Abstract
The thiol group of cysteine (Cys) residues, often present in the active center of the protein, is of particular importance to protein function, which is significantly determined by the redox state of a protein’s environment. Our knowledge of different thiol-based oxidative posttranslational modifications (oxiPTMs), which compete for specific protein thiol groups, has increased over the last 10 years. The principal oxiPTMs include S-sulfenylation, S-glutathionylation, S-nitrosation, persulfidation, S-cyanylation and S-acylation. The role of each oxiPTM depends on the redox cellular state, which in turn depends on cellular homeostasis under either optimal or stressful conditions. Under such conditions, the metabolism of molecules such as glutathione, NADPH (reduced nicotinamide adenine dinucleotide phosphate), nitric oxide, hydrogen sulfide and hydrogen peroxide can be altered, exacerbated and, consequently, outside the cell’s control. This review provides a broad overview of these oxiPTMs under physiological and unfavorable conditions, which can regulate the function of target proteins.
Keywords: Persulfidation, S-cyanylation and S-acylation, S-glutathionylation, S-nitrosation, S-sulfenylation
Introduction
Once synthesized in ribosomes, proteins can undergo numerous posttranslational modifications (PTMs) involving chemical changes in specific amino acid residues, which are mediated by enzymatic or nonenzymatic additions of certain chemical groups. These additional regulatory mechanisms, many of which have a very significant impact on cellular signaling, affect the chemical properties of target proteins and, consequently, their spatial conformation, stability, folding properties, subcellular location and biological function (Navrot et al. 2011, Friso and van Wijk 2015).
This highly diverse range of PTMs includes phosphorylation, ubiquitination, SUMOylation, γ-carboxylation, poly(ADP-ribosyl)ation, acetylation, redox modification, methylation, glycosylation, acylation, alkylation, hydroxylation, nitration and nucleotide addition, among others (Vu et al. 2018, Arefian et al. 2021, Gough and Sadanandom 2021, Péter et al. 2021, Wang et al. 2021c). The UniProtKB/Swiss-Prot databases have identified over 450 different PTMs (Conibear 2020, Zhang and Zeng 2020, Wang et al. 2021b), which demonstrates the complexity of cellular proteomes. The identification of a single PTM in a specific protein requires specific experimental techniques, which, in many cases, involve complex technical protocols, including chemoselective reactions, in order to label specific amino acid residues, combined with tandem mass spectrometry analyses (Chuh and Pratt 2015, Shortreed et al. 2015, Aslebagh et al. 2019). Additionally, although developed to identify protein PTMs in different databases (Li and Tang 2016, Audagnotto and Dal Peraro 2017, Xie et al. 2018), bioinformatic tools need to be corroborated by experimental techniques.
This review provides a comprehensive and updated overview of the major oxidative posttranslational modifications (oxiPTMs), which affect thiol groups of protein cysteine residues and their functioning in plant cells.
Overview of the thiol-based oxiPTMs: a mechanism of protein regulation
Thiol (-SH) groups of cysteine (Cys) residues, which are involved in the protein’s active center and folding, are essential for the functioning and regulation of many proteins (Poole 2015, Ulrich and Jakob 2019). These thiol groups can be deprotonated to a negatively charged thiolate (Cys-S−), resulting in enhanced reactivity. Furthermore, the oxidation of the thiol group can involve either one- or two-electron oxidation events, leading to the formation of thiyl radicals (Cys-S•) and sulfenic acids (Cys-SOH), respectively (Trujillo et al. 2016, Turell et al. 2020). Under cellular oxidant conditions, sulfenic acid (-SOH) is oxidized to sulfinic acid (-SO2H) and then to sulfonic acid (-SO3H), the latter being an irreversible process, which usually triggers the inactivation of the target protein (Fig. 1).
Fig. 1.
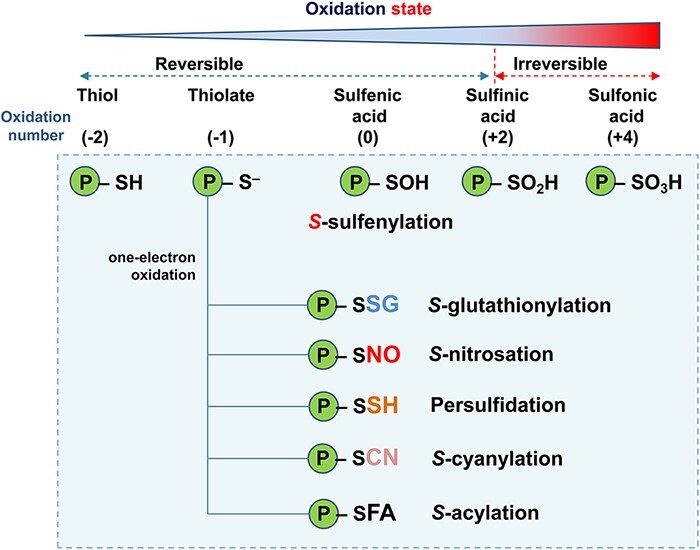
Outline of the main thiol-based oxiPTMs. The upper side of the panel indicates the oxidation states of sulfur (S) in proteins, which can be from thiol (−2) to sulfonic acid (+4). Under cellular oxidant conditions, the oxidation from sulfinic acid to sulfonic acid could take place, the latter being an irreversible process. The principal reversible oxiPTMs result from the interaction between thiolate with either hydrogen peroxide (H2O2; S-sulfenylation); glutathione (GSH; S-glutathionylation), nitric oxide (NO; S-nitrosation), hydrogen sulfide (H2S; persulfidation), cyanide (HCN; S-cyanylation) or fatty acid (FA; S-acylation) are also displayed in the lower side of the panel.
On the other hand, it should be noted that the acid dissociation constant (pKa) of the thiol group is typically close to the physiological pH (7.0–7.4). The thermodynamics and kinetics of protein thiol oxidation in vivo are affected by thiol acidity, which varies according to the protein microenvironment in a specific subcellular location and protein folding (Roos et al. 2013, Zhang et al. 2021). This is particularly important when the cellular redox state is susceptible to modification under stressful conditions during which the generation of oxidant molecules is uncontrollably boosted and initiates a cascade of cellular signals (Chung et al. 2013, Castro et al. 2021). In Arabidopsis, under oxidative stress conditions, the peroxidatic Cys of the chloroplast 2-Cys peroxiredoxin (2-Cys Prx) can be oxidized to sulfinic acid causing its inactivation; however, the activity is restored when this Cys is reduced back to sulfenic acid by a sulfiredoxin enzyme (Iglesias-Baena et al. 2010), a process in which NADPH thioredoxin reductase C appears to be involved (Puerto-Galán et al. 2015).
The change in the redox state of a specific thiol group can either increase or decrease the biological function of the target protein (Table 1). Proteins in any cellular context are prone to be exposed to different potential agents, which can oxidize or reduce the thiol group. The final impact will therefore depend on the accessibility of these potential thiol targets to different potential oxidants or reducing molecules such as certain free radical species, including hydroxyl (•OH), peroxyl (ROO•), superoxide (O2•−), nitric oxide (•NO) and nitrogen dioxide (•NO2), as well as nonradical molecules such as hydrogen peroxide (H2O2), peroxynitrite (ONOO−) and hydrogen sulfide (H2S). Furthermore, certain pair molecules like reduced/oxidized glutathione [GSH/glutathione disulfide (GSSG)] and NADPH/NADP+ (reduced and oxidized nicotinamide adenine dinucleotide phosphate, respectively), which are involved in numerous metabolic pathways either as substrates or cofactors and whose reduced/oxidized ratio can be significantly affected under nitro-oxidative conditions, also play an important role in cellular redox homeostasis (Møller et al. 2020, Vogelsang and Dietz 2020, Corpas et al. 2021b). Additionally, the involvement of other thiol-based redox compounds such as S-nitrosothiols (SNO), polysulfide (H2Sn), thioredoxins (Trxs), glutaredoxins (Grxs) and peroxiredoxins (Prxs) also needs to be taken into account (Rouhier et al. 2015, Knuesting and Scheibe 2018, Benchoam et al. 2020, Kimura 2021, Sánchez-Guerrero et al. 2021, Takata et al. 2021). This complex redox regulation under physiological and stress conditions has mostly been described in chloroplasts (Yoshida et al. 2018, Liebthal et al. 2020, Yokochi et al. 2021) and mitochondria (da Fonseca-Pereira et al. 2019, Martí et al. 2020). Thus, chloroplastic Prx II E is involved in peroxynitrite reductase activity, which is inhibited by S‐nitrosation (Romero-Puertas et al. 2007), while mitochondrial PrxII F through S-nitrosation prevents the thermal aggregation of citrate synthase (Camejo et al. 2015).
Table 1.
Number of identified plant proteins susceptible to undergo oxiPTMs according to proteomic analyses in different plant species and organs
| oxiPTM | No of identified proteins | Plant species/organ | Reference |
|---|---|---|---|
| S-sulfenylation | 1,394a | Arabidopsis cell culture treated with H2O2 | Huang et al. 2019 |
| 132 b | Arabidopsis plastids | De Smet et al. 2019 | |
| S-glutathionylation | 79c | Arabidopsis cell culture | Dixon et al. 2005 |
| 25d | Wheat seedlings | Gietler et al. 2016 | |
| S-nitrosation | 63e | Arabidopsis cell exposed to the NO donor S-nitrosoglutathione | Lindermayr et al. 2005 |
| 52e | Arabidopsis leaves of plants exposed to NO gas | ||
| 46f | Endogenous S-nitrosated proteins in Arabidopsis cells | Fares et al. 2011 | |
| 44f | Endogenous S-nitrosated proteins in Arabidopsis plantlets | Puyaubert et al. 2014 | |
| 32e | Populus × canescens | Vanzo et al. 2014 | |
| 926g | Arabidopsis | Hu et al. 2015 | |
| 402 | Peanut root tips | Pan et al. 2021 | |
| 35h | Arabidopsis guard cells treated with flg22 | Lawrence et al. 2020 | |
| Persulfidation | 106i 2015 |
Arabidopsis leaves | Aroca et al. 2015, Aroca et al. 2017 |
| S-cyanylation | 163 | Arabidopsis leaves | García et al. 2019 |
| S-acylation | 600 2643j |
Arabidopsi s | Hemsley et al. 2013; Kumar et al. 2020 |
| 450 k | Poplar cell culture | Srivastava et al. 2016 |
BTD-based probe.
YAP1C probe.
GS-biotin-labeling studies, 2D-PAGE followed by MALDI-MS.
Immublot probe with anti-GSH antibodies and identified by MALDI-TOF and LC–MS/MS.
BSM and LC–MS/MS.
BSM and labeling with isotope-coded affinity tags.
Site-specific nitrosoproteomic approach.
Iodo tandem mass tag™ labeling.
Modified BSM.
Acyl resin-assisted capture assay.
Acyl-Biotin Exchange method.
Other molecules with antioxidant capacity, such as ascorbate and melatonin, are also involved in this pool of regulatory molecules (Tan et al. 2015, Zechmann 2018, Aghdam et al. 2021). All these elements broaden the network of interactions, which are difficult to decipher if a holistic analysis is not used to examine all the complex interactions. The oxiPTM mechanism for the regulation of signaling protein functions requires that: (i) the oxiPTM-promoted change in protein function is caused by cellular stimuli; (ii) the modification be rapid and be mediated by an enzyme and (iii) the modification be specific and reversible (Shelton et al. 2005). In addition, the effectiveness of these PTMs can vary depending on such factors as the accessibility of the potential target Cys’ thiol group to each molecule incorporated. For example, NO and H2S, being smaller than GSH (reduced glutathione), are expected to access a larger number of Cys residues. Nevertheless, the proteins corresponding to each oxiPTM, whose number and specificity are expected to increase with the development of new techniques, have been identified using proteomic analysis (Alcock et al. 2018, Shi and Carroll 2020, Wang et al. 2021b). Fig. 1 shows the principal oxiPTMs examined in this review.
S-sulfenylation
S-sulfenylation involves the reaction of H2O2 with redox-sensitive Cys in proteins to form cysteine sulfenic acid (Cys-SOH), as well as the reversible covalent addition of one oxygen atom to the thiol group. If the thiol oxidation state persists over time, two or three oxygen atoms can be added to generate an irreversible covalent addition until sulfonic acid is formed (Fig. 1). This generally results in protein degradation or inactivation and is usually associated with nonfunctional proteins and stressful conditions (Filipovic et al. 2018).
With the aid of a dimedone-based DYn-2 probe, initial in vitro studies of protein S-sulfenylation, which used the human colon carcinoma (RKO) cell line treated with 500 µM H2O2, identified over 778 S-sulfenylated proteins following detection with liquid chromatography–tandem mass spectrometry (LC–MS/MS) (Yang et al. 2014). These S-sulfenylated proteins, approximately 92% of which contained only one or two S-sulfenylated residues, include protein kinases, phosphatases, acetyltransferases, deacetylases and deubiquitinases. This suggests that S-sulfenylation regulates these proteins and may mediate other additional PTMs (Yang et al. 2014).
In Arabidopsis thaliana cell cultures treated with 100 µM H2O2, an initial analysis of sulfenome identified roughly 100 potential S-sulfenylated proteins (Waszczak et al. 2014). Later, 1394 proteins susceptible to S-sulfenylation were identified using a more reactive 1-(pent-4-yn-1-yl)-1 H-benzo[c][1,2]thiazin-4(3H)-one 2,2-dioxide (BTD) based probe (Fu et al. 2019) than the DYn-2 probe to detect S-sulfenylated residues (Huang et al. 2019), which had previously identified 200 S-sulfenylated proteins (Akter et al. 2015) (Table 2). These proteins included mitogen-activated protein kinase 4 (MAPK4), which was S-sulfenylated at Cys181, leading to a decrease in kinase activity (Huang et al. 2019). A more in-depth analysis of Arabidopsis cells treated with 1 mM H2O2, using sulfenic acid yeast ACTIVATOR PROTEIN 1 containing single redox-active Cys598 (YAP1C) trapping technology tagged to the plastids, identified 132 S-sulfenylated proteins (De Smet et al. 2019). More recently, a new noninvasive strategy using a disulfide-linked peptide reporter has been implemented to identify S-sulfenylated proteins in Arabidopsis cells (Wei et al. 2020).
Table 2.
Representative examples of plant proteins that undergo several oxiPTMs
Computational prediction.
Proteomic identification.
Activity in vitro assay.
These studies using cell cultures treated with high concentrations of H2O2 have provided a large number of candidate proteins for S-sulfenylation. However, more in-depth analyses need to be carried out on other plant species under both physiological and stressful conditions in order to evaluate the role of S-sulfenylation.
S-glutathionylation
Glutathione (GSH; γ-L-glutamyl-L-cysteinyl-glycine) is a nonprotein thiol compound, which, along with ascorbate, is the most abundant soluble antioxidant present in plant cells (Foyer and Noctor 2011). With its pKa of roughly 8–9 and at physiological pH, the -SH group of GSH is highly protonated. Under optimal physiological conditions, although mainly found in a reduced form of GSH, in oxidative stress environments, free glutathione is oxidized to glutathione disulfide (GSSG) (Airaki et al. 2011; Diaz-Vivancos et al. 2015). GSH is required in a diverse range of detoxification pathways including the ascorbate-glutathione cycle for H2O2 regulation, the glyoxalase pathway for methylglyoxal (MG) detoxification (Kharbech et al. 2020, Dorion et al. 2021), and also for the biosynthesis of phytochelatins, which are a group of low molecular weight polypeptides involved in heavy metal and metalloid detoxification (Gupta et al. 2013, Rodríguez-Ruiz et al. 2019a, Bhat et al. 2021). Some evidence indicates that stress-induced reductions in the GSH/GSSG ratio in different cellular systems promote the formation of mixed disulfide bridges between glutathione and protein thiols. S-glutathionylation, a reversible PTM (Fig. 1), occurring spontaneously between glutathione and a protein thiolate, regulates protein function in different subcellular compartments (Shelton et al. 2005, Mailloux and Treberg 2016, Zhang et al. 2018, Kalinina and Novichkova 2021). This nonenzymatic interaction can be through several mechanisms: (i) protein thiol (P-SH) is oxidized by reactive oxygen species (ROS) to a sulfenic acid (P-SOH) which then rapidly reacts with GSH to form P-SSG; or (ii) exchange of thiol-disulfide between P-SH with glutathione disulfide (GSSG) (Zhang et al. 2018). There is also another proposed mechanism mediated enzymatically by a glutaredoxin (GRX) where P-SH is oxidized to a thiyl radical (PS•), which then quickly reacts with GSH to form a thiyl radical glutathionyl intermediate (PSSG•−), which then reacts with O2 to form PSSG (Beer et al. 2004). Another example in which S-glutathionylation is mediated by a GRX has been described in the Arabidopsis brassinosteroid insensitive 1-associated receptor-like kinase 1 (BAK1), in which this PTM causes the inhibition of its kinase activity (Bender et al. 2015). Under certain adverse environmental conditions, S-glutathionylation can prevent protein Cys overoxidation, which causes its inactivation through the formation of sulfonic residues (Zaffagnini et al. 2007, 2012, Gurrieri et al. 2019). For example, 2-Cys Prx undergoes deglutathiolation in the presence of sulfiredoxin in pea chloroplasts, which does not occur in the mitochondrial Prx IIF. Thus, it has been suggested that glutathionylation/deglutathionylation is associated with changes in redox state during plant development or in response to stress conditions (Calderón et al. 2017).
S-glutathionylated proteins have been detected using techniques such as the use of GSH antibodies and labeling with the aid of 35S radiolabeling and biotinylation (Ito et al. 2003, Gao et al. 2009). Thus, in Arabidopsis cell cultures, approximately 79 S-glutathionylated proteins have been identified using a combination of GSH biotin labeling, 2D-PAGE and matrix-assisted laser desorption/ionization-mass spectrometry (MALDI-MS) (Dixon et al. 2005). In wheat seedlings, 25 S-glutathionylated proteins were identified using 2-D electrophoresis, immunoblot tests, anti-GSH antibodies, MALDI-TOF (time-of-flight) and LC–MS/MS (Gietler et al. 2016) (Table 2).
As mentioned above, given its antioxidant properties, interest in the involvement of GSH in S-glutathionylation processes has been growing in plant research. For example, GSH regulates ethylene biosynthesis in Arabidopsis by modulating 1-aminocyclopropane-1-carboxylate oxidase (ACO) activity through S-glutathionylation. With the aid of in silico docking analysis, Cys63 has been identified as the best candidate for S-glutathionylation of ACO (Datta et al. 2015). In Arabidopsis, S-glutathionylation can trigger the inhibition of chloroplastic NADP-glyceraldehyde-3-phosphate dehydrogenase (AtGAPA1) at Cys149 (Zaffagnini et al. 2007). S-glutathionylation has also been found to facilitate the insoluble aggregation of S-glutathionylated AtGAPA1, which appears to be irreversible. On the other hand, the aggregation process has been reported to be halted by Trx h1 (Zaffagnini et al. 2019). Another target protein for S-glutathionylation, α-amylase 3 (AtAMY3), which catalyzes the cleaving of α-1,4-glucosidic bonds in starch, is involved in the response to osmotic stress and stomatal opening and is also regulated by Trxs (Gurrieri et al. 2019). A pKa analysis of catalytic cysteine residues Cys499 and Cys587 suggests that one of these residues, which are susceptible to S-glutathionylation, can be deprotonated. This mechanism prevents overoxidation under stress conditions when high H2O2 content is present (Gurrieri et al. 2019). Cytosolic NADPH-generating isocitrate dehydrogenase (NADP-ICDH) in Arabidopsis has also been shown to be S-glutathionylated at Cys363, which is located outside the active center. Although it does not directly affect NADP-ICDH activity, S-glutathionylation appears to mediate inhibition caused by S-nitrosation in the presence of S-nitrosoglutathione (GSNO) under in vitro conditions (Niazi et al. 2019). S-glutathionylation appears to play a major role in certain organelles. Accordingly, in an analysis of nine photosynthetic species from streptophyte algae to angiosperms, Müller-Schüssele et al. (2021) have identified 364 proteins susceptible to undergo S-glutathionylation, of which 151 have a plastid location.
S-nitrosation
Nitric oxide (•NO) is a free radical with signaling functions found in higher plants. It mediates a wide variety of plant processes ranging from seed germination to fruit ripening and is involved in mechanisms of response to biotic and abiotic stresses, either directly or through its interaction with other growth regulators (González-Gordo et al. 2019, 2020a, Kolbert et al. 2019, Mishra et al. 2021, Corpas et al. 2022a, 2022b). Much of this regulation is carried out through NO-derived PTMs such as tyrosine nitration (Corpas et al. 2021a), metal nitrosylation and S-nitrosation (Corpas et al. 2020, Gupta et al. 2020). S-nitrosation, previously referred to as S-nitrosylation, is a covalent reaction involving one-electron oxidation of thiol groups. Thus, the presence of the thiol group enables GSH to react with NO to generate S-nitrosoglutathione (GSNO), regarded as a low molecular S-nitrosothiol, which can mediate transnitrosation reactions (Corpas et al. 2013). GSNO content is controlled by GSNO reductase (GSNOR), an enzyme that is inhibited by S-nitrosation (Sakamoto et al. 2002, Leterrier et al. 2011, Guerra et al. 2016, Tichá et al. 2017).
Most research on S-nitrosation, which is one of the most studied oxiPTMs, has been carried out in A. thaliana. Thus, using the biotin switch method (BSM), 63 candidates for S-nitrosation have been identified in Arabidopsis cells treated with the NO donor GSNO (Lindermayr et al. 2005). In the same study, using Arabidopsis plants exposed to NO gas, 52 S-nitrosated proteins were detected in leaves. Using an alternative technique based on a combination of BSM and labeling with isotope-coded affinity tags, a total of 46 endogenous proteins, which appeared to be S-nitrosated, were identified in Arabidopsis cells (Fares et al. 2011). With the aid of a similar approach, 44 endogenous S-nitrosated proteins, 11 of which were overnitrosated under cold stress conditions, were also identified in Arabidopsis plantlets (Puyaubert et al. 2014). The number of Arabidopsis S-nitrosated proteins identified was subsequently extended to 926 with the aid of a site-specific nitrosoproteomic approach (Hu et al. 2015). More recently, the S-nitrosoproteome in Arabidopsis guard cells has been studied in response to the bacterial peptide flagellin using an iodo tandem mass tag labeling technique, which enabled 35 S-nitrosated proteins to be identified (Lawrence et al. 2020).
A study of poplar leaves has identified 172 S-nitrosated proteins, under ozone stress conditions; 32 S-nitrosated proteins were differentially affected, 9 of which showed a higher degree of S-nitrosation, with lower S-nitrosation observed in 23 of these proteins; this suggests the presence of a de-nitrosation mechanism to regulate these proteins (Vanzo et al. 2014). These authors, who specifically studied phenylalanine ammonia-lyase 2 involved in lignin biosynthesis, observed that, under ozone stress, enzyme activity increased due to de-nitrosation. On the other hand, an S-nitrosoproteome analysis of the root tips of peanut plants under aluminum stress, causing programmed cell death, has identified 402 S-nitrosated proteins, which closely correlated with an increase in GSNO content as a consequence of the inhibition of GSNOR activity by S-nitrosation (Pan et al. 2021). A site-specific nitrosoproteomic study of tomato plants under sodium alkaline stress has identified 334 S-nitrosated proteins in 425 different S-nitrosated loci. These proteins were involved in a wide range of metabolic processes, such as NO homeostasis and ROS metabolisms, as well as Ca2+, ethylene and mitogen-activated protein kinase (MAPK) signaling. In this study, potential key target proteins, including ACO, ascorbate peroxidase (APX) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), were selected (Gong and Shi 2019). In silico analyses have been used to complement the study of these phenomena, while different bioinformatic platforms have also been developed for these purposes, although it will eventually be necessary to experimentally confirm all the theoretical data reported (Kolbert and Lindermayr 2021).
Persulfidation
H2S is a gasotransmitter that is endogenously generated as part of the sulfur and Cys metabolism (González-Gordo et al. 2020b). This molecule in plant cells has been shown to be involved in a myriad of physiological and stressful processes through multiple interactions with phytohormones and other signaling molecules, including H2O2, NO and melatonin (Corpas 2019, Corpas and Palma 2020). At the protein level, H2S mediates a PTM called persulfidation, previously known as S-sulfhydration, which involves H2S interactions with a thiol group of susceptible Cys (Aroca et al. 2015, Aroca et al. 2018, 2021a, 2021b, Wang et al. 2021a).
H2S is a gas, but in an aqueous solution, it is dissociated to hydrosulfide (HS–) and sulfide (S2–) anions according to the following reaction: H2S(aqueous solution) ↔ HS− + H + ↔ S2− + 2 H+. As H2S has an estimated pKa value of 6.8 at 37°C, the hydrosulfide anion (HS-) predominates at the physiological pH of 7.4. Using a modified BSM combined with LC–MS/MS analysis, a total of 106 putative persulfidated proteins were initially identified in Arabidopsis leaves (Aroca et al. 2015). Later, using the tag-switch method, the number of persulfidated proteins identified increased to 2015 in the same plant species (Aroca et al. 2017) (Table 2). Like S-glutathionylation, persulfidation has been considered a protection mechanism that generates resistance to irreversible protein oxidation (Pantaleno et al. 2021).
Analysis of the oxiPTM persulfidation, whose importance in plant physiology has been demonstrated with the aid of new methodological approaches, has highlighted the role of H2S under physiological and stressful conditions (Kouroussis et al. 2019, Fu et al. 2020, Zhao et al. 2020). A comparative analysis of protein persufidation, S-glutathionylation and S-nitrosation has shown that persulfidation plays a more prominent role than the other two oxiPTMs in Arabidopsis (Aroca et al. 2018). The physiological importance of persulfidation has been demonstrated in processes such as ROS metabolism regulation in which certain antioxidant enzymes such as APX and catalase have been reported to be persulfidated (Table 1) (Aroca et al. 2015, Corpas et al. 2019). In tomato (Solanum lycopersicum) plants, the H2S-producing enzyme L-cysteine desulfhydrase (SlLCD1), a nuclearly encoded isozyme, is involved in the regulation of fruit ripening (Hu et al. 2020). Persulfidation is also involved in Arabidopsis stomatal closure through the persulfidation of different proteins in the cascade of signals, including SnRK2.6 (Chen et al. 2020), ABAI4 (Zhou et al. 2021), RBOHD and the L-cysteine desulfhydrase (Shen et al. 2020), with the latter two proteins playing a role in the generation of O2•- and H2S, respectively. Furthermore, autophagy is regulated through the persulfidation of ATG4 (Laureano-Marín et al. 2020) and ATG18a (Aroca et al. 2021b).
S-cyanylation
Cyanide (HCN), which inhibits mitochondrial cytochrome oxidase and, consequently, the respiratory pathway, is known to have a negative impact on cellular metabolism and was used as a poison during the First and Second World Wars, as well as a further chemical terrorism weapon. HCN can also inhibit other key metalloproteins such as the antioxidant enzymes copper-zinc superoxide dismutase and catalase (Corpas et al. 1998).
Endogenous HCN is part of the cell metabolism as a result of cyanogenic glycosides hydrolysis by β-glycosidases and α-hydroxynitrile lyase (Arenas-Alfonseca et al. 2018, Cressey and Reeve 2019, Gotor et al. 2019) and is also released as a co-product of ethylene biosynthesis (Ansari et al. 2019). Cyanide is mainly detoxified by mitochondrial β-cyanoalanine synthase (CAS-C1) in the following reaction: L-cysteine + HCN → β-cyano-L-alanine + H2S (Machingura et al. 2016). HCN is associated with several physiological regulatory functions such as seed germination, nitrate assimilation and root growth, as well as a co-product of ethylene biosynthesis; HCN is also accumulated as a defense mechanism against plant pathogens and herbivores (Miller and Conn 1980, Siegień and Bogatek 2006, Zidenga et al. 2017). More recently, HCN has gained more prominence as a mediator of the PTM S-cyanylation due to its interaction with thiol groups (Fig. 1). In human blood and plasma, HCN has been observed to interact with albumin disulfide, as well as with heavy and light IgG chains (Fasco et al. 2007, 2011). Given that HCN does not react with free sulfhydryl groups, it has been suggested that S-cyanylation occurs in free Cys residues from GSH or other small molecules such as mixed disulfides.
In higher plants, our limited knowledge of S-cyanylation mainly comes from a pioneer study of Arabidopsis wild-type plants and a CAS-C1 knockout mutant. This proteomic analysis of Arabidopsis roots and leaves identified 163 proteins susceptible to S-cyanylation (García et al. 2019). As HCN detoxification by CAS-C1 also generates H2S, the correlation between both these molecules, which mediate S-cyanylation and persulfidation, raises new questions about their mutual interactions as a mechanism of metabolic regulation (García et al. 2019). Enzymes that are targeted by S-cyanylation include APX and NADP-ICDH (Table 1) that participate also in the regulation of the levels of H2O2 and NADPH, respectively.
S-acylation
This reversible PTM, frequently known as S-palmitoylation, enables palmitate (C16 fatty acid) to be added to the thiol group of a specific Cys from soluble or peripheral membrane proteins through a thioester bond (Hurst and Hemsley 2015, Li and Qi 2017). This reaction is catalyzed by a family of protein S-acyl transferases containing a Asp-His-His-Cys motif in a Cys-rich domain (Batistič 2012, Yuan et al. 2013, Hemsley 2013, 2020). S-acylation increases the hydrophobicity of target proteins, which facilities their attachment to the membrane and, consequently, trafficking regulation (Hemsley 2020) and can be reversed by thioesterases (Zheng et al. 2019).
Proteomic analyses of Arabidopsis using a biotin switch isobaric tagging for relative and absolute quantification-based approach have identified 600 putative S-acylated proteins (Hemsley et al. 2013) and, in poplar cells, several hundred potential S-acylated proteins (Srivastava et al. 2016) (Table 1). Representative proteins that illustrate the importance of S-acylation include Ca2+-dependent protein kinases, calcineurin-B-like proteins, MAPKs, receptor-like kinases, integral membrane transporters, ATPases and soluble N-ethylmaleimide-sensitive factor-activating protein receptors (Zheng et al. 2019). A more recent proteomic analysis of Arabidopsis using an acyl resin-assisted capture assay has expanded the list of potential S-acylated proteins to 2643 (Kumar et al. 2020). The importance of protein S-acylation has been highlighted in meiotic Arabidopsis, which is involved in the development of male and female sporophyte reproductive structures and associated gametophytes (Li et al. 2019).
Table 1 shows the number of identified proteins susceptible to all the oxiPTMs in higher plants described above.
How oxiPTMs are integrated in the metabolism of plant cells?
As previously described, nucleophilic thiol groups of protein Cys residues facilitate redox PTMs, which modify the function of the affected proteins. Research has mainly focused on specific oxiPTMs through the identification of potential targets with the aid of proteomic and LC–MS analyses adapted to each PTM. Each modification is evaluated in relation to its specific functional effect (an increase, decrease, or no effect on purified proteins) usually under in vitro conditions. This key step in the analysis needs to evaluate the effect of each oxiPTM under all cellular conditions, as specific thiol groups can be targeted by several competing PTM-promoting molecules according to their specificity, concentration, microenvironmental conditions and subcellular location, which will finally determine their role in the process.
In higher plants, protein S-nitrosation is the most studied oxiPTM, with available information mainly obtained from Arabidopsis plants. However, other oxiPTMs are attracting increasing attention given the interactions among them and the signaling properties of H2O2, NO and H2S involved in regulating the final function of target proteins. Table 2 shows different proteins regulated by several oxiPTMs. Examples of multiply regulated proteins include NADP-GAPDH, NADP-ICDH and NADP-malic enzyme, which are involved in generating NADPH (Hildebrandt et al. 2015, Niazi et al. 2019, Corpas et al. 2021b), a key molecule for the maintenance of redox homeostasis. On the other hand, biomolecules such as GSH, Prxs and Trxs, which, in turn, buffer cellular redox status, are also involved in regulating oxiPTMs. All these elements provide a detailed picture of complex redox equilibria, which finally determine the effect on the target protein.
Recently, there has been increasing interest in the potential physiological role of the S-nitrosothiol, thionitrous acid (HSNO), in cellular redox regulation, as a consequence of the interplay between NO and H2S (Cortese-Krott et al. 2015, Nava et al. 2016, Chen et al. 2019, Marcolongo et al. 2019, Marozkina and Gaston 2020). The detection of this compound in biological systems whose chemistry is more complex than expected has been a major challenge. In plants, very little is known about HSNO, which, to our knowledge, has only been studied when applied exogenously to alfalfa plants under drought conditions using NOSH-aspirin, which simultaneously releases NO and H2S to generate HSNO. Despite the beneficial effect of this S-nitrosothiol, little information is available concerning any potential oxiPTMs (Antoniou et al. 2020).
Summary and future perspectives
Interest in the different oxiPTMs has been growing of late due to their major physiological role in a wide range of higher plant processes. Over the last 10 years, significant advances have been made in this field with the development of appropriate technical approaches to specifically identify each redox modification. The methodology most commonly used involves proteomic analysis combined with chemoselective probes and mass spectrometry techniques (Yang et al. 2016, Shi and Carroll 2020, Zhang et al. 2021). Fig. 2 shows a comparative analysis of proteins identified, which could be targeted by the principal oxiPTMs of cysteine thiols in the model plant A. thaliana, with a combination of three PTMs, persulfidation, S-sulfenylation and S-nitrosation, affecting over 690 proteins (Aroca et al. 2018).
Fig. 2.
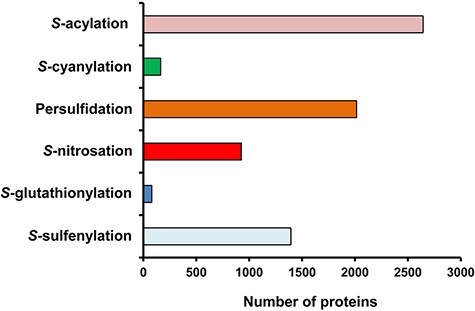
Comparative analysis of the number of proteins identified that could be potentially targeted by one of the main thiol-based oxiPTMs in the model plant Arabidopsis thaliana. These PTMs include 2643 proteins for S-acylation (Kumar et al. 2020), 163 for S-cyanylation (García et al. 2019), 2015 for persulfidation (Aroca et al. 2017), 926 for S-nitrosation (Hu et al. 2015), 79 for S-glutathionylation (Dixon et al. 2005) and 1394 for S-sulfenylation (Huang et al. 2019)
Although some oxiPTMs have been studied in the algal model Chlamydomonas (Berger et al. 2016, De Mia et al. 2019), most studies have been carried out on Arabidopsis plants. Our knowledge of these oxiPTMs, therefore, needs to be extended to other plant species, especially those of agronomic interest under adverse environmental conditions, for crop improvement and biotechnological purposes. This is crucial given that signaling molecules, such as H2O2, NO and H2S, and molecules with antioxidant capacity, such as GSH, are directly involved in these mechanisms. Although cross talk between the different oxiPTMs clearly exists, our knowledge concerning this phenomenon remains limited, and more research will need to be carried out in order to boost the potential of these molecular events.
Funding
European Regional Development Fund cofinanced grant from the Spanish Ministry of Science and Innovation (PID2019-103924GB-I00); Plan Andaluz de Investigación, Desarrollo e Innovación (PAIDI 2020) (P18-FR-1359); Junta de Andalucía (group BIO192), Spain to F.J.C. and J.M.P..
Contributor Information
Francisco J Corpas, Department of Biochemistry, Cell and Molecular Biology of Plants, Group of Antioxidants, Free Radicals and Nitric Oxide in Biotechnology, Food and Agriculture, Estación Experimental del Zaidín (Spanish National Research Council, CSIC), C/ Professor Albareda, 1, Granada 18008, Spain.
Salvador González-Gordo, Department of Biochemistry, Cell and Molecular Biology of Plants, Group of Antioxidants, Free Radicals and Nitric Oxide in Biotechnology, Food and Agriculture, Estación Experimental del Zaidín (Spanish National Research Council, CSIC), C/ Professor Albareda, 1, Granada 18008, Spain.
Marta Rodríguez-Ruiz, Department of Biochemistry, Cell and Molecular Biology of Plants, Group of Antioxidants, Free Radicals and Nitric Oxide in Biotechnology, Food and Agriculture, Estación Experimental del Zaidín (Spanish National Research Council, CSIC), C/ Professor Albareda, 1, Granada 18008, Spain.
María A Muñoz-Vargas, Department of Biochemistry, Cell and Molecular Biology of Plants, Group of Antioxidants, Free Radicals and Nitric Oxide in Biotechnology, Food and Agriculture, Estación Experimental del Zaidín (Spanish National Research Council, CSIC), C/ Professor Albareda, 1, Granada 18008, Spain.
José M Palma, Department of Biochemistry, Cell and Molecular Biology of Plants, Group of Antioxidants, Free Radicals and Nitric Oxide in Biotechnology, Food and Agriculture, Estación Experimental del Zaidín (Spanish National Research Council, CSIC), C/ Professor Albareda, 1, Granada 18008, Spain.
Disclosures
The authors have no conflicts of interest to declare.
References
- Abat J.K., Mattoo A.K. and Deswal R. (2008) S-nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata ribulose-1,5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS J. 275: 2862–2872. [DOI] [PubMed] [Google Scholar]
- Airaki M., Sánchez-Moreno L., Leterrier M., Barroso J.B., Palma J.M. and Corpas F.J. (2011) Detection and quantification of S-nitrosoglutathione (GSNO) in pepper (Capsicum annuum L.) plant organs by LC-ES/MS. Plant Cell Physiol. 52: 2006–2015. [DOI] [PubMed] [Google Scholar]
- Aghdam M.S., Mukherjee S., Flores F.B., Arnao M.B., Luo Z. and Corpas F.J. (2021) Functions of melatonin during postharvest of horticultural crops. Plant Cell Physiol. pcab175. 10.1093/pcp/pcab175. [DOI] [PubMed] [Google Scholar]
- Akter S., Huang J., Bodra N., De Smet B., Wahni K., Rombaut D., et al. (2015) DYn-2 Based identification of Arabidopsis sulfenomes. Mol. Cell. Proteom. 14: 1183–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock L.J., Perkins M.V. and Chalker J.M. (2018) Chemical methods for mapping cysteine oxidation. Chem. Soc. Rev. 47: 231–268. [DOI] [PubMed] [Google Scholar]
- Ansari M.W., Kaushik S., Bains G., Tula S., Joshi B., Rani V., et al. (2019) Cyanide produced with ethylene by ACS and its incomplete detoxification by β-CAS in mango inflorescence leads to malformation. Sci. Rep. 9: 18361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou C., Xenofontos R., Chatzimichail G., Christou A., Kashfi K. and Fotopoulos V. (2020) Exploring the potential of nitric oxide and hydrogen sulfide (NOSH)-releasing synthetic compounds as novel priming agents against drought stress in Medicago sativa plants. Biomolecules 10: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arefian M., Bhagya N. and Prasad T.S.K. (2021) Phosphorylation-mediated signalling in flowering: prospects and retrospects of phosphoproteomics in crops. Biol. Rev. Camb. Philos. Soc. 96: 2164–2191. [DOI] [PubMed] [Google Scholar]
- Arenas-Alfonseca L., Gotor C., Romero L.C. and García I. (2018) Role of mitochondrial cyanide detoxification in Arabidopsis root hair development. Plant Signal. Behav. 13: e1537699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca A., Benito J.M., Gotor C. and Romero L.C. (2017) Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. J. Exp. Bot. 68: 4915–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca A., Gotor C. and Romero L.C. (2018) Hydrogen sulfide signaling in plants: emerging roles of protein persulfidation. Front. Plant Sci. 9: 1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca Á., Serna A., Gotor C. and Romero L.C. (2015) S-sulfhydration: a cysteine posttranslational modification in plant systems. Plant Physiol. 168: 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca A., Yruela I., Gotor C. and Bassham D.C. (2021a) Persulfidation of ATG18a regulates autophagy under ER stress in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 118: e2023604118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca A., Zhang J., Xie Y., Romero L.C. and Gotor C. (2021b) Hydrogen sulfide signaling in plant adaptations to adverse conditions: molecular mechanisms. J. Exp. Bot. 72: 5893–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslebagh R., Wormwood K.L., Channaveerappa D., Wetie A.G.N., Woods A.G. and Darie C.C. (2019) Identification of posttranslational modifications (PTMs) of proteins by mass spectrometry. Adv. Exp. Med. Biol. 1140: 199–224. [DOI] [PubMed] [Google Scholar]
- Audagnotto M. and Dal Peraro M. (2017) Protein post-translational modifications: in silico prediction tools and molecular modeling. Comput. Struct. Biotechnol. J. 15: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistič O. (2012) Genomics and localization of the Arabidopsis DHHC-cysteine-rich domain S-acyltransferase protein family. Plant Physiol. 160: 1597–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer S.M., Taylor E.R., Brown S.E., Dahm C.C., Costa N.J., Runswick M.J., et al. (2004) Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: implications for mitochondrial redox regulation and antioxidant DEFENSE. J. Biol. Chem. 279: 47939–47951. [DOI] [PubMed] [Google Scholar]
- Begara-Morales J.C., López-Jaramillo F.J., Sánchez-Calvo B., Carreras A., Ortega-Muñoz M., Santoyo-González F., et al. (2013) Vinyl sulfone silica: application of an open preactivated support to the study of transnitrosylation of plant proteins by S-nitrosoglutathione. BMC Plant Biol. 13: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begara-Morales J.C., Sánchez-Calvo B., Chaki M., Valderrama R., Mata-Pérez C., López-Jaramillo J., et al. (2014) Dual regulation of cytosolic ascorbate peroxidase (APX) by tyrosine nitration and S-nitrosylation. J. Exp. Bot. 65: 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchoam D., Semelak J.A., Cuevasanta E., Mastrogiovanni M., Grassano J.S., Ferrer-Sueta G., et al. (2020) Acidity and nucleophilic reactivity of glutathione persulfide. J. Biol. Chem. 295: 15466–15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender K.W., Wang X., Cheng G.B., Kim H.S., Zielinski R.E. and Huber S.C. (2015) Glutaredoxin AtGRXC2 catalyses inhibitory glutathionylation of Arabidopsis BRI1-associated receptor-like kinase 1 (BAK1) in vitro. Biochem. J. 467: 399–413. [DOI] [PubMed] [Google Scholar]
- Berger H., De Mia M., Morisse S., Marchand C.H., Lemaire S.D., Wobbe L., et al. (2016) A light switch based on protein S-nitrosylation fine-tunes photosynthetic light harvesting in Chlamydomonas. Plant Physiol. 171: 821–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat J.A., Ahmad P. and Corpas F.J. (2021) Main nitric oxide (NO) hallmarks to relieve arsenic stress in higher plants. J. Hazard. Mater. 406: 124289. [DOI] [PubMed] [Google Scholar]
- Calderón A., Lázaro-Payo A., Iglesias-Baena I., Camejo D., Lázaro J.J., Sevilla F., et al. (2017) Glutathionylation of pea chloroplast 2-Cys Prx and mitochondrial Prx IIF affects their structure and peroxidase activity and sulfiredoxin deglutathionylates only the 2-Cys Prx. Front. Plant Sci. 8: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camejo D., Ortiz-Espín A., Lázaro J.J., Romero-Puertas M.C., Lázaro-Payo A., Sevilla F., et al. (2015) Functional and structural changes in plant mitochondrial PrxII F caused by NO. J. Proteomics 119: 112–125. [DOI] [PubMed] [Google Scholar]
- Castro B., Citterico M., Kimura S., Stevens D.M., Wrzaczek M. and Coaker G. (2021) Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nat. Plants 7: 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Jia H., Wang X., Shi C., Wang X., Ma P., et al. (2020) Hydrogen sulfide positively regulates abscisic acid signaling through persulfidation of SnRK2.6 in guard cells. Mol. Plant 13: 732–744. [DOI] [PubMed] [Google Scholar]
- Chen W., Matsunaga T., Neill D.L., Yang C.T., Akaike T. and Xian M. (2019) Rational design of a dual-reactivity-based fluorescent probe for visualizing intracellular HSNO. Angew. Chem., Int. Ed. Engl. 58: 16067–16070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuh K.N. and Pratt M.R. (2015) Chemical methods for the proteome-wide identification of posttranslationally modified proteins. Curr. Opin. Chem. Biol. 24: 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Wang S.B., Venkatraman V., Murray C.I. and Van Eyk J.E. (2013) Cysteine oxidative posttranslational modifications: emerging regulation in the cardiovascular system. Circ. Res. 112: 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear A.C. (2020) (2020) Deciphering protein post-translational modifications using chemical biology tools. Nat. Rev. Chem. 4: 674–695. [DOI] [PubMed] [Google Scholar]
- Corpas F.J. (2019) Hydrogen sulfide: a new warrior against abiotic stress. Trends Plant Sci. 24: 983–988. [DOI] [PubMed] [Google Scholar]
- Corpas F.J., Alché J.D. and Barroso J.B. (2013) Current overview of S-nitrosoglutathione (GSNO) in higher plants. Front. Plant Sci. 4: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas F.J., González-Gordo S., Cañas A. and Palma J.M. (2019) Nitric oxide and hydrogen sulfide in plants: which comes first? J. Exp. Bot. 70: 4391–4404. [DOI] [PubMed] [Google Scholar]
- Corpas F.J., González-Gordo S. and Palma J.M. (2020) Nitric oxide: a radical molecule with potential biotechnological applications in fruit ripening. J. Biotechnol. 324: 211–219. [DOI] [PubMed] [Google Scholar]
- Corpas F.J., González-Gordo S. and Palma J.M. (2021a) Protein nitration: a connecting bridge between nitric oxide (NO) and plant stress. Plant Stress 2: 100026. [Google Scholar]
- Corpas F.J., González-Gordo S. and Palma J.M. (2021b) Nitric oxide and hydrogen sulfide modulate the NADPH-generating enzymatic system in higher plants. J. Exp. Bot. 72: 830–847. [DOI] [PubMed] [Google Scholar]
- Corpas F.J., González-Gordo S. and Palma J.M. (2022a) NO source in higher plants, an unresolved issue. Trends Plant Sci. 27: 116–119. [DOI] [PubMed] [Google Scholar]
- Corpas F.J., González-Gordo S., Rodríguez-Ruiz M., Muñoz-Vargas M.A. and Palma J.M. (2022b) Nitric oxide and hydrogen sulfide share regulatory functions in higher plant events. BioCell 46: 1–5. [Google Scholar]
- Corpas F.J. and Palma J.M. (2020) H2S signaling in plants and applications in agriculture. J. Adv. Res. 24: 131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas F.J., Sandalio L.M., del Río L.A. and Trelease R.N. (1998) Copper-zinc superoxide dismutase is a constituent enzyme of the matrix of peroxisomes in the cotyledons of oilseed plants. New Phytol. 138: 307–314. [DOI] [PubMed] [Google Scholar]
- Cortese-Krott M.M., Kuhnle G.G., Dyson A., Fernandez B.O., Grman M., DuMond J.F., et al. (2015) Key bioactive reaction products of the NO/H2S interaction are S/N-hybrid species, polysulfides, and nitroxyl. Proc. Natl. Acad. Sci. U.S.A. 112: E4651–E4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressey P. and Reeve J. (2019) Metabolism of cyanogenic glycosides: a review. Food Chem. Toxicol. 125: 225–232. [DOI] [PubMed] [Google Scholar]
- da Fonseca-Pereira P., Daloso D.M., Gago J., de Oliveira Silva F.M., Condori-Apfata J.A., Florez-Sarasa I., et al. (2019) The mitochondrial thioredoxin system contributes to the metabolic responses under drought episodes in Arabidopsis. Plant Cell Physiol. 60: 213–229. [DOI] [PubMed] [Google Scholar]
- Datta R., Kumar D., Sultana A., Hazra S., Bhattacharyya D. and Chattopadhyay S. (2015) Glutathione regulates 1-aminocyclopropane-1-carboxylate synthase transcription via WRKY33 and 1-aminocyclopropane-1-carboxylate oxidase by modulating messenger RNA stability to induce ethylene synthesis during stress. Plant Physiol. 169: 2963–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mia M., Lemaire S.D., Choquet Y. and Wollman F.A. (2019) Nitric oxide remodels the photosynthetic apparatus upon s-starvation in Chlamydomonas reinhardtii. Plant Physiol. 179: 718–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet B., Willems P., Fernandez-Fernandez A.D., Alseekh S., Fernie A.R., Messens J., et al. (2019) In vivo detection of protein cysteine sulfenylation in plastids. Plant J. 97: 765–778. [DOI] [PubMed] [Google Scholar]
- Diaz-Vivancos P., de Simone A., Kiddle G. and Foyer C.H. (2015) Glutathione-linking cell proliferation to oxidative stress. Free Radic. Biol. Med. 89: 1154–1164. [DOI] [PubMed] [Google Scholar]
- Dixon D.P., Skipsey M., Grundy N.M. and Edwards R. (2005) Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol. 138: 2233–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorion S., Ouellet J.C. and Rivoal J. (2021) Glutathione metabolism in plants under stress: beyond reactive oxygen species detoxification. Metabolites 11: 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares A., Rossignol M. and Peltier J.B. (2011) Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochem. Biophys. Res. Commun. 416: 331–336. [DOI] [PubMed] [Google Scholar]
- Fasco M.J., Hauer C.R.I.I.I., Stack R.F., O’hehir C., Barr J.R. and Eadon G.A. (2007) Cyanide adducts with human plasma proteins: albumin as a potential exposure surrogate. Chem. Res. Toxicol. 20: 677–684. [DOI] [PubMed] [Google Scholar]
- Fasco M.J., Stack R.F., Lu S., Hauer C.R. 3rd, Schneider E., Dailey M., et al. (2011) Apr 18;Unique cyanide adduct in human serum albumin: potential as a surrogate exposure marker. Chem. Res. Toxicol. 24: 505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovic M.R., Zivanovic J., Alvarez B. and Banerjee R. (2018) Chemical biology of H2S signaling through persulfidation. Chem. Rev. 118: 1253–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C.H. and Noctor G. (2011) Ascorbate and glutathione: the heart of the redox hub. Plant Physiol. 155: 2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G. and van Wijk K.J. (2015) Posttranslational protein modifications in plant metabolism. Plant Physiol. 169: 1469–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Liu K., Ferreira R.B., Carroll K.S. and Yang J. (2019) Proteome-wide analysis of cysteine S-sulfenylation using a benzothiazine-based probe. Curr. Protoc. Protein Sci. 95: e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Liu K., He J., Tian C., Yu X. and Yang J. (2020) Direct proteomic mapping of cysteine persulfidation. Antioxid. Redox Signal. 33: 1061–1076. [DOI] [PubMed] [Google Scholar]
- Gao X.H., Bedhomme M., Veyel D., Zaffagnini M. and Lemaire S.D. (2009) Methods for analysis of protein glutathionylation and their application to photosynthetic organisms. Mol. Plant 2: 218–235. [DOI] [PubMed] [Google Scholar]
- García I., Arenas-Alfonseca L., Moreno I., Gotor C. and Romero L.C. (2019) HCN regulates cellular processes through posttranslational modification of proteins by S-cyanylation. Plant Physiol. 179: 107–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietler M., Nykiel M., Orzechowski S., Fettke J. and Zagdańska B. (2016) Proteomic analysis of S-nitrosylated and S-glutathionylated proteins in wheat seedlings with different dehydration tolerances. Plant Physiol. Biochem. 108: 507–518. [DOI] [PubMed] [Google Scholar]
- Gong B. and Shi Q. (2019) Identififying S-nitrosylated proteins and unraveling S-nitrosoglutathione reductase-modulated sodic alkaline tolerance in Solanum lycopersicum L. Plant Physiol. Biochem. 142: 84–93. [DOI] [PubMed] [Google Scholar]
- González-Gordo S., Bautista R., Claros M.G., Cañas A., Palma J.M. and Corpas F.J. (2019) Nitric oxide-dependent regulation of sweet pepper fruit ripening. J. Exp. Bot. 70: 4557–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Gordo S., Palma J.M. and Corpas F.J. (2020b) Appraisal of H2S metabolism in Arabidopsis thaliana: In silico analysis at the subcellular level. Plant Physiol. Biochem. 155: 579–588. [DOI] [PubMed] [Google Scholar]
- González-Gordo S., Rodríguez-Ruiz M., Palma J.M. and Corpas F.J. (2020a) superoxide radical metabolism in sweet pepper (Capsicum annuum L.) fruits is regulated by ripening and by a NO-enriched environment. Front. Plant Sci. 11: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotor C., García I., Á A., Laureano-Marín A.M., Arenas-Alfonseca L., Jurado-Flores A., et al. (2019) Signaling by hydrogen sulfide and cyanide through post-translational modification. J. Exp. Bot. 70: 4251–4265. [DOI] [PubMed] [Google Scholar]
- Gough C. and Sadanandom A. (2021) Understanding and exploiting post-translational modifications for plant disease resistance. Biomolecules 11: 1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra D., Ballard K., Truebridge I. and Vierling E. (2016) S-Nitrosation of conserved cysteines modulates activity and stability of s-nitrosoglutathione reductase (GSNOR). Biochemistry 55: 2452–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta D.K., Huang H.G. and Corpas F.J. (2013) Lead tolerance in plants: strategies for phytoremediation. Environ. Sci. Pollut. Res. Int. 20: 2150–2161. [DOI] [PubMed] [Google Scholar]
- Gupta K.J., Kolbert Z., Durner J., Lindermayr C., Corpas F.J., Brouquisse R., et al. (2020) Regulating the regulator: nitric oxide control of post-translational modifications. New Phytol. 227: 1319–1325. [DOI] [PubMed] [Google Scholar]
- Gurrieri L., Distefano L., Pirone C., Horrer D., Seung D., Zaffagnini M., et al. (2019) The thioredoxin-regulated α-amylase 3 of Arabidopsis thaliana is a target of S-glutathionylation. Front. Plant Sci. 10: 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley P.A. (2020) S-acylation in plants: an expanding field. Biochem. Soc. Trans. 48: 529–536. [DOI] [PubMed] [Google Scholar]
- Hemsley P.A., Weimar T., Lilley K.S., Dupree P. and Grierson C.S. (2013) A proteomic approach identifies many novel palmitoylated proteins in Arabidopsis. New Phytol. 197: 805–814. [DOI] [PubMed] [Google Scholar]
- Hildebrandt T., Knuesting J., Berndt C., Morgan B. and Scheibe R. (2015) Cytosolic thiol switches regulating basic cellular functions: GAPDH as an information hub? Biol. Chem. 396: 523–537. [DOI] [PubMed] [Google Scholar]
- Holzmeister C., Fröhlich A., Sarioglu H., Bauer N., Durner J. and Lindermayr C. (2011) Proteomic analysis of defense response of wildtype Arabidopsis thaliana and plants with impaired NO– homeostasis. Proteomics 11: 1664–1683. [DOI] [PubMed] [Google Scholar]
- Hu J., Huang X., Chen L., Sun X., Lu C., Zhang L., et al. (2015) Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol. 167: 1731–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K., Zhang X., Yao G., Rong Y., Ding C., Tang J., et al. (2020) A nuclear-localized cysteine desulhydrase plays a role in fruit ripening in tomato. Hortic. Res. 7: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Willems P., Wei B., Tian C., Ferreira R.B., Bodra N., et al. (2019) Mining for protein S-sulfenylation in Arabidopsis uncovers redox-sensitive sites. Proc. Natl. Acad. Sci. U.S.A. 116: 21256–21261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst C.H. and Hemsley P.A. (2015) Current perspective on protein S-acylation in plants: more than just a fatty anchor? J. Exp. Bot. 66: 1599–1606. [DOI] [PubMed] [Google Scholar]
- Iglesias-Baena I., Barranco-Medina S., Lázaro-Payo A., López-Jaramillo F.J., Sevilla F. and Lázaro J.J. (2010) Characterization of plant sulfiredoxin and role of sulphinic form of 2-Cys peroxiredoxin. J. Exp. Bot. 61: 1509–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Iwabuchi M. and Ogawa K. (2003) The sugar-metabolic enzymes aldolase and triose-phosphate isomerase are targets of glutathionylation in Arabidopsis thaliana: detection using biotinylated glutathione. Plant Cell Physiol. 4: 655–660. [DOI] [PubMed] [Google Scholar]
- Kalinina E. and Novichkova M. (2021) Glutathione in protein redox modulation through S-glutathionylation and S-nitrosylation. Molecules 26: 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharbech O., Sakouhi L., Ben Massoud M., Jose Mur L.A., Corpas F.J., Djebali W., et al. (2020) Nitric oxide and hydrogen sulfide protect plasma membrane integrity and mitigate chromium-induced methylglyoxal toxicity in maize seedlings. Plant Physiol. Biochem. 157: 244–255. [DOI] [PubMed] [Google Scholar]
- Kimura H. (2021) Hydrogen sulfide (H2S) and polysulfide (H2Sn) Signaling: the First 25 years. Biomolecules 11: 896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesting J. and Scheibe R. (2018) Small molecules govern thiol redox switches. Trends Plant Sci. 23: 769–782. [DOI] [PubMed] [Google Scholar]
- Kolbert Z., Barroso J.B., Brouquisse R., Corpas F.J., Gupta K.J., Lindermayr C., et al. (2019) A forty year journey: the generation and roles of NO in plants. Nitric Oxide 93: 53–70. [DOI] [PubMed] [Google Scholar]
- Kolbert Z. and Lindermayr C. (2021) Computational prediction of NO-dependent posttranslational modifications in plants: current status and perspectives. Plant Physiol. Biochem. 167: 851–861. [DOI] [PubMed] [Google Scholar]
- Kouroussis E., Adhikari B., Zivanovic J. and Filipovic M.R. (2019) Measurement of protein persulfidation: improved tag-switch method. Methods Mol. Biol. 2007: 37–50. [DOI] [PubMed] [Google Scholar]
- Kumar M., Carr P. and Turner S. (2020) An atlas of Arabidopsis protein S-acylation reveals its widespread role in plant cell organisation of and function. BioRxiv. 10.1101/2020.05.12.090415. [DOI] [PubMed] [Google Scholar]
- Laureano-Marín A.M., Á A., Pérez-Pérez M.E., Yruela I., Jurado-Flores A., Moreno I., et al. (2020) Abscisic acid-triggered persulfidation of the Cys protease ATG4 mediates regulation of autophagy by sulfide. Plant Cell 32: 3902–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence S.R. 2nd, Gaitens M., Guan Q., Dufresne C. and Chen S. (2020) S-Nitroso-proteome revealed in stomatal guard cell response to flg22. Int. J. Mol. Sci. 21: 1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leterrier M., Barroso J.B., Valderrama R., Palma J.M. and Corpas F.J. (2012) Cytosolic NADP-isocitrate dehydrogenase in Arabidopsis leaves and roots. Biol. Plant 56: 705–710. [Google Scholar]
- Leterrier M., Chaki M., Airaki M., Valderrama R., Palma J.M., Barroso J.B., et al. (2011) Function of S-nitrosoglutathione reductase (GSNOR) in plant development and under biotic/abiotic stress. Plant Signal. Behav. 6: 789–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Tang H. (2016) Computational Methods in Mass Spectrometry-Based Proteomics. In Shen, B., Tang, H. and Jiang, X. Edited by Translational Biomedical Informatics. Advances in Experimental Medicine and Biology. Vol. 939 pp. 63–89. Springer, Singapore. [DOI] [PubMed] [Google Scholar]
- Li Y., Li H.J., Morgan C., Bomblies K., Yang W. and Qi B. (2019) Both male and female gametogenesis require a fully functional protein S-acyl transferase 21 in Arabidopsis thaliana. Plant J. 100: 754–767. [DOI] [PubMed] [Google Scholar]
- Li Y. and Qi B. (2017) Progress toward understanding protein S-acylation: prospective in plants. Front. Plant Sci. 8: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebthal M., Schuetze J., Dreyer A., Mock H.P. and Dietz K.J. (2020) Redox conformation-specific protein-protein interactions of the 2-cysteine peroxiredoxin in Arabidopsis. Antioxidants (Basel) 9: 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C., Saalbach G. and Durner J. (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 137: 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machingura M., Salomon E., Jez J.M. and Ebbs S.D. (2016) The β-cyanoalanine synthase pathway: beyond cyanide detoxification. Plant Cell Environ. 39: 2329–2341. [DOI] [PubMed] [Google Scholar]
- Mailloux R.J. and Treberg J.R. (2016) Protein S-glutathionlyation links energy metabolism to redox signaling in mitochondria. Redox Biol. 8: 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcolongo J.P., Venâncio M.F., Rocha W.R., Doctorovich F. and Olabe J.A. (2019) NO/H2S “Crosstalk” Reactions. The role of thio-nitrites (SNO-) and perthionitrites (SSNO-). Inorg. Chem. 58: 14981–14997. [DOI] [PubMed] [Google Scholar]
- Marozkina N. and Gaston B. (2020) An update on thiol signaling: S-Nitrosothiols, hydrogen sulfide and a putative role for thionitrous Acid. Antioxidants (Basel) 9: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí M.C., Jiménez A. and Sevilla F. (2020) thioredoxin network in plant mitochondria: cysteine S-posttranslational modifications and stress conditions. Front. Plant Sci. 11: 571288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.M. and Conn E.E. (1980) Metabolism of hydrogen cyanide by higher plants. Plant Physiol. 65: 1199–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra V., Singh P., Tripathi D.K., Corpas F.J. and Singh V.P. (2021) Nitric oxide and hydrogen sulfide: an indispensable combination for plant functioning. Trends Plant Sci. 26: 1270–1285. [DOI] [PubMed] [Google Scholar]
- Møller I.M., Igamberdiev A.U., Bykova N.V., Finkemeier I., Rasmusson A.G. and Schwarzländer M. (2020) Matrix redox physiology governs the regulation of plant mitochondrial metabolism through posttranslational protein modifications. Plant Cell 32: 573–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Schüssele S.J., Bohle F., Rossi J., Trost P., Meyer A.J. and Zaffagnini M. (2021) Plasticity in plastid redox networks: evolution of glutathione-dependent redox cascades and glutathionylation sites. BMC Plant Biol. 21: 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Vargas M.A., González-Gordo S., Cañas A., López-Jaramillo J., Palma J.M. and Corpas F.J. (2018) Endogenous hydrogen sulfide (H2S) is up-regulated during sweet pepper (Capsicum annuum L.) fruit ripening. In vitro analysis shows that NADP-dependent isocitrate dehydrogenase (ICDH) activity is inhibited by H2S and NO. Nitric Oxide 81: 36–45. [DOI] [PubMed] [Google Scholar]
- Muñoz-Vargas M.A., González-Gordo S., Palma J.M. and Corpas F.J. (2020) Inhibition of NADP-malic enzyme activity by H2S and NO in sweet pepper (Capsicum annuum L.) fruits. Physiol. Plant. 168: 278–288. [DOI] [PubMed] [Google Scholar]
- Nava M., Martin-Drumel M.A., Lopez C.A., Crabtree K.N., Womack C.C., Nguyen T.L., et al. (2016) spontaneous and selective formation of HSNO, a crucial intermediate linking H2S and nitroso chemistries. J. Am. Chem. Soc. 138: 11441–11444. [DOI] [PubMed] [Google Scholar]
- Navrot N., Finnie C., Svensson B. and Hägglund P. (2011) Plant redox proteomics. J. Proteomics 74: 1450–1462. [DOI] [PubMed] [Google Scholar]
- Niazi A.K., Bariat L., Riondet C., Carapito C., Mhamdi A., Noctor G., et al. (2019) Cytosolic isocitrate dehydrogenase from Arabidopsis thaliana is regulated by glutathionylation. Antioxidants (Basel) 8: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L., Yu J., Liao W., Xie J., Yu J., Lv J., et al. (2019) Proteomic investigation of S-nitrosylated proteins during NO-induced adventitious rooting of cucumber. Int. J. Mol. Sci. 20: 5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma J.M., Mateos R.M., López-Jaramillo J., Rodríguez-Ruiz M., González-Gordo S., Lechuga-Sancho A.M., et al. (2020) Plant catalases as NO and H2S targets. Redox Biol. 34: 101525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C., Li X., Yao S., Luo S., Liu S., Wang A., et al. (2021) S-nitrosated proteomic analysis reveals the regulatory roles of protein S-nitrosation and S-nitrosoglutathione reductase during Al-induced PCD in peanut root tips. Plant Sci. 308: 110931. [DOI] [PubMed] [Google Scholar]
- Pantaleno R., Scuffi D. and Garcıa-Mata C. (2021) Hydrogen sulphide as a guard cell network regulator. New Phytol. 230: 451–456. [DOI] [PubMed] [Google Scholar]
- Péter C., Nagy F. and Viczián A. (2021) SUMOylation of different targets fine-tunes phytochrome signaling. New Phytol. 232: 1201–1211. [DOI] [PubMed] [Google Scholar]
- Poole L.B. (2015) The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 80: 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puerto-Galán L., Pérez-Ruiz J.M., Guinea M. and Cejudo F.J. (2015) The contribution of NADPH thioredoxin reductase C (NTRC) and sulfiredoxin to 2-Cys peroxiredoxin overoxidation in Arabidopsis thaliana chloroplasts. J. Exp. Bot. 66: 2957–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puyaubert J., Fares A., Rézé N., Peltier J.B. and Baudouin E. (2014) Identification of endogenously S-nitrosylated proteins in Arabidopsis plantlets: effect of cold stress on cysteine nitrosylation level. Plant Sci. 215-216: 150–156. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ruiz M., Aparicio-Chacón M.V., Palma J.M. and Corpas F.J. (2019a) Arsenate disrupts ion balance, sulfur and nitric oxide metabolisms in roots and leaves of pea (Pisum sativum L.) plants. Environ. Exp. Bot. 161: 143–156. [Google Scholar]
- Rodríguez-Ruiz M., González-Gordo S., Cañas A., Campos M.J., Paradela A., Corpas F.J., et al. (2019b) Sweet pepper (Capsicum annuum L.) fruits contains an atypical peroxisomal catalase that is modulated by reactive oxygen and nitrogen species. Antioxidants 8: 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Puertas M.C., Campostrini N., Mattè A., Righetti P.G., Perazzolli M., Zolla L., et al. (2008) Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics 8: 1459–1469. [DOI] [PubMed] [Google Scholar]
- Romero-Puertas M.C., Laxa M., Mattè A., Zaninotto F., Finkemeier I., Jones A.M.E., et al. (2007) S‐nitrosylation of peroxiredoxin II E promotes peroxynitrite‐mediated tyrosine nitration. Plant Cell 19: 4120–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos G., Foloppe N. and Messens J. (2013) Understanding the pK(a) of redox cysteines: the key role of hydrogen bonding. Antioxid. Redox Signal. 18: 94–127. [DOI] [PubMed] [Google Scholar]
- Rouhier N., Cerveau D., Couturier J., Reichheld J.P. and Rey P. (2015) Involvement of thiol-based mechanisms in plant development. Biochim. Biophys. Acta 1850: 1479–1496. [DOI] [PubMed] [Google Scholar]
- Sakamoto A., Ueda M. and Morikawa H. (2002) Arabidopsis glutathione-dependent formaldehyde dehydrogenase is an S-nitrosoglutathione reductase. FEBS Lett. 515: 20–24. [DOI] [PubMed] [Google Scholar]
- Sánchez-Guerrero A., Nadal M., Florez-Sarasa I., Ribas-Carbó M., Vallarino J.G., De Brasi-Velasco S., et al. (2021) Decreased levels of thioredoxin o1 influences stomatal development and aperture but not photosynthesis under non-stress and saline conditions. Int. J. Mol. Sci. 22: 1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton M.D., Chock P.B. and Mieyal J.J. (2005) Glutaredoxin: role in reversible protein s-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid. Redox Signal. 7: 348–366. [DOI] [PubMed] [Google Scholar]
- Shen J., Zhang J., Zhou M., Zhou H., Cui B., Gotor C., et al. (2020) Persulfidation-based modification of cysteine desulfhydrase and the NADPH Oxidase RBOHD controls guard cell abscisic acid signaling. Plant Cell 32: 1000–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. and Carroll K.S. (2020) Activity-based sensing for site-specific proteomic analysis of cysteine oxidation. Acc. Chem. Res. 53: 20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortreed M.R., Wenger C.D., Frey B.L., Sheynkman G.M., Scalf M., Keller M.P., et al. (2015) Global identification of protein post-translational modifications in a single-pass database search. J. Proteome Res. 14: 4714–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegień I. and Bogatek R. (2006) Cyanide action in plants—from toxic to regulatory. Acta Physiol. Plant. 28: 483–497. [Google Scholar]
- Srivastava V., Weber J.R., Malm E., Fouke B.W. and Bulone V. (2016) Proteomic analysis of a poplar cell suspension culture suggests a major role of protein S-acylation in diverse cellular processes. Front. Plant Sci. 7: 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata T., Jung M., Matsunaga T., Ida T., Morita M., Motohashi H., et al. (2021) Methods in sulfide and persulfide research. Nitric Oxide 116: 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D.X., Manchester L.C., Esteban-Zubero E., Zhou Z. and Reiter R.J. (2015) Melatonin as a potent and inducible endogenous antioxidant: synthesis and metabolism. Molecules 20: 18886–18906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanou G., Job C., Rajjou L., Arc E., Belghazi M., Diamantidis G., et al. (2009) Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J. 60: 795–804. [DOI] [PubMed] [Google Scholar]
- Tichá T., Lochman J., Činčalová L., Luhová L. and Petřivalský M. (2017) Redox regulation of plant S-nitrosoglutathione reductase activity through post-translational modifications of cysteine residues. Biochem. Biophys. Res. Commun. 494: 27–33. [DOI] [PubMed] [Google Scholar]
- Trujillo M., Alvarez B. and Radi R. (2016) One- and two-electron oxidation of thiols: mechanisms, kinetics and biological fates. Free Radic. Res. 50: 150–171. [DOI] [PubMed] [Google Scholar]
- Turell L., Zeida A. and Trujillo M. (2020) Mechanisms and consequences of protein cysteine oxidation: the role of the initial short-lived intermediates. Essays Biochem. 64: 55–66. [DOI] [PubMed] [Google Scholar]
- Ulrich K. and Jakob U. (2019) The role of thiols in antioxidant systems. Free Radic. Biol. Med. 140: 14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzo E., Ghirardo A., Merl-Pham J., Lindermayr C., Heller W., Hauck S.M., et al. (2014) S-Nitroso-proteome in poplar leaves in response to acute ozone stress. PLoS One 9: e106886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelsang L. and Dietz K.J. (2020) Regulatory thiol oxidation in chloroplast metabolism, oxidative stress response and environmental signaling in plants. Biochem. J. 477: 1865–1878. [DOI] [PubMed] [Google Scholar]
- Vu L.D., Gevaert K. and De Smet I. (2018) Protein language: post-translational modifications talking to each other. Trends Plant Sci. 23: 1068–1080. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang Y., Lv Q., Wang L., Du J., Bao F., et al. (2017) Nitric oxide modifies root growth by S-nitrosylation of plastidial glyceraldehyde-3-phosphate dehydrogenase. Biochem. Biophys. Res. Commun. 488: 88–94. [DOI] [PubMed] [Google Scholar]
- Wang P., Fang H., Gao R. and Liao W. (2021a) Protein persulfidation in plants: function and mechanism. Antioxidants (Basel) 10: 1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Zhang Q., Li S., Cheng B., Xue H., Wei Z., et al. (2021b) iCysMod: an integrative database for protein cysteine modifications in eukaryotes. Brief. Bioinf. 22: bbaa400. [DOI] [PubMed] [Google Scholar]
- Wang W., Li A., Zhang Z. and Chu C. (2021c) Posttranslational modifications: regulation of nitrogen utilization and signaling. Plant Cell Physiol. 62: 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszczak C., Akter S., Eeckhout D., Persiau G., Wahni K., Bodra N., et al. (2014) Sulfenome mining in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 111: 11545–11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B., Willems P., Huang J., Tian C., Yang J., Messens J., et al. (2020) Identification of sulfenylated cysteines in Arabidopsis thaliana proteins using a disulfide-linked peptide reporter. Front. Plant Sci. 11: 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Luo X., Li Y., Chen L., Ma W., Huang J., et al. (2018) DeepNitro: prediction of protein nitration and nitrosylation sites by deep learning. Genomics Proteomics Bioinf. 16: 294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Carroll K.S. and Liebler D.C. (2016) The expanding landscape of the thiol redox proteome. Mol. Cell. Proteom. 15: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Mu J., Chen L., Feng J., Hu J., Li L., et al. (2015) S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol. 167: 1604–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Gupta V., Carroll K.S. and Liebler D.C. (2014) Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat. Commun. 5: 4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokochi Y., Fukushi Y., Wakabayashi K.I., Yoshida K. and Hisabori T. (2021) Oxidative regulation of chloroplast enzymes by thioredoxin and thioredoxin-like proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 118: e2114952118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Hara A., Sugiura K., Fukaya Y. and Hisabori T. (2018) Thioredoxin-like2/2-Cys peroxiredoxin redox cascade supports oxidative thiol modulation in chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 115: E8296–E8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Zhang S., Sun M., Liu S., Qi B. and Li X. (2013) Putative DHHC-cysteine-rich domain S-acyltransferase in plants. PLoS One 8: e75985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun B.W., Feechan A., Yin M., Saidi N.B., Le Bihan T., Yu M., et al. (2011) S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478: 264–268. [DOI] [PubMed] [Google Scholar]
- Zaffagnini M., Bedhomme M., Lemaire S.D. and Trost P. (2012) The emerging roles of protein glutathionylation in chloroplasts. Plant Sci. 18: 86–96. [DOI] [PubMed] [Google Scholar]
- Zaffagnini M., Marchand C.H., Malferrari M., Murail S., Bonacchi S., Genovese D., et al. (2019) Glutathionylation primes soluble glyceraldehyde-3-phosphate dehydrogenase for late collapse into insoluble aggregates. Proc. Natl. Acad. Sci. U.S.A. 116: 26057–26065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M., Michelet L., Marchand C., Sparla F., Decottignies P., Le Maréchal P., et al. (2007) The thioredoxin-independent isoform of chloroplastic glyceraldehyde-3-phosphate dehydrogenase is selectively regulated by glutathionylation. FEBS J. 274: 212–226. [DOI] [PubMed] [Google Scholar]
- Zechmann B. (2018) Compartment-specific importance of ascorbate during environmental stress in plants. Antioxid. Redox Signal. 29: 1488–1501. [DOI] [PubMed] [Google Scholar]
- Zhang J., Ye Z.-W., Singh S., Townsend D.M. and Tew K.D. (2018) An evolving understanding of the S-glutathionylation cycle in pathways of redox regulation. Free Radic. Biol. Med. 120: 204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Gaffrey M.J., Li X. and Qian W.J. (2021) Characterization of cellular oxidative stress response by stoichiometric redox proteomics. Am. J. Physiol., Cell Physiol. 320: C182–C194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. and Zeng L. (2020) Crosstalk between ubiquitination and other post-translational protein modifications in plant immunity. Plant Commun. 1: 100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Zhang J., Zhou M., Zhou H., Gotor C., Romero L.C., et al. (2020) Current approaches for detection of hydrogen sulfide and persulfidation in biological systems. Plant Physiol. Biochem. 155: 367–373. [DOI] [PubMed] [Google Scholar]
- Zheng L., Liu P., Liu Q., Wang T. and Dong J. (2019) Dynamic protein S-acylation in plants. Int. J. Mol. Sci. 20: 560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Zhang J., Shen J., Zhou H., Zhao D., Gotor C., et al. (2021) Hydrogen sulfide-linked persulfidation of ABI4 controls ABA responses through the transactivation of MAPKKK18 in Arabidopsis. Mol. Plant 14: 921–936. [DOI] [PubMed] [Google Scholar]
- Zidenga T., Siritunga D. and Sayre R.T. (2017) Cyanogen Metabolism in Cassava Roots: impact on protein synthesis and root development. Front. Plant Sci. 8: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]


