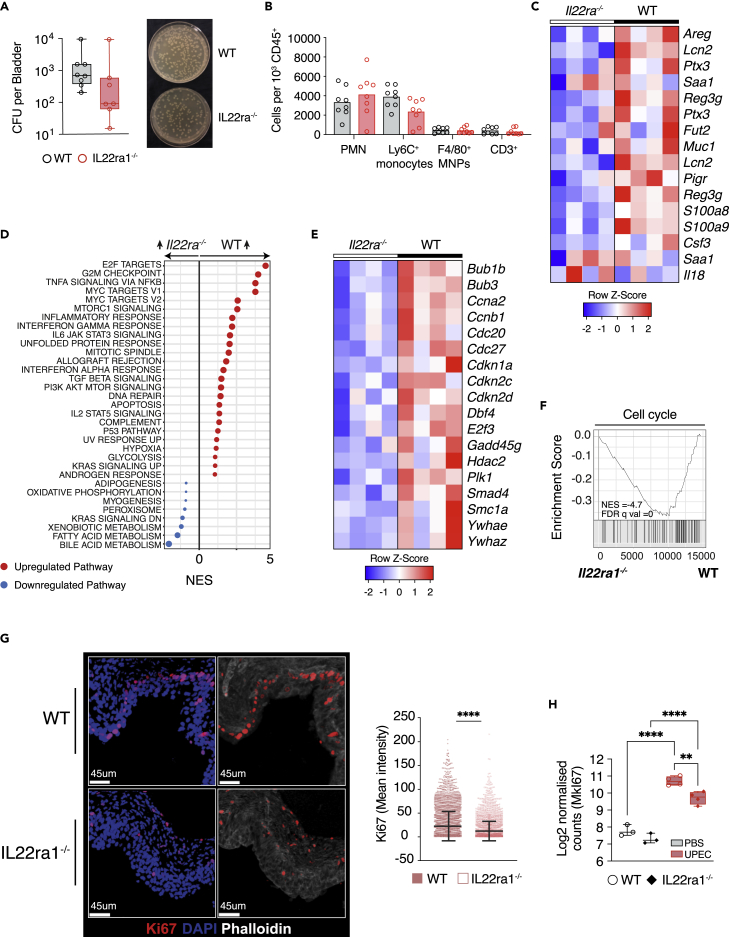
Figure 4.
IL22 promotes epithelial AMP production in the bladder post infection
(A and B) (A) Colony-forming units per bladder and (B) quantification of cells counts (per 1 × 103 CD45+ cells) for indicated subsets 24 h after challenge in C57BL/6N (gray) or IL22ra−/− (red) mice infected with UTI89 (left panel) (Table S1) and corresponding image of bacterial growth on agar plates; 1:30 dilution (right panel). N = 7-8 mice per group.
(C) Heatmap of AMPs from RNA sequencing of bladders infected with UTI89 in C57BL/6N (n = 4) or IL22ra−/− (n = 4) mice 24 h after challenge. Data represent four biological replicates per group (IL22ra−/− and C57BL/6N).
(D) Gene set enrichment analysis (www.gsea-msigdb.org/gsea) of the differential expression from (C) against hallmarks pathways. Only significant pathways (false discovery rate [FDR] q value < 0.05) are plotted. Red dots indicate positive enrichment and blue negative, the size of the dot is inversely correlated with the FDR q value and the position indicates the normalized enrichment score (NES).
(E) Heatmap of cell cycle gene transcripts from data in (C).
(F) Enrichment plot from GSEA for “cell cycle” pathways from the Kegg database.
(G) Representative confocal image of infected bladders from C57BL/6N or IL22ra−/− mice 24 h post infection (left) (red, Ki67; blue, DAPI and white, phalloidin) and quantification of Ki67 mean intensity fluorescence; each square (red filled, C57BL/6N; red open, IL22ra−/−) represents a cell surface (right). Mean ± SEM is shown.
(H) Mki67 Log normalized counts from data in (C). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 by Mann-Whitney test (A, G), one-way ANOVA with Šídák’s multiple comparisons test (B, H), and two-way ANOVA with Dunn’s multiple comparisons test (C, E). All bladders used were from female mice unless otherwise stated.