Significance
A critical component for appendage regeneration is the presence of nerves in the wound. The regenerative effect of nerves is achieved by a threshold amount of nerve endings; however, the molecular mechanism underlying this phenomenon remains largely unknown. Here, we used forward genetic screening to identify a mutant displaying two temperature-sensitive and seemingly unrelated defects: locomotion and regeneration of amputated fins. A causative gene was defined as scn8ab, which encodes the neuronal, voltage-gated sodium channel Nav1.6. Mutation of scn8ab led to disrupted axon regrowth and patterning during the early phase of regeneration, resulting in hypo-innervation and impaired regeneration. This research uncovers a factor critical for innervation in the injured appendage and offers a perspective on the functions of Nav1.6.
Keywords: zebrafish, regeneration, innervation, scn8a, forward genetics
Abstract
Teleost fishes and urodele amphibians can regenerate amputated appendages, whereas this ability is restricted to digit tips in adult mammals. One key component of appendage regeneration is reinnervation of the wound area. However, how innervation is regulated in injured appendages of adult vertebrates has seen limited research attention. From a forward genetics screen for temperature-sensitive defects in zebrafish fin regeneration, we identified a mutation that disrupted regeneration while also inducing paralysis at the restrictive temperature. Genetic mapping and complementation tests identify a mutation in the major neuronal voltage-gated sodium channel (VGSC) gene scn8ab. Conditional disruption of scn8ab impairs early regenerative events, including blastema formation, but does not affect morphogenesis of established regenerates. Whereas scn8ab mutations reduced neural activity as expected, they also disrupted axon regrowth and patterning in fin regenerates, resulting in hypoinnervation. Our findings indicate that the activity of VGSCs plays a proregenerative role by promoting innervation of appendage stumps.
Regenerative capacity is heterogenous across animal species. Teleost fishes and urodele amphibians possess a remarkable ability to regenerate damaged appendages, spinal cords, hearts, and brains, but adult mammals exhibit comparatively limited regenerative capacity (1–3). The central question in the field remains: What genes instruct regeneration? One strategy to select regeneration-associated candidate genes is to dissect the roles of genes expressed differentially upon injury, as these genes are candidates for possessing function during regeneration. In another common strategy, genes with known roles in embryonic development are pursued, as they are candidates for reactivation upon injury to facilitate regeneration (4). However, reverse genetic approaches can fail to detect important candidates if they do not display differential expression upon injury or if they are not known to play developmental roles. As an alternative approach, forward genetics can be employed to identify essential or unexpected genetic factors that would otherwise be challenging to uncover. Forward genetic strategies to regeneration have been previously employed with adult zebrafish fins, which regenerate upon injury, leading to the discovery of novel mutations influencing regeneration (8–10). Here, we conducted a large-scale forward genetics screen with adult zebrafish and isolated mutants exhibiting regeneration defects.
The dependance of appendage regeneration on intact innervation has been studied since the 1820s, when Tweedy J. Todd demonstrated that salamander limbs that had been denervated after wound healing exhibited impaired limb regeneration (11–14). Studies of regenerating teleost fish fins and mouse digit tips have also demonstrated nerve dependance (12–15). Strikingly, deviation of nerves to an injury on the surface of a limb can lead to the development of an accessory limb in salamanders (12, 18). Early studies by Marcus Singer and colleagues indicated that limb regeneration is determined in part by the total number of nerves present near the amputation plane and their cumulative neurotrophic effects (19, 20). Many candidate neurotropic factors, including fibroblast growth factors, bone morphogenic proteins, neuregulin-1, oncostatin M, platelet-derived growth factor, and anterior gradient protein, have been recently reported to regulate blastema formation (16, 21–23). The regenerative effects of these factors likely require a sufficient nerve density threshold, yet, to our knowledge, the molecular mechanisms explaining a quantitative requirement of nerve fibers have not been explored.
Results
Forward Genetics Screening Identifies a Temperature-Sensitive Mutant Defective in Locomotion and Fin Regeneration.
To identify essential factors mediating adult tissue regeneration, we conducted an unbiased forward genetics screen combined with a temperature-shift assay (SI Appendix, Fig. S1A). We mutagenized male fish with ethylnitrosourea (ENU), an alkylating agent and point mutagen, and generated F3 families, which were expected to be homozygous for ENU-induced mutations (SI Appendix, Fig. S1A). Because regeneration genes often play crucial developmental roles, we reasoned that mutations in regeneration genes may affect embryonic development, thereby causing developmental lethality (8–10). As previous screening approaches circumvented this problem and successfully isolated mutants (9, 10), we applied the same screening strategy to identify temperature-sensitive mutations (SI Appendix, Fig. S1B). To do this, F3 families were raised at 26 °C (a permissive temperature) for at least 2 mo and then shifted to 33 °C (a restrictive temperature) immediately after caudal fin amputation. We then quantified fin regeneration at 7 days post-amputation (dpa) and identified fin regeneration–impaired mutants.
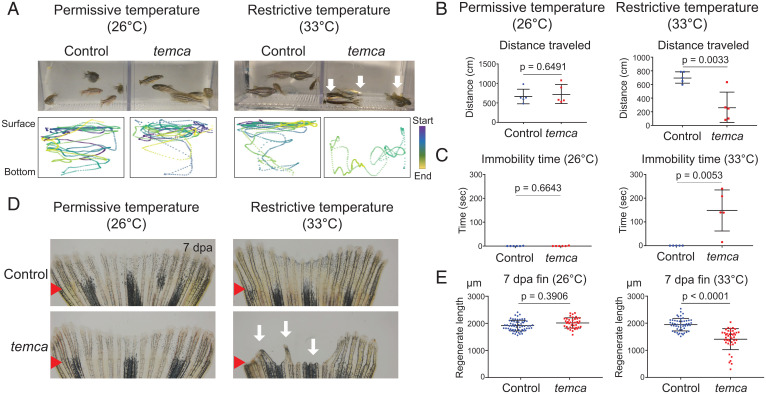
Unexpectedly, our screening isolated one mutant (pd291) exhibiting temperature-sensitive locomotion and fin-regeneration defects (Fig. 1, SI Appendix, Fig. S1 C–E, and Movies S1–S4). The locomotion defects had complete genetic linkage with impaired fin regeneration. The pd291 mutants sank to the bottom of their aquaria and displayed little or no movement when transferred from 26 °C to the restrictive temperature of 33 °C. Movement was regained when the temperature was allowed to decrease to 29 °C, indicating that this defect is reversible (Fig. 1 A–C and SI Appendix, Fig. S1 C and D). Because the restrictive temperature triggered cataplexy-like behavior, characterized by a sudden loss of muscle tone and strength (24), we named this mutant temperature-triggered cataplexy (temca). We quantified swimming behavior of wild-type siblings and temca mutants at 26 °C or 33 °C using the automated tracking systems idTracker (25) and Tracktor (26). The average distance traveled by temca mutants was reduced by 65% at 33 °C compared with controls (Fig. 1 A and B) and was defined by long periods of immobility (Fig. 1C and SI Appendix, Fig. S1C). The swimming activity of temca mutants was normal at 26 °C (Fig. 1 A–C and SI Appendix, Fig. S1D). To quantify regenerative ability, we measured the lengths of regenerates from wild-type and temca siblings at 7 dpa. The temca mutants displayed normal regeneration at 26 °C, but regenerated structures were truncated by 25% at 33 °C (Fig. 1 D and E). Fin rays of temca exhibited a range of phenotypes, including complete blockage, reduced length, or patterning defects (SI Appendix, Fig. S1E). Swimming disability of temca mutants showed 100% penetrance, whereas fin-regenerative defects exhibited ∼50 to 75% penetrance.
Fig. 1.
The temca mutants display temperature-sensitive locomotion and fin-regeneration defects. (A) Swimming behavior of control (wild-type siblings) and temca at 26 °C (Left) and 33 °C (Right). (Bottom) Representative 1-min swimming trajectories of individual fish. Color code indicates video frame, and the distance between dots indicates speed. (B and C) Quantification of 3-min swimming behavior of control and temca mutants at 26 °C (Left) and 33 °C (Right). (B) Average distance traveled (n = 5). (C) Duration of immobility at the bottom of swimming tanks (n = 5). (D) Representative whole-mount images of control and temca 7 dpa fins at 26 °C (Left) and 33 °C (Right). White arrows indicate impaired fin regenerates of temca mutants. Red arrows indicate amputation plane. (E) Quantification of fin-regenerate lengths at 7 dpa at 26 °C (n = 62 and 39 for control and temca, respectively) and 33 °C (n = 51 and 43 for control and temca, respectively). Data are presented as mean ± SD. Student’s unpaired two-tailed t test.
temca Encodes the VGSC scn8ab.
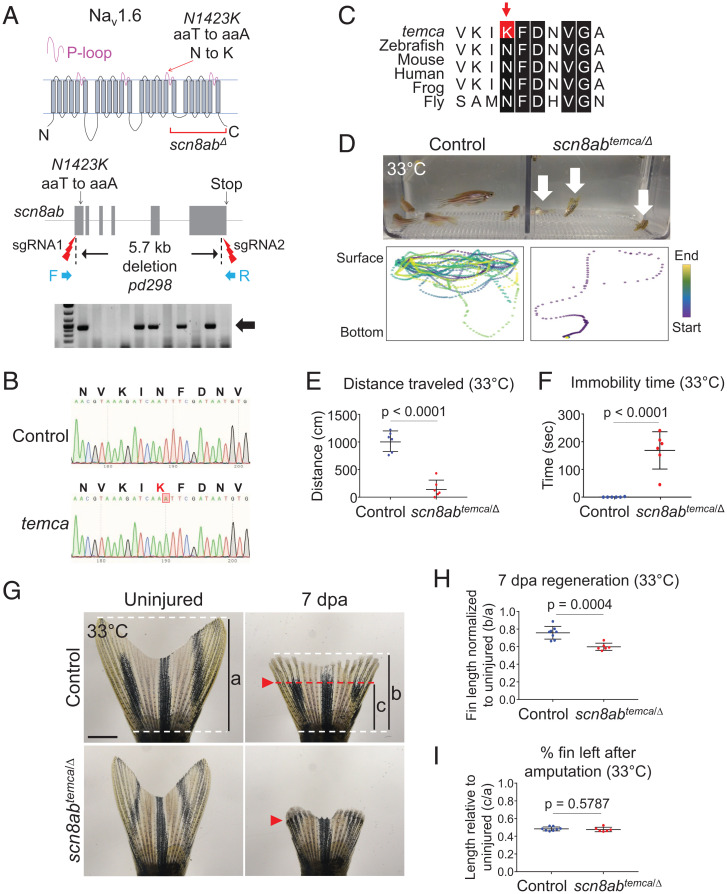
To identify the molecular basis of temca, we scored paralysis phenotypes in genetic mapping crosses and performed whole-exome sequencing with genomic DNA extracted from wild-type siblings and temca mutants. Linkage analysis was first performed with the mapping tool SNPtrack, which identifies mutant-specific single nucleotide polymorphisms (SNPs) and regions of homozygosity (27). SNPtrack analysis yielded a single ∼27.5-Mb peak on chromosome 6 with high homozygosity in temca mutants (SI Appendix, Fig. S2A). We raised progenies generated from crossing of heterozygotes at 26 °C and performed phenotyping at 33 °C to isolate 198 homozygous mutants. Restriction fragment length polymorphism analysis with these homozygotes defined the temca mutation within an 801-kb region (SI Appendix, Fig. S2A). Mutant SNPs present in this region were filtered against an SNP database established from other mutants isolated from the same screening and SNPfisher (28). After filtering, we identified a nonsynonymous (T to A) mutation causing an asparagine (N) to lysine (K) amino acid change at position 1423 of scn8ab, which encodes the voltage-gated sodium channel (VGSC) alpha subunit Nav1.6 (scn8abpd291 or scn8abtemca) (Fig. 2 A–C). N1423 is a highly conserved residue in the pore loop of Nav1.6, which folds toward the channel pore and impacts channel activity (29, 30) (Fig. 2 A and C). Our analyses, therefore, indicate that the temca phenotype likely results from an N1423K mutation in scn8ab.
Fig. 2.
temca Encodes the VGSC alpha subunit, scn8ab. (A, Top) Schematic of Scn8ab (Nav1.6) protein structure. Red arrow points to temca mutation in pore-loop (P-loop) of the third domain. Deleted region of scn8ab deletion allele (scn8abΔ) is shown. (Middle) sgRNA target sites are depicted for 5.7-kb deletion of scn8ab and primers used to confirm deletion. (Bottom) A representative DNA gel image indicating scn8abΔ is shown. (B) Sanger sequencing chromatograms from control (wild-type siblings) and temca. The temca mutation within the scn8ab coding sequence causes an asparagine (N) to lysine (K) change. (C) Amino acid sequence alignment across distant species. Black boxes mark conserved residues. Red box and arrow indicate temca N to K mutation. (D–H) Complementation analysis for temca and scn8abΔ. (D) Swimming behavior of control (scn8ab+/Δ) and scn8ab compound heterozygotes (scn8abtemca/Δ) at 33 °C. (Bottom) Representative 1-min swimming trajectories of individual fish. (E and F) Quantification of 3-min swimming behavior at 33 °C. (E) Average distance traveled (n = 6). (F) Duration of immobility at the bottom of swimming tanks (n = 6). (G–I) Regeneration in scn8abtemca/Δ normalized to uninjured fin lengths. (G) Representative whole-mount fin images of 7 dpa control and scn8abtemca/Δ at 33 °C. (H) Fin lengths at 7 dpa (b in G) normalized to uninjured fin length (a in G). The graph indicates the value of “b/a”. (I) Position of amputation plane. Lengths from base to amputation plane (c in G) were divided by uninjured fin lengths (a in G). The graph indicates the value of “c/a”. The amputation plane was ∼50% of caudal fins in control and scn8abtemca/Δ. n = 8 and 6 for control and scn8abtemca/Δ, respectively. Data are presented as mean ± SD. Student’s unpaired two-tailed t test.
To assess the causative nature of the N1423K mutation by complementation analysis, we generated a loss-of-function allele by deleting 5.7 kb of scn8ab (scn8abpd298 or scn8abΔ) (Fig. 2A and SI Appendix, Fig. S2B). The scn8abΔ allele lacks a portion of the third domain and the entire fourth domain of Nav1.6, which we speculate renders the protein nonfunctional. We then crossed scn8abΔ/+ heterozygotes with scn8abtemca homozygotes and raised their progeny at 26 °C. Analysis of swimming activity using size-matched adults revealed that the average distance traveled by scn8abtemca/Δ adults was reduced by 84% at 33 °C compared with heterozygous siblings (scn8abtemca/+), and periods of immobility were increased (Fig. 2 D–F and Movie S5). The swimming activity of scn8ab compound heterozygotes was normal at 26 °C (SI Appendix, Fig. S2 C and D). With respect to fin regeneration at adult stages, size-matched scn8ab compound heterozygotes regenerated fins normally at 26 °C (SI Appendix, Fig. S2 E and G); however, lengths of regenerated fin rays of size-matched scn8ab compound heterozygotes were reduced by 37% at 33 °C compared with heterozygous siblings (SI Appendix, Fig. S2 F and H). Normalized regenerate lengths of 7 dpa temca fins with uninjured fins also indicated a 21% reduction (Fig. 2 G–I). Our complementation test demonstrates that temca is a conditional allele of scn8ab required for mobility and fin regeneration.
scn8ab Is Dispensable During Embryogenesis and Acquires Functionality in Mature Animals.
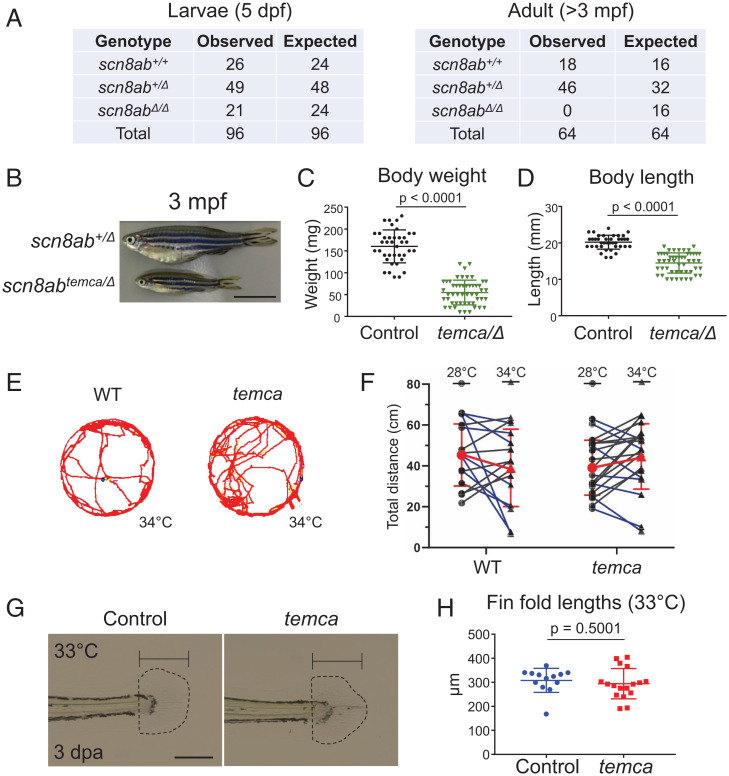
While genotyping progeny generated by crossing scn8abΔ/+ heterozygotes, we noticed that scn8abΔ/Δ homozygotes reached swimming stage (5 days post-fertilization [dpf]) but failed to survive to adulthood (Fig. 3A). Notably, compound scn8ab heterozygous (scn8abtemca/Δ) juveniles and adults were smaller than scn8abtemca/+ siblings, indicating that temca is an scn8ab hypomorphic allele and that scn8abΔ results in a null allele at the permissive temperature (Fig. 3 B-D). To determine when scn8ab is required for mobility, we transferred temca mutants to 33 °C at various developmental stages and monitored swimming ability. Erratic swimming behavior of temca mutants was first observed at ∼2 months post-fertilization (mpf) (Movies S6 and S7), indicating that scn8ab activity becomes vital during the juvenile stage.
Fig. 3.
scn8ab Is dispensable during early development. (A) Number of zebrafish with scn8ab+/+, scn8ab+/Δ, and scn8abΔ/Δ genotypes scored at 5 dpf (Left) and at 3 mpf (Right). The observed genotypic ratio matched the expected ratio in 5 dpf larvae from scn8abΔ/+ in-crosses, but no scn8abΔ homozygotes recovered at adult stages (>3 mpf). (B) Representative whole-mount images of control (heterozygous sibling) and temca at 3 mpf. (C and D) Quantification of body weight (C) and body length (D) at 3 mpf (n = 41 and 53 for scn8abtemca/+ and scn8abtemca/Δ, respectively). (E) Control (heterozygotes) and temca 6 dpf larvae swimming tracks at 34 °C. (F) Quantification of swimming behavior of 6 dpf control and temca larvae (n = 16 and 20 for control and temca, respectively). Two-way ANOVA, temperature [F(1,33) = 0.01992, P = 0.889]; genotype [F(1,33) = 0.005127, P = 0.943]; genotype × temperature interaction [F(1,33) = 4.119, P = 0.051]. (G) Representative images of control (heterozygotes) and temca regenerated fin folds 3 dpa at 33 °C. (H) Quantification of 3 dpa larvae fin fold–regenerate lengths at 33 °C (n = 13 and 17 for control and temca, respectively). Scale bars, 1 cm in B, 200 µm in G. (C, D, and H) Data are presented as mean ± SD by Student’s unpaired two-tailed t test.
To further explore the functionality of scn8ab at the early developmental stage, we evaluated swimming and regenerative abilities of temca larvae. Analysis of swimming behavior of 6 dpf larvae at the restrictive temperature revealed no difference in the total distance traveled between temca mutants and their wild-type siblings (Fig. 3 E and F and SI Appendix, Fig. S3 A and B). To assess regenerative ability, we amputated fin folds of wild-type and temca larvae at 3 dpf, transferred the larvae to 33 °C, and measured the regenerate lengths at 3 dpa. Fin-fold regeneration of temca mutants was similar to that of their heterozygous wild-type siblings (Fig. 3 G and H). We also analyzed locomotion and regenerative ability in homozygous scn8ab deletion mutant larvae (scn8abΔ/Δ) and found no significant defects in swimming behavior or fin-fold regeneration at 33 °C compared with control siblings (SI Appendix, Fig. S3 C–E). Thus, our data indicate that scn8ab is dispensable during immature zebrafish developmental stages.
scn8ab Mutation Leads to Abnormal Neuronal Function in the Adult Brain.
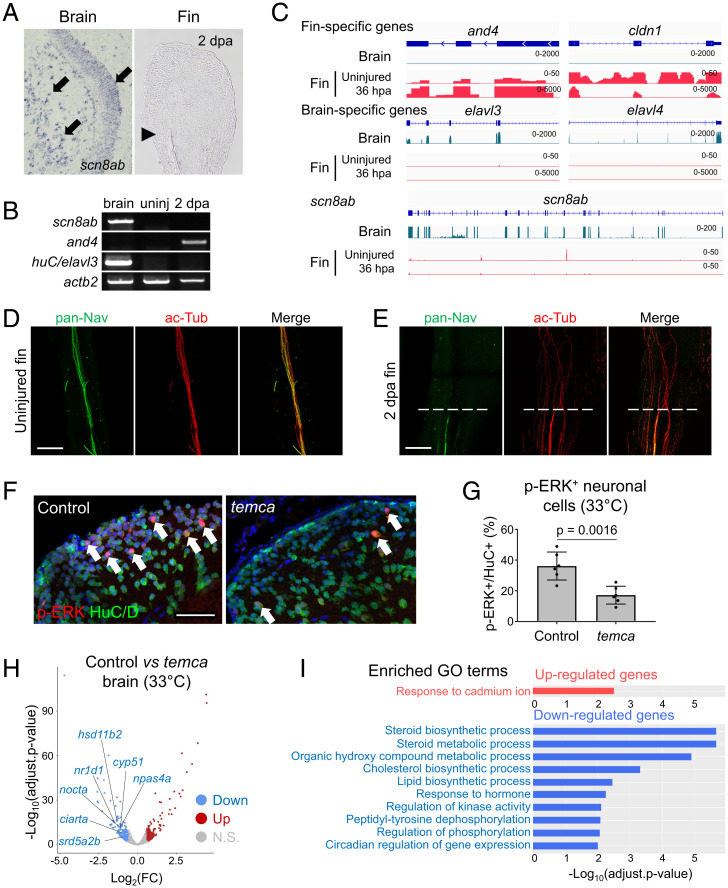
Mammalian SCN8A/NaV1.6 is present primarily in neurons (31, 32), where it is localized to the axon initial segment and the nodes of Ranvier (33, 34). To determine the spatial expression pattern of scn8ab, we performed in situ hybridization, identifying strong scn8ab transcript signals in brain neurons but no detectable expression in regenerating fin tissues (Fig. 4A and SI Appendix, Fig. S4). We also examined scn8ab transcript levels using alternative assays, including RT-PCR and RNA sequencing (RNA-seq) analyses, both of which also failed to detect scn8ab transcript expression in fin tissues (Fig. 4 B and C and SI Appendix, Fig. S5). As the epitope of the available pan-VGSC antibody (pan-Nav Ab) is entirely intact in Scn8ab (SI Appendix, Fig. S6 A and B), we performed whole-mount immunostaining to determine the spatial expression of VGSCs, including Scn8ab, in the fin. Notably, pan-Nav Ab labeled nerves, but we observed little to no expression in other cell types in uninjured and 2 dpa fins (Fig. 4 D and E). These results suggest that VGSCs are present in fin neurons. While we cannot detect scn8ab transcript, it is clear that family members are expressed in fin axons, which might include Scn8ab at levels not detectable by RNA-seq.
Fig. 4.
scn8ab Regulates neuronal activity in the brain. (A) Representative section in situ hybridization of scn8ab transcripts in brain and 2 dpa fins of wild-type fish. Black arrows indicate scn8ab expression. Black arrowhead marks amputation plane. (B) RT-PCR showing scn8ab expression in the zebrafish brain but not in uninjured or regenerating (2 dpa) fins. huC/elavl3 Is used as a brain-specific gene; and4 is a representative gene for regenerating fins; and actb2 is used as a loading control. (C) RNA-seq genome-browser tracks of fin-specific genes (and4 and cldn1), brain-specific genes (elavl3 and elavl4), and scn8ab. Brain and uninjured and 36 h postamputation (hpa) fin profiles of controls were used. (D and E) Confocal, whole-mount, immuno-stained image against pan-Nav antibody of uninjured and 2 dpa wild-type fins. Dashed white lines mark amputation plane. (F) Representative images of brain phosphorylated ERK (p-ERK; red) and HuC/D (green) immunostaining in control (heterozygous siblings) and temca placed at 33 °C for 2 h prior to dissection. White arrows indicate p-ERK+/HuC/D+ double-stained neurons. (G) Quantification of p-ERK+ neurons in 300 × 300 µm2 dorsal telencephalon region (n = 6). Data are presented as mean ± SD. Student’s unpaired two-tailed t test. (H) RNA-seq volcano plot displaying significantly reduced genes in temca placed at 33 °C for 1 d before brain collection. (I) Top Gene Ontology terms for genes up-regulated and down-regulated in temca brains. (D–F) Scale bars, 50 µm. ac-Tub, acetylated α-tubulin; Adjust, adjusted.
To investigate whether neural activity in the brain was altered in the temca mutants, we assessed ERK phosphorylation as a fast readout of brain activity (35–37). The temca mutants and heterozygous siblings were shifted to 33 °C for 2 h before brain dissection and p-ERK staining. The temca brains displayed a 52% decrease in the number of p-ERK+ neurons compared with controls (Fig. 4 F and G), indicative of impaired neuronal activity. Bulk RNA-seq analysis of whole-brain tissues dissected from control and temca fish placed at 33 °C for 1 d identified 649 genes (n = 369 and 280 for up and down-regulated genes, respectively) with significantly changed expression levels (adjusted P [P-adj] < 0.05; fold change > 1.5) (Fig. 4H and Dataset S1), including genes influencing brain function, such as npas4a and fosab (38, 39), circadian clock genes (nr1d1, nocta, and ciarta) (40–42), and steroid biosynthesis genes (hsd11b2, cyp51, and srd5a2b) (43). Gene Ontology analysis of up-regulated genes indicated enrichment in responses to cadmium ions, whereas the down-regulated genes were enriched for steroid/sterol/cholesterol biosynthesis/metabolic processes, and circadian regulation terms (Fig. 4I). Consistent with Gene Ontology analysis, gene-set enrichment (GSE) analysis (44) demonstrated significant reductions in components related to circadian rhythm and steroid biosynthetic processes in temca mutants, as well as genes associated with ribosome-related processes and translation (SI Appendix, Fig. S7). These analyses revealed abnormal neuronal function in temca mutants.
Inactivation of scn8ab Affects Early Fin Regeneration Events.
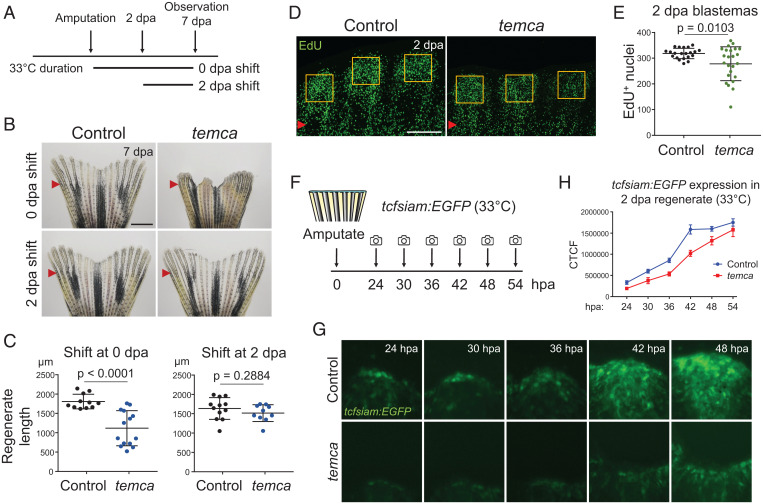
To understand the defects in fin regeneration caused by the temca mutation, we shifted animals from 26 °C to 33 °C immediately after fin amputation to impair scn8ab function either during blastema formation or at 2 dpa to disrupt scn8ab activity after the blastema has been formed (Fig. 5A). Whereas an immediate temperature shift reduced the lengths of regenerating temca fins, shifting at 2 dpa had minimal effects on the ultimate size and shape of regenerates (Fig. 5 B and C). Blastema cell-cycling indices were reduced at 2 dpa of temca mutants (Fig. 5 D and E), and expression of a tcfsiam:EGFP reporter, an in vivo Wnt biosensor that labels proliferative proximal blastema cells (45, 46), was delayed and reduced at early time points (Fig. 5 F–H). RNA-seq analysis of uninjured fins collected at 1 d after 33 °C treatment showed similar transcriptome profiles between control (heterozygotes) and temca fins (n = 4 and 7 significantly down- and up-regulated genes in temca uninjured fins, respectively; P-adj < 0.05; fold change > 1.5) (SI Appendix, Fig. S8A and Dataset S2). In contrast, RNA-seq analysis of 2 dpa temca and control fin regenerates identified 337 genes with significant changes in expression levels (P-adj < 0.05; fold change > 1.5), including 135 and 202 genes with increased and decreased expression, respectively (SI Appendix, Fig. S8A and Dataset S3). Significantly down-regulated genes in temca mutants included several transcription factors that are known to mediate the early regenerative or injury response (SI Appendix, Fig. S8A). Genes with reduced RNA levels were enriched for Gene Ontology terms related to circadian rhythm, regulation of cell cycle, and extracellular matrix organization, whereas up-regulated genes did not yield any significantly enriched terms (SI Appendix, Fig. S8B). GSE analysis similarly indicated reduced expression of DNA replication and cell-cycle gene sets (SI Appendix, Fig. S8 C–E). These results suggest that early regeneration events, including blastema formation, are dependent on scn8ab function.
Fig. 5.
Early fin regeneration is impaired in temca mutants. (A) Schematic of temperature shift assay. (B) Representative whole-mount images of control (heterozygotes) and temca fin regenerates at 7 dpa shifted to 33 °C right after amputation (0 dpa shift) or at 2 dpa (2 dpa shift). (C) Quantification of fin regenerate lengths at 7 dpa (n = 12, 14, 12, and 11 for 0 dpa shift control and temca and for 2-dpa shift control and temca, respectively). (D) Representative EdU-staining images of control and temca fin regenerates at 2 dpa. (E) Quantification of EdU+ cells in blastema area of 2 dpa fin regenerates (n = 8). (F) Schematic showing amputation and imaging time points of tcfsiam:EGFP transgenic reporter line. (G) Representative whole-mount images of tcfsiam:EGFP control (heterozygotes) and temca fin regenerates at 24, 30, 36, 42, and 48 h postamputation (hpa) at 33 °C. (H) Quantification of tcfsiam:EGFP signal intensity (n = 10 and 9 for control and temca, respectively). Red arrowheads indicate amputation planes. Scale bars, 1 mm in B; 150 µm in D. Data are presented as mean ± SD. Student’s unpaired two-tailed t test.
The temca Mutation Impairs Reinnervation.
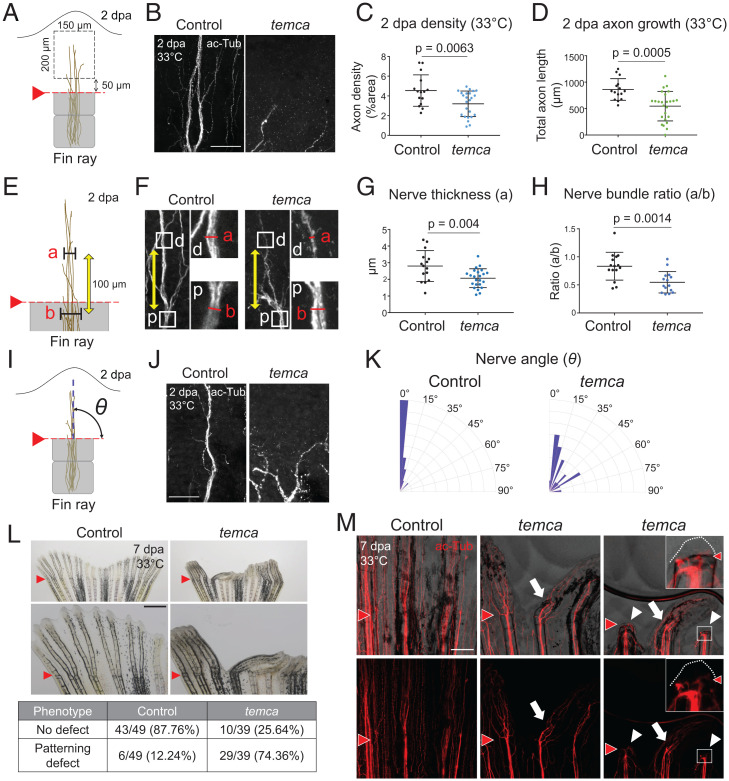
Innervation of appendages is a key early regenerative event (12, 47), and scn8ab plays roles in neurons (29, 48), prompting us to explore whether scn8ab influences nerve regeneration in fins. In assessing innervation during fin regeneration, we observed that nerves near the amputation plane underwent Wallerian degeneration (49), an active process of retrograde degeneration of nerve endings, by 1 dpa (SI Appendix, Fig. S9A). By 2 dpa, at the onset of blastema formation, significant numbers of axons had re-emerged in the regenerates, indicating that reinnervation precedes blastema formation (Fig. 6 A–D). Nerve fibers congregate at the center of fin rays to form a nerve bundle with an average diameter of 2.8 µm in control animals. The diameter of this bundle is similar in regenerated tissue to that below the amputation plane (an 83% nerve bundle ratio in controls) (Fig. 6 E–H). This thick nerve bundle runs vertically along the proximal–distal axis in the regenerates. By contrast, we found that innervation of temca mutant fins exhibited multiple defects. Axon density in temca mutants was reduced by 30% compared with that of their heterozygous siblings, indicating impaired axon regrowth (Fig. 6 A–D). In some regenerating temca fins where axon regrowth occurred, patterning defects were evident. The diameter of nerve bundles in the regenerated portion of temca fins was noticeably thinner (1.5-µm diameter on average) (Fig. 4 E–H), and their direction of regrowth was less consistent, indicative of misguided innervation (Fig. 4 I–K). These results suggest that scn8ab disruption results in the absence of innervation or hypoinnervation prior to or during blastema formation.
Fig. 6.
The temca mutation causes innervation defects in regenerating fins. (A) Schematic of method for assessing axon density and total axon length in 2 dpa fin regenerates. (B) Representative images of acetylated α-tubulin (ac-Tub) staining in 2 dpa control (heterozygotes) and temca fin regenerates at 33 °C. (C and D) Quantification of axon density (C) and total axon length (D) in a 150 × 200 µm2 area of control and temca 2-dpa fin regenerates at 33 °C. (E) Schematic of method for measuring axon bundle thickness and ratio in 2 dpa fin rays. (F–H) Representative images of ac-Tub staining (F) and quantification of axon bundle thickness (G) and ratio (H) in control and temca 2 dpa fin regenerates at 33 °C. (I) Schematic depicting angle of nerve bundle regrowth in 2 dpa fin regenerates. (J and K) Representative images of ac-Tub staining (J) and rose plots depicting orientation of regenerated nerve bundles (K) in control and temca 2 dpa fin regenerates at 33 °C (n = 8 and 12 for control and temca, respectively). (L) Representative whole-mount images of control and temca 7 dpa fins at 33 °C. (M) Representative images of ac-Tub staining in control and temca 7 dpa fin regenerates at 33 °C. White arrows indicate misshaped fin rays having misguided nerve bundles. White arrowheads indicate fin rays with complete blockage of regeneration. Insets display an enlarged version of the area in the white boxes. White dotted lines mark the distal tip of epidermis. Scale bars, 50 µm in B and J; 1.5 mm and 500 µm in L Top and Bottom, respectively; 250 µm in M. Data are presented as mean ± SD. Student’s unpaired two-tailed t test.
To determine how early innervation defects contribute to fin regeneration, we assessed 7 dpa regenerates (Fig. 6 L and M and SI Appendix, Fig. S9C). While all control fins regrew, we observed that 15% of temca fins displayed completely blocked fin regeneration with lack of innervation (Fig. 6M, Inset). Due to the hypomorphic nature of temca, many temca fin rays slightly regenerated, but the majority of these fins exhibited patterning defects (Fig. 6L and SI Appendix, Figs. S1E and S9B). These misshaped fin regenerates likely grew in the same direction as the misguided nerve bundles, and their axon densities were lower than that of controls (SI Appendix, Fig. S9C). Overall, our experiments indicate that scn8ab is essential for fin regeneration by facilitating reinnervation through the regulation of axon regrowth and guidance.
Discussion
SCN8A/NaV1.6 controls neural activity, and SCN8A mutations in humans and mice cause epilepsy and movement defects (29, 50–52). Our study indicates that scn8ab in zebrafish brains plays a similar role to modulate neural activity and influence swimming behavior. In addition, we demonstrate that impaired scn8ab function leads to defective appendage regeneration, ostensibly through reduced axon regrowth and misguided innervation. Given the fact that scn8ab is a main modulator of neural activity, we speculate that scn8ab-mediated electrical signals in fin neurons are crucial for axon regeneration upon fin amputation. Multiple studies in developing embryos have provided evidence that neuronal activity can affect neurite development and further establishment of neural networks (53, 54). For instance, knockdown of scn8aa in developing zebrafish embryos leads to aberrant axonal trajectories and delayed growth of secondary motor neurons (53). Decreased neural activity in developing zebrafish larvae increases the incidence of pathfinding errors in primary motor neurons (54). Our data suggest that the voltage-gated channel activity of Scn8ab is required for nerve reconstruction and for nerves to reach a sufficient threshold number in zebrafish fins, which enable normal blastema formation. According to the neurotrophic-factor model of nerve dependence, reduced nerve density below a certain threshold (hypoinnervated appendages) results in decreased secretion of proregenerative factors from nerves or nerve-associated cells, leading to impairment of blastema formation and proliferation (19, 55–58). Once the blastema forms, however, scn8ab-dependent neural function is likely dispensable for blastema maintenance and outgrowth in the late phases of regeneration.
The scn8ab transcript is undetectable in fin tissues, as scn8ab messenger RNA (mRNA) may not be transported to the distal peripheral axons or the transported scn8ab mRNA level may be highly restricted. However, our pan-Nav staining demonstrates the presence of VGSCs in the nerves within fins. Our analysis indicated that the pan-Nav Ab can detect most Scna/Nav homologs in zebrafish (SI Appendix, Fig. S6B). These VGSCs, including Scn8ab, are known to be primarily expressed in the neurons and muscle (59, 60), and caudal fin muscle is mainly located at the base but not other areas of fins. These facts suggested that scn8ab expressed in neurons rather than other cell types influences fin regeneration. We observed that some fin nerves are not labeled by pan-Nav Ab staining, suggesting heterogeneity of peripheral nerves based on the expression level of VGSCs. Pan-Nav–negative neurons may be distinct subtypes that may not have abundant levels of VGSCs. It will be important to identify which types of neurons are majorly modulated by scn8ab during fin regeneration.
Our transcriptome analysis indicated up-regulation of immune response–related genes in the regenerating fins of temca mutants (SI Appendix, Fig. S7 C–E). As peripheral neurons interact with immune cells in various contexts (61), scn8ab can modulate neuronal activity upon injury to regenerate axons, which coordinate immune cell responses at the wound area. At the early phase of appendage regeneration, infiltration of neutrophils and macrophages to injury sites is one key event for successful regeneration (62, 63). It is interesting to note that the ablation of macrophages impairs fin regeneration and patterning (63), a similar phenotype to that of temca mutants. Identifying molecular regulators mediating reinnervation and neuro-immune response will be an important next step.
Previous work in salamanders suggested that VGSCs contribute to regeneration after blastema formation, as overexpression of Scn5a in blastema cells caused patterning defects (64). In addition, a recent study of paralytic (para), which encodes the sole VGSC alpha-subunit gene in Drosophila, reported the roles of this VGSC in neuroblast proliferation (65). Further investigation of Scn8a mutant animals during regeneration across tissues and species can broaden our understanding of the mechanisms and targets governing the nerve dependence of tissue regeneration.
Materials and Methods
Zebrafish Maintenance and Procedures.
Wild-type or transgenic male and female zebrafish of the outbred Ekkwill (EK) or WIK strains ranging up to 18 mo of age were used for all zebrafish experiments. The water temperature was maintained at 26 °C for animals, unless otherwise indicated. Work with zebrafish was performed in accordance with Duke University and University of Wisconsin-Madison guidelines. Animals were anesthetized in 0.02% tricaine until gill movement stopped. For caudal-fin amputations, 50% of the fin was removed using a razor blade. Fin regenerate lengths were measured starting from the amputation plane to the distal tips of the third to fifth fin rays in both the dorsal and ventral caudal fin lobes, using Zeiss ZEN software. For regeneration normalized to uninjured fin (SI Appendix, Fig. S2 G and H), fin rays 3 to 5 were measured from the base of the fin to the tip of 7 dpa fins and divided by the length of the corresponding uninjured fin. To amputate the fin fold of larvae, we used a surgical blade (no. 10) to transect the proximal region of the pigment gap that marks next to the circulatory loop of the caudal vein at 3 dpf and allowed them to regenerate for 3 d. At 3 dpa, fin folds were imaged, and regenerate lengths were measured from the edge of the blood vessel to the caudal tip of the fin fold, using ZEN software.
Generation of scn8ab Deletion Line by CRISPR/Cas9.
To generate scn8ab deletion allele (pd298), CRISPR single-guide RNA (sgRNA) target sites were designed using the CRISPRscan web tool (https://www.crisprscan.org/) (66). sgRNAs were synthesized by a cloning-free method as described in ref. 67. Briefly, primers containing T7 promoter, sgRNA target, and partial sgRNA scaffold sequences were annealed with the universal sgRNA 3′ scaffold primer, and the annealed oligos were filled using the thermocycler to generate sgRNA templates. sgRNAs were synthesized using the MEGAshortscript T7 Transcription Kit (Invitrogen, AM1354) and purified by the RNA Purification Kit (Zymogen, R1016) according to the manufacturer's instructions. An sgRNA pair targeting scn8ab (each 12 to 15 ng/µL) was mixed and coinjected with Cas9 protein (0.5 µg/µL; PNABio, CP01) into the one-cell–stage embryos. Genomic DNA was extracted from one to two embryos at 2 dpf for genotyping. Deletion was detected by PCR using primers listed in SI Appendix, Table S1. F0 clutches were raised to adulthood, and founders were screened with F1 progenies. The founder was outcrossed with wild-type animals to generate heterozygous animals.
Mutagenesis, Exome Sequencing, Genetic Mapping, and Positional Cloning.
EK males were mutagenized by incubating with fish water containing 3 mM ENU for 1 h/wk for 4 wk (68). Mutagenized males were mated to females of the WIK strain to generate F1 families (SI Appendix, Fig. S1A). A total of 373 F3 families (a total of 9,740 individual fish) were generated for identifying temperature-sensitive fin-regeneration mutants. F3 fish were raised at the permissive temperature (26 °C) for at least 2 mo. After caudal fin amputation, they were shifted to the restrictive temperature (33 °C) and screened individually at 7 dpa for regeneration defects, using a dissecting microscope. Putative mutant families were out-crossed again to either EK or WIK strains to generate F4 heterozygotes, which were in-crossed to generate F5 families. F5 families were used for secondary screening to validate the fin-regeneration phenotype. Secondary screening validated a total of 16 mutants from 37 putative F3 families.
For exome sequencing, genomic DNA was isolated from pools of homozygous mutants and wild-type siblings, using the Puregene Core Kit (Qiagen). Library construction and exome capture were performed at the Duke Center for Genomic and Computational Biology using an Agilent early-access SureSelect XT Zebrafish exome kit. Sequencing was performed using an Illumina HiSeq2000 with 100-bp paired-end runs. A total of 38,006,214 and 41,005,368 paired-end reads were collected for the wild-type sibling and mutant pools, respectively. Exome-sequence files were further analyzed using SNPTrack (genetics.bwh.harvard.edu/snptrack/) (27) to identify SNPs of wild-type sibling and mutants. SNPs were filtered against our SNP dataset generated from other mutant families selected from the same screening and SNPfisher (28). For genetic mapping, heterozygous fish were crossed to generate pools of wild-type siblings and homozygous mutants. We designed primers to genotype individual SNPs using restriction fragment length polymorphism assay and identified a nonsynonymous SNP found in the scn8ab genomic region. All primers and restriction enzyme information are available in SI Appendix, Table S1. To design a genotyping method of scn8abtemca, we searched for restriction enzymes to cut the mutated region and found that ApoI cuts the mutated DNA but not wild-type DNA. Primers for genotyping were 5′-TGTAGGTTGTGGTGAACGCT-3′ and 5′-GCTCAATCCACAGTGGTAGTCA-3′. ApoI digested PCR products of wild-type DNA to generate 63-, 65-, and 313-bp fragments. ApoI digested PCR products of scn8abtemca to generate 63-, 65-, 67-, and 246-bp fragments.
Immunostaining and Histology.
For whole-mount fin immunostaining, fins were fixed with 4% paraformaldehyde (PFA) overnight at 4 °C. After fixation, fins were washed several times in phosphate-buffered saline (PBS) at room temperature. Fins were then dehydrated in serial dilutions of methanol and PBS for 10 min each. After that, fins were incubated in 20% dimethyl sulfoxide (DMSO) and methanol at 4 °C for 30 min and rehydrated in a series of methanol and 0.3% Triton X-100 in PBS (PBST0.3) dilutions. Fins were then permeabilized with acetone for 20 min at 4 °C and washed three times with PBST0.3 at room temperature. Fins were transferred to blocking solution (5% goat serum, 1% bovine serum albumin, 1% DMSO, and 0.3% Triton X-100) for 1 h at room temperature and treated with primary antibody overnight at 4 °C. The next day, fins were washed and incubated with secondary antibodies at 4 °C overnight. The primary and secondary antibodies used in this study were as follows: anti-acetylated α-tubulin (rabbit; #5335, Cell Signaling Technology; 1:500), anti–pan-VGSC (mouse; #S8809, Millipore Sigma; 1:500), anti–phospho-ERK1/2 (rabbit; #9101, Cell Signaling Technology; 1:250), anti–HuC/D (mouse; clone 16A11, #A21271, Invitrogen; 1:100), Alexa Fluor 488 (mouse, rabbit, and goat, A11029, A11034, and A11055, respectively, Life Technologies; 1:500), and Alexa Fluor 594 (mouse and rabbit; A11032 and A11037, respectively, Life Technologies; 1:500). For making frozen sections, fins and brains were fixed with 4% PFA overnight at 4 °C, then washed with 0.1% Tween-20 in PBS 3 times for 10 min. Tissues were then embedded in 1.5% agarose with 5% sucrose and incubated in 30% sucrose overnight at 4 °C. Frozen blocks were sectioned at 14 µm in a cryostat.
Imaging.
Whole-mount fin images were acquired using an AxioZoom stereo fluorescence microscope (Zeiss) at the time points indicated. For imaging Wnt responsive cells, Tg(7xTCFXla.Siam:EGFP)ia4 (tcfsiam:EGFP) fish were used, and images were processed using the Zen software (Zeiss) (69). Whole-mount 2 dpa fin innervation images were acquired using a Nikon A1R-s confocal laser scanning microscope and processed using the NIS-Elements software (Nikon). The 7 dpa fin innervation images were captured using a Keyence BZ-X800 fluorescence microscope and analyzed using Keyence software. Image stitching was automatically processed using NIS-Elements or Keyence software during image acquisition. Maximum projections were obtained using 30 and 20 slices every 1 µm for 2 and 7 dpa fin innervation images, respectively. Further image processing was carried out manually using Photoshop or FIJI/ImageJ software. Regenerate fin lengths were measured and quantified by averaging fin rays 3, 4, and 5 from the dorsal and ventral lobes of the fin.
In situ hybridization on cryosections of 4% PFA–fixed fins was performed as previously described (70). To generate digoxigenin-labeled probes for scn8ab, we used the 3′ untranslated region of scn8ab cDNA, which does not show conservation with other scn genes (60). Primer sequences of scn8ab, fgf20a, and fn1b are listed in SI Appendix, Table S1.
EdU Incorporation Assay.
EdU incorporation assays were done as previously described (46). Briefly, 10 µL of EdU solution (TCI chemical, E1057; 10 mM in PBS) was injected intraperitoneally 40 min prior to collection of fin regenerates, which were then fixed in 4% PFA overnight at 4 °C. After fixation, fins were washed once with TBS for 10 min and incubated in freshly made EdU staining solution (1 mM CuSO4, 50 mM ascorbic acid, 10 mM florescent azide, and 100 mM Tris buffer, pH 8.0) for 60 min at room temperature. Fins were washed several times with 0.2% Triton X-100 in TBS and costained with DAPI.
Analysis of Swimming Trajectories.
Swimming trajectories of adult zebrafish were analyzed using the free tracking-software tools idTracker and Tracktor (25, 26). For quantification of distances traveled, single adult zebrafish were placed in a 1.5-L tank and allowed to acclimate for 5 min, after which they were filmed for 4 min. Videos were then run on idTracker (25) to calculate distances traveled or in Tracktor (26) to generate swimming trajectory plots, changing tracking parameters as necessary to detect only one animal throughout the video and minimize background noise. Total distances traveled were calculated using MATLAB from the coordinates generated by idTracker.
For larval zebrafish, spontaneous swimming was recorded, and total distance moved was quantified over 3-min periods at 28 °C and 34 °C. We placed 6 dpf larvae individually in 22-mm–diameter wells of 12-well plates and allowed them to acclimate for at least 1-h. Spontaneous movement was then recorded for each larva at both 28 °C and 34 °C. Larvae were allowed to acclimate for 15 min between temperature changes and recordings. The starting temperature was alternated so that 28 °C or 34 °C was tested first in equal proportions. Video was recorded from above using a MotionPro Y4 high-speed camera (Integrated Design Tools) with a 50-mm macro lens (Sigma Corp. of America) at 1,000 frames/s. Larvae were illuminated from above with a mounted light-emitting diode light (MCWHL5 6500 K LED, powered by LEDD1B driver; Thorlabs) and below with an infrared light source (IR Illuminator CM-IR200B; C&M Vision Technologies). Swimming trajectories were analyzed and total distance moved was quantified using the free tracking software tool Flote (71).
RNA Isolation, RT-PCR, and Sequencing.
RNA was isolated from uninjured and regenerated fins and brain tissues using Tri-Reagent (ThermoFisher). Complementary DNA (cDNA) was synthesized from 300 ng to 1 μg of total RNA using an NEB ProtoScript II first-strand cDNA synthesis kit (catalog no. E6560). For RT-PCR, HuC/elavl3 was used for the brain-specific marker; and4 was used for regenerating fin tissue markers; and actb2 was used as a loading control. The sequences of the primers used for RT-PCR are listed in SI Appendix, Table S1.
For RNA-seq, total RNA was prepared from two biological replicate pools of uninjured and 36 h postamputation caudal-fin samples of heterozygote siblings and temca mutants at 33 °C. Two biological replicate pools of brain samples of heterozygote siblings and temca mutants were dissected 1 d after 33 °C incubation. Generation of mRNA libraries and sequencing were performed at the Biotechnology Center at University of Wisconsin-Madison using an Illumina NovaSeq with 150-bp paired-end runs. Adapter sequences were trimmed by Cutadapt. Sequences were aligned to the zebrafish genome (genome assembly GRCz11, Ensembl gene annotation release 104) using HISAT2. Differentially regulated transcripts were identified using featureCounts and DESeq2. The accession number for transcriptome datasets is GSE186729.
To determine the scn8ab transcript level by RNA-seq analysis, we used RNA-seq profiles generated by this study and datasets generated by other studies. These include PRJEB39269 [3 dpf, 5.5 mpf, and 30 mpf uninjured brain (72)]; PRJNA307985 [uninjured and 4 dpa adult fin (70)]; PRJNA669701 [uninjured adult fin, anterior cut edge of 2 dpa adult fin, and posterior cut edge of 2 dpa adult (73)]; PRJNA523009 [uninjured and 4 dpa adult fins (74)]; PRJNA517717 [microdissected distal and proximal regenerating fin tissues from 4 dpa adult fins (75)]; PRJNA612515 [uninjured and 2 dpa adult fins (76)]; and PRJNA559885 [uninjured and 1-dpa adult fins (77)]. RNA-seq profiles were aligned to the zebrafish genome (genome assembly GRCz11, Ensembl gene annotation release 104) using HISAT2. Aligned RNA-seq profiles were visualized using the Integrative Genomics Viewer (Broad Institute).
Data Collection and Statistics.
All statistical analyses were performed using GraphPad Prism 7. Significance was calculated using unpaired two-tailed Student’s t tests or two-way ANOVA with P < 0.05 used to denote statistical significance. Clutchmates were randomized into different treatment groups for each experiment. No animal or sample was excluded from the analysis unless the animal died during the procedure. Sample sizes were chosen on the basis of previous publications and experiment types and are indicated in each figure legend or in Materials and Methods. No statistical methods were used to predetermine sample size. For expression patterns, at least five fish from each transgenic line were examined. We pooled 6 fins and 3 brains of each group for RNA purification and subsequent RT-qPCR or RNA-seq. Sample sizes, statistical tests, and P values are indicated in the figures or their legends.
Supplementary Material
Acknowledgments
We thank the University of Wisconsin, Madison, School of Medicine and Public Health Biomedical Research Model Services staff and Duke University zebrafish facility staff for zebrafish care; the University of Wisconsin Biotechnology Center DNA Sequencing Facility (RRID:SCR_017759) for providing RNA-sequencing services; Francesco Argenton for transgenic animals; Nutishia Lee, Elaine Zheng, and Madeline Sullivan for contributions to experiments; Deneen Wellik and Emery Bresnick for discussion of the research; and Jingli Cao and J.K. laboratory members for comments on the manuscript. This work was supported by NIH Grants R35 GM 137878 (to J.K.) and R01 HD 105033 (to K.D.P.); NIH–University of Wisconsin Carbone Cancer Center Support Grant P30 CA 014520 (to J.K.); National Institute of General Medical Sciences Training grant T32 GM 007133 (to D.O.-M.); a Stem Cell and Regenerative Medicine Center Research Training Award (to D.O.-M.); National Heart Lung and Blood Institute Training Grant T32 HL 007936 (to I.J.B.); and American Heart Association Predoctoral Fellowship 827904 (to I.J.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200342119/-/DCSupplemental.
Data and Materials Availability
New sequencing data have been deposited in GEO under accession code GSE186729 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE186729) (78). Previously published data were used for this work, including PRJEB39269 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJEB39269) (79), PRJNA307985 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA307985 and (72)) (80), PRJNA669701 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA669701 and (70)) (81), PRJNA523009 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA523009 and (73)) (82), PRJNA517717 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA517717 and (75)) (83), PRJNA612515 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA612515 and (76)) (84), and PRJNA559885 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA559885 and (77)) (85).
References
- 1.Sehring I. M., Weidinger G., Recent advancements in understanding fin regeneration in zebrafish. Wiley Interdiscip. Rev. Dev. Biol. 9, e367 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Payzin-Dogru D., Whited J. L., An integrative framework for salamander and mouse limb regeneration. Int. J. Dev. Biol. 62, 393–402 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Aztekin C., et al. , Identification of a regeneration-organizing cell in the Xenopus tail. Science 364, 653–658 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerber T., et al. , Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 362, eaaq0681 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C. H., Poss K. D., Regeneration genetics. Annu. Rev. Genet. 51, 63–82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y. T., et al. , Genetic reprogramming of positional memory in a regenerating appendage. Curr. Biol. 29, 4193–4207.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tseng T. L., et al. , The RNA helicase Ddx52 functions as a growth switch in juvenile zebrafish. Development 148, dev199578 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson S. L., Weston J. A., Temperature-sensitive mutations that cause stage-specific defects in zebrafish fin regeneration. Genetics 141, 1583–1595 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poss K. D., Nechiporuk A., Hillam A. M., Johnson S. L., Keating M. T., Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development 129, 5141–5149 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Chen C. H., et al. , Transient laminin beta 1a induction defines the wound epidermis during zebrafish fin regeneration. PLoS Genet. 11, e1005437 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Todd T., On the process of reproduction of the members of the aquatic salamander. Q J Sci Lit Arts 16, 84–96 (1823). [Google Scholar]
- 12.Farkas J. E., Monaghan J. R., A brief history of the study of nerve dependent regeneration. Neurogenesis (Austin) 4, e1302216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka E. M., The molecular and cellular choreography of appendage regeneration. Cell 165, 1598–1608 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Simões M. G., et al. , Denervation impairs regeneration of amputated zebrafish fins. BMC Dev. Biol. 14, 49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carr M. J., et al. , Mesenchymal precursor cells in adult nerves contribute to mammalian tissue repair and regeneration. Cell Stem Cell 24, 240–256.e9 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Johnston A. P., et al. , Dedifferentiated Schwann cell precursors secreting paracrine factors are required for regeneration of the mammalian digit tip. Cell Stem Cell 19, 433–448 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Meda F., et al. , Nerves control redox levels in mature tissues through Schwann cells and hedgehog signaling. Antioxid. Redox Signal. 24, 299–311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Endo T., Gardiner D. M., Makanae A., Satoh A., The accessory limb model: An alternative experimental system of limb regeneration. Methods Mol. Biol. 1290, 101–113 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Singer M., The influence of the nerve in regeneration of the amphibian extremity. Q. Rev. Biol. 27, 169–200 (1952). [DOI] [PubMed] [Google Scholar]
- 20.Dinsmore C. E.; American Society of Zoologists, A History of Regeneration Research: Milestones in the Evolution of a Science (Cambridge University Press, Cambridge, UK, ed. 1, 1991), 228 p. [Google Scholar]
- 21.Satoh A., Makanae A., Nishimoto Y., Mitogawa K., FGF and BMP derived from dorsal root ganglia regulate blastema induction in limb regeneration in Ambystoma mexicanum. Dev. Biol. 417, 114–125 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Farkas J. E., Freitas P. D., Bryant D. M., Whited J. L., Monaghan J. R., Neuregulin-1 signaling is essential for nerve-dependent axolotl limb regeneration. Development 143, 2724–2731 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Kumar A., Godwin J. W., Gates P. B., Garza-Garcia A. A., Brockes J. P., Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318, 772–777 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheung J., Ruoff C. M., Mignot E., “Chapter 8 - Central nervous system hypersomnias” in Sleep and Neurologic Disease, Miglis M., Ed. (Academic Press, 2017). [Google Scholar]
- 25.Pérez-Escudero A., Vicente-Page J., Hinz R. C., Arganda S., de Polavieja G. G., idTracker: Tracking individuals in a group by automatic identification of unmarked animals. Nat. Methods 11, 743–748 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Sridhar V. H., Roche D. G., Gingins S., Tracktor: Image- based automated tracking of animal movement and behaviour. Methods Ecol. Evol. 10, 815–820 (2019). [Google Scholar]
- 27.Leshchiner I., et al. , Mutation mapping and identification by whole-genome sequencing. Genome Res. 22, 1541–1548 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butler M. G., et al. , SNPfisher: Tools for probing genetic variation in laboratory-reared zebrafish. Development 142, 1542–1552 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meisler M. H., et al. , SCN8A encephalopathy: Research progress and prospects. Epilepsia 57, 1027–1035 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanders S. J., et al. , Progress in understanding and treating SCN2A-mediated disorders. Trends Neurosci. 41, 442–456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burgess D. L., et al. , Mutation of a new sodium channel gene, Scn8a, in the mouse mutant ‘motor endplate disease’. Nat. Genet. 10, 461–465 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Schaller K. L., Caldwell J. H., Developmental and regional expression of sodium channel isoform NaCh6 in the rat central nervous system. J. Comp. Neurol. 420, 84–97 (2000). [PubMed] [Google Scholar]
- 33.Caldwell J. H., Schaller K. L., Lasher R. S., Peles E., Levinson S. R., Sodium channel Na(v)1.6 is localized at nodes of ranvier, dendrites, and synapses. Proc. Natl. Acad. Sci. U.S.A. 97, 5616–5620 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osorio N., et al. , Persistent Nav1.6 current at axon initial segments tunes spike timing of cerebellar granule cells. J. Physiol. 588, 651–670 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Randlett O., et al. , Whole-brain activity mapping onto a zebrafish brain atlas. Nat. Methods 12, 1039–1046 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao Y. J., Ji R. R., c-Fos and pERK, which is a better marker for neuronal activation and central sensitization after noxious stimulation and tissue injury? Open Pain J. 2, 11–17 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancedda L., et al. , Patterned vision causes CRE-mediated gene expression in the visual cortex through PKA and ERK. J. Neurosci. 23, 7012–7020 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klarić T., Lardelli M., Key B., Koblar S., Lewis M., Activity-dependent expression of neuronal PAS domain-containing protein 4 (npas4a) in the developing zebrafish brain. Front. Neuroanat. 8, 148 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hudson A. E., Genetic reporters of neuronal activity: c-Fos and G-CaMP6. Methods Enzymol. 603, 197–220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delaunay F., Thisse C., Marchand O., Laudet V., Thisse B., An inherited functional circadian clock in zebrafish embryos. Science 289, 297–300 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Kawai M., et al. , Nocturnin: A circadian target of Pparg-induced adipogenesis. Ann. N. Y. Acad. Sci. 1192, 131–138 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goriki A., et al. , A novel protein, CHRONO, functions as a core component of the mammalian circadian clock. PLoS Biol. 12, e1001839 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storbeck K. H., et al. , Steroid metabolome analysis in disorders of adrenal steroid biosynthesis and metabolism. Endocr. Rev. 40, 1605–1625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wehner D., et al. , Wnt/β-catenin signaling defines organizing centers that orchestrate growth and differentiation of the regenerating zebrafish caudal fin. Cell Rep. 6, 467–481 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Kang J., Nachtrab G., Poss K. D., Local Dkk1 crosstalk from breeding ornaments impedes regeneration of injured male zebrafish fins. Dev. Cell 27, 19–31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar A., Brockes J. P., Nerve dependence in tissue, organ, and appendage regeneration. Trends Neurosci. 35, 691–699 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Makinson C. D., et al. , Regulation of thalamic and cortical network synchrony by Scn8a. Neuron 93, 1165–1179.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waller A., Experiments on the frog, and observations of the alterations produced thereby in the structure of their primitive fibers. Philos. Trans. R. Soc. Lond. 140, 423–429 (1850). [Google Scholar]
- 50.Smith M. R., Smith R. D., Plummer N. W., Meisler M. H., Goldin A. L., Functional analysis of the mouse Scn8a sodium channel. J. Neurosci. 18, 6093–6102 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larsen J., et al. ; EuroEPINOMICS RES Consortium CRP, The phenotypic spectrum of SCN8A encephalopathy. Neurology 84, 480–489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gardella E., et al. , The phenotype of SCN8A developmental and epileptic encephalopathy. Neurology 91, e1112–e1124 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Pineda R. H., et al. , Knockdown of Nav1.6a Na+ channels affects zebrafish motoneuron development. Development 133, 3827–3836 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Plazas P. V., Nicol X., Spitzer N. C., Activity-dependent competition regulates motor neuron axon pathfinding via PlexinA3. Proc. Natl. Acad. Sci. U.S.A. 110, 1524–1529 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singer M., On the nature of the neurotrophic phenomenon in urodele limb regeneration. Am. Zool. 18, 829–841 (1978). [Google Scholar]
- 56.Lehrberg J., Gardiner D. M., Regulation of axolotl (Ambystoma mexicanum) limb blastema cell proliferation by nerves and BMP2 in organotypic slice culture. PLoS One 10, e0123186 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brockes J. P., Mitogenic growth factors and nerve dependence of limb regeneration. Science 225, 1280–1287 (1984). [DOI] [PubMed] [Google Scholar]
- 58.Brockes J. P., Kintner C. R., Glial growth factor and nerve-dependent proliferation in the regeneration blastema of Urodele amphibians. Cell 45, 301–306 (1986). [DOI] [PubMed] [Google Scholar]
- 59.de Lera Ruiz M., Kraus R. L., Voltage-gated sodium channels: Structure, function, pharmacology, and clinical indications. J. Med. Chem. 58, 7093–7118 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Novak A. E., et al. , Embryonic and larval expression of zebrafish voltage-gated sodium channel alpha-subunit genes. Dev. Dyn. 235, 1962–1973 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Chu C., Artis D., Chiu I. M., Neuro-immune interactions in the tissues. Immunity 52, 464–474 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li L., Yan B., Shi Y. Q., Zhang W. Q., Wen Z. L., Live imaging reveals differing roles of macrophages and neutrophils during zebrafish tail fin regeneration. J. Biol. Chem. 287, 25353–25360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petrie T. A., Strand N. S., Yang C. T., Rabinowitz J. S., Moon R. T., Macrophages modulate adult zebrafish tail fin regeneration. Development 141, 2581–2591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sousounis K., Erdogan B., Levin M., Whited J. L., Precise control of ion channel and gap junction expression is required for patterning of the regenerating axolotl limb. Int. J. Dev. Biol. 64, 485–494 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Piggott B. J., et al. , Paralytic, the Drosophila voltage-gated sodium channel, regulates proliferation of neural progenitors. Genes Dev. 33, 1739–1750 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreno-Mateos M. A., et al. , CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12, 982–988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Varshney G. K., et al. , High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 25, 1030–1042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trevarrow B., Techniques for optimizing the creation of mutations in zebrafish using N-ethyl-N-nitrosourea. Lab Anim. (NY) 40, 353–361 (2011). [DOI] [PubMed] [Google Scholar]
- 69.Moro E., et al. , In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev. Biol. 366, 327–340 (2012). [DOI] [PubMed] [Google Scholar]
- 70.Kang J., et al. , Modulation of tissue repair by regeneration enhancer elements. Nature 532, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burgess H. A., Granato M., Sensorimotor gating in larval zebrafish. J. Neurosci. 27, 4984–4994 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Demirci Y., et al. , Comparative transcriptome analysis of the regenerating zebrafish telencephalon unravels a resource with key pathways during two early stages and activation of Wnt/β-catenin signaling at the early wound healing stage. Front. Cell Dev. Biol. 8, 584604 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cao Z., et al. , Calcineurin controls proximodistal blastema polarity in zebrafish fin regeneration. Proc. Natl. Acad. Sci. U.S.A. 118, e2009539118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee H. J., et al. , Regenerating zebrafish fin epigenome is characterized by stable lineage-specific DNA methylation and dynamic chromatin accessibility. Genome Biol. 21, 52 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stewart S., et al. , Transpositional scaling and niche transitions restore organ size and shape during zebrafish fin regeneration. bioRxiv [Preprint] (2019). 10.1101/606970. Accessed 02 October 2019. [DOI]
- 76.Thompson J. D., et al. , Identification and requirements of enhancers that direct gene expression during zebrafish fin regeneration. Development 147, dev191262 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang W., et al. , Changes in regeneration-responsive enhancers shape regenerative capacities in vertebrates. Science 369, eaaz3090 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.D. Osorio-Méndez et al., Voltage-gated sodium channel scn8a is required for innervation and regeneration of amputated adult zebrafish fins. GSE186729. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE186729. Deposited 27 October 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.D. Osorio-Méndez et al., Voltage-gated sodium channel scn8a is required for innervation and regeneration of amputated adult zebrafish fins. PRJEB39269. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJEB39269. Deposited 7 October 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.D. Osorio-Méndez et al., Voltage-gated sodium channel scn8a is required for innervation and regeneration of amputated adult zebrafish fins. PRJNA307985. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA307985. Deposited 5 January 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.D. Osorio-Méndez et al., Voltage-gated sodium channel scn8a is required for innervation and regeneration of amputated adult zebrafish fins. PRJNA669701. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA669701. Deposited 19 October 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.D. Osorio-Méndez et al., Voltage-gated sodium channel scn8a is required for innervation and regeneration of amputated adult zebrafish fins. PRJNA523009. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA523009. Deposited 18 February 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.D. Osorio-Méndez et al., Voltage-gated sodium channel scn8a is required for innervation and regeneration of amputated adult zebrafish fins. PRJNA517717. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA517717. Deposited 29 January 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.D. Osorio-Méndez et al., Voltage-gated sodium channel scn8a is required for innervation and regeneration of amputated adult zebrafish fins. PRJNA612515. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA612515. Deposited 13 March 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.D. Osorio-Méndez et al., Voltage-gated sodium channel scn8a is required for innervation and regeneration of amputated adult zebrafish fins. PRJNA559885. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA559885. Deposited 12 August 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
New sequencing data have been deposited in GEO under accession code GSE186729 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE186729) (78). Previously published data were used for this work, including PRJEB39269 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJEB39269) (79), PRJNA307985 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA307985 and (72)) (80), PRJNA669701 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA669701 and (70)) (81), PRJNA523009 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA523009 and (73)) (82), PRJNA517717 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA517717 and (75)) (83), PRJNA612515 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA612515 and (76)) (84), and PRJNA559885 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA559885 and (77)) (85).