Abstract
Glycine betaine is accumulated in cells living in high salt concentrations to balance the osmotic pressure. Glycine sarcosine N-methyltransferase (GSMT) and sarcosine dimethylglycine N-methyltransferase (SDMT) of Ectothiorhodospira halochloris catalyze the threefold methylation of glycine to betaine, with S-adenosylmethionine acting as the methyl group donor. These methyltransferases were expressed in Escherichia coli and purified, and some of their enzymatic properties were characterized. Both enzymes had high substrate specificities and pH optima near the physiological pH. No evidence of cofactors was found. The enzymes showed Michaelis-Menten kinetics for their substrates. The apparent Km and Vmax values were determined for all substrates when the other substrate was present in saturating concentrations. Both enzymes were strongly inhibited by the reaction product S-adenosylhomocysteine. Betaine inhibited the methylation reactions only at high concentrations.
Bacteria in saline habitats adapt to their environment by accumulating high intracellular concentrations of small organic compounds in order to achieve osmotic balance. These compounds have been termed “compatible solutes” because of their compatibility with cellular metabolism (3). Glycine betaine (called betaine hereafter) is among the most important compatible solutes.
The production of betaine from simple carbon sources is typical of halophilic and halotolerant phototrophic eubacteria but rare among heterotrophic eubacteria (30). However, many microorganisms are able to accumulate betaine from the growth medium or synthesize it from choline by oxidation. Also, many plants accumulate or synthesize betaine in response to salinity or drought (27). In plants, betaine is synthesized from choline, which is a product of the successive methylation reactions of phospho or phosphatidyl derivatives of ethanolamine. Choline is oxidized to betaine in two steps, with betaine aldehyde as the intermediate (5, 9).
We have previously shown that betaine synthesis in the extremely halophilic anaerobic bacterium Ectothiorhodospira halochloris proceeds via the threefold methylation of glycine in the N position. The reactions are catalyzed by two enzymes with partly overlapping substrate specifities. Glycine sarcosine methyltransferase (GSMT) catalyzes the methylation steps from glycine to sarcosine (N-monomethylglycine) and from sarcosine to dimethylglycine. Sarcosine dimethylglycine methyltransferase (SDMT) catalyzes the steps from sarcosine to dimethylglycine and from dimethylglycine to betaine. S-Adenosylmethionine (AdoMet) acts as the methyl group donor in the reactions (24).
Glycine methyltransferases have been isolated from different mammalian origins (26). However, these enzymes are specific for glycine; sarcosine is not methylated further (11). To our knowledge, sarcosine methyltransferases have never been described before. In this study, we have expressed these two novel enzymes in Escherichia coli, purified them, and characterized some of their enzymatic properties.
A possible application of GSMT and SDMT is in enhancing the stress tolerance of plants. Betaine is a well-known protectant of cells and enzymes against various stresses (6, 27). During the past decade, there has been considerable interest in the genetic engineering of drought- and salt-tolerant plants. A well-established approach has been the introduction of choline oxidation into plants which do not synthesize betaine (20). As we have shown with E. coli, the glycine methylation pathway also has potential in improving the stress tolerance of heterologous organisms (35) and could therefore be considered an alternative for choline oxidation.
MATERIALS AND METHODS
Chemicals.
Chemicals were purchased from Sigma-Aldrich unless otherwise indicated.
Methyltransferase activity assays.
A modification of a previously reported methyltransferase activity assay (4) was used. After the methylation of glycine, sarcosine, or dimethylglycine, the unreacted methyl group donor S-adenosyl-l-[methyl-14C]methionine is adsorbed to charcoal, and the remaining methylation products are determined by scintillation counting. The reaction mixture typically contained 25 μl of substrate (1 M glycine or 100 mM sarcosine in GSMT assays and 320 mM sarcosine or 200 mM dimethylglycine in SDMT assays), 25 μl of 0.6 M Tris-HCl (pH 7.4), 25 μl of 8 mM S-adenosyl-l-methionine iodide salt (with 91 nCi of S-adenosyl-l-[methyl-14C]methionine [Amersham Pharmacia Biotech]), and 25 μl of enzyme sample. The reaction mixture was incubated at 37°C for 25 min, the reaction was stopped by adding 75 μl of charcoal suspension (133 g/liter in 5% [wt/vol] trichloroacetic acid), and the mixture was incubated for 10 min at 0°C. After centrifugation for 10 min, 75 μl of the supernatant was removed for assay in a liquid scintillation counter (model 1410; Wallac, Turku, Finland). The enzyme activity is calculated as micromoles of methyl groups transferred per minutes.
The effect of pH on the activity of the purified enzymes was determined by using the following buffers: 125 mM potassium phosphate (pH 4.9 to 6.2), 125 mM triethanolamine (pH 5.8 to 8.3), and 125 mM Tris-HCl (pH 8.0 to 9.0). The pH values were measured at 37°C from the reaction mixtures.
The effects of CaCl2, MgSO4, EDTA, and p-chloromercuribenzoate were tested by incubating the enzymes for 15 min at room temperature with 2.7 mM metal ions, 13.3 mM EDTA, or 1.33 mM p-chloromercuribenzoate. The enzyme reactions were started by adding the substrate (1/4 of the final volume), and the reaction mixtures were incubated for 25 min at 37°C.
The substrate specificity was studied at 37°C using the radiometrical methyltransferase assay described previously (25). In this assay, the unreacted AdoMet is precipitated with phosphotungstic acid and adenosine. The substrates used at 25 mM were ethanolamine (Fluka), monomethylethanolamine (Fluka), and 14 of the common l-amino acids (alanine, asparagine, aspartate, cysteine, glutamate, glutamine, isoleucine, leucine, methionine, phenylalanine, proline, serine, threonine, and valine). The composition of the reaction mixture and the specific radioactivity of AdoMet used at 1 mM were the same as those given above.
Construction of clones expressing GSMT and SDMT.
The GSMT and SDMT gene fragments were amplified by PCR with insertion of NcoI at the 5′ end and BglII at the 3′ end. The primers used were 5′-CGG ACC ATG GAT ACG ACT ACT GAG CAG-3′ and 5′-GCG CAG ATC TTC AGT CCT CCT CCC GAT ATT CCT-3′ for GSMT and 5′-GCA TGC CAT GGC GAC GCG CTA CGA CGA TCA A-3′ and 5′-GCT CAG ATC TTC ACC CTT TGC GGA AGT AAA AGA TAC-3′ for SDMT. The template of the PCRs was the plasmid used for the sequencing of the “betaine operon” of E. halochloris (24). The amplified PCR fragments were purified with the QIAquick DNA purification kit (Qiagen) and cloned into NcoI/BglII-cut pQE-60 expression vectors (Qiagen). The resulting plasmids, pGSMT and pSDMT, were transformed into E. coli XL-1 Blue MRF′ as described previously (8). The gene sequences of the clones showing GSMT or SDMT activity were verified at the Institute of Biotechnology (Helsinki, Finland) by sequencing both DNA strands of the genes.
Expression of GSMT and SDMT and preparation of cell extracts.
Fresh Luria-Bertani broth (750 ml) supplemented with 200 mg of ampicillin/liter was inoculated with 250 ml of overnight culture and grown for 30 min at 37°C and 200 rpm. Isopropyl-β-d-thiogalactopyranoside was added to 1 mM final concentration, and the culture was grown for a further 5 h. The cells were separated by centrifugation (10 min; 1,500 × g), washed once with 20 mM Tris-HCl (pH 7.5), and suspended in 22 ml of 20 mM Tris-HCl (pH 7.5) supplemented with 1 mM phenylmethylsulfonyl fluoride and 2 mM dithiothreitol. The cells were disrupted with a Labsonic 2000 U (B. Braun, Melsungen, Germany) sonicator in 5-ml batches with maximum power. The cell suspension was sonicated for 5 min in 1-min pulses with intermittent cooling. The cell debris was removed by centrifugation at 31,000 × g at 4°C for 30 min.
Purification of GSMT.
Ammonium sulfate was added to the cell extract to achieve 25% saturation, and the solution was incubated for 45 min on ice. The suspension was centrifuged at 31,000 × g for 20 min at 0°C, and the supernatant was applied to a Butyl Sepharose 4 FF (Amersham Pharmacia Biotech) column (1.5 by 12 cm) preequilibrated with 25% saturated ammonium sulfate in 20 mM Tris-HCl, pH 7.5. The column was washed with 100 ml of buffer and eluted with a linear gradient of 25 to 0% saturated ammonium sulfate (250 ml). The active fractions (45 ml) were pooled and concentrated by ultrafiltration (Centriplus 30; Amicon) to 10 ml. The concentrated sample was diluted to 100 ml and applied to a DEAE Sepharose FF (Amersham Pharmacia Biotech) column (1 by 9 cm) preequilibrated with 20 mM Tris-HCl, pH 7.5. The column was washed with 15 ml of buffer and eluted with a linear NaCl gradient from 0 to 1 M NaCl (150 ml). The active fractions were pooled and concentrated by ultrafiltration (Centriplus 30 [Amicon]; Ultrafree MC 10,000 NMWL [Millipore]). In both chromatographic purification steps, GSMT eluted in approximately the middle of the gradient. The purified enzyme was supplemented with 2 mM dithiothreitol, divided into aliquots, and stored at −80°C.
Purification of SDMT.
Ammonium sulfate fractionation at 40% saturation was carried out as described above. The supernatant from this purification step was diluted to 25% saturated ammonium sulfate and applied to a Phenyl Sepharose column (high-sub) (Amersham Pharmacia Biotech) (1.5 by 12 cm) preequilibrated with 25% saturated ammonium sulfate in 20 mM Tris-HCl, pH 7.5. The column was washed with 100 ml of buffer and eluted with a linear gradient of 25 to 0% saturated ammonium sulfate (250 ml). The active fractions were pooled, purified further with DEAE, concentrated, and stored as for GSMT. In both chromatographic purification steps, SDMT eluted in approximately the middle of the gradient.
HPLC analysis of the reaction products.
The reaction products of the radiometrical assays were analyzed by high-performance liquid chromatography (HPLC) using an Aminex HPX-87C column (Bio-Rad) at 80°C with deionized water (0.6 ml/min) as the eluent. The specific radioactivity of AdoMet in the reaction mixtures was from 2 to 13 times higher than in the normal assays. The injection volumes were from 40 to 75 μl. In order to detect the methylated products, 140-μl fractions were collected and analyzed by liquid scintillation counting as described above.
Other methods.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out using 12% polyacrylamide gels according to the standard protocol (16). Protein concentrations were determined using Bio-Rad protein assay reagent with bovine serum albumin as a protein standard. Initial-velocity data were analyzed by nonlinear least-squares regression using the program DYNAFIT (15). The molecular weights of GSMT and SDMT were estimated by analytical gel filtration with a Superose 12-HR-30 (Amersham Pharmacia Biotech) column according to the instructions given by the manufacturer. Metals were analyzed by inductively coupled plasma emission spectrometry using an AtomScan 16 (Thermo Jarell Ash Corp., Franklin, Mass.) instrument at Danisco Cultor Innovation Center (Kirkkonummi, Finland).
RESULTS AND DISCUSSION
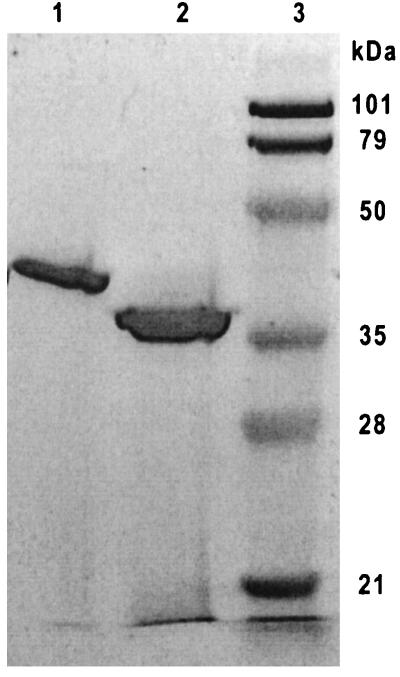
Since it proved to be extremely difficult to purify sufficient amounts of native GSMT and SDMT for characterization, they were produced in E. coli. The expression levels of the enzymes were high. As estimated from SDS-PAGE, GSMT and SDMT represented roughly 30 to 50% of the total protein in the cell extracts. Both recombinant enzymes were purified to homogeneity by ammonium sulfate fractionation, hydrophobic interaction chromatography, and ion-exchange chromatography (Fig. 1). No impurities could be detected with SDS-PAGE either when the enzyme was overloaded or when less enzyme was loaded on the gel. The yields of the purified recombinant enzymes were 29 and 19 mg/liter of culture for GSMT and SDMT, respectively. The molecular masses estimated from the gel were 42 and 36 kDa for GSMT and SDMT, respectively. The value for GSMT differs considerably from the value of 31 kDa calculated from the amino acid sequence. However, the molecular mass of native GSMT previously determined by SDS-PAGE was 38 kDa (24), which is also far higher than the calculated value and reasonably close to the value determined for recombinant GSMT in this study. The determined value for SDMT does not differ significantly from the calculated value of 32 kDa. Estimates for the molecular masses of the enzymes obtained by analytical gel filtration were 40 kDa for GSMT and 25 kDa for SDMT. The results suggest that both recombinant enzymes are monomers.
FIG. 1.
SDS-PAGE of purified GSMT and SDMT. Lane 1, GSMT; lane 2, SDMT; lane 3, molecular mass marker. The proteins were purified by ammonium sulfate fractionation, hydrophobic-interaction chromatography, and ion-exchange chromatography as described in Materials and Methods.
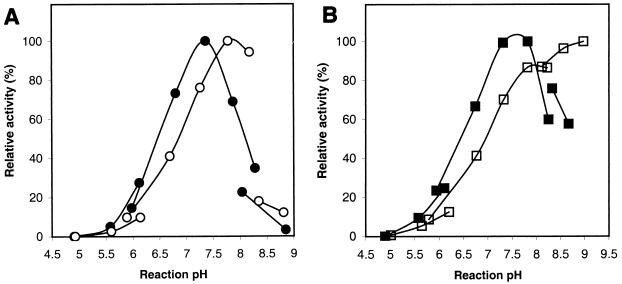
Determination of pH optima.
The effects of pH on the activities are shown in Fig. 2A for GSMT and in Fig. 2B for SDMT. The maximal activities obtained in the range of the triethanolamine buffer used were around pH 7.4 on glycine and around 7.9 on sarcosine with GSMT and around 8.0 on sarcosine and around 7.6 on dimethylglycine with SDMT. The pH optimum of SDMT appears to depend on the buffer used. With Tris-HCl as the buffer, the optimum for the sarcosine activity of SDMT was near pH 9.0, which differs from the one obtained with the triethanolamine buffer. The assays were carried out with 25 mM glycine, 25 mM sarcosine (GSMT), 80 mM sarcosine (SDMT), or 25 mM dimethylglycine and 1 mM AdoMet as the substrates. The results suggest that the enzyme system would work optimally near physiological pH.
FIG. 2.
Effect of pH on methyltransferase activities in 125 mM potassium phosphate (approximately in the range from pH 4.9 to 6.2), 125 mM triethanolamine (approximately in the range from pH 5.8 to 8.3), and 125 mM Tris-HCl (approximately in the range from pH 8.0 to 9.0). (A) GSMT with 25 mM glycine (●) and 25 mM sarcosine (○) as the substrates. (B) SDMT with 80 mM sarcosine (□) and 25 mM dimethylglycine (■) as the substrates.
Substrate specifity.
GSMT has strict specifity for glycine and sarcosine, and SDMT has strict specificity for sarcosine and dimethylglycine as the methyl group acceptors. Neither ethanolamine, monomethylethanolamine, nor any of the l-amino acids (alanine, asparagine, aspartate, cysteine, glutamate, glutamine, isoleucine, leucine, methionine, phenylalanine, proline, serine, threonine, or valine) tested at 25 mM were N methylated by GSMT or SDMT. The results suggest that the enzymes do not catalyze the formation of other betaines (glutamate betaine and proline betaine) that have been reported to be components of the compatible solute pools of cyanobacteria (18) and marine algae (19). The results also indicate that choline, a well-known precursor of betaine, is not formed from ethanolamine by these enzymes. High specifity has been reported for the glycine N-methyltransferases from rat (25) and rabbit livers (11) as well.
Analysis of cofactors.
No evidence of cofactors was found. Incubating GSMT or SDMT with 2.7 mM Ca2+ or Mg2+ or 13.3 mM EDTA had no significant effect on any of the activities. Similar results have also been reported for the mammalian glycine methyltransferases (11). Furthermore, no Mn, Co, or Zn and only insignificant traces of Ca and Mg were detected when purified GSMT and SDMT were analyzed by inductively coupled plasma emission spectroscopy.
In the UV-visible spectra (200 to 800 nm) of GSMT and SDMT, the only peaks were at 280 nm and below 230 nm. Besides these peaks, characteristic of all proteins, no other peaks indicating the presence of cofactors could be detected.
Effect of p-chloromercuribenzoate acid on enzymatic activities.
p-Chloromercuribenzoate acid (1.33 mM) inhibited >95% of the GSMT activities on glycine and sarcosine but only 23% of the SDMT activities on sarcosine and dimethylglycine. With both enzymes, the inhibition was completely counteracted by the addition of dithiothreitol (5.3 mM). The results suggest that the SH groups play an important role in the enzyme reactions catalyzed by GSMT. According to the amino acid sequence, both enzymes have two cysteine residues (24).
Inhibition of enzymatic activities.
The reaction product S-adenosylhomocysteine (AdoHcy) is known to be a strong competitive inhibitor of many methyltransferases (e.g., see references 11, 29, and 34). The concentrations of AdoHcy that cause 50% inhibition are presented in Table 1. The results indicate that GSMT and SDMT are also very susceptible to inhibition by AdoHcy and that its efficient removal is required for betaine synthesis to continue in living cells. This can be achieved by S-adenosylhomocysteine hydrolase-catalyzed hydrolysis of AdoHcy to adenosine and homocysteine (31). The inhibitory effects of dimethylglycine on GSMT and of glycine on SDMT were also studied. These compounds were not methylated by the enzymes under the conditions used. As shown in Table 1, dimethylglycine, the product of the methylation of sarcosine, was a relatively poor inhibitor of GSMT. Also, glycine inhibited the activities of SDMT only at high concentrations. At 50 mM it had no effect on sarcosine activity but inhibited 27% of dimethylglycine activity. At 200 mM, it inhibited 13% of the sarcosine activity and 56% of the dimethylglycine activity. The results indicate that the affinity of glycine to SDMT is low.
TABLE 1.
Inhibition of GSMT and SDMT by AdoHcy and dimethylglycinea
| Enzyme | Methyl group acceptor (mM) | Concn inhibiting 50% (mM)
|
|
|---|---|---|---|
| AdoHcy | Dimethylglycine | ||
| GSMT | Glycine (250) | 0.5 | 38 |
| Sarcosine (25) | 0.4 | 49 | |
| SDMT | Sarcosine (80) | 0.5 | NDb |
| Dimethylglycine (50) | 2.3 | ND | |
All reaction mixtures contained 2 mM AdoMet as the methyl group donor.
ND, not determined.
In phototrophically growing E. halochloris, the intracellular betaine concentrations vary from about 0.7 to 1.6 mol/kg of water at a salinity range from 120 to 240 g/kg of water (7). The intracellular K+ and Na+ concentrations (0.33 M K+ and 0.66 M Na+) remain fairly constant at these salinities (33). The relative activities of GSMT and SDMT when betaine and KCl (0.33 M)-NaCl (0.66 M) were present in the reaction mixtures are shown in Table 2. KCl-NaCl inhibited all methylation steps to various degrees. Betaine is known to protect many enzymes from the perturbing effects of salts and other stress factors (6). However, betaine did not relieve the inhibiting effects of KCl-NaCl on GSMT or SDMT but inhibited the reactions in high concentrations. At 2 M, it inhibited all enzyme activities, both with KCl-NaCl present and with it absent.
TABLE 2.
Effects of betaine, KCl, and NaCl on GSMT and SDMT activitiesa
| Enzyme | Methyl group acceptor (mM) | Relative activity (%)
|
|||||
|---|---|---|---|---|---|---|---|
| No additions | 0.5 M betaine | 2 M betaine | 0.33 M KCl + 0.66 M NaCl | 0.33 M KCl + 0.66 M NaCl + 0.5 M betaine | 0.33 M KCl + 0.66 M NaCl + 2 M betaine | ||
| GSMT | Glycine (250) | 100 | 100 | 41 | 20 | 19 | 7 |
| Sarcosine (25) | 100 | 120 | 30 | 34 | 39 | 11 | |
| SDMT | Sarcosine (80) | 100 | 120 | 29 | 68 | 76 | 21 |
| Dimethylglycine (50) | 100 | 130 | 31 | 75 | 74 | 18 | |
All reaction mixtures contained 2 mM AdoMet as the methyl group donor.
Without KCl-NaCl, 0.5 M betaine had no effect on the first methylation step and a small activating effect on the later steps of the pathway. The concentration of 0.5 M is higher than the reported levels of betaine in salt-stressed plants and plant cell organelles (27), which indicates that in the concentrations naturally found in plants, GSMT and SDMT would not be inhibited by betaine.
Sibley and Yopp (31) have proposed that the regulation of betaine synthesis in the halophilic cyanobacterium Aphanothece halophytica is based on the (putative) strong inhibitory effect of AdoHcy on the methyltransferases catalyzing betaine biosynthesis in this strain. Briefly, according to this model, betaine synthesis is regulated by the different effects of various intracellular K+ and betaine concentrations on the hydrolytic or synthetic activity of AdoHcy hydrolase. The results presented in Table 2 suggest that salts and betaine can also directly affect the activities of the methyltransferases. The regulation of the methylation pathway is presumably very complicated, and the data presented here do not allow a detailed analysis of it.
Kinetic properties.
GSMT catalyzes the reaction sequence glycine-sarcosine-dimethylglycine, and SDMT catalyzes the sequence sarcosine-dimethylglycine-betaine. In all the two-substrate reactions, AdoMet acts as the methyl group donor. The apparent kinetic parameters for both substrates of every reaction step were determined with the other substrate present in excess. GSMT and SDMT displayed Michaelis-Menten kinetics for their substrates. The sigmoidal rate behavior with respect to AdoMet reported for the mammalian glycine-methyltransferases was not detected in any of the initial velocity patterns. The apparent Km and Vmax values determined are shown in Table 3.
TABLE 3.
Kinetic parameters of GSMT and SDMT
| Enzyme | Substrate with varied concn | Substrate with fixed concn (mM) | Apparent Km (mM) | Vmaxa (μmol · min−1 · mg−1) |
|---|---|---|---|---|
| GSMT | Glycine | AdoMet (4) | 18 ± 2 | 1.1 |
| AdoMet | Glycine (250) | 0.42 ± 0.04 | 1.0 | |
| Sarcosine | AdoMet (4) | 2.3 ± 0.2 | 0.15 | |
| AdoMet | Sarcosine (250) | 0.28 ± 0.04 | 0.12 | |
| SDMT | Sarcosine | AdoMet (2.5) | 6.1 ± 1.0b | 1.3b |
| AdoMet | Sarcosine (125) | 0.21 ± 0.02 | 1.1 | |
| Dimethylglycine | AdoMet (2.25) | 4.9 ± 0.9 | 7.4 | |
| AdoMet | Dimethylglycine (125) | 0.16 ± 0.01 | 6.1 |
Errors were less than 10%.
Rough estimate. See Results and Discussion.
As presented in Table 3, in every methylation step, the Vmax values determined for both substrates are close to each other. The initial velocity data fitted reasonably well with the hyperbolic curves with both high and low substrate concentrations, a fact that is also indicated by the standard errors presented in Table 3. However, we cannot completely rule out the possibility of some minor substrate inhibition resulting from the saturating concentrations of the cosubstrate used.
We have previously purified native GSMT from E. halochloris and reported its specific activities on glycine and sarcosine. These results were obtained with 25 mM glycine, sarcosine, or dimethylglycine and 1 mM AdoMet as the substrates (24). According to the results presented in Table 3, some of the substrates were not present in saturating concentrations in these assays. This appears to be especially true for the activities on glycine. Reexamining the linearity of the product formation in the enzymatic reactions revealed that our previous results were not obtained in the strictly linear range. With the activity on glycine, there is also the possibility that the strong inhibition by AdoHcy distorted the results, because the AdoMet concentration was close to its Km value.
Nevertheless, the specific activities of recombinant GSMT and SDMT were determined under the same conditions used earlier in order to compare the activities with our previous results. The specific activities of purified GSMT were 0.16 μmol min−1 mg−1 on glycine and 0.075 μmol min−1 mg−1 on sarcosine, and those of purified SDMT were 0.68 μmol min−1 mg−1 on sarcosine and 3.4 μmol min−1 mg−1 on dimethylglycine.
The specific activities of recombinant GSMT were 3.3 and 2.5 times lower than those reported for native GSMT, with glycine and sarcosine, respectively, as the substrates (24). The deviations of the product formation rates from linearity do not explain the differences between the specific activities. The clones were verified by sequencing, and therefore it is also highly unlikely that the lower activity would be the result of a mutation during the PCR amplification. One possibility is that the low specific activity of recombinant GSMT is a result of incorrect folding of the enzyme due to differences between the intracellular conditions for E. coli and those for E. halochloris. However, taking into account the error margins, the ratios of the activities on different substrates correspond reasonably well to our previous results for native GSMT and also for His6-tagged GSMT and SDMT (24).
Our experiences with the expression of GSMT and SDMT in Saccharomyces cerevisiae have been similar to those with E. coli. In S. cerevisiae, the activities of GSMT were extremely low compared to those of SDMT, although high levels of both enzymes (identified by Western blotting) were synthesized (A. Nyyssölä, unpublished results). It would therefore seem likely that the heterologous expression of GSMT in plants would also present difficulties.
Since both enzymes catalyze two successive steps, it was necessary to discover whether the second methylation step would distort the results of the initial velocity determinations of the first step. Using the apparent Km and Vmax values for the sarcosine step of GSMT, it was estimated that the methylation of the newly formed sarcosine had only a negligible effect on the initial velocity determinations with glycine as the substrate. This was also confirmed by HPLC analyses (see Materials and Methods) of reactions with 4 mM AdoMet and 250 mM glycine, 4 mM AdoMet and 9 mM glycine, and 1 mM AdoMet and 25 mM glycine as substrates. In these reactions, the amounts of dimethylglycine formed were only 3 to 7% of the total products (sarcosine plus dimethylglycine). The reaction times were such that the final product concentrations were considerably higher than those in any of the initial velocity determinations.
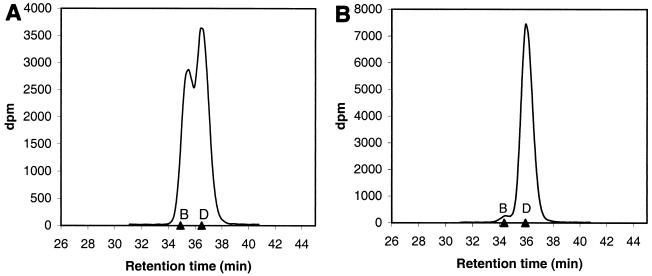
The situation is different with the SDMT activity on sarcosine. As shown in the HPLC analysis of the reaction mixture (Fig. 3A), the newly formed dimethylglycine is readily methylated to betaine with 4 mM sarcosine and 4 mM AdoMet as the substrates. Consequently, reliable values for the apparent Km and Vmax for the sarcosine activity with a fixed AdoMet concentration cannot be obtained by the methods used in this study.
FIG. 3.
HPLC analysis of radioactive methylation products with SDMT. The radioactive products were determined from fractions collected from the eluent as described in Materials and Methods. (A) Reaction with 4 mM sarcosine as the substrate. (B) Reaction with 80 mM sarcosine as the substrate. The methyl group donor S-adenosyl-l-[methyl-14C]methionine was used at 4 mM in both reactions. The retention times are shown by triangles on the x axis (D, dimethylglycine; B, betaine). The retention times in panels A and B are not comparable.
However, with 4 mM AdoMet and an excess of sarcosine (80 mM) as substrates, the amount of betaine formed was negligible (Fig. 3B). Also, with 0.11 and 1 mM AdoMet at this sarcosine concentration, only barely detectable amounts of betaine were formed (data not shown). The results indicate that when saturating concentrations of sarcosine are used, the reaction catalyzed by SDMT stops at the dimethylglycine stage. It would seem plausible that under these conditions sarcosine competitively inhibits the further methylation of the newly formed dimethylglycine to betaine. Because of this phenomenon, we were able to determine the apparent kinetic parameters for the AdoMet activity with a saturating concentration of sarcosine as the cosubstrate (Table 3).
Although the initial velocities of SDMT are unreliable when determined with less than a saturating concentration of sarcosine, the apparent kinetic parameters with sarcosine as the variable substrate were estimated. With the shortest possible reaction times, the apparent Km and Vmax determined were 6.1 mM and 1.3 μmol min−1 mg−1, respectively (Table 3). The Vmax is close to the value determined with the sarcosine concentration fixed and the AdoMet concentration variable, most likely because of the inhibition of the second step by high concentrations of sarcosine. From the estimate of the apparent Km for sarcosine, nothing more can be concluded than that it appears to be of the same order of magnitude as the Km for dimethylglycine of SDMT.
The apparent Km value of 18 mM determined for glycine was significantly greater than the ones determined for the other substrates and also greater than those reported for the mammalian glycine methyltransferases (ranging from 0.13 mM for rat to 11 mM for pig) (25, 26). However, the apparent Vmax of the glycine methylation step is six times greater than the value for the rat glycine methyltransferase (25). Also, the apparent Km for AdoMet for the glycine step (0.42 mM) is higher than that for the other steps and higher than the S0.5 values (the substrate concentration at which the reaction rate is half of its maximal value) of the mammalian glycine methyltransferases (around 0.3 mM for human, rabbit, and pig and 0.05 mM for rat) (26). The apparent Km values for AdoMet decrease somewhat in the successive methylation steps. This suggests that AdoMet is more efficiently used when the reaction to betaine proceeds.
Possible applications of heterologous expression of GSMT and SDMT.
The glycine methylation pathway has many possible applications. Betaine is widely used as an additive in the feed industry. Thus, transgenic plants producing high concentrations of betaine would have a better nutritional value and could therefore be used directly in feed without the supplementation of betaine. As already mentioned, the stress-relieving potential of betaine is not limited to plants. Many industrial microbes suffer from environmental stress under processing conditions. For example, the high substrate concentrations used in fermentation media can cause osmotic stress. The glycine methylation pathway could therefore also be used in improving the stress tolerance of commercially important microbes in agriculture and industry (T. Reinikainen, A. Nyyssölä, and J. Kerovuo, March 2000, U.S. patent application 09/137, 434).
The most interesting possibility, however, is the application of GSMT and SDMT in the genetic engineering of stress-tolerant plants. Drought, salinity, and low temperatures are among the most important environmental factors limiting plant productivity (2). The stress-relieving effects of betaine on plants and its stabilizing effects on plant macromolecules have been widely reported (13, 14, 21, 27, 36). Many important crops, such as rice, potato, tomato, and tobacco, do not synthesize betaine, and therefore, introducing betaine synthesis into these plants has been a well-established target for metabolic engineering (20). So far, the strategy has been the utilization of the choline oxidation pathway. Enzymes catalyzing the oxidation of choline to betaine have been introduced into many plants (1, 10, 12, 17, 22, 28). However, the levels of betaine in the transgenic plants have been significantly lower than those in plants that naturally accumulate it, although in some cases improved stress tolerance has been reported.
It is believed that the reason for the low betaine contents of the transgenic plants is the limited supply of choline (22). Presumably, the reactions of choline metabolism in non-betaine-producing plants form a rigid metabolic network, which makes it difficult to direct choline to betaine synthesis (23). In higher plants, the ethanolamine for choline synthesis is produced from glycine via serine (32). Bypassing the glycine-choline pathway by the introduction of GSMT and SDMT would make it possible to directly engineer all three methylation reactions of betaine synthesis and conceivably to avoid the difficulties associated with the choline oxidation pathway.
It remains to be seen how efficient the glycine methylation pathway is against choline oxidation. We hope that the information on the basic characteristics of GSMT and SDMT presented in this study will facilitate future work on introducing these methyltransferases into plants. In addition to E. halochloris, there are many other halophilic bacteria synthesizing betaine de novo (30), and it is reasonable to assume that the majority of them use the glycine methylation pathway as well. Consequently, there are most likely abundant alternatives to the GSMT and SDMT of E. halochloris in nature.
ACKNOWLEDGMENTS
We are grateful to Markku Saloheimo and VTT Biotechnology for letting us use their scintillation counter. We also thank Xiaoyan Wu, Niklas von Weymarn, and especially Martti Marjamaa for their kind help with the analyses.
This work was supported by the Research Foundation of Helsinki University of Technology and by Danisco Cultor.
REFERENCES
- 1.Alia, Hayashi H, Sakamoto A, Murata N. Enhancement of the tolerance of Arabidopsis to high temperatures by genetic engineering of the synthesis of glycinebetaine. Plant J. 1998;16:155–162. doi: 10.1046/j.1365-313x.1998.00284.x. [DOI] [PubMed] [Google Scholar]
- 2.Boyer J S. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- 3.Brown A D. Microbial water stress. Bacteriol Rev. 1976;40:803–846. doi: 10.1128/br.40.4.803-846.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cook R J, Wagner C. Glycine N-methyltransferase is a folate binding protein of rat liver cytosol. Proc Natl Acad Sci USA. 1984;81:3631–3634. doi: 10.1073/pnas.81.12.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datko A H, Mudd S H. Enzymes of phosphatidylcholine synthesis in Lemna, soybean, and carrot. Plant Physiol. 1988;88:1338–1348. doi: 10.1104/pp.88.4.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galinski E A. Compatible solutes of halophilic eubacteria: molecular principles, water-solute interaction, stress protection. Experientia. 1993;49:487–496. [Google Scholar]
- 7.Galinski E A, Trüper H G. Betaine, a compatible solute in the extremely halophilic phototrophic bacterium Ectothiorhodospira halochloris. FEMS Microbiol Lett. 1982;13:357–360. [Google Scholar]
- 8.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 9.Hanson A D, Rhodes D. 14C tracer evidence for synthesis of choline and betaine via phosphoryl base intermediates in salinized sugarbeet leaves. Plant Physiol. 1983;71:692–700. doi: 10.1104/pp.71.3.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi H, Alia, Mustardy L, Deshnium P, Ida M, Murata N. Transformation of Arabidopsis thaliana with the codA gene for choline oxidase; accumulation of glycinebetaine and enhanced tolerance to salt and cold stress. Plant J. 1997;12:133–142. doi: 10.1046/j.1365-313x.1997.12010133.x. [DOI] [PubMed] [Google Scholar]
- 11.Heady J E, Kerr S J. Purification and characterization of glycine N-methyltransferase. J Biol Chem. 1973;248:69–72. [PubMed] [Google Scholar]
- 12.Huang J, Hirji R, Adam L, Rozwadowski K L, Hammerlindl J K, Keller W A, Selvaraj G. Genetic engineering of glycinebetaine production toward enhancing stress tolerance in plants: metabolic limitations. Plant Physiol. 2000;122:747–756. doi: 10.1104/pp.122.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jolivet Y, Larher F, Hamelin J. Osmoregulation in halophytic higher plants: the protective effect of glycine betaine against the heat destabilization of membranes. Plant Sci Lett. 1982;25:193–201. [Google Scholar]
- 14.Krall J P, Edwards G E, Andreo C S. Protection of pyruvate, Pi dikinase from maize against cold lability by compatible solutes. Plant Physiol. 1989;89:280–285. doi: 10.1104/pp.89.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Lilius G, Holmberg N, Bülow L. Enhanced NaCl stress tolerance in transgenic tobacco expressing bacterial choline dehydrogenase. Bio/Technology. 1996;14:177–180. [Google Scholar]
- 18.Mackay M A, Norton R S, Borowitzka L J. Organic osmoregulatory solutes in cyanobacteria. J Gen Microbiol. 1984;130:2177–2191. [Google Scholar]
- 19.Mason T G, Blunden G. Quaternary ammonium and tertiary sulphonium compounds of algal origin as alleviators of osmotic stress. Botanica Marina. 1989;32:313–316. [Google Scholar]
- 20.McCue K F, Hanson A D. Drought and salt tolerance: towards understanding and application. Trends Biotechnol. 1990;8:358–362. [Google Scholar]
- 21.Murata N, Mohanty P S, Hayashi H, Papageorgiou G C. Glycinebetaine stabilizes the association of extrinsic proteins with the photosynthetic oxygen-evolving complex. FEBS Lett. 1992;296:187–189. doi: 10.1016/0014-5793(92)80376-r. [DOI] [PubMed] [Google Scholar]
- 22.Nuccio M L, Russell B L, Nolte K D, Rathinasabapathi B, Gage D A, Hanson A D. The endogenous choline supply limits glycine betaine synthesis in transgenic tobacco expressing choline monooxygenase. Plant J. 1998;16:487–496. doi: 10.1046/j.1365-313x.1998.00316.x. [DOI] [PubMed] [Google Scholar]
- 23.Nuccio M L, Rhodes D, McNeil S D, Hanson A D. Metabolic engineering of plants for osmotic stress resistance. Curr Opin Plant Biol. 1999;2:128–134. doi: 10.1016/s1369-5266(99)80026-0. [DOI] [PubMed] [Google Scholar]
- 24.Nyyssölä A, Kerovuo J, Kaukinen P, von Weymarn N, Reinikainen T. Extreme halophiles synthesize betaine from glycine by methylation. J Biol Chem. 2000;275:22196–22201. doi: 10.1074/jbc.M910111199. [DOI] [PubMed] [Google Scholar]
- 25.Ogawa H, Fujioka M. Purification and properties of glycine N-methyltransferase from rat liver. J Biol Chem. 1982;257:3447–3452. [PubMed] [Google Scholar]
- 26.Ogawa H, Gomi T, Fujioka M. Mammalian glycine N-methyltransferases. Comparative kinetic and structural properties of the enzymes from human, rat, rabbit and pig livers. Comp Biochem Physiol. 1993;106B:601–611. doi: 10.1016/0305-0491(93)90137-t. [DOI] [PubMed] [Google Scholar]
- 27.Rhodes D, Hanson A D. Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:357–384. [Google Scholar]
- 28.Sakamoto A, Alia, Murata N. Metabolic engineering of rice leading to biosynthesis of glycinebetaine and tolerance to salt and cold. Plant Mol Biol. 1998;38:1011–1019. doi: 10.1023/a:1006095015717. [DOI] [PubMed] [Google Scholar]
- 29.Schneider W J, Vance D E. Conversion of phosphatidylethanolamine to phosphatidylcholine in rat liver. Partial purification and characterization of the enzymatic activities. J Biol Chem. 1979;254:3886–3891. [PubMed] [Google Scholar]
- 30.Severin J, Wohlfarth A, Galinski E A. The predominant role of recently discovered tetrahydropyrimidines for the osmoadaptation of halophilic eubacteria. J Gen Microbiol. 1992;138:1629–1638. [Google Scholar]
- 31.Sibley M H, Yopp J H. Regulation of S-adenosylhomocysteine hydrolase in the halophilic cyanobacterium Aphanothece halophytica: a possible role in glycinebetaine biosynthesis. Arch Microbiol. 1987;149:43–46. [Google Scholar]
- 32.Stewart G R, Larher F. Accumulation of amino acids and related compounds in relation to environmental stress. In: Stumpf P K, Conn E E, editors. The biochemistry of plants. Vol. 5. New York, N.Y: Academic Press; 1988. pp. 609–636. .. [Google Scholar]
- 33.Trüper H G, Galinski E A. Biosynthesis and fate of compatible solutes in extremely halophilic phototrophic eubacteria. FEMS Microbiol Rev. 1990;75:247–254. [Google Scholar]
- 34.Upmeier B, Gross W, Köster S, Barz W. Purification and properties of S-adenosyl-l-methionine:nicotinic acid-N-methyltransferase from cell suspension cultures of Glycine max L. Arch Biochem Biophys. 1988;262:445–454. doi: 10.1016/0003-9861(88)90396-7. [DOI] [PubMed] [Google Scholar]
- 35.von Weymarn N, Nyyssölä A, Reinikainen T, Leisola M, Ojamo H. Improved osmotolerance of recombinant Escherichia coli by de novo glycine betaine biosynthesis. Appl Microbiol Biotechnol. 2001;55:214–218. doi: 10.1007/s002530000515. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y, Aspinall D, Paleg L G. Protection of membrane integrity in Medicago sativa L. by glycinebetaine against the effects of freezing. J Plant Physiol. 1992;140:541–543. [Google Scholar]