Abstract
Non-starter lactic acid bacteria (NSLAB) were isolated from 12 Italian ewe cheeses representing six different types of cheese, which in several cases were produced by different manufacturers. A total of 400 presumptive Lactobacillus isolates were obtained, and 123 isolates and 10 type strains were subjected to phenotypic, genetic, and cell wall protein characterization analyses. Phenotypically, the cheese isolates included 32% Lactobacillus plantarum isolates, 15% L. brevis isolates, 12% L. paracasei subsp. paracasei isolates, 9% L. curvatus isolates, 6% L. fermentum isolates, 6% L. casei subsp. casei isolates, 5% L. pentosus isolates, 3% L. casei subsp. pseudoplantarum isolates, and 1% L. rhamnosus isolates. Eleven percent of the isolates were not phenotypically identified. Although a randomly amplified polymorphic DNA (RAPD) analysis based on three primers and clustering by the unweighted pair group method with arithmetic average (UPGMA) was useful for partially differentiating the 10 type strains, it did not provide a species-specific DNA band or a combination of bands which permitted complete separation of all the species considered. In contrast, sodium dodecyl sulfate-polyacrylamide gel electrophoresis cell wall protein profiles clustered by UPGMA were species specific and resolved the NSLAB. The only exceptions were isolates phenotypically identified as L. plantarum and L. pentosus or as L. casei subsp. casei and L. paracasei subsp. paracasei, which were grouped together. Based on protein profiles, Italian ewe cheeses frequently contained four different species and 3 to 16 strains. In general, the cheeses produced from raw ewe milk contained a larger number of more diverse strains than the cheeses produced from pasteurized milk. The same cheese produced in different factories contained different species, as well as strains that belonged to the same species but grouped in different RAPD clusters.
Non-starter lactic acid bacteria (NSLAB) usually increase from a low number in fresh curd to dominate the microflora of mature cheese (36). In contrast to starters, NSLAB tolerate the hostile environment of cheese during ripening; this environment typically is characterized by 32 to 39% moisture, 4 to 6% salt in moisture, pH 4.9 to 5.3, 5 to 13°C, and a deficiency of nutrients (15, 48). Mesophilic lactobacilli predominate in the NSLAB community, although pediococci and micrococci may also be found (6, 11, 15). Lactobacillus casei subsp. casei, L. casei subsp. pseudoplantarum, Lactobacillus paracasei subsp. paracasei, and Lactobacillus plantarum are the most frequently isolated taxa, but other facultatively and obligately heterofermentative species of lactobacilli are also found (18, 28). Adventitious mesophilic lactobacilli are usually present because of postpasteurization contamination but may also constitute part of the raw milk microflora and survive pasteurization (48).
The role of NSLAB in ripening has not been resolved satisfactorily yet, although inclusion of adjunct cultures of some strains of NSLAB or use of raw milk during cheese manufacturing increases the level of free amino acids, peptides, and free fatty acids, which leads to enhanced flavor intensity and accelerates cheese ripening (4, 14, 31, 32, 34). A comparison of the proteolytic and lipolytic enzymes of starters and NSLAB by quadratic response surface methodology (19) showed greater adaptation to cheese-like conditions by several enzymes of mesophilic lactobacilli. Further application of the same mathematical methodology showed that adaptation of peptidase activities to cheese-like conditions depends on the species and strain of mesophilic lactobacilli (20). The relative abundance of certain species and, especially, the heterogeneity of NSLAB strains in cheese may determine the relationships between NSLAB and cheese flavor. Very few data on the typing of lactobacilli in food ecosystems are available (11).
Most of the Italian ewe cheeses are semihard or hard Pecorino-like cheeses. They are produced at industrial or semi-industrial plants, have different national and international markets, and are produced by different technologies, but they all have typical features which depend on local and regional traditions. The indigenous microbial contents of cheeses, which are selected by the raw milk and cheese-making environment and technology, could be considered some of the main factors in determining the typical cheese features. Although Italian ewe cheeses use thermophilic lactic acid bacterial starters, heterogeneous NSLAB constitute a great part of the indigenous microbial community. Studies of NSLAB diversity may be helpful for (i) differentiating cheeses, (ii) establishing the effects of selected technological parameters on specific differences in the microbial flora, (iii) developing a monitoring system for studying the microflora dynamics in mixed-population fermentations, (iv) evaluating the real contributions of species and strains to cheese ripening, and, in general, (v) obtaining information about the diversity of a large adventitious population. Such information could allow selection of the most suitable strains to introduce as adjunct starters in pasteurized milk cheeses in order to reproduce more closely the flavor of raw milk cheeses or to accelerate cheese ripening.
Randomly amplified polymorphic DNA (RAPD) analysis is a PCR-based method which requires a short time compared with other genetic methods, provides good levels of discrimination, and is applicable to large numbers of strains (49). Moreover, the resolving power of this method can be easily enhanced by increasing the number of primers used to randomly amplify the bacterial genome (46). RAPD analysis has been used to estimate the diversity of Lactobacillus strains in the Centre National de Recherches Zootechniques collection (46), to type strains of L. plantarum (31), Lactococcus lactis isolated from raw milk used to produce Camembert (33), and dairy propionibacteria (40), to establish the correct nomenclature and classification of strains of L. casei subsp. casei (10), and to study the population of NSLAB in mature commercial cheese (11).
Analysis of cell wall protein profiles has been used to study and compare several strains of lactobacilli (1, 38, 50) and to differentiate the thermophilic lactobacilli present in natural or selected starters used to produce several Italian cheeses (16). In particular, Gatti et al. (16) reported that extraction of cell wall proteins, followed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) separation, has been found to be a reliable and rapid way to characterize large numbers of strains and to relate differences in cell wall protein profiles of strains to adaptation to different ecological niches and cheese technology.
To our knowledge, no previous studies have used phenotypic, genotypic, and cell wall protein analyses to characterize a large number of ewe cheese-related bacteria.
In this paper we describe phenotypic, genotypic (RAPD-PCR analysis), and cell wall protein characterization of NSLAB isolated from several Italian ewe cheeses.
MATERIALS AND METHODS
Cheese samples.
Twelve ewe cheeses, corresponding to six different types of cheese, were supplied by the main Italian producers (Table 1). The names of the producers are not included in Table 1. The cheeses were mainly Pecorino-like cheeses which are produced in several regions in central and southern Italy. All of the cheeses were analyzed when they were ready for market. Standard procedures were used to analyze cheeses for fat (Soxhlet method using diethyl ether), protein (micro-Kjeldahl method), salt (24), and moisture (25). The pH was measured by using a cheese slurry prepared from 20 g of cheese in 12 g of water (2).
TABLE 1.
Technological and chemical characteristics of Italian ewe cheeses
| Factory | Cheese | Type of milk | Curd cooking temp (°C)a | Ripening period | NSLAB concn (log CFU/g) | pH | Moisture content (%) | Salt in moisture (g of NaCl/100 g of H2O) |
|---|---|---|---|---|---|---|---|---|
| I | Pecorino Umbro (A)b | Pasteurized | 40–45 | 45 days | 8.2 | 5.2 | 40.3 | 4.2 |
| II | Pecorino Umbro (A) | Pasteurized | 40–45 | 45 days | 7.3 | 5.4 | 41.7 | 4.6 |
| III | Pecorino Umbro (A) | Pasteurized | 40–45 | 45 days | 7.7 | 5.1 | 38.2 | 5.7 |
| I | Pecorino Sardo (B) | Raw | 40–45 | 45 days | 8.4 | 5.3 | 38.5 | 4.7 |
| II | Pecorino Sardo (B) | Raw | 40–45 | 45 days | 7.9 | 5.2 | 40.1 | 5.2 |
| III | Pecorino Sardo (B) | Raw | 48–52 | 6 months | 6.8 | 5.2 | 32.3 | 8.6 |
| I | Pecorino Toscano (C) | Pasteurized | 40–45 | 30 days | 7.8 | 5.4 | 39.5 | 4.5 |
| II | Pecorino Toscano (C) | Pasteurized | 40–45 | 30 days | 7.2 | 5.0 | 37.1 | 4.9 |
| I | Pecorino Romano (D) | Raw | 48–52 | 10 months | 6.2 | 5.2 | 32.6 | 7.8 |
| II | Pecorino Romano (D) | Raw | 48–52 | 10 months | 5.5 | 5.1 | 33.2 | 8.2 |
| I | Fossa (E) | Raw | 52–55 | 6 months | 7.6 | 5.1 | 31.2 | 6.5 |
| I | Canestrato Pugliese (F) | Raw | 70–75 | 3 months | 7.8 | 5.0 | 32.4 | 7.3 |
All the cooking treatments were for 2 min except for the treatment for Canestrato Pugliese cheese (70 to 75°C for ca. 30 s).
The letters in parentheses were used to distinguish cheeses (see Table 2).
NSLAB isolation and enumeration.
Twenty grams of cheese was diluted in 180 ml of a 2% sodium citrate solution and homogenized in a Stomacher Lab-Blender 400 (PBI International, Milan, Italy). Serial dilutions were made in 0.25× Ringer's solution and plated on Lactobacillus-selective agar (Becton Dickinson and Co., Cockeysville, Md.). After incubation under microaerophilic conditions for 72 h at 30°C, 20 isolated colonies were randomly selected from each of a duplicate set of countable Lactobacillus-selective agar plates for each cheese. The isolates were propagated in MRS broth (Difco Laboratories, Detroit, Mich.) and purified. All isolates were checked for the catalase reaction and examined microscopically before stock preparations were prepared.
Group differentiation of mesophilic Lactobacillus isolates.
Four hundred gram-positive, catalase-negative, nonmotile, randomly isolated bacterial rods were first clustered on the basis of growth at 15 and 45°C, CO2 production from glucose, NH3 production from arginine, and esculin hydrolysis. About 30% of the isolates belonging to each group were then used for phenotypic, genotypic, and cell wall protein analyses.
Phenotypic characterization.
In addition to the preliminary assays, presumptively mesophilic lactobacilli were characterized for sugar fermentation by using the API 50 CHL system (bioMerieux, Marcy-l'Etoile, France) as recommended by the manufacturer. The APILAB Plus version 4.0 program (bioMerieux) was used to analyze the fermentation profiles obtained with the identification strips. Additional assays, such as assays for growth responses with and without Tween 80 in the culture medium, isomers of lactate, and ethanol production, were also performed as described in the species descriptions published by Kandler and Weiss (29) and Hammes and Vogel (22). As described by Fitzsimons et al. (11), the pH difference in MRS broth lacking acetate, citrate, and Tween 80 and in MRS broth lacking acetate and citrate after incubation for 48 h at 30°C was used to determine whether Tween 80 was required for growth.
Isomers of lactate and ethanol production were determined enzymatically (Boehringer Mannheim, Milan, Italy).
Ten American Type Culture Collection and Deutsche Sammlung von Mikroorganismen type strains (Table 2) were also included in this and the following analyses.
TABLE 2.
Characteristics fo the NSLAB type strains and isolates from Italian ewe cheeses
| RAPD clustera | Cell wall protein clusterb | Factory | Cheesec | Strain | |
|---|---|---|---|---|---|
| 1 | 3 | L. plantarum ATCC 14917T | |||
| 1 | 3 | I | A | L. plantarum S67 | |
| 8 | 3 | I | A | L. plantarum BL3 | |
| 1 | 3 | II | A | L. plantarum S1 | |
| 8 | 3 | II | A | L. plantarum 9H7 | |
| 8 | 3 | III | A | L. plantarum 12H4 | |
| 8 | 3 | I | B | L. plantarum 1HE | |
| 2 | 3 | I | B | L. plantarum 2HL | |
| 8 | 3 | II | B | L. plantarum 3H4 | |
| 2 | 3 | II | B | L. plantarum 83E | |
| 8 | 3 | II | B | L. plantarum 96E | |
| 2 | 3 | III | B | L. plantarum 4H1 | |
| 8 | 3 | III | B | L. plantarum 4H2 | |
| 8 | 3 | III | B | L. plantarum 4H5 | |
| 2 | 3 | III | B | L. plantarum 4H10 | |
| 8 | 3 | I | C | L. plantarum 14H3 | |
| 1 | 3 | I | C | L. plantarum 14H4 | |
| 8 | 3 | I | D | L. plantarum 6L | |
| 8 | 3 | I | D | L. plantarum 6 C | |
| 8 | 3 | I | D | L. plantarum 6 D | |
| 1 | 3 | II | D | L. plantarum 144 | |
| 1 | 3 | II | D | L. plantarum BV2 | |
| 1 | 3 | II | D | L. plantarum AC4 | |
| 1 | 3 | II | D | L. plantarum CL3 | |
| 1 | 3 | II | D | L. plantarum AH9 | |
| 1 | 3 | II | D | L. plantarum 361 | |
| 1 | 3 | II | D | L. plantarum 440 | |
| 1 | 3 | II | D | L. plantarum 422 | |
| 1 | 3 | II | D | L. plantarum BA1 | |
| 1 | 3 | II | D | L. plantarum BA2 | |
| 1 | 3 | I | E | L. plantarum F13 | |
| 8 | 3 | I | E | L. plantarum F16 | |
| 8 | 3 | I | E | L. plantarum 11H3 | |
| 8 | 3 | I | E | L. plantarum 11H4 | |
| 1 | 3 | I | E | L. plantarum 11H5 | |
| 1 | 3 | I | E | L. plantarum 11H6 | |
| 1 | 3 | I | F | L. plantarum 563A | |
| 1 | 3 | I | F | L. plantarum 592A | |
| 2 | 3 | I | F | L. plantarum 3397 | |
| 2 | 3 | I | F | L. plantarum 366 | |
| 11 | 6 | L. paracasei ATCC 25302T | |||
| scd | sc | L. paracasei subsp. tolerans DSM 20058T | |||
| 11 | 6 | III | A | L. paracasei 12H1 | |
| 11 | 6 | III | A | L. paracasei 12H8 | |
| 11 | 6 | II | B | L. paracasei 1HB | |
| 11 | 6 | II | B | L. paracasei 2HF | |
| 11 | 6 | II | B | L. paracasei 2H9 | |
| 11 | 6 | III | B | L. paracasei 4H3 | |
| 11 | 6 | III | B | L. paracasei 4H6 | |
| 11 | 6 | III | B | L. paracasei 15H4 | |
| 11 | 6 | II | C | L. paracasei 13H3 | |
| 11 | 6 | II | D | L. paracasei S14 | |
| 11 | 6 | I | F | L. paracasei 203 | |
| 11 | 6 | I | F | L. paracasei 364 | |
| 11 | 6 | I | F | L. paracasei 221 | |
| 11 | 6 | I | F | L. paracasei 137 | |
| 11 | 6 | I | F | L. paracasei 280A | |
| 8 | 8 | L. brevis ATCC 14869T | |||
| 1 | 8 | I | B | L. brevis 57E | |
| 1 | 8 | I | B | L. brevis 1HA | |
| 7 | 8 | I | B | L. brevis 1HC | |
| 8 | 8 | I | B | L. brevis 2HB | |
| 8 | 8 | I | B | L. brevis 1H1 | |
| 8 | 8 | II | B | L. brevis 68E | |
| 8 | 8 | III | B | L. brevis 95E | |
| 8 | 8 | I | D | L. brevis 8H1B | |
| 2 | 8 | II | D | L. brevis 102 | |
| 8 | 8 | II | D | L. brevis 265 |
| 2 | 8 | II | D | L. brevis 127 |
| 2 | 8 | II | D | L. brevis 129 |
| 2 | 8 | I | E | L. brevis F1 |
| 7 | 8 | I | E | L. brevis F31 |
| 8 | 8 | I | E | L. brevis F21 |
| 8 | 8 | I | F | L. brevis 109 |
| 2 | 8 | I | F | L. brevis 83A |
| 8 | 8 | I | F | L. brevis 265A |
| 2 | 8 | I | F | L. brevis 111 |
| 5 | 10 | L. curvatus ATCC 25601T | ||
| 5 | 10 | I | A | L. curvatus P14 |
| 5 | 10 | I | A | L. curvatus D54 |
| 5 | 10 | I | A | L. curvatus P402 |
| 5 | 10 | I | A | L. curvatus A63 |
| 5 | 10 | I | A | L. curvatus 1CF |
| 11 | 10 | I | B | L. curvatus 1HD |
| 11 | 10 | I | B | L. curvatus 58E |
| 5 | 10 | I | C | L. curvatus 14H10 |
| 5 | 10 | II | C | L. curvatus 13H5 |
| 11 | 10 | I | E | L. curvatus F5 |
| 5 | 10 | I | E | L. curvatus F19 |
| scd | 3 | L. pentosus ATCC 8041T | ||
| 8 | 3 | III | A | L. pentosus 12H2 |
| 8 | 3 | III | A | L. pentosus 12H5 |
| 2 | 3 | III | A | L. pentosus 12H6 |
| 8 | 3 | I | B | L. pentosus 1HL |
| 8 | 3 | I | B | L. pentosus 2HG |
| 8 | 3 | I | C | L. pentosus 14H9 |
| sc | 1 | L. fermentum ATCC 14931T | ||
| 2 | 1 | I | A | L. fermentum D13 |
| 1 | 1 | II | D | L. fermentum 93 |
| 4 | 1 | II | D | L. fermentum 10A |
| sc | 1 | II | D | L. fermentum 31L |
| 4 | 1 | I | E | L. fermentum F44 |
| 3 | 1 | I | F | L. fermentum 94A |
| 3 | 1 | I | F | L. fermentum 65H |
| sc | 6 | L. casei ATCC 393T | ||
| 11 | 6 | L. casei ATCC 334Te | ||
| 9 | 6 | III | A | L. casei M16 |
| 9 | 6 | III | A | L. casei M2 |
| 9 | 6 | III | A | L. casei K63 |
| sc | 6 | II | B | L. casei S18 |
| sc | 6 | II | B | L. casei V7 |
| 10 | 6 | II | C | L. casei B44 |
| 10 | 6 | II | C | L. casei AF4 |
| 9 | 4 | I | A | L. casei subsp. pseudoplantarum 159A |
| 9 | 4 | I | A | L. casei subsp. pseudoplantarum 134L |
| 9 | 4 | I | A | L. casei subsp. pseudoplantarum 109V |
| 9 | 4 | II | C | L. casei subsp. pseudoplantarum 115K |
| 6 | 7 | L. rhamnosus ATCC 7469T | ||
| 6 | 7 | III | B | L. rhamnosus 15H3 |
| sc | 2 | II | A | 9H6 N.I. |
| 8 | 9 | I | B | 61E N.I. |
| 5 | 2 | II | B | 3H1 N.I. |
| 11 | 5 | II | B | 1H2 N.I. |
| 5 | 2 | II | B | 3H6 N.I. |
| 5 | 2 | II | B | 3H8 N.I. |
| 9 | 9 | II | C | 55V N.I. |
| 11 | 5 | I | E | F18 N.I. |
| sc | 5 | I | E | F29 N.I. |
| sc | 2 | I | F | 476 N.I. |
| sc | 5 | I | F | 433 N.I. |
| 11 | sc | I | F | 590 N.I. |
| sc | 9 | I | F | 21M N.I. |
| 11 | 5 | I | F | 14L N.I. |
Genotypic characterization.
Mesophilic Lactobacillus isolates were characterized genotypically by RAPD-PCR analysis. Genomic DNAs from identified strains were extracted as described by De Los Reyes-Gàvilan et al. (9) from 2-ml samples of overnight cultures growth in MRS broth at 30°C. The final concentration of lysozyme used for cell lysis was 2 mg/ml. The concentration and purity of DNA were assessed by determining the optical densities at 260 and 280 nm, as described by Sambrook et al. (41); the DNA concentration of each sample was adjusted to 25 ng/μl. One microliter of each DNA (25 ng/μl) in a 25-μl PCR mixture was sufficient to give reproducible results.
Ten primers (Life Technologies, Milan, Italy) with arbitrarily chosen sequences were tested at a final concentration of 1 μM. The sequences were as follows: P1, 5′ ACGCGCCCT 3′ (27); P2, 5′ ATGTAACGCC 3′ (26); P3, 5′ CTGCGGCAT 3′ (11); P4, 5′ CCGCAGCGTT 3′ (11); P5, 5′ TGCTCTGCCC 3′ (46); P6 5′ GTCCACACGG 3′ (46); P7, 5′ AGCAGCGTGG 3′ (30); P8, 5′ CGTACAGGCT 3′; P9, 5′ TCACCGTCGC 3′; and P10, 5′ ACTGGCTCCG 3′. Each reaction mixture contained each 2′-deoxynucleoside 5′-triphosphate at a concentration of 200 μM, 1 μM primer, 1.5 mM MgCl2, 1.25 U of Taq DNA polymerase (Life Technologies), 2.5 μl of PCR buffer, 25 ng of DNA, and enough sterile bidistilled water to bring the volume to 25 μl. The PCR program comprised 45 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 35°C, and extension for 2 min at 72°C; the cycles were preceded by denaturation at 94°C for 4 min and were followed by extension at 72°C for 5 min (40).
PCR products (15 μl) were separated by 4 h of electrophoresis at 120 V on a 1.5% (wt/vol) agarose gel (Gibco BRL, Life Technologies, Milan, Italy), and the DNA was detected by UV transillumination after staining with ethidium bromide (0.5 μg/ml). The molecular sizes of the amplified DNA fragments were estimated by comparison with a 123-bp ladder DNA (Gibco BRL, Life Technologies).
Photographs of RAPD-PCR gels were obtained with a high-performance charge-coupled device camera (Cohu, Inc., San Diego, Calif.) and were scanned by using a Scanject IIcx scanner (Hewlett-Packard Co., Palo Alto, Calif.). Electrophoretic profiles in the form of densitometric curves were compared with GelCompar 4.0 software (Applied Maths, Kortrijk, Belgium). Three series of RAPD-PCR profiles were combined to obtain a unique dendrogram. Pairwise comparisons of band patterns were evaluated by calculating an index of genetic similarity by using the simple matching coefficient (44). A cluster analysis was carried out with similarity estimates by using the unweighted pair group method with arithmetic average (UPGMA), from which a dendrogram showing the relationships between Lactobacillus isolates was obtained. The analysis was performed by using the NTSYS.PC package, version 1.8 (39). The reproducibility of RAPD fingerprints was determined from triplicate loadings of independent, triplicate RAPD reaction mixtures prepared from nine strains on three gels, and cluster analysis was performed as described above.
Cell wall protein characterization.
Cell wall protein was extracted by using a slightly modified version of the method of Gatti et al. (16). Twenty-four-hour-old cells of mesophilic lactobacilli cultivated in MRS broth (Difco) were harvested, washed twice in 0.05 M Tris-HCl (pH 7.5) containing 0.1 M CaCl2, and resuspended in 1 ml of the same buffer at an A600 of 10.0. After centrifugation at 8,000 × g for 5 min, cell wall proteins were extracted from the pellets with 1.0 ml of extraction buffer (pH 8.0) containing 0.01 M EDTA, 0.01 M NaCl, and 2% (wt/vol) SDS. Suspensions were stored at room temperature for 60 min, heated at 100°C for 5 min, and centrifuged at 11,600 × g for 10 min at 4°C. The supernatants were analyzed by SDS-PAGE with a Phast system (Pharmacia, Uppsala, Sweden) and stained with Comassie blue (23). Densitometry of SDS-PAGE gels was performed with GelCompar 4.0 software (Applied Maths), and the Rf values of individual proteins were calculated to compare the protein profiles of the strains. The 70 kit molecular weight protein standard (molecular weight range, 14,300 to 66,000; 54 μg of total protein) in addition to β-galactosidase (molecular weight, 116,000; 8 μg of protein) and α2-macroglobulin (molecular weight, 170,000; 6 μg of protein) was used (Sigma Chemical Co., St. Louis, Mo.). The reproducibility of SDS-PAGE was estimated by loading two independent, triplicate cell wall protein extraction preparations from nine strains on two gels, and cluster analysis was performed as described above. The relative error (E) for each band in each gel was calculated as follows: E = [(Rf − Rfm)/Rfm] × 100, where Rf is the distance between the top of the separating gel and a protein band and Rfm is the mean Rf for the band obtained in different gels. Clustering of the resultant profiles was performed as described above for genotypic characterization.
RESULTS
Cheese characteristics.
The main characteristics of the 12 ewe cheeses from which NSLAB were isolated are shown in Table 1. These cheeses are the most important Italian ewe cheeses, and most were supplied by different factories. Some cheeses were produced from raw milk, and others were produced from pasteurized milk; the curd was cooked for 2 min at 40 to 55°C except for Canestrato cheese (70 to 75°C for ca. 30 s). The cheeses were produced by using thermophilic natural lactic acid bacterial starters, and the ripening time ranged from 45 days to 10 months depending on the type of cheese (fresh, semihard, or hard).
All the cheeses contained high numbers of NSLAB, which varied from 5.5 to 8.2 log CFU/g. The pH values differed slightly and ranged between 5.0 and 5.4. The moisture and salt in moisture values varied greatly from 31.2 to 44.5% and from 4.0 to 10.2%, respectively, depending on the ripening period and cheese size. The protein content ranged from 25 to 32%, and the fat content ranged from 33 to 38% (data not shown).
Phenotypic characterization.
Four hundred presumptive Lactobacillus isolates (gram-positive, catalase-negative, nonmotile rods) that were randomly isolated from the 12 Italian ewe cheeses were differentiated into preliminary groups based on growth at 15 and 45°C, CO2 production from glucose, NH3 production from arginine, and esculin hydrolysis (22, 45). About 30% of the isolates belonging to each of the preliminary groups (a total of 123 isolates) were characterized further.
Phenotypic characterization based on sugar fermentation assays, the physiological assays described above, and other biochemical assays (growth responses with and without Tween 80 in the culture medium, isomers of lactate and ethanol production) resulted in identification of 32% of the isolates as L. plantarum isolates, 15% as Lactobacillus brevis isolates, 12% as L. paracasei subsp. paracasei isolates, 9% as Lactobacillus curvatus isolates, 6% as Lactobacillus fermentum isolates, 6% as L. casei subsp. casei isolates, 5% as Lactobacillus pentosus isolates, 3% as L. casei subsp. pseudoplantarum isolates, and 1% as Lactobacillus rhamnosus isolates (22, 29, 45). The percentage of identification obtained by the identification computer program (APILAB Plus) was always more than 99%. Eleven percent of the isolates were not identified. A UPGMA dendrogram based on phenotypic similarities is not shown. Except for Pecorino Umbro AII, Pecorino Toscano CI, and Pecorino Romano DI cheeses, which contained only one to three species, all of the cheeses contained four NSLAB species (Table 2). Except for Pecorino Toscano cheese produced in factory II, L. plantarum isolates were found in all of the ewe cheeses. Except for Fossa cheese, L. paracasei subsp. paracasei isolates were present in all of the cheese types; some exceptions depended on the factory. The same type of cheese (e.g., Pecorino Umbro) produced with the same technology showed some variation in the composition of NSLAB species which was related to the factory. Technology also influenced the microbial composition. L. brevis isolates were found mainly in cheeses produced with a high temperature during curd cooking, a high concentration of salt in moisture, and a long ripening time (e.g., Pecorino Sardo cheese from factory III, Pecorino Romano cheese, and Fossa cheese). The few isolates of L. casei subsp. pseudoplantarum were detected only in fresh ewe cheeses.
Phenotypic characterization identified the 10 type strains as expected.
Genotypic characterization.
Primers P2, P3, P5, P6, P8, P9, and P10 did not give PCR products or gave very few bands, despite extended annealing times at a low temperature. Fitzsimons et al. (11) obtained species-specific bands when they used primer P2 for characterization of some Lactobacillus strains isolated from Cheddar cheese; nevertheless, in our PCR conditions, primer P2 gave only a limited number of non-species-specific bands. P1, P4, and P7 generated the greatest pattern diversity and were selected for genotypic characterization. The importance of using several primers to improve the RAPD resolving power, especially when RAPD patterns generated with single primers are very similar, has been described by other authors (47). The reproducibility of the RAPD fingerprints was assessed by comparing the PCR products obtained with primers P1, P4, and P7 and DNA prepared from three separate cultures of the same strain. Nine strains were studied, and the patterns for the same strain were 93% similar, indicating that the reproducibility of the technique under the conditions used was high (data not shown).
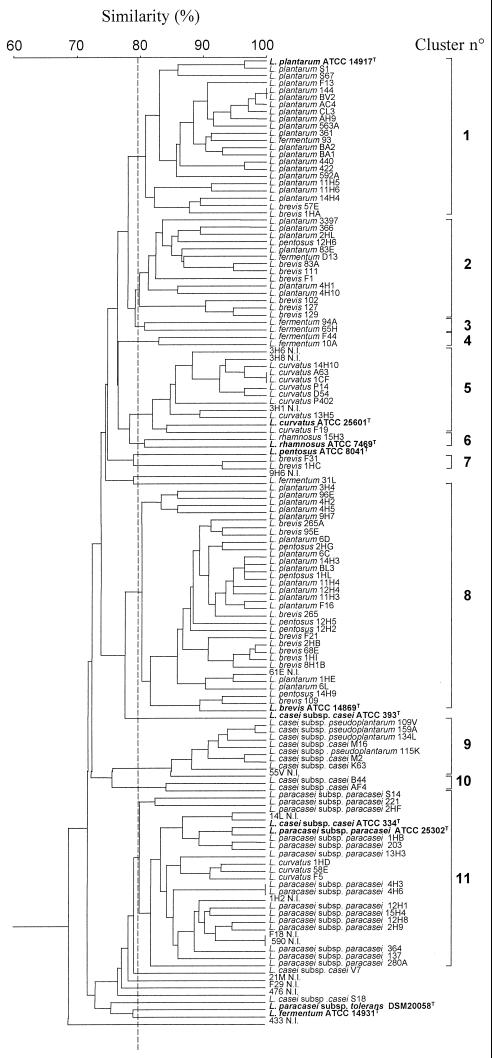
The combined RAPD profiles generated with primers P1, P4, and P7 for 123 isolates from 12 cheeses and 10 type strains produced the UPGMA dendrogram shown in Fig. 1. At 80% similarity, the type strains of the species were separated. The only exceptions were L. casei subsp. casei ATCC 334T and L. paracasei subsp. paracasei ATCC 25302T, which joined the same cluster at a similarity level greater than 95%. Due to the high level of genetic relatedness to L. casei subsp. casei and L. paracasei subsp. paracasei, ATCC 334 was previously proposed as a neotype strain for L. casei along with rejection of the name L. paracasei (8). Type strain L. casei ATCC 393T did not belong to any group and joined the clusters containing L. casei subsp. casei, L. casei subsp. pseudoplantarum, and L. paracasei subsp. paracasei isolates at a similarity level of about 72%. DNA-DNA hybridizations revealed high levels of relatedness among strains of L. casei subsp. casei and related subspecies, all of which differed from type strain ATCC 393T (3).
FIG. 1.
Dendrogram, obtained from combined RAPD patterns with three primers, of Lactobacillus isolates from ewe cheeses and type strains. A cluster analysis was conducted with similarity estimates by using UPGMA.
Above 80% similarity, the mesophilic lactobacilli isolated from the cheeses were grouped into 11 clusters (clusters 1 to 11) (Fig. 1). Twelve isolates, which produced a unique RAPD pattern, were not included in any cluster. Details of each cluster are given in Table 2. Several small clusters contained isolates that belong to unique species (e.g., clusters 3, 4, 6, 7, and 10). Cluster 5 contained several L. curvatus strains together with three unidentified isolates. The major clusters (clusters 1, 2, 8, and 11) contained isolates of two other species in addition to numerous isolates belonging to unique species. In particular, cluster 1 contained most of the L. plantarum isolates together with type strain ATCC 14917T and isolates phenotypically identified as L. brevis 57E and 1HA and L. fermentum 93. L. plantarum and L. brevis isolates were also present in clusters 2 and 8. The phylogenetic tree based on 16S rRNA sequences for the L. casei-Pediococcus group showed ca. 93% similarity between these two taxa (43). L. plantarum isolates, which accounted for the highest percentage of cheese isolates (32%), were characterized by great RAPD pattern variability since they were included in three different clusters, clusters 1, 2, and 8. DNA-DNA hybridizations showed that L. plantarum strains were not homogeneous (7). Cluster 9 included only L. casei subsp. casei, L. casei subsp. pseudoplantarum, and one phenotypically unidentified isolate. All the L. paracasei subsp. paracasei isolates, grouped in cluster 11, which also included three isolates of L. curvatus, some unidentified isolates, and the type strain L. casei subsp. casei ATCC 334T.
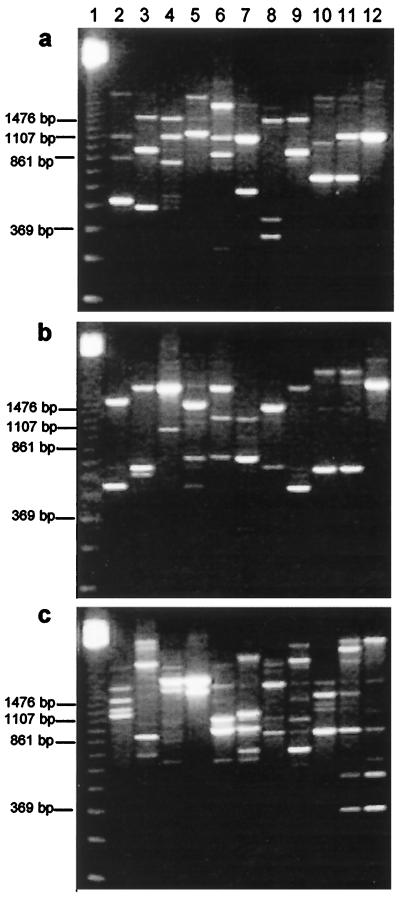
The different RAPD patterns which show the representative fingerprint for each of the 11 clusters are shown in Fig. 2. Primers P1, P4, and P7 amplified DNA and produced bands at 250 to 4,200 bp. Primer P1 produced specific bands for individual clusters (e.g., ca. 490, 450, and 300 to 1,800 bp for clusters 1, 2, and 5, respectively), but none of the three primers used was useful for obtaining species-specific bands.
FIG. 2.
RAPD patterns obtained with primers P1 (a), P4 (b), and P7 (c), showing the representative fingerprints of the 11 clusters. Lane 1, DNA molecular size standards (123-bp ladder DNA; Gibco BRL, Life Technologies); lanes 2 to 12, clusters 1 to 11, respectively.
Cell wall protein characterization.
The reproducibility of SDS-PAGE was estimated by duplicate loading of independent, triplicate cell wall protein extracts from nine strains on two gels. The relative error for each band in each gel was less than 1% (21). Based on preliminary assays, the resolving power of SDS-PAGE was best when 12% acrylamide was used (data not shown).
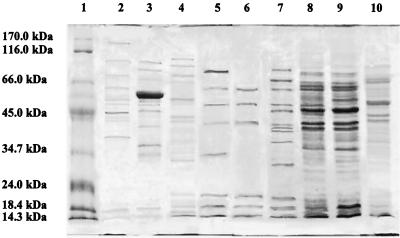
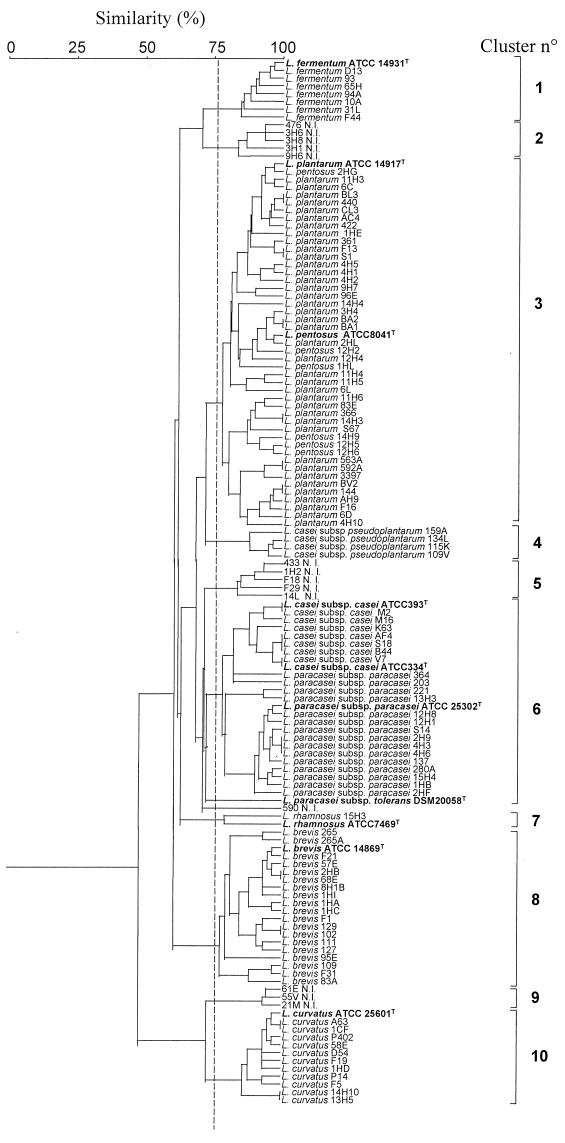
Representative SDS-PAGE cell wall protein profiles for the nine Lactobacillus species and subspecies isolated from cheeses are shown in Fig. 3. Two protein bands at molecular masses of ca. 50 and 18 kDa were produced by all of the isolates. Only L. fermentum and L. casei subsp. pseudoplantarum produced species-specific bands at ca. 123 and 30 kDa, respectively. L. curvatus produced four well-defined protein bands at ca. 53, 43, 41, and 31 kDa in addition to several other bands in the 92- to 66-kDa range. A similar cell wall protein profile was reported for L. curvatus isolates from sausages (42). L. casei subsp. casei and L. paracasei subsp. paracasei produced similar patterns that were characterized mainly by three proteins at 61, 50, and 41.6 kDa; L. plantarum and L. pentosus also behaved similarly, producing six well-defined bands at molecular masses ranging from 65 to 35 kDa. Our analyses showed that most of the species studied were characterized by specific protein profiles which produced the UPGMA dendrogram shown in Fig. 4. When a similarity level of ca. 77% was used, the type strains and the cheese isolates grouped into 10 clusters which, with few exceptions, separated all of the isolates belonging to the different species. Clusters 1, 4, 7, 8, and 10 contained all of the isolates and related type strains of L. fermentum, L. casei subsp. pseudoplantarum, L. rhamnosus, L. brevis, and L. curvatus, respectively. L. plantarum and L. pentosus were grouped together in cluster 3, and L. casei subsp. casei and L. paracasei subsp. paracasei were grouped together in 6. This is not surprising because L. plantarum and L. pentosus strains can be taxonomically distinguished only by the large 16S-23S rRNA spacer region sequence (22) and because of the high level of genetic relatedness between L. casei subsp. casei and L. paracasei subsp. paracasei (8). Except for strain 590, the unidentified isolates were grouped together in clusters 2, 5, and 9. Further details concerning each cluster are given in Table 2.
FIG. 3.
SDS-PAGE patterns of cell wall proteins from Lactobacillus isolates. Lane 1, standard proteins (see Materials and Methods); lane 2, L. fermentum 65H; lane 3, L. brevis 68E; lane 4, L. curvatus 1HD; lane 5, L. casei subsp. casei M16; lane 6, L. paracasei subsp. paracasei 4H3; lane 7, L. casei subsp. pseudoplantarum 115K; lane 8, L. plantarum 144; lane 9, L. pentosus 2HG; lane 10, L. rhamnosus 15H3.
FIG. 4.
Dendrogram, obtained from SDS-PAGE patterns of cell wall proteins, of Lactobacillus isolates from ewe cheeses and type strains. A cluster analysis was conducted with similarity estimates by using UPGMA.
DISCUSSION
NSLAB were isolated from 12 Italian ewe cheeses representing six different types of cheese, which in several cases were produced in different factories. Pecorino Umbro, Pecorino Sardo, Pecorino Toscano, Pecorino Romano, Fossa, and Canestrato cheeses are the Italian ewe cheeses with the greatest market popularity (12, 17, 18) and differ with respect to several technological and chemical characteristics (Table 1). The concentrations of NSLAB cells in the cheeses ranged from 5.5 to 8.2 log CFU/g, values which correspond to the values usually found in cheeses during ripening (11, 13). Phenotypic characterization of 123 catalase-negative, nonmotile, rod-shaped isolates identified nine Lactobacillus species and subspecies which accounted for various percentages of the total. Most of the 12 cheeses contained four different species of NSLAB (Table 2). L. plantarum isolates (32% of the total) were found in 11 of the 12 ewe cheeses. L. paracasei subsp. paracasei isolates (12% of the total) were found in all the cheese types except Fossa cheese, but variations in the percentage depended on the factory of origin. Fossa cheese is a very typical cheese characterized by 3 months of ripening in an underground pit (Fossa) (18). L. paracasei subsp. paracasei has always been found at the highest concentrations in Cheddar cheeses (11, 28). The few L. casei subsp. pseudoplantarum isolates were found only in fresh ewe cheeses, and L. brevis isolates were found mainly in cheeses with very restrictive ripening conditions. In contrast, Pecorino Sardo cheeses ripened for different periods contained the same species after 45 days or 6 months of ripening (the only exception was L. rhamnosus). Other authors (11, 35) have observed changes in the composition of the NSLAB populations in Cheddar cheese which depended on the age of the cheese.
Although phenotypic tests provide some evidence of metabolic capabilities, there are some problems, such as a lack of reproducibility and a lack of discriminatory power. Designation of certain neotype strains based only on phenotypic characteristics gave confused results which were resolved only by using molecular techniques (10). In an effort to corroborate the biochemical classification, we also used genotypic and cell wall protein analyses. To our knowledge, this is the first study which combined characterization of NSLAB based on two different molecular techniques in addition to phenotypic analysis.
Other studies (11, 33, 40, 46, 47) have used RAPD analysis to differentiate strains belonging to a limited number of species of lactobacilli, lactococci, and propionibacteria. Fitzsimons et al. (11) were the first workers to use this technique to study NSLAB from Cheddar cheeses. The isolates from Cheddar cheeses were mainly L. paracasei subsp. paracasei, and only a few isolates of L. plantarum, L. curvatus, and L. brevis were found. A combination of two primers was used, and even if characteristic profiles were obtained with both primers, RAPD analysis with one of the two primers gave species-specific DNA bands. Fitzsimons et al. (11) concluded that their technique was useful for rapidly classifying large numbers of NSLAB occurring in cheese and allowing studies of this community. We used RAPD analysis to differentiate isolates belonging to nine NSLAB species or subspecies in addition to several phenotypically unidentified isolates. Although the combination of three primers used in this study (primers P1, P4, and P7) was useful for partially differentiating 10 type strains, we did not find a species-specific DNA band or a combination of bands which permitted complete separation of all the species considered (Fig. 1 and 2 and Table 2).
The resolving power of RAPD analysis, however, is less than that of protein profiling since the latter technique resolved more the reference isolates at the strain level (5, 11, 37, 42). Indeed, we obtained a combination of cell wall protein profiles which were species specific and could resolve NSLAB in the presence of many species (Fig. 3 and 4 and Table 2). Only isolates that were phenotypically identified as L. plantarum and L. pentosus isolates or L. casei subsp. casei and L. paracasei subsp. paracasei isolates, were grouped together. Although grouped together, the last two species were correctly separated within the same cluster. Gatti et al. (16) found species-specific proteins for Lactobacillus helveticus and Lactobacillus delbrueckii, but within L. delbrueckii there was no discrimination between L. delbrueckii subsp. lactis and L. delbrueckii subsp. bulgaricus when the same electrophoretic technique was used (16). Under our conditions, RAPD analysis did not discriminate between L. plantarum and L. pentosus, but L. paracasei subsp. paracasei isolates were separated from L. casei subsp. casei. As shown in the dendrogram in Fig. 4, the NSLAB species showed different degrees of overall similarity. When the variability of the two strains of L. rhamnosus was not included, the protein profiles of the L. plantarum-L. pentosus group, L. brevis, and the L. casei subsp. casei-L. paracasei subsp. paracasei group were the most heterogeneous. In spite of the lower resolving power of the method, the L. plantarum-L. pentosus group and L. brevis were also found to be the most heterogeneous groups when RAPD analysis was used (Fig. 1). Fitzsimons et al. (11) found the greatest strain diversity in Cheddar cheese for the subspecies L. paracasei subsp. paracasei. However, greater genotypic differentiation observed in a certain species could be the result of a higher number of strains used for comparison and, therefore, of the increased probability of encountering more distantly related taxonomic units (40).
Based on a level of homology greater than 77%, the limit used to differentiate species by protein profiling, and an arbitrarily range from 90 to 100%, strains of the same species which occur in different subclusters may be regarded as more dissimilar strains. Based on this criterion, Italian ewe cheeses (30 to 35 isolates were examined from each cheese) contained different numbers of strains, which ranged from 3 to 16. In Cheddar cheese the average number of strains per cheese was found to be seven (11). Except for Pecorino Romano cheese from factory I, all the cheeses produced from raw ewe milk contained a larger number of different strains (8 to 16 strains) than cheeses produced from pasteurized milk (3 to 7 strains). In particular, Fossa and Canestrato cheeses contained 13 and 16 strains, respectively, belonging to L. plantarum, L. fermentum, L. brevis, L. curvatus (only in Fossa cheese), and L. paracasei subsp. paracasei (only in Canestrato cheese) in addition to several unidentified strains. Based on RAPD analysis and considering the strains included in different clusters more dissimilar (Fig. 1 and Table 2), the microbial diversity of ewe cheeses seemed to decrease. Except for Pecorino Umbro AII, Pecorino Toscano CI, and Pecorino Romano DI, the Italian ewe cheeses contained four to seven NSLAB strains. Protein profiling showed that the same cheese produced in different factories (e.g., Pecorino Sardo and Pecorino Romano cheeses) contained not only some different species but also some strains which belonged to the same species and clustered differently. In contrast, isolates from the same cheese frequently grouped together in the same cluster (e.g., L. plantarum AC4, CL3, 440, and 422 from Pecorino Romano DII cheese; L. plantarum 4H5, 4H2, and 4H1 from Pecorino Sardo BIII cheese; and L. brevis 109 and F31 and L. brevis 1HA and 1HC from Canestrato and Pecorino Sardo BI cheeses, respectively). Also, the majority of strains isolated from Cheddar cheeses made in the same factory grouped together in the same cluster (11).
Based on characterization of a large number of NSLAB species by phenotypic, genotypic, and cell wall protein analyses, the following conclusions can be drawn: (i) although 10 primers were screened and 3 of them were used, RAPD analysis did not completely resolve classification of NSLAB; (ii) cell wall protein profiling seems to be a more appropriate tool for classifying NSLAB; (iii) in some cases, such as discrimination between L. casei subsp. casei and L. paracasei subsp. paracasei, RAPD analysis helped improve the resolving power of protein profiling; (iv) RAPD analysis and particularly protein profiling provided useful information about the diversity of NSLAB in cheeses; and (v) Italian ewe cheeses are characterized by a very heterogeneous NSLAB flora which is influenced by geographical and technological factors, which may be responsible for cheese diversity.
The results of the present work could represent a useful tool for nonrandom selection of NSLAB for use as adjunct cultures in pasteurized milk cheese making in order to improve and standardize product quality.
REFERENCES
- 1.Boot H J, Kolen C P, Pot B, Kersters K, Pouwels P H. The presence of two S-layer-protein-encoding genes is conserved among species related to Lactobacillus acidophilus. Microbiology. 1996;142:2375–2384. doi: 10.1099/00221287-142-9-2375. [DOI] [PubMed] [Google Scholar]
- 2.British Standard Institution. Chemical analysis of cheese. Part 5. Determination of pH value. British Standard 770. Milton Keyes, United Kingdom: British Standard Institution; 1976. [Google Scholar]
- 3.Collins M D, Phillips B A, Zanoni P. Deoxyribonucleic acid homology studies of Lactobacillus casei, Lactobacillus paracasei sp. nov., subsp. paracasei and subsp. tolerans, and Lactobacillus rhamnosus sp. nov., comb. nov. Int J Syst Bacteriol. 1989;39:105–108. [Google Scholar]
- 4.Corsetti A, Gobbetti M, Smacchi E, De Angelis M, Rossi J. Accelerated ripening of Pecorino Umbro cheese. J Dairy Res. 1998;65:631–642. [Google Scholar]
- 5.Costas M, Pot B, Vandamme P, Kersters K, Owen R J, Hill L R. Interlaboratory comparative study of the numerical analysis of one-dimensional sodium dodecyl sulphate-polyacrylamide gel electrophoretic protein patterns of Campylobacter strains. Electrophoresis. 1990;11:467–474. doi: 10.1002/elps.1150110606. [DOI] [PubMed] [Google Scholar]
- 6.Dacre J C. A note on pediococci in New Zealand Cheddar cheese. J Dairy Res. 1958;25:414–417. [Google Scholar]
- 7.Dellaglio F, Bottazzi V, Vescovo M. Deoxyribonucleic acid homology among Lactobacillus species of the subgenus Streptobacterium Orla-Jensen. Int J Syst Bacteriol. 1975;25:160–172. [Google Scholar]
- 8.Dellaglio F, Dicks L M T, Du Tolt M, Torriani S. Designation of ATCC 334 in place of ATCC 393 (NCDO 161) as the neotype strain of Lactobacillus casei subsp. casei and rejection of the name Lactobacillus paracasei. Request for an opinion. Int J Syst Bacteriol. 1991;41:341–342. [Google Scholar]
- 9.De Los Reyes-Gavilàn C G, Limsowtin G K Y, Tailliez P, Séchaud L, Accolas J P. A Lactobacilus helveticus-specific DNA probe detects restriction fragment length polymorphisms. Appl Environ Microbiol. 1992;58:3429–3432. doi: 10.1128/aem.58.10.3429-3432.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dicks L M T, Du Plessis E M, Dellaglio F, Lauer E. Reclassification of Lactobacillus casei subsp. casei ATCC 393 and Lactobacillus rhamnosus ATCC 15820 as Lactobacillus zeae nom. rev., designation of ATCC 334 as the neotype of L. casei subsp. casei, and rejection of the name Lactobacillus paracasei. Int J Syst Bacteriol. 1996;46:337–340. doi: 10.1099/00207713-46-1-337. [DOI] [PubMed] [Google Scholar]
- 11.Fitzsimons N A, Cogan T M, Condon S, Beresford T. Phenotypic and genotypic characterization of non-starter lactic acid bacteria in mature cheddar cheese. Appl Environ Microbiol. 1999;65:3418–3426. doi: 10.1128/aem.65.8.3418-3426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox P F, Guinee T P. Italian cheeses. In: Fox P F, editor. Cheese: chemistry, physics and microbiology. Vol. 2. London, United Kingdom: Elsevier Applied Science; 1987. p. 251. [Google Scholar]
- 13.Fox P F, McSweeney P L H, Lynch C M. Significance of non-starter lactic acid bacteria in Cheddar cheese. Aust J Dairy Technol. 1998;53:5383–5389. [Google Scholar]
- 14.Franklin J G, Sharpe M E. The incidence of bacteria in cheesemilk and Cheddar cheese and their association with flavour. J Dairy Res. 1963;30:87–99. [Google Scholar]
- 15.Fryer T F, Sharpe M E. Pediococci in Cheddar cheese. J Dairy Res. 1966;33:325–331. [Google Scholar]
- 16.Gatti M, Fornasari E, Neviani E. Cell-wall protein profiles of dairy thermophilic lactobacilli. Lett Appl Microbiol. 1997;25:345–348. doi: 10.1046/j.1472-765x.1997.00235.x. [DOI] [PubMed] [Google Scholar]
- 17.Gobbetti M, Corsetti A, Smacchi E, De Angelis M, Rossi J. Microbiology and biochemistry of Pecorino Umbro cheese during ripening. Ital J Food Sci. 1997;9:111–126. [Google Scholar]
- 18.Gobbetti M, Folkertsma B, Fox P F, Corsetti A, Smacchi E, De Angelis M, Rossi J, Kilcawley K, Cortini M. Microbiology and biochemistry of Fossa (pit) cheese. Int Dairy J. 1999;9:763–773. [Google Scholar]
- 19.Gobbetti M, Lanciotti R, De Angelis M, Corbo M R, Massini R, Fox P F. Study of the effects of temperature, pH, NaCl and aw on the proteolytic and lipolytic activities of cheese-related lactic acid bacteria by quadratic response surface methodology. Enzyme Microb Technol. 1999;25:795–809. [Google Scholar]
- 20.Gobbetti M, Lanciotti R, De Angelis M, Corbo M R, Massini R, Fox P F. Study of the effects of temperature, pH and NaCl on the peptidase activities of non-starter lactic acid bacteria (NSLAB) by quadratic response surface methodology. Int Dairy J. 1999;9:865–875. [Google Scholar]
- 21.Gómez-Zavaglia A, Abraham A, Giorgeri S, De Antoni G. Application of polyacrylamide gel electrophoresis and capillary gel electrophoresis to the analysis of Lactobacillus delbrueckii whole-cell proteins. J Dairy Sci. 1999;82:870–877. [Google Scholar]
- 22.Hammes W P, Vogel R. The genus Lactobacillus. In: Wood B J B, Holzapfel W H, editors. The genera of lactic acid bacteria. London, United Kingdom: Blackie Academic and Professional; 1995. p. 19. [Google Scholar]
- 23.Heukeshoven J, Dernik R. Increased sensitivity for Coomassie staining of sodium dodecyl sulfate-polyacrylamide gels using PhastSystem development unit. Electrophoresis. 1988;9:60–61. doi: 10.1002/elps.1150090112. [DOI] [PubMed] [Google Scholar]
- 24.International Dairy Federation. Cheese and processed cheese. Determination of chloride content: potentiometric titration method. Standard 88. Brussels, Belgium: International Dairy Federation; 1979. [Google Scholar]
- 25.International Dairy Federation. Determination of the total solids content (cheese and processed cheese). Standard 4A. Brussels, Belgium: International Dairy Federation; 1982. [Google Scholar]
- 26.Jayarao B M, Oliver S P. Polymerase chain reaction-based DNA fingerprinting for identification of Streptococcus and Enterococcus species isolated from bovine milk. J Food Prot. 1994;57:240–245. doi: 10.4315/0362-028X-57.3.240. [DOI] [PubMed] [Google Scholar]
- 27.Johansson M L, Quednau M, Molin G, Ahrné S. Randomly amplified polymorphic DNA (RAPD) for rapid typing of Lactobacillus plantarum strains. Lett Appl Microbiol. 1995;21:155–159. doi: 10.1111/j.1472-765x.1995.tb01030.x. [DOI] [PubMed] [Google Scholar]
- 28.Jordan K N, Cogan T M. Identification and growth of non-starter lactic acid bacteria in Irish Cheddar cheese. Ir J Agric Food Res. 1993;32:47–55. [Google Scholar]
- 29.Kandler O, Weiss N. Regular, non-sporing Gram-positive rods. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 2. Baltimore, Md: Williams and Wilkins Co.; 1986. p. 1208. [Google Scholar]
- 30.Kangfu Y, Pauls K P. Optimization of the PCR program for RAPD analysis. Nucleic Acids Res. 1992;20:2606. doi: 10.1093/nar/20.10.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lane C N, Fox P F. Contribution of starter and added lactobacilli to proteolysis in Cheddar cheese during ripening. Int Dairy J. 1996;6:715–728. [Google Scholar]
- 32.Lynch C M, McSweeney P L H, Fox P F, Cogan T M, Drinan F B. Manufacture of cheddar cheese with and without adjunct lactobacilli under controlled microbiological conditions. Int Dairy J. 1996;6:851–867. [Google Scholar]
- 33.Mangin I, Corroler D, Reinhardt A, Gueguen M. Genetic diversity among dairy lactococcal strains investigated by polymerase chain reaction with three arbitrary primers. J Appl Microbiol. 1999;86:514–520. doi: 10.1046/j.1365-2672.1999.00699.x. [DOI] [PubMed] [Google Scholar]
- 34.McSweeney P L H, Fox P F, Lucey J A, Jordan K N, Cogan T M. Contribution of the indigenous microflora to the maturation of Cheddar cheese. Int Dairy J. 1993;3:613–634. [Google Scholar]
- 35.Naylor J, Sharpe M E. Lactobacilli in Cheddar cheese. II. Duplicate cheeses. J Dairy Res. 1958;25:421–430. [Google Scholar]
- 36.Peterson S D, Marshall R T. Non-starter lactobacilli in Cheddar cheese: a review. J Dairy Sci. 1990;73:1393–1410. [Google Scholar]
- 37.Pot B, Hertel C, Ludwig W, Descheemaeker P, Kersters K, Schleifer K H. Identification and classification of Lactobacillus acidophilus, L. gasseri and L. johnsonii strains by SDS-PAGE and rRNA-targeted oligonucleotide probe hybridization. J Gen Microbiol. 1993;139:513–517. doi: 10.1099/00221287-139-3-513. [DOI] [PubMed] [Google Scholar]
- 38.Reniero R, Morelli L, Callegari M L, Sommi P, Bottazzi V. Surface proteins in enteric lactobacilli. Ann Microbiol Enzimol. 1990;40:83–91. [Google Scholar]
- 39.Rohlf F J. NTSYS.PC. Numerical taxonomy and multivariate analysis system, version 1.8. New York, N.Y.: Applied Biostatistics Inc.; 1993. [Google Scholar]
- 40.Rossi F, Torriani S, Dellaglio F. Identification and clustering of dairy propionibacteria by RAPD-PCR and CGE-REA methods. J Appl Microbiol. 1998;85:956–964. doi: 10.1111/j.1365-2672.1998.tb05259.x. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Samelis J, Tsakalidou E, Metaxopoulos J, Kalantzopoulos G. Differentiation of Lactobacillus sake and Lact. curvatus isolated from naturally fermented Greek dry salami by SDS-PAGE of whole-cell proteins. J Appl Bacteriol. 1995;78:157–163. doi: 10.1111/j.1365-2672.1995.tb03165.x. [DOI] [PubMed] [Google Scholar]
- 43.Schleifer K H, Ludwig W, Amann R, Hertel C, Ehrmann M, Köhler W, Krause A. Phylogenetic relationships of lactic acid bacteria and their identification with nucleic acid probes. In: Novel G, Le Querler J F, editors. Les bacteries lactiques. Actes du Colloque LACTIC 91. Caen, France: Centre du Publications de l'Université de Caen; 1992. pp. 23–32. [Google Scholar]
- 44.Sokal R R, Michener C D. A statistical method for evaluating systematic relationship. Univ Kans Sci Bull. 1958;38:1409–1438. [Google Scholar]
- 45.Spicher G, Schröeder R. The microflora of sourdough. IV. Communication: bacterial composition of sourdough starters. Genus Lactobacillus Beijerinck. Z Lebensm Unters Forsch. 1978;167:342–354. doi: 10.1007/BF01415931. [DOI] [PubMed] [Google Scholar]
- 46.Tailliez P, Quénée P, Chopin A. Estimation de la diversité parmi les souches de la colection CNRZ: application de la RAPD à un groupe de lactobacilles. Lait. 1996;76:147–158. [Google Scholar]
- 47.Tailliez P, Tremblay J, Ehrlich S D, Chopin A. Molecular diversity and relationship within Lactococcus lactis, as revealed by randomly amplified polymorphic DNA (RAPD) Syst Appl Microbiol. 1998;21:530–538. doi: 10.1016/S0723-2020(98)80065-9. [DOI] [PubMed] [Google Scholar]
- 48.Turner K W, Lawrence R C, Levriere J. A microbiological specification for milk for aseptic cheese making. N Z J Dairy Sci Technol. 1986;21:249–254. [Google Scholar]
- 49.Vincent D, Roy D, Mondou F, Déry C. Characterization of bifidobacteria by random DNA amplification. Int J Food Microbiol. 1998;43:185–193. doi: 10.1016/s0168-1605(98)00109-3. [DOI] [PubMed] [Google Scholar]
- 50.Yasui T, Yoda K, Kamiya T. Analysis of S-layer proteins of Lactobacillus brevis. FEMS Microbiol Lett. 1995;133:181–186. doi: 10.1111/j.1574-6968.1995.tb07881.x. [DOI] [PubMed] [Google Scholar]