Abstract
Purpose
Variability in cytochrome P450 3A4 (CYP3A4) metabolism is mainly caused by non-genetic factors, hence providing a need for accurate phenotype biomarkers. Although 4β-hydroxycholesterol (4βOHC) is a promising endogenous CYP3A4 biomarker, additional investigations are required to evaluate its ability to predict CYP3A4 activity. This study investigated the correlations between 4βOHC concentrations and hepatic and intestinal CYP3A4 protein expression and ex vivo microsomal activity in paired liver and jejunum samples, as well as in vivo CYP3A4 phenotyping (midazolam) in patients with a wide body weight range.
Methods
The patients (n = 96; 78 with obesity and 18 normal or overweight individuals) were included from the COCKTAIL-study (NCT02386917). Plasma samples for analysis of 4βOHC and midazolam concentrations, and liver (n = 56) and jejunal (n = 38) biopsies were obtained. The biopsies for determination of CYP3A4 protein concentration and microsomal activity were obtained during gastric bypass or cholecystectomy. In vivo CYP3A4 phenotyping was performed using semi-simultaneous oral (1.5 mg) and intravenous (1.0 mg) midazolam.
Results
4βOHC concentrations were positively correlated with hepatic microsomal CYP3A4 activity (ρ = 0.53, p < 0.001), and hepatic CYP3A4 concentrations (ρ = 0.30, p = 0.027), but not with intestinal CYP3A4 concentrations (ρ = 0.18, p = 0.28) or intestinal microsomal CYP3A4 activity (ρ = 0.15, p = 0.53). 4βOHC concentrations correlated weakly with midazolam absolute bioavailability (ρ = − 0.23, p = 0.027) and apparent oral clearance (ρ = 0.28, p = 0.008), but not with systemic clearance (ρ = − 0.03, p = 0.81).
Conclusion
These findings suggest that 4βOHC concentrations reflect hepatic, but not intestinal, CYP3A4 activity. Further studies should investigate the potential value of 4βOHC as an endogenous biomarker for individual dose requirements of intravenously administered CYP3A4 substrate drugs.
Trial registration
Clinical.Trials.gov identifier: NCT02386917.
Supplementary information
The online version contains supplementary material available at 10.1007/s00228-022-03336-9.
Keywords: 4β-Hydroxycholesterol, CYP3A4, Midazolam pharmacokinetics, Drug metabolism, Proteomics
Introduction
The cytochrome P450 (CYP) 3A subfamily, consisting mainly of CYP3A4 and the polymorphic CYP3A5 [1], plays a significant role in the metabolism of 30–50% of clinically used drugs [2, 3]. Due to its abundant expression in both the liver and small intestine [4], CYP3A4 contributes significantly to the first-pass and systemic metabolism of substrate drugs. Hence, CYP3A4 is an important determinant for oral bioavailability and systemic clearance and thereby systemic drug exposure. There is substantial inter-individual variability in CYP3A4 activity, mainly due to environmental factors, disease state, and drug-drug interactions, leading to differences in dose requirements between patients. Conditions such as obesity and non-alcoholic fatty liver disease (NAFLD) are associated with a lower expression and activity of CYP3A4, and several studies suggest that body weight is inversely associated with CYP3A4 activity [5–10]. So far, genetic factors seem to be of limited importance for the substantial variability in CYP3A4 phenotype [11, 12]. Thus, non-genetic biomarkers are warranted to study individual variability in CYP3A4 activity in humans.
The current gold standard method to assess in vivo CYP3A4 activity is midazolam, a selective CYP3A4 phenotypic probe drug in vivo with a short half-life [13–16]. However, since midazolam is a medium-to-high extraction ratio drug, systemic clearance may be affected by changes in protein binding, hepatic blood flow, and intrinsic clearance (CLint), the latter representing metabolic capacity [17–19]. Previous studies have suggested that midazolam pharmacokinetics may be influenced by an increased hepatic blood flow in patients with obesity [20, 21]. Thus, in selected patient populations, the use of midazolam for CYP3A4 phenotyping may be challenging as it does not necessarily reflect CYP3A4 activity. 4β-hydroxycholesterol (4βOHC), a cholesterol metabolite mainly produced by CYP3A4, is proposed as a promising endogenous biomarker for CYP3A4 [22, 23]. There are practical advantages with an endogenous biomarker such as 4βOHC; it is non-invasive, and only a single blood sample is required, making it more convenient for measuring CYP3A4 activity than traditional pharmacokinetic studies with CYP3A4 probe drugs. To assess its suitability as a marker for CYP3A4 phenotype, several studies have investigated the correlation between plasma concentrations of 4βOHC and apparent oral clearance (CL/F) or systemic clearance of midazolam, but the results are conflicting [24–27]. The current literature suggests that 4βOHC concentrations reflect inter-individual variability in CYP3A4 activity and have mainly been suggested as a marker for CYP3A4 induction [28–30]. Still, the long elimination half-life may limit the ability to observe acute changes in CYP3A4 activity [31]. Also, the contribution of intestinal CYP3A4 in the formation of 4βOHC remains unclear [32]. The potential use of 4βOHC as a biomarker for individualized oral dosing of CYP3A4 substrates has thus been debated [33–35].
Additional investigations are required before 4βOHC can be accepted as a validated biomarker. As part of the primary endpoint in the COCKTAIL study [32], we evaluated 4βOHC as an endogenous biomarker for CYP3A4 activity by investigating the correlations between 4βOHC and different CYP3A4 metrics: (1) CYP3A4 expression and (2) ex vivo microsomal CYP3A4 activity in paired liver and jejunum biopsies and (3) absolute bioavailability, apparent oral clearance, and systemic clearance of midazolam in vivo.
Methods
Patients
In total, 96 patients from the open-label, three-armed COCKTAIL-study (NCT02386917) were included in the present analysis [36, 37]. The study population included 78 patients with severe obesity (BMI -> 40 or 35–40 kg/m2 combined with at least one obesity-related comorbidity) scheduled for weight loss treatment with Roux-en-Y-gastric bypass (RYGB) (n = 38) or non-surgical calorie restriction (n = 40), and 18 mainly normal to overweight individuals scheduled for cholecystectomy (BMI 18.5–30 kg/m2). The patients with severe obesity were subjected to a 3-week low-energy diet (< 1200 kcal/day) before the study investigation, whereas the normal to overweight individuals were not subjected to any defined diet beforehand. Medications, including statins, and/or other substances that are known to alter CYP3A4 activity/midazolam pharmacokinetics were discontinued at least seven half-lives before the investigational day, and none of the patients used any CYP3A4 inducers or time-dependent inhibitors. The study was approved by the Regional Committee for Medical and Health Research Ethics (2013/2379/REK) and complied with the Declaration of Helsinki. All patients signed a written informed consent before study participation.
Study investigation
Blood samples for determination of plasma 4βOHC concentrations and a 24-h pharmacokinetic investigation using semi-simultaneous oral and intravenous dosing of midazolam were collected from all patients. From 10:00 p.m. before the investigation, patients abstained from food, drink (except water), and drugs. On the investigational day, patients first met for blood sampling before 1.5 mg oral midazolam syrup was administered, followed by 1.0 mg intravenous midazolam 4 h later. Blood samples for determination of midazolam plasma concentrations were collected from a peripheral venous catheter before and at 0.25, 0.5, 1, 1.5, 2, 3, 4, 4.25, 2.5, 5, 5.5, 6, 8, 10, 12, 23, and 24 h following oral dosing. The pharmacokinetic investigation has been described in more detail previously [37]. In vivo midazolam pharmacokinetic data were available in 92 patients. The day after the study investigation, paired liver and jejunum biopsies were obtained during RYGB (n = 38), and liver biopsies were obtained during cholecystectomy (n = 18), as previously described [9, 38].
Body composition was measured with Inbody 720, Body Composition Analyzer (Biospace, Korea). The NAFLD liver fat score was calculated as suggested by Kotronen et al. [39]. Standard clinical chemistry analyses were performed in fresh blood samples at the Department of Laboratory Medicine, Vestfold Hospital Trust, Tønsberg, Norway. Plasma concentrations of high-sensitivity C-reactive protein (hs-CRP) were measured using immunoturbidimetry (Advia Chemistry XPT systems, Siemens) at Fürst Medical Laboratory, Oslo, Norway. Blood samples for determination of 4βOHC and midazolam concentrations were drawn in K2-EDTA vacutainer tubes on ice and centrifuged for 10 min at 4 °C (1,800 g). Plasma was separated into Cryovials and frozen within 1 h at − 70 °C until analysis.
Bioanalytical assays
4βOHC
4βOHC plasma concentrations were determined by a previously described assay [40] with an added filtration step [41]. In short, 4βOHC was de-esterified from fatty acids by ethanolic sodium methoxide and isolated from plasma by liquid–liquid extraction with hexane. Extracts were evaporated by nitrogen and reconstituted in methanol before filtration. After methanol reconstitution, the samples were frozen at − 20 °C for 15 min, and filtered in specialized Eppendorf filter tubes (Costar Spin-x HPLC Micro Centrifuge Filter, 0,2 µM Nylon Filter) through centrifugation (2,000 g for 6 min at 2 °C). The filtered extract was used for UPLC-MS/MS analysis. An Acquity ultra-performance liquid chromatograph (UPLC) followed by a Micromass Quattro micro tandem mass spectrometry (MS) detector (Waters, Milford, MA) was used for quantitative analysis. Chromatographic separation was achieved on a BEH C18 column RP-shield (1.7 μm, 1 × 100 mm) from Waters with a mobile phase of water and methanol. After atmospheric pressure chemical ionization, detection was obtained with multiple reaction monitoring at m/z 385.25 -> 367.45 (4βOHC) and m/z 392.30 -> 374.50 (4βOHC-D7; internal standard). The lower limit of quantification (LLOQ) was 10 ng/mL. Intra‐ and interday imprecision and inaccuracy were < 15% at 10 ng/mL and < 4% at 644 ng/mL (n = 6) [40]. All samples were stored at − 70 °C between sampling and analysis.
Midazolam
The plasma concentrations of midazolam were determined by liquid–liquid sample extraction followed by a previously validated UHPLC-MS/MS method [42]. The sample preparation in short, 50 ng/mL internal standard (deuterated midazolam (MDZ-d6)), was added to a 5-mL Eppendorf tube, evaporated to dryness by nitrogen gas followed by subsequent addition of 250 µL of sample plasma. Aqueous ammonia (0.5 M) was added to the tubes in a 1:1 ratio (250 µL) and agitated prior to the addition of the extracting solvent (1.0 mL of ethyl acetate). The sample was mixed for 10 min (Invitrogen Hulamixer, Thermo Fischer, Scientific Inc) prior to being centrifuged at 2,500 g for 10 min at room temperature (Heraeus Megafuge 16R-centrifuge, Thermo Fisher Scientific Inc). The extraction was carried out twice before the organic phase was transferred to a new Eppendorf tube, evaporated to dryness under nitrogen (60 °C), and subsequently reconstituted in 50 µL 5% ACN in 0.05 M ammonium acetate buffer (pH 4.4). The sample was mixed and transferred to micro vials before UHPLC-MS/MS analysis.
The UHPLC-MS/MS method was carried out in two labs due to limited capacity. The instrument in lab 1 was a Thermo Fisher Scientific Vanquish HPLC coupled to a Thermo Fisher Accucore (C18 2.1 × 50 mm) analytical column and a Thermo Fisher Scientific Altis QqQ MS. In lab 2, a Waters (Waters, Milford, MA) Acquity UPLC with a BEH C18 RP-shield (1.7 μm, 1 × 100 mm) analytical column coupled to a Micromass Quattro micro tandem MS was used. The chromatography was carried out with a flow rate of 0.4 mL/min and 0.2 mL/min and gradient elution for lab 1 and lab 2, respectively. Mobile phase A was 5% acetonitrile in 10 mM ammonium formate, and mobile phase B was 90% acetonitrile and 10% methanol. Positive electrospray ionization and selected reaction monitoring were performed with compound-tuned conditions. For both labs, acquisition was carried out by the MRM transitions m/z 326.1 -> m/z 249.1 and m/z 326.1 -> m/z 291.2 for midazolam, and m/z 332.1 -> m/z 252.1 and m/z 332.1 -> m/z 203.1 for midazolam-d6 in positive mode.
Inter-laboratory cross-validation was performed using clinical study samples, and the bias was within ± 12.5%. Calibrators and quality control (QC) samples were prepared in blank plasma and analyzed in each analytical run. Eight calibrators in the range of 0.1 to 20 ng/mL were applied, and back-calculated values of calibrators within 80 to 120% were accepted. LLOQ was 0.1 ng/mL, and the upper limit of quantification (ULOQ) was 20 ng/mL. Samples with midazolam concentrations above ULOQ were diluted in blank plasma and reanalyzed. Dilution integrity with dilution factors of 1/2, 1/5, 1/10, 1/20, and 1/50 was established; mean accuracy ranged from 88.9 to 103.8%, and the imprecision was < 4.5%. Within-series and between-series performance were assessed with the resulting coefficient of variation < 12.3%, and the mean accuracy ranged from 99.3 to 104.3%.
Liver and jejunum biopsies
The biopsies were transferred into individual cryotubes, snap-frozen in liquid nitrogen directly upon sampling, and stored at − 80 °C until analysis.
CYP3A4 protein quantification
Proteins were extracted from liver and jejunum biopsies in an SDS-containing (2% w/v) lysis buffer and quantified as previously described [43]. In short, samples were processed with the multi-enzyme digestion filter-aided sample preparation protocol, using LysC and trypsin [44]. Proteomics analysis was performed with Q Exactive HF or Q Exactive HF-X. MS data were processed with MaxQuant (version 1.6.10.43) [45], using the human UniProtKB. Spectral raw intensities were normalized with variance stabilization [46] and were subsequently used to calculate the protein concentrations using the total protein approach [47].
Preparation of microsomes and ex vivo CYP3A4 activity assay
Methods for quantifying CYP3A4 activity in liver and jejunum biopsies have been described previously [38]. In brief, liver and jejunum biopsies were prepared with a Potter–Elvehjem homogenizer, and differential centrifugation was used to isolate microsomal fractions. Midazolam was used as a probe drug and the formation of the metabolite 1′-hydroxymidazolam as a probe reaction for CYP3A4 activity. Activity incubation was carried out at 37 °C for 20 min, and midazolam was added in eight different concentrations. The incubation was terminated using ice-cold acetonitrile containing internal standard. Quantification of 1′-hydroxymidazolam was performed using a triple quadrupole mass spectrometer coupled to an ACQUITY UPLC I-class system (Waters XevoTM TQ-S; Waters Corporation, Milford, MA) equipped with an electrospray ionization source as previously described [38].
Genotype analyses
CYP3A4 and CYP3A5 variant alleles were analyzed using Taqman-based real-time polymerase chain reaction assays implemented for routine pharmacogenetic analyses at the Center for Psychopharmacology, Diakonhjemmet Hospital, Norway. CYP3A4, the reduced function allele*22 (rs35599367), and CYP3A5, the null allele *3 (rs776746), were included in the analysis. All alleles were in Hardy–Weinberg equilibrium.
Pharmacokinetic calculations
In vivo absolute bioavailability, apparent oral clearance, and systemic clearance of midazolam from the pharmacokinetic investigation were determined using a previously developed population pharmacokinetic model [37]. Briefly, the modeling was performed using the non-parametric adaptive grid approach implemented in Pmetrics (version 1.5.2) for R (version 3.6.2) [48, 49]. A catenary three-compartment model with absorption lag-time and first-order elimination from the central compartment described the data adequately. The enzyme kinetic parameters from the ex vivo CYP3A4 activity investigation were determined using untransformed data and GraphPad prism 7 by fitting the reaction velocity versus the substrate concentration data to the Michaelis–Menten model or the substrate inhibition model, as previously described [38]. The Michaelis constant (Km) values were adjusted for the fraction of unbound drug in microsomes, which was predicted from the physicochemical properties of the substrates and microsomal protein concentration using the Simcyp prediction tool (https://members.simcyp.com/account/tools/fumic). The unbound CLint (CLint,u) was calculated from the ratio of maximum velocity (Vmax) to the unbound Km. Due to limited capacity, the hepatic and jejunum microsomal CLint,u were determined in 20 RYGB patients.
Data and statistical analysis
Neither 4βOHC nor most of the other variables passed the normality tests (including the Shapiro–Wilk test and visual inspection of plots), and non-parametric statistical tests were therefore used in the statistical analyses. Spearman’s rank-order correlation test was used to describe the rank-based measure of association. Wilcoxon rank-sum test was used to compare patients with obesity with normal to overweight individuals. A p value < 0.05 was considered statically significant. Data are presented as median difference (95% confidence interval (CI)) if not otherwise stated. Demographic data are presented as mean ± standard deviation (SD). All statistical analyses were performed using R for Windows (version 4.1.2) [49].
Results
Patient characteristics
Patient characteristics at the investigational day are given in Table 1. The patients had a mean age of 46 ± 11 years, 70% were women, and 98% were Caucasian. The majority (94%) were genotypic normal CYP3A4 metabolizers (CYP3A4 *1/*1), and 10% also expressed functional CYP3A5 (CYP3A5 *1/*3). In line with the inclusion criteria, mean BMI was higher in patients with obesity compared with normal to overweight individuals. Patients with obesity had lower serum total cholesterol compared with the normal to overweight individuals (− 0.60 mmol/L [95% CI: − 1.0, − 0.00]) but higher NAFLD liver fat score (2.8 [95% CI: 1.9, 3.6]) and hs-CRP (2.1 mg/L [95% CI: 1.1, 3.9]). Twenty percent of the patients received treatment with cholesterol-lowering drugs (mainly statins) (patients with obesity = 19, normal to overweight = 0).
Table 1.
Patient characteristics at the study investigation. Data are described as mean ± SD or absolute numbers (%)
|
Patients with obesity n = 78 |
Normal to overweight individuals n = 18 |
|
|---|---|---|
| Age (years) | 47 ± 10 | 42 ± 15 |
| Sex (male/female) | 26/52 | 3/15 |
| Body weight (kg) | 121 ± 22 | 71 ± 11 |
| BMI (kg/m2) | 41 ± 5.8 | 25 ± 3.5 |
| Total cholesterol (mmol/L) | 3.9 ± 0.95 | 4.4 ± 0.87 |
| Triglycerides (mmol/L) | 1.3 ± 0.59 | 0.93 ± 0.47 |
| LDL (mmol/L) | 2.4 ± 0.82 | 2.5 ± 0.77 |
| HDL (mmol(L) | 0.96 ± 0.20 | 1.5 ± 0.36 |
| hs-CRP (mg/L) | 5.7 ± 6.0 | 2.5 ± 3.8 |
| ALT (U/L) | 39 ± 21 | 22 ± 15 |
| NAFLD liver fat score | 1.1 ± 2.0 | − 1.7 ± 1.1 |
| CYP3A4 genotype (likely phenotype) | ||
| *1/*1 (normal metabolizer) | 74 (95%) | 16 (89%) |
| *1/*22 (intermediate metabolizer) | 4 (5%) | 2 (11%) |
| CYP3A5 genotype (likely phenotype) | ||
| *1/*3 (intermediate metabolizer) | 7 (9%) | 3 (17%) |
| *3/*3 (poor metabolizer) | 71 (91%) | 15 (83%) |
ALT alanine aminotransferase, BMI body mass index, CYP cytochrome P450, HDL high-density lipoproteins, hs-CRP high-sensitivity C-reactive protein, LDL low-density lipoproteins, NAFLD non-alcoholic fatty liver disease
Correlations between 4βOHC and CYP3A4 metrics
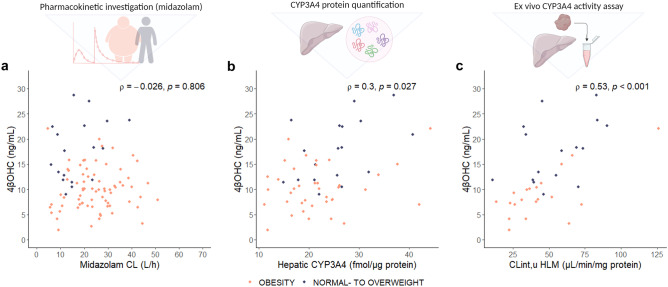
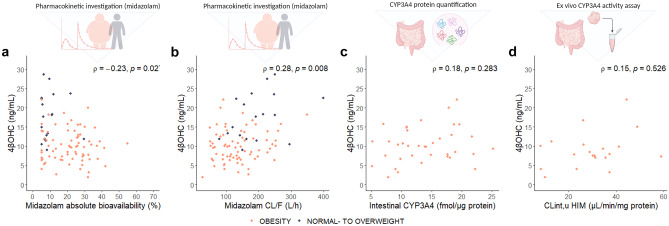
4βOHC concentrations were positively correlated with both hepatic CYP3A4 concentrations (ρ = 0.30, p = 0.027) and hepatic microsomal CLint,u (ρ = 0.53, p < 0.001), but not with systemic midazolam clearance (ρ = − 0.03, p = 0.81) (Fig. 1). The lack of correlation between 4βOHC concentrations and systemic midazolam clearance was also shown when the patients with obesity and the normal to overweight individuals were analyzed separately (Supplementary Fig. S1). Hepatic microsomal activity was significantly correlated with hepatic CYP3A4 concentrations (ρ = 0.51, p = 0.001), but no correlation was observed between systemic midazolam clearance and hepatic microsomal CLint,u (Supplementary Fig. S2). 4βOHC concentrations were not significantly correlated with intestinal CYP3A4 concentrations or intestinal microsomal CLint,u (Fig. 2c and d). However, 4βOHC plasma concentrations showed an inverse correlation with absolute bioavailability of midazolam (ρ = − 0.23, p = 0.027), and a positive correlation with apparent oral clearance (ρ = 0.28, p = 0.008) (Fig. 2a and b). Absolute values of the different CYP3A metrics are reported in Supplementary Table S1.
Fig. 1.
Hepatic CYP3A4 metrics. Correlation between (a) systemic midazolam clearance (CL) and 4βOHC concentrationsa, (b) hepatic CYP3A4 concentrationb and 4βOHC concentrations, and (c) clearance intrinsic for midazolam 1’-hydroxylation in human liver microsomesc and 4βOHC concentrations. Spearman’s rho (ρ) is the correlation coefficient, and the p value is from the Spearman rank correlation analysis. aAvailable in 92 patients (patients with obesity = 74, normal to overweight individiuals = 18). bAvailable in 56 patients (patients with obesity = 38, normal to overweight individuals = 18). cAvailable in 36 patients (RYGB = 20, normal to overweight individuals = 16). Abbreviations: CLint,u, clearance intrinsic unbound; CYP, cytochrome P450; HLM, human liver microsomes; 4βOHC, 4-beta hydroxycholesterol
Fig. 2.
Intestinal CYP3A4 metrics. Correlation between (a) midazolam absolute bioavailabilitya and 4βOHC concentrations, (b) apparent oral midazolam clearance (CL/F)a and 4βOHC concentrations, (c) jejunum CYP3A4 concentrationb and 4βOHC concentrations, and (d) clearance intrinsic for midazolam 1′-hydroxylation in human intestinal microsomesc and 4βOHC concentrations. Spearman’s rho (ρ) is the correlation coefficient, and the p value is from the Spearman rank correlation analysis. aAvailable in 92 patients (patients with obesity = 74, normal to overweight individuals = 18). bAvailable in 37 RYGB-patients. cAvailable in 20 RYGB-patients. Abbreviations: CLint,u, clearance intrinsic unbound; CYP, cytochrome P450; HIM, human intestinal microsomes; 4βOHC, 4-beta hydroxycholesterol
In the patients with obesity, neither 4βOHC concentrations, systemic midazolam clearance, nor absolute bioavailability changed during the 3-week low-energy diet period (Supplementary Fig. S3). However, due to the low-fat low-energy diet and weight loss, the total cholesterol was lowered in this period. All correlations were thus repeated using the 4βOHC/cholesterol ratio (4βOHC/C). The results from these analyses showed a similar pattern (Supplementary Figs. S4 and S5).
Impact of genotype and patient characteristics on 4βOHC
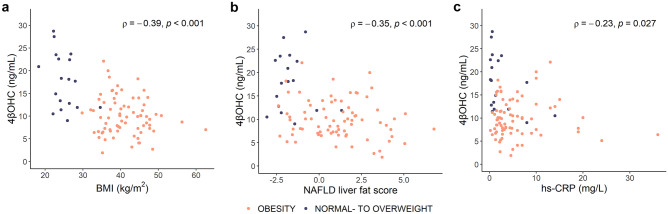
An analysis of 4βOHC concentrations based on genotype did not indicate elevated 4βOHC in the individuals expressing CYP3A5 *1/*3 compared with CYP3A5 *3/*3 expressors, neither in normal to overweight individuals (p = 0.95) nor in patients with obesity (p = 0.33) (Supplementary Fig. 6a and b). However, 4βOHC concentrations were lower in patients with obesity expressing CYP3A4 *1/*22 compared with CYP3A4 *1/*1 (p = 0.024), whereas in the normal to overweight individuals, the difference did not reach statistical significance (p = 0.12) (Supplementary Fig. S6c and d). BMI was inversely correlated with 4βOHC concentrations: ρ = − 0.39 (p < 0.001) (Fig. 3a), and median 4βOHC was 1.9-fold higher in normal to overweight individuals compared with patients with obesity (median difference 7.4 ng/mL [95% CI: 4.4, 11]) (Supplementary Fig. S7a). Both NAFLD liver fat score and hs-CRP were inversely correlated with 4βOHC: ρ = − 0.35 (p < 0.001) and ρ = − 0.23 (p = 0.027), respectively (Fig. 3b and c). Absolute bioavailability and systemic clearance of midazolam were 2.8-fold and 1.6-fold higher in the patients with obesity compared with the normal to overweight individuals (13% [95% CI: 5.6, 18] and 7.7 L/h [95% CI: 2.3, 13], respectively) (Supplementary Fig. S7b and c). There was no sex difference in 4βOHC concentrations in the study population (median difference between females and males: 1.8 ng/mL [95% CI: − 0.30, 3.9]).
Fig. 3.
Clinical variables and 4βOHC. Correlation between (a) BMI and 4βOHC concentrations (n = 96), (b) NAFLD liver fat score and 4βOHC concentrations (n = 95), and (c) hs-CRP and 4βOHC concentrations (n = 96). Spearman’s rho (ρ) is the correlation coefficient, and the p value is from the Spearman rank correlation analysis. Abbreviations: BMI, body mass index; hs-CRP, high-sensitivity C-reactive protein; NAFLD, non-alcoholic fatty liver disease; 4βOHC, 4-beta hydroxycholesterol
Discussion
In this study, a comprehensive investigation of 4βOHC as an endogenous biomarker for CYP3A4 activity was performed. To our knowledge, this is the first study where 4βOHC has been compared with three other CYP3A4 metrics: intestinal and hepatic microsomal CYP3A4 activity and protein expression in paired liver and jejunum samples, as well as absolute bioavailability, apparent oral clearance, and systemic clearance of midazolam in vivo. The main findings were that 4βOHC concentrations were positively correlated with hepatic microsomal CYP3A4 activity and hepatic CYP3A4 expression. Equally important, this study also provides evidence of a negligible contribution from intestinal CYP3A4 to the formation of 4βOHC.
The moderate correlation between 4βOHC concentrations and hepatic microsomal activity (CLint,u) supports that 4βOHC concentrations reflect individual CYP3A4 metabolic capacity and provides evidence that 4βOHC is a suitable marker for hepatic CYP3A4 phenotype. However, there was no correlation between 4βOHC concentrations and systemic clearance of midazolam, the gold standard method to assess CYP3A4 activity. The correlation between these two metrics has been investigated in multiple studies previously, with conflicting findings varying from no correlation as in this study to a weak or moderate correlation [24–27, 50, 51]. Even though systemic clearance of midazolam currently is the preferred method for CYP3A4 phenotyping, it may have some important limitations in special populations, e.g., patients with obesity, as shown in the present study. Surprisingly, we observed no correlation between hepatic microsomal activity (CLint,u) and in vivo systemic midazolam clearance, suggesting that systemic midazolam clearance does not solely reflect CYP3A4 activity in the present population. In contrast to hepatic microsomal CLint,u, in vivo systemic midazolam clearance may also be influenced by differences in hepatic blood flow given its medium to high extraction ratio [17–19] which may be especially relevant in patients with chronic diseases or in case of simultaneous physiological alterations. We have previously hypothesized that hepatic blood flow may have a more pronounced impact on systemic midazolam clearance than CYP3A4 activity in patients with obesity due to higher liver blood flow in this patient population compared with normal-weight individuals [20]. Brill et al. have also suggested that blood flow alterations seem to influence midazolam pharmacokinetics in patients with obesity [21]. This may at least partly explain why patients with obesity had lower 4βOHC concentrations in combination with higher systemic midazolam clearance compared with normal to overweight individuals, and also the lack of correlation between 4βOHC and systemic midazolam clearance in the present study. 4βOHC may therefore be an especially relevant biomarker to include in midazolam-based CYP3A4 phenotyping studies in specific patient populations where physiological factors such as altered blood flow may challenge the interpretation of the results. Additionally, there are some methodological considerations with the ex vivo CYP3A activity assay that should be taken into consideration. Microsomal activity in small liver tissue samples may not be representative of the CYP3A4 activity across the whole liver [52], the unbound fraction was estimated, and it was not accounted for variability in microsomal protein per gram liver and microsomal recovery.
Information regarding the role of intestinal CYP3A4 in 4βOHC formation has been lacking [32, 53], and uncertainty about the intestinal contribution seems to be an important limitation with 4βOHC phenotyping. We showed that neither intestinal CYP3A4 expression nor intestinal microsomal CYP3A4 activity (CLint,u) was correlated with 4βOHC concentrations. Even though there was a weak, but significant correlation between 4βOHC concentrations and (1) midazolam absolute bioavailability, and (2) apparent oral clearance, but no correlation with systemic clearance, these results indicate that intestinal CYP3A4 has a negligible role in the formation of 4βOHC. There have been some indications of a possible contribution from intestinal CYP3A4 in the formation of 4βOHC [40, 54]. However, a more recent study indicates that intestinal CYP3A4 is unlikely to be of significant importance for the 4βOHC formation [55], which is in line with our findings. Hence, it seems that 4βOHC is mainly appropriate for hepatic CYP3A4 phenotyping, but the relatively small sample size in our study should be kept in mind when interpreting these findings.
In this study, we also report a negative correlation between BMI and 4βOHC, and that patients with obesity have significantly lower 4βOHC concentrations compared with normal to overweight individuals. Previous studies have also found an inverse correlation between BMI and 4βOHC/C, and body weight and 4βOHC/C [10, 25]. Intestinal and hepatic CYP3A4 expression as well as CLint,u using midazolam as a probe drug are also reported to be negatively correlated with BMI [8, 9]. This supports that patients with obesity have a lower CYP3A4 activity compared with normal weight individuals [7]. The normal to overweight individuals in this study had similar values of 4βOHC compared to reported values for corresponding Caucasians in other studies [10, 23, 51, 56]. Lower levels of 4βOHC were also observed by Gravel et al. in patients with a mean BMI of ~ 29 kg/m2, although the authors suggested that this was due to type 2 diabetes, not obesity [51], and by Woolsey et al. in patients with a mean BMI of ~ 33 kg/m2 and NAFLD [5]. CYP3A4 activity seems to be suppressed during inflammation [57, 58], and in agreement with this, we also found a weak inverse correlation between 4βOHC concentrations and the inflammation marker hs-CRP. Björkhem-Bergman et al. also reported a similar correlation in a study with patients with increased susceptibility to respiratory infections [59]. Furthermore, and in agreement with a previous study in patients with NAFLD and obesity [5], we observed a weak inverse correlation between NAFLD liver fat score and 4βOHC concentrations, suggesting decreased CYP3A4 activity in patients with NAFLD.
Most of the cholesterol formation to 4βOHC is accounted for by CYP3A4 metabolism, whereas the contribution from CYP3A5 is more ambiguous [12, 22, 23, 60, 61]. Hole et al. found no significant effect of CYP3A5 genotype on 4βOHC concentrations in a similar study population as ours and suggested that CYP3A5 has a limited role in the formation of 4βOHC in vivo [12]. In agreement with this, we did not observe any significant difference in 4βOHC concentrations between individuals expressing functional (CYP3A5*1/*3) and non-functional CYP3A5 (CYP3A5*3/*3). Nevertheless, we do not believe that the heterozygote CYP3A5*1 carriers in our study population (~ 10%) have impacted the interpretation of the results in this study to any significant degree. This is supported by the fact that there were no statistically significant differences between the heterozygote CYP3A5*1 carriers and the homozygote CYP3A5*3 carriers in any of the other metrics investigated. Also, there was a tendency of lower 4βOHC concentrations in CYP3A4*22 carriers (reduced function), but the interpretation is challenged by the low number of individuals carrying this allele. Overall, these results are in line with previous studies [12, 25, 50] and support that 4βOHC primarily seems to be a biomarker for CYP3A4 phenotype.
The major strength of this study includes the comprehensive investigation of three different CYP3A4 metrics in relation to 4βOHC, all obtained from the same individuals at the same time point. The patients with obesity were subjected to a 3-week low-energy diet before the study investigation, which may have influenced the results. However, as neither 4βOHC concentrations, absolute bioavailability, nor systemic clearance of midazolam changed from before to after the diet, a change in the CYP3A4 phenotype during this short period seems unlikely. Nevertheless, in addition to the fact that 20% of the patients used cholesterol-lowering drugs, cholesterol levels also decreased following the diet, which led to a significant increase in 4βOHC/C in the patients with obesity. Several studies report the 4βOHC/C ratio to account for abnormal or changing cholesterol levels. However, the concentration of 4βOHC is less than 0.002% of total cholesterol [53]. We decided to report 4βOHC concentrations, as most individuals had normal cholesterol levels, but analyses using 4βOHC/C yielded similar results. Also, Diczfalusy et al. have previously shown that variability in cholesterol levels only explains 9% of the variability in 4βOHC concentrations [23]. Another limitation includes not adjusting for multiple testing, which may increase the likelihood of type 1 errors. Additionally, the results in this study may not be applicable in other populations, as the majority of the study population were patients with obesity.
In summary, this comprehensive study showed that 4βOHC plasma concentrations reflect hepatic, but not intestinal CYP3A4 activity well. Hence, 4βOHC may be a valuable biomarker for individualized dosing of intravenously administered CYP3A4 substrate drugs. Additionally, 4βOHC is a valuable supplement to traditional phenotyping with probe drugs such as midazolam given its easy implementation and complementary information when other factors, such as disease state or simultaneous physiological alterations, are expected to influence the pharmacokinetics of CYP3A4 probe drugs.
Supplementary information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank the participants, the surgical staff, and the study personnel working on the COCKTAIL study at Vestfold Hospital Trust. The authors also thank the Swedish Research Council, approval numbers 5715 and 01951 (C.W., T.B.A., and P.A.), for supporting the proteomics analyses.
Author contribution
J.H., A.Å., S.A., C.K., T.B.A., H.C., E.S., R.S., and R.J. conceived and designed the COCKTAIL-study. A.Å., E.M., K.H., B.M.W., K.E.K., and I.R. designed the present research. I.R., V.K., L.K.J., K.H., B.M.W., J.K.H, P.A., and C.W. performed the research. K.E.K. and I.R. analyzed the data. K.E.K. and I.R. wrote the manuscript. All authors contributed to critically reviewing the manuscript, and gave their final approval for submission.
Funding
Open access funding provided by University of Oslo (incl Oslo University Hospital). The authors would like to thank the following: Vestfold Hospital Trust, Norway; Department of Pharmacy, University of Oslo, Norway; and AstraZeneca, Sweden.
Data availability
Access to data collected from this study, including anonymized individual-participant data, may potentially be made available following publication upon e-mail request to the corresponding author. After approval of a proposal, data will be shared with investigators whose proposed use of the data has been approved by the COCKTAIL steering committee, according to the consent given by the participants and Norwegian laws and legislations.
Declarations
Ethics approval
The study was approved by the Regional Committee for Medical and Health Research Ethics (2013/2379/REK) and complied with the Declaration of Helsinki.
Conflict of interest
C. Karlsson, S. Andersson, and R. Jansson-Löfmark are AstraZeneca employees and own shares in AstraZeneca, while C. Wegler and T.B. Andersson are former AstraZeneca employees. K.E. Kvitne, I. Robertsen, K. Hole, B.M. Wollmann, L.K. Johnson, J.K. Hertel, R. Sandbu, P. Artursson, E. Molden, E. Skovlund, H. Christensen, V. Krogstad, J. Hjelmesæth, and A. Åsberg have no conflict of interest to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Rendic S, Guengerich FP. Survey of human oxidoreductases and cytochrome P450 enzymes involved in the metabolism of xenobiotic and natural chemicals. Chem Res Toxicol. 2015;28(1):38–42. doi: 10.1021/tx500444e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guengerich FP. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Paine MF, et al. The human intestinal cytochrome P450 “pie”. Drug Metab Dispos. 2006;34(5):880–886. doi: 10.1124/dmd.105.008672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woolsey SJ, et al. CYP3A activity and expression in nonalcoholic fatty liver disease. Drug Metab Dispos. 2015;43(10):1484–1490. doi: 10.1124/dmd.115.065979. [DOI] [PubMed] [Google Scholar]
- 6.Kolwankar D, et al. Association between nonalcoholic hepatic steatosis and hepatic cytochrome P-450 3A activity. Clin Gastroenterol Hepatol. 2007;5(3):388–393. doi: 10.1016/j.cgh.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 7.Brill MJ, et al. Midazolam pharmacokinetics in morbidly obese patients following semi-simultaneous oral and intravenous administration: a comparison with healthy volunteers. Clin Pharmacokinet. 2014;53(10):931–941. doi: 10.1007/s40262-014-0166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulvestad M, et al. Impact of OATP1B1, MDR1, and CYP3A4 expression in liver and intestine on interpatient pharmacokinetic variability of atorvastatin in obese subjects. Clin Pharmacol Ther. 2013;93(3):275–282. doi: 10.1038/clpt.2012.261. [DOI] [PubMed] [Google Scholar]
- 9.Krogstad V, et al. Correlation of body weight and composition with hepatic activities of cytochrome P450 enzymes. J Pharm Sci. 2021;110(1):432–437. doi: 10.1016/j.xphs.2020.10.027. [DOI] [PubMed] [Google Scholar]
- 10.Hole K, et al. Elevated 4β-hydroxycholesterol/cholesterol ratio in anorexia nervosa patients. Pharmacol Res Perspect. 2018;6(5):e00430. doi: 10.1002/prp2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein K, Zanger UM. Pharmacogenomics of cytochrome P450 3A4: recent progress toward the “Missing Heritability” problem. Front Genet. 2013;4:12. doi: 10.3389/fgene.2013.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hole K, et al. Impact of genetic and nongenetic factors on interindividual variability in 4β-hydroxycholesterol concentration. Eur J Clin Pharmacol. 2017;73(3):317–324. doi: 10.1007/s00228-016-2178-y. [DOI] [PubMed] [Google Scholar]
- 13.Heizmann P, Eckert M, Ziegler WH (1983) Pharmacokinetics and bioavailability of midazolam in man. Br J Clin Pharmacol 16 Suppl 1(Suppl 1):43s-49s [DOI] [PMC free article] [PubMed]
- 14.de Jonge H, et al. Impact of CYP3A5 genotype on tacrolimus versus midazolam clearance in renal transplant recipients: new insights in CYP3A5-mediated drug metabolism. Pharmacogenomics. 2013;14(12):1467–1480. doi: 10.2217/pgs.13.133. [DOI] [PubMed] [Google Scholar]
- 15.Yu KS, et al. Effect of the CYP3A5 genotype on the pharmacokinetics of intravenous midazolam during inhibited and induced metabolic states. Clin Pharmacol Ther. 2004;76(2):104–112. doi: 10.1016/j.clpt.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Kharasch ED, et al. Influence of CYP3A5 genotype on the pharmacokinetics and pharmacodynamics of the cytochrome P4503A probes alfentanil and midazolam. Clin Pharmacol Ther. 2007;82(4):410–426. doi: 10.1038/sj.clpt.6100237. [DOI] [PubMed] [Google Scholar]
- 17.Thummel KE, et al. Oral first-pass elimination of midazolam involves both gastrointestinal and hepatic CYP3A-mediated metabolism. Clin Pharmacol Ther. 1996;59(5):491–502. doi: 10.1016/S0009-9236(96)90177-0. [DOI] [PubMed] [Google Scholar]
- 18.Klotz U, Ziegler G. Physiologic and temporal variation in hepatic elimination of midazolam. Clin Pharmacol Ther. 1982;32(1):107–112. doi: 10.1038/clpt.1982.133. [DOI] [PubMed] [Google Scholar]
- 19.Rogers JF, et al. An evaluation of the suitability of intravenous midazolam as an in vivo marker for hepatic cytochrome P4503A activity. Clin Pharmacol Ther. 2003;73(3):153–158. doi: 10.1067/mcp.2003.23. [DOI] [PubMed] [Google Scholar]
- 20.Kvitne KE, et al. Short- and long-term effects of body weight loss following calorie restriction and gastric bypass on CYP3A-activity - a non-randomized three-armed controlled trial. Clin Transl Sci. 2022;15(1):221–233. doi: 10.1111/cts.13142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brill MJ, et al. Semiphysiologically based pharmacokinetic model for midazolam and CYP3A mediated metabolite 1-OH-midazolam in morbidly obese and weight loss surgery patients. CPT Pharmacometrics Syst Pharmacol. 2016;5(1):20–30. doi: 10.1002/psp4.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bodin K, et al. Metabolism of 4 beta -hydroxycholesterol in humans. J Biol Chem. 2002;277(35):31534–31540. doi: 10.1074/jbc.M201712200. [DOI] [PubMed] [Google Scholar]
- 23.Diczfalusy U, et al. 4Beta-hydroxycholesterol is a new endogenous CYP3A marker: relationship to CYP3A5 genotype, quinine 3-hydroxylation and sex in Koreans. Swedes and Tanzanians Pharmacogenet Genomics. 2008;18(3):201–208. doi: 10.1097/FPC.0b013e3282f50ee9. [DOI] [PubMed] [Google Scholar]
- 24.Shin KH, et al. Evaluation of endogenous metabolic markers of hepatic CYP3A activity using metabolic profiling and midazolam clearance. Clin Pharmacol Ther. 2013;94(5):601–609. doi: 10.1038/clpt.2013.128. [DOI] [PubMed] [Google Scholar]
- 25.Woolsey SJ, et al. Relationships between endogenous plasma biomarkers of constitutive cytochrome P450 3A activity and single-time-point oral midazolam microdose phenotype in healthy subjects. Basic Clin Pharmacol Toxicol. 2016;118(4):284–291. doi: 10.1111/bcpt.12492. [DOI] [PubMed] [Google Scholar]
- 26.Tomalik-Scharte D, et al. Plasma 4beta-hydroxycholesterol: an endogenous CYP3A metric? Clin Pharmacol Ther. 2009;86(2):147–153. doi: 10.1038/clpt.2009.72. [DOI] [PubMed] [Google Scholar]
- 27.Björkhem-Bergman L, et al. Comparison of endogenous 4β-hydroxycholesterol with midazolam as markers for CYP3A4 induction by rifampicin. Drug Metab Dispos. 2013;41(8):1488–1493. doi: 10.1124/dmd.113.052316. [DOI] [PubMed] [Google Scholar]
- 28.Bodin K, et al. Antiepileptic drugs increase plasma levels of 4beta-hydroxycholesterol in humans: evidence for involvement of cytochrome p450 3A4. J Biol Chem. 2001;276(42):38685–38689. doi: 10.1074/jbc.M105127200. [DOI] [PubMed] [Google Scholar]
- 29.Josephson F, et al. CYP3A induction and inhibition by different antiretroviral regimens reflected by changes in plasma 4beta-hydroxycholesterol levels. Eur J Clin Pharmacol. 2008;64(8):775–781. doi: 10.1007/s00228-008-0492-8. [DOI] [PubMed] [Google Scholar]
- 30.Kanebratt KP, et al. Cytochrome P450 induction by rifampicin in healthy subjects: determination using the Karolinska cocktail and the endogenous CYP3A4 marker 4beta-hydroxycholesterol. Clin Pharmacol Ther. 2008;84(5):589–594. doi: 10.1038/clpt.2008.132. [DOI] [PubMed] [Google Scholar]
- 31.Diczfalusy U et al (2009) 4beta-hydroxycholesterol as an endogenous marker for CYP3A4/5 activity. Stability and half-life of elimination after induction with rifampicin. Br J Clin Pharmacol 67(1):38–43 [DOI] [PMC free article] [PubMed]
- 32.Penzak SR, Rojas-Fernandez C. 4β-Hydroxycholesterol as an endogenous biomarker for CYP3A Activity: literature review and critical evaluation. J Clin Pharmacol. 2019;59(5):611–624. doi: 10.1002/jcph.1391. [DOI] [PubMed] [Google Scholar]
- 33.Neuhoff S, Tucker GT. Was 4β-hydroxycholesterol ever going to be a useful marker of CYP3A4 activity? Br J Clin Pharmacol. 2018;84(7):1620–1621. doi: 10.1111/bcp.13538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gjestad C et al (2018) Gjestad et al. reply to ‘Was 4β-hydroxycholesterol ever going to be a useful marker of CYP3A4 activity?’ by Neuhoff and Tucker. Br J Clin Pharmacol 84(7):1624–1625 [DOI] [PMC free article] [PubMed]
- 35.Kuypers DRJ, Vanhove T. Kuypers and Vanhove reply to ‘Was 4β-hydroxycholesterol ever going to be a useful marker of CYP3A4 activity?’ by Neuhoff and Tucker. Br J Clin Pharmacol. 2018;84(7):1622–1623. doi: 10.1111/bcp.13592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hjelmesaeth J, et al. Impact of body weight, low energy diet and gastric bypass on drug bioavailability, cardiovascular risk factors and metabolic biomarkers: protocol for an open, non-randomised, three-armed single centre study (COCKTAIL) BMJ Open. 2018;8(5):e021878. doi: 10.1136/bmjopen-2018-021878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kvitne KE et al (2021) Short- and long-term effects of body weight loss following calorie restriction and gastric bypass on CYP3A-activity - a non-randomized three-armed controlled trial. Clin Transl Sci [DOI] [PMC free article] [PubMed]
- 38.Krogstad V, et al. A comparative analysis of cytochrome P450 activities in paired liver and small intestinal samples from patients with obesity. Drug Metab Dispos. 2020;48(1):8–17. doi: 10.1124/dmd.119.087940. [DOI] [PubMed] [Google Scholar]
- 39.Kotronen A, et al. Prediction of non-alcoholic fatty liver disease and liver fat using metabolic and genetic factors. Gastroenterology. 2009;137(3):865–872. doi: 10.1053/j.gastro.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 40.Gjestad C, et al. 4β-hydroxycholesterol correlates with dose but not steady-state concentration of carbamazepine: indication of intestinal CYP3A in biomarker formation? Br J Clin Pharmacol. 2016;81(2):269–276. doi: 10.1111/bcp.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Størset E, et al. The CYP3A biomarker 4β-hydroxycholesterol does not improve tacrolimus dose predictions early after kidney transplantation. Br J Clin Pharmacol. 2017;83(7):1457–1465. doi: 10.1111/bcp.13248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egeland EJ et al (2020) Chronic inhibition of CYP3A is temporarily reduced by each hemodialysis session in patients with end-stage renal disease. Clin Pharmacol Ther [DOI] [PubMed]
- 43.Wegler C et al (2022) Drug disposition protein quantification in matched human jejunum and liver from donors with obesity. Clin Pharmacol Ther [DOI] [PMC free article] [PubMed]
- 44.Wiśniewski JR, Mann M. Consecutive proteolytic digestion in an enzyme reactor increases depth of proteomic and phosphoproteomic analysis. Anal Chem. 2012;84(6):2631–2637. doi: 10.1021/ac300006b. [DOI] [PubMed] [Google Scholar]
- 45.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11(12):2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 46.Huber W, et al. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.S96. [DOI] [PubMed] [Google Scholar]
- 47.Wiśniewski JR, Rakus D. Multi-enzyme digestion FASP and the ‘Total Protein Approach’-based absolute quantification of the Escherichia coli proteome. J Proteomics. 2014;109:322–331. doi: 10.1016/j.jprot.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 48.Neely MN, et al. Accurate detection of outliers and subpopulations with Pmetrics, a nonparametric and parametric pharmacometric modeling and simulation package for R. Ther Drug Monit. 2012;34(4):467–476. doi: 10.1097/FTD.0b013e31825c4ba6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R Foundation for Statistical Computing (2018) R: a language and environment for statistical computing. Vienna, Austria
- 50.Vanhove T, et al. Comparative performance of oral midazolam clearance and plasma 4β-hydroxycholesterol to explain interindividual variability in tacrolimus clearance. Br J Clin Pharmacol. 2016;82(6):1539–1549. doi: 10.1111/bcp.13083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gravel S, et al. use of 4β-hydroxycholesterol plasma concentrations as an endogenous biomarker of CYP3A activity: clinical validation in individuals with type 2 diabetes. Clin Pharmacol Ther. 2019;106(4):831–840. doi: 10.1002/cpt.1472. [DOI] [PubMed] [Google Scholar]
- 52.Wegler C, et al. Influence of proteome profiles and intracellular drug exposure on differences in CYP activity in donor-matched human liver microsomes and hepatocytes. Mol Pharm. 2021;18(4):1792–1805. doi: 10.1021/acs.molpharmaceut.1c00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mao J, et al. Perspective: 4β-hydroxycholesterol as an emerging endogenous biomarker of hepatic CYP3A. Drug Metab Rev. 2017;49(1):18–34. doi: 10.1080/03602532.2016.1239630. [DOI] [PubMed] [Google Scholar]
- 54.Gjestad C, et al. 4β-Hydroxycholesterol level significantly correlates with steady-state serum concentration of the CYP3A4 substrate quetiapine in psychiatric patients. Br J Clin Pharmacol. 2017;83(11):2398–2405. doi: 10.1111/bcp.13341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gjestad C, et al. Effect of grapefruit juice intake on serum level of the endogenous CYP3A4 metabolite 4β-hydroxycholesterol-an interaction study in healthy volunteers. Aaps j. 2019;21(4):58. doi: 10.1208/s12248-019-0330-1. [DOI] [PubMed] [Google Scholar]
- 56.Mannheimer B, et al. No impact of vitamin D on the CYP3A biomarker 4β-hydroxycholesterol in patients with abnormal glucose regulation. PLoS ONE. 2015;10(4):e0121984. doi: 10.1371/journal.pone.0121984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jover R, et al. Down-regulation of human CYP3A4 by the inflammatory signal interleukin-6: molecular mechanism and transcription factors involved. Faseb j. 2002;16(13):1799–1801. doi: 10.1096/fj.02-0195fje. [DOI] [PubMed] [Google Scholar]
- 58.Morgan ET, et al. Regulation of drug-metabolizing enzymes and transporters in infection, inflammation, and cancer. Drug Metab Dispos. 2008;36(2):205–216. doi: 10.1124/dmd.107.018747. [DOI] [PubMed] [Google Scholar]
- 59.Björkhem-Bergman L, et al. Serum levels of 25-hydroxyvitamin D and the CYP3A biomarker 4β-hydroxycholesterol in a high-dose vitamin D supplementation study. Drug Metab Dispos. 2013;41(4):704–708. doi: 10.1124/dmd.113.051136. [DOI] [PubMed] [Google Scholar]
- 60.Nitta SI, et al. Evaluation of 4β-hydroxycholesterol and 25-hydroxycholesterol as endogenous biomarkers of CYP3A4: study with CYP3A-humanized mice. Aaps j. 2018;20(3):61. doi: 10.1208/s12248-018-0186-9. [DOI] [PubMed] [Google Scholar]
- 61.Gebeyehu E, et al. Sex and CYP3A5 genotype influence total CYP3A activity: high CYP3A activity and a unique distribution of CYP3A5 variant alleles in Ethiopians. Pharmacogenomics J. 2011;11(2):130–137. doi: 10.1038/tpj.2010.16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Access to data collected from this study, including anonymized individual-participant data, may potentially be made available following publication upon e-mail request to the corresponding author. After approval of a proposal, data will be shared with investigators whose proposed use of the data has been approved by the COCKTAIL steering committee, according to the consent given by the participants and Norwegian laws and legislations.