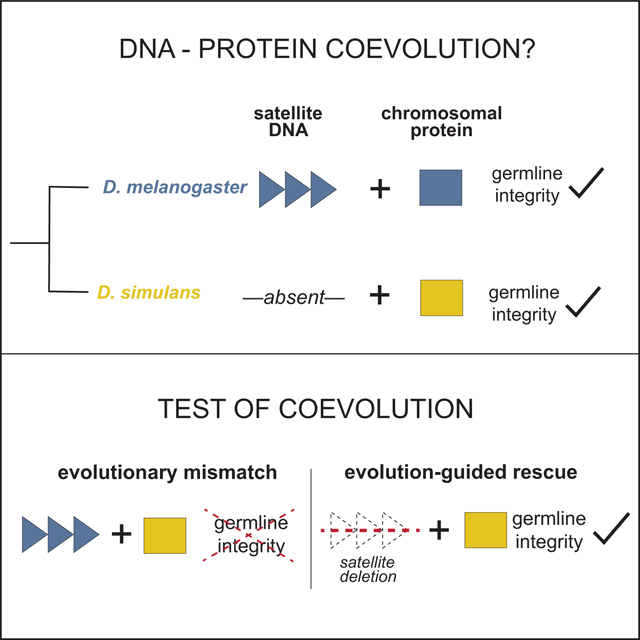
SUMMARY
Satellite DNA spans megabases of eukaryotic sequence and evolves rapidly.1–6 Paradoxically, satellite-rich genomic regions mediate strictly conserved, essential processes like chromosome segregation and nuclear structure.7–10 A leading resolution to this paradox posits that satellite DNA and satellite-associated chromosomal proteins coevolve to preserve these essential functions.11 We experimentally test this model of intra-genomic coevolution by conducting the first evolution-guided manipulation of both chromosomal protein and DNA satellite. The 359bp satellite spans an 11Mb array in Drosophila melanogaster that is absent from its sister species, Drosophila simulans.12–14 This species-specific DNA satellite colocalizes with the adaptively evolving, ovary-enriched protein, Maternal Haploid (MH)–the Drosophila homolog of Spartan.15 To determine if MH and 359bp coevolve, we swapped the D. simulans version of MH (“MH[sim]”) into D. melanogaster. MH[sim] triggers ovarian cell death, reduced ovary size, and loss of mature eggs. Surprisingly, the D. melanogaster mh null mutant has no such ovary phenotypes15, suggesting that MH[sim] is toxic in a D. melanogaster background. Using both cell biology and genetics, we discovered that MH[sim] poisons oogenesis through a DNA damage pathway. Remarkably, deleting the D. melanogaster-specific 359bp satellite array completely restores mh[sim] germline genome integrity and fertility, consistent with a history of coevolution between these two fast-evolving loci. Germline genome integrity and fertility are also restored by overexpressing Topoisomerase II (Top2), suggesting that MH[sim] interferes with Top2-mediated processing of 359bp. The observed 359bp-MH[sim] cross-species incompatibility supports a model under which seemingly inert repetitive DNA and essential chromosomal proteins must coevolve to preserve germline genome integrity.
Keywords: DNA satellite, coevolution, maternal haploid, 359bp, Spartan, Topoisomerase II
Graphical Abstract
eTOC BLURB
Rapid evolution of DNA repeats is thought to trigger rapid evolution of proteins that package and process DNA repeats. Brand and Levine genetically manipulate both protein and DNA satellite to define the molecular players engaged in this intra-genomic coevolution and to reveal the chromosome biology preserved by this coevolution.
RESULTS AND DISCUSSION
DNA satellite-enriched genomic regions evolve rapidly and yet support strictly conserved nuclear functions, including chromosome segregation, chromosome tethering, and telomere integrity.1–10 A classic resolution to this paradox posits that DNA satellite-associated proteins evolve adaptively to mitigate deleterious proliferation of DNA satellite sequence variants.11 Repeated bouts of DNA satellite evolution and chromosomal protein adaptation result in exquisitely coevolved satellites and satellite-associated proteins. This model of coevolution predicts pervasive incompatibilities between satellite DNA and chromosomal proteins from closely related species: adaptively evolving chromosomal proteins from one species should fail to package or process DNA satellites from another.11,16,17
Evidence for this coevolution model has emerged from engineering “evolutionary mismatches” between the adaptively evolving chromosomal protein(s) of one species and the DNA satellite landscape of a close relative. Under one approach, a diverged chromosomal protein is introduced into a closely related species, generating an evolutionary mismatch between the manipulated protein and one or more DNA satellites.17–20 Consistent with disrupted DNA satellite:chromosomal protein coevolution, the naïve protein typically perturbs a satellite-mediated function, such as chromosome segregation or nuclear organization.17,19,20 In these cases, however, the incompatible DNA satellites are unknown. A second approach crosses sister species to generate evolutionary mismatches between chromosomal proteins and DNA satellites in hybrid progeny. Consistent with disrupted DNA satellite:chromosomal protein coevolution, interspecies hybrid inviability has been linked to satellite-rich genomic loci.21,22 In these systems, however, the incompatible chromosomal proteins are unknown. To date, there are no cases of experimental identification of both chromosomal protein and satellite engaged in coevolution.
To experimentally probe both sides of the coevolution model, we searched for a rapidly evolving DNA satellite associated with an adaptively evolving chromosomal protein. In Drosophila melanogaster, the 359bp satellite spans an 11Mb array at the base of the X chromosome.12,13 Close relatives of D. melanogaster, including D. simulans and D. erecta, lack this X-linked satellite array.14 Instead, these species have shorter arrays of “359bp-like” sequence dispersed throughout heterochromatin and euchromatin.23–25 Such extreme lineage-restriction to D. melanogaster makes this DNA satellite array an ideal locus for testing the coevolution model.
On the protein side, we identified from the literature Maternal Haploid (MH), an ovary-enriched protein that is maternally provisioned to the embryo, colocalizes with the 359bp satellite, and supports genome integrity.15,26,27 Embryos of mh null mothers suffer paternal chromosome mis-segregation at the very first mitosis, suggesting that the maternally-provisioned MH prepares the otherwise inert, sperm-deposited paternal chromosomes for participation in embryonic mitosis.15,26 Most of these embryos arrest around the first mitotic division. A smaller fraction develop beyond the first division, cycling only the “maternal haploid” complement of chromosomes until arrest prior to hatching.15,26,28 The mechanism by which MH primes paternal chromosomes for embryonic mitosis is not known; however, the human homolog of MH, called Spartan, is a well-characterized protease.29–31 Spartan resolves DNA-protein crosslinks that block DNA replication, chromatin remodeling, and DNA repair.30,32 In Drosophila, MH may play an analogous role in DNA-protein crosslink resolution during paternal chromosome processing.
If the D. melanogaster-specific 359bp proliferation triggered mh to innovate, we should detect evidence of positive selection at mh between D. melanogaster and D. simulans. To determine if mh evolves adaptively, we conducted a McDonald-Kreitman test33 using polymorphism within D. melanogaster and D. simulans populations and divergence between D. melanogaster and D. simulans (2.5 million years diverged34). This comparison revealed an excess of nonsynonymous fixations, consistent with a history of adaptive evolution (Figure 1A, Table S1). The dynamic evolution of the 359bp satellite and adaptive evolution of a 359bp-associated protein, MH, raises the possibility that mh recurrently evolves to preserve a biological function compromised by 359bp satellite proliferation.
Figure 1. MH evolves adaptively to preserve female fertility.
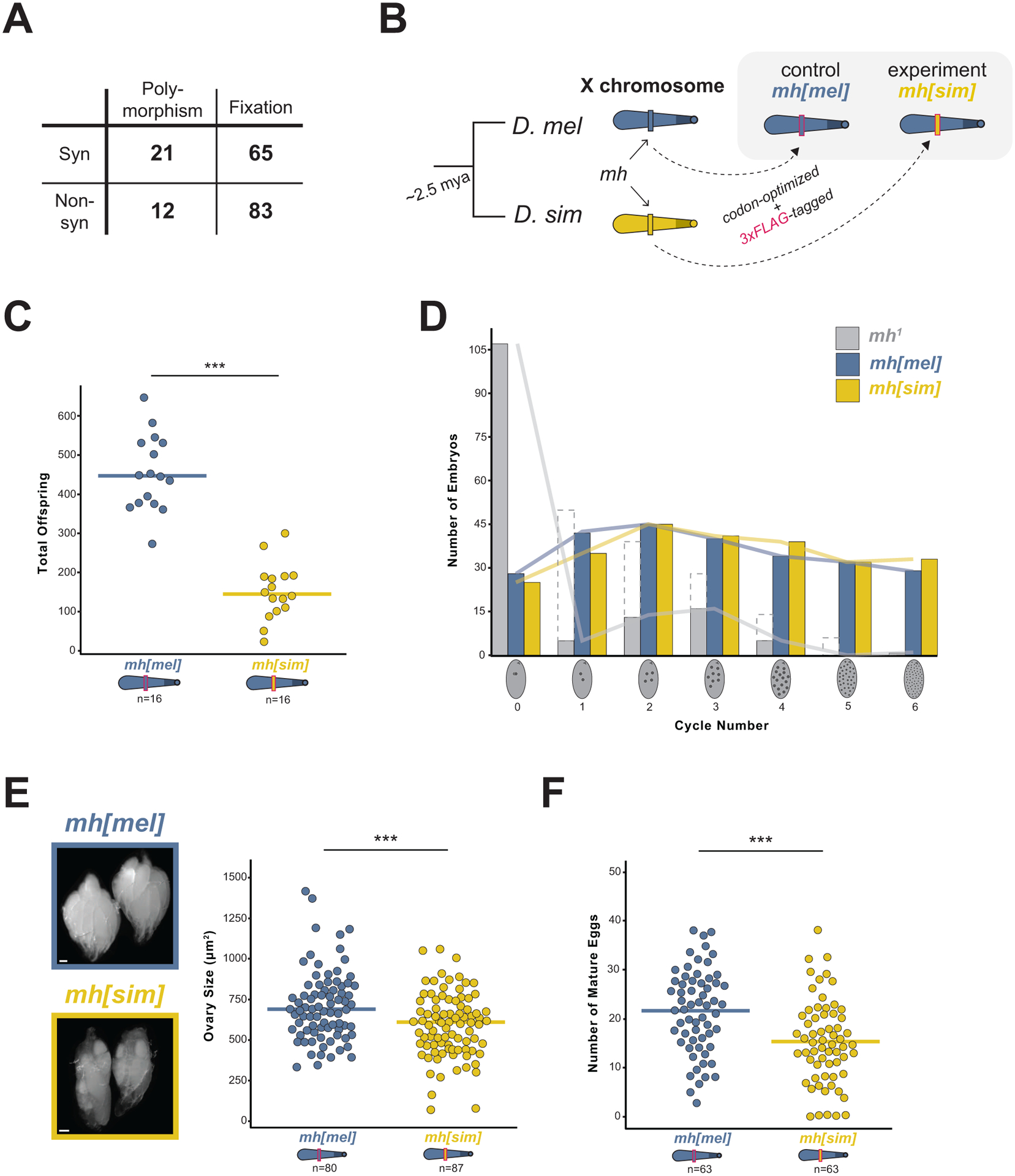
(A) Counts of synonymous and nonsynonymous polymorphic and fixed sites within and between D. melanogaster and D. simulans; χ2 test, p = 0.04. (B) Swap strategy: the D. melanogaster (“mel”, blue) or D. simulans (“sim,” yellow) mh coding sequence, codon-optimized for D. melanogaster and 3xFLAG-tagged, replaced the native mh gene on the X chromosome. (C) Total offspring from mh[mel] or mh[sim] females crossed to wildtype (w1118) males. (D) Frequency distribution of embryos at increasing mitotic cycle numbers collected for 70 minutes from mh1, mh[mel], and mh[sim] females. Dashed bars from mh1 females correspond to embryos undergoing mitotic catastrophe likely triggered at the first mitosis. Solid gray bars representing mh1-derived embryos greater than cycle 0 are presumed maternal haploid. (E) Representative images and ovary size estimates from mh[mel] and mh[sim] females. (F) Number of mature eggs per ovary pair from mh[mel] and mh[sim] females. (t-test: “***” = p < 0.001, scale bar = 100μm)
To test the possibility of MH:359bp coevolution, we first conducted an evolution-guided manipulation of mh to generate an “evolutionary mismatch” between protein and satellite. We used CRISPR/Cas9 to integrate into the native mh locus of D. melanogaster either a 3xFLAG-tagged mh coding sequence from D. melanogaster (our control fly, “mh[mel]”) or a 3xFLAG-tagged mh coding sequence from D. simulans (our experimental fly, “mh[sim]”, Figure 1B). Both the D. melanogaster and the D. simulans coding sequences were codon-optimized for D. melanogaster. We observed equivalent expression of the two transgenes (Figure S1A).
The mh null mutant phenotype in the early embryo motivated our prediction that an evolutionary mismatch between the D. simulans mh and the D. melanogaster 359bp X-linked array would disrupt the first mitotic division. We reasoned that in its native D. simulans background, MH[sim] efficiently processes all D. simulans paternal chromosomes. In a D. melanogaster background, we predicted that MH[sim] would process most D. melanogaster chromosomes but fail to recognize and process the D. melanogaster-specific 359bp array, triggering mis-segregation of the paternal X-chromosome. This defect would result in reduced female fertility and a dearth of female progeny. We discovered that mh[sim] females produced significantly fewer progeny than control mh[mel] females (Figure 1C); however, contrary to our prediction, the progeny sex ratio did not deviate from 50/50 (Figure S1B). These data suggest that paternal 359bp is not uniquely vulnerable to the presence of MH[sim] during the first mitotic division. Consistent with this inference, we observed that mh[sim] completely rescues the first mitotic division: embryos from mh[mel] and mh[sim] mothers show equivalent, normal distributions of embryonic stages from a 70-minute collection (Figure 1D). In contrast, embryos produced by mh null mothers typically arrest during the first division (Figure 1D). Moreover, we observed no evidence of elevated maternal haploid embryos from mh[sim] mothers (Figure S1C). These data suggest that mh[sim] does not phenocopy the mh null early embryonic phenotype.
To uncover an alternative source of the mh[sim] fertility defect, we looked at the developmental stage just before the first embryonic mitosis: oogenesis. Although mh is highly expressed during oogenesis, previous reports suggested that mh null mutation alone yields no ovary phenotype.15,26 We similarly detected no difference in ovary size or mature egg number of mh null mothers compared to heterozygous controls (Figure S1D,E). In contrast, mh[sim] ovaries are significantly smaller than mh[mel] ovaries and are depleted of the most mature egg stages (Figure 1E,F). This unexpected mh[sim] ovary phenotype, combined with the complete rescue of the first embryonic division by mh[sim], suggests that mh[sim] does not behave as a loss-of-function allele. Instead, MH[sim] might be toxic.
To explore the possibility that MH[sim] is toxic, we first asked if MH[sim] localizes aberrantly in the ovary. We visualized MH[mel] and MH[sim] by staining ovaries with anti-FLAG. We discovered that MH[mel] localized primarily in the earliest stages of oogenesis (the germarium, Figure 2A,B). MH[sim] localized in these cell types as well as on the nurse cell nuclei of later stage egg chambers (Figure 2A,B). The aberrant persistence of MH[sim] during oogenesis, combined with compromised mh[sim] ovary development, raised the possibility that MH mislocalization alone might be toxic. To test this hypothesis, we used the UAS/GAL4 system to overexpress MH[mel] in the female germline (driver nos-Gal4-VP16). In ovaries overexpressing MH[mel], we indeed observed elevated levels and aberrant localization of the protein in later stage egg chambers (Figure S2A). Nevertheless, these females gave rise to abundant progeny (Figure S2B), suggesting that mislocalization alone cannot explain the compromised ovary development of mh[sim] females. In contrast, overexpression of MH[sim] resulted in an absence of mature eggs (Figures S2C,D). Consequently, these females were completely sterile (Figure S2B). These data suggest that MH[sim] – which functions normally in its native D. simulans genome – is toxic to oogenesis in D. melanogaster. This toxicity appears to be dose-dependent: heterozygous mh[mel]/mh[sim] females give rise to progeny counts similar to mh[mel] homozygotes (Figure S2E).
Figure 2. MH[sim] poisons oogenesis through a DNA damage pathway.
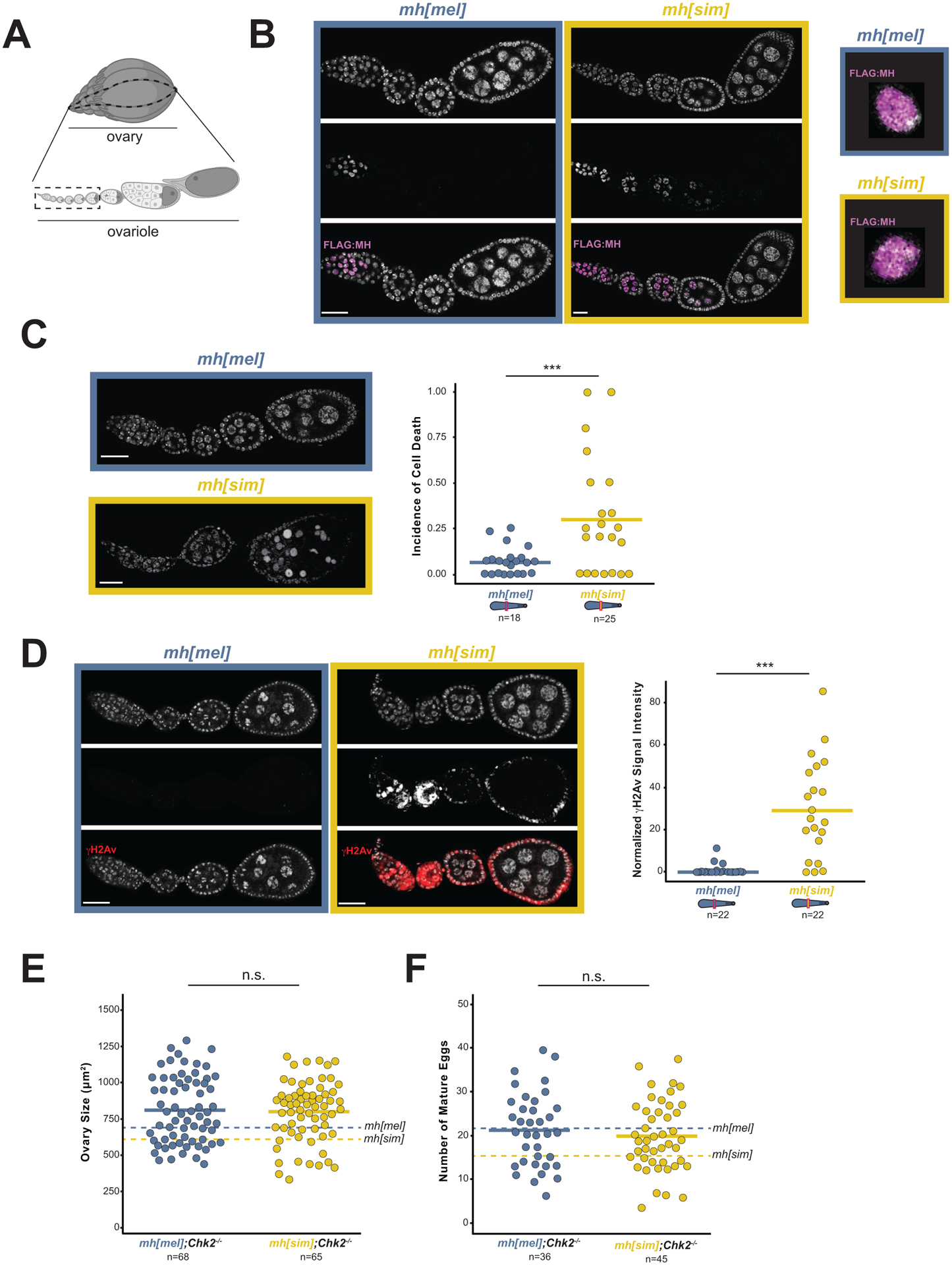
(A) Diagram of a Drosophila ovary (above) and a single ovariole (below) with the germline stem cells in the germarium at the anterior (left) position and the mature eggs at the posterior position (right). The dashed box shows the developmental stages shown in images 2B-2D (Created using BioRender.com). (B) mh[mel] and mh[sim] ovaries stained with anti-FLAG to visualize MH localization (left). Merged images of single nuclei from the germarium (*) show no MH foci on the DNA (right). (C) Incidence of cell death captured by the fraction of ovarioles with condensed nuclei (arrowheads) in mh[mel] and mh[sim] ovaries. (D) γH2Av signal in mh[mel] and mh[sim] ovaries and the quantification of normalized fluorescent signal intensity. Note that the expected γH2Av-positive cells in the germarium in mh[mel] are absent under the imaging parameters used but are indeed present, see Figure S3A. (E) Ovary size estimates from mh[mel]; Chk2−/− and mh[sim]; Chk2−/− females. (F) Number of mature eggs per ovary pair from mh[mel]; Chk2−/− and mh[sim]; Chk2−/− females. In panels E and F, dotted lines correspond to mh[mel] and mh[sim] averages reported in Figure 1E and 1F, respectively. (t-test: “***” = p < 0.001, “n.s.” p > 0.05, scale bar = 25μm)
To study the cell biological basis of this block to oogenesis, we turned back to ovaries of females expressing the CRISPR-introduced mh[mel] or mh[sim] transgene under the native promoter (Figure 1B). We observed an excess of hyper-condensed nuclei in mh[sim] ovaries, consistent with elevated cell death (Figure 2C35,36). A classic trigger of cell death is the accumulation of DNA damage.37 To visualize DNA damage, we stained mh[mel] and mh[sim] ovaries for the double-strand break marker, γH2Av.38,39 We observed elevated DNA damage signaling in mh[sim] ovaries (Figure 2D, S3A). This phenotype further distinguishes mh[sim] from mh null ovaries – mh null ovaries show no evidence of elevated DNA damage (Figure S3B). To address the hypothesis that MH[sim] compromises oogenesis through a DNA repair pathway, we combined mh[sim] with a null mutation in a DNA damage checkpoint gene. The gene, Chk2 (also known as mnk), normally blocks egg production in the presence of DNA damage.40,41 Chk2−/− ovaries bypass this checkpoint, allowing a female to make mature but damaged eggs in the presence of elevated DNA damage. We discovered that Chk2−/− restores mh[sim] ovaries to mh[mel]-like ovary size and mh[mel]-like egg production (Figure 2E,F). However, the mh[sim];Chk2−/− females are sterile while mh[mel]];Chk−/− females retain fertility (Figure S3C). These data suggest that MH[sim] compromises oogenesis by triggering DNA damage.
Applying these phenotypic data to the coevolution model, we hypothesized that MH[sim]-induced DNA damage depends on the 11Mb array of 359bp satellite in D. melanogaster. Under this model, MH[sim]-specific residues are incompatible with 359bp. Removing 359bp should restore germline genome integrity and fertility of mh[sim] females. To directly test this prediction, we took advantage of a fly strain that lacks the 11Mb array of X-linked 359bp satellite (Figure 3A42). We recombined this 359bp deletion, called Zygotic hybrid rescue (Zhr) onto both the mh[mel] and the mh[sim] X chromosomes (Figure 3B). If MH[sim]-induced toxicity depends on the presence of the 359bp expansion, mh[sim],Zhr females should have minimal DNA damage and recover fertility. Remarkably, the 359bp deletion completely restores the DNA damage marker, γH2Av, to wildtype (low) levels (Figure 3C). Consistent with restored germline genome integrity of mh[sim] females, we observed no difference in ovary size and no difference in egg production between mh[mel] and mh[sim] females that lack 359bp (Figure 3D,E). Finally, the 359bp deletion completely restores mh[sim] fertility to mh[mel] levels (Figure 3F). These data reveal that MH[sim] toxicity depends on 359bp, consistent with a history of coevolution between these two fast-evolving components of the Drosophila genome.
Figure 3. The 359bp satellite deletion rescues mh[sim] genome integrity and fertility.
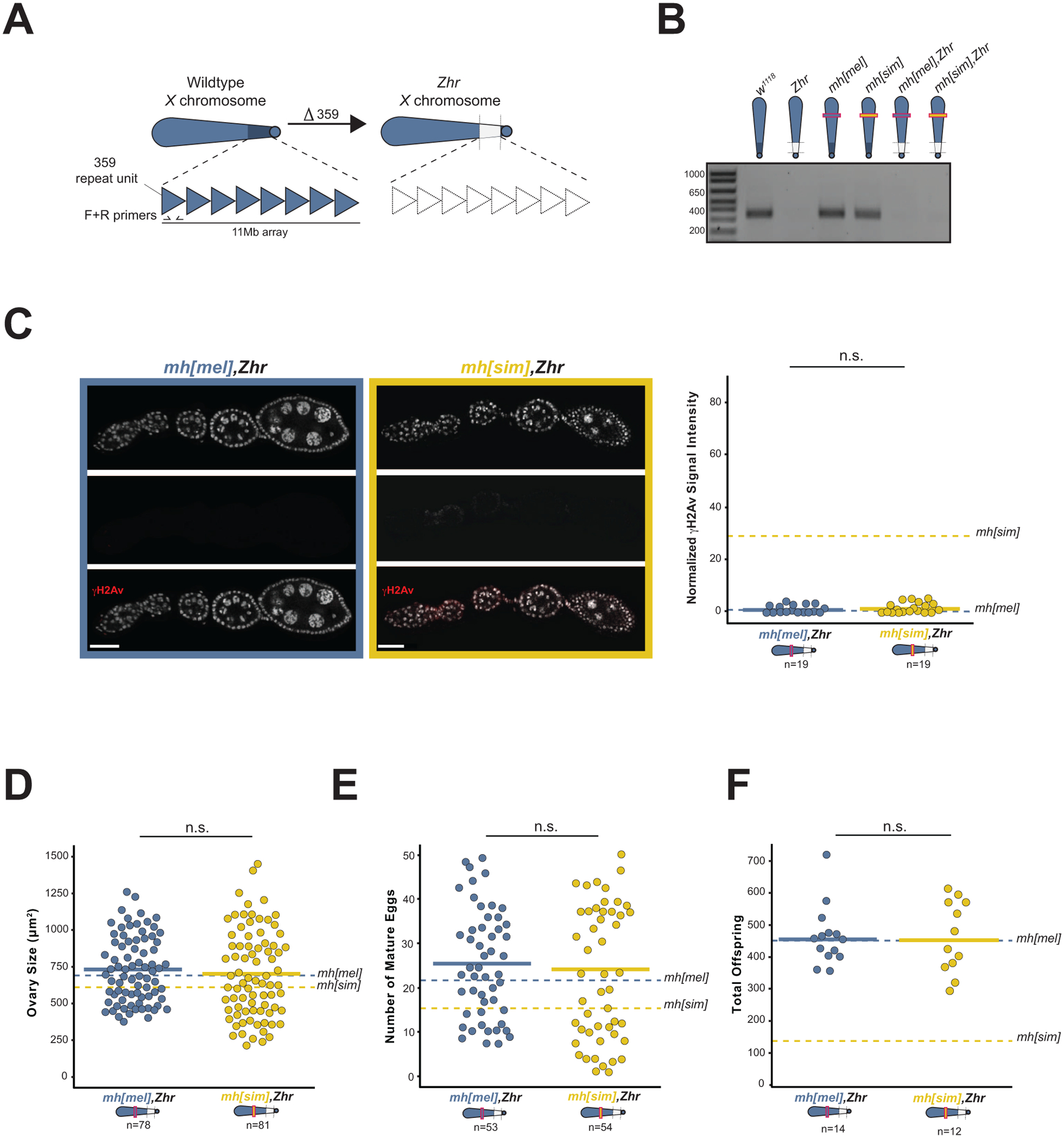
(A) The Zhr X chromosome lacks the 11Mb pericentromeric 359bp satellite array. (B) A 10-cycle PCR distinguishes between wildtype 359bp copy number and the 359bp deletion (Zhr) and validates the recombined mh[mel],Zhr and recombined mh[sim],Zhr X chromosomes. (C) γH2Av signal in mh[mel],Zhr and mh[sim],Zhr ovaries and the quantification of normalized fluorescent signal intensity. Dotted lines correspond to mh[mel] and mh[sim] averages reported in Figure 2D. (D) Ovary size of mh[mel],Zhr and mh[sim],Zhr females. (E) Number of mature eggs per ovary pair from mh[mel],Zhr and mh[sim],Zhr females. (F) Progeny counts from mh[mel],Zhr and mh[sim],Zhr females crossed to wildtype (w1118) males. In panels D, E, and F, dotted lines correspond to mh[mel] and mh[sim] averages reported in Figure 1E, 1F, and 1C, respectively. (t-test: “n.s.” p > 0.05, scale bar = 25μm)
The observed 359bp-dependent toxicity, rather than loss-of-function, suggests that MH[sim] may interfere with the preservation of 359bp integrity. To define a molecular basis for this interference, we used well-characterized MH homologs as guides. The MH homologs in worm (DVC-1) and human (Spartan) use the conserved Spartan metalloprotease domain to repair DNA-protein crosslinks. A major substrate of Spartan/DVC-1-directed repair is Topoisomerase II (Top2).31,43,44 Top2 transiently crosslinks with DNA as it resolves torsional stress and DNA entanglements. Spartan/DVC-1 degrades Top2 when these crosslinks become irreversible and threaten various DNA transactions, including DNA replication, chromatin remodeling, and repair.45–47 In D. melanogaster, Top2 specifically cleaves 359bp48 and resolves DNA entanglements involving 359bp during female meiosis.49 Moreover, Top2 and mh genetically interact in the ovary and colocalize in the embryo.15 We hypothesized that MH[sim] interferes with Top2 resolution of 359bp entanglements during oogenesis.
This interference model predicts that Top2 is limiting in the presence of MH[sim]. To test this prediction, we reduced Top2 using a heterozygous loss-of-function mutant and overexpressed Top2 in the ovary using the UAS/GAL4 system in an mh[mel] or mh[sim] background. Reduction of Top2 exacerbates mh[sim]-dependent subfertility (Figure 4A) while Top2 overexpression in the ovary completely rescues mh[sim] fertility (Figure 4B). The rescued mh[sim] ovaries also showed restored genome integrity (Figure 4C). Excess Top2 appears to mitigate MH[sim] interference. Combined with the literature on DNA-Top2 crosslink resolution by MH homologs DVC-1 and Spartan, these data raise the possibility that MH[sim] over-actively clears Top2:359bp associations that otherwise resolve 359bp entanglements in the female germline. Persistent DNA entanglements would trigger the observed DNA damage that blocks oogenesis progression (Figure 4D). Intriguingly, a sliding window analysis of D. melanogaster-specific mh evolution revealed a striking enrichment of elevated dN/dS in multiple regions of the C-terminus (Figure S4A). The C-terminus of Spartan mediates both its recruitment to chromatin50,51 and its self-cleavage activity.30,52 Lineage-specific evolution of either recruitment to chromatin or autoregulation could modulate MH activity at 359bp.
Figure 4. MH[sim] may interfere with Top2 processing of 359bp entanglements.
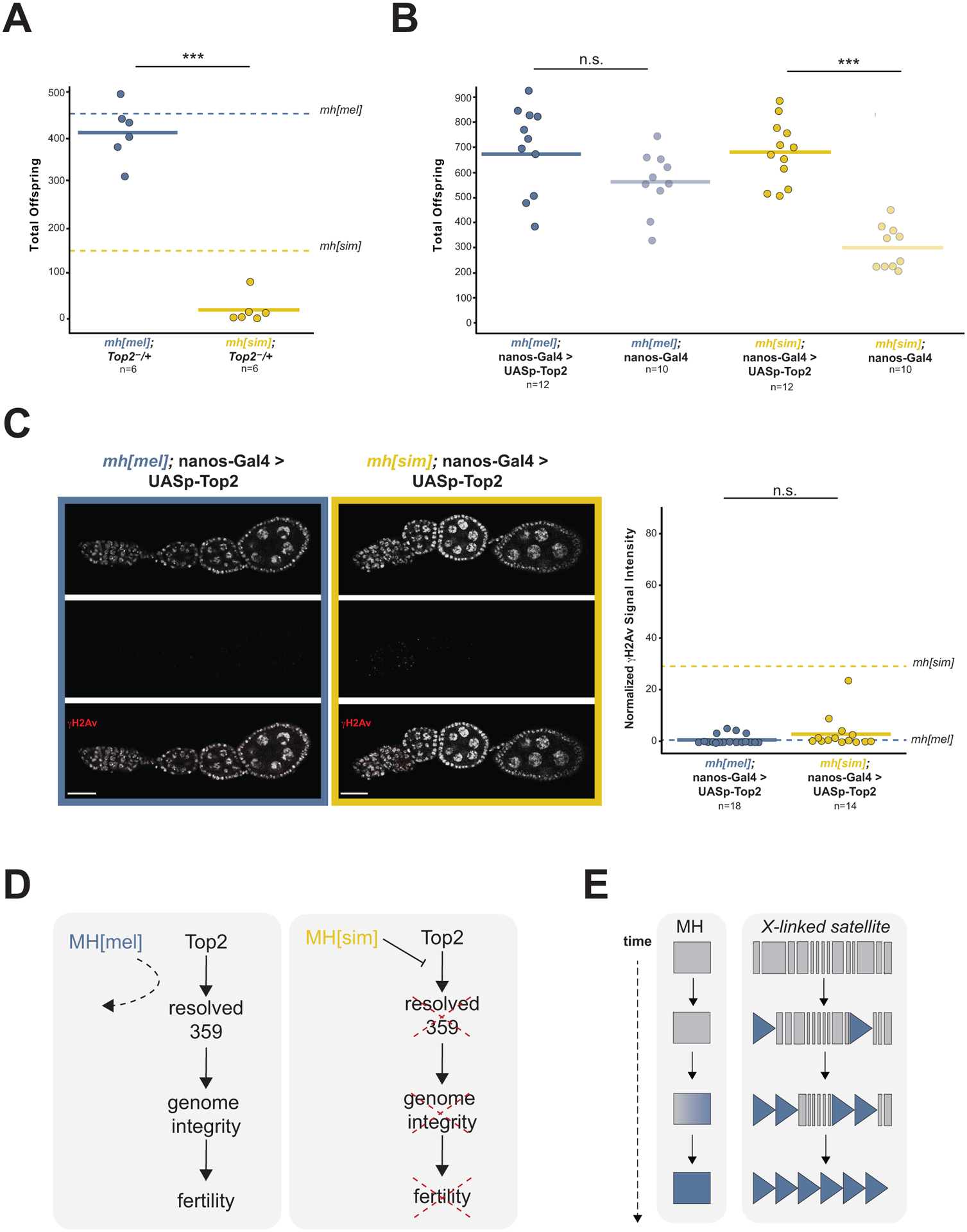
(A) Progeny counts from mh[mel]; Top2−/+ and mh[sim]; Top2−/+ females crossed to wildtype (w1118) males. Dotted lines correspond to mh[mel] and mh[sim] averages reported in Figure 1C. (B) Progeny counts from nos-Gal4-VP16 (female germline GAL4) driven mh[mel]; UASp-Top2 or mh[sim]; UASp-Top2 females crossed to wildtype (w1118) males. (C) γH2Av signal from ovaries of nos-Gal4-VP16 driven mh[mel]; UASp-Top2 or mh[sim]; UASp-Top2 females and quantification of normalized fluorescent signal intensity. Dotted lines correspond to mh[mel] and mh[sim] averages reported in Figure 2D. (D) Model of MH[sim] interference with Top2 processing of 359bp entanglements. These entanglements threaten genome integrity and ultimately, fertility. MH[mel], in contrast, has no measurable function in the ovaries, suggesting that it avoids interfering with 359bp processing by Top2. (E) Model of MH evolution tracking 359bp satellite proliferation. (t-test: “***” = p < 0.001, “n.s.” p > 0.05, scale bar = 25μm)
Our model is motivated in part by the observation that repeat-rich genomic regions, and especially the 11Mb array of 359bp, is uniquely vulnerable to DNA entanglements.21,49,53 If 359bp is so deleterious, how could it have proliferated? DNA satellites can behave selfishly, gaining a transmission advantage from one generation to the next.54,55 We suspect that such non-Mendelian segregation led to 359bp proliferation, triggering MH to evolve adaptively along the D. melanogaster lineage (Figure 4E). We lack sufficient power to detect such lineage-specific adaptive evolution under a McDonald-Kreitman test framework (Table S1); however, a sliding window dN/dS analysis between the reconstructed ancestral mh sequence and either mh[mel] or mh[sim] revealed a highly significant enrichment of codons with elevated dN/dS along the D. melanogaster branch (FET, p < 0.0001, Figure S4A, Table S2). This finding is consistent with 359bp proliferation leading to positive selection on mh. However, we cannot formally rule out the possibility that a selection pressure distinct from 359bp proliferation shaped mh[mel] adaptive evolution. Under this alternative model, the D. melanogaster version of MH evolved first, releasing constraint on 359bp copy number. Most likely, both selection and loss of constraint operate cyclically.
Regardless of the force(s) that promoted 359bp proliferation, the 359bp:MH system offers two important elaborations of the classic model of intra-genomic coevolution.11,56–58 This canonical model posits that chromosomal proteins evolve adaptively to recognize and process novel satellite repeat variants. Under this model, the mismatched mh[sim] allele should fail to perform an mh function; that is, act as a loss-of-function allele. Instead, we demonstrate that mh[sim] is toxic, suggesting that mh[mel] evolved adaptively to avoid interfering with 359bp processing. The canonical coevolution model also envisioned coevolution sculpting specifically a DNA-protein interface. However, MH lacks a sequence-specific DNA binding domain26,51, rejecting the possibility that MH evolves to reduce 359bp sequence recognition. MH adaptive evolution instead likely tracks Top2, and more specifically, Top2-359bp crosslinks. This speculative model suggests that 359bp:MH coevolution is indirect: MH tracks Top2 evolution and Top2 evolution tracks 359bp evolution. Under this model, Top2 should evolve adaptively. To test this possibility, we implemented a McDonald-Kreitman test on Top2 alleles from D. melanogaster and D. simulans. We discovered that Top2 indeed evolves adaptively between these sister species (Figure S4B). Future research will test this model of 359bp-triggered evolution of the protein:protein interaction interface between MH and Top2.
359bp-mediated toxicity to oogenesis highlights the catastrophic functional consequences of DNA satellite evolution. Importantly, 359bp-mediated toxicity is also apparent in D. melanogaster-D. simulans hybrid embryos: a distinct, unmapped gene on D. simulans chromosome 259–62 interacts deleteriously with 359bp to cause embryonic chromosome mis-segregation, genome instability, and lethality.21,42,63 This interspecies hybrid dysfunction in the embryo, together with the 359bp:mh[sim] toxicity in the ovary reported here, suggests that recurrent bouts of coevolution not only shape essential genome functions within species but also can trigger hybrid incompatibilities between species.
STAR METHODS
(see separate Word document for STAR METHODS Table)
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Monoclonal anti-FLAG M2 | Sigma-Aldrich | Cat#F3165 |
| Mouse Monoclonal 12G10 anti-αTubulin | Developmental Studies Hybridoma Bank | Cat#12G10 anti-alpha-tubulin, RRID:AB_1157911 |
| Mouse Monocolonal anti-Histone 2A Gamma Variant, Phosphorylated | Developmental Studies Hybridoma Bank | Cat#UNC93–5.2.1, RRID:AB_261807 |
| Chicken Polyconal anti-Green Fluorescent Protein | Aves Labs | Cat# GFP-1010, RRID:AB_230731 |
| Chemicals, peptides, and recombinant proteins | ||
| Phusion High-Fidelity DNA Polymerase | NEB | Cat#M0530 |
| RIPA buffer | Cell Signaling Technology | Cat#9806 |
| Protease Inhibitor Cocktail | Roche | Cat#11873580001 |
| PMSF | Cell Signaling Technology | Cat#8553 |
| Benzonase | Sigma-Aldrich | Cat#E1014 |
| ProLong Gold Antifade Mountant with DAPI | ThermoFisher Scientific | Cat#P36931 |
| Critical commercial assays | ||
| APAgene GOLD Genome Walking Kit | Bio S&T, Inc | BT901-RT |
| Kwikquant Western Blot Detection Kit | Kindle Biosciences | Cat#R1004 |
| Deposited data | ||
| mh alleles from D. simulans | This manuscript | OL546458..64 |
| Top2 alleles from D. simulans | This manuscript | OL156853..59 |
| Experimental models: Organisms/strains | ||
| yw; nos-Cas9(II-attP40) | Best Gene, Inc | N/A |
| yw; PBac[y + -attP-9A]VK00018 | Bloomington Drosophila Stock Center | BDSC:9736 |
| Gal4::VP16-nos | Bloomington Drosophila Stock Center | BDSC:64277 |
| Zhr 1 | Bloomington Drosophila Stock Center | BDSC:25140 |
| mnk l6 | N. Phadnis | N/A |
| Top2 17−6 | P. Geyer | N/A |
| w 1118 | MTL laboratory | N/A |
| mh 1 | Bloomington Drosophila Stock Center | BDSC:7130 |
| P{gcid.EGFP.cidIII}.2 | K. McKim, Schuh et al.75 | N/A |
| Oligonucleotides | ||
| Primers to amplify and sequence mh alleles from D. simulans, see Table S3 | This manuscript | N/A |
| Primers to screen mh CRISPR transformants, see Table S3 | This manuscript | N/A |
| Primers to screen for PCR-introduced mutations in the cloned mh UAS constructs, see Table S3 | This manuscript | N/A |
| Primers to detect native mh locus, see Table S3 | This manuscript | N/A |
| Primers to screen for 359-bp deletion (Zhr), see Table S3 | Rosic et al.9 | N/A |
| Primers for Genome Walking, see Table S3 | This manuscript | N/A |
| Primers to amplify and sequence Top2 alleles from D. simulans, see Table S3 | This manuscript | N/A |
| Recombinant DNA | ||
| Plasmid: pBFv-U6.2 | Addgene | Addgene#138400 |
| Plasmid: pBFv-U6.2B | Addgene | Addgene#138401 |
| Plasmid: 3xFLAG:mh[mel] and 3xFLAG:mh[sim] HDR plasmids | This manuscript | N/A |
| Plasmid: pUASp-attB | Drosophila Genomics Resource Center | DGRC#1358 |
| Software and algorithms | ||
| Geneious v 11.1.5 | Biomatters | https://www.geneious.com/ |
| FIJI | ImageJ2 | https://imagej.net/software/fiji/ |
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mia Levine (m.levine@sas.upenn.edu).
Materials availability
All reagents generated in this study are available upon request to the lead contact.
Data availability
The mh and Top2 alleles from D. simulans have been deposited in GenBank and are publicly available as of the date of this publication. Accession numbers are listed in the key resources table. All other data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
We maintained Drosophila melanogaster stocks on standard cornmeal food at 24°C. Fly stocks used in this study are listed in the key resources table.
METHOD DETAILS
Population genetic and molecular evolution analyses
We conducted population genetic analysis of mh using multiple alleles from both D. melanogaster and D. simulans. We obtained nine D. melanogaster mh alleles (coordinates X:15472804-15475400, dmel r6.4) from lines collected in Lyon, France.64 We amplified seven D. simulans mh alleles from lines collected in Nairobi, Kenya (Accession OL546458-OL546464).65 Importantly, a duplication event occurred along the D. simulans lineage, resulting in a full-length copy of the mh ortholog, and a tandem partial duplicate.66,67 To specifically amplify the full length mh ortholog, we designed primers that anneal to unique genomic sequence only (Table S3). We then prepared genomic DNA and conducted PCR amplification followed by Sanger sequencing using standard protocols. We aligned the sequences in Geneious using the Geneious Alignment algorithm with default settings (Geneious v11.1.5, Biomatters, Auckland, New Zealand) and confirmed alignment quality by eye. We performed a McDonald-Kreitman test33 with the D. melanogaster and D. simulans mh coding sequences. We performed lineage-specific McDonald-Kreitman tests with the D. yakuba mh coding sequence as an outgroup to polarize mutations along the D. melanogaster and D. simulans linages.
Similarly, we conducted population genetic analysis of Top2 using multiple alleles from both D. melanogaster and D. simulans. We obtained nine D. melanogaster Top2 alleles (coordinates 2L:19447365-19453490, dmel r6.4) from lines collected in Lyon, France.64 We amplified and Sanger sequenced seven D. simulans Top2 alleles from lines collected in Nairobi, Kenya (Accession OL156853-OL156859).65 As described above, we aligned the sequences in Geneious, confirmed alignment quality by eye, and performed a McDonald-Kreitman test.33
We calculated pairwise dN/dS between D. melanogaster and D. simulans mh alleles using a window size of 200bp (step size = 20bp) in the software package, DnaSP.68 We reconstructed the ancestral mh sequence of D. melanogaster and D. simulans using the codeML package in PAML69,70 and repeated this pairwise calculation between the extant D. melanogaster and D. simulans mh alleles and the reconstructed ancestral mh allele.
Fly stock construction
Constructing gene swaps
We used CRISPR/Cas9 to generate D. melanogaster flies that encode a transgenic D. melanogaster allele or a D. simulans allele of mh, integrated into the native location. We first generated a U6 promoter-driven guide RNA construct by cloning sgRNAs flanking the coding sequence of mh (5’: GGATTGGCCCAGGATCAACA, 3’: CGTGGAGAGCTTCTGCCGCG) into pBFv-U6.2 and pBFv-U6.2B backbones. We shuttled the 3’ sgRNA into pBFv-U6.2 to create a dual sgRNA vector (University of Utah Mutagenesis Core). In parallel, we constructed homology directed repair (HDR) plasmids encoding one kilobase homology arms 5’ and 3’ of their respective guide RNAs. Between the homology arms we synthesized a codon-optimized (for D. melanogaster) mh coding sequence from either D. melanogaster or D. simulans (GenScript, Piscataway, NJ). We N-terminally tagged each sequence with 3xFLAG along with a linker sequence (GGTGGTTCATCA). We injected the dual sgRNA vector and a single HDR plasmid into the Cas9-expressing line, yw; nos-Cas9(II-attP40) (BestGene Inc, Chino Hills, CA).
We crossed single males, injected as embryos, to FM7 (X-chromosome balancer) females. We screened F1 females to identify positive transformants using forward primer 5’-AAGTGTCGCGCTATTTCACC-3’ and reverse primer 5’-TCACCGTCATGGTCTTTGTAGTCCAT-3’. We then backcrossed the positive F1 females to FM7 males and self-crossed the balanced F2 progeny to generate lines homozygous for either mh[mel] or mh[sim] allele. To confirm that the introduced alleles encoded the expected sequence, we amplified the entire region from homozygous flies using primers that anneal outside of the homology arms (5’-AATGGATTTCGGCAAATGAG-3’, 5’-GTCGTTGTAGGAGCCCATGT-3’) and then sequenced across the entire region. We also designed primers that amplified the native mh locus (5’-GGCCCTGCTCATATCGTATC-3’, 5’-AAGAACCTTACTGCGTGCAAC-3’) to confirm that our final genotypes were true replacements. Finally, we confirmed that the transgenic alleles were introduced into only the endogenous mh location by performing inverse PCR using the APAgene GOLD Genome Walking Kit (Bio S&T, Inc, Montreal, Canada) following the manufacturer’s instructions. The gene-specific primers for use in combination with the provided degenerate random tagging primers can be found in Table S3.
Constructing UAS-mh and UAS-Top2 lines
We used the ΦC31 integrase-mediated transgenesis system to introduce into the same landing site mh from D. melanogaster or D. simulans downstream of an “upstream activating sequence” or “UAS”71. Using the HDR plasmids as a template (see above), we PCR-amplified the 3xFLA-Gtagged mh coding sequence (either D. melanogaster or D. simulans) using Phusion High-Fidelity DNA Polymerase (NEB, Ipswich, MA). We cloned the resulting PCR products into NotI/XbaI sites of the pUASp-attB vector (Drosophila Genomics Resource Center, Bloomington, IN). We confirmed the absence of PCR-introduced mutations in the cloned UASp-mh[mel] and UASp-mh[sim] alleles by direct Sanger sequencing of the constructs (Table S3). We introduced the constructs into D. melanogaster yw; PBac[y+-attP-9A]VK00018 flies, which have an attP transgene landing site at cytological position 75A10 on chromosome 3L (BestGene Inc, Chino Hills, CA). We next made each transgene homozygous. To overexpress the transgenic alleles, we crossed these stocks to Gal4::VP16-nos (BDSC #64277), which drives germline expression of transgenes downstream of UAS.
Similarly, we used the ΦC31 integrase-mediated transgenesis system to introduce Top2 from D. melanogaster downstream of an UAS promoter.71 We synthesized a codon-optimized, D. melanogaster Top2 coding sequence (Twist, South San Francisco, CA). We N-terminally tagged each sequence with 3xHA along with a linker sequence (GGTGGTTCATCA). We introduced the constructs into D. melanogaster yw; PBac[y+-attP-9A]VK00018 flies (see above, BestGene Inc, Chino Hills, CA). We next constructed either mh[mel]; UASp-Top2[mel] or mh[sim]; UASp-Top2[mel] stocks using balancer chromosomes. To overexpress the transgenic Top2 allele, we crossed either mh[mel]; UASp-Top2[mel] or mh[sim]; UASp-Top2[mel] males to either mh[mel]; Gal4::VP16-nos or mh[sim]; Gal4::VP16-nos females, respectively.
Zhr rescue stocks
To generate stocks that encode both the X-linked mh-transgene and the X-linked 359bp satellite deletion (Zhr1, BDSC #25140), we first generated trans-heterozygote females. We crossed these trans-heterozygote females to FM7 males and used PCR to assay individual recombinant male progeny for the presence of both the mh transgene and Zhr. We detected the mh transgenes with forward primer 5’-AAGTGTCGCGCTATTTCACC-3’ and reverse primer 5’-TCACCGTCATGGTCTTTGTAGTCCAT-3’. To detect the Zhr mutation (i.e., 359bp satellite deletion), we used forward primer 5’-TATTCTTACATCTATGTGACC-3’ and reverse primer 5’-GTTTTGAGCAGCTAATTACC-3’.9 Performing a 10-cycle PCR at an annealing temperature of 52°C yields a band only in the presence of the 11Mb 359bp satellite array (Figure 3B). We backcrossed males positive for both the mh transgene and Zhr mutation to FM7 females to generate a permanent stock.
Additional stocks
We generated heterozygous mh[mel]/mh[sim] females by crossing mh[mel] females to mh[sim] males.
We used a +/FM7; +/CyO stock to generate flies encoding both the mh transgene at the native locus (chromosome X) and the Chk−/− (mnk) mutation (chromosome 2). The mnkl6 stock41 was a gift from N. Phadnis.
To generate heterozygous Top2 hypomorph females, we also used a +/FM7; +/CyO stock to construct flies encoding both the mh transgene at the native locus (chromosome X) and a heterozygous Top217–6/CyO mutation (chromosome 2). The Top217–6 stock72 was a gift from P. Geyer.
Immunoblotting
To assay 3xFLAG MH protein abundance in the ovary, we dissected 20 ovary pairs in 1X PBS and ground the material in RIPA buffer (Cell Signaling Technology, Danvers, MA), Protease Inhibitor Cocktail (Roche, Basel, Switzerland), and 2X PMSF (Cell Signaling Technology, Danvers, MA). To promote solubility, we incubated the lysate in benzonase (Sigma Aldrich, St. Louis, MO) for 1hr at 4C. We used 20μg of lysate and probed with 1:10,000 anti-FLAG (M2, Sigma Aldrich, St. Louis, MO) or 1:1000 anti-αTubulin (Developmental Studies Hybridoma Bank, Iowa City, IA) and 1:1000 anti-mouse HRP secondary antibodies (Kindle Biosciences, Greenwich, CT). We exposed blots with Kwikquant Western Blot detection kit and imaged with a Kwikquant imager (Kindle Biosciences, Greenwich, CT).
Fertility assays
Female fertility
To assay female fertility, we first aged virgin females 3–5 days. For each replicate vial, we crossed four virgin females to four w1118 males. We conducted crosses on molasses food at 24°C. UAS-Gal4 crosses were reared at 25°C. We flipped the parents onto new food every three days over the course of nine days and counted all progeny that emerged. No viability differences across assayed genotypes were noted.
Ovary size and mature egg counts
To determine the number of mature eggs and ovary size from focal genotypes, we first dissected ovary pairs in 1X PBS and imaged at 8X magnification with a Leica DFC7000 T camera. We quantified the area of each ovary pair using the polygon tool in FIJI73 to define the borders of the tissue. We then calculated the area (μm2) within these boundaries using the “Measure” selection in FIJI. After imaging, we counted the number of eggs that contain elongated dorsal appendages (stages 13 and 14).
Immunofluorescence
We conducted immunofluorescence on ovaries following the protocol described in.74 We stained ovaries with anti-FLAG (1:3000, M2, Sigma Aldrich, St. Louis, MO) and anti-γH2Av (1:1000, a gift from R. S. Hawley). We mounted ovaries with ProLong Gold Antifade Reagent with DAPI (Thermo Fisher Scientific, Waltham, MA). We imaged slides at 63X magnification on a Leica TCS SP8 Four Channel Spectral Confocal System. For each experiment, we used the same imaging parameters across genotypes.
We conducted immunofluorescence on embryos collected in a 0–70 minute window from mh[mel], mh[sim], or mh1 females crossed to males homozygous for P{gcid.EGFP.cid}III.275, a gift from K. McKim. We followed the protocol described in76 to fix and stain the embryos with anti-GFP (1:1000, Aves Labs, Tigard, OR). We mounted and imaged the embryos as described above.
Analysis of cytological data
Cell death quantification
To quantify the incidence of cell death, we mounted fixed whole ovaries (as described above) with ProLong Gold Antifade Reagent with DAPI (Thermo Fisher Scientific, Waltham, MA) and imaged at 63X magnification on a Leica TCS SP8 Four Channel Spectral Confocal System using the tile scanning and merging feature. We identified the number of ovarioles that contained egg chambers with >1 condensed, signal-saturated nurse cell nuclei. We then divided this number by the total number of ovarioles present in each ovary to determine the fraction of cell death incidence in mh[mel] and mh[sim] ovaries.
Immunofluorescence quantification
To quantify the average fluorescence of γH2Av in ovaries, we used the polygon tool in FIJI73 to define the borders of a representative stage four egg chamber. We quantified the fluorescent signal intensity using the “Measure” tool in FIJI, which calculates the mean pixels within these boundaries. We normalized the fluorescent signal intensity of mh1, mh[mel], and mh[sim] to the mean intensity signal of the mh[mel]. Similarly, the fluorescent signal intensity of mh[mel],Zhr and mh[sim],Zhr was normalized to the mean intensity signal of mh[mel],Zhr. Finally, the fluorescent signal intensity of nos-Gal4-VP16 driven mh[mel]; UASp-Top2[mel] and mh[sim]; UASp-Top2[mel] was normalized to the mean intensity signal of nos-Gal4-VP16 driven mh[mel]; UASp-Top2[mel].
QUANTIFICATION AND STATISTICAL ANALYSIS
We analyzed population genetic analyses using a χ2 test and molecular evolution analyses using a Fisher’s Exact test, otherwise we used t-tests. We carried out all statistical analyses using the R software (www.R-project.org).
Supplementary Material
HIGHLIGHTS.
The Drosophila homolog of Spartan, Maternal Haploid (MH), evolves adaptively
The D. simulans MH is toxic to oogenesis in its sister species, D. melanogaster
D. simulans MH toxicity is triggered by a D. melanogaster-specific satellite array
Overexpression of Top2 mitigates this D. melanogaster-D. simulans incompatibility
ACKNOWLEDGEMENTS
We thank Isabella Farkas and Courtney Christopher for technical assistance, and the University of Utah Mutation Generation and Detection Core. We also thank the Levine Lab, M. Patel, N. Phadnis, A. Das, and D. Dudka for feedback on the manuscript and the Levine Lab, P. Geyer, H. Malik, R.S. Hawley, and M. Buszczak for discussions about the project. This work was supported by a Shurl and Kay Curci Foundation fellowship from the Life Sciences Research Foundation to C.L.B. and National Institutes of Health (NIH) NIGMS grant R35GM124684 to M.T.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Cechova M, Harris RS, Tomaszkiewicz M, Arbeithuber B, Chiaromonte F, and Makova KD (2019). High satellite repeat turnover in great apes studied with short- and long-read technologies. Mol Biol Evol 36, 2415–2431. 10.1093/molbev/msz156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jagannathan M, Warsinger-Pepe N, Watase GJ, and Yamashita YM (2017). Comparative Analysis of Satellite DNA in the Drosophila melanogaster Species Complex. G3 (Bethesda) 7, 693–704. 10.1534/g3.116.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kipling D, Ackford HE, Taylor BA, and Cooke HJ (1991). Mouse minor satellite DNA genetically maps to the centromere and is physically linked to the proximal telomere. Genomics 11, 235–241. 10.1016/0888-7543(91)90128-2. [DOI] [PubMed] [Google Scholar]
- 4.Round EK, Flowers SK, and Richards EJ (1997). Arabidopsis thaliana centromere regions: genetic map positions and repetitive DNA structure. Genome Res 7, 1045–1053. 10.1101/gr.7.11.1045. [DOI] [PubMed] [Google Scholar]
- 5.Rudd MK, and Willard HF (2004). Analysis of the centromeric regions of the human genome assembly. Trends Genet 20, 529–533. 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Mefford HC, and Trask BJ (2002). The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet 3, 91–102. 10.1038/nrg727. [DOI] [PubMed] [Google Scholar]
- 7.McKinley KL, and Cheeseman IM (2016). The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol 17, 16–29. 10.1038/nrm.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jagannathan M, Cummings R, and Yamashita YM (2018). A conserved function for pericentromeric satellite DNA. Elife 7. 10.7554/eLife.34122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosic S, Kohler F, and Erhardt S (2014). Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J Cell Biol 207, 335–349. 10.1083/jcb.201404097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoeftner S, and Blasco MA (2009). A ‘higher order’ of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J 28, 2323–2336. 10.1038/emboj.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henikoff S, Ahmad K, and Malik HS (2001). The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293, 1098–1102. 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 12.Lohe AR, Hilliker AJ, and Roberts PA (1993). Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics 134, 1149–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brutlag DL (1980). Molecular arrangement and evolution of heterochromatic DNA. Annu Rev Genet 14, 121–144. 10.1146/annurev.ge.14.120180.001005. [DOI] [PubMed] [Google Scholar]
- 14.de Lima LG, Hanlon SL, and Gerton JL (2020). Origins and Evolutionary Patterns of the 1.688 Satellite DNA Family in Drosophila Phylogeny. G3 (Bethesda) 10, 4129–4146. 10.1534/g3.120.401727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang X, Cao J, Zhang L, Huang Y, Zhang Q, and Rong YS (2017). Maternal Haploid, a Metalloprotease Enriched at the Largest Satellite Repeat and Essential for Genome Integrity in Drosophila Embryos. Genetics 206, 1829–1839. 10.1534/genetics.117.200949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferree PM, and Prasad S (2012). How can satellite DNA divergence cause reproductive isolation? Let us count the chromosomal ways. Genet Res Int 2012, 430136. 10.1155/2012/430136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jagannathan M, and Yamashita YM (2021). Defective Satellite DNA Clustering into Chromocenters Underlies Hybrid Incompatibility in Drosophila. Mol Biol Evol 38, 4977–4986. 10.1093/molbev/msab221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayes JJ, and Malik HS (2009). Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science 326, 1538–1541. 10.1126/science.1181756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumon T, Ma J, Akins RB, Stefanik D, Nordgren CE, Kim J, Levine MT, and Lampson MA (2021). Parallel pathways for recruiting effector proteins determine centromere drive and suppression. Cell 184, 4904–4918 e4911. 10.1016/j.cell.2021.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maheshwari S, Tan EH, West A, Franklin FCH, Comai L, and Chan SWL (2015). Naturally occurring differences in CENH3 affect chromosome segregation in zygotic mitosis of hybrids. PLoS Genet 11, e1004970. 10.1371/journal.pgen.1004970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferree PM, and Barbash DA (2009). Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol 7, e1000234. 10.1371/journal.pbio.1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibeaux R, Acker R, Kitaoka M, Georgiou G, van Kruijsbergen I, Ford B, Marcotte EM, Nomura DK, Kwon T, Veenstra GJC, and Heald R (2018). Paternal chromosome loss and metabolic crisis contribute to hybrid inviability in Xenopus. Nature 553, 337–341. 10.1038/nature25188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abad JP, Agudo M, Molina I, Losada A, Ripoll P, and Villasante A (2000). Pericentromeric regions containing 1.688 satellite DNA sequences show anti-kinetochore antibody staining in prometaphase chromosomes of Drosophila melanogaster. Mol Gen Genet 264, 371–377. 10.1007/s004380000331. [DOI] [PubMed] [Google Scholar]
- 24.Losada A, and Villasante A (1996). Autosomal location of a new subtype of 1.688 satellite DNA of Drosophila melanogaster. Chromosome Res 4, 372–383. 10.1007/BF02257273. [DOI] [PubMed] [Google Scholar]
- 25.Sproul JS, Khost DE, Eickbush DG, Negm S, Wei X, Wong I, and Larracuente AM (2020). Dynamic Evolution of Euchromatic Satellites on the X Chromosome in Drosophila melanogaster and the simulans Clade. Mol Biol Evol 37, 2241–2256. 10.1093/molbev/msaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delabaere L, Orsi GA, Sapey-Triomphe L, Horard B, Couble P, and Loppin B (2014). The Spartan ortholog maternal haploid is required for paternal chromosome integrity in the Drosophila zygote. Curr Biol 24, 2281–2287. 10.1016/j.cub.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Svetec N, Cridland JM, Zhao L, and Begun DJ (2016). The Adaptive Significance of Natural Genetic Variation in the DNA Damage Response of Drosophila melanogaster. PLoS Genet 12, e1005869. 10.1371/journal.pgen.1005869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loppin B, Berger F, and Couble P (2001). Paternal chromosome incorporation into the zygote nucleus is controlled by maternal haploid in Drosophila. Dev Biol 231, 383–396. 10.1006/dbio.2000.0152. [DOI] [PubMed] [Google Scholar]
- 29.Kihara S, Matsuzawa Y, Kubo M, Nozaki S, Funahashi T, Yamashita S, Sho N, and Tarui S (1989). Autoimmune hyperchylomicronemia. N Engl J Med 320, 1255–1259. 10.1056/NEJM198905113201906. [DOI] [PubMed] [Google Scholar]
- 30.Stingele J, Bellelli R, Alte F, Hewitt G, Sarek G, Maslen SL, Tsutakawa SE, Borg A, Kjaer S, Tainer JA, et al. (2016). Mechanism and Regulation of DNA-Protein Crosslink Repair by the DNA-Dependent Metalloprotease SPRTN. Mol Cell 64, 688–703. 10.1016/j.molcel.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaz B, Popovic M, Newman JA, Fielden J, Aitkenhead H, Halder S, Singh AN, Vendrell I, Fischer R, Torrecilla I, et al. (2016). Metalloprotease SPRTN/DVC1 Orchestrates Replication-Coupled DNA-Protein Crosslink Repair. Mol Cell 64, 704–719. 10.1016/j.molcel.2016.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weickert P, and Stingele J (2022). DNA-Protein Crosslinks and Their Resolution. Annu Rev Biochem. 10.1146/annurev-biochem-032620-105820. [DOI] [PubMed] [Google Scholar]
- 33.McDonald JH, and Kreitman M (1991). Adaptive protein evolution at the Adh locus in Drosophila. Nature 351, 652–654. [DOI] [PubMed] [Google Scholar]
- 34.Garrigan D, Kingan SB, Geneva AJ, Andolfatto P, Clark AG, Thornton KR, and Presgraves DC (2012). Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res 22, 1499–1511. 10.1101/gr.130922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerr JF, Wyllie AH, and Currie AR (1972). Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 26, 239–257. 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCall K (2004). Eggs over easy: cell death in the Drosophila ovary. Dev Biol 274, 3–14. 10.1016/j.ydbio.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 37.Roos WP, Thomas AD, and Kaina B (2016). DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer 16, 20–33. 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 38.Jang JK, Sherizen DE, Bhagat R, Manheim EA, and McKim KS (2003). Relationship of DNA double-strand breaks to synapsis in Drosophila. J Cell Sci 116, 3069–3077. 10.1242/jcs.00614. [DOI] [PubMed] [Google Scholar]
- 39.Madigan JP, Chotkowski HL, and Glaser RL (2002). DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res 30, 3698–3705. 10.1093/nar/gkf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakhrat A, Pritchett T, Peretz G, McCall K, and Abdu U (2010). Drosophila Chk2 and p53 proteins induce stage-specific cell death independently during oogenesis. Apoptosis 15, 1425–1434. 10.1007/s10495-010-0539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, Golic KG, Rio DC, and Rubin GM (2004). Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol 24, 1219–1231. 10.1128/MCB.24.3.1219-1231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawamura K, Yamamoto MT, and Watanabe TK (1993). Hybrid lethal systems in the Drosophila melanogaster species complex. II. The Zygotic hybrid rescue (Zhr) gene of D. melanogaster. Genetics 133, 307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dokshin GA, Davis GM, Sawle AD, Eldridge MD, Nicholls PK, Gourley TE, Romer KA, Molesworth LW, Tatnell HR, Ozturk AR, et al. (2020). GCNA Interacts with Spartan and Topoisomerase II to Regulate Genome Stability. Dev Cell 52, 53–68 e56. 10.1016/j.devcel.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez-Mosqueda J, Maddi K, Prgomet S, Kalayil S, Marinovic-Terzic I, Terzic J, and Dikic I (2016). SPRTN is a mammalian DNA-binding metalloprotease that resolves DNA-protein crosslinks. Elife 5. 10.7554/eLife.21491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gale KC, and Osheroff N (1992). Intrinsic intermolecular DNA ligation activity of eukaryotic topoisomerase II. Potential roles in recombination. J Biol Chem 267, 12090–12097. [PubMed] [Google Scholar]
- 46.Morimoto S, Tsuda M, Bunch H, Sasanuma H, Austin C, and Takeda S (2019). Type II DNA Topoisomerases Cause Spontaneous Double-Strand Breaks in Genomic DNA. Genes (Basel) 10. 10.3390/genes10110868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pommier Y, Sun Y, Huang SN, and Nitiss JL (2016). Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol 17, 703–721. 10.1038/nrm.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kas E, and Laemmli UK (1992). In vivo topoisomerase II cleavage of the Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. EMBO J 11, 705–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hughes SE, and Hawley RS (2014). Topoisomerase II is required for the proper separation of heterochromatic regions during Drosophila melanogaster female meiosis. PLoS Genet 10, e1004650. 10.1371/journal.pgen.1004650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Centore RC, Yazinski SA, Tse A, and Zou L (2012). Spartan/C1orf124, a reader of PCNA ubiquitylation and a regulator of UV-induced DNA damage response. Mol Cell 46, 625–635. 10.1016/j.molcel.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reinking HK, Kang HS, Gotz MJ, Li HY, Kieser A, Zhao S, Acampora AC, Weickert P, Fessler E, Jae LT, et al. (2020). DNA Structure-Specific Cleavage of DNA-Protein Crosslinks by the SPRTN Protease. Mol Cell 80, 102–113 e106. 10.1016/j.molcel.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao S, Kieser A, Li HY, Reinking HK, Weickert P, Euteneuer S, Yaneva D, Acampora AC, Gotz MJ, Feederle R, and Stingele J (2021). A ubiquitin switch controls autocatalytic inactivation of the DNA-protein crosslink repair protease SPRTN. Nucleic Acids Res 49, 902–915. 10.1093/nar/gkaa1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferree PM, Gomez K, Rominger P, Howard D, Kornfeld H, and Barbash DA (2014). Heterochromatin position effects on circularized sex chromosomes cause filicidal embryonic lethality in Drosophila melanogaster. Genetics 196, 1001–1005. 10.1534/genetics.113.161075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chmátal L, Gabriel SI, Mitsainas GP, Martínez-Vargas J, Ventura J, Searle JB, Schultz RM, and Lampson MA (2014). Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Current biology: CB 24, 2295–2300. 10.1016/j.cub.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmátal L, Yang K, Sullivan BA, Schultz RM, Lampson MA, and Black BE (2017). Expanded Satellite Repeats Amplify a Discrete CENP-A Nucleosome Assembly Site on Chromosomes that Drive in Female Meiosis. Current biology: CB 27, 2365–2373.e2368. 10.1016/j.cub.2017.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malik HS, and Henikoff S (2001). Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157, 1293–1298. 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malik HS, and Henikoff S (2002). Conflict begets complexity: the evolution of centromeres. Curr Opin Genet Dev 12, 711–718. 10.1016/s0959-437x(02)00351-9. [DOI] [PubMed] [Google Scholar]
- 58.Vermaak D, Bayes JJ, and Malik HS (2009). A surrogate approach to study the evolution of noncoding DNA elements that organize eukaryotic genomes. J Hered 100, 624–636. 10.1093/jhered/esp063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carracedo MC, Asenjo A, and Casares P (2000). Location of Shfr, a new gene that rescues hybrid female viability in crosses between Drosophila simulans females and D. melanogaster males. Heredity (Edinb) 84 (Pt 6), 630–638. 10.1046/j.1365-2540.2000.00658.x. [DOI] [PubMed] [Google Scholar]
- 60.Gerard PR, and Presgraves DC (2012). Abundant genetic variability in Drosophila simulans for hybrid female lethality in interspecific crosses to Drosophila melanogaster. Genet Res (Camb) 94, 1–7. 10.1017/S0016672312000031. [DOI] [PubMed] [Google Scholar]
- 61.Orr HA (1996). The unexpected recovery of hybrids in a Drosophila species cross: a genetic analysis. Genet Res 67, 11–18. 10.1017/s0016672300033437. [DOI] [PubMed] [Google Scholar]
- 62.Sawamura K, Taira T, and Watanabe TK (1993). Hybrid lethal systems in the Drosophila melanogaster species complex. I. The maternal hybrid rescue (mhr) gene of Drosophila simulans. Genetics 133, 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sawamura K, and Yamamoto MT (1993). Cytogenetical localization of Zygotic hybrid rescue (Zhr), a Drosophila melanogaster gene that rescues interspecific hybrids from embryonic lethality. Mol Gen Genet 239, 441–449. 10.1007/BF00276943. [DOI] [PubMed] [Google Scholar]
- 64.Pool JE, Corbett-Detig RB, Sugino RP, Stevens KA, Cardeno CM, Crepeau MW, Duchen P, Emerson JJ, Saelao P, Begun DJ, and Langley CH (2012). Population Genomics of sub-saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet 8, e1003080. 10.1371/journal.pgen.1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rogers RL, Cridland JM, Shao L, Hu TT, Andolfatto P, and Thornton KR (2014). Landscape of standing variation for tandem duplications in Drosophila yakuba and Drosophila simulans. Mol Biol Evol 31, 1750–1766. 10.1093/molbev/msu124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chakraborty M, Chang CH, Khost DE, Vedanayagam J, Adrion JR, Liao Y, Montooth KL, Meiklejohn CD, Larracuente AM, and Emerson JJ (2021). Evolution of genome structure in the Drosophila simulans species complex. Genome Res 31, 380–396. 10.1101/gr.263442.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Castillo DM, Kean CM, McCormick B, Natesan S, and Barbash DA (2022). Testing the Drosophila maternal haploid gene for functional divergence and a role in hybrid incompatibility. bioRxiv. 10.1101/2022.03.23.485453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rozas J, Sanchez-DelBarrio JC, Messeguer X, and Rozas R (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497. 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 69.Yang Z (1997). PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13, 555–556. 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 70.Yang Z (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24, 1586–1591. msm088 [pii] 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 71.Venken KJ, He Y, Hoskins RA, and Bellen HJ (2006). P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747–1751. 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 72.Hohl AM, Thompson M, Soshnev AA, Wu J, Morris J, Hsieh TS, Wu CT, and Geyer PK (2012). Restoration of topoisomerase 2 function by complementation of defective monomers in Drosophila. Genetics 192, 843–856. 10.1534/genetics.112.144006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McKim KS, Joyce EF, and Jang JK (2009). Cytological analysis of meiosis in fixed Drosophila ovaries. Methods Mol Biol 558, 197–216. 10.1007/978-1-60761-103-5_12. [DOI] [PubMed] [Google Scholar]
- 75.Schuh M, Lehner CF, and Heidmann S (2007). Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol 17, 237–243. 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 76.Levine MT, Vander Wende HM, and Malik HS (2015). Mitotic fidelity requires transgenerational action of a testis-restricted HP1. Elife 4, e07378. 10.7554/eLife.07378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mh and Top2 alleles from D. simulans have been deposited in GenBank and are publicly available as of the date of this publication. Accession numbers are listed in the key resources table. All other data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


