Abstract
The human immunodeficiency virus/acquired immunodeficiency syndrome (HIV/AIDS) epidemic still exists as a major global public health burden, especially in the middle- and low-income countries. Antiretroviral therapy (ART) remains a sole option to reduce the mortality and morbidity associated with this disease as no approved vaccine candidates are available. About 67% of the people living with HIV (PLHIV) have received the ART in 2019 worldwide. As a consequence of increased ART regimes, the prevalence of drug resistance mutations (DRM) also has been escalating and it would become a significant barrier in achieving the United Nations Programme on HIV/AIDS goal of eliminating HIV by 2030. So far, nucleoside reverse transcriptase inhibitors (NRTI), non-nucleoside reverse transcriptase inhibitors (NNRTI), and protease inhibitor-(PI) associated DRM have been reported across the globe with a considerable escalation in the annual prevalence rate of pretreatment NNRTI DRM. Conversely, NRTI-associated DRM is still under 5%, with a few scattered reports of significant increase from few countries such as southern and eastern Africa. Likewise, in India, the propositions of NRTI and NNRTI-associated DRM have increased since the commencement of the nationwide ART program in 2004. In agreement to the global trend, M1841/V, a type of NNRTI, remains as a dominant DRM among PLHIV. In this review, we tried to collate various mechanisms of DRM in PLHIV. In addition, patterns of HIV DRM in India and their future challenges on drug-related mutations have been discussed.
Keywords: Drug resistance mutations, nonnucleoside reverse transcriptase inhibitors, nucleoside reverse transcriptase inhibitors, people living with human immunodeficiency virus, protease inhibitor
Introduction
The human immunodeficiency virus (HIV) is classified under the genus Lentivirus within the family Retroviridae.[1] Based on its genetic characterization and antigens, it is further subdivided into HIV-1 and HIV-2. However, HIV-1 is relatively more pathogenic and is responsible for the majority of HIV infections around the globe. The first case of HIV infection was reported in 1981 to the Centre for Disease Control, and later in 1983, this was termed as Acquired Immune Deficiency Syndrome (AIDS). As per the United Nations Programme on HIV/AIDS (UNAIDS) data, around 76 million people have been infected since the start of the epidemic and 32.7 million people have died worldwide.[2] In 2019 alone, 1.7 million new infections were recorded and 690,000 died. And as of now, 38 million people are living with HIV (PLHIV). In India, with 2.1 million PLHIV, there are on an average 87,000 new infections and 69,000 deaths, annually.[3] In order to accelerate the process of control and subsequent elimination of HIV/AIDS, the National Strategic Plan has laid strong emphasis on design, development, and implementation of tailored interventions considering context-specific to local evidence. In 1987, zidovudine, a reverse transcriptase inhibitor, was approved by the Food and Drug Administration, USA to treat this disease. Since this first anti-HIV drug, multiple medicines have been validated for HIV treatment. These include nucleoside reverse transcriptase inhibitors (NRTI) and non-nucleoside reverse transcriptase inhibitors (NNRTI). These two major categories of anti-HIV drugs target the reverse transcriptase enzyme thus, blocking the synthesis of proviral DNA. Another category is protease inhibitors (PI), these compounds target HIV protease, which is crucial for the viral maturation process. Subsequently, antiretroviral therapy (ART) was introduced, which is a combination of medicines from NRTI, NNRTI, and PI drug classes, targeting different biological mechanisms in HIV.[4]
At initial stages of the epidemic (during 2004), the WHO guidelines stated that ART should be initiated at CD4 cell count of 200 cells/μl considering the greater chances of acquiring opportunistic infections when the count falls below 200 cells/μl. However, these guidelines have been updated regularly. In 2017, India adopted WHO recommendations of initiating ART “regardless of WHO clinical stage and at any CD4 cell count.” Ever since its inception, ART has impacted both mortality and morbidity worldwide in PLHIV.[5] However, success rate of ART relies on susceptibility of virus and adherence to therapy, failure of which leads to unavoidable emergence of drug resistance. The drug resistance mutations (DRM) which impact the effect of all these anti-HIV drugs are observed in individuals undergoing ART. In addition, these reported DRMs are frequently observed in naïve individuals who have not taken any ART regimes. This indicates the spread of HIV DRM at the community level.[6]
Elements like excessive variability and high evolution rate in HIV favors emergence of DRM. Besides hindering the efficacy of ART, the spread of drug-resistant strains is another adverse effect of DRMs.[7] According to WHO Guidelines <5% DRM prevalence is categorized as low, 5%-15% as moderate and >15% as high prevalence, which signifies the grade of surveillance programs required for monitoring primary HIV-DRM. This review discusses the HIV drug-resistant mutations and their molecular mechanisms. In addition, we analyzed the prevalence and patterns of HIV DRMs in India and their future challenges.
Methodology
This review was aimed to provide the pattern of HIV drug-resistant mutations in India. For compiling this review we used a combination of web-accessible data along with manual curation to survey. An initial comparison among many open-access search engines showed that most search results are similar regardless of the search engine used. Therefore, Google Scholar and PubMed were selected as the primary literature search engines. Databases were searched in November 2020 with the keyword “HIV drug resistant mutations” and “India” and retrieved 229 documents for the period of 1988–2020. Search terms were used in combination to systematically search titles, abstracts, and keywords. From this initial set of articles, 195 were excluded from the study, and 34 alone were taken for the analysis. The study includes original articles containing the epidemiological work on HIV DRM from various regions of India. The excluded articles fall under different categories; 24 documents (Books, Reviews, Systemic reviews, etc.,) 18 (Duplications), 143 (Nonrelevant articles). The nonrelevant articles include those from other country, other diseases, computer modeling studies, docking studies, basic research, etc. The literature survey and article selection criteria are detailed in the PRISMA flow diagram [Figure 1].
Figure 1.
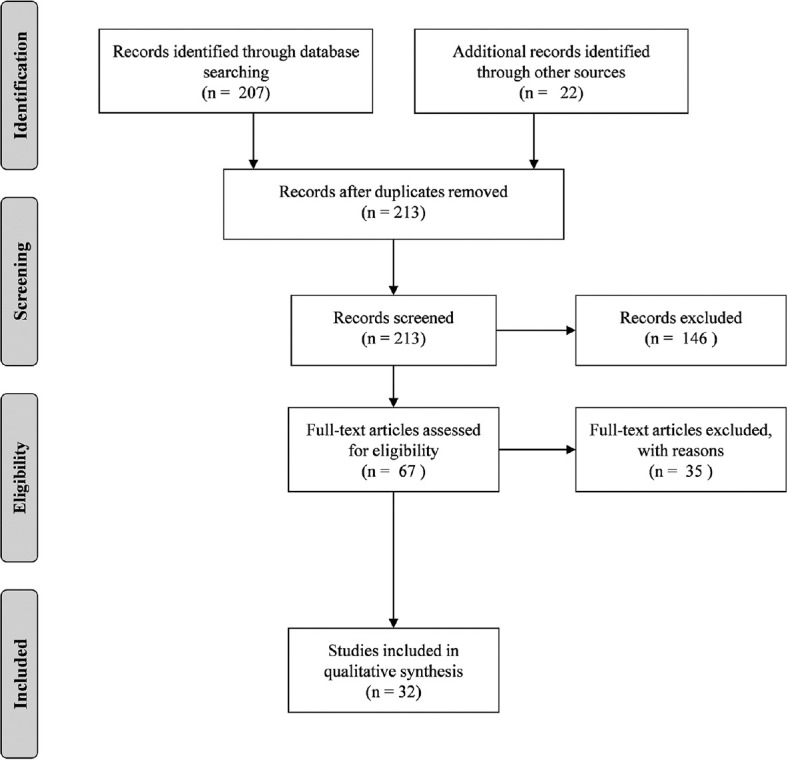
PRISMA flow diagram. Presentation of the procedure of literature searching and selection with number of articles at each stage
Human Immunodeficiency Virus Drug Resistance Mechanism
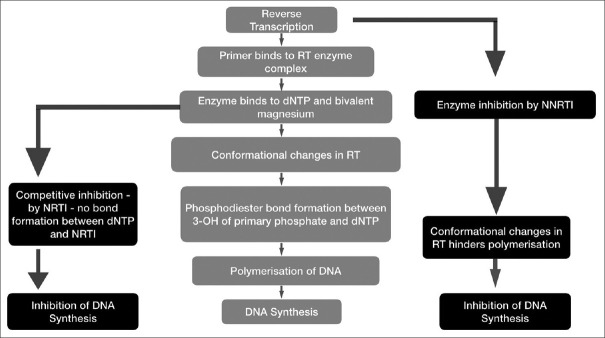
In recent years a great progress in regard to ART for HIV is evinced, but its effects are considerably influenced by DRMs [Table 1]. Most of the anti-HIV drugs target reverse transcriptase enzyme which is a multifunctional enzyme responsible for RNA-dependent DNA synthesis. NRTIs and NNRTIs are anti-HIV drugs which were designed to hinder reverse transcription mechanisms [Figure 2]. NRTIs such as zidovudine, stavudine, abacavir, and lamivudine being dNTP analogs. This competitively inhibits reverse transcriptase enzyme, absence of 3’-OH in NRTI leads to obstruction of phosphodiester bond formation. NNRTIs such as nevirapine, efavirenz, etravirine, on the other hand, follows different mechanism, as they block DNA synthesis by allosterically binding with p66 subunit of reverse transcriptase enzyme leading to conformational changes transcended by restriction of polymerase catalytic triad affecting DNA polymerization.[8]
Table 1.
Common human immunodeficiency virus drug-resistant mutations
| Anti HIV drug category | DRM mutation | Location of mutation | Mechanism of action |
|---|---|---|---|
| NRTI | M184V, K65R, Q151, Y115F, K219R, M41L | Active site of RT | Reduced affinity due to competitive inhibition |
| TAMs K70R, D67N, L210W | Enhancement of primer unblocking | ||
| NNRTI | K103N, Y181C, G190A, A98G, VI79D/E, K101E | Hydrophobic pocket of p66 subunit | Reduced affinity due to allosteric inhibition |
| Protease Inhibitors | V28S, V82L, N83D, 185V, N88D | HIV protease enzyme | Competitive inhibition-blocks viral maturation |
HIV: Human immunodeficiency virus, DRM: Drug resistance mutations, RT: Reverse transcriptase, NRTI: Nucleoside reverse transcriptase inhibitors, NNRTI: Non-nucleoside reverse transcriptase inhibitors
Figure 2.
Mechanism of anti-HIV drugs NRTIs and NNRTIs
Nucleoside Reverse Transcriptase Inhibitors Associated Mutations
NRTI-associated mutations give rise to a large number of effects with regard to drug susceptibilities, replication capacity of virus, and biochemistry of reverse transcriptase enzyme.[9] Point mutations mentioned in Table 1 were observed to be induced after ART, which cause decline in affinity of NRTI to RT enzyme leading to lower incorporation of NRTIs. M184, a mutation located on codon 184 of RT enzyme causes more than 100-fold resistance and develops rapidly in individuals who receive lamivudine monotherapy; this mutation alone is capable of rendering lamivudine therapy ineffective.
The second drug resistance mechanism involves phosphorolytic removal of NRTI from 3 terminus of primer, the process being known as “primer unblocking.” Mutations which have an amplifying effect on primer unblocking are Thymidine Analogue Mutations such as K70R and D67N. This can be enhanced by selective drugs like zidovudine and stavudine as they are thymidine analog. While researchers believed that pyrophosphorolysis as a primary mechanism of resistance to zidovudine and stavudine, the process is not drug-specific in the way that discriminatory mutations tend to be. Hence, these pyrophosphorolysis-enhancing mutations can confer reduced susceptibility to all of the NRTIs. Thus, certain drugs are developing resistance quickly and effectively to some NRTI than others.[10]
Nonnucleoside Reverse Transcriptase Inhibitors Associated Mutations
NNRTI-associated mutations are located on a hydrophobic, which usually emerge when patients are administered NNRTI monotherapies, due to incomplete viral suppression. This indicates NNRTI-associated mutations are caused by viruses selected from a mutant preexisting population. Nucleotide substitutions that affect the susceptibility to NNRTIs are usually present on three regions between codons 98–108, 79–190, and 225–236.
Among NNRTI DRM mutations mentioned in Table 1, K103N and Y181C are common which lead to virological failure. These mutations occur in the enzyme pocket targeted by NNRTI, thus reducing the affinity of these compounds toward reverse transcriptase enzyme.[11] K103N present on codon 103 causes 20–50-fold resistance to NNRTIs, enough for virological failure. Another mutation V106A, causes more than 30-fold resistance to NNRTI-Nevirapine. Recent evidences show the differential effects of genetic background on both the type and degree of HIV DRM. Owing to the polymorphic nature of the hydrophobic pocket of HIV reverse transcriptase, it confers a differential degree of resistance among different subtypes and strains.[12]
Protease Inhibitors Associated Mutations
HIV protease, a homodimeric aspartyl protease plays a vital role in viral maturation by cleaving viral Gag and Gag-Pol polyprotein precursors transitioning them into mature, functional viral enzymes and structural proteins.[13] This transition is essential for production of infectious viral particles. The fact that HIV protease is crucial for HIV infection makes it a favorable target for drug design. PIs such as Ritonavir, Saquiunavir, and Indinavir were approved to be used as a part of highly active ART (HAART) by FDA.[14]
PIs competitively inhibit HIV protease significantly, thereby ceasing the viral maturation process, leading to a reduction in the number of infectious viral particles [Figure 3]. However, high mutation rate due to the absence of proof-reading activity and dynamic viral replication along with potential dual infection and treatment interruption contributes to the development of drug-resistant mutation. A few common HIV drug-resistant PI mutations observed in the Indian population are V28S and V82L.[15] Even though naturally occurring DRMs arise with high frequency in untreated patients they are not significant enough as they rarely cross the threshold of detectable level. Almost all clinically relevant DRMs arise as a consequence of selective drug pressure.[16]
Figure 3.
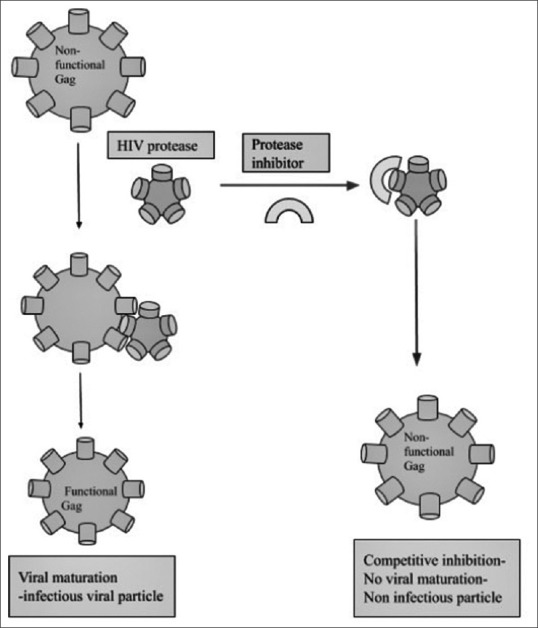
HIV Protease Inhibitor mechanism
The Current Status of Human Immunodeficiency Virus Drug Resistance Mutations in India
Joint UNAIDS and National AIDS Control Organization (NACO) estimated that the prevalence of HIV/AIDS in India is about 0.3%, which is still above the global average of 0.2%.[2,17] To evade the adverse effects of HIV and AIDS in India, free ART was launched in 2004. This is the second-largest ART program globally and the program achieved 54% decrease in the AIDS-associated mortality after a decade of their initiation. Thus, every newly infected patient receives ART that comprises of 2 NRTIs and 1 NNRTI drug. The PI is reserved for second-line treatment if the first-line treatments like NRTIs and NNRTIs fail due to drug resistance.[17] The emergence and spread of DRM is a major threat to the aim of NACO in combating the HIV epidemic in India. The list of the prevalence of HIV DRMs in India are given in Table 2. In order to identify the frequency of DRM among HIV strains circulating across the states, genotyping techniques are in practice. Karade et al.[18] reviewed the DRM status in India by analyzing the 1046 HIV genomic sequences published between 2004 and 2014. The proportion of HIV sequences with any DRM, any NRTI DRM, and any NNRTI DRM was 78.39%, 68.83%, and 73.13%, respectively. Among the NRTI DRM, M184V and K65R were the most predominant mutations found across India. In the case of NNRTI, DRM, K103N, and Y181 C are the prevalent among the reported HIV DRM. Interestingly, the rising trend of K65R mutation was noticed among the Indian population from 2004 to 2014. However, M1841/V continued to be the dominant DRM throughout this study period.
Table 2.
List of human immunodeficiency virus drug resistance mutations in India
| Study name | Study period | Study region | Significant findings of the study | Type of mutations observed |
|---|---|---|---|---|
| Chauhan et al.[23] | September 2014 to June 2016 | North-Western India | Study population: 37 (ART individuals) 22.2% drug resistance observed among FSW and 14.3% was observed among MSM |
K219R, L74V, K219N, and Y181C |
| Thirunavukarasu et al.[24] | March 2010 to March 2014 | South India (Salem, Tamil Nadu) | Study population: 213 (VF patients: 23) NRTI associated mutations: 87% (20/23) NNRTI associated mutations: 91% (21/23) |
I135R/T/V/X, L178 I/M, M184V/I, D67N, K70R, and K103N |
| Ambike et al.[25] | January 2014 to August 2014 | Maharashtra | Study population: 213 (treatment naïve individuals) Mutations associated. with RT and PI were observed in 9.37% (3 of 32) |
M184V, V111I, K65R and Q151M1 |
| Nesakumar et al.[26] | April 2011 to September 2012 | Chennai | Study population: 213 (treatment naïve individuals) TDR was found to be 11.3% (6/53) and NNRTI associated mutations in 8.3% (4/53) |
K101E, Y188C/Y, K103N, |
| Azam et al.[27] | 2010 and 2011 | Aligarh | Study population: 54 (treatment naïve individuals). All patients had at least two secondary mutations that are associated with resistance to PIs |
H69K, I93L, I15V, L19I, M36I, R41K, L63P, and L89M |
| Lakhikumar Sharma et al.[28] | Not stated | Manipur | Study population: 37 (ART individuals).37%, 29% and 7% individuals harbour DRM at the target sites of NNRTI, NRTI and PI respectively | M184V, T215Y, M41L and V108I, H221Y, M46I and I47V |
| Kannangai et al.[29] | 2009 and 2012 | South India | Study population: 127 (treatment naïve individuals) 2.4% people have TDRMs, NNRTI PI and NRTI associated mutations were not found |
K101E, Y181C and G190A |
| Dinesha et al.[30] | Not stated | South India | Study population: 167 (ART individuals) 90.4% (151/167) people had DRMs. NRTI, NNRTI, and dual mutations (NRTI and NNRTI) were seen in 123 (73.7%), 147 (88%), and 119 individuals (71.3%) respectively |
M184IV, K65R, K103NS, V106AM and Y181CIV |
| Azam et al.[31] | 2010 to 2012 | Aligarh | Study population: 103 (ART individuals) 28% (29/103) people have DRM. Out of this 14 (13.6%) mutations at NRTI and 15 (14.6%) mutations at NNRTI |
K219R, E44D, A98G, K103R, V106M, V108I, E138A, Y181C, G190A and T215Y |
| Azam et al.[32] | 2010 to 2012 | Aligarh | Study population: 102 (59-ART and 43-treatment naïve individuals) DRM was observed in 64% (66/102) of the individuals |
D30 N, M46I, G16E, K20R, M36I, D60E, I62V, L63P, I64M, H69K, T74A/S, V77I, V82I, I85V, L89M, I93L, M36I/L/V, H69K, and L89M |
| Rajesh et al.[33] | July 2007 and March 2008 | Madurai | Study population: 26 (ART, pregnant women) NNRTI were observed in 33% (12/26) of the individuals |
Y181C, K103N and Y188C. |
| Saravanan et al.[34] | 2008 and 2009 | Chennai | Study population: 107 (ART individuals) | M46I, I54 V/A, V82A, L90M, K103N, Y181C, and G190A |
| Karade et al.[35] | January 2014 and April 2015 | Pune | Study population: 52 (treatment naïve individuals) 3.8% (2/52) have NNRTI associated mutations |
V106M, K103N |
| Sharma et al.[36] | August 2016 and September 2017 | Manipur | Study population: 44 (14-treatment naïve, 30 ART individuals) 35.7% (5/14) had NRTIs, NNRTIs, and PIs associated mutations. 50% (15/30) had NRTIs, NNRTIs and/or PIs |
M184V, M184I, M41L, D67N, T69D, K70G/R, Y115F, T215N and K219R/Q, A98G, K103N, V179D/E, Y181C, G190A/R, K101E, V106A/M, V108I, Y181F, H221Y and M230I |
| Sinha et al.[37] | April 2006 to August 2008 | New Delhi | Study population: 68 (treatment naïve individuals) 2.9 (2/68) had NRTIs associated mutation observed. No NNRTIs associated mutation were observed. Minor PIs mutation also observed |
M184V, K20R, M36I, and H69K |
| Choudhury et al.[38] | Not Stated | New Delhi | Study population: 44 (17-ART and 27-treatment naïve individuals) Naïve individuals: 25.9 and 85.18% of NRTI and NNRTI associated mutations were found ART individuals: NRTI and NNRTI mutations were observed in 78.7 and 80.6% respectively |
I135T, I135M, E138A, R211K, M184V |
| Sen et al.[39] | June 2004 to December 2005 | Pune | Study population: 44 (50-ART and 25-treatment naïve individuals) No DRM was observed in the Treatment naïve individuals At least one RT mutations were observed in more than 80% (29/44) of ART individuals |
T39A, E203D/K, H208Y, D218E, K122E, M184V, K103N/S and Y181C |
| Gupta et al.[40] | January 2005-April 2005 | Mumbai | Study population: 51 (ART individuals) 96.1% people had resistance to at least one drug used. In that, 92.2% (47/51) had NRTI resistance, 62.7% (32/51) had NNRTI resistance, and 7.8% (4/51) had PI resistance |
M184V, M41L, D67N, K70R, L210W, T215F/Y, V75I, M46I, G48V, I54V, G73S, V82A/F, I84V, L90M, A98G, K101E/P, K103N/S, V106M, V108I, Y181C/I, Y188L, G190A/S/E, F227L, M230L |
| Karade et al.[41] | June and August 2014 | Pune | Study population: 80 (ART individuals) 79.75% (63/80) of individuals had at last one type of RT associated mutations The prevalence of DRMs were 58.75%, 78.75%, and 1.25% for NRTIs, NNRTIs, and protease inhibitors (PIs), respectively |
M184V/I, K103N, L89M, V77I, L63P, H69K/R/Q, M36I, K20I/M/R/T, G16E, K65R and L10V/I |
| Ekstrand et al.[42] | Not stated | Bangalore | Study population: 92 (ART individuals) 86% individuals had at least one RT mutations. NRTI resistance mutations and NNRTI resistance mutations were observed in 68% and 72% individuals respectively |
M184V/MV/I/IM, E44D/DE/A/K, L74V, V75M, V118I, T69D/DN, Y181C/CY/I/V, K103N/KN/R, K101E/EK/Q/KQ, G190A/AG, V108I, V106M/MV |
| Thorat et al.[43] | August 2007 and February 2009 | Kakinada | Study population: 47 (ART women individuals) No NRTI and PI associated mutations were found. Only one patient had NNRTI associated mutation |
K101E |
| Neogi et al.[44] | November 2009 and February 2010 | Bangalore | Study population: 21 (ART individuals) Only 9.56% (2/21) individuals had NNRTI associated mutations |
E138A and L210LS |
| Lall et al.[45] | Not stated | Pune | Study population: 40 (treatment naïve individuals) 10% (4/40) of the individuals had RT associated mutations |
V82A, M41L, D67N, M184V, and A98G |
NRTIs: Nucleoside reverse transcriptase inhibitors; NNRTI: Non-NRTI; PI: Protease inhibitor; TDRM: Transmitted drug resistance-associated mutations; ART: Antiretroviral therapy; DRM: Drug resistance mutations, RT: Reverse transcriptase, FSW: Female Sex Workers, MSM: Man who Sex with Man.
Although ART regimes do not depend upon HIV subtypes, subtype C, which is a predominant subtype in India may develop certain DRM under drug pressure. Thus, systematic sequencing may be practiced to develop a subtype C-specific HIV drug resistance database to overcome the treatment failure. According to recent reports, associated co-infections in HIV patients also can enhance DRM in the Indian population.[7] Among 115 patients analyzed, 13.3% DRM was found in HIV + TB + and 7.5% DRM was found in HIV + TB-. For instance, 10.6%, 4%, and 2.6% of NNRTIs, NRTIs, and PI DRM were respectively found in HIV + TB + patients. On the contrary, only NNRTIs DRM (1<) was found in HIV + TB-. Thus, Tuberculosis (TB) co-infection may enhance the emergence of HIV DRM. However, whether TB alone is responsible for this higher prevalence needs extensive research with scientific methodologies.
Future Challenges for India
In resource-limited countries like India, providing HIV genotyping to all patients with suspected treatment failure is a challenging task. To overcome this, the detection of the most common DRM has been suggested as a cost-effective method. However, this may mislead the evaluation of the emerging DRMs and the changing trends of other uncommon DRM that already exists. Also, different detection techniques used to detect DRM may lead to discrepant results that ultimately affect the treatment protocols.[19] For instance, the detection rate of K65R DRM will be higher in case of allele-specific PCR, however, the same will be negative for Sanger sequencing. It is worth mentioning that HIV drug-resistant mutation in India depends mostly on conventional Sanger sequencing.[20] So, ensuring the implementation of cutting-edge detection technologies is need of the hour to control and eliminate the HIV epidemics.
It is known that patients in low- and middle-income countries, both acquired and transmitted DRM originate in the first-line ART drugs.[21] However, the predominance of transmitted DRMs can be temporarily impaired by employing new classes of drugs. As per the WHO guidelines in 2016, most of the countries have attained the WHO's 10% transmitted DRM threshold.[22] Thus, switching to a new first-line NNRTI named dolutegravir is in progress, and implementing this strategy may be beneficial for both public health response and appropriate amendments in the policy. Overall, the development of routine HIV genotyping and appropriate database for submission is essential along with the updating in the first line ART as per the WHO to estimate the actual rate and mitigate the HIV DRM in India.
Conclusion
Despite the implementation of effective ART, which declined the mortality rate from 1.9 million in 2006 to 0.95 in 2017, the rapid emergence of HIV DRM would possibly reverse the morality trend across the globe. The first step in the prevention strategy involves the identification of infected populations, for which effective HIV DRM surveillance strategies that include point-of-care testing must be developed worldwide. This would minimize the chance of their dissemination among the high-risk sex networks. Current population genotyping methods for the surveillance of resistance are high cost and low throughputs. Thus, the implementation of cutting-edge technologies like next-generation sequencing combined with simpler sample collection and storage matrices has considerable potential to broaden global surveillance and patient monitoring for HIV DRM. For the long term, WHO has initiated a new global action plan to mitigate and prevent HIV-associated DRM. As factors that influence dissemination of DRM varies between countries, regional data are essential for both public health response and appropriate amendments in the policy. Overall, scale-up of clinical measurements, frequent updating in the ART regimens, and execution of effective strategies like prompt counseling and pretreatment education to improve patient adherence to ART probably would help end the AIDS epidemic by 203045.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Fanales-Belasio E, Raimondo M, Suligoi B, Buttò S. HIV virology and pathogenetic mechanisms of infection: A brief overview. Ann Ist Super Sanita. 2010;46:5–14. doi: 10.4415/ANN_10_01_02. [DOI] [PubMed] [Google Scholar]
- 2.UNAIDS. 2019. Archived from the Original on December 4, 2019. [Last accessed on 2019 Dec 21]. https://www.unaids.org/en/resources/documents/2019/2019- UNAIDS-data.
- 3.National AIDS Control Organization (NACO) and National Institute of Medical Statistics (ICMR). India HIV Estimations 2017. Technical Report. [Last accessed on 2021 Dec 24]. Available from: http://naco.gov.in/sites/default/files/Fact%20 Sheets.pdf .
- 4.Kemnic TR, Gulick PG. Stat Pearls. Treasure Island (FL): Stat Pearls Publishing; 2020. HIV Antiretroviral Therapy. Available from: https://www.ncbi.nlm.nih.gov/books/NBK513308/ [PubMed] [Google Scholar]
- 5.World Health Organization. 2nd ed. Geneva: World Health Organization; 2017. [Lastaccessed on 2020 October 11]. Guidelines for Managing Advanced HIV Disease and Rapid Initiation of Antiretroviral Therapy. Recommendations for Public Health Approach; p. 422. Available from: https://www.who.int/hiv/pub/toolkits/ advanced-HIV-disease-policy/en/ [PubMed] [Google Scholar]
- 6.Marcelin AG. Resistance to nucleoside reverse transcriptase inhibitors. In: Geretti AM, editor. Antiretroviral Resistance in Clinical Practice. Ch. 1. London: Mediscript; 2006. [Last accessed on2021 October 11]. Available from: https://www.ncbi.nlm.nih. gov/books/NBK2241/ [PubMed] [Google Scholar]
- 7.Sinha S, Gupta K, Khan NH, Mandal D, Kohli M, Das BK, et al. Higher frequency of HIV-1 drug resistance and increased nucleoside reverse transcriptase inhibitor mutations among the HIV-1 positive antiretroviral therapy-naïve patients coinfected with Mycobacterium tuberculosis compared with only HIV infection in India. Infect Dis (Auckl) 2018;11:1–7. doi: 10.1177/1178633718788870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clutter DS, Jordan MR, Bertagnolio S, Shafer RW. HIV-1 drug resistance and resistance testing. Infect Genet Evol. 2016;46:292–307. doi: 10.1016/j.meegid.2016.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller V, Larder BA. Mutational patterns in the HIV genome and cross-resistance following nucleoside and nucleotide analogue drug exposure. Antivir Ther. 2001;6(Suppl 3):25–44. [PubMed] [Google Scholar]
- 10.Charpentier C, Dwyer DE, Lecossier D, Clavel F, Hance AJ. Antiviral Therapy. Vol. 7. 2-4 Idol Lane, London EC3R 5DD, England: Int Medical Press Ltd; 2002. Co-evolution and competition of viral populations with distinct resistance genotypes in patients failing treatment with protease inhibitors; p. S55. [Google Scholar]
- 11.Mackie N. Resistance to non-nucleoside reverse transcriptase inhibitors. In: Geretti AM, editor. Antiretroviral Resistance in Clinical Practice. Ch. 2. London: Mediscript; 2006. [Last accessed on 2021 Dec 24]. Avaialble from: https://www.ncbi.nlm.nih. gov/books/NBK2249/ [PubMed] [Google Scholar]
- 12.Pokorná J, Machala L, Rezáčová P, Konvalinka J. Current and novel inhibitors of HIV protease. Viruses. 2009;1:1209–39. doi: 10.3390/v1031209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinberg MA, Brenner BG. The impact of HIV genetic polymorphisms and subtype differences on the occurrence of resistance to antiretroviral drugs. Mol Biol Int. 2012;2012:1–10. doi: 10.1155/2012/256982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kempf DJ, Marsh KC, Fino LC, Bryant P, Craig-Kennard A, Sham HL, et al. Design of orally bioavailable, symmetry-based inhibitors of HIV protease. Bioorg Med Chem. 1994;2:847–58. doi: 10.1016/s0968-0896(00)82036-2. [DOI] [PubMed] [Google Scholar]
- 15.Dutta N, Nandi S, Guha S, Saha M K. Emergence of HIV drug-resistant mutations in East Indian population after failure of first-line antiretroviral therapy. HIV AIDS Rev. 2017;16:258–64. [Google Scholar]
- 16.Perelson AS, Ribeiro RM. Modeling the within-host dynamics of HIV infection. BMC Biol. 2013;11:96. doi: 10.1186/1741-7007-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National AIDS Control Organization (NACO) and National Institute of Medical Statistics (ICMR). India HIV Estimations 2015. Technical Report NACO. Technical Report 2015. 2013. [Last accessed on 2021 October 12]. Available from: http:// www.nacoonline.org .
- 18.Karade S, Chaturbhuj DN, Sen S, Joshi RK, Kulkarni SS, Shankar S, et al. HIV drug resistance following a decade of the free antiretroviral therapy programme in India: A review. Int J Infect Dis. 2018;66:33–41. doi: 10.1016/j.ijid.2017.10.020. [DOI] [PubMed] [Google Scholar]
- 19.Toni TA, Brenner BG, Asahchop EL, Ntemgwa M, Moisi D, Wainberg MA. Development of an allele-specific PCR for detection of the K65R resistancemutation in patients infected with subtype C human immunodeficiency virustype 1. Antimicrob Agents Chemother. 2010;54:907–11. doi: 10.1128/AAC.01080-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varghese V, Wang E, Babrzadeh F, Bachmann MH, Shahriar R, Liu T, et al. Nucleic acidtemplate and the risk of a PCR-induced HIV-1 drug resistance mutation. PLoS One. 2010;5:e10992. doi: 10.1371/journal.pone.0010992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scherrer AU, von Wyl V, Yang WL, Kouyos RD, Böni J, Yerly S, et al. Emergence of acquired HIV-1 drug resistance almost stopped in Switzerland: A 15-year prospective cohort analysis. Clin Infect Dis. 2016;62:1310–7. doi: 10.1093/cid/ciw128. [DOI] [PubMed] [Google Scholar]
- 22.World Health Organization. Recommendations for Public Health Approach. 2nd ed. Geneva: World Health Organization; 2016. [Last accessed on 2021 November 15]. Consolidated Guidelines on the Use of Antiretroviral Drugs for Treating HIV Infection; p. 422. Available from: http://www.who.int/hiv/pub/arv/arv-2016/ en/ [Google Scholar]
- 23.Chauhan CK, Lakshmi PV, Sagar V, Sharma A, Arora SK, Kumar R. Primary HIV drug resistance among recently infected cases of HIV in North-West India. AIDS Res Treat. 2019;1525646:1–8. doi: 10.1155/2019/1525646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thirunavukarasu D, Udhaya V, Iqbal HS, Umaarasu T. Patterns of HIV-1 drug-resistance mutations among patients failing first-line antiretroviral treatment in South India. J Int Assoc Provid AIDS Care. 2016;15:261–8. doi: 10.1177/2325957415603508. [DOI] [PubMed] [Google Scholar]
- 25.Ambike SS, Thakar MR, Patil AA, Gangakhedkar RR, Kurle SN. Partial pol sequences from drug naive HIV-2 infected individuals from Maharashtra, India. AIDS Res Hum Retroviruses. 2019;35:505–8. doi: 10.1089/AID.2018.0282. [DOI] [PubMed] [Google Scholar]
- 26.Nesakumar M, Haribabu H, Cheedarla N, Karunaianantham R, Kailasam N, Sathyamurthi P, et al. Transmitted HIV-1 drug resistance in a treatment-naive cohort of recently infected individuals from Chennai, India. AIDS Res Hum Retroviruses. 2019;35:775–9. doi: 10.1089/AID.2019.0022. [DOI] [PubMed] [Google Scholar]
- 27.Azam M, Malik A, Rizvi M, Rai A. Zero prevalence of primary drug resistance-associated mutations to protease inhibitors in HIV-1 drug-naive patients in and around Aligarh, India. J Infect Dev Ctries. 2014;8:079–85. doi: 10.3855/jidc.3480. [DOI] [PubMed] [Google Scholar]
- 28.Lakhikumar Sharma A, Ramsing Singh T, Ranjana Devi K, Shanjukumar Singh L. Prevalence of drug resistance associated mutations among the anti retroviral therapy exposed HIV-1 infected individuals in Manipur, Northeast India. Curr HIV Res. 2016;14:360–70. doi: 10.2174/1570162x14666160401131426. [DOI] [PubMed] [Google Scholar]
- 29.Kannangai R, David S, Sundaresan VC, Sachithanandham J, Mani M, Abraham OC, et al. Frequency of transmitted drug resistance mutations among treatment-naive HIV-1-infected individuals at a tertiary care centre in South India. Mol Diagn Ther. 2015;19:273–5. doi: 10.1007/s40291-015-0160-5. [DOI] [PubMed] [Google Scholar]
- 30.Dinesha TR, Gomathi S, Boobalan J, Sivamalar S, Solomon SS, Pradeep A, et al. Genotypic HIV-1 drug resistance among patients failing tenofovir-based first-line HAART in South India. AIDS Res Hum Retroviruses. 2016;32:1234–6. doi: 10.1089/AID.2016.0110. [DOI] [PubMed] [Google Scholar]
- 31.Azam M, Malik A, Rizvi M, Rai A. Trends of drug-resistance-associated mutations in the reverse transcriptase gene of HIV type 1 isolates from North India. Arch Virol. 2014;159:719–25. doi: 10.1007/s00705-013-1889-y. [DOI] [PubMed] [Google Scholar]
- 32.Azam M, Malik A, Rizvi M, Singh S, Gupta P, Rai A. Emergence of drug resistance-associated mutations in HIV-1 subtype C protease gene in north India. Virus Genes. 2013;47:422–8. doi: 10.1007/s11262-013-0961-8. [DOI] [PubMed] [Google Scholar]
- 33.Rajesh L, Ramesh K, Hanna LE, Narayanan PR, Swaminathan S. Emergence of drug resistant mutations after single dose nevirapine exposure in HIV-1 infected pregnant women in south India. Indian J Med Res. 2010;132:509–12. [PMC free article] [PubMed] [Google Scholar]
- 34.Saravanan S, Vidya M, Balakrishnan P, Kantor R, Solomon SS, Katzenstein D, et al. Viremia and HIV-1 drug resistance mutations among patients receiving second-line highly active antiretroviral therapy in Chennai, Southern India. Clin Infect Dis. 2012;54:995–1000. doi: 10.1093/cid/cir967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karade S, Patil AA, Ghate M, Kulkarni SS, Kurle SN, Risbud AR, et al. Limited HIV pretreatment drug resistance among adults attending free antiretroviral therapy clinic of Pune, India. AIDS Res Hum Retroviruses. 2016;32:377–80. doi: 10.1089/AID.2015.0277. [DOI] [PubMed] [Google Scholar]
- 36.Sharma AL, Singh TR, Singh LS. Antiretroviral resistance, genotypic characterization and origin of Human Immunodeficiency Virus among the infected wives of Intravenous drug users in Manipur. Sci Rep. 2018;8:1–2. doi: 10.1038/s41598-018-33636-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinha S, Ahmad H, Shekhar RC, Kumar N, Dar L, Samantaray JC, et al. Prevalence of HIV drug resistance mutations in HIV type 1 isolates in antiretroviral therapy naive population from northern India. AIDS Res Treat. 2012;2012:905823. doi: 10.1155/2012/905823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choudhury SD, Chaudhury AK, Kalra R, Andrabi R, Wig N, Biswas A, et al. Antiretroviral drug resistance mutations in the reverse transcriptase gene of HIV-1 isolates from Northern Indian patients: A follow-up study. Arch Virol. 2010;155:563–9. doi: 10.1007/s00705-010-0605-4. [DOI] [PubMed] [Google Scholar]
- 39.Sen S, Tripathy SP, Chimanpure VM, Patil AA, Bagul RD, Paranjape RS. Human immunodeficiency virus type 1 drug resistance mutations in peripheral blood mononuclear cell proviral DNA among antiretroviral treatment-naive and treatment-experienced patients from Pune, India. AIDS Res Hum Retroviruses. 2007;23:489–97. doi: 10.1089/aid.2006.0221. [DOI] [PubMed] [Google Scholar]
- 40.Gupta A, Saple DG, Nadkarni G, Shah B, Vaidya S, Hingankar N, et al. One-, two-, and three-class resistance among HIV-infected patients on antiretroviral therapy in private care clinics: Mumbai, India. AIDS Res Hum Retroviruses. 2010;26:25–31. doi: 10.1089/aid.2009.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karade SK, Ghate MV, Chaturbhuj DN, Kadam DB, Shankar S, Gaikwad N, et al. Cross-sectional study of virological failure and multinucleoside reverse transcriptase inhibitor resistance at 12 months of antiretroviral therapy in Western India. Medicine. 2016;95:e4886. doi: 10.1097/MD.0000000000004886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ekstrand ML, Shet A, Chandy S, Singh G, Shamsundar R, Madhavan V, et al. Suboptimal adherence associated with virological failure and resistance mutations to first-line highly active antiretroviral therapy (HAART) in Bangalore, India. Int Health. 2011;3:27–34. doi: 10.1016/j.inhe.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thorat SR, Chaturbhuj DN, Hingankar NK, Chandrasekhar V, Koppada R, Datkar SR, et al. Surveillance of transmitted HIV type 1 drug resistance among HIV type 1-positive women attending an antenatal clinic in Kakinada, India. AIDS Res Hum Retroviruses. 2011;27:1291–7. doi: 10.1089/AID.2011.0036. [DOI] [PubMed] [Google Scholar]
- 44.Neogi U, Prarthana BS, Gupta S, D'souza G, De Costa A, Kuttiatt VS, et al. Naturally occurring polymorphisms and primary drug resistance profile among antiretroviral-naive individuals in Bangalore, India. AIDS Res Hum Retroviruses. 2010;26:1097–101. doi: 10.1089/aid.2010.0092. [DOI] [PubMed] [Google Scholar]
- 45.Lall M, Gupta RM, Sen S, Kapila K, Tripathy SP, Paranjape RS. Profile of primary resistance in HIV-1-infected treatment-naive individuals from Western India. AIDS Res Hum Retroviruses. 2008;24:987–90. doi: 10.1089/aid.2008.0079. [DOI] [PubMed] [Google Scholar]