Abstract
COVID-19 infection is associated with a significant fatality rate in individuals suffering from severe acute respiratory distress syndrome (ARDS). Among the several possibilities, inhibition of hypoxia-inducible factor prolyl hydroxylase-2 or prolyl hydroxylase domain-containing protein 2 (PHD2) in a hypoxia-independent way is a prospective therapeutic target for the treatment of ARDS. Vadadustat, Roxadustat, Daprodustat, Desidustat, and Enarudustat are the available clinical trial inhibitors. This study is proposed to focus on the repurposing of FDA-approved drugs as effective PHD2 inhibitors. This computational study utilises e-pharmacophore hypothesis generation from the native ligand–protein complex (PDB ID: 5OX6) based on XP visualiser information. The hypothesis containing five essential features (AAANR) was incorporated for FDA database screening, followed by Glide XP molecular docking and Prime MM-GBSA binding free energy calculations. Top scored ligands were investigated and Fenbufen was identified as an effective PHD-2 inhibitor by comparing with the native co-crystal ligand (Vadadustat). The manual lead optimisation of the Fenbufen structure was adopted to improve inhibitory potency, by increasing the binding affinity and protein–ligand stability. The newly designed compounds B and C showed additional binding interactions, excellent docking scores, binding free energy, and an acceptable range of ADME properties. Also, Fenbufen and compound C owned preferable protein–ligand stability during MD simulation when compared with the co-crystallised clinical trial ligand. Based on our findings, we deduce that Fenbufen can be proposed as an effective repurposable candidate as its structural modification showed a remarkable improvement in PHD2 inhibition.
Graphical abstract
Supplementary information
The online version contains supplementary material available at 10.1007/s11224-022-02012-z.
Keywords: COVID-19, Acute respiratory distress syndrome (ARDS), Hypoxia-inducible factor prolyl hydroxylase-2 (PHD-2) inhibitors, Fenbufen, e-Pharmacophore modelling, Prime MM-GBSA, ADME prediction, MD simulations
Introduction
Acute respiratory distress syndrome (ARDS) is a severe acute lung injury [ALI] manifestation that is characterised by the sudden development of substantial hypoxemia and pulmonary infiltrates [1]. Increased pulmonary vascular permeability, epithelial cell destruction, and the start of oedema are all related to ARDS [2]. The significant fatality rate in COVID-19 patients is associated with ARDS [3]. The proportion of COVID-19 patients diagnosed with ARDS ranges from 20 to 67%, with patients on mechanical ventilation accounting for 100% [4]. Endothelial cells serve as an important barrier in controlling compartmentalisation between the vascular and interstitial regions, according to cellular physiology. In ARDS, a breakdown of endothelial barrier integrity allows for significant fluid leakage from the vascular bed into the air space [5]. Mechanical and protective mechanical ventilation ensures vascular resistance and reduces alveolar fluid loss [6]. Endothelial damage may be exacerbated by the degree of alveolar pressure and ventilator variability [7]. Because ARDS has a significant mortality rate, new treatments are required. Currently, only a few pharmaceutical therapies are being studied to prevent endothelial cell injury and fluid extravasation into the alveolar space [8].
A molecular pathway study of endothelial barrier function decline identifies prolyl hydroxylase domain-2 (PHD2) as a key participant. When PHD2 is inactivated in a hypoxia-independent way, HIF-2 is activated, which increases the production of vascular endothelial protein tyrosine phosphatase (VEPTP), which promotes the dephosphorylation of VE-cadherin, hence promoting adherent junction integrity and barrier function. Furthermore, because the VE-cadherin gene is not sensitive to hypoxia [9], it is possible that inhibiting PHD2 in a hypoxia-independent way may tighten adherent junction integrity and prevent the loss of barrier function.
Japan has begun a clinical study for COVID-19-associated ARDS utilising the PHD2 inhibitor Vadadustat [10]. Based on the aforementioned collected knowledge, we used in-silico repurposing methodologies for FDA-approved medications in this work to find an effective PHD2 inhibitor.
Materials and methods
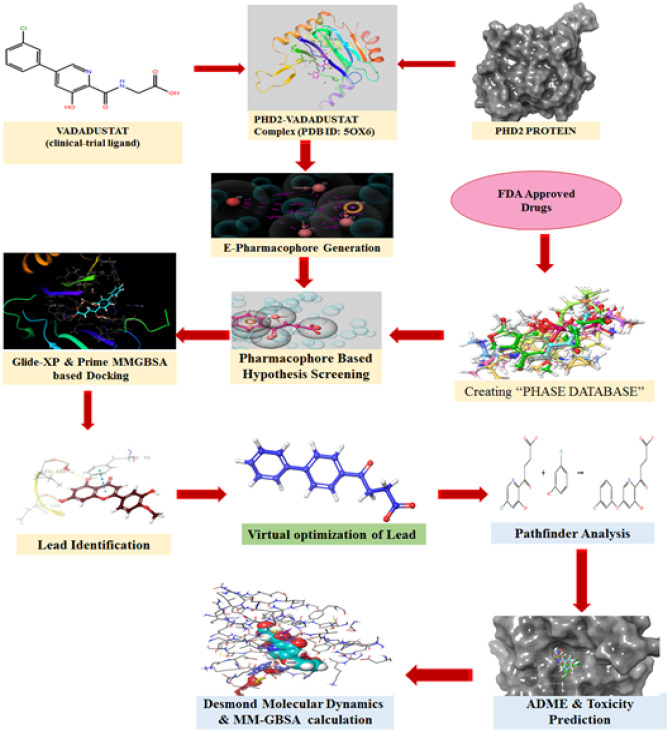
In silico simulations of protein preparation, ligand preparation, grid generation, molecular docking, and molecular mechanics were carried out in the Maestro11.9.011modeling package provided by Schrödinger, LLC, New York, NY, 2019–1, installed in a Dell OptiPlex 3060 with a processor Intel Core i7-8700, Ubuntu 16.04 LTS, graphics GeForce GT 730/PCle/SSE2, and a 64-bit operating system energetically optimised structure-based pharmacophore generation and molecular dynamics were carried out in the Maestro12.4modeling package provided by Schrödinger, LLC, New York, NY, 2020–2, installed in a Dell Precision 7820 with an Intel Xeon(R) Gold 6130 processor, Kernel GNOME Version 3.28.2CentOS Linux 7, Graphics Quadro P5000/PCle/SSE2, and 64-bit OS.
The energetically optimised structure-based pharmacophore (e-pharmacophore), is both a ligand and structure-based approach to obtain structurally diverse active ligands from the FDA-approved drug dataset. The 3-dimensional crystal structure of human PHD2 protein (PDB ID: 5OX6) co-crystallised with a potent inhibitor (Vadadustat) was obtained from the Protein Data Bank (PDB). Simultaneously, the list of FDA-approved drug candidates was collected from the Drugbank database. Initially, the obtained raw protein (5OX6) was pre-processed and energetically minimised with the assistance of the OPLS3e force field to get a refined protein structure using the “Protein Preparation Wizard” panel of GLIDEv7.7 Module provided by the Schrodinger suite (Schrödinger, LLC, New York, NY, 2019–1).
In addition, the reference inhibitor Vadadustat was extracted and produced from the co-crystallised protein using GLIDEv7.7’s “Ligprep” settings (Schrödinger, LLC, New York, NY, 2019–1). The generated ligands were then re-docked in Extra precision mode (XP-docking algorithm) with previously prepared PHD2 protein and the top-ranking docked posture was recovered for further hypothesis creation. The docking methodology was confirmed by computing the RMSD values by superimposing reference ligand (Vadadustat) re-docked conformations with the original co-crystallised conformation of a ligand. The reward and penalty areas of the reference ligand were visualised using the XP visualiser programme in the GLIDEv7.7 module (Schrödinger, LLC, New York, NY, 2019–1).
The fundamental chemical properties necessary for the creation of e-pharmacophore hypotheses were chosen based on the Glide XP descriptor information. Using the PHASE v3.8 module of the Schrodinger suite (Schrödinger, LLC, New York, NY, 2020–2), a customised e-pharmacophore hypothesis including five critical chemical characteristics (AAARN) that enhance ligand binding was constructed using accessible protein–ligand complexes. The PHASE v3.8 module was used to generate a phase database for database screening of the previously stated FDA medications (Schrödinger, LLC, New York, NY, 2019–1). Using the phase database, the derived pharmacophore hypothesis was efficiently evaluated for shape-based similarity. This hypothesis screening allows the selection of ligands from the phase database comprising a list of FDA-authorised medications that have comparable chemical properties to the reference ligand.
To identify the potentially interacting drug candidates from the database, the compounds obtained after database screening were docked with the active pocket of the minimised protein (5OX6) using the same XP docking mode (GLIDEv7.7) as that of the reference ligand. The compounds which possessed better docking scores and protein interaction were selected for further analysis of binding free energy calculation, pharmacokinetic & toxicity profile, and protein–ligand stability predictions. MM-GBSA tool in the Prime v3.5 module (Maestro11.9.011, Schrödinger, LLC, New York, NY, 2019–1) was utilised to calculate the binding free energy of selected ligands [11].
Experimentally, more relevant absorption, distribution, metabolism, and excretion properties of selected drug candidates were predicted theoretically using QikProp v5.4, a more precise rapid software package incorporated in the Schrodinger suite (Maestro11.9.011, Schrödinger, LLC, New York, NY, 2019–1). Additionally, Qikprop was used to check Lipinski’s rule-of-five and Jorgensen’s rule-of-three infringements to determine whether a compound is drug-like [12].
Molecular dynamics
The ligands scrutinised from the above interpretations were subjected to analyse the stability, RMSD, RMSF, and interaction from the protein–ligand complex in a more relevant physiological environment by running MD simulations using the OPLS_2005 force field in DESMOND V5.2 module of Schrodinger suite (Schrödinger, LLC, New York, NY, 2020–2) [13]. Firstly, the system builder was constructed by placing the protein–ligand complex inside the orthorhombic boundary box containing TIP3P water as a solvent model. The protein was again minimised and recalculated to add more suitable counterions to neutralise the charges of the system. Additionally, 0.15 M NaCl salt concentration was fixed in the system, 20 Å away from the ligand to simulate background salt at the physiological condition. The above-prepared system was incorporated into the workspace to start an MD simulation. Molecular dynamics were run with an NPT ensemble at 300-K temperature and 1.01325 bar atmospheric pressure for 100 ns using the OPLS_2005 force field. The structure frames computed for every 50 ps were saved in trajectory during molecular dynamics; a total of 1000 frames were saved. The generated MD results were further analysed through the simulation interaction analysis tool in the DESMOND V5.2 module [14, 15].
Results and discussions
e-Pharmacophore modelling
Pharmacophore modelling is an integral process in lead discovery and optimisation [16]. Among, three different pharmacophore modelling approaches, e-pharmacophore serves as a potential tool in retrieving more diverse actives than conventional ligand and structure-based pharmacophore modelling approaches [17]. In addition, it also allows excluded volume occupied by the receptor and utilises Glide XP scoring functions to accurately characterise the ligand–protein interaction.
Before developing an e-pharmacophore hypothesis for PHD2 inhibitors, the binding site analysis and the interaction patterns of clinical trial candidates (Vadadustat, Roxadustat, Daprodustat, and Enarodustat) with amino acid residues of PHD2 receptor were closely examined (Fig S1). The binding site analysis showed more conserved amino acid residues at Tyr-303, Tyr-329, Arg-383, and Tyr-310. The metal ion Fe2+ is essential for ligand binding and it acts as a co-factor and mediates metal co-ordinations with His-313, His-374, and Asp-315 amino acid residues. It was also inferred that salt bridge interaction with Arg-383 and H-bond interactions at Asp-254, Tyr-303, Tyr-329, and Arg-383 are important contributing interactions with inhibitors in a clinical trial.
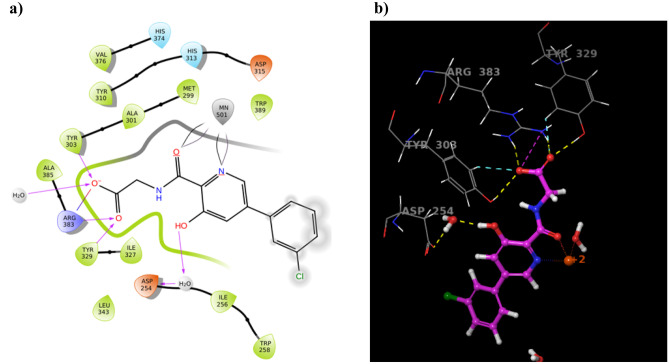
For an e-pharmacophore generation, 3D coordinates of X-ray crystallographic structure of PHD2 with Vadadustat (PDB ID: 5OX6) were selected and subjected to Protein Preparation Wizard. The native interactions were closely analysed by using “XP visualiser” to identify the key features, essential for rewards and penalties for the effective generation of hypotheses. The features essential for inhibitor activity for Vadadustat: (a) the carboxyl group represented hydrophobically packed correlated H-bonds withTyr-303, Tyr-329, and Arg-383 amino acid residues, (b) H-bonding with Asp-254 amino acid residue, (c) presence of two-electron negative groups for interaction with Fe2+ ion, (d) Aromatic hydrophobic enclosure reward helps in stabilisation of ligand binding pocket (Fig. 1). Based on these collective insights, the five features of pharmacophore (three hydrogen bond acceptors (AAA), one negatively charged group (N), and one aromatic ring (A)) were selected using the “Phase module” (Fig. 2).
Fig. 1.
The native two-dimensional (a) and three-dimensional (b) interactions of clinical trial ligand (Vadadustat) after XP molecular docking with PHD2 Protein (PDB: 50X6)
Fig. 2.
Generation of e-pharmacophore hypothesis for PHD2 inhibitors. The five features were mainly selected from the Vadadustat compound. The features A1, A2, and A5 (Pink spheres) are H-bond acceptors, N9 (Red spheres) for a negatively charged ionisable group, and R10 (orange torus) for an aromatic ring
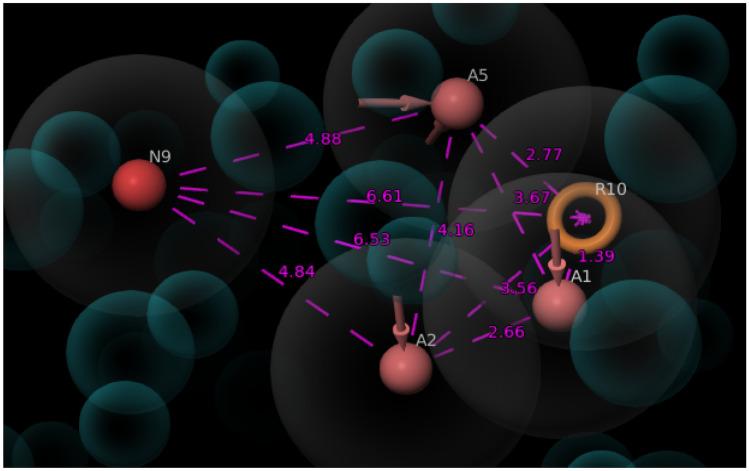
The protein–ligand complex was imported and manual selection mode was opted for creating receptor-based excluded volume cell by keeping radii size and radii scoring function as default features. The receptor atom surface with 2 Å are of ligand surface was ignored and the shell thickness was set as 5 Å. The generated e-pharmacophore hypothesis (Fig. 3) was used for further database screening.
Fig. 3.
Three-dimensional (3D) spatial arrangement of selected features (AAANR) with intersite distances. The intersite distances between the selected features are expressed in angstrom (Å)
Database screening
For hypothesis screening, 2D structures of the FDA-approved drugs from “Drug Bank” was downloaded and subjected to “Phase Database Creation”. We created a phase database for the obtained FDA drugs. During database creation, the ligands were subjected to its preparation and filtration. In ligand preparation, the essential ligand protonation states were generated at PH of 7.0 ± 2.0 using Epik including metal binding states. In ligand filtration, the prepared ligands were filtered based on the Qikprop (ADME) properties, Lipinski’s rule, and reactive fictional groups. The desired ligands obtained from the above database creation were incorporated for pharmacophore hypothesis screening by fixing a minimum of 3 pharmacophore features as matching criteria. The structurally diverse 596 FDA drugs having features similar to that of Vadadustat were obtained which shows that the generated pharmacophore model has the ability to retrieve compounds that inhibit the PHD2 enzyme.
Molecular docking
Initially. the accuracy of the docking programme was examined by calculating the RMSD value of the superimposition of re-docked (Vadadustat) ligands (Fig. 4). The RMSD value was found to be less than 1 (0.9816) which confirmed that further molecular docking could predict a better intermolecular framework between the prepared PHD2 protein (5OX6) and screened FDA drugs.
Fig. 4.
Validation of the docking protocol by re-docking the Vadadustat conformation and calculating the RMSD. a Superimposed pose of Vadadustat conformations with native structure. b The active binding pocket of PHD2 protein occupied by the Vadadustat conformations
Molecular docking was performed to differentiate actives and in-actives from the above obtained 596 screened ligands. It was carried out in Extra Precision Mode (XP-docking) which provided a more sophisticated scoring function and weeded out false positives more effectively. As a result of XPdocking, 126 active ligands were found to be docked with the active binding site of previously minimised PHD2 protein. Then, scoring and binding interactions of the top 10 ligands were analysed manually (Table 1).
Table 1.
Docking results of top-ranked 10 ligands of FDA-approved drugs for Hit identification and drug repurposing
| Compound name | Docking score (kcal/mol) | Glide g score | MM-GBSA binding score (kcal/mol) | Amino acid residue | Nature of interaction |
|---|---|---|---|---|---|
| Mycophenolic acid | −13.743 | −13.823 | −8.89 | Tyr-303 | H-bond |
| Arg-383 | H-bond, salt bridge | ||||
| Tyr-329 | H-bond | ||||
| Dinoprostone | −13.319 | −13.319 | −18.46 | Tyr-303 | H-bond |
| Arg-383 | H-bond, salt bridge | ||||
| Tyr-329 | H-bond | ||||
| Asp-315 | H-bond | ||||
| Fenbufen | −11.764 | −11.764 | −20.64 | Fe2+ | Metal co-ordination |
| Tyr-303 | H-bond | ||||
| Tyr-329 | H-bond | ||||
| Arg-383 | H-bond, salt bridge | ||||
| His-313 | Pi-Pi stacking | ||||
| Amino hippuric acid | −11.629 | −11.629 | −6.00 | Tyr-303 | H-bond |
| Tyr-329 | H-bond | ||||
| Arg-383 | H-bond, salt bridge | ||||
| His-313 | Pi-Pi stacking | ||||
| Calcium gluconate | −11.202 | −11.202 | 11.02 | Fe2+ | Metal co-ordination |
| Tyr-303 | H-bond | ||||
| Tyr-329 | H-bond | ||||
| Arg-383 | H-bond, salt bridge | ||||
| Pyridoxine | −8.774 | −8.774 | −8.42 | Fe2+ | Metal co-ordination |
| Asp-254 | salt bridge | ||||
| Sodium glycerophosphate | −8.768 | −8.811 | 12.16 | Fe2+ | Metal co-ordination |
| Tyr-303 | H-bond | ||||
| Tyr-329 | H-bond | ||||
| Arg-383 | H-bond, salt bridge | ||||
| Gemfibrozil | −7.730 | −7.730 | −12.74 | Fe2+ | Metal co-ordination |
| Tyr-310 | H-bond | ||||
| Bromfenac | −7.528 | −7.528 | −24.60 | Fe2+ | Metal co-ordination |
| Arg-322 | Pi-cation | ||||
| Trp-258 | Pi-Pi stacking | ||||
| Vilanterol | −7.033 | −7.033 | −37.97 | Arg-322 | H-bond |
| Asp-254 | H-bond | ||||
| Gln-239 | H-bond | ||||
| Leu-240 | H-bond | ||||
| Tyr-310 | Pi-cation |
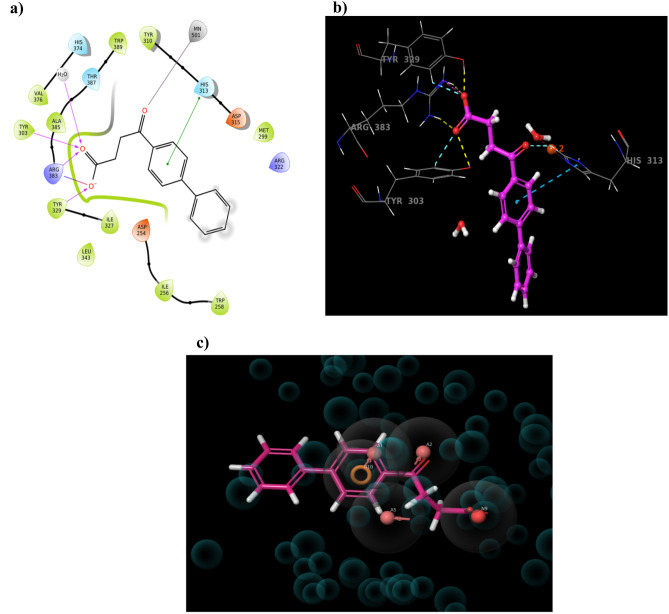
After visual inspection, it was found that the drug Fenbufen possessed a better docking score (−11.764 kcal/mol). Fenbufen is a non-steroidal anti-inflammatory drug that acts by preventing cyclooxygenase from producing prostaglandins which can cause inflammation. It exhibited the most favourable protein interactions−the oxygen functionality of Fenbufen showed metal co-ordination with Fe2+ ion, the end carboxylic acid group made H-bond interaction with Tyr-303, Tyr-329, and Arg-383and also salt bridge interaction with Arg-383. Additionally, the aromatic ring of Fenbufen formed Pi-Pi staking interaction with the imidazole ring of His-313 amino acid residue (Fig. 5). Based on the knowledge of existing PHD2 inhibitors and active site requirements we assumed that repositioning of Fenbufen may produce better inhibition of the PHD2 enzyme.
Fig. 5.
Extra precision (XP) Docking results of PHD2 protein with Fenbufen compound. a, b Represent the 2D and 3D interaction pattern of Fenbufen with PHD2 protein respectively. c Represents the Fenbufen aligned with the generated hypothesis
Manual lead optimisation and ADME/toxicity analysis
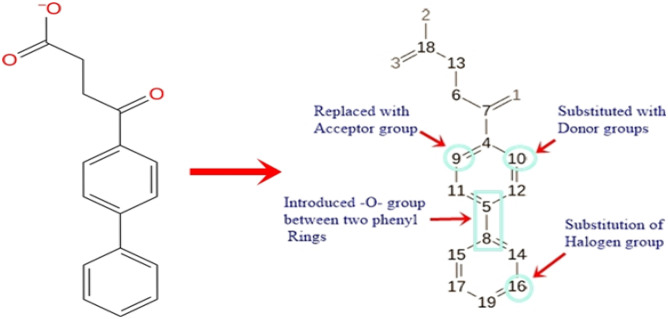
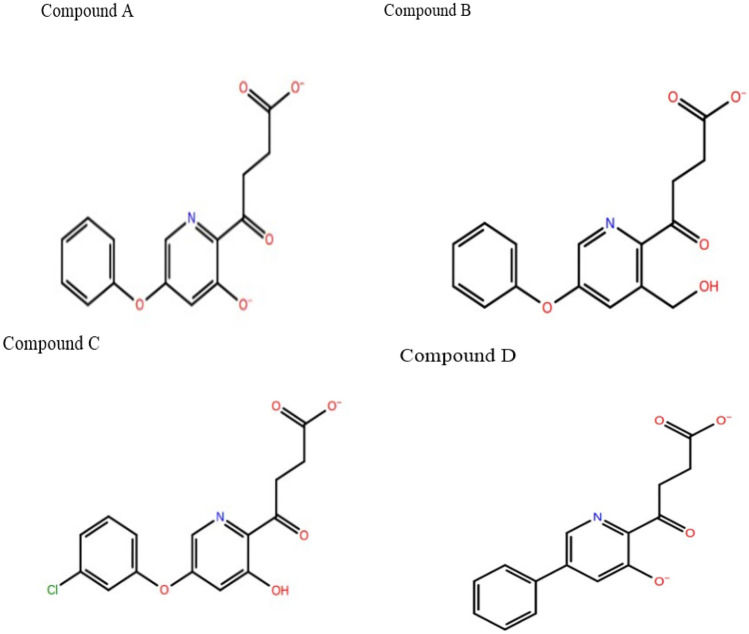
It is assumed that the inhibitory potency of PHD2 inhibitors is enhanced by increasing the binding affinity. So, the manual lead optimisation on the Fenbufen structure was performed to design new compounds with more binding affinity and additional interaction with protein amino acid residues. Fenbufen was selected as the lead molecule and we modified and/or introduced some essential chemical features based on site map analysis of the binding pocket (Fig S2). Briefly, we substituted donor groups (-OH and -CH3OH) at C-10 position, replacing the carbon atom at C-9 position with acceptor groups (˗N = group), introducing the acceptor group (-O-) between the two phenyl rings and substituting the more electronegative halogen groups (-Cl) at C-16 position (Fig. 6) and successfully made four new chemical structures. The structure of newly designed compounds A, B, C, and D is shown in Fig. 7.
Fig. 6.
Manual lead optimisation of Fenbufen structure
Fig. 7.
The structure of newly designed compounds A, B, C, and D from manual lead optimisation of Fenbufen
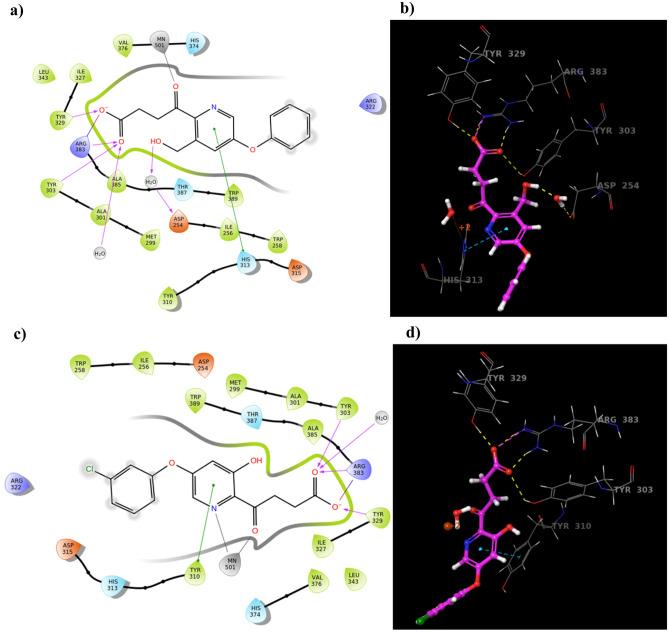
The newly designed structures were incorporated into ligand preparation (ligprep) followed by Glide XP molecular docking with minimised PHD2 protein. Obviously, all four compounds showed a good docking score, among which compounds B and C possessed additional binding interactions with His-313, Asp-254, and Tyr-310 amino acid residues as compared to the Fenbufen lead compound (Table 2 and Fig. 8).
Table 2.
Docking results of manually optimised lead compounds
| Compound name | Docking score (kcal/mol) | Glide g score (kcal/mol) | MM-GBSA binding score (kcal/mol) | Interaction residues | Nature of interactions | Interaction distance (Å) | No. of synthetic pathways (pathfinder) |
|---|---|---|---|---|---|---|---|
| Fenbufen | −11.764 | −11.819 | −20.64 | Fe2+ | Metal co-ordination | 2.31 | 8 |
| His-313 | Pi-Pi stacking | 5.34 | |||||
| Tyr-303 | H-bond | 2.56 | |||||
| Tyr-329 | H-bond | 1.66 | |||||
| Arg-383 | H-bond | 1.70 | |||||
| Salt bridge | 2.78 | ||||||
| Compound A | −14.268 | −14.607 | −16.83 | Fe2+ | Metal co-ordination | 2.01 | 3 |
| Tyr-329 | H-bond | 2.83 | |||||
| Tyr-303 | H-bond | 1.82 | |||||
| Arg-383 | H-bond | 2.02 | |||||
| Salt bridge | 2.31 | ||||||
| Compound B | −13.611 | −13.611 | −29.90 | Fe2+ | Metal co-ordination | 2.17 | 2 |
| Arg-383 | H-bond | 2.21 | |||||
| Tyr-303 | Salt bridge | 1.90 | |||||
| Tyr-329 | H-bond | 2.49 | |||||
| His-313 | H-bond | 1.65 | |||||
| Asp-254 | Pi-Pi stacking | 3.45 | |||||
| Water bridge | 1.75 | ||||||
| Compound C | −11.908 | −12.691 | −36.38 | Fe2+ | Metal co-ordination | 2.26 | 3 |
| Tyr-303 | H-bond | 3.27 | |||||
| Tyr-329 | H-bond | 2.64 | |||||
| Arg-383 | H-bond | 2.87 | |||||
| Tyr-310 | Salt bridge | 2.75 | |||||
| Pi-Pi stacking | 4.87 | ||||||
| Compound D | −15.664 | −16.205 | −13.67 | Fe2+ | Metal co-ordination | 2.19 | 8 |
| Tyr-329 | H-bond | 1.90 | |||||
| Tyr-303 | H-bond | 2.62 | |||||
| Arg-383 | H-bond | 1.89 | |||||
| Salt bridge | 1.01 |
Fig. 8.
The molecular docking results of compound B and compound C with PHD2 protein. a, c Represent the 2D molecular interactions of compounds B and C with PHD2 protein. b, d Represents the 3D binding orientations of compounds B and C with amino acid residues
Additionally, the synthetic feasibility of the above newly designed structures was evaluated using the Pathfinder tool provided by the Schrodinger suite (Maestro11.9.011, Schrödinger, LLC, New York, NY,2019–1). Pathfinder utilised a reaction-based enumeration tool and provided a streamlined approach to evaluate the different pathways involved in the synthesis of newly designed compounds. Interestingly, all four compounds showed more than two retrosynthetic pathways in synthetically accessible chemical space (Table 2). Furthermore, the ADME and Toxicity properties of Vadadustat, Fenbufen, and compounds A, B, C, and D were estimated through Qikprop Analysis and the results are shown in (Table 3). The compounds A, B, C, and D showed an acceptable range of pharmacokinetic properties and toxicity profile as compared to the Vadadustat and Fenbufen compounds.
Table 3.
ADME and Toxicity properties of selected compounds by Qikprop analysis
| Compound namea | Molecular weight (g/mol)b | QPlogo/wc | QPlogSd | QPlog HERGe | QPlogBBf | #metab ± g | QPlogKhsah | %human oral absorptioni |
|---|---|---|---|---|---|---|---|---|
| Vadadustat | 306.70 | 2.75 | −4.31 | −3.55 | −1.696 | 3 | −0.131 | 66.68 |
| Fenbufen | 254.28 | 3.19 | −4.89 | −3.48 | −1.054 | 2 | −0.169 | 81.72 |
| Compound A | 287.27 | 2.16 | −2.78 | −3.22 | −1.638 | 4 | −0.402 | 67.672 |
| Compound B | 301.29 | 1.79 | −2.65 | −3.49 | −1.598 | 4 | −0.619 | 67.63 |
| Compound C | 321.71 | 2.64 | −3.64 | −3.40 | −1.573 | 4 | −0.299 | 69.37 |
| Compound D | 271.27 | 2.22 | −3.02 | −3.30 | −1.595 | 4 | −0.326 | 67.21 |
aLigand name
bMolecular weight of the compound (acceptable range: 130–725 g/mol)
cpredicted octanol/water partition co-efficient logP (acceptable range: −2.0 to 6.5)
dPredicted aqueous solubility; S in Mol/L (acceptable range: −6.5 to 0.5)
epredicted IC50 value for blockage of HERG K+ channels (acceptable range: below −5.0)
fpredicted blood brain barrier (BBB) permeability (acceptable range: −3.0 to 1.2)
gnumber of likely metabolic reactions (range: 1 to 8)
hpredicted Human serum albumin binding (acceptable range: −1.5 to 1.5)
ipercentage of human oral absorption (< 25% is poor and > 80% is high)
Molecular mechanics-general born surface area estimation
The prime molecular mechanics-general born surface area (MM-GBSA) panel can be employed to estimate the ligand binding energies and strain energies for the selected compounds [18]. The low-energy binding poses of previously docked ligands like Vadadustat, Fenbufen, and Compounds A, B, C, and D with PHD2 protein were rescored based on the calculation of total binding free energy using prime MMGBSA technology provided by the Schrodinger suite.
We found that MM-GBSA ΔG bind score of Compound A, B, C, and D were −16.83, −29.90, −36.38, and −13.67 kcal/mol, respectively (Table 2). Also, the MM-GBSA ΔG bind score of Vadadustat and Fenbufen were −30.58 and −20.64 kcal/mol respectively. Altogether, compound B and Cs possessed the lowest MM-GBSA score than the Fenbufen compound. Collectively, the MM-GBSA estimation suggested that compounds B and C formed a more stable complex with PHD2 protein and its stability was analysed through the MD simulation package.
Molecular dynamics
The MD simulation is the fundamental computational tool for capturing dynamic events of the protein–ligand complex [19]. It is performed to analyse their movements at the molecular and atomistic level to understand the ligand-induced conformational changes of the active site [13]. The ligand-receptor interactions obtained from molecular docking may be less convincing due to the lack of receptor flexibility. Hence, we preferred molecular dynamics simulation which favours both ligand and receptor flexibility and also mimics a more realistic physiological environment. During MD simulations, the dynamic trajectory frames of the protein–ligand complex were recorded for every 50 ps which gave more insight into the stability of the complex as well as the contribution of key amino acid residue interactions.
To predict the stability and flexibility of the PHD2 protein complexed with Vadadustat; Fenbufen, compound B, and compound C were submitted to the molecular simulation process for 100 ns using the OPLS_2005 force field.
MD analysis of clinical trial PHD2 inhibitor Vadadustat
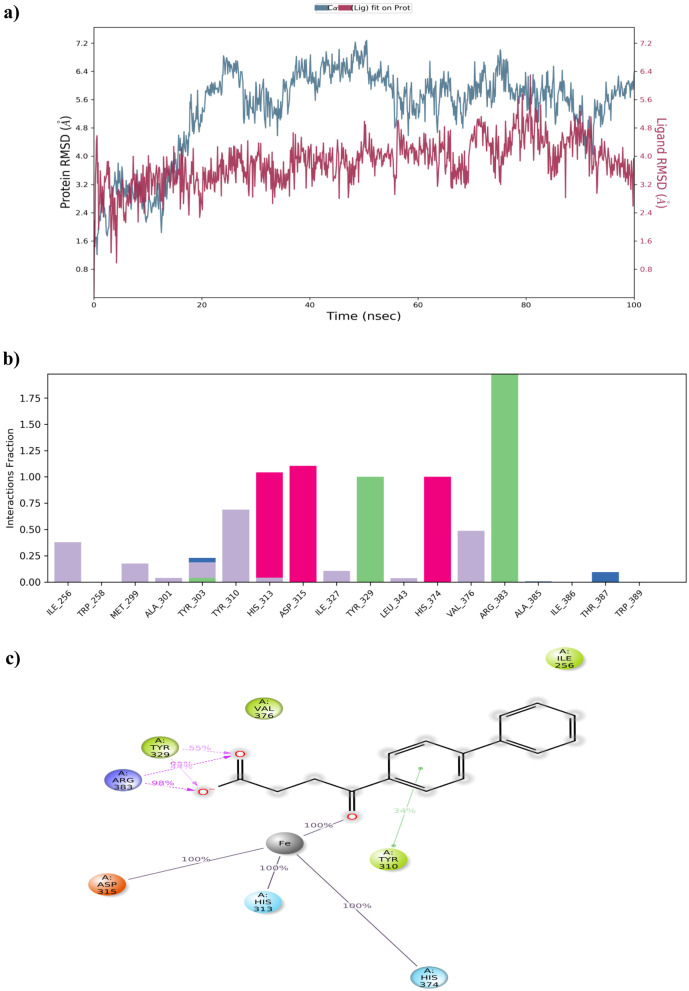
Firstly, the MD simulation for the PHD2-Vadadustat complex was performed. Initially, the RMSD of the protein–ligand complex had deviated until 30 ns. After 30 ns, it was noted that the complex attained equilibrium and maintained its stability within the range of 0.5 to 4.5 Å throughout the simulation time (Fig. 9a). Subsequently, the interactions between the protein and ligand were perceived. Generally, protein–ligand contacts were categorised into 4 types, namely H-bond, hydrophobic, ionic, and water bridges. The interaction types of the PHD2-Vadadustat complex and its percentage levels are shown in Fig. 9b and c. In the case of Vadadustat, Arg-383, Tyr-329, and Tyr-303 amino acid, residues formed 100%, 94%, and 34% of H-bond interaction with PHD2, respectively. Additionally, it showed 100% metal co-ordination interaction with Fe2+ ion, 32% Pi-Pi stacking interaction with Trp-389 residue, and also 62% and 32% water bridge interactions with Asp-254 and Tyr-303 amino acid residues for the whole MD simulation respectively.
Fig. 9.
MD simulation for PHD2-Vadadustat complex. a The protein–ligand RMSD histogram. b protein–ligand contact fractions during the whole molecular dynamic simulation. c 2D molecular interaction fingerprints of PHD2-Vadadustat complex
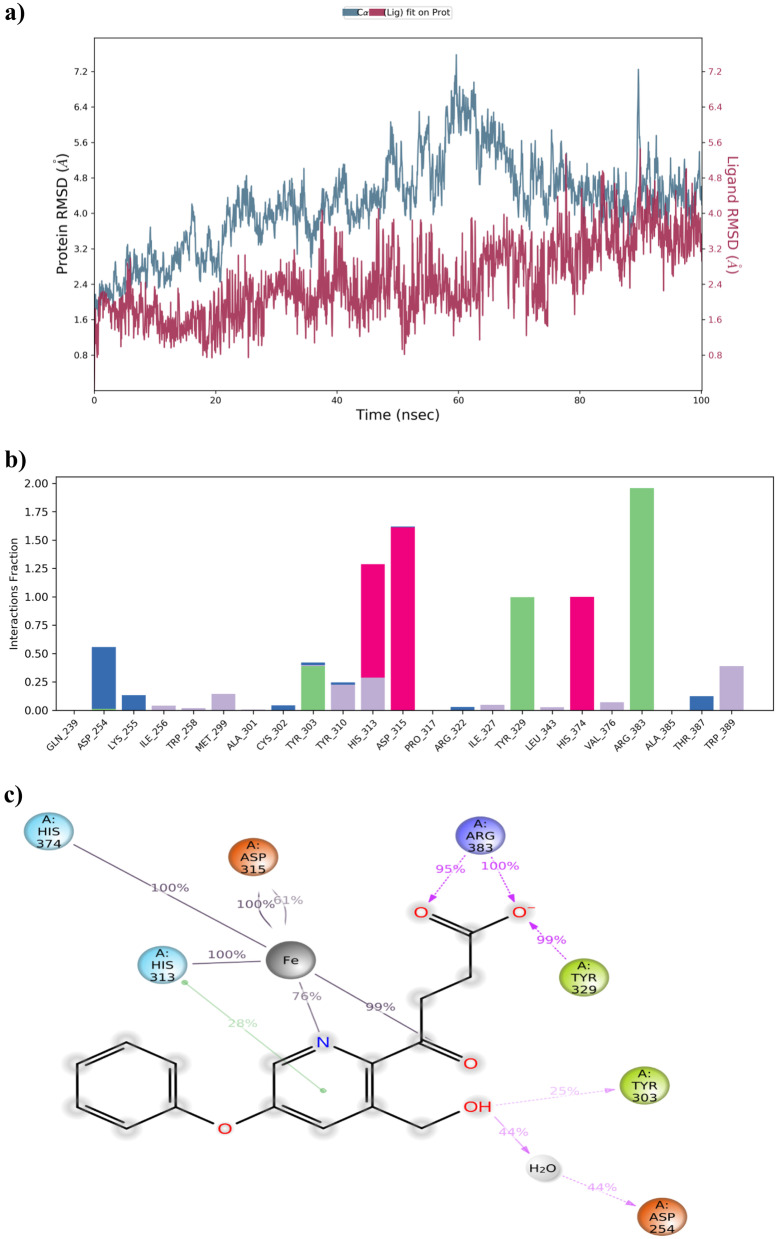
MD simulation analysis of Fenbufen
The MD result of the PHD2-Fenbufen complex was closely monitored. It was noted that protein–ligand RMSD of the complex continued to be within the range of 0.9 to 6.6 Å with slight fluctuations over the period of 100 ns. The interaction patterns of the protein–ligand complex were observed (Fig. 10). The protein–ligand contacts and interaction fingerprints of complex showed similar interactions of Vadadustat. Here, both amino acid residues Arg383 and Tyr329 formed a 98% hydrogen bond interaction with the end carboxylic acid group of Fenbufen. Besides, it also maintained 100% metal co-ordination with Fe2+and 34% Pi-Pi stacking interaction with Tyr-310 throughout the simulation period. Figure 10b and c show protein–ligand contacts and their molecular interactions of Fenbufen respectively.
Fig. 10.
MD simulation for PHD2-Fenbufen complex. a The protein–ligand RMSD histogram. b protein–ligand contact fractions during the whole molecular dynamic simulation. c 2D molecular interaction fingerprints of PHD2-Fenbufen complex
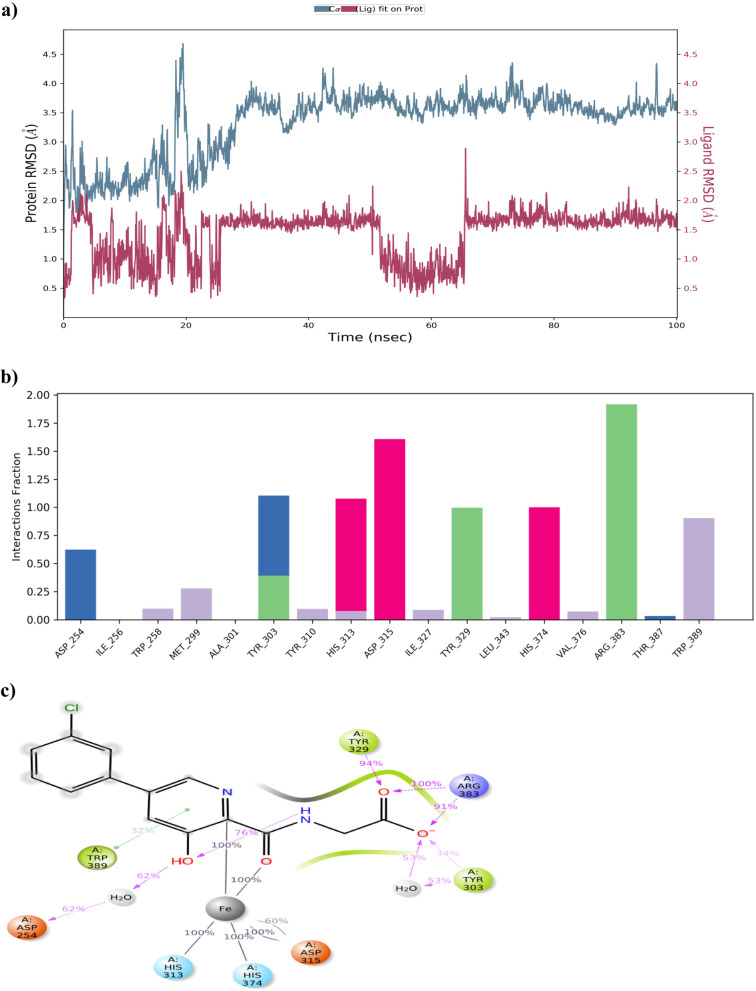
MD simulations of manually optimised lead compounds
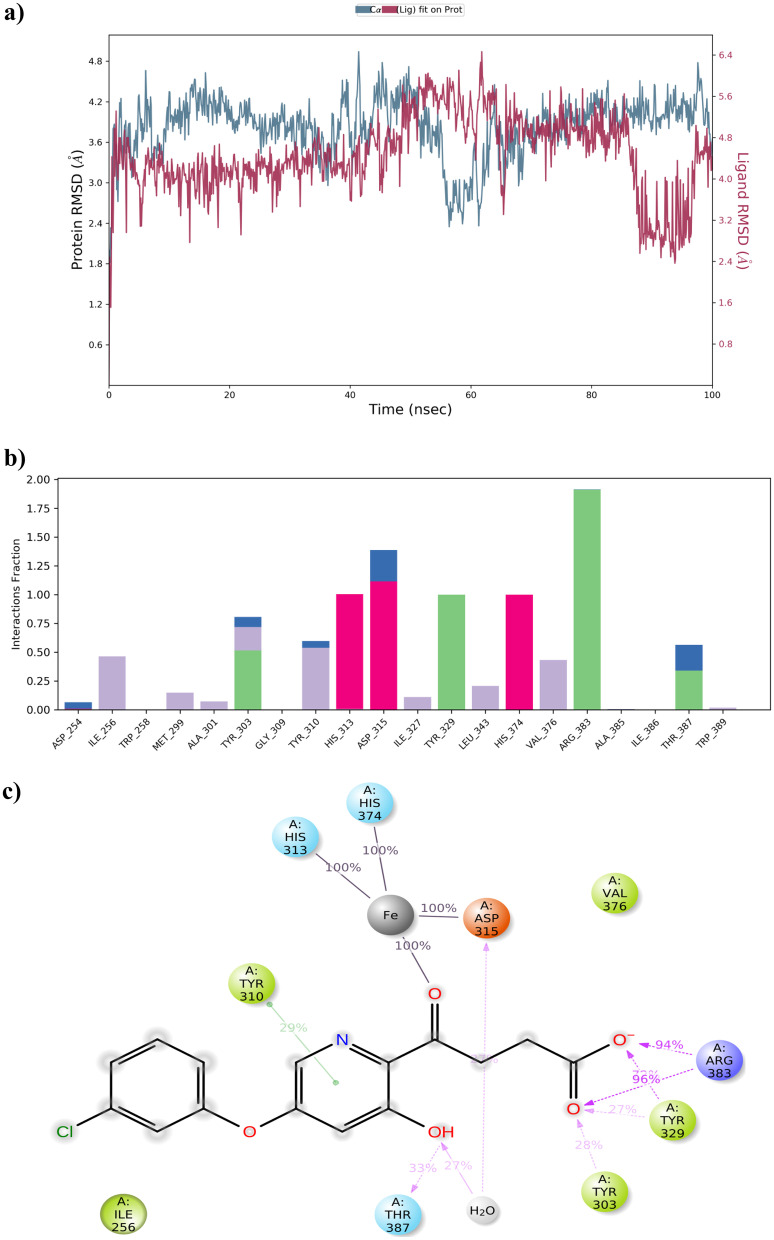
Based on the findings retrieved from MM-GBSA analysis, the better performed compounds, namely compound B and compound C, were exposed to MD simulation. The protein–ligand RMSD of compound B and compound C with PHD2 was visualised manually and the results are depicted in Figs. 11a and 12a. The stability of both compound B and compound C with PHD2 protein was maintained within the range of 0.8 to 7.00 Å and 2.4 to 6.00 Å aside. It was found to be fascinating that both manually optimised new structures B and C did not show more conformational changes. Simultaneously, protein–ligand interaction was monitored during the entire simulation process. All the protein–ligand contacts, as well as the ligand–protein interactions of both compound B and compound C are summarised in Figs. 11b, c and 12b, c.
Fig. 11.
MD simulation for PHD2-compound B complex. a The protein–ligand RMSD histogram. b Protein–ligand contact fractions during the whole molecular dynamic simulation. c 2D molecular interaction fingerprints of PHD2-compound B complex
Fig. 12.
MD simulation for PHD2-compound C complex. a The protein–ligand RMSD histogram. b Protein–ligand contact fractions during the whole molecular dynamic simulation. c 2D molecular interaction fingerprints of the PHD2-compound C complex
Regarding compound B, Tyr-329 and Arg-383 amino acid residues formed 99% and 100% of H-bond interaction with PHD2 respectively. Also, 44% of water bridge interaction was noted with Asp-254 amino acid residue. Additionally, 28% of Pi-Pi stacking with His-313 and 100% of metal co-ordination with Fe2+ were formed. On the other hand, compound C possessed 94%, 96% and 33% H-bond interactions with Arg-383, Tyr-329, and Thr-387 amino acid residues respectively. The metal co-ordination (100%) with Fe2+ was maintained all over the MD simulation.
The overall insight from MD simulations indicated that the compound C-PHD2 complex was more stable and possessed additional binding interactions compared to other complexes like Vadadustat, Fenbufen, and compound B with PHD2 which implied that compound C had a better binding affinity towards the PHD2 protein and might have recommended biological activity than Vadadustat.
Conclusion
In this study, we attempted to identify a reprovable FDA drug, which showed effective PHD2 inhibitory activity, through pharmacophore-based virtual screening and molecular docking approaches for ARDS treatment. Due to the fact that FDA-approved drugs are abundant in number with unique chemical diversity, they were considered one of the most prominent assets for overall drug discovery and development with the advantage of that once identified for drug repurposing, it is not essential to take them for preclinical test again.
Our work aimed to give a new purpose of PHD2 inhibition to some well-established FDA-approved drugs. We adopted in silico molecular modelling methods concurrently with appropriate refinement to spot target-specific PHD2 inhibitors. Here, we proposed that Fenbufen, a non-steroidal anti-inflammatory drug can be effectively repurposed for PHD2 inhibition. Also, manually optimised leads from Fenbufen, i.e., compound B and compound C could act as potential PHD2 inhibitors. These findings were evidenced with the interpretation results of molecular interactions responsible for the effective binding of ligands to the active site of PHD.
Furthermore, based on the knowledge gained from the above observations of the combination of pharmacophore modelling, molecular docking, molecular dynamics, and manual inspection, it can be deduced that, one of the manually optimised lead compound C possessed more binding affinity and protein–ligand complex stability than compound B, Fenbufen, and Vadadustat. These results can be used as a starting point for the commencement of further in vitro testing and in vivo evaluation.
It can be concluded that the appropriate use of combined in silico methods in the drug discovery process can enhance the process of hit identification and lead optimisation and enable the confirmation of their potentiality to serve as scaffolds for the establishment of new beneficial agents than the traditional approach by oneself.
Supplementary information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank PSGCP for the Schrodinger support.
Abbreviations
- ARDS
Acute respiratory distress syndrome
- ALI
Acute lung injury
- COVID-19
Novel coronavirus 2019
- FDA
Food and Drug Administration
- PHD2
Prolyl hydroxylase domain-containing protein 2
- HIF-2α
Hypoxia-inducible factor 2α
- VEPTP
Vascular endothelial protein tyrosine phosphatase
- MM-GBSA
Molecular mechanics-general born surface area
- MD
Molecular dynamics
- RMSD
Root mean square deviation
- RMSF
Root mean square fluctuation
Author contribution
JC conceived the idea of the project. JC and JB performed the computational simulation and analysed the data. JB wrote the original manuscript. JC proofread the manuscript. All the authors have read and approved the manuscript submission.
Data availability
All data generated or analysed during this study are included in this article.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Welker C, Huang J, Gil IJ, Ramakrishna H. Acute respiratory distress syndrome update, with coronavirus disease 2019 Focus. J Cardiothorac Vasc Anesth. 2021;36:1188–1195. doi: 10.1053/j.jvca.2021.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball L, Silva PL, Giacobbe DR, Bassetti M, Zubieta-Calleja GR, Rocco PR, Pelosi P. Understanding the pathophysiology of typical acute respiratory distress syndrome and severe COVID-19. Expert Rev Respir Med. 2022;16(4):437–446. doi: 10.1080/17476348.2022.2057300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X, Ma X. Acute respiratory failure in COVID-19: is it ‘typical’ ARDS? Crit Care. 2020;24:198. doi: 10.1186/s13054-020-02911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang B, Zhou X, Qiu Y, Song Y, Feng F, Feng J, et al. Clinical characteristics of 82 cases of death from COVID-19. PLoS ONE. 2020;15:e0235458. doi: 10.1371/journal.pone.0235458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singleton, Lennon (2012) Acute lung injury regulation by hyaluronan. J Allergy Ther Suppl 4. 10.4172/2155-6121.s4-003 [DOI] [PMC free article] [PubMed]
- 6.Baron A, Matthay M, Teboul L, Bein T, Schultz M, Magder S, et al. Experts’ opinion on management of hemodynamics in ARDS patients: focus on the effects of mechanical ventilation. Intensive Care Med. 2016;42:739–749. doi: 10.1007/s00134-016-4326-3. [DOI] [PubMed] [Google Scholar]
- 7.Petrucci N, Iacovelli W (2004) Ventilation with lower tidal volumes versus traditional tidal volumes in adults for acute lung injury and acute respiratory distress syndrome. Cochrane Database Syst Rev CD003844. 10.1002/14651858.cd003844.pub2 [DOI] [PubMed]
- 8.Silva P, Rocco P, Pelosi P. FG-4497: a new target for acute respiratory distress syndrome? Expert Rev Respir Med. 2015;9:405–409. doi: 10.1586/17476348.2015.1065181. [DOI] [PubMed] [Google Scholar]
- 9.Gong H, Rehman J, Tang H, Wary K, Mittal M, Chatturvedi P, et al. HIF2α signaling inhibits adherens junctional disruption in acute lung injury. J Clin Invest. 2015;125:652–664. doi: 10.1172/jci77701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rishikesh S, Susan P, Leontia B, Shabnam S, Brian S, Ajit C. Evaluation of drug interaction with multiple doses of Rabeprazole, a proton pump inhibitor, on the pharmacokinetics of Vadadustat. Am J Kidney Dis. 2020;75:P631. doi: 10.1053/j.ajkd.2020.02.330. [DOI] [Google Scholar]
- 11.LLC S (2014) Prime, version 3.5. New York.
- 12.No Title Schrödinger Release 2020–1: qikprop Schrödinger, LLC, New York, NY, 2020.
- 13.Schrödinger Release. Desmond molecular dynamics system. Schrödinger LLC. 2019.
- 14.Cutinho P, Shankar R, Anand A, Roy J, Mehta C, Nayak U, et al. Hit identification and drug repositioning of potential non-nucleoside reverse transcriptase inhibitors by structure-based approach using computational tools (part II) J Biomol Struct Dyn. 2020;38:3772–3789. doi: 10.1080/07391102.2019.1663263. [DOI] [PubMed] [Google Scholar]
- 15.Rehman M, Alajmi M, Hussain A, Rather G, Khan MA. High-throughput virtual screening, molecular dynamics simulation, and enzyme kinetics identified ZINC84525623 as a potential inhibitor of NDM-1. Int J Mol Sci. 2019;20:819. doi: 10.3390/ijms20040819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang S. Pharmacophore modeling and applications in drug discovery: Challenges and recent advances. Drug Discov Today. 2010;15:444–450. doi: 10.1016/j.drudis.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Kumar A, Rathi E, Kini S. E-pharmacophore modelling, virtual screening, molecular dynamics simulations and in-silico ADME analysis for identification of potential E6 inhibitors against cervical cancer. J Mol Struct. 2019;1189:299–306. doi: 10.1016/j.molstruc.2019.04.023. [DOI] [Google Scholar]
- 18.Balaji B, Ramanathan M. Prediction of estrogen receptor β ligands potency and selectivity by docking and MM-GBSA scoring methods using three different scaffolds. J Enzyme Inhib Med Chem. 2012;27:832–844. doi: 10.3109/14756366.2011.618990. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Lamim Ribeiro J, Tiwary P. Machine learning approaches for analyzing and enhancing molecular dynamics simulations. Curr Opin Struct Biol. 2020;61:139–145. doi: 10.1016/j.sbi.2019.12.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this article.