Abstract
The novel COVID-19 pandemic, has effectively turned out to be one of the deadliest events in modern history, with unprecedented loss of human life, major economic and financial setbacks and has set the entire world back quite a few decades. However, detection of the COVID-19 virus has become increasingly difficult due to the mutating nature of the virus, and the rise in asymptomatic cases. To counteract this and contribute to the research efforts for a more accurate screening of COVID-19, we have planned this work. Here, we have proposed an ensemble methodology for deep learning models to solve the task of COVID-19 detection from chest X-rays (CXRs) to assist Computer-Aided Detection (CADe) for medical practitioners. We leverage the strategy of transfer learning for Convolutional Neural Networks (CNNs), widely adopted in recent literature, and further propose an efficient ensemble network for their combination. The DenseNet-201 architecture has been trained only once to generate multiple snapshots, offering diverse information about the extracted features from CXRs. We follow the strategy of decision-level fusion to combine the decision scores using the blending algorithm through a Random Forest (RF) meta-learner. Experimental results confirm the efficacy of the proposed ensemble method, as shown through impressive results upon two open access COVID-19 CXR datasets — the largest COVID-X dataset, as well as a smaller scale dataset. On the large COVID-X dataset, the proposed model has achieved an accuracy score of 94.55% and on the smaller dataset by Chowdhury et al., the proposed model has achieved a 98.13% accuracy score.
Keywords: COVID-19, Deep learning, Blending, Ensemble, Classifier fusion, Chest X-ray
1. Introduction
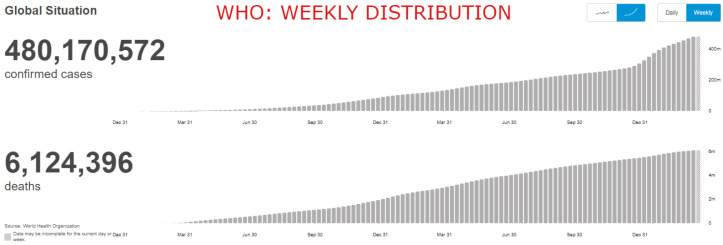
The Novel Coronavirus disease (COVID-19) has massively impacted the world and has brought it to a standstill since its first emergence in December, 2019. The COVID-19 or SARS-CoV-2 is a severe acute respiratory syndrome which spreads via droplets containing the virus produced as a result of coughing or sneezing of a COVID-19 infected person. As of 25th March, , seen below in Fig. 1, the number of people affected by the virus has increased at an astonishing rate and according to the official World Health Organization (WHO) website [1], there have been 480,170,572 confirmed cases of COVID-19, including 6,124,396 deaths worldwide.
Fig. 1.
Weekly distribution of covid positive cases and death counts (worldwide) as of 25th March,2022 [1].
One of the primary ways to curb the spread of the virus is to identify the infected individual and prevent other healthy individuals from coming into their vicinity. The most effective way of screening infected patients is through the Reverse Transcription Polymerase Chain Reaction (RT-PCR) which can detect SARS-CoV-2 ribonucleic acid (RNA) from respiratory specimens obtained via nasopharyngeal or oropharyngeal swabs. But the test involves a lot of labor, takes plenty of time and the results often proved to be inconsistent as mentioned in [2].
A reliable and quick alternative screening method is analyzing chest radiography images e.g., chest X-ray (CXR) or computed tomography (CT) images using deep learning(DL) with the help of CADe which saves both time and labor, thus speeding up the detection process.
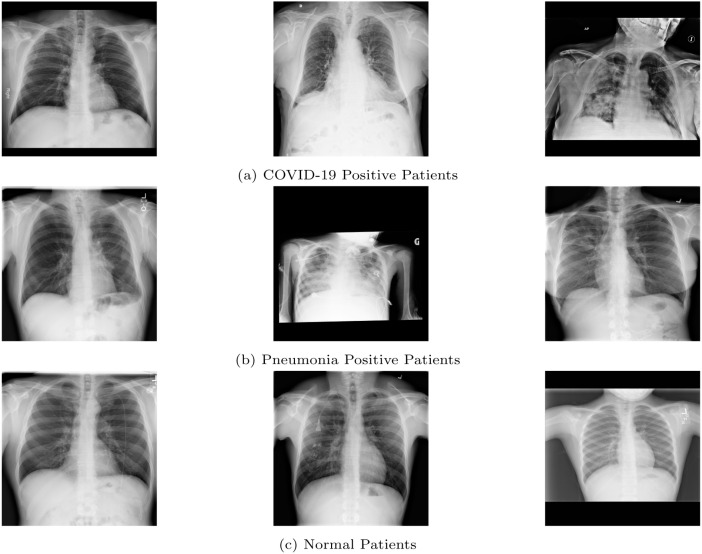
In the field of biomedics, CADe has proved to be very effective as evident from the works of [3], [4], [5]. It finds its use in the detection of pulmonary disorders, coronary artery disease, Alzheimer’s disease, pneumonia and other such diseases. The sample CXR images for COVID-19, pneumonia and normal patients are shown in Fig. 2.
Fig. 2.
Sample images of CXRs for all three classes taken from the COVID-X dataset.
In our paper, we have used deep learning techniques in CADe along with CNNs which are trained by transfer learning followed by ensembling the CNNs using blending which reduces the computational time by a great extent as well as increases the accuracy of the predictions by a considerable amount thus helping the medical community at large.
1.1. Motivation and contributions
In early 2021, there was a significant surge in the research and development related to vaccine production. AstraZeneca, Pfizer, Sputnik, Covishield, and Covaxin are, to name a few, some of the popularly used vaccines. However, as the COVID-19 virus keeps mutating into various other forms, it becomes more challenging for the vaccines to neutralize them. As of 17 March 2022, a total of 10,925,055,390 vaccine doses have been administered [1]. Due to the mutations compiled with the ever increasing population, it is hard to effectively supply vaccines as well as vaccinate all developing nations. Most of the African nations have dangerously low vaccination rates [6] which could potentially cause more loss of human lives. Therefore, it is of acute importance that alternative methods to diagnose COVID-19 virus in patients be developed. It is quite evident from Fig. 1, that even after 1 year since the discovery of vaccines, the number of COVID-19 cases are ever increasing, further highlighting the need for research in this domain.
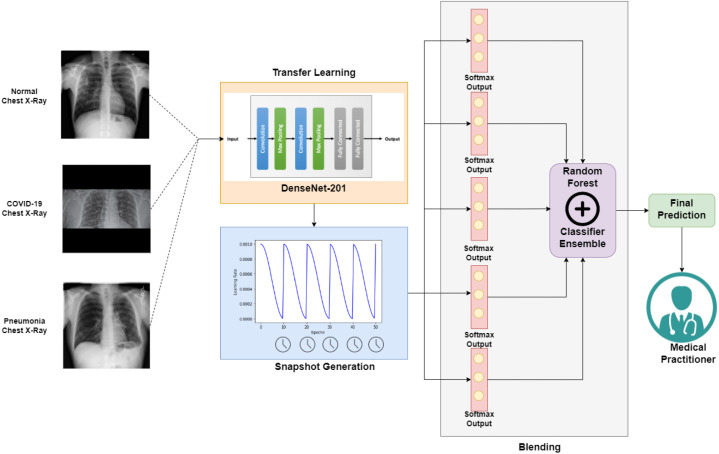
The use of deep learning techniques in CADe along with CNNs, a type of deep learning model, have especially shown outstanding results in the field of image classification due to its higher ability to extract and learn the low-level features and patterns in images and later using the gained knowledge for classifying the images as is evident from the works [7], [8], [9]. Hence, in our paper, a CADe framework has been proposed where the individual predictions of the CNN models generated while training only one state-of-the-art CNN model have been ensembled together, thereby classifying CXRs with greater predication ability. The steps of the proposed work are summarized in Fig. 3. Deep learning based techniques have also been used to predict the number of new cases and death-rate using time series data. As seen in the work [10], the authors have used a comparative study between forecasting methods and are predicting new cases and death rate one, three and seven-day ahead during a span of 100 days.
Fig. 3.
Schematic diagram of our proposed methodology which consists of: (I) Acquisition and Preprocessing of input CXRs, (II) Transfer Learning upon the DenseNet-201 CNN architecture, (III) Generation of multiple Snapshots with Cosine Annealing (Section 3.4.2) with only one training phase, and (IV) Ensemble of classifiers using blending algorithm with RF meta-learner to yield prediction, available for medical practitioners.
Researchers have often turned to transfer learning during cases of low availability of data, where the weights of the deep learning network is provided from a state-of-the-art model’s previous training cycle though in a different domain. It has been utilized previously in the works [11], [12] and more recently in COVID-19 CADe in the works [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23].
The ensembling approach has been explored in the work [24]. Stacking after ensembling has been explored in the work [21]. Although there are approaches using ensembling for combined prediction, the instances of such works to accurately classify and detect COVID-19 from CXRs, are not abundant. Also, the ensembling of several CNN models involve training the models separately which is very time consuming and requires a lot of computational effort, which we solve through a more efficient strategy.
Taking all of the aforementioned facts into consideration, in this paper, we have made the following contributions:
-
•
We utilize the strategy of transfer learning on our CNN based model (DenseNet-201) to predict and generate decision scores upon training on the CXRs.
-
•
We apply the process of cosine annealing based learning rate schedule, thereby generating different snapshots of the CNN, each having learned diverse information from the CXRs. Hence, we generate base model snapshots for ensembling by training the CNN model only once.
-
•
We use the snapshots to generate decision scores, provided as input to a RF meta-learner through the blending ensemble algorithm.
-
•
We achieve satisfactory results on a large COVID-19 CXR dataset of 15471 samples as well as a smaller dataset of 2905 samples.
2. Related work
2.1. COVID-19 detection
There exist two primary sources for generating medical images and thereby using them as a dataset for COVID-19 CADe methods. The first one is the CT scan and the second one is the CXR.
Ensemble learning strategies are a powerful tool for CADe which provides better accuracies in terms of image classification and have been previously utilized successfully in other domains such as detection of Tuberculosis [3], [25] and also in cardiomegaly classification [26].
Such ensembling approach has been further extended in the COVID-19 detection domain as well, which is evident from the work of [24] where they had implemented the approach of majority vote on classical machine learning models using texture features extracted from the CXR images.
For CADe, CNN based classifiers are usually used for image classification. In the work of [27], the authors had proposed a parallel-dilated CNN based COVID-19 detection system to extract radiological features from CXRs. CNN based deep feature extraction using CXRs had also been proposed in the work of [28]. The authors of [29] adapted Darknet-19 CNN architecture to work on CXRs while [30] utilized CNN to explore the network design using generative synthesis, a machine-driven exploration strategy. Similarly, the authors of [31] have employed the MobileNetV2 architecture and integrated it into Capsule Networks to construct a fully automated and lightweight model. The model, termed as MobileCaps is capable of classifying COVID-19 CXR images, and predicting severity scores based on the RALE scoring technique. COVID-NET CT-2 proposed by the authors of [32], in 2021 is another state-of-the-art implementation of Deep Neural Network based COVID-19 detection based on COVIDX [33] dataset. A COFE-Net deep-learning based model, proposed by the authors of the work [34], achieved an accuracy score of 96.39% on multi-class classification of COVID-19 from CXR images.
When working with CNNs, the principle of transfer learning has been utilized very often. The architecture and saved weights of state-of-the-art CNN classifiers on the ImageNet benchmark provides the users with higher accuracies while classifying images with CNNs and hence has developed into a popular choice as depicted in the work of [13]. The authors of [16] utilized the ResNet-50 architecture and performed data augmentation to increase the variations in their data set. [17] used an Xception architecture based CNN transfer learning method. The multi-stage cascaded disease classification problem was solved in the work of [18] utilizing the power of transfer learning with CNNs. SqueezeNet CNN architecture with transfer learning were utilized in the work of [20]. The authors of [14] analyzed a large number of state-of-the-art CNN models with transfer learning along with image augmentation to enhance the limited number of X-ray samples.
There are several methods proposed by authors where transfer learning with CNNs has been implemented along with the strategy of ensembling thereby providing better classification rates, combining various features. VGG architecture based CNNs with transfer learning had been ensembled with the empirical decision-level fusion strategy of stacking in the work of [21]. A similar approach was adapted by [23] where a capsule-based network had been used. The authors of [22] used transfer learning based CNN and combined that with ensembling several empirical fusion schemes which were pruned for optimal hyper-parameters and finding the best result. In [35], the authors have cross-examined the effectiveness of ensemble strategy with DNNs for COVID-19 detection. A CNN and SVM fusion strategy proposed by the authors of [36]has showcased an accuracy score of 99.02% on CXRs for COVID-19 detection.
More work, in the domain of multi-class COVID-19 detection would be useful, as research in the domain is lacking compared to binary-classification of CXR samples. In [37], the authors proposed a single layer-based (SLB) and a feature fusion based (FFB) composite system to automate the detection of COVID-19 in X-ray images using deep features. Similarly, in [38], the authors have proposed an ensemble of pre-trained Deep-CNN models and tested it for multi-class classification on X-ray images in order to attain state-of-the-art results.
Apart from CXRs, CT scans have also been used as a primary source of dataset as is evident from the work of [39], [40]. In the work [41], the authors have proposed a semi-supervised learning based approach and utilized GANs to reach a 99.8% accuracy score. A DNN based approach on CT scans is highlighted in the work [42], where the authors have proposed DNN-GFE and achieved an accuracy score of 96.71%. A CNN-Autoencoder fusion model has shown promising results and achieved 96.05% accuracy score. In the work done in [43] however due to unavailability of data, 200 deceased cases were synthetically generated to avoid an imbalanced dataset and matched the number of recovered cases, which can raise questions regarding the models capabilities in a real world scenario. In the work [44] the authors have utilized a fuzzy, rank-based fusion on decision scores using the Gompertz function to achieve accuracy scores of 98.83%. In the work [45], the authors achieved a 98.93% accuracy score using the Sugeno fuzzy integral ensemble of four pre-trained deep learning models on CT scan images. The authors of the work [46] utilized GoogleNet and ResNet and proposed a hybrid meta-heuristic feature selection algorithm, based Golden Ratio Optimizer to achieve an accuracy score of 99.15% on CT scan dataset. ET-NET, an ensemble of three transfer learning based models proposed by the authors of the work [47] has achieved an accuracy of 97.81%. However, the high cost of machinery involved along with the enhanced difficulty of computational models to extract and interpret features from such data-dense images as obtained from CT scans have rendered them as an infeasible choice for our model. Moreover, CT scans expose the subject to much higher radiation levels, as compared to normal X-rays and require skilled professionals to operate the already expensive machinery, which can be a major challenge in most of the developing countries across the world. Hence, in our paper, we have utilized the CXRs as the source of our datasets.
2.2. Snapshot ensembling
In the work [48], the authors have introduced the concept of warm restarts in gradient-based optimization, as a modification of the commonly used Stochastic Gradient Descent (SGD) algorithm. In the work [49], the authors have further proposed an ensemble strategy of the models generated in the interim training process described in [48]. They have suggested that there is a significant diversity between the local minima found at each cycle, and this diversity can be exploited through an appropriate ensemble approach. The authors of the work [50] proposed the use of a DL based improved Snapshot Ensemble technique for efficient COVID-19 chest X-ray classification, to take advantage of the transfer learning technique using the ResNet-50 model. EDL-COVID [51], an ensemble DL model was generated by combining multiple snapshot models of COVID-Net, by employing a proposed weighted averaging ensembling method that is aware of different sensitivities of DL models on different classes types. The paper [52] proposes an ensemble of CNNs called ECOVNet, based on EfficientNet (for feature extraction) and model snapshots to detect COVID-19 from chest X-rays. Moreover, the results of this literature suggest that the soft ensemble of the proposed ECOVNet model snapshots outperforms the other state-of-the-art methods.
2.3. Random forest classifier
Random Forest (RF) [53], proposed in 2001, is a widely successful method for classification via the ensembling of decision trees. They have been applied for classifier combination in applications such as bearing fault diagnosis [54], solar power forecasting [55] and hyper-spectral image classification [56] to name a few. Even for applications related to COVID-19, RF classifier finds use in the methods [57], [58]. Class imbalance is a huge problem when dealing with Covid-19 datasets, due to the lack of adequate labeled, high quality, Covid positive X-ray samples. Using a Boosted RF Classifier as in the works of [59] can not only aid in Covid-19 detection on imbalanced dataset, but can also greatly reduce the requirement of computational resources as in the alternatively used Snapshot Ensembling methods. A proof-of concept of RF based COVID-19 detection system can be seen in the work of [60], where they have proposed a web based solution, called Heg.IA, to optimize the diagnosis of COVID-19. The system aims to support decision-making regarding diagnosis of COVID-19 and indicates the necessity (even severity e.g. regular ward, semi-ICU or ICU) of hospitalization based on the results of the patients’ blood tests. Medical professionals have participated in the development of this system, and it can help professionals make a decision based on an easy-to-use system, when there is a lack of testing kits.
3. Proposed method
3.1. Data acquisition
Two datasets have been used in this paper.
3.1.1. Wang et al. ’s dataset (COVID-X)
The first one is COVID-X that we have obtained from the work of [33], which is the largest open access COVID-19 X-ray dataset, at the time of the experimental study and consists of CXR images. Five different repositories of chest X-ray scans had been merged to create this dataset. It consists of three different classes of scans — COVID-19 positive patients, pneumonia infected patients, and normal patients. The distribution of data used in this work is shown in Table 1. All of the data used being medical images, and no form of data augmentation has been performed on this dataset as that would imply synthetic or artificial manufacturing of real-world patient data which may be an impractical representation of the human body parameters. In more recent times, the authors of [33] have released a new training dataset with over 30,000 CXR images collected from over 16,400 patients. The dataset consists of 16,490 positive COVID-19 images from over 2,800 patients.
The train set is further split into a train set and a holdout set in the ratio 8:2 which is shown in Table 1.
Table 1.
Class-wise distribution of CXR samples in the COVID-X dataset.
| Phase | COVID-19 | Pneumonia | Normal | Total |
|---|---|---|---|---|
| Train | 375 | 4367 | 6373 | 11115 |
| Test | 100 | 594 | 885 | 1579 |
| Holdout | 93 | 1091 | 1593 | 2777 |
| Total | 568 | 6052 | 8851 | 15471 |
3.1.2. Chowdhury et al. ’s dataset
The second dataset is obtained from the work [14] consisting of CXR images. This data set also consists of three different classes of scans — COVID-19 positive patients, pneumonia infected patients, and normal patients. The dataset is first split into train and test sets in the ratio 89:11 as done in the work [14]. After that, the train set is further split into train and holdout sets in the ratio 7:3. The data distribution for each set is shown in Table 2.
Table 2.
Class-wise distribution of CXR samples in the COVID dataset by Chowdhury et al.
| Phase | COVID-19 | Pneumonia | Normal | Total |
|---|---|---|---|---|
| Train | 136 | 838 | 833 | 1807 |
| Test | 25 | 149 | 147 | 321 |
| Holdout | 58 | 358 | 361 | 777 |
| Total | 219 | 1345 | 1341 | 2905 |
3.2. Pre-processing
The ImageNet dataset has been used to train the CNN models. The ImageNet input images are of dimension 224 × 224. However, the CXRs used in the first dataset are of varying dimensions, while the second dataset are of compatible dimensions.
Subsequently, the CXRs of the first dataset are made of compatible dimension. Hence, at the boundaries of the images, black borders are added to ensure that the CXRs are of required dimension.
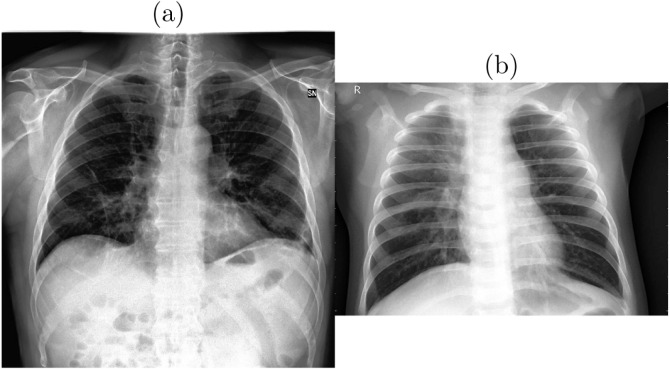
It is of acute importance that, the model is able to correctly distinguish between COVID and Pneumonia cases, as it would be disastrous if the model misclassifies COVID patients as pneumonia. An example of it can be seen in Fig. 4, where the model might falsely classify the COVID case as pneumonia, thereby increasing the need for better pre-processing and data augmentation strategies to aid in model classification.
Fig. 4.
Sample chest X-ray scan images taken from COVID-X dataset [30] showing: (a) COVID-19 positive and (b) Pneumonia cases.
3.3. Blending
Deep learning models have a stochastic nature, and there is possibility for diverse information to be captured from the various local minima in the training process. In this situation, an ensemble strategy may be used for information fusion. Hence, the blending algorithm is used for classifier combination as a form of decision-level fusion. The algorithm of our ensemble strategy is shown in Algorithm 1.
The blending algorithm consists of two stages. The first stage is a traditional training phase which is used for generation of decision scores. The second stage is a training phase of a meta-learner, used to combine the decision scores from multiple sources, which are regarded as input features. It is to be noted that the training data utilized in the first stage are separate from the holdout data used for training the meta-learner in the second stage. This is to ensure that overfitting of the meta-learner model does not take place.
3.4. Base classifiers
As the first part of the blended ensembling method, we have used the strategy of transfer learning to fine-tune pre-trained CNN model which was originally trained on the ImageNet Large Scale Visual Recognition Challenge (ILSVRC) dataset [61], to classify COVID-19 from input CXR images. The CNN that we used in this paper has the pre-trained convolutional blocks and the weights of the standard DenseNet-201 CNN architecture. The process of extracting the low-level features of the CXRs during the training phase is accelerated by the saved weights of the pre-trained convolutional blocks of the DenseNet-201.
The classifier is pre-trained on the benchmark ImageNet dataset. Hence, even if the dataset is small sized as in case of our second dataset, described in Table 2, the shared weights obtained from the DenseNet-201 helps to efficiently make classifications on the new dataset, as it is enhanced by knowledge mined from the ImageNet dataset.
The architecture of conventional CNN models has a feed forward mechanism where the knowledge gained in the previous layer is passed on to the next layer. The DenseNet proposed in the work [62] is a massive improvement over the conventional CNN model as it learns from the output of all the previous layers and uses them as an input to the current layer thus generating its output. The vanishing gradient problem is thus treated and the model also trains for less epochs, thereby lessening the training time.
3.4.1. Architecture
The architecture begins with the conventional convolution and pooling layers followed by three sequences of dense blocks and transition layers. These layers are followed by a dense block and classification layer which finally gives us the output. In each of the dense block, each layer receives inputs from all previous layers thus improving the efficiency of the model as well as the accuracy of classification. The difference of DenseNet with other such models lies in the number of convolutional layers in the third and fourth dense blocks. There are a total of 201 layers in the network, thus the model is called as DenseNet-201.
The architecture of the classifier following the feature extractors has two fully connected dense layers, each consisting of 256 neurons, followed by a softmax layer. Dropout with a probability of 50% has been deployed between the two dense layers to account for complex co-adaptations in the data. The softmax layer which is the final layer of our model provides the necessary output and classifies the input image into one of the three classes.
3.4.2. Cosine annealing
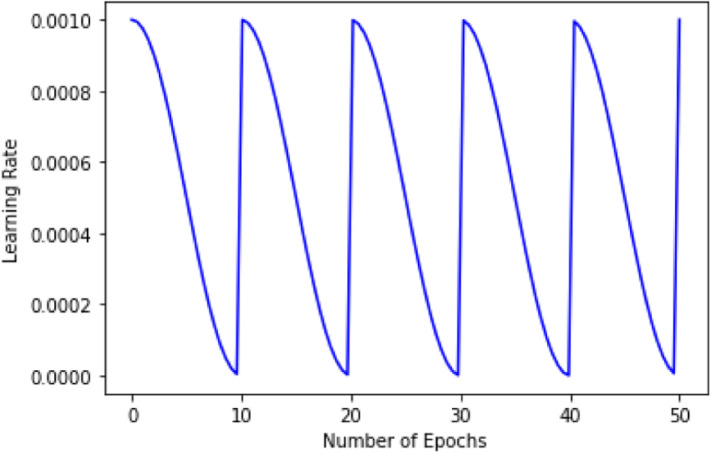
An important hyperparameter for optimization in deep learning based models is the learning rate. In fact, SGD has been shown to require a learning rate annealing schedule to converge to a good minimum in the first place. We have used SGD with Warm Restart (SGDR) technique as proposed in the work [48] with cosine annealing. In this technique, the learning rate is initialized with a high value and is scheduled to decrease. Hence, we start with a high value for the learning rate and with each batch, we get closer and closer to the global minimum. The learning rate decreases along the cosine function.
| (1) |
Eq. (1) obtained from the works of [48], [49] is for the cosine annealing based learning rate schedule, where is the learning rate at epoch , is the maximum or starting learning rate, is the total number of epochs, is the number of cycles, mod is the modulo operation, and square brackets indicate a floor operation.
We perform this for a particular number of epochs, at the end of which the model converges to the local minima. After that, the learning rate shoots up rapidly once again. But this stimulated restart of the learning process does not take place with a new set of small random numbers as the weight, but good weights obtained from the previous learning process is reused thus providing a “warm restart” instead of a cold one. The purpose of the high initial learning rate after a restart is to essentially pull the parameters out of the minimum to which they previously converged and then to converge to a different local minima. This periodic aggressive annealing thus helps the model to rapidly converge to a new and better solution. Fig. 5 shows the learning rate schedule used in our experiments.
Fig. 5.
Shows a cyclic learning rate while following the cosine function providing a warm restart after every 10 epochs.
3.4.3. Generating and ensembling snapshots
Snapshot ensembles are a recent technique that uses warm restarts when training a single model. At the heart of snapshot ensembling is an optimization process which visits several local minima before converging to a final solution i.e., the global minima. As soon as the model converges to a local minima after a number of epochs, we save a snapshot of the model containing the weights, as mentioned in the work [49] . Hence from a single training process, an ensemble of diverse models are produced using the snapshot ensembling technique. A collection of these snapshots are later used as the base models in the blending technique mentioned in Section 3.3.
The success of ensembling relies on the diversity of the individual models in the ensemble. Snapshot ensembling ensures this through cosine annealing in the training process which enables the model to converge to a different local minimum after every restart.
3.5. Meta-learner
Decision Trees are the primary building blocks of the RF model. A RF has a large number of individual decision trees that operate as an ensemble. Each individual tree in the RF gives a prediction and the class with the most number of predictions is the final prediction of the entire model. A large number of relatively uncorrelated models (each Decision Tree) operating as a single unit outperforms any individual Decision Tree model, thereby leading to the robust nature of RF classifier.
After generating the snapshots, we have used them to predict the outcomes on the holdout set, therefore generating decision scores. The outputs in the form of concatenated decision scores serve as the input to the next model. We have utilized the RF classifier to be our meta-learner as part of the blending technique mentioned in Section 3.3. After this meta-learner is trained only on data originating from the holdout set, we are ready to generate the test predictions on our pipeline.
4. Results and analysis
We have trained the DenseNet-201 model on both the COVID-X dataset by Wang et al. and also on the dataset by Chowdhury et al. thereby generating 5 snapshots and later ensembled them using RF classifier model. The accuracies on the two datasets obtained are 94.55% and 98.13%, respectively. The performances of our proposed method and its implementation process has been discussed in the following subsections.
4.1. Implementation
We have utilized the TensorFlow library in Python for implementation of the base classifiers. Our CNN model is trained with the Stochastic Gradient Descent (SGD) algorithm with the momentum value as 0.9 and starting learning rate as 0.001. The model is trained for 50 epochs and snapshots of the model i.e. the model weights are stored at an interval of 10 epochs thus generating 5 snapshot models which are used as the base models in the blending technique mentioned previously.
We have utilized the SciKit-Learn library in Python for implementation of the meta-learner. The hyperparameters for the models were experimentally determined.
COVID-X dataset [33].
For our RF classifier model, we have kept the values of i.e., the number of decision trees in our RF to be 900, i.e., the minimum number of data points placed in a node before the node is split to be 5 and i.e., the minimum number of data points allowed in a leaf node to be 1. We have also kept the value of i.e., the maximum number of levels in each decision tree to be 4.
Chowdhury et al. ’s dataset [14].
For this dataset, we have kept the values of i.e., the number of decision trees in our RF to be 750, i.e., the minimum number of data points placed in a node before the node is split to be 2 and i.e., the minimum number of data points allowed in a leaf node to be 1. We have also kept the value of i.e., the maximum number of levels in each decision tree to be unbounded.
4.2. Comparison of results
Table 3 shows comparison with other methods that use the same dataset from [30] as the proposed work, while Table 4 shows results on the dataset from the work [14]. Table 5 shows comparisons with methods outside these two datasets.
Table 3.
Comparison with state-of-the-art methods on the COVID-X dataset [30].
| Method | Data distribution | Accuracy % |
|---|---|---|
| COVID-Net (2020) [30] | 358 COVID-19, 5538 Pneumonia, 8066 Normal | 93.3 |
| ECOVNet (2020) [52] | 589 COVID-19 , 6053 Pneumonia, 8851 Normal | 96.00 |
| COVID-ResNet (2020) [16] | 68 COVID-19, 931 Bact. Pneumonia, 660 Viral Pneumonia, 1203 Normal | 96.23 |
| COVID-CAPS (2020) [23] | Not specified | 98.3 |
| COVIDiagnosis-Net (2020) [20] | 76 COVID-19, 4290 Pneumonia, 1583 Normal | 98.26 |
| EDL-COVID (2021) [51] | 100 COVID-19, 594 Pneumonia, 885 Normal | 95 |
| COVID-Net CT-2 S (2021) [32] | Not specified | 97.9 |
| COVID-NET CT-2 L (2021) [32] | Not specified | 98.1 |
| Blended ensembling | 568 COVID-19, 6052 Pneumonia, 8851 Normal | 94.55 |
Table 4.
Comparison with state-of-the-art methods on the dataset by Chowdhury et al. [14].
| Method | Data distribution | Accuracy% |
|---|---|---|
| CNN + SVM (2020) [28] | 219 COVID-19, 1345 Pneumonia, 1341 Normal | 98.97 |
| Stacked VGG Ensemble (2020) [21] | 219 COVID-19, 1345 Pneumonia, 1341 Normal | 97.4 |
| PDCOVIDNet (2020) [27] | 219 COVID-19, 1345 Pneumonia, 1341 Normal | 96.58 |
| Blended ensembling | 219 COVID-19, 1345 Pneumonia, 1341 Normal | 98.13 |
Table 5.
Comparison with state-of-the-art methods on multi-class classification.
| Method | Data distribution | Accuracy % |
|---|---|---|
| Transfer Learning (on Dataset 1 in cited paper) (2020) [13] | 224 COVID-19, 700 Pneumonia, 504 Normal | 93.48 |
| Transfer Learning (on Dataset 2 in cited paper) (2020) [13] | 224 COVID-19, 714 Pneumonia, 504 Normal | 94.72 |
| DarkCovidNet (2020) [29] | 127 COVID-19, 500 Pneumonia, 500 Normal | 87.02 |
| Majority Voting ML (2020) [24] | 782 COVID-19, 782 Pneumonia, 782 Normal | 93.41 |
| DenseNet-201 (2020) [14] | 423 COVID-19, 1485 Pneumonia, 1579 Normal | 97.94 |
| VGG16 (2020) [15] | 142 COVID-19, 142 Pneumonia, 142 Normal | 95.88 |
| CovXNet (2020) [19] | 305 COVID-19, 2780 Bact. Pneumonia, 1493 Viral Pneumonia, 1583 Normal | 90.2 |
| CNN + SVM (2021) [36] | 77 COVID-19, 256 Normal | 99.02 |
| Cascaded CNNs (2020) [18] | 69 COVID-19, 79 Bact. Pneumonia, 79 Viral Pneumonia, 79 Normal | 99.9 |
| CoroNet (on Dataset 1 in cited paper) (2020) [17] | 284 COVID-19, 657 Pneumonia, 310 Normal | 95.0 |
| CoroNet (on Dataset 2 in cited paper) (2020) [17] | 157 COVID-19, 500 Pneumonia, 500 Normal | 90.21 |
| Pruned Weighted Average (2020) [22] | 313 COVID-19, 8792 Pneumonia, 7595 Normal | 99.01 |
| FFB3 (2021) [37] | 125 COVID-19, 500 pneumonia, 500 no-finding, and | 87.64 |
| Deep-CNN (2021) [38] | 2161 COVID-19, 2022 pneumonia, and 5863 normal chest | 92.63 |
| ResNet-50 and AlexNet (2021) [63] | 3,616 COVID-19, 1,345 pneumonia, 10,192 normal, and 6,012 lung opacity | 95 |
| Blended ensembling on COVID-X | 568 COVID-19, 6052 Pneumonia, 8851 Normal | 94.55 |
| Blended ensembling on Chowdhury et al. dataset[14] | 219 COVID-19, 1345 Pneumonia, 1341 Normal | 98.13 |
From the comparison, we note that for methods which have been reported using the same as well as different datasets, the proposed method is able to outperform or achieve comparable performance with most of the methods, and achieves impressive accuracy. It can be noted that there are very few state-of-the-art methods that are able to outperform the proposed method and the margin for that is small. Due to the lack of a standardized benchmark dataset, all past methods cannot directly be compared. Even so, the proposed method has been validated to achieve impressive performance with a considerably larger number of CXR samples than all compared methods, in the case of the dataset COVID-X. Overall, we can safely comment that the results are competitive and the proposed method is technically sound and robust.
4.3. Analysis
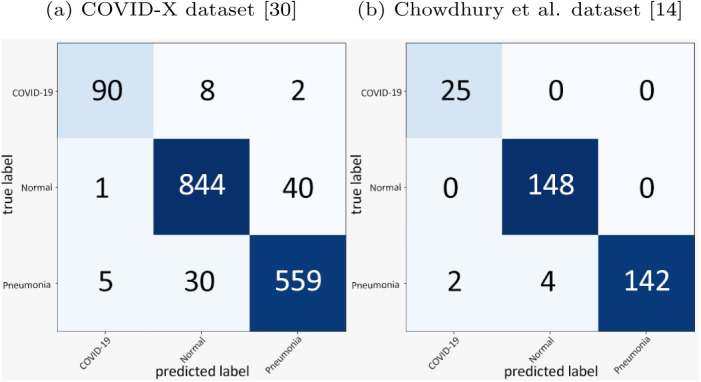
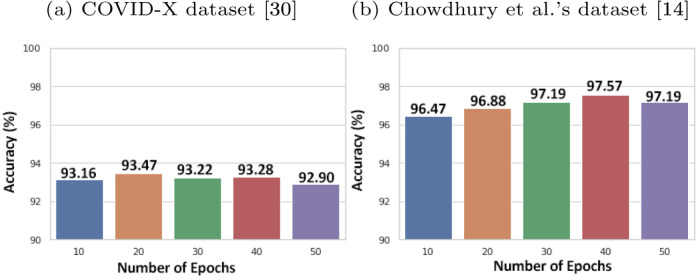
The confusion matrices generated by the prediction of the blended ensembling method for the 3-class classification problem on the COVID-X dataset [33] and the Chowdhury et al. dataset [14] are shown in Fig. 7. Fig. 6 shows the performances of the snapshots or the base models generated at an interval of every 10 epochs on the test sets of each of the datasets. Table 6 shows the class-wise performance of the proposed method on each of the two datasets.
Fig. 7.
Confusion Matrices.
Fig. 6.
Performance of base CNN classifiers.
Table 6.
Recall (Sensitivity), Precision (Positive Predictive Value), and F1-Score for 3-class classification.
| (a) COVID-X dataset [30] | |||
| Metric (%) | COVID-19 | Pneumonia | Normal |
| Recall | 90.00 | 94.10 | 95.36 |
| Precision | 93.75 | 93.01 | 95.69 |
| F1-Score | 91.83 | 93.55 | 95.52 |
| (b) Dataset [14] | |||
| Metric (%) | COVID-19 | Pneumonia | Normal |
| Recall | 100.00 | 95.94 | 100.00 |
| Precision | 92.59 | 100.00 | 97.36 |
| F1-Score | 96.15 | 97.93 | 98.66 |
From a close observation of the confusion matrices in Fig. 7, we observe that our proposed method can classify images into its correct class with a high accuracy. Furthermore, from Fig. 6, we can observe the initial performances of the base models on each of the two datasets. After applying our proposed blended ensembling method, we obtain a clear spike in the accuracy of the prediction as depicted in the confusion matrices which hint at the success of our proposed method.
5. Conclusion
In this paper, we have proposed a method to effectively detect COVID-19 from CXRs. Initially, transfer learning has been used on the state-of-the-art CNN model DenseNet-201, which has been pre-trained on the ImageNet dataset. This training phase has been utilized to generate five snapshots of the model using cosine annealing technique, each extracting and learning different low-level features of the dataset. Next, the predictions of these snapshots have been ensembled via blending to train a RF meta-learner which, in turn, provides more accurate and reliable prediction upon combining the features of each of the base models (i.e., snapshots of DenseNet-201). The results of our proposed method upon the large COVID-X dataset as well as Chowdhury et al.’s dataset indicate that blending the initial decision scores can lead to impressive accuracy, hence more reliable predictions.
Since the results of our method are experimental in nature, it involves finding the optimal hyperparameters for our model. This empirical approach may need manual intervention (i.e., some experimentation) to fine tune the parameters to perfection which may be considered as a possible limitation of our approach. We intend to make the selection process more efficient with an optimization algorithm, which will be a part of our future work. We would also like to extend our proposed method to other areas of healthcare involving other forms of medical imaging apart from CXRs like CT scans to detect other diseases as well where it can increase the efficiency of predictions, thereby boosting the testing rate of patients and benefiting the medical community.
CRediT authorship contribution statement
Avinandan Banerjee: Software, Methodology, Formal analysis, Writing – original draft. Arya Sarkar: Data curation, Software, Methodology, Writing – original draft. Sayantan Roy: Resources, Visualization, Writing – original draft. Pawan Kumar Singh: Writing – review & editing, Resources, Visualization, Project administration, Investigation, Funding acquisition. Ram Sarkar: Conceptualization, Validation, Resources, Data curation, Writing – review & editing, Investigation, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
We would like to thank the Centre for Microprocessor Applications for Training, Education and Research (CMATER) research laboratory of the Computer Science and Engineering Department, Jadavpur University, Kolkata, India for providing us the infrastructural support.
References
- 1.2022. WHO coronavirus (COVID-19) dashboard. URL https://covid19.who.int/ (Accessed 25 March 2022) [Google Scholar]
- 2.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lakhani P., Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284(2):574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 4.Wang P., Liu X., Berzin T.M., Brown J.R.G., Liu P., Zhou C., Lei L., Li L., Guo Z., Lei S., et al. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): a double-blind randomised study. Lancet Gastroenterol. Hepatol. 2020;5(4):343–351. doi: 10.1016/S2468-1253(19)30411-X. [DOI] [PubMed] [Google Scholar]
- 5.Golan R. Science; 2018. Deepcade: A deep learning architecture for the detection of lung nodules in ct scans. [Google Scholar]
- 6.2022. Tracking coronavirus vaccinations around the world. URL https://www.nytimes.com/interactive/2021/world/covid-vaccinations-tracker.html (Accessed 15 March 2022) [Google Scholar]
- 7.Goel T., Murugan R., Mirjalili S., Chakrabartty D.K. OptCoNet: an optimized convolutional neural network for an automatic diagnosis of COVID-19. Appl. Intell. 2020:1–16. doi: 10.1007/s10489-020-01904-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahlol A.T., Yousri D., Ewees A.A., Al-Qaness M.A., Damasevicius R., Abd Elaziz M. COVID-19 image classification using deep features and fractional-order marine predators algorithm. Sci. Rep. 2020;10(1):1–15. doi: 10.1038/s41598-020-71294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang W., Yao J., Chen A., Lv Q., Zanin M., Liu J., Wong S., Li Y., Lu J., Liang H., et al. Early triage of critically ill COVID-19 patients using deep learning. Nature Commun. 2020;11(1):1–7. doi: 10.1038/s41467-020-17280-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayoobi N., Sharifrazi D., Alizadehsani R., Shoeibi A., Gorriz J.M., Moosaei H., Khosravi A., Nahavandi S., Chofreh A.G., Goni F.A., et al. Time series forecasting of new cases and new deaths rate for COVID-19 using deep learning methods. Results Phys. 2021;27 doi: 10.1016/j.rinp.2021.104495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu Y., Lin H., Meng J., Wei X., Guo H., Zhao Z. Deep transfer learning for modality classification of medical images. Information. 2017;8(3):91. [Google Scholar]
- 12.Ravishankar H., Sudhakar P., Venkataramani R., Thiruvenkadam S., Annangi P., Babu N., Vaidya V. Deep Learning and Data Labeling for Medical Applications. Springer; 2016. Understanding the mechanisms of deep transfer learning for medical images; pp. 188–196. [Google Scholar]
- 13.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chowdhury M.E., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Al-Emadi N., et al. 2020. Can AI help in screening viral and COVID-19 pneumonia? arXiv preprint arXiv:2003.13145. [Google Scholar]
- 15.A. Makris, I. Kontopoulos, K. Tserpes, COVID-19 detection from chest X-Ray images using Deep Learning and Convolutional Neural Networks, in: 11th Hellenic Conference on Artificial Intelligence, 2020, pp. 60–66.
- 16.Farooq M., Hafeez A. 2020. Covid-resnet: A deep learning framework for screening of covid19 from radiographs. arXiv preprint arXiv:2003.14395. [Google Scholar]
- 17.Khan A.I., Shah J.L., Bhat M.M. Coronet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karar M.E., Hemdan E.E.-D., Shouman M.A. Cascaded deep learning classifiers for computer-aided diagnosis of COVID-19 and pneumonia diseases in X-ray scans. Complex Intell. Syst. 2020:1–13. doi: 10.1007/s40747-020-00199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahmud T., Rahman M.A., Fattah S.A. CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. 2020;122 doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ucar F., Korkmaz D. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnostic of the Coronavirus disease 2019 (COVID-19) from X-Ray images. Med. Hypotheses. 2020 doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boddeda R., Deepak V S., Patel S.A., et al. 2020. A novel strategy for COVID-19 classification from chest X-ray images using deep stacked-ensembles. arXiv preprint arXiv:2010.05690. [Google Scholar]
- 22.Rajaraman S., Siegelman J., Alderson P.O., Folio L.S., Folio L.R., Antani S.K. 2020. Iteratively Pruned deep learning ensembles for COVID-19 detection in chest X-rays. arXiv preprint arXiv:2004.08379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. 2020. Covid-caps: A capsule network-based framework for identification of covid-19 cases from x-ray images. arXiv preprint arXiv:2004.02696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandra T.B., Verma K., Singh B.K., Jain D., Netam S.S. Coronavirus disease (COVID-19) detection in chest X-Ray images using majority voting based classifier ensemble. Expert Syst. Appl. 2020;165 doi: 10.1016/j.eswa.2020.113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajaraman S., Antani S.K. Modality-specific deep learning model ensembles toward improving TB detection in chest radiographs. IEEE Access. 2020;8:27318–27326. doi: 10.1109/access.2020.2971257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Islam M.T., Aowal M.A., Minhaz A.T., Ashraf K. 2017. Abnormality detection and localization in chest x-rays using deep convolutional neural networks. arXiv preprint arXiv:1705.09850. [Google Scholar]
- 27.Chowdhury N.K., Rahman M.M., Kabir M.A. PDCOVIDNet: a parallel-dilated convolutional neural network architecture for detecting COVID-19 from chest X-ray images. Health Inf. Sci. Syst. 2020;8(1):1–14. doi: 10.1007/s13755-020-00119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nour M., Cömert Z., Polat K. A novel medical diagnosis model for COVID-19 infection detection based on deep features and Bayesian optimization. Appl. Soft Comput. 2020 doi: 10.1016/j.asoc.2020.106580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linda Wang Z.Q.L., Wong A. 2020. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. arXiv:2003.09871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pawan S.J., Sankar R., Prabhudev A.M., Mahesh P.A., Prakashini K., Das S.K., Rajan J. 2021. MobileCaps: A lightweight model for screening and severity analysis of COVID-19 chest X-Ray images. arXiv. [Google Scholar]
- 32.Gunraj H., Sabri A., Koff D., Wong A. 2021. COVID-Net CT-2: Enhanced deep neural networks for detection of COVID-19 from chest CT images through bigger, more diverse learning. arXiv preprint arXiv:2101.07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L., Wong A. 2020. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-Ray images. arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banerjee A., Bhattacharya R., Bhateja V., Singh P.K., Sarkar R., et al. COFE-Net: An ensemble strategy for computer-aided detection for COVID-19. Measurement. 2022;187 doi: 10.1016/j.measurement.2021.110289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asgharnezhad H., Shamsi A., Alizadehsani R., Khosravi A., Nahavandi S., Sani Z.A., Srinivasan D., Islam S.M.S. Objective evaluation of deep uncertainty predictions for covid-19 detection. Sci. Rep. 2022;12(1):1–11. doi: 10.1038/s41598-022-05052-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharifrazi D., Alizadehsani R., Roshanzamir M., Joloudari J.H., Shoeibi A., Jafari M., Hussain S., Sani Z.A., Hasanzadeh F., Khozeimeh F., et al. Fusion of convolution neural network, support vector machine and sobel filter for accurate detection of COVID-19 patients using X-ray images. Biomed. Signal Process. Control. 2021;68 doi: 10.1016/j.bspc.2021.102622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozcan T. A new composite approach for COVID-19 detection in X-ray images using deep features. Appl. Soft Comput. 2021;111 doi: 10.1016/j.asoc.2021.107669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhardwaj P., Kaur A. A novel and efficient deep learning approach for COVID-19 detection using X-ray imaging modality. Int. J. Imaging Syst. Technol. 2021;31(4):1775–1791. doi: 10.1002/ima.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun L., Mo Z., Yan F., Xia L., Shan F., Ding Z., Song B., Gao W., Shao W., Shi F., et al. Adaptive feature selection guided deep forest for covid-19 classification with chest ct. IEEE J. Biomed. Health Inf. 2020 doi: 10.1109/JBHI.2020.3019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaban W.M., Rabie A.H., Saleh A.I., Abo-Elsoud M. A new COVID-19 patients detection strategy (CPDS) based on hybrid feature selection and enhanced KNN classifier. Knowl.-Based Syst. 2020;205 doi: 10.1016/j.knosys.2020.106270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alizadehsani R., Sharifrazi D., Izadi N.H., Joloudari J.H., Shoeibi A., Gorriz J.M., Hussain S., Arco J.E., Sani Z.A., Khozeimeh F., et al. Uncertainty-aware semi-supervised method using large unlabeled and limited labeled COVID-19 data. ACM Trans. Multimedia Comput. Commun. Appl. (TOMM) 2021;17(3s):1–24. [Google Scholar]
- 42.Joloudari J.H., Azizi F., Nodehi I., Nematollahi M.A., Kamrannejhad F., Mosavi A., Hassannatajjeloudari E., Alizadehsani R. EasyChair; 2021. DNN-GFE: A Deep Neural Network Model Combined with Global Feature Extractor for COVID-19 Diagnosis Based on CT Scan Images: Tech. rep. [DOI] [PubMed] [Google Scholar]
- 43.Khozeimeh F., Sharifrazi D., Izadi N.H., Joloudari J.H., Shoeibi A., Alizadehsani R., Gorriz J.M., Hussain S., Sani Z.A., Moosaei H., et al. Combining a convolutional neural network with autoencoders to predict the survival chance of COVID-19 patients. Sci. Rep. 2021;11(1):1–18. doi: 10.1038/s41598-021-93543-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kundu R., Basak H., Singh P.K., Ahmadian A., Ferrara M., Sarkar R. Fuzzy rank-based fusion of CNN models using Gompertz function for screening COVID-19 CT-scans. Sci. Rep. 2021;11(1):1–12. doi: 10.1038/s41598-021-93658-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kundu R., Singh P.K., Mirjalili S., Sarkar R. COVID-19 detection from lung CT-scans using a fuzzy integral-based CNN ensemble. Comput. Biol. Med. 2021;138 doi: 10.1016/j.compbiomed.2021.104895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dey A., Chattopadhyay S., Singh P.K., Ahmadian A., Ferrara M., Senu N., Sarkar R. MRFGRO: a hybrid meta-heuristic feature selection method for screening COVID-19 using deep features. Sci. Rep. 2021;11(1):1–15. doi: 10.1038/s41598-021-02731-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kundu R., Singh P.K., Ferrara M., Ahmadian A., Sarkar R. ET-NET: an ensemble of transfer learning models for prediction of COVID-19 infection through chest CT-scan images. Multimedia Tools Appl. 2022;81(1):31–50. doi: 10.1007/s11042-021-11319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loshchilov I., Hutter F. 2016. Sgdr: Stochastic gradient descent with warm restarts. arXiv preprint arXiv:1608.03983. [Google Scholar]
- 49.Huang G., Li Y., Pleiss G., Liu Z., Hopcroft J.E., Weinberger K.Q. 2017. Snapshot ensembles: Train 1, get m for free. arXiv preprint arXiv:1704.00109. [Google Scholar]
- 50.Annavarapu C.S.R., et al. Deep learning-based improved snapshot ensemble technique for COVID-19 chest X-ray classification. Appl. Intell. 2021;51(5):3104–3120. doi: 10.1007/s10489-021-02199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang S., Wang C., Nie J., Kumar N., Zhang Y., Xiong Z., Barnawi A. EDL-COVID: ensemble deep learning for COVID-19 case detection from chest x-ray images. Ieee Trans. Ind. Inf. 2021;17(9):6539–6549. doi: 10.1109/TII.2021.3057683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chowdhury N.K., Kabir M.A., Rahman M., Rezoana N., et al. 2020. Ecovnet: An ensemble of deep convolutional neural networks based on efficientnet to detect covid-19 from chest x-rays. arXiv preprint arXiv:2009.11850. [Google Scholar]
- 53.Breiman L. Random forests. Mach. Learn. 2001;45(1):5–32. [Google Scholar]
- 54.Xu G., Liu M., Jiang Z., Söffker D., Shen W. Bearing fault diagnosis method based on deep convolutional neural network and random forest ensemble learning. Sensors. 2019;19(5):1088. doi: 10.3390/s19051088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abuella M., Chowdhury B. Random forest ensemble of support vector regression models for solar power forecasting. 2017 IEEE Power & Energy Society Innovative Smart Grid Technologies Conference; ISGT; IEEE; 2017. pp. 1–5. [Google Scholar]
- 56.Wang A., Wang Y., Chen Y. Hyperspectral image classification based on convolutional neural network and random forest. Remote Sens. Lett. 2019;10(11):1086–1094. [Google Scholar]
- 57.Tang Z., Zhao W., Xie X., Zhong Z., Shi F., Liu J., Shen D. 2020. Severity assessment of coronavirus disease 2019 (COVID-19) using quantitative features from chest CT images. arXiv preprint arXiv:2003.11988. [Google Scholar]
- 58.Alqudah A., Qazan S., Alquran H., Qasmieh I., Alqudah A. COVID-19 detection from x-ray images using different artificial intelligence hybrid models. Jordan J. Electr. Eng. 2020;6(6):168. [Google Scholar]
- 59.Iwendi C., Bashir A.K., Peshkar A., Sujatha R., Chatterjee J.M., Pasupuleti S., Mishra R., Pillai S., Jo O. COVID-19 patient health prediction using boosted random forest algorithm. Front. Public Health. 2020;8 doi: 10.3389/fpubh.2020.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barbosa V.A.d.F., Gomes J.C., de Santana M.A., de Lima C.L., Calado R.B., Bertoldo Junior C.R., Albuquerque J.E.d.A., de Souza R.G., de Araújo R.J.E., Mattos Junior L.A.R., et al. COVID-19 rapid test by combining a random forest-based web system and blood tests. J. Biomol. Struct. Dyn. 2021:1–20. doi: 10.1080/07391102.2021.1966509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., et al. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115(3):211–252. [Google Scholar]
- 62.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 63.Senan E.M., Alzahrani A., Alzahrani M.Y., Alsharif N., Aldhyani T.H. Automated diagnosis of chest X-Ray for early detection of COVID-19 disease. Comput. Math. Methods Med. 2021;2021 doi: 10.1155/2021/6919483. [DOI] [PMC free article] [PubMed] [Google Scholar]