Significance
Long noncoding RNAs (lncRNA) play a fundamental role in the process of X-chromosome inactivation. Here we show that these kind of RNAs are also involved in the choice of which chromosome, paternal or maternal, is going to be inactivated in female mice. We identify the lncRNA Lppnx as the driving force for the action of the so-called X-controlling element (Xce). Although the Xce had been described 40 y ago, its localization on the X chromosome together with its molecular action remain elusive. With the results presented here, we shed light on the specific localization of the Xce as well as on the underlying molecular mechanisms how the Xce influences the choice of which chromosome is going to be inactivated.
Keywords: X chromosome inactivation, X-controlling element, pluripotency factors, female mouse embryo, noncoding RNA
Abstract
X chromosome inactivation (XCI) is the process of silencing one of the X chromosomes in cells of the female mammal which ensures dosage compensation between the sexes. Although theoretically random in somatic tissues, the choice of which X chromosome is chosen to be inactivated can be biased in mice by genetic element(s) associated with the so-called X-controlling element (Xce). Although the Xce was first described and genetically localized nearly 40 y ago, its mode of action remains elusive. In the approach presented here, we identify a single long noncoding RNA (lncRNA) within the Xce locus, Lppnx, which may be the driving factor in the choice of which X chromosome will be inactivated in the developing female mouse embryo. Comparing weak and strong Xce alleles we show that Lppnx modulates the expression of Xist lncRNA, one of the key factors in XCI, by controlling the occupancy of pluripotency factors at Intron1 of Xist. This effect is counteracted by enhanced binding of Rex1 in DxPas34, another key element in XCI regulating the activity of Tsix lncRNA, the main antagonist of Xist, in the strong but not in the weak Xce allele. These results suggest that the different susceptibility for XCI observed in weak and strong Xce alleles results from differential transcription factor binding of Xist Intron 1 and DxPas34, and that Lppnx represents a decisive factor in explaining the action of the Xce.
Many metazoa, when undergoing sex differentiation, adopt dosage compensation systems which through varied biological approaches, ensure the equalization of the transcription dosage of the sex chromosomes in the different sexes (1, 2). In flies, for example, hyper-transcription of the X chromosome doubles the level of X chromosome transcriptional activity in males to achieve levels comparable to that ensured by the two X chromosomes present in females (3). In contrast, in mammals, inactivation of one of the two X chromosome present in females occurs, a process known as X chromosome inactivation (XCI) (4–7). In female mice, two forms of XCI exist. In imprinted XCI (iXCI) the exclusive inactivation of the male-derived X chromosome is observed as opposed to random XCI (rXCI), in which cells of the epiblast choose to inactivate either the male or female inherited X (8).
In mouse preimplantation embryos iXCI begins gradually at the two-cell stage and by the morula stage this process is complete, with each cell containing an inactivated and silenced paternal X chromosome. At the late blastocyst stage, this process is reversed within cells of the inner cell mass, which will give rise to the embryo proper, while cells from extraembryonal tissues such as the trophectoderm and primitive endoderm maintain the silencing of the paternal X.
This results at the time of implantation, in cells of the epiblast carrying two fully active X chromosomes. Between days 5.5 and 7.5 in their turn, cells of the epiblast undergo random XCI with the exact timepoint of XCI of each cell depending on the germ layer to which it belongs (9).
By day 9.5 the process of rXCI is essentially complete, with the female embryo consisting of a mosaic of cells carrying either paternal or maternal derived inactivated X chromosomes. This pattern persists in the soma throughout postnatal life as the cell passes on the X which has been inactivated to its daughter cells through mitosis. As the timepoint of rXCI in the early postimplantation embryo coincides with the formation of the three germ layers, it has been proposed that the rXCI process is tightly coupled to the process of differentiation (10). This notion is supported by studies on embryonic stem cells (ES) which have been exploited as an in-vitro model for the study of rXCI. In female pluripotent ES cells both X chromosomes are equally transcribed. Early in the in vitro differentiation the initiation of inactivation of a single X occurs in a random fashion (11).
At the molecular level, XCI in the mouse has been shown to be highly dependent on the expression of two long-noncoding RNAs (lncRNAs), Xist and its antisense counterpart Tsix (12, 13). While in undifferentiated cells Xist and Tsix are equally transcribed, at the onset of differentiation Xist is up-regulated and Tsix down-regulated from the same X chromosome. In the course of differentiation Xist starts to spread over the whole chromosome, attracting histone-modifying complexes like Polycomb repressive complex 1/2 (PRC1/2) and DNA methyltransferases, which will lead to the complete transcriptional silencing and the formation of a heterochromatic environment visible as the Barr body (14).
Genetically, Xist/Tsix localize to a region called the X inactivation center (XIC). This region, extending over roughly 500 kb, is both required and sufficient to drive XCI. The region contains additional lncRNAs such as Ftx, Xite, and Jpx, which modulate Xist or/and Tsix expression (15–17). Studies using ES cells have shown that pluripotency factors are involved in XCI, suggesting again the likelihood of tight and mechanistically meaningful coupling between differentiation and XCI. Part of the generic role of these factors involves the direct blocking of Xist and promotion of Tsix transcription, with the key pluripotency factors Oct4, Sox2, and Nanog having been shown to bind to Intron1 of Xist and block its transcription (18). Conversely, Rex1, Klf4, and c-myc bind to a minisatellite repeat, DxPas34, upstream of the Tsix gene to promote its transcription (19). While genetic deletion of DxPas34 leads to ectopic up-regulation of Xist, overexpression of Rex1 inhibits Xist up-regulation and XCI. Rex1 plays a crucial role in the initiation and fine-tuning of XCI through its interaction with RNF12, an X-linked ubiquitin ligase which is up-regulated at the onset of differentiation and processes the proteasomal degradation of Rex1 (20). RNF12 is also controlled by other pluripotency factors (21).
The initiation of rXCI is thought to require the recognition of the X chromosome (counting) and the decision of which chromosome will be inactivated (choice). Although the choice of which X chromosome will be inactivated in somatic tissues is theoretically random, it has been shown that alleles of the X-controlling element (Xce) can significantly bias the ratio of inactivation between the Xs present in the female (22, 23). Four alleles have been identified among standard inbred mouse strains and inbred wild mouse, according to their behavior in various XCI crosses: Xcea (129Sv, C3H, CBA), Xceb (BALB/C, C57Bl6, DBA), Xcec (Pgk1a, Mus castaneus) and Xced (Mus spretus) with allelic strength ranging from “weak, Xcea < intermediate, Xceb < strong, Xcec < very strong, Xced (24–27). In mice, heterozygous for the Xce, the chromosome carrying the weaker allele is more likely to be inactivated, leading to the skewed XCI ratio. For example, 129Sv/Pgk1a heterozygous females show a 75:25 ratio, meaning that some 25% of the cells carry an inactivated Pgk1a-derived X chromosome while 75% of cells had inactivated the 129Sv-derived X (28). Xce homozygous animals, irrespective of the allele carried, do not exhibit such skewing (29).
Although the Xce was first described nearly four decades ago, the identity and precise genomic localization remains elusive. Initially, the Xce was located between Eda (Tabby) and Atp7a (Mottled), spanning roughly 9 Mb and encompassing the XIC with Xist/Tsix (30–32). That Xist is part of, or even identical to the Xce locus, was disproved several years later (33). Several more recent attempts have been undertaken to identify the genomic loci driving skewed XCI without answering the question whether one or several genomic loci are implicated (33–35).
Here we report a further approach to determine a possible candidate for the Xce. Analyzing previous mapping data which localized the Xce candidate region between DxPas28 and DxPas41 encompassing 80 kb and two protein coding genes (Cdx4 and Chic1), we found a transcriptionally active locus which had gone unreported. Further experiments revealed that the resulting transcript, which we named Lppnx, is mainly localized in the nucleus and that its expression is restricted to the inner cell mass (ICM) of blastocysts in male as well as in female ES cells. Deletion of the putative promoter region in ES cells and mice disrupts expression of Lppnx and does not affect pluripotency or viability. However, upon differentiation we detect elevated expression of Xist in female as well as in male cells compared to WT cells. In addition, when Lppnx-deficient 129Sv animals (weak Xce) were crossed with Pgk1a mice (strong Xce), we detected a shift toward expression of the strong allele of Xist as well as a shift in the allelic ratio of X-linked gene expression, indicating that Lppnx may act as a negative regulator of Xist and thus influence the choice of which X will be inactivated. By analyzing high-order chromatin (Hi-C) data, we established that the Lppnx locus interacts with Xist Intron1, an important regulator of Xist transcription. Deletion of Xist Intron1 in 129Sv Lppnx-deficient male ES cells rescues the expected Xce phenotype , indicating that Lppnx acts via Xist intron 1. In addition, ChIP experiments reveal that in Lppnx-deficient ES cells Oct4 and Rex1 occupancy of Intron1 is decreased, showing a possible regulatory network in which Lppnx modulates the occupancy of pluripotency factors at Xist Intron1 and hence, Xist transcription upon differentiation.
Results
Mapping of the Xce Candidate Region and Its Genetic Elements.
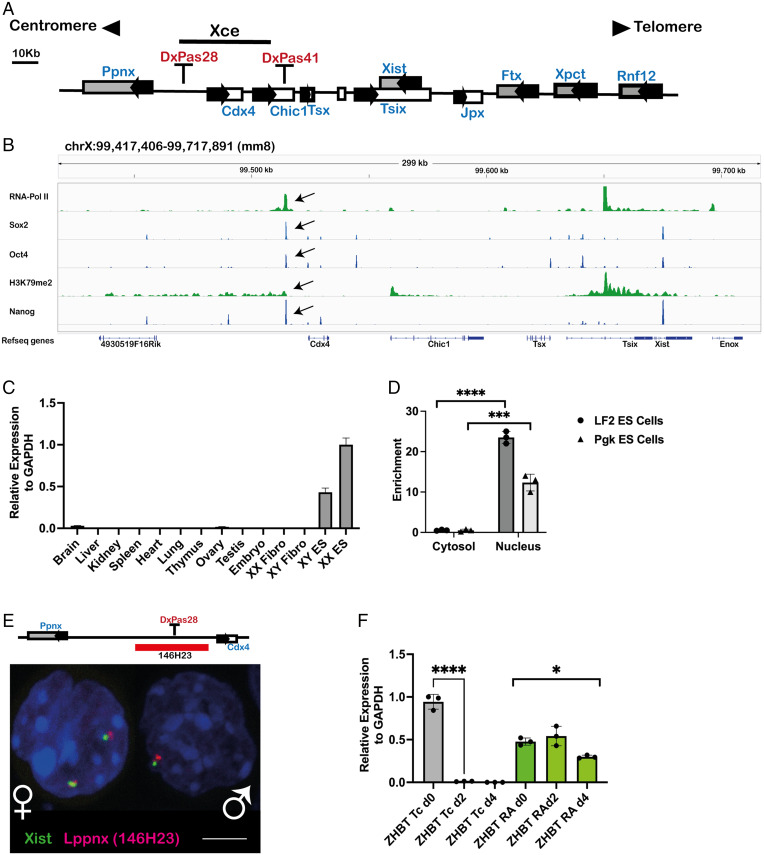
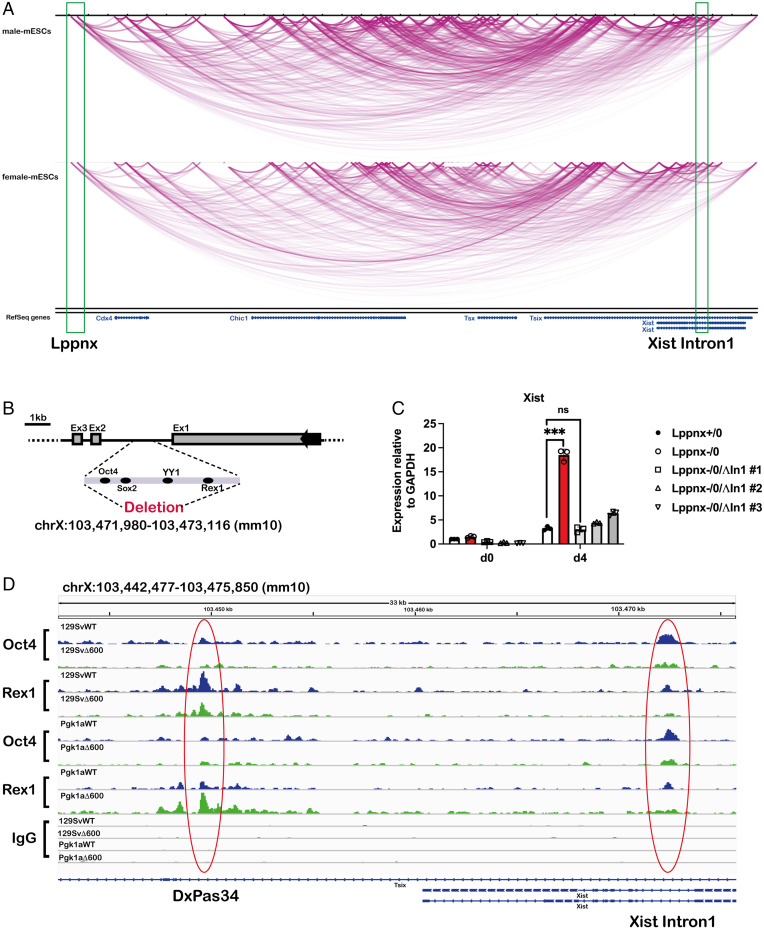
In a previous study (33) and subsequent follow-up studies, we identified a roughly 80 kb element 3′ to Xist between the microsatellite repeats DXPas28 and 41, as a potential Xce candidate (Fig. 1A). In addition to the previously characterized protein coding genes Cdx4 and Chic1, we identified another transcript originating from the candidate region (SI Appendix, Fig. S1A). Analysis of published ChIP-seq data of ES cells allowed us to identify a putative promoter region of 600 bp harboring binding sites for the pluripotency factors Oct4, Sox2, and Nanog as well as PolII, indicating active transcription (Fig. 1B). This is supported by the observation of H3K79me2 marks in the gene body, a consequence of active transcription (36).
Fig. 1.
A long noncoding RNA is transcribed from the predicted X-controlling element. Schematic view of the X-inactivation center with the predicted Xce indicated by a black bar (A). ChIPseq analysis shows a clear signal, marked with a black arrow, for the pluripotency factors Oct4, Nanog and Sox2 within the Xce. RNA-Pol II and H3K79m2 indicate initiation and maintenance of active transcription (B). The transcript identified in B is mainly expressed in mouse male and female ES cells but not in differentiated cells, as exemplified by organ analysis of adult mice. Relative expression levels of the transcript were measured by qRT-PCR in mouse organs, embryos, and cell lines. XX Fibro: Female fibroblast cell line, XY Fibro: Male fibroblast cell line. XX ES: Female embryonic stem cell line XY ES: Male embryonic stem cell line. (C). Fractionation of cytosol and nuclei in two ES cell lines, LF2 and Pgk, reveals the localization of the transcript to be mainly in the nucleus, indicating a bona fide lncRNA transcript. Enrichment in the two compartments was measured using qRT-PCR. Two-sample t test: P < 0.0001(LF2), P = 0.0006 (Pgk) (D). Visualization of the transcript in the nucleus of male and female ES cells using RNA-FISH. The position of the BAC used as FISH probe is highlighted as a red bar (E). The expression of the transcript depends on Oct4. Oct4-deficient ES cell line (ZHBT) which carries a tetracycline-repressible Oct4 transgene was differentiated either in the presence (RA) or absence (TC) of Oct4. In the absence of Oct4 (Tc) the transcript is rapidly down-regulated during differentiation, whereas the maintenance of Oct4 during differentiation (RA) maintains also the transcription. Transcript levels are shown relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Two-sample t test: P < 0.0001. One-way ANOVA: P = 0.0143 (F).
A Long Noncoding RNA within the Xce Expressed in ES Cells and in the ICM of Blastocysts.
Using primers binding to the first exon of the unknown transcript, we analyzed various tissues and cell lines to elucidate its expression pattern. Apart from weak expression in ovary and brain, we did not find transcripts in adult mouse organs nor in male or female fibroblast cell lines, whereas male and female ES cells showed a robust signal (Fig. 1C). This indicates that expression of the transcript is likely restricted to undifferentiated stem cells. In line with this conclusion, using an RNA–fluorescent in situ hybridization (FISH) approach we found a comparable signal in the ICM of expanded blastocysts indicating expression in the mouse preimplantation embryos (SI Appendix, Fig. S1B). To address whether the transcript encodes a protein we fractionated ES cell extracts into nuclei and cytosolic fractions. We found the vast majority of transcripts in the nucleus, indicating a likely noncoding RNA (Fig. 1D). This result is supported by RNA-FISH experiments showing a clear signal in the nucleus of both male and female ES cells (Fig. 1E). Given its restricted expression to ES cells and the early embryo, we were interested to explore the eventual dependency of its expression on pluripotency factors. To this end we made use of an Oct4-deficient ES cell line which carries a tetracycline-repressible Oct4 transgene (37). Differentiation of these ES cells in the presence of tetracycline (i.e., in the absence of Oct4) leads to a rapid down-regulation of the transcript, whereas in the presence of Oct4 its expression was maintained during differentiation (Fig. 1F). These results indicate that the transcript is down-regulated during the differentiation of ES cells and that Oct4 is required for its expression.
In summary, we show evidence that inside the predicted Xce region is localized the origin of a novel putative long noncoding RNA, which is expressed in male and female ES cells as well as in the ICM of late blastocysts. Since during differentiation the transcript is down-regulated, its expression pattern is compatible with the notion that pluripotency factors are required/involved in its expression. Given that the predicted genomic region of the transcript intermingles with the previously defined and published Ppnx gene, we named the transcript long noncoding RNA in the Ppnx gene, Lppnx.
Lppnx Is Differentially Expressed in Hybrid ES Cells during Differentiation.
The choice of which chromosome will be inactivated is made very early upon differentiation. Since we had shown Lppnx is down-regulated during differentiation (Fig. 1F), we took a closer look of the exact timepoint and allele specificity of Lppnx expression during differentiation of ES cells using single cell RT-qPCR on hybrid 129Sv/Pgk1a female ES cells. The use of allele-specific primers allowed the expression of Lppnx on the respective (129 or Pgk1a) X chromosome to be followed. In the pluripotent state these cells express distinct allele-specific levels of Lppnx: in a majority of cells, Lppnx from the 129Sv X chromosome is more highly expressed than from the Pgk1a alelle (SI Appendix, Fig. S2, d0). This ratio reverses by day 1 of differentiation; that is, by the time that choice of the X to be inactivated is established, with higher numbers of cells expressing from the strong (Pgk1a) Lppnx allele (SI Appendix, Fig. S2, d1). By day 2 most of the cells had silenced Lppnx. Interestingly, at day 1 the percentage of cells showing predominant expression from the Pgk1a (strong) allele at 61% compared to 32% from the 129Sv (weak) allele approximates inversely the X-linked gene expression ratio found later in heterozygous Xce crosses (75 vs. 25%). This may indicate that Lppnx expression is linked to Xist expression and involved in the choice of which X is to be inactivated.
Deletion of the Putative Promoter Sequence of Lppnx Abolishes the Expression of Lppnx RNA in ES Cells and Embryos.
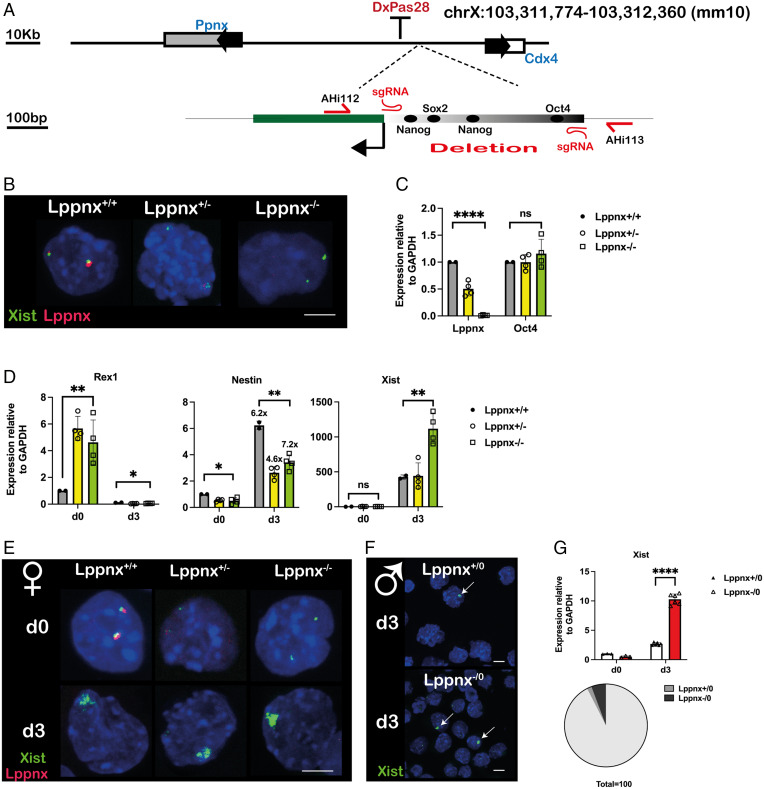
To support the idea of a putative promoter region both required and sufficient for the expression of Lppnx, we deleted this element in 129Sv mice using CRISPR/Cas9 (Fig. 2A and SI Appendix, Fig. S1 C and D). Phenotypically, female and male mice carrying either homo- or hemizygous deletions do not show obvious abnormalities. In addition, female ES cells generated from crosses of these mice are morphologically normal and exhibit normal levels of the ES cell pluripotency marker Oct4 (Fig. 2B and C). However, upon differentiation, Xist levels were found to be two- to three-fold higher compared to WT ES cells (Fig. 2D and E). Male ES cells carrying the deletion similarly exhibited increased Xist levels and, as a consequence, increased Xist foci after differentiation (Fig. 2F and G). Interestingly, the levels of the pluripotency marker Rex1 are moderately higher in undifferentiated hetero and homozygous female ES cells, whereas differentiation capacity indicated by the induction of the neuronal marker Nestin is not affected by the deletion of Lppnx (Fig. 2D). In summary, the deletion of the predicted promoter region leads to a complete silencing of Lppnx transcription and to increased levels of Xist after differentiation of such male and female ES cells.
Fig. 2.
Deletion of the putative promoter sequence of Lppnx in 129Sv mice. CRISPR/Cas9 mediated genome editing was used to delete the 595-bp promoter fragment in mice. Schematic view of the position of the sgRNAs and the primer used for subsequent genotyping of the founder animals (A). FISH analysis of female ES cells generated from LppnxΔ600 mice show the absence of a signal for Lppnx indicating that the predicted promoter fragment drives Lppnx expression (B). Lppnx expression is down-regulated in heterozygous ES cells and absent in homozygous cells carrying the deletion of the putative promoter region. Two-sample t test: P < 0.0001 (comparing WT with homozygous cells). The absence of Lppnx expression does not affect pluripotency in these ES cells as shown by a comparison of Oct4 expression levels. One-way ANOVA: P = 0.4998 (C). Female ES cells homo- and heterozygous for the deletion of Lppnx show different levels of key markers compared to WT cells upon differentiation. Cells were analyzed at day 3 after onset of differentiation (see Materials and Methods section). Rex1 levels are elevated in both, homozygous and heterozygous undifferentiated cells. Two-sample t test: P = 0.0023. After differentiation Rex is down-regulated in all three lines. One-way ANOVA: P = 0.008. Nestin was used as a differentiation marker. The degree of differentiation was calculated as fold change in expression of Nestin. The fold change is given above the bars in d3. All three lines show comparable fold changes during differentiation. Two-sample t test: P = 0.0242 (d0) P = 0.0032 (d3). Xist expression is markedly increased after differentiation in homozygous cells. d0: One-way ANOVA: P = 0.4959. d3:Ttwo-sample t test between WT and homozygous deletion P = 0.0064 (D). RNA-FISH analysis of differentiated female ES cells shows an increased intensity of Xist associated clouds upon Lppnx depletion, in agreement with the analysis in D (E). Accordingly, male ES cells deficient for Lppnx express higher Xist levels after differentiation as shown by qRT-PCR and the elevated number of Xist clouds indicated by arrows (F, G). Two-sample t test: P < 0.0001. The increase of Xist clouds in ES cells deficient for Lppnx compared to WT cells is visualized in a pie chart (G).
The Phenotypic Effect of the Deletion of the Entire 80-kb Xce Candidate and the 600-bp Lppnx Promoter Are Very Similar.
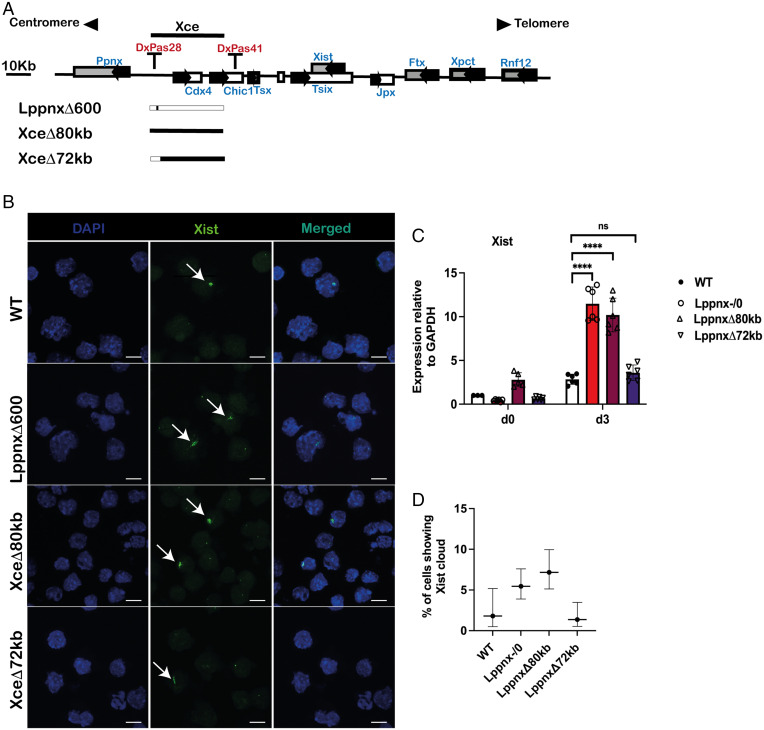
We have shown that the deletion of Lppnx leads to increased Xist levels in 129Sv male and female ES cells during differentiation. Given the robust phenotype shown in male ES cells (Xist RNA level and Xist foci), we decided to compare this phenotype with a deletion of the entire Xce candidate region and finally with the entire Xce except the Lppnx region. Using CRISPR/Cas9 we introduced the deletions in WT 129Sv ES cells (Fig. 3A). Upon differentiation we observed that the 80-kb and the 600-bp lines show a comparable phenotype in terms of Xist levels and foci. In contrast, the deletion of the protein coding genes Cdx4 and Chic1 does not have any effect on Xist levels after differentiation (Fig. 3B–D). From this result, we conclude that Lppnx is the driving factor in the formation of the observed phenotype.
Fig. 3.
Comparison of the number of Xist clouds in male ES cells. Schematic view of the X inactivation center and the CRISPR mediated deletion. 129Sv male ES cells were used to introduce a deletion of the entire predicted Xce (XceΔ80kb) or a deletion which leaves intact the Lppnx region (LppnxΔ72kb). These cell lines were compared with a WT and the previously shown LppnxΔ600 (corresponding to Lppnx-/0) cell line (A). After differentiation XceΔ80kb and LppnxΔ600 show elevated numbers of Xist clouds whereas LppnxΔ72kb and WT show comparable levels (B). qRT-PCR analysis of the cells in B showed elevated Xist expression levels in XceΔ80kb and LppnxΔ600 but not in LppnxΔ72kb compared to WT. Two-sample t test: P < 0.0001 (C). Quantification of Xist clouds shown in B are given in percentage of Xist clouds within the different cell lines. Remarkably, only XceΔ80kb and LppnxΔ600 show elevated ratio of clouds, whereas LppnxΔ72kb and WT cells show comparable levels. Percentages are given with an upper and lower level and are shown with a CI of 95% (Wilson/Brown) (D).
Heterozygous Deletion of Lppnx in a Xce Heterozygous Background Influences the Choice of the X Chromosome To Be Inactivated.
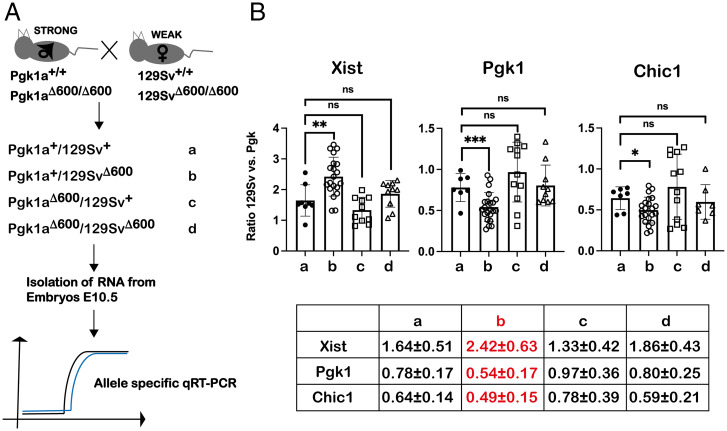
The Xce locus is associated with the introduction of bias in choice of which X will be inactivated. To test the influence of Lppnx on this, we crossed mice heterozygous for Xce carrying a deletion of or not, of Lppnx. As a readout we isolated E10.5 female embryos and performed X chromosome allele-specific RT-qPCR to calculate a quantitative ratio for XCI (Fig. 4A). F1 embryos derived from crossings of 129Sv (Xcea) and HPRT-Pgk1a (Xcec) showed a calculated expression ratio for Xist of 1.64 (Fig. 4B). Conversely, expression ratios of two representative X-linked genes were 0.64 and 0.78, respectively. The deletion of Lppnx in 129Sv embryos shifted the Xist allelic ratio to 2.42, whereas the ratios of X-linked genes decreased to 0.54 and 0.49, respectively (Fig. 4B). These data suggest that the deletion of Lppnx on an Xcea haplotype background influences the choice process in XCI in Xce heterozygous embryos. Moreover, it suggests that Lppnx expression may prevent X inactivation by repressing Xist up-regulation (Fig. 4B).
Fig. 4.
Deletion of Lppnx promotes a shift in X-linked expression ratio in heterozygous embryos. Schematic view of the breeding strategy used to analyze X-linked expression ratios. From heterozygous Xce crosses between Xcea (weak, 129Sv) and Xcec (strong, Pgk1a), indicated from a–d, E10.5 female embryos were isolated. Δ600 corresponds to the 600-bp promoter deletion shown before. We used allele-specific qRT-PCR to define the X-linked expression ratio as follows: 2Ct(Pgk)-Ct(129) (A). Reciprocal crosses of Xcea with Xcec with or without a depletion of Lppnx show a significant shift of Xist ratio as well as of X-linked genes. Pgk1 and Chic1 in E10.5 embryos only in Xcea-LppnxΔ600/XcecWT embryos, whereas all other combinations are comparable to WT. Two-sample t test Xist: P = 0.0048, Pgk1: P = 0.0042, Chic1: P = 0.0379. Each dot in the plots represents the value of a single embryo measured in three technical replicates. Expression ratios are summarized in a table, only crossing b (marked in red) shows significant difference to WT (B).
The Deletion of Lppnx in a Xcec Strain Shows Only Weak Effects.
Given the effect described above we expected to observe a similar phenotype when deleting Lppnx on a Xcec carrying mouse strain. However, analysis of Hprt-Pgk−/129Sv+ embryos carrying such deletions failed to show any significant effect on Xist ratios compared to WT (Fig. 4B). This would suggest that Lppnx is either not active or only weakly active in Xcec-carrying strains. Intriguingly, the ratio in double-knockout (ko)) embryos showed almost identical values to that of WT littermates. If the deletion of Lppnx had no effect on the Xcec strain, then the observed ratio would be expected to be similar to that of heterozygous embryos carrying a deletion on the 129Sv X, rather than having values similar to those of WT. These data were confirmed by the observations of male Xcec ES cells after differentiation. The deletion of Lppnx leads to an up-regulation of Xist and to an increase of Xist foci; however, both to a much weaker degree compared to male Xcea ES cells (SI Appendix, Fig. S3).
The Lppnx Promoter Region Interacts with Xist Intron1.
Higher order chromatin interactions are known to play a crucial role in XCI (38). We analyzed published Hi-C data for possible interactions of the Lppnx locus with elements of the XIC which might help to explain the observed phenotypes using an epigenome browser (epigenomegateway.wustl.edu/browser/). The visualization of interactions within the XIC and the predicted Xce locus revealed a possible connection between the loci of Lppnx and Xist Intron1 (XI1) in both male and female ESCs (Fig. 5A). It has been shown that this Intron contains binding sites for pluripotency factors, playing a crucial role in XCI (18). Interestingly, Intron 1 seems to be dispensable for XCI in vivo and in vitro but it may be involved in the choice of which X is inactivated in female cells (39).
Fig. 5.
The Lppnx locus may interact with Xist Intron1 in mouse ES cells and controls the loading of Oct4 and Rex1 in XI1. Screenshot of a Hi-C analysis showing a part of the X inactivation center including Xist and Lppnx. The green rectangles indicate the transcription start site of Lppnx and Xist Intron1 showing a possible interaction between these two regions in both, male and female ES cells (A). Schematic view of the pluripotency factor binding region of Xist intron 1 which was deleted in 129Sv and 129SvΔ600 male ES cells, respectively, using CRISPR (B). Elevated Xist expression observed in 129SvΔ600 male ES cells (Lppnx-/0) after differentiation is completely reversed after the deletion of Xist Intron1. Three independent lines (#1, #2, #3) are shown. Two-sample t test P = 0.001 (C). Lppnx regulates the loading of pluripotency factors in Xist Intron1 and DxPas34. ChIPseq experiments show that upon deletion of Lppnx the loading of Oct4 and Rex1 is reduced at Xist Intron1 (red circle, Right) in male 129Sv and Pgk1a ES cells. In contrast, Rex1 binding at DxPas34, a regulatory region of Tsix, is reduced in Lppnx-deficient 129Sv ES cells, whereas in Pgk1a cells Rex1 binding is increased (red circle, Left) (D).
Deletion of the Pluripotency Factor Binding Region of Xist Intron1 Rescues the Phenotype in Lppnx KO Male ES Cells.
We decided to use CRISPR mediated ablation of the pluripotency factor binding region of Xist Intron1 in Lppnx deficient 129Sv male ES cells, based on data obtained from (18), to test whether this has any effect on the elevation of Xist levels observed in these cells (Fig. 5B). Elevated Xist levels are observed in Lppnx ko cells as reported earlier; however, analysis of three independent clones carrying a Lppnx/Xist Intron1 double ko showed a complete rescue of the phenotype leading to Xist levels after differentiation comparable to those of wildtype (WT) cells (Fig. 5C).
Lppnx Controls the Loading of Pluripotency Factors at Xist Intron1 and DxPas34.
As mentioned, transcription factors bound to XI1 modulate Xist expression and probably choice during XCI. We analyzed the loading of three key factors on XI1. Oct4 and Rex1 have been shown to repress Xist expression (18), whereas YY1acts as a transcriptional activator of Xist (40). We used ChIP-seq to analyze the loading of these factors in WT and Lppnx ko ES cells. We found that upon Lppnx depletion, the loading of Oct4 and Rex1 at XI1 decreased markedly in Xcea as well as in Xcec ES cells, whereas YY1 binding remained unchanged (Fig. 5D and SI Appendix, Fig. S4A). These data may indicate that Lppnx controls the level of bound transcription factors which determine the onset of Xist up-regulation and hence the choice of which X will be inactivated. Interestingly, the transcription factor Rex1 evolved by retro-position from the YY1 gene. Rex1 binds targets divergent from YY1, but the binding motifs of Rex1 and YY1 show similarity in their core region (41). Considering this, YY1 and Rex1 may compete for binding on XI1, although YY1 binding seemed unchanged.
Similar observations can be carried out at the microsatellite repeat DxPas34, an important control region for the expression of Tsix (42). While on both Xce backgrounds binding of Oct4 decreased slightly upon the deletion of Lppnx, Rex1 showed a very different and divergent pattern: while in Xcea cells there was a decrease in loading, cells from Xcec showed increased loading for Rex1 upon Lppnx depletion (Fig. 5D). This result may explain differences in the phenotype of the Lppnx ko in Xcea and Xcec ES cells. Increased binding of Rex1 at DxPas34 in Xcec ES cells would maintain Tsix transcription and hence compensate for the loss of Oct4 in XI1. In contrast, the low level of Rex1 at DxPas34 in Xcea cells leads to decreased Tsix transcription and together with decreased Oct4 level at XI1, to an enhanced activation of Xist transcription.
Discussion
The most widely discussed and shared idea about how the Xce influences choice in XCI is that this genetic region serves to bind a series of autosomal factors which in turn would regulate the main defined players in XCI, Tsix and Xist (43, 44). In the present study, we define the Xce as an 80-kb spanning region between the microsatellite repeats DxPas28 and DxPas41. We identified Lppnx, a lncRNA originating from within this region, as a novel candidate for the Xce locus and suggest a mechanism involving modulation of the occupancy of pluripotency factors at Xist Intron1 and DxPas34, two key regions in XCI. Our analysis suggests that different binding capacities of pluripotency factors at Xist Intron1 and DxPas34 influence Xist up-regulation and most likely the choice in XCI, a hypothesis which has been presented recently (39). Our results extend this idea by showing that Lppnx can modulate the binding of these factors in a background-dependent manner. In particular, Oct4 and Rex1 showed decreased binding at Intron1 upon deletion of Lppnx. Since Oct4 was shown to inhibit Xist up-regulation, this reduced binding could well explain the increased expression of Xist after differentiation on an Xcea background (18). This effect is counteracted in Xcec ES cells by increased binding of Rex1 at DXPas34, which in turn maintains Tsix transcription and inhibits the Xist up-regulation (19).
Whereas the Xcea allele studied here (129Sv strain) is highly susceptible to the lack of Lppnx, the Xcec strain (HPRT-Pgk1a) shows a weaker phenotype upon the ablation of Lppnx. In both strains, Lppnx modulates Xist expression most likely by controlling the occupancy of Oct4 and Rex1 (and possibly other factors) in Xist Intron1. Lppnx expression itself is highly dependent on the presence of Oct4, linking Lppnx expression to pluripotency and XCI and forming a feedback loop between Lppnx and Xist (SI Appendix, Fig. S4B). This association may explain why in previous studies it has been supposed that the Xce might be identical to Xist (33). Our analysis shows clearly not only that Xce is genetically different from Xist, but also provides a likely mechanism for how Xce acts as a Xist repressor with the requirement of Xist Intron1.
The question of how Lppnx modulates the binding of transcription factors at a molecular level remains to be elucidated. Emerging results from previous studies stress the possible role of lncRNAs as molecular scaffolds (45, 46). For Lppnx to act as a scaffold, binding pluripotency factors for XI1 would require an interaction or proximity between the Lppnx and Xist locus. Analysis of published Hi-C and 5C data suggests that indeed both loci do interact, underlining the importance of topologically associating domains in long-range chromatin interaction in many biological phenotypes (38, 47). The zinc-finger protein CTCF and the cohesion complex are the main players in the establishment of both, long- and short-range chromatin interaction (48). While both the Lppnx promoter and Xist Intron1 contain binding sites for both factors, the extensive genetic heterogeneity between Xcea and Xcec strains might lead to modulation of binding and hence different chromosomal conformations. The same is true for pluripotency binding sites in Xist Intron1 and the Lppnx promoter regulating the level of Lppnx expression (SI Appendix, Fig. S4B).
Recently it was reported that the promoter region of the noncoding RNA Linx (LinxP) acts as a cis-regulatory element and long-range silencer to influence the choice of X chromosome to be inactivated independent of its transcriptional activity (49). We believe that Lppnx and Linx are mostly identical concerning their positions on the X chromosome. However, we think that both transcription and the lncRNA itself play an important role in the interpretation of the results shown here. For one to fully understand the function of a lncRNA, it is important to identify its interacting partners, and in recent years lncRNA-protein interactions have gained increased attention. We analyzed CLIP-Seq data from mouse embryonic stem cells and found that Oct4 binds to Lppnx in these cells, whereas YY1 shows a neglectable signal (SI Appendix, Fig. S5, blue boxed region) (50). Interestingly, both Oct4 and YY1 seem to bind to Tsix but not to Xist (SI Appendix, Fig. S5, red boxed region). These data may indicate that upon a certain proximity between the Lppnx and the Xist/Tsix locus, the Lppnx RNA provides a pool of Oct4 (and other factors) to the regulatory regions in the Xist/Tsix locus. Hence, lack of Lppnx RNA leads to decreased Oct4 occupancy in important regulatory regions like Xist Intron1 and DxPas34. These results indicate that the binding of important transcription factors in XCI can be directly regulated by Lppnx. The quantitative binding of Oct4 (and other factors) to Lppnx strongly depends on the sequence of the RNA. Therefore, we analyzed possible splice variants of Lppnx in Xcea and Xcec ES cells, which may explain the differential loading of these factors to the prospective binding sites. Indeed, we found splice variants predominantly occurring in Xcea but not in Xcec and vice versa (SI Appendix, Fig. S6). This finding brings another level of complexity into the action of Lppnx, suggesting that different splice variants result in differential loading of the transcription factors.
Our results and the model that results suggest that the action of Xce is quite complex, in agreement with the rather quantitative and variable effects of Xce action on X-inactivation ratios observed in in vivo experimental typing systems. Indeed, characterization of unknown Xce alleles historically has been based on extended testing of progeny from crosses involving various known Xce tester strains, precisely to deal with this variability. The characterization of Lppnx as shown here could facilitate substantially the categorization of Xce strains. However, additional efforts will be needed to determine its role within the complex network acting through the Xce. This suggests the question of whether Lppnx acts in concert with additional factors so far not identified. Future research will center on questions concerning the possible binding of Oct4/Rex1 to Lppnx, the role of different isoforms of Lppnx and their implications for differential factor binding, and a more extensive exploration of individual Xce allele variability and its mechanistic implications.
Materials and Methods
Generation of Lppnx KO Mice.
To generate a transgenic mouse line with the promoter of Lppnx deleted we used CRISPR (clustered regularly interspaced short palindromic repeats) -Cas9 to target this region in the mouse zygote. For a more detailed description please refer to SI Appendix, SI Methods.
CRISPR-Mediated Genome Editing in ES Cells.
Using https://horizondiscovery.com/en/ordering-and-calculation-tools/crispr-design-tool, we identified suitable target sequences with minimal off target effects for sgRNA synthesis. Corresponding oligonucleotides (listed in SI Appendix, Table S1) were then cloned into pSpCas9(BB)-2A-GFP (PX458, a gift from Feng Zhang, addgene #48138) (51) using the BbsI site. The plasmids were then transfected in ES cells (Xfect, Takara #631317) followed by fluorescence activated cell sorting of green fluorescent protein–positive cells. Single clones were analyzed by PCR with primer pairs listed in SI Appendix, Table S1.
Generation of ES Cell Lines from Blastocysts and ES Cell Differentiation.
Flushed E3.5 blastocysts were cultured for 12–24 h in KSOM media (Invitrogen). Before hatching they were then transferred in gelatinized 96-well dishes containing normal cell culture media supplemented with 2i, Lif, and 10% serum (52). After several days round colonies were obtained and expanded by continuous dissociation.
ES cell differentiation was carried out as previously described (53). Briefly, 1 million cells were plated on a 6-cm dish coated with laminin. After 6 h Fgf2 was added to the cells. The medium was replaced every day and cells were harvested after 3 d of differentiation.
Nuclei Extraction of ES Cells.
Cells were grown until confluency, trypsinized, and washed with phosphate-buffered saline. The cell pellet was resuspended in a hypotonic buffer (10 mM Hepes, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.05% Nonidet P-40, +protease inhibitors) and incubated 20 min on ice. After centrifugation, supernatant and the nuclei were subjected to RNA isolation.
RNA Isolation and cDNA Synthesis.
RNA from tissue, embryos, and cells was isolated using the Qiagen RNAesy kit according to manufacturer instructions. cDNA synthesis was carried out using 1 µg of total RNA and the SuperScript III First-Strand Synthesis System (Invitrogen#18080-051). Residual genomic DNA was removed by applying a suitable DNase (Thermo #EN0771).
RNA FISH on ES Cells and Preimplantation Embryos.
RNA FISH on ES cells was carried out according to published protocols (54) with the following modification. ES cells were fixed in ice-cold paraformaldehyde for 5 min before being spotted on glass slides. RNA FISH on blastocysts was performed following published protocols (55).
Z-stacks were captured on a Leica SP5 confocal microscope (Leica Microsystems, Germany) and images were prepared with Fiji/ImageJ software.
Single-Cell Gene Expression Analysis.
Single-cell gene expression analysis was performed as described previously (56) and as recommended by Fluidigm https://www.fluidigm.com/area-of-interest/single-cell-analysis/single-cell-analysis-with-microfluidics# (www.fluidigm.com/single-cell-expression.html; South San Francisco, CA, USA). For a more detailed description please refer to SI Appendix, SI Methods.
qRT-PCR Using Allele-Specific Primer and Calculation of Expression Ratios.
The solution from cDNA synthesis was diluted 1:40 and from this 5 µL was mixed with 2× QPCRBIO SYGREEN MIX LO-ROX reagent (PCR BIO) for every single reaction. The sequences of primer are shown in SI Appendix, Table S1. Expression ratios have been defined as following: 2Ct(Pgk)-Ct(129). Statistical analysis was performed using JMP and Prism software.
Chromatin Immunoprecipitation and Sequencing (ChIP-Seq).
This protocol was adapted from a CUT&RUN Protocol from EpiCypher (https://www.epicypher.com/content/documents/protocols/cutana-cut&run-protocol.pdf) and is based on the use of magnetic beads. For a more detailed description please refer to SI Appendix, SI Methods.
ChIP-Seq Data Processing.
Data were processed on Galaxy. Adapters were removed with Trimmomatic v0.36.6 and sequences having an average quality below 20 were eliminated. Quality statistics were verified with Fastqc v0.69. Reads were aligned to mm10 using Bowtie2 v2.3.4.2 using default parameters. Bam files were ordered by coordinates with SortSam v2.7.1.1. Only reads mapping to the main chromosomes were kept. Unmapped and nonprimary reads were removed with Samtools v1.1.2 (“Filter SAM or BAM, output SAM or BAM”). Duplicated reads were removed with picards tools v2.7.1.0 (MarkDuplicatesWithMateCigar). Bigwig files were generated with bamCoverage v2.5.7.0 normalizing to 1× coverage on mm10 and extending reads to 150 bp.
Hi-C Data.
Using an epigenome browser (epigenomegateway.wustl.edu/browser/) we analyzed published Hi-C data obtained from male and female ESC (38). The GEO accession number for the datasets are GSM873934 (male-mESCs) and GSM873927 (female-mESCs).
CLIP-Seq Data.
Data sets were downloaded using the Geo database (https://www.ncbi.nlm.nih.gov/geo/) with the respective accession numbers: Oct4 (GSE68196) and YY1 (GSM1665564). Integrative Genomics Viewer (IGV, Broad Institute) was used to visualize the data.
Supplementary Material
Acknowledgments
We thank Jamie Hackett and Matthieu Boulard from EMBL Rome for critical reading of the manuscript and valuable comments; Neil Humphreys and Neil Dear from the Transgenic Facility of EMBL Rome for generating multiple knock-out mouse lines; Giulia Bolasco and Monica Di Giacomo for guiding us through analytical microscopy; and Philip Hublitz and James Sawitzke for advice in cloning transgenic vectors. The bio-informatic analysis was carried out by Nicolas Descostes.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118182119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Mank J. E., The W, X, Y and Z of sex-chromosome dosage compensation. Trends Genet. 25, 226–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ercan S., Mechanisms of x chromosome dosage compensation. J Genomics 3, 1–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuroda M. I., Hilfiker A., Lucchesi J. C., Dosage compensation in Drosophila-a model for the coordinate regulation of transcription. Genetics 204, 435–450 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Payer B., Lee J. T., X chromosome dosage compensation: How mammals keep the balance. Annu. Rev. Genet. 42, 733–772 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Leeb M., Wutz A., Mechanistic concepts in X inactivation underlying dosage compensation in mammals. Heredity 105, 64–70 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Heard E., Disteche C. M., Dosage compensation in mammals: Fine-tuning the expression of the X chromosome. Genes Dev. 20, 1848–1867 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Brockdorff N., Turner B. M., Dosage compensation in mammals. Cold Spring Harb. Perspect. Biol. 7, a019406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Namekawa S. H., Payer B., Huynh K. D., Jaenisch R., Lee J. T., Two-step imprinted X inactivation: Repeat versus genic silencing in the mouse. Mol. Cell. Biol. 30, 3187–3205 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan S. S., Williams E. A., Tam P. P., X-chromosome inactivation occurs at different times in different tissues of the post-implantation mouse embryo. Nat. Genet. 3, 170–174 (1993). [DOI] [PubMed] [Google Scholar]
- 10.Monk M., Harper M. I., Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature 281, 311–313 (1979). [DOI] [PubMed] [Google Scholar]
- 11.Schulz E. G., et al. , The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell 14, 203–216 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Kay G. F., et al. , Expression of Xist during mouse development suggests a role in the initiation of X chromosome inactivation. Cell 72, 171–182 (1993). [DOI] [PubMed] [Google Scholar]
- 13.Lee J. T., Lu N., Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 99, 47–57 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Sado T., Brockdorff N., Advances in understanding chromosome silencing by the long non-coding RNA Xist. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20110325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun S., et al. , Jpx RNA activates Xist by evicting CTCF. Cell 153, 1537–1551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogawa Y., Lee J. T., Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol. Cell 11, 731–743 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Chureau C., et al. , Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum. Mol. Genet. 20, 705–718 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Navarro P., et al. , Molecular coupling of Xist regulation and pluripotency. Science 321, 1693–1695 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Navarro P., et al. , Molecular coupling of Tsix regulation and pluripotency. Nature 468, 457–460 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Gontan C., et al. , RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature 485, 386–390 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Navarro P., Moffat M., Mullin N. P., Chambers I., The X-inactivation trans-activator Rnf12 is negatively regulated by pluripotency factors in embryonic stem cells. Hum. Genet. 130, 255–264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cattanach B. M., Isaacson J. H., Controlling elements in the mouse X chromosome. Genetics 57, 331–346 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cattanach B. M., Williams C. E., Evidence of non-random X chromosome activity in the mouse. Genet. Res. 19, 229–240 (1972). [DOI] [PubMed] [Google Scholar]
- 24.Cattanach B. M., Pollard C. E., Perez J. N., Controlling elements in the mouse X-chromosome. I. Interaction with the X-linked genes. Genet. Res. 14, 223–235 (1969). [DOI] [PubMed] [Google Scholar]
- 25.West J. D., Chapman V. M., Variation for X chromosome expression in mice detected by electrophoresis of phosphoglycerate kinase. Genet. Res. 32, 91–102 (1978). [DOI] [PubMed] [Google Scholar]
- 26.Johnston P. G., Cattanach B. M., Controlling elements in the mouse. IV. Evidence of non-random X-inactivation. Genet. Res. 37, 151–160 (1981). [DOI] [PubMed] [Google Scholar]
- 27.Cattanach B. M., Rasberry C., Identification of the Mus spretus Xce allele. Mouse Genome 89, 565–566 (1991). [Google Scholar]
- 28.de La Casa-Esperón E., et al. , X chromosome effect on maternal recombination and meiotic drive in the mouse. Genetics 161, 1651–1659 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krietsch W. K., Fehlau M., Renner P., Bücher T., Fundele R., Expression of X-linked phosphoglycerate kinase in early mouse embryos homozygous at the Xce locus. Differentiation 31, 50–54 (1986). [DOI] [PubMed] [Google Scholar]
- 30.Cattanach B. M., Perez J. N., Parental influence on X-autosome translocation-induced variegation in the mouse. Genet. Res. 15, 43–53 (1970). [DOI] [PubMed] [Google Scholar]
- 31.Cattanach B. M., Papworth D., Controlling elements in the mouse. V. Linkage tests with X-linked genes. Genet. Res. 38, 57–70 (1981). [DOI] [PubMed] [Google Scholar]
- 32.Cattanach B. M., Rasberry C., Evans E. P., Burtenshaw M., Further Xce linkage data. Mouse News Lett. 83, 165 (1989). [Google Scholar]
- 33.Simmler M. C., Cattanach B. M., Rasberry C., Rougeulle C., Avner P., Mapping the murine Xce locus with (CA)n repeats. Mamm. Genome 4, 523–530 (1993). [DOI] [PubMed] [Google Scholar]
- 34.Calaway J. D., et al. , Genetic architecture of skewed X inactivation in the laboratory mouse. PLoS Genet. 9, e1003853 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chadwick L. H., Pertz L. M., Broman K. W., Bartolomei M. S., Willard H. F., Genetic control of X chromosome inactivation in mice: Definition of the Xce candidate interval. Genetics 173, 2103–2110 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen A. T., Zhang Y., The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 25, 1345–1358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niwa H., Miyazaki J., Smith A. G., Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24, 372–376 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Nora E. P., et al. , Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minkovsky A., et al. , The pluripotency factor-bound intron 1 of Xist is dispensable for X chromosome inactivation and reactivation in vitro and in vivo. Cell Rep. 3, 905–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makhlouf M., et al. , A prominent and conserved role for YY1 in Xist transcriptional activation. Nat. Commun. 5, 4878 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J. D., Faulk C., Kim J., Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX1. Nucleic Acids Res. 35, 3442–3452 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen D. E., et al. , The DXPas34 repeat regulates random and imprinted X inactivation. Dev. Cell 12, 57–71 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Lyon M. F., Possible mechanisms of X chromosome inactivation. Nat. New Biol. 232, 229–232 (1971). [DOI] [PubMed] [Google Scholar]
- 44.Rastan S., Non-random X-chromosome inactivation in mouse X-autosome translocation embryos—Location of the inactivation centre. J. Embryol. Exp. Morphol. 78, 1–22 (1983). [PubMed] [Google Scholar]
- 45.Spitale R. C., Tsai M. C., Chang H. Y., RNA templating the epigenome: Long noncoding RNAs as molecular scaffolds. Epigenetics 6, 539–543 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ribeiro D. M., et al. , Protein complex scaffolding predicted as a prevalent function of long non-coding RNAs. Nucleic Acids Res. 46, 917–928 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dekker J., Heard E., Structural and functional diversity of topologically associating domains. FEBS Lett. 589, 2877–2884 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghirlando R., Felsenfeld G., CTCF: Making the right connections. Genes Dev. 30, 881–891 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galupa R., et al. , A conserved noncoding locus regulates random monoallelic Xist expression across a topological boundary. Mol. Cell 77, 352–367.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sigova A. A., et al. , Transcription factor trapping by RNA in gene regulatory elements. Science 350, 978–981 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ran F. A., et al. , Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ying Q. L., et al. , The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Attia M., Rachez C., Avner P., Rogner U. C., Nucleosome assembly proteins and their interacting proteins in neuronal differentiation. Arch. Biochem. Biophys. 534, 20–26 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Chaumeil J., Okamoto I., Heard E., X-chromosome inactivation in mouse embryonic stem cells: Analysis of histone modifications and transcriptional activity using immunofluorescence and FISH. Methods Enzymol. 376, 405–419 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Namekawa S. H., Lee J. T., Detection of nascent RNA, single-copy DNA and protein localization by immunoFISH in mouse germ cells and preimplantation embryos. Nat. Protoc. 6, 270–284 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dubois A., et al. , Spontaneous reactivation of clusters of X-linked genes is associated with the plasticity of X-inactivation in mouse trophoblast stem cells. Stem Cells 32, 377–390 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.