Abstract
Cytoglobin is a hemoprotein widely expressed in fibroblasts and related cell lineages with yet undefined physiological function. Cytoglobin, as other heme proteins, can reduce nitrite to nitric oxide (NO) providing a route to generate NO in vivo in low oxygen conditions. In addition, cytoglobin can also bind lipids such as oleic acid and cardiolipin with high affinity. These two processes are potentially relevant to cytoglobin function. Little is known about how specific amino acids contribute to nitrite reduction and lipid binding. Here we investigate the role of the distal histidine His81(E7) and several surface residues on the regulation of nitrite reduction and lipid binding. We observe that the replacement of His81(E7) greatly increases heme reactivity towards nitrite, with nitrite reduction rate constants of up to 1100 M−1s−1 for the His81Ala mutant. His81(E7) mutation causes a small decrease in lipid binding affinity, however experiments on the presence of imidazole indicate that His81(E7) does not compete with the lipid for the binding site. Mutations of the surface residues Arg84 and Lys116 largely impair lipid binding. Our results suggest that dissociation of His81(E7) from the heme mediates the formation of a hydrophobic cavity in the proximal heme side that can accommodate the lipid, with important contributions of the hydrophobic patch around residues Thr91, Val105, and Leu108, whereas the positive charges from Arg84 and Lys116 stabilize the carboxyl group of the fatty acid. Gain and loss-of-function mutations described here can serve as tools to study in vivo the physiological role of these putative cytoglobin functions.
Keywords: Cytoglobin, nitrite reduction, nitric oxide, protein-lipid interaction
1. Introduction
Cytoglobin (Cygb) is a heme globin first described in 2001 with yet undefined physiological function [1–3]. Expression of Cygb is widespread in mammalian tissues, with higher expression levels observed in fibroblasts and fibroblast-related cell lineages [4–6]. The sequence and structure of Cygb shows high similarity with other mammalian globins, including neuroglobin (Ngb), myoglobin and hemoglobin. Like Ngb, Cygb has a six-coordinated heme, with distal (E7) and proximal (F8) histidines coordinating the heme iron. The distal histidine is in a dynamic equilibrium between iron-bound and iron-unbound conformations, with the equilibrium poised towards the iron-bound species but allowing binding of gaseous ligands and other small molecules to the heme iron. Nevertheless, the preferred six-coordinated conformation confers a very different reactivity to Cygb and Ngb as compared to five-coordinated globins like myoglobin and hemoglobin [5]. Phylogenetic reconstructions indicate that Cygb most probably evolved from a six-coordinate globin ancestor, and is itself a common ancestor to five-coordinate globins such as myoglobin and hemoglobin [7]. Interestingly, some Cygbs such as zebrafish Cygb-1 do present significant five-coordination although that does not seem to apply to other fish Cygb-1 proteins [8–10].
Cygb can bind lipids such as oleic acid and cardiolipin with high affinity (KD ≈ 1μM) [11, 12]. This process appears to be regulated by the thiol oxidation state [11, 13] and may have emerged recently in Cygb evolution [8]. The effects on peroxidase activity and the specificity towards anionic phospholipids suggest that a protein-lipid interaction could play a regulatory effect on Cygb function, reminiscent of other protein-lipid interactions like the cardiolipin-mediated activation of cytochrome c on apoptosis [14]. The biological significance of that phenomenon in Cygb is yet unknown [12].
Previous work also purports a possible role of Cygb in nitric oxide (NO) metabolism. Through its NO deoxygenation activity; Cygb can oxidize NO to nitrate at near diffusion-limited rates, as observed for other globins [15–17]. This reaction appears to be supported by Cytochrome b5 reductase 3, Cytochrome b5 and NADH [16–18] and can impact the maintenance of blood pressure in the blood vessels [18, 19]. Conversely, Cygb can reduce nitrite to NO and this can constitute a route to generate NO in vivo in low oxygen conditions [8, 20–22]. In addition, Cygb may act as a superoxide dismutase, further impacting NO bioavailability by decreasing superoxide concentrations [23].
These two processes are potentially relevant to Cygb function. It is clear that a disulfide bond in the surface of Cygb can modulate heme reactivity in these two reactions [11, 13, 21, 24]. However, the role of the distal histidine or other mutations in these processes is unknown. In this work, we use site-directed mutagenesis techniques to investigate the mechanisms regulating nitrite reduction and lipid binding. Previous work on Ngb indicates that the replacement of the distal histidine by residues that cannot provide a sixth ligand to the heme iron leads to a constitutive five-coordinate structure with increased reactivity towards nitrite [25, 26]. Given the significant structural similarity between Ngb and Cygb, it is expected that these mutations will also increase nitrite reduction rates, although that has not been investigated. A deeper understanding of the factors regulating nitrite reduction in Cygb is necessary in order to develop the tools to study the relevance of this reaction for Cygb physiological function in cell culture and animal models.
We have also identified some putative binding pockets for lipid binding to Cygb [12]. Based on Molecular Dynamics simulations, we have noted that the formation of the disulfide bond between Cys38 and Cys83 can pull the E helix away from the heme and weaken the interaction of the distal histidine –His81(E7)– with the heme, and also create a novel hydrophobic pocket in the protein. Models indicate that residues Arg84, Thr91 and Lys116 can interact with the bound lipids [12]. Alternatively, another possible binding site involves the distal side of the heme, and this location would be very sensitive to mutations of the His81 residue. In addition, a third possible site is formed by the hydrophobic surface patch that includes Val105 and Leu108. In order to determine which one of the sites is a more likely binding site, we performed mutations of all the indicated residues and characterized their lipid binding properties. We hypothesize that selective mutation of these sites can identify the most relevant binding spots by changes in the dissociation constant values. We expect that a more detailed knowledge of the lipid binding site of Cygb can also guide further efforts to establish the physiological relevance of this reaction.
Our results confirm the role of the distal histidine on nitrite reduction and show that Cygb can achieve high rates of nitrite reduction with rate constants up to 1100 M−1s−1 for the H81A mutant. Distal histidine mutations generate a gradual increase in rate constants roughly consistent with the size of the new side chain. Although the distal histidine mutations do modify lipid binding properties, imidazole replacement experiments indicate that the lipid binding site does not involve the heme distal site. The protein surface seems to contribute part of the binding energy, with the charges at Arg84 and Lys116 being critical for the stabilization of the carboxylic group of the fatty acid. These observations open the possibility to modifications of Cygb that impact specifically nitrite reduction and/or lipid binding. These can serve as tools for future in vivo studies to establish the possible physiological role of those putative Cygb functions.
2. Materials and Methods
2.1. Reagents and protein preparation –
Reagents were obtained from Sigma unless stated otherwise.
2.2. Protein expression and purification –
Plasmids for the generation of Cygb mutants were generated using standard molecular biology techniques. The wild-type (WT) human Cygb protein was expressed using the pET28-HsaCgb plasmid as described [8, 17]. The mutations H81A, H81Q, H81L, H81W, R84E, T91D, V105Y, L108Q and K116E were introduced using the Quikchange site-directed mutagenesis kit (Stratagene, La Jolla, CA) using adequate primers. Purification of the His-tagged proteins from SoluBL21 cells (Genlantis, San Diego, CA) with Ni-NTA agarose columns was performed as reported for the human WT Cygb protein [8, 17]. His-tagged Ngb H64L was expressed and purified as previously described [26].
2.3. Determination of autoxidation rates –
The autoxidation of the ferrous-dioxygen complexes was studied at 37 °C in sodium phosphate buffer 100 mM, pH 7.4 as previously described [25]. Briefly, the ferrous-dioxygen (FeII-O2) complexes were prepared by reduction of the globins with excess dithionite (to form the reduced deoxy form of the protein) followed by gel filtration through a Sephadex G25-column (PD10, GE Healthcare) equilibrated with aerobic buffer. As the protein is eluted in the column and dithionite is removed from the sample, the deoxy globin binds oxygen from the buffer to yield a final elution sample containing mostly the ferrous-dioxygen form. These concentrated protein samples were mixed with prewarmed buffer at 37 °C and the spectral changes were monitored for 30 to 60 minutes in a Cary-50 spectrophotometer (Agilent Technologies. Palo Alto, CA). For Cygb mutants R84E and K116E, the reaction rates were too fast to be determined by this method as substantial autoxidation occurs as the protein is eluted from the G25 column. Instead, the deoxy species (FeII) was prepared inside an anaerobic glovebox (Coy Laboratory Products, Grass Lake, MI). In this method, the ferrous complexes (FeII) are prepared by mixing the proteins with sodium dithionite and the samples were run through a Sephadex G25-column (PD10, GE Healthcare) equilibrated with anaerobic buffer to remove excess dithionite. Ferrous protein samples were then mixed with oxygen saturated buffer (4:1 v/v ratio). This generates quantitatively the ferrous dioxygen species (FeII-O2) and the spectral changes associated with the decay of this complex to the ferric form (FeIII) are monitored for 1000 seconds (1 spectrum/s) in an Agilent HP8453 spectrophotometer (Agilent Technologies. Palo Alto, CA).
2.4. Nitrite reduction experiments –
Nitrite reduction experiments were performed in 100 mM sodium phosphate buffer, pH 7.4, at 37 °C. The reactions were followed in the presence of 2.5 mM sodium dithionite as described previously [8, 25, 27]. The presence of excess dithionite generates quantitatively the ferrous heme species, reduces ferric species formed during the reaction back to the ferrous form, and consumes oxygen avoiding side reactions between nitrite and ferrous dioxygen heme. In these conditions, ferrous heme reacts with nitrite producing NO and ferric heme. The ferric species is reduced to ferrous heme by dithionite whereas the NO binds to ferrous heme until all the ferrous heme is bound to NO. Samples of Cygbs (2–10 μM) were mixed with different concentrations of nitrite (typically between 1 and 10 mM, except for distal histidine mutants which were reacted with nitrite concentrations in the 250 μM-2 mM range) and the spectral changes were monitored in the 350–700 nm range. Absorbance changes were fit to single exponential equation as described [8, 25, 27].
2.5. Lipid and imidazole binding experiments –
The binding of sodium oleate or sodium octanoate to the globins was assessed by UV-Vis spectroscopy as described previously [8, 11]. Binding experiments were carried out in 100 mM sodium phosphate buffer, pH 7.4 at 25 °C. Sodium oleate and octanoate stocks (50 mM) were prepared in water and stored at −20 °C. Dilutions of sodium oleate/octanoate in sodium phosphate buffer were prepared before use. Globin spectra were acquired in a Cary-50 spectrophotometer (Agilent Technologies. Palo Alto, CA). The binding spectra were obtained by adding increasing amounts of the sodium oleate stocks (200 μM-5 mM) or sodium octanoate stocks (2 mM-50 mM) to a solution of the globin in the ferric state (5–15 μM) to a final concentration of ≈ 60–1500 μM oleate or ≈ 60–7200 μM octanoate. As increasing amounts of the lipid are added, spectral changes in the heme spectra are noticeable. Differences between the highest and lowest points of the difference spectra were plotted vs the concentration of lipid. The data was fitted to a binding model for a 1:1 stoichiometry. From the equilibrium constant and the Beer-Lambert equation, the absorbance change at a given wavelength can be written as:
| (1) |
Where Δε is the difference in the extinction coefficient between the initial, unbound species and the protein-ligand complex; P is the total concentration of protein; L is the total concentration of ligand, and KD is the dissociation constant of the protein-ligand complex.
The experiments to determine imidazole binding to wild-type Cygb and His81 mutants were performed in a similar fashion but instead of oleic acid, imidazole stock solutions were used. Spectra were obtained by adding increasing amounts of an imidazole stock solution (1 mM −100 mM) to a solution of the globin in the ferric state (5–15 μM) to yield a final concentration of ≈100–15000 μM imidazole. The KD for imidazole was calculated using equation 1 as described for the oleate binding experiments.
2.6. Statistical Analysis –
Each experiment was performed at least in triplicate and values are representative of two or more independent determinations using different batches of protein purified separately. Data were analyzed using Origin 8.0 (OriginLab Corporation, Northampton, MA) and expressed as mean ± standard deviation of the mean.
3. Results
3.1. Mutant selection -
For the distal histidine mutations, we generated the homologous mutations to those that we previously studied in Ngb [25] in order to compare the relative effects of these alterations in a similar globin fold. For the mutations relative to lipid binding our criteria were as follows – first, residues with side chains that can stabilize the negative charge at the carboxyl group of the fatty acid were mutated to negatively-charged side chain amino acids to promote charge repulsion (T91D, R84E, K116E). In the case of the residues that form a hydrophobic patch in the Cygb surface (V105Y and L108Q) we replaced the side chains with other amino acids observed in structurally-related proteins such as zebrafish Cygb1 or human Ngb which do not show appreciable lipid binding [8]. Protein expression of the wild type protein and mutants were performed as described protein [8, 17]. There were no significant differences in stability or yield for the mutants studied.
3.2. Spectral properties -
The mutations performed on surface residues (R84E, T91D, V105Y, L108Q, K116E) did not cause significant changes in the spectra of their ferrous or ferric species. However, the mutations in the distal histidine did cause changes in the protein spectra, as expected from the removal of the sixth heme ligand. The visible spectra of the wild type Cygb and distal histidine mutants are shown in Figure 1. In the oxidized heme state (FeIII), the mutants H81L and H81Q show spectra with maxima around 500 nm and 630 nm, contrasting with the peak around 535 nm for the wild type Cygb. This spectrum of H81L and H81Q resembles the ferric form of five coordinate globins such as Hb or Mb, with a predominant aquo-Met species [28]. In the case of H81A and H81W, the spectra are more similar to the wild type Cygb ferric form, and more consistent with a six-coordinated heme. The nature of the sixth ligand is unclear but the absorbance shoulder around 540 and 580 nm, more noticeable in H81W, is consistent with a hydroxyl group bound to the heme, as observed in the alkaline spectrum of wild-type Cygb [11], and consistent with the hydroxy met species [28]. The spectrum of the ferrous (FeII) heme is similar for all the Cygb mutants, showing a single peak with maxima around 560 nm instead of the double peak of the wt Cygb with maxima at 520 nm and 560 nm. These spectra are similar to other five-coordinated globins like Hb and Mb and indicate that the heme coordination in the ferrous form is five-coordinated instead of the six-coordinated wt Cygb species.
Figure 1. Spectral properties of wild type Cytoglobin and distal histidine (His81) mutants.
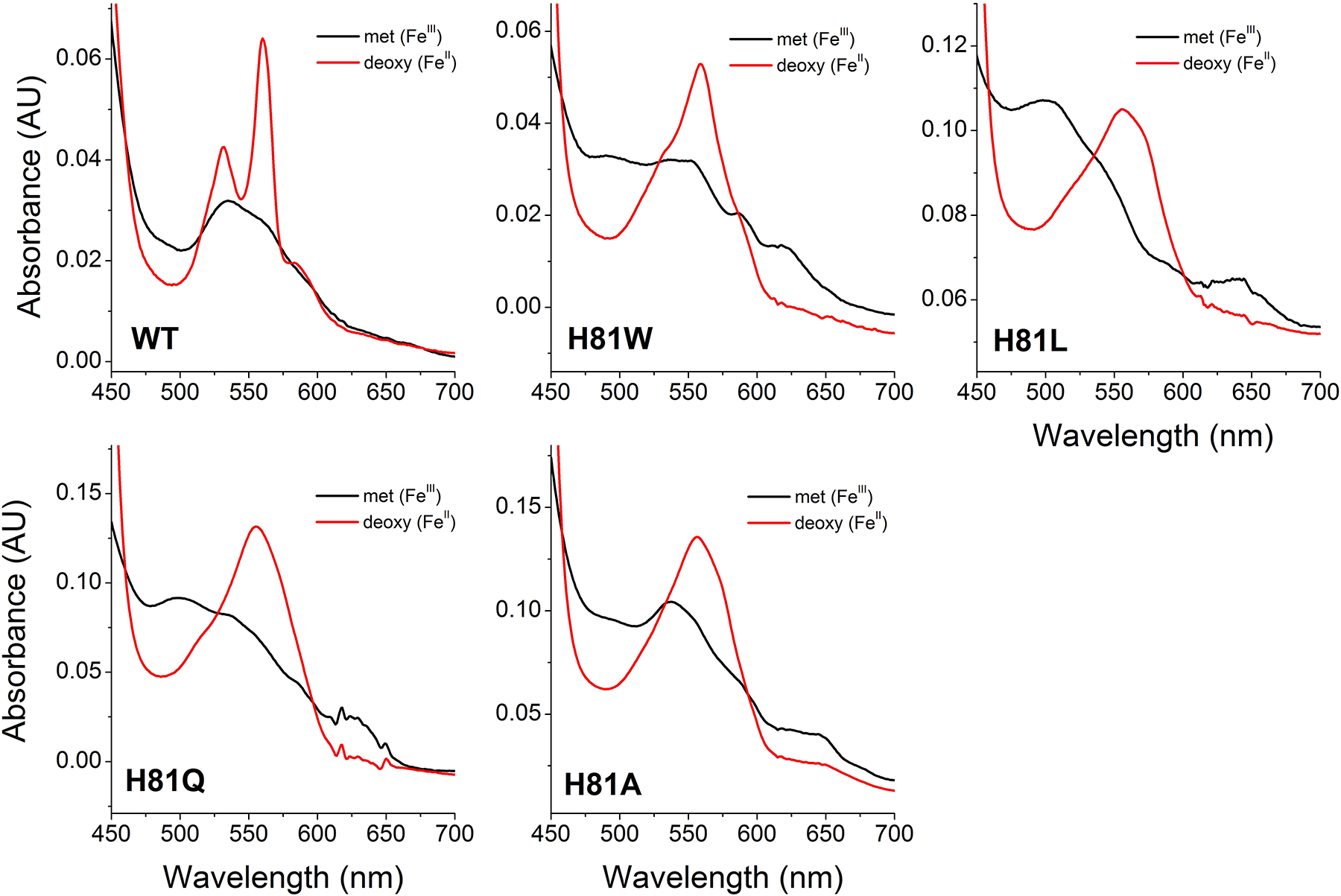
The spectra of the ferric (FeIII) and deoxy ferrous (FeII) species are shown in black and red respectively.
The spectra of the Cygb mutants H81V and H81Y have been reported [29]. In the ferrous form, both mutants show a single peak around 560 nm indicative of the five-coordinate state, as observed for the Cygb mutants in this work. The ferric form of H81V shows noticeable maxima at 550 and 590, probably indicating a population of OH− bound heme as observed in H81W, and to a lesser extent, in H81A.
We have studied the effects of the homologous mutations in Ngb [25, 26]. In that case, the ferric six-coordination is more conserved, with H64A, H64L and H64W mutants showing spectra similar to the six-coordinate wild-type Ngb, and only the H64Q mutant showing peaks around 500 and 630 nm typical of the five coordinate ferric heme. [25, 26] Moreover, the H64A and H64W mutants, show deoxy spectra with some six-coordinate properties, indicating that a sixth ligand –probably a solvent molecule– is stabilized in the heme pocket [25]. Altogether, the spectral data available for distal histidine mutants of Cygb and Ngb suggests that the six-coordination is stronger in Ngb than in Cygb. This is good agreement with the measured binding affinity for the distal histidine, showing a 5-fold preference in Ngb versus Cygb [30, 31]. This situation is also consistent with the proposed evolution of the Ngb and Cygb proteins, where Cygb lies in between the six-coordinate Ngb and the five-coordinate hemoglobins and myoglobins [7], and Cygb can transition to a five coordinate state with minor sequence changes, as observed in the five-coordinate native zebrafish Cygb-1 [8].
3.3. Autoxidation of cytoglobin mutants –
Ferrous Cygb can react with molecular oxygen to form a ferrous heme dioxygen complex (FeII-O2) similarly to oxygen carrier proteins such as Hb or Mb. However, the heme dioxygen complex of wild type Cygb is remarkably unstable in comparison with the analogous species of Hb or Mb, and decays with formation of ferric Cygb with a half-life of 2.6 minutes for the WT protein [8]. We determined the autoxidation rates for all the Cygb mutants generated in this work. The autoxidation rates depend mainly on the heme redox potential and our previous work in Ngb indicates that for six-coordinated globins, these rates are very sensitive to mutations around the heme pocket [25] but usually not altered by changes far from the heme moiety [27]. Thus, we used the autoxidation rates as a surrogate for changes in redox potential. Our hypothesis -in particular for the surface mutations, R84E, T91D, V105Y, L108Q, and K116E- is that mutants with autoxidation rates and nitrite reduction rates similar to the wild type protein will not have significant changes in heme reactivity, and the differences in lipid binding will be related to specific protein-ligand interactions and not to differences in the heme environment or redox potential. Our calculated rates are shown in Table 1, and the spectral changes for the reaction of the Cygb mutants are shown in Figure 2. For the mutations in the protein surface, in most cases we did not observe large variations in the autoxidation rates, as expected (Table 1). However, it is noteworthy that the mutant R84E, shows a roughly 10-fold increase in the rate constant as compared to WT. This indicates some unforeseen effects of the mutation on the stability of the FeII-O2 complex. Some of the mutations in the distal histidine did cause significant changes in the observed rates of autoxidation, as also noted in the equivalent Ngb mutants (Table 1) [25]. Unlike the WT protein, His81 mutants did show the formation of a clear ferrous-oxy species at the beginning of the experiment, noted by the peaks with maxima around 545 and 580 nm. This species is most abundant for the slower autoxidizing mutants, H81A and H81Q (Figure 2). In general, we observed that the distal histidine mutation did decrease the rates of autoxidation, a trend also observed in the homologous Ngb mutants (Table 1). Trends for Cygb and Ngb distal His (E7) mutants were very similar with the exception of H81L mutant, which did not alter the autoxidation rate in Cygb although it did cause a 20-fold decrease in Ngb.
Table 1.
Autoxidation rates of wild-type and distal histidine mutants of cytoglobin and neuroglobin.
| Cytoglobin | Rate (min−1) | Neuroglobin | Rate (min−1) |
|---|---|---|---|
| Wild-type | 0.270 ± 0.020a | Wild-type | 0.23 ± 0.03b |
| H81A | 0.094 ± 0.038 | H64A | 0.066 ± 0.005b |
| H81Q | 0.019 ± 0.005 | H64Q | 0.010 ± 0.002b |
| H81L | 0.314 ± 0.043 | H64L | 0.010 ± 0.004 |
| H81W | 0.135 ± 0.034 | H64W | 0.076 ± 0.006b |
| R84E | 2.362 ± 0.359c | ||
| T91D | 0.062 ± 0.005 | ||
| V105Y | 0.107 ± 0.018 | ||
| L108Q | 0.116 ± 0.051 | ||
| K116E | 0.366 ± 0.057 |
Values determined at 37°C in 100 mM sodium phosphate, pH 7.4. All values from this work except
from reference [8] and
from reference [25].
Rate constant for the fast phase; R84E decay is best fit to a two-exponential equation; the rate constant for the slower phase is 0.303 ± 0.037 min−1. The fast phase accounts for 73 ± 12% of the decay.
Figure 2. Autoxidation of wild type Cytoglobin and distal histidine (His81) mutants.
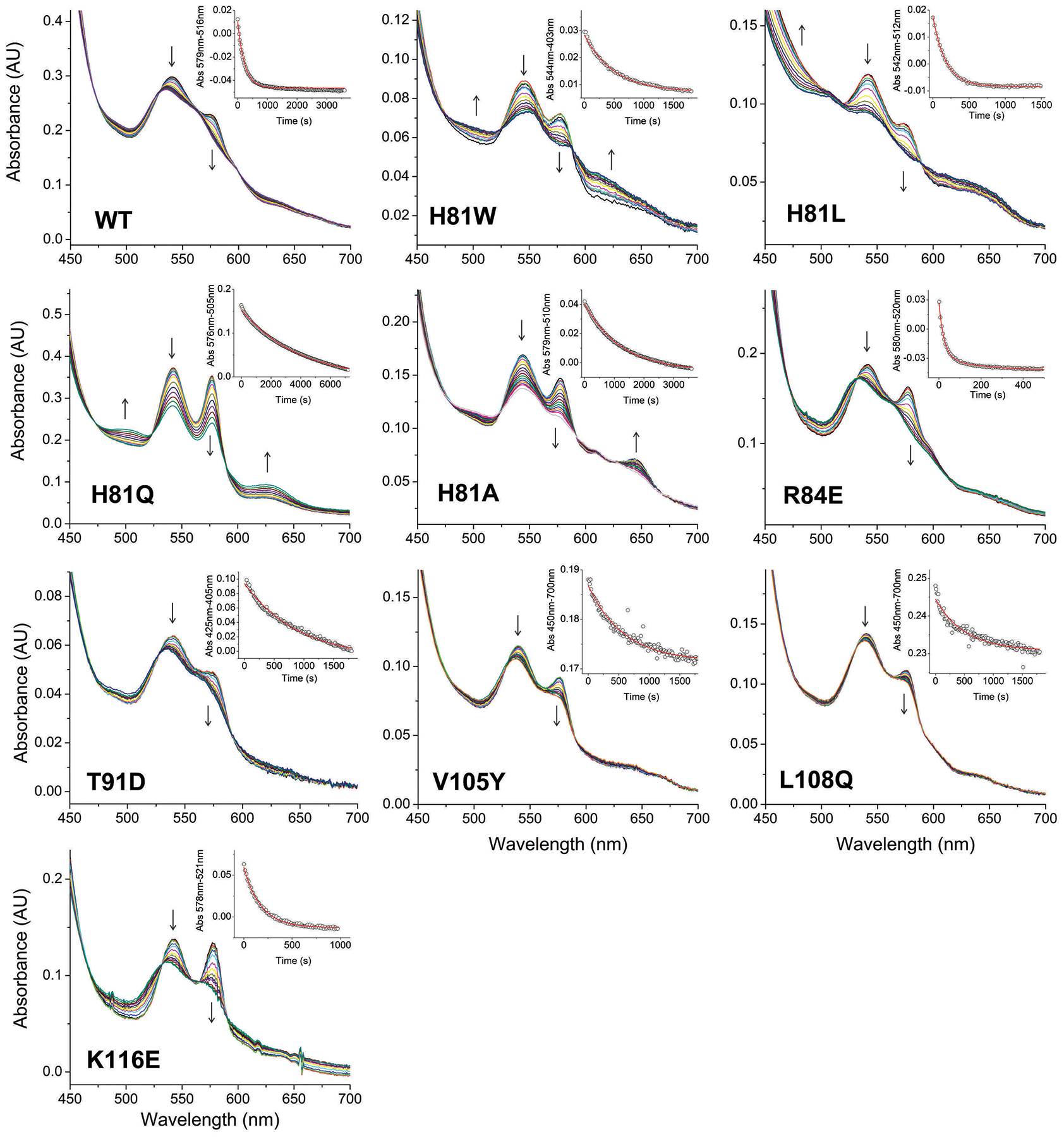
Each plot shows selected spectra during the time course of autoxidation for wild type and the mutants H81W, H81L, H81Q and H81A. Arrows indicate the direction of the absorbance change. Insets show the fitting of the decay to a single exponential equation.
3.4. Nitrite reduction by cytoglobin mutants –
The reduction of nitrite to NO by heme proteins is an important reaction in physiology as it constitutes a pathway that can circumvent the decrease in activity of nitric oxide synthases in low oxygen conditions [22, 32–34]. Relevant generation of NO from nitrite in vivo has been shown for Hb and Mb [35–37]. Cygb and Ngb can also catalyze nitrite reduction to NO [8, 25, 26, 38], although the importance of this reaction for their physiological function is still unknown.
Previous work in Ngb has stressed the role of six-coordination on nitrite reduction rates [25]. We have shown that the replacement of the distal histidine by leucine or glutamine increases the reaction rates of human Ngb by 2000-fold [26]. Mutagenesis studies indicate that the reaction rate for Ngb mutants appears to correlate with the distal pocket accessibility and not with the protein redox potential or autoxidation rates [25]. To further study the effect of the distal residue on nitrite reduction rates and compare the effect of Cygb and Ngb heme environments, we determined the nitrite reduction rates of the homologous His(E7) mutation in Cygb. As the changes in nitrite reduction rates also provide information about the accessibility of the heme pocket, we also determined the rates for the mutations putatively involved in lipid binding to determine if these changes alter the heme pocket properties.
As noted above, Cygb has a disulfide bond that can adjust the reactivity of the heme group by regulating the strain of the bond between the heme iron and the distal histidine [11, 13, 21, 24, 31]. It has been shown that in the presence of the intramolecular disulfide bond, nitrite reduction rates are significantly faster than for the reduced thiol species (32.3 M−1s−1 vs 0.63 M−1s−1 [21]). The nitrite reduction experiments in this work were conducted under excess dithionite and thus, we expect the disulfide bond to be reduced to free thiols. These are the same conditions used in other experiments compared here [8, 25]. We do not expect a significant impact of the thiol redox state on the nitrite reactivity of the distal histidine mutants as the effect of the disulfide bond, at least for the ferric protein, is largely removed in the His81 mutants [24].
Nitrite reduction was studied in the presence of excess dithionite as previously described [8, 25, 27]. In these conditions, the reaction of nitrite with ferrous Cygb produces NO and a ferric Cygb via the formation of a ferric-hydroxyl or ferric-NO species [26]. Excess dithionite reduces the ferric Cygb species and NO binds to the ferrous Cygb so the final species of the reaction is NO-bound ferrous Cygb. The nitrite reduction rate constants for the WT and Cygb mutants, and the corresponding values for the human Ngb mutants are shown in Table 2. Representative reactions as followed by UV-visible spectroscopy are shown in Supplemental Figure 1. With small differences, the trend for Cygb is strikingly similar to the results observed in Ngb. The reaction rate constant for Cygb mutants increases as the side chain volume becomes smaller, in the order H81W < H81L/H81Q < H81A (Table 2). It should be noted that even although the H81W mutant introduces an increase in volume size, the mutation removes the sixth ligand of the heme iron and thus leave a five-coordinated heme with increased accessibility to external ligands like nitrite. The most remarkable difference with Ngb is the lesser effect of the H81L mutation, with a rate about 25% of the Ngb H64L, whereas the other Cygb proteins have values within 2-fold of the corresponding Ngb protein.
Table 2.
Nitrite reductase rates of selected wild-type globins and mutants .
| Protein | Protein | Reaction rate constant (M−1s−1) |
|---|---|---|
| Hemoglobin | T-State | 0.12a |
| R-State | 6.0a | |
| Myoglobin | Wild-type | 5.6 ± 0.6b |
| H64A | 1.8 ± 0.3b | |
| F43H | 1.4 ± 0.1c | |
| F43H/H64A | 49.8 ± 1.5c | |
| Neuroglobin | Wild-type | 0.52 ± 0.19d |
| H64W | 7.6 ± 1.3d | |
| H64L | 302 ± 55 | |
| H64Q | 387 ± 21d | |
| H64A | 1120 ± 140d | |
| Cytoglobin | Wild-type | 1.14 ± 0.07e |
| H81W | 10.7 ± 2.1 | |
| H81L | 76 ± 16 | |
| H81Q | 210 ± 44 | |
| H81A | 1100 ± 100 | |
| R84E | 0.90 ± 0.30 | |
| T91D | 1.24 ± 0.18 | |
| V105Y | 3.76 ± 0.85 | |
| L108Q | 0.66 ± 0.18 | |
| K116E | 0.65 ± 0.14 |
We also studied the effect of the mutations targeting lipid binding on nitrite reductase rates. As indicated above, nitrite reduction is sensitive to the heme pocket and we did not expect significant changes. Confirming this hypothesis, most mutations did not change significantly the calculated rate constants (Table 2) although there was a 3-fold increase in the value for the V105Y mutation.
3.5. Lipid binding to wild-type and mutant Cygbs –
The binding of oleic acid to wild-type Cygb and mutants was determined by difference spectroscopy. All the experiments were conducted on ferric protein with intact intramolecular disulfide Cys38-Cys83 bonds as previously reported [12]. Cygb with the reduced thiols shows little or no binding to lipids [11, 12]. Oleic acid binding to mutants other than His(E7) showed spectral changes similar to those observed for the WT protein [8, 11]. Spectral changes observed for these mutants are shown in Supplemental Figure 2. The difference spectra show the largest changes around the Soret peak, with maxima around 400nm and minima around 425nm. These changes, along with the changes in the 500–700nm range, are consistent with a weakening of the distal histidine-heme interaction and a more five-coordinated state, consistent with previous observations [8, 11]. All these mutants (R84E, T91D, V105Y, L108Q, K116E) showed a weaker binding than Cygb WT as indicated by the higher KD value, but although T91D, V105Y and L108Q showed moderate increases (2 to 7 -fold, changes for V105Y were not statistically different from WT, Student’s t-test P=0.09), the mutations K116E and R84E caused increases in KD of >30-fold (Table 3). This suggests that the side chains of R84 and K116 play a role in the binding of the fatty acid by interacting with its carboxy moiety and this interaction is suppressed when the charge of these side chains is reversed.
Table 3.
Fatty acid binding parameters of wild-type cytoglobin and mutants.
| Cytoglobin | Oleic acid | Octanoic acid |
|---|---|---|
| KD (μM) | KD (μM) | |
| Wild-type | 2.3 ± 1.0a | 1590 ± 740 |
| H81A | 24 ± 2 | 1520 ± 890 |
| H81Q | 46 ± 8 | 250 ± 10 |
| H81L | 36 ± 13 | 950 ± 80 |
| H81W | 32 ± 10 | 140 ± 10 |
| R84E | 170 ± 90 | n.d. |
| T91D | 5.7 ± 1.4 | n.d. |
| V105Y | 4.8 ± 3.2 | n.d. |
| L108Q | 14 ± 3 | n.d. |
| K116E | 62 ± 23 | 185000 ± 13000 |
Dissociation constants determined at 25°C in 100 mM sodium phosphate, pH 7.4. All values from this work except
from [8].
In the case of the His(E7) mutants, the binding of oleic acid induces spectral changes different from those observed on the Cygb WT protein (Figure 3). This is not unexpected as the event causing the spectral changes in the WT protein, the dissociation of the distal histidine, is no longer possible. In a somehow puzzling way, the changes for H81A, H81L, and H81Q show an opposite change to that observed for the WT protein, with the spectra apparently changing to a more six-coordinated state as the lipid binds. The absorbance increase in the 530–550nm range is consistent with a hydroxy moiety being the sixth ligand [28]; thus we hypothesize that oleic acid binding is stabilizing the interaction of the ferric heme with a hydroxy group thus generating a six-coordinate spectrum. With the exception of the H81L mutant, which showed a 4-fold increase in KD, all the other His(E7) mutants showed large (10- to 30-fold) increases in the KD value. Taken in conjunction with the spectral changes, it seems possible that the His(E7) mutants are binding the oleic acid in a different binding site than the WT protein.
Figure 3. Oleate binding to wild type Cytoglobin and distal histidine (His81) mutants determined by differential spectroscopy.
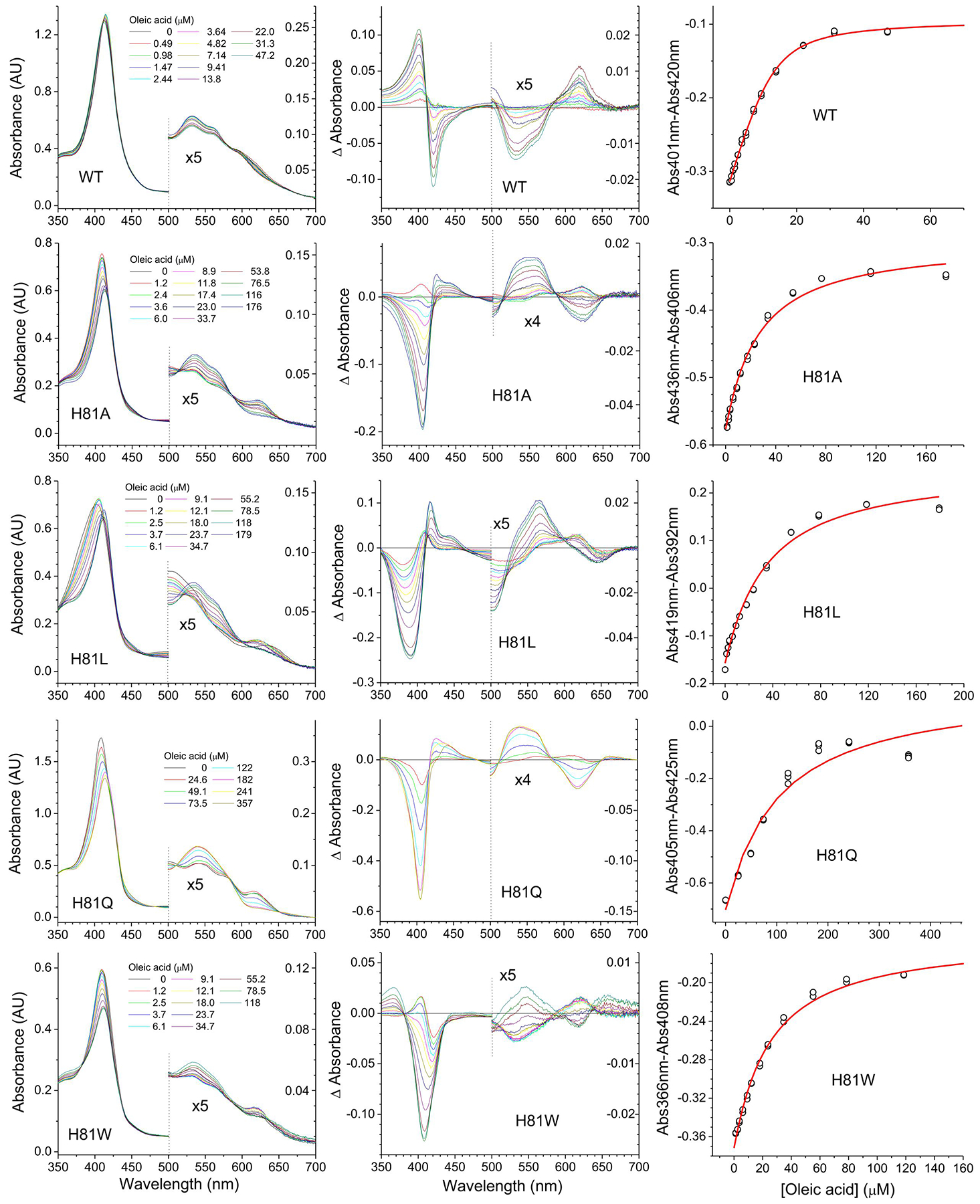
Cytoglobins (5–10 μM) were mixed with increasing concentrations of oleic acid. Left panels indicate the absorbance changes upon addition of increasing amounts of oleate. The concentrations of oleate are indicated in each panel. Middle panels show the difference spectra -subtracting the initial spectrum from each spectrum. Right panels show the fit to determine the apparent KD values by fitting the absorbance differences at the wavelengths showing the largest absorbance change versus the concentration of oleate. The red line denotes the fit of the data to Equation 1. The 500nm-700nm range in the left and middle panels is enlarged to show the spectral changes in detail.
3.6. Binding of octanoic acid to wild type cytoglobin and distal histidine mutants –
There is little known about the lipid properties that promote the binding to cytoglobin. To our knowledge, only the binding of oleic acid or phospholipids containing several oleic acid chains has been reported [11, 12]. Octanoic acid has only eight carbons and no double bonds, unlike the larger oleic acid (18:1 cis-9) so it presents a much shorter hydrophobic moiety than oleic acid whereas the carboxyl group is identical. We do not expect octanoic acid to be a physiological substrate for Cygb, but we used it as a tool to determine the contribution to the binding of the fatty acid chain versus the carboxylic acid, and to help elucidate the differences between His81 mutants and WT Cygb. The observed spectral changes for the binding of octanoic acid and the determined KD values are shown in Supplementary Figure 3 and Table 3. For WT Cygb and K116E it is apparent that the difference spectra are almost identical to those elicited by oleic acid, indicating that octanoic acid is also able to induce the six-coordinate to five-coordinate transition seen for oleic acid, albeit with a much weaker interaction that requires a ≥1000-fold higher concentration of ligand. K116E affinity is much lower than that of the Cygb WT (Table 3), in good agreement with the trend observed for oleic acid, and suggesting that the carboxylate binding has a higher relevance in the binding of octanoic acid relative to oleic acid.
Notably, the difference spectra for the His81 mutants in the presence of octanoic acid are different from those observed with oleic acid, although very similar to those shown for Cygb WT with both oleic and octanoic acid (Supplementary figure 3). In all cases, the difference spectra points to an increase in five coordination, with the oleic acid weakening the binding of the sixth heme-coordinating molecule, most probably water. The binding constants were in all cases lower than for Cygb WT, indicating a higher affinity towards octanoic acid in the mutants. The causes of these discrepancies between the effect of oleic acid and octanoic acid in the His81 mutants are uncertain. It is apparent that the His81 mutants can accommodate the short hydrophobic chain of the octanoate better than Cygb WT; however why this does not also translate into a higher affinity for the oleic acid is unclear.
3.7. Further examination of lipid binding to distal histidine mutants –
As discussed above, the spectral changes observed during lipid binding to Cygb WT –and Cygb mutants that conserve the distal Histidine– are consistent with the shift from a six-coordinate to a five-coordinate heme species. This causes a significant change in the UV-visible spectrum that can be easily detected [11, 12]. However, in the case of the His(E7) mutants the initial species are already five-coordinate (although they seem to preserve some six-coordinate character, probably due to the stabilization of a hydroxy group), so the spectral changes are not as pronounced. In addition, based on our previous Molecular Dynamics studies, the distal heme pocket could also serve as a lipid binding site when the distal histidine moves away from the heme (e.g. in phospholipids with more than one lipid chain such as phosphatidic acid or cardiolipin [12]).
In order to clarify the situation of this set of mutants, we used imidazole as an auxiliary probe to mimic the presence of the distal histidine. Imidazole is structurally identical to the side chain of histidine, and like histidine, it can bind to most heme proteins with a vacant binding site, albeit usually with low affinity [40]. Thus, we conducted new experiments in the presence of imidazole and compared the lipid binding to these mutants in the presence and absence of imidazole.
Firstly, we studied the binding of imidazole to WT Cygb and the distal histidine mutants (Supplemental Figure 4, Table 4). Imidazole binding to Cygb WT is weak, with small absorbance changes and a KD value around 1 mM, comparable to the value observed for other globins [40]. In the case of the His(E7) mutants, the binding affinity was much higher, with KD values between 7 and 24 μM (Table 4). This increase in affinity has been also noted in myoglobin His(E7) mutants as well [40]. The spectral changes observed, with an absorbance increase in the 530–550 nm range, are consistent with the binding of a sixth ligand and a low spin ferric heme (Supplemental Figure 4) [28].
Table 4.
Imidazole binding parameters of wild-type cytoglobin and distal histidine mutants.
| Cytoglobin | KD (μM) |
|---|---|
| Wild-type | 820 ± 150 |
| H81A | 10.6 ± 6.2 |
| H81Q | 15.3 ± 11.0 |
| H81L | 23.5 ± 4.4 |
| H81W | 7.3 ± 0.9 |
Then we studied the binding of oleic acid to the His(E7) mutants in the presence of several saturating concentrations of imidazole (Figures 4–5). The spectral changes for the binding of oleic acid in the presence of imidazole (Figure 4) indicate in all cases a shift towards a more 5-coordinated state.
Figure 4. Oleate binding to Cytoglobin distal histidine mutants in the presence of imidazole determined by differential spectroscopy.
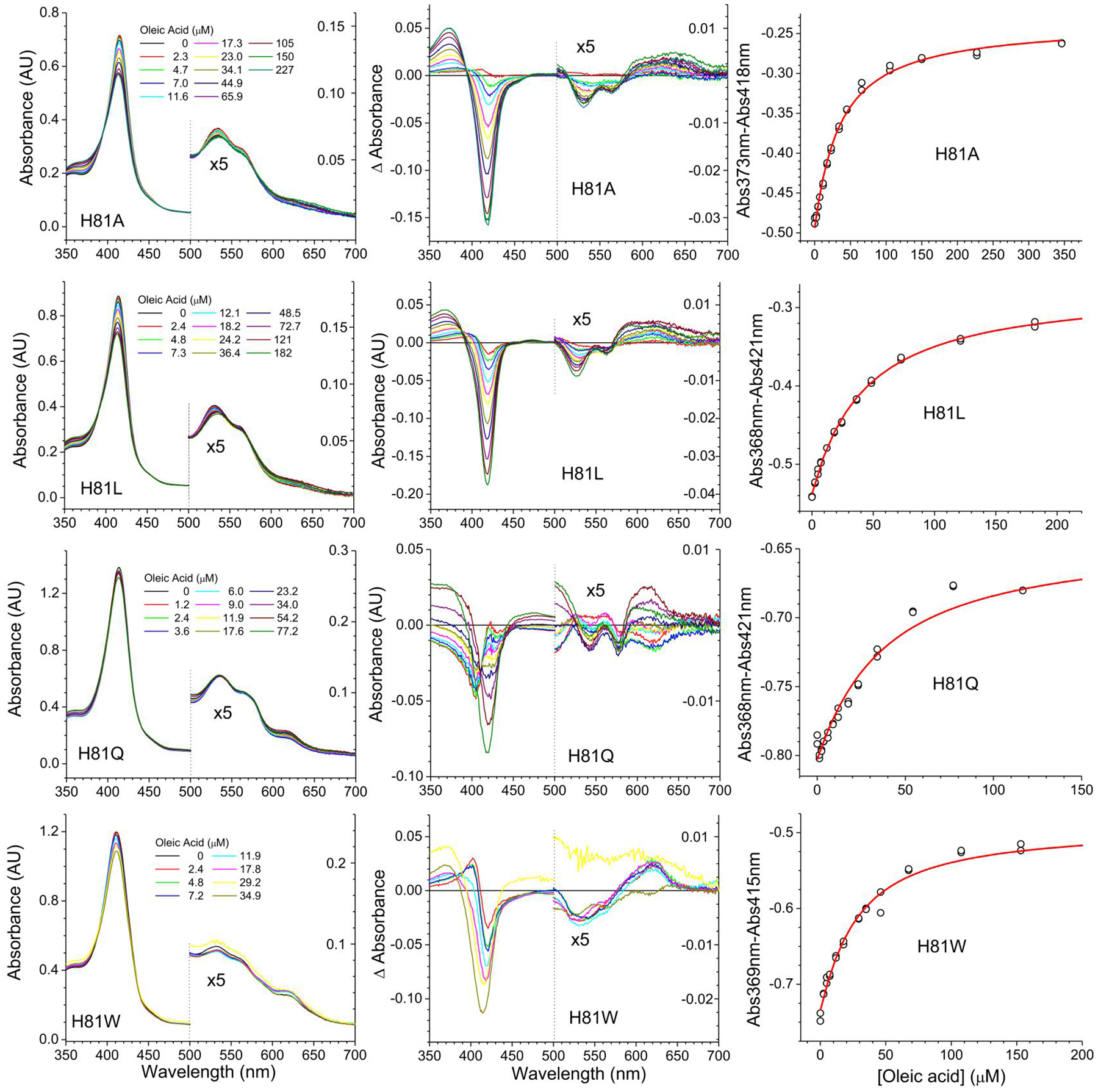
The binding of oleate was monitored in the presence of 250 μM Imidazole. Left panels indicate the absorbance changes upon addition of increasing amounts of oleate. The concentrations of oleate are indicated in each panel. Middle panels show the difference spectra -subtracting the initial spectrum from each spectrum. Right panels show the fit to determine the apparent KD values by fitting the absorbance differences at the wavelengths showing the largest absorbance change versus the concentration of oleate. The red line denotes the fit of the data to Equation 1. The 500 nm-700 nm range in the left and middle panels is enlarged to show the spectral changes in detail.
Figure 5. Oleate binding to Cytoglobin distal histidine mutants in the presence of different concentrations of imidazole determined by differential spectroscopy.
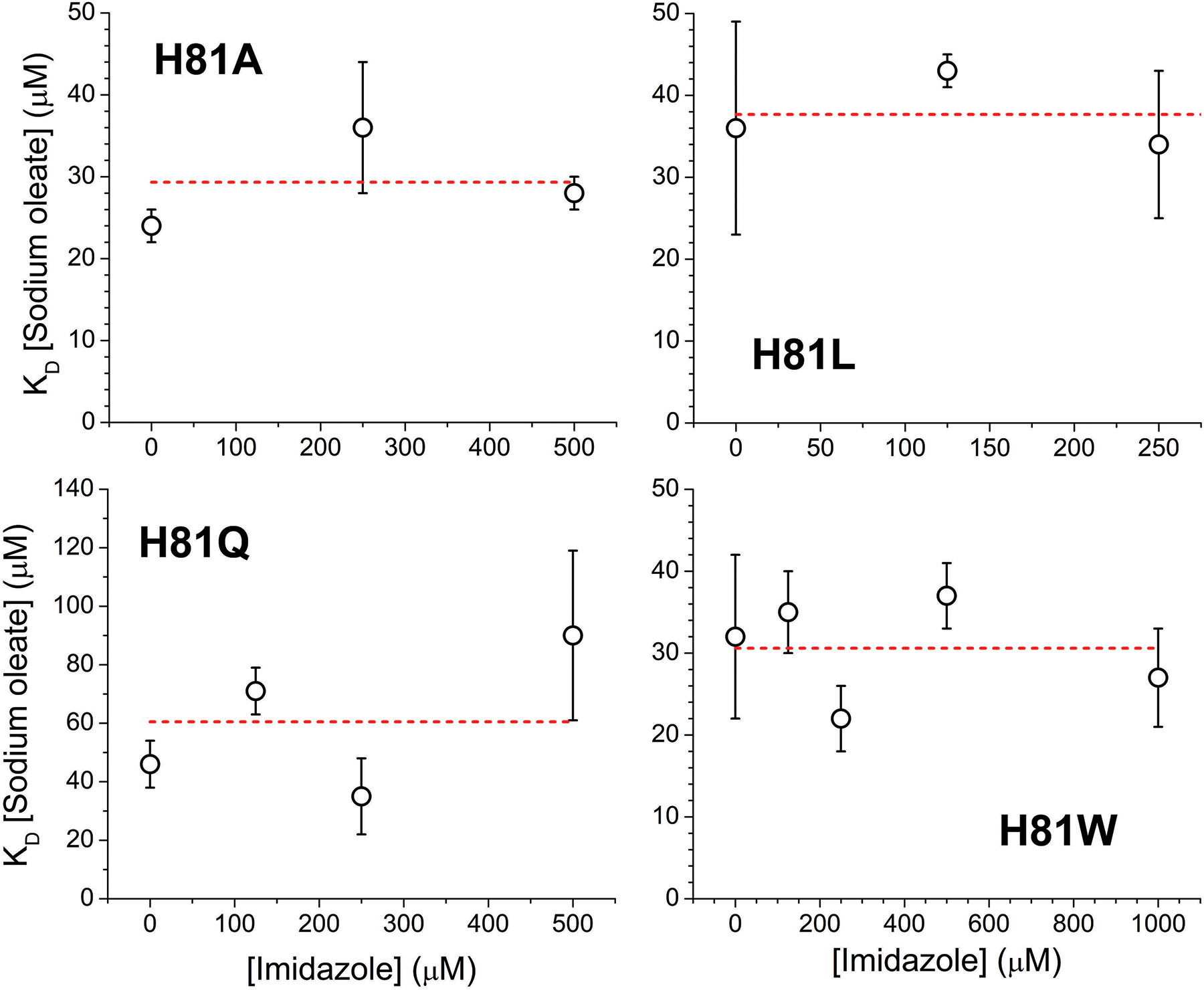
The observed dissociation constants at selected imidazole concentrations are indicated. The dashed red line denotes the linear fit of the observed KD values (slope set to zero).
Although the trend is consistent with the changes observed in the WT protein, it should be noted that in the case of the native protein this shift is due to a weakening of the His81-heme interaction, this is attributed to the lipid stabilizing a displacement of the E-helix [12]; however in the case of the His81 mutants with imidazole bound; the movement of the E-helix does not necessarily impose a strain to the imidazole-heme interaction, as the imidazole in not covalently bound to the protein backbone as in the case of the native His81 residue. Thus, we hypothesize that the apparent weakening of the imidazole-heme interaction is consistent with a movement of the E-helix, that although does not break the imidazole-heme interaction, removes a number of stabilizing interactions between the imidazole group and the distal pocket, decreasing the overall imidazole-heme binding stability.
As discussed above, computational models indicate that the hydrophobic moiety of the oleic acid could occupy the distal heme site when the histidine is not coordinating the heme iron [12]; if that was the case, we would expect that imidazole would compete for the same binding site, and therefore would increase the apparent KD for oleic acid as the concentration of imidazole increases. Our data does not indicate a significant change of oleic acid binding in the presence of imidazole (Figure 5); although a slight upwards trend is observed for the H81Q mutant. Thus, we conclude that the oleic acid is not binding in the distal heme pocket.
4. Discussion
Cytoglobin has been subject to extensive research for the last 20 years, and yet its physiological functions remain unclear. Here, we focus on the structural determinants for two processes of potential relevance to Cygb function: nitrite reduction and lipid binding.
As the mutation of the distal histidine cause large increases in nitrite reduction in Ngb, a closely related six-coordinate globin, we investigated whether these changes also regulate nitrite reduction in Cygb. Our results indicate that the regulation of nitrite reduction rates in Cygb follows very similar patterns to those observed for Ngb [25]. Removal of His81(E7) allows for easier access of nitrite to the heme iron, with nitrite reduction rates increasing as the side chain size decreases (H81W<H81Q/L<H81A). The nitrite reduction rate constants increase up to 1000-fold higher than Cygb WT for the His81Ala mutant. The fast nitrite reduction by H81Q, H81L and H81A makes them interesting mutations to study in a physiological context where nitrite reduction could be relevant to Cygb function. It is remarkable that in the case of five-coordinate globins like hemoglobin and myoglobin, the wild-type rates are similar to those observed in six-coordinated globins, even though the heme pocket is already more accessible to ligands. However, removal of the distal histidine in these proteins does not increase nitrite reduction rates (Table 2). In fact, is possible that the distal histidine in five-coordinated globin may actually help to stabilize the heme-nitrite complex by providing stabilizing contacts with nitrite. Studies on myoglobin indicate that placement of a histidine side chain instead of Phe43(CD1) can provide stabilizing interactions and increase nitrite reduction rates, especially when combined with the increased heme access provided by the H64A mutation (Table 2) [39].
In addition to ligand access to the active site, another factor that can be related to the increased nitrite reduction rates observed for the His81(E7) is the possible stabilization of the O-nitrito binding mode for nitrite, as opposed to the N-nitro binding mode. There is evidence that distal ligands can modulate the binding of nitrite in either conformation, as shown in myoglobin [41, 42] and cytochrome c nitrite reductase [43]. The stabilization of the O-nitrito mode can contribute to increased nitrite reduction rates as shown in myoglobin mutants [44]. The binding mode of nitrite to Ngb and Cygb is unknown but it is possible that binding of the nitrite via the oxygen atom can contribute to the fast nitrite reduction in Ngb and Cygb distal mutants.
The autoxidation experiments showed that most mutants introduce only moderate changes in the rate as compared to WT Cygb. His81 mutants show slower autoxidation rates (except H81L) in a similar fashion to the response observed in Ngb His64(E7) mutants. Unexpectedly, R84E showed a very fast oxidation of its ferrous-dioxygen complex. Arg84 and Lys116 are both located in close proximity to the heme-7-propionate group, and in principle could have similar effects on the heme electronics. Modifications of the propionate groups have been shown to introduce changes in myoglobin ligand binding, with up to 3-fold changes in oxygen dissociation rates [45, 46]. Similar effects can be seen in hemoglobin, related to hydrogen bond networks extending from the heme propionate groups to the heme iron [47]. However, both propionate groups are not equivalent, and changes around the heme-6-propionate seem to have a larger effect on heme properties than those involving the heme-7-propionate. In myoglobin, Arg45(CD3) interacts with the heme-6-propionate and also, via a water molecule, with the distal His64(E7). [46] This hydrogen bond network is not present in the Cygb distal pocket, where Gln62, the Cygb residue equivalent to myoglobin Arg45, adopts a different orientation. Alternatively, the heme-7-propionate could be involved in ligand access to the active site, as described for cytochrome P450cam [48]. Although Lys116 and Arg84 side chains are located at a similar distance of the propionate, it is plausible that Arg84, being located in the distal side, would have a larger effect on ligand entry, perhaps even independently of any interaction with the heme-7-propionate. In that regard, in the crystal structure of WT Cygb the side chain of Arg84 can show alternate confirmations and may directly interact with the heme distal ligand [49]. To further reinforce this observation, in the crystal structure of the H81Q mutant the Arg84 side chain forms a hydrogen bond with the mutant Gln81 side chain [50]. It is possible that Arg84 only interacts with the heme ligand in some circumstances. We do not detect a significant difference in the R84E nitrite reduction rates as compared to WT Cygb; however, it has been postulated that Arg84 can play a role in the reactions of Cygb with ascorbate or superoxide [23, 51]. The involvement of Arg84 on Cygb heme reactivity deserves further study.
We have previously proposed a lipid binding mode where oleic acid binds in a hydrophobic pocket in the heme proximal side, with the lipid carboxy group exposed to the solvent and interacting with Arg84, Lys116 or Thr91. Alternate models suggest that the hydrophobic part of the lipid could bind in the distal heme pocket, competing with the distal histidine [12]. In addition, some of our models predicted another binding site where the hydrophobic surface formed by Val105 and Leu108 accommodates the aliphatic chain. Our mutations target separately these three binding models and can be divided into three groups -mutations around a hydrophobic surface patch (V105Y, L108Q); mutations of tentative carboxylate binding residues in the surface (R84E, T91D, K116E); and distal histidine (His81) mutations.
The mutations V105Y and L108Q cause modest, but noticeable decreases on lipid binding affinity. These changes are larger than expected from our initial model where the hydrophobic portion of the lipid was completely buried inside the protein, but more consistent with the hydrophobic patch around Val105, Leu108 and Ala112 lining the hydrophobic cavity opening (Figure 6). Also T91D, in the inner side of this pocket, has a small effect on oleate binding suggesting that the interactions of Thr91 with the lipid are not critical for binding.
Figure 6. Tentative model of oleic acid binding to Cytoglobin.
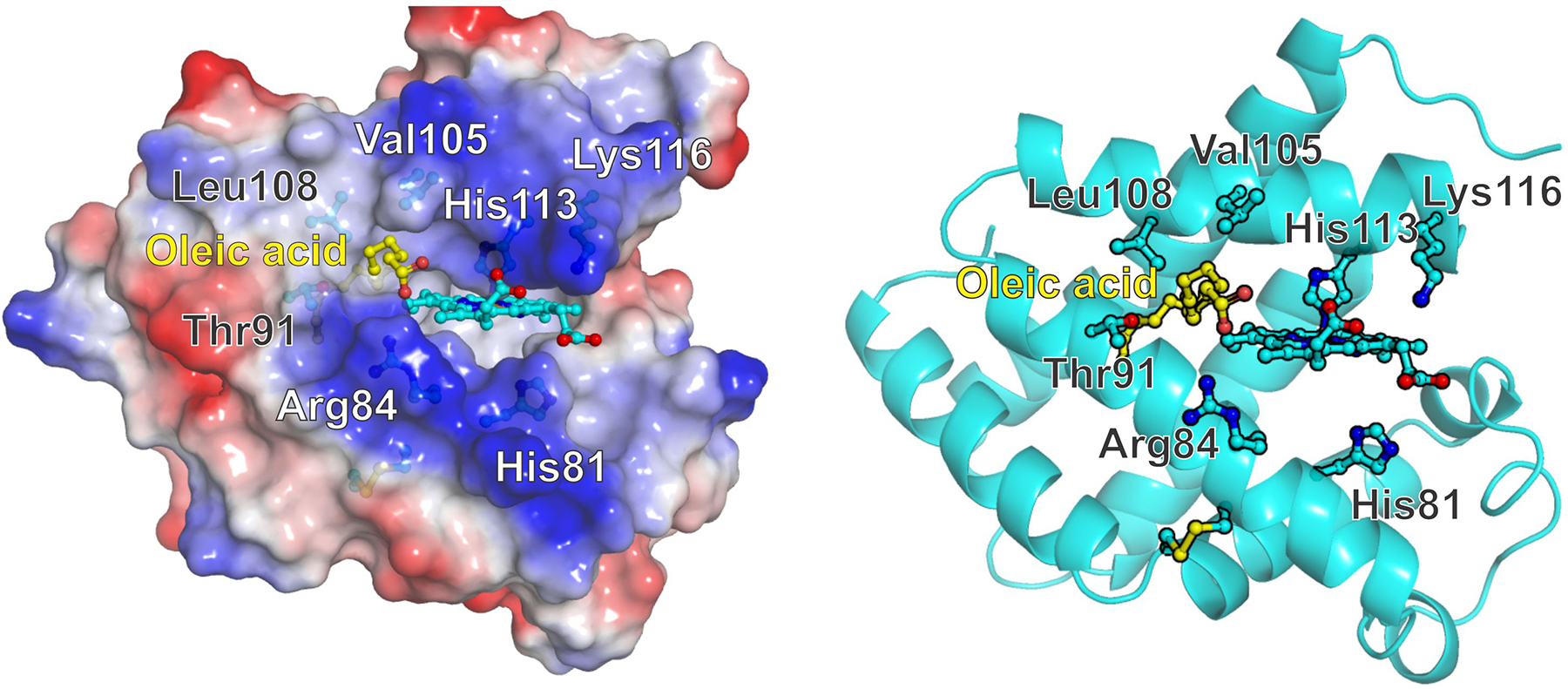
Left, surface potential view indicating the hydrophobic region around residues Val105 and Leu108 and the positive charges contributed by Arg84 and Lys116 sidechains. Right, Cartoon view showing the side residues studied in this work. The formation of the disulfide bridge between Cys38 and Cys83 promotes a movement of the E helix -containing His81- that opens a hydrophobic cavity in the proximal side of the heme. The cavity opening is surrounded by residues Arg 84, Thr91, Val105, Leu108 and Ala 112. The hydrophobic moiety of the fatty acid occupies the hydrophobic cavity whereas the carboxylate can interact with Arg84. Protein structures and electrostatic surface potentials were generated with PyMOL [52].
As noted above, T91D has minimal effect on lipid binding and it is unlikely to bind the carboxylate group. However, mutations of the surface residues Arg84 and Lys116 largely impair lipid binding, indicating that these residues are involved in stabilizing interactions with the carboxylate group of the lipid.
His81(E7) mutation does alter moderately lipid binding affinity, however experiments in the presence of imidazole indicate that His81(E7) does not compete with the lipid for the binding site, as we would expect a significant change in affinity to displace the imidazole. These experiments also strongly suggest that even in the absence of imidazole, oleate does not occupy the distal pocket. This is also consistent with the higher affinity of WT Cygb towards oleic acid as compared to the His81(E7) mutants; if the lipid binding required dissociation of the distal histidine from the heme, we would expect all the His81 mutants, lacking the distal ligand, to show higher affinities than WT Cygb.
We did not study the effect of the disulfide bond in the context of His81 mutations, however it is possible that the formation of the disulfide bond is no longer a requirement for lipid binding in these mutants, as suggested by our experiments on cyanide binding [24].
Our results are consistent with the proposed mechanism indicating that dissociation of His81(E7) from the heme, triggered by the Cys38-Cys83 disulfide bond, mediates the formation of a hydrophobic cavity in the proximal heme side that can accommodate the lipid. These are largely based on the observation that formation of the disulfide bond is a necessary condition for lipid binding [11, 12]. Whereas our initial models proposed a model where almost the complete fatty acid could be inserted in the protein, with the carboxylate interacting with Thr91 [12], the present results support a model where the hydrophobic portion of the lipid is bound in a more shallow cavity, lined by residues Arg 84, Thr91, Val105, Leu108 and Ala 112, and the carboxylate moiety is not bound inside the cavity but stays in the protein surface, free to interact with the solvent and residues Arg84 and possibly Lys116 (Figure 6). Although in this model the side chain of Lys116 appears to be too far from oleic acid to interact directly, the effect of the K116E mutation could be transduced by the repulsion of the heme carboxylate towards Arg84; as the side chain of Arg84 interacts with the heme carboxylate it would limit its ability to stabilize the oleic acid carboxylate group. The presence of positive charges at both Arg84 and Lys116 positions provides the main energetic contributions to the stability of the Cygb-lipid complex; both R84E and K116E mutations can serve as loss-of-function mutations for lipid binding. As these mutations have different effects on the autoxidation rates, they can serve to target lipid binding separately or in conjunction with reactions dependent on the oxygen complex stability (e.g. NO deoxygenation).
Altogether, we present a detailed exploration of the mechanisms regulating nitrite reduction and lipid binding in Cygb. We expect that these observations can help to generate modified Cygbs with specific changes in nitrite reduction and/or lipid interaction properties that can help guide to elucidate the contribution of nitrite reduction and protein-lipid interactions to Cygb function in cellular systems.
Supplementary Material
Acknowledgements
We dedicate this work to Qin Tong, who passed away during the preparation of this manuscript. S.J.K., N.I.M. and T.N.A. were part of the First Experiences in Research program at the University of Pittsburgh, Dietrich School of Arts and Sciences. We thank Mark T. Gladwin for funding support, encouragement and helpful discussions.
Funding
This work was supported by funding from the Vitalant (formerly Institute for Transfusion Medicine) and the Hemophilia Center of Western Pennsylvania (to Mark T. Gladwin, University of Pittsburgh), National Institutes of Health [Grant numbers P01 HL103455 and T32 HL110849 to Mark T. Gladwin, and Grant number R01 HL125886 to Mark T. Gladwin and J.T]. A.W.D. is supported by National Institutes of Health [Grant number T32 HL110849].
Abbreviations:
- Cygb
cytoglobin
- K D
dissociation constant
- Ngb
neuroglobin
- WT
wild-type
Footnotes
Appendix A. Supplementary Data
Supplementary figures 1–4 including the reaction of wild type Cytoglobin and mutants with nitrite and the interaction of cytoglobin with oleic acid, octanoic acid and imidazole are provided.
Declaration of competing interest
The authors declare the following competing financial interest(s): A.W.D. and J.T. are co-inventors of provisional and pending patents for the use of recombinant cytoglobin, neuroglobin, and other heme-based molecules as antidotes for carbon monoxide poisoning. Globin Solutions, Inc., has licensed this technology. A.W.D. is a consultant of Globin Solutions, Inc. J.T. is a shareholder and officer of Globin Solutions, Inc.
REFERENCES
- 1.Kawada N, Kristensen DB, Asahina K, Nakatani K, Minamiyama Y, Seki S, Yoshizato K, Characterization of a stellate cell activation-associated protein (STAP) with peroxidase activity found in rat hepatic stellate cells. J Biol Chem 276 (27) (2001) 25318–25323. [DOI] [PubMed] [Google Scholar]
- 2.Trent JT 3rd, Hargrove MS, A ubiquitously expressed human hexacoordinate hemoglobin. J Biol Chem 277 (22) (2002) 19538–19545. [DOI] [PubMed] [Google Scholar]
- 3.Burmester T, Ebner B, Weich B, Hankeln T, Cytoglobin: a novel globin type ubiquitously expressed in vertebrate tissues. Mol Biol Evol 19 (4) (2002) 416–421. [DOI] [PubMed] [Google Scholar]
- 4.Mathai C, Jourd’heuil FL, Lopez-Soler RI, Jourd’heuil D, Emerging perspectives on cytoglobin, beyond NO dioxygenase and peroxidase. Redox Biol 32 2020) 101468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keppner A, Maric D, Correia M, Koay TW, Orlando IMC, Vinogradov SN, Hoogewijs D, Lessons from the post-genomic era: Globin diversity beyond oxygen binding and transport. Redox Biol 37 2020) 101687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burmester T Hankeln T, Function and evolution of vertebrate globins. Acta Physiol (Oxf) 211 (3) (2014) 501–514. [DOI] [PubMed] [Google Scholar]
- 7.Blank M Burmester T, Widespread occurrence of N-terminal acylation in animal globins and possible origin of respiratory globins from a membrane-bound ancestor. Mol Biol Evol 29 (11) (2012) 3553–3561. [DOI] [PubMed] [Google Scholar]
- 8.Corti P, Ieraci M, Tejero J, Characterization of zebrafish neuroglobin and cytoglobins 1 and 2: Zebrafish cytoglobins provide insights into the transition from six-coordinate to five-coordinate globins. Nitric Oxide 53 2016) 22–34. [DOI] [PubMed] [Google Scholar]
- 9.Cuypers B, Vermeylen S, Hammerschmid D, Trashin S, Rahemi V, Konijnenberg A, De Schutter A, Cheng CC, Giordano D, Verde C, De Wael K, Sobott F, Dewilde S, Van Doorslaer S, Antarctic fish versus human cytoglobins - The same but yet so different. J Inorg Biochem 173 2017) 66–78. [DOI] [PubMed] [Google Scholar]
- 10.Giordano D, Pesce A, Vermeylen S, Abbruzzetti S, Nardini M, Marchesani F, Berghmans H, Seira C, Bruno S, Javier Luque F, di Prisco G, Ascenzi P, Dewilde S, Bolognesi M, Viappiani C, Verde C, Structural and functional properties of Antarctic fish cytoglobins-1: Cold-reactivity in multi-ligand reactions. Comput Struct Biotechnol J 18 2020) 2132–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeder BJ, Svistunenko DA, Wilson MT, Lipid binding to cytoglobin leads to a change in haem co-ordination: a role for cytoglobin in lipid signalling of oxidative stress. Biochem J 434 (3) (2011) 483–492. [DOI] [PubMed] [Google Scholar]
- 12.Tejero J, Kapralov AA, Baumgartner MP, Sparacino-Watkins CE, Anthonymuthu TS, Vlasova II, Camacho CJ, Gladwin MT, Bayir H, Kagan VE, Peroxidase Activation of Cytoglobin by Anionic phospholipids: Mechanisms and Consequences. BBA-Mol. Cell Biol. L 1861 (5) (2016) 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beckerson P, Wilson MT, Svistunenko DA, Reeder BJ, Cytoglobin ligand binding regulated by changing haem-co-ordination in response to intramolecular disulfide bond formation and lipid interaction. Biochem J 465 (1) (2015) 127–137. [DOI] [PubMed] [Google Scholar]
- 14.Kagan VE, Bayir HA, Belikova NA, Kapralov O, Tyurina YY, Tyurin VA, Jiang J, Stoyanovsky DA, Wipf P, Kochanek PM, Greenberger JS, Pitt B, Shvedova AA, Borisenko G, Cytochrome c/cardiolipin relations in mitochondria: a kiss of death. Free Radic Biol Med 46 (11) (2009) 1439–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Tong J, Zweier JR, Follmer D, Hemann C, Ismail RS, Zweier JL, Differences in oxygen-dependent nitric oxide metabolism by cytoglobin and myoglobin account for their differing functional roles. FEBS J 280 (15) (2013) 3621–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amdahl MB, Petersen EE, Bocian K, Kaliszuk SJ, DeMartino AW, Tiwari S, Sparacino-Watkins CE, Corti P, Rose JJ, Gladwin MT, Fago A, Tejero J, The Zebrafish Cytochrome b5/Cytochrome b5 Reductase/NADH System Efficiently Reduces Cytoglobins 1 and 2: Conserved Activity of Cytochrome b5/Cytochrome b5 Reductases during Vertebrate Evolution. Biochemistry 58 (29) (2019) 3212–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amdahl MB, Sparacino-Watkins CE, Corti P, Gladwin MT, Tejero J, Efficient Reduction of Vertebrate Cytoglobins by the Cytochrome b5/Cytochrome b5 Reductase/NADH System. Biochemistry 56 2017) 3993–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilangovan G, Khaleel SA, Kundu T, Hemann C, El-Mahdy MA, Zweier JL, Defining the reducing system of the NO dioxygenase cytoglobin in vascular smooth muscle cells and its critical role in regulating cellular NO decay. J Biol Chem 296 2021) 100196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, El-Mahdy MA, Boslett J, Varadharaj S, Hemann C, Abdelghany TM, Ismail RS, Little SC, Zhou D, Thuy LT, Kawada N, Zweier JL, Cytoglobin regulates blood pressure and vascular tone through nitric oxide metabolism in the vascular wall. Nat Commun 8 2017) 14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Hemann C, Abdelghany TM, El-Mahdy MA, Zweier JL, Characterization of the mechanism and magnitude of cytoglobin-mediated nitrite reduction and nitric oxide generation under anaerobic conditions. J Biol Chem 287 (43) (2012) 36623–36633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reeder BJ Ukeri J, Strong modulation of nitrite reductase activity of cytoglobin by disulfide bond oxidation: Implications for nitric oxide homeostasis. Nitric Oxide 72 2018) 16–23. [DOI] [PubMed] [Google Scholar]
- 22.Dent MR, DeMartino AW, Tejero J, Gladwin MT, Endogenous Hemoprotein-Dependent Signaling Pathways of Nitric Oxide and Nitrite. Inorg Chem 60 (21) (2021) 15918–15940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zweier JL, Hemann C, Kundu T, Ewees MG, Khaleel SA, Samouilov A, Ilangovan G, El-Mahdy MA, Cytoglobin has potent superoxide dismutase function. Proc Natl Acad Sci U S A 118 (52) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeMartino AW, Amdahl MB, Bocian K, Rose JJ, Tejero J, Gladwin MT, Redox sensor properties of human cytoglobin allosterically regulate heme pocket reactivity. Free Radic Biol Med 162 2021) 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tejero J, Sparacino-Watkins CE, Ragireddy V, Frizzell S, Gladwin MT, Exploring the mechanisms of the reductase activity of neuroglobin by site-directed mutagenesis of the heme distal pocket. Biochemistry 54 (3) (2015) 722–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiso M, Tejero J, Basu S, Azarov I, Wang X, Simplaceanu V, Frizzell S, Jayaraman T, Geary L, Shapiro C, Ho C, Shiva S, Kim-Shapiro DB, Gladwin MT, Human neuroglobin functions as a redox-regulated nitrite reductase. J Biol Chem 286 (20) (2011) 18277–18289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tejero J, Negative surface charges in neuroglobin modulate the interaction with cytochrome c. Biochem Biophys Res Commun 523 (3) (2020) 567–572. [DOI] [PubMed] [Google Scholar]
- 28.Rots MJ Zandstra PJ, Influence of the R----T transition on the optical absorption and magnetic circular dichroism spectra of methemoglobin fluoride, aquomethemoglobin, and hydroxymethemoglobin. Biochemistry 23 (5) (1984) 844–851. [DOI] [PubMed] [Google Scholar]
- 29.Beckerson P, Svistunenko D, Reeder B, Effect of the distal histidine on the peroxidatic activity of monomeric cytoglobin [v1; ref status: indexed, http://f1000r.es/4xg] F1000Research 4:87 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dewilde S, Kiger L, Burmester T, Hankeln T, Baudin-Creuza V, Aerts T, Marden MC, Caubergs R, Moens L, Biochemical characterization and ligand binding properties of neuroglobin, a novel member of the globin family. J Biol Chem 276 (42) (2001) 38949–38955. [DOI] [PubMed] [Google Scholar]
- 31.Hamdane D, Kiger L, Dewilde S, Green BN, Pesce A, Uzan J, Burmester T, Hankeln T, Bolognesi M, Moens L, Marden MC, The redox state of the cell regulates the ligand binding affinity of human neuroglobin and cytoglobin. J Biol Chem 278 (51) (2003) 51713–51721. [DOI] [PubMed] [Google Scholar]
- 32.Lundberg JO, Weitzberg E, Gladwin MT, The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat Rev Drug Discov 7 (2) (2008) 156–167. [DOI] [PubMed] [Google Scholar]
- 33.Gladwin MT, Schechter AN, Kim-Shapiro DB, Patel RP, Hogg N, Shiva S, Cannon RO 3rd, Kelm M, Wink DA, Espey MG, Oldfield EH, Pluta RM, Freeman BA, Lancaster JR Jr., Feelisch M, Lundberg JO, The emerging biology of the nitrite anion. Nat Chem Biol 1 (6) (2005) 308–314. [DOI] [PubMed] [Google Scholar]
- 34.Tejero J, Shiva S, Gladwin MT, Sources of Vascular Nitric Oxide and Reactive Oxygen Species and Their Regulation. Physiol Rev 99 (1) (2019) 311–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Z, Shiva S, Kim-Shapiro DB, Patel RP, Ringwood LA, Irby CE, Huang KT, Ho C, Hogg N, Schechter AN, Gladwin MT, Enzymatic function of hemoglobin as a nitrite reductase that produces NO under allosteric control. J Clin Invest 115 (8) (2005) 2099–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cosby K, Partovi KS, Crawford JH, Patel RP, Reiter CD, Martyr S, Yang BK, Waclawiw MA, Zalos G, Xu X, Huang KT, Shields H, Kim-Shapiro DB, Schechter AN, Cannon RO 3rd, Gladwin MT, Nitrite reduction to nitric oxide by deoxyhemoglobin vasodilates the human circulation. Nat Med 9 (12) (2003) 1498–1505. [DOI] [PubMed] [Google Scholar]
- 37.Shiva S, Huang Z, Grubina R, Sun J, Ringwood LA, MacArthur PH, Xu X, Murphy E, Darley-Usmar VM, Gladwin MT, Deoxymyoglobin is a nitrite reductase that generates nitric oxide and regulates mitochondrial respiration. Circ Res 100 (5) (2007) 654–661. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Follmer D, Zweier JR, Huang X, Hemann C, Liu K, Druhan LJ, Zweier JL, Characterization of the function of cytoglobin as an oxygen-dependent regulator of nitric oxide concentration. Biochemistry 51 (25) (2012) 5072–5082. [DOI] [PubMed] [Google Scholar]
- 39.Wu LB, Yuan H, Gao SQ, You Y, Nie CM, Wen GB, Lin YW, Tan X, Regulating the nitrite reductase activity of myoglobin by redesigning the heme active center. Nitric Oxide 57 2016) 21–29. [DOI] [PubMed] [Google Scholar]
- 40.Mansy SS, Olson JS, Gonzalez G, Gilles-Gonzalez MA, Imidazole is a sensitive probe of steric hindrance in the distal pockets of oxygen-binding heme proteins. Biochemistry 37 (36) (1998) 12452–12457. [DOI] [PubMed] [Google Scholar]
- 41.Yi J, Heinecke J, Tan H, Ford PC, Richter-Addo GB, The distal pocket histidine residue in horse heart myoglobin directs the O-binding mode of nitrite to the heme iron. J Am Chem Soc 131 (50) (2009) 18119–18128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang B, Shi Y, Tejero J, Powell SM, Thomas LM, Gladwin MT, Shiva S, Zhang Y, Richter-Addo GB, Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets. Biochemistry 57 (32) (2018) 4788–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lockwood CW, Burlat B, Cheesman MR, Kern M, Simon J, Clarke TA, Richardson DJ, Butt JN, Resolution of key roles for the distal pocket histidine in cytochrome C nitrite reductases. J Am Chem Soc 137 (8) (2015) 3059–3068. [DOI] [PubMed] [Google Scholar]
- 44.Tse W, Whitmore N, Cheesman MR, Watmough NJ, Influence of the heme distal pocket on nitrite binding orientation and reactivity in Sperm Whale myoglobin. Biochem J 478 (4) (2021) 927–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harada K, Makino M, Sugimoto H, Hirota S, Matsuo T, Shiro Y, Hisaeda Y, Hayashi T, Structure and ligand binding properties of myoglobins reconstituted with monodepropionated heme: functional role of each heme propionate side chain. Biochemistry 46 (33) (2007) 9406–9416. [DOI] [PubMed] [Google Scholar]
- 46.Carver TE, Olson JS, Smerdon SJ, Krzywda S, Wilkinson AJ, Gibson QH, Blackmore RS, Ropp JD, Sligar SG, Contributions of residue 45(CD3) and heme-6-propionate to the biomolecular and geminate recombination reactions of myoglobin. Biochemistry 30 (19) (1991) 4697–4705. [DOI] [PubMed] [Google Scholar]
- 47.Ramos-Santana BJ Lopez-Garriga J, Tyrosine B10 triggers a heme propionate hydrogen bonding network loop with glutamine E7 moiety. Biochem Biophys Res Commun 424 (4) (2012) 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayashi T, Harada K, Sakurai K, Shimada H, Hirota S, A role of the heme-7-propionate side chain in cytochrome P450cam as a gate for regulating the access of water molecules to the substrate-binding site. J Am Chem Soc 131 (4) (2009) 1398–1400. [DOI] [PubMed] [Google Scholar]
- 49.Sugimoto H, Makino M, Sawai H, Kawada N, Yoshizato K, Shiro Y, Structural basis of human cytoglobin for ligand binding. J Mol Biol 339 (4) (2004) 873–885. [DOI] [PubMed] [Google Scholar]
- 50.Gabba M, Abbruzzetti S, Spyrakis F, Forti F, Bruno S, Mozzarelli A, Luque FJ, Viappiani C, Cozzini P, Nardini M, Germani F, Bolognesi M, Moens L, Dewilde S, CO rebinding kinetics and molecular dynamics simulations highlight dynamic regulation of internal cavities in human cytoglobin. PLoS One 8 (1) (2013) e49770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gardner AM, Cook MR, Gardner PR, Nitric-oxide dioxygenase function of human cytoglobin with cellular reductants and in rat hepatocytes. J Biol Chem 285 (31) (2010) 23850–23857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeLano WL, The PyMOL molecular graphics system. 2002).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


