Abstract
The family of Adenosine Deaminases Acting on RNA (ADARs) regulate global gene expression output by catalyzing Adenosine-to-Inosine (A-to-I) editing of double-stranded RNA and through interacting with RNA and other proteins. ADARs play important roles in development and disease, including an increasing connection to cancer progression. ADAR1 has demonstrated a largely pro-oncogenic role in a growing list of cancer types, and its function in cancer has been attributed to diverse mechanisms. Here we review existing literature on ADAR1 biology and function, its roles in human disease including cancer, and summarize known cancer-associated phenotypes and mechanisms. Lastly, we discuss implications and outstanding questions in the field, including strategies for targeting ADAR1 in cancer.
Keywords: ADARs, RNA editing, epitranscriptomics, cancer
RNA modifications in human biology and disease
Post-transcriptional modifications to RNA have emerged as a widespread phenomenon and are described as the epitranscriptome of the cell. The field of epitranscriptomics (see Glossary), spurred on by advances in genomic sequencing and analysis, has vastly expanded and complicated our understanding of how the cell regulates its gene expression output. We now understand that over 100 naturally-occurring RNA modifications exist, providing an additional layer of information and regulation to a transcript’s sequence and structure. The majority of these are linked to structural RNAs, such as rRNAs and tRNAs, but several are found more broadly in mRNAs as well. In humans, the most predominant mRNA modifications studied include adenosine to inosine (A-to-I), cytidine to uridine (C-to-U), N6-methyladenosine (m6A), pseudouridine (Ψ), and 5-methylcytosine (5mC)[1].
The epitranscriptome describes a diverse group of RNA alterations. Base editing (A-to-I, C-to-U) involves irreversible deamination reactions to directly alter base pairing preferences, whereas methylation and other modifications (m6A, 5mC, Ψ) regulate RNA processing and stability via association with proteins and/or structural changes. RNA methylation (m6A, 5mC) is notable for involving writer, reader, and eraser proteins that allow for reversible and dynamic regulation[1], [2]. There is also interplay between different modifications, such as m6A inhibiting A-to-I editing in cis, which may contribute to the general lack of overlap between these marks[3].
Regardless of mechanism, these modifications have been demonstrated to play significant roles in human disease, as alterations to the enzymes responsible are linked to cancer, genetic birth defects, cardiovascular disease, neurological disorders, and more[1]. The m6A mark is known to regulate RNA metabolism including translation, microRNA (miRNA) processing, and splicing; consequently METTL proteins that catalyze m6A are implicated in cancers including AML, breast cancer, and glioblastoma via regulation of cancer gene stability[3]. The m5C methyltransferase NSUN2 is associated with defects in memory and learning, and its expression correlates with breast cancer and development[1].
RNA modifications represent yet another layer of regulation on global gene expression output, which can then have important implications for rewiring the cancer cell. Here, we focus on the inosine mark and one of its catalyzing enzymes, ADAR1, and examine their impact on cancer.
ADAR family members, isoforms & domains
The family of Adenosine Deaminases Acting on RNA (ADARs) was first discovered accidentally as the enzyme responsible for “denaturing” dsRNA in Xenopus laevis embyros and inadvertently thwarting RNAi experiments[4]. In more than two decades since its discovery, we have come to appreciate ADARs (previously called DRADA or DSRAD), and more broadly the field of RNA modifications, as a major regulator of the cell’s gene expression output. And, given the power of these modifications to alter the global transcriptome, dysregulation of these processes is increasingly implicated in human disease, especially in the process of malignant transformation to the cancer state.
ADARs are a group of enzymes that bind double-stranded RNA (dsRNA) as homodimers and catalyze the hydrolytic deamination of an adenosine nucleotide to form inosine. Adenosine-to-inosine (A-to-I) editing occurs only at regions of dsRNA and alters the base pair preference from guanosine to cytidine, thus functionally acting as an A-to-G mutation. There are three family members: ADAR1 (also called ADAR), ADAR2 (ADARB1), and ADAR3 (ADARB2). ADAR1 is ubiquitously expressed in nearly all tissues and contains two isoforms generated from alternate promoter usage. The p110 isoform is largely nuclear, while the p150 isoform is interferon-inducible and can be found in the nucleus and cytoplasm. ADAR2 has more targeted expression to tissues like the brain, lungs, & arteries, and is responsible for the especially high editing rates found in neuronal tissues. ADAR3 is brain-specific and has yet to demonstrate detectable editing activity, although it may suppress the activity of other ADARs in the brain[5].
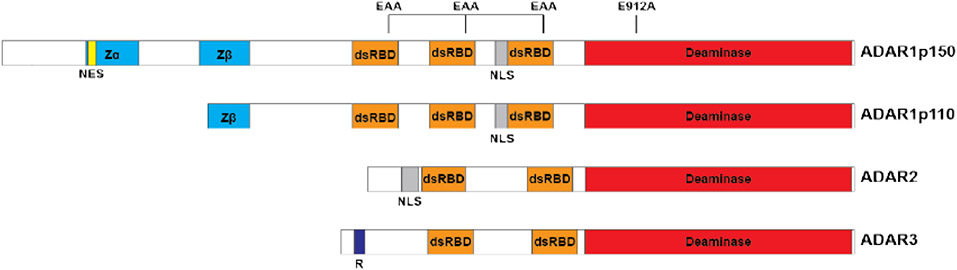
Both ADAR1 and ADAR2 have similar domain structures that are critical for their enzymatic activity (Figure 1). Beginning from the C-terminus, the catalytic deaminase domain is essential for editing activity. Point mutations at a key glutamate residue, required for flipping the targeted adenine base out of a dsRNA duplex, are sufficient to inactivate the enzyme (E912A for ADAR1, E396A for ADAR2)[4]. Each ADAR contains two or three dsRNA binding domains (dsRBDs) that include a key KKxxK motif; mutation to EAxxA abolishes RNA binding activity, and consequently also editing activity. All ADARs contain a nuclear localization signal (NLS), but the ADAR1p150 isoform also contains a nuclear export signal (NES) that allows it to shuttle between the nucleus and cytoplasm. The p150 isoform contains an additional Z-DNA/RNA binding domain at its N-terminus, which may contribute to distinct binding preferences between the two isoforms. A recent analysis found that over half of ADAR1 editing sites were exclusively edited by ADAR1p150, possibly as a result of additional binding capabilities conferred by the Zα domain, although there was no significant difference in motifs surrounding p150-specific and p150/p110 sites[6].
Figure 1. Schematic of the domain structure of the family of human ADARs.
ADAR1 is composed of two isoforms, p150 and p110, which share identical sequence with the exception of additional N-terminal sequence for p150, including an additional Zα domain and the NES which allows for both cytoplasmic and nuclear localization. Both contain three dsRBDs that mediate dsRNA binding and homodimerization, both of which are critical for enzymatic activity. Mutation of the KKxxK dsRBD motif to EAxxA abolishes dsRNA binding and thus editing activity. The E912A point mutation in ADAR1p150 (E617A for p110, E396A for ADAR2) disrupts catalytic deaminase activity. ADAR3 is thought to be catalytically inactive, and contains a unique arginine-rich domain that confers ability to bind single-stranded RNA. Abbreviations: NES, nuclear export signal; dsRBD, dsRNA binding domain; NLS, nuclear localization signal; R, arginine-rich domain; Zα/β, Z-DNA/RNA binding domains.
A-to-I edits are predominantly found in noncoding regions and are most prevalent at Alu repeat elements interspersed throughout the human genome. When two similar Alu repeats are inverted in close proximity, transcription of this locus generates regions of strong complementarity that can form double-stranded RNA (dsRNA). Global analyses to identify A-to-I edit sites generally find that the large majority (80%+) overlap with Alu repeat elements, the largest source of endogenous dsRNA in the human cell[7]. Most A-to-I edit sites are generally found in introns and 3'UTRs, with 1% or fewer occurring in coding exons[8]-[10]. However rare, editing within coding regions does occur, and such examples were the focus of much early research into ADAR downstream mechanisms (e.g. GRIA2, AZIN1).
Analyses of ADAR editing preferences generally indicate nearest neighbor preferences for any base but G one nucleotide upstream (−1) and for G one nucleotide downstream (+1) (most commonly UAG), as well as a general depletion for A in the bases surrounding the edit[9]. Given the similarity in edit motif, there is significant overlap of editing targets for ADAR1 and ADAR2, with the subcellular location and expression level of ADARs dictating much of targeting preference, with the exception of p150-specific targeting noted above[10]. Algorithms for predicting editing based on sequence motif have shown limited utility (~75% accuracy for ADAR1) due to contributions from the deaminase domain, RBDs, and RNA structure in determining the likelihood of editing[11]. Thus, sequencing remains the most accurate and informative method for A-to-I editing detection.
ADAR mutant mouse models
The study of ADAR-knockout (KO) mouse models has contributed greatly to our understanding of the biology of ADARs and the relative contributions of different family members. Adar-KO is lethal at embryonic day 12.5, with mice displaying elevated IFN signatures and failed liver development[12]. Analysis of chimeric Adar-KO/WT mice demonstrated that Adar-null embryonic stem cells could contribute to development of murine brain, gonads, heart, kidney, and lung, but not bone marrow, liver, spleen, or thymus[12]. The embryonic lethality of Adar loss (or catalytic inactivation) is overcome by a second KO of Mavs or MDA5/Ifih1 (and delayed to E15.5 by Ifnar1 or Stat1 KO), indicating critical roles for dsRNA sensing pathway activation and the resulting innate immune response. However, all double KO mice still die shortly after birth, due to additional organ deficiencies not caused by MAVS pathway activation[13]-[15]. ADAR1p150-specific KO is also embryonic lethal but rescued by Mavs KO, implicating this isoform as the major mediator of MDA5/MAVS pathway lethality, whereas p110 contributed to organ abnormalities[14]. In agreement, later work found that ADAR1p110-specific KO mice survive to birth and displayed no activation of the MDA5/MAVS pathway[16]. However, these mice have high neonatal lethality rates that are completely rescued by crossing with catalytically inactive Adar, indicating that they are caused by an RNA-editing independent function of ADAR1p110. Adar KO in adult mice also led to a lethal MAVS-mediated inflammatory response and disruption of tissue homeostasis, confirming critical roles for Adar beyond embryonic development [14]. In short, Adar appears to play two key roles in development and homeostasis in mice: the editing-dependent action of ADAR1p150 in preventing activation of the MDA5/MAVS pathway, and the editing-independent role(s) of ADAR1p110 in maintaining organ development and homeostasis.
Adarb1-KO mice survive to birth but die within three weeks, and are rescued by knock-in of a single recoding Q/R edit site (with endogenous ~100% editing frequency) in the AMPA receptor Gria2[17]. Strikingly, a four-way mutant mouse (AdarE861A/E861A, MDA5/Ifih1-null, Adarb1-null, Gria2R/R) survives to adulthood and lacks any gross abnormalities, suggesting that aside from the Gria2 edit site and preventing MAVS activation, A-to-I editing is not essential for life[18]. However, a mouse model with targeted disruption of a recoding edit site in Flna demonstrates precursor signs of cardiac disease and defects in vascular cells, indicating that single edit sites may still influence biology in a subtler or more tissue-specific manner[19]. Finally, Adarb2-KO mice are grossly normal but display cognitive defects in learning and memory, and global A-to-I editing is essentially unchanged[20].
While KO mouse models allow for the study of whole-organism dynamics and remain a powerful tool, care should be taken in directly transferring conclusions from mouse to humans. Primates have over an order of magnitude higher editing rates than mice at conserved sites, largely attributed to the evolution of increased editing to accommodate primate-specific Alu repeat elements[21]. Especially given the relative ease of redirecting ADAR targeting by altering sequence or structure, it is conceivable that this conserved mechanism could be repurposed over evolutionary time to yield distinct functions in humans over mice. Additionally, the requirements for ADARs and their activities in normal homeostasis and development may not reflect dynamics in disease states – and thus a single edit site may still play an important role in cancer progression, even if it is not required for the maintenance of overall health and survival, as was suggested by the viability of the four-way mutant mouse model.
ADARs in human disease
In humans, mutations in ADAR1 have been linked to several type I interferonopathies, or diseases of dysregulated type I interferon homeostasis, in line with its observed role in suppressing IFN signaling. ADAR1 mutations are associated with Aicardi-Goutières syndrome (AGS), an autoimmune disorder characterized by aberrantly high interferon levels and various cranial abnormalities[22]. AGS-associated ADAR1 mutations cluster within the deaminase domain or the Z-DNA binding domains, which combined with the common pathways of nucleic acid processing and sensing regulated by other AGS-associated genes (IFIH1, RNASEH2A/B/C, etc.) implicates the A-to-I editing activity and masking of self-dsRNA of ADAR1 in the molecular pathophysiology of this disease[22]. Dyschromatosis symmetrica hereditaria (DSH) is an autosomal dominant inherited skin disorder characterized by hypo- and hyper-pigmentation of the face, hands and feet[23], [24]. Haploinsufficiency caused by mutations in ADAR1 is thought to cause DSH, and interestingly the mutations in DSH and AGS patients generally do not overlap.
Aside from discrete genetic perturbations in Mendelian diseases, disruptions in ADARs and A-to-I editing levels have also been associated with a wide variety of cancers, as well as several neurological and immunological diseases. ADAR2, being predominantly expressed in the brain, is commonly found to have reduced editing in brain disorders, including glioblastoma multiforme, Alzheimer’s disease, ALS, schizophrenia, and bipolar disorder[25]-[30]. ADAR1 mutations contribute to the cognitive defects of AGS by inducing a neuroinflammatory state and resulting encephalopathy[31]. Both ADAR1 and ADAR2 edit multiple recoding sites in the serotonin receptor 5HT2CR to fine-tune its activity and downstream signaling in the brain[32].
Unsurprisingly given its connection to IFN signaling, ADAR1 has also demonstrated roles in regulating the virus-host interaction, either as a proviral or antiviral mechanism. Generally these roles involve A-to-I editing of viral genomes themselves or of host factors that regulate the antiviral response, although some editing-independent roles have also been described[33].
RNA editing dynamics have also been studied in the cardiovascular system, which also tends to have high A-to-I editing levels. ADAR1 editing of CTSS increases its expression via HuR-mediated stabilization, and increased CTSS expression is associated with atherosclerotic vascular diseases[34]. ADAR2 has also been implicated in cardiovascular disease through a recoding edit site in FLNA which, when disrupted, results in hypertension and cardiac remodeling in mice[19].
ADAR1 and cancer
Increased ADAR1 expression and/or activity has been implicated in many cancers, including hepatocellular carcinoma (HCC), non-small-cell lung cancer (NSCLC), gastric cancer, chronic myeloid leukemia (CML), esophageal squamous cell carcinoma (ESCC), colorectal cancer, oral squamous cell carcinoma, pancreatic cancer, multiple myeloma, cervical cancer, thyroid cancer, and breast cancer[7], [35]-[45]. Conversely, ADAR1 downregulation was associated with metastatic melanoma and kidney cancers[46], [47]. We have summarized prior literature that experimentally confirmed a role for ADAR1 in cancer hallmarks, including whether ADAR1 perturbation was tested in vivo and any downstream ADAR1 targets mediating cancer phenotypes (Table 1).
Table 1.
ADAR1 of cancer hallmarks
| ADAR1 promoting cancer | ||||
|---|---|---|---|---|
| Cancer | Hallmarks | In vivo | Genes involved | Ref. |
| Hepatocellular carcinoma | Growth, colony formation, invasion/migration | Y[35] | AZIN1[48], CircARSP91[72], ITGA2[73] | [35], [48], [72], [73] |
| Thyroid cancer | Growth, colony formation, invasion/migration | Y[45] | miR-200b-3p & ZEB1[45], CDK13[50] | [45], [50] |
| Cervical cancer | Growth | Y[74] | [63], [74] | |
| Breast cancer | Growth, colony formation, migration/invasion | Y[65], [75] | FLNB, miR-27a-5p, miR-4485-3p[51], DHFR[56] | [56], [65], [75]-[78] |
| Chronic myeloid leukemia | Proliferation, self-renewal | Y[38], [52], [57], [79] | let-7[52], miR-26a and MDM2 & miR-155[57] | [38], [52], [57], [79] |
| Non-small cell lung cancer | Growth, metastasis, colony formation | Y[36], [80] | NEIL1 & miR-381-3p[36], FAK[81], AZIN1[82], CX3CL1[80] | [36], [80]-[82] |
| Gastric cancer | Growth & metastasis, colony formation, migration/invasion | Y[37], [83] | mTOR[83], miR-302a-3p & IRF9[84], circ0004872 & miR-224[85] | [37], [83]-[85] |
| Esophageal squamous cell carcinoma | Growth, colony formation, migration/invasion | Y[39], [86, p.2] | AZIN1[39] | [39], [86] |
| Colorectal cancer | Growth, stemness, migration/invasion* | Y[40]* | AZIN1[40], [87] | [40], [87] |
| Oral squamous cell carcinoma | Growth, colony formation, migration/invasion, stemness | Y[41] | DICER1[41] | [41] |
| Pancreatic cancer | Growth, colony formation, migration/invasion | Y[42] | c-Myc[42], circNEIL3 & miR-432-5p[88] | [42], [88] |
| Multiple myeloma | Growth, colony formation, self-renewal | Y[43], [49] | GLI1[43], NEIL1[49] | [43], [49], [89] |
| Prostate cancer | Growth | Y[90] | PCA3 & PRUNE2[90] | [90], [91] |
| Glioma | Growth, colony formation | Y[92] | CDK2[92] | [92], [93] |
| Melanoma | Growth | N | miR-149-3p & GSK3A | [94] |
| ADAR1 suppressing cancer | ||||
| Cancer | Hallmarks | In vivo | Genes involved | Ref. |
| Melanoma | Growth & metastasis, invasion, T-cell killing | Y[46], [69], [95], [96] | miR-455-5p & CPEB1[95], miR-222 & ICAM1[97], miR-378a-3p & PARVA[98], ITGB3[96], [99] | [46], [69], [95]-[99] |
| Breast cancer | Migration/invasion* | Y* | GABRA3 | [100] |
indicates that phenotypes were only assessed by perturbing the target, not ADAR1 itself
ADAR mechanisms in cancer
ADARs can function to regulate the transcriptome in several distinct ways, and this is reflected in the diversity of ADAR1 targets and mechanisms that have been implicated in its pro-oncogenic effects. Most direct are effects mediated by A-to-I editing, which can alter RNA structure, binding motifs, coding sequence, and more to regulate the target. However, editing-independent roles have also been demonstrated for ADARs, as they can also function as an RNA-binding protein independently from catalytic activity. Such mechanisms are usually demonstrated by rescue of a given phenotype by the catalytic point mutant E912A (Figure 1).
Coding edits
Early mechanistic work on ADARs focused on the tantalizing, though rare, occurrences where editing resulted in direct recoding of a protein. One landmark study characterized a recoding edit site in antizyme inhibitor 1 (AZIN1) which resulted in an S367G substitution, thus altering the structure and localization of the protein to confer a greater pro-oncogenic function for the protein in HCC[48]. The K242R recoding edit in the DNA repair enzyme NEIL1 results in altered lesion specificity and less efficient DNA repair, which in multiple myeloma cells contributed to increased proliferation and colony formation[49]. The CDK13 Q103R edit site promotes increased growth, colony formation, and invasion in thyroid cancer and reduces the nuclear speckle localization of this splicing factor[50]. Filamin B editing yields an M2293V mutation that reduces nuclear localization and EMT-suppressive activity to promote invasion in triple-negative breast cancer[51].
microRNA mechanisms
ADARs can also bind and/or edit the hairpin intermediates of miRNAs. The primary miRNA (pri-miRNA) transcript is cleaved by nuclear Drosha/DGCR8 to yield the precursor (pre-miRNA), which is exported to the cytoplasm, cleaved by Dicer, and the mature miRNA loaded into the RNA-Induced Silencing Complex (RISC)[4]. ADARs have been demonstrated to regulate this process via A-to-I editing, dsRNA binding, or even RNA-independent direct protein interactions. An edit site in the seed region of miR-376a-5p yielded distinct binding preferences between the edited and unedited miRNA that resulted in opposing effects on invasion and migration in glioblastoma[25]. A-to-I editing of pri-let-7d was shown to inhibit processing to its mature form, thus promoting self-renewal capacity of CML progenitors to therapy-resistant leukemia stem cells[52]. Additionally, several studies have demonstrated a direct interaction between Dicer and ADAR1, independent of both catalytic and RNA binding activity of ADAR1, which can promote the processing of pro-oncogenic miRNAs[41], [53], [54].
3'UTR mechanisms
Given that the 3'UTR represents a large fraction of editing sites, it is unsurprising that ADAR1 targeting of the 3'UTR is another common mechanism for regulating gene expression in cancer. Editing of 3'UTRs can create or destroy miRNA binding sites to alter mRNA stability of important cancer genes, as several studies have demonstrated[55]-[57]. Binding of the 3'UTR of targets allowed ADAR1 to exclude Staufen1 binding and subsequent decay of anti-apoptotic genes, thus promoting stress-induced survival of cells in an editing-independent manner[58].
Splicing mechanisms
ADAR1-mediated splicing regulation can occur via edit sites that create/destroy splice signals, as well as binding competition with splice factors. Editing of HNRPLL in multiple cancer types was shown to promote exon 12A inclusion via SRSF1 recruitment, which may promote the expression of pro-growth genes like CCND1[59]. Editing-independent binding of ADARs was shown to regulate access of splicing machinery to multiple transcripts in ESCC, altering exon inclusion and function of several cancer-relevant targets[60]. Changes to global splicing patterns upon ADAR1 dysregulation has been documented in cancer cells, although only a limited number can be attributed to editing in cis, suggesting other mechanisms are responsible for the majority of alterations[61]. ADAR1 has also been shown to target trans splicing factors, such as CDK13 editing in thyroid cancer, which can then exert a larger influence on global splicing than individual edit sites[50].
R-loop edits
Several recent papers have demonstrated roles for A-to-I editing in the maintenance of genome integrity via editing of DNA:RNA hybrids. ADAR2-KD cells were found to have an impaired DNA damage response due to defects in clearing DNA:RNA hybrids[62]. ADAR1p110 was critical for proliferation of a subset of telomerase-activated cancer cells due to a requirement to edit telomeric variant R-loops to promote clearance and genomic stability[63]. Although these telomeric variants constitute a cancer-specific phenomenon, the authors also observed telomeric instability in Adar-null mouse embryonic fibroblasts. Unless a broader mechanism of R-loop clearance that is editing-independent exists, it is unclear how the editing-incompetent AdarE861A/E861A, MDA5/Ifih1−/−, Adarb1−/−, Gria2R/R mouse would have escaped such phenotypes. While the interplay between ADARs and the DNA damage response is not entirely understood, this suggests an additional pathway by which ADAR inhibition could constitute an effective therapeutic strategy.
As these examples illustrate, the multifunctional potential of ADARs allows for diverse possible mechanisms for transcriptome regulation, which can then contribute to cancer development. Thus it is critically important to assess the global transcriptome via unbiased sequencing within a given cancer context to understand how ADAR1 exerts its context-dependent effects on gene expression. Additionally, the potential impact of these mechanisms in a normal context is rarely assessed (the availability of primary cells being a common limitation) but represents an important consideration for understanding ADAR biology in full.
ADARs and immune signaling
As established in mouse models, ADAR1 and A-to-I editing play an essential role in suppressing IFNs and preventing an aberrant innate immune response, mediated by polymerizing of MDA5 on dsRNA and activation of MAVS. The interferon-inducible p150 isoform has been implicated as the major culprit in this pathway, presumably due to the editing of p150-specific targets it can access through its cytoplasmic localization and/or additional Z-DNA/RNA binding domain. Surprisingly, in ADAR1p110-null, ADAR2-null mice, only 2% of A-to-I edit sites in the brain are preserved by p150 activity and yet this is sufficient to prevent activation of MDA5[16]. Thus, it seems increasingly likely that MDA5 is sensitive to only a small subset of A-to-I editing targets, although the identity of these targets is yet unknown.
In addition to its role suppressing the MDA5/MAVS axis, suppression of PKR is another well-recognized facet to ADAR1 immune regulation. PKR is another antiviral dsRNA sensor that upon activation triggers translational shutdown via eIF2α phosphorylation and apoptosis. Loss of ADAR1 is generally associated with increased PKR phosphorylation, and baseline PKR expression has been hypothesized as a marker, or at least a requirement, for ADAR1 sensitivity [64]-[67].
Unsurprisingly, ADAR1 has also demonstrated roles in regulating antitumor immunity and synergizing with checkpoint blockade. In a screen for sensitizers to T-cell killing, ADAR1 emerged as a top hit in several murine cancers[68]. ADAR1-KO cells implanted in immune-competent mice had a strong synergistic response with anti-PD-1 treatment, overcoming several mechanisms of anti-PD-1 resistance and coinciding with increased levels of tumor-killing T and NK cells as well as immune cell-recruiting cytokines and IFNs[69]. This effect was dependent on Ifnar2 and Ifngr1 loss or STAT1 loss, indicating a requirement for IFN sensing, as well as for at least one functional dsRNA sensor (PKR or MDA5). Most compellingly, ADAR1-KO cells with antigen presenting defects (B2m-null were still sensitized to immunotherapy, suggesting the inflammatory tumor environment caused by ADAR1 loss is sufficient to restore antitumor immunity independent of CD8+ T-cell recognition.
However, the role of ADAR1 in cancer remains complex and context-dependent and consists of more than just its immune regulatory functions alone. Studies of ADAR1 perturbation in vitro or in immune-compromised mice have demonstrated roles in growth and metastasis, outside of roles in activating the antitumor immune response (Table 1). Also, rescue of ADAR1-loss-mediated growth defects in vitro by suppression of PKR or MDA5/MAVS are context-dependent and often partial, suggesting there are additional cell-intrinsic mechanisms regulating growth[65], [66].
Concluding Remarks and Future Perspectives
The diverse functions of ADAR1 in the cell complicate our understanding of its role in cancer. Between RNA binding, RNA editing, and protein interactions mediating its activity, ADAR1 exerts wide-ranging control over the global transcriptome. Yet, as the above examples have made clear, certain targets can cut through the noise to significantly impact cancer dynamics. In addition to regulating cancer mechanisms through specific gene targets, roles in suppressing immunostimulatory dsRNAs and, more recently, promoting genome integrity via R-loop clearance have emerged as additional angles to the story of ADAR1 in cancer.
While our understanding of ADAR biology has rapidly expanded, several key gaps remain that are relevant to the cancer context (see Outstanding questions). Unraveling these mysteries has major therapeutic potential to inform nuanced design of candidate drugs and offer better therapeutic precision. Depending on the mechanisms at play in a given cancer context, individual functional domains or isoforms could be targeted. Editing and/or p150-specific inhibition could be leveraged to induce an interferon response and cell death, and even synergize with other immunotherapies [69]. Precise suppression of a specific editing event could be achieved by altering the dsRNA structure using ASOs [70], [71]. Inducing telomere instability by p110-specific targeting was a proposed therapeutic strategy for telomerase-positive cancers, and we speculate that the broader role for p110 in the DNA damage response could lead to synergy with drugs in this pathway, such as PARP inhibition [62], [63]. Ultimately, a more complete ADAR1 targeting strategy may generate the most broad and robust response, but with increased risk for side effects in normal tissues. Finding the right balance for an optimal therapeutic window will be key to the development of anti-ADAR1 therapeutics.
Outstanding Questions Box:
What is the identity of the RNA(s) responsible for activating MDA5/MAVS and PKR pathways? Emerging data suggests a key role for the ADAR1p150 isoform, possibly due to the unique binding properties of its additional Z-DNA/RNA binding domain. This would have major implications both for ADAR1-associated interferonopathies as well as targeting ADAR1 immune suppression in cancer.
How could recently uncovered roles for ADAR1 in regulating genome integrity synergize with DNA damaging agents?
If ADAR1 is ubiquitously expressed and its loss is generally deleterious in normal tissues as well, what is the potential therapeutic window for developing inhibitors to treat cancer and possibly other diseases?
Highlights Box:
The epitranscriptome represents an emerging novel layer of gene regulation that goes awry in cancer. Many RNA modifying enzymes have been implicated in cancer, including the Adenosine-to-Inosine (A-to-I) editors called ADARs.
ADARs are multifunctional proteins that regulate the transcriptome via A-to-I editing, RNA binding, and direct protein interactions. All of these functions have been implicated in cancer development in diverse tissue contexts. Here we focus on the predominantly oncogenic role of ADAR1.
In addition to mechanisms mediated by a specific target, ADAR1 also regulates interferon signaling via dsRNA sensors, which translates to an immunosuppressive function in the cancer context. Recently, a role in resolving R-loops has been proposed to regulate genome integrity and senescence, and thus another cancer hallmark.
Acknowledgements
We acknowledge the NCI Outstanding Investigator Award (R35CA232105) and funding from the Ludwig Center at Harvard to FJS.
Glossary Box:
- Alu repeat elements
Short Interspersed Repeat Elements (SINEs) present in over 1 million copies in our genome due to retrotransposon activity. Alu elements in an inverse repeat orientation are responsible for the large majority of endogenous dsRNA, and thus are a major target of A-to-I editing.
- Checkpoint blockade
A therapeutic strategy inhibiting one of several suppressive receptor-ligand interactions on T cells (checkpoints) in order to stimulate immune activity. Prominent examples include PD-1/PD-L1 and CTLA4/CD80 & CD86.
- Epitranscriptomics
The collection of post-transcriptional RNA modifications that can alter the base pairing and/or activity of the modified RNA.
- MDA5/MAVS pathway
An innate immunity pathway involving the sensing of long stretches of cytoplasmic dsRNA by MDA5 (part of the RIG-I-like receptor family). Binding and oligomerization of MDA5 on dsRNA allows for activation of mitochondrially-associated MAVS, which results in downstream type I interferon signaling via TRAFs.
- microRNAs
A class of short ~22nt noncoding RNAs that direct Argonaute-mediated silencing of coding transcripts via sequence homology. The seed region (positions 2-7) is particularly critical and requires perfect complementarity with the target.
- PKR
Protein kinase R, encoded by the eukaryotic translation initiation factor 2-alpha kinase 2 (EIF2AK2) gene, is another important component of the innate immunity pathway. PKR is activated by binding dsRNA and mediates translational suppression via eIF2α phosphorylation as well as cell death.
- Z-DNA/RNA
Double-stranded DNA or RNA that is in the left-handed coil conformation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, and Novoa EM, “The RNA modification landscape in human disease,” RNA, vol. 23, no. 12, pp. 1754–1769, Dec. 2017, doi: 10.1261/rna.063503.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Xue C, Zhao Y, and Li L, “Advances in RNA cytosine-5 methylation: detection, regulatory mechanisms, biological functions and links to cancer,” Biomark. Res, vol. 8, no. 1, p. 43, Sep. 2020, doi: 10.1186/s40364-020-00225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lian H, Wang Q-H, Zhu C-B, Ma J, and Jin W-L, “Deciphering the Epitranscriptome in Cancer,” Trends Cancer, vol. 4, no. 3, pp. 207–221, Mar. 2018, doi: 10.1016/j.trecan.2018.01.006. [DOI] [PubMed] [Google Scholar]
- [4].Nishikura K, “A-to-I editing of coding and non-coding RNAs by ADARs,” Nat. Rev. Mol.Cell Biol, vol. 17, no. 2, pp. 83–96, Dec. 2015, doi: 10.1038/nrm.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tan MH et al. , “Dynamic landscape and regulation of RNA editing in mammals,” Nature, vol. 550, no. 7675, p. nature24041, Oct. 2017, doi: 10.1038/nature24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sun T et al. , “Decoupling expression and editing preferences of ADAR1 p150 and p110 isoforms,” Proc. Natl. Acad. Sci, vol. 118, no. 12, p. e2021757118, Mar. 2021, doi: 10.1073/pnas.2021757118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Paz-Yaacov N et al. , “Elevated RNA Editing Activity Is a Major Contributor to Transcriptomic Diversity in Tumors,” Cell Rep., vol. 13, no. 2, pp. 267–276, Oct. 2015, doi: 10.1016/j.celrep.2015.08.080. [DOI] [PubMed] [Google Scholar]
- [8].Bahn JH, Lee J-H, Li G, Greer C, Peng G, and Xiao X, “Accurate identification of A-to-I RNA editing in human by transcriptome sequencing,” Genome Res., vol. 22, no. 1, pp. 142–150, Jan. 2012, doi: 10.1101/gr.124107.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Picardi E, Manzari C, Mastropasqua F, Aiello I, D’Erchia AM, and Pesole G, “Profiling RNA editing in human tissues: towards the inosinome Atlas,” Sci. Rep, vol. 5, p. 14941, Oct. 2015, doi: 10.1038/srep14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang IX, So E, Devlin JL, Zhao Y, Wu M, and Cheung VG, “ADAR Regulates RNA Editing, Transcript Stability, and Gene Expression,” Cell Rep., vol. 5, no. 3, pp. 849–860, Nov. 2013, doi: 10.1016/j.celrep.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Eggington JM, Greene T, and Bass BL, “Predicting sites of ADAR editing in double-stranded RNA,” Nat. Commun, vol. 2, p. ncomms1324, May 2011, doi: 10.1038/ncomms1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hartner JC, Schmittwolf C, Kispert A, Müller AM, Higuchi M, and Seeburg PH, “Liver Disintegration in the Mouse Embryo Caused by Deficiency in the RNA-editing Enzyme ADAR1,” J. Biol Chem, vol. 279, no. 6, pp. 4894–4902, Feb. 2004, doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- [13].Mannion NM et al. , “The RNA-Editing Enzyme ADAR1 Controls Innate Immune Responses to RNA,” Cell Rep., vol. 9, no. 4, pp. 1482–1494, Nov. 2014, doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, and Stetson DB, “Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-organ Development,” Immunity, vol. 43, no. 5, pp. 933–944, Nov. 2015, doi: 10.1016/j.immuni.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liddicoat BJ et al. , “RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself,” Science, vol. 349, no. 6252, pp. 1115–1120, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kim JI et al. , “RNA editing at a limited number of sites is sufficient to prevent MDA5 activation in the mouse brain,” PLOS Genet., vol. 17, no. 5, p. e1009516, May 2021, doi: 10.1371/journal.pgen.1009516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Higuchi M et al. , “Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2,” Nature, vol. 406, no. 6791, pp. 78–81, Jul. 2000. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- [18].Chalk AM, Taylor S, Heraud-Farlow JE, and Walkley CR, “The majority of A-to-I RNA editing is not required for mammalian homeostasis,” Genome Biol., vol. 20, no. 1, p. 268, Dec. 2019, doi: 10.1186/s13059-019-1873-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jain M et al. , “RNA editing of Filamin A pre-mRNA regulates vascular contraction and diastolic blood pressure,” EMBO J., p. e94813, Aug. 2018, doi: 10.15252/embj.201694813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mladenova D et al. , “Adar3 Is Involved in Learning and Memory in Mice,” Front. Neurosci, vol. 12, p. 243, 2018, doi: 10.3389/fnins.2018.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li JB and Church GM, “Deciphering the functions and regulation of brain-enriched A-to-I RNA editing,” Nat. Neurosci, vol. 16, no. 11, p. 1518, Nov. 2013, doi: 10.1038/nn.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rice GI et al. , “Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature,” Nat. Genet, vol. 44, no. 11, pp. 1243–1248, Sep. 2012, doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kono M, Suganuma M, Ito Y, Ujiie H, Morimoto K, and Akiyama M, “Novel ADAR1 mutations including a single amino acid deletion in the deaminase domain underlie dyschromatosis symmetrica hereditaria in Japanese families,” Int. J. Dermatol, vol. 53, no. 3, pp. e194–e196, Mar. 2014, doi: 10.1111/j.1365-4632.2012.05765.x. [DOI] [PubMed] [Google Scholar]
- [24].Liu Q et al. , “Two novel mutations and evidence for haploinsufficiency of the ADAR gene in dyschromatosis symmetrica hereditaria,” Br. J. Dermatol, vol. 154, no. 4, pp. 636–642, Apr. 2006, doi: 10.1111/j.1365-2133.2006.07133.x. [DOI] [PubMed] [Google Scholar]
- [25].Choudhury Y et al. , “Attenuated adenosine-to-inosine editing of microRNA-376a*promotes invasiveness of glioblastoma cells,” J. Clin. Invest, vol. 122, no. 11, pp. 4059–4076, Nov. 2012, doi: 10.1172/JCI62925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Maas S, Patt S, Schrey M, and Rich A, “Underediting of glutamate receptor GluR-B mRNA in malignant gliomas,” Proc. Natl Acad. Sci, vol. 98, no. 25, pp. 14687–14692, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gaisler-Salomon I et al. , “Hippocampus-specific deficiency in RNA editing of GluA2 in Alzheimer’s disease,” Neurobiol. Aging, vol. 35, no. 8, pp. 1785–1791, Aug. 2014, doi: 10.1016/j.neurobiolaging.2014.02.018. [DOI] [PubMed] [Google Scholar]
- [28].Hideyama T et al. , “Profound downregulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons,” Neurobiol. Dis, vol. 45, no. 3, pp. 1121–1128, Mar. 2012, doi: 10.1016/j.nbd.2011.12.033. [DOI] [PubMed] [Google Scholar]
- [29].Akbarian S, Smith MA, and Jones EG, “Editing for an AMPA receptor subunit RNA in prefrontal cortex and striatum in Alzheimer’s disease, Huntington’s disease and schizophrenia,” Brain Res., vol. 699, no. 2, pp. 297–304, Nov. 1995, doi: 10.1016/0006-8993(95)00922-D. [DOI] [PubMed] [Google Scholar]
- [30].Kubota-Sakashita M, Iwamoto K, Bundo M, and Kato T, “A role of ADAR2 and RNA editing of glutamate receptors in mood disorders and schizophrenia,” Mol Brain, vol. 7, p. 5, Jan. 2014, doi: 10.1186/1756-6606-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Guo X et al. , “Aicardi-Goutières syndrome-associated mutation at ADAR1 gene locus activates innate immune response in mouse brain,” J. Neuroinflammation, vol. 18, no. 1, p. 169, Jul. 2021, doi: 10.1186/s12974-021-02217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Werry TD, Loiacono R, Sexton PM, and Christopoulos A, “RNA editing of the serotonin 5HT2C receptor and its effects on cell signalling, pharmacology and brain function,” Pharmacol. Ther, vol. 119, no. 1, pp. 7–23, Jul. 2008, doi: 10.1016/j.pharmthera.2008.03.012. [DOI] [PubMed] [Google Scholar]
- [33].Samuel CE, “Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral,” Virology, vol. 411, no. 2, pp. 180–193, Mar. 2011, doi: 10.1016/j.virol.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Stellos K et al. , “Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation,” Nat. Med, vol. 22, no. 10, pp. 1140–1150, Oct. 2016, doi: 10.1038/nm.4172. [DOI] [PubMed] [Google Scholar]
- [35].Chan THM et al. , “A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma,” Gut, vol. 63, no. 5, pp. 832–843, May 2014, doi: 10.1136/gutjnl-2012-304037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Anadón C et al. , “Gene amplification-associated overexpression of the RNA editing enzyme ADAR1 enhances human lung tumorigenesis,” Oncogene, vol. 35, no. 33, pp. 4407–4413, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chan THM et al. , “ADAR-Mediated RNA Editing Predicts Progression and Prognosis of Gastric Cancer,” Gastroenterology, vol. 151, no. 4, pp. 637–650.e10, Oct. 2016, doi: 10.1053/j.gastro.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jiang Q et al. , “ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia,” Proc. Natl. Acad. Sci, vol. 110, no. 3, pp. 1041–1046, Jan. 2013, doi: 10.1073/pnas.1213021110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Qin Y-R et al. , “Adenosine-to-Inosine RNA Editing Mediated by ADARs in Esophageal Squamous Cell Carcinoma,” Cancer Res., vol. 74, no. 3, pp. 840–851, Feb. 2014, doi: 10.1158/0008-5472.CAN-13-2545. [DOI] [PubMed] [Google Scholar]
- [40].Shigeyasu K et al. , “AZIN1 RNA editing confers cancer stemness and enhances oncogenic potential in colorectal cancer,” JCI Insight, vol. 3, no. 12, Jun. 2018, doi: 10.1172/jci.insight.99976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Liu X et al. , “ADAR1 promotes the epithelial-to-mesenchymal transition and stem-like cell phenotype of oral cancer by facilitating oncogenic microRNA maturation,” J. Exp. Clin. Cancer Res, vol. 38, no. 1, p. 315, Jul. 2019, doi: 10.1186/s13046-019-1300-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sun Y et al. , “The aberrant expression of ADAR1 promotes resistance to BET inhibitors in pancreatic cancer by stabilizing c-Myc,” Am. J. Cancer Res, vol. 10, no. 1, pp. 148–163, Jan. 2020. [PMC free article] [PubMed] [Google Scholar]
- [43].Lazzari E et al. , “Alu-dependent RNA editing of GLI1 promotes malignant regeneration in multiple myeloma,” Nat. Commun, vol. 8, no. 1, p. 1922, Dec. 2017, doi: 10.1038/s41467-017-01890-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chen Y, Wang H, Lin W, and Shuai P, “ADAR1 overexpression is associated with cervical cancer progression and angiogenesis,” Diagn. Pathol, vol. 12, no. 1, p. 12, Jan. 2017, doi: 10.1186/s13000-017-0600-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ramírez-Moya J, Baker AR, Slack FJ, and Santisteban P, “ADAR1-mediated RNA editing is a novel oncogenic process in thyroid cancer and regulates miR-200 activity,” Oncogene, vol. 39, no. 18, pp. 3738–3753, Apr. 2020, doi: 10.1038/s41388-020-1248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nemlich Y et al. , “MicroRNA-mediated loss of ADAR1 in metastatic melanoma promotes tumor growth,” J. Clin. Invest, vol. 123, no. 6, pp. 2703–2718, Jun. 2013, doi: 10.1172/JCI62980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Han L et al. , “The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers,” Cancer Cell, vol. 28, no. 4, pp. 515–528, Oct. 2015, doi: 10.1016/j.ccell.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chen L et al. , “Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma,” Nat. Med, vol. 19, no. 2, pp. 209–216, Jan. 2013, doi: 10.1038/nm.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Teoh PJ et al. , “Aberrant hyperediting of the myeloma transcriptome by ADAR1 confers oncogenicity and is a marker of poor prognosis,” Blood, vol. 132, no. 12, pp. 1304–1317, Sep. 2018, doi: 10.1182/blood-2018-02-832576. [DOI] [PubMed] [Google Scholar]
- [50].Ramírez-Moya J, Miliotis C, Baker AR, Gregory RI, Slack FJ, and Santisteban P, “An ADAR1-dependent RNA editing event in the cyclin-dependent kinase CDK13 promotes thyroid cancer hallmarks,” Mol. Cancer, vol. 20, no. 1, p. 115, Sep. 2021, doi: 10.1186/s12943-021-01401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Baker AR et al. , “Transcriptome profiling of ADAR1 targets in triple-negative breast cancer cells reveals mechanisms for regulating growth and invasion,” Mol. Cancer Res, (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zipeto M et al. , “ADAR1 Activation Drives Leukemia Stem Cell Self-Renewal by Impairing Let-7 Biogenesis,” Cell Stem Cell, vol. 19, no. 2, pp. 177–191, Aug. 2016, doi: 10.1016/j.stem.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Qi L et al. , “An RNA editing/dsRNA binding-independent gene regulatory mechanism of ADARs and its clinical implication in cancer,” Nucleic Acids Res., vol. 45, no. 18, pp. 10436–10451, Oct. 2017, doi: 10.1093/nar/gkx667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ota H et al. , “ADAR1 Forms a Complex with Dicer to Promote MicroRNA Processing and RNA-Induced Gene Silencing,” Cell, vol. 153, no. 3, pp. 575–589, Apr. 2013, doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Nakano M, Fukami T, Gotoh S, Takamiya M, Aoki Y, and Nakajima M, “RNA Editing Modulates Human Hepatic Aryl Hydrocarbon Receptor Expression by Creating MicroRNA Recognition Sequence * J. Biol. Chem, vol. 291, no. 2, pp. 894–903, Jan. 2016, doi: 10.1074/jbc.M115.699363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Nakano M, Fukami T, Gotoh S, and Nakajima M, “A-to-I RNA Editing Up-regulates Human Dihydrofolate Reductase in Breast Cancer,” J. Biol. Chem, vol. 292, no. 12, pp. 4873–4884, Mar. 2017, doi: 10.1074/jbc.M117.775684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Jiang Q et al. , “Hyper-Editing of Cell-Cycle Regulatory and Tumor Suppressor RNA Promotes Malignant Progenitor Propagation,” Cancer Cell, vol. 35, no. 1, pp. 81–94.e7, Jan. 2019, doi: 10.1016/j.ccell.2018.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sakurai M et al. , “ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay,” Nat. Struct. Mol Biol, vol. 24, no. 6, pp. 534–543, Jun. 2017, doi: 10.1038/nsmb.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chen Y-T et al. , “Tumor-associated intronic editing of HNRPLL generates a novel splicing variant linked to cell proliferation,” J. Biol Chem, vol. 293, no. 26, pp. 10158–10171, Jun. 2018, doi: 10.1074/jbc.RA117.001197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Tang SJ et al. , “Cis- and trans-regulations of pre-mRNA splicing by RNA editing enzymes influence cancer development,” Nat. Commun, vol. 11, no. 1, p. 799, Feb. 2020, doi: 10.1038/s41467-020-14621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Solomon O et al. , “Global regulation of alternative splicing by adenosine deaminase acting on RNA (ADAR),” RNA, vol. 19, no. 5, pp. 591–604, May 2013, doi: 10.1261/rna.038042.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jimeno S et al. , “ADAR-mediated RNA editing of DNARNA hybrids is required for DNA double strand break repair,” Nat. Commun, vol. 12, no. 1, p. 5512, Sep. 2021, doi: 10.1038/s41467-021-25790-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Shiromoto Y, Sakurai M, Minakuchi M, Ariyoshi K, and Nishikura K, “ADAR1 RNA editing enzyme regulates R-loop formation and genome stability at telomeres in cancer cells,” Nat. Commun, vol. 12, no. 1, p. 1654, Dec. 2021, doi: 10.1038/s41467-021-21921-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Chung H et al. , “Human ADAR1 Prevents Endogenous RNA from Triggering Translational Shutdown,” Cell, vol. 172, no. 4, pp. 811–824.e14, Feb. 2018, doi: 10.1016/j.cell.2017.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kung C-P et al. , “Evaluating the therapeutic potential of ADAR1 inhibition for triple-negative breast cancer,” Oncogene, vol. 40, no. 1, Art. no. 1, Jan. 2021, doi: 10.1038/s41388-020-01515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Gannon HS et al. , “Identification of ADAR1 adenosine deaminase dependency in a subset of cancer cells,” Nat. Commun, vol. 9, no. 1, p. 5450, Dec. 2018, doi: 10.1038/s41467-018-07824-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Liu H et al. , “Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss,” Nat. Med, p. 1, Dec. 2018, doi: 10.1038/s41591-018-0302-5. [DOI] [PubMed] [Google Scholar]
- [68].Lawson KA et al. , “Functional genomic landscape of cancer-intrinsic evasion of killing by T cells,” Nature, vol. 586, no. 7827, Art. no. 7827, Oct. 2020, doi: 10.1038/s41586-020-2746-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ishizuka JJ et al. , “Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade,” Nature, Dec. 2018, doi: 10.1038/s41586-018-0768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Mizrahi RA, Schirle NT, and Beal PA, “Potent and Selective Inhibition of A-to-I RNA Editing with 2′- O -Methyl/Locked Nucleic Acid-Containing Antisense Oligoribonucleotides,” ACS Chem. Biol, vol. 8, no. 4, pp. 832–839, Apr. 2013, doi: 10.1021/cb300692k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Tay DJT et al. , “Targeting RNA Editing of Antizyme Inhibitor 1: a Potential Oligonucleotide-Based Antisense Therapy for Cancer,” Mol Ther., vol. 0, no. 0, May 2021, doi: 10.1016/j.ymthe.2021.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Shi L et al. , “Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma,” Cell Death Dis., vol. 8, no. 11, p. e3171, Nov. 2017, doi: 10.1038/cddis.2017.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Yu J, Zhang C, Yu Q, Yu H, and Zhang B, “ADAR1 p110 Enhances Adhesion of Tumor Cells to Extracellular Matrix in Hepatocellular Carcinoma via Up-Regulating ITGA2 Expression,” Med Sci. Monit. Int. Med. J. Exp. Clin. Res, vol. 25, pp. 1469–1479, Feb. 2019, doi: 10.12659/MSM.911944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wang H, Hou Z, Wu Y, Ma X, and Luo X, “p150 ADAR1 isoform involved in maintenance of HeLa cell proliferation,” BMC Cancer, vol. 6, no. 1, p. 282, Dec. 2006, doi: 10.1186/1471-2407-6-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Morales F et al. , “Increase in ADAR1p110 activates the canonical Wnt signaling pathway associated with aggressive phenotype in triple negative breast cancer cells,” Gene, vol. 819, p. 146246, Apr. 2022, doi: 10.1016/j.gene.2022.146246. [DOI] [PubMed] [Google Scholar]
- [76].Fumagalli D et al. , “Principles Governing A-to-I RNA Editing in the Breast Cancer Transcriptome,” Cell Rep., vol. 13, no. 2, pp. 277–289, Oct. 2015, doi: 10.1016/j.celrep.2015.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Sagredo EA et al. , “ADAR1 Transcriptome editing promotes breast cancer progression through the regulation of cell cycle and DNA damage response,” Biochim. Biophys. Acta BBA -Mol. Cell Res, p. 118716, Apr. 2020, doi: 10.1016/j.bbamcr.2020.118716. [DOI] [PubMed] [Google Scholar]
- [78].Li X et al. , “Upregulation of ADAR Promotes Breast Cancer Progression and Serves as a Potential Therapeutic Target,” J. Oncol, vol. 2021, p. e2012903, Sep. 2021, doi: 10.1155/2021/2012903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Steinman RA et al. , “Deletion of the RNA-editing enzyme ADAR1 causes regression of established chronic myelogenous leukemia in mice,” Int. J. Cancer, vol. 132, no. 8, pp. 1741–1750, 2013, doi: 10.1002/ijc.27851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Wu M, Jin M, Cao X, Qian K, and Zhao L, “RNA editing enzyme adenosine deaminases acting on RNA 1 deficiency increases the sensitivity of non-small cell lung cancer cells to anlotinib by regulating CX3CR1-fractalkine expression,” Drug Dev. Res, vol. n/a, no. n/a, pp. 1–11, Jul. 2021, doi: 10.1002/ddr.21861. [DOI] [PubMed] [Google Scholar]
- [81].Amin EM et al. , “The RNA-editing enzyme ADAR promotes lung adenocarcinoma migration and invasion by stabilizing FAK,” Sci Signal, vol. 10, no. 497, p. eaah3941, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hu X et al. , “RNA editing of AZIN1 induces the malignant progression of non-small-cell lung cancers,” Tumor Biol., vol. 39, no. 8, p. 1010428317700001, Aug. 2017, doi: 10.1177/1010428317700001. [DOI] [PubMed] [Google Scholar]
- [83].Dou N, Yu S, Ye X, Yang D, Li Y, and Gao Y, “Aberrant overexpression of ADAR1 promotes gastric cancer progression by activating mTOR/p70S6K signaling,” Oncotarget, vol. 7, no. 52, pp. 86161–86173, Nov. 2016, doi: 10.18632/oncotarget.13354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Jiang L et al. , “ADAR1 Suppresses Interferon Signaling in Gastric Cancer Cells by MicroRNA-302a-Mediated IRF9/STAT1 Regulation,” Int. J. Mol. Sci, vol. 21, no. 17, Aug. 2020, doi: 10.3390/ijms21176195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Ma C et al. , “Circular RNA hsa_circ_0004872 inhibits gastric cancer progression via the miR-224/Smad4/ADAR1 successive regulatory circuit,” Mol Cancer, vol. 19, no. 1, p. 157, Nov. 2020, doi: 10.1186/s12943-020-01268-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Wu Z et al. , “Reprogramming of the esophageal squamous carcinoma epigenome by SOX2 promotes ADAR1 dependence,” Nat. Genet, vol. 53, no. 6, Art. no. 6, Jun. 2021, doi: 10.1038/s41588-021-00859-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Takeda S et al. , “Activation of AZIN1 RNA editing is a novel mechanism that promotes invasive potential of cancer-associated fibroblasts in colorectal cancer,” Cancer Lett., vol. 444, pp. 127–135, Mar. 2019, doi: 10.1016/j.canlet.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Shen P et al. , “CircNEIL3 regulatory loop promotes pancreatic ductal adenocarcinoma progression via miRNA sponging and A-to-I RNA-editing,” Mol Cancer, vol. 20, no. 1, p. 51, Mar. 2021, doi: 10.1186/s12943-021-01333-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Teoh PJ, Chung T-H, Chng PYZ, Toh SHM, and Chng WJ, “IL6R-STAT3-ADAR1 (P150) interplay promotes oncogenicity in multiple myeloma with 1q21 amplification,” Haematologica, vol. 105, no. 5, Art. no. 5, May 2020, doi: 10.3324/haematol.2019.221176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Salameh A et al. , “PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3,” Proc. Natl. Acad. Sci, vol. 112, no. 27, pp. 8403–8408, jul. 2015, doi: 10.1073/pnas.1507882112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Li X et al. , “Double-stranded RNA-specific adenosine deaminase-knockdown inhibits the proliferation and induces apoptosis of DU145 and PC3 cells by promoting the phosphorylation of H2A.X variant histone,” Oncol. Lett, vol. 22, no. 5, p. 764, Nov. 2021, doi: 10.3892/ol.2021.13025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Tassinari V et al. , “ADAR1 is a new target of METTL3 and plays a pro-oncogenic role in glioblastoma by an editing-independent mechanism,” Genome Biol., vol. 22, no. 1, p. 51, Jan. 2021, doi: 10.1186/s13059-021-02271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Yang B et al. , “PTBP1 induces ADAR1 p110 isoform expression through IRES-like dependent translation control and influences cell proliferation in gliomas,” Cell. Mol. Life Sci, vol. 72, no. 22, pp. 4383–4397, Nov. 2015, doi: 10.1007/s00018-015-1938-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Yujie Ding MM, Shi X, Ji J, and Su Y, “ADAR1p150 regulates the biosynthesis and function of miRNA-149* in human melanoma,” Biochem. Biophys. Res. Commun, vol. 523, no. 4, pp. 900–907, Mar. 2020, doi: 10.1016/j.bbrc.2019.12.110. [DOI] [PubMed] [Google Scholar]
- [95].Shoshan E et al. , “Reduced adenosine-to-inosine miR-455–5p editing promotes melanoma growth and metastasis,” Nat. Cell Biol, vol. 17, no. 3, pp. 311–321, Mar. 2015, doi: 10.1038/ncb3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Nemlich Y, Besser MJ, Schachter J, and Markel G, “ADAR1 regulates melanoma cell invasiveness by controlling beta3-integrin via microRNA-30 family members,” Am. J. Cancer Res, vol. 10, no. 8, pp. 2677–2686, Aug. 2020. [PMC free article] [PubMed] [Google Scholar]
- [97].Galore-Haskel G et al. , “A novel immune resistance mechanism of melanoma cells controlled by the ADAR1 enzyme,” Oncotarget, vol. 6, no. 30, pp. 28999–29015, Aug. 2015, doi: 10.18632/oncotarget.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Velazquez-Torres G, et al. , “A-to-I miR-378a-3p editing can prevent melanoma progression via regulation of PARVA expression,” Nat. Commun, vol. 9, no. 1, p. 461, Jan. 2018, doi: 10.1038/s41467-018-02851-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Nemlich Y et al. , “ADAR1-mediated regulation of melanoma invasion,” Nat. Commun, vol. 9, no. 1, Dec. 2018, doi: 10.1038/s41467-018-04600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Gumireddy K et al. , “The mRNA-edited form of GABRA3 suppresses GABRA3-mediated Akt activation and breast cancer metastasis,” Nat. Commun, vol. 7, p. 10715, Feb. 2016, doi: 10.1038/ncomms10715. [DOI] [PMC free article] [PubMed] [Google Scholar]