Abstract
In contrast to gram-negative bacteria, little is known about the mechanisms by which gram-positive bacteria degrade the toxic metabolic intermediate methylglyoxal (MG). Clostridium beijerinckii BR54, a Tn1545 insertion mutant of the NCIMB 8052 strain, formed cultures that contained significantly more (free) MG than wild-type cultures. Moreover, BR54 was more sensitive to growth inhibition by added MG than the wild type, suggesting that it has a reduced ability to degrade MG. The single copy of Tn1545 in this strain lies just downstream from gldA, encoding glycerol dehydrogenase. As a result of antisense RNA production, cell extracts of BR54 possess significantly less glycerol dehydrogenase activity than wild-type cell extracts (H. Liyanage, M. Young, and E. R. Kashket, J. Mol. Microbiol. Biotechnol. 2:87–93, 2000). Inactivation of gldA in both C. beijerinckii and Clostridium difficile gave rise to pinpoint colonies that could not be subcultured, indicating that glycerol dehydrogenase performs an essential function in both organisms. We propose that this role is detoxification of MG. To our knowledge, this is the first report of targeted gene disruption in the C. difficile chromosome.
Methylglyoxal (MG), a toxic electrophile, interacts with negatively charged nucleophilic centers of cellular macromolecules in both eukaryotic and prokaryotic cells (16, 18). It is a common intermediate in the nonenzymatic glycation of proteins (Maillard reaction), where it forms adducts with arginine, lysine, and cysteine (24, 29, 31, 42). MG is a major precursor of advanced glycation end products which appear to contribute to the pathologies associated with age and the complications of diabetes (4, 39). MG also reacts with guanine, as well as adenine and cytosine, in nucleic acids and is mutagenic in bacteria (20, 25). MG inhibits bacterial growth, and at higher concentrations it is bactericidal.
MG is synthesized in Escherichia coli and other organisms during normal glycolysis by conversion from dihydroxyacetone phosphate (DHAP) with MG synthase (summarized in reference 19). Although toxic, MG is thought to play an important role in bacterial growth under conditions in which there may be excessive accumulation of sugar phosphates (14). Accumulation of toxic levels of sugar phosphates is prevented by diversion of carbon flow via the MG bypass. This is a risky but effective maneuver and allows bacteria to survive within the limits imposed by the toxicity of the electrophile. The MG bypass is used by Clostridium sphenoides under phosphate limitation conditions to convert DHAP to MG (40). The enzymes of the MG bypass, including MG synthase and MG reductase, have been found in cell extracts (40).
Detoxification of MG is essential for cells. In bacteria, this is best understood in E. coli and probably occurs by similar reactions in other gram-negative bacteria (for reviews see references 12, 14, 16, and 39). In E. coli MG detoxification is effected primarily by a glutathione-dependent glyoxalase system. E. coli mutants with deficient glyoxalase enzymes are highly sensitive to MG, while MG-resistant mutants have been reported that have increased glyoxylase I and II activities (28). Additional glutathione-independent MG detoxification pathways seem to play a minor role in E. coli.
Little, if anything, is known about the mechanisms that protect cells against electrophiles in gram-positive bacteria. These organisms are generally thought to lack glutathione (10), and KefB and KefC activities, which can protect E. coli against MG toxicity (13), are not present. Bacillus stearothermophilus is a notable exception, since it possesses MG synthase and appears to have evolved or acquired the glyoxalase I-glyoxalase II pathway (5).
In this paper we show that glycerol dehydrogenase activity is essential for cellular viability of two clostridial species, Clostridium beijerinckii and Clostridium difficile. Our evidence suggests that this enzyme plays an important role in MG detoxification in these bacteria.
MATERIALS AND METHODS
Growth of bacteria.
C. beijerinckii NCIMB 8052 and C. difficile CD37 (a gift from A. P. Roberts, Eastman Dental Centre, London, United Kingdom) were routinely grown under anaerobic conditions at 37°C in medium T.5 containing 0.5% (wt/vol) glucose, as described previously (21). Strain BR54 was generated by mutagenesis of C. beijerinckii NCIMB 8052 with Tn1545 (23) and was selected on agar plates containing medium T.5 supplemented with erythromycin (25 μg/ml) and 1-butanol (12 g/liter) (21). Strain BR54 cells were grown in medium T.5 supplemented with 25 μg of erythromycin per ml. E. coli cells were routinely grown in Luria-Bertani (LB) medium (34) supplemented with the following antibiotics, as required: ampicillin (100 μg/ml), tetracycline (12.5 μg/ml), and kanamycin (50 μg/ml).
Northern blots.
Heat-activated spores of C. beijerinckii NCIMB 8052 and its mutant, BR54, were used to inoculate medium T.5 for overnight growth; erythromycin (25 μg/ml) was added to the BR54 culture. Samples (3 ml) of each overnight culture were inoculated into 200 ml of medium T (21) with 2% (wt/vol) glycerol and grown overnight to a final optical density of 0.3 to 0.4 (10). RNA was extracted as described previously (10) by using an RNeasy kit (Qiagen Inc., Santa Clarita, Calif.) according to the manufacturer's directions. The RNA was treated with RNase-free DNase (Stratagene, La Jolla, Calif.) and repurified on an RNeasy column, and total RNA was electrophoresed according to the manufacturer's instructions. A 32P-labelled probe was prepared by random primer labelling of the entire gldA gene obtained by PCR amplification with primers GLDF and GLDR (Table 1) and with wild-type chromosomal DNA as the template. Hybridizations were carried out by using standard methods.
TABLE 1.
Primer sequences and coordinates
| Primer | Sequence (5′→3′)a | Coordinatesb |
|---|---|---|
| BRE1 | GCAGTATCTTCTTTTGAAGTAGCAGCC | 1175–1149 |
| GLDAF2 | catgtctagaATTGCACCAAGTAAATACA | 181–199 |
| GLDAR2 | gatcctcgagTTAATGTATCATAGCATAG | 799–781 |
| GDHF6 | tctcggtaccGAAAATCAATTAACTGTTCTCATT | 409–432 |
| GLDR | acatccatggGTCCTAATCTGTTAGCTGT | 1245–1229 |
| GLDR1 | acattctagaGTCCTAATCTGTTAGCTGT | 1245–1229 |
| GLDF | actaggtaccAAGCTTCATCATCACCTA | 1–18 |
| GDHF4 | AGCTCCAGTTCGTCTTTTAGTATCT | 642–660 |
| GDHR4 | CACAACAGGAAGATTTGCATAG | 501–481 |
| GDH3 | GCTTGAGCTGTAATAGTAGTAGTACCA | 764–744 |
| GDHR3 | GAAAATCAATTAACTGTTCTCATTGG | 409–434 |
| CDF1 | atgctctagaTTAATGATGTGGGAAAAGAAGTAT | 51–76 |
| CDR1 | gcctctcgagTGGGCAAGTGCATCAAATC | 590–572 |
| CDF11 | aattatctagaGATTTGATGCACTTGCCCA | 572–590 |
| GLCDR | attatctcgagTTATAAAGACTAATGGTAC | 1131–1112 |
| CDF5 | GATTTGATGCACTTGCCCA | 572–590 |
| CDR5 | ATACTTCTTTTCCCACATCATTAA | 76–53 |
| MTL1 | GAATTCGAGCTCGGTACCCGGGGA | |
| MTL2 | TCCCCGGGTACCGAGCTCGAATTC | |
| ECGLDF1 | ctctggatccGCATTTGGCACTACTCATC | 4–24 |
| ECGLDR1 | agtaggatccATATTCGGGTTATTTGGCAA | 486–465 |
| ECGLDR2 | ATTTTCCAGAACCAGCTG | 900–878 |
Mismatched bases are indicated by lowercase letters, and introduced restriction sites are in italics.
The positions of primers are given in relation to the DNA segments encompassing C. beijerinckii and C. difficile gldA in Fig. 1.
C. beijerinckii gldA mutants.
Experiments designed to disrupt gldA in wild-type strain C. beijerinckii NCIMB 8052 were carried out as follows. The desired portions of the gldA gene were PCR amplified from wild-type DNA with appropriate primers derived from the DNA sequence (Fig. 1). The PCR products (see below) were cloned into plasmid pMTL31 (Apr Emr) (43), which is not able to replicate in gram-positive bacteria. The recombinant plasmids containing the desired insertions were electroporated into E. coli HB101 harboring the IncP helper plasmid R702 (Kmr Apr) (43). This strain has all the trans-acting proteins needed to mobilize other plasmids containing IncP oriT, including pMTL31 and pCTC1.
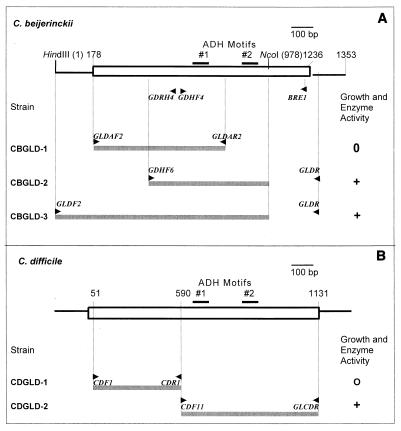
FIG. 1.
(A) Inactivation of gldA in C. beijerinckii NCIMB 8052. At the top is a diagrammatic representation of a 1,353-bp DNA fragment encompassing the gldA coding sequence (box). The gray bars represent the fragments used to construct strains CBGLD-1, CBGLD-2, and CBGLD-3. (B) Inactivation of gldA in C. difficile CD37. At the top is a physical map of gldA, and the DNA fragments used to generate strains CDGLD-1 and CDGLD-2 are represented by gray bars.
Plasmid pCBGLD-1, containing an internal fragment of the coding sequence of C. beijerinckii gldA, was constructed by cloning the XbaI- and XhoI-trimmed PCR product obtained with primers GLDAF2 and GLDAR2 into pMTL31. Plasmid pCBGLD-2, also containing an internal fragment of the coding sequence of C. beijerinckii gldA, was constructed by cloning the KpnI- and NcoI-trimmed PCR product obtained with primers GDHF6 and GLDR into pMTL31. Plasmid pCBGLD-3, containing a fragment of C. beijerinckii gldA including the 5′ end of the coding sequence, was constructed by cloning the KpnI- and NcoI-trimmed PCR product obtained with primers GLDF2 and GLDR into pMTL31. Note that the inserts in pCBGLD-2 and pCBGLD-3 are truncated at an internal Ncol site within gldA (Fig. 1).
C. beijerinckii NCIMB 8052 wild-type cells were mated with E. coli HB101 harboring R702 and the constructed pCBGLD plasmids (43) by using a procedure based on that described by Williams et al. (43). Medium T.5 (50 ml) was inoculated with 1.0 ml of a C. beijerinckii NCIMB 8052 culture grown overnight in the same medium from heat-activated spores. The cultures were grown anaerobically at 37°C to the mid-exponential phase. Donor cells (E. coli HB101 cells containing plasmid R702 and the desired pCBGLD plasmid) were grown aerobically to the mid-exponential phase in LB medium supplemented with ampicillin (100 μg/ml) and kanamycin (50 μg/ml). Both donor and recipient cultures were pelleted in an anaerobic chamber, washed three times with medium T.5, and suspended in 0.5 ml of medium T.5. In separate tubes 10-, 20-, 30-, and 40-μl aliquots of the donor suspension were mixed with 40, 30, 20, and 10 μl of the recipient suspension, respectively. Each mixture of cells was placed separately on a spot on an RCM (Oxoid, Unipath Ltd., Basingstoke, United Kingdom) plate and incubated overnight at 37°C. The cells in each spot, which represented independent mating events, were harvested and suspended separately in 1 ml of medium T.5. Samples (100 μl) were plated on CBM plates (30) containing erythromycin (25 μg/ml). Individual colonies were restreaked on the same medium to obtain single colonies.
Plasmid integration in strain CBGLD-1 was verified by direct PCR amplification using freshly appearing pinpoint colonies obtained after primary transconjugants were restreaked and divergently oriented primers GDHF4 and GDHR4.
The KpnI- and XbaI-trimmed PCR product obtained with primers GLDF and GLDR1, which included 177 bp of DNA flanking the 5′ end (Fig. 1), was cloned into replicative plasmid pCTC1 (Apr Emr) (43), which contains oripAMβ1 conferring autonomous replication in clostridia. The presence of the insert was verified in plasmid preparations obtained from a C. beijerinckii transconjugant by PCR performed with primers GDH3 and GDHR3, which yielded a product of the expected size (ca. 360 bp).
Disruption of C. difficile gldA.
The C. difficile gldA gene was identified by comparison with C. beijerinckii gldA. A BLAST analysis (2) was carried out by using amino acid translation of the gldA nucleotide sequence of C. beijerinckii NCIMB 8052 as the query. The C. difficile nucleotide sequence database (Sanger Centre Pathogen Sequencing Unit) contains an open reading frame whose translated amino acid sequence is similar to that of C. beijerinckii gldA (26% identity and 44% similarity). The approach used was similar to that used for gene disruption in C. beijerinckii. An internal fragment of gldA was amplified from C. difficile DNA with primers CDF1 and CDR1 and was cloned into pMTL31 to obtain plasmid pCDGLD-1. As a control, a fragment containing the 3′ end of gldA was amplified from C. difficile DNA with primers CDF11 and GLCDR and was cloned into pMTL31 to obtain plasmid pCDGLD-2. Integration of plasmids pCDGLD-1 and pCDGLD-2 into the C. difficile genome generated strains CDGLD-1 and CDGLD-2, respectively. Plasmid integration in strain CDGLD-1 was verified by direct PCR amplification using freshly appearing pinpoint colonies with divergently oriented primers CDF5 and CDR5. For verification of plasmid integration in strain CDGLD-2, direct PCR amplification using freshly appearing colonies was carried out with primer CDF1 and vector-specific primer MTL2.
Disruption of gldA in E. coli.
Gene disruption was carried out with a fragment of the E. coli gldA gene lacking the translation start codon and the two alcohol dehydrogenase (ADH) signature motifs. The insert was generated by PCR amplification with primers ECGLDF1 and ECGLDR1, cloned into pKO3 (22), and transformed into strains XL-1 Blue (Stratagene) and Lin 204 (= Coli Genetic Stock Center strain 5995). The latter strain lacks both glycerol kinase and an aerobic glycerol-3-phosphate dehydrogenase (35). PCR amplification with the vector-specific primer pKO3-L (22) and the gldA-specific primer ECGLDR2 yielded a 0.9-kb product, as expected, confirming that integration had taken place.
MG measurement.
Free MG concentrations were determined by a modification of the methods of Barros et al. (3) and Cordeiro and Ponces Freire (9), based on the method of McLellan et al. (26). Reversibly bound MG (24) concentrations were determined by incubating the samples with 2,3-dimethylquinoxaline for 24 h according to the method of Chaplen et al. (7) in order to permit MG to dissociate from soluble cellular components. Samples were acidified with 0.1 volume of 5 M perchloric acid to prevent nonenzymatic formation of MG from cellular triose phosphates. The acidified samples were incubated for 30 min at room temperature with 1,2-diaminobenzene (920 μM) plus dimethylquinoxaline (500 nM) as an internal standard. Preliminary experiments confirmed that formation of quinoxaline from pure MG, which was monitored over time at 336 nm, was almost complete after 6 min. The mixtures were applied to solid-phase extraction cartridges containing 500 mg of Accubond C18 sorbent (J&W Scientific, Folsom, Calif.), washed with 20 mM ammonium phosphate (pH 2.3), and eluted with 2.5 ml of methanol. The methanol was evaporated under N2 at 40°C, and the samples were redissolved in elution buffer consisting of 40% (vol/vol) 25 mM ammonium formate (pH 3.4) and 60% (vol/vol) methanol. High-performance liquid chromatography was carried out with a LiChroshper RP-18 column (250 by 4 mm; Hewlett-Packard Co., Wilmington, Del.) at a rate of 0.2 ml/min by using a high-performance liquid chromatography system (Dionex Corp., Sunnyvale, Calif.) and UV detection at 320 nm (Waters 484 tunable absorbance detector; Millipore Corp., Milford, Mass.). Experiments demonstrated that there was a straight-line increase in A320 from 0 to at least 10 μM MG.
Enzyme assays.
GldA activities in crude extracts of C. beijerinckii cells were measured as described previously (23). MG reductase activity was assayed by measuring the decrease in absorbance at 340 nm during MG-dependent oxidation of NADH at 37°C under aerobic conditions. The reaction mixtures consisted of 100 mM MES (morpholinoethanesulfonic acid) buffer (pH 6.5), 100 mM KCl, 3 to 5 mg of protein of crude C. beijerinckii cell extract (23), 0.1 mM NADH, 1 mM MG, and enough water to bring the total volume to 1.0 ml.
RESULTS
Cultures of BR54 contain significantly more free MG than wild-type cultures.
Owing to its lower GldA activity (23), strain BR54 was expected to accumulate higher levels of MG than the wild type. This was found to be the case. The concentrations of free MG were 74 ± 58 nM (mean ± standard deviation; n = 15 for all values presented here) in wild-type cultures and 132 ± 68 nM in mutant BR54 cultures. Thus, the free MG concentration was low in both cultures but was significantly higher in the mutant cultures (P = 0.019), a difference consistent with the hypothesis that there is less MG detoxification in the mutants. The concentrations of MG reversibly bound to soluble components were the same in the two types of culture (P = 0.67); they were 451 ± 94 nM in the wild type and 427 ± 197 nM in the mutant. The MG concentrations were the same in cell cultures and in supernatants of pelleted cells, indicating that both forms of MG moved readily from the cells to the growth medium.
C. beijerinckii BR54 is more sensitive to added MG than the wild type.
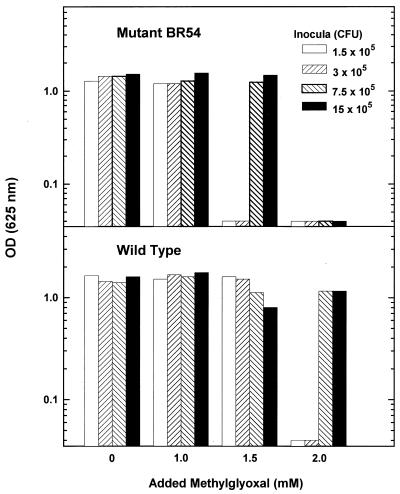
Since strain BR54 possesses only 25% of the GldA activity of the parental strain (23), we expected that the mutants would be more sensitive to added MG than the wild type. The mutants were unable to grow with 1.5 mM added MG when the medium (5 ml) was inoculated with 1.5 × 105 or 3 × 105 CFU, whereas the wild type grew equally well with and without added MG (Fig. 2). When higher inoculum densities were used, both the mutant and the wild type grew in the presence of 1.5 mM MG. When 2 mM MG was added, however, the mutant was unable to grow, and only cultures which received larger inocula (>7.5 × 105 CFU) of the wild type were able to grow. Presumably, only when sufficient GldA was present in the culture to detoxify enough of the added MG could growth be initiated.
FIG. 2.
Effect of MG on growth of wild-type strain C. beijerinckii NCIMB 8052 and mutant BR54. Cultures (5 ml) in medium T.5 were inoculated with 10 to 100 μl (1.5 × 105 to 15 × 105 CFU) of cells grown overnight from activated spores. MG was added at zero time, and the cultures were incubated for 2 days at 37°C in an anaerobic chamber. OD (625 nm), optical density at 625 nm.
gldA is an essential gene in C. beijerinckii NCIMB 8052.
The gldA gene of C. beijerinckii NCIMB 8052 encodes glycerol dehydrogenase. The enzyme activities measured in cell extracts were 2 to 3 orders of magnitude lower than those previously reported for extracts from other clostridia that are able to utilize glycerol as a fermentable carbon source (23). The low enzyme activity is consistent with the inability of C. beijerinckii NCIMB 8052 to use glycerol as a sole fermentable carbon source. Thus, it is unlikely that glycerol dehydrogenase functions in a major catabolic pathway in this organism.
Further experiments were carried out to determine whether glycerol dehydrogenase performs an important function in cellular metabolism in C. beijerinckii by inactivating the cognate gene, gldA. Integrational plasmids carrying internal fragments of the gldA coding sequence were employed for these experiments (Fig. 1A). They were constructed by inserting the desired portions of gldA into pMTL31, which lacks the ability to replicate in gram-positive bacteria (43). The resulting plasmids were mobilized into wild-type C. beijerinckii from an E. coli donor, and transconjugants were selected with the appropriate antibiotic resistance marker. Homologous integration by a Campbell-like mechanism of plasmid pCBGLD-1 containing an internal segment of gldA lacking both the translational start codon and ADH signature motif 2 resulted in transconjugants that showed only limited growth. Only very small pinpoint colonies were formed, and no viable cells were recovered by restreaking these small colonies after 2 days.
To verify that integration had taken place, the pinpoint transconjugant colonies were restreaked within 1 day of their appearance. The new colonies were used directly for PCR amplification with divergently oriented primers GDH4 and GDHR4 (Fig. 1). Only when gene disruption resulting from plasmid integration had taken place could PCR amplification yield a product, which contained the integrated vector and portions of gldA. A product of the expected size, approximately 4.7 kb, was obtained (data not shown), demonstrating that gene disruption had indeed taken place. To ensure that this product was not derived from extracellular plasmid DNA contaminating the colonies, PCR amplification was also carried out with a vector-specific primer, MTL-1, and a primer (BRE1) specific for the 3′ end of gldA, which is not present in pGLDA-1. A PCR product of the expected size (965 bp) was obtained, confirming that integration had taken place.
In another similar experiment, plasmid pCBGLD2 was employed to disrupt gldA in order to generate, after integration, two incomplete copies of gldA. One of these copies lacked 251 bp from the C terminus, whereas the other lacked 231 bp from the N terminus. Both partial copies retained the two iron-containing ADH signature motifs, ADH_IRON_1 and ADH_IRON_2 (PROSITE motifs PS00913 and PS00060). Surprisingly, transconjugants such as CBGLD-2 harboring this integrated plasmid were viable, indicating that they produced a functional glycerol dehydrogenase. This enzyme potentially could have arisen from either the copy of gldA lacking the 3′ end (transcribed from the gldA promoter) or the copy lacking the 5′ end (possibly transcribed by a vector promoter). Since strain CBGLD-1 contained one almost complete copy of gldA, lacking only the 5′ noncoding sequences and the start codon, and this strain failed to make GldA, we inferred that the functional copy in strain CBGLD-2 was the one that was truncated at the NcoI site (Fig. 1A). These experiments indicated that the 87 C-terminal amino acids of GldA are not required for enzyme activity.
Two additional strains were constructed as controls. As expected, in strain CBGLD-3, integration of a plasmid containing a larger fragment encompassing both ADH signature motifs and 177 bp of DNA upstream of gldA but truncated at the NcoI recognition site (Fig. 1) resulted in a wild-type phenotype. This strain expressed gldA at the same level as the wild type. The fact that enzyme activity was not increased in strain CBGLD-3 suggests that the 177 bp of DNA upstream from the start codon does not encompass the gldA promoter. The entire gldA gene plus 177 bp of DNA adjacent to the 5′ end as well as the dispensable C terminus, obtained with primers GLDF and GLDR1, was cloned into replicative plasmid pCTC1 (43). The resultant plasmid, pCBGLD-4, was mobilized into C. beijerinckii, and cell extracts of the resulting strain had the same glycerol dehydrogenase activity as the wild type. This again indicates that the 177 bp of DNA preceding the start codon does not contain the gldA promoter, since otherwise the substantial increase in gene copy number should have resulted in enhanced enzyme activity.
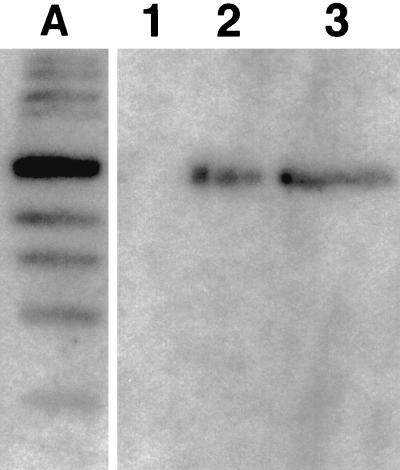
Expression of C. beijerinckii gldA was detected by Northern hybridization in both the wild-type and mutant BR54 strains (Fig. 3). The ca. 3-kb size of the transcripts confirmed that the gldA promoter is not located immediately adjacent to the gene. There may be one or more additional open reading frames located between the promoter region and gldA.
FIG. 3.
Northern blot of RNA prepared from wild-type strain C. beijerinckii NCIMB 8052 (lane 2) and mutant BR54 (lane 3) probed with a DNA fragment containing the gldA gene. A single band at approximately 3 kb was produced by both samples. The molecular size markers in lane A ranged from 7 to 0.5 kb, and the 3.0-kb band was the darkest. Three lanes have been omitted between the marker lane and empty lane 1.
Is there an essential role for glycerol dehydrogenase in E. coli?
Glycerol dehydrogenase specific activity is low in E. coli (35), and the role of this enzyme is unknown. E. coli gldA up-promoter mutants exhibit greatly increased glycerol dehydrogenase activity and are able to grow on glycerol as a sole carbon source (17, 41). We used E. coli as a control, since an essential role for GldA was not expected because there are two or more mechanisms for detoxification of MG in this organism (12, 14, 16, 38). Using the pKO3 vector (22), we were able to isolate gldA mutants derived from two strains of E. coli, XL-1 Blue and Lin 204. Gene disruption was carried out with a fragment of the E. coli gldA gene lacking the translation start codon and the two ADH signature motifs. The E. coli strains with disrupted gldA grew as well as the wild type in LB medium, demonstrating that in contrast to C. beijerinckii, gldA is not an essential gene in E. coli.
C. difficile gldA mutants.
We used wild-type C. difficile strain CD37, a nontoxigenic strain whose genome sequence is nearing completion (http://www.sanger.ac.uk/Projects/C_difficile/). Gene disruption was carried out by the methods used with C. beijerinckii. The segment of C. difficile DNA whose translated amino acid sequence is similar to the C. beijerinckii gldA encoded amino acid sequence was obtained from the Sanger Centre database. Primers were designed based on this sequence so that the PCR amplification product lacked the translational start codon and both ADH iron motifs. The PCR fragment was cloned into the pMTL31 vector, and gene disruption was carried out as described above. C. difficile transconjugants (e.g., CDGLD-1) formed pinpoint colonies on CBM medium, and they could not be subcultured after 2 days. We concluded that gldA performs an essential function in this organism, as it does in C. beijerinckii. In the control, plasmid integration did not disrupt gldA (CDGLD-2 in Fig. 1), and the colonies had a wild-type phenotype. Integration of the relevant plasmids in strains CDGLD-1 and CDGLD-2 was verified by PCR amplification with primers that were expected to yield a product only if the integrated plasmid was present in the transconjugant strain. In strain CDGLD-1 divergently oriented primers CDF5 and CDR5 yielded the expected ca. 4.4-kb PCR product. The vector-specific primer, MTL-2, and primer CDF1 were used with strain CDGLD-2, yielding the expected ca. 5.2-kb PCR product (data not shown).
DISCUSSION
Our gene disruption experiments demonstrated that glycerol dehydrogenase plays an essential role in cellular metabolism in both C. beijerinckii and C. difficile. The role of this enzyme in C. beijerinekii is not in central pathways of sugar catabolism, in which there are high metabolite fluxes, since the observed activity is too low. Moreover, these bacteria cannot use glycerol as a sole fermentable carbon source. The enzyme is therefore likely to participate in another pathway whose metabolic flux is lower than that of a primary fermentative pathway.
Our hypothesis is that GldA plays a role in MG detoxification in these clostridia. This hypothesis is supported by several independent pieces of evidence. (i) C. beijerinckii mutant BR54 with reduced expression of gldA contains higher levels of unbound MG than its parent. (ii) BR54 is more sensitive to added MG than the wild type, which is consistent with lower GldA activity. (iii) Inactivation of gldA in either C. beijerinckii or C. difficile is lethal, as expected if the enzyme plays a cardinal role in MG detoxification. Presumably, the limited growth of gldA transconjugants obtained with these two organisms was due to residual levels of glycerol dehydrogenase. When the remaining enzymatic activity was diluted by growth to a level that was insufficient to detoxify the MG generated by the cells, the cells were killed. (iv) The electrophoretic patterns of acid-precipitable proteins from the two strains differ (data not shown), which is consistent with different degrees of protein glycation in the mutant and the wild type.
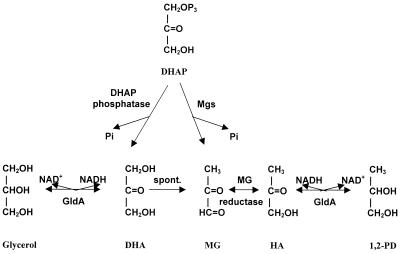
The role proposed for GldA in MG detoxification is illustrated in Fig. 4. In this model MG synthesis from DHAP is catalyzed by a MG synthase similar to that recently identified in Clostridium acetobutylicum (15). In addition, spontaneous conversion of dihydroxyacetone (DHA) to MG has been reported (33). Degradation of MG by glyoxalases does not occur to a significant extent, presumably because of the absence of glutathione in the organisms (11). The essential nature of GldA activity in the clostridia studied is consistent with a minor role for, or even the complete absence of, other MG detoxification systems. In contrast, a gldA knockout mutant of E. coli, which possessed the glyoxalase-linked MG detoxification systems (12, 14, 16), was viable.
FIG. 4.
Hypothetical pathways for synthesis and detoxification of MG in C. beijerinckii and C. difficile. DHAP is converted either to MG by MG synthase (Mgs), releasing inorganic phosphate (Pi), or to DHA by a cellular phosphatase. DHA either is reduced to glycerol by glycerol dehydrogenase (GldA) or is converted spontaneously to MG. The latter is detoxified by reduction to HA by a hypothetical MG reductase; HA is reduced to 1,2-propanediol (1,2-PD) by glycerol dehydrogenase.
Cellular MG levels can be reduced by several mechanisms, including conversion to nontoxic compounds by pathways involving GldA (Fig. 4). In addition, conversion of MG to hydroxyacetone (HA) may be catalyzed by a MG reductase such as that mentioned by Misra et al. (27). MG reductase activities have been reported or have been postulated to occur in E. coli and other bacteria (1, 6, 27). We have found that this activity also occurs in C. beijerinckii, as indicated by an MG-dependent NADH oxidase activity of 0.07 U/mg of protein in crude cell extracts (H. Liyanage and E. R. Kashket, unpublished data). It is characteristic of these oxidoreductases that there are many of them and that they differ considerably in different bacteria (viz., there is a large number of microbial ADHs) (8, 32).
The two reactions catalyzed by GldA are postulated to occur in the physiological direction, that is, NADH oxidation and substrate reduction. This is the likely direction rather than NAD reduction, because of the reducing cellular environment in the anaerobes and because the internal pH is less than 7 (37), near the optimum pH (about pH 6) of NADH oxidation (36; unpublished data). In contrast, the optimal pH for GldA-mediated NAD reduction is pH 10.0 (36, 41). GldA, thus, has a twofold function: to reduce DHA to glycerol and to reduce HA to 1,2-propanediol, thereby decreasing the cellular MG concentration. The result is that MG is detoxified by the diversion of DHAP to DHA, which in turn is reduced to glycerol, thus pulling the DHAP-to-DHA reaction. Similarly, the reduction of HA to 1,2-propanediol is catalyzed by GldA (36), including the C. beijerinckii enzyme (data not shown), and this too tends to reduce cellular levels of MG. A reduction in GldA activity, as occurs in the BR54 mutant, therefore results in higher cellular MG levels and increased sensitivity to added MG.
Finally, to our knowledge, this is the first report of targeted gene disruption in C. difficile. The methods which we employed to study this were adapted from those currently used with other clostridia (44), and they pave the way for functional analysis of virulence determinants in this important pathogen.
ACKNOWLEDGMENTS
This work was supported by the Cooperative State Research, Education, and Extension Service, U.S. Department of Agriculture, under agreement 97-35504-5143.
Sequence data for C. difficile was produced by the Sanger Centre Pathogen Sequencing Unit and can be obtained from http://www.sanger.ac.uk/Projects/C_difficile. We thank E. C. C. Lin and Lewis Wray for valuable discussions.
REFERENCES
- 1.Altaras N E, Cameron D C. Metabolic engineering of a 1,2-propanediol pathway in Escherichia coli. Appl Environ Microbiol. 1999;65:1180–1185. doi: 10.1128/aem.65.3.1180-1185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaumleffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barros A, Rodriguez J A, Almeida P J, Oliva-Teles M T. Determination of glyoxal, methylglyoxal, and diacetyl in selected beer and wine, by HPLC with UV spectrophotometric detection after derivatization with o-phenylenediamine. J Liq Chromatogr Rel Technol. 1999;22:2061–2069. [Google Scholar]
- 4.Brownlee M, Cerami A, Vlassara H. Advanced glycosylation end products in tissue and the biochemical basis of diabetic complications. N Engl J Med. 1988;318:1315–1321. doi: 10.1056/NEJM198805193182007. [DOI] [PubMed] [Google Scholar]
- 5.Burke R M, Tempest D W. Growth of Bacillus stearothermophilus on glycerol in chemostat culture: expression of an unusual phenotype. J Gen Microbiol. 1990;136:1381–1385. doi: 10.1099/00221287-136-7-1381. [DOI] [PubMed] [Google Scholar]
- 6.Cameron D C, Altaras N E, Hoffman M L, Shaw A J. Metabolic engineering of propanediol pathways. Biotechnol Prog. 1998;14:116–125. doi: 10.1021/bp9701325. [DOI] [PubMed] [Google Scholar]
- 7.Chaplen F W R, Fahl W E, Cameron D C. Evidence of high levels of methylglyoxal in cultured Chinese hamster ovary cells. Proc Natl Acad Sci USA. 1998;95:5533–5538. doi: 10.1073/pnas.95.10.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J S. Alcohol dehydrogenase: multiplicity and relatedness in the solvent-producing clostridia. FEMS Microbiol Rev. 1955;17:263–273. doi: 10.1111/j.1574-6976.1995.tb00210.x. [DOI] [PubMed] [Google Scholar]
- 9.Cordeiro C, Ponces Freire A. Methylglyoxal assay in cells as 2-methyl-quinoxaline using 1,2-diaminobenzene as derivatizing reagent. Anal Biochem. 1996;234:221–224. doi: 10.1006/abio.1996.0076. [DOI] [PubMed] [Google Scholar]
- 10.Evans V J, Liyanage H, Ravagnani A, Young M, Kashket E R. Truncation of peptide deformylase reduces the growth rate and stabilizes solvent production in Clostridium beijerinckii NCIMB 8052. Appl Environ Microbiol. 1998;64:1780–1795. doi: 10.1128/aem.64.5.1780-1785.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fahey R C, Brown W C, Adams W B, Worsham M B. Occurrence of glutathione in bacteria. J Bacteriol. 1978;133:1126–1129. doi: 10.1128/jb.133.3.1126-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson G P. Protective mechanisms against toxic electrophiles in Escherichia coli. Trends Microbiol. 1999;7:242–247. doi: 10.1016/s0966-842x(99)01510-3. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson G P, McLaggan D, Booth I R. Potassium channel activation by glutathione-S-conjugates in Escherichia coli: protection against methylglyoxal is mediated by cytoplasmic acidification. Mol Microbiol. 1995;17:1025–1038. doi: 10.1111/j.1365-2958.1995.mmi_17061025.x. [DOI] [PubMed] [Google Scholar]
- 14.Ferguson G P, Totemeyer S, MacLean M J, Booth I R. Methylglyoxal production in bacteria: suicide or survival? Arch Microbiol. 1998;170:209–219. doi: 10.1007/s002030050635. [DOI] [PubMed] [Google Scholar]
- 15.Huang K, Rudolph F B, Bennett G N. Characterization of methylglyoxal synthase from Clostridium acetobutylicum ATCC 824 and its use in the formation of 1,2-propanediol. Appl Environ Microbiol. 1999;65:3244–3247. doi: 10.1128/aem.65.7.3244-3247.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue Y, Kimura A. Methylglyoxal and regulation of its metabolism in microorganisms. Adv Microb Physiol. 1995;37:177–227. doi: 10.1016/s0065-2911(08)60146-0. [DOI] [PubMed] [Google Scholar]
- 17.Jin R Z, Tang J C-T, Lin E C C. Experimental evolution of a novel pathway for glycerol dissimilation in Escherichia coli. J Mol Evol. 1983;19:429–436. doi: 10.1007/BF02102318. [DOI] [PubMed] [Google Scholar]
- 18.Kalapos M P. Methylglyoxal in living organisms: chemistry, biochemistry, toxicology and biological implications. Toxicol Lett. 1999;110:145–175. doi: 10.1016/s0378-4274(99)00160-5. [DOI] [PubMed] [Google Scholar]
- 19.Karp P D, Riley M, Paley S M, Pellegrini-Toole A, Krummenacker M. EcoCyc: encyclopedia of Escherichia coli genes and metabolism. Nucleic Acids Res. 1999;27:55–58. doi: 10.1093/nar/27.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasai H, Kumeno K, Yamaizumi Z, Nishimura S, Nagao M, Fujita Y, Sugimura T, Nukaya H, Kosuge T. Mutagenicity of methylglyoxal in coffee. Jpn J Cancer Res. 1982;73:681–683. [PubMed] [Google Scholar]
- 21.Kashket E R, Cao Z-Y. Isolation of a degeneration-resistant mutant of Clostridium acetobutylicum NCIMB 8052. Appl Environ Microbiol. 1993;59:4198–4202. doi: 10.1128/aem.59.12.4198-4202.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Link A J, Phillips D R, Church G M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli. Application to open reading frame characterization. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liyanage H, Young M, Kashket E R. Butanol tolerance of Clostridium beijerinckii NCIMB 8052 associated with down-regulation of gldA by antisense RNA. J Mol Microbiol Biotechnol. 2000;2:87–93. [PubMed] [Google Scholar]
- 24.Lo T W C, Westwood M E, McLellan C, Selwood T, Thornalley P J. Binding and modification of proteins by methylglyoxal under physiological conditions. A kinetic and mechanistic study with Nα-acetylarginine, Nα-acetylcysteine, Nα-acetyllysine, and bovine serum albumin. J Biol Chem. 1994;269:32299–32305. [PubMed] [Google Scholar]
- 25.Marnett L J, Hurd H K, Hollstein M C, Levin D E, Esterbauer H, Ames B N. Naturally occurring carbonyl compounds are mutagens in Salmonella tester strain TA104. Mutat Res. 1985;148:25–34. doi: 10.1016/0027-5107(85)90204-0. [DOI] [PubMed] [Google Scholar]
- 26.McLellan A C, Phillips S A, Thornalley P J. The assay of methylglyoxal in biological systems by derivatization with 1,2-diamino-4,5-dimethoxybenzene. Anal Biochem. 1992;206:17–23. doi: 10.1016/s0003-2697(05)80005-3. [DOI] [PubMed] [Google Scholar]
- 27.Misra K, Banerjee A B, Ray S, Ray M. Reduction of methylglyoxal in Escherichia coli K12 by an aldehyde reductase and alcohol dehydrogenase. Mol Cell Biochem. 1996;156:117–124. doi: 10.1007/BF00426333. [DOI] [PubMed] [Google Scholar]
- 28.Murata K, Tani K, Kato J, Chibata I. Excretion of glutathione by methylglyoxal-resistant Escherichia coli. J Gen Microbiol. 1980;120:545–547. doi: 10.1099/00221287-120-2-545. [DOI] [PubMed] [Google Scholar]
- 29.Nagaraj R H, Shipanova I N, Faust F M. Protein cross-linking by the Maillard reaction. Isolation, characterization, and in vivo detection of a lysine-lysine cross-link derived from methylglyoxal. J Biol Chem. 1996;271:19338–19345. doi: 10.1074/jbc.271.32.19338. [DOI] [PubMed] [Google Scholar]
- 30.O'Brien R W, Morris J G. Oxygen and the growth and metabolism of Clostridium acetobutylicum. J Gen Microbiol. 1971;68:307–318. doi: 10.1099/00221287-68-3-307. [DOI] [PubMed] [Google Scholar]
- 31.Oya T, Hattori N, Mizuno Y, Miyata S, Maeda S, Osawa T, Uchida K. Methylglyoxal modification of proteins. J Biol Chem. 1999;274:18492–18502. doi: 10.1074/jbc.274.26.18492. [DOI] [PubMed] [Google Scholar]
- 32.Reid M F, Fewson C A. Molecular characterization of microbial alcohol dehydrogenases. Crit Rev Microbiol. 1994;20:13–56. doi: 10.3109/10408419409113545. [DOI] [PubMed] [Google Scholar]
- 33.Riddle V, Lorenz F W. Nonenzymic, polyvalent anion-catalyzed formation of methylglyoxal as an explanation of its presence in physiological systems. J Biol Chem. 1968;243:2718–2724. [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.St. Martin E J, Freedberg W B, Lin E C C. Kinase replacement by a dehydrogenase for Escherichia coli glycerol utilization. J Bacteriol. 1977;131:1026–1028. doi: 10.1128/jb.131.3.1026-1028.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang C-T, Ruch F E, Jr, Lin E C C. Purification and properties of a nicotinamide adenine dinucleotide-linked dehydrogenase that serves an Escherichia coli mutant for glycerol catabolism. J Bacteriol. 1979;140:182–187. doi: 10.1128/jb.140.1.182-187.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terracciano J S, Kashket E R. Intracellular conditions required for initiation of solvent production by Clostridium acetobutylicum. Appl Environ Microbiol. 1986;52:86–91. doi: 10.1128/aem.52.1.86-91.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thornalley P J. The glyoxalase system: new developments toward functional characterization of a metabolic pathway fundamental to biological life. Biochem J. 1990;269:1–11. doi: 10.1042/bj2690001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thornalley P J, Westwood M, Lo T W, McLellan A C. Formation of methylglyoxal-modified proteins in vitro and in vivo and their involvement in AGE-related processes. Contrib Nephrol. 1995;112:24–31. doi: 10.1159/000424089. [DOI] [PubMed] [Google Scholar]
- 40.Tran-Dinh K, Gottschalk G. Formation of d(−)-1,2-propanol and d(−)-lactate from glucose by Clostridium sphenoides under phosphate limitation. Arch Microbiol. 1985;142:87–92. [Google Scholar]
- 41.Truninger V, Boos W. Mapping and cloning of gldA, the structural gene of the Escherichia coli glycerol dehydrogenase. J Bacteriol. 1994;176:1796–1800. doi: 10.1128/jb.176.6.1796-1800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uchida K, Khor O T, Oya T, Osawa T, Yasuda Y, Miyata T. Protein modification by a Maillard reaction intermediate methylglyoxal. FEBS Lett. 1997;410:313–318. doi: 10.1016/s0014-5793(97)00610-8. [DOI] [PubMed] [Google Scholar]
- 43.Williams D R, Young D I, Young M. Conjugative plasmid transfer from Escherichia coli to Clostridium acetobutylicum. J Gen Microbiol. 1990;136:819–826. doi: 10.1099/00221287-136-5-819. [DOI] [PubMed] [Google Scholar]
- 44.Young D I, Evans V J, Jefferies J R, Jennert K C B, Phillips Z E V, Ravagnani A, Young M. Genetic methods in clostridia. Methods Microbiol. 1999;29:191–207. [Google Scholar]