Abstract
Caveolae are plasma membrane invaginations that play important roles in both endocytosis and membrane tension buffering. Typical caveolae have invaginated structures with a high-density caveolin assembly. Membrane sculpting proteins, including PACSIN2 and EHD2, are involved in caveolar biogenesis. PACSIN2 is an F-BAR domain-containing protein with a membrane sculpting ability that is essential for caveolar shaping. EHD2 is also localized at caveolae and involved in their stability. However, the spatial relationship between PACSIN2, EHD2, and caveolin has not yet been investigated. We observed the single-molecule localizations of PACSIN2 and EHD2 relative to caveolin-1 in three-dimensional space. The single-molecule localizations were grouped by their proximity localizations into the geometric structures of blobs. In caveolin-1 blobs, PACSIN2, EHD2, and caveolin-1 had overlapped spatial localizations. Interestingly, the mean centroid of the PACSIN2 F-BAR domain at the caveolin-1 blobs was closer to the plasma membrane than those of EHD2 and caveolin-1, suggesting that PACSIN2 is involved in connecting caveolae to the plasma membrane. Most of the blobs with volumes typical of caveolae had PACSIN2 and EHD2, in contrast to those with smaller volumes. Therefore, PACSIN2 and EHD2 are apparently localized at typically sized caveolae.
Introduction
Caveolae are flask-shaped plasma membrane invaginations that are abundant in several cell types found in muscle, epithelial, and adipose tissues [1–3]. Caveolae play dual roles at the plasma membrane, as an endocytic apparatus and a membrane reservoir for buffering membrane tension. During endocytosis, the caveolar invagination is pinched off to form endocytic vesicles, while in tension buffering it is flattened to provide extra surface area to increase the membrane surface [1,4,5].
Caveolae are composed of a unique set of proteins and lipids. The caveolar membrane is rich in cholesterol, similar to the lipid rafts at the plasma membrane, where several receptors and signaling proteins are reportedly concentrated [6–9]. Caveolae are also a platform for signaling proteins that are regulated by the caveolar endocytic function. The structural caveolar proteins comprise caveolins and cavins [10,11]. Caveolin exists as three isoforms, and the caveolin-1 and caveolin-3 amino acid sequences are almost identical [12,13]. Caveolin-1 is ubiquitously expressed, while caveolin-3 is predominantly expressed in muscle. Mutations associated with diseases such as muscular dystrophy have been identified in caveolin-3 [14,15], consistent with the role of caveolae in the tension buffering of muscle cells [16]. There are four cavin isoforms, and they are essential for caveolae [11,17–20]. Cavins associate with caveolins and generate the characteristic striations on the caveolar surface, as observed by electron microscopy [21–24].
The endocytosis of caveolae is mediated by dynamin [25], as in clathrin-mediated endocytosis. The invaginated membrane of clathrin-coated pits is mainly produced by BAR domain proteins [26,27], which directly generate membrane curvatures and recruit structural proteins for membrane remodeling, including dynamin and Wiskott–Aldrich syndrome family proteins [28]. Dynamin mediates the pinching of invaginations to form vesicles, in cooperation with the actin cytoskeleton [29]. The BAR domains are divided into the BAR, N-BAR, and I-BAR domain subfamilies [30,31]. Among them, the F-BAR domain-containing protein PACSIN (Syndapin) is involved in caveolae [32–34]. Three isoforms of PACSIN have been described. PACSIN3 is a muscle-specific isoform, and its knockout results in caveolar biogenesis abnormalities [35]. PACSIN2 is a ubiquitous isoform involved in caveolae formation and endocytosis [34]. PACSIN1 is brain-specific, and its role in caveolae has not yet been clarified [34]. Importantly, PACSIN2 has membrane deforming ability, which is altered by the cholesterol content of the membrane, implying the important role of PACSIN2 in caveolar homeostasis [36]. Indeed, PACSIN2 is stably localized at caveolae, presumably at the neck of caveolar invaginations [33,34,37]. Furthermore, PACSINs have NPF sequences that bind to the EHD2 protein, which is localized at and stabilizes caveolae, presumably by mediating actin cytoskeleton anchoring [32,38,39]. Importantly, dynamin is recruited to caveolae only when PACSIN2 disappears, suggesting the regulation of dynamin binding to PACSIN2 during endocytosis [40].
Approximately 150 caveolin-1 molecules have been detected in mature caveolae [37,41]. Due to this abundance of caveolin-1, typical caveolae appear to have a certain caveolin-1 density, which can be measured by the nearest neighbor distance (NND) between caveolin-1s in single-molecule localization microscopy (SMLM), a method that can determine the coordinates of molecules at an accuracy equivalent to the protein size; that is, ~10 nm in the plane parallel to the focal plane [42]. Using SMLM data projected onto a two-dimensional plane, the membrane deformation can be monitored by the density estimations of caveolin-1 [42] or the distances between caveolin-1 and caveolae-localized molecules [43]. Furthermore, by introducing a cylindrical lens, the localization depth measurement; that is, the three-dimensional determination of the coordinates, could be achieved with an accuracy of ~50 nm in the depth direction [44,45]. The three-dimensional coordinates of caveolin-1 localization can be grouped into blob-shaped geometrical structures, which were previously classified into the typical caveolae or other caveolin-1 clusters [46,47]. In this study, we used three-dimensional SMLM to examine PACSIN2 and EHD2 localizations relative to caveolin-1. The PACSIN2 and EHD2 coordinates mostly overlapped with the caveolin-1 coordinates in the caveolin-1 blobs of caveolar volume, which are thought to correspond to typical caveolae. However, PACSIN2 was ~5 nm closer to the plasma membrane than EHD2 and caveolin-1, suggesting its role in connecting caveolae to the plasma membrane. Most of the blobs with volumes of typical caveolae had PACSIN2 and EHD2, whereas those with smaller volumes did not. Therefore, PACSIN2 and EHD2 are apparently localized at typically sized caveolae.
Results and discussion
First, we observed the single-molecule localizations in antibody-stained HeLa cells. SMLM uses total internal reflection to illuminate the fluorophore, and thus the observation is limited to the plasma membrane neighboring the glass surface on which the cells attach. HeLa cells were labeled with antibodies against caveolin-1 and PACSIN2 or EHD2. Two kinds of antibodies for each protein were used, Caveolin-1 (7C8) + Caveolin-1 (3238), EHD2 (G-3) + Caveolin-1 (3238), EHD2 (11440-1-AP) + Caveolin-1 (7C8), PACSIN2 (SAB-1402538) + Caveolin-1 (3238), and PACSIN2 (Senju) + Caveolin-1 (7C8). These antibodies were visualized with secondary antibodies that were doubly labeled with Alexa 647 combined with Cy3 or Alexa 405. The activation of Alexa 405 or Cy3 by the excitation light was converted to the activation of Alexa 647, enabling the observation of the two labels at the same wavelength for Alexa 647. Therefore, the wavelength aberration was negligible between the observations of the two labels. The Alexa 647 signals that were associated with the activation Alexa 405 or Cy3 signals were considered for the analysis, to avoid the non-specific observations. To enable three-dimensional observations, a cylindrical lens was utilized to examine the signal depths by observing the deformation of the SMLM spherical signal according to the distance to the focal plane. The typical images reconstructed from the coordinates showed the proximity localizations of clustered signals of PACSIN2 and caveolin-1, as well as EHD2 and caveolin-1 (Fig 1).
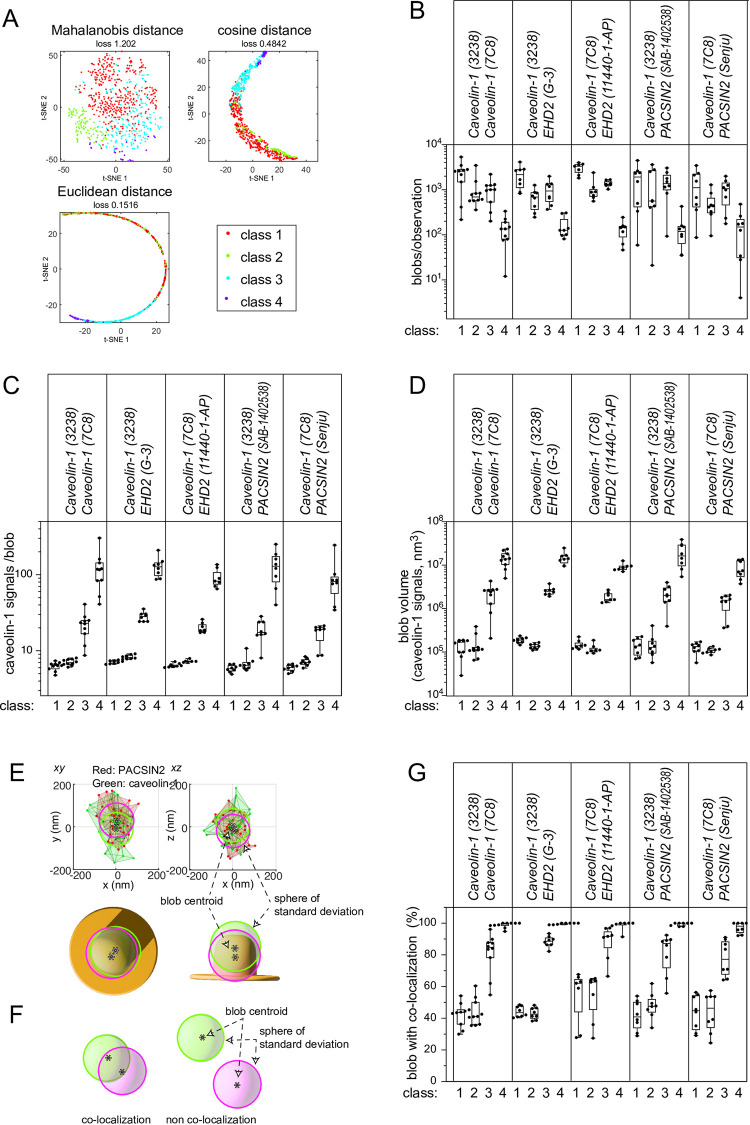
Fig 1. SMLM analyses of PACSIN2, EHD2, and caveolin-1.
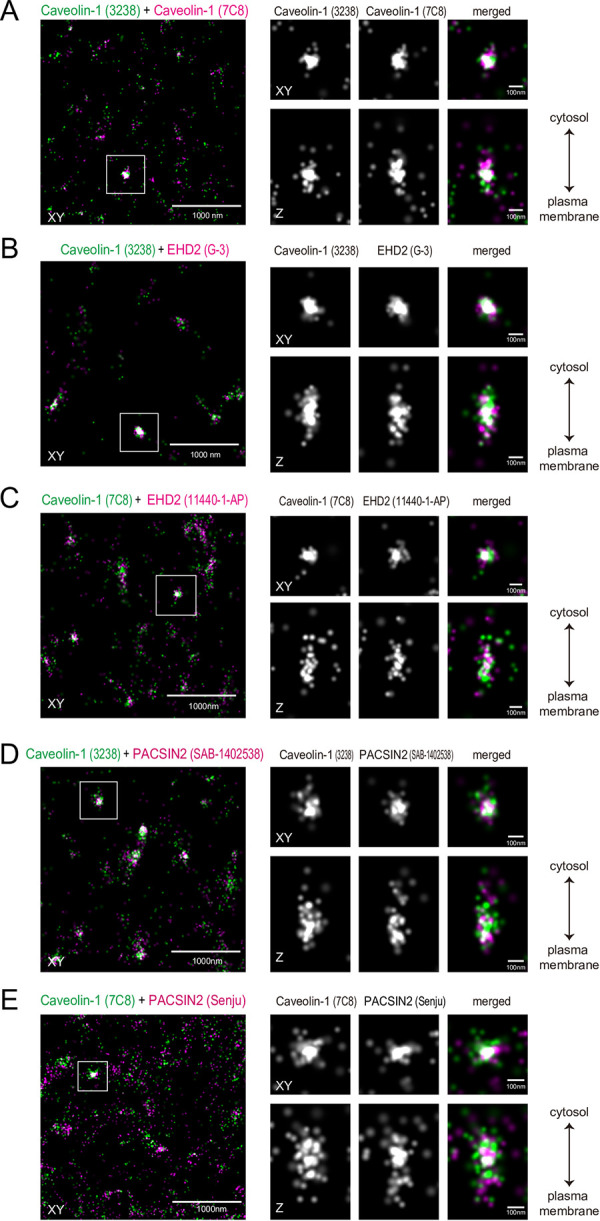
Representative reconstituted image of caveolin-1 (green) with those of PACSIN2 or EHD2 (magenta) in HeLa cells. The merged image of the focal plane (XY) is shown on the left. The box indicates the region for the enlarged images on the right, and contains one blob composed of caveolin-1 presumably corresponding to a caveola, with the projection to show the signal distribution to the Z-direction for each caveolin-1, PACSIN2, or EHD2 image. The combinations of antibodies are: (A) Caveolin-1 (7C8) + Caveolin-1 (3238), (B) Caveolin-1 (3238) + EHD2 (G-3), (C) Caveolin-1 (7C8) + EHD2 (11440-1-AP), (D) Caveolin-1 (3238) + PACSIN2 (SAB-1402538), and (E) Caveolin-1 (7C8) + PACSIN2 (Senju).
The single-molecule localizations of caveolin-1 within an 80 nm distance from each other were grouped into four classes by the SuperResNet software, according to the number of signals, area, shape, and so on [46,47]. The clustering below 80 nm was considered to be reasonable because caveolae are typically 60–100 nm in diameter [10,48], and the number of observations of caveolin-1 was 4–15 within 100 nm of caveolae with this antibody labeling, according to our previous estimations for caveolar shapes [42]. The spatial distributions of these groups of signals were mostly spherical, and hence they are called blobs. The mapping of the blobs by t-distributed stochastic neighbor embedding (t-SNE) by Mahalanobis distance, cosine distance, and Euclidean distance resulted in the clear separation of the blobs, suggesting robust classification (Fig 2A). The number of blobs was consistent between observations (Fig 2B). The estimated size of caveolin-1 blobs in each class was also similar between observations, suggesting the accurate grouping of the signals (S1 Fig). As reported in the development of this method [47], the class 1 and class 2 blobs contained fewer caveolin-1 localizations and smaller volumes, while the class 3 blobs had volumes equivalent to those of typical caveolae (Fig 2C and 2D). The class 3 blobs typically contained abundant ~20 caveolin-1 signals. The volumes of the class 3 blobs were approximately ten times larger than those of the class 1 and 2 blobs, and were similar to the volumes of 60–100 nm spheres, strongly suggesting that class 3 corresponded to typical caveolae. The class 4 blobs contained superabundant localizations of ~100 signals, and thus might represent caveolae rosettes or clusters [49,50].
Fig 2. Clustering of caveolin-1 into blobs and co-localization with PACSIN2 or EHD2.
(A) Typical clustering results of caveolin-1 signals of blobs from a cell into groups according to proximity by SuperResNet and their projections by t-SNE with Mahalanobis distance, cosine distance, and Euclidean distance. (B) The number of each class of caveolin-1 blobs per observation. (C) The average number of caveolin-1 signals from a blob of each class per observation. (D) The average volume of each class of caveolin-1 blobs per observation. In (B-D), the dot represents an average from an observation, which typically contained a cell. N = 6–10 observations for each combination of antibodies. (E) A caveolin-1 blob containing PACSIN2. Each signal of PACSIN2 and caveolin-1 is shown as a dot, and the signals within 80 nm of each other are connected by lines. The centroids of the caveolin-1 and PACSIN2 signals are illustrated by *, with spheres based on the radii of the standard deviations of caveolin-1 and PACSIN2. (F) The definition of colocalization by the overlap of the spheres of standard deviations. (G) Percentages of the colocalizations of blobs. The percentages of caveolin-1 blobs colocalized with PACSIN2 and EHD2 are shown for each antibody combination. The co-staining of two caveolin-1 antibodies indicated the blobs with two co-localized caveolin-1 antibodies.
To estimate the colocalization of the two antibody labels, the spatial distribution of signals was estimated by comparing the distance between the centroids of the two antibody blobs to the standard deviations of the signal coordinates of blobs (Fig 2E). The colocalization was determined by the overlap of the spatial distributions of the two kinds of signals; i.e., the centroid distance below the sum of each standard deviation (Fig 2F). Approximately half of the class 1 and class 2 blobs of caveolin-1 had signals from another antibody against caveolin-1, PACSIN2, and EHD2 (Fig 2G). Within each class, the blobs with colocalization had larger volumes and signals than those without colocalization (S2 Fig). Therefore, the number of molecules to be detected is related to the volume of the blobs and the number of observed signals, suggesting that the class 1 and 2 blobs might have an insufficient number of molecules for colocalization detection or might not have colocalization. Such difficulty in analyzing small structures would probably result from the antibody labeling, where only a small fraction of the proteins can be visualized, as has been shown that 1–8 signals per caveola by electron microscopic analysis by using various caveolin-1 antibodies [51]. Most of the class 3 and class 4 caveolin-1 blobs had the signals from another antibody to caveolin-1, PACSIN2, and EHD2, suggesting that caveolae with a typical size had PACSIN2 and EHD2.
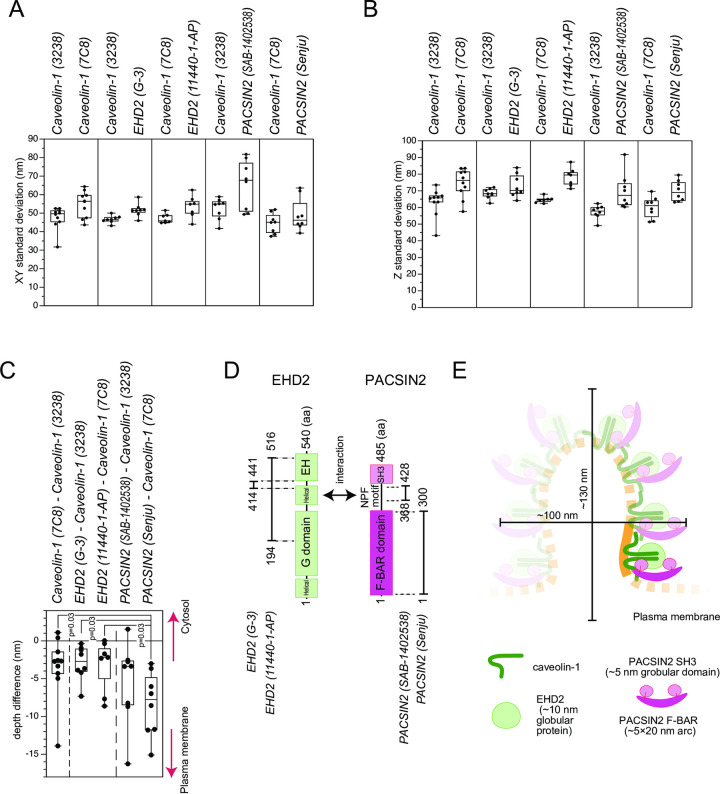
We next examined the spatial distribution of the signals; i.e., the shape, of the class 3 blobs. The standard deviations in the XY plane, parallel to the glass surface or plasma membrane, and the Z direction in depth were similar for the caveolin-1 signals between various antibody combinations (Fig 3A and 3B). The standard deviations of the PACSIN2 and EHD2 signals did not appear to be largely different from those of caveolin-1. Therefore, PACSIN2, EHD2, and caveolin-1 had largely overlapped spatial localizations in caveolin-1 blobs.
Fig 3. The geometry of colocalization in the blobs.
(A) Standard deviations of the blobs of each antibody staining in the XY plane, the plane parallel to the lens surface or coverslip of the cell attachment; i.e., the plasma membrane. (B) Standard deviations of the blobs of each antibody staining in the Z or depth direction. In (A, B), the pairs of antibodies used for co-staining are shown side-by-side. The dot represents an average from an observation, which typically contained a cell. N = 6–10 observations for each combination of antibodies. (C) The difference in the blob centroid depths of the indicated antibodies used for co-staining. A negative value indicates closer localization to the plasma membrane. The dot represents an average from an observation, which typically contained one cell. N = 6–10 observations for each combination of antibodies. The statistical difference in the depth of the PACSIN2 antibodies was evaluated by one-way ANOVA with post-hoc Holm-Bonferroni analysis and the p-values were indicated. (D) Domain diagrams of PACSIN2 and EHD2. The antigens for the antibodies, as well as the interaction sites between PACSIN2 and EHD2, are also illustrated. (E) A model of PACSIN2 F-BAR domain localization in caveolae. The PACSIN2 F-BAR domain, SH3 domain, EHD2, and caveolin-1 are illustrated in an approximate scale. The size of the caveola is obtained from ~2 times of the standard deviations. The PACSIN2 F-BAR domain is located close to the plasma membrane, but PACSIN2, EHD2, and caveolin-1 mostly overlap each other, resulting in the hypothetical unit of PACSIN2, EHD2, and presumably caveolin-1 in caveolae. The shaded part is hypothetical.
Next, we assessed the relative differences in the depth distributions by the distance of the centroid of caveolin-1 signals of the class 3 blobs to that of the associated PACSIN2 and EHD2 blobs. The negative distance in the z direction indicated a localization closer to the plasma membrane, i.e., the glass on which the cells were attached. The distances of the EHD2 centroids by the two kinds of antibodies to that of caveolin-1 were similar to each other and also to the centroid determined with another caveolin-1 antibody, indicating that the EHD2 localization was similar to that of caveolin-1 (Fig 3C). The monoclonal PACSIN2 antibody (SAB-1402538) recognizes the region before the SH3 domain of PACSIN2 that interacts with EHD2 (Fig 3D) [39]. The PACSIN2 centroid determined with the monoclonal antibody did not have a significant difference in depth from the EHD2 and caveolin-1 centroids (Fig 3C), which is consistent with the interaction of the region before the SH3 domain of PACSIN2 with EHD2. The polyclonal PACSIN2 antibody (Senju) recognizes the F-BAR domain, which binds to membrane [34]. Interestingly, the PACSIN2 centroid identified by this polyclonal antibody to the F-BAR domain was 5–10 nm closer to the plasma membrane than the EHD2 centroid (Fig 3C). The F-BAR domain of PACSIN2 is an arc-like rod of ~20 nm in length with a width of 5 nm [52]. The SH3 domains are globular domains with a diameter of ~5 nm [53], and EHD2 is also a globular protein of ~10 nm diameter [54]. Therefore, the PACSIN2 F-BAR domain was localized closer to the plasma membrane at a one protein distance, suggesting the role of PACSIN2 in connecting caveolae to the plasma membrane. Combined with the overall overlaps of PACSIN2, EHD2, and caveolin-1 localizations, these proteins would form a unit of PACSIN2, EHD2, and caveolin-1, in which the F-BAR domain of PACSIN2 faces the plasma membrane (Fig 3E).
This SMLM analysis suggested that PACSIN2 and EHD2 are localized in typical caveolae. We previously reported that PACSIN2 is localized to caveolae throughout the caveolar life cycle [32–34]. However, PACSIN2 appeared to tubulate membrane upon endocytosis and cholesterol depletion. The TIRF analysis indicated that PACSIN2 and caveolin-1 were co-localized, from the appearance of caveolin-1 at the plasma membrane, and the colocalization with dynamin occurred only at the last moment of endocytosis [40]. Upon cholesterol removal, PACSIN2 binding to the membrane was strengthened, supporting the production of membrane tubules, presumably for endocytosis [36]. Accordingly, caveolae with abundant cholesterol would exhibit a weaker affinity to PACSIN2, to prevent the elongation of caveolae for endocytosis. Therefore, the localizations of PACSIN2 and EHD2 at the entire caveola, with the exposure of PACSIN2 at the putative neck region, a boundary between the plasma membrane and the caveolar main body, would provide the potential for the extension of the caveola to such tubular structures upon endocytosis and cholesterol depletion.
Materials and methods
HeLa cell culture
HeLa cells were cultured as described previously [34] in Dulbecco’s modified Eagle’s medium (Nacalai), supplemented with 10% fetal calf serum, 63 μg/ml benzylpenicillin potassium, and 100 μg/ml streptomycin.
Antibodies
The anti-PACSIN2 rabbit polyclonal antibody (Senju) was affinity-purified from the serum of rabbits immunized with the F-BAR domain of PACSIN2 [34]. The anti-EHD2 rabbit polyclonal antibody (11440-1-AP, Proteintech), the mouse monoclonal anti-caveolin-1 (7C8, Santa Cruz Biotechnology, sc-53564, 1:100), the rabbit polyclonal anti-caveolin-1 (Cell Signaling, #3238, 1:200), the mouse monoclonal anti-PACSIN2 (Sigma, SAB-1402538, 1:100), and the mouse monoclonal anti-EHD2 (G-3, Santa Cruz, sc-515458, 1:100) antibodies were purchased.
STORM observation and analysis
The three-dimensional STORM setup with a cylindrical lens for depth measurement was purchased from Nikon and modified based on previous reports [42,44,45]. Dye preparation, secondary antibody labeling, and cell staining for STORM imaging (Nikon) were performed according to the manufacturer’s protocols, using combinations of Alexa Fluor 405 + Alexa Fluor 647 or Cy3 + Alexa Fluor 647 [44,45]. Alexa Fluor 405 and Cy3 are the activator dyes, and Alexa Fluor 647 is the reporter dye. The ratio of the activator dye: the reporter dye: antibody is 2–3: 0.6–1: 1. HeLa cells were cultured on Lab-Tek II chambered cover glasses (Nunc) that were pre-cleaned with 1M KOH for 1 hr. They were fixed in 3% paraformaldehyde (WAKO) + 0.1% glutaraldehyde (TAAB, electron microscopy grade) in HEPES-buffered saline (30 mM Hepes, pH 7.4, 100 mM NaCl, 2 mM CaCl2) for 10 min at room temperature, reduced with 0.1% NaBH4 in PBS for 7 min, and blocked in blocking buffer (3% BSA + 0.2% Triton X-100 in PBS) for 1 hr at room temperature. The cells were then stained with a 1:100 dilution of the primary antibodies in the blocking buffer for at least 1 hr at room temperature. After washing with wash buffer (0.05% Triton X-100, 0.2% BSA in PBS), the cells were incubated with secondary antibodies for 1 hr at room temperature and then washed. Finally, the cells were post-fixed with 3% paraformaldehyde + 1% glutaraldehyde in HEPES-buffered saline for 10 min at room temperature, and then stored in PBS at 4°C.
For image acquisition, the cells were soaked in 50 mM Tris-HCl (pH 8.0), 10 mM NaCl, and 10% glucose supplemented with cystamine, glucose oxidase, and catalase, according to the manufacturer’s instructions. An N-STORM (Nikon) super-resolution microscope equipped with a 100×/1.49 objective lens (Apo TIRF 100× Oil DIC N2, Nikon) and an EMCCD camera (iXon Du-897, ANDOR) was used for imaging. One image for the activation laser (405 or 561 nm) and three sequential images for the reporter laser (647 nm) were obtained for 10,000 cycles (total 40,000 images) and analyzed with the NIS-Elements AR 4.60.00 software provided by Nikon.
The signals upon the reporter laser irradiation following the observation by the activation laser irradiation were considered to be the specific signals and analyzed further. The coordinates that were at almost identical positions (<20 nm) in the continuous observations were eliminated, because the Alexa dye emits signals multiple times [55–57]. The coordinates were converted into the VISP format by the ChriSTORM ImageJ plugin [58–60], as described in the Supplementary Data 1 and 2. In all figures, each dot represents one signal. The clustering of the caveolin-1 signals was performed by SuperResNet [46,47], where >4 caveolin-1 signals within 80 nm, a value determined as a 20% smaller size of caveolin-1, were connected to a cluster. The clusters were divided into 4 classes according to the SuperResNet analysis of caveolin-1 [46,47], by the numbers of signals, shape parameters, and so on. Typically, thousands of blobs were identified per cell. The clusters of caveolin-1 signals exhibited blob structures. The volume of the class 3 blob corresponded to the volume of a sphere with a 60–100 nm diameter, a typical size of a caveola, which ranges from 0.9–4 × 106 nm3. The PACSIN2 and EHD2 signals were also grouped into clusters by SuperResNet. The overlaps of the signal distribution of the signals were considered for co-localization; i.e., the centroid distance below the sum of each standard deviation (Fig 2F). The center of mass of the clusters was considered for the depth difference. These calculations were performed with MATLAB. For each blob, the standard deviations in the XY and Z axes and the depth differences between the centroids of clusters of each antibody labeling were examined and then averaged per each observation, which typically contains a cell. The averages of the observations were then plotted in each Figure. In Fig 3C, the statistical evaluation was examined by One-way ANOVA with post-hoc Holm-Bonferroni analysis.
Supporting information
The average number of caveolin-1 blobs per observation, the XY standard deviation, and the Z standard deviation for each class and each combination of antibodies are shown. The dot represents an average from an observation, which typically contained one cell. N = 6–10 observations for each combination of antibodies. The combinations of antibodies are as follows: a: Caveolin-1 (7C8) + Caveolin-1 (3238), b: Caveolin-1 (3238) + EHD2 (G-3), c: Caveolin-1 (7C8) + EHD2 (11440-1-AP), d: Caveolin-1 (3238) + PACSIN2 (SAB-1402538), and e: Caveolin-1 (7C8) + PACSIN2 (Senju).
(PDF)
The volume of a caveolin-1 blob and the average number of caveolin-1 signals per blob per observation, obtained for each class for each combination of antibodies, shown with or without the colocalization of the two stains. The dot represents an average from an observation, which typically contained one cell. N = 6–10 observations for each combination of antibodies described in S1 Fig. w: Caveolin-1 blobs with the colocalization; wo: Caveolin-1 blobs without the colocalization.
(PDF)
(ZIP)
(ZIP)
Acknowledgments
We thank the laboratory members for fruitful discussions.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by JSPS (https://www.jsps.go.jp/english/)(KAKENHI, JP 20H03252, JP20KK0341, JP21H05047) to SS and by JSPS (JP20K06625) to TN, and JST (https://www.jst.go.jp/EN/) CREST (JPMJCR1863) to SS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Parton RG, McMahon K-A, Wu Y. Caveolae: Formation, dynamics, and function. Current Opinion in Cell Biology. 2020;65:8–16. doi: 10.1016/j.ceb.2020.02.001 [DOI] [PubMed] [Google Scholar]
- 2.Hubert M, Larsson E, Lundmark R. Keeping in touch with the membrane; protein- and lipid-mediated confinement of caveolae to the cell surface. Biochemical Society transactions. 2020;48(1):155–63. Epub 2020/02/13. doi: 10.1042/BST20190386 ; PubMed Central PMCID: PMC7054752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelkmans L, Zerial M. Kinase-regulated quantal assemblies and kiss-and-run recycling of caveolae. Nature. 2005;436(7047):128–33. doi: 10.1038/nature03866 [DOI] [PubMed] [Google Scholar]
- 4.Del Pozo MA, Lolo F-N, Echarri A. Caveolae: Mechanosensing and mechanotransduction devices linking membrane trafficking to mechanoadaptation. Current Opinion in Cell Biology. 2021;68:113–23. doi: 10.1016/j.ceb.2020.10.008 [DOI] [PubMed] [Google Scholar]
- 5.Nassoy P, Lamaze C. Stressing caveolae new role in cell mechanics. Trends in Cell Biology. 2012;22(7):381–9. doi: 10.1016/j.tcb.2012.04.007 [DOI] [PubMed] [Google Scholar]
- 6.Örtegren U, Karlsson M, Blazic N, Blomqvist M, Nystrom FH, Gustavsson J, et al. Lipids and glycosphingolipids in caveolae and surrounding plasma membrane of primary rat adipocytes. European journal of biochemistry. 2004;271(10):2028–36. Epub 2004/05/07. doi: 10.1111/j.1432-1033.2004.04117.x . [DOI] [PubMed] [Google Scholar]
- 7.Razani B, Woodman SE, Lisanti MP. Caveolae: from cell biology to animal physiology. Pharmacol Rev. 2002;54(3):431–67. Epub 2002/09/12. doi: 10.1124/pr.54.3.431 . [DOI] [PubMed] [Google Scholar]
- 8.Smart EJ, Graf GA, McNiven MA, Sessa WC, Engelman JA, Scherer PE, et al. Caveolins, liquid-ordered domains, and signal transduction. Mol Cell Biol. 1999;19(11):7289–304. Epub 1999/10/19. doi: 10.1128/MCB.19.11.7289 ; PubMed Central PMCID: PMC84723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gabor KA, Stevens CR, Pietraszewski MJ, Gould TJ, Shim J, Yoder JA, et al. Super resolution microscopy reveals that caveolin-1 is required for spatial organization of CRFB1 and subsequent antiviral signaling in zebrafish. PLoS One. 2013;8(7):e68759. Epub 20130709. doi: 10.1371/journal.pone.0068759 ; PubMed Central PMCID: PMC3706321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothberg KG, Heuser JE, Donzell WC, Ying Y-S, Glenney JR, Anderson RGW. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68(4):673–82. doi: 10.1016/0092-8674(92)90143-z [DOI] [PubMed] [Google Scholar]
- 11.Hill MM, Bastiani M, Luetterforst R, Kirkham M, Kirkham A, Nixon SJ, et al. PTRF-Cavin, a Conserved Cytoplasmic Protein Required for Caveola Formation and Function. Cell. 2008;132(1):113–24. doi: 10.1016/j.cell.2007.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Z, Scherer PE, Okamoto T, Song K, Chu C, Kohtz DS, et al. Molecular cloning of caveolin-3, a novel member of the caveolin gene family expressed predominantly in muscle. J Biol Chem. 1996;271(4):2255–61. doi: 10.1074/jbc.271.4.2255 . [DOI] [PubMed] [Google Scholar]
- 13.Way M, Parton RG. M-caveolin, a muscle-specific caveolin-related protein. FEBS Lett. 1995;376(1–2):108–12. doi: 10.1016/0014-5793(95)01256-7 . [DOI] [PubMed] [Google Scholar]
- 14.Minetti C, Sotgia F, Bruno C, Scartezzini P, Broda P, Bado M, et al. Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nat Genet. 1998;18(4):365–8. doi: 10.1038/ng0498-365 WOS:000072755500020. [DOI] [PubMed] [Google Scholar]
- 15.McNally EM, de Sa Moreira E, Duggan DJ, Bonnemann CG, Lisanti MP, Lidov HG, et al. Caveolin-3 in muscular dystrophy. Hum Mol Genet. 1998;7(5):871–7. Epub 1998/05/23. doi: 10.1093/hmg/7.5.871 . [DOI] [PubMed] [Google Scholar]
- 16.Sinha B, Koster D, Ruez R, Gonnord P, Bastiani M, Abankwa D, et al. Cells respond to mechanical stress by rapid disassembly of caveolae. Cell. 2011;144(3):402–13. Epub 2011/02/08. doi: 10.1016/j.cell.2010.12.031 ; PubMed Central PMCID: PMC3042189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu L, Pilch PF. A critical role of cavin (polymerase I and transcript release factor) in caveolae formation and organization. J Biol Chem. 2008;283(7):4314–22. Epub 20071203. doi: 10.1074/jbc.M707890200 . [DOI] [PubMed] [Google Scholar]
- 18.Bastiani M, Liu L, Hill MM, Jedrychowski MP, Nixon SJ, Lo HP, et al. MURC/Cavin-4 and cavin family members form tissue-specific caveolar complexes. J Cell Biol. 2009;185(7):1259–73. Epub 20090622. doi: 10.1083/jcb.200903053 ; PubMed Central PMCID: PMC2712963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen CG, Bright NA, Howard G, Nichols BJ. SDPR induces membrane curvature and functions in the formation of caveolae. Nat Cell Biol. 2009;11(7):807–14. Epub 20090614. doi: 10.1038/ncb1887 ; PubMed Central PMCID: PMC2712677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMahon KA, Zajicek H, Li WP, Peyton MJ, Minna JD, Hernandez VJ, et al. SRBC/cavin-3 is a caveolin adapter protein that regulates caveolae function. Embo j. 2009;28(8):1001–15. Epub 20090305. doi: 10.1038/emboj.2009.46 ; PubMed Central PMCID: PMC2683698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohan J, Morén B, Larsson E, Holst MR, Lundmark R. Cavin3 interacts with cavin1 and caveolin1 to increase surface dynamics of caveolae. Journal of Cell Biology. 2015;128(5):979–91. doi: 10.1242/jcs.161463 [DOI] [PubMed] [Google Scholar]
- 22.Stoeber M, Schellenberger P, Siebert CA, Leyrat C, Helenius A, Grunewald K. Model for the architecture of caveolae based on a flexible, net-like assembly of Cavin1 and Caveolin discs. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(50):E8069–E78. Epub 2016/11/12. doi: 10.1073/pnas.1616838113 ; PubMed Central PMCID: PMC5167190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Ariotti N, Rae J, Liang H, Tillu V, Tee S, et al. Caveolin-1 and cavin1 act synergistically to generate a unique lipid environment in caveolae. Journal of Cell Biology. 2021;220(3):e202005138. doi: 10.1083/jcb.202005138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovtun O, Tillu VA, Jung W, Leneva N, Ariotti N, Chaudhary N, et al. Structural insights into the organization of the cavin membrane coat complex. Dev Cell. 2014;31(4):405–19. Epub 2014/12/03. doi: 10.1016/j.devcel.2014.10.002 . [DOI] [PubMed] [Google Scholar]
- 25.Henley JR, Krueger EW, Oswald BJ, McNiven MA. Dynamin-mediated internalization of caveolae. J Cell Biol. 1998;141(1):85–99. Epub 1998/05/16. doi: 10.1083/jcb.141.1.85 ; PubMed Central PMCID: PMC2132718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simunovic M, Evergren E, Callan-Jones A, Bassereau P. Curving Cells Inside and Out: Roles of BAR Domain Proteins in Membrane Shaping and Its Cellular Implications. Annu Rev Cell Dev Biol. 2019;35:111–29. Epub 2019/07/25. doi: 10.1146/annurev-cellbio-100617-060558 . [DOI] [PubMed] [Google Scholar]
- 27.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438(7068):590–6. Epub 2005/12/02. doi: 10.1038/nature04396 . [DOI] [PubMed] [Google Scholar]
- 28.Suetsugu S, Gautreau A. Synergistic BAR-NPF interactions in actin-driven membrane remodeling. Trends Cell Biol. 2012;22(3):141–50. Epub 2012/02/07. doi: 10.1016/j.tcb.2012.01.001 . [DOI] [PubMed] [Google Scholar]
- 29.Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nature reviews Molecular cell biology. 2012;13(2):75–88. Epub 2012/01/12. doi: 10.1038/nrm3266 ; PubMed Central PMCID: PMC3519936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suetsugu S, Toyooka K, Senju Y. Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin Cell Dev Biol. 2010;21(4):340–9. Epub 2009/12/08. doi: 10.1016/j.semcdb.2009.12.002 . [DOI] [PubMed] [Google Scholar]
- 31.Daumke O, Roux A, Haucke V. BAR domain scaffolds in dynamin-mediated membrane fission. Cell. 2014;156(5):882–92. Epub 2014/03/04. doi: 10.1016/j.cell.2014.02.017 . [DOI] [PubMed] [Google Scholar]
- 32.Hansen CG, Howard G, Nichols BJ. Pacsin 2 is recruited to caveolae and functions in caveolar biogenesis. Journal of cell science. 2011;124(Pt 16):2777–85. Epub 2011/08/03. doi: 10.1242/jcs.084319 . [DOI] [PubMed] [Google Scholar]
- 33.Koch D, Westermann M, Kessels MM, Qualmann B. Ultrastructural freeze-fracture immunolabeling identifies plasma membrane-localized syndapin II as a crucial factor in shaping caveolae. Histochemistry and cell biology. 2012;138(2):215–30. Epub 2012/06/22. doi: 10.1007/s00418-012-0945-0 . [DOI] [PubMed] [Google Scholar]
- 34.Senju Y, Itoh Y, Takano K, Hamada S, Suetsugu S. Essential role of PACSIN2/syndapin-II in caveolae membrane sculpting. Journal of cell science. 2011;124(12):2032–40. doi: 10.1242/jcs.086264 WOS:000291048000009. [DOI] [PubMed] [Google Scholar]
- 35.Seemann E, Sun M, Krueger S, Tröger J, Hou W, Haag N, et al. Deciphering caveolar functions by syndapin III KO-mediated impairment of caveolar invagination. Elife. 2017;6. Epub 2017/12/06. doi: 10.7554/eLife.29854 ; PubMed Central PMCID: PMC5716666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gusmira A, Takemura K, Lee SY, Inaba T, Hanawa-Suetsugu K, Oono-Yakura K, et al. Regulation of caveolae through cholesterol-depletion-dependent tubulation mediated by PACSIN2. Journal of cell science. 2020;133(19). doi: 10.1242/jcs.246785 WOS:000581511400012. [DOI] [PubMed] [Google Scholar]
- 37.Ludwig A, Howard G, Mendoza-Topaz C, Deerinck T, Mackey M, Sandin S, et al. Molecular composition and ultrastructure of the caveolar coat complex. PLOS Biology. 2013;11(8):e1001640. Epub 2013/09/10. doi: 10.1371/journal.pbio.1001640 ; PubMed Central PMCID: PMC3754886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoeber M, Stoeck IK, Hänni C, Bleck CKE, Balistreri G, Helenius A. Oligomers of the ATPase EHD2 confine caveolae to the plasma membrane through association with actin. Embo Journal. 2012;31(10):2350–64. doi: 10.1038/emboj.2012.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morén B, Shah C, Howes MT, Schieber NL, McMahon HT, Parton RG, et al. EHD2 regulates caveolar dynamics via ATP-driven targeting and oligomerization. Molecular Biology of the Cell. 2012;23(7):1316–29. doi: 10.1091/mbc.E11-09-0787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Senju Y, Rosenbaum E, Shah C, Hamada-Nakahara S, Itoh Y, Yamamoto K, et al. Phosphorylation of PACSIN2 by protein kinase C triggers the removal of caveolae from the plasma membrane. Journal of cell science. 2015;128(15):2766–80. doi: 10.1242/jcs.167775 WOS:000359782100005. [DOI] [PubMed] [Google Scholar]
- 41.Walser Piers J, Ariotti N, Howes M, Ferguson C, Webb R, Schwudke D, et al. Constitutive Formation of Caveolae in a Bacterium. Cell. 2012;150(4):752–63. doi: 10.1016/j.cell.2012.06.042 [DOI] [PubMed] [Google Scholar]
- 42.Tachikawa M, Morone N, Senju Y, Sugiura T, Hanawa-Suetsugu K, Mochizuki A, et al. Measurement of caveolin-1 densities in the cell membrane for quantification of caveolar deformation after exposure to hypotonic membrane tension. Sci Rep. 2017;7(1):7794. Epub 2017/08/12. doi: 10.1038/s41598-017-08259-5 ; PubMed Central PMCID: PMC5552771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang L, Scarlata S. Super-resolution Visualization of Caveola Deformation in Response to Osmotic Stress. J Biol Chem. 2017;292(9):3779–88. Epub 20170117. doi: 10.1074/jbc.M116.768499 ; PubMed Central PMCID: PMC5339760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317(5845):1749–53. Epub 2007/08/19. doi: 10.1126/science.1146598 ; PubMed Central PMCID: PMC2633025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319(5864):810–3. Epub 2008/01/05. doi: 10.1126/science.1153529 ; PubMed Central PMCID: PMC2633023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khater IM, Liu Q, Chou KC, Hamarneh G, Nabi IR. Super-resolution modularity analysis shows polyhedral caveolin-1 oligomers combine to form scaffolds and caveolae. Sci Rep. 2019;9(1):9888. Epub 2019/07/10. doi: 10.1038/s41598-019-46174-z ; PubMed Central PMCID: PMC6614455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khater IM, Meng F, Wong TH, Nabi IR, Hamarneh G. Super Resolution Network Analysis Defines the Molecular Architecture of Caveolae and Caveolin-1 Scaffolds. Sci Rep. 2018;8(1):9009. Epub 2018/06/15. doi: 10.1038/s41598-018-27216-4 ; PubMed Central PMCID: PMC5998020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parton RG, Simons K. The multiple faces of caveolae. Nature reviews Molecular cell biology. 2007;8(3):185–94. Epub 2007/02/24. doi: 10.1038/nrm2122 . [DOI] [PubMed] [Google Scholar]
- 49.Del Pozo MA, Lolo FN, Echarri A. Caveolae: Mechanosensing and mechanotransduction devices linking membrane trafficking to mechanoadaptation. Curr Opin Cell Biol. 2021;68:113–23. Epub 20201111. doi: 10.1016/j.ceb.2020.10.008 . [DOI] [PubMed] [Google Scholar]
- 50.Sinha B, Köster D, Ruez R, Gonnord P, Bastiani M, Abankwa D, et al. Cells respond to mechanical stress by rapid disassembly of caveolae. Cell. 2011;144(3):402–13. doi: 10.1016/j.cell.2010.12.031 ; PubMed Central PMCID: PMC3042189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujimoto T, Kogo H, Nomura R, Une T. Isoforms of caveolin-1 and caveolar structure. Journal of cell science. 2000;113(19):3509–17. doi: 10.1242/jcs.113.19.3509 [DOI] [PubMed] [Google Scholar]
- 52.Shimada A, Takano K, Shirouzu M, Hanawa-Suetsugu K, Terada T, Toyooka K, et al. Mapping of the basic amino-acid residues responsible for tubulation and cellular protrusion by the EFC/F-BAR domain of pacsin2/Syndapin II. FEBS Lett. 2010;584(6):1111–8. Epub 2010/03/02. doi: 10.1016/j.febslet.2010.02.058 . [DOI] [PubMed] [Google Scholar]
- 53.Rao Y, Ma Q, Vahedi-Faridi A, Sundborger A, Pechstein A, Puchkov D, et al. Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation. Proc Natl Acad Sci U S A. 2010;107(18):8213–8. Epub 2010/04/21. doi: 10.1073/pnas.1003478107 ; PubMed Central PMCID: PMC2889545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoernke M, Mohan J, Larsson E, Blomberg J, Kahra D, Westenhoff S, et al. EHD2 restrains dynamics of caveolae by an ATP-dependent, membrane-bound, open conformation. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(22):E4360–E9. doi: 10.1073/pnas.1614066114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sengupta P, Jovanovic-Talisman T, Skoko D, Renz M, Veatch SL, Lippincott-Schwartz J. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat Methods. 2011;8(11):969–75. Epub 20110918. doi: 10.1038/nmeth.1704 ; PubMed Central PMCID: PMC3400087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fricke F, Beaudouin J, Eils R, Heilemann M. One, two or three? Probing the stoichiometry of membrane proteins by single-molecule localization microscopy. Sci Rep. 2015;5:14072. Epub 20150911. doi: 10.1038/srep14072 ; PubMed Central PMCID: PMC4642553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Annibale P, Vanni S, Scarselli M, Rothlisberger U, Radenovic A. Quantitative photo activated localization microscopy: unraveling the effects of photoblinking. PLoS One. 2011;6(7):e22678. Epub 20110726. doi: 10.1371/journal.pone.0022678 ; PubMed Central PMCID: PMC3144238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leterrier C, Potier J, Caillol G, Debarnot C, Rueda Boroni F, Dargent B. Nanoscale Architecture of the Axon Initial Segment Reveals an Organized and Robust Scaffold. Cell Rep. 2015;13(12):2781–93. Epub 20151217. doi: 10.1016/j.celrep.2015.11.051 . [DOI] [PubMed] [Google Scholar]
- 59.Ovesný M, Křížek P, Borkovec J, Svindrych Z, Hagen GM. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30(16):2389–90. Epub 20140425. doi: 10.1093/bioinformatics/btu202 ; PubMed Central PMCID: PMC4207427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.El Beheiry M, Dahan M. ViSP: representing single-particle localizations in three dimensions. Nat Methods. 2013;10(8):689–90. doi: 10.1038/nmeth.2566 . [DOI] [PubMed] [Google Scholar]