Summary
The intestinal epithelium comprises the body’s largest surface exposed to viruses. Additionally, the gut epithelium hosts a large population of intraepithelial T lymphocytes, or IELs, although their role in resistance against viral infections remains elusive. By fate-mapping T cells recruited to the murine intestine, we observed an accumulation of newly recruited CD4+ T cells after infection with murine norovirus CR6 and adenovirus type-2 (AdV), but not reovirus. CR6- and AdV-recruited intraepithelial CD4+ T cells co-expressed Ly6A and chemokine receptor CCR9, exhibited T helper 1 and cytotoxic profiles and conferred protection against AdV in vivo and in an organoid model in an IFN-γ-dependent manner. Ablation of the T cell receptor (TCR) or the transcription factor ThPOK in CD4+ T cells prior to AdV infection prevented viral control, while TCR ablation during infection did not impact viral clearance. These results uncover a protective role for intraepithelial Ly6A+CCR9+CD4+ T cells against enteric adenovirus.
Graphical Abstract
eTOC Blurb
It is known that a large number of T cells accumulate in the gut epithelium, but it remained unclear whether they play a role against enteric virus infections. Using fate-mapping models, Parsa et al. report that distinct enteric viruses induce the recruitment of CD4+ T cells to the gut epithelium and that these cells control viral replication via IFN-γ secretion.
Introduction
The highly specialized intestinal immune system is charged with maintaining tolerance to harmless stimuli from commensal bacteria and food, while providing protective immunity against pathogens and epithelial cancers (Florsheim et al., 2021; Honda and Littman, 2016; McDonald et al., 2018; Tanoue et al., 2016). Intraepithelial lymphocytes (IELs) comprise a large T cell population located at the critical interface between the intestinal lumen and the core of the body. IELs can provide a first line of immunity in mice and humans, while balancing tolerance and defense (Bilate et al., 2020; Cervantes-Barragan et al., 2017; Edelblum et al., 2015; Hoytema van Konijnenburg et al., 2017; Jabri and Sollid, 2009; Sujino et al., 2016). Among the IEL populations, CD4+ T cells are key players in intestinal homeostasis, finely tuning responses at the level of antigen recognition and functional differentiation. In the intestinal epithelium (IE) and underlying lamina propria (LP), tissue adapted pro-inflammatory, regulatory (Treg) and intraepithelial (CD8αα+ CD4 IEL) CD4+ T cells coordinate immunity and tolerance to diverse intestinal stimuli (Cervantes-Barragan et al., 2017; Honda and Littman, 2016; McDonald et al., 2018; Sujino et al., 2016). Both LP and IE CD4+ T cells are antigen-experienced and co-express markers of activation, gut-homing and tissue residency, including CD44, CD69, CD103 and CD8αα homodimers (Masopust and Soerens, 2019).
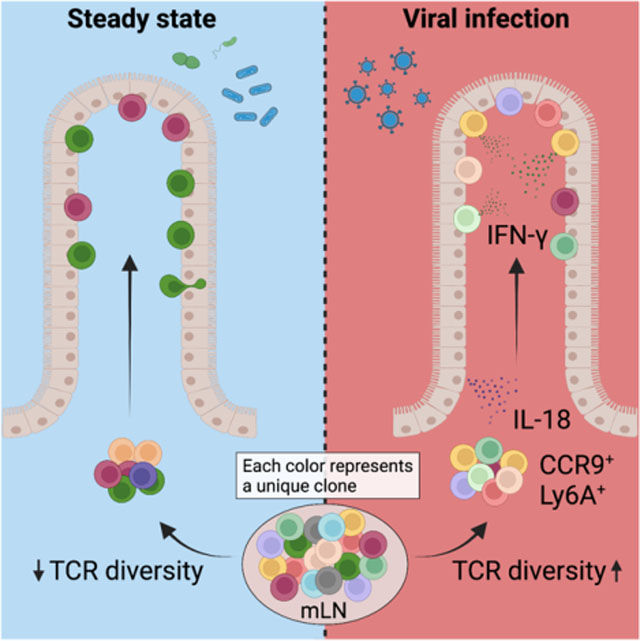
Enteric viruses, such as norovirus, rotavirus and adenovirus, are among the most common causes of acute gastroenteritis in humans and are responsible for morbidity and mortality worldwide (Iliev and Cadwell, 2021; Wilhelmi et al., 2003). These enteric viruses can infect intestinal epithelial cells and myeloid cells in the LP, provoking robust innate and adaptive immune responses (Iliev and Cadwell, 2021); however, a functional role for intestinal T cells in resistance to enteric viruses is not well established. We sought to address the functional role of intestinal T cells in the response to enteric viral infections by developing a T cell fate-mapping strategy to identify and characterize IE T cell dynamics during enteric viral infection. We observed specific recruitment and accumulation of CCR9+Ly6A+CD4+ T cells, expressing a T helper 1 (Th1) and cytotoxic profile, in the epithelium after murine norovirus (MNV) CR6 and murine adenovirus-2 (AdV) infections. Virus-recruited CD4+ T cells displayed increased T cell receptor (TCR) diversity while ablation of the TCR or the CD4-lineage defining transcription factor T-Helper-Inducing POZ/Krueppel-Like Factor (ThPOK) in CD4+ T cells prior to AdV infection prevented viral control. In summary, we found that intraepithelial CD4+ T cells protect against enteric virus infection in a ThPOK- and IFN-γ-dependent manner.
Results
Enteric virus infection leads to distinct T cell dynamics
To characterize T cell dynamics in the IE during enteric viral infections, we infected mice with four different viruses, chronic MNV (CR6), acute MNV (CW3) (Strong et al., 2012), reovirus T1L (T1L) (Bouziat et al., 2017) and AdV (Wilson et al., 2017). CR6 infects intestinal tuft cells in the IE to establish long-lasting chronic infection (Graziano et al., 2021; Wilen et al., 2018), while CW3 and T1L infect multiple cell types including myeloid and stromal cells in the LP (Grau et al., 2017; Graziano et al., 2021). AdV exclusively infects and replicates in the IE (Takeuchi and Hashimoto, 1976) (Figure S1A). We have previously shown that during steady state, CD4+ T cells enter the IE as CD103− effector T cells and gradually acquire an IEL phenotype with progressive expression of CD103 and CD8αα (Bilate et al., 2020; London et al., 2021). To understand T cell recruitment dynamics to the site of enteric infections, we first quantified the frequencies of CD103− T cells in the IE 10 days post infection by flow cytometry. We observed that AdV infection resulted in an increase of CD4+CD103− T cells whereas T1L infection led to an increase in CD8αβ+CD103− T cells in the IE (Figure 1A, B). To independently assess peripheral T cell recruitment to the IE during viral infections, we developed a fate-mapping approach. We crossed SellCreERT2 (Merkenschlager et al., 2021) with Rosa26CAG-LSL-tdTomato (iSellTomato) to permanently label naïve T cells (CD62L+) as Tomato+ upon tamoxifen treatment, enabling a strategy that enriched for “ex-naïve” (CD62L−Tomato+) pathogen-specific T cells that migrated to the intestine (Figure 1C, S1B and S1C). Analysis of tamoxifen-treated iSellTomato mice 10 days post infection revealed a significant increase of recently recruited (Tomato+) CD4+ T cells post AdV and CR6 infections, whereas T1L and CW3 infections led to a preferential recruitment of CD8αβ+ T cells (Figure 1D, E and Figure S1D). Additionally, although all tested enteric viruses led to a significant recruitment of CD4+ T cells to the LP, CW3, CR6 and T1L preferentially recruited CD8αβ+ T cells to this compartment (Figure S1E and S1F).
Figure 1. Distinct intraepithelial T cell dynamics post enteric viral infections.
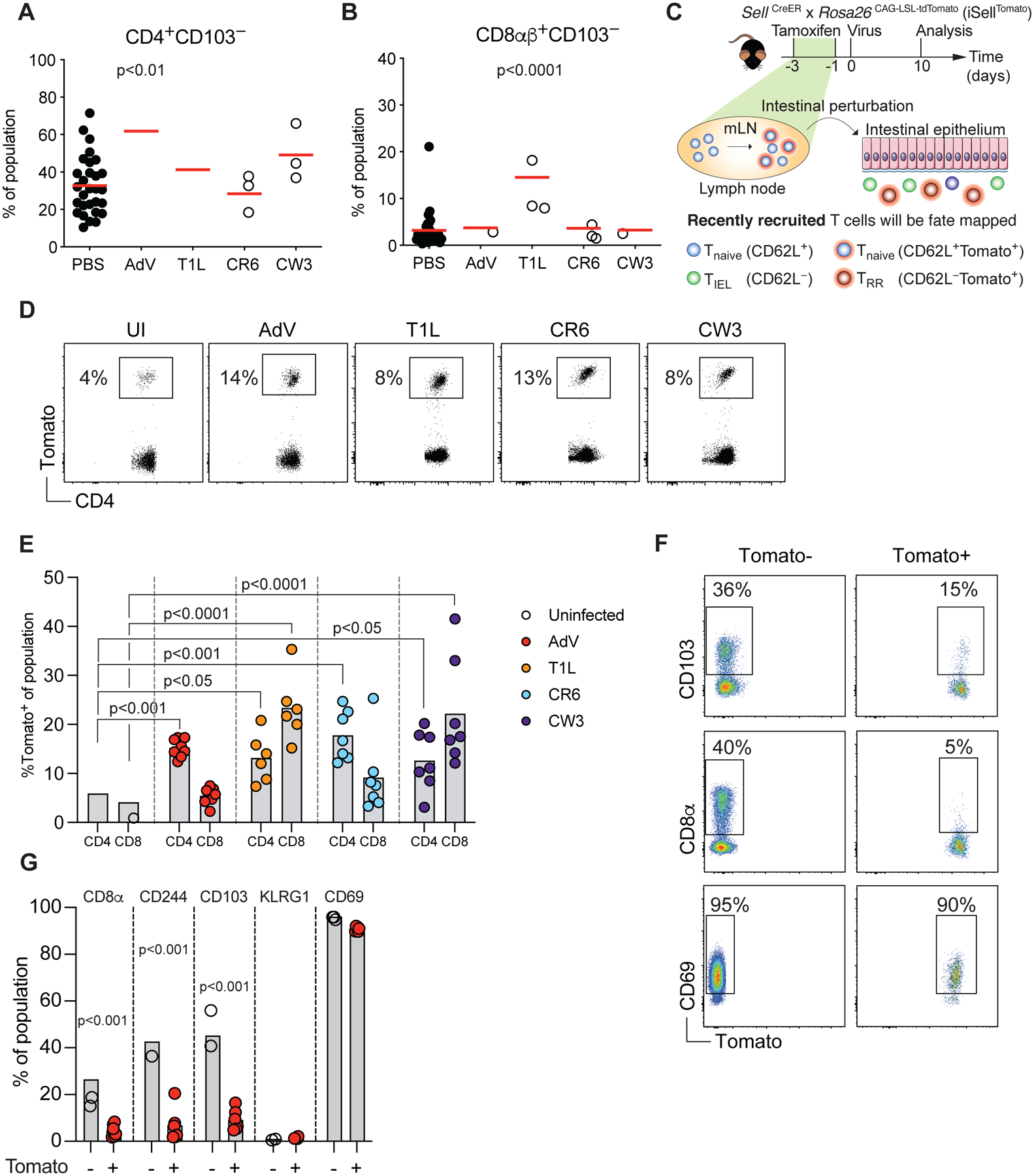
(A-B) B6 mice were orally infected with 107 infectious units (i.u.) of murine adenovirus-2 (AdV), 108 plaque forming units (pfu) of reovirus T1L, 3×106 pfu of either murine norovirus (MNV) CR6 or CW3. CD4+CD103− (A) or CD8αβ+CD103− (B) small intestine intraepithelial lymphocytes (IEL) were analyzed 10 days post infection among total CD4 or CD8αβ T cells, respectively. (C) Experimental overview of the T cell fate-mapping model. iSellTomato mice were treated orally with tamoxifen 1 and 3 days prior to viral infection, RR: recently recruited. (D-E) iSellTomato mice were orally infected with 107 i.u. of AdV, 108 pfu of T1L, or 3×106 pfu of either CR6 or CW3, small intestine TCRβ+CD4+CD62L− and TCRβ+CD8αβ+CD62L− IELs were analyzed for tomato expression 10 days post infection. (D) Representative dot plots of tomato expression among CD4+ IELs. (E) Frequencies of tomato expression among small intestine CD4+ or CD8+ IELs. (F-G) iSellTomato mice were infected with 107 i.u. of AdV and expression of CD103, CD8α and CD69 were analyzed among TCRβ+CD4+CD62L−Tomato+ or Tomato− cells. (F) Representative surface expression of markers as indicated among Tomato+ and Tomato− T cells. (G) Frequencies of Tomato+ or Tomato− cells expressing the indicated markers. Data are expressed as mean of individual mice in A and B (n = 6 for AdV, n = 5 for T1L, n = 12 for CR6, of two independent experiments, n = 3 for CW3, of one experiment), in E (n = 8 for AdV, n = 6 for CW3 and n = 7 for CR6 and T1L, of two independent experiments) and in G (n = 6 of two independent experiments). p values are as indicated, one-way ANOVA plus Bonferroni test in A and E, Student’s t-test in G. See also Figure S1.
We then focused on AdV infection as this virus preferentially recruited CD4+ T cells and exclusively infects the intestinal epithelium. As expected based on a prior study, newly recruited CD4+ T cells in response to AdV infection did not express typical IEL markers such as CD103, CD8αα or CD244 (Reis et al., 2013), but the majority of Tomato+ T cells expressed CD69 indicating recent activation, or early tissue-residency (Faria et al., 2017) (Figure 1F, G and S1G). Using a fate-mapping strategy we revealed that the dynamics of recently recruited T cells differ between enteric viral infections.
Recruited intraepithelial CD4+ T cells display a mixed Th1 and T cytotoxic profile
Next, we analyzed the gene expression profiles of newly recruited IE and LP CD4+ T cells from AdV-infected mice. Unbiased clustering of the gene expression data and principal component analysis segregated CD4+Tomato+ T cells from infected mice in comparison to uninfected mice (Figure 2A, B). CD4+Tomato+ T cells from AdV infected mice displayed tissue specific gene expression such as Ccl5 and Itga1 in the IE, and Fasl and IL12rb2 in the LP, indicating tissue adaptation and some degree of compartmentalization (London et al., 2021). CD4+Tomato+ T cells from both compartments of AdV-infected mice displayed expression of inflammation- and gut residency-associated genes (Ccr9, Ly6a, Lztfl1 and Fyco1), with a mixed Th1 (Prdm1, Tbx21, Id2 and Cxcr6) and T cytotoxic (CTL) (Runx3, Nkg7, Gzma and Gzmk) profiles (Figure 2A, C and D). Overall, these results suggest that intestinal CD4+ T cells recruited upon AdV infection display mixed Th1 and CTL profiles.
Figure 2. Transcriptional analysis of epithelial-recruited CD4+ T cells post AdV-infection.
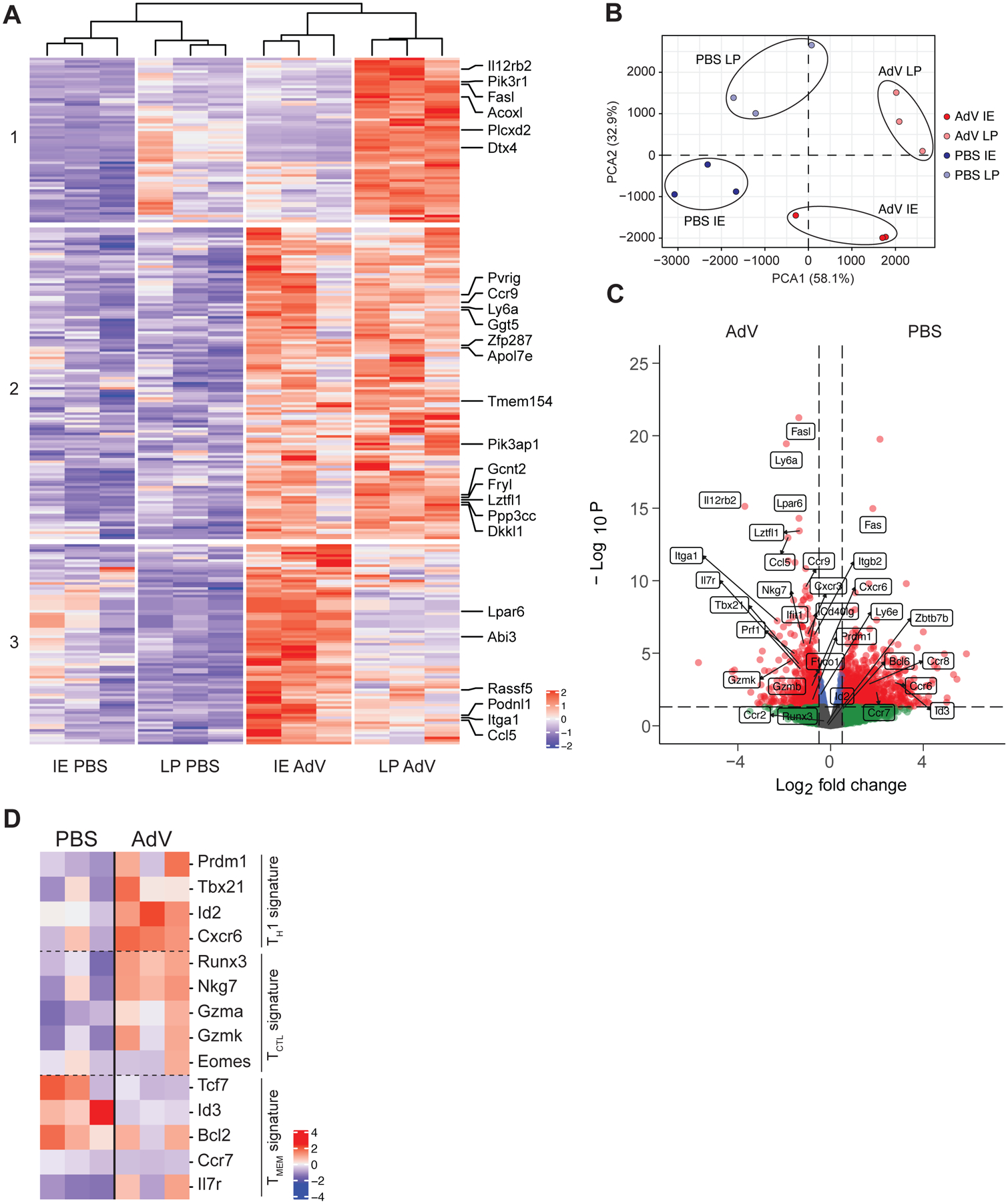
iSellTomato mice were infected with AdV or treated with PBS vehicle control, and CD4+CD62L−Tomato+ T cells were sorted from small intestine lamina propria (LP) or intestinal epithelium (IE) 10 days post infection for bulk RNAseq. (A) Heatmap clustering of the most significant differentially expressed genes represented by normalized Z score in LP and IE of AdV-infected and PBS treated control mice. Top 25 genes are annotated. FDR < 0.05. (B) Principal component analysis of sorted T cells in IE and LP compartment of AdV-infected and control mice. (C) Volcano plot visualizing gene fold change (X-axis) versus p value (Y-axis) between T cells from AdV-infected and control mice for both IE and LP combined. Dashed line on y-axis denotes p value of 0.05 and dashed line on x-axis denotes fold change of ± 2. (D) Transcriptional signatures represented by normalized Z score of T cells between AdV-infected and control mice for both IE and LP combined. n = 3 mice per group from one experiment. Data are expressed as means of individual mice.
Our transcriptional analysis indicated a heterogenous profile of the recruited CD4+ IE T cells post enteric AdV infection. To initially assess their functional profile, we infected tamoxifen-treated iSellTomato mice and characterized by flow cytometry CD4+Tomato+ T cells 10- and 30-days post AdV infection by using markers such as Ly6A and CCR9, enriched in our RNAseq data. As expected, we detected only a few fate-mapped CCR9+Ly6A+CD4+ T cells in the IE and LP compartments in naïve uninfected mice. In contrast, AdV-infection significantly increased the recruitment of CCR9+Ly6A+CD4+ T cells and granzyme B (GzmB) expression (Figure 3A). Additional cell surface markers with transcripts increased in the RNAseq analysis and associated with gut tissue recruitment during AdV-infection include CXCR6 and LFA-1 (CD11a/CD18) for the intraepithelial compartment, and CXCR3 for the LP. By day 30 post infection, no differences were observed in these surface markers between control and AdV-infected mice, while CCR9+Ly6A+, CXCR3+ and LFA-1 remained increased in CD4+Tomato+ T cells isolated from the LP of AdV-infected mice, suggesting different retention or replacement dynamics between these compartments (Figure S2A and S2B).
Figure 3. AdV- and CR6-recruited IE CD4+ T cells are enriched for CCR9+Ly6A+ cells and acquire a Th1 and cytotoxic profile.
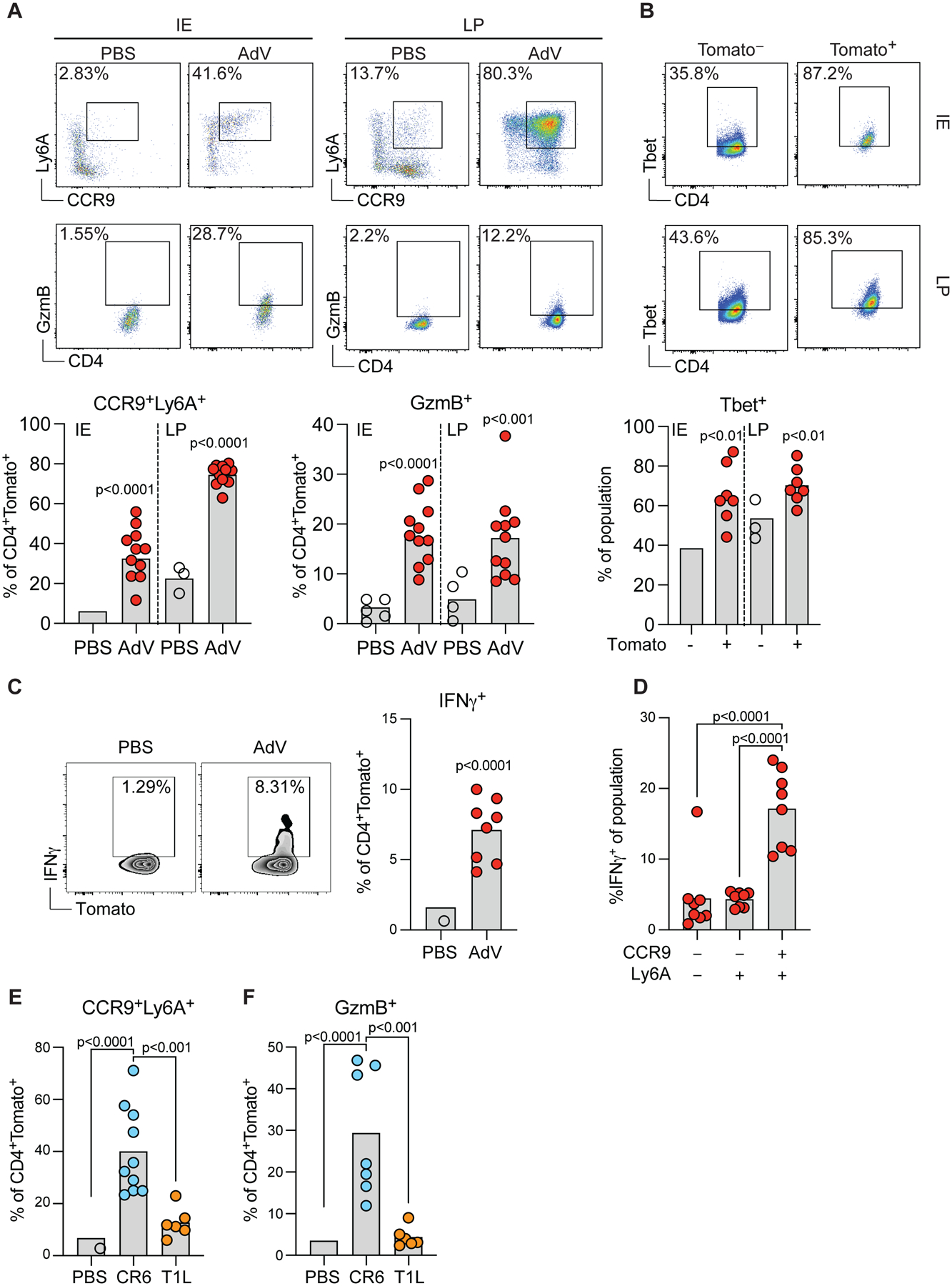
iSellTomato mice were infected with AdV or treated with PBS vehicle only, and small intestine CD4+CD62L−Tomato+ T cells were analyzed 10 days post infection (A-D). (A) Representative plots (top) of marker expression and population frequencies (bottom) as indicated among CD4+CD62L−Tomato+ T cells in LP or IE compartments. (B) Representative plots (top) and frequencies (bottom) of T-bet expression among Tomato+ or Tomato− cells in IE and LP. (C) Representative plots (left) and frequencies (right) of IFN-γ production among CD4+CD62L−Tomato+ T cells in the IE. (D) Frequencies of IFN-γ production among CCR9−Ly6A−, Ly6A+CCR9− and CCR9+Ly6A+ cells within CD4+Tomato+ T cells. (E-F) iSellTomato mice were infected with CR6 or T1L, or treated with PBS vehicle only, and CD4+CD62L−Tomato+ T cells was analyzed for Ly6A, CCR9 (E) and GzmB (F) expression 10 days post infection among CD4+CD62L−Tomato+ cells. Data are expressed as means of individual mice, for A (n = 10 for control and n = 11 for AdV, of two independent experiments), for B (n = 7 of two independent experiments), for C and D (n = 8 of two independent experiments), for E (n = 6–10, two independent experiments), and for F (n = 6–10, two independent experiments). p values are as indicated, Student’s t-test in A, B and C. One-way ANOVA plus Bonferroni test in D, E and F. See also Figure S2, S3 and S4.
The majority of recruited CD4+Tomato+ in AdV-infected mice expressed the transcription factor Tbx21 (T-bet) (Figure 3B), associated with TH1 phenotype as well as with IEL differentiation (Reis et al., 2014). Accordingly, we detected increased IFN-γ expression in CD4+Tomato+ IE T cells from AdV-infected mice in comparison to CD4+Tomato+ IE T cells isolated from uninfected mice (Figure 3C). CCR9+Ly6A+ T cells were the main IFN-γ producers within recruited Tomato+ CD4+ T cells (Figure 3D). Because we observed that different enteric viruses give rise to distinct T cell responses in the IE compartment (see Figure 1E above), we infected tamoxifen-treated iSellTomato mice with CR6 or T1L to determine if these viruses could also induce CCR9+Ly6A+CD4+ T cells. In agreement with differences observed in the CD4 T cell recruitment between these viruses, CR6, but not T1L infection, led to increased CCR9+Ly6A+CD4+ T cells (Figure 3E) with similar GzmB expression as observed in AdV-recruited CD4+Tomato+ T cells (Figure 3F). To address whether other enteric pathogens also induce CCR9 and Ly6A expression in newly recruited CD4+ T cells, we orally infected iSellTomato mice with Listeria monocytogenes 10403S-inlA. Listeria-specific IE CD4+Tomato+ T cells upregulated Ly6A but failed to upregulate CCR9 (Figure S3A, B and C), pointing to pathogen-associated recruiting programs (Kiner et al., 2021). These results reveal that CR6 and AdV infections induce gut recruitment of CD4+ T cells expressing CCR9, Ly6A, and an overall Th1/CTL phenotype.
Recruited intraepithelial CD4+ T cells are clonally diverse and temporally TCR-dependent for controlling AdV-replication in vivo
To determine the role of the T cell receptor (TCR) in controlling AdV viral replication, we first extracted the TCR CDR3 sequences for both the α and the β chain from our bulk RNAseq data. Analysis of the CDR3 sequences suggested an increased number of reads for unique CDR3 sequences in CD4+ T cells recruited during acute viral infection in comparison to CD4+ T cells recently recruited in naïve mice (Figure 4A). We have recently shown that CD4+ T cells in the IE undergo clonal selection and reduction in TCR diversity as cells differentiate from conventional CD103− CD4+ T cells into differentiated CD103+CD8αα+ CD4+IELs (Bilate et al., 2020). To further investigate the TCR repertoire of AdV-recruited IE CD4+ T cells, we infected tamoxifen-treated iSellTomato mice with AdV and sequenced the TCRβ chain of single cell sorted Tomato+ CD4+ T cells, 10- and 30-days post-infection (AdV) or post-PBS injection (PBS). As shown earlier, most of the Tomato+ CD4+ T cells from AdV-infected mice were Ly6A+, whereas Tomato+ CD4+ T cells in uninfected mice were mostly Ly6A− (Figure 4B). Analysis of newly recruited (Tomato+) Ly6A− CD4+ T cells from uninfected mice also confirmed our previous observations, revealing clonal expansions with reduced TCR diversity and increase in CD103 expression in most of the clones at day 30 [Figure 4B–F; TCR diversity was assessed by the diversity 50 index (D50), indexes varied from least diverse; 0, to most diverse; 0.5]. However, contrary to expectations for T cell responses to pathogens, newly recruited (Tomato+) IE CD4+ T cells from AdV-infected mice, regardless of Ly6A expression, did not show reduced TCR diversity and higher clonal expansions. Instead, CD4+ T cells from AdV-infected mice displayed similar or even less clonal expansion than control mice and the Ly6A− population showed significantly higher diversity at day 10 post-infection (Figure 4C–F). Furthermore, recruited CD4+Tomato+ T cells in AdV-infected mice displayed limited IEL differentiation, as assessed by CD8αα and CD103 expression at day 30 post-infection, and remained similarly diverse (Figure 4B and Figure S4A).
Figure 4. Clonally diverse Ly6A+CD4+ T cells are recruited to the IE compartment 10- and 30-days post AdV-infection.
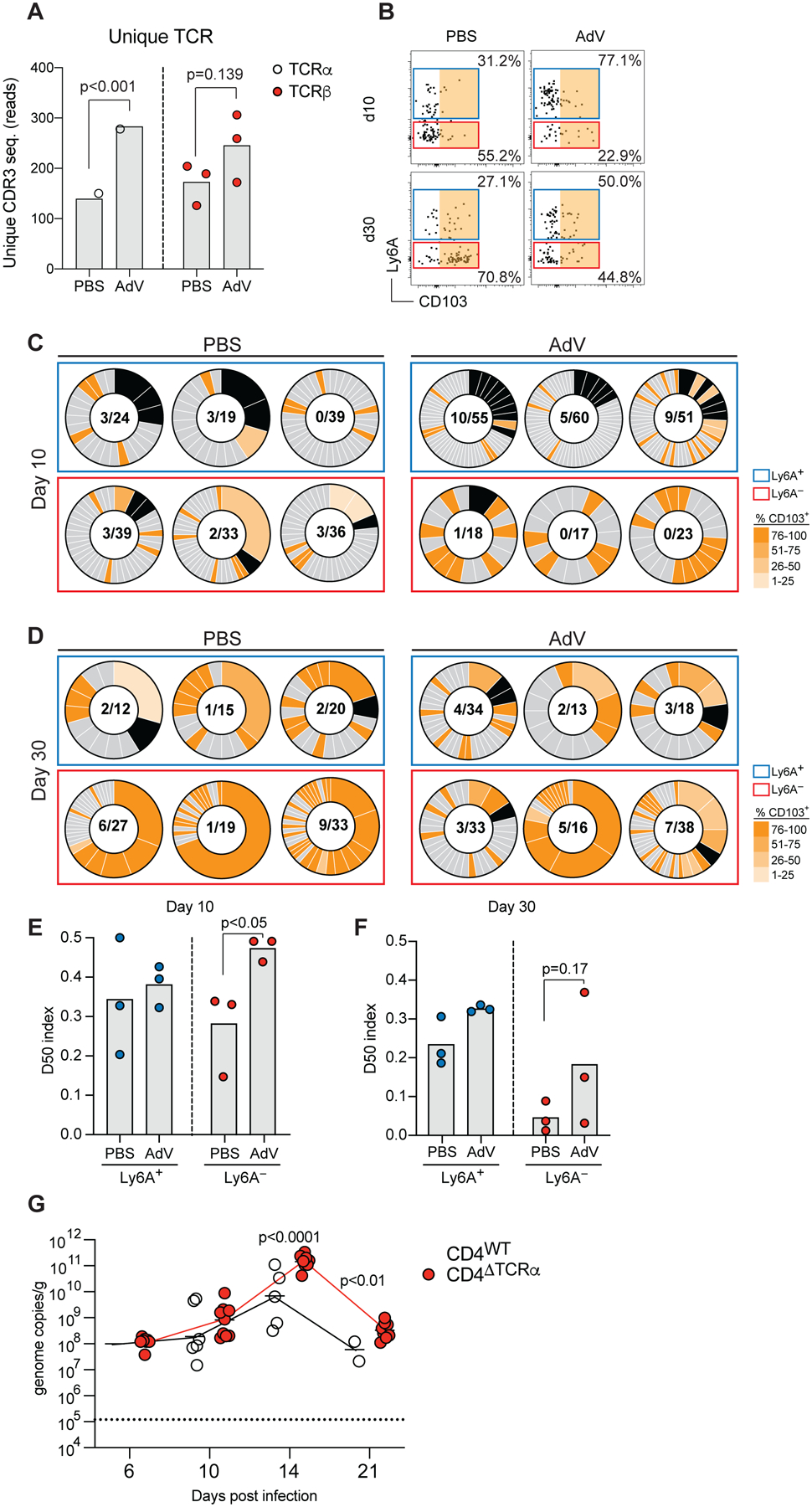
(A) Unique TCRα and TCRβ CDR3 sequence reads were extracted from bulk RNAseq data in figure 2. (B-F) iSellTomato mice were infected with AdV or treated with PBS vehicle control only, and small intestine CD4+CD62L−Tomato+ T cells were single-cell sorted at 10- and 30-days post infection for TCRβ-seq. (B) Ly6A and CD103 expression analysis of CD4+CD62L+Tomato+ T cells indexed and sorted for analysis in B and C. Colored gates represent indexed analysis in B and C. Orange area indicates CD103+ gate for C and D. Single cell TCRβ analysis at day 10 (C) and day 30 (D) post AdV-infection. Box colors represent gate color in A. Black pie regions represent expanded clones, grade of orange represents level of CD103 expression per clone indexed in A. Nominator represents total number of expanded clones, denominator represent total number of sequenced clones. (E-F) D50 analysis of CD4+Tomato+Ly6A+ and Ly6A− at day 10 (E) and day 30 (F) post AdV infection. (G) Viral genome copies of AdV per gram of feces of AdV-infection over time of iCD4ΔTCRα mice, dashed line represents limit of detection. Data are expressed as mean of individual mice, for A (n = 3 per group and timepoint, 1 experiment), for B-F (n = 3 per group and timepoint, 1 experiment), and for G (n = 8–12, two independent experiments). Significant p values as indicated, Student’s t-test in A, E, F and G. See also Figure S4.
To initially investigate possible anti-viral functions of gut recruited CD4+ T cells, we first assessed a role for TCR expression by these cells. In a recent study, we found that intraepithelial CD4+ T cells require TCR expression during their differentiation from peripheral precursors, but not necessarily for maintenance of their epithelial program, or function in a model of bacterial infection (Bilate et al., 2020). We infected mice in which we inducibly deleted TCRα in CD4+ T cells (Cd4CreERxTracfl/fl, iCD4ΔTCRα) immediately before AdV-infection. We found increased AdV titers in the stool of iCD4ΔTCRα mice, pointing to a role for the TCR in CD4+ T cell control of viral replication in the intestine (Figure 4G). However, analogous to findings obtained in our previous study (Bilate et al., 2020), excising TCRα from CD4+ T cells 6 days post infection did not impact anti-viral mechanisms during primary, or memory responses (Figure S4B, C). Hence, recruited IE CD4+ T cells post AdV infection retained clonal diversity and the activation marker Ly6A with limited expression of CD103. In contrast, recruited CD4+ T cells in uninfected mice, had reduced clonal diversity and increased CD103 expression over time. Furthermore, our data suggest that TCR signaling is required during CD4+ T cell activation but removal of the TCR post infection does not impact anti-viral mechanisms.
AdV- and CR6-recruited IE CD4+ T cells clear viruses in intestinal organoid cultures
To further examine the ability of virus-recruited CD4+ T cells to regulate viral replication, we developed an intestinal organoid (Sato et al., 2009) and T cell co-culture system in vitro. To visualize ongoing AdV infection and replication in the organoids, we used a replication competent mouse adenovirus-2 encoding green fluorescent protein (GFP) fused to the minor capsid protein XI (AdV-GFP) (Wilson et al., 2017). AdV-GFP-infected organoids displayed GFP signal as early as 6 hours post-infection; when cultures were continuously monitored for up to 48 hours (Figure 5A, B and Suppl. Movie 1). We sorted CD4+Tomato+ IE T cells isolated from tamoxifen-treated iSellTomato mice infected with AdV, CR6 or T1L, 10 days post infection and co-cultured them with AdV-GFP infected organoids (Figure 5C). CD4+Tomato+ IE T cells isolated from either AdV- or CR6-infected mice, but not cells isolated from T1L-infected mice, controlled AdV-GFP replication in intestinal organoids, suggesting that the differentiation of CCR9+Ly6A+CD4+ T cells is associated with cross-protection against enteric viruses (Figure 5D, E and Figure S4D), data that indirectly reinforces the observations that continuous TCR expression by virus-recruited CD4+ T cells may not be required to regulate viral replication. To independently examine the role of TCR engagement in our organoid model, we co-cultured CD4+Tomato+ IE T cells from AdV- and CR6-infected mice with organoids generated from mice deleted of major histocompatibility complex II (MHCII). Consistent with a TCR-independent function, both AdV- and CR6-derived CD4+ T cells controlled AdV-GFP replication in MHCII-deficient organoid cultures (Figure S4E). To specifically address a functional role by recruited Tomato+CD4+ T cells in controlling AdV replication, we sorted Tomato+CCR9+Ly6A+, Tomato+CCR9−Ly6A−, Tomato−CCR9+Ly6A+ and Tomato−CCR9−Ly6A− IE CD4+ T cells from tamoxifen-treated iSellTomato mice infected with AdV 10 days post infection and co-cultured them with AdV-GFP infected organoids. We found that Tomato+CCR9+Ly6A+CD4+ T cells had significantly higher capacity for controlling viral replication in the organoids in comparison to other T cell populations (Figure 5F).
Figure 5. Newly recruited IE CD4+ T cells show anti-viral activity in AdV-infected intestinal organoid co-cultures.
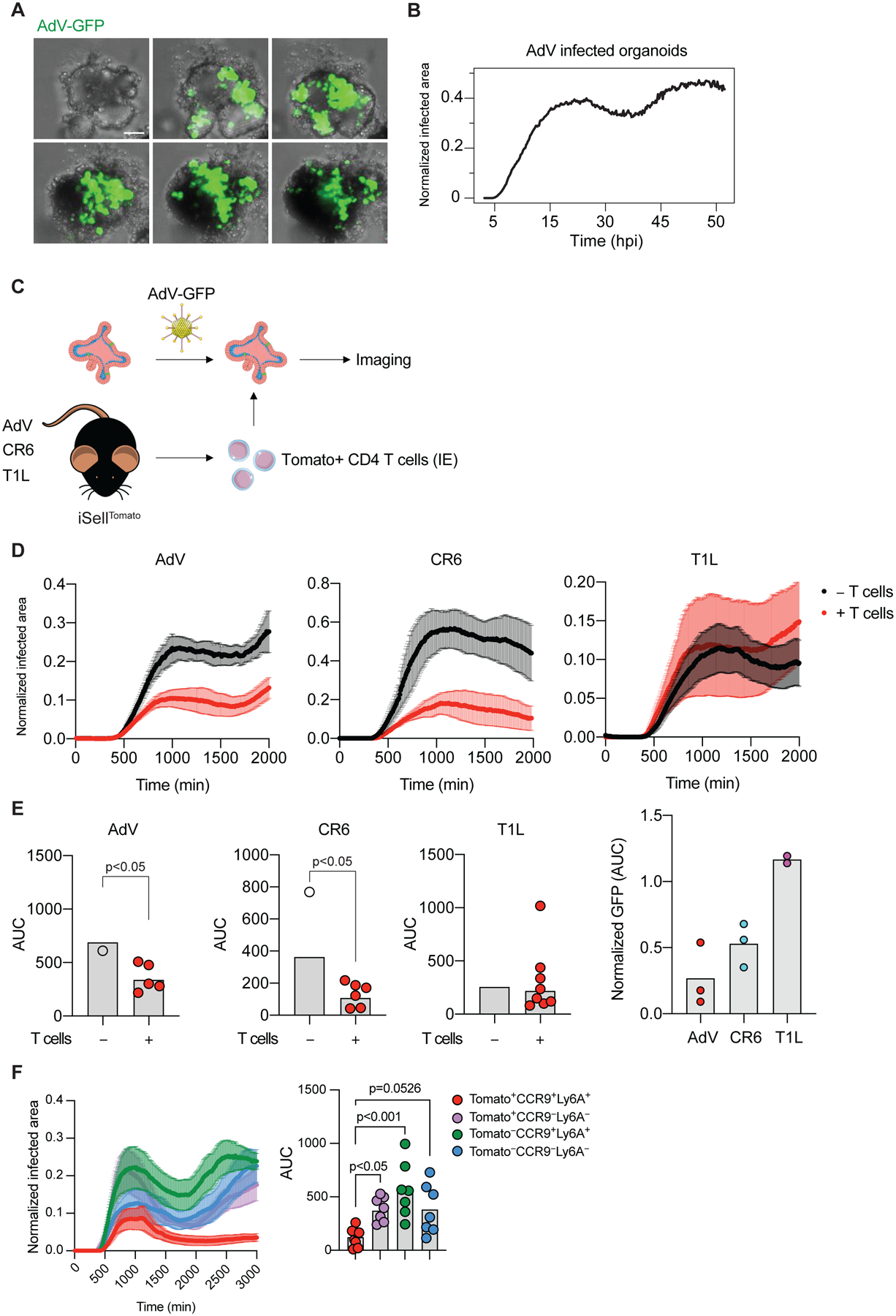
(A-B) Small intestinal organoids were infected with 104 i.u. of AdV-GFP and imaged for 48 hours. (A) Representative images of organoids infected with AdV-GFP (green) at the indicated times post infection (hours, top left corner). (B) GFP expression area over time normalized to the area of organoids. (C-E) iSellTomato mice were infected with different viruses, small intestine CD4+Tomato+ IE T cells were then sorted and co-cultured with 104 i.u. AdV-GFP infected organoids. (C) Experimental overview. (D) Representative graphs of GFP expression area from AdV-GFP infected organoids normalized to the area of the organoids over time with (red) or without (black) CD4+Tomato+ IE T cells derived from mice infected with AdV, CR6 or T1L 10 days post infection. Data are expressed as mean ± SEM of individual experiment. (E) Summarized area under the curve (AUC) for D. Data points for individual organoids in E (left). Pooled data from 2–3 experiments (right). (F) iSellTomato mice were infected with AdV, CD4+Tomato+CCR9+Ly6A+, CD4+Tomato+CCR9−Ly6A−, CD4+Tomato−CCR9+Ly6A+ and CD4+Tomato−CCR9−Ly6A− IE T cells were then sorted and co-cultured with 104 i.u. AdV-GFP infected organoids. Data are expressed as mean of individual organoid in E (except right figure of pooled data) and F. n = 5–10 organoids of two independent experiments A-E, one experiment in F. p values as indicated, Student’s t-test in E or one-way ANOVA plus Bonferroni test in F.
We have previously correlated changes in T cell motility with anti-pathogen responses by IELs and detected preferential displacement of IELs to regions containing pathogens using live intravital microscopy (Hoytema van Konijnenburg et al., 2017). Longitudinal imaging analysis of the organoid and T cell co-cultures showed cellular interactions between CD4+ IE T cells derived from AdV infected mice with AdV-infected epithelial cells (Figure 6A).
Figure 6. Newly recruited IE CD4+ T cells interact with AdV-infected epithelial cells in intestinal organoids.
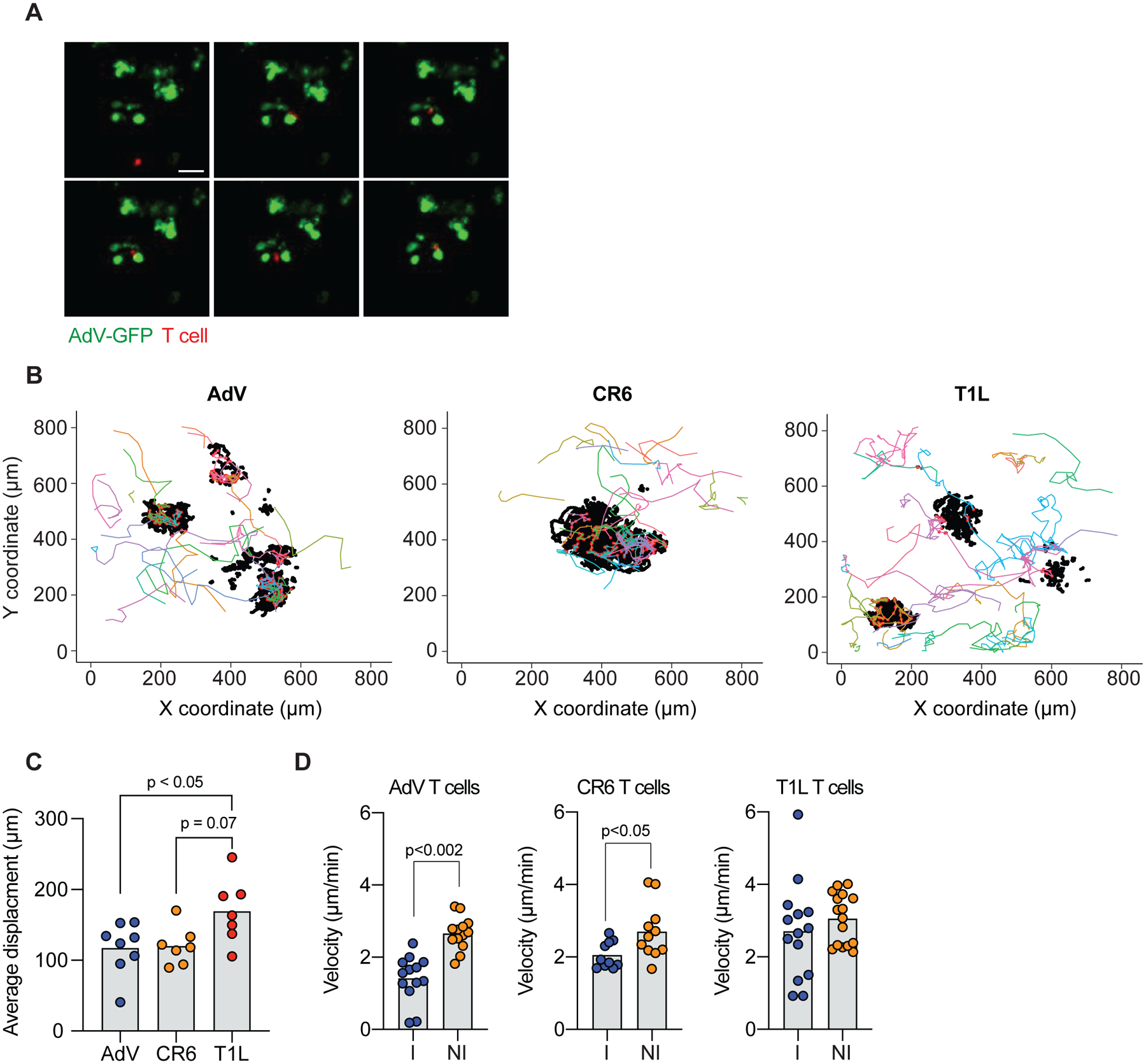
(A-D) Small intestine CD4+Tomato+ IE T cells derived from AdV, CR6 or T1L infected iSellTomato mice were co-cultured with AdV-GFP infected organoids and tracked over 48 ± 2 hours. (A) Representative images of AdV-GFP infected organoids (green) and IE CD4+Tomato+ T cells (red) derived from AdV-infected mice. Arrows indicate tracked T cells, top left corner indicates time post infection in minutes. (B) Tracking was obtained in 3 dimensions (x, y, and z); figure shows only x and y. Each colored line represents a unique T cell track, black dots represent GFP+ epithelial cells and red dots represent T cell-organoid interaction points (<20 μm radius distance). (C) Average displacement distance of tracked Tomato+ T cells co-cultured with AdV-infected organoids. (D) Velocity of tracked tomato+ T cells, interacting T cells (I) within 20 μm radius of GFP+ cells, or non-interacting (NI) T cells outside the 20 μm radius of GFP+ cells. Data are expressed as mean of individual organoid in C (n = 7–8, two independent experiments). Data are expressed as average of pooled T cell tracks in D (n = 10–16 of two independent experiments). p values as indicated, One-way ANOVA plus Bonferroni test in C, Student’s t-test in D.
Tracking of CD4+Tomato+ IE T cells from AdV- or CR6-infected mice suggested preferential migration of T cells towards the organoids, which contrasted with CD4+ T cells isolated from T1L-infected mice (Figure 6B, C and Suppl. Movies 2–4). Moreover, quantifying cellular velocity revealed that CD4+Tomato+ T cells, derived from AdV- or CR6-infected mice, interacting (<20 μm distance) with GFP+ epithelial cells slowed down in comparison to non-interacting T cells (>20 μm distance) from the same mice, an occurrence not observed in CD4+Tomato+ IE T cells derived from T1L-infected mice (Figure 6D). Therefore, the observed cross-protection by CD4+Tomato+ IE T cells derived from AdV- or CR6-infected mice correlates with distinct behavior and proximity to AdV-infected epithelial cells.
ThPOK-dependent CCR9+Ly6A+CD4+ IE T cells cross-protect against an enteric virus
Upon arrival to the intestinal epithelium, peripheral CD4+ T cells gradually lose the helper T cell lineage-defining transcription factor ThPOK while upregulating the CD8 T cell lineage-defining transcription factor Runt-Related Transcription Factor 3 (Runx3), and T-bet (London et al., 2021; Mucida et al., 2013; Reis et al., 2013; Sujino et al., 2016). This transition is accompanied by the loss of hallmarks associated with T helper subsets, including RORγt or FOXP3 expression, and acquisition of IEL markers including CD244 and CD8αα homodimers (Mucida et al., 2013; Reis et al., 2013). Of note, while we had observed upregulation of granzyme B and IFN-γ by recruited CD4+ T cells during viral infection, which are characteristics of IEL differentiation, these cells do not display ThPOK downregulation or express CD8αα (see Figure 1G and Figure S4A above). To determine the role of transcription factors associated with IEL differentiation or CD4 T cell identity in the function and differentiation of gut-recruited CD4 T cells during enteric virus infection, we inducibly and specifically targeted Zbtb7b (iCD4ΔThPOK), Tbx21 (iCD4ΔTbet) or Runx3 (iCD4ΔRunx3) (Mucida et al., 2013; Reis et al., 2014; Reis et al., 2013) in CD4+ T cells upon tamoxifen administration prior to infection. AdV-infected iCD4ΔThPOK mice displayed significantly reduced viral clearance compared to wild-type (WT) infected controls (Figure 7A), while iCD4ΔRunx3 mice presented delayed clearance but similar control by day 21 post-infection (Figure 7B). iCD4ΔTbet showed no differences in viral clearance, compared to WT controls (Figure 7C). Consistent with a functional role for CCR9+Ly6A+CD4+ T cells in viral control, AdV-infected iCD4ΔThPOK mice did not show recruitment of these cells to the IE and LP compartment 10 days post infection, while iCD4ΔTbet and iCD4ΔRunx3 mice displayed significant accumulation similar to control mice (Figure 7D and S5A). Furthermore, CCR9+Ly6A+CD4+ IE T cells from AdV-infected iCD4ΔRunx3 and iCD4ΔThPOK mice showed reduced IFN-γ production when compared to infected control or iCD4ΔTbet mice (Figure 7E and S5B), suggesting that unlike typical Th1 cells, CCR9+Ly6A+CD4+ T cell IFN-γ production is Tbet-independent.
Figure 7. ThPOK-dependent CCR9+Ly6A+ CD4+ T cells show anti-adenoviral function in vivo.
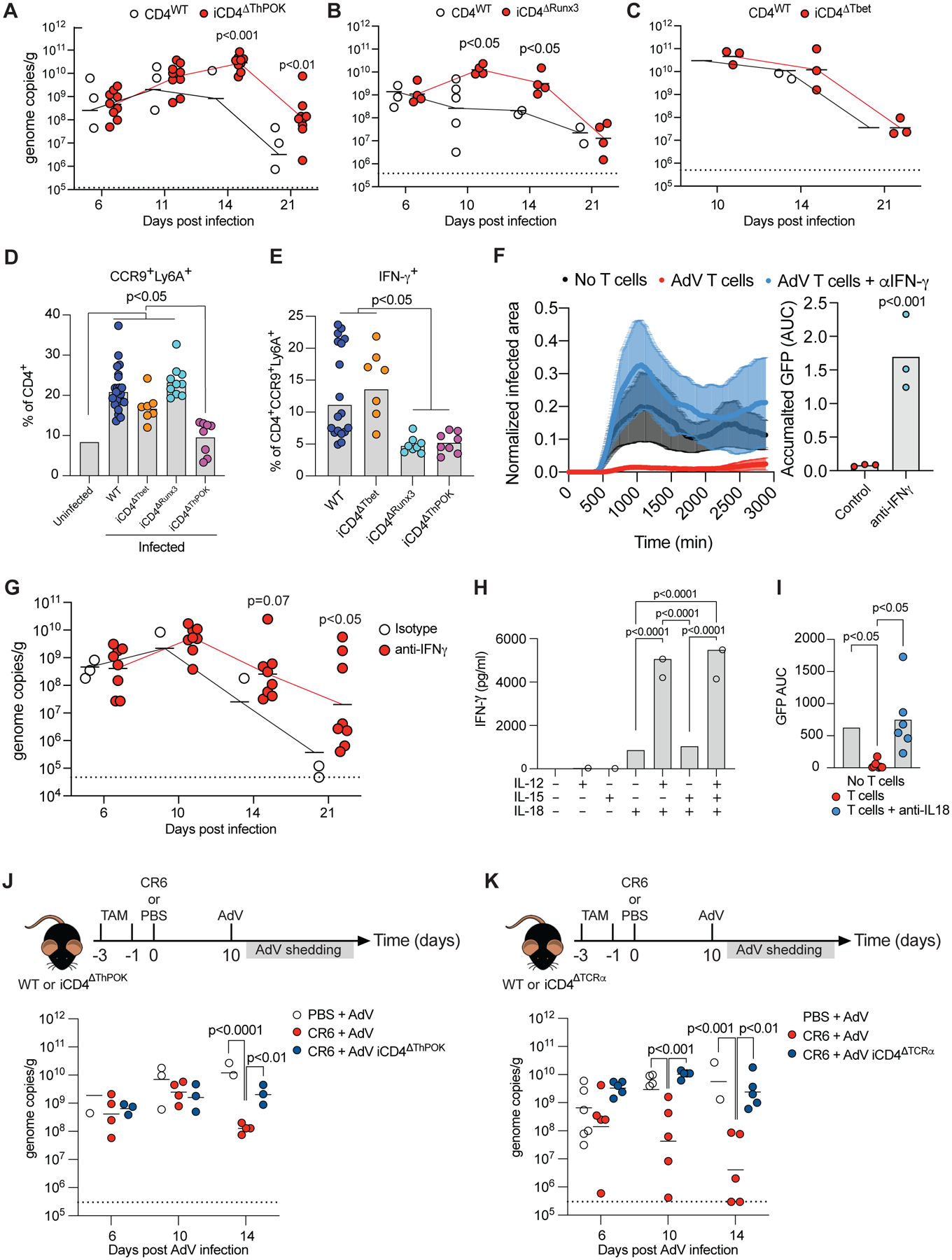
(A-C) AdV genome copies per gram of feces over time of post-infection (A) iCD4ΔThPOK, (B) or iCD4ΔRunx3 (C) iCD4ΔTbet mice. Dashed line represents limit of detection. (D-E) Mice were infected with AdV and small intestine IE T cells were analyzed 10 days post infection. (D) Frequency of CCR9+Ly6A+ cells among CD4+ T cells. (E) IFN-γ production among CCR9+Ly6A+CD4+ T cells. (F, I) iSellTomato mice were infected with AdV and small intestine CD4+Tomato+ T cells were sorted at day 10 post infection. T cells was co-cultured with AdV-GFP infected organoids and imaged with or without anti-IFN-γ (F) or anti-IL-18 (I) blocking antibodies. Normalized infected area over time imaged (left) and accumulated GFP protein expression (right) in indicated conditions. (G) AdV genome copies per gram of feces over time in mice treated with IFN-γ blocking antibody or isotype control at day 8, 10, 12, 14, 16 and 18 post infection. (H) iSellTomato mice were infected with AdV and small intestine CD4+Tomato+ T cells were sorted at day 10 post infection. T cells was cultured and stimulated with IL-12, IL-15 and IL-18 as noted in figure for 24 hours. (J-K) WT, iCD4ΔThPOK or iCD4ΔTCRα mice were infected with CR6 (or PBS as control) and infected with AdV 10 days later. AdV stool shedding was measured post AdV-infection. Dashed line represents limit of detection. AdV genome copies per gram of stool of iCD4ΔThPOK (I) or iCD4ΔTCRα (J) mice relative to WT controls over time. Data are expressed as mean of individual mice in A, B, C, D, E, G, J and K, and F and I is expressed as mean of individual organoids, for A (n = 7–9, two independent experiments), for B (n = 4–5, one experiment), for C (n = 3–5, one experiment), for D and E (n = 7–18, two independent experiments), for G (n = 6–8, two independent experiments), for H (n = 2–3, representative of two independent experiments), for I (n = 6–9, one experiment), for J (n = 3–5, one experiment), for K (n = 5–10, two independent experiments). p values as indicated, one-way ANOVA plus Bonferroni test in D, E, H, I, J (log10 transformed) and K (log10). Student t-test in A (log10), B (log10), C (log10), F and G (log10). See also Figure S5.
To determine if the enhanced IFN-γ production we observed by virus-recruited CD4+ T cells mediates the control of AdV infection, we added neutralizing anti-IFN-γ antibodies in the co-culture organoid system. CD4+Tomato+ IE T cells were sorted from AdV-infected iSellTomato mice 10 days post infection and co-cultured with AdV-GFP infected organoids with or without of anti-IFN-γ. Sorted Tomato+ T cells inhibited viral replication in the control condition, but not in the presence of neutralizing anti-IFN-γ, indicating that IFN-γ derived from virus-recruited CD4+ T cells exerts an important function in controlling AdV replication (Figure 7F). Intraepithelial CD4+Tomato+ T cells isolated from CR6-infected mice controlled AdV replication in a similar fashion (Figure S5C and S5D). Conversely, exogenous IFN-γ added to AdV-infected WT organoids readily suppressed virus replication, an effect not observed in Ifngr1−/− organoids (Figure S5E). In the same line, we observed impaired viral clearance upon coculturing of IE CD4+Tomato+ T cells from AdV-infected mice with Ifngr1−/− organoids (Figure S5F). Finally, in vivo treatment of AdV-infected mice with blocking anti-IFNγ antibodies significantly reduced viral clearance (Figure 7G).
Previous studies reported that a combination of the cytokine IL-18 with IL-12 or IL-15 can synergistically augment IFN-γ production by T cells, potentially without TCR stimulation (Tominaga et al., 2000; Yoshimoto et al., 1998). We observed increased Il12rb2 and ll15ra transcripts, but not Il2rb or Il18r1, by AdV-recruited IE and LP CD4+ T cells (Figure S5G). However, recruited IE and LP CD4+ Tomato+ T cells showed increased IL-18R protein expression in AdV-infected 10 and 30 days post infection compared to CD4+Tomato+ T cells recruited during steady state (Figure S5H), paralleling a preferential expression of IL-18R by CD103−CD4+ IELs when compared to CD103+ CD4+ IELs at steady state, suggesting that CD4 IEL differentiation follows a diverging pathway from IL-18R signaling (Figure S5I). Therefore, we investigated whether these cells produce IFN-γ upon IL-18 stimulation independently of TCR engagement. Sorted intraepithelial CD4+Tomato+ T cells from AdV-infected iSellTomato mice were stimulated ex vivo with IL-12 and/or IL-15 in the presence, or not, of IL-18. While IL-18 stimulation alone induced a modest IFN-γ secretion by CD4+Tomato+ IE T cells, this effect was significantly enhanced upon combination with IL-12 (Figure 7H). After confirming that intestinal organoids produce IL-18 (Figure S5J), we assessed whether IL-18 is necessary for T cell-mediated viral clearance in co-cultures of sorted IE CD4+ Tomato+ from AdV-infected iSellTomato mice and AdV-GFP infected organoids in the presence, or not, of blocking anti-IL-18 antibodies. While sorted IE CD4+ Tomato+ T cells efficiently inhibited viral replication, IL-18 blockage reversed this effect (Figure 7I). Moreover, in vivo treatment of AdV-infected mice with blocking anti-IL-18 antibodies also significantly reduced viral clearance (Figure S5K).
Finally, we addressed whether the cross-protective activity observed for CR6-recruited CD4+ IE T cells in the AdV-infected organoid system could be recapitulated in vivo. Tamoxifen-treated iCD4ΔThPOK or wild-type control mice were infected with CR6 or vehicle, and 10 days later, infected with AdV. WT mice infected with CR6 prior to AdV-infection displayed accelerated AdV clearance compared to vehicle-treated mice, reinforcing the possibility of cross-protection. This phenomenon was dependent on ThPOK expression by CD4+ T cells as tamoxifen-treated CR6-infected iCD4ΔThPOK mice did not show reduction in stool AdV titers post infection (Figure 7J). Additionally, as observed during primary AdV infection, initial engagement of the TCR on CD4+ T cells during the primary CR6-infection was also necessary for cross-protection against AdV, as CR6-infected iCD4ΔTCRα mice displayed increased AdV titers in the stool when compared to wild-type control mice (Figure 7K). Furthermore, excising TCRα from CD4+ T cells 6 days post CR6-infection did not impact cross-protection by the recruited CD4+ T cells against AdV infection (Figure S5L). Hence, ThPOK-dependent CCR9+Ly6A+CD4+ T cells induced by AdV or CR6 are recruited to the IE compartment, acquiring an increased IFN-γ production that controls AdV viral replication in vivo and in vitro.
Discussion
By developing a mouse genetic strategy allowing for the fate-mapping of newly recruited polyclonal T cells to the intestinal tissue, our study uncovered an important role of CD4+ IELs in the protection against enteric adenovirus infection. The fate-mapping approach also revealed that while MNV (CR6) and AdV, viruses with tropism for the epithelium, induced a robust CD4+ T cell recruitment to the epithelium, CW3 and Reovirus T1L, viruses that predominantly infect cells in the LP, preferentially recruited CD8αβ T cells to both LP and IE compartments. After initial priming in the gut-draining LNs (Esterhazy et al., 2019), newly recruited T cells are further exposed to gut tissue signals in the LP, a site with much higher density of myeloid cell populations than the intestinal epithelium (London et al., 2021). This stepwise process probably plays an important role in the differentiation of CCR9+Ly6A+CD4+ T cells, which are exposed to TCR- and cytokine-signals, including IL-12 and IL-18, before even reaching the epithelium. Such intra-tissue specialization, directly associated with the site of insult, highlights distinct lymphocyte differentiation strategies, as recently demonstrated during steady state (London et al., 2021) and infection (Kiner et al., 2021). Nonetheless, whether newly recruited LP versus IE CD4+ T cells play complementary or redundant roles during infections remains to be determined.
Upon arriving in the IE compartment at steady state, recruited peripheral CD103− CD4+ T cells gradually acquire an IEL phenotype, progressively acquiring CD103 and CD8αα expression (Bilate et al., 2020; London et al., 2021). During this process, a progressive loss of ThPOK allows for increased imprinting by other transcription factors including Runx3 and T-bet, resulting in acquisition of a cytotoxic machinery (London et al., 2021; Mucida et al., 2013; Reis et al., 2014; Reis et al., 2013). As virus-induced cytotoxic CD4+ effector T cells have previously been identified in several murine models (Brien et al., 2008; Hou et al., 1992; Stuller et al., 2010) as well as human viral infections (Swain et al., 2012; van Leeuwen et al., 2004; Zaunders et al., 2004). IEL differentiation would presumably facilitate response to viruses (Mucida et al., 2013; Reis et al., 2013). While we observed preferential epithelial recruitment of CD4+ T cells in the context of AdV or chronic MVN infections, acquisition of an CTL-like IEL program was prevented and CD4+ T cells instead differentiated into IFN-γ-producing CCR9+Ly6A+ cells functionally dependent on continuous ThPOK expression. ThPOK has been previously shown to modulate Th1 phenotype during effector differentiation (Vacchio et al., 2014). Additionally, a role for ThPOK in the “functional fitness” of CD4+ T cells during responses to systemic LCMV was recently reported (Ciucci et al., 2019), supporting a role for this transcription factor in anti-viral CD4 T cell responses.
In contrast to the IEL differentiation pathway (London et al., 2021), we observed that AdV-recruited IE CD4+ T cells acquired a mixed Th1 and CTL effector phenotype with most of the cells expressing CD69, resembling some characteristics associated to tissue resident memory (TRM) though lacking CD103 expression (Mackay et al., 2013; Masopust and Soerens, 2019). Furthermore, a recent study reported that CD103− TRM had increased proliferative potential and enhanced function but could also readily modulate their phenotype upon relocation, in contrast to CD103+ TRM (Christo et al., 2021). The distinct differentiation of the fate-mapped cells suggests that the lack of CD103 expression and IEL differentiation is due to a cell-intrinsic regulation. It is possible that their TGF-β signaling is impaired (Konkel et al., 2011; Reis et al., 2013) or that pathways upregulated by these cells counteract Runx3-mediated IEL differentiation. Runx3, which has been previously associated with CTL (Lotem et al., 2013), IEL (Reis et al., 2013) and TRM (Milner et al., 2017) differentiation, is not required for upregulation of CCR9 or Ly6A by virus-recruited CD4+ T cells, but regulates IFN-γ production by these cells. Our data overall suggest a major role for IFN-γ, rather than CTL activity, in the anti-viral activity of these cells. Hence, although the differentiation of IE-recruited CD4+ T cells upon virus infection appears to follow a distinct path from TRM and IEL CD4+ T cells, it could be related to CTL CD4+ T cell responses observed in other tissues, a possibility that requires additional investigation supported by our observations reported here.
Another characteristic of peripheral T cell recruitment to the IE compartment is clonal expansion and progressive loss of TCR diversity as they undergo IEL differentiation during homeostasis (Bilate et al., 2020). In contrast to these steady state observations, virus-recruited IE CD4+ T cells displayed a clonally diverse TCR population, which is comparable to what has been described for peripheral CD4+ T cells upon LCMV infection (Khatun et al., 2021). Nevertheless, corresponding to our observations under steady state (Bilate et al., 2020), while TCR expression was required for initial differentiation and function of IE-recruited CD4+ IELs, TCR removal post infection did not impact the capacity of virus-recruited CD4+ T cells to control viral replication. This observation, in addition to reinforcing the notion that IE T cells may depend less on TCR engagement to exert their function than peripheral T cells (Bilate et al., 2020), may explain their ability to cross protect against unrelated viruses in vivo and in our organoid model. The capacity of recruited CD4+ T cells to secrete IFN-γ in response to IL-18 in combination to IL-12 or IL-15, independent of TCR engagement, is similar to what has been described to memory CD8+ T cells (Ariotti et al., 2014) and could represent a general mechanism co-opted by IELs to confer broad protection in the epithelium. Conversely, cross-protection to unrelated pathogens has been previously reported to skin-resident CD8αβ+ T cells upon herpes simplex virus infection (Ariotti et al., 2014).
A relevant point in our studies is that functional IE-recruited CD4+ IELs were primarily found in a chronic viral model, raising the possibility that targeting strategies to enhance the function of this population may benefit viral control. Primary or secondary (cross-protection) viral clearance data indicated the functional relevance of transcription factors associated with the development of CCR9+Ly6A+CD4+ T cells, or their capacity to secrete IFN-γ, observations related to previous reports describing a crucial role for this cytokine in the human adenovirus replication (Mistchenko et al., 1989). Additionally, germ-free mice display reduced numbers of IFN-γ-producing CD4+ T cells the intestine, a phenotype that can be rescued by MNV-CR6 infection (Kernbauer et al., 2014). Our study indicates that enteric viral infections promote distinct T cell responses with intra-tissue specialization within the intestine, with previously unappreciated characteristics of TCR repertoire and requirements, surface markers and functional adaptation.
Limitations of the study
Our study primarily focused on understanding the functional role of intestinal CD4+ T cells during murine adenovirus-2 infection. While our data indicate that MNV-CR6 induced CCR9+Ly6A+CD4+ T cells cross-protect against AdV-infection, we have not addressed the opposite as MNV-CR6 is known to induce a lifelong chronic infection that cannot be cleared by cell mediated or humoral adaptive immunity (Nice et al., 2015). Because our data suggests that CD4+ T cells recruited by L. monocytogenes, share similarities virus-recruited T cells, yet do not upregulate CCR9, future studies are needed to determine whether additional pathogens can trigger analogous TCR-independent T cell effector function. Further investigation is also needed to determine if similar mechanisms described in this study occur during infection with other enteric viruses. Moreover, follow up studies should address whether similar pathways exist beyond the intestine and if this information can be translated to human virus infections.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Daniel Mucida (mucida@rockefeller.edu).
Materials Availability
All animal strains used in this study are available from The Jackson Laboratory or were provided by the indicated investigators. All mice are available upon request, but due to the complexity of the crosses, availability may be limited. This study did not generate any new unique reagents.
Data and Code Availability
The raw IE and LP T cell RNA sequencing data, IE TCR sequencing data and processed files reported in this study have been deposited in GEO and are publicly available as of the date of publication. The accession number is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC anti-mouse IL-18Ra (clone: A17071D) | Thermo Fisher Scientific | Cat# 157905, RRID:AB_2860734 |
| Alexa Fluor 700 anti-mouse CD4 (clone: RM4–5) | BD Biosciences | Cat# 557956, RRID:AB_396956 |
| APC anti-mouse IFNg (clone: XMG1.2) | Thermo Fisher Scientific | Cat# 25-7311-82, RRID:AB_469680 |
| BV605 anti-mouse CD8a (clone: 53–6.7) | BioLegend | Cat# 100744, RRID:AB_2562609 |
| BV711 anti-mouse CD8b (clone: H35–17.2) | BD Biosciences | Cat# 740761, RRID:AB_2740424 |
| APC anti-mouse CD8b (clone: H35–17.2) | Thermo Fisher Scientific | Cat# 17-0083-81, RRID:AB_657760 |
| PE-Cy7 anti-mouse CD45 (clone: 30-F11) | Thermo Fisher Scientific | Cat# 25-0451-82, RRID:AB_2734986 |
| eFluor 450 anti-mouse granzyme B (clone: NGZB) | Thermo Fisher Scientific | Cat# 48-8898-82, RRID:AB_11149362 |
| BV421 anti-mouse CD103 (clone: 2E7) | BioLegend | Cat# 121421, RRID:AB_10900074 |
| eFluor 450 anti-mouse CD62L (clone: MEL-14) | Thermo Fisher Scientific | Cat# 48-0621-82, RRID:AB_1963590 |
| FITC anti-mouse CD69 (clone: AB_465120) | Thermo Fisher Scientific | Cat# 11-0691-85, RRID:AB_1134101 |
| APC anti-mouse TCRb (clone: H57–597) | Thermo Fisher Scientific | Cat# 17-5961-83, RRID:AB_469482 |
| APCef780 anti-mouse TCRb (clone: H57–597) | Thermo Fisher Scientific | Cat# 47-5961-82, RRID:AB_1272173 |
| PerCP-eFluor710 anti-mouse TCRgd (clone: GL-3, GL3) | Thermo Fisher Scientific | Cat# 46-5711-82, RRID:AB_2016707 |
| BV421 anti-mouse CCR9 (clone: CW-1.2) | BD Biosciences | Cat#: 565412 RRID: AB_2739223 |
| FITC anti-mouse Ly6A (clone: D7) | Biolegend | Cat# 108106, RRID:AB_313342 |
| Alexa Fluor 647 anti-mouse CD244.2 (clone: eBio244F4) | Thermo Fisher Scientific | Cat# 51-2441-82, RRID:AB_657870 |
| V450 anti-mouse/human Tbet (clone: O4–46) | BD Biosciences | Cat#: 561312 RRID:AB_10611714 |
| APC anti-mouse CD5 (clone: 53–7.3) | Thermo Fisher Scientific | Cat#: 17-0051-81 RRID: AB_469330 |
| Aqua fluorescent reactive dye | TheromoFisher Scientific | Cat# 34966 |
| PE or APC TL (T3b) Tetramer | NIH tetramer facility | Not applicable |
| PE or APC LLO Tetramer | NIH tetramer facility | Not applicable |
| Anti-mouse IFNg (clone: XMG1.2) | BioXcell | Cat#: BE0055 RRID: AB_1107694 |
| Anti-mouse IL-18 (clone: YIGIF74–1G7) | BioXcell | Cat#: BE0237 RRID: AB_2687719 |
| Rat IgG1 (clone: HRPN) | BioXCell | Cat#: BE0088 RRID: AB_1107775 |
| Rat IgG2a (clone: 2A3) | BioxCell | Cat#: BE0089 RRID: AB_1107769 |
| Bacterial and Virus Strains | ||
| Murine Adenovirus-2 | Provided by J. Smith | Wilson et al., 2017 |
| Listeria monocytogenes 10403S-inlA expressing full length OVA | Provided by L. Lefrançois | Sheridan et al., 2014 |
| Reovirus T1L | Provided by B. Jabri | Bouziat et al., 2017 |
| Murine norovirus CW3 | Provided by K. Cadwell | Kernbauer et al, 2014 |
| Murine norovirus CR6 | Provided by K. Cadwell | Kernbauer et al, 2014 |
| Murine Adenovirus-2-GFP | Provided by J. Smith | Wilson et al., 2017 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| Ionomycin | Sigma-Aldrich | Cat# I0634 |
| GolgiStop with Monensin | BD Biosciences | Cat# 554724 |
| Dithiothreitol (DTT) | Sigma-Aldrich | Cat# 10197777001 |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | Cat#: P8139 |
| EDTA | ThermoFisher Scientific | Cat# AM9260G |
| Percoll | GE Healthcare | Cat# 17-0891-01 |
| Streptomycin Sulphate | MP Biomedicals | Cat# 0219454125 |
| Oxford Listeria selective agar base | Sigma Aldrich | Cat# 1070040500 |
| Oxford Listeria selective supplement | Sigma Aldrich | Cat# 1070060010 |
| TCL buffer | Qiagen | Cat# 1031576 |
| RNAClean XP beads | Agentcourt | Cat# A63987 |
| Maxima H- reverse transcriptase (RT) | ThermoFisher Scientific | Cat# EP0751 |
| Betaine Solution | Millipore Sigma | Cat# B0300 |
| RNAsin Plus RNase Inhibitor | ThermoFisher Scientific | Cat# PRN2615 |
| CytoFix/CytoPerm Fixation/Permeabilization Solution Kit | BD Biosciences | Cat# 554714 |
| Saponin | Sigma Aldrich | Cat# 47036 |
| RPMI 1650 | Gibco | Cat# 21870076 |
| Fetal Bovine Serum | Sigma | Cat# F0926 |
| Pen/Strep | Gibco | Cat# 10378016 |
| L-glutamine | Gibco | Cat# 25030–081 |
| Sodium Pyruvate | Gibco | Cat# 11360070 |
| Non-essential amino acids | Gibco | Cat# 11140050 |
| 2-mercaptoethanol | Sigma | Cat# M3148 |
| HEPES | Gibco | Cat# 15630–080 |
| Corn Oil | Sigma-Aldrich | Cat# C8267 |
| R-spondin | R&D | Cat# 4645-RS |
| Noggin | Peprotech | Cat# 250–38 |
| EGF | Peprotech | Cat# 315–09 |
| IL-2 | R&D | Cat# 402-ML |
| IL-15 complex | ThermoFisher Scientific | Cat# 16-8156-82 |
| IL-7 | R&D | Cat# 407-ML |
| IL-18 | R&D | Cat# 9124-IL |
| IFN-g | R&D | Cat# 485-MI |
| Matrigel | ThermoFisher Scientific | Cat# CB40230A |
| Critical Commercial Assays | ||
| eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set | Thermo Fisher Scientific | Cat# 00-5523-00 |
| LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit | Thermo Fisher Scientific | Cat# L34966 |
| Miseq Reagent Kit v2 (500 cycles) | Illumina | Cat# MS-103–1003 |
| Nextera XT DNA Library Preparation Kit (24 samples) | Illumina | Cat# FC-131–1024 |
| QIAamp Fast DNA Stool Mini Kit | Qiagen | Cat# 51604 |
| PowerSYBR Green | Applied Biosystems | Cat# 4368577 |
| IFN-γ ELISA | Thermo Fisher Scientific | Cat# 88-7314-88 |
| IL-18 ELISA | Thermo Fisher Scientific | Cat# 88-50618-88 |
| Deposited Data | ||
| Bulk RNA-Seq | This paper | GEO: GSE196417 |
| Single cell TCR seq | This paper | GEO: GSE196417 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Mouse: Sell-CreER | Provided by M. Nuzzensweig | Merkenschlager et al., 2021 |
| Mouse: B6.129S2-H2dlAb1-Ea/J | Jackson Laboratory | Jax: 003584 |
| Mouse: ThpokeGFP: B6.129P2(Cg)-Zbtb7btm2Litt/J | Jackson Laboratory | Jax: 027663 |
| Mouse: Rosa26lsl-tdTomato: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato) Hze/J | Jackson Laboratory | Jax: 007914 |
| Mouse: Tracf/f | Provided by A. Rudensky, (MSKCC) | Bilate et al., 2020 |
| Mouse: Runx3f/f | Jackson Laboratory | Jax: 008773 |
| Mouse: Tbx21f/f | Jackson Laboratory | Jax: 022741 |
| Mouse: Zbtb7bf/f | Jackson Laboratory | Jax: 009369 |
| Mouse: Ifngr1−/− | Jackson Laboratory | Jax: 003288 |
| Oligonucleotides | ||
| Biotinylated-T22VN used for Bulk RNAseq RT/cDNA synthesis: 5’- Bio-TTTTTTTTTTTTTTTTTTTTTTVN - 3’ | Integrated DNA Technologies | Not applicable |
| All TCR and barcode primers used for scTCRseq are found in Table S1. | Integrated DNA Technologies | Bilate et al., 2020 |
| Paired-end (PE) P1 used for scTCRseq by Miseq 5’-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT -3’ | Integrated DNA Technologies | Han et al., 2014 |
| PE P2 used for scTCRseq by Miseq 5’-AAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCT -3’ | Integrated DNA Technologies | Han et al., 2014 |
| Adenovirus FW 5’-GTCCGATTCGGTACTACGGT-3’ | Integrated DNA Technologies | Gounder, A.P et al., 2016 |
| Adenovirus RV 5’-GTCAGACAACTTCCCAGGGT-3’ | Integrated DNA Technologies | Gounder, A.P et al., 2016 |
| Recombinant DNA | ||
| Software and Algorithms | ||
| GraphPad Prism 9.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo v 10 | BD Biosciences | https://www.flowio.com/solutions/flowio |
| FACSDiva | BD Biosciences | https://www.bdbiosciences.com/en-us/instruments/research-instruments/research-software/flow-cytometry-acquisition/facsdiva-software |
| IMGT | Brochet et al., 2008 | imgt.org/HighV-QUEST |
| R | Not applicable | https://cran.r-project.org/ |
| PANDASEQ | Masella et al., 2012 | https://github.com/neufeld/pandaseq |
| sleuth (v0.30) package for R | Pimentel et al., 2017 | https://github.com/pachterlab/sleuth |
| kallisto (v0.46) software | Bray et al., 2016 | https://github.com/pachterlab/kallisto |
| Immunarch (v0.6.5) package | Not applicable | 10.5281/zenodo.3893991) |
| ImageJ | Not applicable | https://imagej.nih.gov/ij/ |
| TrackMate 6.0.2 | Tinevez et al., 2017 | https://imagej.net/plugins/trackmate/ |
| QuantStudio 3 RT PCR System | Applied Biosystems | https://www.thermofisher.com/us/en/home/life-science/pcr/real-time-pcr/real-time-pcr-instruments/quantstudio-systems.html |
| Other | ||
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Animal care and experimentation were consistent with NIH guidelines and were approved by the Institutional Animal Care and Use Committee at the Rockefeller University. B6.129S2-H2dlAb1-Ea/J (Jax: 003584), Rosa26CAG-LSL-tdTomato-WPRE (007914), Zbtb7bfl/fl (009369), Cd4Cre-ERT2 (022356), Ifngr1−/− (003288) and Tbx21fl/fl (022741) mice were purchased from Jackson Laboratories and housed in our facility. Tracf/f mice were kindly provided by A. Rudensky (MSKCC). Runx3fl/fl (008773) mice were provided by T. Egawa (Washington University in St. Louis). SellCre-ERT2 mice were provided by M. Nussenzweig (Merkenschlager et al., 2021). Several of these lines were interbred in our facilities to obtain the final strains described elsewhere in the text. Genotyping was performed according to the protocols established for the respective strains by Jackson Laboratories or by donor investigators. Mice were maintained at the Rockefeller University animal facility under specific pathogen-free (SPF) conditions. Both male and female littermates were used, age 7–12 weeks old.
Oral infection with Listeria monocytogenes
L. monocytogenes 10403S-inlA strain expressing full-length OVA (Lm-OVA) were grown overnight in brain heart infusion media. Lm-OVA was provided by L. Lefrançois. Mice were infected with 109 colony forming units (CFU) 24h after oral treatment with 20 mg of Streptomycin (Sigma-Aldrich) diluted in water. At day 9 post-infection, intraepithelial lymphocytes were harvested and stained with MHCII-restricted LLO-tetramer (NIH) and analyzed by flow cytometry.
METHOD DETAILS
Oral infection with viruses
WT mouse adenovirus-2(AdV) and AdV-GFP [previously called “MAdV-2.IXeGFP (Wilson et al., 2017)] were propagated in the mouse rectal carcinoma CMT-93 cell line (ATCC CCL-223), purified, and quantified as previously described (Gounder et al., 2016). Mice were infected with 107 infectious units of WT AdV in 100 ul PBS by oral gavage. Reovirus T1L (T1L) was provided by T. Dermody (University of Pittsburgh) and propagated in L929 cell line, purified and quantified as previously described (Kobayashi et al., 2010). T1L. Mice were infected with 108 plaque forming units (pfu) of T1L in 100 ul PBS by oral gavage. Murine norovirus (MVN) CW3 and CR6 were provided by K. Cadwell (New York University) and was propagated in RAW264.7 cell line, purified and quantified as previously described (Kernbauer et al., 2014). Mice were infected with 3×106 pfu of MNV CW3 or CR6 in 100 ul PBS by oral gavage.
Tamoxifen Treatment
Tamoxifen (Sigma) was dissolved in corn oil (Sigma) and 10% ethanol, shaking at 37°C for 30 min-1 h. Two doses of Tamoxifen (5 mg/dose) were administered to mice via oral gavage at 50 mg/mL, 3 days and 1 day before viral infection.
Adenovirus fecal shedding
1 or 2 stool pellets were collected at the days post infection as indicated in the figure legends and frozen at −80°C. On the day of processing, stool samples were weighed and processed with QIAamp Fast DNA Stool Mini Kit (Qiagen) for DNA extraction following the manufacturer’s instructions. Samples were measured for DNA content by NanoDrop (Thermo Scientific). RT-qPCR was performed using PowerSYBR Green (Applied Biosystems) with AdV specific primers; FW: 5’-GTCCGATTCGGTACTACGGT-3’; RV: 5’-GTCAGACAACTTCCCAGGGT-3’, at an annealing temperate of 55°C and for 40 cycles on a QuantStudio 3 RT PCR System (Applied Biosystems). Genomic copies were determined by correlation to an AdV DNA standard and normalized to DNA input and stool weight. Stool from uninfected mice was used as a negative control.
Isolation of intestinal T cells
Small intestine intraepithelial and lamina propria lymphocytes were isolated as previously described (Reis et al., 2013). Briefly, small intestines were harvested and washed in PBS and 1 mM dithiothreitol (DTT) followed by 30 mM EDTA. Intraepithelial cells were recovered from the supernatant of DTT and EDTA washes. Lymphocytes from lamina propria were obtained after collagenase digestion of the tissue. Mononuclear cells were isolated by gradient centrifugation using Percoll. Single-cell suspensions were then stained with fluorescently labeled antibodies for 25 min at 4° C prior to downstream flow cytometry (analysis or sorting) as specified in figure legends.
Staining Strategy
The following gating strategy was utilized to examine CD4+ T cells: single live lymphocytes (based on size and live/dead fixable dye Aqua stain), CD45+, TCRγδ−, TCRβ+, CD8βlow/−, CD4+. For analysis of iSellTomato, the following gating strategy was added: CD62L− and Tomato+. For single-cell sorting of cells subjected to scTCRseq the following gating strategy was used: single live lymphocytes, CD45+, TCRγδ−, TCRβ+, CD4+, CD62L−, Tomato+, Ly6a+/−, CD103+/−, CD8α+/−. For sorting of cells subjected to bulk RNaseq we used single live lymphocytes CD45+, TCRγδ−, TCRβ+, CD8β−, CD4+, Tomato+ and CD62L−. For sorting of T cells subjected to organoid co-cultures, we gated on single live lymphocytes CD45+ TCRγδ−, CD8β−, CD62L−, MHCII−, CD11b−, CD11c−, CD19−, NK1.1−, Tomato+ and CD5+.
In vivo IFN-γ or IL-18 blocking
Mice were injected with 500 μg of anti-IFN-γ (XMG1.2, BioXCell) or Rat IgG1 (HRPN, BioXCell) intraperitoneally at day 8, 10, 12, 14, 16 and 18 post infection. Mice were injected with 100 μg of anti-IL-18 (YIGIF74–1G7, BioxCell) or Rat IgG2a (2A3, BioxCell) intraperitoneally at day 9, 10, 11, 12 and 13 post infection.
In vitro T cell cultures
iSellTomato mice were infected with AdV and CD45+CD62L−CD4+Tomato+ IE T cells were sorted 10 days post infection. Sorted T cells were cultured in RPMI 1640 (Gibco), 10% FBS (Sigma), 1% Pen/Strep (Gibco), 1% L-glutamine (Gibco), 1% Sodium Pyruvate (Gibco), 2% Non-essential Amino Acids (Gibco), 2.5% 1M HEPES (Gibco), 50μM 2-Mercaptoethanol (Sigma), 10 ng/ml recombinant murine IL-2 (R&D) and 5 ng/ml recombinant murine IL-7 (R&D) at 10–20.000 cells/well in a 96-well round bottom plate (Corning). T cells were stimulated with 10 ng/ml recombinant murine IL-12 (R&D), 10 ng/ml recombinant murine IL-15/IL-15Rα complex (ThermoFisher) and 10 ng/ml recombinant murine IL-18 (R&D). Supernatants were collected 24-hour post stimulation and IFN-γ was measured with IFN-γ ELISA (Invitrogen) by following manufacturer’s instructions.
Intracellular Staining and Flow Cytometry
For analysis of cytokine secretion, total mononuclear cells isolated from the epithelium were plated in 48-well plates and incubated at 37° C with 100 ng/mL phorbol 12-myristate 13-acetate (PMA, Sigma), 200 ng/mL ionomycin (Sigma) and 2mM monensin (BD Biosciences) for 4 hours. Intracellular staining for IFN-γ and granzyme B was conducted in Perm/Wash buffer after fixation and permeabilization in Fix/Perm buffer (BD Biosciences, USA) according to kit instructions. Flow cytometry data were acquired on an LSR-II flow cytometer (Becton Dickinson,USA) and analyzed using FlowJo 10 software package (Tri-Star, USA).
Organoids and co-cultures
Organoids were established from crypts isolated from adult mouse small intestine and maintained as described previously (Rogoz et al., 2015; Sato et al., 2009). For infection and co-cultures, organoids were removed from the Matrigel by gently pipet off the top media layer and 500 μl of cold culture media containing 10% FBS (Sigma F0926) was added to each well to dissolve the Matrigel. Organoids was washed with cold culture media containing 10% FBS and 30–40 organoids were infected with 104 i.u. of AdV-GFP in 500 μl cold PBS for 20 min on ice. Inoculum was discarded and infected organoids was washed with 15 ml of T-cell culture medium (TCM) [RPMI 1640, 10% FBS, 1% Pen/Strep (Gibco), 1% L-glutamine (Gibco), 1% Sodium Pyruvate (Gibco), 2% Non-essential Amino Acids (Gibco), 2.5% 1M HEPES (Gibco), 50μM 2-Mercaptoethanol (Sigma)]. Then, working on ice, 30 μl of 2–3×104 sorted CD4+Tomato+ T cells in TCM was carefully added to 30 ul of 8–12 infected organoids in 30 μl TCM, next we added 40 ul of ice cold Matrigel. 100 μl of the T cell/organoid/Matrigel mixture was immediately added to a preheated (37°C) 96-well culture plate with glass bottoms (MatTek). The culture plate was incubated for 20–30 min in 37°C to let the Matrigel polymerize. Next, we added 200 μl of TCM containing 50 ng/ml recombinant murine EGF, 100 ng/ml recombinant murine Noggin, 500 ng/ml recombinant human R-spondin, 10 ng/ml recombinant murine IL-2 (R&D), 5 ng/ml recombinant murine IL-7 (R&D) and 10 ng/ml recombinant murine IL-15/IL-15Rα complex (ThermoFisher) on top of the polymerized Matrigel. For IFN-γ neutralizing experiments, we added 10 μg/ml of anti-IFN-γ (XMG1.2, BD Biosciences). For IFN-γ stimulation, we added 10 ng/ml of IFN-γ (R&D). For IL-18 neutralizing experiments, we added 50 μg/ml of anti-IL-18 (YIGIF74–1G7, BioXcell)
Organoid analysis and T cell tracking
Live imaging was performed on the CellVoyager (Yokagawa/Olympus) spinning disk confocal microscope at 37°C and with 5% CO2. Co-cultures were imaged in 96-well plates with glass bottoms (MatTek). 10 to 12 z-stacks of 1.52 μm step-size were acquired every 6–7 min for 48 hours. GFP signal above background per organoid was used to measure the GFP expression area for each z-stack and time point. Organoid area was determined with brightfield images for each z-stack and time point and used to normalize the GFP expression area for each respective z-stack and time point. For T cell tracking, hyperstacks were made of each z-stack, time point and channel. T cell (Tomato+) and infected epithelial (GFP+) tracking was obtained with TrackMate 6.0.2 (Tinevez et al., 2017) on hyperstacked images and over time, cell diameter was set at 14 μm to obtain optimal tracking. Tomato+ T cell tracking 3D coordinates was analyzed together with GFP+ infected epithelial 3D coordinates. If a Tomato+ track point was within 20μm radius of a GFP+ track point in any dimension at a specific given time, that track point was considered as an interacting point. A minimum of 8 sequential interacting points in a specific T cell track were used to calculate the cell average velocity. Reconstruction of images were done using a Python script and the skimage v.0.19 package. Image quantifications and modulations were made in ImageJ 2.1 and all image calculations were made in R studio 1.2.5.
Single-Cell TCR Sequencing
Single cells were index-sorted using a FACS Aria into 96-well plates containing 5μL of lysis buffer (TCL buffer, QIAGEN 1031576) supplemented with 1% β-mercaptoethanol) and frozen at −80° C prior to RT-PCR. RNA and RT-PCRs for TCRβ were prepared as previously described (Dash et al., 2011). PCR products for TCRβ were multiplexed with barcodes and submitted for MiSeq sequencing (Han et al., 2014) using True Seq Nano kit (Illumina). Fastq files were de-multiplexed and paired-end reads were assembled at their overlapping region using the PANDASEQ (Masella et al., 2012) and FASTAX toolkit. Demultiplexed and collapsed reads were assigned to wells according to barcodes. Fasta files from MiSeq sequences were then aligned and analyzed on IMGT (http://imgt.org/HighV-QUEST) (Brochet et al., 2008). Cells with identical TCRβ CDR3 nucleotide sequences were considered as the same clones.
Bulk RNA-seq Library Preparation
Sorted cells (800 cells) were lysed in a guanidine thiocyanate buffer (TCL buffer, QIAGEN) supplemented with 1% β-mercaptoethanol. RNA was isolated by solid-phase reversible immobilization bead cleanup using RNAClean XP beads (Agentcourt, A63987), reversibly transcribed, and amplified as described (Trombetta et al., 2014). Uniquely barcoded libraries were prepared using the Nextera XT kit (Illumina) following the manufacturer’s instructions. Sequencing was performed on an Illumina NextSeq500 for a total yield of 400M reads.
Bulk RNA-seq Analysis
Raw fastq files were processed by using the mouse transcriptome (gencode M23) with the kallisto (v0.46) software (Bray et al., 2016). Analysis of transcript quantification was performed at the gene level by using the sleuth (v0.30) package for R (Pimentel et al., 2017). In short, we modeled batch effect and our experimental design using the sleuth_fit function and detected differentially expressed genes between all groups by the likelihood ratio test (LRT). To determine significantly expressed genes between group pairs, we used the wald-test function. All downstream analysis was made using genes below adjusted p value of 0.05. TCRα and TCRβ CDR3 sequences were reconstructed in silico using MixCR software (Bolotin et al., 2015) and the extracted sequences were analyzed with the Immunarch package (v0.6.5) for R (10.5281/zenodo.3893991).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Analyses
Statistical analysis was carried out using GraphPad Prism v.9. Flow cytometry analysis was carried out using FlowJo software. Data in graphs show mean and p values < 0.05 were considered significant. Repertoire diversity was analyzed by the Diversity 50 (D50). Diversity 50 (D50) was calculated using Excel version 16 as the fraction of dominant clones that account for the cumulative 50% of the TCRβ CDR3s identified. GraphPadPrism v.9 was used for graphs and Adobe Illustrator 2020 used to assemble and edit figures.
Supplementary Material
Supplementary video 1. (Related to Figure 5 and Figure 6) Small intestine organoids were infected with 104 i.u. of AdV-GFP and imaged for 48 hours.
Supplementary video 2. (Related to Figure 5 and Figure 6) Small intestine organoids were infected with 104 i.u. of AdV-GFP and co-cultured with CD4+Tomato+ IELs from AdV-infected mice. Co-cultures was imaged for 50 hours.
Supplementary video 3. (Related to Figure 5 and Figure 6) Small intestine organoids were infected with 104 i.u. of AdV-GFP and co-cultured with CD4+Tomato+ IELs from CR6-infected mice. Co-cultures was imaged for 50 hours.
Supplementary video 4. (Related to Figure 5 and Figure 6) Small intestine organoids were infected with 104 i.u. of AdV-GFP and co-cultured with CD4+Tomato+ IELs from T1L-infected mice. Co-cultures was imaged for 49 hours.
Highlights.
Viral infection results in the recruitment of intraepithelial Ly6A+CCR9+CD4+ T cells
IL-18 activates recruited intestinal CD4+ T cells in the absence of TCR-stimulation
Recruited CD4+ T cells control viral replication via IFN-γ in a TCR-independent manner
Acknowledgments
We are grateful to A. Rogoz, H. Lugo and S. Gonzalez for exceptional animal care, mouse colony management and genotyping; Yasmeen Khan, the Genomics Core, the Bio-Imaging Resource Center (RRID:SCR_017791), and additional Rockefeller University employees for continuous assistance. We thank the NIH tetramer facility for providing the LLO tetramer. We thank A. Bilate for providing advice on the TCR sequencing and editing the manuscript, A. Lockhart for editing the manuscript and all the members of the Mucida, Victora (Rockefeller) and Lafaille (NYU) labs for fruitful discussions. Graphical abstract was created with BioRender.com.
This work was supported by the Black Family Metastasis Center, the Burroughs Wellcome Fund PATH Award, the Mathers Foundation, the Pershing Square Foundation, the FASI/FARE Consortium and National Institute of Health grants R21AI144827, R01DK113375 and R01DK093674, and the Howard Hughes Medical Institute (D.M.). R.P. was supported by the Swedish Research Council and The Sweden-America Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity statement
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. While citing references scientifically relevant for this work, we also actively working to promote gender balance in our reference list.
References
- Ariotti S, Hogenbirk MA, Dijkgraaf FE, Visser LL, Hoekstra ME, Song JY, Jacobs H, Haanen JB, and Schumacher TN (2014). T cell memory. Skin-resident memory CD8(+) T cells trigger a state of tissue-wide pathogen alert. Science 346, 101–105. 10.1126/science.1254803. [DOI] [PubMed] [Google Scholar]
- Bilate AM, London M, Castro TBR, Mesin L, Bortolatto J, Kongthong S, Harnagel A, Victora GD, and Mucida D (2020). T Cell Receptor Is Required for Differentiation, but Not Maintenance, of Intestinal CD4(+) Intraepithelial Lymphocytes. Immunity 53, 1001–1014 e1020. 10.1016/j.immuni.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, and Chudakov DM (2015). MiXCR: software for comprehensive adaptive immunity profiling. Nature methods 12, 380–381. 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- Bouziat R, Hinterleitner R, Brown JJ, Stencel-Baerenwald JE, Ikizler M, Mayassi T, Meisel M, Kim SM, Discepolo V, Pruijssers AJ, et al. (2017). Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science 356, 44–50. 10.1126/science.aah5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nature biotechnology 34, 525–527. 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Brien JD, Uhrlaub JL, and Nikolich-Zugich J (2008). West Nile virus-specific CD4 T cells exhibit direct antiviral cytokine secretion and cytotoxicity and are sufficient for antiviral protection. Journal of immunology 181, 8568–8575. 10.4049/jimmunol.181.12.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet X, Lefranc MP, and Giudicelli V (2008). IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic acids research 36, W503–508. 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J, Cortez VS, Caparon MG, Donia MS, Gilfillan S, et al. (2017). Lactobacillus reuteri induces gut intraepithelial CD4(+)CD8alphaalpha(+) T cells. Science 357, 806–810. 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christo SN, Evrard M, Park SL, Gandolfo LC, Burn TN, Fonseca R, Newman DM, Alexandre YO, Collins N, Zamudio NM, et al. (2021). Discrete tissue microenvironments instruct diversity in resident memory T cell function and plasticity. Nature immunology 22, 1140–1151. 10.1038/s41590-021-01004-1. [DOI] [PubMed] [Google Scholar]
- Ciucci T, Vacchio MS, Gao Y, Tomassoni Ardori F, Candia J, Mehta M, Zhao Y, Tran B, Pepper M, Tessarollo L, et al. (2019). The Emergence and Functional Fitness of Memory CD4(+) T Cells Require the Transcription Factor Thpok. Immunity 50, 91–105.e104. 10.1016/j.immuni.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash P, McClaren JL, Oguin TH 3rd, Rothwell W, Todd B, Morris MY, Becksfort J, Reynolds C, Brown SA, Doherty PC, and Thomas PG (2011). Paired analysis of TCRalpha and TCRbeta chains at the single-cell level in mice. The Journal of clinical investigation 121, 288–295. 10.1172/JCI44752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelblum KL, Singh G, Odenwald MA, Lingaraju A, El Bissati K, McLeod R, Sperling AI, and Turner JR (2015). gammadelta Intraepithelial Lymphocyte Migration Limits Transepithelial Pathogen Invasion and Systemic Disease in Mice. Gastroenterology 148, 1417–1426. 10.1053/j.gastro.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterhazy D, Canesso MCC, Mesin L, Muller PA, de Castro TBR, Lockhart A, ElJalby M, Faria AMC, and Mucida D (2019). Compartmentalized gut lymph node drainage dictates adaptive immune responses. Nature 569, 126–130. 10.1038/s41586-019-1125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria AMC, Reis BS, and Mucida D (2017). Tissue adaptation: Implications for gut immunity and tolerance. J Exp Med 214, 1211–1226. 10.1084/jem.20162014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florsheim EB, Sullivan ZA, Khoury-Hanold W, and Medzhitov R (2021). Food allergy as a biological food quality control system. Cell 184, 1440–1454. 10.1016/j.cell.2020.12.007. [DOI] [PubMed] [Google Scholar]
- Gounder AP, Myers ND, Treuting PM, Bromme BA, Wilson SS, Wiens ME, Lu W, Ouellette AJ, Spindler KR, Parks WC, and Smith JG (2016). Defensins Potentiate a Neutralizing Antibody Response to Enteric Viral Infection. PLoS Pathog 12, e1005474. 10.1371/journal.ppat.1005474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau KR, Roth AN, Zhu S, Hernandez A, Colliou N, DiVita BB, Philip DT, Riffe C, Giasson B, Wallet SM, et al. (2017). The major targets of acute norovirus infection are immune cells in the gut-associated lymphoid tissue. Nat Microbiol 2, 1586–1591. 10.1038/s41564-017-0057-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano VR, Alfajaro MM, Schmitz CO, Filler RB, Strine MS, Wei J, Hsieh LL, Baldridge MT, Nice TJ, Lee S, et al. (2021). CD300lf Conditional Knockout Mouse Reveals Strain-Specific Cellular Tropism of Murine Norovirus. J Virol 95. 10.1128/JVI.01652-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A, Glanville J, Hansmann L, and Davis MM (2014). Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nature biotechnology 32, 684–692. 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, and Littman DR (2016). The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84. 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- Hou S, Doherty PC, Zijlstra M, Jaenisch R, and Katz JM (1992). Delayed clearance of Sendai virus in mice lacking class I MHC-restricted CD8+ T cells. Journal of immunology 149, 1319–1325. [PubMed] [Google Scholar]
- Hoytema van Konijnenburg DP, Reis BS, Pedicord VA, Farache J, Victora GD, and Mucida D (2017). Intestinal Epithelial and Intraepithelial T Cell Crosstalk Mediates a Dynamic Response to Infection. Cell 171, 783–794 e713. 10.1016/j.cell.2017.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, and Cadwell K (2021). Effects of Intestinal Fungi and Viruses on Immune Responses and Inflammatory Bowel Diseases. Gastroenterology 160, 1050–1066. 10.1053/j.gastro.2020.06.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabri B, and Sollid LM (2009). Tissue-mediated control of immunopathology in coeliac disease. Nature reviews. Immunology 9, 858–870. nri2670 [pii] 10.1038/nri2670. [DOI] [PubMed] [Google Scholar]
- Kernbauer E, Ding Y, and Cadwell K (2014). An enteric virus can replace the beneficial function of commensal bacteria. Nature 516, 94–98. 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatun A, Kasmani MY, Zander R, Schauder DM, Snook JP, Shen J, Wu X, Burns R, Chen YG, Lin CW, et al. (2021). Single-cell lineage mapping of a diverse virus-specific naive CD4 T cell repertoire. J Exp Med 218. 10.1084/jem.20200650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiner E, Willie E, Vijaykumar B, Chowdhary K, Schmutz H, Chandler J, Schnell A, Thakore PI, LeGros G, Mostafavi S, et al. (2021). Gut CD4(+) T cell phenotypes are a continuum molded by microbes, not by TH archetypes. Nature immunology 22, 216–228. 10.1038/s41590-020-00836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Ooms LS, Ikizler M, Chappell JD, and Dermody TS (2010). An improved reverse genetics system for mammalian orthoreoviruses. Virology 398, 194–200. 10.1016/j.virol.2009.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel JE, Maruyama T, Carpenter AC, Xiong Y, Zamarron BF, Hall BE, Kulkarni AB, Zhang P, Bosselut R, and Chen W (2011). Control of the development of CD8alphaalpha+ intestinal intraepithelial lymphocytes by TGF-beta. Nature immunology 12, 312–319. 10.1038/ni.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London M, Bilate AM, Castro TBR, Sujino T, and Mucida D (2021). Stepwise chromatin and transcriptional acquisition of an intraepithelial lymphocyte program. Nature immunology 22, 449–459. 10.1038/s41590-021-00883-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotem J, Levanon D, Negreanu V, Leshkowitz D, Friedlander G, and Groner Y (2013). Runx3-mediated transcriptional program in cytotoxic lymphocytes. PloS one 8, e80467. 10.1371/journal.pone.0080467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, Vega-Ramos J, Lauzurica P, Mueller SN, Stefanovic T, et al. (2013). The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nature immunology 14, 1294–1301. 10.1038/ni.2744. [DOI] [PubMed] [Google Scholar]
- Masella AP, Bartram AK, Truszkowski JM, Brown DG, and Neufeld JD (2012). PANDAseq: paired-end assembler for illumina sequences. BMC bioinformatics 13, 31. 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D, and Soerens AG (2019). Tissue-Resident T Cells and Other Resident Leukocytes. Annual review of immunology 37, 521–546. 10.1146/annurev-immunol-042617-053214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald BD, Jabri B, and Bendelac A (2018). Diverse developmental pathways of intestinal intraepithelial lymphocytes. Nature reviews. Immunology 18, 514–525. 10.1038/s41577-018-0013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager J, Finkin S, Ramos V, Kraft J, Cipolla M, Nowosad CR, Hartweger H, Zhang W, Olinares PDB, Gazumyan A, et al. (2021). Dynamic regulation of TFH selection during the germinal centre reaction. Nature 591, 458–463. 10.1038/s41586-021-03187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, Wang D, Getzler AJ, Nguyen T, Crotty S, et al. (2017). Runx3 programs CD8(+) T cell residency in non-lymphoid tissues and tumours. Nature 552, 253–257. 10.1038/nature24993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistchenko AS, Diez RA, and Falcoff R (1989). Inhibitory effect of interferon-gamma on adenovirus replication and late transcription. Biochem Pharmacol 38, 1971–1978. 10.1016/0006-2952(89)90496-6. [DOI] [PubMed] [Google Scholar]
- Mucida D, Husain MM, Muroi S, van Wijk F, Shinnakasu R, Naoe Y, Reis BS, Huang Y, Lambolez F, Docherty M, et al. (2013). Transcriptional reprogramming of mature CD4(+) helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nature immunology 14, 281–289. 10.1038/ni.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, Diamond MS, and Virgin HW (2015). Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science 347, 269–273. 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, and Pachter L (2017). Differential analysis of RNA-seq incorporating quantification uncertainty. Nature methods 14, 687–690. 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- Reis BS, Hoytema van Konijnenburg DP, Grivennikov SI, and Mucida D (2014). Transcription Factor T-bet Regulates Intraepithelial Lymphocyte Functional Maturation. Immunity 41, 244–256. 10.1016/j.immuni.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis BS, Rogoz A, Costa-Pinto FA, Taniuchi I, and Mucida D (2013). Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4(+) T cell immunity. Nature immunology 14, 271–280. 10.1038/ni.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogoz A, Reis BS, Karssemeijer RA, and Mucida D (2015). A 3-D enteroid-based model to study T-cell and epithelial cell interaction. Journal of immunological methods 421, 89–95. 10.1016/j.jim.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, and Clevers H (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. nature07935 [pii] 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Strong DW, Thackray LB, Smith TJ, and Virgin HW (2012). Protruding domain of capsid protein is necessary and sufficient to determine murine norovirus replication and pathogenesis in vivo. J Virol 86, 2950–2958. 10.1128/JVI.07038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuller KA, Cush SS, and Flano E (2010). Persistent gamma-herpesvirus infection induces a CD4 T cell response containing functionally distinct effector populations. Journal of immunology 184, 3850–3856. 10.4049/jimmunol.0902935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujino T, London M, Hoytema van Konijnenburg DP, Rendon T, Buch T, Silva HM, Lafaille JJ, Reis BS, and Mucida D (2016). Tissue adaptation of regulatory and intraepithelial CD4(+) T cells controls gut inflammation. Science 352, 1581–1586. 10.1126/science.aaf3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SL, McKinstry KK, and Strutt TM (2012). Expanding roles for CD4(+) T cells in immunity to viruses. Nature reviews. Immunology 12, 136–148. 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi A, and Hashimoto K (1976). Electron microscope study of experimental enteric adenovirus infection in mice. Infection and immunity 13, 569–580. 10.1128/iai.13.2.569-580.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T, Atarashi K, and Honda K (2016). Development and maintenance of intestinal regulatory T cells. Nature reviews. Immunology 16, 295–309. 10.1038/nri.2016.36. [DOI] [PubMed] [Google Scholar]
- Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, and Eliceiri KW (2017). TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90. 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Tominaga K, Yoshimoto T, Torigoe K, Kurimoto M, Matsui K, Hada T, Okamura H, and Nakanishi K (2000). IL-12 synergizes with IL-18 or IL-1beta for IFN-gamma production from human T cells. International immunology 12, 151–160. 10.1093/intimm/12.2.151. [DOI] [PubMed] [Google Scholar]
- Trombetta JJ, Gennert D, Lu D, Satija R, Shalek AK, and Regev A (2014). Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Current protocols in molecular biology 107, 4 22 21–17. 10.1002/0471142727.mb0422s107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacchio MS, Wang L, Bouladoux N, Carpenter AC, Xiong Y, Williams LC, Wohlfert E, Song KD, Belkaid Y, Love PE, and Bosselut R (2014). A ThPOK-LRF transcriptional node maintains the integrity and effector potential of post-thymic CD4+ T cells. Nature immunology 15, 947–956. 10.1038/ni.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen EM, Remmerswaal EB, Vossen MT, Rowshani AT, Wertheim-van Dillen PM, van Lier RA, and ten Berge IJ (2004). Emergence of a CD4+CD28− granzyme B+, cytomegalovirus-specific T cell subset after recovery of primary cytomegalovirus infection. Journal of immunology 173, 1834–1841. 10.4049/jimmunol.173.3.1834. [DOI] [PubMed] [Google Scholar]
- Wilen CB, Lee S, Hsieh LL, Orchard RC, Desai C, Hykes BL Jr., McAllaster MR, Balce DR, Feehley T, Brestoff JR, et al. (2018). Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science 360, 204–208. 10.1126/science.aar3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmi I, Roman E, and Sanchez-Fauquier A (2003). Viruses causing gastroenteritis. Clin Microbiol Infect 9, 247–262. 10.1046/j.1469-0691.2003.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SS, Bromme BA, Holly MK, Wiens ME, Gounder AP, Sul Y, and Smith JG (2017). Alpha-defensin-dependent enhancement of enteric viral infection. PLoS Pathog 13, e1006446. 10.1371/journal.ppat.1006446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto T, Takeda K, Tanaka T, Ohkusu K, Kashiwamura S, Okamura H, Akira S, and Nakanishi K (1998). IL-12 up-regulates IL-18 receptor expression on T cells, Th1 cells, and B cells: synergism with IL-18 for IFN-gamma production. Journal of immunology 161, 3400–3407. [PubMed] [Google Scholar]
- Zaunders JJ, Dyer WB, Wang B, Munier ML, Miranda-Saksena M, Newton R, Moore J, Mackay CR, Cooper DA, Saksena NK, and Kelleher AD (2004). Identification of circulating antigen-specific CD4+ T lymphocytes with a CCR5+, cytotoxic phenotype in an HIV-1 long-term nonprogressor and in CMV infection. Blood 103, 2238–2247. 10.1182/blood-2003-08-2765. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary video 1. (Related to Figure 5 and Figure 6) Small intestine organoids were infected with 104 i.u. of AdV-GFP and imaged for 48 hours.
Supplementary video 2. (Related to Figure 5 and Figure 6) Small intestine organoids were infected with 104 i.u. of AdV-GFP and co-cultured with CD4+Tomato+ IELs from AdV-infected mice. Co-cultures was imaged for 50 hours.
Supplementary video 3. (Related to Figure 5 and Figure 6) Small intestine organoids were infected with 104 i.u. of AdV-GFP and co-cultured with CD4+Tomato+ IELs from CR6-infected mice. Co-cultures was imaged for 50 hours.
Supplementary video 4. (Related to Figure 5 and Figure 6) Small intestine organoids were infected with 104 i.u. of AdV-GFP and co-cultured with CD4+Tomato+ IELs from T1L-infected mice. Co-cultures was imaged for 49 hours.
Data Availability Statement
The raw IE and LP T cell RNA sequencing data, IE TCR sequencing data and processed files reported in this study have been deposited in GEO and are publicly available as of the date of publication. The accession number is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC anti-mouse IL-18Ra (clone: A17071D) | Thermo Fisher Scientific | Cat# 157905, RRID:AB_2860734 |
| Alexa Fluor 700 anti-mouse CD4 (clone: RM4–5) | BD Biosciences | Cat# 557956, RRID:AB_396956 |
| APC anti-mouse IFNg (clone: XMG1.2) | Thermo Fisher Scientific | Cat# 25-7311-82, RRID:AB_469680 |
| BV605 anti-mouse CD8a (clone: 53–6.7) | BioLegend | Cat# 100744, RRID:AB_2562609 |
| BV711 anti-mouse CD8b (clone: H35–17.2) | BD Biosciences | Cat# 740761, RRID:AB_2740424 |
| APC anti-mouse CD8b (clone: H35–17.2) | Thermo Fisher Scientific | Cat# 17-0083-81, RRID:AB_657760 |
| PE-Cy7 anti-mouse CD45 (clone: 30-F11) | Thermo Fisher Scientific | Cat# 25-0451-82, RRID:AB_2734986 |
| eFluor 450 anti-mouse granzyme B (clone: NGZB) | Thermo Fisher Scientific | Cat# 48-8898-82, RRID:AB_11149362 |
| BV421 anti-mouse CD103 (clone: 2E7) | BioLegend | Cat# 121421, RRID:AB_10900074 |
| eFluor 450 anti-mouse CD62L (clone: MEL-14) | Thermo Fisher Scientific | Cat# 48-0621-82, RRID:AB_1963590 |
| FITC anti-mouse CD69 (clone: AB_465120) | Thermo Fisher Scientific | Cat# 11-0691-85, RRID:AB_1134101 |
| APC anti-mouse TCRb (clone: H57–597) | Thermo Fisher Scientific | Cat# 17-5961-83, RRID:AB_469482 |
| APCef780 anti-mouse TCRb (clone: H57–597) | Thermo Fisher Scientific | Cat# 47-5961-82, RRID:AB_1272173 |
| PerCP-eFluor710 anti-mouse TCRgd (clone: GL-3, GL3) | Thermo Fisher Scientific | Cat# 46-5711-82, RRID:AB_2016707 |
| BV421 anti-mouse CCR9 (clone: CW-1.2) | BD Biosciences | Cat#: 565412 RRID: AB_2739223 |
| FITC anti-mouse Ly6A (clone: D7) | Biolegend | Cat# 108106, RRID:AB_313342 |
| Alexa Fluor 647 anti-mouse CD244.2 (clone: eBio244F4) | Thermo Fisher Scientific | Cat# 51-2441-82, RRID:AB_657870 |
| V450 anti-mouse/human Tbet (clone: O4–46) | BD Biosciences | Cat#: 561312 RRID:AB_10611714 |
| APC anti-mouse CD5 (clone: 53–7.3) | Thermo Fisher Scientific | Cat#: 17-0051-81 RRID: AB_469330 |
| Aqua fluorescent reactive dye | TheromoFisher Scientific | Cat# 34966 |
| PE or APC TL (T3b) Tetramer | NIH tetramer facility | Not applicable |
| PE or APC LLO Tetramer | NIH tetramer facility | Not applicable |
| Anti-mouse IFNg (clone: XMG1.2) | BioXcell | Cat#: BE0055 RRID: AB_1107694 |
| Anti-mouse IL-18 (clone: YIGIF74–1G7) | BioXcell | Cat#: BE0237 RRID: AB_2687719 |
| Rat IgG1 (clone: HRPN) | BioXCell | Cat#: BE0088 RRID: AB_1107775 |
| Rat IgG2a (clone: 2A3) | BioxCell | Cat#: BE0089 RRID: AB_1107769 |
| Bacterial and Virus Strains | ||
| Murine Adenovirus-2 | Provided by J. Smith | Wilson et al., 2017 |
| Listeria monocytogenes 10403S-inlA expressing full length OVA | Provided by L. Lefrançois | Sheridan et al., 2014 |
| Reovirus T1L | Provided by B. Jabri | Bouziat et al., 2017 |
| Murine norovirus CW3 | Provided by K. Cadwell | Kernbauer et al, 2014 |
| Murine norovirus CR6 | Provided by K. Cadwell | Kernbauer et al, 2014 |
| Murine Adenovirus-2-GFP | Provided by J. Smith | Wilson et al., 2017 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| Ionomycin | Sigma-Aldrich | Cat# I0634 |
| GolgiStop with Monensin | BD Biosciences | Cat# 554724 |
| Dithiothreitol (DTT) | Sigma-Aldrich | Cat# 10197777001 |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | Cat#: P8139 |
| EDTA | ThermoFisher Scientific | Cat# AM9260G |
| Percoll | GE Healthcare | Cat# 17-0891-01 |
| Streptomycin Sulphate | MP Biomedicals | Cat# 0219454125 |
| Oxford Listeria selective agar base | Sigma Aldrich | Cat# 1070040500 |
| Oxford Listeria selective supplement | Sigma Aldrich | Cat# 1070060010 |
| TCL buffer | Qiagen | Cat# 1031576 |
| RNAClean XP beads | Agentcourt | Cat# A63987 |
| Maxima H- reverse transcriptase (RT) | ThermoFisher Scientific | Cat# EP0751 |
| Betaine Solution | Millipore Sigma | Cat# B0300 |
| RNAsin Plus RNase Inhibitor | ThermoFisher Scientific | Cat# PRN2615 |
| CytoFix/CytoPerm Fixation/Permeabilization Solution Kit | BD Biosciences | Cat# 554714 |
| Saponin | Sigma Aldrich | Cat# 47036 |
| RPMI 1650 | Gibco | Cat# 21870076 |
| Fetal Bovine Serum | Sigma | Cat# F0926 |
| Pen/Strep | Gibco | Cat# 10378016 |
| L-glutamine | Gibco | Cat# 25030–081 |
| Sodium Pyruvate | Gibco | Cat# 11360070 |
| Non-essential amino acids | Gibco | Cat# 11140050 |
| 2-mercaptoethanol | Sigma | Cat# M3148 |
| HEPES | Gibco | Cat# 15630–080 |
| Corn Oil | Sigma-Aldrich | Cat# C8267 |
| R-spondin | R&D | Cat# 4645-RS |
| Noggin | Peprotech | Cat# 250–38 |
| EGF | Peprotech | Cat# 315–09 |
| IL-2 | R&D | Cat# 402-ML |
| IL-15 complex | ThermoFisher Scientific | Cat# 16-8156-82 |
| IL-7 | R&D | Cat# 407-ML |
| IL-18 | R&D | Cat# 9124-IL |
| IFN-g | R&D | Cat# 485-MI |
| Matrigel | ThermoFisher Scientific | Cat# CB40230A |
| Critical Commercial Assays | ||
| eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set | Thermo Fisher Scientific | Cat# 00-5523-00 |
| LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit | Thermo Fisher Scientific | Cat# L34966 |
| Miseq Reagent Kit v2 (500 cycles) | Illumina | Cat# MS-103–1003 |
| Nextera XT DNA Library Preparation Kit (24 samples) | Illumina | Cat# FC-131–1024 |
| QIAamp Fast DNA Stool Mini Kit | Qiagen | Cat# 51604 |
| PowerSYBR Green | Applied Biosystems | Cat# 4368577 |
| IFN-γ ELISA | Thermo Fisher Scientific | Cat# 88-7314-88 |
| IL-18 ELISA | Thermo Fisher Scientific | Cat# 88-50618-88 |
| Deposited Data | ||
| Bulk RNA-Seq | This paper | GEO: GSE196417 |
| Single cell TCR seq | This paper | GEO: GSE196417 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Mouse: Sell-CreER | Provided by M. Nuzzensweig | Merkenschlager et al., 2021 |
| Mouse: B6.129S2-H2dlAb1-Ea/J | Jackson Laboratory | Jax: 003584 |
| Mouse: ThpokeGFP: B6.129P2(Cg)-Zbtb7btm2Litt/J | Jackson Laboratory | Jax: 027663 |
| Mouse: Rosa26lsl-tdTomato: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato) Hze/J | Jackson Laboratory | Jax: 007914 |
| Mouse: Tracf/f | Provided by A. Rudensky, (MSKCC) | Bilate et al., 2020 |
| Mouse: Runx3f/f | Jackson Laboratory | Jax: 008773 |
| Mouse: Tbx21f/f | Jackson Laboratory | Jax: 022741 |
| Mouse: Zbtb7bf/f | Jackson Laboratory | Jax: 009369 |
| Mouse: Ifngr1−/− | Jackson Laboratory | Jax: 003288 |
| Oligonucleotides | ||
| Biotinylated-T22VN used for Bulk RNAseq RT/cDNA synthesis: 5’- Bio-TTTTTTTTTTTTTTTTTTTTTTVN - 3’ | Integrated DNA Technologies | Not applicable |
| All TCR and barcode primers used for scTCRseq are found in Table S1. | Integrated DNA Technologies | Bilate et al., 2020 |
| Paired-end (PE) P1 used for scTCRseq by Miseq 5’-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT -3’ | Integrated DNA Technologies | Han et al., 2014 |
| PE P2 used for scTCRseq by Miseq 5’-AAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCT -3’ | Integrated DNA Technologies | Han et al., 2014 |
| Adenovirus FW 5’-GTCCGATTCGGTACTACGGT-3’ | Integrated DNA Technologies | Gounder, A.P et al., 2016 |
| Adenovirus RV 5’-GTCAGACAACTTCCCAGGGT-3’ | Integrated DNA Technologies | Gounder, A.P et al., 2016 |
| Recombinant DNA | ||
| Software and Algorithms | ||
| GraphPad Prism 9.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo v 10 | BD Biosciences | https://www.flowio.com/solutions/flowio |
| FACSDiva | BD Biosciences | https://www.bdbiosciences.com/en-us/instruments/research-instruments/research-software/flow-cytometry-acquisition/facsdiva-software |
| IMGT | Brochet et al., 2008 | imgt.org/HighV-QUEST |
| R | Not applicable | https://cran.r-project.org/ |
| PANDASEQ | Masella et al., 2012 | https://github.com/neufeld/pandaseq |
| sleuth (v0.30) package for R | Pimentel et al., 2017 | https://github.com/pachterlab/sleuth |
| kallisto (v0.46) software | Bray et al., 2016 | https://github.com/pachterlab/kallisto |
| Immunarch (v0.6.5) package | Not applicable | 10.5281/zenodo.3893991) |
| ImageJ | Not applicable | https://imagej.nih.gov/ij/ |
| TrackMate 6.0.2 | Tinevez et al., 2017 | https://imagej.net/plugins/trackmate/ |
| QuantStudio 3 RT PCR System | Applied Biosystems | https://www.thermofisher.com/us/en/home/life-science/pcr/real-time-pcr/real-time-pcr-instruments/quantstudio-systems.html |
| Other | ||


