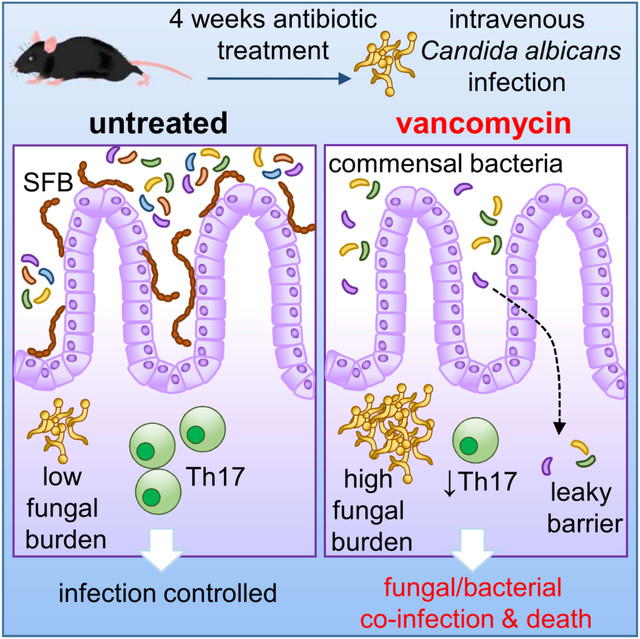
Summary
Antibiotics are a modifiable iatrogenic risk factor for the most common human nosocomial fungal infection, invasive candidiasis, yet the underlying mechanisms remain elusive. We found that antibiotics enhanced susceptibility to murine invasive candidiasis due to impaired lymphocyte-dependent IL-17A- and GM-CSF-mediated antifungal immunity within the gut. This led to non-inflammatory bacterial escape and systemic bacterial co-infection, which could be ameliorated by IL-17A or GM-CSF immunotherapy. Vancomycin alone similarly enhanced susceptibility to invasive fungal infection and systemic bacterial co-infection. Mechanistically, vancomycin reduced gut Th17 cells associated with impaired proliferation and RORγt expression. Vancomycin’s effects on Th17 cells were indirect, manifesting only in vivo in the presence of dysbiosis. In humans, antibiotics were associated with increased risk of invasive candidiasis and death after invasive candidiasis. Our work highlights the importance of antibiotic stewardship in protecting vulnerable patients from life-threatening infections and provides mechanistic insights into a controllable iatrogenic risk factor for invasive candidiasis.
Graphical Abstract
eTOC Blurb
Broad-spectrum antibiotics can underlie invasive candidiasis, but how antibiotics impair antifungal immunity remain elusive. Drummond et al show that antibiotics, particularly vancomycin, cause intestine-specific susceptibility to fungal infection enabling systemic escape of commensal bacteria. These defects map to reduced gut lymphoid IL-17A and GM-CSF responses associated with local microbiome changes.
Introduction
Antibiotics can influence and modulate the immune system, leading to abrogated antimicrobial immunity and altered homeostasis. For example, antibiotic treatment skews the intestinal immune system towards a more regulatory environment, dampening the development of gut Th17 cells by eliminating the commensal bacteria needed to develop these cell types (Ivanov et al., 2008). Recent studies have also shown that antibiotics can exert direct effects on immune cells and may drive phenotypes that extend beyond the gastrointestinal (GI) tract (Erny et al., 2015). Bactericidal antibiotics can also directly disrupt metabolic processes in myeloid cells which limits their ability to kill bacterial pathogens (Kalghatgi et al., 2013; Yang et al., 2017), while a broad-spectrum antibiotic cocktail prevented protective systemic anti-bacterial immunity in vivo by disrupting granulopoeisis in a neonatal sepsis model (Deshmukh et al., 2014). Antibiotics can therefore cause immunological injuries that enhance susceptibility to secondary infections. Identifying which antibiotics carry these potentially harmful side-effects, and the mechanisms underlying these relationships, will be important for informing future antibiotic stewardship strategies and designing immune-based therapies that may circumvent the undesirable effects of antibiotics on the host.
Candidiasis is most often caused by Candida albicans, a common human fungal commensal in the GI tract, or, less frequently, the skin (Fan et al., 2015; Findley et al., 2013). Invasive candidiasis originates from these commensal populations following breach of GI or cutaneous barrier integrity (Lionakis, 2014). GI-derived invasive candidiasis is typically seen in patients with hematological malignancies during chemotherapy-induced mucositis and neutropenia (Lionakis, 2014). In these cases, broad-spectrum antibiotic pre-exposure is a recognized risk factor, operating to expand gut commensal C. albicans populations which then translocate into the bloodstream following barrier integrity disruption and phagocyte depletion (Fan et al., 2015; Koh et al., 2008; Odds et al., 2006; Zhai et al., 2020). Prophylactic fluconazole has dramatically decreased the prevalence of GI-derived invasive candidiasis in these patients (Lionakis, 2014; Pappas et al., 2018). Skin-derived invasive candidiasis has emerged as the most common form of invasive candidiasis in modern hospitals (Ricotta et al., 2020; Strollo et al., 2016). It is typically seen in acutely ill non-neutropenic patients in the intensive care unit, associated with central venous catheter-related breach of the cutaneous barrier. Notably, broad-spectrum antibiotic pre-exposure has also been documented as a risk factor for this form of invasive candidiasis (Keighley et al., 2019; Pappas et al., 2018). However, the mechanisms by which broad-spectrum antibiotics predispose to infection remain unclear as these patients do not typically feature impaired GI barrier integrity to account for fungal translocation.
In this study, we used animal models to analyze how broad-spectrum antibiotics affect systemic immune responses to Candida albicans. We demonstrate that pre-exposure to antibiotics, especially vancomycin, disrupt antifungal immunity in an organ-specific manner and that these effects can be ameliorated by targeted immunotherapy. We also analyzed the relationship between broad-spectrum antibiotic pre-exposure and development of invasive candidiasis in humans using a large health record database.
Results
Broad-spectrum Antibiotics Result in GI Tract-Specific Susceptibility to Invasive Fungal Infection
To determine if antibiotics affect susceptibility to invasive candidiasis, we investigated antibiotic-induced susceptibility to skin-derived invasive candidiasis in mice. Mice are not naturally colonized with C. albicans in their GI tract (Fan et al., 2015; Iliev et al., 2012), and therefore are a suitable model system to evaluate how antibiotics may promote invasive candidiasis and/or perturb antifungal immunity in this setting. Mice treated with a broad-spectrum antibiotic cocktail (ampicillin, metronidazole, neomycin, vancomycin [AMNV]) had significantly higher mortality when infected intravenously with C. albicans (Fig. 1A), an infection route that mimics human skin-derived candidiasis (Lionakis et al., 2011). Susceptibility was not associated with reduced circulating neutrophils, the critical mediators of defense against invasive candidiasis (Lionakis and Levitz, 2017) (Fig. S1A).
Figure 1: Broad-spectrum Antibiotics Promote Mortality Following Systemic C. albicans Challenge.
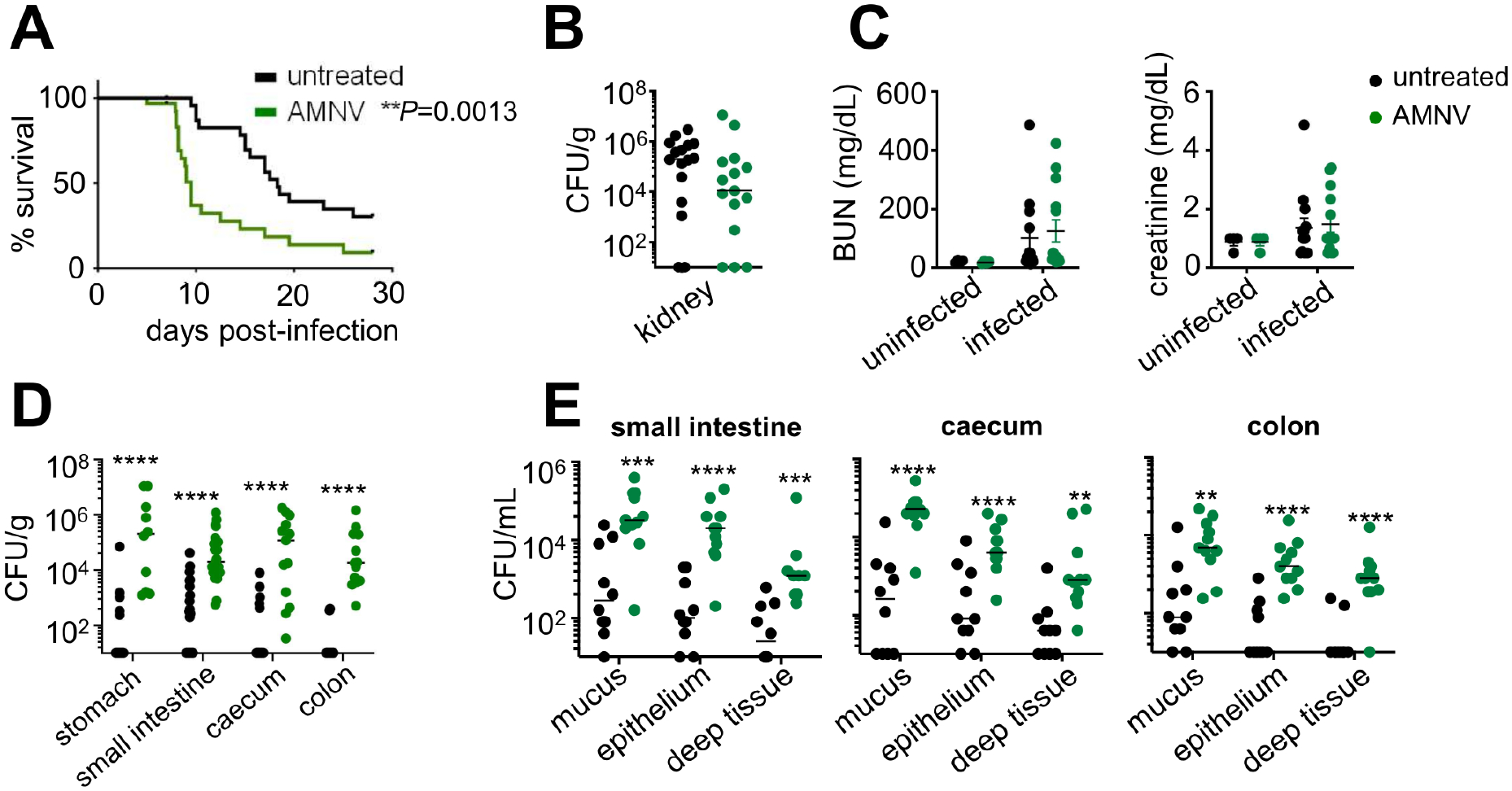
(A) Survival curve of untreated (n=33) and AMNV pre-exposed (n=31) C57BL/6 mice. Data is pooled from 3 independent experiments and analyzed by Log-rank Mantel-Cox test.
(B) Kidney fungal burdens in untreated mice (n=16) and AMNV pre-exposed mice (n=15 animals). Data is pooled from 2 independent experiments.
(C) BUN and creatinine levels in the serum of untreated (n=4 uninfected, n=13 infected) and AMNV pre-exposed (n=4 uninfected, n=14 infected) mice at 4 weeks post-antibiotic treatment (uninfected) or 7 days post-C. albicans infection (infected). Data shown with mean +/− SEM and is pooled from 2 independent experiments.
(D) Fungal burdens of indicated GI tissues (contents removed prior to homogenizing) in untreated mice (stomach n=13; small intestine n=26; caecum/colon n=15) and AMNV pre-exposed mice (stomach n=10; small intestine n=25; caecum/colon n=14) at day 7 post-infection. Data is pooled from 3 (stomach, caecum, colon) or 4 (small intestine) independent experiments and analyzed by Mann Whitney U-test.
(E) Fungal burdens in the mucus, epithelial and deep tissue layers of GI tissues in untreated (n=10) and AMNV pre-exposed (n=11) mice at day 7 post-infection. Data pooled from 2 independent experiments and analyzed by Mann-Whitney U-test. In all panels, each point represents an individual animal. *P<0.05, **P<0.01, ***P<0.005, ****P<0.0001.
Mortality in the mouse model of skin-derived invasive candidiasis is driven by uncontrolled renal fungal growth that causes kidney failure (Lionakis et al., 2011). Unexpectedly, the heightened mortality of antibiotic-treated animals was not due to increased renal fungal proliferation or kidney damage (Fig. 1B–1C), or impaired fungal control in the liver, spleen or brain (Fig. S1B). Instead, we found significantly increased fungal growth across GI tissue compartments in antibiotic pre-exposed mice (Fig. 1D). We asked whether the increased GI fungal burden of antibiotic pre-exposed mice was apparent throughout superficial and deeper gut tissue layers or whether it was only seen in the gut lumen reflective of mouse coprophagic behavior post-infection (Fig. S1C). We found that antibiotic pre-exposed mice had significantly higher fungal burden in all gut tissue layers tested (mucus, intestinal epithelium, deep intestinal tissue), consistent with hematogenously-derived fungal tissue invasion (Fig. 1E). Together, this data shows that broad-spectrum antibiotics promoted GI tract-specific susceptibility to invasive candidiasis in mice.
Commensal Gut Bacteria Disseminate in Antibiotic Pre-exposed Fungal-Infected Mice
Since antibiotic-treated mice had increased GI tissue fungal proliferation, we wondered whether antibiotic pre-exposure compromised gut barrier function. Antibiotics were previously linked to low-level systemic translocation of commensal bacteria (Chakraborty et al., 2018; Knoop et al., 2016; Spiller, 2018; Tulstrup et al., 2015). Strikingly, we cultured large amounts of Gram-positive and Gram-negative bacteria from the spleen of C. albicans-infected antibiotic pre-exposed mice (Fig. 2A). No bacteria were recovered from the spleen of fungus-uninfected antibiotic pre-exposed mice or C. albicans infected mice that did not receive antibiotics, indicating that both antibiotics and systemic fungal challenge are required for systemic bacterial translocation in this model (Fig. 2A). In agreement, we found bacterial translocation in the blood and peritoneal fluid of C. albicans-infected antibiotic pre-exposed mice (Fig. 2B). MALDI-TOF-based identification of these bacteria confirmed that they were GI-derived commensals (Fig. 2C). Histological gut examination indicated that the escape of commensal bacteria in C. albicans-infected antibiotic pre-exposed mice was non-inflammatory (Fig. S2), which has been previously suggested to occur via dendritic cell migration (Knoop et al., 2016). We observed bacterial dysbiosis in AMNV pre-exposed animals (Fig. 2D–2E), which has been linked to disrupted systemic antifungal immunity (Li et al., 2020). Several families that were depleted by AMNV pre-exposure (Bacteroidales, Deferribacterales, Erysipelotrichales, Clostridiales) included species that are major producers of short-chain fatty acids (SCFAs) such as Ruminococcus, Roseburia and Butyricimonas (Fig. S3A). Because these SCFAs promote GI tissue barrier integrity (Kelly et al., 2015), we examined the potential contribution of the loss of these SCFA-producing bacteria to antibiotic-induced susceptibility to invasive candidiasis. We delivered a cocktail of SCFAs (acetate, butryrate, proprionate) to AMNV pre-exposed mice and analyzed the effect on mortality following systemic C. albicans challenge. We found that SCFA supplementation did not rescue antibiotic-induced susceptibility to invasive C. albicans infection (Fig. S3B). Together, we show that broad-spectrum antibiotic pre-exposure causes gut dysbiosis and drives GI tissue fungal proliferation upon systemic fungal challenge, which promotes subsequent bacterial GI tract translocation and systemic bacterial co-infection in a manner that is independent of SCFAs.
Figure 2: Broad-spectrum Antibiotic Pre-exposure Promotes Dissemination of Gut Commensal Bacteria Following Fungal Infection.
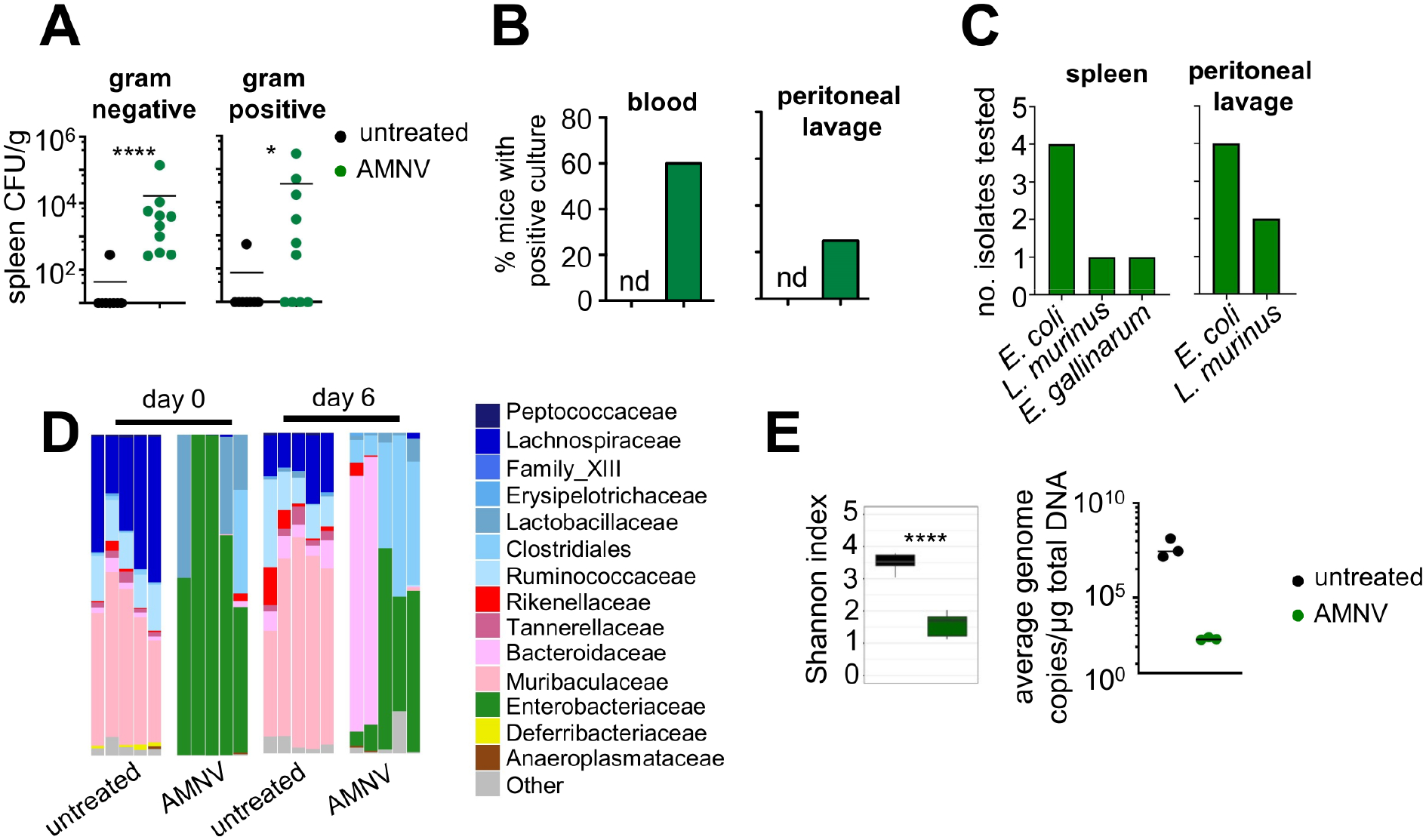
(A) Bacterial burdens in the spleen of untreated (n= 8) and AMNV-preexposed (n= 10) mice at day 7 post-fungal infection. No bacteria were detected in the spleens of AMNV pre-exposed Candida-uninfected or AMNV-unexposed Candida-infected mice. Data is pooled from 2 independent experiments and analyzed by Mann-Whitney U-test.
(B) Percentage of mice with positive bacterial cultures of the blood and peritoneal lavage at day 7 post-fungal infection. Percentages represent combined data from 2 independent experiments with at least 10 mice in each group. nd, not detected.
(C) Bacterial colonies from the spleen or peritoneal lavage fluid of AMNV pre-exposed mice were identified by MALDI-TOF (Escherichia coli, Lactobacillus murinus, Enterococcus gallinarum). Randomly-picked isolates were chosen from at least 6 different animals across 2 independent experiments.
(D) Relative abundance of indicated bacterial families in stools collected from untreated and antibiotic pre-exposed mice at day 0 and day 6 relative to C. albicans challenge. Each bar shows the analysis for an individual mouse and at least 2 mice per group are from an independent experiment.
(E) Left: Shannon index of stool microbiome diversity in mice that were untreated (n=5) and AMNV pre-exposed (n=5) mice at day 6 post-infection. Right: Quantification of total bacterial load in stool samples collected from animals at day 6 post fungal infection. Each point represents an individual animal (n=3 per group). Data from uninfected animals showed similar results.
IL-17A and GM-CSF Protect against Antibiotic-Induced Susceptibility to Invasive Candidiasis
We next aimed to define the molecular and cellular underpinnings of antibiotic-induced dysfunctional GI tract antifungal immunity and barrier integrity. Antibiotics disrupt cytokine production from GI tract-resident CD4 T cells and innate lymphoid cells (ILCs), particularly IL-17A, IL-22, and GM-CSF (Ivanov et al., 2009; Ivanov et al., 2008; Mortha et al., 2014). IL-17A is a critical antifungal molecule at the mucosal barrier as inborn errors of IL-17 immunity cause mucosal candidiasis (Boisson et al., 2013; Lévy et al., 2016; Lionakis and Levitz, 2017; Puel et al., 2011). IL-22 and GM-CSF also protect against C. albicans infection (Aggor et al., 2020; Bichele et al., 2018; Kasahara et al., 2016), and promote intestinal barrier integrity (Gennari et al., 1994; Sonnenberg et al., 2012). We thus examined whether the production of these three cytokines by GI tract-resident lymphocytes was affected by antibiotic treatment before and during fungal infection (Fig. S4) and found significant reductions in the frequencies of IL-17A-, IL-22-, and GM-CSF-producing lymphoid cells upon antibiotic pre-exposure (Fig. 3A–3B). To determine the impact of these antibiotic-induced cytokine perturbations on mouse susceptibility to C. albicans infection, we infected mice deficient in each of these cytokines and examined whether antibiotics accelerated their mortality upon fungal infection, as we observed in wild-type (WT) mice. We found that antibiotic pre-exposed Il22−/− animals had greater mortality post-fungal infection compared to Il22−/− animals that did not receive antibiotics (Fig. 3C), indicating that IL-22 is not required to mediate the susceptibility phenotype in this model. By contrast, mice lacking either IL-17A or GM-CSF exhibited similar susceptibility to fungal infection regardless of antibiotic pre-exposure (Fig. 3C). This indicates that reduced production of IL-17A and GM-CSF, but not IL-22, in antibiotic pre-exposed animals mediates their enhanced susceptibility to subsequent invasive fungal disease.
Figure 3: Broad-Spectrum Antibiotic Pre-exposure Disrupts Lymphocyte-Derived Cytokine Responses within the Gastrointestinal Tract During Invasive Fungal Infection.
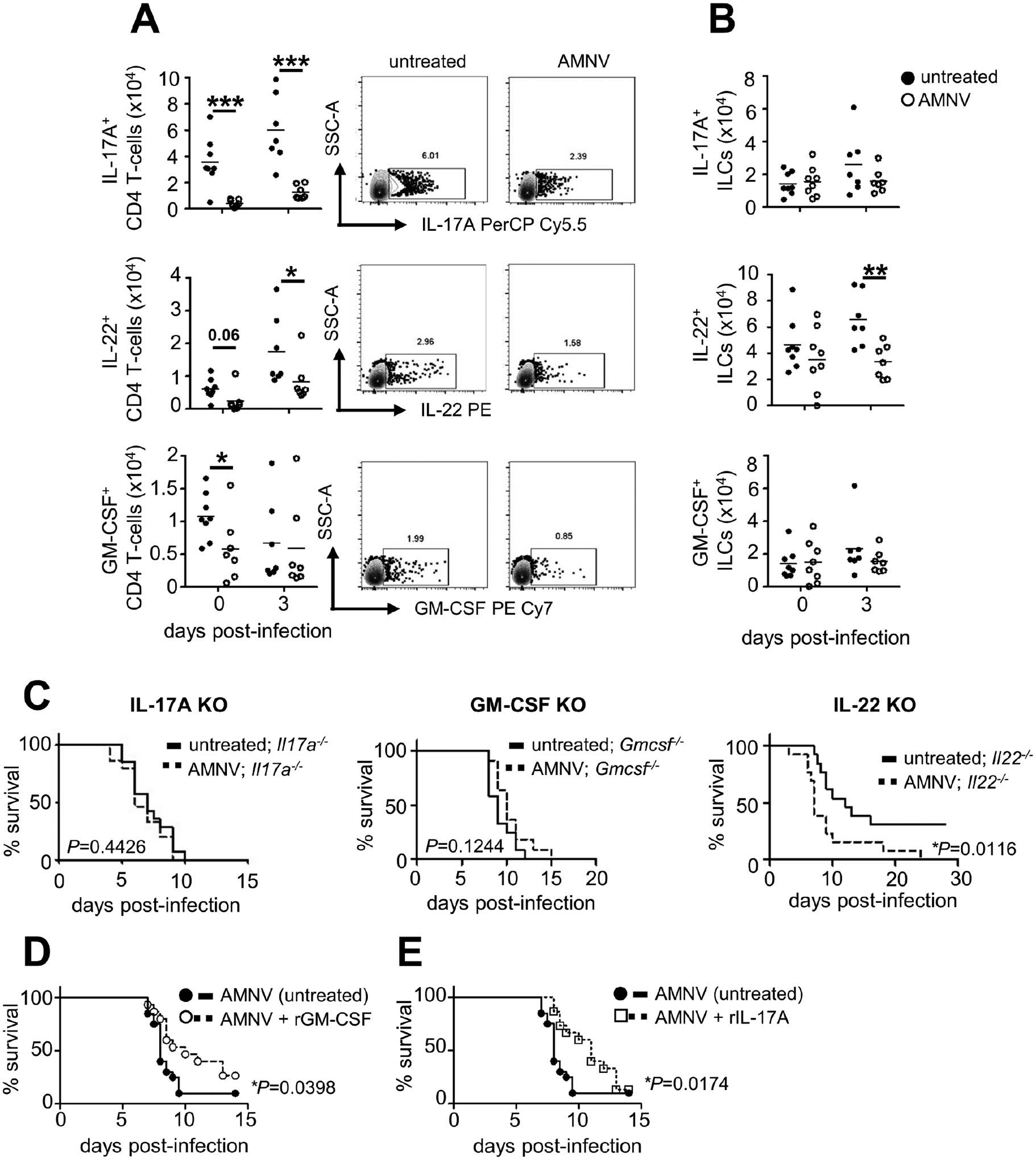
(A and B) (A) Numbers of cytokine-positive CD4+ T cells and (B) ILCs in untreated (day 0 n=8; day 3 n=7) and AMNV pre-exposed (day 0 n=8; day 3 n=7) mice at indicated time points relative to fungal infection. Example FACS plots are gated on CD4+ T cells using the gating strategy shown in Fig. S6. All data shown is pooled from 2 independent experiments; each point represents an individual animal. Data is analyzed by unpaired two-tailed t-tests.
(C) Survival curve of C. albicans-infected Il17a−/− (untreated, n=14; AMNV pre-exposed, n=15), Gmcsf−/− (untreated, n=12; AMNV pre-exposed, n=11) and Il22−/− (untreated, n=13; AMNV pre-exposed, n=13) mice. Data in each curve is from 3 (Il17a−/−) or 2 (Gmcsf−/−, Il22−/−) independent experiments and analyzed by Log rank Mantel-Cox test.
(D and E) (D) Survival curve of C. albicans-infected AMNV pre-exposed WT mice, with or without administration of recombinant GM-CSF, or (E) recombinant IL-17A (untreated n=20 mice, treated n=15 mice). WT mice were pre-exposed to AMNV as in Fig 1A, and 5 μg GM-CSF or 1 μg IL-17A or diluent control was delivered intraperitoneally, using the following dosing strategy: GM-CSF, days −6, −4, −2, 0, +2, +4 and +6 relative to C. albicans infection; IL-17A, −8 hours and +24 hours relative to C. albicans infection. Data in D and E is pooled from 2 independent experiments and analyzed by Log rank Mantel-Cox test. *P<0.05, **P<0.01, ***P<0.005, ****P<0.0001.
As we found that antibiotics reduced the production of GM-CSF and IL-17A in the GI tract, we next explored whether restoring these defects with recombinant cytokine treatment may rescue the susceptibility of antibiotic-treated animals when challenged with C. albicans. Recombinant GM-CSF is FDA-approved for accelerating myeloid reconstitution following myeloablative chemotherapy or hematopoietic stem cell transplantation, and has provided clinical benefit in some patients with invasive fungal (including C. albicans) infections (Dignani et al., 2005; Gavino et al., 2014; Poynton et al., 1998; Rókusz et al., 2001; van de Veerdonk et al., 2012). Thus, we administered GM-CSF to antibiotic pre-exposed C. albicans-infected mice and found that it partially reversed their susceptibility (Fig. 3D). We observed a similar partial rescue with IL-17A administration (Fig. 3E). Our data therefore points to a potential translational avenue for therapeutic intervention to ameliorate antibiotic-induced infection susceptibility in this setting.
Both CD4 T cells and ILCs Promote Protection in the Setting of Antibiotic-Induced Invasive Candidiasis
As both GI tract-resident CD4 T cells and ILCs of antibiotic pre-exposed mice exhibited cytokine production defects (Fig. 3A–3B), we next examined which of these populations’ impairment contributes to susceptibility to systemic fungal infection upon antibiotic pre-exposure. Antibiotic-treated Rag1−/− mice, which lack CD4 T cells (and other αβ and γδ lymphoid cells), had greater mortality following fungal infection compared to Rag1−/− animals that did not receive antibiotics (Fig. 4A), indicating that the antibiotic exposure-induced susceptibility phenotype develops even in the absence of αβ (and γδ) lymphoid cells. By contrast, we found that antibiotic-treated Rag2−/−Ilr2g−/− animals, which also lack ILCs, had similar susceptibility to fungal infection regardless of antibiotic pre-exposure (Fig. 4B). These findings indicate that defects in both innate and adaptive lymphoid populations of antibiotic pre-exposed animals mediate their enhanced susceptibility to subsequent invasive fungal infection. In line with this, non-antibiotic exposed Rag2−/−Ilr2g−/− mice phenocopied WT antibiotic-treated mice at the levels of significantly greater fungal proliferation across the GI tract but not the kidney (Fig. 4C) and systemic escape of commensal bacteria (Fig. 4D), consistent with the notion that innate and adaptive lymphoid cells are necessary to promote fungal control within the GI tract and containment of systemic bacterial translocation. To examine whether broad-spectrum antibiotic pre-exposure is sufficient to compromise innate and adaptive lymphoid cell-mediated protection against invasive candidiasis in this setting, we adoptively transferred CD4 T cells and ILCs into Rag2−/−Ilr2g−/− mice and found that they rescued the susceptibility phenotype (Fig. 4A, 4E). Notably, adoptive transfer of CD4 T cells and ILCs harvested from antibiotic pre-exposed donors into Rag2−/−Il2rg−/− mice significantly blunted their protective effects during invasive candidiasis (Fig. 4E). Together, these studies indicate that broad-spectrum antibiotic pre-exposure impairs lymphocyte-dependent IL-17A- and GM-CSF-mediated antifungal immunity within the GI tract, which is associated with non-inflammatory bacterial translocation, systemic bacterial co-infection, and increased mortality post-fungal challenge.
Figure 4: Defects in CD4+ T cells and ILCs Mediate Broad-spectrum Antibiotic-Induced Susceptibility to Invasive Fungal Infection.
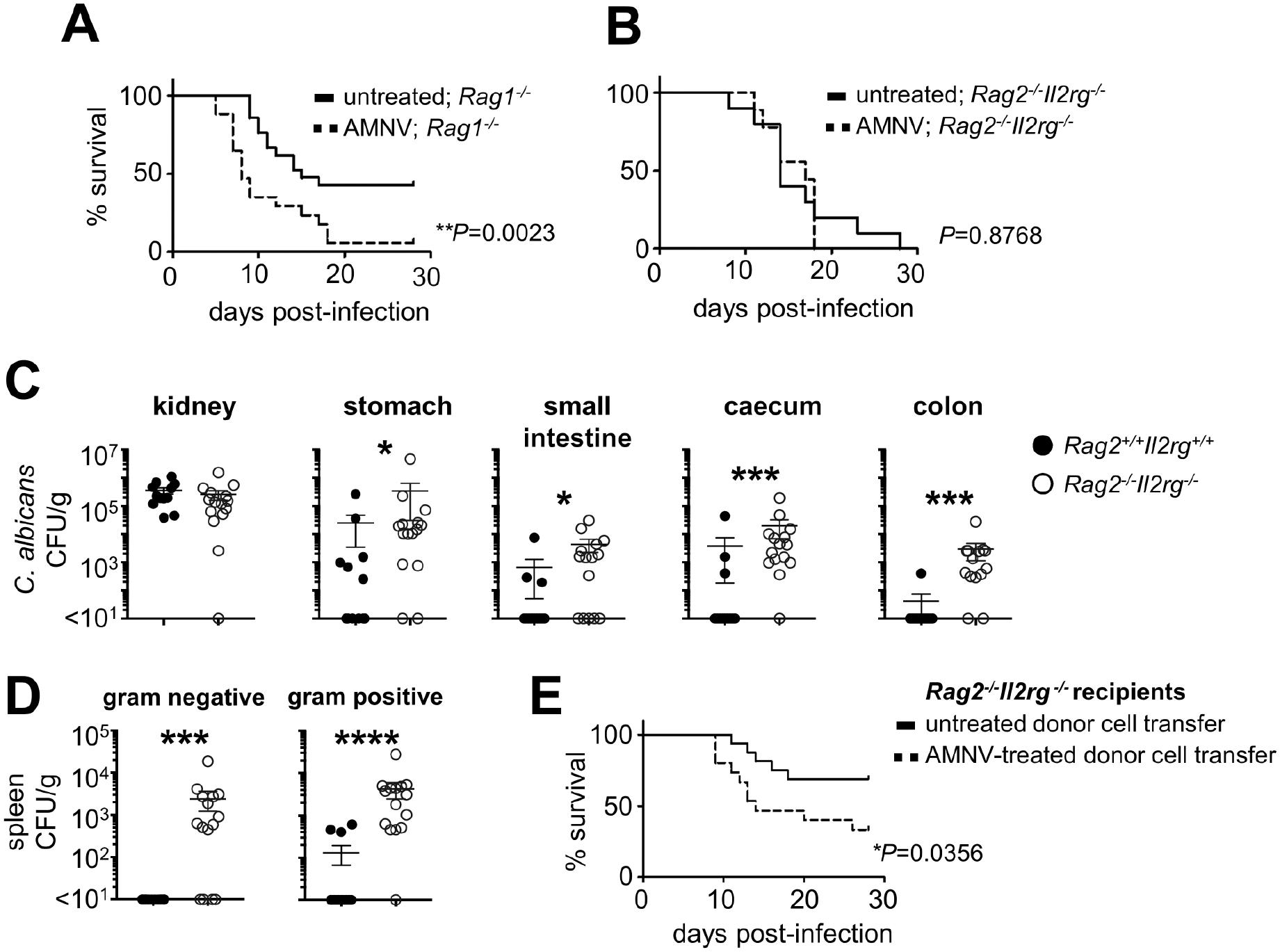
(A) Survival curve of untreated (n=21) and AMNV pre-exposed (n=17) Rag1−/− animals following systemic C. albicans challenge. Data is pooled from 3 independent experiments and analyzed by Log rank Mantel-Cox test.
(B) Survival curve of untreated (n=10) and AMNV pre-exposed (n=9) Rag2−/−Il2rg−/− animals following systemic C. albicans challenge. Data is pooled from 2 independent experiments and analyzed by Log rank Mantel-Cox test.
(C) Fungal burdens within indicated tissues at day 7 post-infection in WT (n=12) and Rag2−/−Il2rg−/− (n=15) mice. Each point represents an individual animal. Data is pooled from 3 independent experiments and analyzed by Mann Whitney U-test. *P<0.05, ***P<0.005.
(D) Bacterial burdens in the spleen of WT (n=12) and Rag2−/−Il2rg−/− (n=15) mice. Each point represents an individual animal. Data is pooled from 3 independent experiments and analyzed by Mann Whitney U-test. ***P<0.005, ****P<0.0001.
(E) Survival curve of C. albicans-infected Rag2−/−Il2rg−/− mice that had received adoptive transfer of lymphocytes from untreated WT mouse donors (n=16 recipients) or AMNV pre-exposed WT mouse donors (n=15 recipients). Data is pooled from 2 independent experiments and analyzed by Log rank Mantel-Cox test.
Vancomycin Pre-exposure Specifically Promotes Susceptibility to Invasive Candidiasis
Different antibiotics can exert specific effects on immune cell function and/or microbiome-dependent immune modulation (Kalghatgi et al., 2013). We therefore sought to examine whether any specific antibiotic was responsible for the enhanced susceptibility to invasive candidiasis of AMNV pre-exposed mice. We single-treated WT mice with each antibiotic and evaluated survival following intravenous C. albicans challenge, as before. Notably, pre-exposure to vancomycin, but not metronidazole or neomycin, promoted markedly increased mortality, whereas ampicillin pre-exposure modestly increased susceptibility (Fig. 5A). Vancomycin has been associated with defective immune responses in mice (Brandl et al., 2008; van Opstal et al., 2016) and humans (Von Drygalski et al., 2007) in other settings, therefore we next focused on vancomycin to understand how it impaired GI tract antifungal immunity.
Figure 5: Vancomycin increases susceptibility to invasive candidiasis.
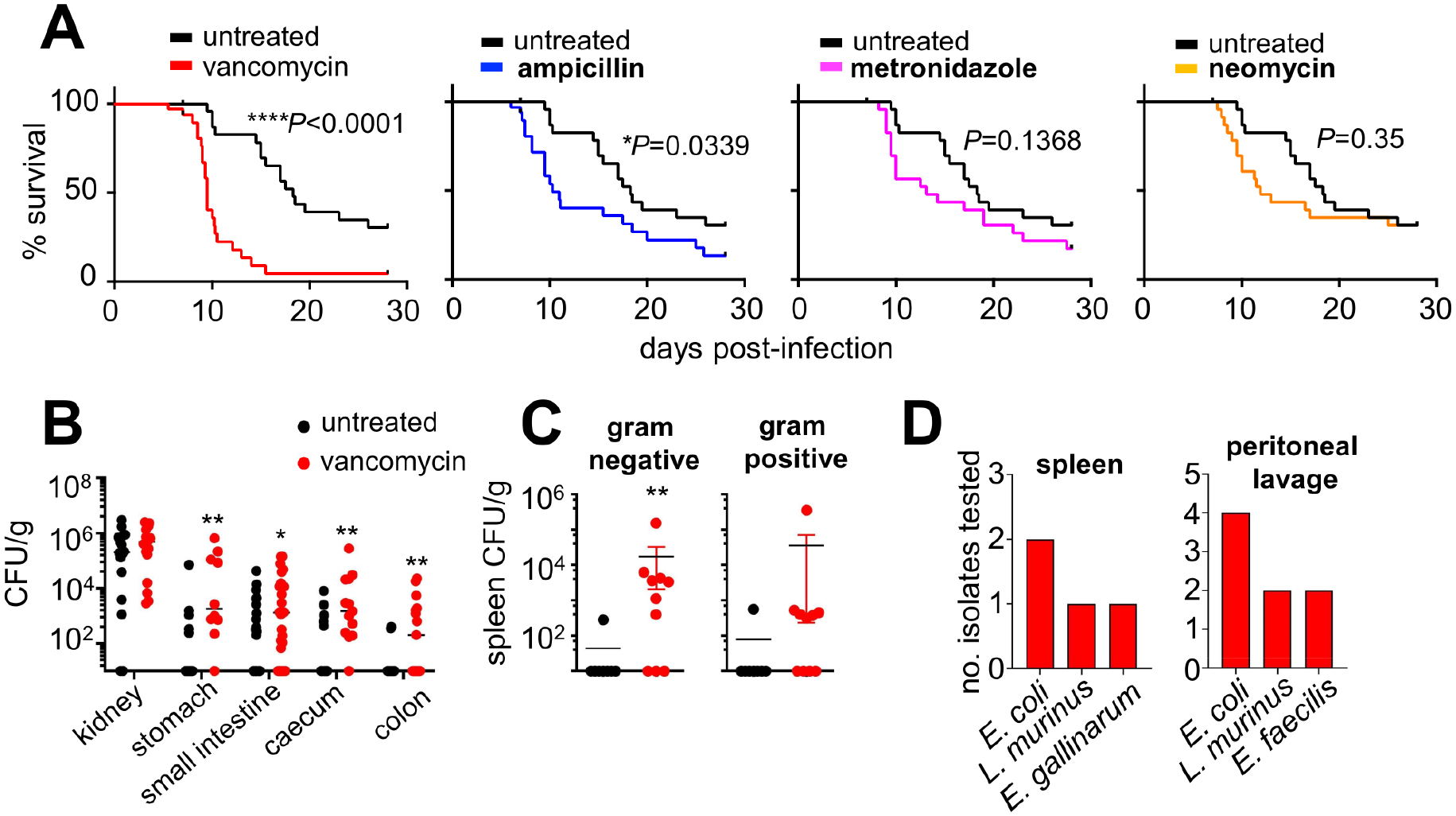
(A) Survival curves of C57BL/6 mice treated with the indicated antibiotics for 4 weeks prior to intravenous infection (n=33 per group), compared with untreated control mice (black lines). Data is pooled from 3 independent experiments and analyzed by Log-rank Mantel-Cox test.
(B) Fungal burdens of the indicated organs in untreated mice (kidney n=16; stomach n=13; small intestine n=26; caecum/colon n=15) and vancomycin pre-exposed mice (kidney n=16; stomach n=10; small intestine n=26; caecum n=13; colon n=15). Data is pooled from 3–4 independent experiments and analyzed by Mann Whitney U-test.
(C) Bacterial burdens in the spleen of untreated (n= 8) and vancomycin pre-exposed (n= 10) mice at day 7 post-fungal infection. Percentages represent combined data from 2 independent experiments with at least 10 mice in each group.
(D) Bacterial colonies from the spleen or peritoneal lavage fluid of vancomycin pre-exposed mice were identified by MALDI-TOF. Randomly-picked isolates were chosen from at least 6 different animals across 2 independent experiments. **P<0.01.
Like AMNV pre-exposed mice, vancomycin pre-exposed mice had impaired control of invasive fungal infection specifically in GI tissues but not kidney (Fig. 5B) and translocated gut commensal bacteria to the spleen and peritoneal fluid (Fig. 5C). Vancomycin-associated susceptibility was evident upon long-term pre-exposure (4 weeks), whereas a 10-day pre-treatment did not enhance susceptibility to invasive candidiasis (Fig. S5A). Moreover, vancomycin-driven susceptibility was specific to invasive fungal infection, since vancomycin pre-exposed mice did not exhibit accelerated mortality when infected systemically with Yersinia pseudotuberculosis (Fig. S5B). Lastly, we examined whether parenteral administration of vancomycin also increased susceptibility to invasive fungal infection. We treated mice every 2 days with intraperitoneal (IP) vancomycin for 4 weeks, followed by intravenous challenge with C. albicans. In contrast to oral vancomycin pre-exposure, IP vancomycin administration did not increase susceptibility to subsequent invasive fungal infection (Fig. S5C). Collectively, our data show that vancomycin pre-exposure selectively enhances susceptibility to invasive candidiasis, which is dependent on long-term oral administration.
Vancomycin Pre-exposure Results in Reduced Proliferation and RORγt Expression of Gut Th17 cells in a Microbiome-Dependent Manner
We next sought to determine whether vancomycin pre-exposure conferred similar defects in intestinal lymphoid cell cytokine production as AMNV pre-exposure did (Fig. 3), focusing primarily on IL-17A. Indeed, we found that vancomycin pre-exposed mice had a significantly reduced frequency of IL-17A+ CD4 T cells in the small intestine lamina propria, whereas the frequency of IL-17A+ ILCs was preserved (Fig. 6A and S6A). To understand the mechanism(s) by which vancomycin pre-exposure might cause the reduction of gut IL-17A+ CD4 T cells, we examined Th17 cell RORγt expression, proliferation, and viability in vivo. Gut IL-17A+ CD4 T cells from vancomycin pre-exposed mice exhibited significantly reduced expression of RORγt relative to untreated controls (Fig. 6B). In addition, we found a significant reduction of proliferating CD4 T cells in the intestine of vancomycin pre-exposed mice (Fig 6C). Ki67+ CD4 T cells produced more IL-17A than IFNγ, and vancomycin pre-exposure specifically reduced proliferating IL-17A+ CD4 T cells, whereas proliferating IFNγ+ CD4 T cells remained unchanged (Fig. 6D). By contrast, we observed no defect in CD4 T cell survival in the intestine of vancomycin pre-exposed mice (Fig. S6B). Moreover, RORγt expression, proliferation, and viability were preserved in intestinal ILCs of vancomycin pre-exposed mice (Fig. S6B–D). Taken together, this data shows that vancomycin pre-exposure reduced gut Th17 cells associated with decreased RORγt expression and proliferation of Th17 cells.
Figure 6: Vancomycin pre-exposure results in Th17 cell defects in the GI tract associated with depletion of Th17-inducing bacteria from the gut microbiome.
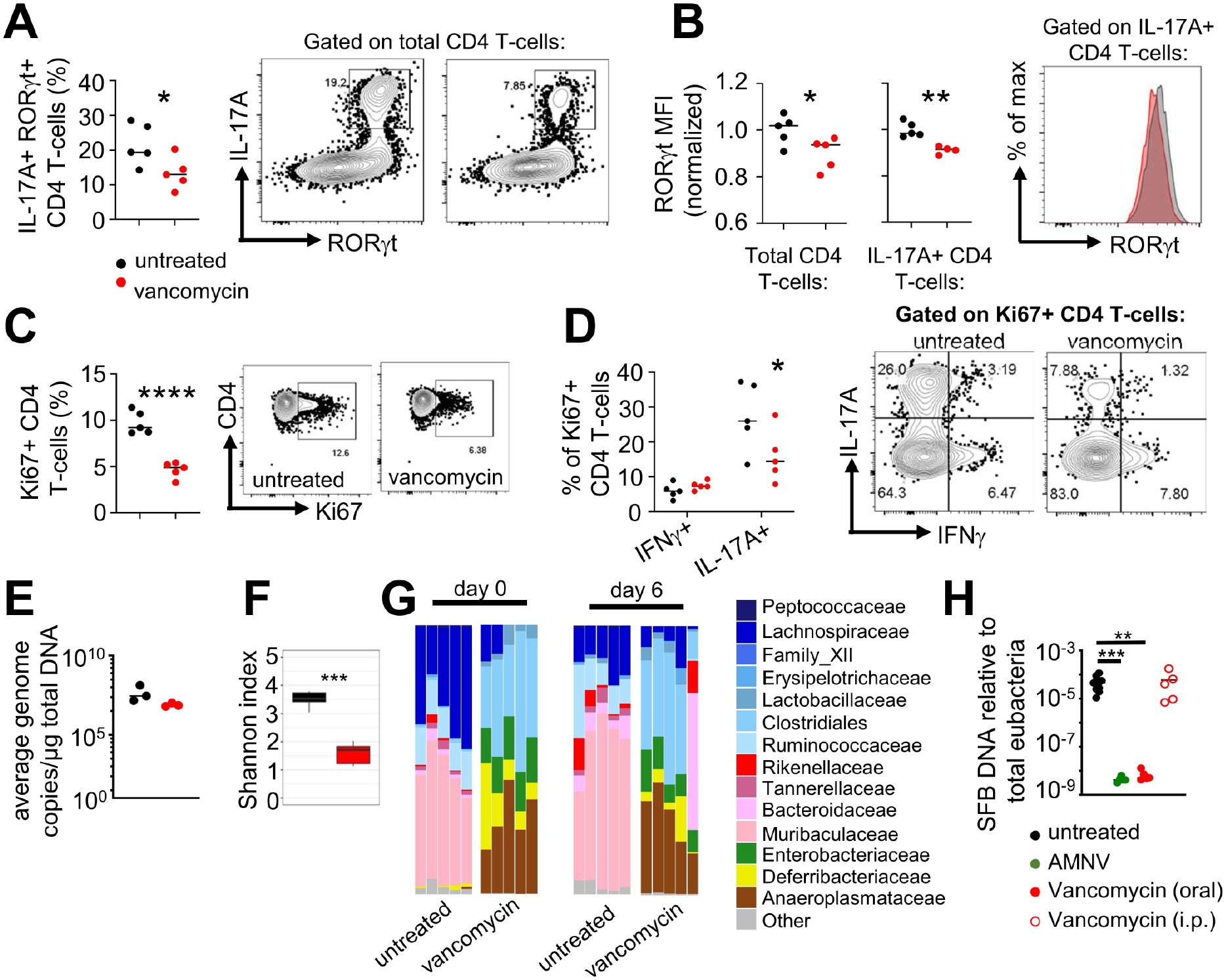
(A) Frequency of IL-17A+ RORγt+ CD4 T cells in the small intestine lamina propria of untreated and vancomycin pre-exposed mice. Representative FACS plots are gated on live CD4 T cell singlets.
(B) Mean fluorescence intensity (MFI) of RORγt in CD4+ T cells (left panel) or IL-17A+ CD4+ T cells (middle panel), normalized to the mean of untreated control mice. Right panel shows representative FACS plot of RORγt expression gated on Th17 cells.
(C) Frequency of Ki67+ CD4 T cells and (D) the proportion of Ki67+ CD4 T cells that were positive for IFNγ or IL-17A. Shown are summary data (left panels) and representative FACS plots (right panels) gated on total CD4 T cells (C) or Ki67+ CD4 T cells (D).
Data in panels A–D are pooled from two independent experiments (n=5 mice per group; each point represents an individual animal) and analyzed using unpaired t-tests. *P<0.05, **P<0.01, ****P<0.0001.
(E) Quantification of total bacterial load in stool samples collected from the indicated groups of animals at day 6 post-fungal infection. Each point represents an individual animal (n=3 per group).
(F) Shannon index of stool microbiome diversity in mice that were untreated (n=5) and vancomycin pre-exposed (n=5) mice at day 6 post-infection.
(G) Relative abundance of indicated bacterial families in stools collected from untreated and vancomycin pre-exposed mice at day 0 and day 6 relative to systemic C. albicans challenge (untreated graphs are also shown Fig 2D). Each bar shows the analysis for an individual mouse and at least 2 mice per group are from an independent experiment.
(H) SFB abundance in the stool as detected by qPCR relative to total eubacteria from untreated (n=10), AMNV pre-exposed (n=5), vancomycin pre-exposed (oral route; n=5), or vancomycin pre-exposed (i.p. route; n=5) uninfected mice. Data analyzed by Kruskal-Wallis with Dunn’s multiple correction. Each dot represents an individual animal.
Next, we asked whether vancomycin acted directly on Th17 cells to induce the immune defects we observed in vivo. We first isolated naïve CD4 T cells from spleen of WT mice and cultured them in vitro under Th17 inducing conditions, in the presence of increasing concentrations of vancomycin. These experiments revealed that vancomycin had no effect on Th17 cell differentiation or proliferation in vitro, since we found similar production of IL-17A by vancomycin-treated CD4 T cells, and no difference in the expression of RORγt or Ki67 (Fig S7A–C). In the same experiments, we measured GM-CSF levels by ELISA upon vancomycin treatment in vitro and found no difference in GM-CSF secretion by T cells in the presence of increasing concentrations of vancomycin (Fig S7C). In an independent line of experiments, we isolated naïve CD4 T cells from the spleen of vancomycin pre-exposed mice and cultured them in vitro under Th17 inducing conditions to determine whether the decreased frequency of IL-17A+ cells we observed in vivo was also evident in vitro. However, we found no defect in the frequency of Th17 cells ex vivo, nor did we detect any impairment in the secretion of GM-CSF by these cells (Fig S7D). Together, these experiments show that vancomycin does not directly impair Th17 cell differentiation or proliferation.
Since our in vitro and ex vivo experiments indicated that the Th17 cell defects only occurred with in vivo exposure to vancomycin, we next examined the microbiome changes occurring in the vancomycin pre-exposed mice which may disrupt microbial-derived signals that influence Th17 cell development. Indeed, it has been shown that specific bacterial species and their products can modulate the number and/or function of gut Th17 cells (Alexander et al., 2022; Atarashi et al., 2015; Ivanov et al., 2009; Martínez-López et al., 2019). We found that vancomycin pre-exposure had no effect on the abundance of total bacteria (Fig 6E), but significantly reduced the diversity of bacterial species (Fig 6F). Specifically, vancomycin pre-exposed animals had significant reductions in the relative abundance of Bacteroides, and expansions in the relative abundance of Anaeroplasma, Deferribacteria and Enterobacteria (Fig 6G). Because segmented filamentous bacteria (SFB) promote Th17 differentiation in the mouse intestine (Ivanov et al., 2009), and vancomycin has activity against SFB (Klaasen et al., 1991), we quantified the amount of SFB in the stool of vancomycin and AMNV pre-exposed animals. We found that the abundance of SFB was significantly reduced in both vancomycin and AMNV pre-exposed animals (Fig 6H). Importantly, we found no reduction in SFB was seen in mice treated with IP vancomycin (Fig 6H) that did not demonstrate increased susceptibility to systemic candidiasis (Fig S5C). Collectively, these results indicate that the reduction of Th17 cells in mice pre-exposed to antibiotics orally may be contributed, at least in part, by the loss of SFB from the microbiota.
Broad-spectrum Antibiotic Exposure is Associated with Susceptibility to and Mortality from Invasive Candidiasis in Humans
To examine the potential human relevance of these findings, we utilized the Cerner HealthFacts database, which includes linked microbiological and clinical data for >60 million patients across the US, to determine whether broad-spectrum antibiotic pre-exposure in hospitalized patients may increase the risk of invasive candidiasis, mortality after invasive candidiasis, and/or development of systemic bacterial co-infection during invasive candidiasis. We extracted records for all hospital encounters from 2009 through 2017 where any Candida species was isolated from blood or other sterile site and identified and evaluated 8,294 patients with invasive candidiasis. Pre-exposure to broad-spectrum antibiotics was defined as having received >7 days of a systemic antibiotic in the 31 days prior to the first positive Candida culture. The mean duration of pre-exposure to broad-spectrum antibiotics in all evaluable patients was 13 days (median, 11 days; range, 8–79 days). Among the patients with invasive Candida infection, the mean duration of pre-exposure to broad-spectrum antibiotics was 14.6 days (median, 12 days; range, 8–52 days). To assess the odds of developing invasive candidiasis, we used logistic regression controlling for patient, encounter, and hospital-level characteristics. Among patients with invasive candidiasis, we calculated the cumulative incidence function of the incidence of bacterial co-infection, considering mortality as a competing risk.
We found that receipt of broad-spectrum antibiotics for >7 days was significantly and independently associated with increased odds of developing invasive candidiasis (OR: 1.13, 95% CI: 1.03 – 1.25), and increased the risk of death following invasive candidiasis (HR: 1.18, 95% CI: 1.02 – 1.36) (Table 1). The receipt of >7 days of broad-spectrum antibiotics exhibited a trend towards an increased risk of bacterial co-infection in individuals with invasive candidiasis not yet experiencing the event (HR: 1.20, 95% CI: 0.90 – 1.60) (Table 1). Although additional studies are required to verify these results, this data suggest that broad-spectrum antibiotics may be a risk factor for developing invasive candidiasis, increased mortality following invasive candidiasis, and increased risk of systemic bacterial co-infection during invasive candidiasis in patients.
Table 1: Risk analysis of broad-spectrum antibiotic pre-exposure in humans.
Adjusted risk ratios for the association between broad-spectrum antibiotic pre-exposure and developing invasive candidiasis (Odds ratio), broad-spectrum antibiotic pre-exposure and 31-day mortality post-invasive Candida infection (hazards ratio), and broad-spectrum antibiotic pre-exposure and developing a systemic bacterial co-infection within 31 days after developing invasive candidiasis (where 31-day mortality was treated as a competing risk, subdistribution hazard ratio). See main text and methods section for total numbers of patients and details of co-variates used. CI, confidence interval.
| Clinical variable | Adjusted risk ratio | Lower CI | Upper CI |
|---|---|---|---|
| Invasive candidiasis | 1.13 | 1.03 | 1.25 |
| Mortality post-invasive candidiasis | 1.18 | 1.02 | 1.36 |
| Bacterial co-infection, post-invasive candidiasis | 1.20 | 0.90 | 1.60 |
Discussion
Antibiotics are a well-known risk factor for invasive candidiasis in humans, yet the mechanisms for this relationship have remained unclear. Here, we show that long-term antibiotic treatment, particularly with vancomycin, enhances susceptibility to invasive candidiasis and drives non-inflammatory escape of gut bacteria resulting in systemic co-infection and death. Using animals deficient in lymphocytes and cytokines associated with mucosal immunity and barrier integrity, we found that antibiotic treatment significantly alters the function of CD4 T cells and ILCs during invasive fungal infection, and that protection against antibiotic-associated mortality is partially dependent on IL-17A and GM-CSF.
Our data revealed that although antibiotic pre-exposed mice had increased mortality following systemic candidiasis, these animals did not have enhanced fungal infection of tissues that typically correlate with mortality (Spellberg et al., 2005; Spellberg et al., 2003). Instead, we found a GI tract-specific susceptibility to fungal infection following antibiotic pre-exposure. Laboratory-bred mice are not naturally colonized with C. albicans (Iliev et al., 2012), and antibiotics are needed to break this colonization resistance which is dependent on HIF1α, LL-37, and certain microbiota (Fan et al., 2015). Penicillin and streptomycin are most often used to permit fungal colonization of laboratory-bred mice, as other antibiotics - including vancomycin, are less able to allow for persistent C. albicans GI colonization (Shankar et al., 2015). While we found evidence that our antibiotic pre-exposed animals did become colonized post-systemic infection, it is important to note that this colonization likely occurred initially from the bloodstream and was maintained by coprophagic behavior. Higher fungal burdens in the stool of intravenously-infected animals has also been observed in mice deficient in DECTIN-1, which are also unable to control fungal invasion of GI tissues (Drummond et al., 2016; Vautier et al., 2012). Mechanisms enabling C. albicans translocation across the intestinal barrier are incompletely understood; recent work implicated hypha formation and secretion of the peptide toxin, Candidalysin, as essential for transcellular migration across enterocyte barriers (Allert et al., 2018). Whether similar mechanisms operate to expel C. albicans from the deep GI tissue into the GI lumen is unknown.
In addition to uncontrolled GI fungal infection, AMNV and vancomycin pre-exposure was associated with systemic escape of gut bacteria. Although we were able to culture gut bacteria from several distal anatomical sites, we did not find evidence of overt intestinal inflammation or generalized permeability defect of the gut barrier in antibiotic pre-exposed mice. Our study adds to the growing body of evidence that antibiotics may induce the translocation of commensal bacteria across the intestinal barrier independently of epithelial cell disruption and tight-junction damage (Chakraborty et al., 2018; Yu et al., 2014). The mechanisms by which this occurs remain incompletely understood, although one study indicated that dissemination of bacteria to the mesenteric lymph nodes requires microbial signaling to colonic goblet cells and CX3CR1+ dendritic cells (Knoop et al., 2016). In these studies, antibiotic pre-exposure caused limited bacterial translocation without morbidity. By contrast, our data reveals that antibiotic-induced translocation of bacteria is significantly exacerbated by invasive fungal infection of the GI tract, which markedly elevated bacterial translocation leading to mortality.
Antibiotics are known to cause disruption to mucosal lymphocytes (Ivanov et al., 2009). IL-17A production by CD4 T cells is especially sensitive to antibiotic treatment, as we confirmed here. IL-17A is a key antifungal cytokine at mucosal sites since patients with IL-17 receptor signaling defects are susceptible to mucosal candidiasis and enhanced C. albicans GI colonization (Puel et al., 2011; Puel et al., 2010). Indeed, we found that IL-17A was required to control C. albicans infection of GI tissues in the setting of antibiotic pre-exposure, and that AMNV and vancomycin treatment significantly reduced the number of intestinal Th17 cells. A recent study by Li et al utilized a similar antibiotic treatment in WT animals and also found that antibiotics promoted mortality following systemic candidiasis, which was associated with reduced IL-17A production since recombinant IL-17A rescued antibiotic-treated animals (Li et al., 2020). However, in that study, antibiotics promoted uncontrolled fungal growth in several organs including the kidney and liver, and dissemination of commensal bacteria was not investigated. It is possible that the discrepancy in renal fungal control between that and our study is due to microbiota differences in the mouse colonies. In our study, both AMNV and vancomycin pre-exposed animals had reductions in Bacteroidetes whereas Li et al found little change in this phylum (Li et al., 2020). Microbiota composition is important for determining immune responses, including to fungi. Indeed, LPS produced by commensal bacteria prevents the development of protective antifungal immunity by limiting neutrophil expansion (Jiang et al., 2014), while GI colonization with C. albicans helps drive Th17 expansion and neutrophil reactivity, which in turn promotes protection against systemic fungal infection (Shao et al., 2019). Therefore, the composition of the commensal microbiota is an important determinant of how IL-17A-dependent responses are regulated which in turn may affect the outcome of systemic candidiasis in mice.
In line with that, we found that vancomycin-induced changes to the microbiome caused significant reductions in the proliferation and RORγt expression of gut Th17 cells. Vancomycin did not directly affect the development of Th17 cells, in line with similar studies that found no direct effect of vancomycin on autoimmune Th17 cells (Almeida et al., 2021). Interestingly, the same study also found no difference in the frequency of infiltrating Th17 cells within the inflamed CNS of vancomycin-treated mice (Almeida et al., 2021), which may indicate that vancomycin exerts tissue-specific effects on Th17 cells that are context-dependent. The commensal microbiota are important for maintaining the proliferative potential of CD4 T cells in mucosal tissues, as antibiotic treatment has been associated with reduced CD4 T cell division (Cording et al., 2013; Dutzan et al., 2018), in line with our findings. In addition, specific bacterial species or their products are required for the maintenance of functional CD4 T cell phenotypes. In particular, SFB drives the differentiation of Th17 cells in the small intestine (Ivanov et al., 2009), a process shown to be indirect and occur following bacterial binding to the intestinal epithelium (Atarashi et al., 2015). Another driver of gut Th17 cells is microbial-derived ATP. Both SFB and bacterial ATP are reduced by antibiotics which has been linked to reduced IL-17A production (Atarashi et al., 2008; Kusu et al., 2013). Furthermore, orally delivered antibiotics have a greater effect on microbiota-dependent immune modulation compared to parenteral antibiotics (Nakamura et al., 2016; Strzępa et al., 2017). Indeed, SFB abundance was reduced in mice pre-exposed to vancomycin orally but not intraperitoneally, although we cannot rule out the possibility that other parenteral routes and/or dosing schemes may also reduce SFB and thus enhance susceptibility to invasive candidiasis. Collectively, these studies may explain our findings that show vancomycin specifically enhanced susceptibility to C. albicans following long-term oral treatment and only indirectly via dysbiosis. Of note, oral administration of vancomycin for Clostridioides difficile infections has been associated with the development of invasive candidiasis in patients (Falcone et al., 2016; Russo et al., 2015).
In addition to IL-17A, we found that GM-CSF production was reduced by antibiotics and that the presence of GM-CSF was required for the development of antibiotic-induced susceptibility during invasive candidiasis. GM-CSF has been shown to have a protective benefit against invasive candidiasis in humans (Dignani et al., 2005; Gavino et al., 2014). We show here that GM-CSF can ameliorate mortality in the setting of antibiotic-associated invasive candidiasis in mice. GM-CSF treatment may be effective at boosting the antifungal activity of myeloid cells (Brown et al., 2017), which were previously shown to be susceptible to antibiotic-induced damage, including with vancomycin (Ha et al., 2014). While we did not observe antibiotic-induced quantitative defects in neutrophils, it is possible that there might be functional defects since neutrophil dysfunction is linked with enhanced susceptibility to invasive candidiasis (Lionakis and Levitz, 2017), and antibiotics have been previously shown to affect neutrophil development (Deshmukh et al., 2014). We previously found that GM-CSF therapy does not enhance neutrophil recruitment to Candida-infected tissues (Drummond et al., 2018), but whether this immunotherapy boosts the functional capacity of these cells, especially in the context of antibiotic treatment, remains unknown. To understand how GM-CSF exerts this protective benefit, future studies will also need to examine the mechanism by which antibiotics disrupt GM-CSF production in the GI tract. Gut bacteria have been shown to regulate GM-CSF production from ILCs and this can be disrupted by treatment with certain antibiotics (Mortha et al., 2014). We found that vancomycin did not have a direct effect on GM-CSF production by CD4 T cells in vitro. Whether vancomycin (or other antibiotics) affect GM-CSF production via other mechanisms is unclear. Moreover, antibiotic pre-exposed mice may have effects beyond IL-17A/GM-CSF that could promote susceptibility to C. albicans infection. Future studies will need to carefully assess the direct and/or indirect mechanisms that specific antibiotics (such as vancomycin and others) perturb cellular antimicrobial immunity.
Lastly, we used a large database of patient health records to determine whether broad-spectrum antibiotic pre-exposure may be associated with the development of (and death from) invasive candidiasis, and whether trans-kingdom infections may occur more frequently in the setting of antibiotic-associated invasive candidiasis. This clinical data corroborated our observations with the mouse model. Patients treated with broad-spectrum antibiotics were more likely to develop and die from invasive candidiasis. We also noted a trend towards developing bacterial co-infections in the setting of antibiotic-associated invasive candidiasis in this dataset. Additional data will be needed to confirm our findings, however bacterial co-infections in patients with systemic C. albicans infections has been reported in observational studies, and this was associated with enhanced mortality and prior antibiotic treatment (Isenmann et al., 2002; Sipsas et al., 2009). Therefore, enhanced vigilance for trans-kingdom systemic infections in antibiotic-treated patients is warranted. Analysis of our patient dataset did not provide the power to examine the role of pre-exposure to different classes of antibiotics, whether oral and/or parenteral, in the development of invasive candidiasis, its mortality, and bacterial co-infection. This will be an important direction of future investigation using larger datasets and prospective study design.
In summary, our data uncover an unexpected mechanistic relationship between administration of broad-spectrum antibiotics and the development of and risk of death from life-threatening trans-kingdom systemic infections and stimulate new avenues of investigation into how antibiotics may affect antimicrobial immunity. This research adds to the growing body of evidence that antibiotics can cause immunological injuries, points towards a potential translational immune-based avenue for intervention, and underscores the importance of antibiotic stewardship, which may help prevent antimicrobial resistance and protect vulnerable patients from life-threatening invasive infections.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Michail Lionakis (lionakism@niaid.nih.gov).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
16S sequencing data have been deposited at NCBI BioProject under the ID number PRJNA812495 and are publicly available as of the date of publication. Access information can also be found in the Key Resource Table.
This paper does not report original code
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Candida albicans maintenance
Candida albicans strain SC5314 was used for all infections in this study. Yeast was serially passaged 3 times in YPD (yeast extract, bacto-peptone and dextrose) broth, grown at 30°C with shaking for 18–24 hours at each passage.
Yersinia pseudotuberculosis culture
WT Yersinia pseudotuberculosis (32777) was grown on MacConkey plates at room temperature for 48 hours. A single colony was picked and grown as a liquid bacterial culture in 2X YT media (Sigma) shaking (220 rpm) at 25°C overnight.
Animals
8–12 week-old female mice were maintained in individually ventilated cages under specific pathogen-free conditions at the National Institutes of Health (Bethesda, MD, USA). Mice were housed in groups of 2–6 per cage, on a 12 hour light/dark cycle at 20–24°C, and had access to food (standard rodent chow) and water ad libitum. Animal studies were performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, under the auspices of protocol LCIM14E approved by the Animal Care and Use Committee of the National Institute of Allergy and Infectious Diseases, or under project license PBE275C33 at the University of Birmingham approved by the local Animal Welfare and Ethical Review Board. WT refers to C57BL/6 (Taconic) or the corresponding littermate/genetically matched controls of the genetically-modified lines; Rag1−/−, Rag2−/−Il2rg−/−, Il17a−/− (all Taconic), Il22−/− (Jackson) and Gmcsf−/− (Charles River). In some experiments, transgenic animals that report on IL-17A production were used. These included Il17aSmart (Price et al., 2012) and Il17aCre animals crossed with Rosa26flSTOP-tdRFP mice (Fiancette et al., 2021); these animals were bred at the Biomedical Services Unit at the University of Birmingham. Animals were euthanized by cervical dislocation following administration of ketamine/xylazine cocktail at defined time-points post-infection (see Figure legends) or when humane endpoints (e.g. hypothermia, weight loss) had been reached, whichever occurred earlier.
Mouse Antibiotic Treatments and Fungal Infections
Ampicillin (1 mg/mL), metronidazole (1 mg/mL), neomycin (1 mg/mL) and vancomycin (0.5 mg/mL) were diluted in autoclaved water and delivered to mice ad libitum as their drinking water for up to 4 weeks, either alone or in combination. Waters were refreshed twice weekly. In some experiments, mice were pre-exposed to vancomycin delivered intraperitoneally at 100 μg every 2 days for 4 weeks. C. albicans yeast cells were washed in PBS, counted, and 0.95 × 105 yeast cells injected intravenously via the lateral tail vein. To control for the background level of sucrose found in the antibiotic cocktail, we treated animals with sucrose in their drinking water and challenged with C. albicans as above. From 2 independent experiments, we found no difference in survival between untreated (n=20; survival rate 20%) and sucrose-treated (n=18; survival rate 22%) mice.
Yersinia pseudotuberculosis Mouse Infection Model
Infection and analysis of mouse survival was carried out as described previously (Collins et al., 2019). Briefly, mice were infected intravenously with 200 colony forming units (CFU) of Y. pseudotuberculosis cultured as described above. Survival (defined as 30% weight loss) was assessed daily.
Mouse Short-Chain Fatty Acid Treatments
Acetate, propionate and butyrate (all from Sigma) were diluted in autoclaved water and delivered to mice ad libitum as their drinking water for at least one week prior to infection (in combination with antibiotics) and continued throughout the infection period. Final concentrations in the drinking water were 40 mM butyrate, 67.5 mM acetate and 25.9 mM propionate (Smith et al., 2013). Waters were refreshed twice weekly.
Mouse Adoptive Transfers
C57BL/6 mice (Taconic) were treated for 4 weeks with AMNV (see above) or control water, and then used as donors for the transfers. Lungs and small intestines were digested as outlined above (lungs were put directly into the same digest buffer as the intestines, after finely mincing), then stained with anti-CD45, anti-CD90.2, anti-CD127 and lineage markers (CD3, CD19, CD11b). Isolated leukocytes from the lung/intestine were pooled and ILCs were FACS-sorted (defined as CD45+CD90.2+CD127+lineage−) into sterile sorting buffer (HBSS supplemented with 2 mM EDTA, 10% FCS, 100 U/mL penicillin, 100 μg/mL streptomycin) using a FACS Aria instrument. CD4+ T-cells were isolated from the spleens of donor mice using the Miltenyi CD4+ T-cell negative selection kit, as per the manufacturer’s instructions. Purified ILCs and CD4+ T-cells were combined in sterile PBS at a ratio of 200:1 (T-cells: ILCs) and 100 μL (2×106 CD4 T-cells, 1×104 ILCs) injected intraperitoneally into Rag2−/−Il2rg−/− recipients. The mice were infected with C. albicans approximately 18 hours after the adoptive transfer of CD4 T-cells and ILCs.
In Vitro Culture Conditions
In all experiments where cells were cultured or stimulated ex vivo, cells were maintained in supplemented media (see specific sections below for details) and cultured at 37°C with 5% CO2. Cultures were checked at least every 2 days for contamination.
Clinical Data
Cerner HealthFacts, a large de-identified electronic health record database from a well-distributed cohort of US academic and community hospitals (Cerner Inc., Kansas City, MO), was used to identify hospital encounters from 2009–2017 where any Candida species was isolated from any body source (excluding stool and respiratory samples). Encounters with more than one Candida spp isolated were excluded. Invasive disease was defined as Candida isolated from blood, sterile abdominal, or other non-abdominal sterile site microbiology test. Bacterial co-infection was assessed in these patients by determining whether they had any positive bacterial microbiology test from blood or a sterile source (including abdominal, respiratory, and urinary tract) 1–31 days after the first positive Candida culture was drawn. Patients with evidence of bacterial infection in the 31 days prior to the first positive Candida culture were excluded. Prior exposure to antibiotics was defined as having received ≥7 days of a broad-spectrum systemic antibiotic (i.e. clindamycin, vancomycin, piperacillin/tazobactam, ceftolozane/tazobactam, ceftriazone, levofloxacin) in the 31 days prior to the first positive Candida culture. In our analysis, there were 116,337 unique inpatient hospitalizations that were positive for Candida (any species), of which 6,681 were excluded due to evidence of bacterial infection prior to Candida infection diagnosis. Of the remaining 110,044 encounters, 7.5% (n=8,294) were invasive Candida infections, and 15% (n=14,737) of patients died or were discharged to hospice. 5.5% (n=6,022) received at least one systemic antibiotic for >7 days.
METHOD DETAILS
Determination of Organ Fungal and Bacterial Burdens
At defined time points post-infection (see Figure legends), animals were euthanized and organs were weighed, homogenized in PBS, and serially diluted before plating onto YPD agar supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin (for fungal burdens), or onto MacConkey and CNA selective agar (Remel) for enumeration of Gram-negative and Gram-positive bacteria, respectively. Colonies were counted after incubation at 37°C for 24–48 hours. Contents of the stomach, intestines and caecum were removed prior to homogenization and plating.
Serological Determination of Renal Function
Terminal blood samples were taken via cardiac puncture and serum obtained by centrifugation at 15,000 rpm for 10 minutes at 4°C. Serum samples were diluted in sterile PBS and analyzed for concentrations of blood urea nitrogen (BUN) and creatinine as markers of renal function at the NIH Clinical Chemistry Laboratory.
Histopathology
Organs were removed from infected mice at the indicated time points and fixed in 10% formalin for 24 hours before embedding in paraffin wax. Tissue sections were stained with either periodic acid-Schiff (PAS) and/or hematoxylin and eosin (H&E).
Determination of Gastrointestinal Fungal Burdens
In some experiments, GI fungal burdens were determined in the different layers of GI tissues. Aseptically removed GI tissues were opened up laterally (to create a flat sheet) and soaked in 10 mL sterile PBS before shaking vigorously to remove contents. Washed tissues were then added to 5 mL of HBSS supplemented with 5% FBS and 25 mM HEPES, shaken vigorously and supernatant stored in a fresh tube (this was designated as ‘mucus layer’). This step was repeated to create 10 mL of ‘mucus layer’, which was centrifuged at 4000rpm for 4 minutes and pellets resuspended in 1 mL sterile PBS prior to plating on YPD plates for determination of fungal burdens. After mucus removal, GI tissues were added to 5mL of pre-warmed HBSS supplemented with 15 mM HEPES, 5 mM EDTA, 10% FBS and 1 mM dithioerythritol. Tissues were vigorously shaken twice and supernatants collected and resuspended into 1 mL sterile PBS, as before. Supernatants from this step were designated as ‘epithelial layer’. Tissues were finally washed twice in sterile PBS (by shaking in 5 mL each time), and then remaining tissue homogenized in 1mL sterile PBS prior to plating (designated ‘deep tissue layer’)(Kang et al., 2020).
Bacterial Identification by MALDI-TOF MS
Individual colonies were isolated from MacConkey and CNA plates and re-streaked onto Sheep Blood agar plates (Remel) to create single cultures. Microbiological identification of isolates was performed using MALDI-TOF MS (Bruker Daltonics). For protein extraction, bacterial colonies in sheep blood agar plate were resuspended in 1 ml 70 % ethanol, vortexed for 1 min, and centrifuged at 13,000 rpm for 2 min. The supernatant was removed completely, and the sample was vortexed for 10 seconds with 50 μl of 70 % formic acid and 50 μl acetonitrile. After a 2 min centrifugation at 13,000 rpm, 1 μl of supernatant was spotted onto the target plate and overlaid with 2 μl of alpha-cyano-4-hydroxycinnamic acid. MALDI-TOF MS analysis was performed using a Microflex LT spectrometer (Bruker Daltonics) and the Biotyper version 4.0.0.1. Manufacturer-recommended cutoff scores of ≥2.0 for species-level identification and ≥1.7 for genus-level identification, and >10 % difference of top score from other genera and species were applied.
Preparation of Genomic DNA from Murine Stool Samples
Microbiome analysis was performed on total DNA extracted from stool samples, collected from mice housed in sterilized empty cages using sterilized forceps. Stool samples were stored dry at −80°C until DNA extraction. Total DNA was extracted from stool samples using the PowerLyzer PowerSoil DNA Extraction Kit (MoBio) as per the manufacturer’s instructions, with the following modifications: Proteinase K (5 mg/mL) was added to the stool sample in the provided lysis buffer (C1) and incubated at 65°C for 10 minutes. Bead-beating was performed on the samples for 2 minutes at 30 Hz using the TissueLyzer (MoBio). Samples were then centrifuged and processed as outlined in the manufacturer’s instructions.
Quantitative PCR for Segmented Filamentous Bacteria
To assess the abundance of Segmented Filamentous Bacteria (SFB) in total genomic DNA from stool samples, qPCR reactions were carried out using 5 ng of total DNA, PerfecTa SYBR Green Fast mix (Quanta Biosciences) and primers specific for SFB (SFB736F: 5’- GACGCTGAGGCATGAGAGCAT - 3’; SFB844R: 5’- GACGGCACGGATTGTTATTCA - 3’) or total eubacteria (UniF340: 5’ - ACTCCTACGGGAGGCAGCAGT - 3’; UniF514: 5’ – TATTACCGCGGCTGCTGGC – 3’), as reported previously (Molloy et al., 2013).
16S Amplicon Sequencing and Analysis
16S rRNA sequencing libraries were prepared based on a previously described strategy(Findley et al., 2013). The V1-V3 region of the 16S rRNA gene was amplified using primers 27F (5’- AGAGTTTGATCCTGGCTCAG) and 534R (5’ -ATTACCGCGGCTGCTGG). Amplicons were generated and processed as previously described (Fadrosh et al., 2014; Jo et al., 2016). Paired-end reads were quality trimmed and joined using FLASH v1.2.11 (https://github.com/dstreett/FLASH2) and parameters of Min overlap: 10, Max mismatch density: 0.25 (Magoč and Salzberg, 2011). Joined pairs were analyzed using the Nephele platform (v.2.15.0, http://nephele.niaid.nih.gov), in particular the DADA2 based pipeline with default parameters. This pipeline de-noised the sequences, removed chimeras and assigned amplicon sequence variants (ASVs) according to the DADA2 algorithm (Weber et al., 2018). Taxonomic assignment was performed with the RDP method against the SILVA v.132 database (Quast et al., 2013). An unrarefied and rarefied biom file with 895 ASVs (40,000 reads/sample) was produced and used for visualization and downstream analyses. Alpha diversity (Shannon) was performed in R with package phyloseq (McMurdie and Holmes, 2013) and phylosmith https://github.com/schuyler-smith/phylosmith.
Isolation and Stimulation of Lamina Propria Leukocytes
The entire small intestine was aseptically removed and contents flushed out with ice-cold PBS. Intestines were opened up laterally and rinsed with ice-cold PBS to wash. Washed intestines were cut into small (1–2 mm) squares and added to 10mL pre-warmed RPMI supplemented with 15% FBS and 5 mM EDTA. Intestine pieces were incubated at 37°C with shaking for 20 minutes, vortexed vigorously for 8 seconds and supernatants removed. 10mL of pre-warmed RPMI supplemented with 5 mM EDTA was added to intestine pieces, vortexed again and supernatants removed. Intestine pieces were washed in PBS and then added to 10 mL digest buffer: RPMI supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 50 μM β-mercaptoethanol, 1 mg/mL collagenase D (Worthington), 1 mg/mL Dispase (Gibco) and 0.2 mg/mL DNAse (Roche). Intestines were incubated in digest buffer at 37°C with shaking for approximately 30 minutes until tissue had disintegrated. Digested intestines were poured through a 40μm filter, spun down and resuspended in RPMI supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin and 50 μM β-mercaptoethanol. In some experiments, intestinal leukocytes were stimulated with PMA (50 ng/mL) and ionomycin (500 ng/mL) in the presence of Brefeldin A (3 μg/mL) for 5 hours at 37°C prior to washing in PBS and staining for flow cytometry analysis.
Flow Cytometry
Isolated leukocytes were resuspended in PBS and stained with Live/Dead stain (Invitrogen) on ice as per manufacturer’s instructions. Fc receptors were blocked with anti-CD16/32 and staining with fluorochrome-labelled antibodies was performed on ice. Labelled samples were acquired immediately or fixed and permeabilized for intracellular staining using the Foxp3 staining kit (eBioscience). Anti-mouse antibodies used in this study were: CD45 (30-F11), CD11b (M1/70), CD11c (N418), MHC Class II (M5/114.15.2), CD3 (145-2C11), CD19 (eBio1D3), F480 (BM8), CD127 (A7R34), IL-17A (TC11–18H10.1), IL-22 (1H8PWSR), Ki67 (SolA15), RORgt (AFKJS-9) all from eBioscience, Ly6G (1A8), Ly6C (AL-21), all from BD Biosciences, and GM-CSF (MP1–22E9), CD90.2 (30-H12) all from Biolegend. Samples were acquired on a BD LSR Fortessa equipped with BD FACSDiva software. Analysis was performed using FlowJo (TreeStar).
In vitro Th17 Cell Differentiation
Naïve CD4 T cells (from spleens of untreated or vancomycin-preexposed mice) were first isolated using the Mojo negative selection kit (Biolegend). These cells were then further purified using FACS (using the BD FACSAria) based on markers CD4+CD44lowCD25− which typically yielded >95% purity. In some experiments, IL-17A reporter animals (Il17aSmart) were used to determine the transcription of IL-17A, in which IL-17A production is detected by staining for the human nerve growth factor receptor (hNGFR) that is under the same transcriptional control as IL-17A (Price et al., 2012). Naïve CD4 T cells were seeded into 96 well round-bottom plates (1×105 cells per well) that had been pre-coated with anti-CD3 (2μg/mL, 145-2C11, Biolegend) and anti-CD28 (10μg/mL, 37.51, BD) for 3 hours at 37°C. T cells were cultured in these pre-coated plates in complete IMDM media (Gibco) supplemented with 10% FBS, 1% Pen/Strep, 1% L-glutamine, 0.1% β-mercaptoethanol, 2ng/mL TGFβ, 25ng/mL IL-6, 10ng/mL IL-1β and 20μg/mL anti-IFNγ (all Biolegend) for 72 hours at 37°C with 5% CO2. Some cultures were further supplemented with vancomycin hydrochloride (Sigma; see figure legends for final concentrations). Culture supernatants were collected and stored at −80°C prior to ELISA-based detection for IL-17A or GM-CSF (DuoSet, RnD Systems). CD4 T-cells were stained with anti-CD4-AlexaFluor700 (RM4.4, Biolegend) and anti-HNGFR-APC (ME20.4, Biolegend) and analyzed by flow cytometry as outlined above.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis of mouse and in vitro data
All data is expressed as mean +/− SEM unless otherwise stated in the figure legends. Statistical analyses were performed using GraphPad Prism 9.0 software. Details of individual tests are included in the figure legends. In general, data was tested for normal distribution by Kolmogorov-Smirnov normality test and analyzed accordingly by unpaired two-tailed t-test or Mann Whitney U-test. In cases where multiple data sets were analyzed, two-way ANOVA was used with Bonferroni correction. In all cases, P values <0.05 were considered significant.
Risk Analysis in Patient Cohort Data
To assess the odds of developing an invasive Candida infection, logistic regression was used controlling for patient, encounter, and hospital-level characteristics. These variables were the patient’s gender and age, whether they had received any parenteral nutrition, had a prior abdominal surgery, or had a central venous catheter, and the maximum Elixhauser Comorbidity Index, as well as the hospital bed size, whether it was a teaching facility, and whether it was urban or rural. The hospital bed sized was used to control for potential differential antibiotic prescribing and treatment practices in various acuity hospital settings. Among those patients with invasive Candida infections, the cumulative incidence function of the incidence of bacterial co-infection, taking into account mortality as a competing risk, was calculated. A Fine-Gray proportional hazards model was used to regress development of a bacterial coinfection on antibiotic pre-exposure using the same definitions and covariates as above, with the additional inclusion of days of antibiotics post-Candida infection. We also assessed the risk of 31-day mortality using a Cox proportional hazards model, with coinfection as a covariate.
Supplementary Material
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| eVolve 605 CD45 Monoclonal Antibody (clone 30-F11) | Thermo Fisher Scientific | Cat# 83-0451-42, RRID:AB_2574712 |
| APC/Cyanine7 anti-mouse CD90.2 (Thy1.2) antibody (clone 53-2.1) | Biolegend | Cat# 140331, RRID:AB_2894662 |
| eFluor 450 CD127 Monoclonal Antibody (Clone A7R34) | Thermo Fisher Scientific | Cat# 48-1271-82, RRID:AB_2016698 |
| APC CD3 antibody (clone 145-2C11) | Thermo Fisher Scientific | Cat# 17-0031-82, RRID:AB_469315 |
| APC CD19 antibody (clone 1D3) | Thermo Fisher Scientific | Cat# 17-0193-82, RRID:AB_1659676 |
| APC CD11b antibody (clone M1/70) | Thermo Fisher Scientific | Cat# 17-0112-82, RRID:AB_469343 |
| APC CD11c antibody (clone N418) | Thermo Fisher Scientific | Cat# 17-0114-82, RRID:AB_469346 |
| eFluor450 MHC class II antibody | Thermo Fisher Scientific | Cat# 48-5321-82, RRID:AB_1272204 |
| PE/Cyanine7 anti-mouse F4/80 antibody (clone BM8) | Thermo Fisher Scientific | Cat# 25-4801-82, RRID:AB_469653 |
| PerCP-Cyanine5.5 IL-17A Monoclonal Antibody (clone TC11-18H10.1) | Biolegend | Cat# 506920, RRID:AB_961384 |
| PE IL-22 Monoclonal Antibody (clone 1H8PWSR) | Thermo Fisher Scientific | Cat# 12-7221-82, RRID:AB_10597428 |
| PE Cy7 anti-mouse Ki67 (clone SolA15) | Thermo Fisher Scientific | Cat#: 25-5698-82, RRID: AB_11220070 |
| APC anti-mouse RORgt (clone AFKJS-9) | Thermo Fisher Scientific | Cat#: 17-6988-82, RRID: AB_10609207 |
| PerCP/Cyanine5.5 anti-mouse Ly-6G antibody (clone 1A8) | BD Biosciences | Cat# 560602, RRID:AB_1727563 |
| AF700 Ly-6C antibody (clone AL-21) | BD Biosciences | Cat# 561237, RRID:AB_10612017 |
| PE/Cyanine7 anti-mouse GM-CSF antibody (clone MP1-22E9) | Biolegend | Cat# 505411, RRID:AB_2721681 |
| Purified anti-mouse CD3 (clone 145-2C11) | Biolegend | Cat#: 100301, RRID:AB_312666 |
| Purified anti-mouse CD28 (clone 37.51) | BD Biosciences | Cat#: 553294, RRID: AB_394763 |
| APC anti-human NFGR (clone ME20.4) | Biolegend | Cat#: 345107, RRID:AB_10639737 |
| Purified anti-mouse IFNg (clone AN-18) | Biolegend | Cat#: 517905, RRID: AB_2810651 |
| Fungal and bacterial strains | ||
| Candida albicans SC5314 | Laboratory of M. Lionakis | ATCC: MYA-2876 |
| Yersinia pseudotuberculosis 32777 | Laboratory of Y. Belkaid | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Ampicillin | Sigma Aldrich | Cat#: A1593 |
| Metronidazole | Sigma Aldrich | Cat#: M1547 |
| Neomycin | VET one | Cat#: 510570 |
| Vancomycin | Fresenius Kabi | Cat#: C28420 |
| Sodium Acetate | Sigma | Cat#: 241245 |
| Sodium Propionate | Sigma | Cat#: P1880 |
| Butyrate | Sigma | Cat#: 303410 |
| DNAse I | Roche | Cat#: 03724778103 |
| Collagenase type IV | Worthington | Cat#: LS004189 |
| Dispase II | Thermo Fisher Scientific | Cat#: 17105041 |
| TGFb | Biolegend | Cat#: 763102 |
| IL-1b | Biolegend | Cat#: 575104 |
| IL-6 | Peprotech | Cat#: 216-16 |
| Deposited data | ||
| 16S Microbiome Data | NCBI BioProject | PRJNA812495 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6Tn | NIAID Taconic contract | N/A |
| Mouse: C57BL/10SgSnAi-[KO]RAG2 | NIAID Taconic contract | Line 103 |
| Mouse: C57BL/10SgSnAi-[KO]RAG2-[KO]gamma c | NIAID Taconic contract | Line 111 |
| Mouse: C57BL/6-[KO]IL17A | NIAID Taconic contract | Line 8434 |
| Mouse: C57BL/6-Il22tm1.1(icre)Stck/J | Jackson Laboratories | 027524 |
| Mouse: GMcsf | Charles River | |
| Mouse: Il17aSmart | Laboratory of D. Withers | N/A |
| Mouse: Il17aCreRosa26flSTOP-tdRFP | Laboratory of D. Withers | N/A |
| Oligonucleotides | ||
| Primer: SFB quantification; SFB736F: 5’- GACGCTGAGGCATGAGAGCAT - 3’; | IDT | N/A |
| Primer: SFB quantification; SFB844R: 5’- GACGGCACGGATTGTTATTCA - 3’ | IDT | N/A |
| Primer: Eubacteria quantification ; UniF340: 5’ - ACTCCTACGGGAGGCAGCAGT - 3’ | IDT | N/A |
| Primer: Eubacteria quantification; UniF514: 5’ – TATTACCGCGGCTGCTGGC – 3’ | IDT | N/A |
| Primers: 16S amplification (forward); 5’- AGAGTTTGATCCTGGCTCAG | IDT | N/A |
| Primer: 16S amplification (reverse); 5’ - ATTACCGCGGCTGCTGG | IDT | N/A |
| Commercial Kits and Assays | ||
| PowerLyzer PowerSoil DNA Extraction Kit | MOBIO | Cat# 1288-100 |
| Mouse GM-CSF DuoSet ELISA | Biotechne | DY415-05 |
| IL-17A ELISA | DY421-05 | |
| Software and algorithms | ||
| FLASH v1.2.11 | Magoć et al., 2011. DOI: 10.1093/bioinformatics/btr507 | https://github.com/dstreett/FLASH2 |
| Nephele v.2.15.0 | Weber et al., 2018. DOI: 10.1093/bioinformatics/btx617 | http://nephele.niaid.nih.gov |
| R package phyloseq | McMurdie and Holmes, 2013. DOI: 10.1371/journal.pone.0061217 | https://github.com/schuyler-smith/phylosmith |
| FlowJo v10.7.2 | TreeStar |
https://www.flowjo.com/solutions/flowjo/downloads
|
| GraphPad Prism v9.2.0 | GraphPad | https://www.graphpad.com/scientificsoftware/prism/ |
Highlights.
Broad-spectrum antibiotics promote susceptibility to invasive fungal infection.
Antibiotics impair control of fungal infection specifically in the intestine.
Commensal gut bacteria escape systemically in antibiotic-treated mice.
Antibiotics impair protective antifungal gut lymphoid IL-17A and GM-CSF responses.
Acknowledgments
This work was supported by the Division of Intramural Research of the National Institute of Allergy & Infectious Disease, National Human Genome Research Institute, the National Cancer Institute, and the NIH Clinical Center. The computational resources of the NIH High-Performance Computation Biowulf Cluster (http://hpc.nih.gov) were used for this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
The authors declare that no conflicts of interest exist.
References
- Aggor FEY, Break TJ, Trevejo-Nuñez G, Whibley N, Coleman BM, Bailey RD, Kaplan DH, Naglik JR, Shan W, Shetty AC, et al. (2020). Oral epithelial IL-22/STAT3 signaling licenses IL-17-mediated immunity to oral mucosal candidiasis. Sci Immunol 5. 10.1126/sciimmunol.aba0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M, Ang QY, Nayak RR, Bustion AE, Sandy M, Zhang B, Upadhyay V, Pollard KS, Lynch SV, and Turnbaugh PJ (2022). Human gut bacterial metabolism drives Th17 activation and colitis. Cell Host & Microbe 30, 17–30.e19. 10.1016/j.chom.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allert S, Förster TM, Svensson C-M, Richardson JP, Pawlik T, Hebecker B, Rudolphi S, Juraschitz M, Schaller M, Blagojevic M, et al. (2018). Candida albicans-Induced Epithelial Damage Mediates Translocation through Intestinal Barriers. mBio 9, e00915–00918. 10.1128/mBio.00915-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida L, Dhillon-LaBrooy A, Castro CN, Adossa N, Carriche GM, Guderian M, Lippens S, Dennerlein S, Hesse C, Lambrecht BN, et al. (2021). RibosomeTargeting Antibiotics Impair T Cell Effector Function and Ameliorate Autoimmunity by Blocking Mitochondrial Protein Synthesis. Immunity 54, 68–83.e66. 10.1016/j.immuni.2020.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, and Takeda K (2008). ATP drives lamina propria TH17 cell differentiation. Nature 455, 808–812. 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, et al. (2015). Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 163, 367–380. 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichele R, Kärner J, Truusalu K, Smidt I, Mändar R, Conti HR, Gaffen SL, Peterson P, Laan M, and Kisand K (2018). IL-22 neutralizing autoantibodies impair fungal clearance in murine oropharyngeal candidiasis model. Euro J Immunol 48, 464–470. 10.1002/eji.201747209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson B, Wang C, Pedergnana V, Wu L, Cypowyj S, Rybojad M, Belkadi A, Picard C, Abel L, Fieschi C, et al. (2013). An ACT1 mutation selectively abolishes interleukin-17 responses in humans with chronic mucocutaneous candidiasis. Immunity 39, 676–686. 10.1016/j.immuni.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, and Pamer EG (2008). Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455, 804–807. http://www.nature.com/nature/journal/v455/n7214/suppinfo/nature07250_S1.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RL, Sequeira RP, and Clarke TB (2017). The microbiota protects against respiratory infection via GM-CSF signaling. Nature Communications 8, 1512. 10.1038/s41467-017-01803-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R, Lam V, Kommineni S, Stromich J, Hayward M, Kristich CJ, and Salzman NH (2018). Ceftriaxone Administration Disrupts Intestinal Homeostasis, Mediating Noninflammatory Proliferation and Dissemination of Commensal Enterococci. Infect Immun 86, e00674–00618. 10.1128/IAI.00674-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N, Han SJ, Enamorado M, Link VM, Huang B, Moseman EA, Kishton RJ, Shannon JP, Dixit D, Schwab SR, et al. (2019). The Bone Marrow Protects and Optimizes Immunological Memory during Dietary Restriction. Cell 178, 1088–1101.e1015. 10.1016/j.cell.2019.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cording S, Fleissner D, Heimesaat MM, Bereswill S, Loddenkemper C, Uematsu S, Akira S, Hamann A, and Huehn J (2013). Commensal microbiota drive proliferation of conventional and Foxp3(+) regulatory CD4(+) T cells in mesenteric lymph nodes and Peyer’s patches. Eur J Microbiol Immunol (Bp) 3, 1–10. 10.1556/EuJMI.3.2013.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O’Leary CE, Oliver PM, Kolls JK, Weiser JN, and Worthen GS (2014). The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med 20, 528–534. 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignani MC, Rex JH, Chan KW, Dow G, deMagalhaes-Silverman M, Maddox A, Walsh T, and Anaissie E (2005). Immunomodulation with interferon-gamma and colony-stimulating factors for refractory fungal infections in patients with leukemia. Cancer 104, 199–204. 10.1002/cncr.21142. [DOI] [PubMed] [Google Scholar]
- Drummond RA, Dambuza IM, Vautier S, Taylor JA, Reid DM, Bain CC, Underhill DM, Masopust D, Kaplan DH, and Brown GD (2016). CD4+ T-cell survival in the GI tract requires dectin-1 during fungal infection. Mucosal Immunol 9, 492–502. 10.1038/mi.2015.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RA, Zahra FT, Natarajan M, Swamydas M, Hsu AP, Wheat LJ, Gavino C, Vinh DC, Holland SH, Mikelis CM, and Lionakis MS (2018). GM-CSF Therapy in Human CARD9 Deficiency. J Allergy Clin Immunol. 10.1016/j.jaci.2018.05.025. [DOI] [PubMed] [Google Scholar]
- Dutzan N, Kajikawa T, Abusleme L, Greenwell-Wild T, Zuazo CE, Ikeuchi T, Brenchley L, Abe T, Hurabielle C, Martin D, et al. (2018). A dysbiotic microbiome triggers TH17 cells to mediate oral mucosal immunopathology in mice and humans. Science translational medicine 10. 10.1126/scitranslmed.aat0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, Keren-Shaul H, Mahlakoiv T, Jakobshagen K, Buch T, et al. (2015). Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci 18. 10.1038/nn.4030 http://www.nature.com/neuro/journal/vaop/ncurrent/abs/nn.4030.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, and Ravel J (2014). An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2, 6. 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone M, Russo A, Iraci F, Carfagna P, Goldoni P, Vullo V, and Venditti M (2016). Risk Factors and Outcomes for Bloodstream Infections Secondary to Clostridium difficile Infection. Antimicrob Agents Chemother 60, 252–257. 10.1128/aac.01927-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D, Coughlin LA, Neubauer MM, Kim J, Kim MS, Zhan X, Simms-Waldrip TR, Xie Y, Hooper LV, and Koh AY (2015). Activation of HIF-1[alpha] and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med 21, 808–814. 10.1038/nm.3871 http://www.nature.com/nm/journal/vaop/ncurrent/abs/nm.3871.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiancette R, Finlay CM, Willis C, Bevington SL, Soley J, Ng STH, Baker SM, Andrews S, Hepworth MR, and Withers DR (2021). Reciprocal transcription factor networks govern tissue-resident ILC3 subset function and identity. Nat. Immunol 22, 1245–1255. 10.1038/s41590-021-01024-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Intramural Sequencing Ctr NIH, et al. (2013). Topographic diversity of fungal and bacterial communities in human skin. Nature 498, 367–370. 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavino C, Cotter A, Lichtenstein D, Lejtenyi D, Fortin C, Legault C, Alirezaie N, Majewski J, Sheppard DC, Behr MA, et al. (2014). CARD9 Deficiency and Spontaneous Central Nervous System Candidiasis: Complete Clinical Remission With GM-CSF Therapy. Clin Infect Dis 59, 81–84. 10.1093/cid/ciu215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennari R, Alexander JW, Gianotti L, Eaves-Pyles T, and Hartmann S (1994). Granulocyte macrophage colony-stimulating factor improves survival in two models of gut-derived sepsis by improving gut barrier function and modulating bacterial clearance. Ann Surg 220, 68–76. 10.1097/00000658-199407000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha YE, Kong K-H, Cho M-H, Kim D-H, Song Y-S, and Yoon S-Y (2014). Vancomycin blocks autophagy and induces interleukin-1β release in macrophages. J Antibiotics 68, 76. 10.1038/ja.2014.112. [DOI] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KD, Quoclinh N, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. (2012). Interactions Between Commensal Fungi and the C-Type Lectin Receptor Dectin-1 Influence Colitis. Science 336, 1314–1317. 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenmann R, Schwarz M, Rau B, Trautmann M, Schober W, and Beger HG (2002). Characteristics of infection with Candida species in patients with necrotizing pancreatitis. World J Surgery 26, 372–376. 10.1007/s00268-001-0146-9. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. (2009). Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell 139, 485–498. 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Frutos R.d.L., Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, and Littman DR (2008). Specific Microbiota Direct the Differentiation of IL-17-Producing T-Helper Cells in the Mucosa of the Small Intestine. Cell Host Microbe 4, 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang TT, Chaturvedi V, Ertelt JM, Xin L, Clark DR, Kinder JM, and Way SS (2014). Commensal enteric bacteria lipopolysaccharide impairs host defense against disseminated Candida albicans fungal infection. Mucosal Immunol. 10.1038/mi.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo JH, Deming C, Kennedy EA, Conlan S, Polley EC, Ng WI, Segre JA, and Kong HH (2016). Diverse Human Skin Fungal Communities in Children Converge in Adulthood. J Invest Dermatol 136, 2356–2363. 10.1016/j.jid.2016.05.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalghatgi S, Spina CS, Costello JC, Liesa M, Morones-Ramirez JR, Slomovic S, Molina A, Shirihai OS, and Collins JJ (2013). Bactericidal Antibiotics Induce Mitochondrial Dysfunction and Oxidative Damage in Mammalian Cells. Sci Trans Med 5, 192ra185. 10.1126/scitranslmed.3006055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B, Alvarado LJ, Kim T, Lehmann ML, Cho H, He J, Li P, Kim B-H, Larochelle A, and Kelsall BL (2020). Commensal microbiota drive the functional diversification of colon macrophages. Mucosal Immunol 13, 216–229. 10.1038/s41385-0190228-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara S, Jhingran A, Dhingra S, Salem A, Cramer RA, and Hohl TM (2016). GM-CSF signaling regulates neutrophil antifungal activity and the oxidative burst during respiratory fungal challenge. J Infect Dis 213, 1289–1298. 10.1093/infdis/jiw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keighley C, Chen SC, Marriott D, Pope A, Chapman B, Kennedy K, Bak N, Underwood N, Wilson HL, McDonald K, et al. (2019). Candidaemia and a risk predictive model for overall mortality: a prospective multicentre study. BMC Infect Dis 19, 445. 10.1186/s12879-019-4065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly Caleb J., Zheng L, Campbell Eric L., Saeedi B, Scholz Carsten C., Bayless Amanda J., Wilson Kelly E., Glover Louise E., Kominsky Douglas J., Magnuson A, et al. (2015). Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host & Microbe 17, 662–671. 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaasen HLBM, Koopman JP, Vollaard EJ, Theeuwes AGM, Van Den Brink ME, Scholten PM, Bakker MH, and Beynen AC (1991). Influence of Antimicrobial Drugs on Segmented Filamentous Bacteria in the Ileum of Mice. Microbial Ecology in Health and Disease 4, 391–397. 10.3109/08910609109140155. [DOI] [Google Scholar]
- Knoop KA, McDonald KG, Kulkarni DH, and Newberry RD (2016). Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut 65, 1100–1109. 10.1136/gutjnl-2014-309059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh AY, Köhler JR, Coggshall KT, Van Rooijen N, and Pier GB (2008). Mucosal Damage and Neutropenia Are Required for Candida albicans Dissemination. PLoS Pathog 4, e35. 10.1371/journal.ppat.0040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusu T, Kayama H, Kinoshita M, Jeon SG, Ueda Y, Goto Y, Okumura R, Saiga H, Kurakawa T, Ikeda K, et al. (2013). Ecto-nucleoside triphosphate diphosphohydrolase 7 controls Th17 cell responses through regulation of luminal ATP in the small intestine. J Immunol 190, 774–783. 10.4049/jimmunol.1103067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy R, Okada S, Béziat V, Moriya K, Liu C, Chai LY, Migaud M, Hauck F, Al Ali A, Cyrus C, et al. (2016). Genetic, immunological, and clinical features of patients with bacterial and fungal infections due to inherited IL-17RA deficiency. Proc Natl Acad Sci U S A 113, E8277–e8285. 10.1073/pnas.1618300114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MM, Li CY, Wu XA, Chen TT, Ren L, Xu BL, and Cao J (2020). Microbiota-driven interleukin-17 production provides immune protection against invasive candidiasis. Critical Care 24, 268, 268. 10.1186/s13054-020-02977-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionakis MS (2014). New insights into innate immune control of systemic candidiasis. Med Mycol 52, 555–564. 10.1093/mmy/myu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionakis MS, and Levitz SM (2017). Host control of fungal infections: lessons from basic studies and human cohorts. Ann Rev Immunol 36, 157–191. 10.1146/annurevimmunol-042617-053318. [DOI] [PubMed] [Google Scholar]
- Lionakis MS, Lim JK, Lee CCR, and Murphy PM (2011). Organ-specific innate immune responses in a mouse model of invasive candidiasis. J Innate Immun 3, 180–199. 10.1159/000321157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč T, and Salzberg SL (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-López M, Iborra S, Conde-Garrosa R, Mastrangelo A, Danne C, Mann ER, Reid DM, Gaboriau-Routhiau V, Chaparro M, Lorenzo MP, et al. (2019). Microbiota Sensing by Mincle-Syk Axis in Dendritic Cells Regulates Interleukin-17 and −22 Production and Promotes Intestinal Barrier Integrity. Immunity 50, 446–461.e449. 10.1016/j.immuni.2018.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, and Holmes S (2013). phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One 8, e61217. 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy MJ, Grainger JR, Bouladoux N, Hand TW, Koo LY, Naik S, Quinones M, Dzutsev AK, Gao JL, Trinchieri G, et al. (2013). Intraluminal containment of commensal outgrowth in the gut during infection-induced dysbiosis. Cell Host Microbe 14, 318–328. 10.1016/j.chom.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, and Merad M (2014). Microbiota-Dependent Crosstalk Between Macrophages and ILC3 Promotes Intestinal Homeostasis. Science 343, 1249288. 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura YK, Metea C, Karstens L, Asquith M, Gruner H, Moscibrocki C, Lee I, Brislawn CJ, Jansson JK, Rosenbaum JT, and Lin P (2016). Gut Microbial Alterations Associated With Protection From Autoimmune Uveitis. Investigative Ophthalmology & Visual Science 57, 3747–3758. 10.1167/iovs.16-19733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC, Davidson AD, Jacobsen MD, Tavanti A, Whyte JA, Kibbler CC, Ellis DH, Maiden MCJ, Shaw DJ, and Gow NAR (2006). Candida albicans strain maintenance, replacement, and microvariation demonstrated by multilocus sequence typing. J Clin Microbiol 44, 3647–3658. 10.1128/jcm.00934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, and Kullberg BJ (2018). Invasive candidiasis. Nat Rev Dis Primers 4, 18026. 10.1038/nrdp.2018.26. [DOI] [PubMed] [Google Scholar]
- Poynton CH, Barnes RA, and Rees J (1998). Interferon gamma and granulocyte-macrophage colony-stimulating factor for the treatment of hepatosplenic candidosis in patients with acute leukemia. Clin Infect Dis 26, 239–240. 10.1086/517077. [DOI] [PubMed] [Google Scholar]
- Price AE, Reinhardt RL, Liang H-E, and Locksley RM (2012). Marking and quantifying IL-17A-producing cells in vivo. PLoS One 7, e39750–e39750. 10.1371/journal.pone.0039750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel A, Cypowyj S, Bustamante J, Wright JF, Liu L, Lim HK, Migaud M, Israel L, Chrabieh M, Audry M, et al. (2011). Chronic Mucocutaneous Candidiasis in Humans with Inborn Errors of Interleukin-17 Immunity. Science 332, 65–68. 10.1126/science.1200439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel A, Daffinger R, Natividad A, Chrabieh M, Barcenas-Morales G, Picard C, Cobat A, Ouachace-Chardin M, Toulon A, Bustamante J, et al. (2010). Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. J Exp Med 207, 291–297. 10.1084/jem.20091983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, and Glöckner FO (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic acids research 41, D590–D596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricotta EE, Lai YL, Babiker A, Strich JR, Kadri SS, Lionakis MS, Prevots DR, and Adjemian J (2020). Invasive candidiasis species distribution and trends, United States, 2009–2017. J Infect Dis, jiaa502. 10.1093/infdis/jiaa502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rókusz L, Liptay L, and Kádár K (2001). Successful treatment of chronic disseminated candidiasis with fluconazole and a granulocyte-macrophage colony-stimulating factor combination. Scand J Infect Dis 33, 784–786. 10.1080/003655401317074671. [DOI] [PubMed] [Google Scholar]
- Russo A, Falcone M, Fantoni M, Murri R, Masucci L, Carfagna P, Ghezzi MC, Posteraro B, Sanguinetti M, and Venditti M (2015). Risk factors and clinical outcomes of candidaemia in patients treated for Clostridium difficile infection. Clin Microbiol Infect 21, 493.e491–494. 10.1016/j.cmi.2014.12.024. [DOI] [PubMed] [Google Scholar]
- Shankar J, Solis NV, Mounaud S, Szpakowski S, Liu H, Losada L, Nierman WC, and Filler SG (2015). Using Bayesian modelling to investigate factors governing antibiotic-induced Candida albicans colonization of the GI tract. Sci. Rep 5. 10.1038/srep08131 http://www.nature.com/srep/2015/150203/srep08131/abs/srep08131.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao T-Y, Ang WXG, Jiang TT, Huang FS, Andersen H, Kinder JM, Pham G, Burg AR, Ruff B, Gonzalez T, et al. (2019). Commensal Candida albicans Positively Calibrates Systemic Th17 Immunological Responses. Cell host & microbe 25, 404–417.e406. 10.1016/j.chom.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipsas NV, Lewis RE, Tarrand J, Hachem R, Rolston KV, Raad II, and Kontoyiannis DP (2009). Candidemia in patients with hematologic malignancies in the era of new antifungal agents (2001–2007). Cancer 115, 4745–4752. 10.1002/cncr.24507. [DOI] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, and Garrett WS (2013). The microbial metabolites, short chain fatty acids, regulate colonic Treg cell homeostasis. Science (New York, N.Y.) 341, 10.1126/science.1241165. 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, Shibata N, Grunberg S, Sinha R, Zahm AM, et al. (2012). Innate Lymphoid Cells Promote Anatomical Containment of Lymphoid-Resident Commensal Bacteria. Science 336, 1321–1325. 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellberg B, Ibrahim AS, Jr JEE, and Filler SG (2005). Mice with Disseminated Candidiasis Die of Progressive Sepsis. J. Infect. Dis 192, 336–343. [DOI] [PubMed] [Google Scholar]
- Spellberg B, Johnston D, Phan QT, Edwards JE, French SW, Ibrahim AS, and Filler SG (2003). Parenchymal organ, and not splenic, immunity correlates with host survival during disseminated candidiasis. Infect Immun 71, 5756–5764. 10.1128/iai.71.10.5756-5764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller RC (2018). Hidden Dangers of Antibiotic Use: Increased Gut Permeability Mediated by Increased Pancreatic Proteases Reaching the Colon. Cell Mol Gastroenterol Hepatol 6, 347–348.e341. 10.1016/j.jcmgh.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strollo S, Lionakis MS, Adjemian J, Steiner CA, and Prevots DR (2016). Epidemiology of Hospitalizations Associated with Invasive Candidiasis, United States, 2002–2012(1). Emerg Infect Dis 23, 7–13. 10.3201/eid2301.161198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzępa A, Majewska-Szczepanik M, Lobo FM, Wen L, and Szczepanik M (2017). Broad spectrum antibiotic enrofloxacin modulates contact sensitivity through gut microbiota in a murine model. Journal of Allergy and Clinical Immunology 140, 121–133.e123. 10.1016/j.jaci.2016.11.052. [DOI] [PubMed] [Google Scholar]
- Tulstrup MV-L, Christensen EG, Carvalho V, Linninge C, Ahrné S, Højberg O, Licht TR, and Bahl MI (2015). Antibiotic Treatment Affects Intestinal Permeability and Gut Microbial Composition in Wistar Rats Dependent on Antibiotic Class. PLoS One 10, e0144854–e0144854. 10.1371/journal.pone.0144854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Veerdonk FL, Kullberg BJ, and Netea MG (2012). Adjunctive immunotherapy with recombinant cytokines for the treatment of disseminated candidiasis. Clin Microbiol Infect 18, 112–119. 10.1111/j.1469-0691.2011.03676.x. [DOI] [PubMed] [Google Scholar]
- van Opstal E, Kolling GL, Moore JH 2nd, Coquery CM, Wade NS, Loo WM, Bolick DT, Shin JH, Erickson LD, and Warren CA (2016). Vancomycin Treatment Alters Humoral Immunity and Intestinal Microbiota in an Aged Mouse Model of Clostridium difficile Infection. J Infect Dis 214, 130–139. 10.1093/infdis/jiw071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vautier S, Drummond RA, Redelinghuys P, Murray GI, MacCallum DM, and Brown GD (2012). Dectin-1 Is not required for controlling Candida albicans colonization of the gastrointestinal tract. Infect Immun 80, 4216–4222. 10.1128/iai.00559-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Drygalski A, Curtis BR, Bougie DW, McFarland JG, Ahl S, Limbu I, Baker KR, and Aster RH (2007). Vancomycin-Induced Immune Thrombocytopenia. N. Eng. J. Med 356, 904–910. 10.1056/NEJMoa065066. [DOI] [PubMed] [Google Scholar]
- Weber N, Liou D, Dommer J, MacMenamin P, Quiñones M, Misner I, Oler AJ, Wan J, Kim L, Coakley McCarthy M, et al. (2018). Nephele: a cloud platform for simplified, standardized and reproducible microbiome data analysis. Bioinformatics 34, 1411–1413. 10.1093/bioinformatics/btx617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Bhargava P, McCloskey D, Mao N, Palsson BO, and Collins JJ (2017). Antibiotic-Induced Changes to the Host Metabolic Environment Inhibit Drug Efficacy and Alter Immune Function. Cell Host Microbe 22, 757–765.e753. 10.1016/j.chom.2017.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu LC-H, Shih Y-A, Wu L-L, Lin Y-D, Kuo W-T, Peng W-H, Lu K-S, Wei S-C, Turner JR, and Ni Y-H (2014). Enteric dysbiosis promotes antibiotic-resistant bacterial infection: systemic dissemination of resistant and commensal bacteria through epithelial transcytosis. Am J Physiol Gastrointest Liver Physiol 307, G824–G835. 10.1152/ajpgi.00070.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai B, Ola M, Rolling T, Tosini NL, Joshowitz S, Littmann ER, Amoretti LA, Fontana E, Wright RJ, Miranda E, et al. (2020). High-resolution mycobiota analysis reveals dynamic intestinal translocation preceding invasive candidiasis. Nat Med 26, 59–64. 10.1038/s41591-019-0709-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S sequencing data have been deposited at NCBI BioProject under the ID number PRJNA812495 and are publicly available as of the date of publication. Access information can also be found in the Key Resource Table.
This paper does not report original code
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


