Abstract
Molecular and physiological analyses were used to study the evolution of the yeast population, from alcoholic fermentation to biological aging in the process of “fino” sherry wine making. The four races of “flor” Saccharomyces cerevisiae (beticus, cheresiensis, montuliensis, and rouxii) exhibited identical restriction patterns for the region spanning the internal transcribed spacers 1 and 2 (ITS-1 and ITS-2) and the 5.8S rRNA gene, but this pattern was different, from those exhibited by non-flor S. cerevisiae strains. This flor-specific pattern was detected only after wines were fortified, never during alcoholic fermentation, and all the strains isolated from the velum exhibited the typical flor yeast pattern. By restriction fragment length polymorphism of mitochondrial DNA and karyotyping, we showed that (i) the native strain is better adapted to fermentation conditions than commercial strains; (ii) two different populations of S. cerevisiae strains are involved in the process of elaboration, of fino sherry wine, one of which is responsible for must fermentation and the other, for wine aging; and (iii) one strain was dominant in the flor population integrating the velum from sherry wines produced in González Byass wineries, although other authors have described a succession of races of flor S. cerevisiae during wine aging. Analyzing all these results together, we conclude that yeast population dynamics during biological aging is a complex phenomenon and differences between yeast populations from different wineries can be observed.
The production of sherry wines comprises two successive processes: first, alcoholic fermentation of must by yeast to produce white wine, and second, biological aging (using the “soleras” system) of the wine under a velum (“flor”) produced by yeast, the so-called flor yeast. All wines made by this special procedure including finos, amontillados, and olorosos) are called sherry wines.
Fermentation of grape juice into wine is a complex microbial reaction. Yeasts are primarily responsible for the alcoholic fermentation of musts, while many vines undergo another fermentation process mediated by lactic acid bacteria (6). Traditionally, wines have been produced by natural fermentation due to the development of yeasts originating from the grapes and winery equipment, although modern enological practices include the inoculation of dry wine yeasts. In Jerez de la Frontera (Andalusia, southern Spain), wineries follow a particular alcohol fermentation process. In these wineries, enologists add the dry wine yeasts to a volume of fresh must equivalent to 1/3 of the total capacity of the fermentation tank. The same volume of fresh must is added 4 to 5 days after fermentation starts (another 1/3 of the total volume of the fermentation tank). After another 4 to 5 days, tanks are filled with fresh must (the last 1/3 volume). The first volume acts as an inoculum for the second portion of fresh must added, and so on. This progressive addition of must during sherry wine making ensures a uniform quality of wines before their biological aging.
When fermentation is completed, wines are supplemented with as much as 15 to 15.5% alcohol before the aging process. Aging is achieved in two phases. The first called “sobretablas,” is a static process in which wines are introduced into oak butts. The second phase, called soleras, is a dynamic system, involving several kinds of oak butts at different aging stages, in which the lowest level contains the oldest wines, and the newest wines are at the top. Twenty percent of the butt volume is left empty to allow the growth of flor yeast. These flor yeasts appear on the surface of the wine forming a thin pellicle (for a review of the whole process, see references 14, 16, and 17). Growth of yeasts on velum surfaces produces important changes in the characteristics of the wine due to the oxidative metabolism of the flor yeasts (17, 18). Various microbiological studies on the fermentation (7, 13, 21, 22, 23) and aging (11, 15, 16) processes in the elaboration of sherry wines have been carried out. However, although some of these studies analyze the dynamics of yeast strains during the specific steps of sherry wine making (18), no complete monitoring of the whole process, from grape must to aged wine has been performed to date.
In this study, the whole elaboration process of sherry wines has been studied using molecular techniques for identification and characterization of yeast species and strains. Yeast population dynamics were studied during alcoholic fermentation, before and after alcohol addition, and during the biological aging of sherry wines in oak butts (sobretablas and soleras). The objectives of this work were (i) to investigate the diversity of Saccharomyces cerevisiae strains involved in the alcoholic fermentation of sherry wines, (ii) to analyze the level of implantation of inoculated strains during sherry wine fermentations, (iii) to study the relationship between the strains present during wine fermentation and the strains involved in velum formation, and (iv) to study the relationship between strains isolated at different stages of biological aging.
MATERIALS AND METHODS
Fermentation assays.
Fermentations were carried out in 30,000-liter tanks. Must was obtained from grapes of the “Palomino fino” variety. Each step of the fermentation process was conducted with 10,000 liters of must. All musts (pH 3.2) were sulfited with 100 ppm of SO2 and rectified with tartaric acid to a level of 4 to 4.5 g/liter. In all cases the sugar content of the must varied from 200 to 230 g/liter.
Sampling.
Two different tanks were selected to monitor alcoholic fermentation, with 2 days' difference between them. Yeast strains were isolated during three different steps of the alcoholic fermentation (referred to below as step 1, step 2, and step 3) representative of the fresh must additions (described in the introduction), and at the end, when the fermentation was completed (step 4). Samples were also taken after the addition of alcohol and during biological aging. Additional samples were aseptically taken from the vela of yeasts growing on the surfaces of fino sherry wines in oak butts. For each of different soleras systems (D and E) two different butts (D1, D2, E1, and E2) were sampled at each level. Soleras system D had four levels, or “criaderas” (from the oldest wine at the 1st criadera to the newest wine at the 4th, and system E had three criaderas. Isolations were made on yeast-peptone-dextrose (YPD) and lysine agar media (during alcoholic fermentation only), after several dilutions in 1‰ saline solution. Plates were incubated at 28°C for 2 to 6 days (9, 10). At each sampling point 50 colonies were picked on YPD petri dishes, and after incubation they were stored at 4°C.
Yeast identification.
Colonies isolated at each sampling point were identified by PCR amplification of the region spanning internal transcribed spacers 1 and 2 (ITS-1, and ITS-2) and the 5.8S rRNA gene (5.8S-ITS region) and subsequent restriction analysis according to the work of Esteve-Zarzoso et al. (4) using DyNAzyme II DNA polymerase (Finnzymes OY, Espoo, Finland). PCR products and restriction fragments were separated on 1.4 and 3% agarose gels, respectively. Cfol, Ddel, HaeIII, and HinfI (Roche Molecular Biochemicals, Mannheim, Germany) were used as restriction endonucleases to identify all yeasts isolated from sherry wines. Fragment lengths were estimated by comparison to a 100-bp ladder (Gibco-BRL, Gaithersburg, Md.). Restriction patterns obtained were compared with those obtained by Esteve-Zarzoso et al. (4) and Fernández-Espinar et al. (5).
Strain differentiation.
Mitochondrial DNA (mtDNA) restriction analysis and karyotyping were used to differentiate strains of S. cerevisiae. Restriction analysis of mtDNA was performed according to the work of Querol et al. (19); HinfI was used as the most suitable restriction endonuclease for differentiation among S. cerevisiae flor and nonflor strains (11, 15, 19). DNA for electrophoretic karyotyping was prepared in agarose plugs as described by Carle and Olson (2). Chromosomal profiles were determined by the contour-clamped homogeneous electric field (CHEF) technique with a DRIII apparatus (Bio-Rad Laboratories, Hercules, Calif.) using standard S. cerevisiae chromosomes as a marker (Bio-Rad Laboratories). Yeast chromosomes were separated on 1% agarose gels in two steps, comprising 60-s pulses for 14 h and then 120-s pulses for 10 h, both at 6 V/cm with an angle of 120°. The running buffer used was 0.5× TBE (45 mM Tris-borate, 1 mM EDTA) cooled at 14°C.
RESULTS
Yeast population dynamics during alcoholic fermentation.
A total of 1,126 colonies were identified. Table 1 shows the identification results and the percentage of colonies corresponding to each species at each step during must fermentation. The species Candida stellata, Dekkera anomala, Hanseniaspora guilliermondii, Hanseniaspora uvarum, Issatchenkia terricola, and S. cerevisiae were isolated at frequencies higher than 2%. Other species, such as Candida incommunis, Candida sorbosa, and Zygosaccharomyces cidri or Z. fermentati, were isolated at very low frequencies and are considered sporadic. We found five different restriction pattern profiles of the 5.8S-ITS region that do not correspond to any of the species included in our database of patterns from more than 132 yeast species isolated from food, including wine fermentations (4, 8). It is possible that these patterns correspond to yeasts from soil contaminants in some cases, or to yeast species infrequently involved in wine making. Yeast identification by molecular techniques agrees with the description of wine yeast diversity obtained by classical techniques (9). At the beginning of the process, the most frequent yeast species are apiculate yeasts, and at the end Saccharomyces species become the main species responsible for fermentation.
TABLE 1.
Evolution of yeast species during alcoholic fermentation of sherry winesa
| Species | Frequency (%) in:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Tank X
|
Tank Y
|
|||||||
| Step 1 | Step 2 | Step 3 | Step 4 | Step 1 | Step 2 | Step 3 | Step 4 | |
| Candida incommunis | 1.5 | |||||||
| Candida sorbosa | 0.81 | |||||||
| Candida stellata | 2.78 | 8.27 | 16.80 | 15.62 | ||||
| Dekkera anomala | 2.40 | |||||||
| Hanseniaspora guilliermondii | 0.56 | 3.20 | 5.74 | |||||
| Hanseniaspora uvarum | 18.40 | |||||||
| Issatchenkia terricola | 2.4 | |||||||
| Metschnikowia pulcherrima | 0.81 | |||||||
| Saccharomyces cerevisiae | 76.11 | 100 | 91.30 | 100 | 46.40 | 100 | 78.60 | 100 |
| Zygosaccharomyces cidri/fermentati | 0.81 | |||||||
| Pattern GB-Bb | 1.60 | |||||||
| Pattern GB-Cb | 19.44 | 0.43 | ||||||
| Pattern GB-Db | 3.20 | |||||||
| Pattern GB-Fb | 1.11 | |||||||
| Pattern GB-Vb | 1.5 | |||||||
Colonies were identified by the method of Esteve-Zarzoso et al. (4).
Restriction patterns not identified.
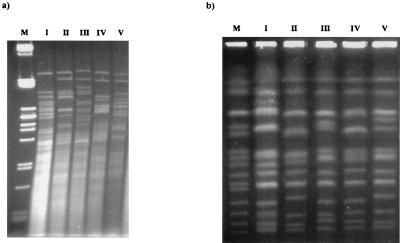
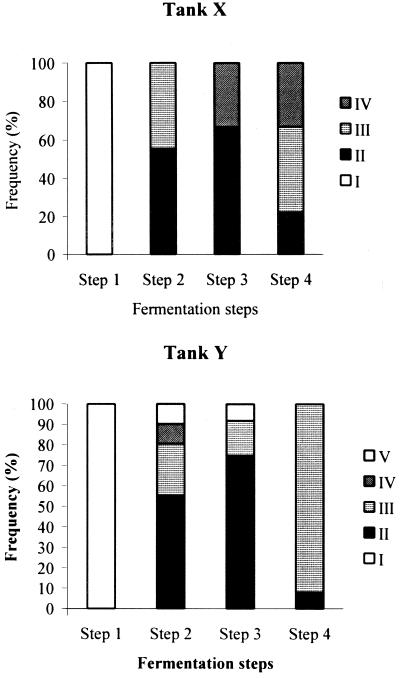
S. cerevisiae population dynamics during alcoholic fermentation and the role of the active dry yeast were studied by means of mtDNA restriction analysis and karyotyping of strains sampled during these processes. A rapid and simple method of Saccharomyces yeast characterization based on mtDNA restriction analysis, has been described for monitoring of wine fermentations (19), and HinfI has been determined to be the restriction endonuclease that recovers the highest mtDNA variability. Using this method with HinfI as the restriction endonuclease, we obtained five different mtDNA restriction patterns from a total of 953 S. cerevisiae colonies isolated throughout alcoholic fermentation and confirmed by electrophoretic karyotyping (Fig. 1). Pattern I corresponds to the inoculated commercial strain, and the other patterns correspond to wild isolates. It is worth noting the high similarities among the four mtDNA restriction patterns (II, III, IV, and V) exhibited by wild S. cerevisiae isolates. The evolution of the five patterns during alcoholic fermentation is shown in Fig. 2. The most interesting result is that the inoculated strain (restriction pattern I) is not responsible for the vinification process. At the beginning of step 2 this strain was replaced by natural strains with other restriction patterns in both fermentation tanks, X and Y (Fig. 2).
FIG. 1.
(a) mtDNA restriction patterns of S. cerevisiae isolates from two fermentation tanks (see Materials and Methods) using the restriction endonuclease HinfI. Lane M corresponds to lambda DNA digested with PstI, used as a marker. (b) Chromosomal profiles of the S. cerevisiae isolates from fermentation tanks. Lane M corresponds to strain YNN295 (Bio-Rad), used as a marker.
FIG. 2.
Growth of yeast strains present in two fermentation tanks (X and Y) during alcoholic fermentation. Strains were characterized by mtDNA restriction analysis, confirmed later by karyotyping (Fig. 1). Pattern I corresponds to the commercial strain.
Yeast population evolution during sobretablas.
Once fermented wines are clarified by natural sedimentation, they are fortified to an alcohol content of 15% and placed in oak butts in sobretablas location. During these procedures samples were taken. After isolation on YPD medium, 47 colonies were identified as described above (4, 5). Yeast colonies isolated were identified as belonging to the species S. cerevisiae (16.65%), Pichia membranaefaciens (53.85%), Pichia anomala (25.35%), and S. cerevisiae flor yeast (4.15%). Pichia species appear at high percentages in this step of the process; this has been described as usual in sherry wine (16).
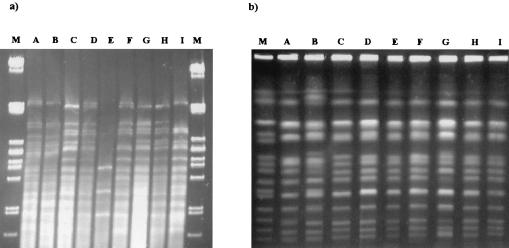
The most important finding for this sherry wine stage is the presence of S. cerevisiae flor strains at a detectable frequency for the first time. These particular strains were molecularly characterized by Fernández-Espinar et al. (5), who showed that they can easily be identified by their specific restriction patterns of the 5.8S-ITS region. These flor yeast patterns are detected only after wines are fortified, never before, probably because they are present at very low frequencies during alcohol fermentation and thus cannot be detected. mtDNA restriction patterns and karyotyping were again used to differentiate Saccharomyces isolates at the strain level. Two different mtDNA restriction patterns were obtained (Fig. 3); pattern A was exhibited by nonflor S. cerevisiae, and pattern B was exhibited by flor strains. However, these patterns appear during the sobretablas process and are not observed during the subsequent biological aging.
FIG. 3.
mtDNA restriction patterns (a) and chromosomal profiles (b) of S. cerevisiae isolates from fortified wine (lanes A and B) and velum (lanes C to I). Lanes M correspond to the size markers described for Fig. 1.
Yeast population evolution during biological aging.
Biological aging is of great importance during the elaboration of sherry wine. Once the wine has been fortified to an alcohol content of 15% (vol/vol), a velum is formed by flor yeasts on the wine surface. Recent studies (14, 21, 22) have shown that the yeasts isolated from the velum, or flor, mainly correspond to various strains of S. cerevisiae (flor strains). To study microbial diversity during biological aging, samples were taken from the vela of two different soleras systems (D and E). A total of 169 different colonies were isolated on YPD medium and identified at the species level by amplification of the 5.8S-ITS region and subsequent restriction analysis using CfoI, HaeIII, and HinfI by the method of Esteve-Zarzoso et al. (4). The same method allowed us to differentiate S. cerevisiae flor strains from other S. cerevisiae strains according to the work of Fernández-Espinar et al. (5). As can be seen in Table 2, S. cerevisiae flor yeasts are the most frequent species growing on velum in both soleras systems, although in some particular butts other, undesirable yeasts, like Dekkera bruxellensis, were found, at even higher frequencies than S. cerevisiae flor yeasts (Table 2, D1, level 1, and E2, level 1). These species have also been isolated in sherry wines during biological aging by Martínez et al. (16), indicating the complex ecological community growing on the velum. The species of the genus Dekkera (Brettanomyces) are typical spoilage yeasts isolated from many fermented beverages, including wines. Ibeas et al. (12) detected Dekkera yeast by nested PCR in barrel-aging sherry wines suspected of Dekkera contamination because of their high acetic acid content. In the present study, Dekkera yeasts have been found at significant levels in sherry wines, indicating that these yeasts may coexist with S. cerevisiae flor yeasts in some butts during normal biological aging.
TABLE 2.
Evolution of yeast species during the biological aging of sherry wines in two soleras systemsa
| Species | Frequency (%) in solera:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D1
|
D2
|
E1
|
E2
|
|||||||||||
| Level 4 | Level 3 | Level 2 | Level 1 | Level 4 | Level 3 | Level 2 | Level 1 | Level 3 | Level 2 | Level 1 | Level 3 | Level 2 | Level 1 | |
| NIb | 33.3 | |||||||||||||
| Candida cantarelli | 91.6 | |||||||||||||
| Dekkera bruxellensis | 13.3 | 8.3 | 66.6 | |||||||||||
| Saccharomyces cerevisiae Jerez | 100 | 53.3 | 100 | 8.3 | 100 | 100 | 100 | 100 | 91.6 | 100 | 100 | 100 | 100 | 33.3 |
Designated D and E. Two different butts were sampled at each level for each system. Level 1 contained the oldest wine. Colonies were identified by the method of Esteve-Zarzoso et al. (4).
NI, restriction patterns not identified.
All the S. cerevisiae strains isolated during biological aging exhibited the typical ribosomal pattern described for S. cerevisiae flor yeast (5). These strains were also characterized by mtDNA restriction analysis and karyotyping (Fig. 3). According to Ibeas et al (11), the digestion of mtDNA with HinfI allowed the differentiation of S. cerevisiae flor yeast at the strain level. Figure 3 shows the seven mtDNA HinfI restriction patterns exhibited by flor strains (lanes C to I). Although restriction patterns obtained from all these strains were similar, with the exception of pattern E, which is the most divergent, some differences among them were observed, mainly due to the gain or loss of one restriction band. As can be seen from Table 3, a general predominance of the strain exhibiting pattern C is observed in the soleras (oldest wines), although in particular oak butts two or more strains may coexist. These results are in accordance with those obtained by Ibeas et al. (11), who also found a predominant strain coexisting with some minority strains in some butts, but alone in other butts. In our study, minority strains exhibiting a special mtDNA restriction pattern characterized by fewer and shorter restriction fragments were found. These strains, corresponding to petite mutants, present the typical ribosomal pattern and karyotype of S. cerevisiae, but a totally different mtDNA restriction analysis (short bands), probably due to the mutagenic effect of the high alcohol content of these wines (11).
TABLE 3.
Frequencies of the mtDNA patterns during the biological aging of sherry wines in two soleras systemsa
| mtDNA pattern | Frequency (%) in solera:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D1
|
D2
|
E1
|
E2
|
|||||||||||
| Level 4 | Level 3 | Level 2 | Level 1 | Level 4 | Level 3 | Level 2 | Level 1 | Level 3 | Level 2 | Level 1 | Level 3 | Level 2 | Level 1 | |
| C | 100 | 50 | 100 | 100 | 100 | 25 | 66.6 | 33.3 | 100 | 100 | ||||
| D | 100 | 50 | 54.5 | 75 | ||||||||||
| E | 9.1 | |||||||||||||
| F | 25 | |||||||||||||
| G | 36.4 | |||||||||||||
| H | 66.6 | |||||||||||||
| I | 100 | 75 | 33.3 | |||||||||||
Designated D and E. Two different butts were sampled at each level for each system. Level 1 contained the oldest wine.
DISCUSSION
Many studies have been conducted to learn more about the non-Saccharomyces yeasts involved in wine making (1), as well as to monitor the evolution of the S. cerevisiae strains during natural and inoculated alcoholic fermentation (19). The Saccharomyces strains responsible for sherry wine aging constitute a special group of wine yeasts, which have been poorly studied at a molecular level. In this sense, only one attempt at the molecular characterization of these yeast strains using mtDNA restriction analysis and electrophoretic karyotyping has been made (11, 15). In the present study, we have shown, for the first time, the evolution of yeast populations from alcoholic fermentation to biological aging in the production of fino sherry wines.
The use of active dry yeasts is of particular interest for the wine industry. There has been considerable controversy over the use of selected pure strains in wine fermentation. It is generally assumed that indigenous yeasts are suppressed by the starter. However studies show that indigenous yeasts can still participate in the fermentation (19, 20) or that only 50% implantation was achieved when fermentation was conducted with some commercial strains (3). In the present study we have shown another example where the native strain is better adapted to fermentation conditions than commercial strains.
Jackson (14) has stated that strains involved in alcoholic fermentation are responsible for velum formation. In this study we show that there are two different populations of S. cerevisiae strains conducting the elaboration of fino sherry wine. One of them is involved in must fermentation, and the other is involved in wine aging. The S. cerevisiae flor strains exhibited 5.8S-ITS region restriction patterns different from those typical of the species S. cerevisiae. These differences can easily be used to differentiate this interesting group of strains (5). They demonstrate that the specific patterns exhibited by flor yeasts are due to the presence of a 24-bp deletion in the ITS-1 region. In the present study, we have demonstrated that this deletion is fixed in flor yeast and that this type of yeast can be isolated from fortified wine. We have never found a Saccharomyces flor yeast pattern among the 953 colonies identified as S. cerevisiae isolated during alcoholic fermentation. This result was confirmed by mtDNA restriction analysis and karyotyping. Flor yeast patterns do not appear during alcoholic fermentation or during sobretablas aging.
In conclusion, we propose using restriction fragment length polymorphism analysis of the 5.8S-ITS region as an alternative to identify wine yeasts, including Saccharomyces flor yeasts. In this work, we demonstrated that in some cases, the inoculated commercial yeast strain is not responsible for the alcoholic fermentation, because it is not adapted to the wine area and cannot compete with the natural flora. Besides, using molecular techniques to characterize yeasts, we demonstrated that the S. cerevisiae strains involved in wine fermentation are different from the strains responsible for biological aging (S. cerevisiae flor yeast). One dominant strain in the flor population integrating the velum was observed in sherry wines produced in González Byass wineries. These results are not in accordance with results published previously by Martínez et al. (16). These authors show a progressive ecological succession of races of S. cerevisiae during wine aging. However, Ibeas et al. (11) observed that a single strain dominates individual barrels and that a dominant strain is stable for two consecutive years. Analyzing all the results together, we conclude that yeast population dynamics during biological aging is a complex phenomenon, and we can observe differences between yeast populations in different wineries.
ACKNOWLEDGMENTS
This work was supported by CICYT grants to A.Q. (ALI96-0457-CO2-01) and F.U. (ALI96-0457-CO2-02). B.E.Z. was the recipient of an FPI fellowship from the Spanish government.
Thanks are due to E. Barrio for critical discussion of the manuscript.
REFERENCES
- 1.Amerine M A, Berg H W, Kunkee R E, Ough C S, Singleton V L, Webb A D. The technology of wine making. 4th ed. Wesport, Conn: AVI Publishing Company, Inc.; 1982. [Google Scholar]
- 2.Carle G F, Olson M V. An electrophoretic karyotype of yeasts. Proc Natl Acad Sci USA. 1985;82:3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esteve-Zarzoso B, Gostíncar A, Bobet R, Uruburu F, Querol A. Selection and molecular characterisation of wine yeasts isolated from the “El Penedès” area (Spain) Food Microbiol. 2000;17:553–562. [Google Scholar]
- 4.Esteve-Zarzoso B, Belloch C, Uruburu F, Querol A. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int J Syst Bacteriol. 1999;49:329–337. doi: 10.1099/00207713-49-1-329. [DOI] [PubMed] [Google Scholar]
- 5.Fernández-Espinar M T, Esteve-Zarzoso B, Querol A, Barrio E. RFLP analysis of the ribosomal internal transcribed spacer and the 5.8S rRNA gene region of the genus Saccharomyces: a fast method for species identification and the differentiation of flor yeasts. Antonie Leeuwenhoek. 2000;78:87–97. doi: 10.1023/a:1002741800609. [DOI] [PubMed] [Google Scholar]
- 6.Fleet G H, Lafon-Lafourcade S, Ribéreau-Gayon P. Evolution of yeasts and lactic acid bacteria during fermentation and storage of Bordeaux wines. Appl Environ Microbiol. 1984;48:1034–1038. doi: 10.1128/aem.48.5.1034-1038.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García Maiquez E. Los microorganismos del Jerez. Microbiol SEM. 1995;11:51–58. [PubMed] [Google Scholar]
- 8.Guillamón J M, Sabaté J, Barrio E, Cano J, Querol A. Rapid identification of wine yeast species based on RFLP analysis of the ribosomal ITS regions. Arch Microbiol. 1998;169:387–392. doi: 10.1007/s002030050587. [DOI] [PubMed] [Google Scholar]
- 9.Heard G M, Fleet G H. Growth of natural yeast flora during the fermentation of inoculated wines. Appl Environ Microbiol. 1985;50:727–728. doi: 10.1128/aem.50.3.727-728.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heard G M, Fleet G H. Evaluation of selective media for enumeration of yeasts during wine fermentation. J Appl Bacteriol. 1986;60:477–481. [Google Scholar]
- 11.Ibeas J I, Lozano I, Perdigones F, Jimenez J. Dynamics of flor yeast populations during the biological aging of sherry wines. Am J Enol Vitic. 1997;48:75–79. [Google Scholar]
- 12.Ibeas J I, Lozano L, Perdigones F, Jimenez J. Detection of Dekkera-Brettanomyces strains in sherry by a nested PCR method. Appl Environ Microbiol. 1996;62:998–1003. doi: 10.1128/aem.62.3.998-1003.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iñigo B, Vazquez D, Arroyo V. Los agentes de la fermentación vínica en la zona de Jerez. Rev Cienc Aplic. 1963;93:296–305. [Google Scholar]
- 14.Jackson R S. Wine science. Principles and applications. San Diego, Calif: Academic Press Inc.; 1994. pp. 338–379. [Google Scholar]
- 15.Martínez P, Codón A C, Pérez L, Beníez T. Physiological and molecular characterization of flor yeasts: polymorphism of flor yeast populations. Yeast. 1995;11:1399–1411. doi: 10.1002/yea.320111408. [DOI] [PubMed] [Google Scholar]
- 16.Martínez P, Pérez Rodriguez L, Benítez T. Evolution of flor yeast population during the biological aging of fino sherry wine. Am J Enol Vitic. 1997;48:160–168. [Google Scholar]
- 17.Martínez de la Ossa E, Caro L, Bonat M, Pérez L, Domeq B. Dry extract in sherry and its evolution in the aging process. Am J Enol Vitic. 1987;38:321–325. [Google Scholar]
- 18.Martínez de la Ossa E, Pérez L, Caro I. Variations of the major volatiles through aging of sherry. Am J Enol Vitic. 1987;38:293–297. [Google Scholar]
- 19.Querol A, Barrio E, Ramón D. Population dynamics of natural Saccharomyces strains during wine fermentation. Int J Food Microbiol. 1994;21:315–323. doi: 10.1016/0168-1605(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 20.Schütz M, Gafner J. Analysis of yeast diversity during spontaneous and induced alcoholic fermentations. J Appl Bacteriol. 1993;75:551–558. [Google Scholar]
- 21.Valcarcel M J, Pérez L, González P, Domeq B. Proceedings IV Jornadas Universitarias de Viticultura y Enología en Jerez. 1987. Efecto del pie de cuba sobre la fermentación dirigida en la zona del Jerez; pp. 160–165. Cadiz, Spain. [Google Scholar]
- 22.Valcarcel M J, Pérez L, González P, Domeq B. Proceedings IV Jornadas Universitarias de Viticultura y Enología en Jerez. 1987. Efecto de la sulfitación sobre las levaduras responsables de la fermentación en la zona del Jerez; pp. 187–195. Cadiz, Spain. [Google Scholar]
- 23.Valcarcel M J, Pérez L, González P, Domeq B. Proceedings V Jornadas Universitarias de Viticultura y Enología en Jerez. 1989. Efecto de la modificación de pH mediante la corrección de la acidez sobre la flora fermentativa en la zona del Jerez; pp. 145–150. Cadiz, Spain. [Google Scholar]