRN3+ xenograft and many BRN3+ MYCNA tumor cells were Ki67+ (Fig. 4 Dvi, Evi, F, and G). The xenograft and MYCNA tumors more widely expressed the retinal ganglion cell-specific GAP43 (SI Appendix, Fig. S9), consistent with its widespread expression in MYCNA and high-MYCN-expressing RB1−/− retinoblastomas (15). Moreover, nearly all NRL+, SOX9+, AP2α+, and BRN3+ cells in xenograft and patient MYCNA tumors coexpressed cone markers RXRγ or TRβ2 (Fig. 5), indicating that they comprised subsets of the RXRγ+ and TRβ2+ cell populations.
Fig. 5.
MYCN-transformed cone precursors coexpress markers of multiple retinal cell types. (A and B) Coimmunostaining of cone markers RXRγ or TRβ2, rod marker NRL, progenitor and Müller cell marker SOX9, amacrine cell marker AP2α, and retinal ganglion cell marker BRN3 in MYCN-transduced retina xenograft (A) and MYCNA patient retinoblastoma (B). Boxes, regions enlarged at right. T, tumor in subretinal space; R, mouse retina. Arrows, cells coexpressing each marker. Scale bar, 40 µm. (C and D) Proportions of cells coexpressing retinal cell markers in explanted retina at 12, 25, and 56 d after MYCN transduction (representative data in SI Appendix, Fig. S6) (C) and in xenograft and patient tumors (representative data in panels A and B) (D) determined by Weka-based segmentation. One xenograft tumor and one MYCNA tumor analyzed; 1,367–12,528 cells examined in two to eight stainings per antibody and sample type.
Notably, the proportions of NRL+, AP2α+, SOX9+, and BRN3+ cells varied, and the proportions that coexpressed RXRγ or TRβ2 increased with different timing in retinal explant cultures and xenografts (Fig. 5 C and D). Few if any AP2α+ and BRN3+ cells coexpressed RXRγ or TRβ2 in explants, but nearly all did so in the xenograft and MYCNA tumor. Likewise, only rare NRL+ cells coexpressed RXRγ in 12 and 25 DIC explants, but most coexpressed RXRγ at 56 DIC and in tumors. Nuclear SOX9 was not coexpressed with RXRγ in 12 DIC explants but was abundant in 25 and 56 DIC explants and tumors. The different timing most likely reflects differentially timed death of rod, amacrine, progenitor, and ganglion cells and appearance of each marker in the N-Myc–overexpressing cone precursors that overtake the retina cultures and tumors.
SOX9 Misexpression Enables N-Myc–Overexpressing Cone Precursor Survival.
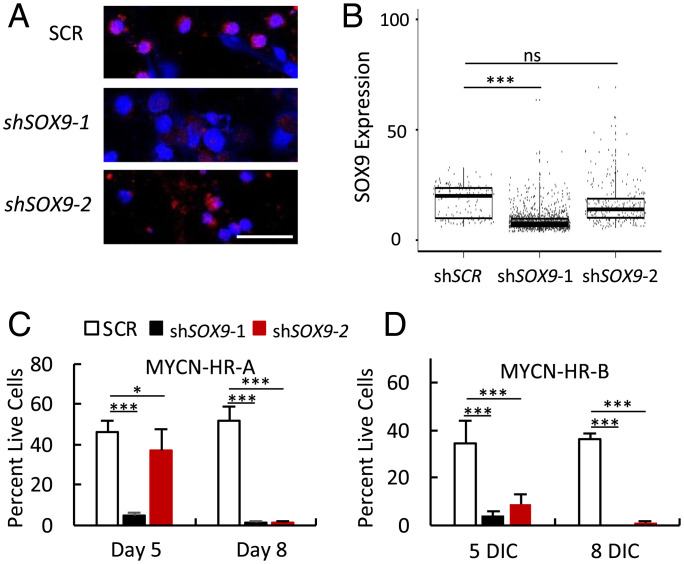
As SOX9 was robustly induced in MYCN-transduced cells and is often upregulated in cancer and associated with poor patient survival (26, 27), we examined the SOX9 role in N-Myc–induced cone precursor proliferation. Toward this end, we segregated aberrantly growing cell foci from two 21-DIC MYCN-transduced retinae (MYCN-HR-A and MYCN-HR-B), after which cultures expanded continuously, with ongoing cell death, for 26 d (MYCN-HR-A) or 35 d (MYCN-HR-B) before cryopreservation. On further expansion, cells were infected with lentiviral vectors expressing either of two SOX9-directed shRNAs or a shSCR control, selected in puromycin, and examined for SOX9 expression by quantitative immunostaining on day 5. In MYCN-HR-A cells, shSOX9-1 downregulated mean SOX9 levels by 45% and impaired cell viability by 90%, while shSOX9-2 decreased expression by 10% and viability by 19% (Fig. 6 A–C). By 8 DIC, both SOX9 shRNAs decreased viability by >97%. Similar results were obtained in MYCN-HR-B cultures (Fig. 6D). Thus, SOX9 has a critical role in N-Myc–overexpressing human cone precursor survival preceding the appearance of MYCN-induced retinoblastoma tumors.
Fig. 6.
SOX9 is essential for N-Myc–overexpressing cone precursor survival. (A) SOX9 immunostaining of early passage MYCN-HR-A cells at 5 d after lentiviral transduction of control (shSCR) or SOX9-directed (shSOX9-1, shSOX9-2) shRNAs. Scale bar, 40 µm. (B) Weka-based quantitation of SOX9 immunostaining. Boxes show interquartile range. Horizontal lines indicate median. (C and D) Percentage live cells at 5 and 8 d after transduction of shSCR or shSOX9 shRNAs in MYCN-HR-A cells (300,000/well) and MYCN-HR-B cells (50,000/well) in triplicate. Error bars, SD. Significance assessed by Student’s t test (*P < 0.05, ***P < 0.0001, ns = not significant).
Discussion
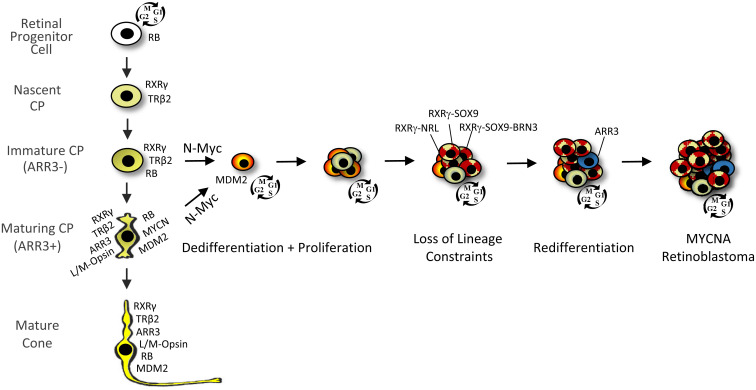
MYCNA retinoblastomas are a unique subtype with a previously undefined cell of origin. Here, we show that in developing human retina, N-Myc overexpression induced cell cycle entry in cells expressing diverse cell type markers but induced robust proliferation and tumorigenesis only in cone precursors. In contrast to the response to pRB loss (8), both immature ARR3– and maturing ARR3+ cone precursors entered the cell cycle and formed proliferating foci (Fig. 7). N-Myc–overexpressing cone precursors downregulated ARR3 and L/M-opsin and lost cell lineage constraints, with rare cells in RXRγ+ foci coexpressing rod marker NRL or amacrine marker AP2α and most RXRγ+ cells coexpressing SOX9. MYCN-transduced retinal xenografts formed tumors resembling explanted retina foci and a MYCNA tumor, with extensive RXRγ and SOX9 coexpression. The rapid and continuous cone precursor proliferation, sustained SOX9 coexpression, and similarity of cone precursor–like masses in vitro, in xenografts, and in patient tumors provide compelling support for a lineage-deconstrained cone precursor origin of the MYCNA retinoblastoma subtype.
Fig. 7.
Immature and maturing cone precursor (CP) responses to N-Myc overexpression. N-Myc–induced proliferation of immature cone precursors, which rapidly upregulate MDM2 (30), as well as maturing cone precursors, which rapidly dedifferentiate, exemplified by loss of ARR3 and L/M-opsin. Proliferation is associated with deconstrained cell lineage boundaries such that rod (NRL), progenitor (SOX9), and ganglion cell (BRN3) markers are coexpressed with cone markers (RXRγ and TRβ2) to varied extents and with varied timing, and with SOX9 having a critical cell survival role. Redifferentiation, exemplified by ARR3 expression, may be observed in nonproliferating cells in N-Myc–overexpressing tumors. Unlike pRB-depleted cone precursors (8), N-Myc–overexpressing cone precursors continuously proliferate without a retinoma stage to yield heterogeneous cancers that coexpress markers of cones and other retinal cell types.
At present, the basis for the cone precursors’ unique sensitivity to N-Myc is unclear. As one possibility, human cone precursors may be predisposed to express N-Myc at higher levels than other retinal cells (Fig. 1F). For example, human cone precursors may enable N-Myc overexpression via an MDM2-mediated increase in N-Myc translation (28) that is sustained by a reciprocal N-Myc induced upregulation of MDM2 (29, 30). However, the cone precursors’ robust N-Myc expression seems unlikely to be the sole basis for their proliferative response, as some RXRγ– cells had high N-Myc yet failed to form foci (Figs. 1 and 2). Our data suggest that the cone precursors’ proliferative response to N-Myc relates to their ability to robustly induce Cyclin D2 and CDK4 expression and pRB phosphorylation (Fig. 3), which are known to mediate Myc-induced cell cycle entry in fibroblasts (25). Why N-Myc induces Cyclin D2 selectively in cone precursors is unclear yet most likely reflects cone precursor chromatin features. Notably, we did not detect N-Myc–induced expression of the TEAD4 master regulator of the MYCNA neuroblastoma transcription network (31), implying that the N-Myc–induced oncogenic program in retinoblastoma differs from that of MYCNA neuroblastoma.
In addition to losing maturation markers, N-Myc–overexpressing cone precursors differed from pRB-depleted cone precursors in that N-Myc induced cell cycle entry in both immature ARR3– and maturing ARR3+ cells. This accords with the notion that pRB-depleted cone precursor cell cycle entry requires a maturation-associated increase in N-Myc and MDM2 (6, 8), which may be fulfilled by ectopic N-Myc and N-Myc–induced MDM2 upregulation (29, 30). Thus, our data suggest that either immature or maturing cone precursors may initiate MYCNA retinoblastoma depending on the in vivo stage when MYCN amplification and overexpression occur. Ectopic N-Myc further differed from pRB loss in that it caused cone precursor dedifferentiation (Fig. 2F), a phenomenon common to other Myc-driven cancers (32). We also identified MYCNA retinoblastoma regions with ARR3+,Ki67– cells (Fig. 4E), implying that the tumor cells can redifferentiate in a conducive microenvironment.
In addition to their loss of maturation markers, N-Myc–overexpressing cone precursors lost cell lineage constraints. Rare cells coexpressed RXRγ with rod or amacrine markers NRL or AP2α in cone precursor foci, xenografts, and MYCNA tumors (Fig. 5D). More striking was the cone precursors’ acquired expression of SOX9 in MYCN-transduced retina, retinal xenografts, and MYCNA tumor cells (Figs. 2A and 5). Whereas SOX9 is normally expressed in retinal progenitor and Müller cells (23), proliferating cone precursors did not widely coexpress other progenitor or Müller cell markers such as VSX2, PROX1, or PAX6 (Fig. 2 and SI Appendix, Figs. S4 and S8 C and D). Xenograft and MYCNA tumors also widely expressed retinal ganglion cell markers BRN3 and GAP43, consistent with high POU4F2 (which encodes BRN3b), GAP43, and other retinal ganglion cell gene expression in MYCNA,RB1+/+ and high-MYCN RB1−/− retinoblastomas (15). Thus, whereas Liu et al. (15) showed that MYC/MYCN activation correlates with cone dedifferentiation and retinal ganglion cell features in subtype 2 retinoblastomas, we here demonstrate that N-Myc overexpression suffices to drive dedifferentiation and lineage infidelity during retinoblastoma genesis. These findings extend evidence of Myc family–induced loss of cell lineage constraints (32, 33) to a human cancer cell of origin.
Another difference between N-Myc–overexpressing and pRB-deficient cone precursors was their lack of a senescence-like retinoma-like stage, evidenced by continuous proliferation and lack of retinoma histology in explanted retinae and rapid tumorigenesis in xenografts (Fig. 4 A–C), in keeping with the earlier appearance of MYCNA tumors in patients (3). Notably, N-Myc induced high p16 expression along with continuous proliferation (Fig. 3E), in contrast to the pRB-deficient cone precursors’ induction of p16 followed by entry into a retinoma-like state (8, 9). The induction of p16 is consistent with p16 expression in some MYCNA neuroblastomas (34) but is still surprising since p16 is expected to inhibit Cyclin D:CDK4/CDK6 kinases that inactivate pRB. As one possibility, N-Myc may induce sufficiently high Cyclin D2:CDK4 to overcome p16 inhibition.
We also found that N-Myc induced expression of retinal progenitor and Müller cell marker SOX9 in cone precursor foci, that SOX9 was expressed throughout the xenograft and MYCNA tumors, and that SOX9 was critical to proliferating cone precursor survival (Figs. 2 and 4–6). SOX9 is overexpressed in diverse human cancers (26, 27) and is linked to cancer cell survival via suppression of proapoptotic genes (35). Like N-Myc, SOX9 may promote tumorigenesis via BMI1 upregulation and induces a wider cancer-related gene expression network (27, 36). SOX9 was previously linked to N-Myc–induced cancers via its overexpression in N-Myc–driven SHH medulloblastoma; however, in that setting SOX9 was inferred to continue its normal expression in neural stem cells (37), whereas in the current study N-Myc induced de novo SOX9 expression in a committed retinal cell type. Although SOX9 was upregulated after the initial N-Myc–induced cone precursor proliferation, it was critical to sustaining the proliferating cone precursor survival (Fig. 6).
In sum, this study reveals the cone precursor origin of N-Myc–overexpressing retinoblastoma and an N-Myc–induced oncogenic trajectory distinct from that initiated by pRB loss (Fig. 7). Whereas pRB-depleted cone precursor proliferation depends on the cone precursors’ intrinsically high N-Myc (5, 6), the far higher N-Myc in MYCN-transduced cone precursors eliminates the need for intrinsically expressed cone maturation–related circuitry and enables dedifferentiation, lineage infidelity, continuous proliferation, and rapid tumorigenesis that bypasses the retinoma stage and circumvents a cell survival barrier via acquired SOX9 expression.
Materials and Methods
Lentivirus Constructs and Production.
BE-GFP-MYCN was produced by inserting MYCN cDNA sequences 734–2,139 (NM_001293228.1, isoform 1), retaining 0 bp of 5′ UTR and 11 bp of 3′UTR, into the SalI and XbaI site of BE-GFP (20) with In-Fusion (Clontech). pLKO.1C-shSOX9-1 and shSOX9-2 were produced by insertion of validated shRNA sequences for shSOX9-1 (5′-GAGCAGCGACGTCATCTCCAA-3′) (38) and shSOX9-2 (5′-CTCCACCTTCACCTACATGAA-3′) (39) into pLKO.1C (40). Virus was produced, concentrated via tangential flow filtration, and stored frozen as described (8).
Retinal Tissue, Culture, and Lentiviral Transduction.
Human fetal samples were collected under Institutional Review Board approved protocols (USC-HS-13-0399 and CHLA-14-2211). Following the patient decision for pregnancy termination, informed consent for donation of the products of conception for research purposes was obtained and samples collected without patient identifiers. Fetal age was determined according to the American College of Obstetrics and Gynecology guidelines (41). Retinae were harvested and cultured on polytetrafluoroethylene cell culture inserts in six-well dishes as described (8). Concentrated virus was thawed on ice and supplemented with media components, and ∼600 µl was used to transduce retinae on the same day as the retina explant (8). Retinae were incubated with 10 µM EdU for 4 h prior to harvest, fixed in 4% paraformaldehyde (PFA) for 10 min, and embedded in OCT sucrose (8).
Immunohistochemical Staining.
We placed 10-µm sections on positively charged slides and stored them at −80 °C. Immunohistochemical staining and imaging on an LSM 710 confocal microscope were performed as described (8) with antibody sources and titers as shown in SI Appendix, Table S1. LSM images were opened in ImageJ, converted to TIFFs, cropped, saved as JPEGs, and copied to PowerPoint.
Trainable Weka Segmentation and Automated Quantitation.
The Weka plugin (21) was obtained via Fiji/ImageJ and used to train the segmentation algorithm with randomly chosen DAPI-stained confocal images. Nuclei and nonnuclei in tightly packed as well as less packed regions were outlined with the freehand tool and added to “nuclei” or “not nuclei” classes. The training data were applied to images to obtain probability maps followed by Watershed segmentation. Based on the segmentation results, more objects were added or removed from the trained classes. This process was continued until adding or removing objects did not further improve segmentation accuracy. The data and classifiers were saved and applied to images to segment nuclei and quantitate integrated density values from other fluorescent channels. All segmented nuclei were included for the analysis. ImageJ macros were made to automate the segmentation and quantitation.
Xenografts.
Mouse experiments were approved and performed according to the guidelines of the Institutional Animal Care and Usage Committees of The Saban Research Institute of Children’s Hospital Los Angeles. Fetal week 16 retinae were dissociated and transduced with BE-MYCN-GFP or BE-GFP vectors. At 17 d after transduction, 100,000 cells were injected into the subretinal space of nonobese diabetic–severe combined immunodeficient mice as described (6). At 63 d after transduction, eyes were harvested, fixed in 4% PFA, embedded in OCT, and cryosectioned.
Cell Culture and Lentiviral Transduction.
Foci were segregated from MYCN-transduced retinae and cultured in RB media (6). Cells were plated in 96-well-plates at 300,000 per well (line A) or 50,000 per well (line B) in triplicate for each timepoint and transduced with pLKO.1C (40) expressing validated SOX9 shRNA1 or the shSCR control (6). Half of the medium was replaced daily. Puromycin selection (1.8 µg/mL) was started at day 3 after lentiviral transduction. Cells were stained with trypan blue and counted in triplicate for each well. The remaining cells were pelleted, attached to glass slides coated with poly-D-lysine (0.01%, Sigma), fixed in 4% PFA, and used for immunostaining.
Supplementary Material
Acknowledgments
We thank Melissa L. Wilson (USC Department of Preventive Medicine) and Family Planning Associates for assistance in obtaining fetal tissue, Angela Ferrario for technical support, the Stem Cell Analytics Core Facility, Animal Care Core Facility, and Imaging Core Facility of The Saban Research Institute of Children’s Hospital Los Angeles, and Zachary Fouladian for critical reading of the manuscript. This study was supported in part by the Larry and Celia Moh Foundation, the Neonatal Blindness Research Fund, the A. B. Reins Foundation, the Knights Templar Eye Foundation, an unrestricted grant to the USC Department of Ophthalmology from Research to Prevent Blindness, and NIH grants P30CA014089 and R01CA137124.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200721119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Sieber O. M., Tomlinson S. R., Tomlinson I. P., Tissue, cell and stage specificity of (epi)mutations in cancers. Nat. Rev. Cancer 5, 649–655 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Schneider G., Schmidt-Supprian M., Rad R., Saur D., Tissue-specific tumorigenesis: Context matters. Nat. Rev. Cancer 17, 239–253 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rushlow D. E., et al. , Characterisation of retinoblastomas without RB1 mutations: Genomic, gene expression, and clinical studies. Lancet Oncol. 14, 327–334 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Ewens K. G., et al. , Phosphorylation of pRb: Mechanism for RB pathway inactivation in MYCN-amplified retinoblastoma. Cancer Med. 6, 619–630 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu X. L., et al. , Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell 137, 1018–1031 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu X. L., et al. , Rb suppresses human cone-precursor-derived retinoblastoma tumours. Nature 514, 385–388 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H., et al. , Human embryonic stem cell-derived organoid retinoblastoma reveals a cancerous origin. Proc. Natl. Acad. Sci. U.S.A. 117, 33628–33638 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh H. P., et al. , Developmental stage-specific proliferation and retinoblastoma genesis in RB-deficient human but not mouse cone precursors. Proc. Natl. Acad. Sci. U.S.A. 115, E9391–E9400 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimaras H., et al. , Loss of RB1 induces non-proliferative retinoma: Increasing genomic instability correlates with progression to retinoblastoma. Hum. Mol. Genet. 17, 1363–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Dimaras H., et al. , Retinoblastoma. Nat. Rev. Dis. Prim. 1, 15021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munier F. L., et al. , Conservative management of retinoblastoma: Challenging orthodoxy without compromising the state of metastatic grace. “Alive, with good vision and no comorbidity”. Prog. Retin. Eye Res. 73, 100764 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Cobrinik D., Retinoblastoma progression. EBioMedicine 2, 623–624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kooi I. E., et al. , Somatic genomic alterations in retinoblastoma beyond RB1 are rare and limited to copy number changes. Sci. Rep. 6, 25264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kooi I. E., et al. , A meta-analysis of retinoblastoma copy numbers refines the list of possible driver genes involved in tumor progression. PLoS One 11, e0153323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J., et al. , A high-risk retinoblastoma subtype with stemness features, dedifferentiated cone states and neuronal/ganglion cell gene expression. Nat. Commun. 12, 5578 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zugbi S., et al. , Clinical, genomic, and pharmacological study of MYCN-amplified RB1 wild-type metastatic retinoblastoma. Cancers (Basel) 12, 2714 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moulin A. P., et al. , Secondary enucleated retinoblastoma with MYCN amplification. Ophthalmic Genet. 42, 354–359 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Polski A., et al. , Variability in retinoblastoma genome stability is driven by age and not heritability. Genes Chromosomes Cancer 59, 584–590 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kooi I. E., et al. , Loss of photoreceptorness and gain of genomic alterations in retinoblastoma reveal tumor progression. EBioMedicine 2, 660–670 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cobrinik D., Francis R. O., Abramson D. H., Lee T. C., Rb induces a proliferative arrest and curtails Brn-2 expression in retinoblastoma cells. Mol. Cancer 5, 72 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arganda-Carreras I., et al. , Trainable Weka Segmentation: A machine learning tool for microscopy pixel classification. Bioinformatics 33, 2424–2426 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Dyer M. A., Livesey F. J., Cepko C. L., Oliver G., Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat. Genet. 34, 53–58 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Poché R. A., Furuta Y., Chaboissier M. C., Schedl A., Behringer R. R., Sox9 is expressed in mouse multipotent retinal progenitor cells and functions in Müller glial cell development. J. Comp. Neurol. 510, 237–250 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee T. C., Almeida D., Claros N., Abramson D. H., Cobrinik D., Cell cycle-specific and cell type-specific expression of Rb in the developing human retina. Invest. Ophthalmol. Vis. Sci. 47, 5590–5598 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Bouchard C., et al. , Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 18, 5321–5333 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguilar-Medina M., et al. , SOX9 stem-cell factor: Clinical and functional relevance in cancer. J. Oncol. 2019, 6754040 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matheu A., et al. , Oncogenicity of the developmental transcription factor Sox9. Cancer Res. 72, 1301–1315 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi D. L., Cobrinik D., MDM2 but not MDM4 promotes retinoblastoma cell proliferation through p53-independent regulation of MYCN translation. Oncogene 36, 1760–1769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slack A., et al. , The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc. Natl. Acad. Sci. U.S.A. 102, 731–736 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran H. N., et al. , Reciprocal induction of MDM2 and MYCN in neural and neuroendocrine cancers. Front. Oncol. 10, 563156 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajbhandari P., et al. , Cross-cohort analysis identifies a TEAD4-MYCN positive feedback loop as the core regulatory element of high-risk neuroblastoma. Cancer Discov. 8, 582–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poli V., et al. , MYC-driven epigenetic reprogramming favors the onset of tumorigenesis by inducing a stem cell-like state. Nat. Commun. 9, 1024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rickman D. S., Schulte J. H., Eilers M., The expanding world of N-MYC-driven tumors. Cancer Discov. 8, 150–163 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Ochiai H., et al. , Bmi1 is a MYCN target gene that regulates tumorigenesis through repression of KIF1Bbeta and TSLC1 in neuroblastoma. Oncogene 29, 2681–2690 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Ma Y., et al. , SOX9 is essential for triple-negative breast cancer cell survival and metastasis. Mol. Cancer Res. 18, 1825–1838 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larsimont J. C., et al. , Sox9 controls self-renewal of oncogene targeted cells and links tumor initiation and invasion. Cell Stem Cell 17, 60–73 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Swartling F. J., et al. , Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell 21, 601–613 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang J., Guo L., Knockdown of SOX9 inhibits the proliferation, invasion, and EMT in thyroid cancer cells. Oncol. Res. 25, 167–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z., et al. , SOX9-PDK1 axis is essential for glioma stem cell self-renewal and temozolomide resistance. Oncotarget 9, 192–204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee S., Cobrinik D., Improved third-generation lentiviral packaging with pLKO.1C vectors. Biotechniques 68, 349–352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettker C. M., et al. , Committee opinion no 700: Methods for estimating the due date. Obstet. Gynecol. 129, e150–e154 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.