Abstract
The microbial ecology of traditional postharvesting processing of vanilla beans (curing) was examined using a polyphasic approach consisting of conventional cultivation, substrate utilization-based and molecular identification of isolates, and cultivation-independent community profiling by 16S ribosomal DNA based PCR-denaturing gradient gel electrophoresis. At two different locations, a batch of curing beans was monitored. In both batches a major shift in microbial communities occurred after short-term scalding of the beans in hot water. Fungi and yeast disappeared, although regrowth of fungi occurred in one batch during a period in which process conditions were temporarily not optimal. Conventional plating showed that microbial communities consisting of thermophilic and thermotolerant bacilli (mainly closely related to Bacillus subtilis, B. licheniformis,, and B. smithii) developed under the high temperatures (up to 65°C) that were maintained for over a week after scalding. Only small changes in the communities of culturable bacteria occurred after this period. Molecular analysis revealed that a proportion of the microbial communities could not be cultured on conventional agar medium, especially during the high-temperature period. Large differences between both batches were observed in the numbers of microorganisms, in species composition, and in the enzymatic abilities of isolated bacteria. These large differences indicate that the effects of microbial activities on the development of vanilla flavor could be different for each batch of cured vanilla beans.
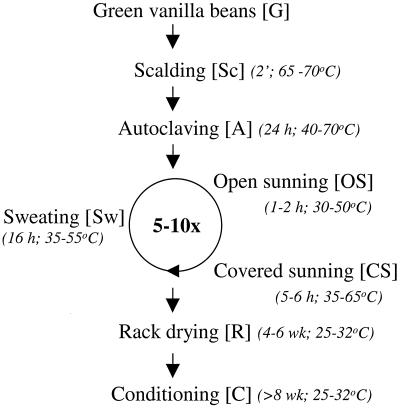
Natural vanilla is the second most valuable flavoring in the food industry ($4,000/kg of natural vanillin [21]) and is derived from the fruits of the tropical orchid Vanilla planifolia. The mature green vanilla beans have no characteristic aroma. Flavor develops during the postharvest processing of the beans (curing). Curing processes differ from country to country, consist of several steps, and are still rather traditional (25). Indonesia is the second largest producer of natural cured vanilla in the world after Madagascar (25). Indonesian curing (see Fig. 1) starts with scalding the beans in hot water (65 to 70°C) for 2 min. After the scalding steps which stops vegetative development and disrupts the cell structures, the drained beans are put in an isolated box for 24 h. During this step, called autoclaving, the beans slowly cool. Subsequently, beans undergo cycles of so-called sunning and sweating, during which beans are exposed to the sun during the daytime and then put in isolated boxes overnight to retain their warmth. A significant part of the aroma, which is thought to be due to bean-derived enzyme activities, is formed during this stage. After 5 to 10 days, depending on weather conditions, the beans are put on racks and dried for a month in a windy, sheltered place to diminish the risk of fungal spoilage. Finally, the beans are put in plastic bags and left to complete the overall vanilla flavor development. This conditioning lasts at least 2 months.
FIG. 1.
Scheme of the postharvesting processing of vanilla beans (curing) in Indonesia. The different steps in the process are shown, along with their approximate durations and temperature ranges (in italic) and the abbreviation (between brackets) used in the text and Fig. 2 to 5.
The lengthy process and the high demand for natural vanilla are the main reasons why traditionally cured vanilla is expensive. People have sought to obtain cheaper natural vanilla-like flavors by using biotechnological processes based on vanilla plant cell cultures (12) or microbial bioconversions (21, 29, 34). However, yields in biotechnological processes are still too low for economic exploitation, and the products also lack the full flavor of natural vanilla (19).
Although many aroma compounds of vanilla have been reported (1, 17, 18), little is known about the processes by which important compounds (vanillin, vanillic acid, p-hydroxybenzaldehyde, p-cresol, 2-phenylalcohol, anisaldehyde, guaiacol, phenyl-acetaldehyde, diacetyl, eugenol, and methyl-cinnamate) are formed during curing. Thermal processes, plant enzyme reactions, and microbial activities are considered important in flavor generation (25). Glucosides, such as glucovanillin, are major aroma precursors in green vanilla beans (35). During the curing process β-glucosidases in the vanilla bean are activated and release aroma compounds from the glucosides. Vanillin is the most important and abundant compound, with concentrations up to 3% (wt/wt). Together with thermal reactions, plant polyphenol oxidisases and peroxidases are assumed to be involved in browning reactions and the production of aroma compounds (25). A microbial contribution to natural vanilla flavor has been suggested but never investigated. Microbial cellulose and hemicellulose degradation in general involves β-glucosidases (8) which, like the plant β-glucosidase, could attack bean glycosides. Degradation of lignin by a wide variety of microorganisms such as white rot fungi, actinomycetes, and some other bacteria also yields aromatic compounds (16). In addition, microbial activities on cell wall compounds release ferulic acid (34) that can be transformed via a large variety of bacteria and fungi into flavor compounds, such as vanillin and guaiacol (29).
Knowledge of the processes contributing to natural vanilla flavor formation will be useful for designing cost-effective processes that yield high-quality vanilla-like flavors. As part of our aim to determine the contribution of microorganisms to vanilla flavor generation, the presence of microorganisms and changes in microbial communities during two curing processes at the largest Indonesian vanilla curing company Djasula Wangi were studied. Microbial communities were determined in a polyphasic approach using conventional plating, identification of dominantly occuring strains, and molecular analysis using 16S/18S ribosomal DNA (rDNA)-based denaturing gradient gel electrophoresis (DGGE) (22). Characteristics that could benefit vanilla flavor were determined for isolated strains.
MATERIALS AND METHODS
Strains.
Type strains Bacillus coagulans DSM1, B. amyloliquefaciens DSM7, B. subtilis DSM10, B. licheniformis DSM13, B. stearothermophilus DSM22, B. thermodenitrificans DSM465, B. thermoglucosidasius DSM2542, B. pallidus DSM3670, B. smithii DSM4216, B. thermocloacae DSM2542, and B. kaustophilus DSM7263, obtained from the Deutsche Sammlung von Microorganismen und Zellkulturen GmbH (DSM), were used for comparison to isolated strains DGGE and Biolog.
Sampling.
Curing was examined at two local branches of the company Djasula Wangi in Indonesia. In June 1998 a curing (ca. 100-kg beans) was followed until the second day of rack drying in Tulungagung, East-Java. Humidity and temperature were continuously measured using Testo 650 equipment (Testo, Almere, The Netherlands). During sunning and rack drying, one sensor was put between the beans and the other was put on top. During sweating one sensor was put in the middle of the jar and the other at the outside. On a regular base, 300-g samples (ca. 30 beans) were withdrawn and immediately stored in sealed plastic bags at 4°C (maximally 7 days) until microbial analysis at the Agricultural University of Bogor, Bogor, Indonesia. Samples from later stages of rack drying and conditioning were sent by normal mail to Bogor. To ensure representative sampling, beans were collected at random directly after the curing process had entered into a next stage, as during each transfer the batch of beans was mixed by workers. In April 1999 a second curing (200 kg) was monitored for two days in Payung, North-Sumatra. Samples were collected as described above and stored at 4°C (maximally 2 days) until microbial analysis in Bogor. After 2 days, 10-kg beans as well as original materials involved in the process, such as cotton cloth, were transferred to Bogor for continuation of the curing process.
Extraction of microorganisms.
A total of 50 g (wet weight) of vanilla beans was cut into 1-cm pieces and put into a 500-ml bottle containing 40 g of glass beads (3 mm in diameter) and 200 ml of 0.85% NaCl salt solution. The bottle was shaken at 200 rpm on a reciprocal shaker, at room temperature, for 1 h and then used for plating and DNA extraction.
Microbial enumeration.
Salt solution containing extracted microorganisms was diluted in a decimal system in 0.85% NaCl. Appropriate dilutions were surface or pour plated. For the enumeration of total microorganisms, normal (3%) and diluted (0.1%) tryptic soy agar (TSA; Difco, Detroit, Mich.) was used. Fungi and yeast were enumerated on potato dextrose agar (PDA; Oxoid, Bastingstoke, United Kingdom) containing 50 μg of tetracycline per ml. Plates were incubated at 30 or 55°C for at most 2 weeks. Numbers were expressed as logarithmic transformed CFU per gram (dry weight) of beans (log CFU/g). The dry weight was determined by cutting beans into 1-cm pieces followed by overnight drying at 100°C.
Isolation and physiological characterization of isolates.
Bacteria or fungi were isolated from plates by picking separate colonies and spreading them on, respectively, 3% TSA or PDA plates. Bacterial strains were characterized based on colony appearance, microscopy, and Gram staining. Identification based on substrate utilization tests was performed with Biolog GN or GP plates (Biolog, Inc., Hayward, Calif.) according to the instructions of the manufacturer. Specific utilization of flavor (precursor) compounds by isolates was tested in Biolog MT plates, using a 250-mg/liter filter-sterilized substrate and the same inoculation procedure as for the Biolog GN and GP plates. Vanillin resistance was determined by incubation for 5 days on TSA plates containing 0, 0.05, 0.1, 0.2, 0.3, 0.4, or 0.5% vanillin (wt/vol). Vanillin, as a 25% (wt/vol) solution in ethanol, was added after sterilization of TSA at 121°C. Protease production was determined on casein-gelatin plates, prepared by dissolving 10 g of casein, 10 g of gelatin, 1 g of tryptic soy broth (TSB) and 15 g of agar per liter of 0.02 M NaOH. The pH was readjusted to 6.5 before sterilization. Positive reactions were scored when a clear halo around the strains was observed. Hydrolysis of hemicellulose, cellulose, and pectin was also determined via plate assays (37) at pH 6.5. For fungal and yeast characterization, PDA and a pH of 5 were used instead of TSA and a pH of 6.5, while the protease assay was performed on skim milk plates (10). A ligninolytic plate assay based on the decoloring of Remazol brilliant blue was used to determine the presumptive abilities of fungal isolates (5). For the determination of peroxidase (9) and β-glucosidase, strains were grown for 3 to 4 days in 2 ml of 0.01% TSB–0.5% xylan–0.5% cellulose. For the β-glucosidase assay, 100 μl of culture or supernatant was mixed with 900 μl of 2.5 mM p-nitrophenyl-β-d-glucopyranoside in 0.1 M phosphate buffer (pH 6.3) and incubated for 4 to 6 h at 37°C. Developed yellow color was measured at 410 nm.
Molecular analysis of isolates and microbial communities.
Microorganisms extracted from the beans were collected via centrifugation for 2 h at 4,500 × g. Cells on the TSA plates with the highest number of separate colonies were collected by suspending the colonies in 0.85% NaCl with a sterile cotton swab and centrifugation for 20 min at 10,000 × g. Cells were washed once with 0.85% NaCl and centrifuged for 20 min at 10,000 × g. Pellets were stored at −30°C and freeze-dried before transport to The Netherlands for molecular analysis. DNA extraction was essentially performed according to the method of Duarte et al. (13). However, pellets were dissolved in 0.8 ml of 120 mM Sodium phosphate buffer (pH 8.0) and mixed with 0.6-g glass beads (0.1 mm in diameter), 100 μl of 20% sodium dodecyl sulfates and 0.7 ml of phenol (pH 8.0). A Biospec bead beater (Techno Lab, Alkmaar, The Netherlands) was used at 4,200 rpm. DNA from cells extracted from the beans was cleaned by one round of Wizard column purification (Promega, Madison, Wis.). For molecular analysis of isolated strains, a colony was transferred to 200 μl of TE (10 mM Tris-HCl, EDTA [pH 8.0]) and 1 μl was used as a template in the PCR. Bacterium-specific PCR was performed in a total volume of 25 μl containing 0.4 μM concentration of primer F341-GC (22), a 0.4 μM concentration of primer R518 (22), a 0.4 mM concentration of deoxynucleoside triphosphates, 10 μg of bovine serum albumin, Expand buffer (Boehringer, Mannheim, Germany), 2.6 U of Expand enzyme, and 1 μl of template. Amplification was performed in a Perkin-Elmer DNA ThermoCycler as follows: 94°C for 4 min, followed by 35 cycles of 94°C for 0.5 min, 54°C for 1 min, and 72°C for 1 min, with a final elongation at 72°C for 5 min. Fragments of 16S rDNA were also amplified using primers U968-GC and L1401 (23), using the same PCR mixture and program. A fungus-specific nested PCR was performed as described by Smit et al. (31). DGGE was performed with the Bio-Rad DCode system. PCR product was loaded onto 1-mm-thick 8% (wt/vol) polyacrylamide (37.5:1 acrylamide-bisacrylamide) gels containing a 40 to 60% linear denaturing gradient for the bacterial PCR and a 25 to 55% gradient for the fungal PCR. A 100% denaturant is defined as 7 M urea and 40% (vol/vol) formamide. Gels were run in 1× TAE buffer (16 mM Tris-HCl [pH 8.0], 0.8 mM sodium-EDTA, 20 mM acetic acid) at 60°C and 70 V for 16 h. Gels were stained in 1× TAE buffer containing 1 μg of ethidium bromide per ml and recorded with a charge-coupled-device camera system (The Imager; Appligen, Illkirch, France).
Cloning and sequencing of 16S rRNA genes of isolates.
PCR primers 8f and 1512r were used to amplify 16S rRNA sequences from isolated strains (14). Products (cleaned with the Qiaquick Rep Purification Kit [Qiagen, Hilden, Germany]) were cloned in Escherichia coli JM109 by using the Promega pGEM-T vector system. Transformants were checked for the correct insert by performing a PCR with pGEM-T specific primers T7 and Sp6 for checking the size of the insert and performing a PCR with F341-GC and R518 to check for the correct banding position in DGGE. Sequencing PCR was carried out with ABI PRISM Dye Termintor Cycle Sequencing Core Kit (Perkin-Elmer), and the purified products were runned in a SEQUAGEL-6 sequence gel (National Diagnostics) in a 373A/DNA Sequencer (Applied Biosystems). Both strands of the 16S rRNA gene were sequenced. Sequences were compared to sequences deposited in the GenBank DNA database by using the BLAST algorithm (2).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the 16S rDNA sequences as determined for the Bacillus isolates mentioned in Table 1 are AF286478 (strain type 1, corresponding to upper band in DGGE), AF286479 (strain type 1, corresponding to lower band in DGGE), AF286482 (strain type 2), AF286486 (strain type 3), AF286481 (strain type 4), and AF286484 (strain type 5).
TABLE 1.
Identities of strains isolated during curing, as determined by three independent methods
| Strain typea | Biolog identification (similarityb) | DGGE identificationc | Closest cultured relative,d GenBank accession no. (% similarity) | Curing stage(s)e at:
|
|
|---|---|---|---|---|---|
| Tulungagung | Payung | ||||
| 1 | B. licheniformis (0.33–0.49) | B. licheniformis (upper bandf) | B. licheniformis, AF276309 (99.0) | A, Sw, R, C | C |
| B. licheniformis, AF276309 (98.1) | |||||
| 2 | B. subtilis (0.29–0.72) | B. subtilis | B. subtilis, Z99104 (97.0) | G, A, Sw, R, C | A, Sw, R, C |
| 3 | B. cereus (0.03–0.16) | NDh | B. firmus, D16268 (96.8) | A, Sw, R | |
| 4 | B. pumilis (0.61) | ND | B. pumilis, AF260751 (98.0) | A, Sw, R | |
| 5 | ND | ND | B. firmus, AJ229200 (97.2) | A, Sw, R | |
| 6 | B. smithii (0.92–0.99g) | B. smithii | ND | A, Sw, R | |
The Type numbers refer to the numbered bands in Fig. 5.
Significant similarities (>0.5) are shown in boldface.
DGGE was performed by using two primer sets: one amplifying the V3 region, the second amplifying the V6-V8 region. Results were compared to DSM type strains.
Determined based on 16S rDNA sequence homology. GenBank accession numbers for 16S rDNA sequences of the isolated strains are mentioned in Materials and Methods.
Abbreviations are as defined in Fig. 2.
B. licheniformis type strain (DSM13) and isolate both showed two bands in DGGE after amplification of the V3 region; only the upper band matched. Clones corresponding to both bands were sequenced.
B. smithii is not in the Biolog database; profiles were compared to that of the type strain DSM4216, incubated at 55°C.
ND, not determined.
RESULTS
Physicochemical changes during vanilla curing.
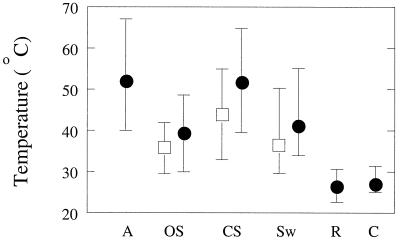
Two curing processes at the company Djasula Wangi were examined, in 1998 at Tulungagung and in 1999 at Payung. The processes were similar and are outlined in Fig. 1. During curing at Tulungagung, the temperature and humidity were monitored until the second day of rack drying. The temperatures during autoclaving and sweating cycles were high and did not drop below 30°C (Fig. 2). When beans were put in thin layers (ca. 3 cm deep) under the sun, the temperature of the beans rose rapidly. The maximum temperature reached 65°C; the averages during open (sun-exposed) and cotton cloth-covered sunning were, respectively, 41 and 52°C. During sunning, a temperature gradient was observed across the bean layer; beans on top of the layer were up to 10°C warmer than the beans at the bottom, which were more shielded from the influence of the sun. During box sweating, the temperature dropped slowly and remained well above the outside temperature (on average, 22°C at night). Beans on the outside lost their temperature much faster than beans in the center of the box. The average temperature during sweating was 39°C; a much higher temperature was observed during autoclaving, when beans had become hot by scalding. Beans were exposed to ambient temperatures (22 to 30°C) during rack drying and conditioning. The humidity during sweating was close to saturation (95 to 100%), while during sunning it varied from 20 to 90% (average, 65%) and during rack drying it was about 75%. Moisture was only slowly lost from the beans; the moisture content of the green beans was 84% (wt/wt) and dropped during sunning and sweating to 75% (Tulungagung, June 1998, ten sweating cycles) and 79% (Payung, April 1999, seven sweating cycles). During rack drying the moisture content decreased to 35% at Tulungagung and to 27% at Payung. The measured pH levels in the bean extracts used for microbial analysis were similar at 4.7 ± 0.2 and 4.8 ± 0.1 for the Tulungagung and Payung curings, respectively. The pH was not affected by the processing steps.
FIG. 2.
Temperature changes during different stages in vanilla curing at Tulungagung. Stages: A, autoclaving; OS, open sunning; CS, closed sunning; Sw, sweating; R, rack drying; C, conditioning (see Fig. 1). Measured minimum and maximum temperatures are indicated by a line; a solid circle indicates the average temperature. For stages in which a temperature gradient is formed, a solid circle represents the average temperature of the beans at the middle of the sweating box (for Sw) or beans directly exposed to sunlight (for OS and CS), while an open box represents the average temperature of beans at the outside of the sweating box or of the underlying beans, respectively.
Changes in fungal and yeast communities during vanilla curing.
Prior to the curing experiments the most suitable method for extracting microorganisms from the beans was determined. Microorganisms were enumerated on TSA after first washing off the microorganisms on the bean surface (by shaking in salt solution in the presence of glass beads), followed by blending. No significant higher numbers of microorganisms were obtained after blending (t test, P > 0.05); thus, the majority of microorganisms was located on the surface of the beans. Liquid from the washed beans could easily be used for collecting cells for DNA isolation, in contrast to the sticky, particle-containing liquid from the blended beans. Therefore, the washing method was used to extract microorganisms from beans throughout this study.
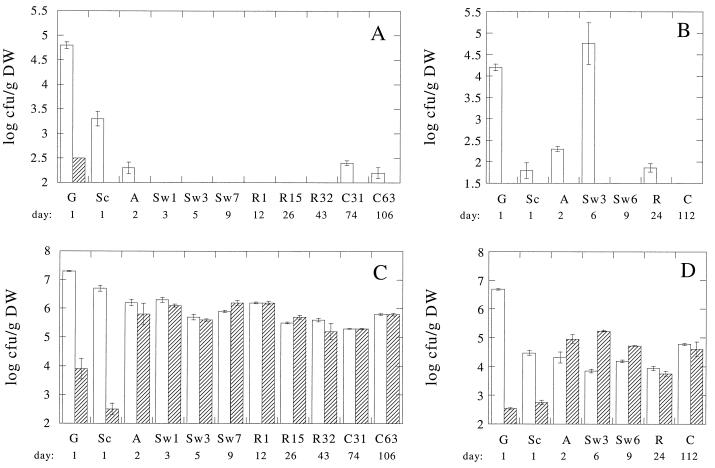
The optimal incubation time for fungi and yeast was 7 days; no further increases in colony forming units were observed with extended incubation. As depicted in Fig. 3A and B, fungi and yeast quickly disappeared during both curing experiments. Yeast was not encountered after scalding. Fungi isolated during sunning and sweating were only able to grow at 30°C but not at the high temperatures prevalent during sunning and sweating. During the Payung curing (Fig. 3B) an increase in fungal numbers was observed between autoclaving and the third cycle of sunning and sweating; this result probably relates to clouded weather conditions which did not allow the beans to heat up much during sunning. When weather conditions improved, the numbers quickly dropped.
FIG. 3.
Changes in microbial numbers during two vanilla curing processes. Numbers were determined at 30°C (open columns) and 55°C (hatched columns) and expressed as the log number per gram (dry weight) of beans. (A) Fungal and yeast numbers at Tulungagung in 1998. (B) Fungal and yeast numbers at Payung in 1999. (C) Bacterial numbers at Tulungagung in 1998. (D) Bacterial numbers at Payung in 1999. Sampling times are indicated by day after the start of the process (lower row), and a letter depicting the stage from which the sample was obtained, followed by a number referring to the number of days the particular stage has been executed at the time of sampling (upper row). Columns: G, green beans; Sc, scalding; A. autoclaving; Sw, sunning-sweating; R, rack drying; C, conditioning (see also Fig. 1). Error bars indicate the standard deviations; all determinations were performed in duplicate.
A variety of fungi and yeast were encountered on the green beans. DGGE of 18S rDNA fragments amplified from yeast isolates revealed five different banding positions for 11 isolates (Fig. 4, yeast isolates), showing considerable diversity in yeast. During curing mainly black and green Aspergillus and Penicillium strains were encountered. Despite the fact that the Aspergillus strains had different morphologies, 18S rDNA PCR fragments of the 15 isolates showed similar mobilities in DGGE (band a in Fig. 4). The 18S rDNA fragments of the three Penicillium isolates had an identical mobility and ended a position lower than that of the Aspergillus isolates in the gel (band b in Fig. 4). Thus, the diversity in fungi isolated during the curing is rather limited. Two isolates from the green beans had an unique position in DGGE (band c in Fig. 4).
FIG. 4.
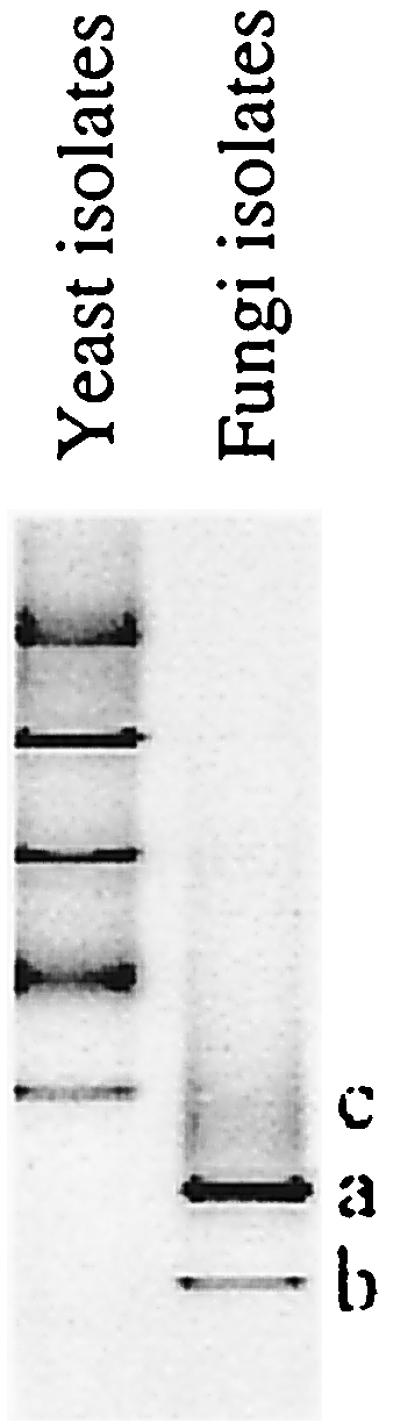
DGGE of 18S rDNA PCR fragments of fungi and yeast cultured from curing beans at Tulungagung and Payung.
DGGE profiling of 18S rDNA PCR fragments obtained from DNA directly extracted from the beans, without intermediate culturing, was attempted but in general failed to give banding patterns in DGGE.
Changes in communities of culturable bacteria during vanilla curing.
The growth of bacteria on TSA was rapid at the incubation temperatures examined (30 and 55°C); after 2 days incubation no new colonies appeared on the plates. For the Tulungagung curing, bacterial numbers were also determined at 45°C, and these were similar to those seen at 30°C (data not shown).
An obvious difference in the total numbers was observed between the two curing processes. At Tulungagung (Fig. 3C), the numbers of microorganisms able to grow at 30 and 55°C were, respectively, about 2 and 1 order of magnitude higher than at Payung (Fig. 3D). On green beans, the numbers of bacteria able to grow at 55°C were much lower than at 30°C in both processes but, after a few cycles of high-temperature sunning and sweating, these numbers increased and became comparable (Tulungagung; Fig. 3C) or higher (Payung; Fig. 3D) than at 30°C. After autoclaving only minor changes in numbers occurred and, even during rack drying and conditioning at a temperature of ca. 30°C, the numbers of thermophilic bacteria remained rather constant.
Severe changes in microbial communities were induced by the scalding and autoclaving steps, as revealed by identification of the isolates based on substrate utilization tests (Biolog system). Isolates from green beans (54 in total) belonged to the genera Pseudomonas, Chyseomonas, Flavimonas, Burkholderia, Enterobacter, Vibrio, Corynebacterium, Bacillus, Staphylococcus, Tsukamurella, Actinomycetes, Leuconostoc, Brevibacterium, Cellumonas, and Rhodococcus. Of these isolates, Actinomycetes and Bacillus strains were able to grow at 55°C. After scalding, only Bacillus strains were cultivated from both processes (179 isolates tested). The high-temperature scalding step evidently has a large influence on the microbial communities. At Tulungagung split beans were also cured. Split beans are not scalded, since this will result in the loss of solids. Instead, they are only exposed to the sun to heat them up. From a sample which was cured for 4 days besides Bacillus strains other bacteria (Xanthomonas, Cellumonas, Vibrio, and Staphylococcus species) were also isolated.
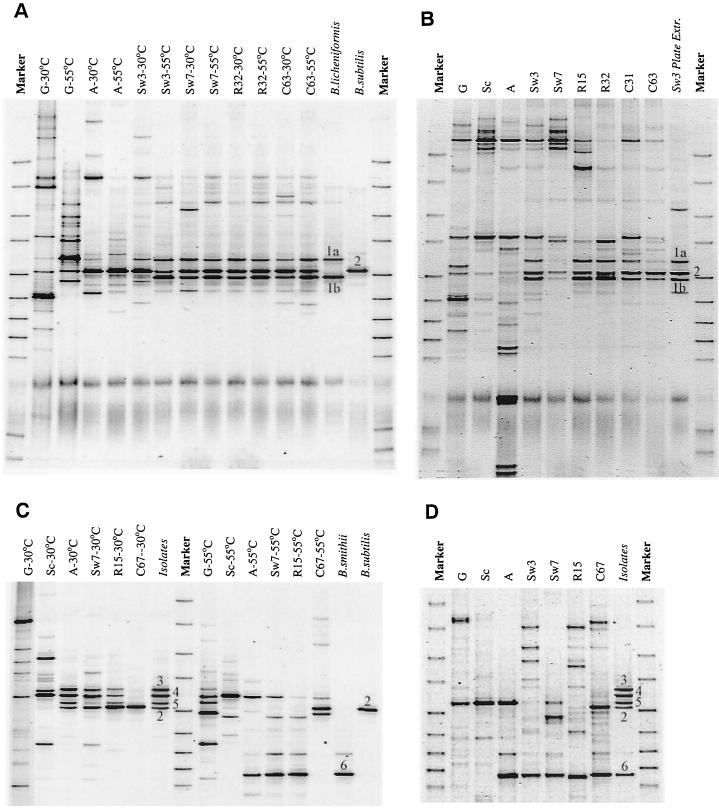
The changes in microbial communities and the decrease in diversity due to scalding and autoclaving were also clearly reflected in 16S rDNA-based DGGE analyses of colonies scrapped from TSA plates (Fig. 5A and C). Green beans (lanes noted with G) contained a more diverse community, as reflected in the number of bands, than did beans during curing, where only a few dominant bands were observed. This agrees well with the Biolog characterization of individual isolates. Culturable microbial communities after scalding differed considerably between the two curings. At the Tulungagung curing a similar DGGE profile consisting of three dominant bands derived from two types of strains (types 1 and 2) was observed for agar plates incubated at 30 and 55°C (Fig. 5A). This was well in line with the fact that comparable numbers of bacilli were found at both temperatures (Fig. 3C). The composition of culturable microbial communities did not change during the curing; only their relative numbers changed (as indicated by the differences in intensity of the individual bands). For the Payung curing (Fig. 5C), a different profile was observed than for the Tulungagung curing. DGGE also revealed that Bacillus strains that developed dominantly on plates incubated at 30°C were different from those growing well at 55°C; for the latter, only one dominant band was observed. Again, this finding agreed well with colony counts; at 55°C higher numbers of bacilli were encountered than at 30°C (Fig. 3D). Furthermore, an obvious change in the microbial community of the Payung curing was observed after rack drying (lane C67 in Fig. 5C), when the profile became comparable to those encountered at Tulungagung. This was confirmed by the identification of isolated strains (Table 1). DGGE profiles of Bacillus strains isolated from the different curing stages (lanes indicated in italics in Fig. 5A and C) matched the dominant bands in DGGE profiles of cells scraped from plates. For the Tulungagung curing two dominant Bacillus species were encountered; for the Payung curing six species were encountered. Only one of the 179 Bacillus isolates revealed a band in DGGE that did not correspond to a dominant band in profiles of colonies extracted from TSA plates.
FIG. 5.
Changes during vanilla curing in DGGE profiles of the amplified V3 region of the bacterial 16S rDNA of microorganisms extracted from TSA plates 3 days after inoculation with an extract of vanilla beans (A and C) and of microbial communities directly isolated from vanilla beans, without intermediate cultivation (B and D). (A and B) Tulungagung experiment. (C and D) Payung experiment. Sampling times are indicated by a letter depicting the stage from which the sample was obtained, followed by a number referring to the number of days the particular stage has been executed at the time of sampling. G, green beans; Sc, scalding; A, autoclaving; Sw, sunning-sweating; R, rack drying; C, conditioning (see also Fig. 1). The temperature refers to the temperature at which the plates were incubated. In italics are indicated the profiles of isolated strains; the numbers refer to different isolates and are explained in the text and Table 1. The blurry bands near the bottom in panels A and B are artifacts of the PCR-DGGE protocol.
Cultivation-independent profiling of bacterial communities.
Effects of culturing media on culturable microbial communities were limited: counts on 3% TSA (heterotrophic) and 0.1% TSA (oligotrophic), as well as DGGE profiles of colonies scraped from the plates, were similar for the Payung experiment (data not shown). The presence of bacteria unable to grow on these diverse media was established by 16S rDNA PCR-DGGE profiling of DNA extracted directly from the beans, without intermediate culturing (Fig. 5B and D), and comparison to the profiles of the culturable microorganisms (Fig. 5A and C, respectively). The presence of not culturable-on-TSA bacteria is especially obvious after autoclaving at Tulungagung (lane A in Fig. 5B). The bands belonging to the culturable microorganisms can hardly be seen in the profile. Assuming an equal PCR efficiency for all DNA templates (although it should be realized that PCR has many pitfalls [36]), this indicates that the actual number of microorganisms is much higher than the 106 CFU/g determined by plating. Only during the last cycles of sunning and sweating did the bands from culturables become dominant, although additional bands remained. For the Payung curing (Fig. 5D), the contribution of nonculturables is relatively lower; the band corresponding to the dominantly culturable thermophilic Bacillus strain is clearly dominant in the profiles. Bands that belong to strains dominantly cultured on plates incubated at 30°C cannot be seen, which relates to the fact that their numbers were much lower than the numbers of the Bacillus strain (type 6) growing at 55°C (Fig. 3D). Interesting is the fact that the latter strain, which was cultured from samples from the sunning-sweating and rack-drying stages (Fig. 5C, lanes A [55°C] to R15 [55°C]), could not be cultivated after conditioning (lane C67), while a band with similar running properties in DGGE remains present throughout the curing, as revealed by analysis of DNA directly extracted from the beans (Fig. 5D, lanes A to C67). The profiles for the Payung curing also showed more time-to-time variation in the composition of the DGGE profiles than those of the Tulungagung curing.
Identification and characterization of isolates.
Identification to the species level and characterization were primarily focused on strains isolated after the scalding step. Strains were profiled in DGGE analyses and compared to profiles of colonies extracted from TSA plates. Identification was performed by substrate utilization-based Biolog identification and 16S rDNA-based molecular identification (sequencing and/or DGGE profiling and comparison to DSM reference strains [using two primer sets, amplifying the V3 and V6-V8 regions of the 16S rRNA genes]). In total, six dominant types of strains were encountered, and their DGGE banding profiles are shown numbered in Fig. 5. All of these strains belonged to the genus Bacillus (Table 1). Biolog identification was of limited use due to the low similarity to profiles in the Biolog database (Table 1), especially for the thermophilic Bacillus strains (only two species in the database). The inability to reliably identify environmental isolates to the species level with Biolog has also been observed by others, and a Biolog identification needs to be confirmed by additional investigations (4, 38). Molecular identification (Table 1) showed that the strains isolated from the Tulungagung curing were related to B. subtilis (type 2) and B. licheniformis (type 1). The later species gave rise to two bands in DGGE when the primers F341-GC and R518 were used to amplify the V3 region, a finding indicative of two different 16S rDNA sequences (Fig. 5). Indeed, three base-pair differences in the V3 region were observed by sequencing. These strains were also isolated from materials used in curing, and two B. subtilis isolates were obtained from green beans. The dominant thermophilic strain (type 6) at Payung was related to B. smithii, with strain type 2 (B. subtilis) present in lower numbers up until rack drying but becoming dominant, together with B. licheniformis (type 1), during conditioning. Isolates only able to grow at 30°C (see Table 2) were related to B. firmus (types 3 and 5) and B. pumilis (type 4). These strains could also be isolated from cotton cloth used in the curing but were not obtained from green beans.
TABLE 2.
Characteristics of isolates from two vanilla curing processes
| Curing station | Strain or samplea | No. of isolates (no. of different strains) | % Strains growing at:
|
% Strains with enzyme activities
|
% Strains with β-glucosidase
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| 30°C | 55°C | Protease | Cellulase | Hemicellulase | Pectinase | Extracellular | Cell bound | |||
| Tulungagung | Green beans | 18 (8) | 100 | 39 | 61 | 61 | 39 | 0 | NDd | ND |
| Type 1 strain | 18 | 100 | 100 | 94 | 100 | 89 | 0 | 0 | 75 | |
| Type 2 strain | 28 | 100 | 100 | 100 | 100 | 100 | 0 | 0 | 75 | |
| Payung | Green bean (30°C)b | 23 (12) | 100 | 8 | 26 | 9 | 17 | 0 | ND | ND |
| Green bean (55°C)b | 13 (4) | 46 | 100 | 38 | 38 | 38 | 0 | 0 | 46 | |
| 30°C isolatesc | 12 (3) | 100 | 0 | 25 | 33 | 17 | 0 | 0 | 17 | |
| Types 1 and 2 | 24 (2) | 100 | 100 | 100 | 96 | 88 | 17 | 0 | 100 | |
| Type 6 strain | 34 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | |
See Table 1.
Temperature indicates the incubation temperature of the plate from which the strains were isolated.
Strain types 3, 4, and 5 which were isolated during the curing.
ND, not determined.
Abilities to degrade vanilla cellular compounds differed largely among both processes and between isolates (Table 2). At Tulungagung, a microbial community (strain types 1 and 2) able to use a large amount of cellular compounds (cellulose, hemicellulose, and/or proteins) developed after scalding. Only pectine was not utilized, although isolates could utilize a degradation product of pectin (polygalacturonic acid) in Biolog MT plates. At Payung the ability to degrade vanilla cell compounds was very limited, since only some isolates able to grow at 30°C produced protease, cellulase, hemicellulase, or pectinase, while the thermophilic B. smithii strains (type 6) did not produce any of these enzymes. Remarkably, the enzyme production abilities of the thermophilic bacteria isolated from the green beans indicated that these strains were able to use complex compounds. Since cellulose and hemicellulose degradation was often observed and in general involves β-glucosidases, a specific test was done for its production and excretion. β-Glucosidase was never excreted (Table 2); however, a large number of isolates showed activity when whole cells were added to the assay, indicating the presence of cell-bound β-glucosidases. Peroxidase activity, another important enzyme for vanilla flavor, was not detected.
To investigate whether bacteria were able to use important aroma compounds of vanilla (vanillic acid, vanillin, p-hydroxybenzaldehyde, p-cresol, 2-phenylalcohol, anisaldehyde, guaiacol, phenyl-acetaldehyde, diacetyl, eugenol, and methyl-cinnamate) as sole sources of carbon, their utilization was tested at a concentration of 250 mg/liter in Biolog MT. Only p-hydroxybenzaldehyde was utilized. In addition, assumed flavor precursors such as leucine, isoleucine, phenylalanine, salicin, coumaric acid, ferulic acid, and catechol were tested. Only leucine, isoleucine, and phenylalanine were utilized. Resistance to vanillin was 0.2 to 0.4% (wt/vol) and did not increase during curing. Isolates from green beans showed a wider range of resistance, i.e., from 0.05 to >0.5%.
Also fungi and yeast were characterized for specific activities. Isolated fungi possessed weak proteolytic, cellulytic, hemicellulytic, and pectinolytic activities. No lignifying activities were detected. A few yeasts possessed very weak proteolytic activities, but no pectinolytic, cellulytic, and hemicellulytic activities were detected.
DISCUSSION
This study describes the microbial ecology of postharvesting processing of vanilla beans. It provides a base for further, more-detailed research on the contribution of microorganisms to vanilla flavor. Considerable numbers of thermotolerant and thermophilic bacilli were detected on vanilla beans undergoing curing. To determine the microbial communities present, microorganisms were first extracted from the beans. A wide variety of physical methods exist to dislodge microorganisms (20). The quantity of microorganisms released can strongly depend on the method used (15, 20). Some methods, such as sonification, destroy part of the microorganisms, while other methods are not powerful enough to release all microorganisms. In general, blending gives the best results (15, 20). In this study it was found that low-speed shaking with glass beads resulted in a similar number of microorganisms released from beans as with high-speed blending. Samples treated with low-speed shaking were easier to deal with; therefore, this method was used throughout the study. As evidenced via the analysis on the green beans, this approach permitted the isolation of a wide variety of microorganisms, although it is possible that a minor fraction of culturable microorganisms remains attached to the beans.
For logistical reasons, samples from the initial stages of the curing process had to be stored at 4°C for some time (at maximum of 2 and 7 days for the curings at Payung and Tulungagung, respectively) before microbial analysis could be conducted. The occurrence of significant microbial growth during cold storage would be expected to decrease diversity and result in the isolation of mainly strains capable of growth at 4°C. Although samples of green beans were stored for the longest time, they revealed the highest diversity. Only 33 and 17% of the isolates from green beans of the Tulungagung and Payung curings, respectively, were capable of slow growth at 4°C. None of the isolates obtained after scalding was able to grow at 4°C (data not shown). Therefore, storage had a minor effect at most.
Thus, the dominance of bacilli in our study is not due to applied methodology (storage, extraction), but the result of considerable growth of thermotolerant and thermophilic bacilli during vanilla curing. The dominance of thermophilic bacilli is not remarkable, since they can sporulate and survive unfavorable conditions such as heat treatment by scalding or nutrient shortage on materials used during curing, such as cotton and boxes. Bacilli are also dominant species in other high-temperature processes, such as composting (6, 7, 32) and cacao fermentation (30). While in these processes microbial fermentation significantly contributed to heat generation, this could not be observed during vanilla curing. The high temperature has to be maintained via the input of heat from hot water scalding and sun and prevention of heat loss via special storage of the beans.
High-temperature counteracts the growth of fungi. Fungus numbers were low during sunning and sweating, and isolates were only able to grow at 30°C, while the temperature during sunning and sweating was in general much higher. Only during curing in unfavorable, cloudy weather conditions was a temporary increase in the numbers observed. The role of fungi in both vanilla curing processes seems negligible and is possibly unwanted. Beans visually overgrown with fungi are discarded in the Indonesian process.
The autoclaving and especially scalding at high temperatures induce major changes in microbial communities. These are the steps where in the microbiology of the process is influenced. The Bacillus strains dominant during curing (related to B. smithii, B. licheniformis, B. subtilis, B. pumilis, and B. firmus) were not present on the green beans in high numbers, while other thermophilic, unidentified species were present but disappeared after scalding. Materials used in the process likely served as natural sources of inoculation, since the dominant species could be isolated from covering materials and boxes. The selection for the dominantly occurring strains is possibly favored by the depletion of oxygen during autoclaving. Beans become brown in the isolated box as the result of chemical and enzymatic oxidation (25). The dominantly occurring B. smithii, B. licheniformis, and B. subtilis are able to grow well under low-oxygen conditions (24).
Strains related to B. subtilis (type 2) and B. licheniformis (type 1) were encountered in both curings and were also the dominant strains isolated from beans from a curing station on Bali, Indonesia (data not shown). In particular, these strains possessed enzymatic activities (proteases, hemicellulases, cellulases, and β-glucosidases) that help them to degrade and consume vanilla cell components. However, the potential to degrade vanilla cell components does not necessarily benefit the natural selection for thermophilic strains possessing these enzymes after scalding. In the Payung curing, thermophilic strains isolated from the green beans and possessing proteases, hemicellulases, cellulases, and β-glucosidases disappeared during curing, while the dominant developing strain (strain type 6, related to B. smithii) did not possess these enzymes. Bacillus types 1 and 2 only became dominant during conditioning. In the Tulungagung curing these strains dominated the curing right from the start of the process.
Other differences were also apparent between the Tulungagung and Payung curings. The numbers of bacilli were much higher during the Tulungagung curing. A comparison of DGGE profiles of bacteria present on the beans and those able to grow on TSA plates revealed that, especially for the Tulunagung curing, a considerable part of the microorganisms developing during autoclaving and sunning-sweating could not be detected by conventional culturing techniques. Several factors likely contributed to the differences in the microbial communities and the numbers between the two curings. (i) The process at Tulungagung was monitored at the end of the curing season, while the curing at Payung was executed at the start of the curing season, when possibly the materials used for curing (covering materials, boxes) contain relatively fewer microorganisms. (ii) Tulungagung is located in the lowlands of Java, 30 m above sea level, while Payung is located in a mountainous area, 750 m above sea level, resulting in differences in temperature and the number of hours of sunlight. Large differences in microbial abundance and populations between different batches and between different producers have also been observed for Indonesian soy sauce production (26–28).
The large differences in microbial abundance, communities, and strain characteristics between the two investigated batches indicate that the effects of microbial activities on the development of vanilla flavor could differ for each batch of cured vanilla beans. Bacilli are known to positively contribute to cacao and several legume food fermentations in Asia and Africa (11, 30). Whether the growth and enzyme activities of bacilli are indeed favorable for vanilla flavor (precursor) formation has yet to be examined, e.g., by comparing flavor profiles of inoculated curings and surface-sterilized controls.
While bacilli were the dominant species that could be cultured, DGGE analysis showed the presence of uncultured species in this food item, especially during the sunning and sweating stage, when a large number of activities related to flavor formation are proposed to occur (25). The presence of unculturable microorganisms in food fermentation was recently also revealed for Mexican pozol (3). Our research supports the remark of Ampe et al. (3) that cultivation-independent characterizations should be included in studies on (possible) food fermentations, since the uncultured species could also influence flavor formation. Molecular identification of dominant members in DGGE profiles can aid in the selection of suitable isolation media, since the phylogenetic positions of bacteria are often consistent with their physiological properties and culture requirements (33).
ACKNOWLEDGMENTS
The financial support by the INDONESTEC program of the Dutch Ministery of Economical Affairs is gratefully acknowledged.
We thank Djasula Wangi, Jakarta, Indonesia, for its cooperation and Mark Dignum, Leiden University, The Netherlands, for supplying samples of cured beans from Bali and a culture of vanilla plant cells.
REFERENCES
- 1.Adedeji B A. Ph.D. thesis. New Brunswick, N.J: Rutgers State University; 1993. [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Ampe F, Ben Omar N, Moizan C, Wacher C, Guyot J P. Polyphasic study of the spatial distribution of microorganisms in Mexican pozol, a fermented maize dough, demonstrates the need for cultivation-independent methods to investigate traditional fermentations. Appl Environ Microbiol. 1999;65:5464–5473. doi: 10.1128/aem.65.12.5464-5473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baillie L W J, Jones M N, Turnbull P C B, Manchee R J. Evaluation of the Biolog system for the identification of Bacillus anthracis. Lett Appl Microbiol. 1995;20:209–211. doi: 10.1111/j.1472-765x.1995.tb00429.x. [DOI] [PubMed] [Google Scholar]
- 5.Bending G D, Read D J. Lignin and soluble phenolic degradation by ectomycorrhizal and ericoid mycorrhizal fungi. Mycol Res. 1997;101:1348–1354. [Google Scholar]
- 6.Blanc M, Marilley L, Beffa T, Aragno M. Rapid identification of heterotrophic, thermophilic, spore-forming bacteria isolated from hot composts. Int J Syst Bacteriol. 1997;47:1246–1248. doi: 10.1099/00207713-47-4-1246. [DOI] [PubMed] [Google Scholar]
- 7.Blanc M, Marilley L, Beffa T, Aragno M. Thermophilic bacterial communities in hot composts as revealed by most probable number counts and molecular (16S rDNA) methods. FEMS Microbiol Ecol. 1999;28:141–149. [Google Scholar]
- 8.Cairney J W G, Burke R M. Fungal enzymes degrading plant cell walls: their possible significance in the ectomycorrhizal symbiosis. Mycol Res. 1994;98:1345–1356. [Google Scholar]
- 9.Chance B, Maehly A C. Assay of catalases and peroxidases. Methods Enzymol. 1956;11:764–766. doi: 10.1002/9780470110171.ch14. [DOI] [PubMed] [Google Scholar]
- 10.Cohen B L. Ammonium repression of extracellular protease in Aspergillus nidulans. J Gen Microbiol. 1972;71:293–299. [Google Scholar]
- 11.Cook P E. Fermented foods as biotechnological resources. Food Res Int. 1994;27:309–316. [Google Scholar]
- 12.Dornenburg H, Knorr D. Production of phenolic flavor compounds with cultured cells and tissues of vanilla species. Food Biotechnol. 1996;10:75–92. [Google Scholar]
- 13.Duarte G F, Rosado A S, Seldin L, Keijzer-Wolters A C, van Elsas J D. Extraction of ribosomal RNA and genomic DNA from soil for studying the diversity of the indigenous bacterial community. J Microbiol Methods. 1998;32:21–29. [Google Scholar]
- 14.Felske A, Wolterink A, van Lis R, Akkermans A D L. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands) Appl Environ Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garland J L, Mills A L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol. 1991;57:2351–2359. doi: 10.1128/aem.57.8.2351-2359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh P, Singh A. Physicochemical and biological treatments for enzymatic/microbial conversion of lignocellulosic biomass. Adv Appl Microbiol. 1993;33:295–333. [Google Scholar]
- 17.Kanisawa T, Tohoro K, Kawakara S. Flavor development in the beans of Vanilla planifolia. In: Kurihara K, Suzuki N, Ogawa H, editors. Olfaction taste. Proceedings of the 11th International Symposium. Tokyo, Japan: Springer; 1994. pp. 268–270. [Google Scholar]
- 18.Klimes I, Lamparsky D. Vanilla volatiles—comprehensive analysis. Int Flavours food Additives. 1976;7:272–291. [Google Scholar]
- 19.Krings U, Berger R G. Biotechnological production of flavours and fragrances. Appl Microbiol Biotechnol. 1998;49:1–8. doi: 10.1007/s002530051129. [DOI] [PubMed] [Google Scholar]
- 20.Lindahl V, Bakken L R. Evaluation of methods for extraction of bacteria from soil. FEMS Microbiol Ecol. 1995;16:135–142. [Google Scholar]
- 21.Muheim A, Lerch K. Towards a high-yield bioconversion of ferulic acid to vanillin. Appl Microbiol Biotechnol. 1999;51:456–461. [Google Scholar]
- 22.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R I, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16 rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Priest F G. Systematics and ecology of Bacillus. In: Sonenshein A I, editor. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 1–26. [Google Scholar]
- 25.Ranadive A S. Vanilla—cultivation, curing, chemistry, technology and commercial products. In: Charalambous G, editor. Developments in food science. Vol. 34. Amsterdam, The Netherlands: Elsevier Science Publishers BV; 1994. pp. 517–577. [Google Scholar]
- 26.Röling W F M, Apriyantono A, van Verseveld H W. Comparison between traditional and industrial soy sauce (kecap) fermentation in Indonesia. J Ferment Bioeng. 1996;81:275–278. [Google Scholar]
- 27.Röling W F M, Timotius K H, Prasetyo A B, Stouthamer A H, van Verseveld H W. Changes in microflora and biochemical composition during the baceman stage of traditional Indonesian kecap (soy sauce) production. J Ferment Bioeng. 1994;77:62–70. [Google Scholar]
- 28.Röling W F M, van Verseveld H W. Characterization of Tetragenococcus halophila populations in Indonesian soy mash (kecap) fermentation. Appl Environ Microbiol. 1996;62:1203–1207. doi: 10.1128/aem.62.4.1203-1207.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosazza J P N, Huang Z, Dostal L, Volm T, Rousseau B. Review: biocatalytic transformations of ferulic acid: an abundant aromatic natural product. J Ind Microbiol. 1995;15:457–471. doi: 10.1007/BF01570016. [DOI] [PubMed] [Google Scholar]
- 30.Schwan R F, Rose A H, Board R G. Microbial fermentation of cocoa beans, with emphasis on enzymatic degradation of the pulp. J Appl Bacteriol. 1995;79:S96–S107. [Google Scholar]
- 31.Smit E, Leeflang P, Glandorf B, van Elsas J D, Wernars K. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:2614–2621. doi: 10.1128/aem.65.6.2614-2621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strom P F. Identification of thermophilic bacteria in solid-waste composting. Appl Environ Microbiol. 1985;50:906–913. doi: 10.1128/aem.50.4.906-913.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teske A, Sigalevich P, Cohen Y, Muyzer G. Molecular identification of bacteria from a coculture by denaturing gradient gel electrophoresis of 16S ribosomal DNA fragments as a tool for isolation in pure cultures. Appl Environ Microbiol. 1996;62:4210–4215. doi: 10.1128/aem.62.11.4210-4215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thibault J F, Asther M, Ceccaldi B C, Couteau D, Delattre M, Duarte J C, Faulds C, Heldt-Hansen H P, Kroon P, Lesage-Meessen L, Micard V, Renard C M G C, Tuohy M, van Hulle S, Williamson G. Fungal bioconversion of agricultural by-products to vanillin. Food Sci Technol Lebensm Wiss. 1998;31:530–536. [Google Scholar]
- 35.Tokoro K, Kawakara S, Amano A, T K, Tudo M. Glucosides in vanilla beans and changes of their contents during maturation. In: Bressiere Y, Thomas A F, editors. Flavour science and technology. New York, N.Y: Wiley Interscience; 1990. pp. 73–76. [Google Scholar]
- 36.von Wintzingerode F, Gobel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 37.Williams S T, Wellington E M H. Actinomycetes. In: Page A L, editor. Methods of soil analysis. Vol. 2. Madison, Wis: American Society of Agronomy; 1982. pp. 781–802. [Google Scholar]
- 38.Wünche L, Babel W. The suitability of the Biolog Automated Microbial Identification System for assessing the taxonomical composition of terrestrial bacterial communities. Microbiol Res. 1996;151:133–143. [Google Scholar]