Abstract
The vascular structure of the tumor microenvironment (TME) plays an essential role in the process of metastasis. In vitro microvascular structures that can be maintained for a long time will greatly promote metastasis research. In this study, we constructed a mimicking breast cancer invasion model based on a microfluidic chip platform, and the maintenance time of the self-assembled microvascular networks significantly improved by culturing with fibroblasts (up to 13 days). Using this model, we quantified the invasion ability of breast cancer cells and angiogenesis sprouts caused by cancer cells, and the intravasation behavior of cancer cells was also observed in sprouts. We found that cancer cells could significantly cause angiogenesis by promoting sprouting behaviors of the self-assembled human umbilical vein endothelial cells, which, in turn, promoted the invasion behavior of cancer cells. The drug test results showed that the drug resistance of the widely used anti-cancer drugs 5-Fluorouracil (5-FU) and Doxorubicin (DOX) in the 3D model was higher than that in the 2D model. Meanwhile, we also proved that 5-FU and DOX had the effect of destroying tumor blood vessels. The anti-angiogenic drug Apatinib (VEGFR inhibitor) enhanced the drug effect of DOX on MDA-MB-231 cells, further proving the promoting effect of angiogenesis on the invasion ability of cancer cells. These results indicate that our model is of great value in reconstructing TME and drug testing in vitro.
I. INTRODUCTION
In 2020, breast cancer surpassed lung cancer, becoming the most common cancer for women and the leading cause of cancer deaths among women worldwide.1 In advanced breast cancer, spread of the cancer throughout the body is caused by the complex transendothelial migration behavior of cancer cells, making it difficult to cure cancer.2 Therefore, the tumor microenvironment (TME) with complex microvascular networks has been identified as one of the driving factors of tumor progression.3–5 As the most abundant component in TME,6 cancer-associated fibroblasts (CAFs) not only produce many components of ECM and the basement membrane7 but also produce a crucial angiogenesis agent–vascular endothelial growth factor (VEGF-A), promoting the development of cancer angiogenesis.8–10 Some researchers have also found that the CAF could enhance drug resistance of cancer cells.11,12 In TME, tumor angiogenesis is the inevitable result of further development of the cancer.13 New blood vessel structures are produced in this process, while also destroying the barrier function of the original blood vessel.14,15 Tumor angiogenesis is also a vital sign, suggesting that tumors turn from benign to malignant.16 Therefore, constructing an in vitro cancer model containing CAFs and self-assembled microvascular networks is critical in investigating the physiological behavior of cancer and testing the accurate preclinical effect of anti-cancer drugs.
Most methods for evaluating cancer treatments were first verified in two-dimensional (2D) monolayer models and then in animal models.17 However, animal models cannot reflect the specific interaction between human cancer cells and blood vessels.18 2D culture lacks the physical cell–cell interactions of tumors,19 although it has an advantage in high-throughput drug screening due to its simple implementation.17 Scientists have gradually realized that simple 2D culture models or animal models are not sufficient enough for complicated tumor metastasis research.20 3D co-culture models have become important methods for complex vascular simulation21 and tumor invasion research,22 although their construction still has challenges. The rapid development of microfluidic chip technology has made it possible to build a 3D human organ model on a chip, and a new term, the organ chip, has been derived.23 In the tumor microenvironment in vitro, the construction of a blood vessel structure is the key. There are two methods for constructing blood vessel structure on chips, one was simply attaching human umbilical vein endothelial cells (HUVECs) to the gel wall for growing,24,25 and another was forming a blood vessel network through the self-assembly behavior of HUVECs.26–29 The latter is the more natural morphogenesis of endothelial cells.30 Kamm et al. introduced several extravasation models of breast cancer containing self-assembled microvascular networks within a bone-mimicking microenvironment18 or within a physiological flow environment.29 Although they provided a usable model for the extravasation study, the stage of tumor development before the formation of circulating tumor cells is more worthy of attention, starting from the perspective of timely treatment of cancer.21,29 Nagaraju et al. constructed a breast cancer cell invasion model containing a self-assembled microvascular network to simulate breast cancer cells’ invasion toward the matrix of the primary tumor.31 This model provided a potential to study the signal transduction in breast cancer cells and self-assembled vessels, but the vascular structure in this model could not be maintained for a long time (only 6 days) because of the lack of necessary stromal cells, such as fibroblasts.7 In addition, this model also ignored the study of vascularization behavior in the process of tumor invasion.
In this study, a mimicking breast cancer invasion model based on microfluidic technology was constructed to study the role of the vascular structure of TME in the process of tumor metastasis. First, fibroblast cells were involved in the TME to explore suitable co-culture strategies to develop and maintain the microvascular networks in vitro. Then, based on the self-assembled microvascular networks, a breast cancer invasion model was constructed to analyze and evaluate tumor metastasis behavior from multiple perspectives, such as cancer cell invasion, tumor angiogenesis, and cancer cell intravasation. Finally, the drug tests were carried out to evaluate the drug resistance differences of the anti-cancer drugs 5-Fluorouracil (5-FU) and Doxorubicin (DOX) between our model and the 2D model. Meanwhile, we also analyzed the effect of the combination of anti-cancer drugs (5-FU and DOX) and anti-angiogenic drug Apatinib (VEGFR inhibitor) on tumor invasion behavior to illustrate the critical role of anti-cancer drug combination strategies in tumor treatment.
II. MATERIALS AND METHODS
A. Device fabrication
The chip structure pattern was designed using Auto CAD software and then using it for manufacturing the mask. According to the standard process, SU-8-2100 (Microchem) was poured on the 3-in. silicon wafer to form a lithography layer (120 μm) at 2500 r/min. Then, the ultraviolet light was shined on the silicon wafer through the mask after the soft bake. The silicon wafer became the mold after postexposure bake, development, rinse, and dry, according to the standard process. Then, polydimethylsiloxane (PDMS, SYLGARD184, Dow Corning) was poured on the mold and was solidified at 80 °C for 1 h. The cured PDMS was cut and punched, and then invisible tapes were used to clean the surface of the PDMS structure, which needed to be bonded. Then, the PDMS structure was bonded to a 24 × 60 mm2 glass sheet with a thickness of 0.2 mm to form the microfluidic chip. After that, the chip was ultrasonically cleaned with 75% alcohol for 10 min and was sterilized by the high temperature and pressure sterilization and the UV irradiation, respectively.
B. Cell culture
Primary human umbilical vein endothelial cells (HUVECs, 8000, ScienCell, <p8) were transduced to stably express fluorescent protein by lentivirus. The HUVECs were cultured in the endothelial growth medium (EGM, CC3162, Lonza) supplemented with 5% fetal bovine serum (10100147, Gibco). The breast cancer cell line MDA-MB-231 cells that were obtained from the Shanghai cell bank of the Chinese Academy of Sciences were cultured in DMEM high glucose medium (C11995500BT, Gibco) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (15140122, Gibco). MDA-MB-231 cells were also infected by lentivirus to express the fluorescent protein for observation of the cell morphology and migration. Primary human mammary cancer-associated fibroblasts (HMCAF, HUM-iCELL-f028, iCELL, <p9) were cultured in a special medium for fibroblasts (PriMed-iCELL-003, iCELL). The above cells were cultured in an incubator at 37 °C, 5% CO2. In the process of cell passaging, cells were trypsinized with 0.25% Trypsin-EDTA (25200072, Gibco) to obtain the cell suspension.
C. Forming pre-existing blood vessels through vasculogenesis
Human fibrinogen (F3879, Sigma) was dissolved in Ca2+ and Mg2+ free DPBS (14190144, Gibco) to prepare a working solution at 8 mg/ml. Thrombin (T4648, Sigma) was dissolved in Ca2+ and Mg2+ free DPBS to obtain a stock solution of 100 U/ml, and then the stock solution was diluted with EGM to make a working solution at 4 U/ml. When loading the cells, HMCAFs were resuspended in thrombin working solution at a cell density of 3 × 106 cells/ml, and then the cell suspension was mixed with a fibrinogen working solution at a ratio of 1:1. The mixture was immediately perfused into the matrix channel of the chip (Fig. 1, Ch2). Waiting for 10 min could ensure the formation of the stable gel in Ch2 and effectively prevent the gel structure in CH2 from causing any damage to the perfusion pressure when loading the HUVERC cells into Ch1. The same method was performed to obtain the stable HUVECs gel in the vessel area of the chip (Fig. 1, Ch1). The fresh medium was replaced into the chips every 24 h.
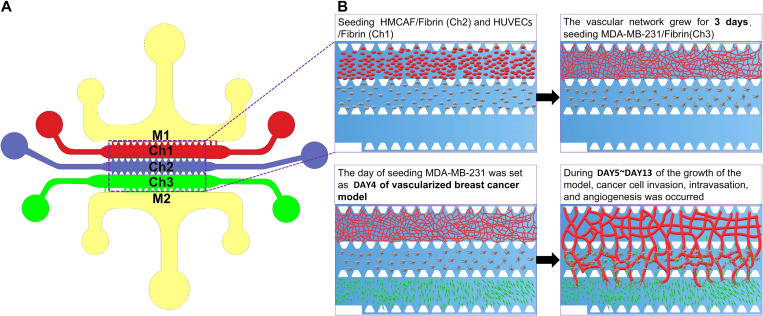
FIG. 1.
The design of the microfluidic chip and schematic diagram of the construction of the cancer model. (a) The materials were perfused in each channel as follows: Ch1: HUVECs + Fibrin gel; Ch2: HMCAF + Fibrin gel; Ch3: EGM (to form the self-assembled microvascular networks) or MDA-MB-231 + fibrin gel (to form a self-assembled vascularized breast cancer model); and M1 and M2: medium channels. (b) We obtained the functional self-assembled microvascular networks through this program and further constructed the vascularized breast cancer model.
D. Seeding MDA-MB-231 gel into the self-assembled blood vessel model
At day 3 of developing the self-assembled microvascular network, the medium in the chip was gently aspirated. The MDA-MB-231 cell-thrombin suspensions were mixed with the prepared fibrinogen working solution, and then the mixture was immediately added into the cancer cell channel (Fig. 1, Ch3) to form 3D tumor tissue. The final cell density of MDA-MB-231 cells was 1 × 107 cells/ml, and this vascularized breast cancer model was cultured for 9–11 days. During this period, the growth behaviors of MDA-MB-231 cells, such as cancer cell invasion, intravasation, angiogenesis, were recorded by the microscope.
E. Perfusibility measurement of the self-assembled blood vessel
In order to test the perfusion performance of the self-assembled blood vessel, 70 kDa FITC-Dextran solution (50 μg/ml, 46945, Sigma) was used. A fluorescence microscope (Dmi8, Leica) was used for continuous observation of the tracers. The tip of a pipet was cut and then was inserted into the one side opening module of the chip. Solid acrylic columns were used to block the other outlets on the chip, except for the opening module on the other side. The prepared solution mixed with tracers was loaded into the tip through a syringe to generate a hydrostatic pressure gradient along the vertical line of the blood vessel development area, providing a driving force for the tracer flowing through the blood vessel.
F. Quantitative analysis of microvascular networks and vascular sprouting
The software of Angiotool was used to quantitatively analyze the three development indicators of blood vessels under the same microscope magnification: vessel percentage area, total vessel length, and the total number of junctions.32 Vessel percentage area was the percentage of the blood vessel area in the total area of the culture region, representing the blood vessel density. The total vessel length and the total number of junctions, respectively, referred to the sum of the vessel length and the sum of the vessel branches in the blood vessel network.32,33 S1 in the supplementary material is a schematic diagram of the analysis of blood vessel network pictures using Angiotool in this article. The solid yellow line represents the outline of the blood vessel, and it could be used to calculate the total coverage area of the blood vessel. The green line represents the skeleton of the blood vessel, and it can be used to calculate the total length of the blood vessel. The blue points represent the vessel branches, and it can be used to analyze the total number of junctions. The values of the vessels’ percentage area under different conditions were divided by the valve of the vessels’ percentage area under the control condition at day 2 to get the relative vessels’ percentage area, facilitating a comparison between the values of different conditions. The relative total vessel length and the relative total number of junctions were obtained under the same procedure.
Software ImageJ was used to quantify the blood vessel diameter, blood vessel sprout area, and average sprout length under the same microscope magnification.34 S2(A) in the supplementary material was a schematic diagram for calculating the angiogenic sprout length of blood vessels using ImageJ. The length of all the dashed green lines was averaged, representing the average length of sprout blood vessels. S2(B) in the supplementary material is a schematic diagram for calculating the angiogenic sprout area of the sprout blood vessel in the yellow dashed box using ImageJ.
G. Immunofluorescence
All liquids are loaded into the chip medium channel. First, excess PBS solution was added into the chips to wash the cells, and then 4% paraformaldehyde was used to perform the fixing process for 20 min at 4 °C. Then, the cells were washed with PBS. After 30 min of permeabilization with 0.1% Triton X-100 solution, the cells were washed again with PBS. A blocking step was then performed using 5% BSA solution (or 4% goat serum solution) for 3 h at room temperature (RT), and then the primary antibody working solution was added into the chip, incubating cells overnight at 4 °C. For vascular barrier markers, mouse monoclonal anti-CD31 (1:100, Ab24590, Abcam) and mouse monoclonal anti-VE-Cadherin (5 μg/ml, 14-1449-82, eBioscience™) were used to detect the expressions of the corresponding proteins. Rabbit monoclonal to anti-FAP (1:100, 66562S, CST) were used to analyze the expression of HMCAF markers. For the cytoskeleton, we used Phalloidin-iFluor 555 conjugate (1:1000, ab176755, Abcam) to incubate cells at RT for 1 h. After incubating cells with the primary antibody, the cells were washed with PBS and were incubated with the corresponding appropriate secondary antibody (A 32731, ThermoFisher; or A 32727, ThermoFisher; or Ab 150113, Abcam) for 1 h according to the added primary antibody species. Then, the cells were washed again with PBS and were incubated with for DAPI (1:500, C1002, Beyotime) for 20 min. Finally, the cells were washed with PBS and were observed under a fluorescence microscope for taking pictures.
H. CCK8 assay and live/dead assay
MDA-MB-231 cells were treated with various concentrations of 5-FU and DOX for 24 or 48 h in a 96-well plate. Then, the cells were incubated in a serum-free medium supplemented with 10% CCK8 solution (Abs 50003, Absin) for 2 h at 37 °C. After shaking the 96-well plate for 5 min, the optical density was measured at 450 nm with a microplate reader.
The Live/Dead Cell Imaging Kit (Molecular Probes, Life Technologies Corporation) was used to evaluate the cell viability of MDA-MB-231 cells treated with 5-FU and DOX for 24 or 48 h on a chip. First, PBS was used to wash the chip for 1–3 min. Then, the cells were incubated with the Live/Dead Cell Imaging Kit for 15–30 min at 37 °C. Finally, we used PBS again to wash out the reagent for 3–5 min and observed the chip under a fluorescent microscope.
I. Statistical analysis
For statistical analysis, all values were obtained from three independent experiments (devices), and the data were expressed as mean ± standard deviation. Values, such as cell number, migration distance, and vessel diameter, were obtained from the fluorescent images by using the image processing and data statistics functions of the ImageJ software and the Angiotool software. Origin software was used to evaluate significant differences by performing unpaired two-tailed t-tests, * means p < 0.05, ** means p < 0.01, *** means p < 0.001, **** means p < 0.0001, and n means the number of independent replicate experiments.
IV. RESULTS AND DISCUSSION
A. Model design and chip design
In order to directly observe tumor vascularization behavior and screen anti-cancer drug lead compounds, we constructed a co-culture model containing vascular network tissue and tumor tissue based on a microfluidic chip. Four types of cells (pri-HUVECs, imm-HUVECs, HMCAF, and MD-MB-231) were used to study microvessel development and tumor vascularization behavior. The structure of this model was improved based on previous research, guaranteeing the communication of juxcrine and paracrine signals between different cells in 3D ECM environment.35 This model was designed to achieve the real-time observation of cancer cell invasion, tumor-related angiogenesis, and subsequently intravasation of cancer cells. The model mainly contains three gel channels (Fig. 1, Ch1–Ch3) for the construction of cell 3D growth environment and two medium channels (Fig. 1, M1–M2) for nutrient supply. The HCMAF fibrinogen solution was added to the middle matrix channel (Fig. 1, Ch2), and the HUVECs’ fibrinogen solution was added to the adjacent side channel 10 min later (Fig. 1, Ch1). These two kinds of cells were co-cultured in the gel for 3 days. During this time, microvascular networks were generated under the stimulation of special medium, and vascularization conditions were similar to the previous works.26–29 Then, the MDA-MB-231 fibrinogen solution was added to the cancer area (Fig. 1, Ch3) at day 3 to finally realize the construction of the vascularized breast cancer model. In the chip, the micro-pillar arrays separated the different channels.
CAFs were very important components in TME6 and even more in tumor margins.36 Many components of the extracellular matrix of cancer are produced by activated CAF,7,37 and it can promote not only tumor angiogenesis8 but also cancer cell invasion.38 Considering such factors, we used human breast cancer-associated fibroblasts (HMCAF) as supporting cells to construct the original TME microvascular networks when selecting the cell types of the matrix channel.
B. Construction of the self-assembled microvascular networks
We used primary HUVECs (pri-HUVECs) as vascular precursor cells to form microvascular networks and compared the growth of immortalized HUVECs (imm-HUVECs) under the same conditions (S3 in the supplementary material). The result intuitively showed that imm-HUVECs did not self-assemble into a blood vessel network, demonstrating the decisive role of primary vascular endothelial cells in the development of microvascular networks.
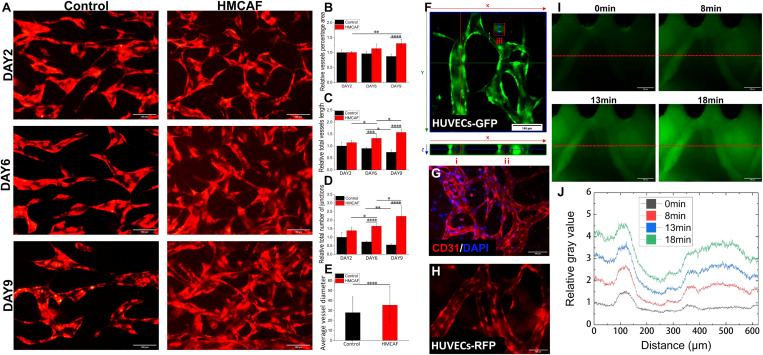
By seeding HMCAF and RFP pri-HUVECs into adjacent channels (Fig. 1, Ch2 and Ch1) to simulate paracrine signals,39 we monitored the self-assembled behaviors of RFP pri-HUVECs and observed the development of microvascular networks for 9 days [Fig. 2(a)]. The results showed that RFP pri-HUVECs had been developed into the microvascular networks at day 2 after perfusion. The microvascular networks formed by RFP pri-HUVECs under the single culture condition gradually faded after culturing for 6–9 days. Then, the values of relative vessel percentage area, relative total vessel length, and relative total number of junctions were quantitatively analyzed [Figs. 2(b)–2(d)]. We found that the values of these three indicators in the HUVECs/HMCAF co-culture group showed a significant upward trend over time, while in the control group (or single culture group), they showed a downward trend. Immunofluorescence staining of the microvascular networks confirmed the robust expression of CD31 [Fig. 2(g)] and the HMCAF marker FAP (S4 in the supplementary material). In addition, we also found that the microvascular networks sprouted in the direction of HMCAF, and then the sprouted vessels developed into larger-diameter vessels [Fig. 2(h)] in the matrix area of HMCAF. These results fully proved that HMCAF cells could help promote the formation and maturation of microvascular networks and maintain the integrity of the structure and function of self-assembled microvascular networks.
FIG. 2.
Vascular function evaluation of self-assembled microvascular networks. (a) The image of blood vessel growth over time under control (HUVECs-RFP single culture) and HMCAF (HUVECs-RFP/HMCAF co-culture) conditions; scale bar: 100 μm. (b)–(d) Under different culture conditions, quantitative results of relative vessel percentage area, relative total vessel length, and relative total number of junctions (compared with the day 2 control group, n = 3). (e) Average vessel diameter of day 6 (n = 3). (f) Confocal cross-sectional view of the blood vessel and the cross-sectional diameter of some blood vessels [(i) 28.4 μm; (ii) 29.8 μm; and (iii) 7.6 μm]; scale bar: 100 μm. (g) CD31 immunofluorescence staining of the self-assembled microvascular networks (CD31, red and DAPI, blue); scale bar: 100 μm. (h) Sprouting vessel morphology in the matrix area of HMCAF (HUVECs-RFP, day 12); scale bar: 100 μm. (i) and (j) The relative fluorescence intensity distribution along the red dotted line at different time points after perfusing the blood vessel with FITC-dextran (70 kDa, green); scale bar: 100 μm.
It is worth mentioning that our microvascular networks or their sprouting structures can be maintained for about 13 days [Fig. 3(a)], while in other studies, they can be maintained at 4–6 days,30,31 which lays the foundation for the construction of complex vascularized tissues and long-term in vitro culture research. Because the microvascular networks were formed in the fibrinogen under a limited space, the maintenance of vascular structure stability in our study was inseparable from the continuous existence of the fibrinogen and the restricted growth of HUVECs. Due to the continuous degradation of fibrinogen in the process of cell metabolism,31 collagen was continuously consumed. Fibroblasts are the main source of the fibrinogen in the matrix.7 Therefore, the co-culture with fibroblasts could effectively maintain the fibrous collagen structure for a longer time, which was one of the main reasons for the long-term maintenance of the vascular structure in our model. On the other hand, the co-culture contributed to the competition between the two cells in both biological nutrients and growth space. The limited space keeps the population numbers of the two types of cells in a stable equilibrium state, which may be another possible reason for the long-term maintenance of the vascular structure.
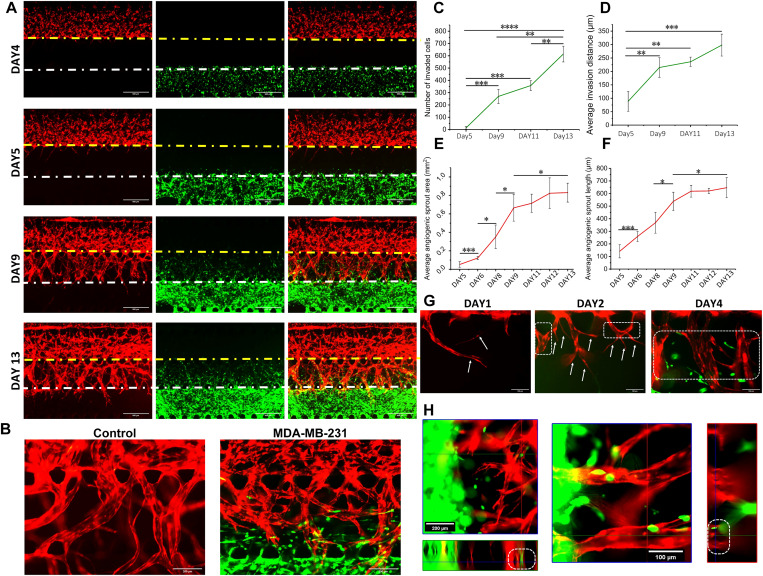
FIG. 3.
Images and analyses of the self-assembled vascularized breast cancer model. (a) The model was imaged on day 4–day 13 under the fluorescence microscope; scale bar: 500 μm. (b) The sprouting vessel morphology under the condition of HUVECs culture alone and HUVECs/MDA-MB-231 co-culture; scale bar: 200 μm. (c) The number of invasions of cancer cells toward the matrix area and (d) the average invasion distance were quantified, taking the pre-existing microvessel region boundary as a starting point. (e) The average sprout area and (f) the average sprout length of the neovascularization toward the matrix region were quantified. (g) The images of angiogenic sprouting on days 5, 6, and 8; white arrow: tip cells; white dashed box: stalk cells; and scale bar: 100 μm. (h) Confocal imaging of the matrix area in the model showed that the cancer cells had performed intravasation (white dashed box); scale bar: 200 (left) and 100 μm (right). HUVECs: with RFP, MDA-MB-231: with GFP, and HMCAF: no fluorescence; (c)–(f): n = 3.
By measuring the average diameter of blood vessels on day 6, we found that the diameters of blood vessels formed by monoculture and co-culture were both at the micrometer level [20–30 μm, Fig. 2(e)], and this was consistent with the diameter of the microvessel networks in human body tissues.40 Next, we observed the cross-sectional view of the blood vessel under the co-culture condition under a confocal microscope, showing that it had a hollow lumen structure [Fig. 2(f)]. After loading the 70 kDa FITC-Dextran solution into the medium channel on day 14 (Fig. 1), the fluorescence intensity in the blood vessel was more potent than the surrounding matrix. Also, the measured fluorescence intensity in the blood vessel was higher than in the surrounding collagen [Fig. 2(j)]. This result meant that the diffusion rate of FITC-Dextran was faster in the sprouting blood vessel than in the surrounding collagen, showing that the original vascular network was perfusable and had a certain blood vessel barrier function.
C. 3D invasion, anglogenesis, and intravasation in self-assembled vascularized breast cancer models
The microvascular area had self-assembled into more physiological microvascular networks after three days of blood vessel development, and then we seeded MDA-MB-231 cells into the chip and set this day as day 4 [Fig. 3(a)]. This operation could exclude the influence of the cancer cells on the vascular development and contributed to the subsequent control analysis. During the culture process [Fig. 3(a)], cancer cells invaded into the matrix area (the white dotted line was the boundary of the cancer cell area and the matrix area), and the blood vessel network started to sprout new blood vessels (the yellow dotted line was the boundary of the microvascular area and the matrix area). The observation and analysis of the process of blood vessel sprouting is of great significance in the blood vessel sprouting research,13,41 and our models could be a very useful platform to meet this need. During sprouting of blood vessels, the tip cells guided the direction of sprouts, and the stalk cells would further undergo vascular anastomosis to form blood vessels with a larger diameter [Figs. 3(a)–3(h)]. In addition, after adding MDA-MB-231 cells to the pre-existing blood vessel network, the morphology of the vessels also changed. On day 13, the blood vessel surface had become rough and tubular structures tended to break because of excessive vascularization [Fig. 3(b)]. In the research of others, they also found that tumor cells could secrete high levels of growth factors to create chaotic blood vessels,42 and this, in turn, exacerbated the progression of tumors. Immature tumor blood vessels also led to hyperosmotic pressure of tumor tissue and, thus, cause difficulty in drug delivery,43 which have become a crucial target of tumor therapy.42 These results also suggested that it was highly necessary to construct the tumor models with the biomimetic blood vessel environment.
Then, through the quantitative analysis over time [Figs. 3(c) and 3(d)], we found that there were significant differences in the number of invaded cells (increased ∼256) and the average invaded distance (increased ∼126 μm) on day 9 compared with day 5. On day 9, the cells had already completed large-scale invasion. From day 9 to day 13, the average invasion distance (increased ∼84 μm) was slower than days 5–9, and there was no significant difference, while the number of invasions increased significantly (about 344). It showed that more cancer cells had invaded, but the range of invasion tended to be stable and concentrated in the HMCAF matrix area. A small number of cancer cells invaded into the pre-existing self-assembled microvascular area (S5 in the supplementary material) on day 13.
The average distance and area of angiogenic sprouts in the matrix area after seeding with MDA-MB-231 cells were analyzed quantitatively [Figs. 3(e) and 3(f)]. According to the results, we found that from day 5 to day 9, both the sprouting length increment and the sprouting area increment were higher than from day 9 to day 13. At day 9, partial angiogenesis had already sprouted into the cancer cell area (S6 in the supplementary material). This meant that days 5–9 were the rapid development stage for tumor-related angiogenesis. During days 9–13, the growth speed of angiogenesis decreased, and angiogenesis was in a phase of slow development. Meanwhile, we imaged the model under a confocal microscope and found that a small number of cancer cells had invaded into the blood vessels [Figs. 3(g) and 3(h) and S-MOVIE1 in the supplementary material] on day 8. This intravasation behavior of cancer cells was a crucial step in cancer metastasis, and after this step, circulating tumor cells were produced, becoming the key to evaluate the progress of tumor metastasis.21 Our model intuitively reflected the biological process from tumor growth to cancer cell intravasation. These results revealed that in the process of tumor metastasis, the invasion of cancer cells to the surrounding stroma and tumor-related angiogenesis were interactive and simultaneous processes. When the new blood vessels overlapped with the invading cancer cells, the intravasation behavior of the cancer cells had already occurred, and this process may happen much earlier than the detection of circulating tumor cells in the blood. At the same time, cancer cells that have invaded into blood vessels at the early stage had the opportunity to obtain more adequate nutrition, and the proliferation of these cells may play a more important role in the process of tumor metastasis. Of course, this guess has yet to be further verified through new understanding in clinical tumor research.
D. Effect of tumor vessels on invasion distance of breast cancer
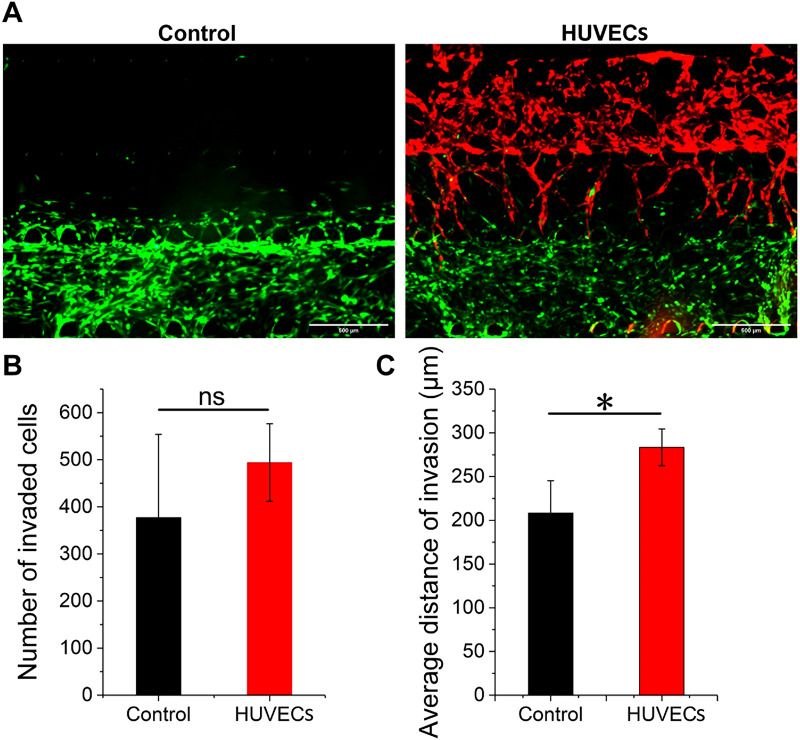
According to the previous analysis of invaded breast cancer cell distances [Figs. 3(c) and 3(d)] and tumor-related angiogenesis [Figs. 3(e) and 3(f)] in the model, we proved the mutual promotion of the cancer cell invasion and tumor-related angiogenesis. Therefore, we set up a control experiment, in which cancer cells grew in the tumor microenvironment without microvascular networks, to further analyze the influence of self-assembled microvascular networks on the invasion ability of cancer cells. According to the invasion imaging of cells on day 12 [Fig. 4(a)] and then a quantitative analysis of the invasion ability of cancer cells [Figs. 4(b) and 4(c)], we found that in the HUVECs group, the number of cancer cells that invaded into channel 2 (Fig. 1, Ch2) was higher than that in the control group (above 31%), but there was no significant difference. In terms of the average invasion distance, the cancer cells in the HUVECs group had stronger invasion ability, indicating that the microvascular networks indeed promoted the invasion of breast cancer cells. This result was consistent with results previously reported in the pancreatic cancer model, and the presence of HUVECs increased the rate of migration of pancreatic cancer cells.24
FIG. 4.
Observation of invasive behavior and evaluation of invasive ability about cancer cells with (HUVECs) and without (control) self-assembled microvascular networks on day 12. (a) The invasion image of MDA-MB-231-GFP cells on day 12; HUVECs: with RFP; HMCAF: no fluorescence; and scale bar: 500 μm. (b) The number of invaded cancer cells and (c) the average invasion distance toward the matrix area (n = 3). Ns: not significant.
E. Drug effect of 5-FU, DOX, and Apatinib on self-assembled vascularized breast cancer models
In our tumor model, it was observed that the invasion behavior of cancer cells and tumor-related angiogenesis together promoted the process of tumor metastasis. In recent years, considering the critical role of tumor blood vessels in cancer metastasis, the combinations of anti-angiogenic drugs and chemotherapeutic drugs became a new tumor treatment strategy.44,45 5-Fluorouracil (5-FU) and Doxorubicin (DOX) are natural anti-tumor lead compounds.46 Apatinib is a common VEGFR inhibitor during drug testing and can effectively inhibit the proliferation,47 migration, and vascular growth of HUVECs for its highly targeted effect on VEGFR-2 to inhibit the phosphorylation and function of VEGF/VEGFR-2 signaling.48,49 Therefore, we used 5-FU, DOX, and Apatinib to treat the self-assembled vascularized breast cancer model to explore suitable and available anti-tumor drug combination strategies.
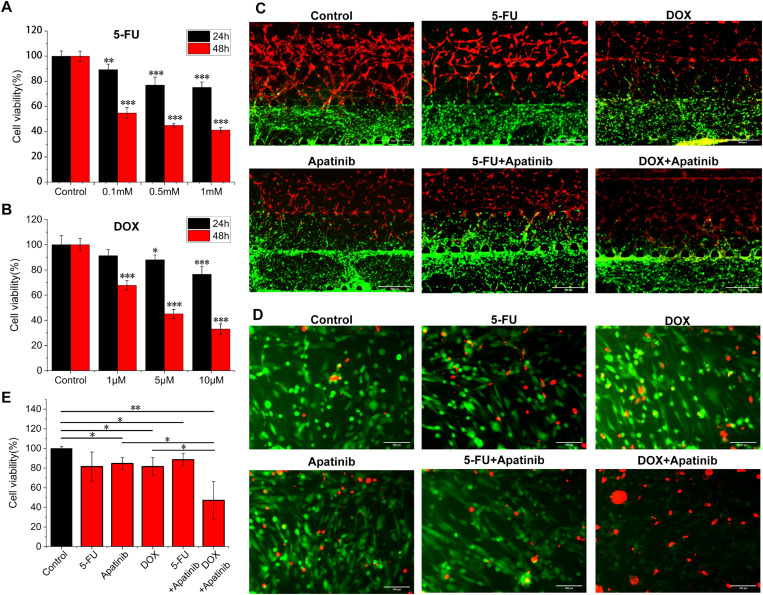
First, we evaluated the cell viability of MDA-MB-231 cells under different concentrations of 5-FU and DOX on a 2D 96-well plate [Figs. 5(a) and 5(b)]. The drug concentrations of two drugs (5-FU: 0.5 mM and DOX: 5 μM), which contributed the same cancer cell viability (about 45%) of under 2D condition for 48 h, were selected to compare the drug effect between 5-FU and DOX in 3D models. The concentration of Apatinib was selected as 10 μM.44 Then, the drugs were uniformly mixed in the medium, and the mixture was loaded into the two medium channels (M1 and M2) simultaneously, enabling a consistent drug action environment. We observed that the blood vessel network was damaged more or less after adding anti-cancer drugs, compared with the control group [Fig. 5(c)]. The damage to the DOX group and DOX/Apatinib collaborative group on the blood vessel network was more than that in the other groups. Moreover, the damage of 5-FU and Apatinib applied together on the blood vessel network was also more than that in 5-FU or Apatinib applied singly. These results verified the inhibitory effect of Apatinib (VEGFR inhibitor) on blood vessels and were also consistent with previous findings that DOX50 and 5-FU46 had a certain vascular toxicity and had the potential to destroy new blood vessels.46
FIG. 5.
Cell viability assay of the vascularization model. The cell viability assay of MDA-MB-231 cells after loading the (a) 5-FU and (b) DOX for 24 and 48 h on the 2D 96-well plates; significant differences were compared with the control group at the same time (n = 3). (c) The merged fluorescence images of the self-assembled vascularized breast cancer model after loading the drug for 48 h; HUVECs (in red): with RFP, MDA-MB-231 (in green): with GFP, HMCAF: no fluorescence, and scale bar: 500 μm. (d) Live–dead assay and (e) cell viability analysis of the MDA-MB-231 cells in the model at day 6 (n = 3); red: dead cells; green: live cells; and scale bar: 100 μm.
Then, we performed a live/dead assay to evaluate the cancer cell viability in the vascularized breast cancer model. From the overall death information of the model, we found that anti-cancer drugs caused varying degrees of killing on all three types of cells (S7 in the supplementary material). We merged the live/dead images [Fig. 5(d)] and calculated the cell viability of the MDA-MB-231 area [Fig. 5(e)]. Since the drug tests were performed for 2 days after the cancer cells were loaded on chip, we thought that the proportion of other cells migrating into the cancer cell gel area was low, and the statistical effect of other cells on the cancer cell viability was not considered. Compared with the cell viability (∼45%) measured at the same concentration in the 2D model, the cell viability of the 5-FU group and the DOX group was about 81%, which was about 36% higher, indicating that the cancer cells in 3D biomimetic models had a stronger drug resistance on 5-FU and DOX. This result was consistent with that of previous studies, which proved that the 3D breast cancer models had lower 5-FU51,52 and DOX51,53 drug sensitivity than the 2D model. Compared with 5-FU (∼81%) and Apatinib (∼85%) used alone, there was no significant difference in cell viability (∼89%) after the combined use. Compared with DOX (∼81%) and Apatinib (∼85%) used alone, the cell viability of combined use (∼47%) was significantly reduced, indicating that the addition of Apatinib significantly enhanced the DOX drug efficiency on MDA-MB-231 cells.
V. CONCLUSIONS
In this work, we developed a biomimetic breast cancer model containing self-assembled microvascular networks and the HMCAF matrix. The decisive role of primary vascular endothelial cells in the development of microvascular networks in vitro has been proved, and the HMCAF cells played an important role in maintaining the structure and function of the microvascular networks for a long time. Therefore, the blood vessel network developed in our work has achieved the morphology and function maintenance for up to 13 days, which was much higher than the reported time of the existing studies. In our self-assembled vascularized breast cancer model, the invasion behavior of tumor cells and the phenomenon of tumor vascularization occurred at the same time, and the results of related quantitative analysis also showed that the two promote each other. We also observed that the intravasation behavior already existed in the early stage of tumor invasion. This suggests that, although these intravasation cancer cells were unable to enter the main blood vessel channel, due to the sufficient blood nutrient supply, their proliferation behavior in blood vessels may play an important role in the process of tumor metastasis. In the drug test, the drug effects were assessed from the aspects of the cancer cell invasion behavior, the vascular endothelial behavior, and cell viability. The results showed that this cancer model had more consistent drug resistance with the human body, compared with the 2D models. Meanwhile, the anti-angiogenic drug Apatinib enhanced the treatment of DOX on breast cancer cells, instead indicating that the combination strategy of anti-cancer drugs and anti-angiogenic drugs can become an important means of cancer treatment. In general, our research provided a more bionic and quantitatively analyzable model for the construction of in vitro breast cancer models and the testing of anti-cancer drugs.
SUPPLEMENTARY MATERIAL
See the supplementary material for the following figures: schematic diagram of quantifying blood vessels and blood vessel sprouting using ImageJ software (S1 and S2); HUVEC growth image and immunofluorescence staining (S3); the picture of HMCAF marker fibroblast activation protein (FAP) immunofluorescence staining (S4); the number of invaded cancer cells corresponding to invasion distance range on day 13 (S5); details of the breast cancer model growth on days 4, 5, 9, and 13 (S6); and the death image of the self-assembled vascularized breast cancer model (on day 8) after drug action for 2 days (S7). MOVIE1 was the 3D confocal image of the vascular structure in the self-assembled vascularized breast cancer model on day 8.
ACKNOWLEDGMENTS
This work was funded by the Project of Basic Research of Shenzhen, China (Nos. JCYJ20180507183655307 and JCYJ20190813143221901). Shengli Mi received the funding. The funders had no role in the study design, data collection, and analysis and in the decision on the publication or preparation of the manuscript.
Contributor Information
Ke Zhang, Email: mailto:zhangkescience@126.com.
Zhichang Du, Email: mailto:202061000118@jmu.edu.cn.
Shengli Mi, Email: mailto:mi.shengli@sz.tsinghua.edu.cn.
AUTHOR DECLARATIONS
Conflict of Interest
The authors have no conflicts to disclose.
Author Contributions
K.Z., Z.D., and T.Y. contributed equally to this work.
Author Contributions
Ke Zhang: Software (equal); Supervision (equal). Zhichang Du: Data curation (equal); Investigation (equal); Visualization (equal); Writing – original draft (lead); Writing – review and editing (lead). Tianying Yuan: Methodology (equal); Project administration (equal). Jiajun Huang: Investigation (equal); Methodology (equal); Software (equal). Xiaoyu Zhao: Data curation (equal); Formal analysis (equal). Shengli Mi: Conceptualization (equal); Data curation (equal); Funding acquisition (equal).
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1.Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., and Bray F., CA Cancer J. Clin. 71(3), 209–249 (2021). 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Harbeck N. and Gnant M., Lancet 389, 1134–1150 (2017). 10.1016/S0140-6736(16)31891-8 [DOI] [PubMed] [Google Scholar]
- 3.Junttila M. R. and de Sauvage F. J., Nature 501, 346–354 (2013). 10.1038/nature12626 [DOI] [PubMed] [Google Scholar]
- 4.Nurmik M., Ullmann P., Rodriguez F., Haan S., and Letellier E., Int. J. Cancer 146, 895–905 (2020). 10.1002/ijc.32193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peela N., Danh T., Saini H., Chu H., Mashaghi S., Ham S. L., Singh S., Tavana H., Mosadegh B., and Nikkhah M., Biomaterials 133, 176–207 (2017). 10.1016/j.biomaterials.2017.04.017 [DOI] [PubMed] [Google Scholar]
- 6.Chen X. M. and Song E. W., Nat. Rev. Drug Discovery 18, 99–115 (2019). 10.1038/s41573-018-0004-1 [DOI] [PubMed] [Google Scholar]
- 7.Kalluri R., Nat. Rev. Cancer 16, 582–598 (2016). 10.1038/nrc.2016.73 [DOI] [PubMed] [Google Scholar]
- 8.De Palma M., Biziato D., and Petrova T. V., Nat. Rev. Cancer 17, 457–474 (2017). 10.1038/nrc.2017.51 [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D. and Coussens L. M., Cancer Cell 21, 309–322 (2012). 10.1016/j.ccr.2012.02.022 [DOI] [PubMed] [Google Scholar]
- 10.Lee S. H., Jeong D., Han Y. S., and Baek M. J., Ann. Surg. Treat. Res. 89, 1–8 (2015). 10.4174/astr.2015.89.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su S., Chen J., Yao H., Liu J., Yu S., Lao L., Wang M., Luo M., Xing Y., Chen F., Huang D., Zhao J., Yang L., Liao D., Su F., Li M., Liu Q., and Song E., Cell 172, 841–856 (2018). 10.1016/j.cell.2018.01.009 [DOI] [PubMed] [Google Scholar]
- 12.Shintani Y., Fujiwara A., Kimura T., Kawamura T., Funaki S., Minami M., and Okumura M., J. Thorac. Oncol. 11, 1482–1492 (2016). 10.1016/j.jtho.2016.05.025 [DOI] [PubMed] [Google Scholar]
- 13.Potente M., Gerhardt H., and Carmeliet P., Cell 146, 873–887 (2011). 10.1016/j.cell.2011.08.039 [DOI] [PubMed] [Google Scholar]
- 14.Bergers G. and Benjamin L. E., Nat. Rev. Cancer 3, 401–410 (2003). 10.1038/nrc1093 [DOI] [PubMed] [Google Scholar]
- 15.Carmeliet P. and Jain R. K., Nature 407, 249–257 (2000). 10.1038/35025220 [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D. and Weinberg R. A., Cell 144, 646–674 (2011). 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 17.Costard L. S., Hosn R. R., Ramanayake H., O’Brien F. J., and Curtin C. M., Acta Biomater. 132(132), 360–378 (2021). 10.1016/j.actbio.2021.01.023 [DOI] [PubMed] [Google Scholar]
- 18.Jeon J. S., Bersini S., Gilardi M., Dubini G., Charest J. L., Moretti M., and Kamm R. D., Proc. Natl. Acad. Sci. U.S.A. 112, 214–219 (2015). 10.1073/pnas.1417115112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montanez-Sauri S. I., Beebe D. J., and Sung K. E., Cell. Mol. Life Sci. 72, 237–249 (2015). 10.1007/s00018-014-1738-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts S., Peyman S., and Speirs V., Adv. Exp. Med. Biol. 1152, 413–427 (2019). 10.1007/978-3-030-20301-6_22 [DOI] [PubMed] [Google Scholar]
- 21.Reymond N., d’Agua B. B., and Ridley A. J., Nat. Rev. Cancer 13, 858–870 (2013). 10.1038/nrc3628 [DOI] [PubMed] [Google Scholar]
- 22.Khuon S., Liang L., Dettman R. W., Sporn P. H. S., Wysolmerski R. B., and Chew T.-L., J. Cell Sci. 123, 431–440 (2010). 10.1242/jcs.053793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sontheimer-Phelps A., Hassell B. A., and Ingber D. E., Nat. Rev. Cancer 19, 65–81 (2019). 10.1038/s41568-018-0104-6 [DOI] [PubMed] [Google Scholar]
- 24.Nguyen D. H. T., Lee E., Alimperti S., Norgard R. J., Wong A., Lee J. J. K., Eyckmans J., Stanger B., and Chen C. S., Sci. Adv. 5, eaav6789 (2019). 10.1126/sciadv.aav6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polacheck W. J., Kutys M. L., Tefft J. B., and Chen C. S., Nat. Protoc. 14, 1425–1454 (2019). 10.1038/s41596-019-0144-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen M. B., Whisler J. A., Froese J., Yu C., Shin Y. J., and Kamm R. D., Nat. Protoc. 12, 865–880 (2017). 10.1038/nprot.2017.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paek J., Park S. E., Lu Q. Z., Park K.-T., Cho M., Oh J. M., Kwon K. W., Yi Y. S., Song J. W., Edelstein H. I., Ishibashi J., Yang W. L., Myerson J. W., Kiseleva R. Y., Aprelev P., Hood E. D., Stambolian D., Seale P., Muzykantov V. R., and Huh D., ACS Nano 13, 7627–7643 (2019). 10.1021/acsnano.9b00686 [DOI] [PubMed] [Google Scholar]
- 28.Kim S., Chung M., Ahn J., Lee S., and Jeon N. L., Lab Chip 16, 4189–4199 (2016). 10.1039/c6lc00910g [DOI] [PubMed] [Google Scholar]
- 29.Hajal C., Ibrahim L., Serrano J. C., Offeddu G. S., and Kamm R. D., Biomaterials 265, 120470 (2021). 10.1016/j.biomaterials.2020.120470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S., Lee H., Chung M., and Jeon N. L., Lab Chip 13, 1489–1500 (2013). 10.1039/c3lc41320a [DOI] [PubMed] [Google Scholar]
- 31.Nagaraju S., Truong D., Mouneimne G., and Nikkhah M., Adv. Healthcare Mater. 7, 1701257 (2018). 10.1002/adhm.201701257 [DOI] [PubMed] [Google Scholar]
- 32.Zudaire E., Gambardella L., Kurcz C., and Vermeren S., PLoS One 6, e27385 (2011). 10.1371/journal.pone.0027385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segarra M., Aburto M. R., Cop F., Llao-Cid C., Hartl R., Damm M., Bethani I., Parrilla M., Husainie D., Schanzer A., Schlierbach H., Acker T., Mohr L., Torres-Masjoan L., Ritter M., and Acker-Palmer A., Science 361, 15 (2018). 10.1126/science.aao2861 [DOI] [PubMed] [Google Scholar]
- 34.Schneider C. A., Rasband W. S., and Eliceiri K. W., Nat. Methods 9, 671–675 (2012). 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du Z., Mi S., Yi X., Xu Y., and Sun W., Biofabrication 10, 034102 (2018). 10.1088/1758-5090/aac70c [DOI] [PubMed] [Google Scholar]
- 36.Quail D. F. and Joyce J. A., Nat. Med. 19, 1423–1437 (2013). 10.1038/nm.3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomasek J. J., Gabbiani G., Hinz B., Chaponnier C., and Brown R. A., Nat. Rev. Mol. Cell Biol. 3, 349–363 (2002). 10.1038/nrm809 [DOI] [PubMed] [Google Scholar]
- 38.Dumont N., Liu B., DeFilippis R. A., Chang H., Rabban J. T., Karnezis A. N., Tjoe J. A., Marx J., Parvin B., and Tlsty T. D., Neoplasia 15, 249–262 (2013). 10.1593/neo.121950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan S. Y., Leung Z. W., and Wu A. R., Small 16, 1905055 (2020). 10.1002/smll.201905055 [DOI] [PubMed] [Google Scholar]
- 40.Braverman I. M., J. Investig. Dermatol. Symp. Proc. 5, 3–9 (2000). 10.1046/j.1087-0024.2000.00010.x [DOI] [PubMed] [Google Scholar]
- 41.Carmeliet P. and Jain R. K., Nature 473, 298–307 (2011). 10.1038/nature10144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viallard C. and Larrivée B., Angiogenesis 20, 409–426 (2017). 10.1007/s10456-017-9562-9 [DOI] [PubMed] [Google Scholar]
- 43.Heldin C. H., Rubin K., Pietras K., and Ostman A., Nat. Rev. Cancer 4, 806–813 (2004). 10.1038/nrc1456 [DOI] [PubMed] [Google Scholar]
- 44.Tang D., Ma J., Chu Z., Wang X., Zhao W., and Zhang Q., Am. J. Transl. Res. 12, 3741–3753 (2020), available at https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7407711. [PMC free article] [PubMed] [Google Scholar]
- 45.Han S., Kim S., Chen Z., Shin H. K., Lee S. Y., Moon H. E., Paek S. H., and Park S., Int. J. Mol. Sci. 21, 2993 (2020). 10.3390/ijms21082993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soultati A., Mountzios G., Avgerinou C., Papaxoinis G., Pectasides D., Dimopoulos M. A., and Papadimitriou C., Cancer Treat. Rev. 38, 473–483 (2012). 10.1016/j.ctrv.2011.09.002 [DOI] [PubMed] [Google Scholar]
- 47.Zhang H. J., Drug Des. Devel. Ther. 9, 6075–6081 (2015). 10.2147/DDDT.S97235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian S., Quan H. T., Xie C. Y., Guo H. Y., Lue F. F., Xu Y. P., Li J., and Lou L. G., Cancer Sci. 102, 1374–1380 (2011). 10.1111/j.1349-7006.2011.01939.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng M. M., Zha J., Zhao H. J., Jia X., Shi Y. F., Li Z. F., Fu G., Yu L., Fang Z. H., and Xu B., Exp. Cell Res. 390, 111934 (2020). 10.1016/j.yexcr.2020.111934 [DOI] [PubMed] [Google Scholar]
- 50.Sawicki K. T., Sala V., Prever L., Hirsch E., Ardehali H., and Ghigo A., Annu. Rev. Pharmacol. Toxicol. 61, 309–332 (2021). 10.1146/annurev-pharmtox-030620-104842 [DOI] [PubMed] [Google Scholar]
- 51.Yan X. J., Zhou L., Wu Z. Z., Wang X., Chen X. Y., Yang F., Guo Y. A., Wu M., Chen Y. Y., Li W. J., Wang J., and Du Y. A., Biomaterials 198, 167–179 (2019). 10.1016/j.biomaterials.2018.05.020 [DOI] [PubMed] [Google Scholar]
- 52.Li W. F., Hu X. Y., Wang S. P., Xing Y., Wang H., Nie Y., Liu T. Q., and Song K. D., Int. J. Biol. Macromol. 130, 166–176 (2019). 10.1016/j.ijbiomac.2019.02.136 [DOI] [PubMed] [Google Scholar]
- 53.Bray L. J., Binner M., Holzheu A., Friedrichs J., Freudenberg U., Hutmacher D. W., and Werner C., Biomaterials 53, 609–620 (2015). 10.1016/j.biomaterials.2015.02.124 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See the supplementary material for the following figures: schematic diagram of quantifying blood vessels and blood vessel sprouting using ImageJ software (S1 and S2); HUVEC growth image and immunofluorescence staining (S3); the picture of HMCAF marker fibroblast activation protein (FAP) immunofluorescence staining (S4); the number of invaded cancer cells corresponding to invasion distance range on day 13 (S5); details of the breast cancer model growth on days 4, 5, 9, and 13 (S6); and the death image of the self-assembled vascularized breast cancer model (on day 8) after drug action for 2 days (S7). MOVIE1 was the 3D confocal image of the vascular structure in the self-assembled vascularized breast cancer model on day 8.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.