Abstract
Native mass spectrometry (nMS) is evolving into a workhorse for structural biology. The plethora of online and offline preparation, separation, and purification methods as well as numerous ionization techniques combined with powerful new hybrid ion mobility and mass spectrometry systems has established the great potential of nMS as a workhorse for structural biology. Fundamental to the progression of nMS has been the development of novel activation methods for dissociating proteins and protein complexes to deduce primary, secondary, tertiary, and quaternary structure through the combined use of multiple MS/MS technologies. This review highlights the key features and advantages of surface collisions (surface-induced dissociation, SID) for probing the connectivity of subunits within protein and nucleoprotein complexes and, in particular, for solving protein structure in conjunction with complementary techniques such as cryo-EM and computational modeling. A focus on several case studies wherein SID provided connectivity maps that were otherwise inaccessible by ‘gold standard’ structural biology techniques, or that agreed with solved crystal or cryo-EM structures, highlights the significant role SID, and more generally nMS, will play in structural elucidation of biological assemblies in the future as the technology becomes more widely adopted.
Graphical Abstract

1. Introduction to native MS
Within the field of structural biology, mass spectrometry (MS) has emerged as a powerful tool for the study of proteins and protein complexes, nucleic acids, and other biological macromolecules.1–5 The advent of electrospray ionization (ESI), along with its lower flow rate variant nanoelectrospray ionization (nESI), transformed the field of mass spectrometry, enabling large non-covalent assemblies to be transferred into the gas-phase, manipulated, and measured by MS and other gas-phase analytical techniques.6 Native MS (nMS) utilizes soft ionization methods along with soft instrument conditions to transfer native proteins from MS-friendly electrolyte solutions (typically ammonium acetate, although ammonium bicarbonate7 and ethylenediammonium diacetate8 have also been employed) to the gas phase without significant restructuring, thereby enabling the study of proteins and protein complexes in their native state.2 This approach has been used to study protein-protein complexes (from dimeric species to even very large assemblies up to MegaDaltons in size9,10), protein-ligand interactions11–13 and protein-RNA/DNA14,15 interactions. nMS is not limited to soluble proteins but has also been used to examine membrane proteins and membrane protein complexes and their interactions with lipids.16–19 nMS offers several advantages over traditional structural biology techniques in that, due to high MS sensitivity, sample consumption is low. Typically, only a few microliters of sample at low micromolar concentrations are needed. Unlike x-ray crystallography, high-quality crystals are not needed for MS analysis, and, in contrast to both crystallography and cryo-electron microscopy, sample preparation for nMS is generally much less extensive since nMS can directly analyze proteins from solution state. Cryo-EM, the gold standard for determination of biomolecular structures, requires fine-tuned conditions to convert samples into solid state (crystal or frozen in ice), and so it is cumbersome relative to nMS and generally unsuitable for high-throughput analyses. nMS is also intrinsically well-suited to the study of heterogeneous samples (e.g. mixtures of nucleic acids, proteins, and different proteoforms), meaning they need not go through the rigorous purification procedures necessary for other techniques. For example, recent studies have shown it is possible to study overexpressed proteins from crude lysates by nMS.20,21 Furthermore, when experiments are performed on high-resolution instruments, even small mass differences, such as posttranslational modifications or cofactor binding,22–24 can be resolved which can provide further insight into structure and therefore function. When coupled to mass analysis, gas-phase ion mobility (IM) provides an orthogonal dimension of separation. In IM ions under the influence of a weak electric field are passed through a pressurized chamber filled with a buffer gas that counteracts the ion motion induced from the electric field gradient. Ions are separated in the IM cell based on their mobility, which is influenced by their mass, size, charge, shape, and flexibility. Indirect information on the conformation of ions can be obtained from IM in the form of a rotationally averaged collision cross section (CCS).25 Ion mobility can detect changes in conformation that are not evident from mass spectra alone and, along with tandem mass spectrometry (MS/MS), augments the native mass spectrometry workflow with powerful capabilities useful for structural biology.26,27
1.1. Dissecting protein complexes with tandem mass spectrometry (MS/MS)
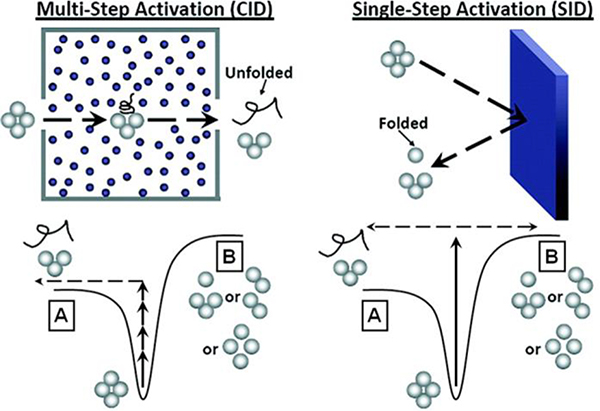
An attractive feature of mass spectrometry is the ability to couple two stages of m/z analysis, i.e., conduct MS/MS. This allows for the isolation of a species at a particular m/z ratio in the first stage and then the activation and subsequent dissociation of in the second stage. The ability to select a species of interest is particularly advantageous for heterogeneous samples in that each protein of interest with a unique m/z can be dissociated selectively, providing information on the individual components. Multiple dissociation methods are useful for nMS, each with advantages and disadvantages. The most common activation technique is collision-induced dissociation (CID), which is available on virtually all modern commercial instruments. During CID analyte ions are accelerated into a neutral collision gas (typically nitrogen or argon) and undergo multiple low-energy collisions with the gas molecules. During each collision a portion of the ion’s kinetic energy is converted into vibrational internal energy, and the stepwise buildup of internal energy during this multi-collision process can eventually result in dissociation if the energy threshold for dissociation is met.28 The products from CID are often reflective of the lowest energy dissociation pathways, which can result from rearrangements (e.g., collapse or unfolding/restructuring of the complex or a single subunit). For protein complexes, CID typically produces highly charged, unfolded monomers and complementary (N-1)mers, regardless of the initial oligomeric state, and is thought to be a result of unfolding or elongation of one of the monomers (see Figure 1).29,30 The charge distribution of fragment ions from CID of a typical protein complex is often described as ‘asymmetric’ in that the fragments do not retain an amount of charge proportional to their mass or native surface area since the restructured monomer retains proportionally ‘too much’ (often ~1/2 of the charge of the precursor), leaving the remaining (N-1)mer charge deficient. CID is a useful tool to confirm the stoichiometry of the protein complex, but due to gas-phase restructuring it can only provide limited information on substructure and subunit connectivity. It should be noted that under certain conditions an alternative CID fragmentation pathway has been observed in which compact monomers or subcomplexes are products of dissociation.31 This pathway remains atypical but it appears to be promoted when protein complexes have low subunit flexibility, higher charge densities, small protein-protein interfaces and fewer salt bridges.31 At low collision energies CID is useful for ejecting proteins or protein complexes from membrane mimetics,16,17 or, when coupled with ion mobility, to intentionally unfold the protein, study stability, and explore the conformational space of the protein.32–34 Low-energy CID is also beneficial for removing extraneous salt and buffer from gas-phase ions prior to mass analysis in order to increase the accuracy of the measured mass of the intact complex or monomers of the complex; although the complex might be restructured, obtaining its accurate mass in the absence of adducts is useful.35,36 ‘Cleaner’ ions can also be produced through the use of submicron capillaries37,38 or addition of solvent vapor additives to the ion source during desolvation.39
Figure 1:
Schematic representations of CID and SID of noncovalent protein complexes with corresponding simplified potential energy diagrams shown at the bottom. In CID (left) protein complexes undergo multiple collisions with the collision gas, which can result in rearrangement/unfolding and ejection of elongated, highly charged monomer and complementary (N-1)mer (A). In SID (right) the high, rapid energy jump can favor a faster, more direct dissociation pathway (B) into folded subunits carrying charge proportional to their mass (surface area), referred to as ‘symmetric charge partitioning’. Reproduced with permission from ref 40. Copyright 2014 American Chemical Society.
An alternative method of dissociation, initially conceived and developed in Graham Cooks’ laboratory,41,42 is surface-induced dissociation (SID).40,43–45 In SID ions are made to collide with a surface. SID is a rapid, approximately single-step, energy deposition process, and dissociation can occur without extensive unfolding (Figure 1). Early SID studies of protein complexes demonstrated that dissociation occurred with more symmetric charge partitioning than CID (i.e. fragments retain charge roughly proportional to their mass and/or surface area), alongside producing a greater variety of subcomplexes consistent with native protein topology. These observations led to the hypothesis that SID could proceed without extensive unfolding,46,47 a hypothesis that was later supported by ion mobility (IM) measurements.48 To rule out unfolding followed by refolding, it was also shown that ligands can be retained in SID when they are not retained by CID. When SID was coupled with IM measurements it was clear that the species of low charge produced in SID were compact, supporting the hypothesis that SID can proceed without unfolding.48,49 However, it is important to note that identical fragments (same stoichiometry and charge state) produced by CID and SID usually have similar CCSs, and so in some cases similar conformations can be sampled by both activation techniques. Because smaller SID fragments (monomers) tend to have fewer charges than those afforded by CID, SID fragments tend to be more compact than CID fragments. Moreover, it has become clear that SID produces fragments that are reflective of the assembly of noncovalent complexes, consistently cleaving the weakest interfaces in protein complexes and so producing structurally informative subcomplexes.40,45,49–53 For this reason SID has emerged as a useful tool in protein structural studies.
Additional dissociation techniques have been applied in nMS studies, including electron-based techniques such as electron capture dissociation (ECD), electron transfer dissociation (ETD) and electron impact dissociation (EID) as well as photon-based techniques including ultraviolet photodissociation (UVPD) and infrared multi-photon dissociation (IRMPD).54,55 The major pathway for these dissociation techniques is covalent backbone fragmentation, contrasting with non-covalent fragmentation of complexes to subcomplexes by collisional activation. Covalent fragmentation using these techniques has proven useful in studying posttranslational modifications (PTMs),56,57 ligand binding sites and conformational changes upon ligand binding,58–60 and protein folding and unfolding.61–63 With UVPD it has also been observed that protein complexes can dissociate non-covalently via both symmetric and asymmetric pathways, with the symmetric pathway increasing in intensity with higher laser power.64,65 The Robinson lab also demonstrated using IRMPD that protein complexes can dissociate asymmetrically or symmetrically, with the symmetric pathway being favored under supercharging conditions for some complexes.66
Clearly tandem mass spectrometry is a powerful tool in nMS and a variety of tools can be employed for activation, depending on the experimental needs. However, alternative complementary methods can also provide structural information. One example is solution disruption, in which subunit interactions are intentionally perturbed in solution, either by changing the ionic strength or pH of the solution or through the addition of organic solvents. Solution disruption experiments have shown that it is possible to disrupt the intact complex, producing structurally relevant subcomplexes,67–69 though alteration of solution conditions may also cause protein denaturation or precipitation. Moreover, as this is a solution-based technique it affects all species present in solution and therefore can be challenging with heterogeneous samples if no liquid-phase separation (e.g. chromatography) is performed beforehand. For heterogeneous samples it is often preferred to transfer them into the gas-phase and then isolate the unique species of interest for further structural interrogation.
1.2. Impact of charge state on SID pathways
A central question for early nMS experiments was “how native is native MS?”. Early protein studies demonstrated that solution conditions (which alter the conformation of the protein) impacted the charge state distribution,70 that non-covalent interactions could be preserved,71 that proteins could be transferred into the gas-phase, that their native structures could be ‘kinetically trapped’ for seconds, and that these ions could be collected while retaining biological function,72,73 providing early evidence that MS could provide structurally relevant information on proteins and their complexes. It is clear from the literature, though, that monomeric proteins can be analyzed under conditions where the structures deviate significantly from “native,”74–78 although this is less likely for protein complexes that are held together by intermolecular protein-protein interfaces. It has also taken the community a while to explore enough systems in depth to discover conditions that best retain native structure. As the community has worked to understand and define the behaviors of monomeric proteins vs. protein complexes, reports have not always pointed out that these systems can show different behaviors. This led to some early, and perhaps lingering, confusion in the broader scientific community about whether nMS does maintain native structure. For example, collapse of monoclonal antibodies in the gas phase has been reported in the literature79,80 in addition to compaction of a range of non-globular proteins.80 These conclusions were drawn from comparisons of experimental ion mobility collision cross sections to cross sections calculated from high-resolution crystal structures.
Fortunately, a large number of cases have appeared where nMS has made significant contributions to structural biology, and so the technique has gained acceptance for characterization of protein complexes. Ion mobility studies of proteins and protein complexes have since demonstrated that the experimental CCS can be in good agreement with the predicted CCS calculated from solved or model structures, providing evidence for native-like proteins in the gas phase.81–84 However, at high charge states some monomeric proteins have been shown to adopt more extended structures, and some protein complexes collapse into compact states.77,81,85 Using basic solution-phase additives, it is possible to reduce the charge states of protein ions, a process known as ‘charge reduction’. Charge reduction better preserves native-like conformations due to reduced intramolecular Coulombic repulsion, and charge-reduced species are generally more stable towards activation/restructuring in the gas phase.86,87 Zhou et. al., demonstrated that charge-reduced precursors produced greater information on the quaternary structure of protein complexes with SID than their ‘normal-charge’ counterparts (in ammonium acetate), which was attributed to suppressed unfolding/elongation/restructuring and better preservation of subunit contacts.49 SID of charge-reduced C-reactive protein (Figure 2a), for example, causes dissociation into a variety of product ions from monomer to tetramer, whereas CID of charge-reduced or normal-charge (24+) CRP simply proceeds in a predictable fashion, ejecting a highly charged monomer from the pentamer (Figure 2b). Compared to surface activation of 24+ CRP (Figure 2c), SID of 18+ CRP yields a greater variety of fragments and suggests an overall cyclic arrangement of the subunits since no intersubunit cleavage is preferred. Given the increased structural information obtained from charge-reduced protein complexes, the majority of SID experiments are performed under charge-reducing conditions, typically through the addition of a small amount of triethylammonium acetate (TEAA) to the protein solution of interest.
Figure 2:
Charge-reduced species have more native-like SID fragmentation patterns than their normal-charge counterparts. (a) SID spectrum of (charge-reduced) 18+ C-reactive protein (CRP) at 1 keV, (b) CID spectrum of CRP 18+ at 3.6 keV, (c) SID spectrum of CRP 24+ (no charge reduction) at 1 keV, and (d) CID spectrum of CRP 24+ at 3.6 keV. Adapted from ref. 49 with permission from the Royal Society of Chemistry.
The majority of nMS experiments have been performed in positive ion mode, and, in the case of SID as discussed above, typically under charge-reduced conditions. However, it should be noted that negative mode can also be used and several studies have shown that the average charge states of protein complexes ionized in negative mode are typically lower than in positive mode without the addition of any additives.88,89 Recent SID studies in negative mode have demonstrated that similar structural information can be obtained in negative mode as in charge-reduced positive mode.90 This is advantageous as solution-phase charge reduction can often result in peak broadening and adduction, decreasing mass accuracy,91 whereas this was not observed in negative mode.90 Negative mode SID therefore could be useful in studies when substructural information is required along with high mass accuracy (e.g proteoform identification or ligand binding studies). However, as the majority of SID experiments have been performed in positive mode this review will focus on positive mode SID.
2. SID for protein structural prediction
A major development in the fields of biological mass spectrometry and structural biology came with the ability to ionize and transmit large molecules.6 Progress started with the study of single proteins, and now, with modern instrumentation, it is possible to study macromolecular complexes weighing several MDa.9,10,70 Given the low sample requirements (μL of sample at μM or lower concentrations), along with the intrinsic ability to study dynamic and heterogenous samples, nMS has emerged as a promising low resolution structural technique. A central question in nMS is how much structural information can be obtained from such studies. As discussed above, the multi-collision process of CID typically produces monomer and (N-1)mer regardless of the starting structure.30,92 While this is undoubtedly useful for proteoform characterization, it can be a limitation for structural studies. It has been reported that for some proteins altering the solution conditions and then subjecting the complexes to CID can result in products more reflective of the starting structure, but so far this appears to be an atypical pathway limited to a small number of cases.31
Early studies of protein complexes demonstrated that SID produced a wider range of subcomplexes, products with more symmetrical charge partitioning, and products without the high degree of unfolding observed in CID, suggesting it could be a useful dissociation technique for structural studies.40,47,49,51 To determine how much structural information could be obtained from SID, studies initially focused on protein complexes of known structure, typically with the use of solution-phase charge reduction86,87 which increases the stability of the precursor and limits unfolding/restructuring, resulting in more structurally informative fragments.49 The observed SID products can be compared to the expected products from analysis of the solved structure. This was achieved using PISA interfacial analysis which allows interfacial information in the form of interface area, number of potential salt bridges, and number of hydrogen bonds to be obtained from high resolution crystal structures.93 This information can then be used to rank the interfaces from strongest to weakest. If dissociation is occurring in a manner consistent with the structure, it would be expected that the weakest (smallest) interfaces would cleave first. For a given homomer, depending on the oligomeric state, there are a number of different ways the subunits can assemble to form a complex, which will result in differences in the interfacial strengths.94 For example, a homotetramer could assemble either in a cyclic manner (i.e. monomer to tetramer) forming a C4 symmetric complex, or through dimerization (i.e. monomer to dimer, dimer to tetramer) forming a complex with D2 symmetry. In a cyclic complex, the interfaces between subunits are equal, and therefore all interfaces could be considered equally likely to break upon activation. Hence it would be predicted that a cyclic tetramer would dissociate to monomer+trimer and two dimers at low collision energy. The cyclic tetramer aquaporin-Z (Figure 3), a membrane protein from E. Coli., has been previously studied with SID and it was found to fragment in a manner consistent with its structure, producing monomer+trimer and dimers.95 In contrast, a D2 tetramer has a stronger monomer-monomer interface and weaker interfaces between the dimers forming the tetramer; therefore, it would be predicted to produce dimers at low collision energy. This expected fragmentation pattern was indeed observed for a series of three soluble D2 tetramers (streptavidin, neutravidin and transthyretin (TTR)), which dissociated to dimers at low SID energies.53 In addition, the experimental collision cross sections of the dimers were consistent with theoretical CCS of the dimers produced via cleavage of the weakest interface. The striking difference in SID spectra for C4 tetramers vs D2 tetramers, as highlighted in Figure 3, clearly demonstrates that SID provides structural information in the form of subunit connectivity. Furthermore, while streptavidin, neutravidin, and transthyretin all have D2 symmetry, they have significant differences with regards to the relative strengths of all interfaces, as shown in Figure 3. It is clear that the interfaces in TTR are more similar to each other (interface A vs. B vs. C), whereas for streptavidin the monomer-monomer interface (interface A) is much stronger than the other interfaces (interfaces B & C). This difference in relative interface strength influences not only the onset energy for dissociation but also the variety of products observed at each energy, with more monomer+trimer being observed for TTR than streptavidin due to the lower barrier to forming this product. The interface area is an important factor to consider for protein complex dissociation, as previous reports have highlighted that it can also influence CID fragmentation of complexes, with smaller interface areas being more likely to fragment via the atypical pathway along with complexes with higher charge density and fewer interfacial salt bridges.31
Figure 3:
SID can distinguish between tetramers of different arrangements. A) PISA interfacial analysis for C4 tetramer aquaporinZ, and D2 tetramers streptavidin, neutravidin and transthyretin. B) Low energy SID for 13+ aquaporin Z, adapted with permission from ref 95 with permission from the Royal Society of Chemistry. C) low energy SID of 11+ streptavidin, adapted with permission from ref. 52.
Predictable fragmentation patterns based on interface analysis are also observed from SID of oligomers containing a greater number of subunits and which have a greater number of potential arrangements. For example, hexamers can exist in many different arrangements including cyclic structures and stacked or planar dimers-of-trimers and trimers-of-dimers. Both the full-length (HFQ102) and truncated (HFQ65) versions of cyclic RNA-binding hexamer HFQ (host factor-I protein) have equal interfaces between subunits, and therefore if dissociation occurs in a manner consistent with its structure it would be expected to dissociate to form all subcomplexes from monomer to pentamer, which is indeed the case when activated by SID.96–98 The hexamer glutamate dehydrogenase (GDH), in contrast, can be described as dimer-of-trimers in which the interfacial areas within the glutamate dehydrogenase trimers are much larger than the interactions between monomers on opposing trimers, and as a result this complex dissociates by SID to form primarily trimers. 50,52,96,97,99 Insulin, on the other hand, is a trimer-of-dimers arrangement that has more equal interfaces between subunits (compared to GDH) and so SID produces dimers and tetramers along with trimers. SID therefore can distinguish between different subunit arrangements.
Table 1 summarizes the topologies that have been explored by SID over the past several years; both homomers and heteromers are represented, with molecular weights ranging from ~25 kDa cytochrome c dimers46,100 up to 801 kDa GroEL 14mers51,96,101 on the homomeric side and heteromers represented from ~65 kDa α2β2 hemoglobin to ~700 kDa 20S proteasomes [(α7β7β7α7)] from several species.102,103 This MS/MS approach has not been limited to soluble proteins; membrane proteins AmtB and Aqp0 also fragment in a manner consistent with their structure by SID.95 As mentioned above, for heteromers the presence of multiple subunits with different structures poses more difficulty in determining the complete connectivity within the complex compared to homomers. Tryptophan synthase is a hetero-4mer with a near-linear αββα arrangement.104–106 The interfacial area, as determined by PISA, between the (red) α and (blue) β subunits is weaker than between the two β in the middle of the complex (1363 vs. 1624 Å2) so that the primary fragmentation channel is loss of α monomer. When the leftover αββ complex is subjected to a second stage of SID, the second α monomer is ejected, leaving the stronger ββ interaction intact.107 Toyocamycin nitrile hydratase (TNH), an (αβγ)2 arrangement, does not have a high-resolution crystal structure for PISA analysis but dissociation into αβγ subcomplexes at low energy and αβ and βαγγ complexes at higher energy is suggestive of its overall topology as a dimer-of-heterotrimers.98,108–110 Hemoglobin [(αβ)2] has also been studied by SID, and dissociates to produce both monomer+trimer and dimers due to the similar interfacial areas that have to be broken to produce these products (1767 Å2 vs 1842 Å2).52 There is an abundance of evidence that, as a result of a surface collision, protein complexes dissociate in a manner that is reflective of their substructure, 45,50–53,109,110 consistently cleaving the weakest interfaces. Those interfaces with the fewest residues, hydrogen bonds, and salt bridges will be cleaved preferentially, especially when activation involves only a limited amount of collision energy that can be internalized and distributed throughout the molecule.44,45,52 At higher collision energy more dissociation pathways are available for the molecule to explore, but even fragments produced via higher energy pathways are reflective of substructure.
Table 1:
Examples of noncovalent protein complexes of varying topologies and their major dissociation pathways. Line thickness represents relative interfacial strength

|
2.1. SID as a constraint in computational modelling
Given that SID has been shown to consistently cleave the weakest interfaces in protein complexes, with the exception of complexes with intertwined inter-protein domains,82,118 and hence can provide structural information consistent with native structure, current research is exploring if SID can be used as a constraint for computational modelling of proteins. Predictive models could then be used to provide increased structural information for protein complexes of unknown structure and could use information from SID along with data from other measurements obtained at low resolution (cryo-EM, SAXS, etc).52,119 Initial studies related the appearance energy (AE) to features of the protein:protein interfaces within the complex, with AE defined arbitrarily as 10% fragmentation to avoid ill-defined low-rise fragmentation onset values and competition with alternative fragmentation pathways.52 Initially a set of eight globular proteins that had a complete complex structure deposited in the PDB were chosen. Energy-resolved mass spectrometry (ERMS) plots were generated for each complex, enabling the experimental AEs to be determined. To determine the predicted AEs, Rosetta’s InterFaceAnalyzer was used to calculate features of the protein:protein interfaces, including interface area, hydrogen bonds, and salt bridges.120 The study found that while individual features could be correlated with experimental AE, a more accurate AE prediction could be obtained when several of these features were combined. Furthermore, predictions could be improved by also considering a rigidity factor (RF), which was based upon the intrasubunit salt bridges, hydrogen bonds, and disulfide bonds, and is thought to account for differences in subunit propensity for partial unfolding upon surface collision. The study found that the best prediction of AE was obtained by having nonzero weights (w) for the number of interacting residues (NRs) and unsatisfied hydrogen bonds (UHBs) at the interface, and for the RF, as shown in equation 1.
| [Eq. 1] |
The model suggested that proteins with a larger interface have a higher AE, while lower flexibility (increased RF) and higher numbers of UHB at the interface both result in lower AEs. Using this relationship a strong correlation was obtained between the predicted and observed AEs, as shown in Figure 4.52 This approach was then expanded to consider an additional two complexes not included in the training set which were predicted to have higher AE than those within the training set. In fact, the correlation improved upon including these complexes. The ability to predict AE from a structure and compare it to an experimental AE is attractive for structure validation/selection, when no solved structure exists but predicted structures are available. Therefore, to test this approach the model was applied to study four complexes for which there are no solved structures; three computationally designed dodecamers and a protein complex for which a homology model exists.82,121 The predicted and experimental AEs for these complexes show good correlation, but not as high as when a solved structure was used (in the training and test sets). This is expected as there may be some variation in interface strength between the model and the experimental structure. However, these results demonstrated that AE could be predicted from structures and compared to experimental AEs, which has promise in the selection of candidate structural models for unknown systems.52 We note here that all of the protein complexes used to date are somewhat globular and that very different structural motifs (elongated coiled coils) might not fit on the same AEprediction line, although an appropriate prediction could be attempted for different structural motifs.
Figure 4:
Predicted AE, based on the initial optimized model as shown in equation 1, shows good correlation to experimental AE. Reproduced with permission from ref 52.
This approach was more recently expanded, demonstrating that SID AE could be used in combination with Rosetta to successfully evaluate protein-protein docking poses.119 Firstly, the SID AE prediction model was improved and then incorporated into a scoring function which combined the RosettaDock122 scoring function with an SID scoring term, quantifying the agreement between the experimental results and structures generated from RosettaDock from substructures. The improved AE prediction model replaced UHB with the hydrophobic surface area (HSA) of the interface (see equation 2), as it was found that poses with low interface RMSDs can have significantly different UHB and hence prediction using this term can be challenging.
| [Eq. 2] |
When the SID scoring term, using this model, was incorporated, it was possible to predict structures with less than 2 Å RMSD from the native structure for 6/9 of the complexes tested, while without the SID constraint this was only possible for 3/9 complexes.119 It was hypothesized that the inclusion of SID helps RosettaDock identify native-like structures based on interface size and hydrophobicity, as the interfaces are scored based on number of interface residues and buried hydrophobic surface area at the interface. However, this approach relied on knowing the correct tertiary structure and hence an approach in which less prior knowledge is required is attractive. Current work has performed docking using input structures from homology models, unbound crystal structures, and bound and perturbed structures instead. In this case both flexible and symmetric docking were performed, and low resolution cryo-EM density maps were included as an additional method of scoring and ranking the structures. Using this approach, and scoring with both cryo-EM and SID, the RMSD100 (normalized version of root mean square deviation) to native was below 4 Å for all 15 structures tested with ensemble docking. Full complex prediction via symmetric docking also benefited from the inclusion of this data, with the RMSD100 of the predicted structure less than 4 Å for 14/15 cases, compared to only 5/15 without the data.
2.2. SID provides structural information for complexes without solved structure
From the study of known structures, it has become clear that SID is a promising tool to study the assembly and subunit connectivity of protein complexes. SID consistently cleaves the weakest interfaces in protein complexes, producing structurally informative subcomplexes. Hence, SID can be a useful tool in the study of protein complexes of unknown structure. This approach shows even more promise when it can be combined with computational approaches, model structures, and/or complementary methods to propose or validate a structural model. This information can be beneficial when protein complexes cannot be solved using traditional techniques or when it is simpler to have the SID data prior to collecting data that require greater effort/time/expense.
A favorable feature of MS in the study of proteins and protein complexes is the ability to separate and select different species in a complex mixture. When samples are heterogeneous, traditional techniques can fail to produce unambiguous or interpretable data, but in nMS the species of interest can be mass selected and individually studied. Heterogeneity can be attributed to the molecule itself (e.g., different proteoforms or different conformations) or heterogeneity can be intentionally introduced (e.g. in assembly/disassembly studies and subunit exchange experiments). Shirzadeh et al. used nMS and SID to provide insight into the disassembly mechanism of transthyretin.114 Transthyretin (TTR) is a homotetramer which has been implicated in amyloidosis, but the mechanism(s) surrounding fibril formation are not fully understood.123 While two models had been previously proposed for TTR tetramer disassembly, subunit exchange experiments suggested differences in the mechanisms proposed, mainly the production of monomer through a dimer intermediate or from the tetramer itself.124,125 TTR is a dimer of dimers and low energy SID produces dimers resulting from cleavage of the weakest interfaces.53 Combining low-energy SID with subunit exchange experiments, greater mechanistic details on the exchange and hence disassembly/assembly mechanism can be obtained.114 For example, if we consider a tetramer containing two tagged (T) and two untagged (U) monomers, there are different ways the monomers could be arranged depending on the disassembly/assembly mechanism, these unique arrangements would be indistinguishable by mass alone but can be distinguished by SID as shown in Figure 5. Using nMS, SID, and subunit exchange over a range of incubation times the authors were able to validate the previously proposed TTR disassembly model in which TTR monomers are produced in a two-step mechanism wherein tetramer dissociates to dimer which further dissociates into monomers.124 However, they were also able to further refine the model and include the addition of parallel and early formation of the heterotetramer with two tags from the dimer assembly. In this case the coupling of SID to subunit exchange experiments was key to providing details on TTR dynamics.114
Figure 5:
SID can distinguish between different arrangements of subunits. A) SID from a sample at 4 °C of a TTR UU/TT tetramer yielding an MS spectrum with equal signal intensity for UU and TT dimers. (B) SID-MS spectrum of an equimolar solution of UU/TT, UT/UT, and UT/TU. Reproduced with permission from Ref 114. Copyright 2019 American Chemical Society.
nMS in combination with SID can be used to study not only protein:protein interactions but also ligand binding, either to determine binding site (as discussed below in the SID in the Orbitrap section) investigate complex stability upon binding, or to explore the interaction of proteins with RNA or DNA.15,53,95,116,126,127 Protein-RNA/DNA interactions are crucial to understand as they facilitate many fundamental biological processes, such as gene expression, RNA splicing, and protein synthesis.128 Generating high resolution structures of these complexes is often not possible, due in part to challenges obtaining homogeneous samples at high enough concentrations for traditional techniques. nMS is a promising alternative characterization method that can handle low concentrations and heterogeneous samples, and hence can be used to provide insight into structure and function. However it should be noted that Mg2+ is often required for appropriate RNA folding and thus for complex formation, which can pose challenges with nMS experiments due to its non-volatile nature.129 Even so, low mM quantities are sometimes sufficient for activity and can be handled by nMS.15,127 One such study utilized nMS and SID to determine the stoichiometry of Pyrococcus furiosus RNaseP, which catalyzes maturation of tRNA. SID of the assembled complex revealed the presence of all four expected subunits; RPP21, RPP29, POP5, and RPP30 and demonstrated that they were bound 1:1 with the catalytic RNaseP RNA. CID on the other hand, was unable to dissociate this complex and determine the stoichiometry.15 In another study the Ebola virus matrix protein VP40 was studied.130 This protein exists in multiple different oligomeric forms in order to perform its function (including an octameric ring structure), and it is thought that host RNA may be necessary for the transformation of VP40 from its structural role to its essential non-structural role in the virus life cycle. By nMS it was observed that the low weight SEC fractions were consistent with the RNA free monomer and dimer while the higher weight SEC yielded a broad unresolvable spectrum, consistent with it binding a variety of RNA. Dissociation of a portion of this unresolvable species with SID yielded dimer, trimer and tetramer and hence it was thought that this fraction corresponds to the ring. Interestingly all subcomplexes contained an additional mass of 4.7 kDa, attributed to a ligand on unknown nature. In addition, incubation of the monomer/dimer with a 35 nucleotide DNA oligo was able to produce the VP40 octameric ring, with two bound oligos, as demonstrated with nMS. nMS was then used to determine oligonucleotide binding and stoichiometry of the protein-RNA complexes for a range of oligonucleotides, and providing insight into this transformer protein.130
In the absence of a solved structure, model structures often exist for protein complexes, either homology models or, in the case of computationally-designed proteins, the designed structure. Validating these models with experimental evidence is often key. Native mass spectrometry, in conjunction with SID in particular, is being utilized to study computationally designed protein complexes, offering the advantage of rapid structural characterization.82,131–134 In one such study, three computationally designed hetero-dodecamers with dihedral point symmetry were characterized with SID-IM.82 These protein assemblies could not be crystallized and therefore alternative methods to confirm the design were required. nMS first confirmed the designed oligomeric state could be observed, then SID experiments were performed to confirm the subunit connectivity. The complexes were all designed such that two trimers were connected by three dimers. SID was performed over a range of energies to build detailed connectivity maps for two of the complexes using low and mid-energy SID the products were consistent with the designed hetero-oligomerization interfaces and consistent with the designed structure. For the third design, the dominant products at low energy SID products deviate from the expected dominant products based on the protein-protein interfaces, however the observed connectivity maps are consistent with the designed structure, the difference in onset may in-part be due to some restructuring in the gas-phase.82
In the absence of a predetermined model structure, nMS and SID can be used to guide model determination, either in combination with docking as discussed in the previous section or in combination with other techniques. One such complex that has benefitted from SID-guided structural determination is toyocamycin nitrile hydratase (TNH) from Streptomyces rimosus.109,110 TNH is a small (~85kDa) heterohexamer comprised of two α subunits, two β subunits and two γ subunits. TNH is a challenging complex to characterize with traditional techniques. It is too large for NMR studies and despite many efforts has proved challenging to study with both X-ray crystallography (no crystals) and cryo-EM. In an initial SID study, it was determined that the subunits were arranged in a dimer of αβγ trimers arrangement.109 More recently, SID was combined with additional MS-based methods of structure determination, as shown In Figure 6.110 Performing SID over multiple energies allowed the construction of a complete connectivity map and enabled ranking of the interfacial strengths; from SID data it was determined that the primary contact between the αβγ trimers is the γ-γ subunits with a weaker contact between the β-β subunits. The CCS of subunits from either solution disruption68,135 or SID were then used as constraints in coarse-grained modelling,136,137 enabling a model to be proposed. This structure could be further refined using homology modelling involving tetrameric nitrile hydratases, enabling possible atomic structures to be proposed, using the SID data, solution disruption, and covalent labeling of surface exposed residues138–140 as constraints. Finally, the structures were further validated using chemical crosslinking,141,142 resulting in three proposed hexamer models.110 This combination of MS-based methods enabled detailed structural information to be obtained on a protein complex that has been challenging to study with traditional techniques. Cryo-EM studies have since subsequently been attempted for this complex, but its small size coupled with its propensity to move in the ice means that only low-resolution maps could be obtained, however two of three MS proposed structures fit the low-resolution envelopes.
Figure 6:
Workflow for characterizing the TNH structure by complementary mass spectrometric tools, reproduced with permission from ref 110. Copyright 2015 American Chemical Society.
Another example in which SID and nMS were utilized to guide modeling is the multicopper oxidase Mnx from Bacillus sp., which has also proved challenging to characterize with traditional techniques. Mnx is known to be comprised of three subunits MnxE, MnxF and MnxG. MnxE and MnxF are very similar in size (both ~12kDa), making them difficult to distinguish in low resolution methods. Furthermore, these two subunits (MnxE/F) do not have homologues of known structure, making homology modeling impossible. The average diameter of the complex could be determined using transmission electron microscopy.117 However, in order to obtain more detailed structural information, alternative methods for structural elucidation had to be considered. The intact complex is ~210 kDa and was thought to be composed of one MnxG subunit and six MnxE/F subunits although it was not clear how many E and F were present or how they were arranged. While collision-induced dissociation could not provide information on the assembly of the complex, SID determined that the complex was composed of a MnxG subunit bound to a MnxE3F3 hexamer. Increasing the SID energy allowed the MnxE3F3 hexamer to be further dissociated; in this case all possible oligomeric states (monomer to pentamer) were observed, consistent with a cyclic hexamer. Based on the observed subcomplexes (e.g., EF dimers, EFE and FEF trimers, etc), it was proposed that the hexamer has an alternating MnxE/MnxF arrangement. The information from SID was then combined with docking of ab initio models of MnxE and MnxF and a homology model of MnxG, allowing a proposed model to be built, as shown in Figure 7, although one should always remember than more than one model can fit the measured CCS values.117 These experiments again highlighted the power of SID coupled with computational modelling to propose a connectivity map in the absence of a high-resolution crystal structure. Low resolution cryoEM maps have subsequently been obtained that are consistent with the SID proposed structure.
Figure 7:
Using data from nMS and SID experiments, along with homology and ab initio models, a structural model for Mnx could be proposed. Reproduced with permission from ref 117.
2.3. Complementarity of SID and high-resolution structural biology techniques
SID has also proven beneficial for complexes that have been studied using traditional techniques but when distinction of subunits is not possible even with high-resolution structures available. One such example is pyridoxine Biosynthesis 1 (PDX1) from Arabidopsis thaliana, of which there are three homologs; PDX1.1, PDX1.2 and PDX1.3. PDX 1.1 and PDX1.3 are catalytically active enzymes while PDX1.2 is inactive.143 However, it has been shown that PDX1.2 forms dodecameric heterocomplexes with either PDX1.1 or PDX1.3.144 Previous studies have sought to solve the structure of these mixed complexes and, while PDX1.2/PDX1.3 heterocomplexes could be crystalized, the positions of the individual proteins could not be pinpointed.145 However, it was proposed that the likely assembly would be a hexamer ring of active PDX1.1 or PDX1.3 stacked on top of an inactive hexamer ring of PDX 1.2.144,145 Recent studies using cell-free expression of PDX heterocomplexes found that a range of different heterododecamers could be observed, and by varying the input DNA ratio (and hence the protein ratios) the full range of co-complexes from 0:12 to 12:0 could be observed.146 These complexes, along with the PDX1.2 dodecamer, were studied with cryo-EM, and while a 3.2Å resolution structure consistent with the stacked ring structure could be reconstructed for PDX1.2, the mixed PDX1.2/1.3 structures posed challenges, due in part to the variability in the stoichiometry of the co-expressed samples. A well-defined two-ring fold could be observed for PDX1.2/1.3 hetero-dodecamer; however, like with the previous crystal structure of PDX1.2/PDX1.3, the two different subunits could not be distinguished in the structure. To provide insight into subunit arrangement, the 6:6 PDX1.2:PDX1.3 complex was studied with SID (each co-complex from 0:12 to 12:0 was fragmented by SID). MS offers an advantage in such studies because even when multiple stoichiometries exist, a single species can be mass-selected and investigated. When dissociated by SID the most abundant product of the 6:6 dodecamer was hexamer, consistent with a dodecamer formed via stacking of two hexamer rings. Interestingly, the hexamers were found to contain both PDX1.2 and PDX1.3 in varying ratios-including 3:3 (dominant), 2:4 and 4:2. This suggests that the 6:6 12mers are formed in either the [3:3 and 3:3] or [2:4 and 4:2] combination of PDX1.2:PDX1.3, as opposed to the stacked homohexamer rings. The SID results suggest that the co-assembly is based on stochastic subunit incorporation at different locations but with a degree of symmetry.146 This study demonstrates that complementing high-resolution techniques such as cryo-EM or X-ray crystallography with nMS and SID can provide increased information on heterogenous complexes.
3. SID instrumentation for native mass spectrometry
SID can be accomplished on virtually any mass spectrometer, from trapping instruments exemplified by quadrupole (or linear) ion traps (QITs),147 electrostatic linear ion traps (ELITs),111,148 Orbitraps,97,99 and FT-ICRs96–98,108,149–151 to transmission configurations (single or multiple quadrupoles152,153 and TOF platforms48,97,112,154–159) and even hybrid configurations utilizing combinations of trapping and transmission ion optics. If SID is performed in a trapping region, which can be a mass analyzer (FT-ICR cell, QIT, ELIT) or a multipole ion trap (e.g., collision cell), then the SID voltages must usually be pulsed as a tandem-in-time MS/MS configuration. In contrast, transmission ion optics necessitate a transmission SID configuration in which the potentials are static but optimized for SID sensitivity. One exception is SID inside a TOF reflectron,160 in which case transmission is achieved even though the surface voltage is pulsed sequentially with the TOF injection voltages.
There are a variety of important choices to consider when designing SID devices and implementing them in new or existing platforms. Utilizing SID to probe noncovalent protein complexes necessitates the evaluation of additional instrument parameters such as the available mass and energy range, pressure, and ion heating (which can cause structural rearrangements76,113). Next, we will discuss general design principles for SID cells, focusing on the effect of SID cell geometry and instrument parameters on the analysis of protein complexes by SID-MS. We then discuss these parameters in conjunction with published SID cell designs that have been successfully utilized to probe the structures of noncovalent protein complexes on Q-IM-TOF, Orbitrap, FT-ICR, and ELIT platforms to evaluate strengths and weaknesses of each design, with a particular emphasis on sensitivity, mass range, energy range, and usefulness/applicability, and how SID and IM couple together to create a workhorse for tertiary and quaternary structure determination. We will conclude this section with a perspective on the future of SID instrumentation for native mass spectrometry guided structural biology.
3.1. General design principles for SID devices
Fundamentally, SID involves collision of a projectile ion with a surface for ion activation to induce unimolecular dissociation sometime later. SID patterns are indicative of the assembly and interfacial interactions of noncovalent protein complexes, enabling the creation of connectivity maps between the individual subunits, as shown in a variety of examples in Table 1.40,45,52
To effect a surface collision, the analyte ion must be accelerated to a particular kinetic energy by raising the voltages on all ion optics preceding the SID surface or similarly by dropping the voltages on the surface and all ion optics behind the surface. It is important that the collision energy, the ion’s kinetic energy at the moment of surface collision, is accurately and precisely defined so that laboratory frame SID collision energies are comparable across experiments and instrument platforms. It is thus important to determine where the ion has its last meaningful collision with background gas molecules so that its effective birth potential can be determined. The voltage difference (ΔV) between the electrode in this region and the SID surface multiplied by the ion charge state (z) is the laboratory frame SID collision energy (ELAB), eq. 3
| [Eq. 3] |
The maximum energy that is available for internalization and redistribution into the ion’s many degrees of freedom is
| [Eq. 4] |
where ECOM is the center-of-mass energy, MN is the mass of the neutral target (gas molecule in CID or surface in SID), and MION is the mass of the analyte projectile ion. It is obvious from this equation that the efficiency of energy transfer to the projectile increases with the mass of the target, i.e., per-collision energy deposition in SID is more efficient than in CID as a first approximation simply because the surface mass is much greater than the ion’s mass. We will expand upon this concept later when we discuss the mechanism for SID of protein complexes.
The ions that collide with the surface must then be extracted off the surface and be transmitted and/or trapped throughout the remainder of the instrument. Fragmentation of a protein complex by SID is usually followed by a combination of ion mobility separation,48 further stages of mass selection and fragmentation,98,110,115 other gas-phase manipulations (e.g. charge manipulation, hydrogen-deuterium exchange, etc.), and mass analysis so that structural information can be maximized in each experiment, providing comprehensive topological maps and ligand-binding information. The SID device geometry and surface material, incidence and scattering angles, ion beam velocity and position distribution, pressure, and collision energy are all important experimental variables to optimize, discussed next in the context of protein complexes.
3.1.1. SID design considerations: general guidelines
A first consideration in the design of an SID cell is whether to use DC-only optics or a combination of rf and DC potentials. For straightforward cases of product ions of a single charge state and similar kinetic energies, DC optics such as Einzel lenses and immersion lenses provide less biased ion transmission than rf optics (multipoles and rf ion traps), which have low-mass cutoffs and a drop-off in transmission at high m/z due to insufficient pseudopotential well depth. Arrangements of DC lenses can act as kinetic energy analyzers but the effect on m/z-dependent ion transmission is less severe. DC optics also yield more predictable ion trajectories than oscillating rf devices, making collision with a surface and ion trajectory modeling more straightforward. However, multipoles and ion traps are excellent ion focusing devices, particularly when filled with a neutral background gas such as helium or nitrogen at low pressure (< 10−2 torr). Efficient collection of ions after SID can be challenging because the analyte projectiles scatter off the surface with a wide range of m/z values, angles, and kinetic energies, but rf optics such as collision cells and multipole ion guides are well suited for trapping and transmission of diverse ion beams. Providing a focused ion beam prior to collision is important for SID sensitivity and can be accomplished with rf multipoles and quadrupoles or immersion lenses and einzel lenses. In summary, it is best to utilize DC optics in the vicinity of the surface to avoid fringing rf fields from multipoles while taking advantage of the focusing capabilities of rf optics in order to maximize sensitivity. Regardless of design, prior to fabrication and testing of a new SID cell, results from ion optics simulations (e.g. SIMION) should be examined in detail in order to iteratively refine the design for optimum transmission.96,97,112,155
A second design consideration is whether the SID voltages should be pulsed or constant and whether SID is accomplished on-axis or off-axis. Pulsed configurations can only be achieved in ion traps such as QITs,147 FT-ICR cells,149,151,161 and ELITs (one such experimental setup is discussed later)111,148 or TOF relectrons160,162 and rely on fast-switching DC power supplies to cause collision with an on-axis surface over a period of 100–101 μs. These configurations are common in FT-ICR experiments but are decreasing in popularity as more versatile hybrid mass spectrometer configurations become more prevalent. The strict timing requirements make these experiments difficult, and so transmission SID configurations have gained in popularity for nMS.
Older transmission SID configurations utilized orthogonal combinations of quadrupoles, electric and magnetic sectors, and time-of-flight analyzers to direct projectiles into a surface mounted at ~45° and subsequently to collect and mass analyze the scattered product ions.152–154,160,163–165 Strictly speaking these configurations could be considered ‘on-axis’ since the ion beam need not be deflected for the initial collision, though scattered ions are collected on the orthogonal axis. In modern hybrid mass spectrometers suitable for native mass spectrometry (FT-ICR, IM-TOF, and Orbitraps) the optics are generally arranged linearly, save for orthogonal injection TOF analyzers and Orbitrap hybrids, and as a result the development of SID devices utilizing off-axis surfaces became necessary so that the collision target is not directly in the ion path.48,96,97,99,108,112,155,156 Usually the surface is mounted such that its normal is perpendicular to the ions’ direction of travel, requiring deflection into the surface and extraction of the scattered beam into the remainder of the instrument. The voltages in transmission SID are static and usually need not be synced with instrument electronics, making implementation easier than pulsed SID. Ideally, voltages are supplied by internal power supplies, but this is often not possible when retrofitting commercial instruments, thus requiring additional external circuitry (and in the case of pulsed SID, timing circuits). SID voltages are preferably controlled in the native instrument software (or simple auxiliary software) with minimal user intervention but options for additional control for advanced users. Transmission SID cells ought to be simple yet efficient. The greater the number of electrodes, the more complex the fabrication, assembly, and installation, which can increase the possibility of alignment errors and device-to-device variability. Moreover, complex configurations can be difficult (for non-experts) to use and tune. Importantly, SID cells that take up substantial volume for installation may require truncation of important elements originating in the instrument, which can compromise mass range, sensitivity, and dynamic range.96,97,108
There are other important device specifications which need be considered, but discussing them in detail is beyond the scope of this text (for further information refer to ref. 166). Surface material and morphology are well known to alter chemical and physical reactions as the projectile interacts with the surface; the internal energy deposition, generally ~15–20%, and kinetic energy retention and dispersion are also dependent upon surface rigidity and morphology.167–170 Self-assembled monolayers (SAMs) of long fluorinated hydrocarbon chains on gold have been the most prominent surface material in the literature171,172 both for small molecules and protein complexes because they increase internal energy deposition and reduce neutralization and other deleterious reactions on the surface, which can otherwise chemically alter analyte ions and their fragments.173 Several studies throughout the literature have shown that for both small molecules and multiply charged ions such as proteins and protein complexes stainless steel surfaces are sufficient SID targets.42,97,112,152–154,174 The extent of neutralization of protein complexes has not been explicitly characterized but it is thought to play only a minor role for these types of ions. Moreover, the effects of surface material on SID fragmentation dynamics and patterns have not been explicitly characterized, an important goal of recent work in the Wysocki laboratory.
3.1.2. Special considerations for native mass spectrometry
Throughout the ~45-year history of SID development, small organic molecules and peptides have been the subject of most studies.175 Only in the mid-2000s, many years after the invention of electrospray ionization6 and the resulting emergence of native mass spectrometry1,78,176,177 was it illustrated that SID could provide valuable topological information when fragmenting protein complexes.46,100 Transporting protein complexes to the gas phase from solution, transmitting them through the mass spectrometer, and fragmenting them has proven much more difficult than for small molecules. Special considerations and instrument modifications have been necessary to accomplish SID (and more broadly, transmission and mass analysis) of such large molecules.
First, almost all instruments rely on high pressure differential vacuum stages, pressurized multipoles, and collision cells for collisional cooling of ions, which serves to narrow the distributions of kinetic energies and positions of the ions produced in the ion source and thereby increase transmission through the remainder of the instrument. Because protein complexes are orders of magnitude larger than small molecules (tens to hundreds of kDa vs. < 1,000 Da), their collisions with low-mass background gas molecules are less effective on a per-collision basis. It has thus been necessary to increase the pressure near the ion source as well as in ion guides and collision cells in order to increase the number of cooling collisions and so transmit complexes more effectively.178,179 Increasing the pressure in collision cells allows more efficient dissociation of large noncovalent protein complexes which are otherwise difficult to fragment if the number of collisions or collision energy is insufficient.180–182 For example, the Robinson group modified a Micromass QTOF II183 and the Heck group modified a Micromass QTOF I for native mass spectrometry, in part, by increasing the pressures near the source and in the collision cell and by dropping the selection quadrupole rf frequency to improve high m/z transmission and isolation.179 The same rationale can be applied to SID of protein complexes. It is imperative that the ion beam is focused and efficiently transmitted from the source to the SID cell as well as collected and trapped/transmitted through the remainder of the instrument after collision. It is preferable to conduct SID in a low pressure region (<10−5 torr) so as to prevent ‘CID contamination’ and to reduce ion scattering, although SID in higher pressure regions is sometimes necessary and effective, but it is also necessary to collisionally cool scattered complexes and fragments after collision because they will retain 5–25% of the collision energy as axial kinetic energy, which must eventually be dissipated.98
On a related note, while increasing pressure improves transmission of large ions through the mass spectrometer, efficient transmission through multipoles and other rf optics can only be accomplished in combination with appropriate rf potentials. In general, rf frequencies need to be lowered and voltages raised in order to provide an ample pseudopotential well for confining high m/z ions radially and thus avoid m/z biases in SID fragmentation spectra.180–182 The mass range of quadrupoles and multipoles is more sensitive to changes in rf voltage or frequency than stacked ring ion guides. The frequencies of quadrupole rf drivers must also be lowered (a more practical choice than raising the maximum voltage, which can cause electrical breakdown) to enable mass selection at high m/z.179,181,183 Ion mobility may also serve as a selection device if it precedes the SID cell.156,184
The discussion of greater rf voltages and dc gradients leads naturally to a concern that excessive ‘heating’ of proteins, protein complexes, and fragments can restructure them. As we will discuss later, SID fragmentation patterns are sensitive to changes in tertiary and quaternary structures of complexes,45,47,113,185 a feature that is attractive for structural characterization and determination of upstream “heating” of the ions, and therefore it is particularly critical to maintain ‘soft’ instrument conditions prior to collision if a native topology map is desired. Proteins can also restructure in rf trapping regions, though the extent depends on many factors including pressure, rf voltage, rf frequency, ion residence time, and device geometry.74,75,186,187 Whether before or after SID, restructuring of macromolecular ions also alters the collision cross sections obtained from ion mobility measurements and thus should be avoided when studying native-like structures.
There are several additional parameters that are important to SID-IM-MS measurements. The energy range and overall mass range of new generations of TOFs, Orbitraps, and FT-ICRs are suitable for native SID-MS, but these considerations are of concern for older instruments or instruments designed primarily for ‘omics’ work. As the analyte mass increases into the MDa range and beyond in the near future, access to greater collision energies will be necessary due to the increased degrees of freedom of these enormous species. Utilization of collision voltages within commercial instruments requires placement of the SID cell just prior to the commercial collision cell99,115,156 (or replacing it with a hybrid SID-CID cell96,108) or prior to the IM cell in Waters Q-IM-TOFs48,155 but is the most sensible choice to achieve the desired 200–300 V voltage range so that even large noncovalent complexes (e.g. human 20S proteasome102,103 and GroEL51,188) can be dissociated by SID.
3.2. SID cells for native mass spectrometry
While the use of SID for analytical MS/MS stretches back to the mid-1980s,42 its application to structural biology studies of native proteins and protein complexes is a more recent effort pioneered by the Wysocki group. SID has been installed into multiple instrument platforms with a wide array of capabilities, from Waters Q-IM-TOF instruments48,97,112,115,155,156 utilizing ion mobility as an orthogonal characterization scheme, to high-resolution Orbitrap Exactives with extended mass ranges,97,99 and an ultrahigh resolution FT-ICR.96–98,108 In this section we will discuss the evolution of SID cells on each platform and the unique advantages each offers in the SID experiment. A special focus on the coupling of SID and IM for protein structural determination will also be presented.
3.2.1. SID cells for IM-TOF instruments
Ion mobility platforms can offer an additional stage of selectivity prior to final mass analysis by separating analyte ions based on their rotationally averaged collision cross sections, a useful mode of characterization for conformationally dynamic molecules like proteins and protein assemblies.189 Ion mobility enables the observation of changes in protein conformation due to gas-phase restructuring (ion heating), interactions with gas-phase or solution-phase reactants, changes in solution conditions (e.g. pH or ionic strength), and binding events with ligands, RNA, DNA, or other proteins.
The Wysocki group installed a 10-lens in-line SID cell in a Waters/Micromass QTOF II mass spectrometer by removing the ion transfer hexapole bridging the collision cell and the TOF chamber.155 After modification, the QTOF II configuration consisted of a nESI source, a quadrupole mass filter, an SID cell, and a CID cell followed by a TOF analyzer. Although this instrument does not have an ion mobility cell, it was a precursor to the Waters Synapt Q-IM-TOF instruments equipped with traveling wave ion mobility cells190 which all utilized nearly identical SID cells.156 Amongst the proteins investigated initially on the QTOF II was the previously discussed toyocamycin nitrile hydratase (TNH), a heterohexameric Co-binding protein complex that consists of two each of three unique subunits (α, 21.2 kDa; β, 10.1 kDa; γ, 11.5 kDa) and which has been resistant to characterization using traditional high-resolution methods, as discussed previously.109 The differences in the CID (Figure 8a) and SID (Figure 8b) fragmentation patterns of the 19+ precursor illustrate the increase in connectivity information between gas-phase collisions and a surface collision even with early prototype SID cells. CID of the 19+ precursor results in ejection of α and β monomers, yet SID generates αβγ heterotrimers as the most abundant fragment ion, as illustrated in Figure 8c. This is consistent with the SID-determined dimer-of-heterotrimers connectivity of the subunits, which would otherwise be unclear from the CID spectrum. We can make further conclusions about the substructure of TNH using additional fragments that are unique to the SID spectrum, as will be discussed in detail later.
Figure 8:
Comparison of (a) CID and (b) SID spectra of 19+ toyocamycin nitrile hydratase heterohexamer (αβγ)2 on a Micromass/Water QTOF II mass spectrometer retrofitted with a Gen 1 SID device. Reproduced from ref. 109. Copyright 2011 American Chemical Society.
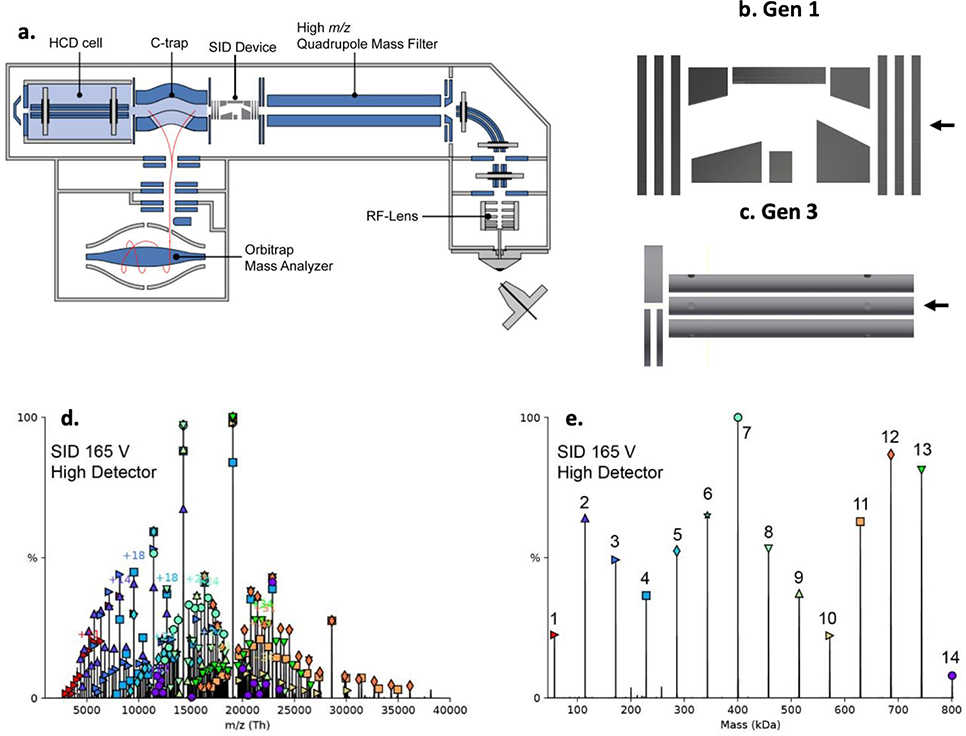
For practical purposes we will discuss the SID cell design in the context of the Waters Synapt G2 platform191 (shown in Figure 9a) which consists of a nESI source, a high pressure traveling wave ion guide (TWIG), a high m/z quadrupole, two TWIGs serving as collision cells on either side of the traveling wave ion mobility cell, and finally a TOF analyzer. SID devices incorporated on the G2 platform are also compatible with the G2-S platform, which incorporates a stepwave ion guide in the front end of the instrument. The ‘Gen 1’ SID cell installed in the QTOF II as well as Waters Synapt G2 and G2-S mass spectrometers is shown in Figure 9b.48,155,156 This SID cell is approximately 3.0 cm in length. Accommodations were made in the Synapt by truncating either the Trap or Transfer TWIG by the corresponding amount. This truncation would normally be deleterious to instrument sensitivity but in this case has little effect due to the enormous ion capacity of the Trap cell. The ‘Gen 1’ SID cell is composed of three primary regions: 1) an immersion lens to focus the precursor ion beam, 2) a surface collision region wherein a series of deflectors guide the analyte ions for collision with the surface and subsequently extract the ions away from the surface, and 3) a product ion collection and focusing region formed by another immersion lens. The electrodes are made of stainless steel while the assembly is held together by PEEK on a stainless-steel mounting bracket. The surface is a glass slide mounted to a stainless-steel holder electrode. The glass slide is covered with a 10 Å layer of titanium and a 1000 Å layer of gold. A self-assembled monolayer (SAM) is created by immersing the gold surface in an ethanolic solution of a fluorinated hydrocarbon, e.g. 2-(perfluorodecyl)ethanethiol.155 In some instances, stainless steel surfaces have been utilized with similar results.97,112
Figure 9:
SID cells for Q-IM-TOF platforms. Schematic diagram of (a) Waters Synapt G2 platform with three integrated SID cells with locations noted. Three generations of SID cells have been installed in the G2, (a) Gen 1,48,156 (b) Gen 2,112 (c), Gen 3.97 Note that the Gen 1 & 2 devices can be located before and after the IM cell, whereas the Gen 3 install location is in the quadrupole chamber prior to the Trap.
The placement of the SID device enables excellent experimental flexibility. Ions generated by nESI can be mass selected by the quadrupole and either stored, or activated (CIU or CID), and thermalized in the Trap TWIG. In mobility-TOF mode, the ions are stored in the Trap during the prior scan’s IM separation (~20 ms), after which they are pulsed into the SID cell where they can be subject to activation via a surface collision or can simply be passed through the cell in transmission mode. Any remaining precursor and fragment ions are then mobility separated in the traveling wave ion mobility cell and can then be activated further in the Transfer TWIG or transmitted to the TOF for detection. The SID collision energy in this SID-IM configuration is defined as the potential difference between the exit of the Trap cell and the surface multiplied by the ion charge state. It is worth noting, though, that the traveling waves during ion ejection from the Trap may also affect the kinetic energy distributions of precursor ions and thereby alter the effective collision energy.
The robustness and usefulness of SID prior to IM allows the relative quantitation of oligomers with the same m/z as well as the monitoring of changes in precursor and fragment ion conformation, which can be compared between ‘hot/harsh’ and ‘cold/soft’ conditions, denaturing solution conditions, and ‘native’ conditions. For example, returning to the TNH example discussed earlier, the QTOF II did not have IM capabilities, so the complexity of the TNH SID spectrum made interpretation and evaluation of fragment ions of low abundance difficult. Figure 10a shows the low-energy (700 eV) SID-IM-MS spectrum of 14+ TNH obtained on a G2-S platform,110 with IM serving as an orthogonal separation mechanism to disambiguate heterotrimer fragments from unfragmented precursor ions, which have the same nominal m/z values. At higher SID collision energy (Figure 10b, 1680 eV), more fragmentation of the hexamer and secondary fragmentation of the heterotrimers is induced, resulting in a substantial increase in topologically informative peaks that are resolved by ion mobility, even for species of low abundance. For example, based on the mobiligram in Figure 10b, we can suggest the following interactions between individual subunits: 1) detection of αβ as an abundant SID fragment suggests a strong interfacial overlap between α and β, 2) observation of γγ suggests a connection between the γ monomers; 3) detection of αγ implies an interaction between these two subunits; 4) observation of βγ suggests an intersubunit interaction; 5) tetramers βαγγ and αββα as complements of αβ and γγ are further indicative of the overall subunit arrangement. The fragmentation patterns from the SID-IM-MS experiment were key in proposing the first connectivity map for TNH. This seminal work provides a glimpse into the usefulness of SID for native mass spectrometry guided structural biology.
Figure 10:
SID-IM reveals the connectivity within a heterohexamer. SID-IM of 14+ TNH on a Waters G2-S fitted with a Gen 1 SID device prior to the IM cell. Shown are low- and high-energy spectra at (a) 700 eV and (b) 1680 eV, respectively. Reproduced from ref. 110. Copyright 2015 American Chemical Society.
In some cases, it may be useful to conduct IM separation prior to SID. For example, if a protein complex is heterogeneous, existing in multiple conformations that have similar charge state distributions, they cannot be disambiguated by the quadrupole or the TOF but may be separated by collision cross section. Zhou et al. placed a Gen 1 SID cell after the IM cell in a Synapt G2 by truncating the Transfer TWIG instead of the Trap.156 Mobility-separated CsI cluster ions, transthyretin, and human serum amyloid P (SAP) oligomers were characterized by SID. Two conformers of the SAP dodecamers were observed and mobility separation allowed distinct SID patterns to be obtained for the less compact and more compact oligomers. Quintyn et. al illustrated an SID-IM-SID configuration taking advantage of the placement of dual SID cells prior to and after the IM cell of a Synapt G2-S platform, discussed in Section 3.2.5.115
Although the Gen 1 SID cell has been installed on several different platforms (Q-IM-TOF, Orbitrap, and FT-ICR), not all instruments have the luxury of being able to accommodate an SID cell 3 cm in length. On the Synapts, truncation of either the Trap or Transfer TWIGs has minimal impact on performance not only because these cells are long to begin with (~13 cm) and thus have high ion capacity, but also because ions are only accumulated in the Trap for a brief amount of time (~20 ms) prior to injection into the ion mobility cell. Even so, it became necessary to reconsider the design of the SID cell with a focus on 1) decreasing the SID cell volume, 2) reducing the number of independent electrodes to simplify tuning, and 3) increasing SID efficiency and thus sensitivity by improving ion collection after surface collision.
These considerations gave rise to ‘Gen 2’ and ‘Gen 3’ SID cells on the Synapt97,112 and FT-ICR96,97 platforms. For Synapt platforms, Stiving et al. designed a shortened and simplified ‘Gen 2’ SID cell consisting of 5 DC-only electrodes and a DC-only ion carpet to improve collection of product ions after collision (Figure 9c).112 The first two electrodes serve as an immersion lens for ion focusing prior to collision, and a single deflector guides ions into the surface for collision. The surface is tilted to improve ease of ion extraction compared to a level surface that is farther off axis. The ion carpet, which consists of a series of concentric ring electrodes connected by a voltage divider, creates a 3D potential well which focuses ions toward the center aperture of the device.192 Overall the device length was ~1.6 cm, a nearly 50% reduction in size. The SID spectra of several model protein complexes were investigated and compared to the Gen 1 SID cell. An improvement in product ion collection at low SID energy was noted, as well as improvements to energy resolved SID curves (smoother transitions between energies) but the device showed lower ion transmission for large complexes and at high SID energies. A similar cell design was briefly adopted on the solariX FT-ICR platform, discussed later.
The requirement for truncation of the Trap TWIG for both the Gen 1 and Gen 2 SID cells as well as the lingering tuning complexity prompted the design and implementation of a much simpler third generation (‘Gen 3’) design, shown in Figure 9d.97 The Gen 3 SID cell is located behind the quadrupole mass filter, taking the place of the pre-installed dynamic range enhancement (DRE) lens, and simplifies the cell design to three electrodes: a deflector to guide ions into the surface, a stainless steel surface, and an extractor. The three electrodes are arranged in a split lens arrangement taking up approximately 3–4 mm along the optical axis, an order of magnitude reduction in size from the Gen 1 cell design. The relocation of the SID cell in front of the trap allows thermalization of SID fragment ions prior to injection into the ion mobility cell without compromising the mass selection or ion mobility capabilities of the instrument. An extensive characterization of the device showed 2–5x improvements to sensitivity at low SID voltages (<45 V), modest sensitivity improvements for large complexes analyzed at high SID voltages, and otherwise similar sensitivities to the Gen 1 configurations. The simplification of the Gen 3 cell led to the first commercialization of SID on Waters’ SELECT SERIES Cyclic IMS (cIMS).193 The cIMS offers improved IM resolution as well as multifunction capabilities for mobility selection and activation in the cIMS array.194
As SID is primarily useful for informing on the stoichiometry and topology of noncovalent protein complexes (quaternary structure),45 it is desirable to combine it with other MS/MS activation techniques that provide complementary information on, for example, primary structure. Although SID can generate primary sequence fragments from peptides and proteins,175 only limited sequence information can be acquired since dominant a, b, and y, but not c and z, fragments are created from backbone cleavages. UVPD, pioneered by the Brodbelt research group for proteins and protein complexes, offers a high sequence coverage alternative activation technique for probing the primary structures of proteins and peptides.56,195,196 The Barran group demonstrated 266 nm UVPD of mobility-selected flavin mononucleotides, peptides, and proteins in the Transfer cell of a Synapt G2-S using a gate-and-trap scheme.197–199 The peptides gramicidin A and melittin had several conformations that exhibited unique dissociation patterns, with the more compact conformers exhibiting fewer cleavages. The Barran group also utilized IM-UVPD to study the unfolding of the proteins ubiquitin, cytochrome C, and myoglobin with a 213 nm laser199 and have more recently combined multiplexed fragmentation strategies (UVPD-IM-CID) to characterize cytochrome C, hemoglobin, and concanavalin A.200 Notably, for this setup UVPD was accomplished prior to IM by irradiating the analytes in the Trap TWIG instead of the Transfer cell. Stiving et al. utilized a gate-and-trap scheme to probe the structures of several model peptides and proteins by UVPD, where insulin chain B, melittin, and conformationally selected species including des-Arg1 and des-Arg9, were irradiated by 193 nm UVPD in the Transfer cell of a Synapt G2-S.201 A future goal of the work is to implement SID-IM-UVPD for complete “complex-down” characterization of noncovalent protein complexes, with SID generating topologically important subcomplexes that can be irradiated by UVPD, or energized by electron-based activation methods, ExD,202–204 to generate sequence fragments that inform on the folding and interfacial interactions between subunits.
3.2.1.1. SID-IM
Now that we have discussed instrumentation for combining SID with ion mobility, we can more precisely describe why they are so often used in conjunction with one another. Ion mobility can distinguish between species of the same m/z but different mobilities, including multiple conformations of the same species or species with different oligomeric states (e.g., a doubly charged dimer and singly charged monomer will have the same nominal mass-to-charge but different mobilities). As discussed above, this feature of ion mobility makes it particularly useful in protein complex SID studies, in which a range of products of different oligomeric states can be observed. In addition, the symmetric charge partitioning observed in SID often results in the oligomers overlapping in the m/z dimension.47 When SID is installed prior to the ion mobility cell, IM can be exploited to help deconvolute and interpret the data. This is particularly advantageous for larger oligomers or systems with many dissociation pathways. One such example is the cyclic undecameric protein, trp RNA-binding attenuation protein (TRAP). Due to the equal interfaces between the subunits, when TRAP is subjected to SID it produces all possible subcomplexes from monomer to decamer (Figure 11).40,97 By using IM after SID, the different oligomers, which overlap in m/z, can be clearly separated and distinguished. Each oligomeric state lies on a “trendline” in the mobilogram as they all have similar compact CCSs and their drift times scale with charge. This has proven useful for many systems including the ~801 kDa 14-mer GroEL, which consists of two stacked heptamer rings. By SID this complex dissociates to the heptamer along with all additional possible products (monomer to 13mer); interpreting the data was made significantly easier through the addition of mobility separation.51
Figure 11:
SID-IM-TOF of the 18+ charge state of holoTRAP 11mer (in 200 mM EDDA with 14 equiv of trp) on a Synapt G2. All oligomeric fragments are observed in mobility space, consistent with the cyclic arrangement of the subunits. Reproduced with permission from Ref 97. Copyright 2020 American Chemical Society.
IM clearly offers advantages for SID data interpretation due to its ability to separate overlapping oligomers at the same m/z, which is also possible using high-resolution MS instead of SID.96,99,108 However IM can also provide increased structural information when the rotationally averaged CCS is determined.81,84,205,206 These experiments can either be performed in tandem on the same instrument, or on different platforms, depending on the experimental design and requirements. One such study by Jacobs et al. investigated the cooperativity of copper binding in the homotetrameric copper-sensitive operon repressor (CsoR) and combined results from high resolution drift tube ion mobility with SID on a separate platform.207 MS titrations revealed that CuI binding is cooperative. From IM the authors found that, interestingly, the holo tetramer was more compact than the apo tetramer, and the level of compaction was dependent on the number of CuI bound with compaction being observed upon the initial CuI binding, with no variation upon the second and third CuI and a final additional compaction upon the fourth CuI binding. SID also showed marked differences with CuI binding. When the tetramer was fully loaded, not only was it more resistant to fragmentation, but instead of fragmenting primarily to monomer as it did in the apo and partially loaded samples, a relatively high proportion of dimer was observed along with the monomer. This change in fragmentation was attributed to the strong, subunit-bridging, coordinate covalent CuI-S bonds.207 In another study Sengupta et al. used an integrated approach coupling computational modelling, biochemical assays, genetic studies, IM, nMS and SID to better understand Salmonella FraB deglycase, showing that a modeled dimer structure with one substrate per monomer had correctly predicted CCS and SID collision onset.121
The ability to perform SID and IM on the same platform allows the CCS of the precursor and products to be determined.40,49 For example, streptavidin is a D2 symmetric tetramer that dissociates to dimers at low SID energy. As the complex has D2 symmetry, as opposed to cyclic symmetry, dimers produced through different interface cleavages would have different arrangements, and hence different CCSs. Using IM, comparing the experimental and theoretical CCS, it was determined that the dimers are produced from cleaving the weakest interfaces, demonstrating that the SID disassembly pathway is the reverse of the protein assembly pathway. 53 IM can also be useful for confirming that a precursor ion is native-like, that is, that it has not been restructured in the gas phase. Membrane proteins, for example, and their complexes must be solubilized in a membrane mimetic; nanodiscs, detergent micelles, bicelles, or amiphols have all been used with nMS. 17,208–210 Membrane proteins are introduced into the gas-phase within the mimetic and then the protein or protein complex can be released using collisional activation. However, increasing the collision energy can cause unfolding, which while useful in studying the stability imparted upon lipid binding,12 may affect fragmentation studies aimed at structure elucidation. Therefore, when performing SID on membrane protein complexes, it is useful to determine that the precursor is native-like under the conditions needed to remove the detergent before performing SID.95 This is important as previous studies have shown pre-activated, rearranged, structures fragment differently with SID than their native-like counterparts as discussed in detail below.113
Combining SID and IM has also proven beneficial in the study of orthologous protein complexes. Orthologous proteins are responsible for equivalent functions in different organisms; however, evolution has caused them to adapt to their individual physiological needs, giving rise to structural diversity. Determining the extent of structural preservation or variation can be challenging, especially when high resolution structures have not been solved. The Sharon lab recently presented a MS approach, including nMS with IM and SID, top-down MS, and denaturing MS (for proteoform determination) to characterize the 20s proteasome from different organisms (rat, rabbit, human, yeast (Saccharomyces cerevisiae), and archaeal (Thermoplasma acidophilum)). The 20s proteasome is a hetero-28mer compromised of four stacked heptameric rings in an α7β7β7α7 arrangement.103 The CCS and conformational spread of the intact complexes were determined and the authors found similar conformational spread between the eukaryotic species studied while the archaeal species had a narrower conformational spread. SID of each proteasome yielded complicated MS/MS spectra, the interpretation of which was aided through mobility separation, as shown in for rabbit 20s proteasome (see SID-IM of rabbit proteasome in Figure 12).103 The rabbit, human and rat proteasomes were observed to dissociate to half proteasome and α7 plus β7β7α7, along with some further fragmentation to monomer and dimer. The archaeal proeteasome, however, exhibited a preference for dissociation to half proteasome while yeast proteasome was significantly harder to fragment. The differences in SID patterns for these orthologous proteins was attributed to differences in the strength of the interfaces between the different subunits. The 20s proteasome from Methanosarcina thermophila has also previously been reported to dissociate to half proteasome and monomers.102 The authors also used collision-induced unfolding (CIU) to assess the stability of the 20s species. In CIU, the collision energy is raised in a stepwise manner and the conformations of the ion ensemble are monitored using IM. The unfolding or restructuring of the ion can be monitored by acquiring mobility data over a range of collision energies, allowing one to build up a CIU “fingerprint” reflective of the protein’s stability and conformational landscape. This technique has been used to study the structure and stability of proteins including antibodies, protein complexes, protein-ligand interactions.32–34,76,211 The Sharon lab used CIU to study the structural stability of the 20s proteasome from different organisms, finding that archaeal 20s proteasome unfolded at lower energies than the other species, but all exhibited the same number of conformational states. Together, the results from this suite of nMS experiments determined that mammalian (rat, rabbit, human) 20s complexes share similar size and stability, while yeast 20s has increased size and stability and archaeal 20s has decreased size and stability.103 This study demonstrated the wealth of structural information that can be obtained from complementary nMS experiments. In addition, the mass spectrometry data revealed a new rat PSMA7 proteoform, that had not been described previously, and involved truncation of the final two amino acids information that could have been easily lost using other techniques including cryoEM in which the termini often cannot be resolved.
Figure 12:
SID-IM of the 43+ and 44+ rabbit 20s proteasome at 150 V. A) full MS (top) and isolation (bottom), B and C) IM-MS analysis of SID products, D-J) extracted spectra for the different regions underlined in panel B. Reproduced with permission from ref 103. Copyright 2020 American Chemical Society.
As discussed above, CIU is emerging as a powerful analytical fingerprinting technique. 32–34,76,211 Recently the Prell lab performed experiments using collisions with a surface to intentionally unfold monomeric proteins (surface-induced unfolding, SIU) and compared these SIU conformations to those observed from CIU.212 Interestingly they found that the same conformer families and unfolding transitions were sampled in both CIU and SIU. While the same conformational families were sampled, the Prell group demonstrated that the intensity of the unfolded forms for some proteins can be higher in SIU than in CIU, consistent with surface collisions imparting greater activation than gas collisions on the timescale of these experiments. SIU was also used to probe energy deposition upon surface collision, by studying ten proteins under native-like conditions with a range of molecular weights from 17–80 kDa; it was found that the energy deposition efficiency in SIU increased with mass, from a low of ~20% to up to ~68%.212 Previous studies into the mechanism and kinetics of SID have been performed, but these typically have focused on small molecules as opposed to protein complexes, and so the relevance of prior mechanistic studies to protein assemblies is currently unclear.43,153,167,213–215
3.2.1.2. IM-SID
The previous section focused on experiments in which SID was performed prior to IM; however, SID is equally appealing after IM separation.156 When SID is conducted after IM, species can be separated or selected by IM and then individually probed by SID. This configuration is useful for distinguishing different protein conformations. 113,156 It is well known that subjecting gas-phase proteins to steep DC gradients or harsh rf voltages in the presence of non-negligible background gas can activate protein complexes, causing them to rearrange, collapse, or unfold.216 Previous SID experiments performed on hemoglobin activated in-source on a QTOF instrument showed deviations from the native structure, with a higher proportion of monomer being observed.47 These results were compared to IM-MS experiments on a separate instrument which demonstrated that the elevated sampling cone voltage (steep dc gradient) caused the protein to undergo activating collisions and unfold as a result. Being able to combine SID and IM on the same platform clearly allows greater information to be obtained in a single experiment. When SID is coupled after IM, different conformations can be separated before dissociation, providing structural information on each conformation individually.113,156 Quintyn et al. studied three protein complexes by SID and CID after intentional activation in the source of a Waters Synapt G2 Q-IM-TOF (see Figure 9).113 One of the complexes studied, C-reactive protein (CRP), showed significant changes in its conformation upon source activation, namely collapse with modest activation energies and then expansion at higher activation energies. However, the CID spectra of these conformations were disappointingly similar despite the change in quaternary structure evident by variations in drift time. On the other hand, when SID was performed (SID-IM) markedly different spectra were obtained for native (low cone voltage) and collapsed or expanded (high cone voltage) species. At low cone voltages the pentamer dissociated to monomer+tetramer and dimer+trimer as complementary pairs expected from a native cyclic penatamer, whereas at high cone voltages monomer+tetramer was the dominant dissociation pathway (concurrent with a decrease in precursor ion depletion compared to the native state, an indication of gas-phase annealing), demonstrating that SID can distinguish between different conformations. When SID was coupled post-mobility (IM-SID), the fragmentation patterns of each conformation could be compared, as shown in. After activating CRP at a cone voltage of 200 V (a,b,c), three conformations could be observed. Based on the different SID patterns of the mobility-separated conformers (d,e,f) it was suggested that the conformational changes were due to the cone-CID-induced unfolding of a monomer from the complex, with the more elongated conformations having more unfolded monomer that resulted in a corresponding increase in the average monomer charge state in the SID spectrum. Although this approach studied conformational changes induced in the gas phase, it is equally applicable to protein complexes that exist naturally in a dynamic set of conformations in solution.
As discussed previously, SID can be coupled both before and after IM on the Waters Synapt. 115 A useful experiment that is enabled when SID is installed in both locations is one in which a first stage of dissociation by low-energy SID produces subcomplexes, which are then separated by IM and subjected to a second stage of SID to produce smaller subcomplexes (often monomers).115 This approach can provide information on how different subunits can assemble to form multisubunit complexes, particularly if those complexes are large or heterogeneous.110 While the majority of work presented in this section focuses on SID coupled with TWIM on the Q-TOF platform, SID has also been coupled with IM on different instrument platforms. Recently, SID has been coupled with trapped ion mobility spectrometry (TIMS) on an FT-ICR instrument. In this case SID is located after IM so that TIMS serves as a useful stage of mobility selection that can be followed by mass selection in a quadrupole and finally SID to explore different conformations of peptides or proteins.184 This approach has been used to study the model tetrameric protein complex streptavidin, pentameric cholera toxin B plus the ligand GM1s (the headgroup of the GM1 ganglioside receptor), and conformationally selected peptides with the same m/z but different sequences (namely bradykinin desArg1 and desArg9). While this combination of technologies is in its early stages, coupling of SID technology to this and other ion mobility platforms is appealing for studying gas-phase conformations of protein assemblies.
3.2.2. SID cells for Orbitraps
Both Gen 199 and Gen 3 (EMR: ref 97; UHMR: unpublished) SID cells have been built and tested on Thermo Scientific Extended Mass Range (EMR) and Ultrahigh Mass Range (UHMR) Exactive series mass spectrometers, respectively, as illustrated by the schematic shown in Figure 14a. Both the EMR and UHMR mass spectrometers offer high-resolution capabilities (EMR: 140,000 @ m/z 200; UHMR: 200,000 @ m/z 400) for resolving ligand-bound species, adducts, or PTMs and have expanded mass ranges (EMR: 350 to 20,000 m/z; UHMR: 350 to 80,000 m/z) ideal for studying macromolecular assemblies. The ion sources on both instruments, but on the UHMR in particular, have been modified to improve desalting and desolvation of large species180,181 although these options should be used with caution to avoid restructuring of native complexes.185 The UHMR, for example, has an ‘in-source trapping’ option wherein ions are collisionally activated in the transfer multipole behind the S-lens in the high pressure region of the instrument near the source. Both instruments have high m/z quadrupoles and higher energy collision-induced dissociation (HCD) cells. The Orbitrap platform excels in analysis of macromolecular assemblies such as antibodies,218 chaperones,180,181,219 proteasomes,180 and viral particles.220
Figure 14:
SID cells for Orbitrap platforms. (a) Schematic of a Thermo Scientific Extended Mass Range (EMR) Orbitrap Exactive platform with SID cell taking the place of a transport multipole, (b) the Gen 1 SID cell design and (c) Gen 3 SID cell design, (d) SID spectrum of GroEL 14mer (stacked 7mer rings) 65+ to 74+ (165 V) obtained on a UHMR equipped with a Gen 1 SID cell, and (e) Unidec222 deconvoluted mass spectrum with 7mer as a prominent fragment. Numbers indicate the oligomeric state that corresponds to each peak in the mass domain. Panel (a) reproduced from ref. 99. Copyright 2019 American Chemical Society. Panels (b) and (c) adapted from ref. 97. Copyright 2020 American Chemical Society. Panels (d) and (e) reproduced from ref. 101 with permission from The Royal Society of Chemistry.
VanAernum et al. designed a 12-lens SID cell to replace the transport octupole preceding the C-trap (Figure 14b). Ions from the nESI source are focused via the S-lens (similar to an ion funnel) and are transported via multipoles to a high m/z quadrupole mass filter (on the UHMR and modified EMR) where precursor ions can be mass selected for MS/MS. These ions can then be transmitted through the SID cell or caused to collide with the surface and collected in the C-trap or HCD cell. Transmitted ions can be activated by CID in the HCD cell (if not doing SID) for dissociation or cleanup/desalting in order to improve apparent resolution, peak shape, and signal-to-noise. After collection in the C-trap, the ions are pulsed into the Orbitrap where their signal transients are measured and then processed by Fourier transform to give a mass spectrum.
The Gen 1 SID cell consists of three regions, 1) a precursor ion focusing region made up of a three-electrode Einzel lens, 2) a surface collision region with multiple deflection and extraction electrodes, and 3) a product ion focusing region which is also an Einzel lens. The cell maintains the transmission capabilities of the commercial instrument and uses 10 independent voltages provided by an external power supply. An 11th voltage, the C-trap offset, must also be supplied externally in order to provide the necessary acceleration voltages within the SID cell and extraction into the C-trap.99 This cell design has been implemented on 3 Orbitraps (1 EMR, 2 UHMRs) in our own laboratory as well as in two other laboratories for beta testing (Russell at Texas A&M and Sharon at the Weizmann Institute; both labs have previously successfully used Gen 1 or Gen 3 SID devices on their Synapt instruments103,114).
The high resolution and sensitivity of the Orbitrap platform are useful for a variety of purposes, although users must be aware that low mass biases occur as resolution settings are increased – low mass ions are favored over high mass ions because of the long path length in the Orbitrap cell and the high velocity with which the ions traverse it, resulting in desynchronizing collisions.221 VanAernum et al. utilized the high resolution to probe isotopic abundances of overlapping dimer and monomer from SID of streptavidin 11+.99 On the Synapt platform ion mobility allows the quantitation of dimers and monomers with the same m/z since they have different arrival times, even when the species are not isotopically resolved in the TOF. On the Orbitrap isotopic resolution enables the deconvolution of the mass spectrum and thus quantitation of dimer and monomer abundance. Moreover, even lower resolution settings are useful for resolving multiple peaks with small mass differences that have, e.g., variable numbers of N-terminal methionines and sodium ions on the streptavidin monomers.
Because of its high sensitivity and resolution combined with the extended mass range, the Orbitrap platform has been utilized in conjunction with SID in several studies over the last few years. Harvey and VanAernum analyzed the 801 kDa tetradecameric GroEL chaperone on a UHMR platform equipped with a Gen 1 SID cell.101 SID of the 65+ through 74+ charge states of the 14mer (Figure 14d) provided an abundance of subcomplexes consistent with those observed by Zhou on a Synapt IM-MS platform, though with more balanced oligomer abundances throughout the mass range due to improved transmission of large complexes through the UHMR.51 The deconvoluted mass spectrum in Figure 14e, in which 7mer is the most abundant fragment ion, is consistent with the known ‘stacked 7mer’ topology of the complex.
As discussed earlier, the high mass resolution of the Orbitrap platform allows small differences such as ligand-binding to be resolved, hence allowing ligand-binding sites to be probed. Busch et al.116 demonstrated that different binding motifs could be probed using SID-MS on an Orbitrap EMR. Pentamers cholera toxin B (CTB) and C-reactive protein (CRP) bind their respective ligands (GM1s ganglioside sugar head group and phosphocholine) in different manners; the binding site for GM1s resides between two adjacent subunits, whereas the PC binding site in CRP is within a single monomer. Note that CRP also binds 10 calcium ions. While CID of the holo-complexes results in ejection of a monomer from the pentamer, with the monomer likely to be restructured, SID produces compact fragments of all oligomeric states up to pentamer. Busch et. al. found that the CRP CID fragments retained both too many and too few ligands. For example, the tetramer fragment should retain 4 ligands based on the binding site being within each subunit but instead retained 2 through 5 PCs (right inset in Figure 15a) while some highly charged monomers retained no ligand (left inset in Figure 15a). After CID of [CTB+5GM1s]18+ the tetramer retained the expected 3, 4, or 5 ligands, consistent with ligand interactions with two adjacent subunits, while the monomers retained no ligand (Figure 15c, inset). SID fragments, on the other hand, retained the ligands more readily and in a manner more consistent with the ligand binding sites. CRP monomers, dimers, and tetramers from SID were observed to be nearly 100% populated by the expected number of ligands (1 per monomer), Figure 15b. CTB fragments formed by SID also retained ligand; monomers, for example, were found to retain up to 2 GM1s ligands (Figure 15d, inset), consistent with ligand binding between subunits rather than within a single monomer.
Figure 15:
SID distinguishes ligand binding locations in pentamers CRP (with phosphocholine, PC) and CTB (with GM1s). CID spectra of (a) 18+ CRP at 2700 eV and (c) 18+ CTB at 2200 eV and corresponding SID spectra of (b) 18+ CRP at 630 eV and (d) 18+ CTB at 605 eV. Reproduced from ref. 116. Copyright 2018 American Chemical Society.
The UHMR Orbitraps are excellent instruments for characterizing heterogeneous samples due, in part, to optimized ion source conditions, high mass range, and powerful CID activation capabilities, as well as their high resolution. Augmentation of the UHMR with SID provides another means to probe heterogeneous proteins and protein complexes by high resolution mass spectrometry. For example, Harvey et al. investigated the structures of nanodiscs comprised of either DMPC or DMPG lipids, or a 50:50 mix of both, as well as nanodiscs containing antimicrobial peptides.101 Nanodiscs are promising membrane mimetics for solubilization of membrane proteins, and have been successfully utilized in nMS experiments.223–226 They consist of monodisperse nanoscale membrane bilayers encapsulated within an engineered membrane protein scaffold and serve as ‘containers’ for small molecules and membrane proteins; these containers can be ‘opened’ in the gas phase through harsh instrumental conditions via collisional activation. Utilizing an SID-equipped UHMR, Harvey et al studied the dissociation patterns of nanodiscs comprised of DMPC and DMPG lipids and found that the CID behavior depended on the lipid composition, but the SID behavior did not. Figure 16a, for example, shows a deconvolved native mass spectrum of a DMPC nanodisc; the wide peak in the mass domain is due to heterogeneity in the number of lipids contained in each nanodisc. By CID, the DMPC nanodiscs were found to shed lipid but otherwise remain intact even at high collision energy (Figure 16b). Collision with a surface, on the other hand, resulted in the shearing of the nanodisc in half, producing half discs consistent with the nanodisc’s topology (Figure 16c). Interestingly, DMPG nanodiscs sheared in half by both CID and SID within the energy range of the instrument, suggesting that the CID behavior, but not SID, depends on the lipid composition.
Figure 16:
(a) Deconvoluted mass spectrum of DMPC nanodiscs. Waterfall plots showing (b) CID and (c) SID spectra of DMPC nanodiscs with increasing collision energy. Reproduced from ref. 101 with permission from the Royal Society of Chemistry.
Recently, a second SID cell design for the Orbitraps has been reported, also termed a ‘Gen 3’ design (instead of Gen 2) due to geometric similarities with the later Synapt and solariX SID cells, and consists of a transfer hexapole with a split exit lens just before the C-trap (Figure 14c).97 Similar to the Synapt design, the exit lens consists of a deflector, a surface, and an extractor arranged in a split lens configuration. Similar transmission and sensitivities were observed compared to the Gen 1 cell but with fewer voltages, reducing tuning complexity. However, the placement of either SID cell prior to the C-trap and substitution of the C-trap offset with an external voltage are not ideal for ease of use and technology dissemination.
The Orbitrap platform continues to offer promising new capabilities for native mass spectrometry. Most recently, single particle charge detection mass spectrometry (CDMS) has been implemented on UHMR platforms, enabling the detection of single ions in the Orbitrap analyzer.220,227–230 Single particle CDMS has demonstrated unparalleled resolution for protein complexes, up to isotopic resolution for a 466 kDa complex.228,230 CDMS additionally can probe and resolve heterogeneous populations with ultrahigh sensitivity and improve sequence coverage from top-down analyses.227,229 In the future CDMS may prove useful in combination with SID for structural biology studies; the combination of unique SID fragmentation and ultrahigh resolution and sensitivity afforded by CDMS should be a powerful tool.
3.2.3. SID cells for FT-ICR
FT-ICR instruments have a rich history in the development of novel SID configurations and the investigation of the kinetics, energetics, and dynamics of fragmentation of peptides and proteins. Much of this pioneering early work was conducted by McLafferty, Laskin, and Futrell.149–151,161,231,231–234 Only recently has the FT-ICR platform been adopted for native mass spectrometry studies of macromolecular assemblies up to 1.6 MDa.22,96–98,108,235–237 High-field (e.g. 15 and 21 T) magnets, particularly those with dynamically harmonized FT-ICR cells, readily achieve resolutions of hundreds of thousands and even millions,238–241 enough for isotopic resolution of many proteins and protein complexes and their fragments. Isotopic resolution is useful for interpreting SID fragmentation spectra that exhibit symmetric charge partitioning, resulting in significant spectral overlap amongst fragment ions. Hybrid FT-ICR instruments (e.g. Bruker solariX) also have auxiliary MS/MS capabilities such as electron transfer dissociation and electron capture dissociation that can determine the primary sequence of proteins and complexes or manipulate the charge states of ions prior to or after dissociation.
There have been several SID cells adopted specifically for native mass spectrometry on the FT-ICR platform. A schematic of a Bruker solariX FT-ICR is given in Figure 17a. The instrument consists of an ESI/nESI ion source, dual ion funnels, a multipole, a quadrupole mass filter, and a collision cell for CID and ion accumulation. The ions are pulsed into the ultrahigh vacuum region of the instrument from the collision cell through focusing lenses and a transport multipole, eventually residing in the FT-ICR cell where the ions are mass analyzed.
Figure 17:
SID cells for FT-ICR platforms. (a) Schematic of the solariX FT-ICR platform, (b) CAD renderings of three generations of hybrid SID-CID cells (which replace the red collision cell), (c) illustration of transmission vs. SID modes, (d) schematic of the ‘Gen 3’ split lens SID design in the front endcap of the Bruker collision cell, (e) mass spectrum of TNH charge-reduced with EDDA, (f) SID spectrum using acceleration voltage of 45 V on the Gen 1 SID-CID cell, (g) zoom-in of the 6+ charge state of the α subunit showing isotopic resolution, and (h) Gen 3 SID spectrum of the mass selected 16+ charge state of TNH, showing an increase in S/N compared to the Gen 1 configuration. Panels (a, adapted), (c), and (e-g) reproduced from ref. 108. Copyright 2017 American Chemical Society. Panels (b) and (d) reproduced from ref. 97. Copyright 2020 American Chemical Society.
The Wysocki group has designed, fabricated, and tested three generations of SID cells for structural biology on the solariX platform (Figure 17b). The first-generation cell (‘Gen 1’) designed by Yan et al. replaced the Bruker collision cell entirely.108 The 6 cm space occupied by the collision cell was instead populated with an assembly consisting of a 3 cm long SID cell with a truncated rectilinear ion trap making up the remainder. The SID cell can be operated in ‘transmission’ mode or in SID mode (Figure 17c). Several protein complexes were studied to assess the coupling of SID with the ultrahigh resolution capabilities of the FT-ICR cell. Streptavidin tetramer, cholera toxin B pentamer, and TNH heterohexamer all exhibited SID profiles like those previously recorded on Q-IM-TOF platforms equipped with SID cells. The ultrahigh resolution was particularly useful for quantifying oligomer abundance in SID spectra of streptavidin (monomer vs. dimer) and cholera toxin B (overlapping monomer, dimer, trimer, and tetramer). Isotopic resolution is also useful for assigning charge states in complex spectra. For example, Figure 17e shows a native mass spectrum of TNH charge-reduced with ethylenediamine diacetate (EDDA). Using the Gen 1 cell, the SID fragmentation pattern (at 45 V) of TNH was obtained (Figure 17f) and had remarkable similarities to those obtained previously (e.g. Figure 8 and Figure 10), with αβγ trimers as the most abundant fragments with modest contributions of complementary αβ and αβγγ. The isotopic resolution allows the assignment of the peak at m/z 3515 as α6+ (spacing of 0.1666 Da) and the unparalleled mass accuracy enables the assignment of the peak at m/z ~3533 as (α+Co+3O)6+ (cobalt plus three cysteine-sulfenate oxygens).
The utility of the ultrahigh resolution was on display when investigating a more heterogeneous complex, 211 kDa multicopper oxidase complex (Mnx), which is now known (based on SID data) to be composed of a hexameric ring containing two distinct subunits (MnxE and MnxF, 12.2 and 11.2 kDa, respectively) arranged in alternating positions and stacked on a larger MnxG subunit (138 kDa).22,117 Using a Gen 1 SID cell tuned in an atypical manner (i.e. to collide ions with an unknown stainless-steel electrode instead of the usual FSAM-Au surface), SID of the native Mnx complex on a 15 T FT-ICR produced a range of oligomers (1mer through 6mer) containing varying numbers of MnxE and MnxF subunits, consistent with a cyclic hexamer structure in which the subunits alternate in their positions, as well as a sole MnG distribution suggesting that the hexameric ring is ‘stacked’ on the larger MnxG subunit (Figure 18a). Several peaks that were thought to be iron-bound species (based on low-resolution data from SID-IM-TOF117 experiments discussed later) were clarified as protein modifications instead. In addition, the stoichiometry of metal binding on different protein subunits was revealed. For example, the peak near m/z 3087 was determined to be MnxE + C6H10O6 rather than MnxE + 2Cu + Fe based on ultrahigh resolution and mass accuracy currently only achievable on the FT-ICR platform (Figure 18b–d). Similarly, the peaks near m/z 3813 were determined to be several overlapping species, MnxF + Cu + C6H10O6, MnxF + Cu + C8H9NO2S, and MnxF + 4Cu (Figure 18e–h).
Figure 18:
Surface-induced dissociation of 211 kDa multicopper oxidase Mnx on an ultrahigh resolution 15 T FT-ICR platform. (a) SID spectrum of Mnx 26+ through 29+ charge states (inset shows precursor ion population), (b) experimental isotopic distribution of peaks near m/z 3087 and comparison to theoretical isotope distributions for (c) MnxE + 2Cu + Fe and (d) MnxE + C6H10O, and (e) experimental isotope distributions observed near m/z 3813 and theoretical isotope distributions for (f) MnxF + Cu + C6H10O6, (f) MnxF + Cu + C8H9NO2S, and (f) MnxF + 4Cu. Adapted from ref. 22. Copyright 2018 American Chemical Society.
Whereas installation of SID cells in the Synapt platform by truncating the Trap or Transfer TWIGs or in the Orbitrap Exactive platform by replacing a transfer multipole had minimal impact on performance, on the solariX truncation of the collision cell (by half) and modification of the rod geometry (from hexapole to rectilinear trap and a change in rod-to-rod spacing) diminished the mass range, sensitivity, and dynamic range of the instrument. As a result, it became imperative to iterate on the SID-CID hybrid cell design to mitigate these losses. Snyder et al. simplified the SID optics in a similar manner as the ‘Gen 2’ Synapt design in order to accommodate a larger collision cell (Figure 17b, ‘Gen 2’).96 In addition, the collision cell geometry was made to match the original hexapole collision cell design. All told, these modifications resulted in an improvement in mass range by 3x (enabling the fragmentation of 330 kDa glutamate dehydrogenase and 801 kDa GroEL by SID) and an improvement in sensitivity and dynamic range by approximately an order of magnitude.
Motivated by a desire to simplify the ion optics even further, a much simpler ‘Gen 3’ SID cell was designed and tested on the solariX.97 The Gen 3 cell utilizes the entrance lens region of the original Bruker collision cell for SID, removing the need for a custom collision cell. Figure 17d shows the three electrodes that make up the SID cell in the front lens of the collision cell. As with the two other Gen 3 designs, after passing through the quadrupole, ions are deflected into the surface for activation and extracted into the collision cell for fragmentation sometime later. In addition to a simplification of the voltage scheme and overall SID assembly, the improvement to S/N using this cell design is evident in the SID spectrum of TNH 16+ in Figure 17h as compared to the Gen 1 cell in Figure 17f, all while retaining similar fragmentation patterns and avoiding CID contamination. Up to this point we have shown four virtually identical SID spectra of TNH in this review, and each was collected on a different platform using a different SID design. We have done so to emphasize that SID produces similar fragmentation patterns regardless of instrument platform. In other words, SID patterns are largely platform agnostic, save for any transmission differences as a function of m/z which are characteristic of the instrument design rather than SID itself.
The ultrahigh resolving power in combination with the increased sensitivity of this design was demonstrated with cholera toxin B pentamer, whose full scan and SID pattern are shown in Figure 19a,b. The 13+ charge state was selected for fragmentation and was analyzed with various transient lengths. Due to symmetric charge partitioning the peak at m/z 5803 has contributions from monomer, dimer, trimer, tetramer, and even charge stripped pentamer. The resolution obtained with (Figure 19c) an 18s transient was approximately 313,000 and with (Figure 19d) a 36s transient was 733,000. These resolutions are not currently achievable with other high-resolution instrumentation. As proteins increase in size, they tend to become more heterogeneous and bind to more nonspecific adducts, providing a motivation for implementing nMS technologies on the FT-ICR platform to resolve isotopes (for highly accurate charge state assignment) and ligand- or adduct-bound species. Care must be taken, however, to consider that larger oligomers tend to decrease in relative abundance as the transient length is increased on both FT-ICR and Orbitrap platforms due to desynchronizing collisions during the transient acquisition, with a greater distortion observed on the Orbitrap.97,99,221
Figure 19:
FT-ICR offers unparalleled resolution for quantifying oligomer abundances from SID. (a) Mass spectrum of cholera toxin B charge-reduced with EDDA and (b) SID fragmentation pattern at collision energy 715 eV. The ultrahigh resolution is demonstrated for the overlap peak at m/z 5803 using (c) an 18s transient or (d) a 36 s transient. Reproduced from ref. 97. Copyright 2020 American Chemical Society.
3.2.4 SID cells for ELIT
The McLuckey group has implemented SID on a custom electrostatic linear ion trap (ELIT), as illustrated in Figure 20a.111,148 Although the ELIT is analogous to an Orbitrap in that static DC potentials confine ions within the device and a mass spectrum is acquired in the form of an image current transient, MS/MS cannot be performed inside an Orbitrap due to the complicated ion motion and arrangement of the electrodes. However, in contrast, Hilger et al. demonstrated that SID could take place inside an ELIT by placing a surface at the back of the reflectron (similar to SID in a TOF reflectron160,162,242–245) and pulsing that surface to an attractive potential for several μs as the ions approach it.148 The ions then briefly undergo collision and the voltages are switched to extract ions back into the ELIT where they are mass analyzed. Hilger demonstrated the analysis of fragment ions of tetraalkylphosphonium salts. Johnson recently reported SID of a protein complex.111 Figure 20b is a native mass spectrum of the dimer triose phosphate isomerase. The 14+ charge state was isolated using mirror switching246 (Figure 20c) and subjected to SID, generating monomers that exhibit symmetric charge partitioning (Figure 20d), indicative of a successful surface collision. Although these results are preliminary, the high resolution of the ELIT and the versatile yet effective isolation and SID capabilities in a simple and compact mass analyzer make the platform a promising avenue for future exploration.
Figure 20:
(a) Illustration of SID in an ELIT, (b) native mass spectrum of triose phosphate isomerase, (c) isolation of the 14+ charge state by mirror switching, and (d) SID spectrum of the 14+ charge state to produce symmetrically charged monomers. Panel (a) adapted from 148. Copyright 2014 American Chemical Society. Panels (b-d) adapted from ref. 111 with written permission from the author.
3.2.5. Tandem SID configurations
So far, we have discussed configurations in which a single stage of SID is used to probe the connectivity of protein complexes, often in combination with ion mobility separations for improved selectivity. While a single stage of dissociation is often sufficient to probe the structures of small and simple protein complexes, it can be insufficient in the case of large or complicated homomers or especially in the case of heteromers. To that end several groups have developed MS3 or pseudo-MS3 technologies for characterizing the structures of proteins and protein complexes. For example, the Robinson group took advantage of the multistage isolation and activation capabilities of an Orbitrap tribrid instrument to identify ligands bound to membrane proteins.182 The Kelleher group demonstrated a ‘complex-down’ pseudo-MS3 approach in which GroEL monomers formed from in-source dissociation on an Orbitrap platform were dissociated into sequence fragments by HCD.247 The Brodbelt group has implemented multi-stage activation workflows combining CID and UVPD for characterizing proteins and protein complexes.248,249 The Sharon group has also taken advantage of the hybrid capabilities of an Orbitrap platform to elucidate the heterogeneity of the yeast homotetrameric FBP1 complex.24
At least one stage of fragmentation in most of these studies was induced through gas-phase collisions; however, as discussed above, CID ejects monomers and can restructure complexes and their fragments. While this is useful when top-down sequencing is required it can be a limitation in structural studies. On the other hand, SID fragments are compact and more informative in terms of deducing quaternary structure (though SID fragments may exhibit compaction or expansion in some cases49,50). Combining SID with CID in most instrument configurations is relatively straightforward because the SID cell is commonly placed prior to a collision cell, enabling SID-CID experiments to be performed. On the Synapt platform, for example, quadrupole isolated precursors can be fragmented by SID and subsequently separated by ion mobility prior to a second stage of activation by CID in the Transfer TWIG (Figure 9). If a second SID device is placed in front of a truncated Transfer cell, then combinations of SID/CID-IM-SID/CID experiments are possible. Quintyn et al. reported a tandem SID configuration (i.e. SID/SID) in which fragments from a first stage of SID were mobility separated and subjected to SID or CID post-IM.107
For example, tryptophan synthase is a heterotetramer with a near-linear αββα composition. A single stage, low-energy SID-IM experiment (Figure 21a) primarily ejects the smaller peripheral α subunit from the complex since the αβ interaction is weaker than the ββ interaction. At higher collision energy (Figure 21b) ejection of the α monomer is still the dominant dissociation pathway but with additional fragments corresponding to β monomer, ββ dimer, and αββ trimer. Fragmentation of the 12+ αββ trimer and 8+ ββ dimer (Figure 21c and d respectively) after mobility separation enables the reconstruction of their mass spectra. The trimer dissociates in an expected manner, with ejection of the smaller-interface/weakly-bound α monomer as the dominant pathway. The ββ dimer has only a single dissociation pathway, cleavage of the noncovalent interactions between the individual subunits. Taken together these results suggest an assembly mechanism wherein the ββ dimer is formed first, followed by association of a single α monomer to form the trimer and finally association of the second α monomer, resulting in a tetrameric species.
Figure 21:
Illustration of SID-IM-SID. (a) SID spectrum of 19+ tryptophan synthase at a collision energy of 570 eV, (b) SID-IM at a higher energy of 1330 eV, (c) SID-IM-SID at 2280 eV (second stage) of 12+ αββ trimer, and (d) 1330 eV SID of the 8+ ββ dimer. The dissociation pathways are illustrated in the insets. Reproduced from ref. 107 with permission from the Royal Society of Chemistry.
Returning to the example of the heterohexamer TNH, which we have previously discussed as being a dimer-of-heterotrimers (αβγ)2 arrangement with particularly strong interactions between the α and β subunits, the SID-IM-SID experiment was utilized to further probe the assembly and interfacial strengths within the heterotrimer substructures.110 In this case low energy SID was used as the first activation stage to produce αβγ trimers, which were then further dissociated in the second stage of SID. Figure 22a and b show energy-resolved mass spectrometry (ERMS) plots for SID-IM-SID of the 8+ and 9+ heterotrimers, respectively. Only minor differences in relative fragment ion abundance can be attributed to charge state. The ejection of the γ subunit is consistent with the αβ interactions being much stronger than interaction of the γ subunit with any of the other subunits. All three types of heterodimers were detected, suggesting that the arrangement of the three subunits in the heterotrimer is not linear but rather that each subunit interacts with every other subunit. The high abundance of the αβ dimer relative to the two other heterodimers is again indicative of the strongest interaction within the heterotrimer. This experiment highlights the utility of SID/SID for probing heteromeric structures, providing greater insight into protein substructure and assembly.
Figure 22:
Illustration of SID-IM-SID for native TNH heterohexamer. SID-IM-SID spectra of heterotrimer αβγ (a) 8+ and (b) 9+ produced from a first stage of SID of the heterohexamer (αβγ)2 on a Synapt G2-S platform equipped with SID cells prior to and after the TWIM cell. Reproduced from ref. 110. Copyright 2015 American Chemical Society.
SID/SID has also been accomplished on the high-resolution solariX (15 T) FT-ICR platform (schematic in Figure 17a).98 The shrinking of the SID cell design (Gen 3) enabled the simultaneous installation of SID in the entrance lenses of both the quadrupole mass filter and the collision cell. The experiments that are possible in this configuration are SID-MS, SID-Q-CID, and SID-Q-SID, where Q represents a stage of mass selection in the quadrupole mass filter. Note that because the first SID cell is prior to the quadrupole, precursor ions cannot be mass selected prior to the first stage of SID. It may be possible to combine this design with trapped ion mobility spectrometry in the entrance funnels of the instrument250 to enable mobility selection prior to SID/SID but this combination has not yet been demonstrated, although trapped ion mobility coupled to SID (TIMS-SID) has recently been illustrated, discussed in more detail in the IM section below.184
The SID/SID configuration on the solariX was evaluated for several model protein complexes, many with cyclic geometry, in order to demonstrate the importance of high resolution (as an alternative to ion mobility) for deconvoluting oligomer overlap in SID spectra of protein complexes.98 Oligomer abundances and charge state distributions were in agreement with those obtained previously on the Synapt platform.115 The unrivaled ultrahigh resolution of the FT-ICR was best showcased in the SID-Q-SID spectra of the SID fragments of homohexamer HFQ65, as shown in Figure 23. When the charge-reduced precursors in (a) were subject to a single stage of SID in the entrance lens of the FT-ICR quadrupole, all types of subcomplexes from monomer to pentamer were detected as is expected for a complex with cyclic subunit arrangement. The subcomplexes produced by the first stage of SID were then mass-selected by the quadrupole and dissociated by a second stage of SID in the entrance lens of the collision cell, as shown in panels (c) through (f). The expected fragments in each case are 1mer through (N-1)mer (where N is the precursor oligomeric state). In these spectra each oligomer has a narrow charge state distribution, only 1 or 2 charge states each, and so the relative abundances of the SID/SID fragments is only clear due to the isotopic resolution afforded by the 15 T FT-ICR analyzer, particularly at m/z 7186. While the dimer 3+ in (c) fragments only to monomer 1+ (1 m/z isotope spacing at m/z 7186) and monomer 2+, the trimer 4+ exhibits both monomer 1+ and dimer 2+ fragments at m/z 7186, the tetramer generates an additional trimer 3+, and the pentamer 7+ also likely produces tetramer 4+ (though it is not resolved under these conditions). This example illustrates the utility of isotopic resolution for determining and understanding SID product ion distributions, much in the same way that ion mobility is also used. Combining isotopic resolution with ion mobility should provide a higher measure of confidence in SID product ion distributions, particularly as ion mobility couples with high-resolution Orbitraps and as the resolution of TOF analyzers continues to increase to compete with FTMS systems.
Figure 23:
Illustration of SID-Q-SID on an ultrahigh resolution 15 T FT-ICR. (a) native mass spectrum of HFQ65 homohexamer charge reduced with TEAA, (b) single stage SID spectrum of the entire charge state distribution in the entrance lens of the FT-ICR quadrupole, and SID-Q-SID of the isolated (c) dimer 3+, (d) trimer 4+, (e) tetramer 5+, and (f) pentamer 7+. The utility of isotopic resolution is evident when comparing the fragment ion peak at m/z 7186 for each subcomplex. Adapted from ref. 98 with permission from Elsevier.
3.3. Perspective on the future of SID instrumentation
Clearly the future is bright for SID; its usefulness for structural biology studies of noncovalent protein complexes is unrivaled and, in combination with other ion activation techniques such as CID, ETD, ECD, IRMPD, and UVPD, it forms a strong foundation for structural elucidation. Usefulness notwithstanding, the dissemination of SID technology to the broader structural biology community and its use as a complementary tool with other structural biology techniques is a critical next step for widespread adoption and acceptance of the technique. Our group has made and continues to make progress with instrument vendors and in dissemination of the technology to other laboratories in the form of beta testing, with several generations of SID cells having been developed (as summarized in Table 2). Gen 1 SID cells have been installed on various Synapt platforms in the Robinson, Barran, McLean, Sobott, Russell,114 Prell,212 and Sharon laboratories.103 Gen 3 SID cells have been installed in Waters Synapt instruments in PNNL,251 Vachet, and Sharon laboratories recently, as well in Waters G1, G2, and Cyclic IMS instruments in our own laboratory. The Gen 3 SID cell is the first (and currently only) commercially available SID variant, having been recently announced by Waters as an option for the SELECT SERIES Cyclic IMS platforms. On the Orbitrap, Gen 1 cells have been installed in one EMR and two UHMR instruments in our own laboratory as well as in UHMRs in the Sharon and Russell laboratories. We continue to work with both large and small vendors to further develop and disseminate SID technology for the Orbitrap platform as a whole. The solariX FT-ICR platform in our laboratory currently utilizes the Gen 3 variant and is available for dissemination to the wider community for beta testing. We continue to work with Bruker for potential incorporation of SID technology on other platforms. The key to success of future commercialization and dissemination is further simplification and simultaneous optimization of the SID cell design in a vendor neutral manner. Miniaturization of the design and incorporation of SID in the most fitting location in each instrument – without compromising instrument performance or capabilities - combined with control and ideally automation of SID in native instrument software are important goals.
Table 2:
Evolution of SID cells for native mass spectrometry guided structural biology. Adapted from ref. 97. Copyright 2020 American Chemical Society.

|
4. Mechanism of SID for protein assemblies
The final section in this review discusses mechanistic aspects of SID, with particular focus on the activation and dissociation of noncovalent protein complexes. We will begin by summarizing the well-characterized mechanistic aspects of SID, particularly in contrast to CID. For example, in the absence of intertwined subunits, SID typically will dissociate a protein complex in a manner consistent with the complex’s assembly and topology; the smallest and weakest interactions between subunits tend to cleave first, leaving the stronger noncovalent and covalent interactions intact in the remaining product ions.45,50–53,109,110 SID spectra also usually exhibit symmetric charge partitioning, wherein fragment ions retain an amount of charge proportional to their mass.46,100 In some cases fragments may retain more charge than they should, which can be indicative of a restructured precursor ion or a monomer that readily unfolds, exposing basic sites for charge localization. Symmetric charge partitioning in SID contrasts with gas-phase collisional activation (CID) which tends to restructure and eject a highly charged monomer from a protein complex, leaving the remaining subcomplex with proportionally less charge.47 This disparity between collisional activation techniques can be readily explained by considering the timescale of activation and number of collisions; SID involves a single, quick collision with a surface while CID involves hundreds to thousands of collisions with small targets (causing a collisional cooling pathway to compete with dissociation), during which restructuring and asymmetric charge partitioning can take place. SID and CID fragments of low or modest charge tend to be compact,48,49,53 whereas highly charged SID and CID product ions tend to be restructured, often unfolded/elongated/expanded. In some cases, multi-subunit SID subcomplex fragments are observed to be compact after surface collision compared to structures that have been ‘clipped’ from the crystal structure of the native complex, consistent with intuitive expectations for ions in the gas phase, which would be expected to stabilize by maximizing intracomplex interactions.49,50 CID and SID product ions of the same charge state generally have similar CCS, and perhaps similar structures, as suggested by IM-MS49,50 and (unpublished) hydrogen deuterium exchange experiments within our own laboratory, suggesting that the charge of the fragment ion, rather than the manner in which it was produced, is more indicative of its structure. Recent work from Jia shows that dimer precursors of two different charge states (e.g. 9+ and 11+ β-lactoglobulin, BLG) showcase radically different gas-phase behaviors by CID vs. SID. The two charge states of BLG states were found to restructure over a broad range of activation energies when subject to collisions with gaseous targets (i.e. CIU was the dominant process), while the same precursor charge states tended to have fragmentation as a dominant pathway by SID even at relatively low collision energies.252 Energy deposition via SID is well known to be more efficient on a per-collision basis than CID, owing to the greater effective mass of the target/surface compared to any realistic ionic projectile.44,50,102,212,215
Despite the substantial evidence for symmetric charge partitioning, efficient energy deposition, and topologically consistent fragmentation, there are many aspects of the dissociation of protein complexes by SID that remain either unexplored or under-characterized. Among them are the energy deposition and partitioning as a function of collision energy and ion structure, optimal incidence and scattering angles, chemical and physical changes at the surface or in the protein complexes themselves induced by collision, the timescale of dissociation, and any transient interactions of the protein complex with the surface (e.g. sticking or surfing along the surface). These will be discussed at the end of this section; first, we begin by reviewing what is known about SID.
4.1. SID is a fast, high-energy deposition event; CID is a multi-step activation process
Given that CID and SID fragmentation patterns are remarkably different (see e.g., Figures 2, 3, 7, 8), with CID consistently ejecting highly charged (often restructured/elongated) monomers to leave (N-1)mers and with SID generating fragments reflective of substructure, it stands to reason that the mechanism of activation for these two techniques has some key differences. One obvious dissimilarity between CID and SID is the mass of the neutral collision target. CID involves collisions with small inert gas atoms or molecules such as He, N2, Ar, Xe, or SF6, all of which have orders of magnitude less mass than even the smallest of noncovalent protein complexes. We can deduce the effect this has on energy deposition from equation 4. If MION >> MN, then eq. 4 can be approximated by
| [Eq. 5] |
in which case each collision of a protein complex with a background gas molecule is remarkably inefficient because MN / MION ~ 0. For example, for the 53 kDa streptavidin tetramer colliding with a single molecule of nitrogen, the maximum energy transferred to the complex in a single collision is ~0.05% of the collision energy! Therefore, if the 10+ charge state is accelerated through a nitrogen bath gas using a 100 V potential difference, the maximum energy transfer per collision is 0.5 eV. For reference, a single hydrogen bond between water molecules has a dissociation threshold of 0.24 eV,253 yet we must consider that noncovalent interactions between protein subunits involve many hydrogen bonds as well as salt bridges and Van der Waals interactions. This simple calculation illustrates the need for many low-efficiency collisions when fragmenting protein complexes by CID and the importance of the target mass toward imparting enough energy for dissociation. For these reasons, several investigators have explored the use of high mass gases such as argon, xenon, or SF6 rather than He or N2 for CID, CIU, and collisional cooling of protein complexes (targets with more degrees of freedom preferred for collisional cooling).34,254 Pressure also plays an important role in CID, as the number of collisions (hundreds to many thousands) that an ion experiences will increase with pressure. During this ‘heating’ process in which the ion has many successive collisions with background gas, not only is the internal energy of the ion increased (and overall internal energy distribution broadened255) the protein can undergo structural changes that can be measured by ion mobility or deduced from the observed charge partitioning. During the long activation process in CID, collisional cooling can compete with ion activation, making CID even less effective for dissociation of large molecules.
In contrast, SID involves collision with a target surface, with the most common surface materials being stainless steel, gold, and SAMs on gold. Ignoring surface composition for a moment, again consider equation 4 except with a surface as the target, in which case MN >> MION and we obtain
| [Eq. 6] |
In other words, through collision with a clean stainless steel target, equation 6 suggests that the energy transfer could be up to ~100% efficient. Note that this would imply that no energy is lost to the surface as heat and the protein would stick to the surface (and hence not be transmitted through the remainder of the instrument for mass analysis). In reality, the surface is not perfectly clean or smooth and does not have an infinite mass. Instead the effective mass of FC12 SAM-covered surfaces has been measured by Futrell and coworkers as ~150 for a CS2+ projectile (MW: 76.1 Da), which corresponds to the CF3CF2CF end group of the SAM utilized in that particular set of experiments.256 The effective mass of the surface was also found to be independent of mass for a series of small peptides (trialanine, tetraalanine, and two pentapeptides).257 Moreover, what is experimentally observed is that when a protein complex collides with the surface, ~5–25% of the collision energy is internalized to be redistributed amongst the bonds of the molecule (based on experiments with small molecules165,255,258), 5–25% of the collision energy remains as residual kinetic energy (allowing the ion to move away from the surface),98 and the remainder is lost to the surface as heat (or motion of the chains for a SAM-coated surface).258 As we will discuss later, the particulars of energy partitioning for protein complexes have only been investigated in a handful of recent studies (whereas energy partitioning is well characterized for small molecules and peptides), and so this is an area of research that needs further exploration. Even so, it is clear that for a given lab frame energy, the greater mass of the surface lends itself to higher energy deposition that doesn’t depend on a series of many low-energy collisions, a key mechanistic difference with CID.44,175,212,215 As determined by Laskin and Futrell for a series of peptides, the efficiency of T->V energy transfer (~15–20%) is determined by the center-of-mass energy rather than the laboratory frame collision energy and is largely independent of collision energy,255 in agreement with molecular dynamics simulations of N-protonated diglycine in which the percentage of collision energy internalized was nearly independent of the collision energy.259 Another factor to consider when comparing SID and CID is that the long time-scale of activation by CID causes collisional cooling to occur as a competing process, whereas the time-scale of interaction between analyte and target in SID is very brief.257 These complications highlight that differences between SID and CID fragmentation patterns cannot be simply explained by efficiency of energy deposition alone, but by a series of contributing factors that include energy deposition, timescale of interaction, number of collisions, and influence of competing processes (cooling).
While there are no direct measurements of internal energy deposition upon collision of a protein complex with a surface, there is an abundance of evidence that SID deposits energy differently than multi-collision CID. First, noncovalent protein complexes consistently dissociate at lower SID collision energies compared to CID; in other words, the threshold for dissociation by SID is lower than CID, consistent with the higher mass target of SID and lack of competing processes (cooling) during activation. The 11+ charge states of the D2 symmetric homotetramers streptavidin, neutravidin, and transthyretin, for example, all dissociate to dimers at an SID collision energy of 330 eV but remain intact after 330 eV CID.53 Some particularly sturdy protein complexes do not dissociate by CID at the maximum CID voltage of commercial mass spectrometers (200–300 V) but fragment readily by SID. For example, phosphorylase B 29+ and 21+ (normal-charge and charge-reduced) do not dissociate by CID 200 V (5800 eV and 4200 eV) on a Synapt platform but begin to fragment to monomers around ~1000 eV via SID.50 Glutamate dehydrogenase hexamer (39+ and 27+) shows similar behavior, with limited, if any, fragmentation by CID up to 200 V (7800 and 5400 eV) but fragmenting to trimers and monomers by SID 130 V and 190 V (~5100 eV). On a UHMR, DMPC nanodiscs were found to shear in half by SID at ~105 V but only lost lipids via CID up to 225 V.101 Charge-reduced GroEL 14mer 50+ was found to fragment by SID at 180 V (9,000 eV) but not by CID at 200 V (10,000 eV).51 Taken together, these results suggest higher energy deposition in SID than CID at the same lab frame collision energy, though the single vs. multi-collision nature of SID vs. CID should also be taken into account when comparing energy deposition and makes direct comparison of collision energies problematic.255 The Prell group studied energy deposition in SIU and CIU for a variety of protein monomers (not complexes). The CIU and SIU fingerprints of bovine serum albumin (66 kDa) in Figure 24a and Figure 24b, respectively, exhibited similar overall transitions between conformers but at drastically different energies.212 Similarly, the SIU and CIU profiles of the N-terminal domain of anthrax lethal factor (LFN10+) in Figure 24c and d showed similar structural transitions regardless of activation method. Overall, using CIU and SIU data for several additional monomeric proteins up to 80 kDa, a correlation between nominal SIU and CIU energy was approximated by the power law ESIU ∝ ECIU0.61 (Figure 24e), implying more efficient energy deposition via surface collisions.
Figure 24:
Comparison of CIU and SIU plots for bovine serum albumin (BSA) 15+ and the N-terminal domain of anthrax lethal factor (LFN10+). (A) CIU OF BSA15+, (b) SIU of BSA15+, (c) CIU of LFN10+, and (d) SIU of LFN10+. Adapted from ref. 212 with permission from The Royal Society of Chemistry.
4.2. Restructuring of fragment ions after SID and CID: symmetric vs. asymmetric charge partitioning
Differences in the manner in which energy is deposited into protein complexes upon collision with a surface (an energy “jump” rather than multistep, restructuring activation) vs. collision with a much smaller gaseous target partially explains why marked differences in fragmentation patterns are observed between CID and SID and also explains dissimilarities in charge partitioning for the two collisional activation techniques. Charge partitioning is intimately linked not only to the charge density of the protein ions, but also to the method of dissociation. CID is well known to eject highly charged monomers from a noncovalent protein complex, leaving an (N-1)mer intact.30,47,51,65,188,260–265 Multiple monomeric subunits can be stripped from a complex through typical collisional activation with target gas (involving multiple low-efficiency collisions), though whether the cleavages occur simultaneously or (more likely) sequentially is unclear.30,266–268 Sequential removal of monomers was suggested by Benesch et al. who observed production of 11mer of TaHSP16.9 12mer at modest CID collision energies with a concurrent disappearance of the 12mer, but at high collision energies the 11mer species decreased in relative abundance concurrent with appearance of a 10mer. It has also been reported that peripheral subunits are preferentially released.40,102,269,270 Wang et al. found that non-peripheral subunits could be released through secondary dissociation after primary removal of peripheral subunits or directly from charge-reduced or elongated protein complexes.270 Song studied dissociation of the heterohexamer TNH (untagged) by CID and SID and observed changes in preferential ejection of monomers via CID as a result of altered charge density on the complex.271 Leney studied two hexameric protein complexes made up of three copies of two monomers of equal mass (which in one case had the same pIs and in a second case had very different pIs).272 The hexamer with monomers of ~equal pIs expelled both kinds of monomers upon CID, but the hexamer composed of monomers of unequal pIs preferentially ejected the lower pI monomer. One possible explanation is an asymmetric distribution of charges on the subunits. Curiously, in-solution dissociation experiments showed that the higher pI monomer was expelled. Regardless of which subunit is released, however, charge is generally partitioned asymmetrically amongst CID fragments. The charge partitioning is governed primarily by the relative surface area of each subunit, which is consistent with restructuring mechanisms in CID in which a monomer unfolds/elongates and hence increases in surface area relative to the rest of the complex, thereby taking proportionally ‘too much’ charge with it.30
In contrast, SID spectra consistently exhibit more symmetric charge partitioning, wherein the fraction of charge retained by subcomplexes is approximately equal to their fraction of the total mass of the precursor ion.46,100 For example, CID of concentration-produced (non-biological) 11+ cytochrome C dimers results in asymmetric charge partitioning, with two distributions of monomers having most abundant charge states of 8+ and 3+ (Figure 25a), whereas in the SID spectrum only a single Gaussian charge state distribution is observed with an average charge of 5.5, or half the charge of the mass-selected precursor (Figure 25b).273 This is true for other dimers as well, for example phosphorylase B dimers with 29+ and 21+ charges fragment by SID to monomers with 14.5+ and 10.5+ charges, on average (Figure 26a,b).50 In fact, this observation is consistent across topologies, from simple dimers50,52,100,111 and cyclic complexes40 to dihedral53 and even heteromeric proteins.22,109,110,117 For dihedral tetramers streptavidin, neutravidin, and transthyretin with 11+ charges, low-energy SID generates dimers with an average of 5.5 charges, in agreement with a symmetric partitioning model.53 SID of glutamate dehydrogenase hexamer produces topologically consistent trimers with half the charge of the precursors (39+ ->19.5+; 27+ -> 13.5+) despite the high collision energy needed for activation (Figure 26c,d).50
Figure 25:
(a) CID and (b) SID spectra of (Cyt c)211+ Reproduced from ref. 46. Copyright 2006 American Chemical Society.
Figure 26:
SID spectra of protein complexes exhibit symmetric charge partitioning. SID spectra of (a) phosphorylase B dimer 29+ at 110 V, (b) phosphorylase B dimer 21+ at 150 V, (c) glutamate dehydrogenase hexamer 39+ at 130 V, and (d) glutamate dehydrogenase hexamer 27+ at 190 V. Adapted from ref. 50 with kind permission from Springer Science & Business Media.
The charge partitioning models for neither SID (symmetric) or CID (asymmetric) are absolute; there are exceptions and curiosities that depend on protein structure, charge density, presence of salt bridges and other noncovalent interactions,31 and activation energy. For example, CID can result in symmetric charge partitioning in some cases. Jurchen et al. studied CID of α-lactalbumin homo- and heterodimers (one subunit oxidized and one reduced) and observed that the 15+ homodimer fragmented to 8+/7+ monomers but the heterodimer dissociated in an asymmetric manner in which the reduced subunit retained 11+ charges and the oxidized form carried ~4+ charges.262 It was also observed that the 17+ dimer of cytochrome C dissociated symmetrically whereas the charge-reduced 13+ dimer dissociated asymmetrically, implying a dependence on precursor ion charge density. Curiously, when the 17+ dimer was transferred to the gas phase and charge-reduced to 13+ (via proton transfer reactions), symmetric charge partitioning was observed. In other words, the structure of the gas-phase 13+ charge state depended on the initial solution conditions which translated directly into differences in fragmentation. Dodds et al. studied the dissociation pathways of model homodimers of bacteriophage N15 Cro, bacteriophage λ Cro, and bacteriophage P22 Arc.118 The N15 Cro dimer exhibited symmetric charge partitioning by CID, whereas λ Cro exhibited asymmetric charge partitioning. SID of both complexes yielded symmetric charge partitioning. While neither dimer contains intrasubunit disulfide bonds or cross-links, there are interfacial electrostatic interactions within the λ Cro complex which increase the energy threshold for subunit loss by CID, leading to monomer unfolding as the primary CID pathway. Jia et al. recently investigated the unfolding and dissociation pathways of beta-lactoglobulin A homodimers in both oxidized (disulfide bonds intact) and reduced forms and observed symmetric charge partitioning in both cases when SID was used for activation but asymmetric partitioning (as a result of increased subunit flexibility) in CID when the disulfides were reduced.274 While SID spectra usually exhibit symmetric charge partitioning, cyclic complexes (e.g. TRAP and HFQ), complexes with cyclic substructures (e.g. stacked rings like GroEL) and particularly ions generated directly from ammonium acetate solutions (normal-charge) have been known to generate highly charged monomers (and sometimes other small oligomers) from both CID and SID.50,51,51,97,99 The mechanism by which the monomers acquire ‘too much’ charge after surface collision is not clear but might be caused by secondary fragmentation of restructured intermediates, monomer restructuring as monomer departs, or by excess charge on the precursor, increasing its propensity to restructure.50,51 Extended interactions (e.g. multiple collisions or ‘skating’ along) with the surface also cannot be ruled out without further fundamental studies.
The origin of asymmetric charge partitioning in CID yet symmetric charge partitioning in SID is best contrasted as a multi-step ‘heating’ activation method (CID) vs. a single high-energy jump activation (SID) (Figure 1).47,175,275 There is both experimental evidence264,264,276 and molecular dynamics simulations263,277–283 that asymmetric charge partitioning is the result of a monomer unfolding during the multi-step CID process. Benesch et al. found a correlation between monomer surface area as a fraction of the total surface area of the monomer and (N-1)mer produced from CID.30 The Konermann group has developed several molecular dynamics simulations that show initial breakage of zwitterionic interactions (salt bridges) through ion heating followed by gradual unfolding of a monomer during the CID process, with simultaneous proton migration to the unfolding monomer in order to redistribute the charge evenly throughout the protein complex as it is restructuring.276,283 Salt-bridges have been considered an important factor in the stabilization of protein-ions in the gas phase, as the low dielectric constant in the vacuum strengthens electrostatic interactions. It should, however, also be noted that consideration of mobile protons are important when discussing the stabilization imparted from salt-bridges in gas-phase protein ions and protein unfolding, with mobile protons being shown recently in simulations to weaken salt bridges during CIU. 284 It was also observed that subunits could ‘compete’ for charge as they each start to unfold, with a single subunit ‘winning’ and separating from the remaining subunits. The excess charge on the unfolded monomer is the direct result of Coulombic repulsion to equalize the charge density throughout the surface of the complex.260 We can also conclude that monomers of low charge should be compact (assuming charge migration is driven by changes in monomer surface area), which is supported by several studies.49,87,285 The Thachuk group developed molecular dynamics simulations to study charge partitioning and structural changes during CID.263,277–281 For the case study of cytochrome c dimers it was determined that when only small structural changes were observed during dissociation, the charges were distributed symmetrically amongst the fragments, whereas asymmetric charge partitioning was observed as a less well-defined transition wherein a monomer unfolds but is temporarily tethered to the folded monomer by its charged N-terminus (Figure 27).281 Generally, charge movement and unfolding were found by Thachuk to be charge-driven by Coulombic repulsion, and that charge distributes across the protein complex so as to maintain the same charge density throughout.263,279
Figure 27:
Monomer orientations during asymmetric dissociation of cytochrome c dimer 10+ at a center-of-mass distance of (a) 6 nm, (b) 9 nm, and (c) 11 nm. The yellow monomer has 8 charges, and the green monomer has 2 charges. Reproduced from ref. 281. Copyright 2010 American Chemical Society.
The slow heating in CID contrasts directly with an approximately single collision with a high-mass surface target. Strictly speaking, it has not been explicitly demonstrated that protein complexes only hit the surface target a single time, but it is certainly reasonable to assume that the number of interactions that a protein complex has with a surface is orders of magnitude lower than the number of collisions with background gas in CID experiments. As a result of the combination of higher energy deposition, providing access to fragmentation pathways otherwise inaccessible by the less efficient (lower target mass), multistep CID, and the ~single-collision nature of the surface collision, monomers cleaving from a complex do not extensively restructure by SID, resulting in symmetric charge partitioning unless dissociation can only occur following unfolding.
Based on the above arguments we might expect all SID fragments to be compact, and while most SID fragments are compact, it is not true for SID fragments of relatively high charge.49,50 Consider, for example, that the highly charged monomers (< m/z 2,000) from SID of TRAP in appear more extended (and hence unfolded) than the monomers of lower charge (> m/z 2,000). Moreover, many fragments have been observed to be compacted or otherwise restructured compared to native crystal structures as judged by collision cross section and gas-phase hydrogen deuterium exchange measurements, suggesting that compact fragments may be rearranged regardless of their origin. This is most prevalent for cyclic structures which would be expected to rearrange following activation/dissociation in order to maximize intersubunit interactions.113
Returning first to the case of the D2 symmetric homotetramers streptavidin, neutravidin, and transthyretin, Quintyn et al. compared the collision cross sections of CID and SID monomers, dimers, and trimers to those calculated from the native crystal structure and found that SID dimers were well in agreement with the native state (~21 nm2), assuming cleavage of the smallest interfacial area.53 Monomers from SID of dimer phosphorylase B also agreed well with values calculated from crystal structure so long as the charge of the monomer was low. In particular, 15+ through 20+ monomers from SID of 29+ dimer appeared unfolded, whereas monomers of lower charge, produced from SID of either the 29+ or 21+ dimer, had CCSs that agreed well with crystal structure.50 SID experiments with membrane proteins as analyte ions had similar results, with dimers and monomers from SID of trimer AmtB exhibiting CCSs consistent with the crystal structure and monomers and dimers from SID of tetramer Aqp0 agreeing with crystal structure.95 However, the trimer CCS agreed better with a rearranged form as is common for cyclic complexes whose subcomplexes rearrange (collapse), presumably to maximize intersubunit interactions and overall stability of the gas-phase subcomplex.
Investigation of the SID and CID products of pentamer C-reactive protein and tetramer concanavalin A revealed two interesting findings.49 First, the collision cross sections of monomers and pentamers originating from CID and SID were identical so long as the charge state of the fragment was also identical between the two methods (Figure 28). This experimental finding suggests that the charge state of the fragment, rather than the dissociation method, is a better indicator of its conformation. While highly charged monomers from both CID and SID were unfolded, monomers of low or modest charge were not, even after CID. The second interesting finding from this study was that, while monomer collision cross sections agreed quite well with those predicted from the crystal structures,48 the CCSs of the dimers, trimers, and tetramers from both CID and SID were not consistent with the ‘native’ arrangement within the original complex and had CCSs smaller than those calculated from ‘collapsed’ configurations in which intersubunit interactions were maximized. This suggests that both CID and SID product ions, even compact ones, may undergo some degree of restructuring (e.g. relocation of polar residues or binding sites to increase intersubunit stability, as suggested by Zhou et al.49) that causes only a modest change in CCS. Similar findings were reported for trimers from SID of both 39+ and 27+ hexamer glutamate dehydrogenase.50 The compact trimers (< 19 charges) appeared collapsed regardless of precursor or fragment ion charge state, whereas trimers with >18 charges, generated exclusively from the 39+ hexamer, were unfolded, suggesting restructuring of the complex to produce the unfolded trimers. Our group has preliminary gas-phase hydrogen deuterium exchange data that also suggest 1) that CID and SID fragments are similar (i.e. have similar HDX rates) regardless of origin so long as their charge states are the same, and 2) that subcomplexes produced by SID can be restructured compared to their alignment in the native complex, which is evidenced by the proportionally lower exchange rates of SID fragments compared to their native, unactivated precursors (e.g. a dimer fragment exchanges less than half as much as a tetramer precursor). Taken together, ion mobility and hydrogen deuterium exchange measurements show that while SID fragments are compact, they are not necessarily ‘native’ and may be restructured in such a way to increase intersubunit interactions and thus stabilize the subcomplexes.
Figure 28:
Comparison of CCSs of subcomplexes produced by (a,c) CID and (b,d) SID of pentamer C-reactive protein and tetramer concanavalin A. While monomer CCSs agree well with crystal structures, the CCSs of larger oligomers suggest collapse/restructuring in both CID and SID. Open symbols are product ions from normal-charge precursors (24+ CRP and 19+ ConA) while filled circles originate from charge-reduced precursors (18+ CRP and 13+ ConA). Reproduced from ref. 49 with permission from The Royal Society of Chemistry.
The prior discussion assumes that subcomplexes restructure after the precursor complex has dissociated, but it is also possible for collapsed or unfolded subcomplexes to be produced from a restructured or charge-stripped precursor, although it might be surprising if the fragments reflect the initial complex topology if there is significant restructuring prior to dissociation. For example, Ma et al. conducted energy-resolved SID studies of phosphorylase B and glutamate dehydrogenase from ammonium acetate (‘normal’ charge) and charge-reducing conditions (with TEAA additive).50 SID of phosphorylase B dimers produced both folded and unfolded monomers as measured by ion mobility, with unfolded monomers being the prevalent fragment under normal-charge conditions and folded monomers preferred under charge-reduced conditions. For the normal-charge precursors (29+), the proportion of unfolded monomer increased rapidly as the amount of unfolded precursor was observed, suggesting that they may, in fact, originate from the unfolded dimer rather than unfolding after dissociation. Two charge states of glutamate dehydrogenase hexamer were also studied (39+ and 27+). Using the same logic as for phosphorylase B, Ma et al. hypothesized that the 39+ hexamer restructures and subsequently dissociates, rationalizing the observation of unfolded trimer and monomer, whereas the charge-reduced 27+ state remains compact at low SID energy but eventually expands slightly and dissociates to give primarily folded trimers. Secondary dissociation of folded trimers was speculatively suggested to rationalize observation of both folded and unfolded monomers. GroEL, an 801 kDa 14mer of a stacked 7mer ring arrangement, has been observed to dissociate by SID (from ammonium acetate charge states, ~71+) to form some hexamers centered around the 15+ charge state as well as highly charged monomers (average ~22+) which may be the result of secondary dissociation of the topologically relevant fragments (~35+ 7mers).51 These fragments were also produced from charge-reduced ~50+ GroEL, though in much lower abundance (with concurrent increase in topologically-relevant 7mer fragment) as in most cases asymmetric charge partitioning and non-native fragmentation is significantly diminished when the precursor charge is reduced.51 Moreover, there is some evidence that large complexes with many charges may fragment from a charge-reduced state after some charge has been stripped away by collision with a surface. If this were the case, then the sum of the charge on the fragments ought to be lower than the total charge of the precursors, as observed for GroEL, where the average charge state of the 7mer SID fragment generated from ~50+ 14mers was ~21+.51 Secondary dissociation and charge symmetry continue to be explored in our and other laboratories in order to improve our understanding of the fragmentation mechanism for noncovalent protein complexes.
4.3. Improving our understanding of SID
Certain aspects of the mechanism of SID of noncovalent protein complexes are better characterized than others. It is clear from the previous sections that charge partitioning, topologically reflective dissociation patterns, and collision cross sections of some representative activated complexes and their fragments are well characterized, but there are several questions that remain unanswered. For example, the optimal incidence and most likely scattering angles of precursor ions and fragments have not been determined for any noncovalent protein complexes. Although it can be reasonably asserted that SID involves fewer collisions with the neutral target than CID, it is not clear whether SID involves only a single collision or perhaps a few collisions. Or rather it may be that under certain conditions a protein complex ‘skates’ across the surface or undergoes a ‘sticky’ collision in which the ion is deposited on the surface (akin to soft landing286) and is later ejected by further primary collisions. The latter might be implied if unfragmented precursor ions are left with less residual kinetic energy than fragments due to thermalization while on the surface.287 However, kinetic energies of scattered complexes and their fragments have not been measured and reported in detail, with only a handful of preliminary measurements having been made,98,288 in part because of uncertainties in the quality of these measurements in instruments designed for analytical mass spectrometry. For example, we have recently measured the residual kinetic energy of streptavidin tetramers (10+ and 11+) as a percentage of collision energy as 18% at SID 450 eV but only 9.4% at SID 850 eV.98 Based on recently acquired data we have determined approximate residual kinetic energies of several other complexes and their fragments (as a percentage of the SID collision energy) after SID with a stainless steel surface to be ~20–30% for ~200 eV collisions, linearly decreasing to ~10% at 1000 eV collision energy.289 That the residual kinetic energy decreases with collision energy either implies more efficient energy deposition (internalization) or, in better agreement with direct dynamics simulations conducted previously with octaglycine, more energy transferred to the surface and lost as heat/excitation of surface coatings or contaminants.290 Regardless, other aspects of SID energy partitioning for noncovalent protein complexes have not been measured, for example, the internal energy deposition and the energy transfer to the surface, particularly as a function of collision energy and protein mass and conformation.
While there is a lack of experimental data characterizing SID energy partitioning for protein complexes, several groups (Laskin & Futrell,151,161,169,231,232,255,257,291 Barnes,290,292–297 Hase,168,170,170,290,296–302 Hanley303–309) have performed extensive experimental and theoretical characterization of energy partitioning with peptides as analytes. Discussion of these studies in detail is beyond the scope of this text but is enumerated briefly here. Much of Laskin & Futrell’s work has been discussed earlier in the paper and focuses on kinetics, energetics, and dynamics of peptide fragmentation by SID on an FT-ICR mass spectrometer.151,161,169,231,232,255,257,291 Key findings include similar peptide fragmentation patterns in both SID and CID,255,257 similar internal energy deposition in the two techniques,255,257 and observation of a (still contested) ‘shattering’ fragmentation mechanism in which dissociation occurs at or very near the surface rather than sometime later away from the surface.291 Hase (and more recently continued by Barnes) developed classical trajectory simulations to study many aspects of SID, including the effect of surface stiffness170 and projectile orientation301 on energy deposition, general energy partitioning in peptide and metal carbonyl SID,259,298–300,302 and importance of shattering fragmentation for small peptides.298–300,310 Barnes has studied the effect of protonation site and conformation on SID,295 reactions of peptide analyte ions with SAM surfaces,293 the role of proton motion in SID,294 and fragmentation and energetics of SID of octaglycine.290 Barnes and Hase have coauthored several excellent review articles describing the utility of chemical dynamics simulations.168,296,297 Hanley laboratory studied internal energy deposition, kinetic energy retention, and activation energies for SID of small molecules,304,306,308,309 shattering during SID,303 and the effect of peptide structure on kinetic energy retention.307 For example, for a series of peptides, linear molecules retained 24% of the incident ion SID energy as kinetic energy, while only 21% and 17% was retained for cyclic dipeptides and a four-peptide ring, respectively.307 It is likely that these results will translate directly into differences in energy partitioning for protein complexes of different shapes, sizes, and flexibility, as recently described by Prell group for monomeric proteins.212
Another mechanistic question needing further experimental investigation is the timescale of dissociation and whether SID of protein complexes can be fully described by an RRKM model or whether a ‘shattering’ mechanism proposed for metal carbonyls and small peptides also contributes.168,291,298,299 An energy partitioning model in which residual kinetic energy is distributed proportionally by fragment mass, which is in agreement with our recent kinetic energy measurements,98 suggests the RRKM model as the dominant dissociation pathway for protein complexes rather than fast dissociation at or near the surface (in which the fragments would have identical kinetic energies rather than identical velocities). The strong similarity of SID and CID spectra of peptides and other small molecules is also consistent with both undergoing typical RRKM type fragmentation and consistent with what Barnes has reported as a decrease in ‘fast fragmentation events’ (shattering) making up ~50% of the total fragment population to <10% as peptides increase in size from 2 to 8 residues, respectively.292 The extent of neutralization on the surface has not been explicitly characterized but it is thought to be minimal for large, multiply charged species. Even so, chemical and physical changes on the surface and in the protein complexes themselves have not been explored in detail. It has recently been suggested, for example, that lipid molecules may be left on the surface after conducting SID of lipid-containing nanodiscs,101
Finally, it is worth investigating further how fragments are formed by either primary vs. secondary dissociation, and determining which structural motifs require unfolding or restructuring to cleave the noncovalent interaction within some complexes. It is not known, when restructuring is required, which structural motifs are most prevalent. We have also observed that some complexes charge strip before dissociating, but the extent of charge stripping and and its correlation with structure or flexibility have not yet been investigated or systematically compared with charge stripping that occurs by CID. Furthermore, as we have already discussed, ion mobility and HDX measurements have suggested some degree of collapse/restructuring for some SID products, even though fragment ions from SID are generally compact. We have been careful to use ‘compact’ rather than ‘native’ to describe SID fragments, as the structures of these subcomplexes are not explicitly known (e.g. no crystal or cryo-EM structures exist for SID fragments). A straightforward and promising approach to determine the structures of fragments after SID and CID is ion soft landing, which was initially conceived in the Cooks’ laboratory73 but has been utilized by Benesch and Robinson to study protein assemblies by electron microscopy after manipulation in the gas phase.311,312 Anggara et. al. recently coupled scanning tunneling microscopy with soft surface collisions to study the conformational landscape of oligosaccharides, further highlighting the utility of such an approach.313 A combination of ion mobility, gas-phase HDX, and computational modeling should also provide insight into morphologies of these collapsed structures.
5. Conclusion & Outlook
Surface-induced dissociation has been applied to protein complexes for almost 15 years and has been coupled with drift tube, traveling wave, and trapped ion mobility for structural studies of protein assemblies. It has been illustrated with multiple device designs on multiple nMS instrument types, including QTOF, FTICR, Orbitraps, and ELITs, with recent efforts to improve SID accessibility and ease-of-use proving fruitful for dissemination to the wider nMS community through collaborations with instrument manufacturers. Through the application of SID to multiple model and real-world (unknown structure) complexes, much has been learned about the characteristics of SID as an especially effective dissociation method for protein complexes, a method that provides information that is not readily accessible by other established activation methods. Although SID has only recently been introduced as a commercial product,193 SID is already playing a role in structure determination of protein and nucleoprotein complexes as more and more collaborators request devices or data. nMS coupled to SID has shown its value as a complementary structural biology tool that is easy to apply, providing important project progress information prior to, or as an alternative to, the use of more cumbersome tools. Computational methods are allowing SID data, sometimes in conjunction with low resolution data from other methods, to provide improved structure prediction or refinement. Surface induced dissociation, coupled with nMS and ion mobility as well as other activation methods, is poised to become an important tool in the structural biology toolbox.
Figure 13:
IM-SID can be used to individually probe different conformations of in-source activated CRP (cone 200 V). Right hand panels show extracted SID spectra (1260 eV) from the highlighted regions in the left hand panel. Cartoon representations of the structure are also shown as inserts on the CCS panel. Reproduced with permission from ref 217. Copyright 2015 American Chemical Society.
Table 3:
Typical features of CID compared to SID of protein complexes and examples of each throughout the text.
| CID | SID | Example | |
|---|---|---|---|
| Collision Target | Gaseous atoms or molecules: He, Ar, Xe, N2, SF6; Target much smaller than projectile protein complex | Surface, typically metal (contaminated with adventitious hydrocarbons in vacuum) or self-assembled monolayers of fluorinated alkanethiols | Figure 1 |
| Structural Information | Confirm stoichiometry; produce highly charged, elongated monomer and (N-1)mer; consecutive loss of additional monomers; increase apparent mass accuracy and resolution | Confirm stoichiometry; Cleave at weaker non-covalent interfaces at lower collision energies, substructure products provide topology information/connectivity map | Figure 2, Figure 8, Figure 16, Table 1 |
| Activation characteristics | Multi-step activation where many very low energy collisions gradually raise internal energy content while restructuring complex | ~single collision for conversion of kinetic to internal energy; energy “jump” because target size is large compared to projectile size; lower kinetic energies required than for CID | Figure 1, Figure 27 |
| Charge partitioning in homomers | High charge on monomer; low charge on (N-1)mer (‘asymmetric charge partitioning’); symmetric charge partitioning is sometimes observed but not typical | Charge partitioning among fragments is proportional to mass of each fragment (‘symmetric charge partitioning’); asymmetric partitioning (particularly among monomers) is atypical but sometimes observed and poorly understood | Figure 2, Figure 8, Figure 15, Figure 25, Figure 26 |
| Ligands | Ligands typically lost from monomer because of monomer elongation | Ligand number on fragments consistent with expectations (1 ligand per monomer if ligand in a binding pocket or ligand number consistent with subunit number if ligand bound between subunits) | Figure 15 |
| Effect of pre-activation (e.g., source CID/activation) | Elongated, highly charged monomer produced with or without source pre-activation | SID spectra change as in-source CID restructures complex (SID products native-like with no pre-activation, more CID-like after CID preactivation) | |
| CCS of products | Monomers elongated; when SID and CID products have the same charge they often have the same CCS | Monomers compact because they generally have lower charge; higher order products compact but not the same as a structure “clipped” from a high resolution method (may restructure/collapse to maximize stability in gas-phase) | Figure 28 |
| Kinetic model of fragmentation | Consistent with RRKM | Consistent with RRKM | N/A |
| Utility for informing computational modeling/docking | Stoichiometry can inform structural models | SID appearance energy can be predicted computationally and can be used to make decision between docking poses | Figure 4, Figure 6, Figure 7 |
6. Acknowledgements
The entire team of the Resource for Native Mass Spectrometry Guided Structural Biology is gratefully acknowledged [NIH grant P41 GM128577, technology development driven by biological problems]. The FT-ICR was purchased with funds from NIH award S10 OD018507. NSF grants [DBI 0244437 and 1455654] funded the early SID instrument development for application to protein complexes and an NIH R01 [GM113658] funded work that improved our understanding of SID of protein complexes. Data analysis software improvements are being funded by an NIH SBIR R44GM133239to Marshall Bern and a combination SID/ECD device is under development with eMSions through SBIR R43GM140749. We thank all former and current Wysocki group members and outstanding vendor, technology, and protein complex collaborators who contributed to the Wysocki group work mentioned in this paper.
7. Abbreviations
- AE
Appearance energy
- CCS
Collision cross section
- CDMS
Charge detection mass spectrometry
- CID
Collision-induced dissociation
- CIU
Collision-induced unfolding
- Cryo-EM
Cryo-electron microscopy
- ECD
Electron capture dissociation
- EDDA
Ethylenediamine diacetate, a charge reducing reagent
- ETD
Electron transfer dissociation
- ExD
Electron activated dissociation
- ELIT
Electrostatic linear ion trap
- ERMS
Energy-resolved mass spectrum (or spectrometry)
- FT-ICR
Fourier transform ion cyclotron resonance
- HCD
Higher energy collisional dissociation (beam-type CID)
- HDX
Hydrogen-deuterium exchange
- IRMPD
Infrared multiphoton dissociation
- m/z
mass-to-charge ratio
- MS/MS
Tandem mass spectrometry
- NMR
Nuclear magnetic resonance
- nMS
Native mass spectrometry
- PTM
Post-translational modification
- Q-IM-TOF
Quadrupole, ion mobility, time-of-flight (mass spectrometer)
- Rf
Radiofrequency (voltage)
- RRKM
Rice-Ramsberger-Kassel-Marcus (theory of reactivity)
- SAM
Self-assembled monolayer
- SEC
Size-exclusion chromatography
- SID
Surface-induced dissociation
- SIU
Surface-induced unfolding
- TEAA
Triethylammonium acetate, a charge reducing reagent
- T->V
Conversion of kinetic energy to internal vibrational energy
- UVPD
Ultraviolet photodissociation
Biographies
8. Biographies
Dalton T. Snyder is a research associate in the Resource for Native Mass Spectrometry Guided Structural Biology (nMS->SB) under Prof. Vicki Wysocki at the Ohio State University. He obtained his B.S. degree in chemistry and applied mathematics from the University of Evansville in 2014 and later earned his PhD in chemistry (with a focus on mass spectrometry) under Prof. R. Graham Cooks at Purdue University in 2018. Since joining the nMS->SB Resource, he has focused on simplification and dissemination of surface-induced dissociation technology for the native mass spectrometry community. His current research interests include gas-phase hydrogen-deuterium exchange of native proteins and protein complexes as well as study of amyloid beta aggregation pathways by native mass spectrometry.
Sophie R. Harvey received her MSci in Forensic and Analytical Chemistry at the University of Strathclyde in 2010 and then in 2014 received her Ph.D from the University of Edinburgh working under the supervision of Professor Perdita Barran, using mass spectrometry and ion mobility-mass spectrometry to study conformationally dynamic proteins. She did her postdoctoral research in the lab of Vicki Wysocki at The Ohio State University using surface induced dissociation, and ion mobility-mass spectrometry, to study soluble and membrane protein complexes. She is currently a senior research associate working in The Ohio State University mass spectrometry and proteomics facility and in the resource for Native Mass Spectrometry Guided Structural Biology. Sophie’s research interests include characterization of soluble and membrane proteins with and without ligands bound, posttranslational modifications and proteomics.
Vicki H. Wysocki received her PhD from Purdue University in 1987, under the direction of Professor Graham Cooks. Following a National Research Council postdoctoral appointment at the US Naval Research Laboratory, she became an Assistant Professor at Virginia Commonwealth University. She joined the University of Arizona in 1996 and eventually served as Chair of the Department of Chemistry and Biochemistry. In 2012 she moved to The Ohio State University where she is an Ohio Eminent Scholar and Director of the Campus Chemical Instrument Center. She is director of the NIH-funded Resource for Native Mass Spectrometry Guided Structural Biology. Professor Wysocki’s research interests include bioanalytical mass spectrometry, peptide fragmentation mechanisms, proteomics and metabolomics, and instrument development for improved dissociation and characterization of non-covalent protein and nucleoprotein complexes.
9. References
- (1).Heck AJ Native Mass Spectrometry: A Bridge between Interactomics and Structural Biology. Nat. Methods 2008, 5, 927–933. [DOI] [PubMed] [Google Scholar]
- (2).Lorenzen K; Duijn E. van. Native Mass Spectrometry as a Tool in Structural Biology. Curr. Protoc. Protein Sci. 2010, 62, 17.12.1–17.12.17. 10.1002/0471140864.ps1712s62. [DOI] [PubMed] [Google Scholar]
- (3).Ben-Nissan G; Sharon M The Application of Ion-Mobility Mass Spectrometry for Structure/Function Investigation of Protein Complexes. Curr. Opin. Chem. Biol. 2018, 42, 25–33. 10.1016/j.cbpa.2017.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Mehmood S; Allison TM; Robinson CV Mass Spectrometry of Protein Complexes: From Origins to Applications. Annu. Rev. Phys. Chem. 2015, 66, 453–474. 10.1146/annurev-physchem-040214-121732. [DOI] [PubMed] [Google Scholar]
- (5).Boeri Erba E; Signor L; Petosa C Exploring the Structure and Dynamics of Macromolecular Complexes by Native Mass Spectrometry. J. Proteomics 2020, 222, 103799. 10.1016/j.jprot.2020.103799. [DOI] [PubMed] [Google Scholar]
- (6).Fenn JB; Mann M; Meng CK; Wong SF; Whitehouse CM Electrospray Ionization for Mass Spectrometry of Large Biomolecules. Science 1989, 246, 64–71. [DOI] [PubMed] [Google Scholar]
- (7).Sterling HJ; Cassou CA; Susa AC; Williams ER Electrothermal Supercharging of Proteins in Native Electrospray Ionization. Anal. Chem. 2012, 84, 3795–3801. 10.1021/ac300468a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Dyachenko A; Gruber R; Shimon L; Horovitz A; Sharon M Allosteric Mechanisms Can Be Distinguished Using Structural Mass Spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 7235–7239. 10.1073/pnas.1302395110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).van Berkel WJ; van den Heuvel RH; Versluis C; Heck AJ Detection of Intact MegaDalton Protein Assemblies of Vanillyl-Alcohol Oxidase by Mass Spectrometry. Protein Sci. 2000, 9, 435–439. 10.1110/ps.9.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).van de Waterbeemd M; Fort KL; Boll D; Reinhardt-Szyba M; Routh A; Makarov A; Heck AJR High-Fidelity Mass Analysis Unveils Heterogeneity in Intact Ribosomal Particles. Nat. Methods 2017, 14, 283–286. 10.1038/nmeth.4147. [DOI] [PubMed] [Google Scholar]
- (11).Pacholarz KJ; Garlish RA; Taylor RJ; Barran PE Mass Spectrometry Based Tools to Investigate Protein–Ligand Interactions for Drug Discovery. Chem. Soc. Rev. 2012, 41, 4335–4355. 10.1039/C2CS35035A. [DOI] [PubMed] [Google Scholar]
- (12).Laganowsky A; Reading E; Allison TM; Ulmschneider MB; Degiacomi MT; Baldwin AJ; Robinson CV Membrane Proteins Bind Lipids Selectively to Modulate Their Structure and Function. Nature 2014, 510, 172–175. 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Eschweiler JD; Kerr R; Rabuck-Gibbons J; Ruotolo BT Sizing Up Protein–Ligand Complexes: The Rise of Structural Mass Spectrometry Approaches in the Pharmaceutical Sciences. Annu. Rev. Anal. Chem. 2017, 10, 25–44. 10.1146/annurev-anchem-061516-045414. [DOI] [PubMed] [Google Scholar]
- (14).Lai LB; Tanimoto A; Lai SM; Chen W-Y; Marathe IA; Westhof E; Wysocki VH; Gopalan V A Novel Double Kink-Turn Module in Euryarchaeal RNase P RNAs. Nucleic Acids Res. 2017, 45, 7432–7440. 10.1093/nar/gkx388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ma X; Lai LB; Lai SM; Tanimoto A; Foster MP; Wysocki VH; Gopalan V Uncovering the Stoichiometry of Pyrococcus Furiosus RNase P, a Multi-Subunit Catalytic Ribonucleoprotein Complex, by Surface-Induced Dissociation and Ion Mobility Mass Spectrometry. Angew. Chem. Int. Ed. Engl. 2014, 53, 11483–11487. 10.1002/anie.201405362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Barrera NP; Di Bartolo N; Booth PJ; Robinson CV Micelles Protect Membrane Complexes from Solution to Vacuum. Science 2008, 321, 243–246. 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- (17).Laganowsky A; Reading E; Hopper JTS; Robinson CV Mass Spectrometry of Intact Membrane Protein Complexes. Nat. Protoc. 2013, 8, 639–651. 10.1038/nprot.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Bolla JR; Agasid MT; Mehmood S; Robinson CV Membrane Protein–Lipid Interactions Probed Using Mass Spectrometry. Annu. Rev. Biochem. 2019, 88, 85–111. 10.1146/annurev-biochem-013118-111508. [DOI] [PubMed] [Google Scholar]
- (19).Keener JE; Zhang G; Marty MT Native Mass Spectrometry of Membrane Proteins. Anal. Chem. 2021, 93, 583–597. 10.1021/acs.analchem.0c04342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Gan J; Ben-Nissan G; Arkind G; Tarnavsky M; Trudeau D; Noda Garcia L; Tawfik DS; Sharon M Native Mass Spectrometry of Recombinant Proteins from Crude Cell Lysates. Anal. Chem. 2017, 89, 4398–4404. 10.1021/acs.analchem.7b00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Vimer S; Ben-Nissan G; Sharon M Direct Characterization of Overproduced Proteins by Native Mass Spectrometry. Nat. Protoc. 2020, 15, 236–265. 10.1038/s41596-019-0233-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zhou M; Yan J; Romano CA; Tebo BM; Wysocki VH; Paša-Tolić L Surface Induced Dissociation Coupled with High Resolution Mass Spectrometry Unveils Heterogeneity of a 211 KDa Multicopper Oxidase Protein Complex. J. Am. Soc. Mass Spectrom. 2018, 29, 723–733. 10.1007/s13361-017-1882-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Skinner OS; Haverland NA; Fornelli L; Melani RD; Do Vale LHF; Seckler HS; Doubleday PF; Schachner LF; Srzentić K; Kelleher NL; Compton PD Top-down Characterization of Endogenous Protein Complexes with Native Proteomics. Nat. Chem. Biol. 2018, 14, 36–41. 10.1038/nchembio.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ben-Nissan G; Belov ME; Morgenstern D; Levin Y; Dym O; Arkind G; Lipson C; Makarov AA; Sharon M Triple-Stage Mass Spectrometry Unravels the Heterogeneity of an Endogenous Protein Complex. Anal. Chem. 2017, 89, 4708–4715. 10.1021/acs.analchem.7b00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Mason EA; Schamp HW Mobility of Gaseous Lons in Weak Electric Fields. Ann. Phys. (N. Y.) 1958, 4, 233–270. 10.1016/0003-4916(58)90049-6. [DOI] [Google Scholar]
- (26).Clemmer DE; Russell DH; Williams ER Characterizing the Conformationome: Toward a Structural Understanding of the Proteome. Acc. Chem. Res. 2017, 50, 556–560. 10.1021/acs.accounts.6b00548. [DOI] [PubMed] [Google Scholar]
- (27).McCabe JW; Hebert MJ; Shirzadeh M; Mallis CS; Denton JK; Walker TE; Russell DH The Ims Paradox: A Perspective on Structural Ion Mobility-Mass Spectrometry. Mass Spectrom. Rev. 40, 280–305. 10.1002/mas.21642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).McLuckey SA Principles of Collisional Activation in Analytical Mass Spectrometry. J. Am. Soc. Mass Spectrom. 1992, 3, 599–614. 10.1016/1044-0305(92)85001-Z. [DOI] [PubMed] [Google Scholar]
- (29).Benesch JLP Collisional Activation of Protein Complexes: Picking up the Pieces. J. Am. Soc. Mass Spectrom. 2009, 20, 341–348. 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- (30).Benesch JLP; Aquilina JA; Ruotolo BT; Sobott F; Robinson CV Tandem Mass Spectrometry Reveals the Quaternary Organization of Macromolecular Assemblies. Chem. Biol. 2006, 13, 597–605. 10.1016/j.chembiol.2006.04.006. [DOI] [PubMed] [Google Scholar]
- (31).Hall Z; Hernández H; Marsh JA; Teichmann SA; Robinson CV The Role of Salt Bridges, Charge Density, and Subunit Flexibility in Determining Disassembly Routes of Protein Complexes. Structure 2013, 21, 1325–1337. 10.1016/j.str.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Tian Y; Lippens JL; Netirojjanakul C; Campuzano IDG; Ruotolo BT Quantitative Collision-Induced Unfolding Differentiates Model Antibody–Drug Conjugates. Protein Sci. 2019, 28, 598–608. 10.1002/pro.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Fantin SM; Parson KF; Niu S; Liu J; Polasky DA; Dixit SM; Ferguson-Miller SM; Ruotolo BT Collision Induced Unfolding Classifies Ligands Bound to the Integral Membrane Translocator Protein. Anal. Chem. 2019, 91, 15469–15476. 10.1021/acs.analchem.9b03208. [DOI] [PubMed] [Google Scholar]
- (34).Gadkari VV; Ramírez CR; Vallejo DD; Kurulugama RT; Fjeldsted JC; Ruotolo BT Enhanced Collision Induced Unfolding and Electron Capture Dissociation of Native-like Protein Ions. Anal. Chem. 2020, 92, 15489–15496. 10.1021/acs.analchem.0c03372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Sobott F; Robinson CV Characterising Electrosprayed Biomolecules Using Tandem-MS—the Noncovalent GroEL Chaperonin Assembly. Int. J. Mass Spectrom. 2004, 236, 25–32. 10.1016/j.ijms.2004.05.010. [DOI] [Google Scholar]
- (36).McKay AR; Ruotolo BT; Ilag LL; Robinson CV Mass Measurements of Increased Accuracy Resolve Heterogeneous Populations of Intact Ribosomes. J. Am. Chem. Soc. 2006, 128, 11433–11442. 10.1021/ja061468q. [DOI] [PubMed] [Google Scholar]
- (37).Panczyk EM; Gilbert JD; Jagdale GS; Stiving AQ; Baker LA; Wysocki VH Ion Mobility and Surface Collisions: Submicrometer Capillaries Can Produce Native-like Protein Complexes. Anal. Chem. 2020, 92, 2460–2467. 10.1021/acs.analchem.9b03666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Xia Z; Williams ER Protein-Glass Surface Interactions and Ion Desalting in Electrospray Ionization with Submicron Emitters. J. Am. Soc. Mass Spectrom. 2018, 29, 194–202. 10.1021/jasms.8b05670. [DOI] [PubMed] [Google Scholar]
- (39).Hopper JTS; Sokratous K; Oldham NJ Charge State and Adduct Reduction in Electrospray Ionization-Mass Spectrometry Using Solvent Vapor Exposure. Anal. Biochem. 2012, 421, 788–790. 10.1016/j.ab.2011.10.034. [DOI] [PubMed] [Google Scholar]
- (40).Zhou M; Wysocki VH Surface Induced Dissociation: Dissecting Noncovalent Protein Complexes in the Gas Phase. Acc. Chem. Res. 2014, 47, 1010–1018. 10.1021/ar400223t. [DOI] [PubMed] [Google Scholar]
- (41).Cooks RG; Terwilliger DT; Ast T; Beynon JH; Keough T Surface Modified Mass-Spectrometry. J Am Chem Soc 1975, 97, 1583–1585. 10.1021/ja00839a056. [DOI] [Google Scholar]
- (42).Mabud MA; Dekrey MJ; Graham Cooks R Surface-Induced Dissociation of Molecular Ions. Int. J. Mass Spectrom. Ion Processes 1985, 67, 285–294. 10.1016/0168-1176(85)83024-X. [DOI] [Google Scholar]
- (43).Cooks RG; Ast T; Pradeep T; Wysocki V Reactions of Ions with Organic Surfaces. Acc. Chem. Res. 1994, 27, 316–323. 10.1021/ar00047a001. [DOI] [Google Scholar]
- (44).Dongre AR; Somogyi A; Wysocki VH Surface-Induced Dissociation: An Effective Tool to Probe Structure, Energetics and Fragmentation Mechanisms of Protonated Peptides. J. Mass Spectrom. 1996, 31, 339–350. . [DOI] [PubMed] [Google Scholar]
- (45).Stiving AQ; VanAernum ZL; Busch F; Harvey SR; Sarni SH; Wysocki VH Surface-Induced Dissociation: An Effective Method for Characterization of Protein Quaternary Structure. Anal. Chem. 2019, 91, 190–209. 10.1021/acs.analchem.8b05071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Jones CM; Beardsley RL; Galhena AS; Dagan S; Cheng G; Wysocki VH Symmetrical Gas-Phase Dissociation of Noncovalent Protein Complexes via Surface Collisions. J. Am. Chem. Soc. 2006, 128, 15044–15045. 10.1021/ja064586m. [DOI] [PubMed] [Google Scholar]
- (47).Beardsley RL; Jones CM; Galhena AS; Wysocki VH Noncovalent Protein Tetramers and Pentamers with “n” Charges Yield Monomers with n/4 and n/5 Charges. Anal. Chem. 2009, 81, 1347–1356. 10.1021/ac801883k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Zhou M; Dagan S; Wysocki VH Protein Subunits Released by Surface Collisions of Noncovalent Complexes: Nativelike Compact Structures Revealed by Ion Mobility Mass Spectrometry. Angew. Chem. Int. Ed. Engl. 2012, 51, 4336–4339. 10.1002/anie.201108700. [DOI] [PubMed] [Google Scholar]
- (49).Zhou M; Dagan S; Wysocki VH Impact of Charge State on Gas-Phase Behaviors of Noncovalent Protein Complexes in Collision Induced Dissociation and Surface Induced Dissociation. Analyst 2013, 138, 1353–1362. 10.1039/C2AN36525A. [DOI] [PubMed] [Google Scholar]
- (50).Ma X; Zhou M; Wysocki VH Surface Induced Dissociation Yields Quaternary Substructure of Refractory Noncovalent Phosphorylase B and Glutamate Dehydrogenase Complexes. J. Am. Soc. Mass Spectrom. 2014, 25, 368–379. 10.1007/s13361-013-0790-y. [DOI] [PubMed] [Google Scholar]
- (51).Zhou M; Jones CM; Wysocki VH Dissecting the Large Noncovalent Protein Complex GroEL with Surface-Induced Dissociation and Ion Mobility-Mass Spectrometry. Anal. Chem. 2013, 85, 8262–8267. 10.1021/ac401497c. [DOI] [PubMed] [Google Scholar]
- (52).Harvey SR; Seffernick JT; Quintyn RS; Song Y; Ju Y; Yan J; Sahasrabuddhe AN; Norris A; Zhou M; Behrman EJ; Lindert S; Wysocki VH Relative Interfacial Cleavage Energetics of Protein Complexes Revealed by Surface Collisions. Proc. Natl. Acad. Sci. U.S.A. 2019, 116, 8143–8148. 10.1073/pnas.1817632116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Quintyn RS; Yan J; Wysocki VH Surface-Induced Dissociation of Homotetramers with D2 Symmetry Yields Their Assembly Pathways and Characterizes the Effect of Ligand Binding. Chem. Biol. 2015, 22, 583–592. 10.1016/j.chembiol.2015.03.019. [DOI] [PubMed] [Google Scholar]
- (54).Zhou M; Lantz C; A. Brown K; Ge Y; Paša-Tolić L; A. Loo J; Lermyte F Higher-Order Structural Characterisation of Native Proteins and Complexes by Top-down Mass Spectrometry. Chem. Sci. 2020, 11, 12918–12936. 10.1039/D0SC04392C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Lermyte F Advanced Fragmentation Methods in Biomolecular Mass Spectrometry; Royal Society of Chemistry, 2020.
- (56).Madsen JA; Boutz DR; Brodbelt JS Ultrafast Ultraviolet Photodissociation at 193 Nm and Its Applicability to Proteomic Workflows. J. Proteome Res. 2010, 9, 4205–4214. 10.1021/pr100515x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Han S-W; Lee S-W; Bahar O; Schwessinger B; Robinson MR; Shaw JB; Madsen JA; Brodbelt JS; Ronald PC Tyrosine Sulfation in a Gram-Negative Bacterium. Nat. Commun. 2012, 3, 1153. 10.1038/ncomms2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Erales J; Gontero B; Whitelegge J; Halgand F Mapping of a Copper-Binding Site on the Small CP12 Chloroplastic Protein of Chlamydomonas Reinhardtii Using Top-down Mass Spectrometry and Site-Directed Mutagenesis. Biochem. J. 2009, 419, 75–82, 4 p following 82. 10.1042/BJ20082004. [DOI] [PubMed] [Google Scholar]
- (59).Li H; Wongkongkathep P; Van Orden SL; Ogorzalek Loo RR; Loo JA Revealing Ligand Binding Sites and Quantifying Subunit Variants of Noncovalent Protein Complexes in a Single Native Top-Down FTICR MS Experiment. J. Am. Soc. Mass Spectrom. 2014, 25, 2060–2068. 10.1021/jasms.8b04675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Cammarata MB; Brodbelt JS Structural Characterization of Holo- and Apo-Myoglobin in the Gas Phase by Ultraviolet Photodissociation Mass Spectrometry. Chem. Sci. 2015, 6, 1324–1333. 10.1039/C4SC03200D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Breuker K; Oh H; Horn DM; Cerda BA; McLafferty FW Detailed Unfolding and Folding of Gaseous Ubiquitin Ions Characterized by Electron Capture Dissociation. J. Am. Chem. Soc. 2002, 124, 6407–6420. 10.1021/ja012267j. [DOI] [PubMed] [Google Scholar]
- (62).Oh H; Breuker K; Sze SK; Ge Y; Carpenter BK; McLafferty FW Secondary and Tertiary Structures of Gaseous Protein Ions Characterized by Electron Capture Dissociation Mass Spectrometry and Photofragment Spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 15863–15868. 10.1073/pnas.212643599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Zhang H; Cui W; Wen J; Blankenship RE; Gross ML Native Electrospray and Electron-Capture Dissociation FTICR Mass Spectrometry for Top-Down Studies of Protein Assemblies. Anal. Chem. 2011, 83, 5598–5606. 10.1021/ac200695d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Morrison LJ; Brodbelt JS 193 Nm Ultraviolet Photodissociation Mass Spectrometry of Tetrameric Protein Complexes Provides Insight into Quaternary and Secondary Protein Topology. J. Am. Chem. Soc. 2016, 138, 10849–10859. 10.1021/jacs.6b03905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Tamara S; Dyachenko A; Fort KL; Makarov AA; Scheltema RA; Heck AJR Symmetry of Charge Partitioning in Collisional and UV Photon-Induced Dissociation of Protein Assemblies. J. Am. Chem. Soc. 2016, 138, 10860–10868. 10.1021/jacs.6b05147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Mikhailov VA; Liko I; Mize TH; Bush MF; Benesch JLP; Robinson CV Infrared Laser Activation of Soluble and Membrane Protein Assemblies in the Gas Phase. Anal. Chem. 2016, 88, 7060–7067. 10.1021/acs.analchem.6b00645. [DOI] [PubMed] [Google Scholar]
- (67).Hernández H; Dziembowski A; Taverner T; Séraphin B; Robinson CV Subunit Architecture of Multimeric Complexes Isolated Directly from Cells. EMBO Rep. 2006, 7, 605–610. 10.1038/sj.embor.7400702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Levy ED; Erba EB; Robinson CV; Teichmann SA Assembly Reflects Evolution of Protein Complexes. Nature 2008, 453, 1262–1265. 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Hernández H; Robinson CV Determining the Stoichiometry and Interactions of Macromolecular Assemblies from Mass Spectrometry. Nat. Protoc. 2007, 2, 715–726. 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- (70).Katta V; Chait BT Observation of the Heme-Globin Complex in Native Myoglobin by Electrospray-Ionization Mass Spectrometry. J. Am. Chem. Soc. 1991, 113, 8534–8535. 10.1021/ja00022a058. [DOI] [Google Scholar]
- (71).Baca M; Kent SBH Direct Observation of a Ternary Complex between the Dimeric Enzyme HIV-1 Protease and a Substrate-Based Inhibitor. J. Am. Chem. Soc. 1992, 114, 3992–3993. 10.1021/ja00036a066. [DOI] [Google Scholar]
- (72).Siuzdak G; Bothner B; Yeager M; Brugidou C; Fauquet CM; Hoey K; Change C-M Mass Spectrometry and Viral Analysis. Chem. Biol. 1996, 3, 45–48. 10.1016/S1074-5521(96)90083-6. [DOI] [PubMed] [Google Scholar]
- (73).Ouyang Z; Takáts Z; Blake TA; Gologan B; Guymon AJ; Wiseman JM; Oliver JC; Davisson VJ; Cooks RG Preparing Protein Microarrays by Soft-Landing of Mass-Selected Ions. Science 2003, 301, 1351–1354. 10.1126/science.1088776. [DOI] [PubMed] [Google Scholar]
- (74).Liu FC; Kirk SR; Bleiholder C On the Structural Denaturation of Biological Analytes in Trapped Ion Mobility Spectrometry – Mass Spectrometry. Analyst 2016, 141, 3722–3730. 10.1039/C5AN02399H. [DOI] [PubMed] [Google Scholar]
- (75).Morsa D; Hanozin E; Eppe G; Quinton L; Gabelica V; De Pauw E Effective Temperature and Structural Rearrangement in Trapped Ion Mobility Spectrometry. Anal. Chem. 2020, 92, 4573–4582. 10.1021/acs.analchem.9b05850. [DOI] [PubMed] [Google Scholar]
- (76).Dixit SM; Polasky DA; Ruotolo BT Collision Induced Unfolding of Isolated Proteins in the Gas Phase: Past, Present, and Future. Curr. Opin. Chem. Biol. 2018, 42, 93–100. 10.1016/j.cbpa.2017.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Clemmer DE; Hudgins RR; Jarrold MF Naked Protein Conformations: Cytochrome c in the Gas Phase. J. Am. Chem. Soc. 1995, 117, 10141–10142. 10.1021/ja00145a037. [DOI] [Google Scholar]
- (78).Chandler SA; Benesch JL Mass Spectrometry beyond the Native State. Curr. Opin. Chem. Biol. 2018, 42, 130–137. 10.1016/j.cbpa.2017.11.019. [DOI] [PubMed] [Google Scholar]
- (79).Hansen K; Lau AM; Giles K; McDonnell JM; Struwe WB; Sutton BJ; Politis A A Mass-Spectrometry-Based Modelling Workflow for Accurate Prediction of IgG Antibody Conformations in the Gas Phase. Angewandte Chemie International Edition 2018, 57, 17194–17199. 10.1002/anie.201812018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Devine PWA; Fisher HC; Calabrese AN; Whelan F; Higazi DR; Potts JR; Lowe DC; Radford SE; Ashcroft AE Investigating the Structural Compaction of Biomolecules Upon Transition to the Gas-Phase Using ESI-TWIMS-MS. J. Am. Soc. Mass Spectrom. 2017, 28, 1855–1862. 10.1007/s13361-017-1689-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Ruotolo BT; Giles K; Campuzano I; Sandercock AM; Bateman RH; Robinson CV Evidence for Macromolecular Protein Rings in the Absence of Bulk Water. Science 2005, 310, 1658–1661. 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- (82).Sahasrabuddhe A; Hsia Y; Busch F; Sheffler W; King NP; Baker D; Wysocki VH Confirmation of Intersubunit Connectivity and Topology of Designed Protein Complexes by Native MS. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 1268–1273. 10.1073/pnas.1713646115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Politis A; Park AY; Hyung S-J; Barsky D; Ruotolo BT; Robinson CV Integrating Ion Mobility Mass Spectrometry with Molecular Modelling to Determine the Architecture of Multiprotein Complexes. PLOS ONE 2010, 5, e12080. 10.1371/journal.pone.0012080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Bernstein SL; Wyttenbach T; Baumketner A; Shea J-E; Bitan G; Teplow DB; Bowers MT Amyloid β-Protein: Monomer Structure and Early Aggregation States of Aβ42 and Its Pro19 Alloform. J. Am. Chem. Soc. 2005, 127, 2075–2084. 10.1021/ja044531p. [DOI] [PubMed] [Google Scholar]
- (85).Li J; Taraszka JA; Counterman AE; Clemmer DE Influence of Solvent Composition and Capillary Temperature on the Conformations of Electrosprayed Ions: Unfolding of Compact Ubiquitin Conformers from Pseudonative and Denatured Solutions. Int. J. Mass Spectrom. 1999, 185–187, 37–47. 10.1016/S1387-3806(98)14135-0. [DOI] [Google Scholar]
- (86).Hall Z; Politis A; Bush MF; Smith LJ; Robinson CV Charge-State Dependent Compaction and Dissociation of Protein Complexes: Insights from Ion Mobility and Molecular Dynamics. J. Am. Chem. Soc. 2012, 134, 3429–3438. 10.1021/ja2096859. [DOI] [PubMed] [Google Scholar]
- (87).Pagel K; Hyung S-J; Ruotolo BT; Robinson CV Alternate Dissociation Pathways Identified in Charge-Reduced Protein Complex Ions. Anal. Chem. 2010, 82, 5363–5372. 10.1021/ac101121r. [DOI] [PubMed] [Google Scholar]
- (88).Heck AJR; Van Den Heuvel RHH Investigation of Intact Protein Complexes by Mass Spectrometry. Mass Spectrom. Rev. 2004, 23, 368–389. 10.1002/mas.10081. [DOI] [PubMed] [Google Scholar]
- (89).Allen SJ; Schwartz AM; Bush MF Effects of Polarity on the Structures and Charge States of Native-Like Proteins and Protein Complexes in the Gas Phase. Anal. Chem. 2013, 85, 12055–12061. 10.1021/ac403139d. [DOI] [PubMed] [Google Scholar]
- (90).Harvey S; VanAernum Z; Wysocki V Surface-Induced Dissociation of Anionic vs Cationic Native-like Protein Complexes. J. Am. Chem. Soc. 2021, 143, 7698–7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Stiving AQ; Jones BJ; Ujma J; Giles K; Wysocki VH Collision Cross Sections of Charge-Reduced Proteins and Protein Complexes: A Database for Collision Cross Section Calibration. Anal. Chem. 2020, 92, 4475–4483. 10.1021/acs.analchem.9b05519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Schwartz BL; Bruce JE; Anderson GA; Hofstadler SA; Rockwood AL; Smith RD; Chilkoti A; Stayton PS Dissociation of Tetrameric Ions of Noncovalent Streptavidin Complexes Formed by Electrospray Ionization. J Am Soc Mass Spectrom 1995, 6, 459–465. 10.1016/1044-0305(95)00191-F. [DOI] [PubMed] [Google Scholar]
- (93).Krissinel E; Henrick K Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- (94).Ahnert SE; Marsh JA; Hernández H; Robinson CV; Teichmann SA Principles of Assembly Reveal a Periodic Table of Protein Complexes. Science 2015, 350. 10.1126/science.aaa2245. [DOI] [PubMed] [Google Scholar]
- (95).Harvey SR; Liu Y; Liu W; Wysocki VH; Laganowsky A Surface Induced Dissociation as a Tool to Study Membrane Protein Complexes. Chem. Commun. 2017, 53, 3106–3109. 10.1039/C6CC09606A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Snyder DT; Panczyk E; Stiving AQ; Gilbert JD; Somogyi A; Kaplan D; Wysocki V Design and Performance of a Second-Generation Surface-Induced Dissociation Cell for Fourier Transform Ion Cyclotron Resonance Mass Spectrometry of Native Protein Complexes. Anal. Chem. 2019, 91, 14049–14057. 10.1021/acs.analchem.9b03746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Snyder DT; Panczyk E; Somogyi A; Kaplan D; Wysocki VH Simple and Minimally Invasive SID Devices for Native Mass Spectrometry. Anal. Chem. 2020, 92, 11195–11203. 10.1021/acs.analchem.0c01657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Snyder DT; Lin Y-F; Somogyi A; Wysocki VH Tandem Surface-Induced Dissociation of Protein Complexes on an Ultrahigh Resolution Platform. Int. J. Mass Spectrom. 2021, 461, 116503. 10.1016/j.ijms.2020.116503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).VanAernum ZL; Gilbert JD; Belov ME; Makarov AA; Horning SR; Wysocki VH Surface-Induced Dissociation of Noncovalent Protein Complexes in an Extended Mass Range Orbitrap Mass Spectrometer. Anal. Chem. 2019, 91, 3611–3618. 10.1021/acs.analchem.8b05605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Wysocki VH; Jones CM; Galhena AS; Blackwell AE Surface-Induced Dissociation Shows Potential to Be More Informative Than Collision-Induced Dissociation for Structural Studies of Large Systems. J. Am. Soc. Mass Spectrom. 2008, 19, 903–913. 10.1016/j.jasms.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Harvey SR; VanAernum ZL; Kostelic MM; Marty MT; Wysocki VH Probing the Structure of Nanodiscs Using Surface-Induced Dissociation Mass Spectrometry. Chem. Commun. 2020, 56, 15651–15654. 10.1039/D0CC05531J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Ma X; Loo JA; Wysocki VH Surface Induced Dissociation Yields Substructure of Methanosarcina Thermophila 20S Proteasome Complexes. Int. J. Mass Spectrom. 2015, 377, 201–204. 10.1016/j.ijms.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Vimer S; Ben-Nissan G; Morgenstern D; Kumar-Deshmukh F; Polkinghorn C; Quintyn RS; Vasil’ev YV; Beckman JS; Elad N; Wysocki VH; Sharon M Comparative Structural Analysis of 20S Proteasome Ortholog Protein Complexes by Native Mass Spectrometry. ACS Cent. Sci. 2020, 6, 573–588. 10.1021/acscentsci.0c00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Schupfner M; Busch F; Wysocki VH; Sterner R Generation of a Stand-Alone Tryptophan Synthase α-Subunit by Mimicking an Evolutionary Blueprint. ChemBioChem 2019, 20, 2747–2751. 10.1002/cbic.201900323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Miles EW Tryptophan Synthase: A Multienzyme Complex with an Intramolecular Tunnel. Chem. Rec. 2001, 1, 140–151. 10.1002/tcr.4. [DOI] [PubMed] [Google Scholar]
- (106).Miles EW The Tryptophan Synthase Α2β2 Complex: A Model for Substrate Channeling, Allosteric Communication, and Pyridoxal Phosphate Catalysis. J. Biol. Chem. 2013, 288, 10084–10091. 10.1074/jbc.X113.463331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Quintyn RS; Harvey SR; Wysocki VH Illustration of SID-IM-SID (Surface-Induced Dissociation-Ion Mobility-SID) Mass Spectrometry: Homo and Hetero Model Protein Complexes. Analyst 2015, 140, 7012–7019. 10.1039/C5AN01095K. [DOI] [PubMed] [Google Scholar]
- (108).Yan J; Zhou M; Gilbert JD; Wolff JJ; Somogyi Á; Pedder RE; Quintyn RS; Morrison LJ; Easterling ML; Paša-Tolić L; Wysocki VH Surface-Induced Dissociation of Protein Complexes in a Hybrid Fourier Transform Ion Cyclotron Resonance Mass Spectrometer. Anal. Chem. 2017, 89, 895–901. 10.1021/acs.analchem.6b03986. [DOI] [PubMed] [Google Scholar]
- (109).Blackwell AE; Dodds ED; Bandarian V; Wysocki VH Revealing the Quaternary Structure of a Heterogeneous Noncovalent Protein Complex through Surface-Induced Dissociation. Anal. Chem. 2011, 83, 2862–2865. 10.1021/ac200452b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Song Y; Nelp MT; Bandarian V; Wysocki VH Refining the Structural Model of a Heterohexameric Protein Complex: Surface Induced Dissociation and Ion Mobility Provide Key Connectivity and Topology Information. ACS Cent. Sci. 2015, 1, 477–487. 10.1021/acscentsci.5b00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Johnson JT Development of an Electrostatic Linear Ion Trap for Tandem Mass Spectrometry thesis, Purdue University Graduate School, 2020. 10.25394/PGS.12015783.v1. [DOI] [Google Scholar]
- (112).Stiving AQ; Gilbert JD; Jones BJ; Wysocki VH A Tilted Surface and Ion Carpet Array for SID. J. Am. Soc. Mass Spectrom. 2020, 31, 458–462. 10.1021/jasms.9b00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Quintyn RS; Zhou M; Yan J; Wysocki VH Surface-Induced Dissociation Mass Spectra as a Tool for Distinguishing Different Structural Forms of Gas-Phase Multimeric Protein Complexes. Anal. Chem. 2015, 87, 11879–11886. [DOI] [PubMed] [Google Scholar]
- (114).Shirzadeh M; Boone CD; Laganowsky A; Russell DH Topological Analysis of Transthyretin Disassembly Mechanism: Surface-Induced Dissociation Reveals Hidden Reaction Pathways. Anal. Chem. 2019, 91, 2345–2351. 10.1021/acs.analchem.8b05066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Quintyn RS; Harvey SR; Wysocki VH Illustration of SID-IM-SID (Surface-Induced Dissociation-Ion Mobility-SID) Mass Spectrometry: Homo and Hetero Model Protein Complexes. Analyst 2015, 140, 7012–7019. 10.1039/C5AN01095K. [DOI] [PubMed] [Google Scholar]
- (116).Busch F; VanAernum ZL; Ju Y; Yan J; Gilbert JD; Quintyn RS; Bern M; Wysocki VH Localization of Protein Complex Bound Ligands by Surface-Induced Dissociation High-Resolution Mass Spectrometry. Anal. Chem. 2018, 90, 12796–12801. 10.1021/acs.analchem.8b03263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Romano CA; Zhou M; Song Y; Wysocki VH; Dohnalkova AC; Kovarik L; Paša-Tolić L; Tebo BM Biogenic Manganese Oxide Nanoparticle Formation by a Multimeric Multicopper Oxidase Mnx. Nat. Commun. 2017, 8, 1–8. 10.1038/s41467-017-00896-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Dodds ED; Blackwell AE; Jones CM; Holso KL; O’Brien DJ; Cordes MHJ; Wysocki VH Determinants of Gas-Phase Disassembly Behavior in Homodimeric Protein Complexes with Related Yet Divergent Structures. Anal. Chem. 2011, 83, 3881–3889. 10.1021/ac2003906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Seffernick JT; Harvey SR; Wysocki VH; Lindert S Predicting Protein Complex Structure from Surface-Induced Dissociation Mass Spectrometry Data. ACS Cent. Sci. 2019, 5, 1330–1341. 10.1021/acscentsci.8b00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Lewis SM; Kuhlman BA Anchored Design of Protein-Protein Interfaces. PLOS ONE 2011, 6, e20872. 10.1371/journal.pone.0020872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Sengupta A; Wu J; Seffernick JT; Sabag-Daigle A; Thomsen N; Chen T-H; Capua AD; Bell CE; Ahmer BMM; Lindert S; Wysocki VH; Gopalan V Integrated Use of Biochemical, Native Mass Spectrometry, Computational, and Genome-Editing Methods to Elucidate the Mechanism of a Salmonella Deglycase. J. Mol. Biol. 2019, 431, 4497–4513. 10.1016/j.jmb.2019.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Gray JJ; Moughon S; Wang C; Schueler-Furman O; Kuhlman B; Rohl CA; Baker D Protein-Protein Docking with Simultaneous Optimization of Rigid-Body Displacement and Side-Chain Conformations. J. Mol. Biol. 2003, 331, 281–299. 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- (123).Westermark P; Sletten K; Johansson B; Cornwell GG Fibril in Senile Systemic Amyloidosis Is Derived from Normal Transthyretin. Proc. Natl. Acad. Sci. U.S.A. 1990, 87, 2843–2845. 10.1073/pnas.87.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Wiseman RL; Green NS; Kelly JW Kinetic Stabilization of an Oligomeric Protein under Physiological Conditions Demonstrated by a Lack of Subunit Exchange: Implications for Transthyretin Amyloidosis. Biochemistry 2005, 44, 9265–9274. 10.1021/bi050352o. [DOI] [PubMed] [Google Scholar]
- (125).Keetch CA; Bromley EHC; McCammon MG; Wang N; Christodoulou J; Robinson CV L55P Transthyretin Accelerates Subunit Exchange and Leads to Rapid Formation of Hybrid Tetramers. J. Biol. Chem. 2005, 280, 41667–41674. 10.1074/jbc.M508753200. [DOI] [PubMed] [Google Scholar]
- (126).Quintyn RS; Zhou M; Dagan S; Finke J; Wysocki VH Ligand Binding and Unfolding of Tryptophan Synthase Revealed by Ion Mobility-Tandem Mass Spectrometry Employing Collision and Surface Induced Dissociation. Int. J. Ion Mobil. Spec. 2013, 16, 133–143. 10.1007/s12127-013-0126-4. [DOI] [Google Scholar]
- (127).Chen L; Tanimoto A; So BR; Bakhtina M; Magliery TJ; Wysocki VH; Musier-Forsyth K Stoichiometry of Triple-Sieve TRNA Editing Complex Ensures Fidelity of Aminoacyl-TRNA Formation. Nucleic Acids Res. 2019, 47, 929–940. 10.1093/nar/gky1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Flores JK; Ataide SF Structural Changes of RNA in Complex with Proteins in the SRP. Front. Mol. Biosci. 2018, 5. 10.3389/fmolb.2018.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Draper DE RNA Folding: Thermodynamic and Molecular Descriptions of the Roles of Ions. Biophys. J. 2008, 95, 5489–5495. 10.1529/biophysj.108.131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Landeras-Bueno S; Wasserman H; Oliveira G; VanAernum ZL; Busch F; Salie ZL; Wysocki VH; Andersen K; Saphire EO Cellular MRNA Triggers Structural Transformation of Ebola Virus Matrix Protein VP40 to Its Essential Regulatory Form. Cell Rep 2021, 35. 10.1016/j.celrep.2021.108986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).VanAernum ZL; Busch F; Jones BJ; Jia M; Chen Z; Boyken SE; Sahasrabuddhe A; Baker D; Wysocki VH Rapid Online Buffer Exchange for Screening of Proteins, Protein Complexes and Cell Lysates by Native Mass Spectrometry. Nat. Protoc. 2020, 15, 1132–1157. 10.1038/s41596-019-0281-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Chen Z; Boyken SE; Jia M; Busch F; Flores-Solis D; Bick MJ; Lu P; VanAernum ZL; Sahasrabuddhe A; Langan RA; Bermeo S; Brunette TJ; Mulligan VK; Carter LP; DiMaio F; Sgourakis NG; Wysocki VH; Baker D Programmable Design of Orthogonal Protein Heterodimers. Nature 2019, 565, 106–111. 10.1038/s41586-018-0802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Chen Z; Kibler RD; Hunt A; Busch F; Pearl J; Jia M; VanAernum ZL; Wicky BIM; Dods G; Liao H; Wilken MS; Ciarlo C; Green S; El-Samad H; Stamatoyannopoulos J; Wysocki VH; Jewett MC; Boyken SE; Baker D De Novo Design of Protein Logic Gates. Science 2020, 368, 78–84. 10.1126/science.aay2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Boyken SE; Benhaim MA; Busch F; Jia M; Bick MJ; Choi H; Klima JC; Chen Z; Walkey C; Mileant A; Sahasrabuddhe A; Wei KY; Hodge EA; Byron S; Quijano-Rubio A; Sankaran B; King NP; Lippincott-Schwartz J; Wysocki VH; Lee KK; Baker D De Novo Design of Tunable, PH-Driven Conformational Changes. Science 2019, 364, 658–664. 10.1126/science.aav7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (135).Marsh JA; Hernández H; Hall Z; Ahnert SE; Perica T; Robinson CV; Teichmann SA Protein Complexes Are under Evolutionary Selection to Assemble via Ordered Pathways. Cell 2013, 153, 461–470. 10.1016/j.cell.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (136).Hall Z; Politis A; Robinson CV Structural Modeling of Heteromeric Protein Complexes from Disassembly Pathways and Ion Mobility-Mass Spectrometry. Structure 2012, 20, 1596–1609. 10.1016/j.str.2012.07.001. [DOI] [PubMed] [Google Scholar]
- (137).Pukala TL; Ruotolo BT; Zhou M; Politis A; Stefanescu R; Leary JA; Robinson CV Subunit Architecture of Multiprotein Assemblies Determined Using Restraints from Gas-Phase Measurements. Structure 2009, 17, 1235–1243. 10.1016/j.str.2009.07.013. [DOI] [PubMed] [Google Scholar]
- (138).Konermann L; Vahidi S; Sowole MA Mass Spectrometry Methods for Studying Structure and Dynamics of Biological Macromolecules. Anal. Chem. 2014, 86, 213–232. 10.1021/ac4039306. [DOI] [PubMed] [Google Scholar]
- (139).Mendoza VL; Vachet RW Probing Protein Structure by Amino Acid-Specific Covalent Labeling and Mass Spectrometry. Mass Spectrom. Rev. 2009, 28, 785–815. 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Mendoza VL; Vachet RW Protein Surface Mapping Using Diethylpyrocarbonate with Mass Spectrometric Detection. Anal. Chem. 2008, 80, 2895–2904. 10.1021/ac701999b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Rappsilber J The Beginning of a Beautiful Friendship: Cross-Linking/Mass Spectrometry and Modelling of Proteins and Multi-Protein Complexes. J. Struct. Biol. 2011, 173, 530–540. 10.1016/j.jsb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (142).Leitner A; Walzthoeni T; Kahraman A; Herzog F; Rinner O; Beck M; Aebersold R Probing Native Protein Structures by Chemical Cross-Linking, Mass Spectrometry, and Bioinformatics*. Mol. Cell. Proteom. 2010, 9, 1634–1649. 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (143).Tambasco-Studart M; Titiz O; Raschle T; Forster G; Amrhein N; Fitzpatrick TB Vitamin B6 Biosynthesis in Higher Plants. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 13687–13692. 10.1073/pnas.0506228102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Moccand C; Boycheva S; Surriabre P; Tambasco-Studart M; Raschke M; Kaufmann M; Fitzpatrick TB The Pseudoenzyme PDX1.2 Boosts Vitamin B6 Biosynthesis under Heat and Oxidative Stress in Arabidopsis*. J. Biol. Chem. 2014, 289, 8203–8216. 10.1074/jbc.M113.540526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (145).Robinson GC; Kaufmann M; Roux C; Martinez-Font J; Hothorn M; Thore S; Fitzpatrick TB Crystal Structure of the Pseudoenzyme PDX1.2 in Complex with Its Cognate Enzyme PDX1.3: A Total Eclipse. Acta Cryst. D 2019, 75, 400–415. 10.1107/S2059798319002912. [DOI] [PubMed] [Google Scholar]
- (146).Novikova IV; Zhou M; Du C; Parra M; Kim DN; VanAernum ZL; Shaw JB; Hellmann H; Wysocki VH; Evans JE Tunable Hetero-Assembly of a Plant Pseudoenzyme-Enzyme Complex. bioRxiv 2020, 717082. 10.1101/717082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (147).Lammert S; Cooks R Surface-Induced Dissociation of Molecular Ions in a Quadrupole Ion Trap Mass Spectrometer. J. Am. Soc. Mass Spectrom. 1991, 2, 487–491. [DOI] [PubMed] [Google Scholar]
- (148).Hilger RT; Santini RE; McLuckey SA Tandem Mass Spectrometry in an Electrostatic Linear Ion Trap Modified for Surface-Induced Dissociation. Anal. Chem. 2014, 86, 8822–8828. 10.1021/ac502143p. [DOI] [PubMed] [Google Scholar]
- (149).Williams ER; Henry KD; McLafferty FW; Shabanowitz J; Hunt DF Surface-Induced Dissociation of Peptide Ions in Fourier-Transform Mass Spectrometry. J. Am. Soc. Mass Spectrom. 1990, 1, 413–416. 10.1016/1044-0305(90)85022-E. [DOI] [PubMed] [Google Scholar]
- (150).Chorush RA; Little DP; Beu SC; Wood TD; McLafferty FW Surface-Induced Dissociation of Multiply-Protonated Proteins. Anal. Chem. 1995, 67, 1042–1046. [DOI] [PubMed] [Google Scholar]
- (151).Laskin J; Denisov EV; Shukla AK; Barlow SE; Futrell JH Surface-Induced Dissociation in a Fourier Transform Ion Cyclotron Resonance Mass Spectrometer: Instrument Design and Evaluation. Anal. Chem. 2002, 74, 3255–3261. [DOI] [PubMed] [Google Scholar]
- (152).Wysocki VH; Ding J-M; Jones JL; Callahan JH; King FL Surface-Induced Dissociation in Tandem Quadrupole Mass Spectrometers: A Comparison of Three Designs. J. Am. Soc. Mass Spectrom. 1992, 3, 27–32. [DOI] [PubMed] [Google Scholar]
- (153).Bier M; Amy J; Cooks R; Syka J; Ceja P; Stafford G A Tandem Quadrupole Mass Spectrometer for the Study of Surface-Induced Dissociation. Int. J. Mass Spectrom. Ion Processes 1987, 77, 31–47. [Google Scholar]
- (154).Schey K; Cooks R; Grix R; Wöllnik H A Tandem Time-of-Flight Mass Spectrometer for Surface-Induced Dissociation. Int. J. Mass Spectrom. Ion Processes 1987, 77, 49–61. [Google Scholar]
- (155).Galhena AS; Dagan S; Jones CM; Beardsley RL; Wysocki VH Surface-Induced Dissociation of Peptides and Protein Complexes in a Quadrupole/Time-of-Flight Mass Spectrometer. Anal. Chem. 2008, 80, 1425–1436. 10.1021/ac701782q. [DOI] [PubMed] [Google Scholar]
- (156).Zhou M; Huang C; Wysocki VH Surface-Induced Dissociation of Ion Mobility-Separated Noncovalent Complexes in a Quadrupole/Time-of-Flight Mass Spectrometer. Anal. Chem. 2012, 84, 6016–6023. 10.1021/ac300810u. [DOI] [PubMed] [Google Scholar]
- (157).Stone E; Gillig KJ; Ruotolo B; Fuhrer K; Gonin M; Schultz A; Russell DH Surface-Induced Dissociation on a MALDI-Ion Mobility-Orthogonal Time-of-Flight Mass Spectrometer: Sequencing Peptides from an “in-Solution” Protein Digest. Anal. Chem. 2001, 73, 2233–2238. [DOI] [PubMed] [Google Scholar]
- (158).Sun W; May JC; Gillig KJ; Russell DH A Dual Time-of-Flight Apparatus for an Ion Mobility-Surface-Induced Dissociation-Mass Spectrometer for High-Throughput Peptide Sequencing. Int. J. Mass Spectrom. 2009, 287, 39–45. [Google Scholar]
- (159).Sun W; May JC; Russell DH A Novel Surface-Induced Dissociation Instrument for Ion Mobility-Time-of-Flight Mass Spectrometry. Int. J. Mass Spectrom. 2007, 259, 79–86. [Google Scholar]
- (160).Williams ER; Fang L; Zare RN Surface Induced Dissociation for Tandem Time-of-Flight Mass Spectrometry. Int. J. Mass Spectrom. Ion Processes 1993, 123, 233–241. [Google Scholar]
- (161).Laskin J; Futrell JH New Approach for Studying Slow Fragmentation Kinetics in FT-ICR: Surface-Induced Dissociation Combined with Resonant Ejection. Int. J. Mass Spectrom. 2015, 378, 160–168. 10.1016/j.ijms.2014.07.029. [DOI] [Google Scholar]
- (162).Gamage CM; Fernández FM; Kuppannan K; Wysocki VH Submicrosecond Surface-Induced Dissociation of Peptide Ions in a MALDI TOF MS. Anal. Chem. 2004, 76, 5080–5091. [DOI] [PubMed] [Google Scholar]
- (163).Schey KL; Durkin DA; Thornburg KR Design and Performance of an In-Line Surface-Induced Dissociation Device in a Four-Sector Mass Spectrometer. J. Am. Soc. Mass Spectrom. 1995, 6, 257–263. [DOI] [PubMed] [Google Scholar]
- (164).Mohammed S; Chalmers MJ; Gielbert J; Ferro M; Gora L; Smith DC; Gaskell SJ A Novel Tandem Quadrupole Mass Spectrometer Allowing Gaseous Collisional Activation and Surface Induced Dissociation. J. Mass Spectrom. 2001, 36, 1260–1268. [DOI] [PubMed] [Google Scholar]
- (165).Shukla AK; Futrell JH A Beam Scattering Instrument for the Dynamics Studies of Surface-Induced Dissociation Processes. Rev. Sci. Instrum. 2003, 74, 168–175. 10.1063/1.1524713. [DOI] [Google Scholar]
- (166).Snyder DT; Harvey SR; Busch F; Wysocki VH Chapter 11:Surface-Induced Dissociation in Biomolecular Mass Spectrometry. In Advanced Fragmentation Methods in Biomolecular Mass Spectrometry; 2020; pp 281–336. 10.1039/9781839161056-00281. [DOI] [Google Scholar]
- (167).Vékey K; Somogyi Á; Wysocki VH Internal Energy Distribution of Benzene Molecular Ions in Surface-induced Dissociation. J. Mass Spectrom. 1995, 30, 212–217. 10.1002/jms.1190300132. [DOI] [Google Scholar]
- (168).Martin Somer A; Macaluso V; Barnes GL; Yang L; Pratihar S; Song K; Hase WL; Spezia R Role of Chemical Dynamics Simulations in Mass Spectrometry Studies of Collision-Induced Dissociation and Collisions of Biological Ions with Organic Surfaces. J. Am. Soc. Mass Spectrom. 2020, 31, 2–24. 10.1021/jasms.9b00062. [DOI] [PubMed] [Google Scholar]
- (169).Yang Z; Hadjar O; Laskin J Effect of the Surface Morphology on the Energy Transfer in Ion–Surface Collisions. Int. J. Mass Spectrom. 2007, 265, 124–129. 10.1016/j.ijms.2007.01.018. [DOI] [Google Scholar]
- (170).Meroueh O; Hase WL Effect of Surface Stiffness on the Efficiency of Surface-Induced Dissociation. Phys. Chem. Chem. Phys. 2001, 3, 2306–2314. 10.1039/B100892G. [DOI] [Google Scholar]
- (171).Winger B; Julian R Jr; Cooks R; Chidsey C Surface Reactions and Surface-Induced Dissociation of Polyatomic Ions at Self-Assembled Organic Monolayer Surfaces. J. Am. Chem. Soc. 1991, 113, 8967–8969. [Google Scholar]
- (172).Wysocki VH; Jones JL; Ding JM Polyatomic Ion/Surface Collisions at Self-Assembled Monolayers Films. J. Am. Chem. Soc. 1991, 113, 8969–8970. [Google Scholar]
- (173).Cyriac J; Pradeep T; Kang H; Souda R; Cooks RG Low-Energy Ionic Collisions at Molecular Solids. Chem. Rev. 2012, 112, 5356–5411. 10.1021/cr200384k. [DOI] [PubMed] [Google Scholar]
- (174).McCormack AL; Jones JL; Wysocki VH Surface-Induced Dissociation of Multiply Protonated Peptides. J. Am. Soc. Mass Spectrom. 1992, 3, 859–862. [DOI] [PubMed] [Google Scholar]
- (175).Wysocki VH; Joyce KE; Jones CM; Beardsley RL Surface-Induced Dissociation of Small Molecules, Peptides, and Non-Covalent Protein Complexes. J. Am. Soc. Mass Spectrom. 2008, 19, 190–208. 10.1016/j.jasms.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (176).Leney AC; Heck AJ Native Mass Spectrometry: What Is in the Name? J. Am. Soc. Mass Spectrom. 2017, 28, 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (177).Ruotolo BT; Robinson CV Aspects of Native Proteins Are Retained in Vacuum. Curr. Opin. Chem. Biol. 2006, 10, 402–408. 10.1016/j.cbpa.2006.08.020. [DOI] [PubMed] [Google Scholar]
- (178).Ruotolo BT; Benesch JLP; Sandercock AM; Hyung S-J; Robinson CV Ion Mobility–Mass Spectrometry Analysis of Large Protein Complexes. Nat. Protoc. 2008, 3, 1139–1152. 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- (179).van den Heuvel RHH; van Duijn E; Mazon H; Synowsky SA; Lorenzen K; Versluis C; Brouns SJJ; Langridge D; van der Oost J; Hoyes J; Heck AJR Improving the Performance of a Quadrupole Time-of-Flight Instrument for Macromolecular Mass Spectrometry. Anal. Chem. 2006, 78, 7473–7483. 10.1021/ac061039a. [DOI] [PubMed] [Google Scholar]
- (180).Rose RJ; Damoc E; Denisov E; Makarov A; Heck AJR High-Sensitivity Orbitrap Mass Analysis of Intact Macromolecular Assemblies. Nat. Methods 2012, 9, 1084–1086. 10.1038/nmeth.2208. [DOI] [PubMed] [Google Scholar]
- (181).Fort KL; Van de Waterbeemd M; Boll D; Reinhardt-Szyba M; Belov ME; Sasaki E; Zschoche R; Hilvert D; Makarov AA; Heck AJ Expanding the Structural Analysis Capabilities on an Orbitrap-Based Mass Spectrometer for Large Macromolecular Complexes. Analyst 2018, 143, 100–105. [DOI] [PubMed] [Google Scholar]
- (182).Gault J; Liko I; Landreh M; Shutin D; Bolla JR; Jefferies D; Agasid M; Yen H-Y; Ladds MJGW; Lane DP; Khalid S; Mullen C; Remes PM; Huguet R; McAlister G; Goodwin M; Viner R; Syka JEP; Robinson CV Combining Native and ‘Omics’ Mass Spectrometry to Identify Endogenous Ligands Bound to Membrane Proteins. Nat. Methods 2020, 17, 505–508. 10.1038/s41592-020-0821-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (183).Sobott F; Hernández H; McCammon MG; Tito MA; Robinson CV A Tandem Mass Spectrometer for Improved Transmission and Analysis of Large Macromolecular Assemblies. Anal. Chem. 2002, 74, 1402–1407. 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- (184).Panczyk EM; Snyder DT; Ridgeway ME; Somogyi Á; Park MA; Wysocki VH Surface-Induced Dissociation of Protein Complexes Selected by Trapped Ion Mobility Spectrometry. Anal. Chem. 93, 5513–5520. 10.1021/acs.analchem.0c05373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (185).Harvey S; VanAernum Z; Wysocki V Surface-Induced Dissociation of Anionic vs Cationic Native-like Protein Complexes. 2021. 10.26434/chemrxiv.13547837.v1. [DOI] [PMC free article] [PubMed]
- (186).Harvey SR; Yan J; Brown JM; Hoyes E; Wysocki VH Extended Gas-Phase Trapping Followed by Surface-Induced Dissociation of Noncovalent Protein Complexes. Anal. Chem. 2016, 88, 1218–1221. 10.1021/acs.analchem.5b03479. [DOI] [PubMed] [Google Scholar]
- (187).Bleiholder C; Liu FC; Chai M Comment on Effective Temperature and Structural Rearrangement in Trapped Ion Mobility Spectrometry: TIMS Enables Native Mass Spectrometry Applications. Anal. Chem. 2020. 10.1021/acs.analchem.0c02052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (188).Sobott F; Robinson CV Characterising Electrosprayed Biomolecules Using Tandem-MS—the Noncovalent GroEL Chaperonin Assembly. Int. J. Mass Spectrom. 2004, 236, 25–32. 10.1016/j.ijms.2004.05.010. [DOI] [Google Scholar]
- (189).Uetrecht C; Rose RJ; Duijn E. van; Lorenzen K; Heck AJR Ion Mobility Mass Spectrometry of Proteins and Protein Assemblies. Chem. Soc. Rev. 2010, 39, 1633–1655. 10.1039/B914002F. [DOI] [PubMed] [Google Scholar]
- (190).Giles K; Pringle SD; Worthington KR; Little D; Wildgoose JL; Bateman RH Applications of a Travelling Wave-Based Radio-Frequencyonly Stacked Ring Ion Guide. Rapid Commun. Mass Spectrom. 2004, 18, 2401–2414. 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- (191).Giles K; Williams JP; Campuzano I Enhancements in Travelling Wave Ion Mobility Resolution. Rapid Commun. Mass Spectrom. 2011, 25, 1559–1566. 10.1002/rcm.5013. [DOI] [PubMed] [Google Scholar]
- (192).Anthony SN; Shinholt DL; Jarrold MF A Simple Electrospray Interface Based on a DC Ion Carpet. Int. J. Mass Spectrom. 2014, 371, 1–7. 10.1016/j.ijms.2014.06.007. [DOI] [Google Scholar]
- (193).SELECT SERIES Cyclic IMS ion mobility mass spectrometer : Waters https://www.waters.com/waters/en_US/SELECT-SERIES-Cyclic-IMS-ion-mobility-mass-spectrometer/nav.htm?cid=135021297&locale=en_US (accessed Jul 6, 2020).
- (194).Giles K; Ujma J; Wildgoose J; Pringle S; Richardson K; Langridge D; Green M A Cyclic Ion Mobility-Mass Spectrometry System. Anal. Chem. 2019, 91, 8564–8573. 10.1021/acs.analchem.9b01838. [DOI] [PubMed] [Google Scholar]
- (195).Shaw JB; Li W; Holden DD; Zhang Y; Griep-Raming J; Fellers RT; Early BP; Thomas PM; Kelleher NL; Brodbelt JS Complete Protein Characterization Using Top-down Mass Spectrometry and Ultraviolet Photodissociation. J. Am. Chem. Soc. 2013, 135, 12646–12651. 10.1021/ja4029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (196).Brodbelt JS Ion Activation Methods for Peptides and Proteins. Anal. Chem. 2016, 88, 30–51. 10.1021/acs.analchem.5b04563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (197).Bellina B; Brown JM; Ujma J; Murray P; Giles K; Morris M; Compagnon I; Barran PE UV Photodissociation of Trapped Ions Following Ion Mobility Separation in a Q-ToF Mass Spectrometer. Analyst 2014, 139, 6348–6351. 10.1039/C4AN01656D. [DOI] [PubMed] [Google Scholar]
- (198).Theisen A; Yan B; Brown JM; Morris M; Bellina B; Barran PE Use of Ultraviolet Photodissociation Coupled with Ion Mobility Mass Spectrometry To Determine Structure and Sequence from Drift Time Selected Peptides and Proteins. Anal. Chem. 2016, 88, 9964–9971. 10.1021/acs.analchem.6b01705. [DOI] [PubMed] [Google Scholar]
- (199).Theisen A; Black R; Corinti D; Brown JM; Bellina B; Barran PE Initial Protein Unfolding Events in Ubiquitin, Cytochrome c and Myoglobin Are Revealed with the Use of 213 Nm UVPD Coupled to IM-MS. J. Am. Soc. Mass Spectrom. 2019, 30, 24–33. 10.1021/jasms.8b05921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (200).Black R; Barkhanskiy A; Ramakers L; Theisen A; Brown J; Bellina B; Trivedi D; Barran P Characterization of Protein Structure with Ion Mobility Mass Spectrometry, Multiplexed Fragmentation Strategies and Data Directed Analysis. Chemrxiv 2020. 10.26434/chemrxiv.13205918.v1. [DOI] [Google Scholar]
- (201).Stiving AQ; Harvey SR; Jones BJ; Bellina B; Brown JM; Barran PE; Wysocki VH Coupling 193 Nm Ultraviolet Photodissociation and Ion Mobility for Sequence Characterization of Conformationally-Selected Peptides. J. Am. Soc. Mass Spectrom. 2020, 31, 2313–2320. 10.1021/jasms.0c00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (202).Williams JP; Morrison LJ; Brown JM; Beckman JS; Voinov VG; Lermyte F Top-Down Characterization of Denatured Proteins and Native Protein Complexes Using Electron Capture Dissociation Implemented within a Modified Ion Mobility-Mass Spectrometer. Anal. Chem. 2020, 92, 3674–3681. 10.1021/acs.analchem.9b04763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (203).Shaw JB; Malhan N; Vasil’ev YV; Lopez NI; Makarov A; Beckman JS; Voinov VG Sequencing Grade Tandem Mass Spectrometry for Top–Down Proteomics Using Hybrid Electron Capture Dissociation Methods in a Benchtop Orbitrap Mass Spectrometer. Anal. Chem. 2018, 90, 10819–10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (204).Fort KL; Cramer CN; Voinov VG; Vasil’ev YV; Lopez NI; Beckman JS; Heck AJR Exploring ECD on a Benchtop Q Exactive Orbitrap Mass Spectrometer. J. Proteome Res. 2018, 17, 926–933. 10.1021/acs.jproteome.7b00622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (205).Gadkari VV; Harvey SR; Raper AT; Chu W-T; Wang J; Wysocki VH; Suo Z Investigation of Sliding DNA Clamp Dynamics by Single-Molecule Fluorescence, Mass Spectrometry and Structure-Based Modeling. Nucleic Acids Res. 2018, 46, 3103–3118. 10.1093/nar/gky125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (206).Cole HL; Kalapothakis JMD; Bennett G; Barran PE; MacPhee CE Characterizing Early Aggregates Formed by an Amyloidogenic Peptide by Mass Spectrometry. Angew. Chem. Int. Ed. Engl. 2010, 122, 9638–9641. 10.1002/ange.201003373. [DOI] [PubMed] [Google Scholar]
- (207).Jacobs AD; Chang F-MJ; Morrison L; Dilger JM; Wysocki VH; Clemmer DE; Giedroc DP Resolution of Stepwise Cooperativities of Copper Binding by the Homotetrameric Copper-Sensitive Operon Repressor (CsoR): Impact on Structure and Stability. Angew. Chem. Int. Ed. Engl. 2015, 54, 12795–12799. 10.1002/anie.201506349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (208).Marty MT; Hoi KK; Gault J; Robinson CV Probing the Lipid Annular Belt by Gas-Phase Dissociation of Membrane Proteins in Nanodiscs. Angew. Chem. Int. Ed. Engl. 2016, 55, 550–554. 10.1002/anie.201508289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (209).Leney AC; McMorran LM; Radford SE; Ashcroft AE Amphipathic Polymers Enable the Study of Functional Membrane Proteins in the Gas Phase. Anal. Chem. 2012, 84, 9841–9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (210).Hopper JTS; Yu YT-C; Li D; Raymond A; Bostock M; Liko I; Mikhailov V; Laganowsky A; Benesch JLP; Caffrey M; Nietlispach D; Robinson CV Detergent-Free Mass Spectrometry of Membrane Protein Complexes. Nat. Methods 2013, 10, 1206–1208. 10.1038/nmeth.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (211).Tian Y; Ruotolo BT Collision Induced Unfolding Detects Subtle Differences in Intact Antibody Glycoforms and Associated Fragments. Int. J. Mass Spectrom. 2018, 425, 1–9. 10.1016/j.ijms.2017.12.005. [DOI] [Google Scholar]
- (212).T. Donor M; M. Mroz A; S. Prell J Experimental and Theoretical Investigation of Overall Energy Deposition in Surface-Induced Unfolding of Protein Ions. Chem. Sci. 2019, 10, 4097–4106. 10.1039/C9SC00644C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (213).Morris MR; Riederer DE; Winger BE; Cooks RG; Ast T; Chidsey CED Ion/Surface Collisions at Functionalized Self-Assembled Monolayer Surfaces. Int. J. Mass Spectrom. Ion Processes 1992, 122, 181–217. 10.1016/0168-1176(92)87016-8. [DOI] [Google Scholar]
- (214).Miller SA; Riederer DE; Cooks RG; Cho WR; Lee HW; Kang H Energy Disposal and Target Effects in Hyperthermal Collisions of Ferrocene Molecular Ions at Surfaces. J. Phys. Chem. 1994, 98, 245–251. 10.1021/j100052a041. [DOI] [Google Scholar]
- (215).DeKrey MJ; Kenttämaa HI; Wysocki VH; Cooks RG Energy Deposition in [Fe(CO)5]+˙ upon Collision with a Metal Surface. J. Mass Spectrom. 1986, 21, 193–195. 10.1002/oms.1210210405. [DOI] [Google Scholar]
- (216).Ruotolo BT; Hyung S-J; Robinson PM; Giles K; Bateman RH; Robinson CV Ion Mobility-Mass Spectrometry Reveals Long-Lived, Unfolded Intermediates in the Dissociation of Protein Complexes. Angew. Chem. Int. Ed. Engl. 2007, 46, 8001–8004. 10.1002/anie.200702161. [DOI] [PubMed] [Google Scholar]
- (217).Quintyn RS; Zhou M; Yan J; Wysocki VH Surface-Induced Dissociation Mass Spectra as a Tool for Distinguishing Different Structural Forms of Gas-Phase Multimeric Protein Complexes. Anal. Chem. 2015, 87, 11879–11886. [DOI] [PubMed] [Google Scholar]
- (218).Rosati S; Rose RJ; Thompson NJ; van Duijn E; Damoc E; Denisov E; Makarov A; Heck AJR Exploring an Orbitrap Analyzer for the Characterization of Intact Antibodies by Native Mass Spectrometry. Angew. Chem. Int. Ed. Engl. 2012, 51, 12992–12996. 10.1002/anie.201206745. [DOI] [PubMed] [Google Scholar]
- (219).McCabe JW; Mallis CS; Kocurek KI; Poltash ML; Shirzadeh M; Hebert MJ; Fan L; Walker TE; Zheng X; Jiang T; Dong S; Lin C-W; Laganowsky A; Russell DH First-Principles Collision Cross Section Measurements of Large Proteins and Protein Complexes. Anal. Chem. 2020. 10.1021/acs.analchem.0c01285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (220).Wörner TP; Snijder J; Bennett A; Agbandje-McKenna M; Makarov AA; Heck AJR Resolving Heterogeneous Macromolecular Assemblies by Orbitrap-Based Single-Particle Charge Detection Mass Spectrometry. Nat. Methods 2020, 1–4. 10.1038/s41592-020-0770-7. [DOI] [PubMed] [Google Scholar]
- (221).Makarov A; Denisov E Dynamics of Ions of Intact Proteins in the Orbitrap Mass Analyzer. J. Am. Soc. Mass Spectrom. 2009, 20, 1486–1495. [DOI] [PubMed] [Google Scholar]
- (222).Marty MT; Baldwin AJ; Marklund EG; Hochberg GKA; Benesch JLP; Robinson CV Bayesian Deconvolution of Mass and Ion Mobility Spectra: From Binary Interactions to Polydisperse Ensembles. Anal. Chem. 2015, 87, 4370–4376. 10.1021/acs.analchem.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (223).Marty MT; Zhang H; Cui W; Blankenship RE; Gross ML; Sligar SG Native Mass Spectrometry Characterization of Intact Nanodisc Lipoprotein Complexes. Anal. Chem. 2012, 84, 8957–8960. 10.1021/ac302663f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (224).Reid DJ; Keener JE; Wheeler AP; Zambrano DE; Diesing JM; Reinhardt-Szyba M; Makarov A; Marty MT Engineering Nanodisc Scaffold Proteins for Native Mass Spectrometry. Anal. Chem. 2017, 89, 11189–11192. 10.1021/acs.analchem.7b03569. [DOI] [PubMed] [Google Scholar]
- (225).Walker LR; Marzluff EM; Townsend JA; Resager WC; Marty MT Native Mass Spectrometry of Antimicrobial Peptides in Lipid Nanodiscs Elucidates Complex Assembly. Anal. Chem. 2019, 91, 9284–9291. 10.1021/acs.analchem.9b02261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (226).Keener JE; Zambrano DE; Zhang G; Zak CK; Reid DJ; Deodhar BS; Pemberton JE; Prell JS; Marty MT Chemical Additives Enable Native Mass Spectrometry Measurement of Membrane Protein Oligomeric State within Intact Nanodiscs. J. Am. Chem. Soc. 2019, 141, 1054–1061. 10.1021/jacs.8b11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (227).Kafader JO; Durbin KR; Melani RD; Des Soye BJ; Schachner LF; Senko MW; Compton PD; Kelleher NL Individual Ion Mass Spectrometry Enhances the Sensitivity and Sequence Coverage of Top Down Mass Spectrometry. J. Proteome Res. 2020. 10.1021/acs.jproteome.9b00797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (228).Kafader JO; Melani RD; Senko MW; Makarov AA; Kelleher NL; Compton PD Measurement of Individual Ions Sharply Increases the Resolution of Orbitrap Mass Spectra of Proteins. Anal. Chem. 2019, 91, 2776–2783. 10.1021/acs.analchem.8b04519. [DOI] [PubMed] [Google Scholar]
- (229).Kafader JO; Melani RD; Durbin KR; Ikwuagwu B; Early BP; Fellers RT; Beu SC; Zabrouskov V; Makarov AA; Maze JT; Shinholt DL; Yip PF; Tullman-Ercek D; Senko MW; Compton PD; Kelleher NL Multiplexed Mass Spectrometry of Individual Ions Improves Measurement of Proteoforms and Their Complexes. Nat. Methods 2020, 17, 391–394. 10.1038/s41592-020-0764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (230).McGee JP; Melani RD; Yip PF; Senko MW; Compton PD; Kafader JO; Kelleher NL Isotopic Resolution of Protein Complexes up to 466 KDa Using Individual Ion Mass Spectrometry. Anal. Chem. 2020, 93, 2723–2727. 10.1021/acs.analchem.0c03282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (231).Laskin J; Bailey TH; Futrell JH Mechanisms of Peptide Fragmentation from Time-and Energy-Resolved Surface-Induced Dissociation Studies: Dissociation of Angiotensin Analogs. Int. J. Mass Spectrom. 2006, 249, 462–472. [Google Scholar]
- (232).Laskin J; Futrell JH Surface-Induced Dissociation of Peptide Ions: Kinetics and Dynamics. J. Am. Soc. Mass Spectrom. 2003, 14, 1340–1347. [DOI] [PubMed] [Google Scholar]
- (233).Zhong W; Nikolaev EN; Futrell JH; Wysocki VH Tandem Fourier Transform Mass Spectrometry Studies of Surface-Induced Dissociation of Benzene Monomer and Dimer Ions on a Self-Assembled Fluorinated Alkanethiolate Monolayer Surface. Anal. Chem. 1997, 69, 2496–2503. 10.1021/ac9612887. [DOI] [PubMed] [Google Scholar]
- (234).Fernandez FM; Wysocki VH; Futrell JH; Laskin J Protein Identification via Surface-Induced Dissociation in an FT-ICR Mass Spectrometer and a Patchwork Sequencing Approach. J. Am. Soc. Mass Spectrom. 2006, 17, 700–709. 10.1016/j.jasms.2006.01.012. [DOI] [PubMed] [Google Scholar]
- (235).Li H; Sheng Y; McGee W; Cammarata M; Holden D; Loo JA Structural Characterization of Native Proteins and Protein Complexes by Electron Ionization Dissociation-Mass Spectrometry. Anal. Chem. 2017, 89, 2731–2738. 10.1021/acs.analchem.6b02377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (236).Campuzano IDG; Nshanian M; Spahr C; Lantz C; Netirojjanakul C; Li H; Wongkongkathep P; Wolff JJ; Loo JA High Mass Analysis with a Fourier Transform Ion Cyclotron Resonance Mass Spectrometer: From Inorganic Salt Clusters to Antibody Conjugates and Beyond. J. Am. Soc. Mass Spectrom. 2020. 10.1021/jasms.0c00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (237).Li H; Nguyen HH; Ogorzalek Loo RR; Campuzano IDG; Loo JA An Integrated Native Mass Spectrometry and Top-down Proteomics Method That Connects Sequence to Structure and Function of Macromolecular Complexes. Nat. Chem 2018, 10, 139–148. 10.1038/nchem.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (238).Smith DF; Podgorski DC; Rodgers RP; Blakney GT; Hendrickson CL 21 Tesla FT-ICR Mass Spectrometer for Ultrahigh-Resolution Analysis of Complex Organic Mixtures. Anal. Chem. 2018, 90, 2041–2047. 10.1021/acs.analchem.7b04159. [DOI] [PubMed] [Google Scholar]
- (239).Hendrickson CL; Quinn JP; Kaiser NK; Smith DF; Blakney GT; Chen T; Marshall AG; Weisbrod CR; Beu SC 21 Tesla Fourier Transform Ion Cyclotron Resonance Mass Spectrometer: A National Resource for Ultrahigh Resolution Mass Analysis. J. Am. Soc. Mass Spectrom. 2015, 26, 1626–1632. 10.1021/jasms.8b05110. [DOI] [PubMed] [Google Scholar]
- (240).Nikolaev EN; Kostyukevich YI; Vladimirov GN Fourier Transform Ion Cyclotron Resonance (FT ICR) Mass Spectrometry: Theory and Simulations. Mass Spectrom. Rev. 2016, 35, 219–258. 10.1002/mas.21422. [DOI] [PubMed] [Google Scholar]
- (241).Kostyukevich YI; Vladimirov GN; Nikolaev EN Dynamically Harmonized FT-ICR Cell with Specially Shaped Electrodes for Compensation of Inhomogeneity of the Magnetic Field. Computer Simulations of the Electric Field and Ion Motion Dynamics. J. Am. Soc. Mass Spectrom. 2012, 23, 2198–2207. 10.1021/jasms.8b04191. [DOI] [PubMed] [Google Scholar]
- (242).Nikolaev EN; Somogyi Á; Smith DL; Gu C; Wysocki VH; Martin CD; Samuelson GL Implementation of Low-Energy Surface-Induced Dissociation (EV SID) and High-Energy Collision-Induced Dissociation (KeV CID) in a Linear Sector-TOF Hybrid Tandem Mass Spectrometer. Int. J. Mass Spectrom. 2001, 212, 535–551. 10.1016/S1387-3806(01)00462-6. [DOI] [Google Scholar]
- (243).de Maaijer-Gielbert J; Beijersbergen JH; Kistemaker PG; Weeding TL Surface-Induced Dissociation of Benzene on a PFPE Liquid Insulator in a Time-of-Flight Mass Spectrometer. Int. J. Mass Spectrom. Ion Processes 1996, 153, 119–128. [Google Scholar]
- (244).Konijn SW; Steenvoorden RJJM; Kistemaker PG; Weeding TL Mass-Resolved Excitation Spectra of Guaiacol and 4-Methylguaiacol and Geometry Assignment of the Minimum Energy Conformations of Guaiacol. J. Phys. Chem. 1994, 98, 5399–5403. 10.1021/j100072a002. [DOI] [Google Scholar]
- (245).Beck RD; St. John P; Alvarez MM; Diederich F; Whetten RL Resilience of All-Carbon Molecules C60, C70, and C84: A Surface-Scattering Time-of-Flight Investigation. J. Phys. Chem. 1991, 95, 8402–8409. 10.1021/j100174a066. [DOI] [Google Scholar]
- (246).Johnson JT; Carrick IJ; Eakins GS; McLuckey SA Mirror Switching for High-Resolution Ion Isolation in an Electrostatic Linear Ion Trap. Anal. Chem. 2019, 91, 8789–8794. 10.1021/acs.analchem.9b00874. [DOI] [PubMed] [Google Scholar]
- (247).Belov ME; Damoc E; Denisov E; Compton PD; Horning S; Makarov AA; Kelleher NL From Protein Complexes to Subunit Backbone Fragments: A Multi-Stage Approach to Native Mass Spectrometry. Anal. Chem. 2013, 85, 11163–11173. 10.1021/ac4029328. [DOI] [PubMed] [Google Scholar]
- (248).Crittenden CM; Herrera CM; Williams PE; Ricci DP; Swem LR; Trent MS; Brodbelt JS Mapping Phosphate Modifications of Substituted Lipid A via a Targeted MS3 CID/UVPD Strategy. Analyst 2018, 143, 3091–3099. 10.1039/C8AN00561C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (249).Mehaffey MR; Xia Q; Brodbelt JS Uniting Native Capillary Electrophoresis and Multistage Ultraviolet Photodissociation Mass Spectrometry for Online Separation and Characterization of Escherichia Coli Ribosomal Proteins and Protein Complexes. Anal. Chem. 2020, 92, 15202–15211. 10.1021/acs.analchem.0c03784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (250).Panczyk E; Snyder D; Ridgeway ME; Somogyi A; Park MA; Wysocki V Surface-Induced Dissociation of Protein Complexes Selected by Trapped Ion Mobility Spectrometry. Chemrxiv 2020. 10.26434/chemrxiv.13477275.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (251).Choi R; Zhou M; Shek R; Wilson JW; Tillery L; Craig JK; Salukhe IA; Hickson SE; Kumar N; James RM; Buchko GW; Wu R; Huff S; Nguyen T-T; Hurst BL; Cherry S; Barrett LK; Hyde JL; Voorhis WCV High-Throughput Screening of the ReFRAME, Pandemic Box, and COVID Box Drug Repurposing Libraries against SARS-CoV2 Nsp15 Endoribonuclease to Identify Small-Molecule Inhibitors of Viral Activity. bioRxiv 2021, 2021.01.21.427657. 10.1101/2021.01.21.427657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (252).Jia M Native Mass Spectrometry for Characterization of Protein:Protein and Protein:RNA Complexes. Dissertation, Ohio State University, Columbus, OH. [Google Scholar]
- (253).Suresh SJ; Naik VM Hydrogen Bond Thermodynamic Properties of Water from Dielectric Constant Data. J. Chem. Phys. 2000, 113, 9727–9732. 10.1063/1.1320822. [DOI] [PubMed] [Google Scholar]
- (254).Reading E; Walton TA; Liko I; Marty MT; Laganowsky A; Rees DC; Robinson CV The Effect of Detergent, Temperature, and Lipid on the Oligomeric State of MscL Constructs: Insights from Mass Spectrometry. Chem. Biol. 2015, 22, 593–603. 10.1016/j.chembiol.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (255).Laskin J; Futrell JH On the Efficiency of Energy Transfer in Collisional Activation of Small Peptides. J. Chem. Phys. 2002, 116, 4302–4310. 10.1063/1.1450544. [DOI] [Google Scholar]
- (256).Shukla AK; Futrell JH; Sen AD Dynamics of the Dissociative and Nondissociative Scattering of Hyperthermal CS 2+ from a Self-Assembled Fluoroalkyl Monolayer Surface on Gold Substrate. J. Chem. Phys. 2003, 118, 11217–11225. [Google Scholar]
- (257).Laskin J; Denisov E; Futrell J Comparative Study of Collision-Induced and Surface-Induced Dissociation. 2. Fragmentation of Small Alanine-Containing Peptides in FT-ICR MS. J. Phys. Chem. B 2001, 105, 1895–1900. [Google Scholar]
- (258).Žabka J; Dolejšek Z; Herman Z Energy Partitioning in Collisions of Slow Polyatomic Ions with Surfaces: Ethanol Molecular Ions on Surfaces Covered by Self-Assembled Monolayers (CF-SAM, CH-SAM, COOH-SAM). J. Phys. Chem. A 2002, 106, 10861–10869. 10.1021/jp014715f. [DOI] [Google Scholar]
- (259).Wang Y; Hase WL; Song K Direct Dynamics Study of N-Protonated Diglycine Surface-Induced Dissociation. Influence of Collision Energy. J. Am. Soc. Mass Spectrom. 2003, 14, 1402–1412. [DOI] [PubMed] [Google Scholar]
- (260).Felitsyn N; Kitova EN; Klassen JS Thermal Decomposition of a Gaseous Multiprotein Complex Studied by Blackbody Infrared Radiative Dissociation. Investigating the Origin of the Asymmetric Dissociation Behavior. Anal. Chem. 2001, 73, 4647–4661. 10.1021/ac0103975. [DOI] [PubMed] [Google Scholar]
- (261).Jurchen JC; Williams ER Origin of Asymmetric Charge Partitioning in the Dissociation of Gas-Phase Protein Homodimers. J. Am. Chem. Soc. 2003, 125, 2817–2826. 10.1021/ja0211508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (262).Jurchen JC; Garcia DE; Williams ER Further Studies on the Origins of Asymmetric Charge Partitioning in Protein Homodimers. J. Am. Soc. Mass Spectrom. 2004, 15, 1408–1415. 10.1016/j.jasms.2004.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (263).Wanasundara SN; Thachuk M Theoretical Investigations of the Dissociation of Charged Protein Complexes in the Gas Phase. J. Am. Soc. Mass Spectrom. 2007, 18, 2242–2253. 10.1016/j.jasms.2007.09.022. [DOI] [PubMed] [Google Scholar]
- (264).Sever AIM; Yin V; Konermann L Interrogating the Quaternary Structure of Noncanonical Hemoglobin Complexes by Electrospray Mass Spectrometry and Collision-Induced Dissociation. J. Am. Soc. Mass Spectrom. 2020. 10.1021/jasms.0c00320. [DOI] [PubMed] [Google Scholar]
- (265).Light-Wahl KJ; Schwartz BL; Smith RD Observation of the Noncovalent Quaternary Associations of Proteins by Electrospray Ionization Mass Spectrometry. J. Am. Chem. Soc. 1994, 116, 5271–5278. 10.1021/ja00091a035. [DOI] [PubMed] [Google Scholar]
- (266).Aquilina JA; Benesch JLP; Bateman OA; Slingsby C; Robinson CV Polydispersity of a Mammalian Chaperone: Mass Spectrometry Reveals the Population of Oligomers in ΑB-Crystallin. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 10611–10616. 10.1073/pnas.1932958100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (267).Lei QP; Cui X; Kurtz DM; Amster IJ; Chernushevich IV; Standing KG Electrospray Mass Spectrometry Studies of Non-Heme Iron-Containing Proteins. Anal. Chem. 1998, 70, 1838–1846. 10.1021/ac971181z. [DOI] [PubMed] [Google Scholar]
- (268).McCammon MG; Hernández H; Sobott F; Robinson CV Tandem Mass Spectrometry Defines the Stoichiometry and Quaternary Structural Arrangement of Tryptophan Molecules in the Multiprotein Complex TRAP. J. Am. Chem. Soc. 2004, 126, 5950–5951. 10.1021/ja0317170. [DOI] [PubMed] [Google Scholar]
- (269).Loo JA; Berhane B; Kaddis CS; Wooding KM; Xie Y; Kaufman SL; Chernushevich IV Electrospray Ionization Mass Spectrometry and Ion Mobility Analysis of the 20S Proteasome Complex. J. Am. Soc. Mass Spectrom. 2005, 16, 998–1008. 10.1021/jasms.8b02446. [DOI] [PubMed] [Google Scholar]
- (270).Wang G; Chaihu L; Tian M; Shao X; Dai R; de Jong RN; Ugurlar D; Gros P; Heck AJR Releasing Nonperipheral Subunits from Protein Complexes in the Gas Phase. Anal. Chem. 2020. 10.1021/acs.analchem.0c02845. [DOI] [PubMed] [Google Scholar]
- (271).Song Y Protein Primary and Quaternary Structure Elucidation by Mass Spectrometry, The Ohio State University, 2015. [Google Scholar]
- (272).Leney AC Subunit PI Can Influence Protein Complex Dissociation Characteristics. J. Am. Soc. Mass Spectrom. 2019, 30, 1389–1395. 10.1021/jasms.8b06060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (273).Jones CM; Beardsley RL; Galhena AS; Dagan S; Cheng G; Wysocki VH Symmetrical Gas-Phase Dissociation of Noncovalent Protein Complexes via Surface Collisions. J. Am. Chem. Soc. 2006, 128, 15044–15045. 10.1021/ja064586m. [DOI] [PubMed] [Google Scholar]
- (274).Jia M; Song Y; Du C; Wysocki V Charge Distributions of the Gas-Phase Dissociation Products of Dimeric Proteins. Submitted. [Google Scholar]
- (275).McLuckey SA; Goeringer DE Special Feature: Tutorial Slow Heating Methods in Tandem Mass Spectrometry. J. Mass Spectrom. 1997, 32, 461–474. [Google Scholar]
- (276).Sciuto SV; Liu J; Konermann L An Electrostatic Charge Partitioning Model for the Dissociation of Protein Complexes in the Gas Phase. J. Am. Soc. Mass Spectrom. 2011, 22. 10.1021/jasms.8b03911. [DOI] [PubMed] [Google Scholar]
- (277).Wanasundara SN; Thachuk M Free Energy Barrier Estimation for the Dissociation of Charged Protein Complexes in the Gas Phase. J. Phys. Chem. A 2009, 113, 3814–3821. 10.1021/jp8094227. [DOI] [PubMed] [Google Scholar]
- (278).Thachuk M; Fegan SK; Raheem N Description and Control of Dissociation Channels in Gas-Phase Protein Complexes. J. Chem. Phys. 2016, 145, 065101. 10.1063/1.4960615. [DOI] [Google Scholar]
- (279).Fegan SK; Thachuk M A Charge Moving Algorithm for Molecular Dynamics Simulations of Gas-Phase Proteins. J. Chem. Theory Comput. 2013, 9, 2531–2539. 10.1021/ct300906a. [DOI] [PubMed] [Google Scholar]
- (280).Csiszar S; Thachuk M Using Ellipsoids to Model Charge Distributions in Gas Phase Protein Complex Ion Dissociation. Can. J. Chem. 2011. 10.1139/v04-154. [DOI] [Google Scholar]
- (281).Wanasundara SN; Thachuk M Toward an Improved Understanding of the Dissociation Mechanism of Gas Phase Protein Complexes. J. Phys. Chem. B 2010, 114, 11646–11653. 10.1021/jp103576b. [DOI] [PubMed] [Google Scholar]
- (282).Sciuto SV; Liu J; Konermann L An Electrostatic Charge Partitioning Model for the Dissociation of Protein Complexes in the Gas Phase. J. Am. Soc. Mass Spectrom. 2011, 22. 10.1021/jasms.8b03911. [DOI] [PubMed] [Google Scholar]
- (283).Popa V; Trecroce DA; McAllister RG; Konermann L Collision-Induced Dissociation of Electrosprayed Protein Complexes: An All-Atom Molecular Dynamics Model with Mobile Protons. J. Phys. Chem. B 2016, 120, 5114–5124. 10.1021/acs.jpcb.6b03035. [DOI] [PubMed] [Google Scholar]
- (284).Konermann L; Aliyari E; Lee JH Mobile Protons Limit the Stability of Salt Bridges in the Gas Phase: Implications for the Structures of Electrosprayed Protein Ions. J. Phys. Chem. B 2021. 10.1021/acs.jpcb.1c00944. [DOI] [PubMed] [Google Scholar]
- (285).Bornschein RE; Niu S; Eschweiler J; Ruotolo BT Ion Mobility-Mass Spectrometry Reveals Highly-Compact Intermediates in the Collision Induced Dissociation of Charge-Reduced Protein Complexes. J. Am. Soc. Mass Spectrom. 2016, 27, 41–49. 10.1021/jasms.8b05128. [DOI] [PubMed] [Google Scholar]
- (286).Miller SA; Luo H; Pachuta SJ; Cooks RG Soft-Landing of Polyatomic Ions at Fluorinated Self-Assembled Monolayer Surfaces. Science 1997, 275, 1447–1450. 10.1126/science.275.5305.1447. [DOI] [Google Scholar]
- (287).de Clercq HL; Sen AD; Shukla AK; Futrell JH Inelastic Ion-Surface Collisions: Scattering and Dissociation of Low Energy Benzene Molecular Cations. Int. J. Mass Spectrom. 2001, 212, 491–504. [Google Scholar]
- (288).Zhou M Incorporation of Surface Induced Dissociation into a Commercial Ion Mobility – Tandem Mass Spectrometer and Application of Mass Spectrometry Methods for Structural Analysis of Non-Covalent Protein Complexes. Dissertation, The Ohio State University, The Ohio State University, 2013. [Google Scholar]
- (289).Snyder D; Lin Y; Somogyi A; Wysocki V Kinetic Energy Distributions of Protein Complexes and Their Fragments after Surface Collision. Submitted.
- (290).Barnes GL; Hase WL Energy Transfer, Unfolding, and Fragmentation Dynamics in Collisions of N-Protonated Octaglycine with an H-SAM Surface. J. Am. Chem. Soc. 2009, 131, 17185–17193. 10.1021/ja904925p. [DOI] [PubMed] [Google Scholar]
- (291).Laskin J; Bailey TH; Futrell JH Shattering of Peptide Ions on Self-Assembled Monolayer Surfaces. J. Am. Chem. Soc. 2003, 125, 1625–1632. 10.1021/ja027915t. [DOI] [PubMed] [Google Scholar]
- (292).Barnes GL; Shlaferman A; Strain M Fast Fragmentation during Surface-Induced Dissociation: An Examination of Peptide Size and Structure. Chem. Phys. Lett. 2020, 754, 137716. 10.1016/j.cplett.2020.137716. [DOI] [Google Scholar]
- (293).Barnes GL; Young K; Yang L; Hase WL Fragmentation and Reactivity in Collisions of Protonated Diglycine with Chemically Modified Perfluorinated Alkylthiolate-Self-Assembled Monolayer Surfaces. J. Chem. Phys. 2011, 134, 094106. 10.1063/1.3558736. [DOI] [PubMed] [Google Scholar]
- (294).Gregg Z; Ijaz W; Jannetti S; Barnes GL The Role of Proton Transfer in Surface-Induced Dissociation. J. Phys. Chem. C 2014, 118, 22149–22155. 10.1021/jp507069x. [DOI] [Google Scholar]
- (295).Shaikh K; Blackwood J; Barnes GL The Effect of Protonation Site and Conformation on Surface-Induced Dissociation in a Small, Lysine Containing Peptide. Chem. Phys. Lett. 2015, 637, 83–87. 10.1016/j.cplett.2015.07.062. [DOI] [Google Scholar]
- (296).Pratihar S; Barnes GL; Laskin J; Hase WL Dynamics of Protonated Peptide Ion Collisions with Organic Surfaces: Consonance of Simulation and Experiment. J. Phys. Chem. Lett 2016, 7, 3142–3150. 10.1021/acs.jpclett.6b00978. [DOI] [PubMed] [Google Scholar]
- (297).Pratihar S; Barnes GL; Hase WL Chemical Dynamics Simulations of Energy Transfer, Surface-Induced Dissociation, Soft-Landing, and Reactive-Landing in Collisions of Protonated Peptide Ions with Organic Surfaces. Chem. Soc. Rev. 2016, 45, 3595–3608. [DOI] [PubMed] [Google Scholar]
- (298).Meroueh SO; Wang Y; Hase WL Direct Dynamics Simulations of Collision-and Surface-Induced Dissociation of N-Protonated Glycine. Shattering Fragmentation. J. Phys. Chem. 2002, 106, 9983–9992. [Google Scholar]
- (299).Park K; Deb B; Song K; Hase WL Importance of Shattering Fragmentation in the Surface-Induced Dissociation of Protonated Octaglycine. J. Am. Soc. Mass Spectrom. 2009, 20, 939–948. 10.1016/j.jasms.2009.02.028. [DOI] [PubMed] [Google Scholar]
- (300).Park K; Song K; Hase WL An Ab Initio Direct Dynamics Simulation of Protonated Glycine Surface-Induced Dissociation. Int. J. Mass Spectrom. 2007, 265, 326–336. 10.1016/j.ijms.2007.03.009. [DOI] [Google Scholar]
- (301).Rahaman A; Zhou JB; Hase WL Effects of Projectile Orientation and Surface Impact Site on the Efficiency of Projectile Excitation in Surface-Induced Dissociation: Protonated Diglycine Collisions with Diamond {111}. Int. J. Mass Spectrom. 2006, 249–250, 321–329. 10.1016/j.ijms.2005.12.020. [DOI] [Google Scholar]
- (302).Bosio SBM; Hase WL Simulations of Energy Transfer in Cr(CO)6+ Surface-Induced Dissociation. Int. J. Mass Spectrom. Ion Processes 1998, 174, 1–9. 10.1016/S0168-1176(97)00286-3. [DOI] [Google Scholar]
- (303).Schultz DG; Hanley L Shattering of SiMe 3+ during Surface-Induced Dissociation. J. Chem. Phys. 1998, 109, 10976–10983. [Google Scholar]
- (304).Burroughs JA; Wainhaus SB; Hanley L Impulsive Excitation of FeCp+2 and SiMe+3 during Surface-induced Dissociation at Organic Multilayers. J. Chem. Phys. 1995, 103, 6706–6715. 10.1063/1.470401. [DOI] [Google Scholar]
- (305).Wainhaus SB; Gislason EA; Hanley L Determination of Activation Energies for Ion Fragmentation by Surface-Induced Dissociation. J. Am. Chem. Soc. 1997, 119, 4001–4007. [Google Scholar]
- (306).Wainhaus SB; Lim H; Schultz DG; Hanley L Energy Transfer and Surface-Induced Dissociation for SiMe 3+ Scattering off Clean and Adsorbate Covered Metals. J. Chem. Phys. 1997, 106, 10329–10336. [Google Scholar]
- (307).Schultz D; Lim H; Garbis S; Hanley L Energy Partitioning in the Surface-induced Dissociation of Linear and Cyclic Protonated Peptides at an Organic Surface. J. Mass Spectrom. 1999, 34, 217–225. [Google Scholar]
- (308).Lim H; Schultz DG; Gislason EA; Hanley L Activation Energies for the Fragmentation of Thiophene Ions by Surface-Induced Dissociation. The Journal of Physical Chemistry B 1998, 102, 4573–4580. [Google Scholar]
- (309).Lim H; Schultz DG; Yu C; Hanley L Relative Dissociation Energies of Protonated Peptides by Electrospray Ionization/Surface-Induced Dissociation. Anal. Chem. 1999, 71, 2307–2317. [DOI] [PubMed] [Google Scholar]
- (310).Macaluso V; Homayoon Z; Spezia R; L. Hase W Threshold for Shattering Fragmentation in Collision-Induced Dissociation of the Doubly Protonated Tripeptide TIK(H +) 2. Phys. Chem. Chem. Phys. 2018, 20, 19744–19749. 10.1039/C8CP02577K. [DOI] [PubMed] [Google Scholar]
- (311).Mikhailov VA; Mize TH; Benesch JLP; Robinson CV Mass-Selective Soft-Landing of Protein Assemblies with Controlled Landing Energies. Anal. Chem. 2014, 86, 8321–8328. 10.1021/ac5018327. [DOI] [PubMed] [Google Scholar]
- (312).Benesch JLP; Ruotolo BT; Simmons DA; Barrera NP; Morgner N; Wang L; Saibil HR; Robinson CV Separating and Visualising Protein Assemblies by Means of Preparative Mass Spectrometry and Microscopy. J. Struct. Biol. 2010, 172, 161–168. 10.1016/j.jsb.2010.03.004. [DOI] [PubMed] [Google Scholar]
- (313).Anggara K; Zhu Y; Delbianco M; Rauschenbach S; Abb S; Seeberger PH; Kern K Exploring the Molecular Conformation Space by Soft Molecule–Surface Collision. J. Am. Chem. Soc. 2020, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]